Abstract
Mutations and single nucleotide polymorphisms of AT-rich interactive domain-containing protein 5B (ARID5B) are involved in the oncogenesis of acute lymphoblastic leukemia (ALL) and treatment outcomes. However, ARID5B expression and clinical significance in ALL remain unclear. We found ARID5B is significantly down-regulated in ALL compared to healthy bone marrow controls. ARID5B also interacts with PHD finger protein 2 (PHF2). Low expression of ARID5B (ARID5Blow) or ARID5B and PHF2 (ARID5BlowPHF2low) is correlated with the markers of cell proliferation and poor prognosis in ALL patients. Ikaros directly regulates ARID5B expression in ALL. Restoring Ikaros function by Casein Kinase II inhibition also promotes ARID5B expression through recruitment of trimethylation of lysine 4 on histone H3 (H3K4me3) at its promoter region. In summary, our data show that aberrant expression of ARID5B and PHF2 is related to leukemic cell proliferation and several poor prognostic markers. Our data indicate ARID5Blow expression, particularly ARID5BlowPHF2low expression, is linked to Ikaros dysfunction and involved in the oncogenic effect of high-risk ALL, which may represent a high-risk subgroup of ALL.
Introduction
The complex of AT-rich interactive domain-containing protein 5B (ARID5B) formed with PHD finger protein 2 (PHF2) induces the demethylation of lysine 9 di-methylation on histone H3 (H3K9me2) to activate the transcription of the target genes1,2. ARID5B is widely expressed throughout the human body. However ARID5B dysfunction appears to be closely linked with leukemia2–10. ARID5B mutations /SNPs (single nucleotide polymorphisms) are reported to be involved in the oncogenesis of acute lymphoblastic leukemia (ALL) and treatment outcome3–10. Reports also showed that ARID5B knockdown impairs cell cycling by up-regulating p21, and contributes to methotrexate (MTX) and 6-mercaptopurine (6-MP) resistance and eventual relapse3–10. We observed that PHF2 expression is down-regulated in ALL cells. Until now, the clinical significance of ARID5B expression has not been determined in ALL patients.
Ikaros, the product of the IKZF1 gene, is not only an essential transcription factor for lymphocyte development but also a key suppressor in leukemogenesis11,12. The profile of Ikaros’ global genomic binding has been identified in ALL cells13–16. Ikaros binding sites are observed at the ARID5B promoter using ChIP-seq. We reported that Casein Kinase II (CK2) inhibition could restore the leukemia suppressor activity of Ikaros and CK2 inhibitors are the activator of the Ikaros function12–15. We demonstrated that once activated, Ikaros regulates the expression of gene targets by histone modification mechanism, and that it can induce transcription activation of its target genes by recruitment of H3K4me3 in ALL13–17. Here, we studied ARID5B expression in patients with ALL and discovered that ARID5Blow expression is linked to the markers of leukemia cell proliferation and that ARID5BlowPHF2low expression is possibly a poor prognostic indicator in patients with ALL. We also show that ARID5Blow expression is closely related with IKZF1 gene deletion in B-ALL. Our data manifest that Ikaros directly modulates ARID5B expression and that restoring Ikaros function in ALL cells from patients promotes ARID5B expression through the acquisition of H3K4me3. Our results identify the oncogenic effects of the ARID5BlowPHF2low expression pattern and its association with Ikaros dysfunction, which may reveal a novel high-risk subgroup of ALL.
Results
Laboratory characteristics in patients with low ARID5B expression
The mRNA level of ARID5B in the adult ALL patients’ bone marrow samples was significantly lower than those in normals (Fig. 1). Similarly, the cohort studies in B-cell ALL (B-ALL) and T-cell ALL (T-ALL) (Fig. S1) showed that ARID5B expression in mRNA levels was significantly lower than that in B cells from healthy controls (Fig. S1). The laboratory features were compared in patients with B-ALL by dividing them into two groups: high ARID5B mRNA levels (ARID5Bhigh) or low ARID5B mRNA levels (ARID5Blow) (Table 1 and Table S1). A significantly higher median percentage of BM blasts (90.0% vs. 84.6%, P = 0.037) and a significantly higher percentage of cases positive for CD34 (CD34+), the stem cell marker (88.8% vs. 37.5%, P = 0.000) or CD33 (CD33+), the myeloid marker (48.5% vs. 25.0%, P = 0.046) were observed in patients with low ARID5B mRNA level compared to that of high level. Similarly, low ARID5B mRNA level in patients was correlated with a higher frequency of cases positive for expression of Ikaros isoform 6 (IK6+), the gene product of the most common IKZF1 deletion isoform (42.5% vs. 20.0%, P = 0.042), and also a lower median hemoglobin (HGB) and platelet (PLT) count compared to patients with high ARID5B expression (Table S1). We discovered that B-ALL patients with low ARID5B expression represented a cohort with a significantly higher percentage of those requiring more than 4 weeks to reach complete remission (CR), a poor prognostic indicator in ALL, (51.4% vs. 16.0%, P = 0.002), as compared to that with high ARID5B expression (Table S1). However, among T-ALL patients, the low and high ARID5B expression groups did not show significantly different representation in the patient cohort (data not shown).
Fig. 1. ARID5B expression in ALL.
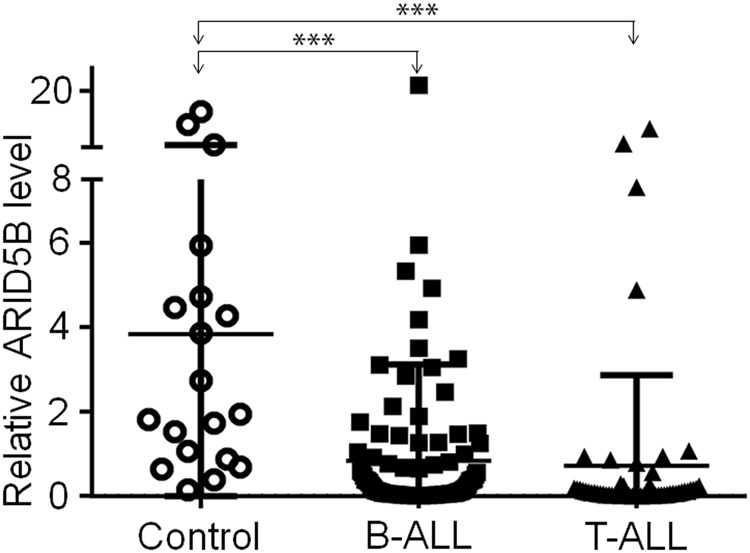
ARID5B expression in B-ALL (N = 123) and T-ALL (n = 57) and normal bone marrow controls (n = 19). ***p < 0.001
Table 1.
Significant correlation of ARID5BlowPHF2low expression with high-risk markers in B-ALL
| Characteristics | ARID5B low PHF2 low | non-ARID5B low PHF2 low | Univariate analyses (Chi-Square Tests) | Multivariate analyses (Multivariate Cox model) | |
|---|---|---|---|---|---|
| P value | P value | HR(95% CI) | |||
| IKZF1 deletion (IK6 expressing) (%) | 49.3 | 15.8 | 0.001 | 0.001 | 0.062 (0.013–0.298) |
| Blasts (%) median (range) bone marrow | 91.2 (59.0–100.0) | 82.4 (28.0–98.0) | 0.000 | 0.038 | 0.005 (0.000–0.742) |
| Extramedullary infiltration (%) spleen | 50.0 | 22.9 | 0.008 | 0.964 | 1.032 (0.264–4.029) |
| Stem cell marker CD34 + (%) | 88.2 | 55.6 | 0.000 | 0.135 | 0.370 (0.100–1.362) |
| Myeloid marker CD33 + (%) | 50.9 | 28.6 | 0.036 | 0.711 | 1.307 (0.317–5.381) |
| Time to CR after treatment is > 4 weeks (%) | 53.0 | 21.2 | 0.003 | 0.002 | 0.132 (0.036–0.478) |
Correlation of ARID5BlowPHF2low expression with clinical features in B-ALL
ARID5B and PHF2 interact with one another1,2. We found that ARID5B mRNA levels were positively correlated with PHF2 expression in the microarray analysis of B-ALL and T-All cohort studies (Fig. S2). We analyzed the co-occurrence of low-level ARID5B and low-level PHF2 expression (ARID5BlowPHF2low) and its association with clinical features (Table S2). ARID5BlowPHF2low expression was correlated to a higher percentage of cases with splenomegaly (50.0% vs. 22.9%, P = 0.008) and a lower PLT count (109/L) (32.0 vs. 58.5, P = 0.020) when compared to patients that were non-ARID5BlowPHF2low (Table S2). Moreover, the percentage of bone marrow blasts, a direct marker of high leukemic cell proliferation, showed significantly higher in ARID5BlowPHF2low than that in none-ARID5BlowPHF2low(91.2% vs. 82.4%, P = 0.000), and multivariate analyses confirmed this result (HR 0.005, 95% CI [0.000, 0.742]; P = 0.038) (Table 1).
We observed the correlation between ARID5BlowPHF2low expression and several poor prognostic markers. A higher percentage of the ARID5BlowPHF2low cases were positive for CD34 (88.2% vs. 55.6%, P = 0.000) or CD33 (50.9% vs. 28.6%, P = 0.036). Importantly, the low expression cohort also showed a significantly higher frequency of Ik6 + cases (49.3% vs. 15.8%, P = 0.001), and a substantially higher percentage of patients with a CR time ≥ 4 weeks when compared to the none-ARID5BlowPHF2low expression cohort and confirmed by multivariable analysis (Table 1).
We looked over the relationship between ARID5B expression and survival. No significant differences were identified in the overall survival (OS) of the patients with ARID5Blow or ARID5BlowPHF2low expression as compared to those in the ARID5Bhigh or none-ARID5BlowPHF2low cohorts, respectively (Fig. S3 and Fig. S4). However, we did observe a trend towards a shortened relapse-free survival (RFS) in patients with ARID5Blow expression, especially the ARID5BlowPHF2low cohort, compared to those with ARID5Bhigh or non-ARID5BlowPHF2low expression, respectively (Fig. S3 and Fig. S4).
The ARID5B expression is regulated by Ikaros in ALL
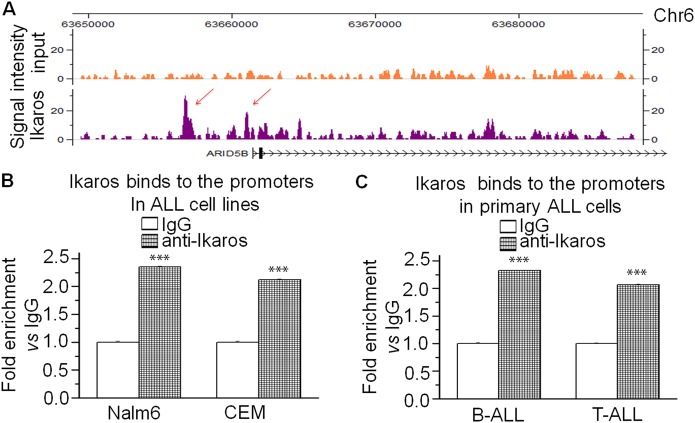
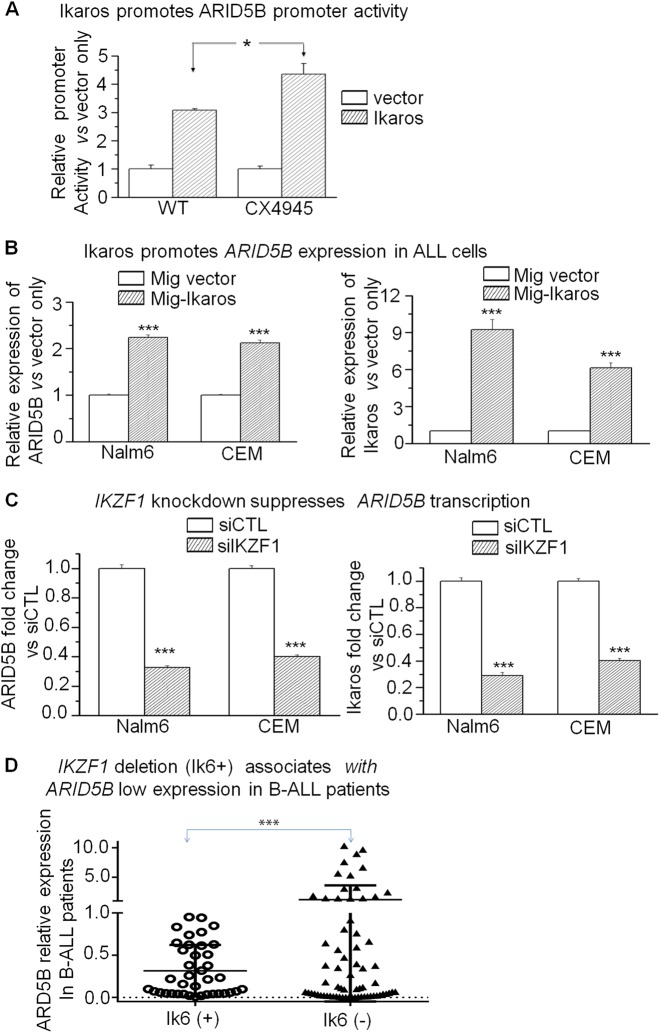
To understand the underlying mechanism of ARID5B low expression in ALL, we studied Ikaros binding sites present in the ARID5B promoter region by ChIP-seq assay, in Nalm6 (Fig. 2a) and primary B-ALL cells (Fig S5)13,14. qChIP assay confirmed Ikaros recruitment at ARID5B promoter in the leukemia cell lines (Fig. 2b) and primary cells (Fig. 2c). These results suggest Ikaros has a direct regulation on ARID5B transcription. We further showed that Ikaros increases promoter activity of ARID5B using the luciferase reporter assay (Fig. 3a). Ikaros transduction of Nalm6 and CEM cells results in the significant increase of ARID5B expression (Fig. 3b). Conversely, efficient Ikaros knockdown significantly decreased ARID5B mRNA level in both of these cell lines (Fig. 3c).
Fig. 2. Ikaros binding sites at ARID5B promoter in B-ALL cells were determined by ChIP-seq (a).
b, c Ikaros binding at ARID5B promoter was validated by qChIP assay in b ALL cell lines and (c) primary ALL cells. Graphed data are the mean ± SD of triplicates representative of one of 3 independent experiments (a, b) or 3 patient samples (c). ***p < 0.001
Fig. 3. Ikaros induces ARID5B expression in ALL.
a The activity of the ARID5B promoter was assessed with transfection of Ikaros or control vector in HEK293 cells with or without the CK2 inhibitor, CX-4945, by luciferase reporter assay; (b) Nalm6 and CEM cells were transduced to express Ikaros (Mig-Ikaros) or with empty vector (Mig vector) and assessed by qPCR for expression of ARID5B. Graphed indicates the relative ARID5B expression; (c) Nalm6 and CEM cells were treated with IKZF1 siRNA (si-IKZF1) or control siRNA (siCTL) and assessed by qPCR for expression of ARID5B. Graphed is the relative expression of ARID5B; (d) Patients that were positive (n = 39) vs. negative (n = 68) for Ik6, the expressed gene product of the IKZF1 deletion, were assessed by qPCR for expression of ARID5B. The ARID5B expression in a–c expresses as the mean ± SD of triplicates representative of one of 3 independent experiments. *p < 0.05, ***p < 0.01
Association of IKZF1 deletion with ARID5B low expression in B-ALL patients
Microarray analysis in B-ALL and T-ALL cohorts18–20 showed the positive correlation of IKZF1 mRNA levels with ARID5B expression (Fig. S6). A significant ARID5B low expression was observed in B-ALL patients that were IK6 + (0.3153 ± 0.0938 vs. 1.2052 ± 0.58441, P = 0.02439) (Fig. 3d), which is consistent with our finding that the ARID5Blow cohort has a significantly higher percentage of IK6 + cases in B-ALL (Table S1). These data reveal the contribution of the IKZF1 genetic defects to low ARID5B expression in B-ALL patients.
CK2 inhibitor CX-4945 promotes ARID5B transcription by enhancing Ikaros activity
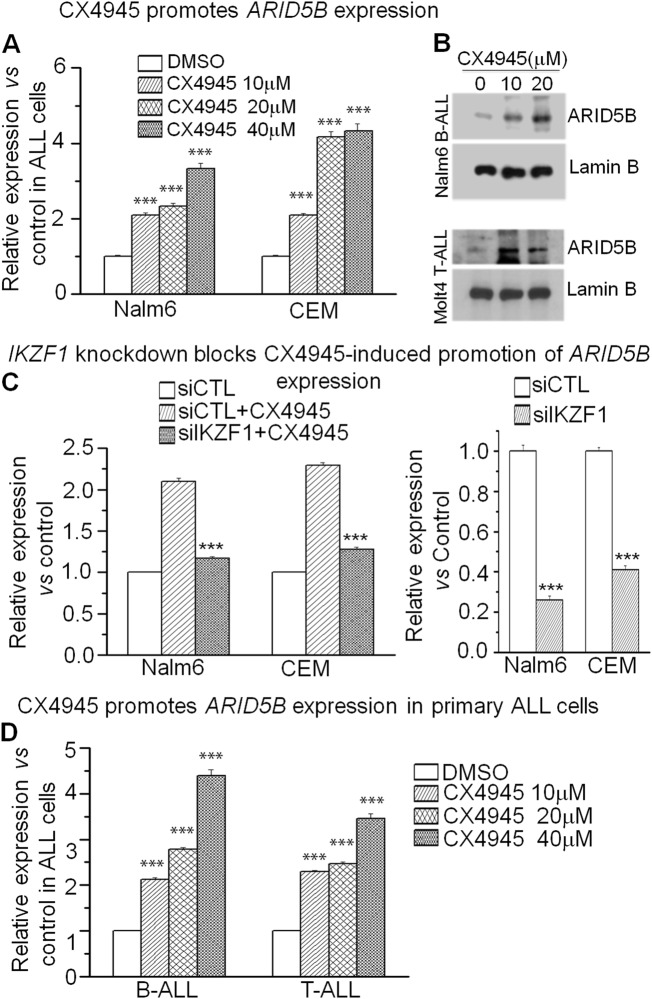
Our previous studies show that the CK2 inhibitor, CX-4945, can restore Ikaros’ tumor suppressor activity13. CX-4945 treatment further improves Ikaros-mediated increase of ARID5B promoter activity when compared to that without treatment (Fig. 3a). Using qPCR, we showed that CX-4945 treatment in Nalm6 and CEM cells enhances ARID5B mRNA level in a dose-dependent manner (Fig. 4a). Western blot data showed that CX-4945 treatment also increases the ARID5B protein level as compared to that of DMSO control in the two cell lines (Fig. 4b). Moreover, Ikaros knockdown significantly attenuates CX-4945-induced increases in the ARID5B mRNA level in ALL cell lines (Fig. 4c). The effect of CX-4945 on ARID5B mRNA levels is also observed in primary B-/T-ALL cells (Fig. 4d). These results indicate that CX-4945 promotes ARID5B transcription by increasing Ikaros function as tumor suppressor in ALL.
Fig. 4. Ikaros dependence on CX-4945 promoting ARID5B expression.
a Treatment with CX-4945 induces an increase in ARID5B expression in Nalm6 and CEM cells; ***p < 0.001 compared to DMSO control. b Protein levels of ARID5B as evaluated by Western blot in the indicated cells that were incubated with different doses (10 μM, 20 μM) of CX-4945 or DMSO control (0) for 48 h. Lamin B was used for loading control. c Effect of Ikaros shRNA knockdown on the CX-4945-induced promotion of ARID5B expression. ***p < 0.01 compared to siCTL + CX4945 group; (d) CX-4945 promotes ARID5B expression in primary ALL cells; ***p < 0.001 compared to the control. Graphed data in A-D represents the mean+/– SD of triplicates representative of one of 3 independent experiments
Increasing Ikaros activity by CK2-inhibition promotes H3K4me3 occupancy at the ARID5B promoter
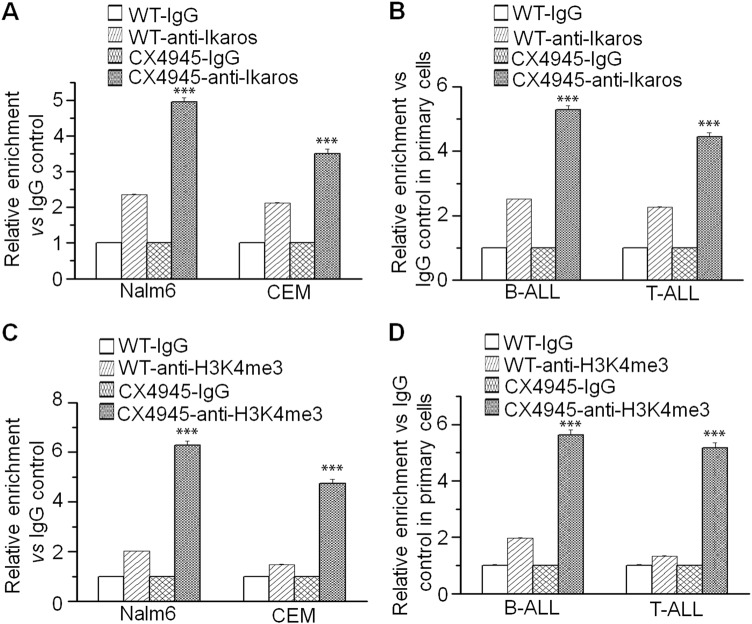
Ikaros regulates target gene expression through histone modification mechanism14. To explored if Ikaros regulates ARID5B expression also via epigenetic mechanisms, we performed ChIP assays to amplify the resulting ARID5B promoter sequences. Our data show that the Ikaros binding to the ARID5B promoter is significantly increased upon CX-4945 treatment not only in Nalm6 and CEM cells (Fig. 5a), but also in primary B-/T-ALL cells (Fig. 5b). CX-4945 treatment also results in the increases of H3K4me3 recruitment at the ARID5B promoter in the cell lines (Fig. 5c), and in the primary cells (Fig. 5d).
Fig. 5. Chromatin switches upon CX-4945 treatment.
Indicated cell lines and primary cells were treated with 10 μM CX-4945 or with DMSO control and evaluated by qChIP for Ikaros binding (a, b) and the H3K4me3 histone mark (c, d) at the ARID5B promoter in the indicated cells. ***p < 0.001 compared to WT-anti-Ikaros control. Graphed data in a–d are the mean+/– SD of triplicates representative of one of 3 independent experiments or 3 patient samples
Discussion
The ARID5B gene product is widely expressed in the human tissue and has been linked to leukemia2–10,21–25. ARID5B mutations /SNPs are linked to the ALL development and adverse treatment outcomes4. Aberrant ARID5B expression halts B-lymphocyte maturation in the developing fetus and contributes to leukemogenesis21. However, the mRNA level of ARID5B in primary ALL and its association with clinical findings have not been reported. Our findings show the correlation of ARID5B expression with a difference in clinical features in ALL. We previously showed that PHF2 is down-regulated in ALL26. We saw that ARID5B and PHF2 expression were positively correlated in ALL and that ARID5BlowPHF2low expression is associated with leukemic cell proliferation (high bone marrow blasts and splenomegaly, low HGB and PLT), as well as a poor prognosis (high percentage of Ik6+, ≥4 weeks to reach CR upon treatment, and CD33+) in B-ALL patients. Next, we showed that Ik6 expression, the most common IKZF1 deletion is significantly linked to ARID5B low expression in B-ALL. We further demonstrated that ARID5B is a direct gene target of Ikaros, the IKZF1 gene product, in ALL. Finally, our study identifies a potential high-risk subgroup of ALL with ARID5BlowPHF2low expression and reveals the oncogenic effect of ARID5BlowPHF2low expression and its correlation with Ikaros dysfunction in ALL.
There have been many reports that SNPs affect gene expression. In addition to reports that ARID5B SNPs increase the risk of ALL, several reports also indicate that both ARID5B and IKZF1 SNPs are positively associated with ALL4–9,22–25,27–29. However, no reports are involved in exploring the relationship between ARID5B SNPs and ARID5B expression. Our data reveal that the IKZF1 genetic defect (Ik6 expression) is associated with ARID5B low expression and that Ikaros directly promotes ARID5B expression. This information also suggests that the association of ARID5B and IKZF1 SNPs with an increased risk of ALL may result from the low expression of ARID5B and IKZF1, although the effects of ARID5B and IKZF1 SNPs on their expression need to be further investigated.
Transcriptional and epigenetic abnormalities are key factors in oncogenesis. The ARID5B-PHF2 complex is involved in the activation of tumor suppressors, such as p53, through its effect on methylation30. Our data shows that the correlation between ARID5BlowPHF2low expression and leukemic cell proliferation, with poor prognostic markers in B-ALL. We also found that restoring Ikaros function by CK2 inhibition could increase ARID5B and PHF2 expression, as well as increase H3K4me3 binding at the promoter region. This data is the first to indicate the regulatory mechanism underlying ARID5B gene expression. It also suggests that targeting transcriptional and epigenetic abnormalities is a potential strategy for developing effective new therapeutics for ALL.
In conclusion, we show that ARID5BlowPHF2low expression is correlated with markers for leukemic cell proliferation and poor outcome. Our results further reveal the effects of ARID5BlowPHF2low expression on ALL oncogenesis and identify a possible subgroup of high-risk ALL with characterization of both ARID5BlowPHF2low expression and Ikaros dysfunction.
Materials and methods
Patient samples and therapies
The 164 bone marrow samples were obtained from patients with ALL, diagnosed at our institutes between 2008 and 2016. All of the patients (107 B-ALL and 57 T-ALL), ages 12–77 years old, were recruited in the cohort study, with diagnoses based on the 2008 revision of the WHO Diagnosis and Classification of ALL. As controls, 19 normal bone marrow samples were used. Following the Declaration of Helsinki, the informed consent was documented by all patients before recruitment.
As previously published (CALLG2008)31, patients received either VDCLP therapy, which consists of Vincristine (V), Daunorubicin (D), Cyclophosphamide (C), L- Asparaginase (L), and Prednisone (P), or CAT therapy, which contains C, Cytarabine (A), Thioguanine (T), high-dose Mitoxantrone (M), and methotrexate/L-Asparaginase (Met/Asp) for induction or early induction. For late consolidation, VDLP or the combined therapy of CVCED (E: Epipodophyllotoxin and D: Dexamethasone), and high-dose Met/Asp, E and A were utilized. Lastly, 6-Mercaptopurine and M were used during maintenance therapy. Imatinib was also added to regimens for patients with Ph (+) ALL starting on day 15 of induction therapy.
The Ethics Committee of Zhongda Hospital Southeast University and the First Affiliated Hospital of Nanjing Medical University, Nanjing, China approved this study.
Cytogenetic and molecular analyses
Ikaros 6 (IK6), the most common expression product from the IKZF1 deletion, was detected as previously described32. Briefly, the isolated genomic DNA with QIAamp DNA Blood Mini Kit (Qiagen, Germantown, MD, USA) was utilized for performing the genomic PCR amplification for detection of IKZF1 deletion on exons 4–7 (△4–7). The flanking deletion breakpoints of IK6 was characterized by direct sequencing of the resulted PCR products. Cytogenetics was also analyzed as described32.
Quantitative Real-time PCR (qPCR) assay
For qPCR of patient samples, the real-time PCR system (StepOne Plus 7500) from Applied Biosystem-Thermofisher (Foster, CA, USA) was utilized. Briefly, cDNA was generated from total RNA (1.0 μg) using SuperScript II first-Strand synthesis kit (Invitrogen, Carlsbad, CA, USA) with poly d(T)20 primers. The genes’ mRNA level was analyzed from the resulting cDNAs on the machine by using the specific primer of each gene. Primers for the qPCR of ARID5B are: Sense: 5′- TCTTAAAGGCAGACCACGCAA −3′, Anti-sense: 5′- TGCCATCGGAATTGTTGTTGG −3′. Primers for qPCR of 18 s rRNA were as previously reported13–15,17,31. Two groups of the cohorts were divided as patients with high or those with low ARID5B expression (4th quartile vs. 1st–3rd quartiles), and SPSS 20.0 was utilized for determination of the cut-off value. ARID5B or PHF2 expression was calculated in the individual sample by a formula as previously described15–17,31–33. The formula was determined from the value of a scatter Ct graph in a serially diluted template standard. ARID5B or PHF2 expression level was normalized to housekeeping gene 18 s rRNA with a formula of ARID5B/18 s rRNA or PHF2/18 s rRNA.
The qPCR assay was also used to analyze ARID5B mRNA levels in the cell lines. Results of drug treatment, Ikaros overexpression, or IKZF1 knockdown were divided by those acquired with housekeeping gene18s rRNA and expressed as fold change over DMSO or vector controls.
Cell culture
The previously described Nalm6 cell line34, is verified by the American Type Culture Collection (ATCC, Manassas, VA). The CCRF-CEM (CEM) and HEK 293 T cell lines were obtained from ATCC. DMEM (Cellgro, Tewksbury, MA, USA), supplemented with 10% FBS and 1% L-glutamine (Cellgro, Tewksbury, MA, USA) was used for culture of HEK 293 T cells; and the 10% FBS (Hyclone, Logon, Utah, USA) supplemented RPMI 1640 medium (Cellgro, Tewksbury, MA, USA) for culturing Nalm6, CEM, and primary human B-/T-ALL cells at 37 °C in a 5% CO2 humidified atmosphere. CX-4945 was obtained from Selleckchem (S2248, Houston, USA). Cells with or without CX-4945 treatment were used for total RNA isolation, as well as western blot.
Plasmid construction and retroviral gene transduction
Human full-length Ikaros (IKZF1) cDNA was cloned into the retroviral vector, MSCV-IRES-GFP (MIG) with BglII and EcoRI site15,34,35. The plasmids were transiently transfected into amphotropic packaging HEK 293 cell lines and the retroviruses were generated and concentrated as described15,34,35. Cells plated on a 24-well plate at 4 × 10E5 cells/well were centrifuged 1400×g in retroviral supernatants plus 12.5 mg/ml polybrene, at 32 °C, for 1 h. The cells were further cultured in fresh media at 37 °C, 5% CO2 incubator for 3 days. The GFP(+) cells were sorted with BD FACS Aria SORP high-performance sorter (BD Biosciences, Sparks, MD, USA), and the sorted cells are cultured for further RNA isolation and ChIP assay.
Luciferase assay
LightSwitch luciferase reporter constructs for promoters of ARID5B were purchased from Active Motif-SwitchGear Genomics (Carlsbad, CA, USA). The transfection-ready promoter plasmid, or pLightSwitch-Rom vector, was transfected with Ikaros in pCDNA3.1 vector or vector only into HEK293 cells and the transient luciferase assay was done with or without 10 μM CX-4945 according to Switchgear Genomics manual by a luminometer as previously described14–17,31–36. Briefly, ARID5B promoter-reporter plasmids and pcDNA3.1-Ikaros or pcDNA3.1 vector were delivered into HEK293 cells in 24-well plates in a 1:3 ratio with the transfection reagent, lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The cells were lysed 24 h after transfection in 100 μl of lysis buffer (Active Motif-SwitchGear Genomics, Carlsbad, CA, USA). Half of the lysate was used for luciferase activity measurement on a GloMax Luminometer (Promega, Madison, WI, USA). The luciferase activity was determined as fold change of the values from the cells transfected with promoter construct relative to ones obtained from pLightSwitch-Rom vector-only control cells. Ikaros effect on the promoter activity was presented as a ratio of Ikaros-induced luciferase activity over that of the vector. The graphed data was the average of triplicates which is one representative of 3 independent experiments.
Western blot assay
Nuclear extracts were isolated by osmotic swelling and homogenization from the cells treated with different doses of CX-4945 and DMSO as controls1,3,14,15,25. Protein concentrations were determined by the quantitative Bradford assay. Total protein (20 μg) of each sample was used for the western blot assay as previously described13,15. ARID5B protein expression was detected with the anti-ARID5B antibody (ab226776, Abcam, Cambridge, MA, USA) and Lamin B was detected by the anti-Lamin B1 antibody (VPA00119, Bio-Rad, USA) as a loading control.
Quantitative chromatin immune precipitation (qChIP)
Chromatin from cells treated with CX-4945 was incubated with antibodies against Ikaros14,15,25. Cells were cross-linked in the 1% formaldehyde solution on ice and the cross-link reaction was ceased with 0.125 M glycine. The chromatin for Ikaros ChIP assay was prepared from 2 × 10E7 Nalm6 or CEM cells or primary leukemia cells (4–10 × 10E6) and fragmented with a Bioruptor (Diagenode, Denville, NJ) to obtain the average DNA size of 400 bp as previously described14,15,25. For ChIP assays, the chromatin was incubated with Dyneabeads-coated affinity-purified rabbit polyclonal anti-Ikaros antibody14,15,25 or normal rabbit IgG (Abcam, ab46540) as the control. The protein/DNA complexes were isolated with a Magnetic separator (Invitrogen, Carlsbad, CA, USA) and extensively washed with RIPA buffer. The ChIP’d DNA was eluted and reversely crosslinked. The resulted samples were further treated with proteinase K digestion, phenol/chloroform extraction, and RNaseA incubation. A QIAquick PCR Purification kit (QIAGEN) was used for recovering the ChIP’d DNAs. Enrichment of Ikaros-bound-ARID5B promoter in the ChIP’d DNA sample vs. that with normal rabbit IgG (ab171870, Abcam, Cambridge, MA, USA) as a control was measured by qPCR with the primers at ARID5B promoter(forward: 5′- GCAGTCGCTGTCCGTTCAA −3′, reverse: 5′- CAAGTGAGCAGTGCACACACA −3′)14,15,25. At least three technical replicates were performed for each assay. The relative Ikaros binding at the ARID5B promoter is expressed as the fold change of Ikaros-bound DNA vs. that of rabbit IgG controls. H3K4me3 qChIP assay was done using the same protocol as Ikaros qChIP, with the anti-H3K4me3 antibody (ab8580, Abcam, Cambridge, MA, USA), except using 1 × 10E7 cells for them as we previously reported14,15,25.
IKZF1 shRNA knockdown
A set of 4 pGFP-V-RS constructs containing unique human Ikaros (ikzf1) 29mer shRNA were purchased from Origene (Rockville, MD, USA). The optimal gene knockdown shRNA plasmid from the 4 constructs was tested and selected using the Neon Transfection System (Invitrogen, Carlsbad, CA, USA) for further studies. After transfection for one day, cells were observed with 80–90% (green cells) transfection efficiency and more than 95% cell viability. The cells incubated with 10 μM CX-4945 or non-treatment DMSO control for 2 days were harvested for total RNA isolation. The cells transfected with a scrambled shRNA (29-mer) vector were used as a control. Ikaros level was evaluated in the cells by qPCR with IKZF1 specific primer as previously reported15,35.
Statistical analyses
Median differences between the groups in the cohort study were tested utilizing a Mann–Whitney U-test. The univariate and multivariate Cox models were used for statistical analysis of frequency differences. The Kaplan-Meier analysis with the log-rank test was utilized to judge the significance for RFS and OS. The date of diagnosis was the initial point for OS, and RFS was started at the time of declared remission to that of patients achieving complete remission (CR). Living patients were counted on for survival at follow up. Data were graphed as mean value ± SEM (standard error of the mean). Analysis of variance (ANOVA) or Student t-test was used to evaluate the statistical significance for comparisons of two groups or comparing multiple groups, respectively.
Electronic supplementary material
Acknowledgements
This work is supported in part by Key Research and Technology Projects in Jiangsu Province (BE2017747); Milstein Medical Asian American Partnership (MMAAP) Foundation Research Project Award in Hematology (2017); The National Natural Science Foundation of China (81770172, 81270613); The Fundamental Research Funds for the Central Universities (2242017K40271, 2242016K40143); The Scientific Research Foundation for the Returned Overseas Chinese Scholars; State Education Ministry (39th); China Postdoctoral Science Foundation (20090461134); Jiangsu Province Key Medical Talents (RC2011077); Special grade of the financial support from China Postdoctoral Science Foundation (201003598); The Six Great Talent Peak Plan of Jiangsu (2010-WS-024) (to Z.G.). This work has also been partially supported by National Institutes of Health (NIH), National Cancer Institute (NCI) grants R01CA209829, (K.J.P. and S.D.) R01CA213912, Hyundai Hope on Wheels Scholar Grant, the Four Diamonds Fund of the Pennsylvania State University College of Medicine (to S.D. and C.S.); Bear Necessities Pediatric Cancer Foundation, Alex’s Lemonade Stand Foundation, and the John Wawrynovic Leukemia Research Scholar Endowment (to S.D.).
Author contributions
Z.G., Q.H., Y.G., Q.G., J.S., G.G., H.S., J.M., J.H., B.C., C.S. and S.D. performed experiments and analyzed data; C.S., Z.G. and S.D. designed and supervised data analysis; C.S., Z.G., S.D., J.S., G.G., L.S. and K.J.P. wrote the manuscript.
Availability of data and materials
In accordance with local health research ethics protocols, the patient datasets for the current study are not publicly accessible; however, it may be available from the corresponding author.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval and informed consent
The written informed consents were provided by all the patients in accordance with the Declaration of Helsinki before enrollment in the study. The Institutional Review Board of Zhongda Hospital Southeast University and the Nanjing Medical University, Nanjing, China, approved the study.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zheng Ge, Phone: +86-25-83262468, Email: Janege879@hotmail.com.
Chunhua Song, Phone: +717-531-1841, Email: csong@pennstatehealth.psu.edu.
Sinisa Dovat, Phone: +717-531-6012, Email: sdovat@pennstatehealth.psu.edu.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41389-018-0095-x).
References
- 1.Baba A, et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat. Cell Biol. 2011;13:668–675. doi: 10.1038/ncb2228. [DOI] [PubMed] [Google Scholar]
- 2.Okuno Y, et al. Novel insights into histone modifiers in adipogenesis. Adipocyte. 2013;2:285–288. doi: 10.4161/adip.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu H, et al. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J. Clin. Oncol. 2012;30:751–757. doi: 10.1200/JCO.2011.38.0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudant J, et al. ARID5B, IKZF1 and non-genetic factors in the etiology of childhood acute lymphoblastic leukemia: the ESCALE study. PLoS ONE. 2015;10:e0121348. doi: 10.1371/journal.pone.0121348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans TJ, et al. Confirmation of childhood acute lymphoblastic leukemia variants, ARID5B and IKZF1, and interaction with parental environmental exposures. PLoS ONE. 2014;9:e110255. doi: 10.1371/journal.pone.0110255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudant J, et al. Are ARID5B and IKZF1 polymorphisms also associated with childhood acute myeloblastic leukemia: the ESCALE study (SFCE)? Leukemia. 2013;27:746–748. doi: 10.1038/leu.2012.244. [DOI] [PubMed] [Google Scholar]
- 7.Linabery AM, et al. ARID5B and IKZF1 variants, selected demographic factors, and childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Leuk. Res. 2013;37:936–942. doi: 10.1016/j.leukres.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peyrouze P, et al. Genetic polymorphisms in ARID5B, CEBPE, IKZF1 and CDKN2A in relation with risk of acute lymphoblastic leukaemia in adults: a Group for Research on Adult Acute Lymphoblastic Leukaemia (GRAALL) study. Br. J. Haematol. 2012;159:599–602. doi: 10.1111/bjh.12063. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, et al. Association of three polymorphisms in ARID5B, IKZF1 and CEBPE with the risk of childhood acute lymphoblastic leukemia in a Chinese population. Gene. 2013;524:203–207. doi: 10.1016/j.gene.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 10.Yang W, et al. ARID5B SNP rs10821936 is associated with risk of childhood acute lymphoblastic leukemia in blacks and contributes to racial differences in leukemia incidence. Leukemia. 2014;24:894–896. doi: 10.1038/leu.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song C, et al. Regulation of Ikaros function by casein kinase 2 and protein phosphatase 1. World J. Biol. Chem. 2011;2:126–131. doi: 10.4331/wjbc.v2.i6.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dovat S, et al. Ikaros, CK2 kinase, and the road to leukemia. Mol. Cell Biochem. 2011;356:201–207. doi: 10.1007/s11010-011-0964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song C, et al. Targeting casein kinase II restores Ikaros tumor suppressor activity and demonstrates therapeutic efficacy in high-risk leukemia. Blood. 2015;126:1813–1822. doi: 10.1182/blood-2015-06-651505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song C, et al. Epigenetic regulation of gene expression by Ikaros, HDAC1 and Casein Kinase II in leukemia. Leukemia. 2016;30:1436–1440. doi: 10.1038/leu.2015.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge Z, et al. Clinical significance of high c-MYC and low MYCBP2 expression and their association with Ikaros dysfunction in adult acute lymphoblastic leukemia. Oncotarget. 2015;6:42300–42311. doi: 10.18632/oncotarget.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gowda C, et al. Regulation of cellular proliferation in acute lymphoblastic leukemia by Casein Kinase II (CK2) and Ikaros. Adv. Biol. Regul. 2017;63:71–80. doi: 10.1016/j.jbior.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, et al. Transcriptional Regulation of JARID1B/KDM5B Histone Demethylase by Ikaros, Histone Deacetylase 1 (HDAC1), and Casein Kinase 2 (CK2) in B-cell Acute Lymphoblastic Leukemia. J. Biol. Chem. 2016;291:4004–4018. doi: 10.1074/jbc.M115.679332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homminga I, et al. Characterization of a pediatric T-cell acute lymphoblastic leukemia patient with simultaneous LYL1 and LMO2 rearrangements. Haematologica. 2012;97:258–261. doi: 10.3324/haematol.2011.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang H, et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115:1394–1405. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harvey RC, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116:4874–4884. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrena S. Aberrant expression of tetraspanin molecules in B-cell chronic lymphoproliferative disorders and its correlation with normal B-cell maturation. Leukemia. 2005;19:1376–1383. doi: 10.1038/sj.leu.2403822. [DOI] [PubMed] [Google Scholar]
- 22.Bhandari P, et al. Association of Genetic Variants in ARID5B, IKZF1 and CEBPE with Risk of Childhood de novo B-Lineage Acute Lymphoblastic Leukemia in India. Asian Pac. J. Cancer Prev. 2016;17:3989–3995. [PubMed] [Google Scholar]
- 23.Gharbi H, et al. Association of genetic variation in IKZF1, ARID5B, CDKN2A, and CEBPE with the risk of acute lymphoblastic leukemia in Tunisian children and their contribution to racial differences in leukemia incidence. Pediatr. Hematol. Oncol. 2016;33:157–167. doi: 10.3109/08880018.2016.1161685. [DOI] [PubMed] [Google Scholar]
- 24.Hsu LI, et al. Association of genetic variation in IKZF1, ARID5B, and CEBPE and surrogates for early-life infections with the risk of acute lymphoblastic leukemia in Hispanic children. Cancer Causes Control. 2015;26:609–619. doi: 10.1007/s10552-015-0550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burmeister T, et al. Germline variants in IKZF1, ARID5B, and CEBPE as risk factors for adult-onset acute lymphoblastic leukemia: an analysis from the GMALL study group. Haematologica. 2014;99:e23–e25. doi: 10.3324/haematol.2013.090720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge Z, et al. Plant homeodomain finger protein 2 as a novel IKAROS target in acute lymphoblastic leukemia. Epigenomics. 2018;10:59–69. doi: 10.2217/epi-2017-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartram T, et al. Childhood acute lymphoblastic leukemia-associated risk-loci IKZF1, ARID5B and CEBPE and risk of pediatric non-Hodgkin lymphoma: a report from the Berlin-Frankfurt-Munster Study Group. Leuk. Lymphoma. 2015;56:814–816. doi: 10.3109/10428194.2014.933479. [DOI] [PubMed] [Google Scholar]
- 28.Lin CY, et al. High-resolution melting analyses for genetic variants in ARID5B and IKZF1 with childhood acute lymphoblastic leukemia susceptibility loci in Taiwan. Blood Cells Mol. Dis. 2014;52:140–145. doi: 10.1016/j.bcmd.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Pastorczak A, et al. Role of 657del5 NBN mutation and 7p12.2 (IKZF1), 9p21 (CDKN2A), 10q21.2 (ARID5B) and 14q11.2 (CEBPE) variation and risk of childhood ALL in the Polish population. Leuk. Res. 2011;35:1534–1536. doi: 10.1016/j.leukres.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 30.Lee KH, et al. PHF2 histone demethylase acts as a tumor suppressor in association with p53 in cancer. Oncogene. 2015;34:2897–2909. doi: 10.1038/onc.2014.219. [DOI] [PubMed] [Google Scholar]
- 31.Ge Z, et al. Targeting High Dynamin-2 (DNM2) expression by restoring Ikaros function in acute lymphoblastic leukemia. Sci. Rep. 2016;6:38004. doi: 10.1038/srep38004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu P, et al. Expression of dominant-negative Ikaros isoforms and associated genetic alterations in Chinese adult patients with leukemia. Ann. Hematol. 2012;91:1039–1049. doi: 10.1007/s00277-012-1415-4. [DOI] [PubMed] [Google Scholar]
- 33.Guo X, et al. Characterization of LEF1 High Expression and Novel Mutations in Adult Acute Lymphoblastic Leukemia. PLoS ONE. 2015;10:e0125429. doi: 10.1371/journal.pone.0125429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campana D, et al. Human B cell development. I. Phenotypic differences of B lymphocytes in the bone marrow and peripheral lymphoid tissue. J. Immunol. 1985;134:1524–1530. [PubMed] [Google Scholar]
- 35.Wang H, et al. Protein phosphatase 1 (PP1) and Casein Kinase II (CK2) regulate Ikaros-mediated repression of TdT in thymocytes and T-cell leukemia. Pediatr. Blood Cancer. 2014;61:2230–2235. doi: 10.1002/pbc.25221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popescu M, et al. Ikaros stability and pericentromeric localization are regulated by protein phosphatase 1. J. Biol. Chem. 2009;284:13869–13880. doi: 10.1074/jbc.M900209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In accordance with local health research ethics protocols, the patient datasets for the current study are not publicly accessible; however, it may be available from the corresponding author.