Abstract
Flightless I (FliI) is a calcium-dependent, actin severing and capping protein that localizes to cell matrix adhesions, contributes to the generation of cell extensions, and colocalizes with Ras. Currently, the mechanism by which FliI interacts with Ras to enable assembly of actin-based cell protrusions is not defined. R-Ras, but not K-ras, H-ras, or N-ras, associated with the leucine-rich region (LRR) of FliI. Mutations of the proline-rich region of R-ras (P202A, P203A) prevented this association. Knockdown of Ras GTPase-activating SH3 domain-binding protein (G3BP1) or Rasgap120 by small interfering RNA inhibited the formation of cell extensions and prevented interaction of R-ras and G3BP1 in FliI wild-type (WT) cells. Pull-down assays using G3BP1 fusion proteins showed a strong association of R-ras with the C-terminus of G3BP1 (amino acids 236–466), which also required the LRR of FliI. In cells that expressed the truncated N-terminus or C-terminus of G3BP1, the formation of cell extensions was blocked. Endogenous Rasgap120 interacted with the N-terminus of G3BP1 (amino acids 1–230). We conclude that in cells plated on collagen FliI-LRR interacts with R-ras to promote cell extension formation and that FliI is required for the interaction of Rasgap120 with G3BP1 to regulate R-ras activity and growth of cell extensions.
INTRODUCTION
Extracellular matrix (ECM) remodeling is crucial for human health and is of central importance in diverse processes in mammals including development, cell differentiation, wound healing, angiogenesis, and tissue homeostasis. Dysregulation of ECM remodeling is associated with congenital defects (e.g., heart valve malformations), fibrosis, and invasive cancers (Bonnans et al., 2014). Fibroblasts contribute to degradative remodeling of the ECM by an extracellular pathway involving matrix metalloproteinases and by an intracellular, phagocytic pathway in which matrix molecules like collagen are degraded in lysosomal compartments (Everts et al., 1996). To enable phagocytosis of collagen, which is the main structural protein of the ECM (Perez-Tamayo, 1978), fibroblasts reorganize subcortical actin filaments to generate plasma membrane extensions that engulf collagen fibrils (Arora et al., 2015). Flightless I (FliI) is a calcium-dependent, actin severing and capping protein that is localized at cell matrix adhesions and is involved in the generation of plasma membrane extensions (Arora et al., 2017). Currently it is unknown how FliI is integrated with key signal transduction pathways to enable the generation of cell extensions.
Flightless I (FliI), originally discovered in Drosophila, is the most evolutionarily conserved member of the gelsolin superfamily of proteins (Claudianos and Campbell, 1995), which are key regulators of actin filament assembly and turnover. FliI comprises six gelsolin-like domains and an N-terminal leucine-rich repeat (LRR) domain (Kopecki and Cowin, 2008). The gelsolin-like domains mediate the severing and capping of actin filaments. The LRR domain (∼400 residues) contains 16 tandem repeats of a 23-amino-acid motif that forms an amphipathic β–α structural unit (Liu and Yin, 1998). The LRR motif is present in a functionally diverse set of proteins that are mainly involved in protein–protein interactions (e.g., decorin binding to collagen) and in signal transduction pathways (e.g., FSH binding to FSH receptors) (Kobe and Deisenhofer, 1995). The LRR domain of FliI protein, which is not present in other gelsolin family members, may enable interactions between FliI and other molecules involved in signal transduction, thereby spatially integrating signaling and actin remodeling functions (Kopecki and Cowin, 2008). Notably, the actin filament-modifying and signaling functions of FliI that are associated with its gelsolin-like and LRR domains are reminiscent of another actin-binding protein, filamin A, which promotes high angle branching of actin filaments but also interacts with over 90 other proteins to mediate a variety of signaling functions (Kim and McCulloch, 2011; Nakamura et al., 2011). While FliI may not interact with as many proteins as does filamin A, associations between the LRR domain of FliI and Ras have been previously observed (Goshima et al., 1999).
The Ras subfamily of GTPases promotes cell adhesion (Hall, 1992) and spreading (Radinsky et al., 1990). Ras subfamily proteins, including H-Ras, K-Ras, N-Ras, and R-Ras, are small monomeric GTPases that contribute to the signaling systems involved in regulation of the actin cytoskeleton (Zhang et al., 1996; Campbell and Der, 2004). In particular, R-ras regulates integrins and invasive processes in cancer (Keely et al., 1999) and cell adhesion formation (Kwong et al., 2003; Self et al., 2001), processes that require tight regulation of actin assembly. R-Ras is structurally distinct from other Ras superfamily proteins because of its proline-rich sequence (residues 199–203) near the C-terminal region (Hansen et al., 2003) that enables interaction with the SH2 and SH3 domains of other proteins to regulate function (Wang et al., 2000). As FliI colocalizes with Ras (Davy et al., 2001), R-ras may be part of a pathway involving FliI that mediates cytoskeletal rearrangements required for cell adhesion and extension formation. Currently, the mechanism by which FliI interacts with Ras to mediate the generation of cell extensions remains undefined.
Ras activity is regulated in part by Ras guanine nucleotide exchange factors (RasGEFs) and by Ras guanine-activating proteins (Rasgaps) (Bos et al., 2007). RasGEFs enhance the catalytic activity of Ras by enabling GDP dissociation from Ras to promote GTP binding (Cherfils and Zeghouf, 2013). Rasgaps inactivate Ras proteins by promoting hydrolysis of the GTP-bound form of Ras to the inactive, GDP-bound protein (Iwashita and Song, 2008). The carboxy-terminal of one prominent Rasgap, Rasgap120, contains a catalytic domain that binds GTP-loaded Ras, promotes GTP hydrolysis, and regulates signal transduction (Pomerance et al., 1996). The amino terminus of Rasgap120 contains a Ca2+-dependent lipid-binding domain, a pleckstrin homology domain, and an SH3 domain that is flanked by two SH2 domains in its amino-terminal domain that are important for triggering downstream signals (Tocque et al., 1997). Rasgap120 is distinguished by its interaction with other regulatory proteins. For example, immunoprecipitation using an amino-terminal construct of Rasgap120 (Parker et al., 1996) lead to the identification of a 68-kDa, Ras GTPase-activating protein SH3 domain-binding protein (G3BP1), which is strongly associated with invasive breast cancers (French et al., 2002). Notably, G3BP1 coimmunoprecipitates with Rasgap120 only in actively growing cells, indicating that the Rasgap120-G3BP1 complex may bind only to the activated form of Ras (Parker et al., 1996).
Here we examined the roles of FliI, the Rasgap120-G3BP1 complex and R-Ras in mediating the formation of plasma membrane extensions that are crucial for ECM remodeling and for localized cell invasion by cancer cells. Our main findings are that the LRR in the N-terminus of FliI interacts with R-ras to promote cell extension formation and that FliI is required for the interaction of Rasgap120 with G3BP1 to regulate R-ras activity.
RESULTS
FliI associates with R-ras
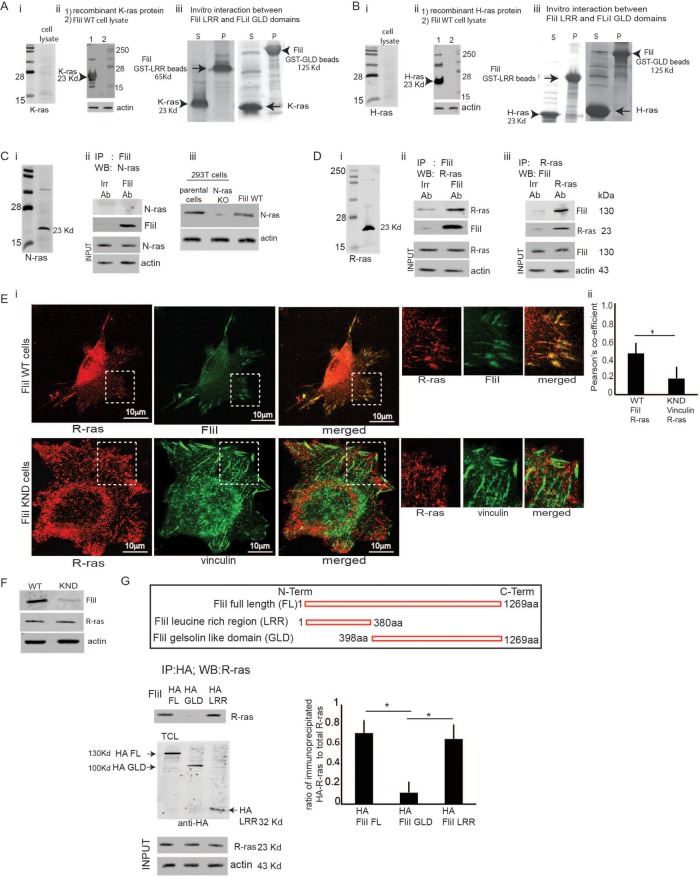
Although it has been suggested that the LRR in the N-terminus of FliI interacts with Ras to effect downstream signaling (Goshima et al., 1999), the mechanism by which FliI interacts with Ras to mediate generation of cell extensions is not defined. We determined the expression of different Ras isoforms in cell lysates from FliI wild-type (WT) cells by Western blotting. Our experiments showed absence of K-ras and H-ras and presence of R-ras and N-ras protein expression (Figure 1, Ai, Bi, Ci, and Di). We probed recombinant K-ras and H-ras proteins to validate the proficiency of K and H-ras antibodies (Figure 1, Aii and Bii). Further in in vitro experiments there was absence of interaction between recombinant K/H-ras and truncated FliI either FliI-LRR or FliI-gelsolin-like domains (GLD) (Figure 1, Aiii and Biii). We looked for an association of FliI with R-ras or N-ras. FliI immunoprecipitates showed no detectable N-ras (Figure 1Cii), while FliI and R-ras clearly associated in coimmunoprecipitation experiments (Figure 1D, ii and iii). We used N-ras knockout (KO) cells and parental 293T cells to show proficiency of the N-ras antibody (Figure 1Ciii).
FIGURE 1:
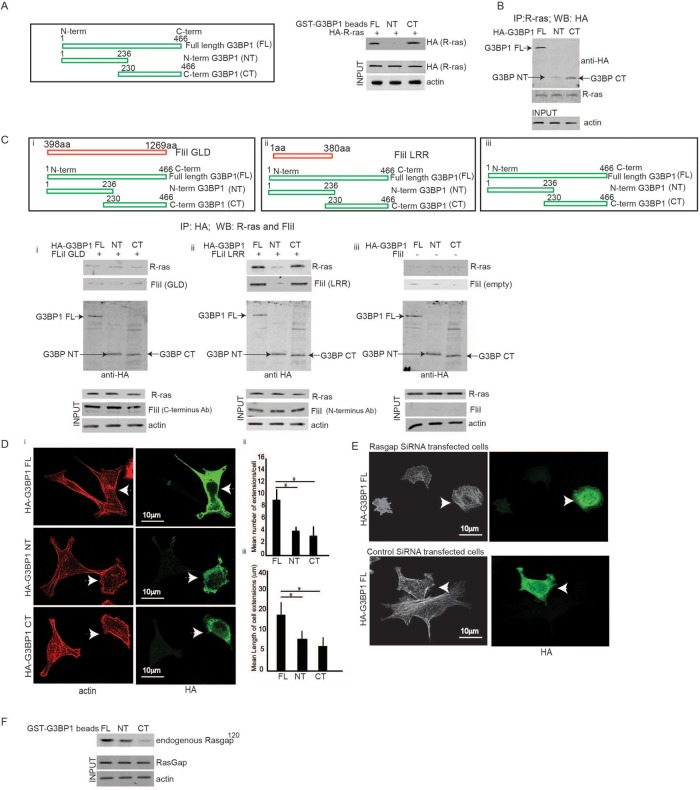
(A) FliI LRR interacts with R-ras. (Ai, Bi, Ci, Di) FliI Wt cell lysates probed for Ras isoforms show protein expression levels of R, K, H, and N-ras. Absence of K-ras and H-ras proteins and presence of K-ras and N-ras proteins. (Aii, Bii) Proficiency of K and H-ras antibodies shown by Western blotting recombinant K-ras and H-ras proteins. (Aiii, Biii) Absence of interaction between recombinant K/H-ras and FliI-LRR or FliI-GLD shown by in vitro experiments. (Cii) In immunoprecipitation assays there is undetectable association of FliI with N-ras or nebulin (irrelevant control antibody). (Ciii) Western blotting of N-ras KO cell lysates and parental 293T cell lysates showing proficiency of the N-ras antibody. (D, ii and iii) Coimmunoprecipitation experiments show that R-ras and FliI associate. These experiments repeated three times. (E, i and ii) Representative images of FliI WT and KND cells incubated with collagen-coated beads showing localization of R-ras at the adhesion sites in FliI WT cells. Bar, 10 μm. (ii) Pearson’s correlation was applied to image pairs of FliI and R-ras in WT cells and vinculin and R-ras in FliI KND cells immunostained cells to estimate colocalization of these proteins using ImageJ. Data in histogram show 40% reduction in colocalization in FliI KND cells. Data reported as mean ± SD and analyzed by ANOVA from observation made on image pairs of 25 cells. (F) FliI WT and KND cells showing equivalent R-ras protein levels. (G) HA antibody immunoprecipitation assay of cells transfected with HA-tagged FliI full-length and N-terminal LRR and C-terminal GLD domains show specific association of FliI-LRR region with R-ras. Data in histogram are from three different experiments and show interaction between R-ras and FliI WT (4.6-fold) (p < 0.05) and FliI LRR (4-fold) (p < 0.05) compared with FliI GLD. Data are reported as mean ± SD, analyzed by ANOVA.
We immunostained for R-ras and FliI to show colocalization of FliI with R-ras in FliI WT cells. Cells plated on collagen showed targeting of FliI and R-ras proteins to the adhesion sites. In contrast, R-ras did not localize to vinculin at adhesion sites in FliI knockdown (KND) cells (Pearson r of FliI/R-ras colocalization coefficient = 55% for FliI WT cells and 15% for FliI KND cells) (Figure 1E, i and ii). There were equivalent expression levels of R-ras in FliI WT and KND cells (Figure 1F).
R-ras interacts with FliI-leucine–rich region
As LRRs in a large number of different proteins are involved in mediating protein–protein interactions (Kobe and Kajava, 2001), we examined the interaction of the LRR of FliI with R-ras (Claudianos and Campbell, 1995). Cells were transfected with hemagglutinin (HA)- tagged, full-length FliI or truncated FliI (either the GLD of the C-terminus or the LRR of the N-terminus). Immunoprecipitation experiments showed strong associations of R-ras with the LRR domain and with full-length FliI, while there was minimal association with the GLDs in the C-terminus of FliI (Figure 1G).
Active R-ras is required for binding to FliI-leucine–rich region
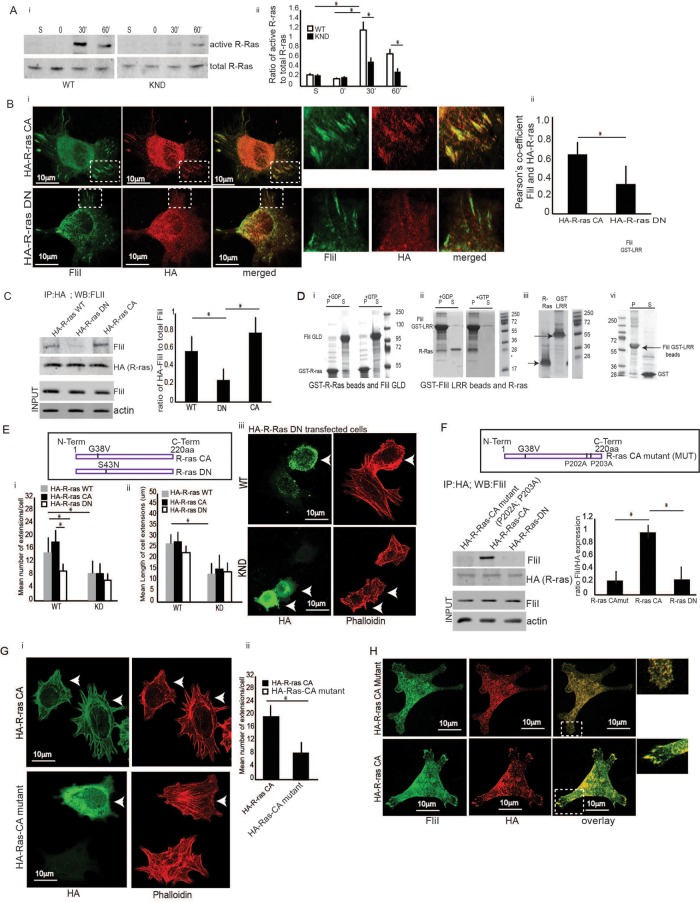
Spreading cells exhibit various types of cell extensions that are regulated by small GTPases, and we anticipated that R-ras is important for the growth of cell extensions (Higashi et al., 2010). Cell lysates from FliI WT and KND cells that had been plated on collagen were collected. Analysis of these lysates showed that spreading FliI WT cells exhibited enhanced R-ras activity at 30 min, whereas FliI KND cells showed weak R-ras activity. Cells in suspension showed no R-ras activity. Collectively, the data indicated that cell spreading on collagen substrates induced R-ras activity and that FliI expression enhanced R-ras activity (Figure 2Ai). Data in the histogram show maximal R-ras activity at 30 min. There is a sixfold increase in FliI WT cells and 1.5-fold increase in FliI KND cells (Figure 2Aii).
FIGURE 2:
Collagen adhesion promotes R-ras activity. (A, i and ii) FliI WT and KND cells plated on collagen-coated tissue culture plates over 0–60 min. Cell lysates analyzed by immunoblot show maximal R-ras activity at 30 min in FliI WT cells, which was delayed and reduced in KND cells. R-ras activity was specific to collagen substrate adhesion as cells in suspension (S) failed to induce R-ras activity. Blots shown are from three independent experiments. (ii) Data in histogram are reported as mean ± SD and analyzed by ANOVA. (Bi) Representative images of cells transfected with constitutively active (CA) R-ras target to the adhesion sites of the collagen substrates and localize with FliI but not in cells transfected with DN R-ras. Bar, 12 μm. (ii) Histogram showing colocalization data analyzed by ImageJ. Pearson’s correlation was applied to images pairs of FliI and R-ras immunostained cells in. Data in the histogram show 62 and 29% colocalization in HA-R-ras CA and HA-R-ras DN cells, respectively. Data reported as mean ± SD and analyzed by ANOVA from observation made on image pairs of 25 cells. (C) In immunoprecipitation experiments with cells transfected with HA-tagged WT, DN, and CA R-ras, there was 3.5-fold (p < 0.05) and 2.5-fold (p < 0.05) increased association of CA and WT R-ras and FliI compared with DN R-ras-transfected cells. Data in histogram are from three different experiments. Data reported as mean ± SD and analyzed by ANOVA. (D) In vitro experiments showing interaction between R-ras and FliI GLD and FliI LRR domains and requirement of GTP/GDP nucleotides for their interaction. i-GST-R-ras (9 μM) or ii-GST-FliI LRR (8 μM) Sepharose beads incubated with 140 μM GTP γS or GDP in buffer containing 50 mM Tris, pH 7.4, 1 mM EDTA, 20 mM NaCl, and 1% Triton X-100 for 10 min at room temperature followed by GTP γS or GDP. After 30 min, samples precleared with 50 μl glutathione-Sepharose before incubation with purified (i) FliI-GLD (12 μM) or (ii) R-ras (10 μM). These experiments showed (i) no interaction between FliI GLD and R-ras in the absence or presence of GTPγS as the GST R-ras beads and purified FliI GLD protein appear separately in pellet (P) and supernatant (S) fractions. The interaction between (ii) FliI LRR and R-ras required the presence of GTPγS as purified R-ras binds to GST FliI LRR beads and appear in the pellet fraction (P). (iii) GST-LRR and R-ras shown independently before incubation. (iv) In control experiments, GST-FliI LRR beads showed no interaction with purified GST (6 μΜ). (i–iv) S, supernatant, P, pellet. These experiments were repeated three times. One set of representative Coomassie-stained SDS–PAGE gels is shown. (Ei) Data presented in histogram from FliI WT cells and KND cells transfected with HA-tagged WT, CA, and DN R-ras plated on collagen substrate for 60 min show twofold more mean number of extensions/cell in CA R-ras (p < 0.05) and WT R-ras (p < 0.05) transfected cells in FliI WT cells as compared with FliI KND cells. FliI WT cells transfected with HA-WT-R-ras and HA-CA-R-ras show twofold higher number of extensions as compared with cells with HA-DN-R-ras (p < 0.05; p < 0.05). (Eii) FliI WT cells show twofold higher mean length of cell extensions (μm) as compared with FliI KND cells (p < 0.05). (Eiii) Confocal images showing absence of cell extensions in cells transfected with HA-DN R-ras in FliI WT and KND cells. (F) Immunoprecipitations with HA antibody of cells transfected with HA-tagged CA R-ras (P202A; P203A) mutations of the prolines (MUT), HA-CA R-ras, and HA-DN R-ras show absence of interaction between mutated R-ras and DN R-ras with FliI. Data showing fourfold decrease in cells transfected with HA-R-ras mutant (p < 0.05) and HA-DN R-ras (p < 0.05). Data in histogram are from three different experiments. Data reported as mean ± SD were analyzed by ANOVA. (Gi) Representative images of cells transfected with HA-tagged CA R-ras MUT and HA CA R-ras cells and plated on collagen. Transfected cells (arrows) immunostained with HA antibody and phalloidin to localize the nontransfected cells. (Gii) Histogram shows a 2.2-fold reduction in the number of extensions per cell in R-ras mutated cells. p < 0.05. Data reported as mean ± SD, analyzed by ANOVA from three experiments. (H) Confocal images showing localization of FliI and R-ras in cells transfected with HA-CA R-ras mutant and HA-CA R-ras. Transfected cells immunostained with HA and FliI antibody show failed targeting of R-ras to growing extensions and localization with FliI in HA CA R-ras MUT cells (P202A; P203A). Observations were recorded from 20 cells each.
Since FliI localizes to adhesions of spreading cells (Mohammad et al., 2012), we transfected cells with hemagglutinin (HA)-tagged WT R-ras, constitutively active R-ras (R-ras CA [G38V) or dominant-negative (DN) R-ras (R-ras [S43N]) and allowed the cells to spread on collagen plates. Adherent, constitutively active R-ras transfected cells that were immunostained for HA and FliI showed colocalization at adhesion sites (Pearson r of FliI/HA-R-ras colocalization coefficient = 61% for HA-R-ras CA cells and 25% for HA-R-ras DN cells) (Figure 2B, i and ii). These results were consistent with immunoprecipitation experiments in which lysates from cells transfected with HA-tagged R-ras WT, constitutively active R-ras (G38V), or dominant-negative (S43N) R-ras were immunoprecipitated with HA antibody. When the HA immunoprecipitates were immunoblotted with a FliI antibody, constitutively active R-ras associated with FliI (Figure 2C).
We conducted in vitro studies, which showed that the interaction between GST-R-Ras beads and purified FliI-GLD was undetectable and was unaffected by the nucleotide state of R-ras (GTP γS or GDP in the buffer; Figure 2Di). In contrast, the interaction of GST-FliI-LRR with purified R-ras required GTP γS, and this interaction occurred only if R-ras was active (Figure 2Dii). GST-LRR and R-ras shown independently before incubation (Figure 2Diii). In control experiments, GST did not associate with GST-FliI-LRR beads (Figure 2Div). These experiments indicated that the LRR region of FliI interacts with active R-ras.
Active R-ras is required for cell extension formation
FliI WT and KND cells were transfected with HA-tagged R-ras CA (G38V) or R-ras DN (S43N), plated on collagen, immunostained with HA antibody, and counterstained with rhodamine phalloidin. In FliI WT cells, there were twofold more extensions/cell when constitutively active R-ras was expressed compared with cells transfected with dominant-negative R-ras. In contrast, FliI KND cells showed no difference in the number of cell extensions in constitutively active R-ras or dominant-negative R-ras-transfected cells. There were 2.5-fold more extensions per cell in FliI WT compared with FliI KND cells that expressed constitutively active R-ras (Figure 2E, i–iii).
Since R-ras differs from other Ras family members because of its distinct proline-rich domain in the C-terminus, we assessed whether the function of the proline-rich region is required for interaction with FliI to promote adhesion and cell extension development. We introduced point mutations (P202A and P203A) in HA-tagged, constitutively active R-ras. In immunoprecipitation assays, lysates from cells transfected with the CA R-ras mutant (P202A, P203A) showed fourfold reductions in the association of FliI compared with CA WT R-ras (Figure 2F).
To determine whether the function of the R-ras proline-rich region was important for cell extension formation, we transfected cells with HA CA R-Ras or with HA-R-ras CA mutants and plated cells on collagen. There was a 2.4-fold reduction in the number of cell extensions/cell in cells with mutant active R-ras, suggesting that the function of the proline-rich sites of R-ras is required for interaction with FliI to induce cell extension formation (Figure 2G, i and ii). These results were extended by immunostaining FliI WT cells transfected with HA-tagged CA R-Ras or with HA-R-ras CA mutants. In cells transfected with the mutant R-ras, R-ras was not recruited to adhesion sites, suggesting that in spreading cells, the association of FliI with R-ras at adhesion sites is required for R-ras recruitment (Figure 2H).
Involvement of Ras GTPase-activating protein SH3 domain-binding protein (G3BP1)
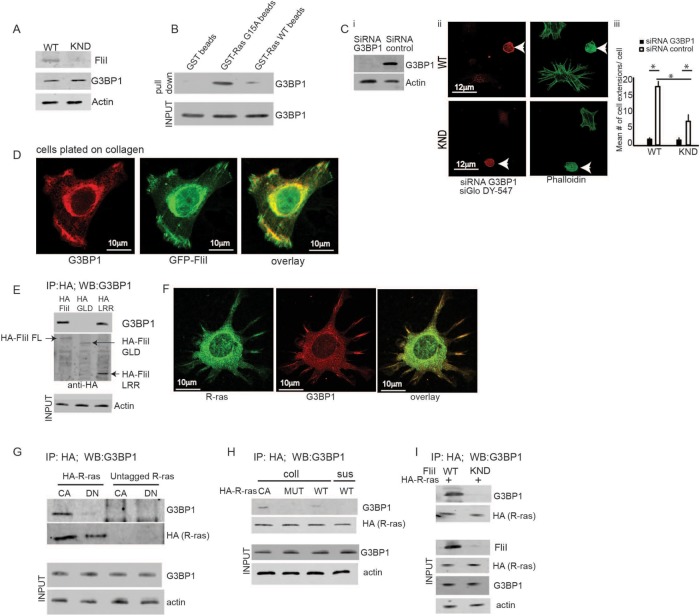
FliI cells previously plated on collagen were screened for GEFs and GAPs that may regulate R-ras during adhesion and the growth of cell extensions. The screen was conducted with a nucleotide-free R-ras (G15A) mutant, which forms stable, high-affinity complexes with active GEFs and GAPs (Cherfils and Chardin, 1999). Mass spectrometry analysis of proteins bound to GST-R-ras G15A beads or GST WT R-ras beads showed enrichment (higher number of bound peptides) of the Ras GTPase-activating protein SH3 domain-binding protein (G3BP1) to the R-ras-G15A beads than to the WT R-ras beads (Table 1). We confirmed the presence of G3BP1 in cell lysates (Figure 3A) in pull-down assays with GST G15A and WT beads (Figure 3B).
TABLE 1:
Mass spectrometry analysis of R-ras G15A mutant and R Ras WT pull down.
| Mus musculus identified protein | Accession number | Molecular weight (kDa) | RRasG15A # of tryptic peptides | RRasWT # of tryptic peptides |
|---|---|---|---|---|
| Ras GTPase-activating protein SH3 domain-binding protein (G3BP1) | P 97855 | 52 | 22 | 1 |
| Ras-related protein Ral-A | P63321 | 24 | 1 | |
| Ras-related protein Rab-1 | Q9D1G1 | 22 | 1 | 7 |
| Ran-specific GTPase-activating protein | P34022 | 24 | 1 | |
| Ras GTPase-activating–like protein | Q9JKF1 | 189 | 2 | 1 |
FIGURE 3:
FliI needed for the assembly of R-ras/G3BP1. (A) FliI WT and KND cells show equivalent expression levels of G3BP1 protein. (B) Pull-down assay shows proteins eluted from GST only, GST RasG15A, and GST Ras WT Sepharose beads incubated with cell lysates. There are increased levels of G3BP1 protein on GST Ras G15A beads. Equal amounts of proteins loaded on beads (INPUT). These experiments repeated four times. (Ci) Knockdown of G3BP1 expression with two different sets of siRNA showing 75 and 72% decrease in protein levels. (Cii, Ciii) Confocal images of FliI WT and KND cells transiently cotransfected with G3BP1 siRNA and fluorescent siRNA (siGloDY-547) show a 2.5-fold difference in development of cell extensions in nontransfected FliI WT and FliI KND cells (p < 0.05). In G3BP1 siRNA-transfected cells there was a sevenfold reduction in FliI WT (p < 0.05) cells and 2.4-fold reduction in FliI KND (p < 0.05) as compared with nontransfected cells. Data in histogram are from three different experiments. Data reported as mean ± SD, analyzed by ANOVA (p < 0.05). (D) Confocal optical sections showing localization of G3BP1 and GFP-FliI to the adhesions sites on collagen substrate. Observations recorded in 25 cells. (E) HA antibody immunoprecipitation of cells transfected with HA-tagged full-length FliI (FL), HA-GLD domain, and HA-LRR domain show association of FliI FL and FliI LRR to G3BP1. Experiment repeated three times. (F) Confocal images showing colocalization of G3BP1 and R-ras at the cell extensions. Observations recorded in 30 cells. (G) Immunoprecipitaion with HA antibody of cells transfected with HA-tagged or control, HA-untagged CA R-ras, and DN-R-ras show interaction between G3BP1 and CA R-ras. (H) Cells transfected with HA-tagged CA-R-ras, CA-R-ras MUT (P202A; P203A), and WT R-ras plated on collagen or in suspension. Cell lysates immunoprecipitated with HA antibody and analyzed by SDS–PAGE gels showing interaction between constitutively active-R-ras and G3BP1 and not between mutant R-ras and G3BP1 or cells in suspension (sus). (I) FliI WT and KND cells were transfected with HA-tagged CA R-ras. Lysates from cells plated on collagen immunoprecipitated with HA antibody showed absence of FliI reduced association between G3BP1 and R-ras. Experiment in G, H, and I repeated three times.
To assess a potential role for G3BP1 in cell extension formation, G3BP1 small interfering RNA (siRNA)-transfected cells were cotransfected with Fluorescent siGlo DY-547 to facilitate identification of transfected cells. Cells transfected with two different sets of G3BP1 siRNA showed 75 and 72% knockdown (Figure 3Ci) (data for the second siRNA are shown in Supplemental Figure 1Bi). Double-labeled cells showed 10-fold fewer numbers of cell extensions in FliI WT cells and 2.5-fold fewer extensions in FliI KND cells compared with control siRNA transfected cells (Figure 3C, ii and iii; see also Supplemental Figure 1B, ii and iii).
As G3BP1 and GFP-FLiI colocalized in cell adhesions and growing extensions (Figure 3D), we examined this relationship between G3BP1 and FliI in more detail in cells that were transfected with truncated forms of HA-tagged FliI (full-length [FL], GLD, LRR) and plated on collagen. In cell lysates immunoprecipitated with HA antibody, there was evidence of a specific interaction between G3BP1and the full-length FliI and with the FliI LRR domain, but there was minimal interaction with the FliI GLD domain (Figure 3E).
Since our data showed an association of FliI with constitutively active (CA) R-ras, we examined the relationship of G3BP1 with R-ras. In immunostaining experiments, cells spreading on collagen showed colocalization of R-ras with G3BP1 in growing cell extensions (Figure 3F) Further, in immunoprecipitation experiments of cells transfected with HA-tagged, constitutively active (CA)-R-ras or DN R-ras, there was evidence of association of G3BP1 with CA-R-ras but negligible association with DN-R-ras (Figure 3G).
In separate experiments, cells transfected with HA-tagged constitutively active R-ras or the constitutively active R-ras mutant (P202A; P203A) were plated on collagen. WT R-ras-transfected cells were maintained in suspension as nonadherent controls. Immunoprecipitation showed evidence for association between constitutively active-R-ras and G3BP1 but not between mutant R-ras and G3BP1 nor of cells in suspension (Figure 3H). To determine whether FliI was required for the association between G3BP1 and R-ras, we transfected HA-tagged CA R-ras into FliI WT and KND cells. Lysates from cells plated on collagen were immunoprecipitated with HA antibody, and these results showed that in the absence of FliI, the association between G3BP1 and R-ras was reduced. These data indicated that at cell adhesions, FliI is required for initiating the interplay between G3BP1 and R-ras (Figure 3I).
Role of Rasgap 120
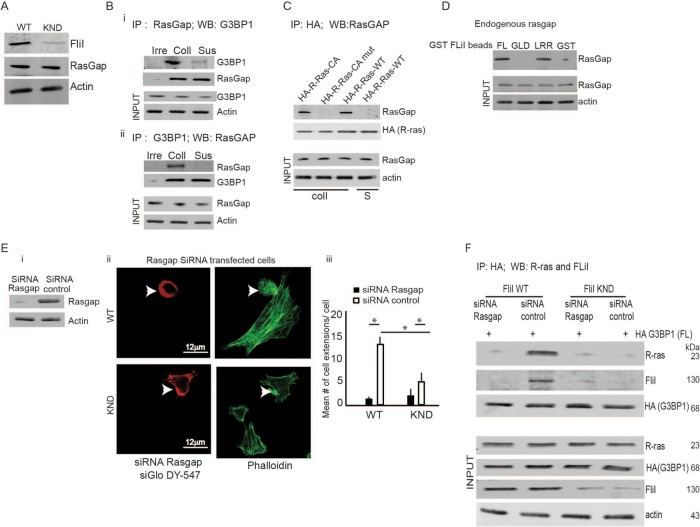
In proliferating and spreading cells, G3BP1 binds tightly to the Rasgap SH3 domain at its N-terminus (Parker et al., 1996), an interaction that mediates Ras downstream signals and acts as an intrinsic Ras effector for regulating cytoskeletal reorganization (Tocque et al., 1997). We explored associations between these signaling molecules and FliI to determine how FliI, as a potential adaptor protein, may promote regulatory processes in cell extension growth. We confirmed that was equivalent expression of Rasgap120 in FliI WT and KND cells (Figure 4A). In coimmunoprecipitation experiments, the association between G3BP1 and Rasgap120 was substrate specific as cells in suspension did not show an association between G3BP1 and Rasgap120 (Figure 4B, i and ii). We also confirmed an association between Rasgap120 and R-ras. FliI WT cells were transfected with HA-tagged constitutively active R-ras or constitutively active R-ras mutant (P202A; P203A) or WT R-ras. Cells were plated on collagen or maintained in suspension and lysates were immunoprecipitated with HA antibody. Immunoblotting showed an association between Rasgap120 and constitutively active-R-ras but not between Rasgap120 and mutant R-ras proteins. These associations were substrate specific as cells in suspension showed no detectable association of R-ras with Rasgap120 (Figure 4C). To establish a relationship between FliI and Rasgap120, we used recombinantFL FliI and the N- and C-terminal domains (GLD and LRR), which were bound to glutathione Sepharose beads and incubated with cell lysates. In pull-down assays, Sepharose bead-bound proteins probed for Rasgap120 showed that endogenous Rasgap120 interacted avidly with full-length FliI and the LRR domain of FliI (Figure 4D).
FIGURE 4:
FIiI and Rasgap are required for G3BP1/R-ras association. (A) FliI WT and KND cells show equivalent expression levels of Rasgap120. (B) Coimmunoprecipitation experiments showing association between G3BP1 and Rasgap120 was substrate specific, as cells in suspension and immunoprecipitation with an irrelevant antibody did not show interaction between G3BP1 and Rasgap120. Experiment repeated three times. (C) Cell lysates immunoprecipitated with HA antibody from FliI cells transfected with HA-tagged constitutively active (CA) R-ras, constitutively active R-ras mutant (P202A; P203A), and WT R-ras plated on collagen or in suspension and analyzed by SDS–PAGE gels showed interaction between CA-R-ras and G3BP1 and not between mutant R-ras and G3BP1. These interactions were substrate specific as cells in suspension (S) showed no interaction. These experiments were repeated four times. (D) Pull-down assay showing proteins eluted from GST only, GST- FL FliI, GST-GLD FliI, and GST-LRR FliI Sepharose beads incubated with lysates. Data showing association between FliI-LRR domain and endogenous Rasgap120. (Ei) Knockdown of Rasgap expression with two different sets of siRNA showing 80 and 76% decrease in protein levels of Rasgap. (Eii, Eiii) Confocal images of FliI WT and KND cells transiently cotransfected with Rasgap siRNA and fluorescent siRNA (siGloDY-547) show 2.4-fold difference in development of cell extensions between nontransfected FliI WT and FliI KND cells (p < 0.05). In Rasgap siRNA-transfected cells, there was 6.25-fold reduction in FliI WT (p < 0.05) cells and 1.6-fold reduction in FliI KND (p < 0.05) compared with nontransfected cells. Data in histogram are from three different experiments. Data reported as mean ± SD, analyzed by ANOVA. (F) FliI WT and KND cells cotransfected with HA-tagged G3BP1 and control siRNA or Rasgap siRNA were immunoprecipitated with anti-HA to detect association of G3BP1with R-ras and FliI. Blots shown are from one of the three experiments repeated.
We examined a potential role for Rasgap120 in cell extension formation. We used two sets of different Rasgap120 siRNAs; in transfected cells there was 80 and 76% knockdown of Rasgap expression (Figure 4Ei; data for the second siRNA are shown in Supplemental Figure 1Ci). Confocal images of FliI WT and KND cells transiently cotransfected with Rasgap120 siRNA and fluorescent siRNA (siGloDY-547) showed a 2.4-fold reduction of cell extensions in FliI KND cells compared with FliI WT cells. In Rasgap120 siRNA-transfected cells there were, respectively, 6.25-fold and 1.6-fold fewer cell extensions compared with control siRNA-transfected cells in WT and KND cells (Figure 4E, i and ii; see also Supplemental Figure 1C, ii and iii). In further experiments, FliI WT and KND cells cotransfected with HA-tagged G3BP1 and Rasgap120 siRNA or with control siRNA were immunoprecipitated with anti-HA antibody to examine associations of G3BP1with R-ras and to assess the roles of Rasgap120 and FliI (Figure 4F). These data indicated that in the absence of FliI or Rasgap120, interaction between G3BP1 and R-ras was attenuated.
Interactions with truncated G3BP1
We examined in more detail which particular regions of G3BP1 are required for its associations with the LRR of FliI, activated R-Ras, and Rasgap120. We used several GST-G3BP1 peptide fusions that were expressed in cells and were examined by pull-down assays or with HA-tagged G3BP1 peptide fusions in transfected cells subjected to immunoprecipitations. To examine the association of R-ras with truncated forms of G3BP1 (FL), G3BP1 residues 1–236 (N-terminal), or G3BP1 residues 230–466 (C-terminal), cells were transfected with HA-tagged R-ras and plated on collagen. Cell lysates were incubated with glutathione beads bound to the various GST G3BP1 truncated forms. Specific protein–protein interactions between glutathione beads with bound G3BP1 peptides and HA-tagged R-ras were detected by immunoblotting with HA antibody. These results showed that there was a robust association between R-ras and the C-terminal domain of G3BP1 (residues 230–466) while there was only a weak association with the N-terminal domain (residues 1–236) (Figure 5A). These results were confirmed in a separate experiment in which cells were transfected with HA-tagged G3BP1 truncated constructs and lysates were immunoprecipitated with R-ras. When the immunoprecipitates were immunoblotted for HA there was evidence of an association of R-ras with full-length G3BP1 and the C-terminal domain (residues 230–466) of G3BP1 (Figure 5B).
FIGURE 5:
C-terminus and N-terminus of G3BP1 required for cell extension formation. (A) Cells transfected with HA-tagged G3BP1 were plated on collagen, and pull-down assays were performed with glutathione beads bound G3BP1 peptides (GST-G3BP1 [FL], GST-G3BP1 amino acids 1–236 [N-terminus], GST-G3BP1 amino acids 230–466 [CT]) and HA-tagged R-ras detected by running the bound proteins on a Western and identified by probing with HA antibody. The results show that the interaction between the R-ras maps to the C-terminal domain of G3BP1 (amino acids 230–466) and full-length G3BP1 and weak interactions detected with N-terminal domain (amino acids 1–236). Blots shown from three experiments. (B) Cells transfected with HA-tagged G3BP1 truncated constructs (FL, NT, and CT) were immunoprecipitated with anti-R-ras and immunoprecipitates separated by Western blotting probed with HA antibody to determine the association of R-ras with specific domain of G3BP1. Experiment repeated three times. (Ci–Ciii) FliI KO fibroblasts transfected with either FliI-GLD, FliI-LRR regions, or empty construct were cotransfected with HA-tagged truncated G3BP1 fusions. Transfected cells plated on collagen and lysates subjected to immunoprecipitation with HA antibody determined the relationship between R-ras and HA-G3BP1 fusion domains in FliI knockout fibroblasts expressing FliI-GLD and FliI LRR domains. These experiments show association between R-ras and full-length G3BP1 and C-terminal domain of G3BP1 in FliI-LRR and weak association in FliI-GLD cells or cells with empty construct. Experiments repeated three times. (Di) Confocal images of HA-tagged G3BP1 (FL, NT, and CT) fusion proteins transfected cells plated on collagen. (Dii) Histogram shows 2.5-fold increase (p < 0.05) in mean number and length of cell extensions in HA-G3BP1 full-length-transfected cells and not in N- or C-terminal truncated domains, suggesting, probably, that G3BP1 interactions at the C-terminal are also required for FliI to regulate cell extension formation. Data reported as mean ± SD and analyzed by ANOVA. (E) Cells cotransfected with Rasgap or control siRNA and HA-tagged G3BP1 that were plated on collagen show cell extensions in transfected (arrows) and nontransfected cells. Observations recorded in 30 cells. (F) Pull-down assay showing association of endogenous Rasgap120 with N-terminus of G3BP1 and weak association with C-terminus.
FliI-LRR region in R-ras signaling
We probed the role of FliI GLD and FliI LRR in G3BP1-R-ras interactions. For these experiments we used cells from FliI conditional KO mice as the shRNA targeted to the FliI GLD domain 1 in FliI KND cells blocked expression of GLD 1-6 if transfected back into these cells. Fibroblast-conditional FliI knockout mice showed equivalent expression levels of G3BP1 and Rasgap120 (Supplemental Figure 1). In three different experiments of similar design, KO fibroblasts transfected with either FliI-GLD, FliI-LRR regions, or an empty construct were cotransfected with HA-tagged truncated G3BP1 fusions (G3BP1 FL, NT, and CT). Transfected cells were plated on collagen and the lysates were analyzed by immunoprecipitation with HA antibody. These data showed an association between R-ras and HA-G3BP1 full-length and C-terminal HA G3BP1 (230aa–466aa) fusion domains in FliI KO fibroblasts that expressed FliI LRR domains. In contrast, there was only a weak association of these same proteins in FliI-GLD cells or in cells with an empty FliI construct (Figure 5C, i–iii). Since interactions between G3BP1 and SH3 domain of Rasgap120 at the N-terminal mediate Ras downstream signaling to regulate cytoskeletal reorganization (Parker et al., 1996), we asked whether the HA-tagged truncated G3BP1 fusion proteins transfected in cells affected the formation of cell extensions. FliI cells plated on collagen exhibit 2.5-fold increases in the mean number and length of cell extensions in HA-G3BP1 full-length-transfected cells but not in N- or C-terminal truncated domains. These data indicated that interactions of the C-terminus and N-terminus of G3BPI are required for FliI to regulate cell extension formation (Figure 5D, i–iii). We next cotransfected cells with Rasgap siRNA or control siRNA and HA-tagged G3BP1. When these cells were plated on collagen, there were very few extensions in the Rasgap siRNA-transfected cells (arrows), suggesting that G3BP1 is required for the function of Rasgap in regulating cytoskeletal organization and forming cell extensions (Figure 5E).
Interactions between Rasgap120 and the NTF2-like motifs within the N-terminal domain of G3BP1 have been reported in mouse tissues (Parker et al., 1996). We mapped endogenous Rasgap120 associations with truncated GST-G3BP1 forms in lysates from FliI cells plated on collagen that were then incubated with glutathione beads bound to G3BP1 peptide fusion proteins. In pull-down assays, endogenous Rasgap120 (identified with Rasgap120 antibody) showed an association of Rasgap120 with the N-terminal domain of G3BP1 but a weak association with the C-terminus (Figure 5F). Collectively, these data show that the N-terminus of G3BP1 associates with Rasgap120 to regulate signal transduction in cell extension growth. This process critically involves the C-terminus of G3BP1 in associating with R-ras and the LRR domain of FliI.
DISCUSSION
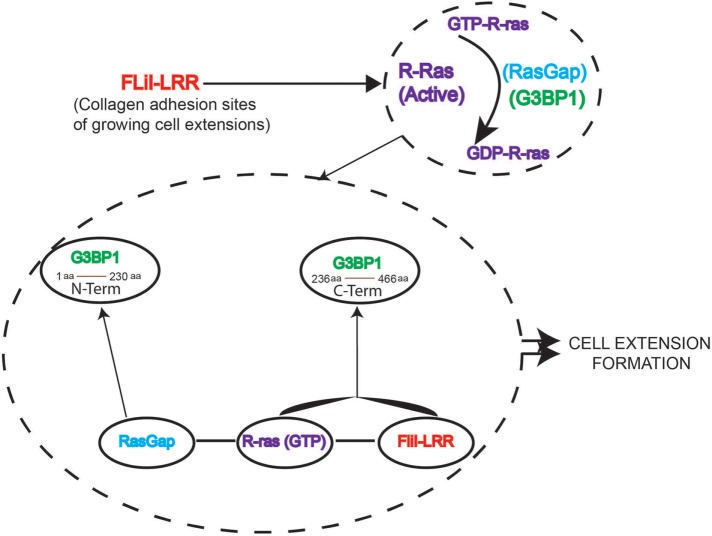
FliI is an actin capping and severing protein that localizes to cell adhesion sites (Mohammad et al., 2012) and regulates the growth of extensions in cells plated on collagen. The GLDs in the C-terminal of FliI interact with nonmuscle myosin IIA to regulate collagen remodeling (Arora et al., 2015). Although the LRR in the N-terminal of FliI has been suggested to interact with Ras for effecting downstream signaling (Goshima et al., 1999), the mechanism by which FliI interacts with Ras to mediate generation of cell extensions is not defined. Here we show that the LRR of FliI interacts with R-ras and promotes association between G3BP1 and Rasgap120 to enable cytoskeletal reorganization and cell extension formation (Figure 6). Earlier reports showed that the N-terminus of G3BP1 (residues 1–229) interacts specifically with the SH3 domain of Rasgap120 (Kennedy et al., 2001; Parker et al., 1996), implicating G3BP1 in Rasgap120 signaling (Pazman et al., 2000). Here we suggest that FliI, by virtue of its multi-domain structure, performs two quite separate functions. The actin-binding domain (homologous to gelsolin family proteins) is involved in actin cytoskeletal rearrangement, while the LRR region regulates signal transduction through its association with R-ras and the C-terminal of G3BP1.
FIGURE 6:
Interplay among R-ras, G3BP1 and Rasgap in FliI-dependent cell extension formation. 1) FliI LRR is required for adhesion-induced R-ras activation and cell extension formation. 2) R-ras and G3BP1 are targeted to developing cell extensions. 3) FliI LRR interacts with G3BP1 and colocalizes with G3BP1 at the growing extensions. 4) FliI LRR interacts with Rasgap and this protein complex of FLiI-Rasgap is required for cell extension formation. 5) Rasgap interacts with N-terminus of G3BP1, which is required for cell extension formation. 6) FliI promotes interaction between R-ras and C-terminus of G3BP1, but cell extension formation also requires the C-terminus and N-terminus of G3BP1.
FliI-LRR domain and R-ras
Unlike other members of the gelsolin family of proteins, the N-terminal half of FliI consists of 16 tandem repeats of a 23-amino-acid motif, which comprises the LRR domain (Liu and Yin, 1998). The LRR in FliI exhibits similar amino acid sequences as LRR domains in other proteins involved in a wide array of cellular functions (Campbell et al., 1993) that include hormone binding, cell adhesion, DNA repair, bacterial virulence, and Ras signal transduction. Nearly 100 proteins containing LRR motifs have been identified (Buchanan and Gay, 1996) and are found in extracellular, cytoplasmic, transmembrane, and nuclear sites. Our data show that FliI acts as an adaptor protein in which its LRR region interacts specifically with R-ras to facilitate the regulatory processes involved in the growth of cell extensions.
Ras proteins (H-ras, N-ras, K-ras, and R-ras) share similar molecular structures, exhibit an ability to hydrolyze guanine nucleotides, and exist in active (GTP-bound) and inactive (GDP-bound) states. However, unlike other Ras proteins, R-ras contains a proline rich motif in its C-terminal that resembles certain SH3 domain-binding sites (Wang et al., 2000), which confer distinct functions on R-ras. In this context, the adaptor protein Nck contains SH3 domains that interact with the proline-rich sequence in the C-terminus of R-ras that mediates integrin activation (Wang et al., 2000). Similarly, signaling through the C-terminus of R-ras is required for focal adhesion formation and integrin activation (Furuhjelm and Peranen, 2003). We found that in response to plating on collagen, FliI localizes to focal adhesions and interacts with activated R-ras (but not H-Ras, N-Ras, or K-Ras). This interaction between the LRR of FliI and the proline-rich sites of active R-ras is evidently involved in the growth of cell extensions because mutations at its C-terminal (proline to alanine at residues 202 and 203 of R-ras) inhibited the growth of cell extensions. R-ras is recruited to the leading edge of migrating cells (Wozniak et al., 2005) where it regulates cell adhesion by affecting the affinity and avidity of integrins (Hansen et al., 2003; Keely et al., 1999). R-ras also controls membrane dynamics by modulating β1 integrin function; this process involves a cycle of membrane protrusion, ruffling, and endocytosis (Conklin et al., 2010). Consistent with these data, we found that the LRR domain of FliI associates with constitutively active R-Ras at growing cell extensions and that inactive R-ras inhibited the growth of extensions.
Role of G3BP1
The functions controlled by small guanine-binding proteins require GEFs and GAPs to regulate extracellular signals and localize cues (Bos et al., 2007). We used a nucleotide-free probe (R-ras G15A) to search for proteins that may be involved in regulation of R-ras activity. Nucleotide-free GTPases, which are intermediates in GDP-GTP exchange reactions and form high-affinity complexes, have been used to identify GAPs and GEFs (Cherfils and Chardin, 1999). With this approach, we used mass spectrometry analysis of proteins eluted from RasG15A to identify G3BP1, which was confirmed by immunoblotting. G3BP1 is implicated in cancer progression through Ras signaling (French et al., 2002) and G3BP1 is frequently overexpressed in human cancers, suggesting its clinical importance (Zhang et al., 2007). The RNA-binding and protein interaction domains of the G3BP1 indicate their involvement in signal-regulated mRNA metabolism (Tourriere et al., 2001). Further, recruitment of G3BP1 and Rasgap120 at filopodial adhesions suggests a possible role of G3BP1 in reorganization of the cytoskeleton and cell migration (Meng et al., 2004). We found that activated R-ras associates with G3BP1 in its C-terminus to promote cell extension growth and that knockdown of G3BP1 prevented the growth of cell extensions. Earlier reports showed that the N-terminal of G3BP1 (residues 1–229) interacts specifically with the SH3 domain of Rasgap120 (Parker et al., 1996), implicating G3BP1 in Rasgap120 signaling (Pazman et al., 2000). In this context, the SH3 domain of Rasgap120 is essential for transducing signals downstream of Ras (Tocque et al., 1997) and is involved in cytoskeletal organization and cell adhesion (Leblanc et al., 1998). Our data, which support a role for FliI in the G3BP1-Rasgap120-R-ras signaling system, indicate that FliI coordinates Ras signaling in cell extension growth and actin cytoskeletal modification at localized sites (Hennig et al., 2015).
Collectively, we have shown that the N-terminus of G3BP1 associates with Rasgap120 to regulate signal transduction for cell extension growth. This process critically involves the C-terminus of G3BP1 in associating with R-ras and the LRR domain of FliI. Therefore, in addition to its role as an actin-binding protein, FliI plays a central role as a critical adaptor protein in R-Ras-mediated cell extension formation in early phases of cell migration.
MATERIALS and METHODS
Reagents
Antibodies to FliI, R-ras, K-ras, H-ras, N-ras, and G3BP1 wer purchased from Abcam. RasGap120 and FliI antibodies were obtained from Santa Cruz. The G3BP1 expression plasmid was obtained from Addgene. Protein G beads and protein Sepharose glutathione beads were purchased from Dynamed. Duolink in Situ Detection Reagents Far Red, Duolink in Situ Probemaker Minus/Plus and Glutathione Sepharose beads were from Sigma Aldrich (Oakville, ON). On-Target siRNA to G3BP1 and Rasgap were purchased from GE Dharmacon (Mississauga, ON). Type 1 bovine collagen was purchased from Advanced BioMatrix (Carlsbad, CA). Transwell, permeable, 24-mm-diameter inserts (8.0-μm pore size; tissue culture-treated polycarbonate membrane) obtained from Costar (Corning). Recombinant K-ras and H-ras proteins were purchased from Abcam. N-ras knockout 293T cell lysate and parental 293T cell lysate were obtained from Novus Biologicals.
Cell culture
Wild-type and KND FliI mouse fibroblasts were cultured at 37°C in complete DMEM containing 10% fetal bovine serum (FBS) and 10% antibiotics (124 U/ml penicillin G, 50 μg/ml gentamicin sulfate, and 0.25 μg/ml Fungizone). Cells were maintained in a humidified incubator containing 95% air and 5% CO2 and passaged with 0.01% trypsin (Life Technologies, Burlington, ON).
Colocalization of proteins
Fibroblasts were plated on glass-bottom microwell dishes (35-mm pertri dishes, 14-mm microwell No. 1.5 coverglass; MatTek Corp.) and incubated for 2.5 h at 37°C. Cells fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min, permeabilized with 0.3% Triton X-100 for 10 min, and blocked with CAS-Block (Life Technologies) for 20 min at room temperature. Cells incubated with appropriate primary and fluorescent secondary antibodies diluted in 0.03% Triton X-100 both for 1 h at 37°C. Confocal microscopy (×40 oil-immersion lens; Leica TCS SP8, Heidelberg, Germany) was used to localize proteins of interest.
Nucleotide-free R-ras and RBD pull downs
Affinity precipitation of exchange factors with the nucleotide-free R-ras mutant (G15A) was performed on cells lysed in 20 mM HEPES (pH 7.6), 150 mM NaCl, 1% Triton X-100, 5 mM MgCl2, 200 mM orthovanadate plus protease inhibitors. Equal amounts of protein from clarified lysates were incubated with 20 μg of purified R-ras (G15A) bound to glutathione-Sepharose beads for 60 min at 4°C. Samples were washed 3× in lysis buffer and processed for SDS–PAGE. For mass spectrometry analysis, gels stained with Coomassie Blue and the bands of interest analyzed by matrix-assisted laser desorption/ionization, TOF 5800 system mass spectrometry (MALDI-TOF-MS). Selected tryptic peptides sequenced by nano-electrospray ionization (ESI)-MS/MS at the UHN Proteomics Facility (Toronto, ON).
Pull-down experiments for active R-ras were performed by lysing adherent or suspended fibroblasts in buffer (50 mM Tris, pH 7.6, 500 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate, 10 mM MgCl2, 200 mM orthovanadate, and protease inhibitors). Lysates were clarified by centrifugation, equalized for total volume and protein concentration, and rotated for 30 min with 30 μg of purified glutathione S-transferase (GST)-RBD bound to glutathione-Sepharose beads. The bead pellets were washed in a buffer solution (50 mM Tris, pH 7.6, 150 mM NaCl, 1% Triton X-100, 10 mM MgCl2, 200 µM orthovanadate, and protease inhibitors), and the proteins were separated by SDS–PAGE.
In vitro experiments
Protein concentrations were determined by running standards on SDS polyacrylamide gels or by the bicinchoninic acid assay (BCA) protein determination method. Pull-down assays were used to assess binding of Ras isoforms and FliI LRR and FliI GLD domains. Recombinant GST-FLiI proteins were bound to glutathione beads, and various concentrations of GST-Sepharose bead bound flightless proteins (0.25–6 μM) were incubated with recombinant Ras isoforms (1 μM) at 23°C for 30 min in the reaction buffer containing 20 mM Tris-HCl, pH 7.5, 20 mM NaCl, 2 mM MgCl2, 0.3 mM CaCl2, and 1 mM dithiothreitol (DTT). Supernatant were removed and pellets consisting of Sepharose beads were washed, and bound proteins were recovered in Laemmli sample buffer and analyzed on SDS–PAGE and stained with Coomassie Blue.
Purification of FliI recombinant proteins
GST-tagged proteins expressed in bacterial expression systems were isolated and purified by adapting earlier methods (Frangioni and Neel, 1993). Briefly, BL21(DE3) cells were transformed with FliI-LRR and FliI GLD constructs. Luria broth (250 ml) containing ampicillin (100 μg/ml) was inoculated at a 1:50 ratio from overnight bacterial culture containing the FliI construct. The culture was grown at 37°C followed by induction with isopropyl β-d-1-thiogalactopyranoside (IPTG) (1 mM) for 3.5 h. Proteins were isolated from inclusion bodies in sodium chloride–Tris–EDTA (STE) buffer (10 mM Tris [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1.5% Sarkosyl, 5 mM DTT) were dialyzed overnight and incubated with glutathione Sepharose beads (Pharmacia) followed by washing three times with STE buffer without sarkosyl.
Immunoblotting
Cell extracts were prepared by scraping cells into lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, and a 1:50 dilution of protease inhibitor cocktail). The homogenate was centrifuged at 10,000 rpm for 4 min and the supernatant separated for analysis. BCA analysis was conducted to ensure that equal amounts of protein were separated on SDS–PAGE gel and transferred to nitrocellulose membranes. Membranes were blocked with 1% bovine serum albumin in Tris-buffered saline (TBS) overnight and incubated with appropriate primary and secondary antibodies diluted in TBS with 0.01% Tween-20 for 1 h at room temperature.
Constructs
Stable FliI KND fibroblast cells were developed using the following oligonucleotides. For the top strand: 5′-gatccGAAGATACACACTATGTTATTCAAGAGATAACATAGTGTGTATCTTCTTTTTTACGCGTg—3′; and for the bottom strand: 5′-aattcACGCGTAAAAAAGAAGATACACACTATGTTATCTCTTGAATAACATAGTGTGTATCTTCg—3′ corresponding to the sense 5′-GAAGATACACACTATGTTA-3′ and antisense 5′-TAACATAGTGTGTATCTTC-3′ for mouse FliI annealed and inserted into an RNAi-Ready pSIREN-RetroQ-DsRed-Express vector (Clontech) at BamHI/EcoRI sites. Insert sequences were confirmed by sequencing. The plasmid was cotransfected with pVSV-G into GP-293 cells for retrovirus production. NIH-3T3 cells were infected with the virus; 2 wk later, the transfected cells were sorted in PBS/0.5% FBS with a Beckman-Coulter Altra flow cytometer/sorter. Cells with strong red fluorescence were cloned by limiting dilution. There was 90% knockdown of FliI expression in these cells (Arora et al., 2015). A FliI WT 3T3 cell line was created by stably transfecting of a scrabble Luciferase RNA sequence 5′-GTGCGTTGCTAGTACCAACTTCAAGAGA-3′.
G3BP1 constructs
For GST-tagged truncated G3BP1 constructs, PCR products containing appropriate G3BP1 fragments were ligated into pGEX-4T-2 (Amersham, Oakville, ON) and transformed into DH5 α-competent Escherichia coli cells (Invitrogen; Burlington, ON). The construct was sequenced (ACGT Corp., Toronto, ON) and transformed into BL21(DE3) competent E. coli cells for GST-tagged-protein expression and purification. Briefly, bacterial cells grown in LB containing 100 μg/ml ampicillin were induced with 0.5 mM IPTG for 3 h at 30°C. To prepare the GST-tagged G3BP1 glutathione beads, bacteria expressing the constructs were pelleted by centrifugation for 30 min at 10,000 × g. The bacterial pellets were suspended in STE buffer (10 mM Tris, pH 8.0, 15 0 mM NaCl, 1 mM EDTA containing 100 μg/ml lysozyme, 5 mM DTT, 1.5% Sarkosyl, 3% Triton X-100, and protease inhibitors). Lysates collected after homogenization and centrifugation were incubated with glutathione beads (Pierce) O/N at 4°C. The beads were washed three times with PBS and used for experiments. HA-tagged G3BP1 constructs were prepared by inserting the appropriated HA-tagged PCR products in pCMV6 plasmid.
Statistical analysis
For all continuous variable data, means and standard errors were computed. For comparisons between two groups, Student’s t test was performed, and for analyses involving more than two groups, analysis of variance (ANOVA) was conducted followed by Tukey’s post hoc test. Statistical significance was set at p < 0.05.
Supplementary Material
Acknowledgments
This work was supported by a Canadian Institute of Health Research operating grant to C.A.M. (MOP-36332). C.A.M. is supported by a Canada Research Chair (Tier1). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations used:
- G3BP1
Ras GTPase-activating protein SH3 domain-binding protein
- GST
glutathione S-transferase
- KND
knockdown
- LRR
leucine-rich repeat
- RBD
Ras-binding domain
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-03-0147) on August 9, 2018.
REFERENCES
- Arora PD, Di Gregorio M, He P, McCulloch CA. (2017). TRPV4 mediates the Ca(2+) influx required for the interaction between flightless-1 and non-muscle myosin, and collagen remodeling. J Cell Sci , 2196–2208. [DOI] [PubMed] [Google Scholar]
- Arora PD, Wang Y, Bresnick A, Janmey PA, McCulloch CA. (2015). Flightless I interacts with NMMIIA to promote cell extension formation, which enables collagen remodeling. Mol Biol Cell , 2279–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnans C, Chou J, Werb Z. (2014). Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol , 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. (2007). GEFs and GAPs: critical elements in the control of small G proteins. Cell , 865–877. [DOI] [PubMed] [Google Scholar]
- Buchanan SG, Gay NJ. (1996). Structural and functional diversity in the leucine-rich repeat family of proteins. Prog Biophys Mol Biol , 1–44. [DOI] [PubMed] [Google Scholar]
- Campbell HD, Schimansky T, Claudianos C, Ozsarac N, Kasprzak AB, Cotsell JN, Young IG, de Couet HG, Miklos GL. (1993). The Drosophila melanogaster flightless-I gene involved in gastrulation and muscle degeneration encodes gelsolin-like and leucine-rich repeat domains and is conserved in Caenorhabditis elegans and humans. Proc Natl Acad Sci USA , 11386–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PM, Der CJ. (2004). Oncogenic Ras and its role in tumor cell invasion and metastasis. Semin Cancer Biol , 105–114. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Chardin P. (1999). GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem Sci , 306–311. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. (2013). Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev , 269–309. [DOI] [PubMed] [Google Scholar]
- Claudianos C, Campbell HD. (1995). The novel flightless-I gene brings together two gene families, actin-binding proteins related to gelsolin and leucine-rich-repeat proteins involved in Ras signal transduction. Mol Biol Evol , 405–414. [DOI] [PubMed] [Google Scholar]
- Conklin MW, Ada-Nguema A, Parsons M, Riching KM, Keely PJ. (2010). R-Ras regulates beta1-integrin trafficking via effects on membrane ruffling and endocytosis. BMC Cell Biol , 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy DA, Campbell HD, Fountain S, de Jong D, Crouch MF. (2001). The flightless I protein colocalizes with actin- and microtubule-based structures in motile Swiss 3T3 fibroblasts: evidence for the involvement of PI 3-kinase and Ras-related small GTPases. J Cell Sci , 549–562. [DOI] [PubMed] [Google Scholar]
- Everts V, van der Zee E, Creemers L, Beertsen W. (1996). Phagocytosis and intracellular digestion of collagen, its role in turnover and remodelling. Histochem J , 229–245. [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG. (1993). Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem , 179–187. [DOI] [PubMed] [Google Scholar]
- French J, Stirling R, Walsh M, Kennedy HD. (2002). The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem J , 223–231. [DOI] [PubMed] [Google Scholar]
- Furuhjelm J, Peranen J. (2003). The C-terminal end of R-Ras contains a focal adhesion targeting signal. J Cell Sci , 3729–3738. [DOI] [PubMed] [Google Scholar]
- Goshima M, Kariya K, Yamawaki-Kataoka Y, Okada T, Shibatohge M, Shima F, Fujimoto E, Kataoka T. (1999). Characterization of a novel Ras-binding protein Ce-FLI-1 comprising leucine-rich repeats and gelsolin-like domains. Biochem Biophys Res Commun , 111–116. [DOI] [PubMed] [Google Scholar]
- Hall A. (1992). Ras-related GTPases and the cytoskeleton. Mol Biol Cell , 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Prior IA, Hughes PE, Oertli B, Chou FL, Willumsen BM, Hancock JF, Ginsberg MH. (2003). C-terminal sequences in R-Ras are involved in integrin regulation and in plasma membrane microdomain distribution. Biochem Biophys Res Commun , 829–838. [DOI] [PubMed] [Google Scholar]
- Hennig A, Markwart R, Esparza-Franco MA, Ladds G, Rubio I. (2015). Ras activation revisited: role of GEF and GAP systems. Biol Chem , 831–848. [DOI] [PubMed] [Google Scholar]
- Higashi T, Ikeda T, Murakami T, Shirakawa R, Kawato M, Okawa K, Furuse M, Kimura T, Kita T, Horiuchi H. (2010). Flightless-I (Fli-I) regulates the actin assembly activity of diaphanous-related formins (DRFs) Daam1 and mDia1 in cooperation with active Rho GTPase. J Biol Chem , 16231–16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashita S, Song SY. (2008). RasGAPs: a crucial regulator of extracellular stimuli for homeostasis of cellular functions. Mol Biosyst , 213–222. [DOI] [PubMed] [Google Scholar]
- Keely PJ, Rusyn EV, Cox AD, Parise LV. (1999). R-Ras signals through specific integrin alpha cytoplasmic domains to promote migration and invasion of breast epithelial cells. J Cell Biol , 1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy D, French J, Guitard E, Ru K, Tocque B, Mattick J. (2001). Characterization of G3BPs: tissue specific expression, chromosomal localisation and rasGAP(120) binding studies. J Cell Biochem , 173–187. [DOI] [PubMed] [Google Scholar]
- Kim H, McCulloch CA. (2011). Filamin A mediates interactions between cytoskeletal proteins that control cell adhesion. FEBS Lett , 18–22. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. (1995). Proteins with leucine-rich repeats. Curr Opin Struct Biol , 409–416. [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. (2001). The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol , 725–732. [DOI] [PubMed] [Google Scholar]
- Kopecki Z, Cowin AJ. (2008). Flightless I: an actin-remodelling protein and an important negative regulator of wound repair. Int J Biochem Cell Biol , 1415–1419. [DOI] [PubMed] [Google Scholar]
- Kwong L, Wozniak MA, Collins AS, Wilson SD, Keely PJ. (2003). R-Ras promotes focal adhesion formation through focal adhesion kinase and p130(Cas) by a novel mechanism that differs from integrins. Mol Cell Biol , 933–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc V, Tocque B, Delumeau I. (1998). Ras-GAP controls Rho-mediated cytoskeletal reorganization through its SH3 domain. Mol Cell Biol , 5567–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YT, Yin HL. (1998). Identification of the binding partners for flightless I, A novel protein bridging the leucine-rich repeat and the gelsolin superfamilies. J Biol Chem , 7920–7927. [DOI] [PubMed] [Google Scholar]
- Meng X, Krishnan J, She Y, Ens W, Standing K, Wilkins JA. (2004). Association of rasGAPSH3 binding protein 1, G3BP1, and rasGap120 with integrin containing complexes induced by an adhesion blocking antibody. J Proteome Res , 506–516. [DOI] [PubMed] [Google Scholar]
- Mohammad I, Arora PD, Naghibzadeh Y, Wang Y, Li J, Mascarenhas W, Janmey PA, Dawson JF, McCulloch CA. (2012). Flightless I is a focal adhesion-associated actin-capping protein that regulates cell migration. FASEB J , 3260–3272. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Stossel TP, Hartwig JH. (2011). The filamins: organizers of cell structure and function. Cell Adh Migr , 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker F, Maurier F, Delumeau I, Duchesne M, Faucher D, Debussche L, Dugue A, Schweighoffer F, Tocque B. (1996). A Ras-GTPase-activating protein SH3-domain-binding protein. Mol Cell Biol , 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazman C, Mayes CA, Fanto M, Haynes SR, Mlodzik M. (2000). Rasputin, the Drosophila homologue of the RasGAP SH3 binding protein, functions in ras- and Rho-mediated signaling. Development , 1715–1725. [DOI] [PubMed] [Google Scholar]
- Perez-Tamayo R. (1978). Pathology of collagen degradation. A review. Am J Pathol , 508–566. [PMC free article] [PubMed] [Google Scholar]
- Pomerance M, Thang MN, Tocque B, Pierre M. (1996). The Ras-GTPase-activating protein SH3 domain is required for Cdc2 activation and mos induction by oncogenic Ras in Xenopus oocytes independently of mitogen-activated protein kinase activation. Mol Cell Biol , 3179–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radinsky R, Flickinger KS, Kosir MA, Zardi L, Culp LA. (1990). Adhesion of Kirsten-ras+ tumor-progressing and Kirsten-ras- revertant 3T3 cells on fibronectin proteolytic fragments. Cancer Res , 4388–4400. [PubMed] [Google Scholar]
- Self AJ, Caron E, Paterson HF, Hall A. (2001). Analysis of R-Ras signalling pathways. J Cell Sci , 1357–1366. [DOI] [PubMed] [Google Scholar]
- Tocque B, Delumeau I, Parker F, Maurier F, Multon MC, Schweighoffer F. (1997). Ras-GTPase activating protein (GAP): a putative effector for Ras. Cell Signal , 153–158. [DOI] [PubMed] [Google Scholar]
- Tourriere H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, Tazi J. (2001). RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol , 7747–7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zou JX, Ek-Rylander B, Ruoslahti E. (2000). R-Ras contains a proline-rich site that binds to SH3 domains and is required for integrin activation by R-Ras. J Biol Chem , 5222–5227. [DOI] [PubMed] [Google Scholar]
- Wozniak MA, Kwong L, Chodniewicz D, Klemke RL, Keely PJ. (2005). R-Ras controls membrane protrusion and cell migration through the spatial regulation of Rac and Rho. Mol Biol Cell , 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HZ, Liu JG, Wei YP, Wu C, Cao YK, Wang M. (2007). Expression of G3BP and RhoC in esophageal squamous carcinoma and their effect on prognosis. World J Gastroenterol , 4126–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Vuori K, Wang H, Reed JC, Ruoslahti E. (1996). Integrin activation by R-ras. Cell , 61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.