Significance
Organisms living in extreme environments are useful for studying the process of adaptation. We studied two distantly related groups that are distributed across a broad elevational gradient on and near the Qinghai-Tibetan Plateau and identified molecular adaptations to increasing elevations. We show that high-elevation adaptation (HEA) emerged soon after a split from low-elevation lineages, and adaptations continue to evolve in species that inhabit increasingly high elevations. Genes related to DNA repair and energy metabolism evolved rapidly, suggesting a crucial role of these genes in HEA. Moreover, we observed common patterns of HEA for similar functions between distantly related lineages, although these functional changes often involved different specific genes.
Keywords: Nanorana, Phrynocephalus, altitudinal gradient, high-elevation adaptation, DNA repair
Abstract
Although many cases of genetic adaptations to high elevations have been reported, the processes driving these modifications and the pace of their evolution remain unclear. Many high-elevation adaptations (HEAs) are thought to have arisen in situ as populations rose with growing mountains. In contrast, most high-elevation lineages of the Qinghai-Tibetan Plateau appear to have colonized from low-elevation areas. These lineages provide an opportunity for studying recent HEAs and comparing them with ancestral low-elevation alternatives. Herein, we compare four frogs (three species of Nanorana and a close lowland relative) and four lizards (Phrynocephalus) that inhabit a range of elevations on or along the slopes of the Qinghai-Tibetan Plateau. The sequential cladogenesis of these species across an elevational gradient allows us to examine the gradual accumulation of HEA at increasing elevations. Many adaptations to high elevations appear to arise gradually and evolve continuously with increasing elevational distributions. Numerous related functions, especially DNA repair and energy metabolism pathways, exhibit rapid change and continuous positive selection with increasing elevations. Although the two studied genera are distantly related, they exhibit numerous convergent evolutionary changes, especially at the functional level. This functional convergence appears to be more extensive than convergence at the individual gene level, although we found 32 homologous genes undergoing positive selection for change in both high-elevation groups. We argue that species groups distributed along a broad elevational gradient provide a more powerful system for testing adaptations to high-elevation environments compared with studies that compare only pairs of high-elevation versus low-elevation species.
Groups of organisms that are broadly distributed with respect to diverse environmental conditions provide an excellent opportunity to study the genetic basis of adaptation. High elevations offer extreme environments with low levels of oxygen, high levels of UV radiation, and cold temperatures, which challenge the survival of many species. Animals that have evolved in high-elevation environments typically exhibit numerous adaptations to counter these environmental challenges, such as altered developmental rates and body sizes, increased pulmonary ventilation, and increased hemoglobin oxygen affinity (1–6). In particular, the Qinghai-Tibetan Plateau and its surrounding slopes constitute a large high-elevation region with a mean elevation exceeding 4,000 m, as well as substantial spatial variation in both oxygen content and solar UV radiation (7), making it an ideal region for study of high-elevation adaptation (HEA).
Most previous studies of the genetic processes of HEA have compared species or populations from high elevations above 3,500 m with those from low elevations to identify sequence variation and/or expression shifts in the high-elevation group (8–15). In these studies, changes in genes involved in energy metabolism and responses to hypoxia have been routinely identified (8–15). However, “high elevation” is not a single point above sea level but, instead, represents a continuum. Despite this fact, studies along a broad elevational continuum are lacking, which has hindered our understanding of the genetic processes that result in HEA. Furthermore, the degree of evolutionary convergence in adaptation (e.g., refs. 16–19) among different species that live at similar elevations is unknown.
Multiple groups, especially of frogs and lizards, are distributed across a large range of elevations on the Qinghai-Tibetan Plateau and its adjacent slopes, and can be used to explore the genetic processes involved in HEA. For example, there are three species of the frog genus Nanorana (Nanorana parkeri, Nanorana liebigii, and Nanorana phrynoides) that occur up to elevations of 4,700 m, 3,500 m, and 2,400 m, respectively (20, 21), in addition to species related to Nanorana that are distributed at relatively low elevations. Studies of N. parkeri have identified several physiological and phenotypic adaptations that help this species cope with hypoxia and cold temperatures, including a slower developmental rate and a smaller body size (4). Similarly, three species of the lizard genus Phrynocephalus (Phrynocephalus erythrurus, Phrynocephalus vlangalii, and Phrynocephalus putjatia) occur at 4,300–5,200 m, 2,200–4,400 m, and 2,200–3,000 m, respectively, and, together with their low-elevation relatives, they are distributed across a broad elevational range. The high-elevation lizards have also evolved some physiological innovations, such as higher capability to carry oxygen (5). Given these broad elevational distributions, these two groups of ectotherms provide excellent models to explore the dynamics and convergent evolution of HEA across an elevational gradient.
Advances in high-throughput sequencing, especially RNA sequencing (RNA-Seq) (22), enable the collection of gene sequences for nonmodel animals. These advances greatly facilitate comparative evolutionary studies (23). We analyzed the transcriptomes of the three high-elevation Nanorana and Phrynocephalus species noted above, as well as two low-elevation (∼1,000 m) relatives of each genus. Multitissue transcriptomic sequencing allowed construction of >7,000 high-quality orthologous genes for both groups. Evolutionary analyses involved the detection of positive selection among individual genes on different branches, inferences of the timing of these changes with respect to changes in elevational distribution, and identification of convergent evolution of functional adaptations and specific genes.
Results and Discussion
Sampling and Sequencing.
For the frog group, four closely related species, including N. parkeri, N. liebigii, N. phrynoides, and Quasipaa spinosa, were collected for RNA-Seq. Among them, N. parkeri is the highest distributed species (specimens were collected at elevations >4,000 m). N. liebigii and N. phrynoides were collected at middle elevations (∼3,000 m and ∼1,800 m, respectively), and Q. spinosa was collected at low elevations (<1,000 m) (Fig. 1A and SI Appendix, Fig. S1). The low-elevation species Odorrana margaretae (24) was collected as an outgroup for comparative analysis.
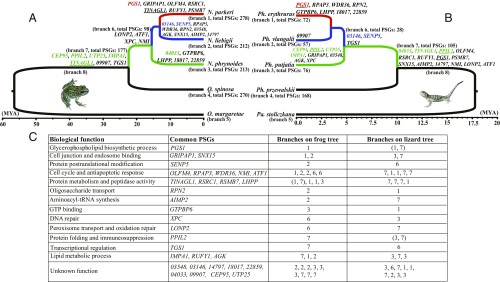
Fig. 1.
Time-scaled phylogenetic trees for frogs (A) and lizards (B) distributed across an elevational gradient of the Qinghai-Tibetan Plateau. Black lineages represent lowland distributions, green lineages represent distributions up to about 2,000 m, blue lineages represent distributions from roughly 2,000–3,500 m, and red lineages represent distributions from about 3,500–4,500 m. The branch number and the number of PSGs are indicated beside each tip or branch of the two trees, and PSGs that evolved in parallel are shown on the respective branches of each tree. (C) All of the parallel PSGs that were identified in both groups of high-elevation species, and their functions (where known), are shown. Parallel PSGs that evolved at the same change in elevational distribution are shown in the respective color of that lineage (green, blue, or red, respectively). Underlined genes showed positive selection on more than one branch of the frog or lizard tree. The functional unknown PSGs are represented by the last five digits of the anole lizard’s ENSEMBL gene identifiers.
Multiple tissues (brain, heart, liver, skin, ovary, and testis) were collected from each species of frog to maximize the number of expressed genes that were examined. N. parkeri, N. liebigii, N. phrynoides, and Q. spinosa had 282,884,179, 197,229,347, 96,028,690, and 287,795,501 high-quality reads, respectively (SI Appendix, Table S1). The final assemblies showed both high-contig N50 values and read-mapping ratios (SI Appendix, Table S1). All of the original RNA-Seq data were deposited into an the open-access Genome Sequence Archive (gsa.big.ac.cn) under accession no. CRA000352.
For each species, we generated a de novo transcriptomic assembly, from which we examined a total of 8,138 orthologous genes among the five frogs. The average mean identity score (MIS) of these gene alignments is 93%, indicating reliable predictions of orthology. Based on the “supergene” alignment that was constructed from concatenations of all of the high-quality orthologous genes, we constructed a maximum likelihood (ML) tree (Fig. 1A). The divergence time at the root was estimated to be 56.5 (29.1–92.8) Mya, which provides an appropriate time interval for identifying episodic positive selection across the group (25).
For the lizard group, all of the transcriptomes of P. przewalskii, P. putjatia, P. vlangalii, and P. erythrurus were obtained from previous studies (13, 14). Paralaudakia stoliczkana, another low-elevation species (occurs at 100–1,700 m; www.zoology.csdb.cn/page/index.vpage) that is closely related to the Phrynocephalus species, was collected (at ∼1,050 m) in this study for outgroup comparison. The full-length transcripts of P. stoliczkana were generated in this study through the PacBio sequencing technology. Using the same procedures used for the frog transcriptomes, a total of 7,288 orthologous genes (MIS = 96%) were analyzed and an ML phylogeny was constructed from the concatenated transcripts of the lizards (Fig. 1B). This phylogeny showed shallower evolutionary divergences for the lizards than the frogs, indicating a later colonization of high elevations in these species.
HEA Is Evident in the Earliest Stages of Elevational Expansion.
Previous studies have shown that the highest elevation lineages (>4,000 m) in each group have many HEAs, especially at the molecular level (14, 26). However, because these studies compared only pairs of high-elevation versus low-elevation species, it is unclear if these adaptations accumulated gradually at increasing elevations or if they evolved at a particular elevational threshold. The two distantly related species groups studied here (Fig. 1 A and B) provide opportunities to address these questions through comparative evolutionary analyses. To address these issues, we examined evidence for positive selection along each of the branches of the two phylogenies.
We first estimated the dN/dS ratio on each branch of the two phylogenies. For both groups, 87–96% of the genes display a dN/dS ratio <1 (Dataset S1), indicating that most genes are undergoing purifying selection. This is true for species at all elevational distributions. To detect episodic positive selection in this general background of purifying selection, we used the branch-site likelihood-based test (25, 27), which has been widely used to find signals of positive selection along specific branches of a phylogenetic tree (8–15). Under this model, we were able to identify genes under positive selection on each branch in the phylogeny (Fig. 1 A and B), and thereby assign genes under selection to particular elevational shifts in distribution. By comparison with the lowland species, we removed genes that were under positive selection in both low and high elevations, and then identified specific genes that were under positive selection at each increase in elevational distribution.
We identified a total of 835 genes under positive selection exclusively on high-elevational lineages of the frog tree and 301 genes under positive selection exclusively on high-elevational lineages of the lizard tree (Fig. 1 A and B). The smaller number of positively selected genes (PSGs) identified in the lizards is not accounted for by their shallower evolutionary divergences compared with the frogs, because lower sequence divergence has little or no correlation with the mean dN/dS ratio (28). Therefore, other biological factors likely account for the fewer PSGs identified in the lizards; one possibility is interspecific hybridization (29), which can dilute signals of positive selection. Nonetheless, we found 32 genes under positive selection on high-elevational lineages of both the frog and lizard trees. These 32 genes are shown along their respective branches in Fig. 1 A and B, and the genes are shown with their associated functions (where known) in Fig. 1C.
In the first stages of HEA on the frog tree (Fig. 1A; green lineages, branches 7 and 3), PSGs are significantly overrepresented by functions such as regulation of endocytosis and exocytosis [gene ontology (GO):0048261, GO:0045056, and GO:0045921], positive regulation of cellular amine metabolic process (GO:0033240), positive regulation of the HIF-1-alpha signaling pathway (GO:1902073), respiratory chain complex IV assembly (GO:0008535), response to radiation and postreplication repair (GO:0009314 and GO:0006301), and vasculogenesis (GO:0001570) (SI Appendix, Tables S2–S5). Each of these functions is closely related to the physiological stresses produced by hypoxia or strong UV radiation (UV). For example, endocytosis could be regulated by many genetic and posttranslational alternations, and such regulation is important for cell survival under hypoxia (30). Based on the enrichment results (SI Appendix, Tables S3 and S5), even the frogs distributed at 2,000 m appear to have evolved multiple adaptations related to endocytosis, including modifications of membrane docking and negative regulation of the p38 MAPK cascade (30). This highlights the potential roles of endocytosis and exocytosis in HEA. Further, biogenic amines, such as dopamine, norepinephrine, and serotonin, act as neurotransmitters; are involved in numerous functional pathways; and are particularly susceptible to the effects of hypoxia (31–33). Adaptations related to cellular biogenic amine metabolic processes are important to maintain physiological homeostasis.
The rapid modification and adaptation of several functions mentioned above, especially the response to radiation, cellular responses to hypoxia, and vasculogenesis, are also observed in other high-elevation animals, such as the snub-nosed monkey (15) and Tibetan antelope (8), suggesting they are major targets of natural selection to deal with cellular stresses associated with high-elevation existence, and thus show convergence in function among different groups of high-elevation animals.
The above results strongly suggest that even frogs at moderate elevations (up to about 2,000 m) have already evolved molecular adaptations to reduced oxygen content and/or increased UV radiation. Similar results were also obtained from the distantly related group of lizards (SI Appendix, Tables S6–S9). On the lizard phylogeny, 178 PSGs were identified on the moderate-elevation (up to ∼2,000 m) lineages (branches 7 and 3). Of these, there are three PSGs (PPIL2, CPLX1, and NPHS2) undergoing positive selection on both branches 7 and 3 on the lizard phylogeny. The 178 lizard PSGs were significantly enriched in GO terms overrepresented by functions like peroxisomal transport (GO:0043574), DNA damage recognition and repair (GO:0000715, GO:0000717, GO:0006303, and GO:0000726), cellular response to oxidative stress (GO:0034599), response to amine (GO:0014075), and cellular carbohydrate biosynthetic and metabolic process (GO:0034637 and GO:0044262), whose modifications are also closely related to adaptations to high elevations and have been widely observed in other high-elevation animals (8–15).
Continued Evolution of HEAs at Increasing Elevations.
Given the early emergence of positive selection consistent with HEA (at elevational expansions to just 2,000 m), we next asked if there was continuing adaptation with increasing elevations or, alternatively, if the adaptations to moderate elevations simply allow expansion of the group to higher elevations? Our results showed that all of the higher elevation species showed positive selection for genes associated with HEA beyond the changes observed in the moderate-elevation species, suggesting a continuous increase in HEA with increasing elevational distribution.
In the frog species with an elevational distribution around 3,000 m (N. liebigii), the identified PSGs were further enriched in categories of carbohydrate transport (GO:0008643), angiogenesis involved in coronary vascular morphogenesis (GO:0060978), heart morphogenesis and development (GO:0003247 and GO:0007507), mitochondrial DNA repair (GO:0043504), and generation of precursor metabolites and energy (GO:0006091) (SI Appendix, Tables S10–S13). Among these functional categories, angiogenesis is well known for responding to hypoxia by increasing the oxygen supply, a function that has also been observed in other high-elevation animals (8, 9, 15). Combining the early adaptations evolved by N. phrynoides, such as the functions associated with vasculogenesis, our results support a similar role of angiogenesis in high-elevation frogs.
The middle-high elevational lizard (P. vlangalii) showed further changes consistent with HEA, with positive selection in genes responsible for nitric oxide-mediated transduction and signaling pathway (GO:0007263 and GO:0038060), cellular response to nitric oxide (GO:0071732 and GO:0071732), cellular glucan metabolic process (GO:0006073), and generation of precursor metabolites and energy (GO:0006091) (SI Appendix, Tables S14–S17). This is consistent with a previous study that focused on the HEAs of P. vlangalii (13).
The increasing adaptation to high elevations continued in the species with the highest elevational distributions (>4,000 m; frog: N. parkeri; lizard: P. erythrurus; branch 1 in Fig. 1 A and B) (for both frogs and lizards). There were continued selective changes, with positive selection in frog genes responsible for regulation of carbohydrate derivative biosynthesis and transport (GO:1901137 and GO:1901264), lipid biosynthesis (GO:0008610), response to oxygen levels (GO:0070482), and cellular response to calcium ions (GO:0071277) (SI Appendix, Tables S18 and S19). The lizard PSGs at this stage mainly function as a response to radiation and DNA damage (GO:0010165, GO:0030330, and GO:0042770), as well as in multiple categories associated with DNA repair (GO:0006281, GO:0000726, and GO:0045002) (SI Appendix, Tables S20 and S21).
By examining the GO terms associated with PSGs from all of the three high-elevation lineages (branches colored green, blue, and red in Fig. 1), we observed several candidate HEA functions that appear to be evolving continuously with increasing elevational expansion in both groups, including functions associated with response to UV and respiratory electron transport chain in frogs (Table 1 and SI Appendix, Table S22) and functions associated with response to radiation and DNA repair in lizards (Table 1 and SI Appendix, Table S23).
Table 1.
Number of PSGs associated with specific GO terms across all three elevational expansions in frogs and lizards
| Taxa | GO ID | P value | Term | Green* | Blue† | Red‡ |
| Frogs | GO:0005996 | 0.038 | Monosaccharide metabolic process | 4 | 2 | 5 |
| GO:0009411 | 0.039 | Response to UV | 1 | 4 | 4 | |
| GO:0046165 | 0.003 | Alcohol biosynthetic process | 2 | 3 | 5 | |
| GO:0043255 | 0.006 | Regulation of carbohydrate biosynthetic process | 5 | 1 | 3 | |
| GO:0022904 | 0.037 | Respiratory electron transport chain | 2 | 1 | 2 | |
| Lizards | GO:0006302 | 0.001 | Double-strand break repair | 4 | 1 | 4 |
| GO:0044262 | 0.001 | Cellular carbohydrate metabolic process | 2 | 2 | 4 | |
| GO:0010212 | 0.021 | Response to ionizing radiation | 2 | 1 | 2 | |
| GO:0006073 | 0.002 | Cellular glucan metabolic process | 1 | 2 | 2 | |
| GO:0000726 | 0.000 | Nonrecombinational repair | 3 | 1 | 2 |
ID, identifier.
Branches 7 and 3 on both phylogenies.
Branches 6 and 2 on both phylogenies.
Branch 1 on both phylogenies.
One interesting finding is the continuous evolution of genes related to UV responses across increasing elevational distributions in frogs (GO:0009411; Table 1 and SI Appendix, Table S22). UV is a major environmental stress factor at high elevations, and UV exposure is especially problematic for exposed amphibian skin, where it can produce oxidant stress and DNA damage. Different genes related to responses to UV exposure appear to be under positive selection in different branches on the tree, suggesting a stepwise adaptation to UV exposure. Some of these genes showed the highest expression levels in the frog skin (the tissue with the highest level of UV exposure); however, the majority of the PSGs showed the highest expression levels in the brain (SI Appendix, Table S24). Another interesting finding is the continuous evolution of the respiratory electron transport chain in frogs (GO:0022904 for ATP generation; SI Appendix, Table S22). Given that the high-elevation Nanorana species have slower development rates (4), both strategies, that of reducing energy consumption and that of increasing energy generation, probably shaped the metabolic adaptations of Nanorana to high elevations.
When we mapped the PSGs onto Kyoto Encyclopedia of Genes and Genomes (KEGG)-annotated pathways, we found additional evidence of continuous evolution of HEA in both groups. For example, at each elevational stage in the frogs, we found multiple PSGs that are involved in the oxidative phosphorylation pathway (with PSGs of SDHA and LHPP on the lineage from low elevation to ∼2,000 m; SDHA and NDUFA6 from ∼2,000 m to ∼3,000 m; and NDUFB9, ATP6AP1, COX4I1, ATP6V0A1, and ATP6V1H from ∼3000 m to ∼4,500 m), further supporting the role of ATP generation (energy metabolism) in HEA. We observed similar patterns in several other pathways, including nucleotide excision repair and the calcium signaling pathway (SI Appendix, Table S25). Although our analysis of the KEGG-annotated pathways in the lizards identified different pathways from those identified in the frogs, these analyses nonetheless supported the role of energy metabolism in HEA. Specifically, we found evidence of positive selection acting on the pathways of glycolysis, fructose and mannose metabolism, and galactose metabolism with increasing elevational stages of the lizards (detailed information is provided in SI Appendix, Table S26).
Convergent Evolution of HEA Between Frogs and Lizards.
Although the two groups studied here (Nanorana and Phrynocephalus) are distantly related, they face similar environmental conditions and challenges at comparable elevations. To assess the degree of convergence in HEA between the two groups, we examined the GO terms associated with the PSGs from both groups at each elevational stage. This represents convergence at the functional level, rather than convergence of specific nucleotide substitutions in genes or amino acid replacements in proteins.
Our analyses identified 103, 35, and 35 GO terms associated with PSGs at elevational expansions to 2,000 m, 3,500 m, and 4,500 m, respectively (SI Appendix, Tables S27–S29). These included a number of functions that have evolved convergently in frogs and lizards (Table 2). For example, the genes GTSE1 and NBN in the frogs and the genes HIPK2 and SMC1A in the lizards are all associated with a response to DNA damage (GO:0042770) at the highest elevation species (branch 1; Table 2). Moreover, the gene PPP1R3B in the frogs and the genes SIRT6, PPIF, and ENSACAG00000024726 in the lizards are all associated with regulation of generation of precursor metabolites and energy (GO:0043467) at the middle-high elevational lineages (branches 6 and 2; Table 2). When we considered the entire pathway leading to the species with the highest elevational distributions (branches 7, 6, and 1 in Fig. 1 A and B, leading to N. parkeri in the frogs and P. erythrurus in the lizards), the PSGs from both groups were associated with many terms related to hypoxia adaptation, such as the glycolytic process, which is a primary energy pathway during hypoxia (Table 2 and SI Appendix, Table S30).
Table 2.
Convergent GO terms between frogs and lizards at comparable elevations
| Branch | GO term | P value | GO term definition |
| 1 | GO:0006109 | 0.002529437 | Regulation of carbohydrate metabolic process |
| GO:0006953 | 0.006084761 | Acute-phase response | |
| GO:0042770 | 0.025243823 | Signal transduction in response to DNA damage | |
| GO:0032355 | 0.043205949 | Response to estradiol | |
| GO:0045216 | 0.044599456 | Cell–cell junction organization | |
| 2 and 6 | GO:0034976 | 0.019758571 | Response to endoplasmic reticulum stress |
| GO:0006457 | 0.026225839 | Protein folding | |
| GO:0043467 | 0.029021199 | Regulation of generation of precursor metabolites and energy | |
| GO:0043547 | 0.029612275 | Positive regulation of GTPase activity | |
| GO:0036119 | 0.046455853 | Response to platelet-derived growth factor | |
| 3 and 7 | GO:0051660 | 0.000308287 | Establishment of centrosome localization |
| GO:0006457 | 0.004496455 | Protein folding | |
| GO:0043547 | 0.012808641 | Positive regulation of GTPase activity | |
| GO:0072524 | 0.013543229 | Pyridine-containing compound metabolic process | |
| GO:0030397 | 0.024392786 | Membrane disassembly | |
| GO:0032846 | 0.034964282 | Positive regulation of homeostatic process | |
| GO:0051591 | 0.034999104 | Response to cAMP | |
| GO:0019318 | 0.03834842 | Hexose metabolic process | |
| GO:0048872 | 0.044892596 | Homeostasis of number of cells | |
| 1, 6, and 7 | GO:0032355 | 0.000587452 | Response to estradiol |
| GO:0051660 | 0.000692194 | Establishment of centrosome localization | |
| GO:0006096 | 0.002883309 | Glycolytic process | |
| GO:0048520 | 0.013978057 | Positive regulation of behavior | |
| GO:0043547 | 0.024198176 | Positive regulation of GTPase activity | |
| GO:0001666 | 0.030533707 | Response to hypoxia | |
| GO:2000352 | 0.035373391 | Negative regulation of endothelial cell apoptotic process |
If all elevational stages are considered, there is additional functional convergence evident in the GO terms associated with PSGs of the frogs and lizards, such as terms associated with energy metabolism, response to UV radiation, and DNA repair (SI Appendix, Tables S22, S23, S25, and S26). However, there are some differences in the DNA repair-related pathways between the two groups. In the frogs, changes in DNA repair functions were evident mainly in the earliest elevational shifts (up to 2,000 m); these mainly included changes to postreplication repair. In contrast, among the lizards, several different types of repair functions exhibited continual evolution along the elevational gradient, including double-strand break repair and nonrecombinational repair.
Although there are a large number of functions showing convergent evolution in the high-elevation frogs and lizards, many of these functions are associated with different sets of PSGs. However, there are 32 PSGs in common between the frogs and lizards (Fig. 1). Among these overlapping PSGs, XPC is involved in the damage recognition of nucleotide excision repair and plays an important role in the early steps of global genome nucleotide excision repair, especially in damage recognition, open complex formation, and repair protein complex formation (34). Other important functions associated with the overlapping PSGs include peroxisomal transport, lipid metabolism, and glycerophospholipid biosynthesis (Fig. 1C).
KEGG pathway analysis further highlights the roles of energy metabolism and DNA repair in HEA. The related energy metabolism pathways associated with the PSGs of both the frogs and lizards include pathways connected to glycolysis, fructose and mannose metabolism, and oxidative phosphorylation (SI Appendix, Table S31). The pathways associated with DNA repair in both groups are connected to nucleotide excision repair, homologous recombination, and nonhomologous end-joining (SI Appendix, Table S31). Moreover, categories related to both energy metabolism and DNA repair had a higher proportion of PSGs and more genetic changes than even the typically fast-evolving spermatogenesis and immune systems (SI Appendix, Fig. S2).
Convergent Functional Adaptation to Hypoxia Involves Different Sets of Genes.
Because many PSGs appeared to be associated with adaptation to hypoxia in both frogs and lizards, we further examined this convergent functional adaption by comparing the frog and lizard branch-specific PSGs with the genes showing significant expression alterations under hypoxia (35). We expected strong changes in expression levels under hypoxia to be a strong indication that the genes are involved in the core hypoxia-responsive pathways (36). Accordingly, we examined all genes from HypoxiaDB (a highly curated database with more than 72,000 entries) that show expression changes under hypoxia (35) and constructed a “reference” list of hypoxia-responsive genes with a series of filtering analyses (Methods). Although the reference genes are from studies of humans, given the functional conservation of many genes across tetrapods, we expected the comparisons to provide useful information about genes involved in hypoxic responses in frogs and lizards. The final reference list of 1,191 genes contained 317 associated GO terms, which cover a wide range of functions, including immunity, neurodevelopment, energetic metabolism, and related aspects (SI Appendix, Table S32). We show the association of hypoxia-associated GO terms with PSGs and particular shifts in elevational distribution on the frog and lizard trees in Fig. 2.
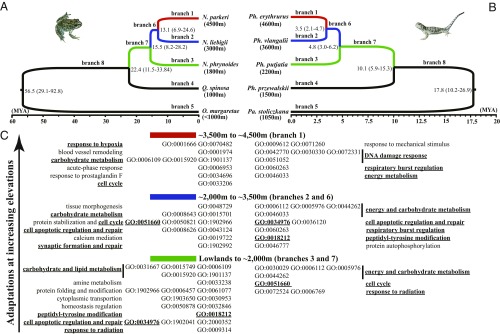
Fig. 2.
Convergent evolution of functions associated with adaptation to hypoxia in high-elevation frogs (A) and lizards (B). The numbers at each node show the estimated time of divergence for the corresponding split in the tree, with 95% confidence limits for the split shown in parentheses. (C) Example GO terms associated with each elevational shift that include functions that have been associated with adaptation to hypoxia. Functions that evolved in parallel in high-elevation lineages of both frogs and lizards are in shown in boldface and are underlined.
We next examined the GO terms between the frog and lizard PSGs and the reference list of hypoxia-responsive genes, and conducted statistical analyses to test whether the frogs and lizards showed more selection among hypoxia-responsive categories than expected at random. We identified 22 hypoxia-responsive categories along the frog branches 7 and 3, a number significantly higher than the mean expected value that was obtained from 1,000 randomly selected gene sets of the same size (22 versus 7.8; P < 2.2e-16), strongly suggesting the emergence of hypoxia adaptation at an elevation of about 2,000 m. However, the lizard PSGs at this stage did not show significantly higher similarity with the hypoxia-associated reference list compared with the mean expected value (7 versus 6.8; P = 0.142). At the elevational expansion to 3,500 m, both groups showed significantly higher similarity with the reference list than the mean expected value (frogs: 10 versus 6.7 with P < 2.2e-16; lizards: 9 versus 5.7 with P < 2.2e-16). The same pattern was observed at the highest elevational expansion (frogs: 9 versus 7.2 with P < 2.2e-16; lizards: 8 versus 5.6 with P < 2.2e-16). Therefore, it appears that much of the adaptation to hypoxia may occur at somewhat higher elevations in lizards than in frogs. Of course, some of the evolution of functions shown in Fig. 2 may also be related to factors other than hypoxia. There are many other ecological and physiological factors that are different among the lizards, including reproductive mode (i.e., oviparity versus viviparity).
Although many functions associated with adaptation to hypoxia evolved in parallel in frogs and lizards (Fig. 2C), the PSGs associated with these functions do not contain any overlap (Table 3). Therefore, there appears to be considerable parallel adaptation to hypoxia, but involving completely different sets of genes.
Table 3.
Branch-associated PSGs related to adaptations to hypoxia (Fig. 2) in high-elevation frogs and lizards
| Branch | Elevational shift | Frog PSGs | Lizard PSGs |
| 1 | ∼3,500 to ∼4,500 m | FMN2, KIAA0196, BMPR2, TYW5, B4GALT7, B3GAT2, IMPDH1, ADCY4, RRM2B, GMDS, GPAA1, ENSACAG00000002886, AP2A1, PPP4R3A, ADIPOR1, GPER1, PLCD1, SCARB1, ALDOA, HIPK2, F7, POSTN, ADAM17, FN1, ITIH4, FLNA | BAG3, RAC1, ENSACAG00000008899, ZRANB3, DSCC1, NBN, GTSE1 |
| 2 and 6 | ∼2,000 to ∼3,500 m | EPHA2, PTK7, BMP4, RXRA, PBX1, COL3A1, STARD13, EZR, SLC26A4, CA12, SPHK1, PTGFR, PPP3CB, SLC37A3, SLC37A2, SLC35F6, UBE4B, IPO9, FLOT2, CRTAP, CEP83, NLRX1, ENSACAG00000025660 | ADK, PPP1R1A, PPP1R3B, JAK2, FYN, CDKL5, AARS, ATF6, RAC1 |
| 3 and 7 | Lowlands to ∼2,000 m | XRCC6, FBXL3, PRKAA1, CLK2, ENSACAG00000013018, COL3A1, MAPK8, MEN1, CCKAR, FGG, TNFAIP3, STX4, SCARB1, ENSACAG00000002091, ENSACAG00000022125, ENSACAG00000009367, SERPINC1, F5, MBD1, ENSACAG00000015535, ENSACAG00000015364, FKBP4, HSPH1, ENSACAG00000017064, HSPG2, COASY, CANT1, MAT2B, TYW5, ALG3, UPP2, FGFR1, EZR, CEP120, PPIL2, PDIA4, PRDX4, CCT4, TBCE, PDIA3, PPP1R15B, PLA2G6, FLOT1, GATA1, THY1, GAS6, KEAP1, TPR, SEMA4D, CTNND1, VEGFA, NOC2L, PLS1 | APOA1BP, PGM2, IMPA1, PPP1R2, PAFAH1B1, NPHS2, MYL6, FRMD1 |
Methods
Transcriptome Sequencing and Assembly.
The project approval (BBCJ-2014-001) was issued by Kunming Institute of Zoology (KIZ). Sample collections followed the animal use protocols approved by the Animal Care and Ethics Committee of KIZ. Total RNA for each sample was extracted using the SV Total RNA Isolation System (Promega). At least three tissues among the brain, heart, liver, skin, ovary, and testes were sampled from each species of Nanorana to allow the isolation of as many expressed genes as possible. All samples were immediately placed in LN2 until RNA extraction. For each tissue, a single paired-end cDNA library was constructed for poly (A)+ RNAs as described in the Illumina manual and sequenced using the HiSeq 2000 sequencing platform with a read length of 101 bp. Each tissue was sequenced with a high depth in the sequencing step. All of the sequencing information for the samples is summarized in SI Appendix, Table S1.
Raw sequencing reads obtained from each tissue were first cleaned with Cutadapt version 1.1.2 (37) to remove potential adapter sequences and low-quality regions with options “-q 20.” For each species, all cleaned reads from the different tissues were combined together and used as a single input for transcript assembly. De novo assembly was generated by Trinity version 20140717 (38) with its default parameters. CD-HIT version 4.6 (39) was then applied on the final assemblies to eliminate redundant contigs, which helped in subsequent homolog prediction. Transrate (40) was used to evaluate the qualities of the de novo transcriptome assemblies based on common statistical indexes, such as average contig length, contig N50 values, and read mapping ratios. The same filtering and assembling methods were used for all samples.
Orthology Determination and Alignment Construction.
We applied a modified reciprocal best hits method (“crb-blast”) to improve the accuracy of ortholog assignments between each assembly and a reference gene dataset (41). The reference dataset was the total protein sequences of Anolis carolinensis, which is available at the ENSEMBL genome database (www.ensembl.org). Before performing BLAST analyses, the coding regions of each assembled contig were determined using TransDecoder version 3.01 (42). This program first identified ORFs that were at least 50-aa long (using TransDecoder.LongOrfs), and then verified those peptides with BLAST or domain hits to the public protein databases (using TransDecoder.Predict). CD-HIT (39) was used again to remove redundant protein sequences, especially when there were more than one ORF predicted for the transcript. The final consensus 1:1 orthologous gene set was then obtained based on the ortholog predictions generated between the query species and the reference gene set.
For each gene, Prank (43) was first used to generate an alignment at the codon level (with options “-F -codon”). The codon alignments were then analyzed by FasParser (44) with the “Trim” function to strap low-quality aligned columns. Final alignments with a length of less than 120 bp were discarded to reduce false-positive findings in subsequent evolutionary analyses. To evaluate the quality of our set of predicted orthologous genes, we estimated the mean identity ratio (MIR) for each alignment with FasParser. Briefly, for each alignment, we first calculated the identity ratio (IR) between each pair of sequences and then obtained the MIR by averaging all of the IR values. Generally, a high MIR was assumed to indicate a more accurate orthologous prediction due to the conservative nature of orthologous genes.
Evolutionary Analyses.
Based on the corresponding topology of the frogs (20, 21) and lizards (45), codeml (version 4.9d) (46) was used to estimate dN, dS, and dN/dS with the free ratio model (“model = 1, NSsites = 0”) to obtain a general evolutionary pattern of selective pressure along the lineages. Mean dN/dS ratios were estimated by excluding genes with very small estimates of dS (<0.001, which would always result in a very large dN/dS). Genes showing evidence of positive selection along each branch were identified with the improved branch-site model (27), during which the targeted branch(es) were individually assigned as the foreground branch and others as background branches. Finally, a likelihood ratio test was used to compare a model (“model = 2, NSsites = 2, omega = 0.5|1.5, fix_omega = 0”) of positive selection on the foreground branch with a null model (“model = 2, NSsites = 2, omega = 1, fix_omega = 1”) where no positive selection occurred on the foreground branch and to calculate a P value.
The divergence time for each ancestral node on the phylogenetic tree was estimated using MCMCtree (46). All alignments were first concatenated to a supergene alignment by FasParser, and the ML tree was then constructed by RAxML version 8.1.15 (47) with a GTR + Γ model of sequence evolution. For divergence time estimates, the calibration times were obtained from the TimeTree project (www.timetree.org/) and/or previous studies (21, 45).
Functional Annotation and Enrichment Analysis.
Functional annotations were obtained using the Trinotate pipeline (trinotate.github.io/). Briefly, we used several methods of functional annotation for each gene set as follows: BLAST+ (48) was used to search for homologies in SwissProt, HMMER (49) was used to search the PFAM protein domain database, SignalP (50) was used to predict protein signal peptides, and tmHMM (51) was used to identify transmembrane domains. All search hits were used for comparison with curated annotation databases (52, 53). Finally, all functional annotation data were then integrated into an SQLite database for fast, efficient searching for terms to create a whole annotation report for each transcriptome assembly.
GO enrichment analyses were conducted with GOSeq (54). For determining the foreground branch-specific enrichments, we additionally applied enrichment analysis using the total PSGs from both foreground and background branches, and then chose the categories exclusively containing the foreground branch PSGs to represent the foreground-specific enrichments. To avoid noise in the enrichments, categories containing more than 500 entries (which are likely to represent very general biological functions) were discarded, after which all of the categories were filtered with REVIGO (55) to remove redundancy.
Construction of the Reference Hypoxia-Responsive Pathways.
All genes showing expression shifts under hypoxia were collected from the HypoxiaDB database (35), as was their expression information. To gain a more precise reference dataset of hypoxia-responsive pathways, these genes were filtered according to the following steps: (i) For genes with quantitative measurements of expression change, there had to be at least a twofold change in expression in at least one study for retention, and (ii) for genes with no quantitative expression shift measurement, the description for the expression change had to include qualitative descriptions such as “dramatically” or “rapidly” affected. After this filtration, 713 strongly up-regulated genes and 478 strongly down-regulated genes were obtained from 3,500 differentially regulated genes.
With these retained genes, functional enrichments were constructed using the GOSeq program (54). We discarded the categories with more than 500 entries to avoid nonspecific functions. To avoid nonmeaningful categories that might be introduced due to the pleiotropy of genes, we also deleted the categories that were also associated with the PSGs of the lowland species (SI Appendix, Tables S33–S36). Further, we removed the redundancy of these categories with the REVIGO method (55). Finally, only categories with a P value lower than 0.01 were retained.
Supplementary Material
Acknowledgments
We thank Peng Shi for helpful discussions; Ke Jiang, Fang Yan, Hong-Man Chen, Jun-Xiao Yang, Xiao-Long Tu, Kai Wang, Bao-Lin Zhang, and Zi-Jie Zhang for great assistance in collections; and Lu Wang, Chun-Ling Zhu, and Li Zhong for help in RNA-Seq. This research was supported by the Strategic Priority Research Program (B) (Grant XDB13020200) of the Chinese Academy of Sciences (CAS); National Natural Science Foundation of China Grants 91431105, 31671326, and 31871275; and the Animal Branch of the Germplasm Bank of Wild Species of the CAS (Large Research Infrastructure Funding). J.C. and Y.-B.S. were supported by the Youth Innovation Promotion Association of the CAS. D.M.H. was funded by the CAS President’s International Fellowship Initiative (Grant 2018VBA0039).
Footnotes
The authors declare no conflict of interest.
Data deposition: All the original RNA-sequencing data reported in this paper have been deposited into the Genome Sequence Archive, gsa.big.ac.cn (accession no. CRA000352).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813593115/-/DCSupplemental.
References
- 1.Ruiz G, Rosenmann M, Veloso A. Respiratory and hematological adaptations to high-altitude in Telmatobius frogs from the Chilean Andes. Comp Biochem Physiol A. 1983;76:109–113. [Google Scholar]
- 2.Altshuler DL, Dudley R. The physiology and biomechanics of avian flight at high altitude. Integr Comp Biol. 2006;46:62–71. doi: 10.1093/icb/icj008. [DOI] [PubMed] [Google Scholar]
- 3.Navas CA, Chauí-Berlinck JG. Respiratory physiology of high-altitude anurans: 55 years of research on altitude and oxygen. Respir Physiol Neurobiol. 2007;158:307–313. doi: 10.1016/j.resp.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Ma X, Lu X, Merila J. Altitudinal decline of body size in a Tibetan frog. J Zool. 2009;279:364–371. [Google Scholar]
- 5.Lu S, et al. Differences in hematological traits between high- and low-altitude lizards (genus Phrynocephalus) PLoS One. 2015;10:e0125751. doi: 10.1371/journal.pone.0125751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Storz JF, Scott GR, Cheviron ZA. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J Exp Biol. 2010;213:4125–4136. doi: 10.1242/jeb.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norsang G, Kocbach L, Stamnes J, Tsoja W. Spatial distribution and temporal variation of solar UV radiation over the Tibetan Plateau. Appl Phys Res. 2011;3:37–46. [Google Scholar]
- 8.Ge RL, et al. Draft genome sequence of the Tibetan antelope. Nat Commun. 2013;4:1858. doi: 10.1038/ncomms2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu Q, et al. The yak genome and adaptation to life at high altitude. Nat Genet. 2012;44:946–949. doi: 10.1038/ng.2343. [DOI] [PubMed] [Google Scholar]
- 10.Qu Y, et al. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun. 2013;4:2071. doi: 10.1038/ncomms3071. [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Wang Y, Zhang Z, He S. Comprehensive transcriptome analysis reveals accelerated genic evolution in a Tibet fish, Gymnodiptychus pachycheilus. Genome Biol Evol. 2014;7:251–261. doi: 10.1093/gbe/evu279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, Qi Y, Bi K, Fu J. Toward understanding the genetic basis of adaptation to high-elevation life in poikilothermic species: A comparative transcriptomic analysis of two ranid frogs, Rana chensinensis and R. kukunoris. BMC Genomics. 2012;13:588. doi: 10.1186/1471-2164-13-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, Qi Y, Fu J. Exploring the genetic basis of adaptation to high elevations in reptiles: A comparative transcriptome analysis of two toad-headed agamas (genus Phrynocephalus) PLoS One. 2014;9:e112218. doi: 10.1371/journal.pone.0112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, et al. Comparative transcriptomic analysis revealed adaptation mechanism of Phrynocephalus erythrurus, the highest altitude lizard living in the Qinghai-Tibet Plateau. BMC Evol Biol. 2015;15:101. doi: 10.1186/s12862-015-0371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L, et al. Genomic analysis of snub-nosed monkeys (Rhinopithecus) identifies genes and processes related to high-altitude adaptation. Nat Genet. 2016;48:947–952. doi: 10.1038/ng.3615. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, et al. Convergent evolution of rumen microbiomes in high-altitude mammals. Curr Biol. 2016;26:1873–1879. doi: 10.1016/j.cub.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Yeaman S, et al. Convergent local adaptation to climate in distantly related conifers. Science. 2016;353:1431–1433. doi: 10.1126/science.aaf7812. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y, et al. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc Natl Acad Sci USA. 2017;114:1081–1086. doi: 10.1073/pnas.1613870114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foote AD, et al. Convergent evolution of the genomes of marine mammals. Nat Genet. 2015;47:272–275. doi: 10.1038/ng.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Che J, et al. Phylogeny of the Asian spiny frog tribe Paini (Family Dicroglossidae) sensu Dubois. Mol Phylogenet Evol. 2009;50:59–73. doi: 10.1016/j.ympev.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Che J, et al. Spiny frogs (Paini) illuminate the history of the Himalayan region and Southeast Asia. Proc Natl Acad Sci USA. 2010;107:13765–13770. doi: 10.1073/pnas.1008415107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilhelm BT, Landry JR. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Gerstein MB, et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512:445–448. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiao L, Yang W, Fu J, Song Z. Transcriptome profile of the green odorous frog (Odorrana margaretae) PLoS One. 2013;8:e75211. doi: 10.1371/journal.pone.0075211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 26.Sun YB, et al. Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proc Natl Acad Sci USA. 2015;112:E1257–E1262. doi: 10.1073/pnas.1501764112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 28.Dos Reis M, Yang Z. Why do more divergent sequences produce smaller nonsynonymous/synonymous rate ratios in pairwise sequence comparisons? Genetics. 2013;195:195–204. doi: 10.1534/genetics.113.152025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noble DWA, Qi Y, Fu J. Species delineation using Bayesian model-based assignment tests: A case study using Chinese toad-headed agamas (genus Phrynocephalus) BMC Evol Biol. 2010;10:197. doi: 10.1186/1471-2148-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Ohh M. Oxygen-mediated endocytosis in cancer. J Cell Mol Med. 2010;14:496–503. doi: 10.1111/j.1582-4934.2010.01016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis JN, Carlsson A. The effect of hypoxia on monoamine synthesis, levels and metabolism in rat brain. J Neurochem. 1973;21:783–790. doi: 10.1111/j.1471-4159.1973.tb07522.x. [DOI] [PubMed] [Google Scholar]
- 32.Eldridge FL, Millhorn DE. Central regulation of respiration by endogenous neurotransmitters and neuromodulators. Annu Rev Physiol. 1981;43:121–135. doi: 10.1146/annurev.ph.43.030181.001005. [DOI] [PubMed] [Google Scholar]
- 33.McNamara MC, Gingras-Leatherman JL, Lawson EE. Effect of hypoxia on brainstem concentration of biogenic amines in postnatal rabbits. Brain Res. 1986;390:253–258. doi: 10.1016/s0006-8993(86)80234-7. [DOI] [PubMed] [Google Scholar]
- 34.Sugasawa K, et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 35.Khurana P, Sugadev R, Jain J, Singh SB. HypoxiaDB: A database of hypoxia-regulated proteins. Database (Oxford) 2013;2013:bat074. doi: 10.1093/database/bat074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khurana P, Tiwari D, Sugadev R, Sarkar S, Singh SB. A comprehensive assessment of networks and pathways of hypoxia-associated proteins and identification of responsive protein modules. Netw Model Anal Health Inform Bioinform. 2016;5:1–13. [Google Scholar]
- 37.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 38.Grabherr MG, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith-Unna R, Boursnell C, Patro R, Hibberd JM, Kelly S. TransRate: Reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 2016;26:1134–1144. doi: 10.1101/gr.196469.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aubry S, Kelly S, Kümpers BM, Smith-Unna RD, Hibberd JM. Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 2014;10:e1004365. doi: 10.1371/journal.pgen.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Löytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun YB. FasParser: A package for manipulating sequence data. Zool Res. 2017;38:110–112. doi: 10.24272/j.issn.2095-8137.2017.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin YT, Brown RP. Species history and divergence times of viviparous and oviparous Chinese toad-headed sand lizards (Phrynocephalus) on the Qinghai-Tibetan Plateau. Mol Phylogenet Evol. 2013;68:259–268. doi: 10.1016/j.ympev.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 46.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 47.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 51.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 52.Powell S, et al. eggNOG v3.0: Orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40:D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.