Abstract
Post-transcriptional addition of poly(A) tails to the 3′ end of RNA is one of the fundamental events controlling the functionality and fate of RNA in all kingdoms of life. Although an enzyme with poly(A)-adding activity was discovered in Escherichia coli more than 50 years ago, its existence and role in prokaryotic RNA metabolism were neglected for many years. As a result, it was not until 1992 that E. coli poly(A) polymerase I was purified to homogeneity and its gene was finally identified. Further work revealed that, similar to its role in surveillance of aberrant nuclear RNAs of eukaryotes, the addition of poly(A) tails often destabilizes prokaryotic RNAs and their decay intermediates, thus facilitating RNA turnover. Moreover, numerous studies carried out over the last three decades have shown that polyadenylation greatly contributes to the control of prokaryotic gene expression by affecting the steady-state level of diverse protein-coding and non-coding transcripts including antisense RNAs involved in plasmid copy number control, expression of toxin–antitoxin systems and bacteriophage development. Here, we review the main findings related to the discovery of polyadenylation in prokaryotes, isolation, and characterization and regulation of bacterial poly(A)-adding activities, and discuss the impact of polyadenylation on prokaryotic mRNA metabolism and gene expression.
This article is part of the theme issue ‘5′ and 3′ modifications controlling RNA degradation’.
Keywords: polyadenylation, mRNA stability, tRNA surveillance, small RNA turnover, gene expression, bacteria
1. Introduction
Polyadenylation refers to an enzymatic process carried out by poly(A) polymerase to add adenine residues to the 3′ extremity of RNAs. Following its discovery, various poly(A) polymerases and their functional homologues were found in a variety of pro- and eukaryotic cells and were partially purified and characterized (reviewed in [1]). These findings and the detection of poly(A) sequences in phylogenetically distant organisms [2] suggested the widespread presence of polyadenylation in pro- and eukaryotic organisms. The poly(A) tails at the 3′ end of mRNA were considered for a long time a characteristic feature of eukaryotic mRNAs (except those encoding metazoan replication-dependent histone mRNAs) and were known to stabilize eukaryotic mRNAs and promote their translation. By contrast, polyadenylation in prokaryotes gained little attention and was neglected until the pioneering work of Nilima Sarkar published in 1992 [3,4].
The polyadenylation story in prokaryotes began in 1962 [5,6], with the partial purification and characterization of Escherichia coli poly(A) polymerase I (PAP I). In the following years, polyadenylated RNAs were detected in Caulobacter crescentus and E. coli and later in Bacillus brevis, Bacillus subtilis, Rhodopseudomonas capsulata and Rhodospirillum rubrum (reviewed in [7,8]). By analysing the length of poly(A) tracts in prokaryotic RNAs bound to oligo(dT) cellulose, it was found that the median length of poly(A) stretches was between 14 and 60 nucleotides and that only a small fraction of total RNA was polyadenylated. Further progress in the study of polyadenylation was achieved after the systematic analysis of poly(A) tracts of E. coli trpA and lpp transcripts [3,9,10]. Then the E. coli pcnB gene, a non-essential gene previously reported to control ColE1 plasmid copy number [11–14], was identified as the gene encoding poly(A) polymerase I [3,15], providing the first evidence of the biological function of polyadenylation in E. coli. Namely, it was shown that polyadenylation promotes the rapid turnover of RNAI, a small antisense RNA complementary to RNAII, which, in turn, serves as a primer for DNA polymerase during replication of ColE1-type plasmids [15–17]. Later, the elucidation of the degradation pathways of both non-coding and protein-coding RNAs demonstrated the involvement of E. coli poly(A) polymerase I in the turnover of several RNAs, thus reinforcing the idea that polyadenylation often destabilizes bacterial transcripts and their decay intermediates. The destabilizing role of polyadenylation is not restricted to bacteria. It was later demonstrated in Archaea and chloroplasts, and in mitochondria and nuclei of diverse eukaryotes including yeasts, plants and humans [18–25].
2. Escherichia coli poly(A) polymerase I and functionally related enzymes able to carry out poly(A) addition in bacteria
A few years prior to the identification of the biological function of the E. coli pcnB gene, it was noted that the product of this gene shares a high sequence similarity with tRNA nucleotidyltransferase [14], an enzyme that catalyses the addition of A and C residues to incomplete CCA termini of tRNAs [26]. Both enzymes are members of the class II polymerase beta-superfamily of nucleotidyltransferases [27] and poly(A) polymerase I, upon its overexpression, can partially compensate for the absence of tRNA nucleotidyltransferase by adding (although with a lower efficiency) a 3'-terminal CCA sequence to defective tRNAs in E. coli [28]. Moreover, it was demonstrated that the high sequence similarity of these enzymes opens up the possibility to swap amino acids within the evolutionarily conserved N-terminal catalytic domain and to generate recombinant polypeptides with altered specificities [29].
The difficulty in distinguishing poly(A) polymerases from other closely related enzymes makes it challenging to identify poly(A) polymerase genes in other bacteria, including Gram-positive species. However, additional genetic tools and functional assays have been used to assure the enzyme's identity. For instance, among three candidates tested for the role of poly(A) polymerase in Streptomyces coelicolor, only polynucleotide phosphorylase (PNPase) was found to be responsible for RNA 3′-tail synthesis in vivo [30]. PNPase, known for its 3′–5′ degradative activity of RNA, also possesses a 3'-terminal oligonucleotide polymerase activity at low inorganic phosphate concentrations. Namely, the enzyme is able to add A, C and U residues to an RNA chain in E. coli in the absence of PAP I [31]. Attempts to identify a poly(A)-adding enzyme in another Gram-positive bacterium, B. subtilis [32], showed that PNPase was not able to carry out polyadenylation of RNA in vivo and also did not identify any other enzyme(s) possessing a poly(A)-adding activity. Therefore, it was proposed that polyadenylation in B. subtilis was likely performed by a still unknown enzyme [30,32].
3. Biochemical properties of Escherichia coli poly(A) polymerase I
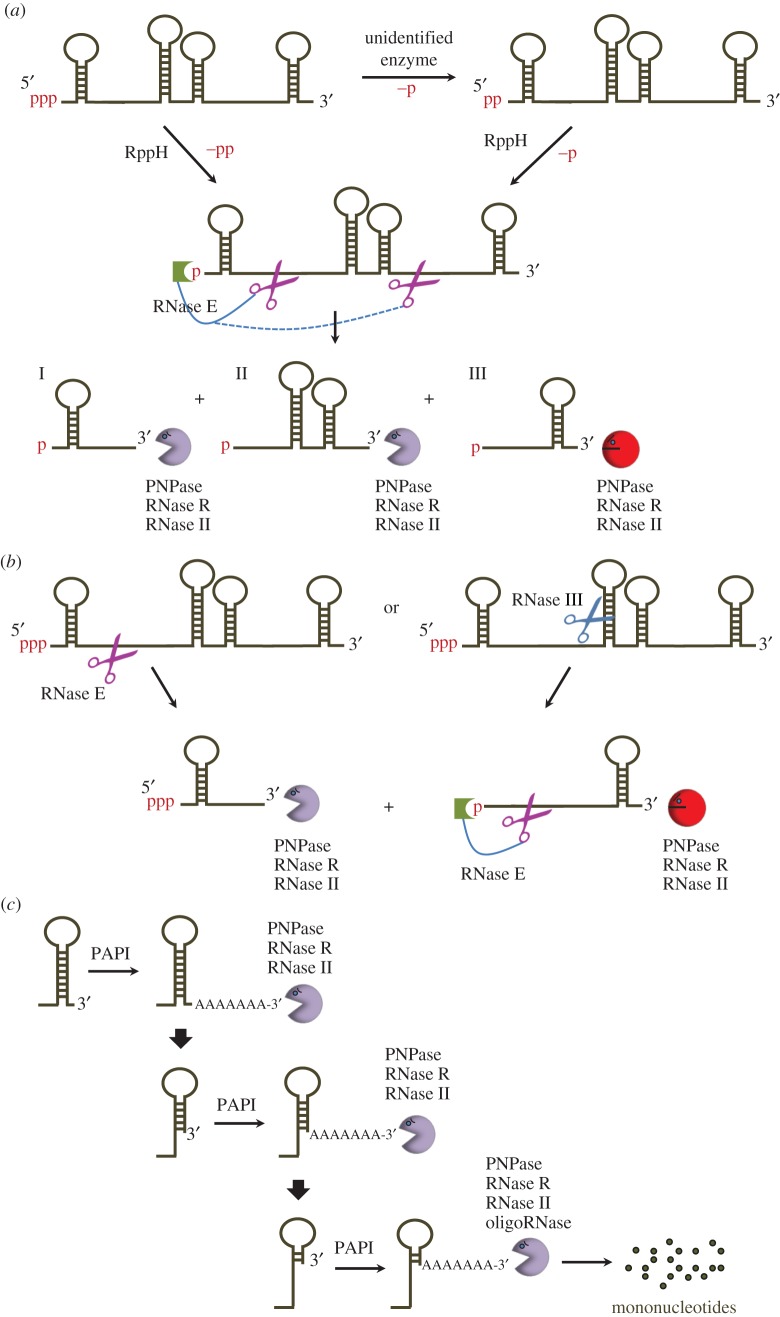
PAP I is a monomer of approximately 53 kDa. The enzyme requires divalent metal ions (i.e. Mg2+ or Mn2+) for activity and it uses ATP as a substrate to add adenosine residues at the 3′ end of RNA molecules. Moreover, in vitro PAP I prefers to carry out polyadenylation of non-structured RNAs and its activity is inhibited by 3′-terminal stem–loop structures [33]. Although CTP can also be used by PAP I, the efficiency of its incorporation is only 5% of that observed for ATP [33,34]. These properties may explain why Cs are occasionally incorporated in poly(A) tails [35]. In vivo poly(A) tails were detected at the 3′ ends of many (but not all) RNA, including primary transcripts, processed RNAs and intermediate products of exonucleolytic degradation [36–39]. However, folding predictions and searches for conserved motifs potentially present in polyadenylated RNA could not reveal any consensus sequence or structure that could serve as a signal for PAP I to begin poly(A) addition [37]. Instead, in vitro studies suggested that PAP I activity is controlled by more general features of RNA. Namely, it was found that unpaired 5′ and 3′ termini, as well as the presence of 5′-monophosphate in substrate RNAs, could greatly increase the rate of poly(A) addition [34,40]. These findings are consistent with the well-documented role of polyadenylation in accelerating the decay of intermediate products of RNA degradation that are produced in the initial steps of RNA decay, i.e. after the removal of 5′-diphosphate by the combined action of RppH and an unknown enzyme [41] followed by the initial RNase E cleavage (figure 1a) or after ‘direct entry cleavages' made by RNase E or RNase III (figure 1b) [43]. As the resulting decay intermediates carry 5′-monophosphate groups and likely possess unstructured 5′ and 3′ termini, they are further targeted for polyadenylation and hence rapid turnover (figure 1c).
Figure 1.
RNA turnover in E. coli and its facilitation by polyadenylation. (a) The principal pathway for RNA turnover in E. coli. RNA decay begins with pyrophosphate removal by the combined action of RppH and an unknown enzyme [41], followed by endonucleolytic cleavage(s) made by the 5′-monophosphate-dependent endoribonuclease, RNase E. The resulting intermediate products such as I, II and III (schematically shown above) are further degraded by the combined action of 3′–5′ exonucleases, primarily including polynucleotide phosphorylase (PNPase), ribonuclease R (RNase R) and ribonuclease II (RNase II). (b) Degradation of some transcripts can involve ‘direct entry’ cleavages made by RNase E or RNase III. The resulting decay intermediates are further degraded by exo- and endonucleases. (c) The PAP I-dependent degradation of structured RNAs in E. coli. Although stable stem–loop structures at the 3′ end of E. coli transcripts and their decay intermediates (e.g. fragment III shown in (a)) per se are resistant to the action of major E. coli exoribonucleases, their decay can be facilitated by polyadenylation. Moreover, repeated cycles of polyadenylation and subsequent 3′–5′ degradation of the polyadenylated species by exoribonucleases ensure their progressive degradation in vivo [42], in turn rendering short oligonucleotides that are further digested by oligoribonuclease (oligoRNase) to yield mononucleotides. (Online version in colour.)
4. Regulation of poly(A) polymerase I level and activity
The intracellular level of E. coli PAP I is usually low, as its overproduction is detrimental to cell growth [13,44]. Previous work has shown that an increase in the abundance of this enzyme beyond its normal level (e.g. by plasmid-dependent overexpression) leads to polyadenylation of mature tRNA. This results in a dramatic decrease in the level of functional tRNAs and a concomitant inhibition of translation, ultimately leading to cell death [45].
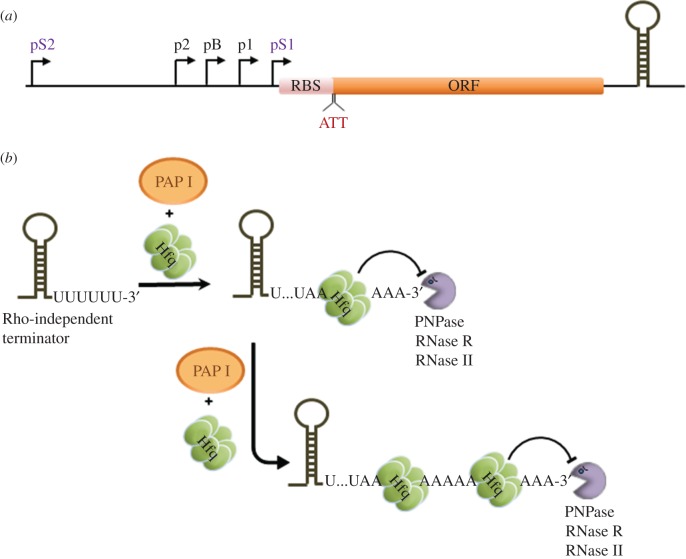
The tight control of PAP I production is exerted at the transcriptional and post-transcriptional levels. Transcription of pcnB mRNA can be initiated from five distinct promoters (figure 2a; [47]). The activities of at least two of them are inversely dependent on the growth rate, which is manifested by an increase in the abundance of PAP I and higher levels of RNA polyadenylation observed in slowly growing bacteria [48,49]. The intricate regulation of pcnB transcription in E. coli was shown to be dependent on two sigma factors, σ70 and σS, responsible for promoter recognition by RNA polymerase, and on ppGpp and DksA, two effectors of the stringent response produced in response to amino acid starvation [46]. Moreover, transcriptional control of pcnB expression in other bacteria seems to also involve additional transcription factors [50]. The use of the non-canonical initiation codon AUU to initiate the translation of pcnB may be another way to limit the availability of PAP I. Moreover, there are indications that PAP I activity could potentially be regulated post-translationally. It was reported that a PAP I variant, with a C-terminal His tag, can be phosphorylated both in vivo and in vitro and that this modification impairs its activity [48]. However, it remains unknown whether phosphorylation of the untagged enzyme occurs in vivo or in vitro and can likewise impact PAP I activity.
Figure 2.
Control of poly(A) polymerase I level and activity in E. coli. (a) Control of E. coli pcnB expression. Transcription of the pcnB gene coding for PAP I is driven by three σ70-dependent (i.e. p1, pB and p2) and two σS-dependent (pS1 and pS2) promoters [46]. Moreover, ppGpp and DksA were found to directly inhibit transcription from pB, pS1 and pS2 and the overall low level of PAP I is maintained owing to lower efficiency of translation caused by the presence of a non-canonical start codon (AUU) within the ribosome-binding site (RBS). (b) Effect of Hfq on poly(A) addition. The RNA chaperone Hfq can bind to poly(A) tails, thereby impeding their exonucleolytic degradation and concomitantly increasing the processivity of the poly(A) addition by PAP I. ORF, open reading frame. (Online version in colour.)
Apart from the regulation of pcnB expression, polyadenylation can be controlled by the RNA chaperone Hfq. The latter was found to bind to poly(A) tails [34] and stimulate their elongation in vitro by increasing the processivity of PAP I (figure 2b; [51]). Moreover, inactivation of Hfq affects recognition and polyadenylation of transcripts terminated by Rho-independent terminators. For instance, the average length, occurrence of polyadenylated transcripts and percentage of poly(A) tails located at the 3′ end of the rpsO Rho-independent transcription terminator were considerably higher in the wild-type strain than in the hfq mutants [51,52]. Therefore, it is possible that oligo(A) tails less than 10 nt in length may be too short to form stable complexes with Hfq, but seem to be long enough to be used by 3′–5′ exoribonucleases which carry out the degradation of structured RNAs [53]. When the tails are longer, Hfq binds tightly to these oligo(A) sequences and may protect polyadenylated RNA from RNase II and PNPase-mediated decay as shown in vitro (figure 2b [54]).
5. Poly(A)-dependent mechanisms and their role in RNA metabolism and gene expression
An increasing number of studies indicate that polyadenylation is broadly used by bacteria to control RNA metabolism (mRNA turnover, stable RNA surveillance and recycling of small RNAs), thereby regulating gene expression at the post-transcriptional level.
(a). Effects of polyadenylation on mRNA and stable RNA metabolism
Polyadenylation of prokaryotic transcripts is best known for its ancillary role in RNA turnover. Although the rate-limiting step in the degradation of prokaryotic mRNA usually involves an endoribonucleolytic cleavage at the 5′ end of the transcript subsequently resulting in its functional inactivation (figure 1a), the remaining steps that are responsible for further fragmentation of the primary cleavage products and their ultimate conversion to nucleotides often involve the combined action of endo- (RNase E/G, RNase III, RNase Y or RNase J1/J2) and exonucleases (e.g. PNPase, RNase II, RNase R and oligoribonuclease) [55]. Moreover, the three major exonucleases (i.e. PNPase, RNase II and RNase R) were shown to play different roles in the PAP I-dependent pathway in E. coli (figure 1c). In contrast to PNPase and RNase R, which initiate and efficiently degrade polyadenylated structured RNAs [56–60], RNase II appears to play a protective role. RNase II is unable to degrade structured RNAs, but because it can eliminate poly(A) tails from polyadenylated decay intermediates, it thereby inhibits their exonucleolytic degradation by PNPase and RNase R [34,61]. In addition to the above mechanism, polyadenylated primary transcripts and their decay intermediates can also be stabilized by Hfq (see above), which binds to their poly(A) tails and thus protects them from exonucleolytic decay [54].
The efficiency of mRNA turnover is often regulated by additional cis- and trans-acting factors differentially affecting the susceptibility of transcripts to the components of the translation apparatus or the RNA decay machine (reviewed in [62]). In vivo studies have shown that stable RNA structures (e.g. transcriptional terminators or repetitive extragenic palindromic (REP) sequences [63,64]) at the 3′ end of decay intermediates or processed transcripts [35,36,65] can confer resistance to degradation by 3′–5′ exonucleases and therefore can protect the adjacent upstream regions from exonucleolytic decay in vivo. Analysis of this resistance mechanism in vitro has revealed that E. coli can employ at least two different strategies to overcome the stabilizing effect of secondary structures.
The first one is associated with the formation of the E. coli degradosome, a multienzyme ribonucleolytic complex, whose major components include RNase E, PNPase, the RNA helicase B (RhlB) and enolase [66,67]. It has been shown that the interaction of RhlB and PNPase in the context of the degradosome enables their functional cooperation in the degradation of structured RNAs [67,68]. Namely, the RNA-unwinding activity of RhlB partly unfolds structured RNAs, yielding single-stranded 3′ ends, thereby facilitating 3′–5′ degradation by exonucleases.
An alternative mechanism involves the action of poly(A) polymerase I (figure 1b). PAP I adds poly(A) tails, thus providing a toe-hold for 3′–5′ exoribonucleases unable to bind in the absence of such single-stranded stretches of nucleotides downstream of stable stem–loop structures. However, once exonucleases become attached to free 3′ ends, they can progressively digest structured RNAs [42,68,69]. As the processivity of exonucleases such as PNPase on highly structured RNA is low, they cannot finish digestion of RNA during a single round owing to their dissociation from substrates [42]. Nevertheless, consecutive cycles of polyadenylation and exonucleolytic degradation are believed to ensure the complete degradation to mononucleotides (figure 1c [42]). In principle, the physical interaction of PAP I and the degradosome detected in vivo [70] suggests that E. coli can likely orchestrate mRNA turnover by coordinating the action of the degradosome- and poly(A)-dependent pathways.
Although polyadenylation of stable RNA precursors in vivo was discovered a long time ago, its biological significance remains largely unknown, in particular for 5S and 23S RNAs [71,72]. As to the contribution of polyadenylation to tRNA metabolism, it is proposed to be linked to the role of PAP I in a quality-control mechanism. The latter helps to eliminate defective tRNAs by targeting them for 3′–5′ exonucleolytic decay [37,73], apparently independent of an interaction between PAP I and the degradosome.
(b). Polyadenylation and its role in the regulation of gene expression
Apart from its contribution to recycling of structured RNA decay intermediates and surveillance of tRNA, polyadenylation also plays an important role in the regulation of gene expression. The regulatory function of polyadenylation can affect the steady-state levels of mRNAs available for translation either positively or negatively (figure 3 and table 1) [37,39,74–77].
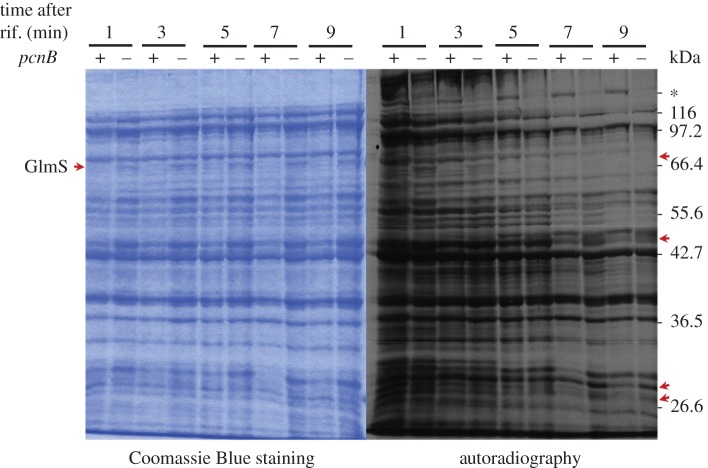
Figure 3.
Effects of PAP I inactivation on functional mRNA stability and protein synthesis in E. coli. The residual capacity of E. coli mRNAs to direct protein synthesis after inhibition of RNA synthesis by rifampicin (rif.) was assessed in the wild-type (pcnB+) and pcnB mutant (pcnB−) strains. Soluble proteins extracted from cells that were treated with rifampicin for 1, 3, 5, 7 and 9 min and pulse-labelled with radioactive methionine as described in [74] were separated on SDS-polyacrylamide gels. The steady-state level of polypeptides and those synthesized de novo were revealed by Coomassie Blue staining and autoradiography, respectively. The position of GlmS and unknown polypeptides overproduced in the mutant strain are indicated by arrowheads, while the position of an unknown protein overproduced in the wild-type strain is indicated by an asterisk.
Table 1.
Role of polyadenylation in global RNA turnover [39]. Indicated are the numbers of genes that were up- or downregulated in the pcnB mutant when compared with the wild-type (wt) strain. Their functional classification is provided according to the Gene Ontology database. The first number represents the number of regulated transcripts with log2 fold change (log2FC) greater than 2, whereas the second one corresponds to the transcripts with log2FC between 1.5 and 2.
| functional categories |
pcnB versus wt |
|
|---|---|---|
| up | down | |
| membrane proteins | 7/7 | 9/12 |
| prophage proteins | 1/4 | 2/0 |
| RNA metabolism | 2/2 | 1 |
| ammonium metabolism | 3/1 | 1/4 |
| motility (flagella proteins) | 0 | 11/15 |
| cell adhesion (fimbriae) | 1/5 | 0 |
| catabolism | 0 | 2/5 |
| miscellaneous | 10/22 | 9/10 |
Besides its apparently direct role in the regulation of mRNA abundance, polyadenylation can also indirectly influence gene expression controlled by numerous cis- and trans-encoded small RNAs. Previous studies have shown that the addition of poly(A) tails has a destabilizing effect on several cis-encoded antisense RNAs known for their role in the control of plasmid replication (e.g. RNAI [17,56] and CopA [78] controlling the copy number of ColE1-type and R1 plasmids, respectively), bacteriophage lambda development (Oop RNA [79]), bacteriophage P4 immunity (CI RNA [80]) and expression of Type I toxin–antitoxin systems (Sok, [81], and others [39]).
Likewise, polyadenylation is known to lead to destabilization of trans-encoded sRNAs. Recent studies have shown that polyadenylation destabilizes the small RNA GlmY involved in the GlmZ-dependent control of glmS expression [82,83]. Besides controlling the level of GlmY, polyadenylation also affects the steady-state levels of RyjA, RybB, SroH [84] and many other sRNAs [39] that become more abundant in pcnB mutant strains. Nevertheless, the actual impact of poly(A) tails on the biological functions of most of these riboregulators remains unknown and merits further analysis.
(c). Perspectives
Despite the significant progress recently achieved in revealing the multifaceted role of polyadenylation in the regulation of prokaryotic RNA metabolism and gene expression, there are still many open questions that have to be addressed in future.
In particular, more work needs to be done to characterize the positive effect of polyadenylation on prokaryotic gene expression. Namely, in contrast to the well-known examples, in which the addition of poly(A) tails to the 3′ end of prokaryotic RNAs or their decay intermediates promotes RNA turnover and reduces the level of functional RNA, the presence of an active poly(A) polymerase in certain cases can enhance the steady-state transcript level (e.g. those expressed from the E. coli flagellar operon [76] and others [39]). The mechanisms, direct or indirect, by which polyadenylation can increase RNA levels and whether this also enhances gene expression are poorly understood and warrant further analysis.
In addition, future studies should reveal more details regarding the complex regulatory networks used by E. coli and other bacteria to coordinate and modulate the action of poly(A) polymerase I and its interaction with known (e.g. the degradosome and Hfq) or still unknown interacting partners to exert post-transcriptional control of gene expression.
Moreover, although the pcnB gene per se is not essential for sustaining E. coli survival and growth under laboratory conditions, the gene might play a more critical role in cell survival in adverse environments. Therefore, it is of particular interest to compare the regulation of PAP I expression and activity along with its impact on gene expression under normal and various stress conditions. This might yield particularly interesting results by testing the effect of major stress factors (such as limitation of iron and nutrients, oxidative stress, etc.) faced by bacterial pathogens in their hosts.
Finally, as the main findings regarding the role of polyadenylation in gene expression and cell physiology were obtained by using E. coli as a model organism, there is little known about the actual mechanisms controlled by polyadenylation in other bacteria. In this regard, the recently reported function of the Pantoea agglomerans YS19 pcnB gene in the indole regulatory pathway [85] found to be involved in host–pathogen interactions is intriguing and clearly demonstrates the merits of using phylogenetically distant organisms to discover new biological functions.
Acknowledgements
The authors are indebted to Jackie Plumbridge for critical reading of the manuscript.
Data accessibility
This article has no additional data.
Authors' contributions
E.H. and V.R.K. wrote the paper.
Competing interests
We have no competing interests.
Funding
This work was supported by the Centre National de la Recherche Scientifique (UMR8261), University Paris-Diderot, ‘Initiative d'Excellence’ programme from the French State (grant ‘DYNAMO’, ANR-11-LABX-0011) (to E.H.), Spanish Ministry of Economy and Competitiveness grant CGL2015-70929-R and IKERBASQUE (Basque Foundation for Science) (to V.R.K.)
References
- 1.Edmonds M, Winters MA. 1976. Polyadenylate polymerases. Prog. Nucleic Acid Res. Mol. Biol. 17, 149–179. ( 10.1016/S0079-6603(08)60069-0) [DOI] [PubMed] [Google Scholar]
- 2.Karpetsky TP, Boguski MS, Levy CC. 1979. Structures, properties, and possible biologic functions of polyadenylic acid. Subcell. Biochem. 6, 1–116. [DOI] [PubMed] [Google Scholar]
- 3.Cao G-J, Sarkar N. 1992. Poly(A) RNA in Escherichia coli: nucleotide sequence at the junction of the lpp transcript and the polyadenylate moiety. Proc. Natl Acad. Sci. USA 89, 7546–7550. ( 10.1073/pnas.89.16.7546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao G-J, Sarkar N. 1992. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl Acad. Sci. USA 89, 10 380–10 384. ( 10.1073/pnas.89.21.10380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.August JT, Ortiz PJ, Hurwitz J. 1962. Ribonucleic acid-dependent ribonucleotide incorporation. I. Purification and properties of the enzyme. J. Biol. Chem. 237, 3786–3793. [PubMed] [Google Scholar]
- 6.Sippel AE. 1973. Purification and characterization of adenosine triphosphate:ribonucleic acid adenyltransferase from Escherichia coli. Eur. J. Biochem. 37, 31–40. ( 10.1111/j.1432-1033.1973.tb02953.x) [DOI] [PubMed] [Google Scholar]
- 7.Sarkar N. 1996. Polyadenylation of mRNA in bacteria. Microbiology 142, 3125–3133. ( 10.1099/13500872-142-11-3125) [DOI] [PubMed] [Google Scholar]
- 8.Sarkar N. 1997. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 66, 173–197. ( 10.1146/annurev.biochem.66.1.173) [DOI] [PubMed] [Google Scholar]
- 9.Karnik P, Taljanidisz J, Sasvari-Szekely M, Sarkar N. 1987. 3'-terminal polyadenylate sequences of Escherichia coli tryptophan synthetase α-subunit messenger RNA. J. Mol. Biol. 196, 347–354. ( 10.1016/0022-2836(87)90695-4) [DOI] [PubMed] [Google Scholar]
- 10.Taljanidisz J, Karnik P, Sarkar N. 1987. Messenger ribonucleic acid for lipoprotein of the Escherichia coli outer membrane is polyadenylated. J. Mol. Biol. 193, 507–515. ( 10.1016/0022-2836(87)90263-4) [DOI] [PubMed] [Google Scholar]
- 11.Lopilato J, Bortner S, Beckwith J. 1986. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol. Gen. Genet. 205, 285–290. ( 10.1007/BF00430440) [DOI] [PubMed] [Google Scholar]
- 12.March JB, Colloms MD, Hart-Davis D, Oliver IR, Masters M. 1989. Cloning and characterization of an Escherichia coli gene, pcnB, affecting plasmid copy number. Mol. Microbiol. 3, 903–910. ( 10.1111/j.1365-2958.1989.tb00239.x) [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Parkinson JS. 1989. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J. Bacteriol. 171, 1254–1261. ( 10.1128/jb.171.3.1254-1261.1989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masters M, March JB, Oliver IR, Collins JF. 1990. A possible role for the pcnB gene product of Escherichia coli in modulating RNA:RNA interactions. Mol. Gen. Genet. 220, 341–344. ( 10.1007/BF00260507) [DOI] [PubMed] [Google Scholar]
- 15.Masters M, Colloms MD, Oliver IR, He L, Macnaughton EJ, Charters Y. 1993. The pcnB gene of Escherichia coli, which is required for ColE1 copy number maintenance, is dispensable. J. Bacteriol. 175, 4405–4413. ( 10.1128/jb.175.14.4405-4413.1993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L, Söderbom F, Wagner EGH, Binnie U, Binns N, Masters M. 1993. PcnB is required for the rapid degradation of RNAI, the antisense RNA that controls the copy number of ColE1-related plasmids. Mol. Microbiol. 9, 1131–1142. ( 10.1111/j.1365-2958.1993.tb01243.x) [DOI] [PubMed] [Google Scholar]
- 17.Xu F, Lin-Chao S, Cohen SN. 1993. The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc. Natl Acad. Sci. USA 90, 6756–6760. ( 10.1073/pnas.90.14.6756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portnoy V, Evguenieva-Hackenberg E, Klein F, Walter P, Lorentzen E, Klug G, Schuster G. 2005. RNA polyadenylation in Archaea: not observed in Haloferax while the exosome polynucleotidylates RNA in Sulfolobus. EMBO Rep. 6, 1188–1193. ( 10.1038/sj.embor.7400571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kudla J, Hayes R, Gruissem W. 1996. Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J. 15, 7137–7146. ( 10.1002/j.1460-2075.1996.tb01105.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupold DS, Caoile AGFS, Stern DB. 1999. Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell 11, 1565–1577. ( 10.1105/tpc.11.8.1565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gagliardi D, Leaver CJ. 1999. Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 18, 3757–3766. ( 10.1093/emboj/18.13.3757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3, e189 ( 10.1371/journal.pbio.0030189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724. ( 10.1016/j.cell.2005.04.029) [DOI] [PubMed] [Google Scholar]
- 24.Wyers F, et al. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121, 725–737. ( 10.1016/j.cell.2005.04.030) [DOI] [PubMed] [Google Scholar]
- 25.Slomovic S, Laufer D, Geiger D, Schuster G. 2005. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol. Cell. Biol. 25, 6427–6435. ( 10.1128/MCB.25.15.6427-6435.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deutscher MP. 1990. Ribonucleases, tRNA nucleotidyltransferase, and the 3′ processing of tRNA. Prog. Nucleic Acid Res. Mol. Biol. 39, 209–240. ( 10.1016/S0079-6603(08)60628-5) [DOI] [PubMed] [Google Scholar]
- 27.Yue D, Maizels N, Weiner AM. 1996. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA 2, 895–908. [PMC free article] [PubMed] [Google Scholar]
- 28.Reuven NB, Zhou Z, Deutscher MP. 1997. Functional overlap of tRNA nucleotidyltransferase, poly(A)polymerase I and polynucleotide phosphorylase. J. Biol. Chem. 272, 33255–33259. ( 10.1074/jbc.272.52.33255) [DOI] [PubMed] [Google Scholar]
- 29.Betat H, Rammelt C, Martin G, Mörl M. 2004. Exchange of regions between bacterial poly(A)polymerase and the CCA-adding enzyme generates altered specificities. Mol. Cell 15, 389–398. ( 10.1016/j.molcel.2004.06.026) [DOI] [PubMed] [Google Scholar]
- 30.Bralley P, Gust B, Chang S, Chater KF, Jones GH. 2006. RNA 3'-tail synthesis in Streptomyces: in vitro and in vivo activities of RNase PH, the SCO3896 gene product and polynucleotide phosphorylase. Microbiology 152, 627–636. ( 10.1099/mic.0.28363-0) [DOI] [PubMed] [Google Scholar]
- 31.Mohanty BK, Kushner SR. 2000. Polynucleotide phosphorylase functions both as a 3'→5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl Acad. Sci. USA 97, 11966–11971. ( 10.1073/pnas.220295997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campos-Guillén J, Bralley P, Jones GH, Bechhofer DH, Olmedo-Alvarez G. 2005. Addition of poly(A) and heteropolymeric 3′ ends in Bacillus subtilis wild-type and polynucleotide phosphorylase-deficient strains. J. Bacteriol. 187, 4698–4706. ( 10.1128/JB.187.14.4698-4706.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yehudai-Resheff S, Schuster G. 2000. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA binding affinities and RNA structure dependence. Nucleic Acids Res. 28, 1139–1144. ( 10.1093/nar/28.5.1139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Folichon M, Allemand F, Régnier P, Hajnsdorf E. 2005. Stimulation of poly(A) synthesis by E. coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 272, 454–463. ( 10.1111/j.1742-4658.2004.04485.x) [DOI] [PubMed] [Google Scholar]
- 35.Mohanty BK, Kushner SR. 1999. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 34, 1094–1108. ( 10.1046/j.1365-2958.1999.01673.x) [DOI] [PubMed] [Google Scholar]
- 36.Haugel-Nielsen J, Hajnsdorf E, Régnier P. 1996. The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J. 15, 3144–3152. ( 10.1002/j.1460-2075.1996.tb00677.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maes A, Gracia C, Hajnsdorf E, Régnier P. 2012. Search for poly(A) polymerase targets in E. coli reveals its implication in surveillance of Glu tRNA processing and degradation of stable RNAs. Mol. Microbiol. 83, 436–451. ( 10.1111/j.1365-2958.2011.07943.x). [DOI] [PubMed] [Google Scholar]
- 38.Régnier P, Marujo PE. 2002. Polyadenylation and degradation of RNA in Escherichia coli. In Translation mechanisms (eds Lapointe J, Brakier-Gingras L), pp. 184–196. New York, NY: Kluwer Academic/Plenum Publishers. [Google Scholar]
- 39.Maes A, Gracia C, Innocenti N, Zhang K, Aurell E, Hajnsdorf E. 2017. Landscape of RNA polyadenylation in E. coli. Nucleic Acids Res. 45, 2746–2756. ( 10.1093/nar/gkw894) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Y, Cohen SN. 2000. Unpaired terminal nucleotides and 5′ monophosphorylation govern 3′ polyadenylation by Escherichia coli poly(A) polymerase I. Proc. Natl Acad. Sci. USA 97, 6415–6420. ( 10.1073/pnas.120173797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luciano DJ, Vasilyev N, Richards J, Serganov A, Belasco JG. 2017. A novel RNA phosphorylation state enables 5′ end-dependent degradation in Escherichia coli. Mol. Cell 67, 44–54. ( 10.1016/j.molcel.2017.05.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coburn GA, Mackie GA. 1996. Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3'-exonucleases dependent on oligoadenylation and RNA secondary structure. J. Biol. Chem. 271, 15776–15781. ( 10.1074/jbc.271.26.15776) [DOI] [PubMed] [Google Scholar]
- 43.Bouvier M, Carpousis AJ. 2011. A tale of two mRNA degradation pathways mediated by RNase E. Mol. Microbiol. 82, 1305–1310. ( 10.1111/j.1365-2958.2011.07894.x) [DOI] [PubMed] [Google Scholar]
- 44.Liu JD, Parkinson JS. 1991. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J. Bacteriol. 173, 4941–4951. ( 10.1128/jb.173.16.4941-4951.1991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohanty BK, Kushner SR. 2013. Deregulation of poly(A) polymerase I in Escherichia coli inhibits protein synthesis and leads to cell death. Nucleic Acids Res. 41, 1757–1766. ( 10.1093/nar/gks1280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nadratowska-Wesolowska B, Slominska-Wojewodzka M, Lyzen R, Wegrzyn A, Szalewska-Palasz A, Wegrzyn G. 2010. Transcription regulation of the Escherichia coli pcnB gene coding for poly(A) polymerase I: roles of ppGpp, DksA and sigma factors. Mol. Genet. Genomics 284, 289–305. ( 10.1007/s00438-010-0567-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jasiecki J, Wegrzyn G. 2006. Transcription start sites in the promoter region of the Escherichia coli pcnB (plasmid copy number) gene coding for poly(A) polymerase I. Plasmid 55, 169–172. ( 10.1016/j.plasmid.2005.10.002) [DOI] [PubMed] [Google Scholar]
- 48.Jasiecki J, Wegrzyn G. 2006. Phosphorylation of Escherichia coli poly(A) polymerase I and effects of this modification on the enzyme activity. FEMS Microbiol. Lett. 261, 118–122. ( 10.1111/j.1574-6968.2006.00340.x) [DOI] [PubMed] [Google Scholar]
- 49.Jasiecki J, Wegrzyn G. 2003. Growth-rate dependent RNA polyadenylation in Escherichia coli. EMBO Rep. 4, 172–177. ( 10.1038/sj.embor.embor733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang XX, Liu YH, Rainey PB. 2010. CbrAB-dependent regulation of pcnB, a poly(A) polymerase gene involved in polyadenylation of RNA in Pseudomonas fluorescens. Environ. Microbiol. 12, 1674–1683. ( 10.1111/j.1462-2920.2010.02228.x) [DOI] [PubMed] [Google Scholar]
- 51.Hajnsdorf E, Régnier P. 2000. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A)polymerase I. Proc. Natl Acad. Sci. USA 97, 1501–1505. ( 10.1073/pnas.040549897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Derout J, Folichon M, Briani F, Dehò G., Régnier P, Hajnsdorf E. 2003. Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res. 31, 4017–4023. ( 10.1093/nar/gkg456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Régnier P, Hajnsdorf E. 2013. The interplay of Hfq, poly(A) polymerase I and exoribonucleases at the 3′ ends of RNAs resulting from Rho-independent termination: a tentative model. RNA Biol. 10, 602–609. ( 10.4161/rna.23664) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Régnier P, Hajnsdorf E. 2003. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 31, 7302–7310. ( 10.1093/nar/gkg915) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hajnsdorf E, Régnier P. 1999. E. coli rpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J. Mol. Biol. 286, 1033–1043. ( 10.1006/jmbi.1999.2547) [DOI] [PubMed] [Google Scholar]
- 56.Xu F, Cohen SN. 1995. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature 374, 180–183. ( 10.1038/374180a0) [DOI] [PubMed] [Google Scholar]
- 57.Hajnsdorf E, Braun F, Haugel-Nielsen J, Régnier P. 1995. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc. Natl Acad. Sci. USA 92, 3973–3977. ( 10.1073/pnas.92.9.3973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hajnsdorf E, Steier O, Coscoy L, Teysset L, Régnier P. 1994. Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams rnb pnp mutant. EMBO J. 13, 3368–3377. ( 10.1002/j.1460-2075.1994.tb06639.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng Z.-F, Deutscher MP. 2005. An important role for RNase R in mRNA decay. Mol. Cell 17, 313–318. ( 10.1016/j.molcel.2004.11.048) [DOI] [PubMed] [Google Scholar]
- 60.Andrade JM, Hajnsdorf E, Régnier P, Arraiano CM. 2009. The poly(A)-dependent degradation pathway of rpsO mRNA is primarily mediated by RNase R. RNA 15, 316–326. ( 10.1261/rna.1197309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marujo PE, Hajnsdorf E, Le Derout J, Andrade R, Arraiano CM, Régnier P. 2000. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA 6, 1185–1193. ( 10.1017/S135583820000073X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaberdin VR, Blasi U. 2006. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol. Rev. 30, 967–979. ( 10.1111/j.1574-6976.2006.00043.x) [DOI] [PubMed] [Google Scholar]
- 63.Newbury SF, Smith NH, Higgins CF. 1987. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell 51, 1131–1143. ( 10.1016/0092-8674(87)90599-X) [DOI] [PubMed] [Google Scholar]
- 64.Newbury SF, Smith NH, Robinson EC, Hiles ID, Higgins CF. 1987. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell 48, 297–310. ( 10.1016/0092-8674(87)90433-8) [DOI] [PubMed] [Google Scholar]
- 65.Goodrich AF, Steege DA. 1999. Roles of polyadenylation and nucleolytic cleavage in the filamentous phage mRNA processing and decay pathways in Escherichia coli. RNA 5, 972–985. ( 10.1017/S1355838299990398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miczak A, Kaberdin VR, Wei CL, Lin-Chao S. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl Acad. Sci. USA 93, 3865–3869. ( 10.1073/pnas.93.9.3865) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Py B, Higgins CF, Krisch HM, Carpousis AJ. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381, 169–172. ( 10.1038/381169a0) [DOI] [PubMed] [Google Scholar]
- 68.Coburn GA, Mackie GA. 1998. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J. Mol. Biol. 279, 1061–1074. ( 10.1006/jmbi.1998.1842) [DOI] [PubMed] [Google Scholar]
- 69.Blum E, Carpousis AJ, Higgins CF. 1999. Polyadenylation promotes degradation of 3'-structured RNA by the Escherichia coli mRNA degradosome in vitro. J. Biol. Chem. 274, 4009–4016. ( 10.1074/jbc.274.7.4009) [DOI] [PubMed] [Google Scholar]
- 70.Carabetta VJ, Silhavy TJ, Cristea IM. 2010. The response regulator SprE (RssB) is required for maintaining poly(A) polymerase I-degradosome association during stationary phase. J. Bacteriol. 192, 3713–3721. ( 10.1128/JB.00300-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Z, Pandit S, Deutscher MP. 1998. Polyadenylation of stable RNA precursors in vivo. Proc. Natl Acad. Sci. USA 95, 12 158–12 162. ( 10.1073/pnas.95.21.12158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Z, Pandit S, Deutscher MP. 1999. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA 5, 139–146. ( 10.1017/S1355838299981669) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Z, Reimers S, Pandit S, Deutscher MP. 2002. RNA quality control: degradation of defective transfer RNA. EMBO J. 21, 1132–1138. ( 10.1093/emboj/21.5.1132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joanny G, Le Derout J, Brechemier-Baey D, Labas V, Vinh J, Regnier P, Hajnsdorf E. 2007. Polyadenylation of a functional mRNA controls gene expression in Escherichia coli. Nucleic Acids Res. 35, 2494–2502. ( 10.1093/nar/gkm120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khemici V, Carpousis AJ. 2004. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 51, 777–790. ( 10.1046/j.1365-2958.2003.03862.x) [DOI] [PubMed] [Google Scholar]
- 76.Maes A, Gracia C, Brechemier D, Hamman P, Chatre E, Lemelle L, Bertin PN, Hajnsdorf E. 2013. Role of polyadenylation in regulation of the flagella cascade and motility in Escherichia coli. Biochimie 95, 410–418. ( 10.1016/j.biochi.2012.10.017) [DOI] [PubMed] [Google Scholar]
- 77.Aiso T, Yoshida H, Wada A, Ohki R. 2005. Modulation of mRNA stability participates in stationary-phase-specific expression of ribosome modulation factor. J. Bacteriol. 187, 1951–1958. ( 10.1128/JB.187.6.1951-1958.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soderbom F, Wagner EG. 1998. Degradation pathway of CopA, the antisense RNA that controls replication of plasmid R1. Microbiology 144, 1907–1917. ( 10.1099/00221287-144-7-1907) [DOI] [PubMed] [Google Scholar]
- 79.Wrobel B, Herman-Antosiewicz A, Szalewska-Palasz S, Wegrzyn G. 1998. Polyadenylation of oop RNA in the regulation of bacteriophage lambda development. Gene 212, 57–65. ( 10.1016/S0378-1119(98)00127-9) [DOI] [PubMed] [Google Scholar]
- 80.Briani F, Del Vecchio E, Migliorini D, Hajnsdorf E, Regnier P, Ghisotti D, Deho G. 2002. RNase E and polyadenyl polymerase I are involved in maturation of CI RNA, the P4 phage immunity factor. J. Mol. Biol. 318, 321–331. ( 10.1016/S0022-2836(02)00085-2) [DOI] [PubMed] [Google Scholar]
- 81.Mikkelsen N Dam, Gerdes K. 1997. Sok antisense RNA from plasmid R1 is functionally inactivated by RNase E and polyadenylated by poly(A) polymerase I. Mol. Microbiol. 26, 311–320. ( 10.1046/j.1365-2958.1997.5751936.x) [DOI] [PubMed] [Google Scholar]
- 82.Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Gorke B. 2008. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 36, 2570–2580. ( 10.1093/nar/gkn091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Urban JH, Vogel J. 2008. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 6, e64 ( 10.1371/journal.pbio.0060064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruiz-Larrabeiti O, Plagaro AH, Gracia C, Sevillano E, Gallego L, Hajnsdorf E, Kaberdin VR. 2016. A new custom microarray for sRNA profiling in Escherichia coli. FEMS Microbiol. Lett. 363, fnw131. ( 10.1093/femsle/fnw131) [DOI] [PubMed] [Google Scholar]
- 85.Li Z, Jiang J, Yu X, Wu C, Shen D, Feng Y. 2017. Poly(A) polymerase I participates in the indole regulatory pathway of Pantoea agglomerans YS19. Microbiology 163, 197–206. ( 10.1099/mic.0.000415) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.