Abstract
Synaptic function and neurotransmitter release are regulated by specific proteins. Cortical neuronal differentiation of human induced pluripotent stem cells (hiPSC) provides an experimental model to obtain more information about synaptic development and physiology in vitro. In this study, expression and secretion of the synaptic proteins, neurogranin (NRGN), growth-associated protein-43 (GAP-43), synaptosomal-associated protein-25 (SNAP-25) and synaptotagmin-1 (SYT-1) were analyzed during cortical neuronal differentiation. Protein levels were measured in cells, modeling fetal cortical development and in cell-conditioned media which was used as a model of cerebrospinal fluid (CSF), respectively. Human iPSC-derived cortical neurons were maintained over a period of at least 150 days, which encompasses the different stages of neuronal development. The differentiation was divided into the following stages: hiPSC, neuro-progenitors, immature and mature cortical neurons. We show that NRGN was first expressed and secreted by neuro-progenitors while the maximum was reached in mature cortical neurons. GAP-43 was expressed and secreted first by neuro-progenitors and its expression increased markedly in immature cortical neurons. SYT-1 was expressed and secreted already by hiPSC but its expression and secretion peaked in mature neurons. SNAP-25 was first detected in neuro-progenitors and the expression and secretion increased gradually during neuronal stages reaching a maximum in mature neurons. The sensitive analytical techniques used to monitor the secretion of these synaptic proteins during cortical development make these data unique, since the secretion of these synaptic proteins has not been investigated before in such experimental models. The secretory profile of synaptic proteins, together with low release of intracellular content, implies that mature neurons actively secrete these synaptic proteins that previously have been associated with neurodegenerative disorders, including Alzheimer's disease. These data support further studies of human neuronal and synaptic development in vitro, and would potentially shed light on the mechanisms underlying altered concentrations of the proteins in bio-fluids in neurodegenerative diseases.
Keywords: Neurogranin, Synaptotagmin-1, SNAP-25, GAP-43, Development, Stem cells
Abbreviations: NRGN, neurogranin; SYT-1, synaptotagmin-1; GAP-43, growth-associated protein-43; (SNAP-25, synaptosomal-associated protein-25; hiPSCs, human induced pluripotent stem cells; LTP, long term potentiation; LDH, lactate dehydrogenase
Highlights
-
•
Human iPSC-derived cortical neurons express and secrete synaptic proteins.
-
•
NRGN, GAP-43 and SNAP-25 are first expressed and secreted by neuro-progenitors.
-
•
SYT-1 is expressed and secreted by stem cells.
-
•
Synaptic proteins are secreted the highest when mature cortical neurons are formed.
1. Introduction
Neurons are connected by synapses that form the functional units in neuronal communication. Because of their ability to undergo various chemical and electrical changes during learning processes, synapses are the most essential components of the connectome in the brain; a synapse serves as a transmission line between the pre- and post-synaptic terminal of two connected units in the connectome (Abbas et al., 2018). Synapses contain specific scaffolding and signaling molecules and receptors that are essential for neurotransmitter release and signal transduction (Eagle et al., 1959). Synapse formation in the human neocortex begins in the third trimester of gestation and continues during the first two postnatal years. Synaptogenesis occurs during the same period, when dendritic and axonal growth and myelination are initiated (Huttenlocher and Dabholkar, 1997). Pre- and post-synaptic proteins interact to promote synaptic development and tuning of cortical circuits (Pinto et al., 2013). In CNS disorders, particularly in neurodegenerative diseases such as Alzheimer's disease (AD), synapses become dysfunctional and undergo degeneration (Blennow et al., 1996; Davies et al., 1987; DeKosky and Scheff, 1990). Several synaptic proteins including neurogranin (NRGN), synaptosomal-associated protein-25 (SNAP-25), growth-associated protein-43 (GAP-43), synaptotagmin-1 (SYT-1) and others can be detected in cerebrospinal fluid (CSF), and have been identified as being released at increased concentrations during neurodegeneration (Blennow, 2004; Brinkmalm et al., 2014; Thorsell et al., 2010; Öhrfelt et al., 2016), while their protein concentrations decreased in the brain (Davidsson and Blennow, 1998). These synaptic proteins are detected in CSF from healthy individuals (albeit at a lower concentration than in AD CSF), showing that they are secreted into the brain interstitial fluid in vivo also in the absence of synaptic injury (Brinkmalm et al., 2014; Thorsell et al., 2010; Öhrfelt et al., 2016).
NRGN (also known as RC3, canarigranin, B-50-immunoreactive C-kinase substrate (BICKS) or p17) is a 78 amino acid postsynaptic protein that is expressed abundantly in the cerebral cortex, hippocampus, amygdala and caudate-putamen in human brain (Represa et al., 1990). It is a neuron-specific protein and its expression is absent in glial cells in the central nervous system (Diez-Guerra, 2010; Gnatenko et al., 2003). NRGN is a calmodulin (CaM)-binding protein and has a role in synaptic plasticity by regulating CaM-mediated signaling and facilitating long term potentiation (LTP) (Zhong et al., 2015). It is suggested to be involved in neuronal growth and differentiation via protein kinase C (PKC) and extracellular signal-regulated kinases 1 and 2 (ERK1/2) signaling pathways (Han et al., 2007).
GAP-43, also known as neuromodulin, B-50, P-57, F1 or pp46, is a presynaptic protein that plays an important role during neuronal development when it is highly expressed in neuronal growth cones (Benowitz and Routtenberg, 1997). It is suggested that primary sensory neurons that lack GAP-43 in their growth cones are deficient in adhesion, spreading, branching and devoid of f-actin (Aigner and Caroni, 1995; Benowitz and Routtenberg, 1997). Furthermore, GAP-43 is suggested to be involved in LTP and long term depression (LTD) and its expression is related to axonal elongation and synaptic sprouting (Denny, 2006; Routtenberg et al., 2000).
Synaptotagmins (SYTs) are a family of synaptic vesicle proteins. The mammalian synaptotagmin family has 15 members. SYT-1 is a pre-synaptic calcium-binding synaptic vesicle protein known to be expressed in cerebral cortex, hippocampus and cerebellum (Chen et al., 2013; Sudhof and Rizo, 2011). It is suggested that SYT-1 has a role in exocytosis and endocytosis, synapse function, behavioral cognition, neuronal polarity, axon differentiation, neuro-development and activity-induced remodeling of synaptic structures (Baker et al., 2015; Inoue et al., 2013, 2015; Sudhof and Rizo, 2011).
SNAP-25 is an essential component of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex; the SNARE complex is needed for several biological processes throughout neuronal development. SNAP-25 helps in synaptic communication by mediating fusion of synaptic vesicles to the plasma membrane during exocytosis and regulating calcium dynamics and neuronal plasticity (Cupertino et al., 2016; Zylbersztejn and Galli, 2011). Both SNAP-25 and SYT-1 interactions are involved in vesicle priming and in neurotransmission (Schupp et al., 2016).
Previously, NRGN, SNAP-25, SYT-1 and GAP-43 expression has been studied in monkeys and rodents during development (Alvarez-Bolado et al., 1996; Capone et al., 1991; Cooper and Gillespie, 2011; Frassoni et al., 2005; Fukuda, 2006; Higo et al., 2006; Mahalik et al., 1992; Murata et al., 2005; Sidor-Kaczmarek et al., 2004) and in human post-mortem brains (Bogdanovic et al., 2000; Furuya et al., 2012). The proteins are also secreted from neurons into the brain interstitial fluid that communicates with the CSF (Blennow et al., 2010). In neurodegenerative diseases such as AD, brain levels of synaptic proteins are commonly reduced, whilst the CSF concentrations are higher (Blennow, 2004; Brinkmalm et al., 2014; Thorsell et al., 2010; Öhrfelt et al., 2016). However, human and rodent brain development varies greatly. Further, it is not possible to study the expression and secretion of synaptic proteins during human fetal brain development.
The possibility to reprogram human somatic cells to pluripotent stem cells has opened ways to develop cell models to study human brain development and physiology (Takahashi et al., 2007). Human induced pluripotent stem cells (hiPSC) were differentiated to cortical neurons using a protocol that follows the same cortical development steps and time frames as observed in human fetal cortex (Shi et al., 2012). During this process, we studied the expression and secretion of NRGN, GAP-43, SYT-1 and SNAP-25. These proteins were sequentially expressed and secreted over a period of at least 150 days, which encompasses the different stages of neuronal development.
2. Material and methods
2.1. Cell culture
Two different hiPSC lines were used in this study, femoral condyle chondrocyte-derived hiPSC (A2B) (Boreström et al., 2014) and fibroblast-derived hiPSC (Con1) (Sposito et al., 2015). A2B hiPSC were cultured on coat-1 in DEF-CS medium (Cellartis, Takara-Bio Europe) while Con1 hiPSC were cultured on Geltrex in complete Essential-8 medium (both from Thermo Fisher Scientific) according to the manufacturer's instructions. All cell cultures were kept in a humidified atmosphere at 5% CO2 and 37 °C.
2.2. Differentiation to cortical neurons
A previously described protocol (Shi et al., 2012), with slight modifications (Bergström et al., 2016), was followed to differentiate hiPSC to cortical neurons. In brief, when hiPSC attained 95–100% confluency, neural induction was initiated by replacing either DEF-CS or complete Essential-8 medium with neural maintenance medium (NMM; a 1:1 mixture of DMEM/F12 and neurobasal medium supplemented with: 1x N2, 1x B27, 50 μM 2-mercaptoethanol, 0.5x non-essential amino acids (all from Thermo Fisher Scientific), 2500 U/mL Penicillin/Streptomycin (GE Healthcare), 10 μg/mL insulin and 0.5 mM sodium pyruvate (all from Sigma Aldrich)) further supplemented with 500 ng/mL mouse Noggin-CF chimera (R&D Systems) and 10 μM SB431542 (Tocris). Cells were maintained in this medium for 10–12 days to attain neuro-epithelial cell morphology followed by passaging with 10 mg/mL dispase (Thermo Fisher Scientific) and plating on laminin-coated plates (surface coverage of 1 μg/cm2 laminin coating solution; Sigma Aldrich) in NMM supplemented with 20 ng/mL FGF-2 (Peprotech) for 4–5 days. Thereafter, cells were further passaged twice using dispase between days 12–22 and with StemPro Accutase® (Thermo Fisher Scientific) between days 25–35 to obtain single cells. On the 35th day of differentiation, immature cortical neurons were seeded on plates pre-coated overnight with poly-L-ornithine (0.01% coating solution; Sigma Aldrich), followed by laminin (as above) for at least 4 h. The cells were seeded at 50,000 cells/cm2 and thereafter maintained for three to seven months in NMM with change of media every second day to achieve mature cortical neurons (Shi et al., 2012).
2.3. Immunocytochemistry
For immunocytochemistry, we followed Shi et al. protocol (Shi et al., 2012). In brief, cells were cultured on 8 well μ-slides and fixed in 4% paraformaldehyde in PBS for 15 min at room temperature (RT) or ice cold methanol at 4 °C. Thereafter, cells were permeabilized for 15 min at RT using 0.3% Triton-X100 (Sigma Aldrich) in PBS, followed by blocking in block buffer (5% donkey serum (Sigma Aldrich) in 0.3% Triton-X100 diluted in PBS). Primary antibodies, OCT-4 (1:400; Cell Signaling, D73G4), NANOG (1:800, Cell Signaling, C30A3), TUJ-1 (1:2000; Abcam ab14545), SV-2 (1:500; DSHB), PSD-95 (1:100; NeuroMab, P78352), KI-67 (1:600; BD Pharmingen™, 550609), PAX-6 (1:600; BioLegend, 901301), nestin (1:50; R&D Systems, MAB1259), CTIP-2 (1:300; Abcam, ab18465), TBR-1 (1:300; Abcam, ab31940), BRN-2 (1:400; Santa Cruz, sc-6029), CUX-1 (1:300; Santa Cruz, sc-13024), GAP-43 (1:1000; Abcam, ab75810), tau (1:1000; Biorbyt, orb175815), NRGN (1:100; Upstate Biotechnologies, 07–425), SNAP-25 (1:400; Sigma Aldrich, S9684-100UL) and SYT-1 antibody (1:200; Synaptic systems, 105,011) were diluted in block buffer and incubated at 4 °C overnight. The next day cells were washed with PBS and incubated with secondary alexa488- and alexa568-conjugates (Life technologies) at RT for 1 h followed by incubation with DAPI (Thermo Fisher Scientific, D3571) for 5 min at RT. The cells were mounted with mounting media (Ibidi) and imaged using either Zeiss LSM700 inverted confocal microscope with 40–63 × objectives using ZEN2000 software (Zeiss) or Nikon confocal microscope A1 with 40–60 × objectives using the Nis Elements software (Nikon). Confocal images were analyzed using ImageJ (NIH).
2.4. Total RNA extraction and cDNA synthesis
For total RNA extraction, the cell layer was washed with pre-warmed Dulbecco's Phosphate-Buffered Saline without calcium and magnesium (DPBS (−/−); Thermo Fisher Scientific), followed by addition of 600 μL of RNeasy Lysis Buffer (Qiagen) supplemented with 4 mM dithiothreitol (Sigma Aldrich) to lyse the cells. The cell lysates were stored at −80 °C until further analysis. Total RNA was extracted and purified using a Qiacube robotic work station (Qiagen), following the RNeasy Mini protocol as per the manufacturer's instructions. Total RNA concentrations were determined on a NanoDrop 2000/2000c spectrophotometer (Thermo Fisher Scientific) followed by dilution of total RNA in RNase-free water to a final concentration of 100 ng/μL cDNA synthesis was performed using the High Capacity cDNA kit with RNase inhibitor (Applied Biosystems). Briefly, total RNA (500 ng) was converted to cDNA in a reaction volume of 20 μL using a single-cycle reaction on a 2720 Thermal Cycler (Applied Biosystems): 25 °C for 10 min, 37 °C for 120 min and 85 °C for 5 min. The cDNA was finally adjusted to a concentration of 0.5 ng/uL before being used in the quantitative PCR reactions.
2.5. Quantitative PCR
Quantitative PCR was performed using TaqMan® Gene Expression Assays with FAM reporter dye in TaqMan® Universal PCR Master Mix with UNG according to the manufacturer's instructions. Quantitative PCR reactions were carried out on Micro-Amp™ 96-well optical microtitre plates on the 7900HT Fast QPCR System (Applied Biosystems) using standard settings. TaqMan® Gene Expression Assays were used for NRGN (Hs00382922_m1), GAP-43 (Hs00967138_m1), SNAP-25 (Hs00938962_m1) and SYT-1 (Hs00194572_m1) (all from Applied Biosystems). Each reaction mixture consisted of 2.5 ng of cDNA and was run in duplicates. Quantitative PCR results were analyzed using the SDS 2.3 software (Applied Biosystems). The relative quantities were calculated following the ΔΔCT method (Livak and Schmittgen, 2001). Average CT:s of RPL-27 (Hs03044961_g1), RPL-30 (Hs00265497_m1) and HPRT-1 (Hs02800695_m1) were used as endogenous controls. All samples were normalized to the endogenous controls; the sample with the highest ratio was determined in each run and all the other samples were then compared to that sample (which was set to 1).
2.6. Western blot
Cells were collected for detecting intracellular protein expression during differentiation and frozen at −80 °C until further analysis. Cells were lysed in Radio-Immunoprecipitation Assay buffer (RIPA buffer; 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS) supplemented with Mini-Complete protease inhibitor cocktail (Roche, Sigma-Aldrich, 04693159001) followed by sonication for 10 min and incubation on ice for 20 min. The lysates were centrifuged at 4000 g for 5 min at +4 °C and the supernatants were transferred to new tubes. Protein concentrations were determined using the Pierce BCA protein assay kit (Thermo Fisher Scientific). Samples were stored at −80 °C until further analysis.
Nu-PAGE loading buffer containing SDS reducing agent was added to aliquots of the protein samples, and after heating to +95 °C for 5 min, samples were either loaded onto 4–12% Bis-tris gels (for NRGN, GAP-43 and SNAP-25) or 3–8% Tris-acetate gels (SYT-1) using XT MES running buffer (all from BioRad Laboratories) or NUPAGE Tris-acetate SDS running buffer (Thermo Fisher Scientific), respectively. The separated proteins were blotted onto a 0.2 μm nitrocellulose membrane (GE Healthcare) using semi-dry technique, followed by blocking in 3% (NRGN) or 5% (GAP-43, SYT-1, SNAP-25) non-fat milk (BioRad Laboratories) and overnight incubation at +4 °C with either anti-NRGN antibody Ab270 (1:5000; gift from Dr. Kuo-Ping Huang (Late)), anti-GAP-43 antibody [EP890Y] (1:100,000; Abcam, ab75810), anti-SYT-1 antibody (1:2000; Synaptic systems, 105,011), and anti-SNAP-25 antibody (1:20,000; Sigma Aldrich, S9684-100UL). After washing, membranes were incubated with horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit secondary antibody (1:40,000 Cell Signaling Technologies) for 1 h at RT. For protein detection, SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific) was used and bands were visualized using ChemiDoc XRS+ (BioRad Laboratories). Membranes were thereafter stripped using Restore stripping buffer (ThermoFisher Scientific) and re-probed with an HRP-conjugated glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (Novus Biologicals). All band intensities were analyzed using ImageJ (NIH); expression of NRGN, GAP-43, SYT-1 and SNAP-25 intensity was correlated to GAPDH. All samples were thereafter related to the sample with highest synaptic protein expression to GAPDH ratio (which was set to 1) in each differentiation batch.
2.7. Cell culture media sample collection
Cell-conditioned media were collected after 48 h of incubation with cells and centrifuged at 360 g for 5 min (to remove cell debris) before the supernatants were transferred to new tubes and stored at −80 °C until further analysis.
2.8. Neurogranin ELISA
NRGN concentration in cell-conditioned media was measured using an in house sandwich ELISA. The Ng36 monoclonal antibody (in house) was used as capturing antibody and was coated on Nunc Maxisorp 96-well microtiter plates at a final concentration of 0.5 μg/mL (100 μL/well) in 50 mM sodium bicarbonate buffer, pH 9.6, overnight at 4 °C. All washes were performed in PBS-T (0.05% Tween20 in PBS (0.01 M phosphate buffer, 0.14 M NaCl, pH 7.4). The remaining protein-binding sites were blocked with 1% bovine serum albumin (BSA) in PBS for 1 h at 20 °C (250 μL/well). Thereafter, plates were washed with PBS-T followed by the addition of calibrators (full length NRGN with concentrations from 7.8 to 1000 pg/mL, blanks, quality control (QC) samples and media samples (100 μL/well)). After washing, biotinylated Ng2 (in house) detection antibody (0.5 μg/mL in PBS-T) was added and incubated for 1 h at RT. After washing, enhanced streptavidin HRP (Enhanced Streptavidin-HRP, 4740 N, Kem En Tech) was diluted 1:20000 in 1% BSA-PBS-T and incubated (100 μL/well) for 30 min at RT followed by three washes. After a final wash, 100 μL of 3,3′,5,5′-tetramethylbenzidine (TMB One Solution, 4380A, Kem En Tech) was added to produce the color reaction. After 20 min incubation in the dark, the reaction was stopped by addition of 100 μL of 0.2 M H2SO4 and the absorbance was measured at 450 nm (reference wavelength 650 nm) using an ELISA plate reader (Vmax, Molecular Devices, USA). A fitted 4-parameter logistic model was used to generate the calibration curve and the blank was included as zero concentration of NRGN.
2.9. GAP-43 ELISA
GAP-43 concentration was measured using an in house sandwich ELISA (Sandelius et al., 2018). Nunc-Immuno Polysorp microwell modules (Thermo Fisher Scientific) were coated with mouse anti-GAP-43 antibody (0.77 μg/mL NM4 ((Fujirebio, Ghent, Belgium)) in 50 mM sodium bicarbonate buffer pH 9.6, overnight at 4 °C. After washing, wells were blocked with PBS-T, 1x Casein (diluted from 10x casein blocking buffer, B6429, Sigma-Aldrich) for 1 h at RT. Thereafter, in house recombinant full-length GAP-43 calibrators (78 pg/mL – 50000 pg/mL), blanks, two folds pre-diluted control samples and media samples in assay diluent (PBS-T, 1% BSA) were co-incubated with a rabbit detector antibody (0.14 μg/mL ABB-135 (Nordic Biosite, Täby, Sweden)) overnight at 4 °C. After additional washes, plates were incubated with anti-rabbit HRP (1:30000, Promega) for 1.5 h. After subsequent washes, wells were incubated for 20 min with TMB (TMB, KemEnTech Diagnostics) in the dark. The color reaction was stopped by addition of 100 μL 0.2 M H2SO4 and the absorbance was read in a SunriseTM micropl ate absorbance reader (Tecan) at 450 nm (650 nm as reference value).
2.10. In-house SNAP-25 and SYT-1 IP-MS assay
The combined SNAP-25 and SYT-1 assay was conducted at the Clinical Neurochemistry Laboratory at the Sahlgrenska University Hospital (Mölndal, Sweden) and consists of immuno-enrichment combined with liquid chromatography/tandem mass spectrometry (LC-MS/MS (Brinkmalm et al., 2014; Fernstrom et al., 2018; Öhrfelt et al., 2016). Briefly, mouse monoclonal antibody 41.1 recognizing SNAP-25 and mouse monoclonal antibody SMI-81R recognizing SYT-1 were used to co-immunoprecipitate both proteins with a KingFisher™ Flex System (Thermo Fisher Scientific), which uses magnetic rods to move particles through the various binding, mixing, washing and elution phases in a 96-well plate format. Immunoprecipitated SNAP-25/SYT-1 together with isotopically labeled protein and peptide standards were subsequently digested with trypsin/Lys-C and analyzed on a quadrupole–orbitrap mass spectrometer Q Exactive (Thermo Fisher Scientific) coupled to an Ultimate 3000 chromatography system (Thermo Fisher Scientific). The instrument was set to acquire scheduled pairs of parallel reaction monitoring (PRM) scans in profile mode allowing simultaneous detection of both the SNAP-25/SYT-1 peptides and the corresponding internal standards (IS). Data acquisition and analysis were performed with Xcalibur software version 2.2 SP1.48 (Thermo Fisher Scientific) and Pinpoint 1.3.0. Levels of SNAP-25tot (amino acids Ac2-16) and SNAP-25aa40 (amino acids 32–40) were calculated by multiplying the ratio of the LC-MS peak areas with the concentration of the corresponding IS.
2.11. Lactate dehydrogenase (LDH) assay
To estimate the viability of the cells, we performed the LDH release assay. hiPSC were differentiated to mature cortical neurons. As a 0% viability control, mature cortical neurons were lysed with 1% Triton X-100 (Sigma Aldrich) (1% v/v to cell-conditioned media). The unlysed (samples) and lysed cortical neurons (controls) were incubated at 37 °C for 30 min to release LDH. Thereafter, the unlysed or lysed cells together with cell-conditioned media were centrifuged at 360×g for 5 min and the supernatants were collected. The freshly collected supernantants (250 μL) were sent to Clinical Chemistry Laboratory at Sahlgrenska University Hospital, Gothenburg, Sweden for analysis. In brief, the LDH assay was performed using the LDH activity test kit as per manufacturer's instructions (Roche Diagnostics Scandinavia AB, 03004732122) on the fully automated analyzer Cobas e 501 module (Roche, Germany). The working range for the assay was 0.167–16.70 μkat/L.
2.12. Statistical analysis
Mean values were compared using two-way analysis of variance (ANOVA) followed by Tukey-Kramer post hoc analysis. Statistical significance was defined as p < 0.05. All statistical analyses were performed using SPSS (IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY: IBM Corp).
3. Results
3.1. Characterization of cells during differentiation from hiPSC towards cortical neurons
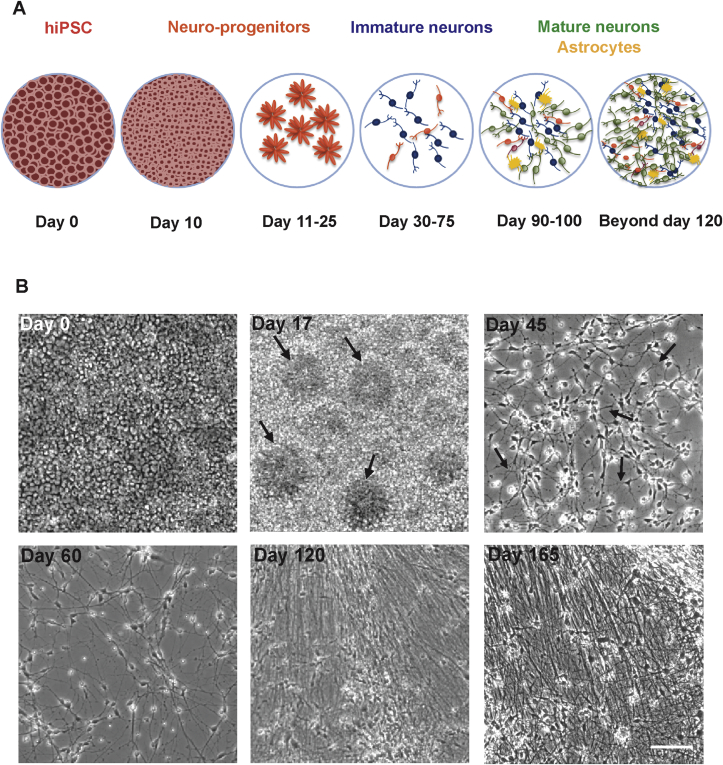
A schematic representation of hiPSC differentiation to cortical neurons is shown in Fig. 1A. The cells differentiated gradually from hiPSC to neuro-progenitors followed by immature and mature cortical neurons. During differentiation, cell morphology was monitored using phase contrast microscopy. Fig. 1B shows the morphological changes occurring during differentiation to cortical neurons. At d0, hiPSC were cultured at 95–100% confluency. The cells formed neural rosettes between d15 to d25 of differentiation (neural rosettes at d17 are shown in Fig. 1B). The contacts between neurites were first observed after d30 of differentiation and more thereafter (d45 and d60 are shown). The neurons were further maintained for two to five months which led to a substantial increase in the neuritic network and formation of thick bundles of neurites. Extensive neuritic networks are shown for d120 and d165 of differentiation.
Fig. 1.
(A) Schematic representation of hiPSC differentiation stages during development to cortical neurons: hiPSC (undifferentiated cells), neuro-progenitors, immature and mature neurons. (B) Cell morphology and neurite network, as observed during hiPSC differentiation to cortical neurons. Phase contrast images at d0 show hiPSC maintained at high density (95–100% confluency); d17 illustrates the formation of rosette structures (arrows); d45 depicts single cells forming neurites (arrows) during early cortical development; d60, neurite connections increase and a network is formed. Further maturation of cortical neurons coincides with establishment of an extensive neuronal network as shown on d120 and d165 of differentiation. Scale bar = 100 μm.
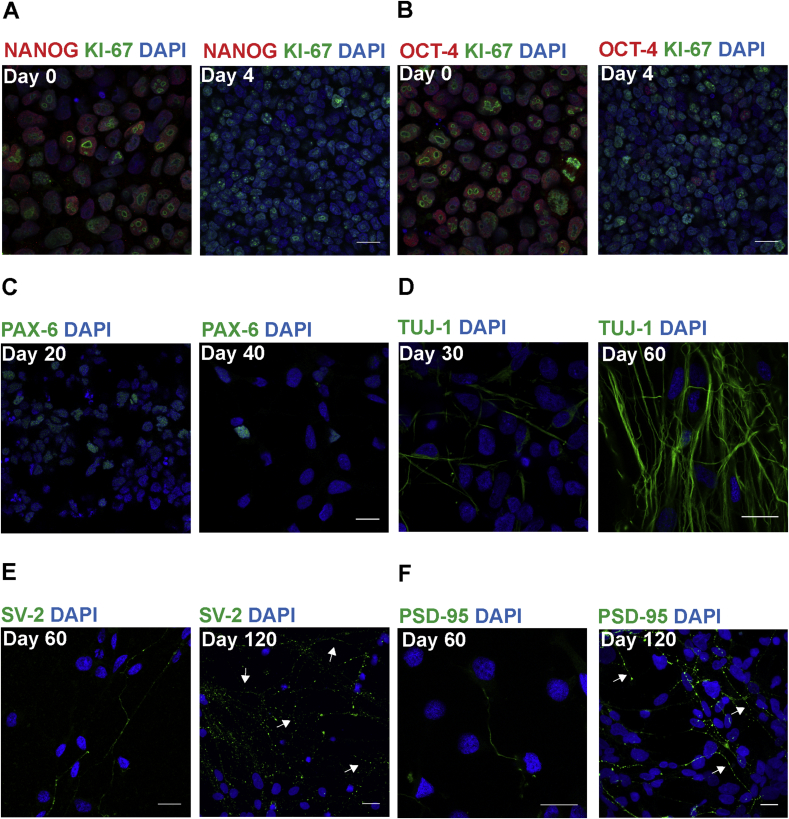
To characterize the neurons, immunocytochemistry was performed during hiPSC differentiation to cortical neurons. Pluripotent markers of stem cells, octamer-binding transcription factor-4 (OCT-4) and homeobox protein NANOG (NANOG) (Fig. 2A and B), were expressed highly on d0 which suggested that these cells are stem cells. NANOG and OCT-4 expression weakened on d4 of differentiation suggesting that these cells loose pluripotency. KI-67, a proliferation marker had similar staining intensities on d0 and d4 of differentiation (showing a typical KI-67 nuclear staining pattern depending on cell cycle stage (Sobecki et al., 2016; Verheijen et al., 1989)), which indicated that these cells have the ability to proliferate during this stage. Nestin, a marker for neuro-progenitors (Supplementary Fig. 2A) was already expressed on d15 of differentiation. Further, paired box protein-6 (PAX-6) (Fig. 2C), another neuro-progenitor marker was highly expressed on d20 which suggested that the cells are neuro-progenitors at this stage. PAX-6 expression weakened on d40 as compared to d20, indicating that neuro-progenitors were decreased during this period. Neuron-specific class III beta-tubulin (TUJ-1) expression increased markedly from d30 to d60 of differentiation (Fig. 2D) indicating neuritic development during differentiation. Further, cortical markers for deep layer neurons, COUP-TF interacting protein-2 (CTIP-2) and T-Box, Brain-1 (TBR-1) were expressed during this period (d40 is shown in Supplementary Fig. 2B and C). Synaptic vesicle glycoprotein-2 (SV-2), a pre-synaptic protein, and post-synaptic density protein-95 (PSD-95) (Fig. 2E and F) were weakly expressed on d60 of differentiation. On d120, SV-2 and PSD-95 expression increased markedly and became punctate, suggesting that synapses matured during this period. Furthermore, markers for upper layer cortical neurons; POU class 3 homeobox-2 (BRN-2) and Cut-Like Homeobox-1 (CUX-1) were first expressed during this stage (d90 and d105 are shown respectively in Supplementary Fig. 2D and E). These stainings indicate the step-wise expression of stem cells, neuro-progenitors, immature and mature cortical neurons during hiPSC differentiation to cortical neurons. In addition, the cells used in this study have been characterized extensively in a previous publication from our group (Bergström et al., 2016), which described the mRNA expression of transcription factors for stem cells, neuro-progenitors and cortical neurons and immunocytochemistry images for glial fibrillary acidic protein (GFAP), an astrocyte marker and the vesicular glutamate transporter 1 (vGLUT1). GFAP expression increased from d60 to d90 while vGLUT1 became punctate on d90 of differentiation (Bergström et al., 2016). Previously, we have also shown that evoked action potentials could first be measured from d100 and onwards (Bergström et al., 2016). This indicates that these neurons have functional synapses around d100 of differentiation.
Fig. 2.
Neuronal and synaptic maturation during differentiation of hiPSC to cortical neurons. Representative images from immunocytochemistry staining during differentiation. (A and B) NANOG and OCT-4 were strongly stained on d0 and the stainings became weak on d4, while KI-67 had similar staining intensities on d0 and d4. (C) PAX-6, a primary neuro-progenitor expression decreased from d20 to d40 of differentiation. (D) Staining for neuron specific tubulin, TUJ-1 was weak on d30 but strong staining was observed on d60. (E) SV-2, a pre-synaptic protein, staining was weak on d60. On d120, a strong punctuate staining was observed (arrows). (F) PSD-95, a post-synaptic density protein, staining was weak on d60. On d120, the staining had intensified and became punctate (arrows). Green or red = protein of interest, blue = nuclei (DAPI), Scale bar = 20 μm.
3.2. Expression and secretion of NRGN during hiPSC differentiation to cortical neurons
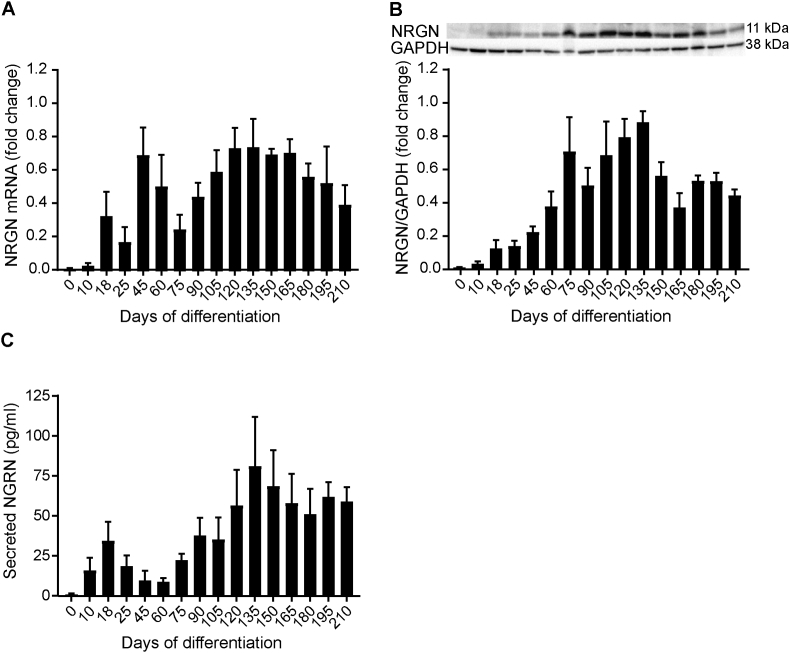
NRGN mRNA expression was first observed on d10 of differentiation followed by a biphasic increase from d18 to d60 (Fig. 3A). After d75, there was a tendency towards a gradual increase in NRGN mRNA expression until d135 of differentiation, when the highest expression was detected. Thereafter, NRGN mRNA expression decreased gradually. Intracellular NRGN protein was first detected on d10 and there was a gradual increase of NRGN expression from d18 to d135. NRGN was expressed at the highest level on d135 as shown in Fig. 3B and Supplementary Fig. 1A. A reduction of NRGN expression was observed after d135, although the levels remained clearly detectable. NRGN secretion to the media was first detected on d10 (Fig. 3C). The NRGN secretion pattern correlated well (Spearman rank correlation coefficient r = 0.64, p = 0.007) with the NRGN mRNA expression as shown in Supplementary Fig. 3. The highest secretion of NRGN was detected on d135 of differentiation. After d135, the secreted levels of NRGN decreased.
Fig. 3.
hiPSC-derived cortical neurons express and secrete NRGN during differentiation. (A) Expression of NRGN mRNA as detected by quantitative PCR; NRGN mRNA was first expressed on d10 at low level. During d18 to d60, there was differential NRGN mRNA expression. When cortical neurons matured, NRGN mRNA levels increased gradually from d90 to d135, when it was expressed the highest. There was a trend towards decreased expression of NRGN mRNA after d165. NRGN mRNA was normalized to the average of HPRT-1, RPL-27 and RPL-30 genes. NRGN mRNA normalized change at each differentiation time point was related to the highest NRGN mRNA expression, which was set to 1. For a summary of assigned significances for the measured changes of NRGN mRNA, see Supplementary Table 1. (B) Intracellular NRGN protein levels were detected by western blot. NRGN was first detected on d10 and its levels increased between d18 and d135. On d135, NRGN levels were the highest; thereafter there was a trend towards decreased NRGN levels. NRGN protein levels were normalized to GAPDH and its fold change in each differentiation was related to the highest NRGN protein level, which was set to 1. Representative bands of NRGN and GAPDH are shown. The summary of assigned significances for the measured changes of NRGN protein level is shown in Supplementary Table 2. (C) Total NRGN secretion was detected by ELISA and NRGN was first secreted on d10. NRGN secretion increased gradually from d90 to d135, when NRGN was secreted the most; thereafter a trend towards decreased secretion was observed. The summary of assigned significances for the changes of secreted NRGN concentrations are summarized in Supplementary Table 3. Bars represent mean±SEM. For intracellular expression of NRGN mRNA and protein, n = 4 (except from d165 to d210 where n = 2) and for secreted NRGN, n = 5 (except from d165 to d210 where n = 3).
3.3. Expression and secretion of GAP-43 during hiPSC differentiation to cortical neurons
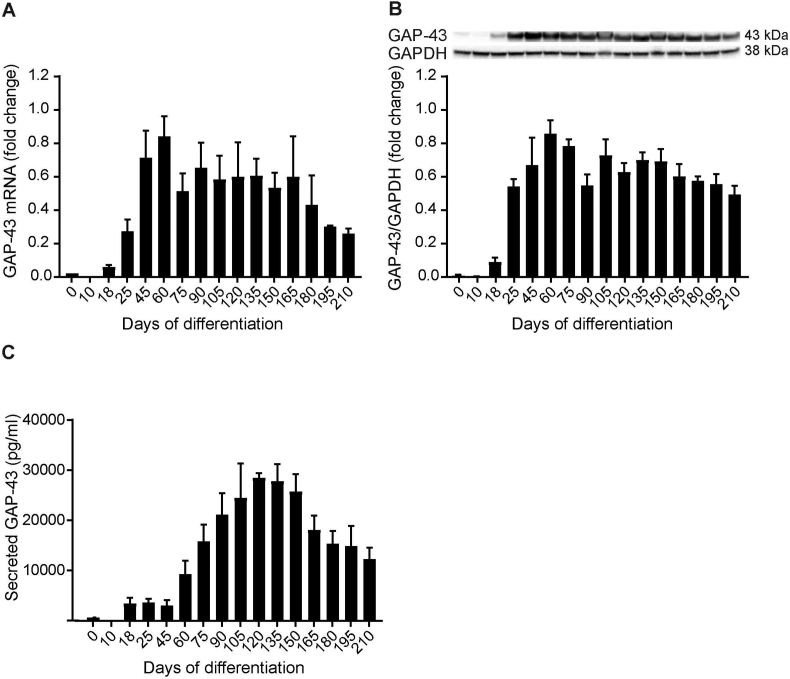
GAP-43 mRNA was first expressed on d18 of differentiation (Fig. 4A). A marked increase in GAP-43 mRNA was observed until d60 of differentiation, when GAP-43 mRNA was expressed at the highest level. Thereafter, the GAP-43 expression decreased. Intracellular GAP-43 protein levels (Fig. 4B and Supplementary Fig. 1B) followed the pattern of GAP-43 mRNA expression (Spearman rank correlation coefficient r = 0.756, p = 0.001). GAP-43 protein levels were stable, with slightly decreased levels of GAP-43 after d165 of differentiation. Secretion of total GAP-43 to media was first noted on d18 of differentiation (Fig. 4C). GAP-43 was secreted at low concentration from d18 to d45. Thereafter, there was a marked increase of secreted GAP-43 until d135. After d135, GAP-43 secretion decreased.
Fig. 4.
GAP-43 mRNA and protein expression during hiPSC differentiation to cortical neurons. (A) Expression of GAP-43 mRNA was detected by quantitative PCR; GAP-43 mRNA was first expressed on d18 of differentiation. A marked increase in GAP-43 mRNA expression was seen between d18 and d60. On d60, GAP-43 mRNA was expressed the highest; thereafter the levels decreased and plateaued until d165 of differentiation. After d165, there was a trend towards decreased expression of GAP-43. GAP-43 mRNA was normalized to the average of HPRT-1, RPL-27 and RPL-30 mRNA. GAP-43 mRNA normalized change at each differentiation time point was related to the highest GAP-43 mRNA expression, which was set to 1. For a summary of assigned significances for the measured changes of GAP-43 mRNA, see Supplementary Table 4. (B) GAP-43 protein levels were detected by western blot and GAP-43 protein was first detected on d18. GAP-43 levels increased gradually between d18 and d60, when it was expressed the highest. Thereafter, GAP-43 levels slightly decreased and plateaued until d165, which was followed by a gradual decrease in GAP-43 levels. GAP-43 levels were correlated to GAPDH and GAP-43 fold change in each differentiation was related to the highest GAP-43 protein level, set to 1. Representative bands of GAP-43 and GAPDH are shown. The summary of assigned significances for the measured changes of GAP-43 protein is shown in Supplementary Table 5. (C) Total GAP-43 secretion was detected by ELISA; GAP-43 was first observed to be secreted on d18. Between d18 and d45, secreted GAP-43 levels plateaued; thereafter there was a trend towards gradual increase of secreted GAP-43 between d60 and d120. On d120, GAP-43 was secreted the highest followed by a gradual decrease in GAP-43 secretion. The summary of assigned significances for the changes of secreted GAP-43 concentrations is summarized in Supplementary Table 6. Bars represents mean±SEM. For intracellular expression of GAP-43 mRNA and protein, n = 4 (except from d165 to d210 where n = 2) and for secreted GAP-43, n = 5 (except from d165 to d210 where n = 3).
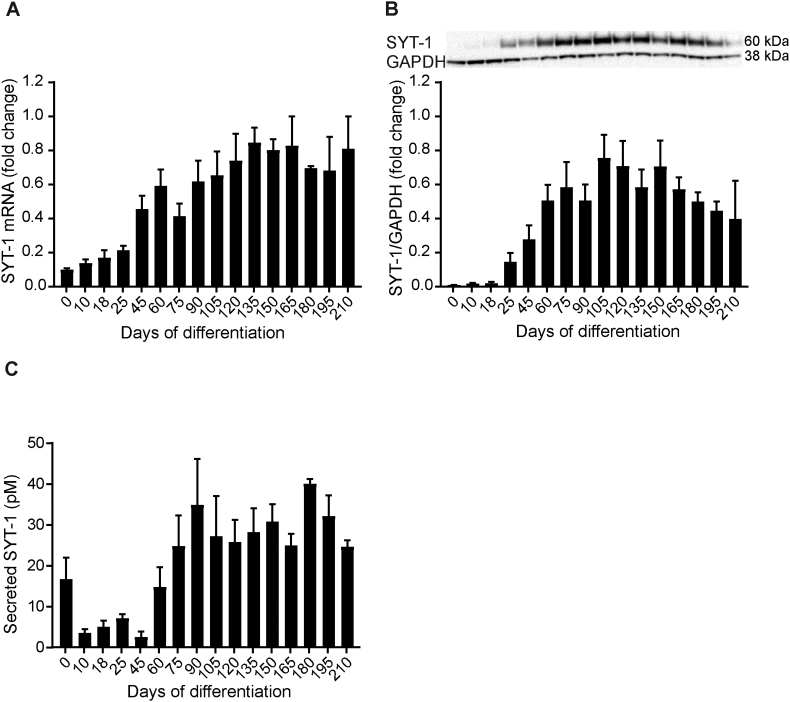
3.4. Expression and secretion of SYT-1 during hiPSC differentiation to cortical neurons
SYT-1 mRNA was expressed throughout the differentiation including d0 (Fig. 5A). There was a gradual increase in the expression of SYT-1 mRNA from d0 to about d165. SYT-1 mRNA was expressed the most during d135 to d165 and on d210. Intracellular expression of SYT-1 protein was first noted on d25 (Fig. 5B and Supplementary Fig. 1C). The SYT-1 protein pattern did not follow SYT-1 mRNA expression during d0 to d18, when SYT-1 protein was undetectable. Thereafter, there was a gradual increase in SYT-1 protein until d105, when protein levels were the highest. After d150, a tendency towards decreased SYT-1 protein was observed. Secreted SYT-1 was monitored by IP followed by PRM mass spectrometry (Fig. 5C). SYT-1 was secreted at relatively high concentrations on d0 compared to d10 through d45. There was a gradual increase in secretion during d60 to d90 with the highest levels seen on d180 of differentiation.
Fig. 5.
SYT-1 mRNA and protein expression during hiPSC differentiation to cortical neurons. (A)SYT-1 mRNA was detected by quantitative PCR and was first expressed on d0. There was a trend towards increased expression of SYT-1 mRNA between d0 and d165. SYT-1 mRNA levels were normalized to the average of HPRT-1, RPL-27 and RPL-30 genes and SYT-1 mRNA fold change in each differentiation was related to the highest SYT-1 mRNA expression, set to 1. For a summary of assigned significances for the measured changes of SYT-1 mRNA, see Supplementary Table 7. (B) SYT-1 protein levels were detected by western blot and were not observed until d18 of differentiation. Thereafter, there was a gradual increase in SYT-1 levels until d105, when expression was the highest. After d150, SYT-1 levels decreased. SYT-1 protein levels were correlated to GAPDH and SYT-1 fold change in each differentiation was related to the highest SYT-1 protein level, set to 1. Representative bands of SYT-1 and GAPDH are shown. The summary of assigned significances for the measured changes of SYT-1 protein is shown in Supplementary Table 8. (C) Secreted SYT-1 was detected by immuno-precipitation (IP) followed by parallel reaction monitoring (PRM) mass spectrometry; the assay targets mainly the SYT-1 C2A calcium binding domain. Fig. 4C show that SYT-1 was secreted at high concentrations on d0. The secreted SYT-1 decreased remarkably from d0 until d45. Thereafter, SYT-1 secretion increased gradually from d60 to d90. After d180, there was a trend towards decreased secretion of SYT-1. The summary of assigned significances for the changes of secreted SYT-1 concentration is summarized in Supplementary Table 9. Bars represent mean±SEM. For intracellular expression of SYT-1 mRNA and protein, n = 4 (except from d165 to d210 where n = 2) and secreted SYT-1, n = 5 (except from d165 to d210 where n = 3).
3.5. Expression and secretion of SNAP-25 during hiPSC differentiation to cortical neurons
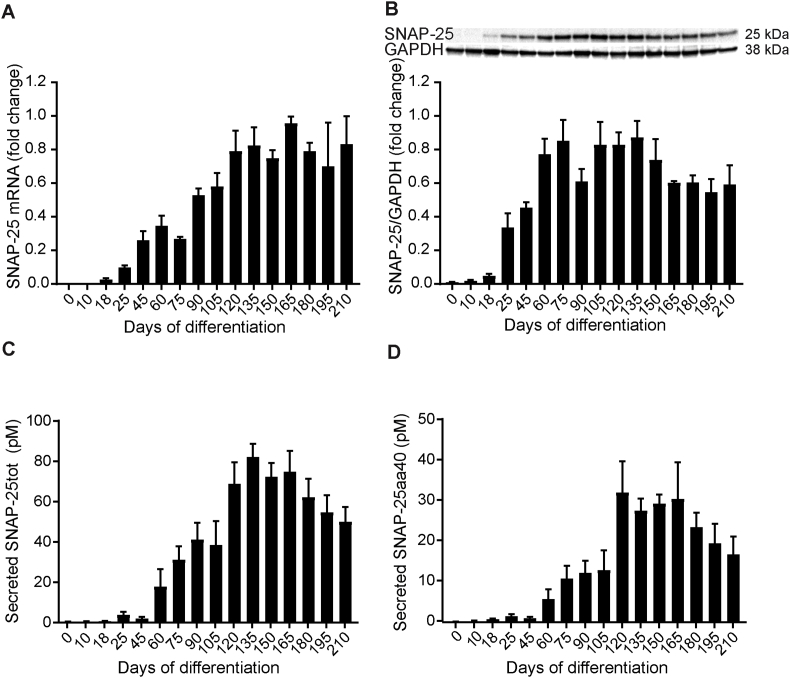
SNAP-25 mRNA was first expressed on d18; thereafter, there was a gradual increase in the expression of SNAP-25 mRNA until d165 (Fig. 6A). Intracellular SNAP-25 protein was first noted on d18 of differentiation. Thereafter, a marked increase in SNAP-25 protein was seen until d75 and after d135 a slight reduction was observed (Fig. 6B and Supplementary Fig. 1D). Secreted SNAP-25 was monitored by IP followed by PRM mass spectrometry. Our method measures the levels of total SNAP-25 (SNAP-25tot) shown in Fig. 6C and SNAP-25 amino acid 40 peptide (SNAP-25aa40) shown in Fig. 6D. SNAP-25tot measures all soluble forms of SNAP-25, whereas SNAP-25aa40 only measures the longer soluble forms (including at least amino acid 40). SNAP-25 was first secreted on d25 at very low concentrations. There was a gradual increase in secreted SNAP-25 during d60 to d135; thereafter, the secreted concentrations plateaued between d135 to d165 followed by a tendency towards decreased secretion of SNAP-25. The highest SNAP-25tot secretion was detected on d135 (Fig. 6C) while the highest SNAP-25aa40 concentrations were detected on d120 of differentiation (Fig. 6D).
Fig. 6.
SNAP-25 mRNA and protein expression during hiPSC differentiation to cortical neurons. (A)SNAP-25 mRNA was detected by quantitative PCR and its expression was first observed on d18. SNAP-25 mRNA expression increased gradually during differentiation until d165, when SNAP-25 mRNA was expressed the highest. SNAP-25 mRNA was normalized to the average of HPRT-1, RPL-27 and RPL-30 mRNA levels. SNAP-25 mRNA fold change in each differentiation was related to the highest SNAP-25 mRNA expression, set to 1. For a summary of assigned significances for the measured changes of SNAP-25 mRNA, see Supplementary Table 10. (B) SNAP-25 protein was first detected on d18 by western blot. SNAP-25 protein levels increased gradually from d18 to d75. After d105, SNAP-25 levels plateaued until d135, followed by reduction in the levels of SNAP-25. SNAP-25 levels were correlated to GAPDH and SNAP-25 fold change in each differentiation was related to the highest SNAP-25 expression, set to 1. Representative bands of SNAP-25 and GAPDH are shown. The summary of assigned significances for the measured changes of SNAP-25 protein is shown in Supplementary Table 11. (C and D) IP followed by PRM mass spectrometry was used to detect secreted SNAP-25; SNAP-25tot (shown in C), and SNAP-25aa40 (shown in D). (C) SNAP-25tot represents all soluble forms of SNAP-25 and was first secreted on d25. The secretion of SNAP-25tot increased markedly from d60 to d135, when it was secreted the highest. Thereafter, the secreted concentrations of SNAP-25tot plateaued until d165 followed by decreased secretion. (D) SNAP-25aa40 measures the longer soluble forms (including at least amino acid 32–40). There was a gradual increase of secreted SNAP-25aa40 between d60 and d105, followed by a marked increase from d105 to d120, when it was secreted the most. After d165, a trend towards decreased secretion of SNAP-25aa40 was observed. The summary of assigned significances for the changes of secreted SNAP-25tot and SNAP-25aa40 concentrations are summarized in Supplementary Table 12 and 13, respectively. Bars represents mean±SEM. For intracellular expression of SNAP-25 mRNA and protein, n = 4 (except from d165 to d210 where n = 2) and for secreted SNAP-25, n = 5 (except from d165 to d210 where n = 3).
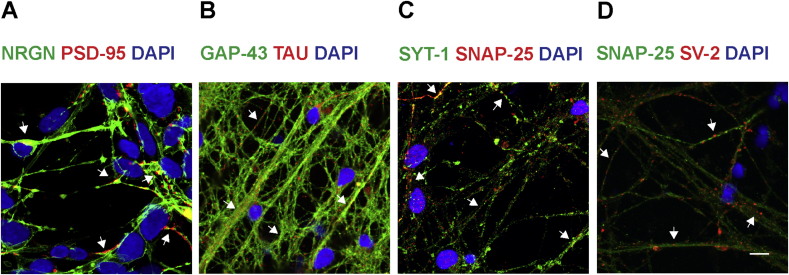
3.6. Localization of synaptic proteins in mature cortical neurons
Synaptic proteins were highly expressed in mature cortical neurons as detected by western blot. Therefore, immunocytochemistry was performed on mature cortical neurons on d120 of differentiation. Synaptic proteins of interest were localized by co-staining with either pre- or post-synaptic protein as shown in Fig. 7. NRGN was co-stained with PSD-95, a dendritic marker; the arrow points at the localization of NRGN and PSD-95 in dendrites (Fig. 7A). A punctate staining for PSD-95 was observed. GAP-43 was co-stained with tau, an axonal protein; the arrows point at the localization of GAP-43 and tau in axons (Fig. 7B). Pre-synaptic proteins, SYT-1 and SNAP-25 were co-stained; they were co-expressed in the same compartment as indicated by arrows (Fig. 7C). A punctate staining for SV-2 and SNAP-25 was observed in the same compartment as indicated by arrows (Fig. 7D).
Fig. 7.
Localization of synaptic proteins in mature corical neurons. Representative images at d120 from immunocytochemistry staining of mature cortical neurons. (A) NRGN (green) and PSD-95 (red), post-synaptic proteins are co-expressed in the same compartment (arrows). (B) GAP-43 (green) and tau (red) are stained together in axons (arrows). (C) SYT-1 (green) and SNAP-25 (red), pre-synaptic proteins are co-stained in the same compartment (arrows). (D) SNAP-25 (green) and SV-2 (red), pre-synaptic proteins are co-expressed in the same compartment (arrows). blue = nuclei (DAPI), Scale bar = 10 μm.
3.7. Release of LDH and GAPDH to cell-conditioned media
To confirm that the synaptic proteins found in cell-conditioned media during neuronal differentiation were secreted, rather than released through damaged cell membranes due to cell death, LDH activity was measured on d135 differentiated cells (Supplementary Fig. 4A), which was the time point when the highest concentrations of synaptic proteins were detected in cell-conditioned media. Cell-conditioned media contained very low LDH activity, 0.285 μkat/L (2.4%; as compared to LDH activity of 11.9 μkat/L in triton-X100 treated cells set to 100% (control)). The limit of detection for the assay was 0.17 μkat/L. Further to support these findings, we performed western blots to detect GAPDH in cell-conditioned media on d0, d18, d60 and d120 of differentiation (Supplementary Fig. 4B). We detected very weak bands on d0, d60 and d120 of differentiation and there were no correlations between secreted synaptic protein levels (NRGN, GAP-43, SYT-1 and SNAP-25) and GAPDH densitometry (arbitrary units) (Correlations for NRGN and GAP-43 are shown in Supplementary Fig. 4C and 4D).
4. Discussion
hiPSC, when undergoing an in vitro differentiation process to induce the same developmental steps as observed during human fetal brain development, provide an opportunity to study molecular changes during human cortical development (Shi et al., 2012). Here, we used this model to study temporal changes in the expression and secretion of a number of synaptic proteins during the differentiation to cortical neurons. We found that NRGN, GAP-43 and SNAP-25 were first expressed and secreted between d10 to d25, when neurite outgrowth was not yet observed. SYT-1 mRNA was expressed and secreted already on d0 of differentiation. NRGN, SNAP-25 and SYT-1 were expressed at the highest levels after d90, when mature synapses were formed while GAP-43 was expressed the highest level already on d60 of differentiation, when neurite outgrowth was intense. For all these synaptic proteins, the highest secretion was observed between d120 and d135, when functional synapses had formed. The expression of synaptic proteins thus followed the timing of development of cortical neurons and network formation.
The hiPSC-derived cortical neurons used in this study are characterized here and also elsewhere (Bergström et al., 2016). On the basis of cell morphology, immunocytochemistry, gene expression and electrophysiological data shown in this study and by Bergström et al. (2016), hiPSC differentiation to cortical neurons can be divided into (i) stem cell stage (d0), (ii) neuro-progenitor stage (d11 to d25), (iii) immature cortical neurons (d30-d75) and (iv) mature cortical neurons (d90 and beyond). During stem cell stage, hiPSC were maintained at 100% confluency and the cells expressed the pluripotency markers, NANOG and OCT-4. At the neuro-progenitor stage, the neural rosette structures appeared and pluripotency genes disappeared, and the markers for neuro-progenitors, nestin and PAX-6 were first expressed. The immature cortical neuronal stage was characterized by neurite outgrowth and formation of first neurite connections followed by neurite network formation. During this stage, markers for deep layer cortical neurons, CTIP-2 and TBR-1 were first expressed. Further, during this stage there was a marked increase in TUJ-1 expression, while synaptic markers, SV-2 and PSD-95 were expressed weakly and the neurons fired low to moderate evoked action potentials. Mature cortical neurons were formed around d90 and beyond. During this period, markers for upper layer cortical neurons, BRN-2 and CUX-1 were first expressed. The neuritic network was further increased and thick bundles of neurites were formed. In addition, the synaptic markers showed punctate localizations. The neuritic network activity was enhanced and the neurons spontaneously fired action potentials suggesting the maturation of functional synapses. The hiPSC differentiation to cortical neurons followed step-wise maturation whereby stem cells appear first followed by their gradual transition to neuro-progenitors. Thereafter, deep-layer neurons were first expressed in immature cortical neurons followed by expression of upper-layer neurons in mature cortical neurons. The stages are summarized in Fig. 1A.
Taken together, increased mRNA expression, intracellular protein levels and secretion of synaptic proteins coincided with the development of neurites and synapses. During neuro-progenitor stage, when neurite outgrowth was absent, NRGN, GAP-43, SNAP-25 and SYT-1 were expressed at low levels. However, expression of the synaptic proteins increased gradually as the neuritic network started to form at the stage of immature neurons. Increased expression of GAP-43 in immature neurons coincided with the formation of neurite connections. Further increases in the expression of NRGN, SNAP-25 and SYT-1 followed the formation of neuritic networks and thick bundles of neurites. Our expression results support an association of NRGN, GAP-43, SNAP-25 and SYT-1 in synaptic sprouting, axonal outgrowth, axonal branching, synaptic plasticity, and synaptogenesis, as suggested elsewhere (Alvarez-Bolado et al., 1996; Baker et al., 2015; Benowitz and Routtenberg, 1997; Diez-Guerra, 2010; Greif et al., 2013; Zhong et al., 2015). Furthermore, a previous study showed that increased expression of synaptic proteins was associated with synapse stabilization (Pinto et al., 2013).
NRGN mRNA was expressed in two phases during early differentiation (neuro-progenitors and immature neurons), thereafter NRGN protein expression increased gradually and peaked in mature neurons. A study on rat telencephalon suggests a bi-phasic NRGN mRNA expression; an early phase and a juvenile phase. In addition, NRGN protein in rats is first expressed on embryonic day 18 and its expression increased markedly at birth (postnatal stage-P1) (Alvarez-Bolado et al., 1996). A study on monkeys also suggests NRGN mRNA expression to be developmentally regulated (Higo et al., 2006). These studies highlight the differential expression of the NRGN gene during early development (corresponding to neuro-progenitors and immature neurons in our model) and suggest NRGN to be expressed the highest in mature neurons.
GAP-43 mRNA expression and intracellular protein levels showed the same profile during neuronal differentiation. The secretion of GAP-43 was delayed in comparison to its intracellular protein levels (by about 4 weeks). The increased levels of GAP-43 protein in immature neurons coincided with neuritic branching, which is in agreement with a possible role of GAP-43 in this process. Our expression results support the increase in GAP-43 protein levels during embryonic brain development in rodent models (Capone et al., 1991; Mahalik et al., 1992).
SYT-1 protein levels were first detected in neuro-progenitors when neither neurite outgrowth nor synaptogenesis were observed. Our finding is in line with a previous report that suggests that SYT-1 is expressed well before the beginning of synaptogenesis both in vivo and in vitro (Greif et al., 2013).
SNAP-25 mRNA expression and protein levels increased gradually during the formation of immature cortical neurons and were highest in mature neurons. One study in developing rat cerebral cortex suggests that SNAP-25 protein is highly expressed on post-natal day 10 (P10) and (P60), after which SNAP-25 levels decrease (Sidor-Kaczmarek et al., 2004). Increased expression of SNAP-25 in mature neurons may reflect the establishment of neuritic networks.
Our novel, sensitive methods to detect NRGN, GAP-43, SYT-1 and SNAP-25 in bio-fluids make the detection of these synaptic proteins in cell-conditioned media possible. The analysis of NRGN, GAP-43, SYT-1, and SNAP-25 secretion to CSF has previously been focused in context of neurodegeneration including AD (Brinkmalm et al., 2014; Sjogren et al., 2001; Thorsell et al., 2010; Öhrfelt et al., 2016). However, the secretion of synaptic proteins from human cellular models during development has not been investigated previously.
The secretory pattern of NRGN followed the NRGN mRNA fold change. NRGN was first secreted by neuro-progenitors. During neuro-progenitor and immature cortical neuronal stages, NRGN secretion was low. As neurons matured and functional synapses had formed, NRGN secretion increased in mature cortical neurons. The secretory pattern of GAP-43 protein suggests that GAP-43 may be required during early development of immature neurons. When cortical neurons mature, GAP-43 may be needed for synaptic stabilization, as suggested by an earlier study (Allegra Mascaro et al., 2013). The SYT-1 expression pattern was puzzling. SYT-1 mRNA was detected already at the hiPSC stage and so was SYT-1 protein secretion. However, intracellular SYT-1 expression was not detectable until more mature differentiation stages. This result suggests that SYT-1 may play distinct roles at early and late differentiation stages. The secretory profile of SNAP-25 showed that SNAP-25 was not detected in stem cells or neuro-progenitors; it was first released when markers for immature neurons were expressed and it peaked when mature synapses were formed. Taken, together, the secretion of synaptic proteins was highest in mature cortical neurons, which coincides with the formation of stable synaptic networks. As we only used one protocol for differentiation of cortical neurons, the results should be confirmed using other protocols.
As no correlations were found between secreted synaptic proteins and GAPDH in cell-conditioned media from stem cells, neuro-progenitors, immature or mature cortical neurons, and LDH activity was low in cell-conditioned media from mature neurons, the secretion of these synaptic proteins was most likely an active process. One possible reason for their secretion could be physiological protein turnover, causing synaptic proteins to be released from synapses once they have executed their actions. It has been reported that general synaptic protein half-life is 99 h (Alvarez-Castelao and Schuman, 2015). The secretion may also reflect synaptic remodeling of the cortical connectome during development. Synaptic proteins may also be secreted as an inherent part of synaptic transmission.
5. Conclusion and significance
This study describes a detailed expression and secretory profile of synaptic proteins in a model reflecting human fetal cortical development. This implicates that increased expression of synaptic proteins reflects an increase in neurite outgrowth followed by formation of synaptic networks. Further, it implicates that mature neurons are more prone to secrete synaptic proteins than neuro-progenitors and immature neurons. This study should facilitate the possibilities to determine the particular role and function of synaptic proteins during cortical development and help dissect the mechanism for synaptic protein release to CSF, as seen in neurodegenerative disease, AD in particular.
Conflicts of interest
HZ has served at advisory boards of Eli Lilly, Roche Diagnostics and Wave, has received travel support from Teva and is a co-founder of Brain Biomarker Solutions in Gothenburg AB, a GU Ventures-based platform company at the University of Gothenburg. KB has served as a consultant or at advisory boards for Alzheon, BioArctic, Biogen, Eli Lilly, Fujirebio Europe, IBL International, Merck, Novartis, Pfizer, and Roche Diagnostics, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB, a GU Ventures-based platform company at the University of Gothenburg. The other authors report no conflicts of interest.
Contributions
FN, LA and HZ designed the study. FN planned and performed neuronal differentiation, collected samples, analyzed data and wrote the manuscript. PB and TS planned and performed neuronal differentiations. FN performed RNA and confocal analysis. FN and BB performed protein analysis. AB planned and performed mass spectrometry experiments and analyzed the data. AS and KH performed ELISA. FN analyzed all secretion data. HZ and KB took part in analyzing and interpreting data. AÖ helped designing experiments to detect SNAP-25. All authors reviewed the manuscript and approved the final version.
Acknowledgements
This work was supported by grants from the Swedish Research Council, the European Research Council, Swedish State Support for Clinical Research (ALFGBG), the Knut and Alice Wallenberg Foundation, Frimurarstiftelsen, Alzheimerfonden, the Parkinson Research Foundation, Stiftelsen Wilhelm och Martina Lundgrens Vetenskapsfond, the Torsten Söderberg Foundation, the Swedish Brain Foundation, and Stiftelsen För Gamla Tjänarinnor. Dr. Kuo-Ping Huang (late) is acknowledged for providing us the anti-NRGN antibody (Ab270).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neuint.2018.10.014.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- Abbas Y., Jeon Y.R., Sokolov A.S., Kim S., Ku B., Choi C. vol. 8. 2018. p. 1228. (Compliance-free, Digital SET and Analog RESET Synaptic Characteristics of Sub-tantalum Oxide Based Neuromorphic Device). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner L., Caroni P. Absence of persistent spreading, branching, and adhesion in GAP-43-depleted growth cones. J. Cell Biol. 1995;128:647–660. doi: 10.1083/jcb.128.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegra Mascaro A.L., Cesare P., Sacconi L., Grasselli G., Mandolesi G., Maco B., Knott G.W., Huang L., De Paola V., Strata P., Pavone F.S. In vivo single branch axotomy induces GAP-43-dependent sprouting and synaptic remodeling in cerebellar cortex. Proc. Natl. Acad. Sci. U.S.A. 2013;110:10824–10829. doi: 10.1073/pnas.1219256110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Bolado G., Rodriguez-Sanchez P., Tejero-Diez P., Fairen A., Diez-Guerra F.J. Neurogranin in the development of the rat telencephalon. Neuroscience. 1996;73:565–580. doi: 10.1016/0306-4522(96)00061-9. [DOI] [PubMed] [Google Scholar]
- Alvarez-Castelao B., Schuman E.M. The regulation of synaptic protein turnover. J. Biol. Chem. 2015;290:28623–28630. doi: 10.1074/jbc.R115.657130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K., Gordon S.L., Grozeva D., van Kogelenberg M., Roberts N.Y., Pike M., Blair E., Hurles M.E., Chong W.K., Baldeweg T., Kurian M.A., Boyd S.G., Cousin M.A., Raymond F.L. Identification of a human synaptotagmin-1 mutation that perturbs synaptic vesicle cycling. J. Clin. Invest. 2015;125:1670–1678. doi: 10.1172/JCI79765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz L.I., Routtenberg A. GAP-43: an intrinsic determinant of neuronal development and plasticity. Trends Neurosci. 1997;20:84–91. doi: 10.1016/s0166-2236(96)10072-2. [DOI] [PubMed] [Google Scholar]
- Bergström P., Agholme L., Nazir F.H., Satir T.M., Toombs J., Wellington H., Strandberg J., Bontell T.O., Kvartsberg H., Holmström M., Boreström C., Simonsson S., Kunath T., Lindahl A., Blennow K., Hanse E., Portelius E., Wray S., Zetterberg H. Amyloid precursor protein expression and processing are differentially regulated during cortical neuron differentiation. Sci. Rep. 2016;6:29200. doi: 10.1038/srep29200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennow K. Cerebrospinal fluid protein biomarkers for Alzheimer's disease. NeuroRx : J. Am. Soc. Exp. NeuroTherap. 2004;1:213–225. doi: 10.1602/neurorx.1.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennow K., Bogdanovic N., Alafuzoff I., Ekman R., Davidsson P. Synaptic pathology in Alzheimer's disease: relation to severity of dementia, but not to senile plaques, neurofibrillary tangles, or the ApoE4 allele. J. Neural. Transm. (Vienna, Austria. 1996;103:603–618. doi: 10.1007/BF01273157. 1996. [DOI] [PubMed] [Google Scholar]
- Blennow K., Hampel H., Weiner M., Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat. Rev. Neurol. 2010;6:131–144. doi: 10.1038/nrneurol.2010.4. [DOI] [PubMed] [Google Scholar]
- Bogdanovic N., Davidsson P., Volkmann I., Winblad B., Blennow K. Growth-associated protein GAP-43 in the frontal cortex and in the hippocampus in Alzheimer's disease: an immunohistochemical and quantitative study. J. Neural. Transm. (Vienna, Austria) 2000;107:463–478. doi: 10.1007/s007020070088. 1996. [DOI] [PubMed] [Google Scholar]
- Boreström C., Simonsson S., Enochson L., Bigdeli N., Brantsing C., Ellerström C., Hyllner J., Lindahl A. Footprint-free human induced pluripotent stem cells from articular cartilage with redifferentiation capacity: a first step toward a clinical-grade cell source. Stem Cells Trans. Med. 2014;3:433–447. doi: 10.5966/sctm.2013-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmalm A., Brinkmalm G., Honer W.G., Frolich L., Hausner L., Minthon L., Hansson O., Wallin A., Zetterberg H., Blennow K., Ohrfelt A. SNAP-25 is a promising novel cerebrospinal fluid biomarker for synapse degeneration in Alzheimer's disease. Mol. Neurodegener. 2014;9:53. doi: 10.1186/1750-1326-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone G.T., Bendotti C., Oster-Granite M.L., Coyle J.T. Developmental expression of the gene encoding growth-associated protein 43 (Gap43) in the brains of normal and aneuploid mice. J. Neurosci. Res. 1991;29:449–460. doi: 10.1002/jnr.490290405. [DOI] [PubMed] [Google Scholar]
- Chen G., Hu T., Wang Z., Li Q., Li J., Jia Y., Xu W.H. Down-regulation of synaptotagmin 1 in cortex, hippocampus, and cerebellum after experimental subarachnoid hemorrhage. Ann. Clin. Lab. Sci. 2013;43:250–256. [PubMed] [Google Scholar]
- Cooper A.P., Gillespie D.C. Synaptotagmins I and II in the developing rat auditory brainstem: synaptotagmin I is transiently expressed in glutamate-releasing immature inhibitory terminals. J. Comp. Neurol. 2011;519:2417–2433. doi: 10.1002/cne.22634. [DOI] [PubMed] [Google Scholar]
- Cupertino R.B., Kappel D.B., Bandeira C.E., Schuch J.B., da Silva B.S., Muller D., Bau C.H., Mota N.R. SNARE complex in developmental psychiatry: neurotransmitter exocytosis and beyond. J. Neural. Transm. (Vienna, Austria) 2016;123:867–883. doi: 10.1007/s00702-016-1514-9. 1996. [DOI] [PubMed] [Google Scholar]
- Davidsson P., Blennow K. Neurochemical dissection of synaptic pathology in Alzheimer's disease. Int. Psychogeriatr. 1998;10:11–23. doi: 10.1017/s1041610298005110. [DOI] [PubMed] [Google Scholar]
- Davies C.A., Mann D.M., Sumpter P.Q., Yates P.O. A quantitative morphometric analysis of the neuronal and synaptic content of the frontal and temporal cortex in patients with Alzheimer's disease. J. Neurol. Sci. 1987;78:151–164. doi: 10.1016/0022-510x(87)90057-8. [DOI] [PubMed] [Google Scholar]
- DeKosky S.T., Scheff S.W. Synapse loss in frontal cortex biopsies in Alzheimer's disease: correlation with cognitive severity. Ann. Neurol. 1990;27:457–464. doi: 10.1002/ana.410270502. [DOI] [PubMed] [Google Scholar]
- Denny J.B. Molecular mechanisms, biological actions, and neuropharmacology of the growth-associated protein GAP-43. Curr. Neuropharmacol. 2006;4:293–304. doi: 10.2174/157015906778520782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Guerra F.J. Neurogranin, a link between calcium/calmodulin and protein kinase C signaling in synaptic plasticity. IUBMB Life. 2010;62:597–606. doi: 10.1002/iub.357. [DOI] [PubMed] [Google Scholar]
- Eagle H., Piez K.A., Fleischman R., Oyama V.I. Protein turnover in mammaliar cell cultures. J. Biol. Chem. 1959;234:592–597. [PubMed] [Google Scholar]
- Fernstrom E., Minta K., Andreasson U., Sandelius A., Wasling P., Brinkmalm A., Hoglund K., Blennow K. 2018. Cerebrospinal Fluid Markers of Extracellular Matrix Remodelling, Synaptic Plasticity and Neuroinflammation before and after Cranial Radiotherapy. [DOI] [PubMed] [Google Scholar]
- Frassoni C., Inverardi F., Coco S., Ortino B., Grumelli C., Pozzi D., Verderio C., Matteoli M. Analysis of SNAP-25 immunoreactivity in hippocampal inhibitory neurons during development in culture and in situ. Neuroscience. 2005;131:813–823. doi: 10.1016/j.neuroscience.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Fukuda M. Distinct developmental expression of synaptotagmin I and IX in the mouse brain. Neuroreport. 2006;17:179–182. doi: 10.1097/01.wnr.0000198435.14142.be. [DOI] [PubMed] [Google Scholar]
- Furuya T.K., Silva P.N., Payao S.L., Bertolucci P.H., Rasmussen L.T., De Labio R.W., Braga I.L., Chen E.S., Turecki G., Mechawar N., Mill J., Smith M.A. Analysis of SNAP25 mRNA expression and promoter DNA methylation in brain areas of Alzheimer's Disease patients. Neuroscience. 2012;220:41–46. doi: 10.1016/j.neuroscience.2012.06.035. [DOI] [PubMed] [Google Scholar]
- Gnatenko D.V., Dunn J.J., McCorkle S.R., Weissmann D., Perrotta P.L., Bahou W.F. Transcript profiling of human platelets using microarray and serial analysis of gene expression. Blood. 2003;101:2285–2293. doi: 10.1182/blood-2002-09-2797. [DOI] [PubMed] [Google Scholar]
- Greif K.F., Asabere N., Lutz G.J., Gallo G. Synaptotagmin-1 promotes the formation of axonal filopodia and branches along the developing axons of forebrain neurons. Develop. Neurobiol. 2013;73:27–44. doi: 10.1002/dneu.22033. [DOI] [PubMed] [Google Scholar]
- Han N.L., Wen J., Lin Q., Tan P.L., Liou Y.C., Sheu F.S. Proteomics analysis of the expression of neurogranin in murine neuroblastoma (Neuro-2a) cells reveals its involvement for cell differentiation. Int. J. Biol. Sci. 2007;3:263–273. doi: 10.7150/ijbs.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo N., Oishi T., Yamashita A., Murata Y., Matsuda K., Hayashi M. Northern blot and in situ hybridization analyses for the neurogranin mRNA in the developing monkey cerebral cortex. Brain Res. 2006;1078:35–48. doi: 10.1016/j.brainres.2006.01.062. [DOI] [PubMed] [Google Scholar]
- Huttenlocher P.R., Dabholkar A.S. Regional differences in synaptogenesis in human cerebral cortex. J. Comp. Neurol. 1997;387:167–178. doi: 10.1002/(sici)1096-9861(19971020)387:2<167::aid-cne1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Inoue Y., Takayanagi M., Sugiyama H. Presynaptic protein synaptotagmin1 regulates the activity-induced remodeling of synaptic structures in cultured hippocampal neurons. J. Neurosci. Res. 2013;91:882–889. doi: 10.1002/jnr.23215. [DOI] [PubMed] [Google Scholar]
- Inoue Y., Kamikubo Y., Ezure H., Ito J., Kato Y., Moriyama H., Otsuka N. Presynaptic protein Synaptotagmin1 regulates the neuronal polarity and axon differentiation in cultured hippocampal neurons. BMC Neurosci. 2015;16:92. doi: 10.1186/s12868-015-0231-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mahalik T.J., Carrier A., Owens G.P., Clayton G. The expression of GAP43 mRNA during the late embryonic and early postnatal development of the CNS of the rat: an in situ hybridization study. Brain Res. Develop. Brain Res. 1992;67:75–83. doi: 10.1016/0165-3806(92)90027-t. [DOI] [PubMed] [Google Scholar]
- Murata Y., Higo N., Oishi T., Yamashita A., Matsuda K., Hayashi M. Developmental changes in the expression of growth-associated protein-43 mRNA in the monkey thalamus: northern blot and in situ hybridization studies. Neuroscience. 2005;136:497–507. doi: 10.1016/j.neuroscience.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Öhrfelt A., Brinkmalm A., Dumurgier J., Brinkmalm G., Hansson O., Zetterberg H., Bouaziz-Amar E., Hugon J., Paquet C., Blennow K. The pre-synaptic vesicle protein synaptotagmin is a novel biomarker for Alzheimer's disease. Alzheimer's Res. Ther. 2016;8:41. doi: 10.1186/s13195-016-0208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto J.G., Jones D.G., Murphy K.M. Comparing development of synaptic proteins in rat visual, somatosensory, and frontal cortex. Front. Neural Circ. 2013;7:97. doi: 10.3389/fncir.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Represa A., Deloulme J.C., Sensenbrenner M., Ben-Ari Y., Baudier J. Neurogranin: immunocytochemical localization of a brain-specific protein kinase C substrate. J. Neurosci.: Off. J. Soc. Neurosci. 1990;10:3782–3792. doi: 10.1523/JNEUROSCI.10-12-03782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routtenberg A., Cantallops I., Zaffuto S., Serrano P., Namgung U. Enhanced learning after genetic overexpression of a brain growth protein. Proc. Natl. Acad. Sci. U.S.A. 2000;97:7657–7662. doi: 10.1073/pnas.97.13.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelius Å., Portelius E., Källén Å., Zetterberg H., Rot U., Olsson B., Toledo J.B., Shaw L.M., Lee V.M.Y., Irwin D.J., Grossman M., Weintraub D., Chen-Plotkin A., Wolk D.A., McCluskey L., Elman L., Kostanjevecki V., Vandijck M., McBride J., Trojanowski J.Q., Blennow K. 2018. CSF GAP-43 Increase Is Alzheimer's Disease-specific and Associated with Tau and Amyloid Pathology. Alzheimer's & Dementia (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupp M., Malsam J., Ruiter M. Interactions between SNAP-25 and synaptotagmin-1 are involved in vesicle priming. Clamping Spontaneous and Stimulating Evoked Neurotransm. 2016;36:11865–11880. doi: 10.1523/JNEUROSCI.1011-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Kirwan P., Livesey F.J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- Sidor-Kaczmarek J., Labuda C., Litwinowicz B., Spodnik J.H., Kowianski P., Dziewiatkowski J., Morys J. Developmental expression of SNAP-25 protein in the rat striatum and cerebral cortex. Folia Morphol. 2004;63:285–288. [PubMed] [Google Scholar]
- Sjogren M., Davidsson P., Gottfries J., Vanderstichele H., Edman A., Vanmechelen E., Wallin A., Blennow K. The cerebrospinal fluid levels of tau, growth-associated protein-43 and soluble amyloid precursor protein correlate in Alzheimer's disease, reflecting a common pathophysiological process. Dement. Geriatr. Cognit. Disord. 2001;12:257–264. doi: 10.1159/000051268. [DOI] [PubMed] [Google Scholar]
- Sobecki M., Mrouj K., Camasses A., Parisis N. vol. 5. 2016. (The Cell Proliferation Antigen Ki-67 Organises Heterochromatin). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sposito T., Preza E., Mahoney C.J., Seto-Salvia N., Ryan N.S., Morris H.R., Arber C., Devine M.J., Houlden H., Warner T.T., Bushell T.J., Zagnoni M., Kunath T., Livesey F.J., Fox N.C., Rossor M.N., Hardy J., Wray S. Developmental regulation of tau splicing is disrupted in stem cell-derived neurons from frontotemporal dementia patients with the 10 + 16 splice-site mutation in MAPT. Hum. Mol. Genet. 2015;24:5260–5269. doi: 10.1093/hmg/ddv246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof T.C., Rizo J. Synaptic vesicle exocytosis. Cold Spring Harbor Perspectiv. Biol. 2011;3 doi: 10.1101/cshperspect.a005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thorsell A., Bjerke M., Gobom J., Brunhage E., Vanmechelen E., Andreasen N., Hansson O., Minthon L., Zetterberg H., Blennow K. Neurogranin in cerebrospinal fluid as a marker of synaptic degeneration in Alzheimer's disease. Brain Res. 2010;1362:13–22. doi: 10.1016/j.brainres.2010.09.073. [DOI] [PubMed] [Google Scholar]
- Verheijen R., Kuijpers H.J., van Driel R., Beck J.L., van Dierendonck J.H., Brakenhoff G.J., Ramaekers F.C. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. II. Localization in mitotic cells and association with chromosomes. J. Cell Sci. 1989;92(Pt 4):531–540. doi: 10.1242/jcs.92.4.531. [DOI] [PubMed] [Google Scholar]
- Zhong L., Brown J., Kramer A. vol. 35. 2015. pp. 7503–7508. (Increased Prefrontal Cortex Neurogranin Enhances Plasticity and Extinction Learning). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylbersztejn K., Galli T. Vesicular traffic in cell navigation. FEBS J. 2011;278:4497–4505. doi: 10.1111/j.1742-4658.2011.08168.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.