Abstract
Aging is the single greatest risk factor for the development of disease. Understanding the biological molecules and mechanisms that modulate aging is therefore critical for the development of health-maximizing interventions for older people. The effect of fats on longevity has traditionally been disregarded as purely detrimental. However, new studies are starting to uncover the possible beneficial effects of lipids working as signaling molecules on health and longevity. These studies highlight the complex links between aging and lipid signaling. In this review we summarize accumulating evidence that points to changes in lipid metabolism, and in particular lipid signaling, as an underlying mechanism for healthy aging.
Keywords: aging, longevity, lipid signaling, nuclear receptors, lipidomics
Aging can be modulated
The discovery that environmental and genetic interventions can increase lifespan in diverse model organisms inspired a revolution in the search for the biological bases of aging [1]. By understanding how aging acts as the major risk factor for age-associated conditions such as cancer, neurodegenerative and metabolic diseases [2], we may learn how to prevent these conditions. Recently, nine different hallmarks of aging were proposed [3], providing a reference framework that allows a better understanding of the mechanisms underlying longevity and healthy aging. These hallmarks interact with each other, especially at the metabolic level [4], but the mechanisms that govern these interactions remain largely unclear. Interestingly, multiple studies have started to pinpoint a pivotal role for diverse lipids in lifespan-extending interventions, suggesting that lipid metabolism may be a key component in healthy aging [5,6]. This review focuses on recent data uncovering how lipid-signaling molecules modulate life-extending interventions, particularly in the nematode worm Caenorhabditis elegans. Most of these lipid signals induce their pro-longevity transcriptional changes through nuclear receptor (NR) transcription factors, while others seem to re-balance lipid homeostasis. Finally, we discuss conservation of these lipids and their effectors, and how they might affect other organisms, including humans.
Nuclear receptors and dauer formation in C. elegans
The ability of organisms to respond to different environmental conditions is fundamental for development and longevity-assurance. Organisms integrate these responses by, for example, the production of hormones and the regulation of NRs. NRs work as molecular switches whose transactivation activity is controlled by the presence of lipophilic hormones produced in response to diverse stimuli [7,8].
Due to its short lifespan and ease for genetic manipulation, C. elegans has been a key model organism in the study of lifespan-extending interventions and the role therein of multiple NRs [9]. The best-studied NR in C. elegans is DAF-12 (dauer formation related gene). daf-12 and around 30 other daf genes control the developmental transition of L1 larvae into either a stress-resistant diapause state, called dauer, or reproductive development (Box 1) [10]. DAF-12 transactivation activity is regulated by its ligands, dafrachronic acids (DA), which are cholesterol-derived molecules whose production is tightly controlled by environmental conditions [11]. In addition to dauer formation, DAF-12 modulates lipid metabolism, developmental timing and lifespan [7]. Lifespan modulation by DAF-12 is complex, since it can promote or repress longevity under low or high temperatures, respectively. Interestingly, modulation of lifespan in response to temperature is independent of DA [12].
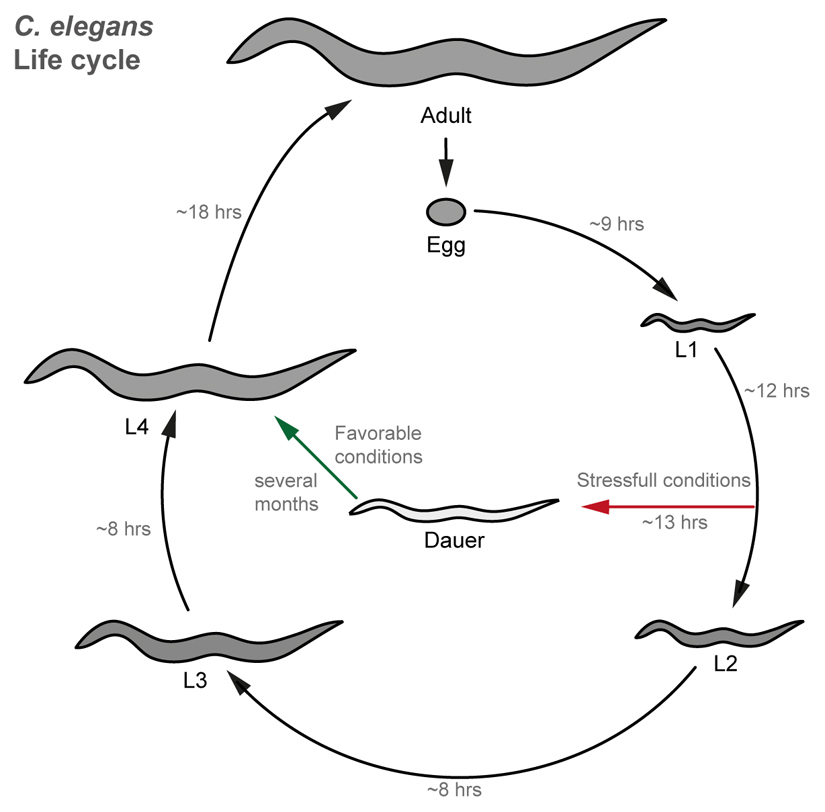
Box 1. Life cycle of C. elegans.
The life cycle of the nematode worm has been widely described (Figure I). Briefly, after embryonic development of the fertilized egg, the worm undergoes four different larval stages (L1-L4), to finally reach adulthood and sexual maturity [93]. At the end of L1, stressful conditions such as overcrowding, high temperatures or starvation, trigger the worm to adopt an alternative developmental pathway called dauer. The dauer worm is able to survive long periods of time under stressful conditions. Once conditions improve, dauer worms resume development at L4 stage to then reach adulthood and reproduce [93].
Figure I. Worm life cycle.
Representation of C. elegans life cycle at 22ºC. Numbers along the arrows represent the hours (hrs) necessary to transition from one stage to the next.
Cholesterol and insulin signaling mediated longevity
Other lifespan-extending interventions also require DAF-12 and its ligand DA. A prominent example is the insulin / insulin-like growth factor signaling (IIS) pathway, an evolutionarily conserved, nutrient-sensing network that modulates a plethora of biological processes, including development and lifespan [13]. IIS reduction extends lifespan in multiple model organisms, and this effect is, in part, mediated by regulating the conserved FOXO transcription factors [14,15]. In C. elegans, mutation of daf-2, the homolog of mammalian insulin-like growth factor (IGF) and insulin receptors, yields long-lived worms, and this longevity requires the presence of DAF-16, the worm FOXO homolog [14]. Interestingly, daf-2 genetically interacts with daf-12 to modulate lifespan [16]. This interaction is intricate, as DAF-12 acts as an anti- or pro-aging factor in daf-2 mutant worms in the presence or absence of DA, respectively [12]. This observation indicates that both DAF-12 and DA are fundamental for reduced IIS induced longevity (Figure 1).
Figure 1. Reduced insulin insulin-like signaling (IIS) extends lifespan in a DAF-16/DAF-12/NSBP-1 dependent manner.
In wild type C. elegans, IIS phosphorylates and negatively regulates NSBP-1, a cholesterol binding protein, and DAF-16 transcription factor. Under reduced IIS, these proteins become dephosphorylated and migrate into the nucleus and interact in a cholesterol-dependent manner to modulate the transcription of pro-longevity genes. Extension of lifespan by lowered IIS is also dependent on DAF-12 and its ligand DA. Worms with reduced IIS cannot maximize lifespan-extension in the absence of DA or excess of cholesterol. I: Insulin; P: Phosphate; C: Cholesterol; DA; Dafachronic Acids
The association between reduced IIS and longevity is regulated not only by DAF-16 but also by multiple other proteins. Among these, the nematode sterol-binding protein 1 (NSBP-1) is especially interesting due to its ability to bind cholesterol, and therefore work as a cholesterol sensor [17]. NSBP-1, much like DAF-16, is phosphorylated under high insulin signaling by AKT, and thereby excluded from the nucleus. Conversely, under reduced IIS, both DAF-16 and NSBP-1 migrate into the nucleus and interact, but only under low cholesterol concentrations (Figure 1). Both proteins regulate the transcription of a small set of genes involved in lipid metabolism and aging [17]. However, it is unclear whether NSBP-1 can regulate additional DAF-16-target genes and whether this mechanism of FOXO regulation is conserved in mammals. Moreover, DAF-12 and DAF-16 induce a negative feedback on DA synthesis upon dauer formation [18], suggesting that cholesterol levels are affected by their interaction, which could have implications for aging modulation, perhaps through NSBP-1. These studies thus show a link between cholesterol metabolism and lifespan.
The Drosophila melanogaster homolog of DAF-12, DHR96, binds cholesterol and modulates lipid metabolism by regulating diverse target genes [19,20]. DHR96 is required for the enhanced xenobiotic detoxification induced by lowered IIS, but not for the lifespan extension, which is unaffected by removal of this gene [21]. The mammalian homolog of daf-12, farnesoid X receptor (FXR), is also involved in cholesterol and lipid metabolism [22]. FXR expression decreases with age-associated endoplasmic reticulum (ER) stress and, as a consequence, plays a prominent role in the development of fatty liver [23]. However, whether FXR plays a role in mammalian lifespan regulation is unknown. In addition to regulating activity of diverse NRs, IIS can also modulate the mevalonate pathway, which in turn produces sterols and isoprenoids [24], adding an extra layer of complexity between IIS and its interaction with cholesterol.
Does dietary restriction promote longevity through lipid signals?
Activity of the IIS network is linked to dietary restriction (DR), which is the most robust lifespan-extending and health-promoting intervention. DR, broadly defined as a reduction in food intake that avoids malnutrition, was first reported over 80 years ago to extend the lifespan of rats [25]. DR has since been shown to increase health and lifespan in a plethora of organisms, including yeast, worms, flies, mice and even primates [25–29]. In addition, DR improves health and has a beneficial effect on risk factors for age-related disorders such as cancer, diabetes and cardiovascular disease in humans [30]. In C. elegans, at least nine different DR regimes exist, with differences in the way that they modulate lifespan [31,32]. However, these regimes can be broadly classified in the three categories discussed below.
Dietary restriction through eat-2 mutation
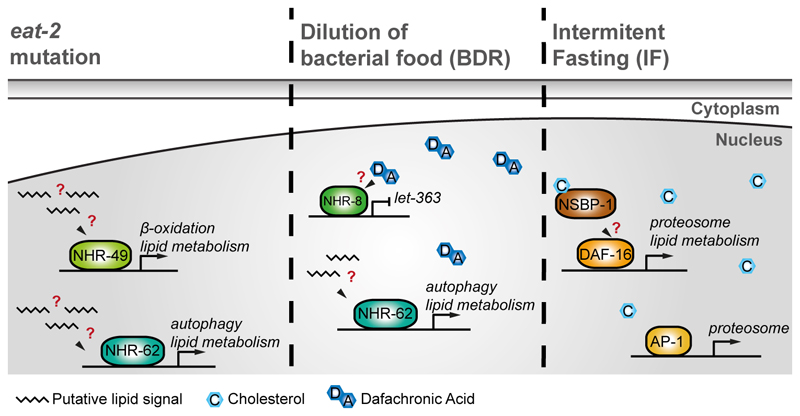
Genetic mutation of eat-2, disturbs an acetylcholine receptor subunit in the pharynx of the worm and reduces pumping and therefore food intake, thereby mimicking DR and extending lifespan by up to 40% [33]. Interestingly, an RNAi screen with eat-2 mutant worms identified nhr-62, a hepatic nuclear factor 4α (HNF4α)-like NR homolog, as a critical factor in DR-induced lifespan-extension (Figure 2) [34]. Other hallmarks of DR, such as the decreased triglycerides and enhanced autophagy also rely, at least partially, on NHR-62-dependent transcriptional regulation. Moreover, NHR-62 controls the expression of genes involved in lipolysis and fatty acid desaturation, thereby implicating lipid metabolism in this form of DR-mediated lifespan extension [34].
Figure 2. Different dietary restriction (DR) paradigms promote longevity in ways that depend on lipid signals.
While different DR interventions in C. elegans have at least partially independent mechanisms for lifespan extension, they all seem to rely on the regulation of transcription factors that act in response to putative lipid signals. Hence, increased lifespan from eat-2 mutation relies on NHR-49 and NHR-62, although their respective lipid ligands (if they have one) remain unknown. NHR-62 is also involved in extension of lifespan by BDR, at least at low food concentrations, and NHR-8 and DA are also required, although the nature of their interaction is unclear. IF relies on DAF-16 and AP-1 to achieve maximum lifespan extension, but whether NSBP-1 may play a role here remains unknown.
In addition to NHR-62, another HNF4α-like homolog, NHR-49, regulates lipid metabolism and lifespan in C. elegans, [35]. This NR is also required for eat-2 lifespan extension, which is reversed by depletion of nhr-49 by RNAi. eat-2 longevity is dependent on regulation of genes associated with β-oxidation by NHR-49 [36]. However, whether any lipid acts as a ligand for either NHR-49 or NHR-62 is still unknown (Figure 2). In mice and flies, HNF-4 regulates lipid metabolism in response to starvation and its ligands are free fatty acids [37–40]. Hence, it is possible that HNF-4 proteins control a gene network that regulates the production of “starvation signals”. These signals would in turn act as ligands for these NRs and be fundamental for lifespan modulation under DR.
Dietary restriction through dilution of bacterial food
Dilution of bacterial food (BDR) was first reported to extend lifespan of C. elegans lifespan almost 40 years ago [41]. BDR worms have increased expression of DAF-9, a critical enzyme in DA biosynthesis, and therefore increased levels of this hormone. Surprisingly, under these conditions, DA, but not its receptor DAF-12, is required for lifespan extension. Instead, DA relies on another NR, NHR-8, which controls cholesterol homeostasis to exert its beneficial effects on lifespan [42,43] (Figure 2). Further, NHR-8 acts upstream of let-363, the worm homolog of mTOR, a prominent metabolic regulator that responds to nutritional inputs [27], which in turn affects germline plasticity. This observation is intriguing, because DR is generally associated with a decrease in fertility, even though longevity and fecundity can be uncoupled in flies [44]. While it is still unclear whether DA binds directly to NHR-8, these results provide a novel link between steroid signaling, mTOR and lifespan under DR [42]. It is important to highlight that BDR modulates lifespan in a cell non-autonomous manner, by regulating the function of the SKN-1 transcription factor, suggesting the existence of one or more “starvation signals” [45]. Furthermore, NHR-62 seems to partially regulate the response to BDR [34], suggesting that this transcription factor is involved in at least two different kinds of DR, eat-2 mutation and BDR.
Dietary restriction through intermittent fasting
An additional DR regime, called intermittent fasting (IF), requires cycles of ad libitum food availability and complete food deprivation. Like the previously described regimes, IF also extends lifespan in diverse organisms such as worms and mice. Intriguingly, mice on IF can consume the same or even more calories than ad libitum fed mice and still reap the beneficial effects of DR [46]. This observation suggests that IF induces some form of “starvation signals” that are more important than the amount of calories eaten. In C. elegans, an IF regime of 2 days on/2 days off food increases lifespan and this depends on at least two transcription factors, DAF-16 and AP-1 [32,47]. The regulation of IF-induced longevity by these two transcription factors seems to be partially explained by expression of E3 ubiquitin ligases, which in turn modulate proteostasis (the regulation of protein synthesis and degradation) in response to “starvation signals” (Figure 2) [47].
Worms are unable to synthesize cholesterol, and its removal from the food medium prevents lifespan-extension induced by IF [48]. This is not a developmental effect, because withdrawal of cholesterol only during adulthood has similar effects. Furthermore, cholesterol deprivation suppresses daf-2-induced longevity, suggesting that cholesterol is required for both reduced IIS and IF-induced longevity [48]. As daf-2 longevity is dependent on DAF-16 regulation by NSBP-1, and NSBP-1 is regulated by cholesterol [17], it is possible that IF-induced lifespan extension is also dependent on the ability of NSBP-1 to bind to cholesterol and DAF-16. Consistent with this, RNAi against daf-16 or nsbp-1 partially suppresses the lifespan extension achieved by IF [32,47,48]. However, it is currently unclear whether cholesterol regulates only DAF-16 subcellular localization, or also its transactivation activity.
Dietary restriction affects lipid metabolism in mammals
In mammals, IF and DR also extend lifespan and have profound effects on energy metabolism, altering fat stores and the production of various hormones such as growth hormone (GH), leptin and adiponectin [49–52]. These hormones are in turn regulated by growth-hormone releasing-hormone (GHRH), a hypothalamic-derived hormone. Mice that lack GHRH are long-lived and have reduced levels of leptin and increased levels of adiponectin. However, the lifespan of this already long-lived GHRH-KO mouse can be further extended by DR, suggesting that DR does not rely on this particular hormone to modulate aging [53].
A meta-analysis of transcriptional changes associated with DR in mice and rats, found that genes associated with hormone signaling and lipid metabolism are highly perturbed [54]. Moreover, recent studies in mice demonstrated that DR affects saturation and elongation of several fatty acids [55,56]. Interestingly, the adiposity changes associated with DR are different from those observed in normal aging, as aging seems to particularly affect phospholipid composition [55]. It is therefore possible that one of the underlying mechanisms modulating lifespan, at least under DR, is the regulation of lipid metabolism, although causality remains to be established.
In humans, multiple studies have established a clear connection between cholesterol levels and mortality [57]. Interestingly, population studies have shown that the offspring of long-lived individuals have healthier cholesterol markers [58], indicating that at least some forms of familial longevity may exert control over cholesterol homeostasis. In addition, short dietary interventions (with 12.5% - 25% caloric restriction) are sufficient to bring cholesterol makers to a healthier state [30,59,60]. These observations further highlight a prominent link between cholesterol homeostasis in longevity. Yet, the possible beneficial effects of cholesterol as a signaling molecule in mammalian aging remain unknown.
Germline ablation signals and longevity
Like DR and reduced IIS, germline ablation is a robust intervention that extends lifespan in a plethora of organisms including worms, flies, mice, rats and even humans [61–65]. Multiple studies suggest that the germline is able to produce signals that partially coordinate reproduction and aging, with lipid metabolism playing a prominent role [5,6]. In C. elegans, germline removal induces fat accumulation, longevity and stress resistance [61,66]. These phenotypes are associated with the regulation of multiple transcription factors, including SKN-1, TCER-1, DAF-16, and NRs DAF-12, NHR-80 and NHR-49 (Figure 3) [61,66–68].
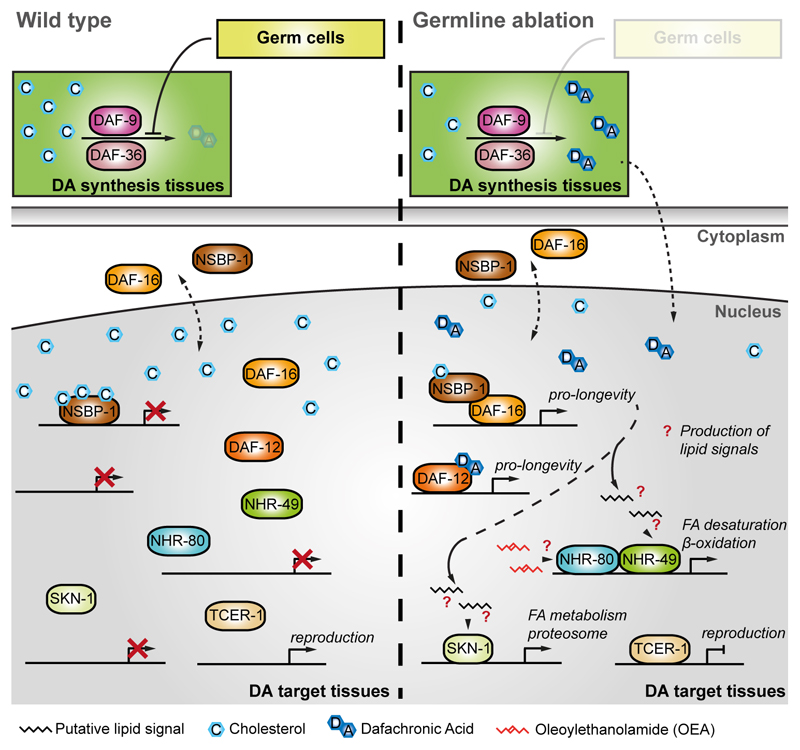
Figure 3. Germline ablation increases lifespan by modulating lipid metabolism.
Germ cells inhibit the production of DA. We propose that, as a consequence, the transcriptional activity of both DAF-16 and DAF-12 is dampened. Conversely, removal of germ cells allows production of DA and transcription of pro-longevity genes by DAF-16 and DAF-12, including genes involved in lipid metabolism. These changes promote the generation of putative lipid signals, which in turn activate downstream factors such as NHR-49, NHR-80 and SKN-1 that promote longevity through different transcriptional programs. However, these putative lipid signals remain unknown. Whether Oleoylethanolamide (OEA) serves as a NHR-80 ligand under germline ablation remains unclear. FA stands for fatty acids.
As with reduced IIS, lifespan extension by germline removal is abolished upon daf-12 mutation and is dependent on the presence of DA [12,61]. Consistently, mutation of daf-9 or daf-36, both of which encode enzymes involved in DA biosynthesis, reduces lifespan in germline-less worms. DA supplementation of daf-9 or daf-36 mutant worms is in turn sufficient to restore germline ablation longevity [69]. This observation suggests that the interaction between DAF-12 and DA is fundamental for lifespan extension by germline removal.
In addition to DAF-12, germline removal longevity also relies on DAF-16 and on the ability of both transcription factors to regulate target genes such as triglyceride lipase and acyl-CoA reductase [70]. These genes are involved in lipid metabolism and are proposed to generate putative lipid signals that would allow lifespan-extension under germline removal [70]. Furthermore, germline-less worms require DAF-12 in the gut for the production of a lipophilic signal, which in turn promotes DAF-16 nuclear localization through a protein called KRI-1[71].
NHR-80, another NR, is a key player in germline removal mediated longevity [67]. NHR-80 modulates the transcription of FAT-6, a desaturase that produces oleic acid, a monounsaturated fatty acid, and this underpins the longevity phenotype [67,72]. However, it is unclear how oleic acid exerts its beneficial effects. Moreover, as NHR-49 and NHR-80 play key roles in DR and germline removal longevity, it is possible that these interventions share at least part of the mechanisms involved in aging modulation.
It was previously shown that germline ablation promotes lipolysis by up-regulating multiple lipases, such as LIPL-4, a homolog of the mammalian lysosome associated lipase (LIPA) [73]. LIPL-4 over-expression in the gut, the organ where C. elegans stores fat, is sufficient to induce longevity [73]. However, germline-less worms have increased fat accumulation, despite the increase in lipid catabolism [74]. This extra accumulated fat appears to come from unconsumed yolk, and from here a signal is produced that activates SKN-1 [75]. SKN-1, homolog to mammalian Nrf2 and member of the cap’n’collar transcription factor family, plays a critical role in regulating genes involved in processes such as proteasome activity and lipid homeostasis [75]. Interestingly, a more recent report suggests that, upon germline ablation, not only lipid catabolism but also lipid anabolism is activated. Enhanced lipid turnover is achieved through DAF-16 and TCER-1, and both proteins are required for germline ablation longevity [76]. This mode of lifespan modulation could be evolutionarily conserved, because enhanced lipid turnover appears to be fundamental for DR-mediated lifespan extension in Drosophila [77,78], and DR also increases lipid turnover in mice [79]. It is therefore possible that both germline ablation and DR have, at least partially, overlapping roles enhancing lipid homeostasis.
Use of lipids in disease treatment and lifespan extension
In C. elegans LIPL-4 appears to also induce lifespan extension by generating the fatty acid oleoylethanolamide (OEA), a monounsaturated fatty acid. OEA works as a novel lysosomal signaling molecule by binding to the lysosomal lipid chaperone LBP-8, inducing its nuclear localization and activating NHR-80 and NHR-49 [80]. The additional observation that OEA supplementation is sufficient to extend lifespan in worms [80] further highlights the critical role lipid signaling may play in extension of lifespan. However, it is currently unknown whether this lipid signal works in a cell-autonomous manner, whether OEA regulates lifespan by modulating gene transcription downstream of NHR-80 and NHR-49. Since these NRs have already been associated with longevity under DR and germline ablation [34,67], it is not farfetched to expect that this mode of longevity control will turn out to be evolutionarily conserved.
In addition to OEA, supplementation of other monounsaturated fatty acids (MUFAs), such as oleic acid and palmitoleic acid, can also extend worm lifespan [81], although not all oleic acid supplementation protocols seem to do so [67,68]. These MUFAs, but not their derived polyunsaturated fatty acids (PUFAs), underpin the long life of histone 3 lysine 4 trimethylation (H3K4me3) deficient worms [81]. Moreover, diets that are rich in MUFAs seem to protect against diabetes and cardiovascular disease in humans [82] Nevertheless, it is currently unclear how these MUFAs exert their beneficial effects. In contrast, ω-6 PUFAs supplementation extends worm lifespan by activating autophagy (Box 2) [83]. This mechanism might also be evolutionarily conserved, because ω-6 and other PUFAs induce autophagy in human cell lines [83,84]. Hence, the relationship between MUFAs, PUFAs and longevity is complex. Higher MUFA:PUFA ratios have been observed in long-lived worms and in the daughters of long lived humans, suggesting higher PUFA levels could be detrimental [85,86]. PUFAs are more prone to oxidation and are thought to increase oxidative damage by further producing free radicals [86]. It is therefore possible that only specific PUFAs have beneficial effects on the organism, and only as long as they do not reach a critical threshold. With a better understanding of the role of lipid metabolism in health during aging comes the opportunity to use this knowledge in the treatment of disease. For example, a recent genetic screen revealed that inhibition of cholesterol biosynthesis by genetic or pharmacological means is sufficient to improve motor performance and lifespan of a mouse model of Rett syndrome [87]. Similarly, a mouse model of Cockayne syndrome was shown to improve its associated phenotypes upon supplementation of beta-hydroxybutirate, a ketone body [88]. These studies underscore the important role that modulation of lipid metabolism may have in the treatment of diverse diseases. Concordantly, medium chain triglyceride (MCT) supplementation is widely used in children as a treatment of drug resistant epilepsy [89]. MCT supplementation induces the body to switch into a ketogenic state, where lipids become the preferred source of energy. Similarly, ketone body supplementation was recently shown to shift the energy source in muscles from glycolysis to a lipid oxidation state [90]. The consequence of this switch was enhanced performance of athletic activity. However, it remains to be determined whether this translates into increased health and lifespan. These observations open the exciting possibility that lipid supplementation can be used not only in the treatment of disease, but also in its prevention and promotion of healthy aging.
Box 2. Autophagy and lipid metabolism.
Autophagy, a highly regulated process that degrades cellular components in response to diverse cues, such as starvation, is highly associated with health and longevity [94]. Autophagy is initiated with the formation of a membrane structure called the autophagosome (AP), which then engulfs molecules and/or organelles to later fuse with lysosomes and degrade the cargo. AP formation is in part regulated by diverse membrane components as it requires specific lipids for its formation [95]. For example, studies in yeast demonstrate that lipids such as phosphatidylinositol 3-phosphate are enriched in endoplasmic reticulum regions that then allow recruitment of proteins involved in AP formation [96–99]. Moreover, lipid homeostasis is fundamental for proper autophagy, since deletion of enzymes responsible for TG and sterol esters completely blocks starvation-induced autophagy [100]. These studies highlight the fundamental role of lipid homeostasis in regulation of autophagy, not only at the level of lipid signals/transcription factors that regulate autophagy-related genes (see text), but also at the level of membrane components.
Lipidomics leads the way
In order to identify and quantify the lipid species relevant for many of the interventions described above [81,85], researchers are taking advantage of the recent developments in the area of lipidomics, i.e. the analysis of lipid metabolites by mass spectrometry techniques [91]. Lipidomics is rapidly opening the doors to a new world of information that points at diverse lipid species with key roles in the regulation of diverse biological processes. For example, a recent and compelling meta-analysis of lipid composition looked at the correlation between diverse lipids within multiple tissues and maximum lifespan in as many as 35 different species including mice, bats and humans [92]. This analysis showed, for example, that structural lipids are more likely to be found in a saturated state in the long-lived species. Although not directly analyzed, this observation also implies a higher MUFA:PUFA ratio. In contrast, energy-related lipids, such as triacyglycerols (TG), were more likely to be in an unsaturated state in the long lived species [92]. Although preliminary, this study highlights the power lipidomic studies could have in the identification of lipid species and their association with longevity.
Concluding remarks and future directions
Recent studies are starting to uncover the prominent link between different lipids, particularly signaling molecules, and aging modulation. Intriguingly, many of the life-promoting interventions discussed here share underlying proteins and lipids critical for lifespan extension. For example, both DAF-12 and DAF-16 are involved in lifespan modulation during reduced IIS and germline ablation [14,61,66–68]. Similarly, NHR-49 and NHR-80 are fundamental for DR and germline ablation induced longevity [34,67]. Moreover, lipid turnover and the accumulation of specific lipid molecules, such as oleic acid, are important for intervention-induced longevity or even normal aging [80,81,85,92]. All these results suggest that lipid metabolism can be affected by multiple life-promoting interventions, albeit at different levels, and this underpins the longevity effects. However, these pioneering studies leave many new questions to be answered (see Outstanding questions). Future studies should address the link between lipid signals, the transcription factors they regulate, and the effects on lipid homeostasis these may have by regulating the expression of enzymes involved in lipid metabolism.
Outstanding Questions.
-
-
Can we use specific lipids as health-promoting and lifespan-extending interventions?
-
-
Are there fasting signals that extend lifespan? And, if so, are they conserved and do they promote healthy longevity under normal conditions?
-
-
Which cells/organs produce the lipid molecules associated with health and longevity?
-
-
Do HNF-4-related transcription factors modulate longevity in organisms other than worms?
-
-
Are supplemented lipids, such as medium chain fatty acids, binding a specific NR or pushing the metabolic machinery towards enhanced lipid turnover that in turn produces fasting signals?
-
-
Is enhanced lipid metabolism (turnover) a conserved mechanism underlying lifespan modulation?
-
-
What other health-promoting and lifespan-extending lipid signals can we identify? And are they associated to known or unknown NRs?
Acknowledgments
We acknowledge funding from the Max Planck Society, the European Research Council (ERC) under the European Union’s Seventh Framework Program (FP7/2007–2013)/ERC grant agreement (268739), the Wellcome Trust UK (098565/Z/12/Z) to L.P., and a Boehringer Ingelheim Fonds fellowship for V.B. We thank Carolina Gomez, Joris Deelen, Maarouf Baghdadi and Javier Moron for helpful comments on the manuscript. We apologize to the authors whose work could not be cited due to length restrictions.
References
- 1.Kenyon C. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 2.Niccoli T, Partridge L. Ageing as a Risk Factor for Disease. Curr Biol. 2012;22:R741–R752. doi: 10.1016/j.cub.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 3.López-Otín C, et al. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.López-Otín C, et al. Metabolic Control of Longevity. Cell. 2016;166:802–821. doi: 10.1016/j.cell.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 5.Hansen M, et al. Reproduction, Fat Metabolism, and Life Span: What Is the Connection? Cell Metab. 2013;17:10–19. doi: 10.1016/j.cmet.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ackerman D, Gems D. The mystery of C. elegans aging: An emerging role for fat: Distant parallels between C. elegans aging and metabolic syndrome? BioEssays. 2012;34:466–471. doi: 10.1002/bies.201100189. [DOI] [PubMed] [Google Scholar]
- 7.Magner DB, Antebi A. Caenorhabditis elegans nuclear receptors: insights into life traits. Trends Endocrinol Metab. 2008;19:153–160. doi: 10.1016/j.tem.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans RM, Mangelsdorf DJ. Nuclear Receptors, RXR, and the Big Bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tissenbaum HA. Using C. elegans for aging research. Invertebr Reprod Dev. 2015;59:59–63. doi: 10.1080/07924259.2014.940470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antebi A, et al. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- 11.Motola DL, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 12.Dumas KJ, et al. Influence of Steroid Hormone Signaling on Life Span Control by Caenorhabditis elegans Insulin-Like Signaling. G3 GenesGenomesGenetics. 2013;3:841–850. doi: 10.1534/g3.112.005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piper MDW, et al. Models of insulin signalling and longevity. Drug Discov Today Dis Models. 2005;2:249–256. [Google Scholar]
- 14.Kenyon C, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 15.Slack C, et al. dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell. 2011;10:735–748. doi: 10.1111/j.1474-9726.2011.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen PL, et al. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheong MC, et al. NSBP-1 mediates the effects of cholesterol on insulin/IGF-1 signaling in Caenorhabditis elegans. Cell Mol Life Sci. 2013;70:1623–1636. doi: 10.1007/s00018-012-1221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penkov S, et al. Integration of carbohydrate metabolism and redox state controls dauer larva formation in Caenorhabditis elegans. Nat Commun. 2015;6 doi: 10.1038/ncomms9060. 8060. [DOI] [PubMed] [Google Scholar]
- 19.Sieber MH, Thummel CS. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 2009;10:481–490. doi: 10.1016/j.cmet.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horner MA, et al. The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis. Genes Dev. 2009;23:2711–2716. doi: 10.1101/gad.1833609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afschar S, et al. Nuclear hormone receptor DHR96 mediates the resistance to xenobiotics but not the increased lifespan of insulin-mutant Drosophila. Proc Natl Acad Sci. 2016;113:1321–1326. doi: 10.1073/pnas.1515137113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao Y, et al. Farnesoid X receptor: a master regulator of hepatic triglyceride and glucose homeostasis. Acta Pharmacol Sin. 2015;36:44–50. doi: 10.1038/aps.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong X, et al. Hepatic steatosis exacerbated by endoplasmic reticulum stress-mediated downregulation of FXR in aging mice. J Hepatol. 2014;60:847–854. doi: 10.1016/j.jhep.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Mullen PJ, et al. The interplay between cell signalling and the mevalonate pathway in cancer. Nat Rev Cancer. 2016;16:718–731. doi: 10.1038/nrc.2016.76. [DOI] [PubMed] [Google Scholar]
- 25.McCay CM, et al. The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. Nutr Burbank Los Angel Cty Calif. 1989;5:155–171. discussion 172. [PubMed] [Google Scholar]
- 26.Mattison JA, et al. Caloric restriction improves health and survival of rhesus monkeys. Nat Commun. 2017;8 doi: 10.1038/ncomms14063. 14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fontana L, Partridge L. Promoting Health and Longevity through Diet: From Model Organisms to Humans. Cell. 2015;161:106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontana L, et al. Extending Healthy Life Span--From Yeast to Humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hine C, et al. Endogenous Hydrogen Sulfide Production Is Essential for Dietary Restriction Benefits. Cell. 2015;160:132–144. doi: 10.1016/j.cell.2014.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heilbronn LK, et al. Effect of 6-Month Calorie Restriction on Biomarkers of Longevity, Metabolic Adaptation, and Oxidative Stress in Overweight Individuals: A Randomized Controlled Trial. JAMA. 2006;295:1539. doi: 10.1001/jama.295.13.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honjoh S, et al. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- 33.Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heestand BN, et al. Dietary Restriction Induced Longevity Is Mediated by Nuclear Receptor NHR-62 in Caenorhabditis elegans. PLoS Genet. 2013;9:e1003651. doi: 10.1371/journal.pgen.1003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Gilst MR, et al. Nuclear Hormone Receptor NHR-49 Controls Fat Consumption and Fatty Acid Composition in C. elegans. PLoS Biol. 2005;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chamoli M, et al. A novel kinase regulates dietary restriction-mediated longevity in Caenorhabditis elegans. Aging Cell. 2014;13:641–655. doi: 10.1111/acel.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palanker L, et al. Drosophila HNF4 Regulates Lipid Mobilization and β-Oxidation. Cell Metab. 2009;9:228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirota K, et al. Hepatocyte nuclear factor-4 is a novel downstream target of insulin via FKHR as a signal-regulated transcriptional inhibitor. J Biol Chem. 2003;278:13056–13060. doi: 10.1074/jbc.C200553200. [DOI] [PubMed] [Google Scholar]
- 39.Hirota K, et al. A Combination of HNF-4 and Foxo1 Is Required for Reciprocal Transcriptional Regulation of Glucokinase and Glucose-6-phosphatase Genes in Response to Fasting and Feeding. J Biol Chem. 2008;283:32432–32441. doi: 10.1074/jbc.M806179200. [DOI] [PubMed] [Google Scholar]
- 40.Hertz R, et al. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 1998;392:512–516. doi: 10.1038/33185. [DOI] [PubMed] [Google Scholar]
- 41.Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 42.Thondamal M, et al. Steroid hormone signalling links reproduction to lifespan in dietary-restricted Caenorhabditis elegans. Nat Commun. 2014;5 doi: 10.1038/ncomms5879. 4879. [DOI] [PubMed] [Google Scholar]
- 43.Magner DB, et al. The NHR-8 Nuclear Receptor Regulates Cholesterol and Bile Acid Homeostasis in C. elegans. Cell Metab. 2013;18:212–224. doi: 10.1016/j.cmet.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grandison RC, et al. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 46.Anson RM, et al. Intermittent fasting dissociates beneficial effects of dietary restriction on glucose metabolism and neuronal resistance to injury from calorie intake. Proc Natl Acad Sci. 2003;100:6216–6220. doi: 10.1073/pnas.1035720100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uno M, et al. A Fasting-Responsive Signaling Pathway that Extends Life Span in C. elegans. Cell Rep. 2013;3:79–91. doi: 10.1016/j.celrep.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 48.Ihara A, et al. Cholesterol regulates DAF-16 nuclear localization and fasting-induced longevity in C. elegans. Exp Gerontol. 2017;87:40–47. doi: 10.1016/j.exger.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 49.Jiang T, et al. Calorie restriction modulates renal expression of sterol regulatory element binding proteins, lipid accumulation, and age-related renal disease. J Am Soc Nephrol JASN. 2005;16:2385–2394. doi: 10.1681/ASN.2004080701. [DOI] [PubMed] [Google Scholar]
- 50.Kuhla A, et al. Lifelong Caloric Restriction Reprograms Hepatic Fat Metabolism in Mice. J Gerontol A Biol Sci Med Sci. 2014;69:915–922. doi: 10.1093/gerona/glt160. [DOI] [PubMed] [Google Scholar]
- 51.Zhu M, et al. Circulating adiponectin levels increase in rats on caloric restriction: the potential for insulin sensitization. Exp Gerontol. 2004;39:1049–1059. doi: 10.1016/j.exger.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 52.Sethi JK, Vidal-Puig AJ. Thematic review series: Adipocyte Biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res. 2007;48:1253–1262. doi: 10.1194/jlr.R700005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun LY, et al. Growth hormone-releasing hormone disruption extends lifespan and regulates response to caloric restriction in mice. eLife. 2013;2 doi: 10.7554/eLife.01098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plank M, et al. A meta-analysis of caloric restriction gene expression profiles to infer common signatures and regulatory mechanisms. Mol Biosyst. 2012;8:1339. doi: 10.1039/c2mb05255e. [DOI] [PubMed] [Google Scholar]
- 55.Miller KN, et al. Aging and caloric restriction impact adipose tissue, adiponectin, and circulating lipids. Aging Cell. 2017 doi: 10.1111/acel.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hahn O, et al. Dietary restriction protects from age-associated DNA methylation and induces epigenetic reprogramming of lipid metabolism. Genome Biol. 2017;18 doi: 10.1186/s13059-017-1187-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prospective Studies Collaboration. Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55 000 vascular deaths. The Lancet. 2007;370:1829–1839. doi: 10.1016/S0140-6736(07)61778-4. [DOI] [PubMed] [Google Scholar]
- 58.Deelen J, et al. Employing biomarkers of healthy ageing for leveraging genetic studies into human longevity. Exp Gerontol. 2016;82:166–174. doi: 10.1016/j.exger.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 59.van de Rest O, et al. Metabolic effects of a 13-weeks lifestyle intervention in older adults: The Growing Old Together Study. Aging. 2016;8:111–124. doi: 10.18632/aging.100877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ravussin E, et al. A 2-Year Randomized Controlled Trial of Human Caloric Restriction: Feasibility and Effects on Predictors of Health Span and Longevity. J Gerontol A Biol Sci Med Sci. 2015;70:1097–1104. doi: 10.1093/gerona/glv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 62.Flatt T, et al. Drosophila germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci. 2008;105:6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cargill SL, et al. Age of ovary determines remaining life expectancy in old ovariectomized mice. Aging Cell. 2003;2:185–190. doi: 10.1046/j.1474-9728.2003.00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drori D, Folman Y. Environmental effects on longevity in the male rat: Exercise, mating, castration and restricted feeding. Exp Gerontol. 1976;11:25–32. doi: 10.1016/0531-5565(76)90007-3. [DOI] [PubMed] [Google Scholar]
- 65.Min K-J, et al. The lifespan of Korean eunuchs. Curr Biol. 2012;22:R792–R793. doi: 10.1016/j.cub.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 66.Alper S, et al. The Caenorhabditis elegans Germ Line Regulates Distinct Signaling Pathways to Control Lifespan and Innate Immunity. J Biol Chem. 2010;285:1822–1828. doi: 10.1074/jbc.M109.057323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goudeau J, et al. Fatty Acid Desaturation Links Germ Cell Loss to Longevity Through NHR-80/HNF4 in C. elegans. PLoS Biol. 2011;9:e1000599. doi: 10.1371/journal.pbio.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ratnappan R, et al. Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of C. elegans. PLoS Genet. 2014;10:e1004829. doi: 10.1371/journal.pgen.1004829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerisch B, et al. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCormick M, et al. New genes that extend Caenorhabditis elegans’ lifespan in response to reproductive signals: New genes that extend C. elegans’ lifespan. Aging Cell. 2012;11:192–202. doi: 10.1111/j.1474-9726.2011.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berman JR, Kenyon C. Germ-Cell Loss Extends C. elegans Life Span through Regulation of DAF-16 by kri-1 and Lipophilic-Hormone Signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 72.Brock TJ, et al. Genetic Regulation of Unsaturated Fatty Acid Composition in C. elegans. PLoS Genet. 2006;2:e108. doi: 10.1371/journal.pgen.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang MC, et al. Fat Metabolism Links Germline Stem Cells and Longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Rourke EJ, et al. C. elegans Major Fats Are Stored in Vesicles Distinct from Lysosome-Related Organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Steinbaugh MJ, et al. Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. eLife. 2015;4 doi: 10.7554/eLife.07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amrit FRG, et al. DAF-16 and TCER-1 Facilitate Adaptation to Germline Loss by Restoring Lipid Homeostasis and Repressing Reproductive Physiology in C. elegans. PLOS Genet. 2016;12:e1005788. doi: 10.1371/journal.pgen.1005788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katewa SD, et al. Peripheral Circadian Clocks Mediate Dietary Restriction-Dependent Changes in Lifespan and Fat Metabolism in Drosophila. Cell Metab. 2016;23:143–154. doi: 10.1016/j.cmet.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Katewa SD, et al. Intramyocellular Fatty-Acid Metabolism Plays a Critical Role in Mediating Responses to Dietary Restriction in Drosophila melanogaster. Cell Metab. 2012;16:97–103. doi: 10.1016/j.cmet.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bruss MD, et al. Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. AJP Endocrinol Metab. 2010;298:E108–E116. doi: 10.1152/ajpendo.00524.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Folick A, et al. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science. 2015;347:83–86. doi: 10.1126/science.1258857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han S, et al. Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature. 2017;544:185–190. doi: 10.1038/nature21686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gillingham LG, et al. Dietary Monounsaturated Fatty Acids Are Protective Against Metabolic Syndrome and Cardiovascular Disease Risk Factors. Lipids. 2011;46:209–228. doi: 10.1007/s11745-010-3524-y. [DOI] [PubMed] [Google Scholar]
- 83.O’Rourke EJ, et al. ω-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 2013;27:429–440. doi: 10.1101/gad.205294.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Niso-Santano M, et al. Unsaturated fatty acids induce non-canonical autophagy. EMBO J. 2015;34:1025–1041. doi: 10.15252/embj.201489363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gonzalez-Covarrubias V, et al. Lipidomics of familial longevity. Aging Cell. 2013;12:426–434. doi: 10.1111/acel.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shmookler Reis RJ, et al. Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging. 2011;3:125–147. doi: 10.18632/aging.100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buchovecky CM, et al. A suppressor screen in Mecp2 mutant mice implicates cholesterol metabolism in Rett syndrome. Nat Genet. 2013;45:1013–1020. doi: 10.1038/ng.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scheibye-Knudsen M, et al. A High-Fat Diet and NAD+ Activate Sirt1 to Rescue Premature Aging in Cockayne Syndrome. Cell Metab. 2014;20:840–855. doi: 10.1016/j.cmet.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang P, et al. Seizure control by ketogenic diet-associated medium chain fatty acids. Neuropharmacology. 2013;69:105–114. doi: 10.1016/j.neuropharm.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cox PJ, et al. Nutritional Ketosis Alters Fuel Preference and Thereby Endurance Performance in Athletes. Cell Metab. 2016;24:256–268. doi: 10.1016/j.cmet.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 91.Yang K, Han X. Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem Sci. 2016;41:954–969. doi: 10.1016/j.tibs.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bozek K, et al. Lipidome determinants of maximal lifespan in mammals. Sci Rep. 2017;7 doi: 10.1038/s41598-017-00037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu PJ. Dauer. WormBook. 2017 doi: 10.1895/wormbook.1.144.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rubinsztein DC, et al. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 95.Shatz O, et al. Complex Relations Between Phospholipids, Autophagy, and Neutral Lipids. Trends Biochem Sci. 2016;41:907–923. doi: 10.1016/j.tibs.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 96.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Graef M, et al. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24:2918–2931. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Young ARJ, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 99.Longatti A, et al. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Velázquez AP, et al. Lipid droplet–mediated ER homeostasis regulates autophagy and cell survival during starvation. J Cell Biol. 2016;212:621–631. doi: 10.1083/jcb.201508102. [DOI] [PMC free article] [PubMed] [Google Scholar]