Abstract
Mitochondrial gene expression in Saccharomyces cerevisiae is responsible for the production of highly hydrophobic subunits of the oxidative phosphorylation system. Membrane insertion occurs cotranslationally on membrane-bound mitochondrial ribosomes. Here, by employing a systematic mass spectrometry–based approach, we discovered the previously uncharacterized membrane protein Mrx15 that interacts via a soluble C-terminal domain with the large ribosomal subunit. Mrx15 contacts mitochondrial translation products during their synthesis and plays, together with the ribosome receptor Mba1, an overlapping role in cotranslational protein insertion. Taken together, our data reveal how these ribosome receptors organize membrane protein biogenesis in mitochondria.
INTRODUCTION
The mitochondrial proteome consists of polypeptides from two genetic sources. Most proteins are encoded in the nucleus and posttranslationally imported into their respective mitochondrial compartments (Neupert, 2015; Wiedemann and Pfanner, 2017). A small subset of proteins, eight in Saccharomyces cerevisiae, is encoded in the mitochondrial genome (Foury et al., 1998). Their genes are transcribed and translated by a genetic system residing in the matrix of the organelle. The mitochondrial ribosomes (mitoribosomes) synthesize, almost exclusively, membrane proteins that are subunits of oxidative phosphorylation complexes. Recent structural data have revealed that mitoribosomes are adapted for the production of hydrophobic polypeptides by a specific makeup of the polypeptide exit tunnel, which is rather apolar to accommodate the hydrophobic translation products (Amunts et al., 2014; Brown et al., 2014; Greber et al., 2014a,b). In addition, mitoribosomes are permanently attached to the inner mitochondrial membrane (Preuss et al., 2001; Ott et al., 2006; Prestele et al., 2009; Pfeffer et al., 2015), in contrast to bacterial ribosomes that are targeted to the membrane by the signal-recognition particle (Bernstein et al., 1989). In yeast mitochondria, cotranslational membrane insertion is mediated in a concerted manner by Oxa1 and Mba1 (Preuss et al., 2001; Ott et al., 2006). Oxa1, which belongs to the conserved YidC/Alb3/Oxa1 family, inserts client proteins into the membrane (He and Fox, 1997; Hell et al., 2001). The peripheral membrane protein Mba1 cooperates with the C-terminal domain of Oxa1 in membrane protein insertion and ribosome–membrane attachment (Ott et al., 2006). The human homologue of Mba1 (Mrpl45) is integrated in the large ribosomal subunit and was speculated to fulfill a similar role in membrane attachment (Greber et al., 2014b). Additionally, Mba1 shuttles the mitochondrial-encoded cytochrome c oxidase subunit 2 (Cox2) to the assembly factor Cox20 (Lorenzi et al., 2016).
In a previous study, we found that the mitochondrial ribosome is organized in large assemblies that we termed MIOREX complexes (Kehrein et al., 2015). These assemblies contain factors responsible for mRNA maturation and turnover, translation, chaperones, proteases, and a number of previously uncharacterized proteins, some of which were later shown to play a role in mitochondrial gene expression (Moda et al., 2016; Rak et al., 2016). Another study used a SILAC (stable isotope labeling of amino acids in cell culture)-based approach (Woellhaf et al., 2016) to identify several factors comigrating with the assembled mitoribosome in sucrose gradients, but the ribosomal subunits with which they interacted remained unknown.
Here, we developed an alternative strategy to further characterize the mitoribosomal interactome. We thereby discovered a previously uncharacterized protein (Mrx15) that interacts with the large ribosomal subunit. Simultaneous absence of Mrx15 and Mba1 provokes a profound respiratory growth defect and decrease in cytochrome c oxidase. We demonstrate that Mba1 and Mrx15 are ribosome receptors with overlapping roles in cotranslational protein insertion, directly interacting with the large ribosomal subunits, nascent polypeptide chains, and the inner membrane.
RESULTS
Identification of mitoribosomal interactors by a systematic mass-spectrometric approach
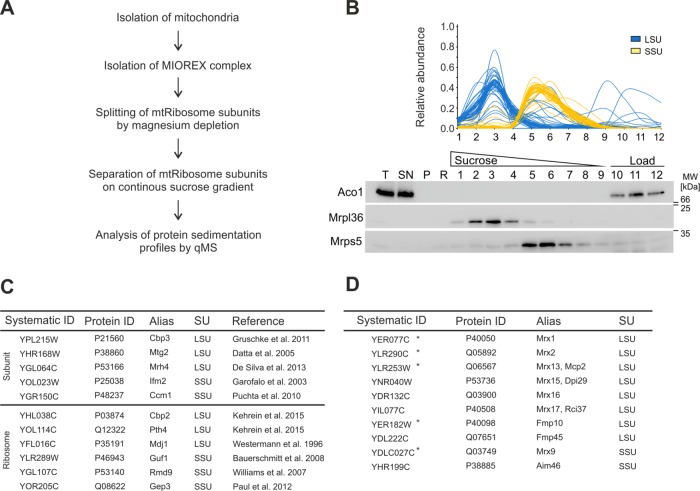
To categorize the extensive interactome of the yeast mitoribosome in the MIOREX complexes (Kehrein et al., 2015), we developed a strategy to isolate mitoribosome subunits and analyze their respective interactomes (Figure 1A). After isolation of MIOREX complexes, ribosomes were split into their subunits by magnesium depletion and separated on a continuous sucrose gradient. Western blotting confirmed that both subunits migrated at distinct peaks (Figure 1B). Next, we repeated the experiment and determined proteins in all fractions by label-free quantitative mass spectrometry (Supplemental Data S1). Proteins from the large subunit (LSU) almost exclusively migrated in fractions 2–4, while proteins of the small subunit (SSU) were found in fractions 4–6 (Figure 1B).
FIGURE 1:
Proteomic profiling of the small and large mitochondrial ribosomal subunits. (A) Strategy to isolate the MIOREX complex with subsequent separation of mitoribosome subunits. (B) After the procedure described in A, the protein abundance of individual fractions from a continuous sucrose gradient was determined by label-free quantitative mass spectrometry. Sedimentation profiles are depicted for the large subunit (blue) and small subunit (yellow). As a control, the experiment was probed for proteins from the large (Mrpl36) and the small (Mrps5) subunit as well as aconitase (Aco1). (C) Top, summary of known interactors of the large and small subunit. These factors showed the same migration behavior as their described ribosomal-interacting subunit. Bottom, summary of known mitoribosome interactors and the interacting subunit found in this study. (D) Summary of known (star) or newly discovered MIOREX components found in this study that comigrated with either the large or the small ribosomal subunit. LSU, large subunit; P, pellet; qMS, quantitative mass spectrometry; R, resuspended; SN, supernatant; SSU, small subunit; T, total.
Next, we cross-referenced our data set with known interactors of the large and small ribosomal subunit. As expected, previously described mitoribosome interactors like Cbp3, Mhr4, and Mtg2 (Figure 1C, top) (Datta et al., 2005; Gruschke et al., 2011; De Silva et al., 2013) comigrated with the LSU. The mitochondrial initiation factor 2 (Ifm1) (Garofalo et al., 2003) and Cmm1 (Puchta et al., 2010) comigrated with the SSU, indicating that we can identify predicted ribosomal interaction partners with our methodology. We next checked for proteins that have been reported to interact with the mitoribosome, but for which the subunit they interact with was unknown (Westermann et al., 1996; Williams et al., 2007; Bauerschmitt et al., 2008; Paul et al., 2012; Kehrein et al., 2015). This allowed us to establish that Cbp2, Pth4, and Mdj1 interact with the LSU, while Rmd9, Gep3, and Guf1 interact with the SSU (Figure 1C, bottom), information likely important to understand their function. For example, the proposed role of Rmd9 in delivering mRNAs to the ribosome (Nouet et al., 2007; Williams et al., 2007) is supported by our finding that it interacts with the SSU.
To identify novel ribosomal interaction partners, we screened the data set for uncharacterized proteins with a predicted or confirmed mitochondrial localization that comigrate with the LSU or SSU (Figure 1D). We thereby identified proteins found in the original MIOREX data set (Kehrein et al., 2015), which were termed Mrx proteins (Figure 1D, star). Consequently, new and uncharacterized ribosomal interaction partners found in this study were named according to this convention (Mrx15–17), as they apparently play a role in expression of mitochondrially encoded proteins. We concluded that this approach allowed us to identify new ribosomal interaction partners and to determine the subunit with which they are interacting.
Mrx15 is a mitochondrial inner membrane protein that interacts with the LSU
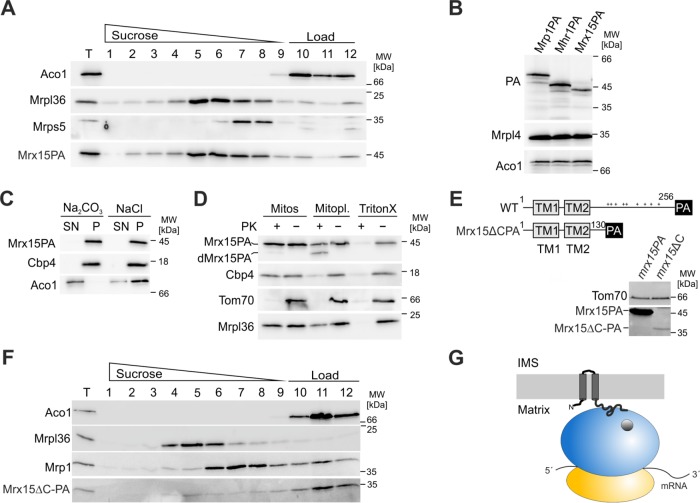
Mrx15 is encoded by the open reading frame YNR040W and shows the same sedimentation profile as the LSU (Figure 1D). Mrx15 is a 29-kDa protein with two predicted transmembrane segments flanked by N- and C-terminal domains. A chromosomally protein A (PA)-tagged variant (Mrx15-PA) quantitatively comigrated with the LSU (Figure 2A) and was present in mitochondria in similar quantities as the mitoribosomal subunits (Figure 2B). To investigate the submitochondrial localization of Mrx15, we performed carbonate extraction and protease protection assays. Mrx15 behaved during carbonate extraction like the integral membrane protein Cbp4 (Figure 2C) (Crivellone, 1994). This is in line with a high-throughput proteomic study that also identified Mrx15 as an integral membrane protein of the inner mitochondrial membrane (Morgenstern et al., 2017). Furthermore, Mrx15 was resistant toward externally added proteinase K when mitochondria were intact, but was partly degraded to a truncated form upon removal of the outer membrane (dMrx15-PA; Figure 2D). Because Mrx15 was detected via its C-terminal PA tag, this shows that the N- and C-termini are facing the mitochondrial matrix. In silico analysis of Mrx15 revealed that it belongs to a family of uncharacterized proteins that are conserved within fungi (Supplemental Figure 1). All members contain two predicted transmembrane segments and a charged C-terminal domain.
FIGURE 2:
Mrx15 is an inner membrane mitochondrial protein that interacts with the large ribosomal subunit. (A) Western blot of mitochondrial extracts containing a chromosomally PA-tagged variant of Mrx15. Mitochondria were lysed in 10 mM EDTA for Mg depletion and separated afterward on a continuous sucrose gradient. Individual fractions were probed for aconitase (Aco1), proteins from the LSU (Mrpl36) and SSU (Mrps5), and PA for Mrx15 detection. (B) Abundancy of PA-tagged variants of the SSU (Mrp1PA), LSU (Mhr1PA), and Mrx15 were compared by Western blotting. Aconitase (Aco1) and Mrpl4 were used as loading controls. (C) Carbonate extraction of mitochondrial extracts. After incubation with either 0.2 M Na2CO3 or 0.2 M NaCl, the membrane and soluble fractions of mitochondrial extracts were separated by high-velocity centrifugation. Supernatant and pellet fractions were probed for membrane (Cbp4) and soluble (Aco1) proteins and PA for Mrx15 detection. (D) Protease protection assay of mitochondria (Mitos), mitoplasts, and lysed mitochondria. Mitochondria, mitoplasts created by hypotonic swelling of mitochondria, and detergent-lysed mitochondria were treated for 20 min with proteinase K (PK). Afterward, a Western blot of each sample was probed for proteins of the outer membrane (Tom70), inner membrane (Cbp4), and the matrix (Mrpl36). The PA antibody was used for detection of the PA-tagged variant of Mrx15. (E) Schematic representation of full-length Mrx15 tagged with PA and a C-terminal truncation variant. Cell extracts of strains grown to log phase containing either variant were probed for PA to determine abundance of each construct. (F) Western blot of mitochondrial extracts containing C-terminally truncated variant of Mrx15. Interaction with the mitoribosome was tested by Mg depletion and separation of ribosomal subunits on a continuous sucrose gradient. Individual fractions were probed for aconitase (Aco1), proteins from the LSU (Mrpl36) and SSU (Mrp1), and PA for Mrx15 detection. (G) Mrx15 inner membrane topology. According to our results, the Mrx15 protein is a component of the inner mitochondrial membrane. The N- and C-termini of the protein face the matrix. The C-terminal domain is necessary for interaction with the large ribosomal subunit. IMS, intermembrane space; Mitopl., mitoplasts; P, pellet; SN, supernatant; T, total; TM, transmembrane segment; WT, wild type.
Next, we tested whether the positively charged, soluble C-terminal domain of Mrx15 is important for the interaction with the mitoribosome. We constructed a mutant in which the C-terminal 126 amino acids were deleted (mrx15ΔC; Figure 2E). This deletion leads to decreased levels of Mrx15ΔC-PA (Figure 2E). Importantly, Mrx15ΔC-PA did not comigrate with the LSU (Figure 2F). Accordingly, the positively charged C-terminal domain is necessary for ribosomal interaction. We concluded that Mrx15 is an integral protein of the inner mitochondrial membrane that interacts with the LSU of the mitoribosome via a C-terminal domain (Figure 2G).
Synthetic genetic interaction between MRX15 and MBA1 suggests a common function
To investigate the function of Mrx15, we created a chromosomal deletion strain and tested the growth of this mutant. The mrx15∆ mutant did not show a growth defect on fermentable and nonfermentable media at 30 or 37°C (Figure 3A). Mba1 is a peripheral inner membrane protein involved in membrane insertion of mitochondrial translation products (Preuss et al., 2001; Ott et al., 2006). It directly interacts with the LSU of the mitoribosome and aligns the tunnel exit for membrane protein insertion (Gruschke et al., 2010; Pfeffer et al., 2015). Because Mba1 and Mrx15 both bind to the membrane and the ribosome, but are not necessary for respiratory growth, we asked whether the combined absence of Mrx15 and Mba1 would result in a synthetic growth phenotype suggesting a common function. As reported previously, the mba1∆ mutant showed only a mild growth defect on a nonfermentable carbon source (Figure 3A) (Ott et al., 2006). Strikingly, the mrx15∆mba1∆ mutant was respiratory deficient at 30 and 37°C, while fermentative growth was not affected. We concluded that loss of Mrx15 is tolerated under the tested conditions but required for respiratory growth in the absence of Mba1, suggesting that both proteins have overlapping functions in the biogenesis of the respiratory chain. Next, we tested whether deletion of the C-terminus of Mrx15 is sufficient to provoke the combined respiratory growth phenotype and found that, while the single mutant did not show a growth phenotype under the tested conditions, cells became respiratory deficient in the mrx15ΔCmba1∆ strain (Figure 2A), thereby demonstrating that the C-terminal domain of Mrx15 is required for ribosome interaction and respiratory growth upon deletion of MBA1.
FIGURE 3:

Deletion of MRX15 together with MBA1 leads to respiratory deficiency and altered membrane attachment. (A) Serial-dilution growth test on full medium fermentable (glucose) and nonfermentable (glycerol) carbon sources of indicated strains. In the mrx15∆C mutant, the terminal 126 amino acids were replaced by a PA tag. In the C-terminal Oxa1 mutant the 71 C-terminal residues were deleted alone or together with MRX15. Cells were grown to logarithmic phase, spotted in 10-fold dilutions, and incubated at 30 and 37°C. (B) Ribosome pelletation at different ionic strengths. The ribosome and attached factors were separated after lysis in different salt conditions by a sucrose cushion and high-velocity centrifugation. The different fractions were probed for a soluble protein (Aco1), a ribosomal marker (Mrpl36), Mba1, and PA for Mrx15 detection. (C) Flotation gradient of mitochondrial extracts from wild-type or mrx15∆mba1∆ strains. Soluble and membrane fractions were separated after freeze–thaw cycles by a sucrose step gradient and high-velocity centrifugation. Fractions were probed for the LSU (Mrpl36, Mrpl4, and Mrpl40), membrane (Cbp4), and soluble proteins (Aco1). P, pellet; SN, supernatant; T, total; WT, wild type.
Mba1 cooperates with Oxa1 in the insertion of mitochondrial translation products into the inner membrane (Preuss et al., 2001). Simultaneous deletion of Mba1 and of 71 C-terminal residues of Oxa1 leads to a severe respiratory growth inhibition combined with an insertion defect of mitochondrially encoded proteins (Ott et al., 2006). We tested whether deletion of the C-terminal tail of Oxa1 in the absence of Mrx15 would impair respiratory growth. However, unlike Mba1, Mrx15 did not show a synthetic genetic interaction with the C-terminal domain of Oxa1 (Figure 3A). This suggests that Mrx15 and the C-terminal ribosome-binding domain of Oxa1 do not have overlapping roles for membrane protein insertion, in contrast to Mrx15 and Mba1, which apparently share a common function.
Next, we investigated the salt sensitivity of the Mrx15-LSU and Mba1-LSU interactions and pelleted ribosomes through a sucrose cushion at different ionic strengths (Figure 3B). At low ionic strength conditions, Mrx15 interacted with the LSU, while Mba1 did not comigrate with the LSU under any of the tested conditions. Consistently, Mba1 comigration with any of the mitoribosomal subunits was not observed in the mass spectrometry data set (Figure 1C). Instead, interaction of Mba1 with the LSU was found through chemical cross-linking or ribosome comigration upon magnesium depletion only, pointing to a rather transient interaction of Mba1 with the mitoribosome (Ott et al., 2006; Gruschke et al., 2010).
Mitoribosomes are permanently associated with the inner mitochondrial membrane (Ott et al., 2006; Prestele et al., 2009; Pfeffer et al., 2015). Based on the high-resolution cryo–electron microscopy (cryo-EM) structure of the LSU and cryo-EM tomography, a second point of membrane attachment was proposed, consisting of a membrane-facing protuberance buildup by mitoribosome-specific proteins and rRNA expansion segments (Amunts et al., 2014; Pfeffer et al., 2015). This is supported by data showing that the mitoribosome stays membrane bound upon deletion of MBA1, either alone or in combination with the C-terminal tail of Oxa1 (Ott et al., 2006). Because we found the Mba1-LSU interaction to be rather weak, we tested whether Mba1 and Mrx15 showed overlapping functions in ribosome–membrane attachment. We therefore followed ribosome–membrane interaction upon simultaneous deletion of MRX15 and MBA1 in a flotation gradient (Figure 3C). In wild-type mitochondria, a fraction of the mitoribosomes cofractionated with the membrane marker Cbp4, indicating membrane interaction. In contrast, only a minor fraction of the mitoribosome in the mrx15∆mba1∆ mutant floated with the membranes. Therefore, we concluded that the interaction of mitoribosomes with the membrane is decreased upon simultaneous deletion of Mba1 and Mrx15.
Absence of Mba1 and Mrx15 leads to complex IV deficiency
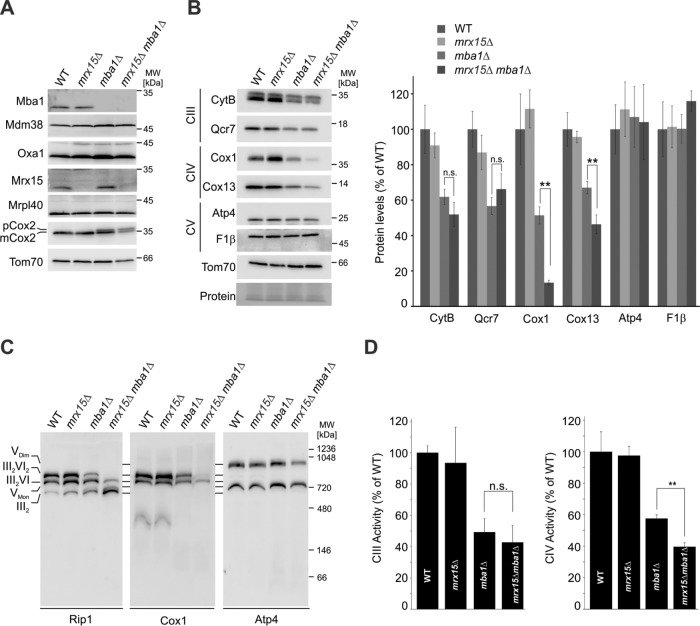
To further characterize the growth defect in the mutants lacking Mrx15, Mba1, or both, we determined steady-state levels of mitochondrial proteins in the strains. Amounts of proteins implicated in mitochondrial protein biogenesis did not change in the strains (Figure 4A). Levels of the cytochrome c oxidase (complex IV) subunit Cox2 were not changed upon loss of Mrx15, but were reduced in the absence of Mba1, with a concomitant accumulation of the precursor form of Cox2 (pCox2), in line with previous data (Preuss et al., 2001; Bauerschmitt et al., 2010). After synthesis and membrane insertion, the Cox2 precursor is processed into a mature from (mCox2) by proteolytic removal of an N-terminal domain by Imp1 and Cox20, thereby generating mature, assembly-competent Cox2 (Nunnari et al., 1993; Jan et al., 2000). Interestingly, simultaneous loss of Mba1 and Mrx15 leads to a further increase in pCox2 levels relative to mCox2.
FIGURE 4:
Mrx15 and Mba1 are required for complex IV accumulation. (A) Steady-state levels of mitochondrial proteins in all indicated strains. Cells were grown in galactose to logarithmic phase, and Western blots probed for proteins necessary for mitochondrial protein biogenesis and complex IV accumulation. (B) Mitochondrial extracts of indicated strains were probed for subunits of complex III (CytB, Qcr7), complex IV (Cox1, Cox13), and complex V (Atp4, F1β). Signal intensities of three independent experiments were quantified and normalized. Significance of observed changes was assessed by a Student’s t test. (C) BN-PAGE of digitonin-solubilized mitochondria from indicated strains. Separated protein complexes were analyzed by Western blotting against subunits of complex III (Rip1), complex IV (Cox1), and complex V (Atp4) (D) Complex III and IV activity measurement. Activity of complex III was followed by measuring cytochrome c reduction, complex IV activity by measuring cytochrome c oxidation at 550 nm. CIII, complex III; CIV, complex IV; CV, complex V; n.s., p > 0.05; **, p ≤ 0.01.
Employing Western blot followed by densitometry analyses, we found that cytochrome bc1 complex (complex III) and complex IV subunits were accumulating normally in the absence of Mrx15, while mitochondria from mba1∆ mutants had diminished amounts of both complexes (Figure 4B). Importantly, upon simultaneous deletion of MRX15 and MBA1, complex IV levels were further reduced, while complex III did not change. ATP synthase (complex V), which also contains mitochondrially encoded subunits, was unchanged in these strains, pointing to a specific defect in accumulating complex IV in the mrx15Δmba1Δ cells.
To confirm this conclusion, we analyzed respiratory chain supercomplex formation and respiratory chain activity. Consistent with the steady-state analyses, blue native PAGE (BN-PAGE) revealed that complex IV abundance in respiratory supercomplexes was lower in the mba1∆ mutant and severely reduced in the double-deletion mutant, where only the smaller isoform of the supercomplexes (III2IV) was detected (Figure 4C, middle). Complex III, the partner of complex IV in respiratory supercomplexes, as revealed by the late-assembling subunit Rip1 (Ndi et al., 2018), was mainly present in the dimeric form, indicating severe reduction of complex IV. ATP synthase was present as a monomer and dimer in all strains, and the mrx15Δmba1Δ cells had slightly reduced levels of the dimer. The specific decrease in complex IV levels upon simultaneous deletion of Mrx15 and Mba1 was also reflected in a significant decrease in complex IV activity, confirming that both Mba1 and Mrx15 are specifically important for the biogenesis of complex IV. In contrast, complex III activity was lower upon MBA1 deletion (Bauerschmitt et al., 2010), but not further decreased in the mrx15Δmba1Δ mutant.
Mrx15 and Mba1 interact with nascent polypeptide chains
Mba1 interacts with nascent polypeptide chains as they emerge from the mitoribosome (Preuss et al., 2001; Ott et al., 2006; Gruschke et al., 2010). Owing to the synthetic genetic interaction and the cofractionation of Mrx15 and Mba1 with the LSU, we hypothesized that Mrx15 could be in contact with nascent chains. To test this, we performed in organello labeling of mitochondrial translation products followed by chemical cross-linking and purification of Mrx15. We chose a cross-linker (DFDNB) with a very short linker length (0.3 nm) that can only covalently connect molecules that are in very close proximity. Autoradiography of the elution fraction showed multiple species with electrophoretic mobilities above 25 kDa (Figure 5A). In a control experiment with mitochondria from wild type, these species were not detected. This indicated that Mrx15 was cross-linked to nascent chains. Mba1 also cross-linked to nascent chains, which is in line with previous results (Preuss et al., 2001; Gruschke et al., 2010). We concluded that Mrx15, like Mba1, is not only in contact with the LSU but directly interacts with nascent chains emerging from the tunnel exit.
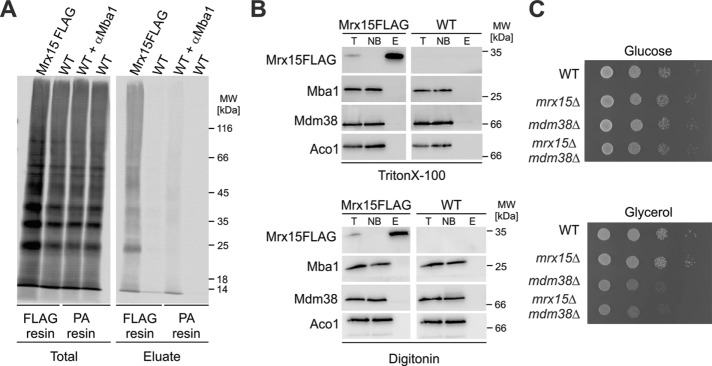
FIGURE 5:
Mrx15 interacts with nascent chains but does not form a stable complex with Mba1. (A) Cross-linking to nascent mitochondrial polypeptide chains. Mitochondrial translation products were radiolabeled in wild-type mitochondria or mitochondria containing a FLAG-tagged variant of Mrx15. After 15 min of labeling, the chemical cross-linker DFDNB was added to covalently link molecules in direct physical contact. The different reactions were stopped, and Mrx15 cross-linking products were purified with a FLAG resin. Mba1 and cross-linked molecules were iummunoprecipitated employing a PA resin and an Mba1 antibody. As a control, the reactions were performed without a FLAG tag or an Mba1 antibody. (B) Purification of FLAG-tagged Mrx15 variants. Mitochondria were lysed in two detergents (Triton X-100, digitonin) and Mrx15 purified by employing a FLAG resin. Western blots of the purification were probed for FLAG, Mba1, Mdm38, and Aco1. (C) Serial-dilution growth test on fermentable (YPD) and nonfermentable (YPG) carbon sources of indicated strains. E, elution; NB, not bound; T, total; WT, wild type.
Mba1 forms a stable complex with Mdm38 that can be purified (Bauerschmitt et al., 2010). We hypothesized that Mrx15 could also physically interact with Mba1 or Mdm38. Therefore, we purified FLAG-tagged variants of Mrx15 from mitochondrial extracts with two different detergents, but no stable complex between these proteins was detected (Figure 5B). Likewise, we did not observe a combined growth defect in the mrx15Δmdm38Δ cells (Figure 5C). Unlike Mdm38 and Mba1, Mrx15 and Mba1 apparently carry out their overlapping functions without forming a stable complex. We concluded that Mrx15, like Mba1, binds to nascent polypeptide chains but does not form a stable complex with Mba1 or Mdm38.
Mrx15 and Mba1 jointly mediate biogenesis of the Cox2 precursor
The direct interaction of Mrx15 with nascent polypeptide chains prompted us to investigate whether the protein together with Mba1 plays a role in protein insertion. Therefore, we radiolabeled mitochondrial translation products in vivo in a pulse–chase experiment (Figure 6A). Mitochondrial protein synthesis and stability were not affected in the mrx15∆ mutant compared with the wild type. As reported previously (Preuss et al., 2001), the MBA1 deletion mutants showed an accumulation of pCox2, and this was enhanced in the mrx15∆mba1∆ mutant. To quantify this effect, we determined the pCox2/mCox2 ratio in both mutants after a 15-min pulse and 90-min chase from five independent experiments (Figure 6B). The pCox2/mCox2 ratio after 15 min of protein synthesis and a 90-min chase was significantly higher in the mrx15∆mba1∆ mutant, revealing an increased accumulation of the pCox2 variant when both Mrx15 and Mba1 were absent. To check whether Mrx15 interacts with specific mitochondrial translation products, we purified Mrx15-containing complexes after in organello labeling of mitochondrial translation products from digitonin lysates. While Mrx15-FLAG could be clearly enriched, only a smear of nascent chains and some mitoribosomes were copurified (Figure 6C). This indicates that Mrx15, in contrast to Mba1, which interacts specifically with Cox2 (Lorenzi et al., 2016), does not bind posttranslationally to mitochondrially encoded proteins. Taken together, these results suggest that the observed complex IV deficiency and the respiratory growth defect in the mrx15∆mba1∆ mutant are caused by hampered Cox2 biogenesis due to disturbances of ribosome–membrane interactions required for efficient cotranslational insertion combined with a specific defect in Cox2 maturation that is carried out by Mba1 (Lorenzi et al., 2016).
FIGURE 6:
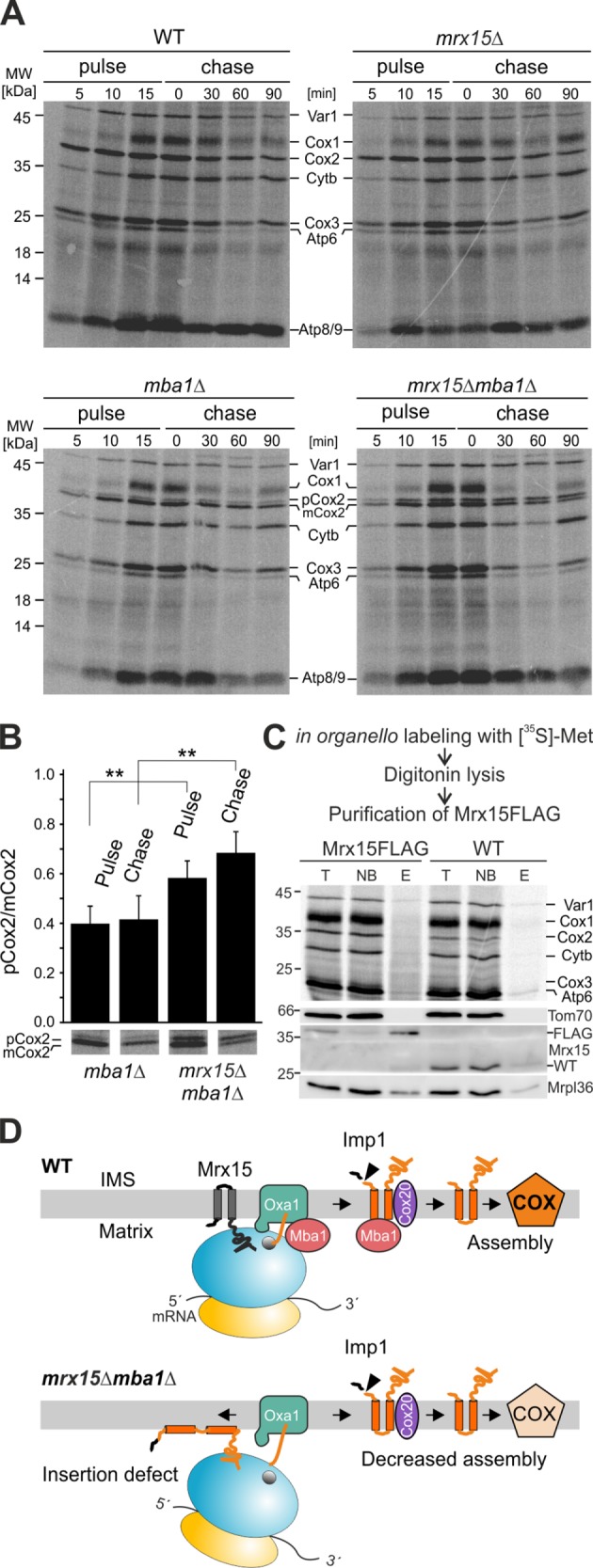
Mrx15 is involved in accumulation of premature Cox2. (A) In vivo radiolabeling of all indicated strains. Mitochondrial protein synthesis of all mutants was followed for 15 min The stability of the synthesized polypeptides was chased for 90 min. (B) Quantification of the pCox2 to mCox2 ratio after 15 min of synthesis and a 90-min chase. Bands of five independent experiments were quantified, and significance between displayed values was assessed by a Student’s t test. (C) Copurification of newly synthesized mitochondrial polypeptides with Mrx15. After in organello labeling of mitochondrial translation products, a FLAG-tagged variant of Mrx15 was purified, and eluates were analyzed by Western blotting and autoradiography. (D) Model of Mrx15 and Mba1 function in Cox2 membrane insertion. In wild-type cells, Mrx15 and Mba1 bind to the LSU and nascent chains to ensure ribosome–membrane attachment and proper membrane insertion. Upon deletion of both factors, membrane insertion is hampered, and pCox2 accumulates. Consequently, this leads to decreased Cox2 maturation, complex IV assembly, and respiratory deficiency. E, elution; IMS, intermembrane space; NB, not bound; T, total; WT, wild type; n.s., p > 0.05; **, p ≤ 0.01.
DISCUSSION
In this study, we identified Mrx15, which helps to tether the mitoribosome to the inner membrane for efficient cotranslational insertion. On the basis of results presented in this study, we propose the following model (Figure 6D): Mba1 and Mrx15 bind to the LSU of the mitoribosome and interact with nascent chains emerging from the exit tunnel. Both proteins tether the LSU to the inner mitochondrial membrane to ensure proper alignment of the LSU with the insertion site. Mba1 and Mrx15 are in proximity to nascent chains to ensure efficient membrane insertion by the insertase Oxa1. Upon completion of Cox2 synthesis, Mba1 dissociates from the mitoribosome to escort the newly synthesized protein toward downstream assembly processes, while Mrx15 remains ribosome bound.
In the simultaneous absence of Mrx15 and Mba1, a large part of the mitoribosomes is liberated from the inner membrane, pointing to a critical function of both proteins as ribosome–membrane tethers. However, Mrx15 and Mba1 greatly differ in their mitoribosome-binding efficiencies. While the integral inner membrane protein Mrx15 stayed ribosome bound in the presence of magnesium and at moderate ionic strength, we did not detect a permanent interaction between the peripheral membrane protein Mba1 and the LSU of the mitoribosome under these conditions. This suggests that Mrx15 is a constant ribosome tether, while Mba1 dynamically interacts with the ribosome. This is in line with a recent study demonstrating a posttranslational function of Mba1 for complex IV biogenesis (Lorenzi et al., 2016), whereby Mba1 shuttles Cox2 from synthesis to its assembly factor Cox20 for maturation and respiratory chain assembly.
Cryo-EM tomography has revealed the organization of mitoribosomes in isolated organelles, where Mba1 occupies a position connecting the tunnel exit with the insertion site. In the absence of Mba1, a specific density was absent, but the general arrangement of the ribosome relative to the inner membrane was unchanged (Pfeffer et al., 2015), suggesting the presence of other ribosome tethers like the Mrx15 protein identified here. In contrast to yeast Mba1, its mammalian homologue mL45 is an integral and firmly bound subunit of the mitoribosomal LSU (Greber et al., 2014b). A recent tomographic study of the human mitoribosome established that mL45 is the single contact site to the inner membrane, while the general architecture of the membrane–mitoribosome contact is conserved between yeast and human (Englmeier et al., 2017). Because homologous proteins of Mrx15 are found only in fungi, it is tempting to speculate that Mrx15 substitutes Mba1 as a permanently bound mitoribosome receptor. Therefore, Mrx15 could occupy a similar binding site on the ribosome as Mba1, ensuring correct mitoribosome–membrane attachment while Mba1 is escorting Cox2 toward respiratory chain assembly.
Permanent membrane attachment of ribosomes appears to be specifically important for mitochondrial gene expression. A recent study of mitoribosome assembly in yeast found that the mitoribosomal LSU is already attached to the membrane during assembly (Zeng et al., 2018). This study found mitoribosome–membrane attachment even upon simultaneous deletion of MBA1, MDM38, and OXA1. Therefore mitoribosome–membrane attachment likely mainly depends on 21S rRNA incorporation into the LSU. This notion is supported by data showing that mutations in 21S rRNA can alter mitoribosome–membrane attachment (Spithill et al., 1978). According to data presented here, mitoribosomes from the double-deletion mutant of MBA1 and MRX15 were still partly membrane attached, suggesting that the 21S rRNA of LSU could connect to the inner mitochondrial membrane via the expansion segment E29 (Pfeffer et al., 2015).
Combined absence of Mba1 and Mrx15 provokes a strong respiratory growth defect, which at least partly reflects the strong complex IV deficiency that exhibits a specific accumulation of the Cox2 precursor. Cox2 precursor accumulation could be caused by a general defect of membrane insertion or the disruption of shuttling newly synthesized Cox2 to the maturation complex consisting of Cox20 and Imp1/2 (Lorenzi et al., 2016). A general insertion defect in the mrx15Δmba1Δ cells that exacerbates the insertion defect of mba1Δ cells would be revealed as a pleiotropic defect in the assembly of all OXPHOS complexes, as has been observed in mutants affected in common processes for synthesis and insertion of the mitochondrially encoded membrane proteins (Altamura et al., 1996; Prestele et al., 2009; Bauerschmitt et al., 2010; Kuzmenko et al., 2016; Ostojic et al., 2016; Suhm et al., 2018). This, however, is not the case in the mrx15Δmba1Δ cells. Furthermore, our data, which indicate that MRX15 does not show a genetic interaction with the C-terminal domain of Oxa1, the insertase, or Mdm38, are in line with a context-specific function Mrx15 exerts together with Mba1 during Cox2 biogenesis. A special aspect of Cox2 biogenesis is that the protein contains by far the largest and most charged soluble domains of all mitochondrially encoded proteins, and these domains need to be translocated over the inner membrane (Herrmann et al., 1995). A disturbed ribosome–membrane contact could specifically impact the translocation of the highly hydrophilic Cox2 domains, particularly when connected with the absence of the shuttling factor Mba1 (Lorenzi et al., 2016) that presumably removes pCox2 from the insertion site.
In addition to the clear energetic advantages of tethering ribosomes to membranes to facilitate cotranslational protein insertion, the firm interaction of the mitoribosome and the inner mitochondrial membrane allows further specialization in how translation is organized. For example, translational activators regulate translation of specific client mRNA in yeast mitochondria (Fox, 2012). This class of proteins is firmly membrane bound and essential for translation in yeast mitochondria. Because of these characteristics, it was suggested that a main function of translational activators is to localize the synthesis of the hydrophobic translation products to the inner membrane (McMullin and Fox, 1993; Fox, 1996). Moreover, by gathering several translational activators controlling synthesis of the three cytochrome oxidase subunits (Naithani et al., 2003), localized assembly sites are established that likely facilitate joining of the subunits. These sites of translation and subsequent biogenesis are not evenly distributed in the inner membrane, as assembly of each OXPHOS complex preferentially occurs at different subcompartments within the inner membrane (Stoldt et al., 2018), highlighting the intricate organization of mitochondrial gene expression.
We have previously found mitoribosomes to interact with many proteins to organize biogenesis of the organellar-encoded proteins. These interactions give rise to large assemblies called MIOREX complexes (Kehrein et al., 2015) that unite many steps of mitochondrial gene expression, including mRNA maturation, translation, assembly, and turnover. Here, employing an alternative strategy to identify and classify components interacting with the mitoribosome provided support for this general concept. Specifically, at least parts of mRNA maturation occur on the LSU, as revealed by its interaction with the COB mRNA splicing factor Cbp2. It remains an exciting task to develop strategies to further categorize the interactome of the yeast mitoribosome and to establish how the different processes are organized on the mitoribosome.
MATERIALS AND METHODS
Yeast strains and growth media
All yeast strains used in this study (Supplemental Table 1) were isogenic to either the wild-type strains W303a or CW04. MRX15 was disrupted with a kanamycin resistance cassette. The MBA1 coding sequence was replaced with an HIS3 selection cassette. FLAG- or PA-tagged variants of Mrx15 were created by replacing the endogenous stop codon with a FLAG sequence or a PA sequence employing a TRP1 or HIS3 selection cassette. Cells were grown at 30°C in YP medium (2% peptone, 1% yeast extract) in the presence of either 2% dextrose, 2% galactose, or 2% glycerol.
Isolation of mitochondria
Mitochondria were isolated as described previously (Gruschke et al., 2011). For experiments analyzed by mass spectrometry, mitochondria were isolated by a sucrose step gradient. The mitochondrial pellet was resuspended in SHE buffer (20 mM HEPES/KOH, pH 7.4, 250 mM sucrose, 1 mM EDTA) and loaded on a step gradient. The layers consisted of 1.75, 0.95, 0.68, and 0.45 M sucrose in 20 mM HEPES/KOH (pH 7.4) and 1 mM EDTA. Mitochondria were spun down (134,000 × g, 1 h, 4°C), recovered from the 1.74–0.95 interface, and resuspended in SH buffer (20 mM HEPES, pH 7.4, 0.6 M sorbitol). At a concentration of 10 mg/ml, mitochondria were snap-frozen in liquid nitrogen.
Sucrose gradients
Isolated mitochondria were lysed (10 mM Tris/HCl, pH 7.4, 10 mM KOAc, 0.5 mM Mg(OAc)2, 10 mM EDTA, 5 mM β-mercaptoethanol, 1% dodecylmaltoside, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1X complete, 0.1 mM spermidine, 5% glycerol), and sucrose gradient experiments were performed as described previously (Kehrein et al., 2015). Collected samples were precipitated by addition of 12% trichloroacetic acid. The resuspended samples were analyzed by SDS–PAGE and Western blotting or mass spectrometry according to published procedures (Chiurillo et al., 2016).
Carbonate extraction and protease protection
Isolated mitochondria (100 µg) were resuspended in 0.1 M Na2CO3 or NaCl and incubated 30 min on ice. Membrane and soluble fractions were separated by centrifugation (100,000 × g, 30 min, 4°C), TCA precipitated, and analyzed by SDS–PAGE and Western blotting.
Mitochondria (100 µg) were incubated in SH buffer or 20 mM HEPES/KOH or lysis buffer (SH buffer with 0.2% Triton X-100) for 30 min at 4°C. Afterward, 0.1 mg/ml proteinase K was added, and the reaction was incubated for 20 min at 4°C. Intact mitochondria or mitoplasts were spun down (25,000 × g, 10 min, 4°C). The supernatant and pellet were analyzed by SDS–PAGE and Western blotting.
Ribosome sedimentation
Isolated mitochondria were lysed in different buffers (10 mM Tris/HCl, pH 7.4, 10/25/100/250 mM KOAc, 5/5/20/50 mM Mg(OAc)2, 0.1 mM spermidine, 5% glycerol, 1% DDM, 1 mM PMSF, 1X complete) for 10 min on ice. The lysate was diluted in one volume of the respective lysis buffer without DDM and cleared by centrifugation (16,000 × g, 10 min, 4°C). The cleared lysate was underlaid with half a volume of sucrose cushion (10 mM Tris/HCl, pH 7.4, 10/25/100/250 mM KOAc, 5/5/20/50 mM Mg(OAc)2, 0.1 mM spermidine, 5% glycerol, 1% DDM, 1 mM PMSF, 1X complete, 1.2 M sucrose) and centrifuged (189,062 × g, 105 min, 4°C). Afterward, the supernatant and pellet fractions were separated and analyzed by Western blotting.
Flotation gradients
Isolated mitochondria were resuspended in buffer A (20 mM Tris/HCl, pH 7.4, 25 mM KOAc, 5 mM Mg(OAc)2, 1 mM PMSF) and treated with three consecutive freeze–thaw cycles. Mitochondria were frozen in liquid nitrogen and thawed at 37°C. Afterward, the reaction was resuspended in 10 volumes of 2.5 M sucrose in 20 mM Tris/HCl (pH 7.4). The suspension was overlaid with one volume of 2.2, 1.5, and 1 M sucrose and centrifuged (40,000 × g, 16 h, 4°C). The gradient was fractionated, and individual fractions were analyzed by Western blotting.
BN-PAGE and Western blotting
Isolated mitochondria (150 µg) were resuspended in BN-PAGE buffer (50 mM Bis-Tris/HCl, pH 7.2, 150 mM NaCl, 2 mM aminohexanoic acid, 1 mM EDTA, 1X complete, 1 mM PMSF, 1% digitonin or 1% DDM, 12% glycerol) and lysed 10 min on ice. The lysate was cleared by centrifugation (16,000 × g, 10 min, 4°C) and NativePAGE 5% G-250 sample additive supplemented to a final concentration of 0.25%. Subsequently, samples were loaded on NativePAGE 3–12% Bis-Tris gels (Thermo Fisher). When the dye front had migrated one-third of the way through the gel, the dark blue cathode buffer was replaced with a light blue cathode buffer. Afterward, separated protein complexes were blotted onto polyvinylidene difluoride membranes.
Complex III and IV activity assay
For respiratory chain activity measurements, 0.5 mg of mitochondria were lysed (50 mM Tris/HCl, pH 7.4, 100 mM KCl, 2.5 MgCl2, 0.1 mM EDTA, 1X complete, 1 mM PMSF, 2% digitonin) for 10 min on ice. Afterward, the lysate was cleared by centrifugation (16,000 × g, 10 min, 4°C) and diluted 50-fold in 50 mM Tris/HCl (pH 7.4).
For complex III activity measurements, KCN (1 mM) and cytochrome c (0.5 mg/ml) were added to the lysate. The reduction of cytochrome c was monitored by measuring the change in absorbance at 550 nm upon addition of quinol (0.08 mg/ml). Activity was calculated by determining the slope of the reaction.
For complex IV activity measurements, antimycin A (0.2 mg/ml) was added to the lysate. The oxidation of cytochrome c was monitored by measuring the change in absorbance at 550 nm upon addition of reduced cytochrome c (0.33 mg/ml). Activity was calculated as for complex III.
Purification of FLAG-tagged Mrx15
Mitochondria from yeast strains expressing FLAG-tagged variants of Mrx15 were lysed for 10 min on ice (10 mM Tris/HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% digitonin or Triton X-100, 1 mM PMSF, 1X complete). Subsequently, samples were diluted 1:1 in dilution buffer (10 mM, Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% digitonin or Triton X-100, 1 mM PMSF, 1x complete), and the lysate was cleared by centrifugation (16,000 × g, 10 min, 4°C). The cleared lysate was incubated with anti-FLAG M2 affinity gel for 2 h at 4°C. The affinity gel was washed three times with dilution buffer. Purified proteins were eluted by incubation for 20 min with dilution buffer supplemented with 0.15 µg/µl 3× FLAG peptide. Samples of all relevant fractions were precipitated by addition of 12% TCA. The resuspended samples were analyzed by SDS–PAGE and Western blotting.
For testing copurification of newly synthesized mitochondrial translation products with Mrx15, a FLAG-tagged variant was purified after in organello labeling (Gruschke et al., 2010). Mitochondria were lysed after the labeling reaction (10 mM Tris/HCl, pH 7.4, 100 mM NaCl, 1 mM EDTA, 2% digitonin, 1 mM PMSF, 1X complete), and the purification was carried out as described earlier.
Miscellaneous methods
Polyclonal antisera against Mrp1 and Mrx15 were generated by injecting rabbits with recombinantly expressed and purified antigens. The antibodies against PA and FLAG were obtained from Sigma. Labeling of mitochondrial translation products and chemical cross-linking of nascent chains were carried out as previously described (Gruschke et al., 2010).
Supplementary Material
Acknowledgments
We thank members of our laboratories for stimulating discussions and Marc Wirth and Ignasi Forne (Ludwig Maximilian University of Munich, Germany) for help with mass spectrometric analyses. This work was supported by grants from the Swedish Research Council, the Carl Tryggers Foundation, and the Knut and Alice Wallenberg Foundation. Proteomic analyses were performed at the Protein Analysis Unit (ZfP) of the Ludwig Maximilians University of Munich, a registered research infrastructure of the Deutsche Forschungsgemeinschaft (DFG; RI-00089).
Abbreviations used:
- BN-PAGE
blue native PAGE
- cryo-EM
cryo–electron microscopy
- LSU
large subunit
- PMSF
phenylmethylsulfonyl fluoride
- SSU
small subunit.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-04-0227) on August 9, 2018.
REFERENCES
- Altamura N, Capitanio N, Bonnefoy N, Papa S, Dujardin G. (1996). The Saccharomyces cerevisiae OXA1 gene is required for the correct assembly of cytochrome c oxidase and oligomycin-sensitive ATP synthase. FEBS Lett , 111–115. [DOI] [PubMed] [Google Scholar]
- Amunts A, Brown A, Bai XC, Llacer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SH, Ramakrishnan V. (2014). Structure of the yeast mitochondrial large ribosomal subunit. Science , 1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerschmitt H, Funes S, Herrmann JM. (2008). The membrane-bound GTPase Guf1 promotes mitochondrial protein synthesis under suboptimal conditions. J Biol Chem , 17139–17146. [DOI] [PubMed] [Google Scholar]
- Bauerschmitt H, Mick DU, Deckers M, Vollmer C, Funes S, Kehrein K, Ott M, Rehling P, Herrmann JM. (2010). Ribosome-binding proteins Mdm38 and Mba1 display overlapping functions for regulation of mitochondrial translation. Mol Biol Cell , 1937–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. (1989). Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature , 482–486. [DOI] [PubMed] [Google Scholar]
- Brown A, Amunts A, Bai XC, Sugimoto Y, Edwards PC, Murshudov G, Scheres SH, Ramakrishnan V. (2014). Structure of the large ribosomal subunit from human mitochondria. Science , 718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurillo MA, Moraes Barros RR, Souza RT, Marini MM, Antonio CR, Cortez DR, Curto MA, Lorenzi HA, Schijman AG, Ramirez JL, da Silveira JF. (2016). Subtelomeric I-SceI-mediated double-strand breaks are repaired by homologous recombination in Trypanosoma cruzi. Front Microbiol , 2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivellone MD. (1994). Characterization of CBP4, a new gene essential for the expression of ubiquinol-cytochrome c reductase in Saccharomyces cerevisiae . J Biol Chem , 21284–21292. [PubMed] [Google Scholar]
- Datta K, Fuentes JL, Maddock JR. (2005). The yeast GTPase Mtg2p is required for mitochondrial translation and partially suppresses an rRNA methyltransferase mutant, mrm2. Mol Biol Cell , 954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva D, Fontanesi F, Barrientos A. (2013). The DEAD box protein Mrh4 functions in the assembly of the mitochondrial large ribosomal subunit. Cell Metab , 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englmeier R, Pfeffer S, Förster F. (2017). Structure of the human mitochondrial ribosome studied in situ by cryoelectron tomography. Structure , 1574–1581. [DOI] [PubMed] [Google Scholar]
- Foury F, Roganti T, Lecrenier N, Purnelle B. (1998). The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae . FEBS Lett , 325–331. [DOI] [PubMed] [Google Scholar]
- Fox TD. (1996). Translational control of endogenous and recoded nuclear genes in yeast mitochondria: regulation and membrane targeting. Experientia , 1130–1135. [DOI] [PubMed] [Google Scholar]
- Fox TD. (2012). Mitochondrial protein synthesis, import, and assembly. Genetics , 1203–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo C, Trinko R, Kramer G, Appling DR, Hardesty B. (2003). Purification and characterization of yeast mitochondrial initiation factor 2. Arch Biochem Biophys , 243–252. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, Ban N. (2014a). The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature , 283–286. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leitner A, Bieri P, Voigts-Hoffmann F, Erzberger JP, Leibundgut M, Aebersold R, Ban N. (2014b). Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature , 515–519. [DOI] [PubMed] [Google Scholar]
- Gruschke S, Gröne K, Heublein M, Holz S, Israel L, Imhof A, Herrmann JM, Ott M. (2010). Proteins at the polypeptide tunnel exit of the yeast mitochondrial ribosome. J Biol Chem , 19022–19028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschke S, Kehrein K, Römpler K, Gröne K, Israel L, Imhof A, Herrmann JM, Ott M. (2011). Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J Cell Biol , 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Fox TD. (1997). Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N and C termini and dependence on the conserved protein Oxa1p. Mol Biol Cell , 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K, Neupert W, Stuart RA. (2001). Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J , 1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Koll H, Cook RA, Neupert W, Stuart RA. (1995). Topogenesis of cytochrome oxidase subunit II—mechanisms of protein export from the mitochondrial matrix. J Biol Chem , 27079–27086. [DOI] [PubMed] [Google Scholar]
- Jan PS, Esser K, Pratje E, Michaelis G. (2000). Som1, a third component of the yeast mitochondrial inner membrane peptidase complex that contains Imp1 and Imp2. Mol Gen Genet , 483–491. [DOI] [PubMed] [Google Scholar]
- Kehrein K, Schilling R, Möller-Hergt BV, Wurm CA, Jakobs S, Lamkemeyer T, Langer T, Ott M. (2015). Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep , 843–853. [DOI] [PubMed] [Google Scholar]
- Kuzmenko A, Derbikova K, Salvatori R, Tankov S, Atkinson GC, Tenson T, Ott M, Kamenski P, Hauryliuk V. (2016). Aim-less translation: loss of Saccharomyces cerevisiae mitochondrial translation initiation factor mIF3/Aim23 leads to unbalanced protein synthesis. Sci Rep , 18749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi I, Oeljeklaus S, Ronsor C, Bareth B, Warscheid B, Rehling P, Dennerlein S. (2016). Ribosome-associated Mba1 escorts Cox2 from insertion machinery to maturing assembly intermediates. Mol Cell Biol , 2782–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullin TW, Fox TD. (1993). COX3 mRNA-specific translational activator proteins are associated with the inner mitochondrial membrane in Saccharomyces cerevisiae. J Biol Chem , 11737–11741. [PubMed] [Google Scholar]
- Moda BS, Ferreira-Júnior JR, Barros MH. (2016). Partial suppression of the respiratory defect of qrs1/her2 glutamyl-tRNA amidotransferase mutants by overexpression of the mitochondrial pentatricopeptide Msc6p. Curr Genet , 607–617. [DOI] [PubMed] [Google Scholar]
- Morgenstern M, Stiller SB, Lubbert P, Peikert CD, Dannenmaier S, Drepper F, Weill U, Hoss P, Feuerstein R, Gebert M, et al. (2017). Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep , 2836–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naithani S, Saracco SA, Butler CA, Fox TD. (2003). Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae . Mol Biol Cell , 324–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndi M, Marin-Buera L, Salvatori R, Singh AP, Ott M. (2018). Biogenesis of the bc1 complex of the mitochondrial respiratory chain. J Mol Biol, DOI: 10.1016/j.jmb.2018.04.036. [DOI] [PubMed] [Google Scholar]
- Neupert W. (2015). A perspective on transport of proteins into mitochondria: a myriad of open questions. J Mol Biol , 1135–1158. [DOI] [PubMed] [Google Scholar]
- Nouet C, Bourens M, Hlavacek O, Marsy S, Lemaire C, Dujardin G. (2007). Rmd9p controls the processing/stability of mitochondrial mRNAs and its overexpression compensates for a partial deficiency of oxa1p in Saccharomyces cerevisiae . Genetics , 1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Fox D, Walter P. (1993). A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science , 1997–2004. [DOI] [PubMed] [Google Scholar]
- Ostojic J, Panozzo C, Bourand-Plantefol A, Herbert CJ, Dujardin G, Bonnefoy N. (2016). Ribosome recycling defects modify the balance between the synthesis and assembly of specific subunits of the oxidative phosphorylation complexes in yeast mitochondria. Nucleic Acids Res , 5785–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Prestele M, Bauerschmitt H, Funes S, Bonnefoy N, Herrmann JM. (2006). Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J , 1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MF, Alushin GM, Barros MH, Rak M, Tzagoloff A. (2012). The putative GTPase encoded by MTG3 functions in a novel pathway for regulating assembly of the small subunit of yeast mitochondrial ribosomes. J Biol Chem , 24346–24355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Woellhaf MW, Herrmann JM, Forster F. (2015). Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat Commun , 6019. [DOI] [PubMed] [Google Scholar]
- Prestele M, Vogel F, Reichert AS, Herrmann JM, Ott M. (2009). Mrpl36 is important for generation of assembly competent proteins during mitochondrial translation. Mol Biol Cell , 2615–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M, Leonhard K, Hell K, Stuart RA, Neupert W, Herrmann JM. (2001). Mba1, a novel component of the mitochondrial protein export machinery of the yeast Saccharomyces cerevisiae . J Cell Biol , 1085–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta O, Lubas M, Lipinski KA, Piatkowski J, Malecki M, Golik P. (2010). DMR1 (CCM1/YGR150C) of Saccharomyces cerevisiae encodes an RNA-binding protein from the pentatricopeptide repeat family required for the maintenance of the mitochondrial 15S ribosomal RNA. Genetics , 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Su CH, Xu JT, Azpiroz R, Singh AM, Tzagoloff A. (2016). Regulation of mitochondrial translation of the ATP8/ATP6 mRNA by Smt1p. Mol Biol Cell , 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spithill TW, Trembath MK, Lukins HB, Linnane AW. (1978). Mutations of the mitochondrial DNA of Saccharomyces cerevisiae which affect the interaction between mitochondrial ribosomes and the inner mitochondrial membrane. Mol Gen Genet , 155–162. [DOI] [PubMed] [Google Scholar]
- Stoldt S, Wenzel D, Kehrein K, Riedel D, Ott M, Jakobs S. (2018). Spatial orchestration of mitochondrial translation and OXPHOS complex assembly. Nat Cell Biol , 528–534. [DOI] [PubMed] [Google Scholar]
- Suhm T, Kaimal JM, Dawitz H, Peselj C, Masser AE, Hanzen S, Ambrozic M, Smialowska A, Bjorck ML, Brzezinski P, et al. (2018). Mitochondrial translation efficiency controls cytoplasmic protein homeostasis. Cell Metab , 1309–1322 e1306. [DOI] [PubMed] [Google Scholar]
- Westermann B, Gaume B, Herrmann JM, Neupert W, Schwarz E. (1996). Role of the mitochondrial DnaJ homolog Mdj1p as a chaperone for mitochondrially synthesized and imported proteins. Mol Cell Biol , 7063–7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N. (2017). Mitochondrial machineries for protein import and assembly. Annu Rev Biochem , 685–714. [DOI] [PubMed] [Google Scholar]
- Williams EH, Butler CA, Bonnefoy N, Fox TD. (2007). Translation initiation in Saccharomyces cerevisiae mitochondria: functional interactions among mitochondrial ribosomal protein Rsm28p, initiation factor 2, methionyl-tRNA-formyltransferase and novel protein Rmd9p. Genetics , 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woellhaf MW, Sommer F, Schroda M, Herrmann JM. (2016). Proteomic profiling of the mitochondrial ribosome identifies Atp25 as a composite mitochondrial precursor protein. Mol Biol Cell , 3031–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R, Smith E, Barrientos A. (2018). Yeast mitoribosome large subunit assembly proceeds by hierarchical incorporation of protein clusters and modules on the inner membrane. Cell Metab , 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.