Abstract
Whole transcriptome analysis plays an essential role in deciphering genome structure and function, identifying genetic networks underlying cellular, physiological, biochemical and biological systems and establishing molecular biomarkers that respond to diseases, pathogens and environmental challenges. Here, we review transcriptome analysis methods and technologies that have been used to conduct whole transcriptome shotgun sequencing or whole transcriptome tag/target sequencing analyses. We focus on how adaptors/linkers are added to both 5′ and 3′ ends of mRNA molecules for cloning or PCR amplification before sequencing. Challenges and potential solutions are also discussed. In brief, next generation sequencing platforms have accelerated releases of the large amounts of gene expression data. It is now time for the genome research community to assemble whole transcriptomes of all species and collect signature targets for each gene/transcript, and thus use known genes/transcripts to determine known transcriptomes directly in the near future.
Keywords: Next generation sequencing, PolyA+ RNAs, PolyA− RNAs, Circular RNAs, RNA methylation, 5′ ends and 3′ ends
Introduction
Whole transcriptome analysis aims at capturing both coding and non-coding RNA and quantifying gene expression heterogeneity in cells, tissues, organs and even a whole body. This analysis is also important because it provides the first steps toward functional characterization and annotation of genes/genomes previously revealed by DNA sequencing [1]; builds blueprints for reconstruction of genetic interaction networks to understand cellular functions, growth/development and biological systems [2]; produces molecular fingerprints of disease processes and prognoses to pinpoint potential targets for drug discovery and diagnostics [3, 4], and offers opportunities to examine the relationship between host and pathogen for novel strategies that can be used for therapeutic and prophylactic intervention [5]. For example, cellular function, growth and cycling pathways are among the most important gene networks contributing to age-related degeneration in tendons of older humans [6], while enrichment of activating and repressive histone modifications represented the major sex-dimorphic signatures [7]. Evidence has also shown that alternative polyadenylation site usage preferences have functional relevance. When differentiated cells are used to generate induced pluripotent stem cells, for example, global 3′UTR (untranslated regions), shortening events occur [8]. In addition to broad 3′UTR shortening, intronic polyA sites can be substantially induced under cell proliferation [9]. The same situation also applies to T-cells during their activation: 86 % of genes expressed short 3′UTR isoforms following an immune response [10]. Therefore, whole transcriptome analysis provides a foundation to explore regulatory pathways and genetic networks that control both qualitative and quantitative phenotypes important to agriculture and human medicine.
No doubt, the rapid development of next generation sequencing (NGS) methods and technologies has made it possible to conduct large scale whole transcriptome sequencing projects. Since 2005, at least five NGS platforms have dominated the market, including the Roche 454 GS FLX(+) system, Applied Biosystems SOLiD (supported oligonucleotide ligation and detection) and Ion Proton/PGM/Chef systems now owned by Life Technologies (Grand Island, NY); Solexa GA (Genome Analyzer)/HiSeq/MiSeq/NextSeq developed by Illumina (San Diego, CA); and PacBio RSII system made by Pacific Biosciences (Menlo Park, CA) [11, 12]. None of these platforms rely on Sanger sequencing. While SOLiD uses sequencing by ligation, all other systems employ sequencing by synthesis [12]. Among these five NGS platforms, only PacBio system uses single molecules as templates for sequencing, while others must conduct either bridge amplification (Illumina platforms) or emulsion PCR amplification (all three Life Technologies platforms) for preparation of “clusters” of same templates for sequencing. The read lengths are also quite variable among these platforms: up to 75 bp (paired end), 300 bp (overlapping paired end), 400 bp (bidirectional), 700 bp (paired end) and 8500 bp produced by SOLiD, Illumina, Ion Torrent, 454 and PacBio systems, respectively [12]. Furthermore, the number of reads per NGS run can range from 1 million to 5000 million with the machine running time varying from 8 h to 11 days, depending on the platforms [13].
In the present review, we classify whole transcriptome analysis with sequencing methods and technologies into four categories: (1) whole transcriptome shotgun sequencing (WTSS), (2) whole transcriptome target/tag sequencing with restriction digestion, (3) whole transcriptome target/tag sequencing without restriction digestion, and (4) other developments. These methods and technologies rely on the library preparation. As such, we will mainly review: (1) what serves as library starting material, such as total RNAs, polyA+ RNAs or polyA− RNAs, (2) how adaptors or linkers are added to both 5′ and 3′ ends of RNA molecules, such as through cDNA synthesis or ligation, and (3) how products are amplified for sequencing, such as via cloning or PCR amplification. We also discuss the challenges associated with library preparation and potential options or solutions that can be used to address these difficulties, biases or challenges. In brief, we review the advantages and disadvantages of each method so that readers are completely informed as they decide what method(s) meet their research objectives.
Whole transcriptome shotgun sequencing
EST (expressed sequence tag) sequencing
ESTs are single pass reads derived from cDNA libraries with random selection. EST libraries can be prepared from single or multiple sources of tissue(s), organ(s) or cell type(s) to meet various research goals. Relatively large scale EST sequencing projects were started in the early 1990s. In 1991, for example, Adams and coworkers [14] reported over 600 ESTs derived from randomly selected human brain cDNA (complementary DNA) clones with more than half (337 ESTs) representing new genes at that time. Ten years later, publicly available ESTs were used to build the TIGR (The Institute for Genomic Research) gene indices for 21 species [15, 16]. Now, gene indices can be found for over 150 species at ftp://occams.dfci.harvard.edu/pub/bio/tgi/data/. ESTs have served as the “unigene” resources at the National Center for Biotechnology Information (NCBI) to infer approximate expression patterns by tissue, age, and health status for more than 140 species (http://www.ncbi.nlm.nih.gov/unigene). ESTs have also been used to examine exclusively expressed genes, co-expressed genes, house-keeping genes and genes that are differentially expressed due to different conditions or diseases [17–21].
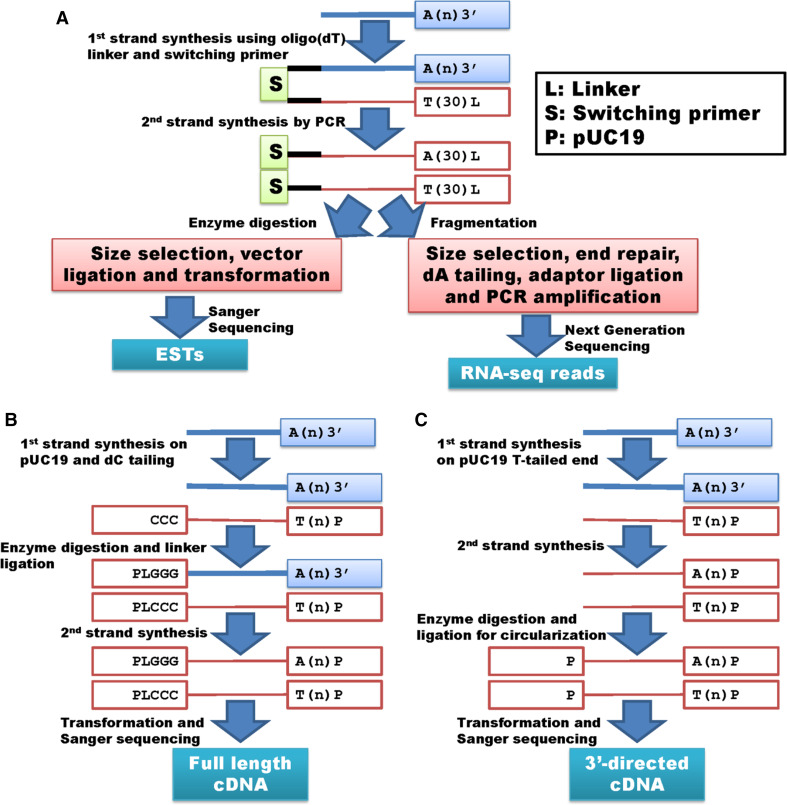
As most eukaryotic mRNAs have a poly(A) tail at their 3′ ends, EST library preparation often uses oligo(dT) primers (typically 20 nucleotides in length) to initiate reverse transcription. During the process, a switching primer is also incorporated in the first-strand cDNA synthesis to form DNA–RNA hybrids. Subsequently, the product is amplified with primers derived from the known adaptor/linker sequences, followed by restriction digestion, fractionation and cloning [22] (Fig. 1a). Cloning involves ligation with vectors and transformation into E. coli for replication. Finally, the clones are randomly picked for Sanger sequencing. In addition to EST sequencing with random selection, protocols have been also developed to prepare libraries for full-length cDNA and 3′-directed cDNA sequencing [23–25]. As shown in Fig. 1b, the conventional full-length cDNA library preparation involves six steps: (1) first-strand cDNA synthesis using plasmid (pUC19) primers that have an overhang of dTs; (2) extension of first-strand cDNA by adding an oligo(dC) tail using terminal deoxynucleotidyl transferase; (3) digestion with HindIII to produce a sticky end for step; (4) ligation with oligo(dG) DNA linkers to form circular DNAs; followed by (5) second-strand synthesis and subsequent transformation into E. coli for cloning collection and sequencing [23].
Fig. 1.
Whole transcriptome shotgun sequencing. a Lists both EST and RNA-seq procedures, while b, c demonstrate library preparations for full-length cDNA and 3′-directed cDNA sequencing. EST, full-length cDNA and 3′-directed cDNA are all cloning-based approaches using Sanger sequencing so that the adaptors are provided by the cloning vectors. However, the adaptors used in RNA-seq library preparation depend on the next generation sequencing platforms. Certainly other derivatives exist for whole transcriptome shotgun sequencing or RNA-seq
Preparation of 3′-directed cDNA uses only pUC-19 based vector primers (Fig. 1c). First-strand cDNA is synthesized in the same manner as described for full-length cDNA described above, but without addition of oligo(dC) tails. After second-strand cDNA synthesis, the constructs are cleaved with both BamHI and MboI and ligated for plasmid re-circularization [24]. The products are introduced into E. coli for clone selection and sequencing. Okubo and colleagues [24] found that the 3′-directed cDNA library proportionally represents the original mRNA population. In particular, the uniqueness of 3′ sequences allows for gene assignments and provides “signatures” for global profiling of gene expression [26]. Gautheret and colleagues [27] assembled 164,704 3′ end ESTs into 15,325 clusters, which were then used to characterize alternative polyadenylation in human. Alternatively, Beaudoing and Gautheret [28] used mRNA sequences as queries to predict alternative polyadenylation against ESTs and investigate tissue biases in 3′ end usages.
A large scale genome wide map of polyA sites using ESTs was first described by Tian and co-workers [29]. The authors used both BLAST and MegaBLAST suites with default settings to align ESTs to genomes. For polyA site determination, they required unaligned sequences at either 5′ or 3′ termini of the EST that had stretches of 8 or more Ts and As, respectively. Using sequences that met these requirements, 29,283 poly(A) sites were identified in 13,942 human genes and 16,282 poly(A) sites were located in 11,155 mouse genes with ESTs or cDNA support [29]. We recently explored the publicly available EST data and collected initial information on polyA sites in X. tropicalis using the PASA (Program to Assemble Spliced Alignments) tool, which was originally developed at The Institute for Genomic Research in 2002 (http://pasa.sourceforge.net/). We downloaded 1,271,480 X. (Silurana) tropicalis ESTs, assembled some public RNA-seq data representing different tissues and embryos at different developmental stages (deposited at NCBI/SRA), and used the reference genome (xtropicalis 7.1) as input data for PASA analysis, which identified 51,659 polyA sites. Using coordinates of gene models as references, we assigned 46,617 polyA sites to 13,250 genes in X. tropicalis. Among those genes, 2749 (21 %) had only one polyA site, while the remaining genes (79 %) contained more than one polyA site.
RNA-seq
Morin and colleagues [30] constructed four libraries for whole transcriptome shotgun sequencing using randomly primed cDNA and massively parallel short-read sequencing on an Illumina Genome Analyzer I. The initial steps for the library preparation were similar to those described above for EST library preparation (Fig. 1a). Basically, both modified oligo(dT) and template-switching primers were used to produce full-length single-stranded cDNAs containing the complete 5′ end of the mRNA and universal priming sequences for end-to-end PCR amplification. The amplified products were fragmented and size-selected for 100–300 bp, which were then end-repaired, dA-tailed and ligated with the Illumina sequencing adaptors (Fig. 1a). PCR was performed again using Illumina’s genomic DNA primer set for cluster generation and sequencing on the Illumina Cluster Station and Illumina Genome Analyzer I. The whole transcriptome shotgun sequencing technique, now known as the RNA-Seq method has dramatically shaped the landscape of whole transcriptome profiling [31–33]. RNA-seq can detect transcript levels, can also reveal splicing isoforms and expressed polymorphisms.
Smibert and coworkers [34] developed a so-called strand-specific RNA-seq method to enrich fragments associated with the polyA site regions. Briefly, oligo-dT selection was used to isolate polyA+ RNA, which was subsequently fragmented. Ends of the fragmented poly polyA+ RNA were then repaired by treatment with phosphatase and polynucleotide kinase. A primer complementary to the 3′ linker was used to reversely transcribe RNA adaptors (5′ and 3′) that were sequentially ligated to the RNA. Each strand-specific RNA-seq library (1 ng) was then re-amplified with 10 cycles of PCR using two primers complementary to the 5′ adaptor and 3′ adaptor with an additional six T residues at the 3′ end, respectively. A second round of PCR with 15 cycles was performed using the same 5′ adaptor primer, but also included a new 3′ primer complementary to the adaptor with an added 5′ extension that contained a 6 nt index sequence and a sequence complementary to the flow cell primer. The libraries were quantitated, and 10–12 libraries were pooled together and sequenced on an Illumina HiSeq 2000 using paired-end 100 bp and 6 bp index read chemistry [34].
On the other hand, RNA-seq reads can also be used for genome wide profiling of polyA sites for discovery of alternative polyadenylation. For example, Schlackow and colleagues [35] sampled S. pombe RNA-seq data and selected reads with a minimum of five consecutive adenine (A) residues [poly(A) tail] at their ends as candidates for polyA site reads. The authors then painted these candidate reads on the chromosome to estimate the “correctness” based on the similarity and coverage. When a sequence mapped to multiple places in the genome, the RNA-seq read with the closest correspondence or closest downstream from an ORF was selected. After discarding potential internal priming polyA sites, the team was able to identify 3′UTRs for nearly 90 % of the yeast genes and re-annotated 3′UTRs of 4535 genes, including extensive examples of alternative polyadenylation and heterogeneity.
Challenges and potential solutions
As discussed above, construction of libraries for both EST and RNA-seq projects are similar, but sequences are produced on different platforms. ESTs are usually determined by Sanger sequencing, which requires a cloning step to produce identical templates for the sequencing reaction. In comparison, RNA-seq is performed in a massive parallel manner on next generation sequencing platforms. Advantages of high throughputs, large data outputs and relatively low costs have led RNA-seq to gradually dominate the field, which has almost replaced conventional EST sequencing in recent years.
There are several challenges associated with RNA-seq analysis. The methodological challenge includes fragmentation bias, length bias and transcriptome composition bias [32, 36]. When RNA is fragmented, the library preparation favors the internal transcript body, while depleting the transcript ends by producing shorter fragments [32]. The short 5′ and 3′ ends fragment can easily get lost during the size selection steps of library preparation [37]. As a consequence, RNA-seq lacks uniform coverage for the whole gene region, implying that RNA-seq data cannot be used to accurately determine both transcription start and end sites. On the other hand, the number of reads per gene should depend on expression abundance, transcript lengths and degradation process. When genes are expressed at a similar level, longer transcripts would produce more reads than shorter ones, resulting in gene length bias [36]. Defects in nonsense mediated decay pathways can decrease the decay of the aberrant RNA molecules [38], influencing the number of reads for a specific mRNA. Transcriptome composition bias occurs when one or a few transcripts in a given sample are expressed at extremely high levels, thereby downplaying the number of reads collected for other transcripts [36]. To correct these biases, several bioinformatics tools have been developed to adjust gene expression level based on (1) a probability weighting function (estimation of differentially expressed changes as a function of its transcript length) [39]; (2) each gene’s test statistic (using the square root of transcript length) [40] or (3) a likelihood (simultaneous estimation of bias parameters and expression levels using the likelihood framework) [41]. All claimed that these corrections and adjustments can significantly improve the results or outcomes, as the adjusted data are highly correlated with qRT-PCR validation [39], overlap with microarray data [40] or are more consistent with known biology [41].
RNA-seq often requires 10–20 times more reads than a typical tag or target sequencing method [42], thus creating challenges in bioinformatics analysis and computational methods due to large data storage, retrieval and processing [32]. Therefore, it is impossible to use spreadsheet software for data processing [43]. No doubt, RNA-seq can provide reads for annotation of known genes, assembly of novel transcripts and compilation of potential splicing forms within a gene or transcript. However, a recent study found that reconstruction of full-length isoforms of genes/transcripts using short reads presents a challenging task as well [44]. When unguided transcript reconstruction was performed using short reads, the authors revealed that valid isoforms were assembled for roughly half of expressed genes on average (H. sapiens mean 41 %, maximum 61 %; D. melanogaster mean 55 %, maximum 73 %; C. elegans mean 50 %, maximum 73 %). In addition, RNA-seq cannot sufficiently detect genes/transcripts with low levels of expression [43].
Although enrichment of 3′ end fragments from strand-specific RNA-seq libraries [34] helps to overcome some bioinformatics challenges, the process remains inefficient due to the many steps involved in library preparation and difficulties in direct RNA–RNA ligation [32]. Removing the gene specific biases and reducing the computational complexities associated with RNA-seq should rely on development of tag/target transcriptome analysis approaches, which will be discussed below in detail. Assembly of RNA-seq reads for discovery of novel transcripts and compilation of transcriptional isoforms might be replaced in the near future by the so-called Iso-seq method. This assay uses the long read lengths of SMRT® Sequencing technology to generate full-length transcripts (http://pacb.com/applications/isoseq/index.html). Two research teams have found that the method has many advantages, such as single molecule sequencing without amplification or fragmentation, single read for entire exon–intron structure, high coverage of all splice sites of the original transcripts, discovery of novel splicing forms and detection of allele specific isoforms [45, 46].
Whole transcriptome tag/target sequencing with restriction digestion
SAGE (serial analysis of gene expression)
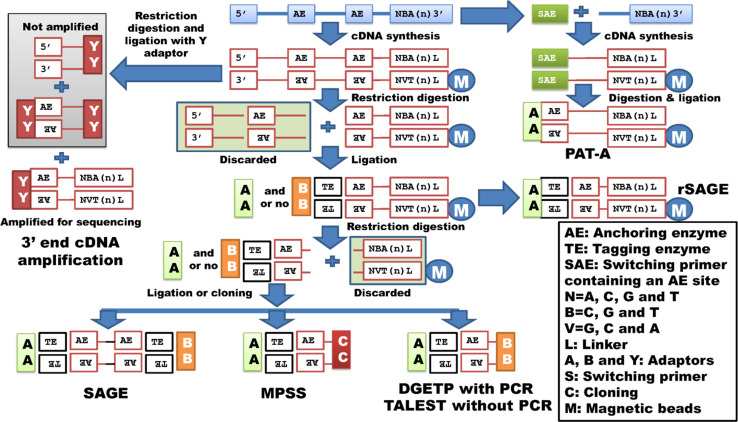
The SAGE technique might be the first true whole transcriptome analysis with sequencing method developed to produce a snapshot of the transcripts in biological samples of interest with collection of small tags that correspond to genes/transcripts [47]. In conventional SAGE analysis, magnetic beads coated with polyT tails are used to capture polyA+ RNA, which are then converted into cDNA using reverse transcriptase. Next, cDNA molecules are digested with the anchoring enzymes (NlaIII and DpnII, for example) and then ligated with adaptors containing the tagging enzyme site (BsmFI, for example) that can be subsequently digested to produce small tags (Fig. 2). Two tags are combined into a di-tag by ligation. The di-tags are further glued together to form long concatamers, which are subsequently cloned and copied millions of times and sequenced. The data are processed to count the small sequence tags for transcriptome analysis. Obviously, it is difficult to assign the short lengths of tags to genes/transcripts. As such, the LongSAGE (MmeI as the tagging enzyme) and SuperSAGE (EcoP15I as the tagging enzyme) methods have further improved the technique by increasing the tag size up to 21–26 bp in length [48, 49]. Furthermore, Spinella et al. [50] developed a method very similar to SAGE, called “TALEST,” or “tandem arrayed ligation of expressed sequence tags.” This assay employed an oligonucleotide adapter containing a type IIs restriction enzyme site to facilitate the generation of short (16 bp) ESTs of fixed position in the mRNA. This process involved cloning and sequencing without PCR at all stages of the assay (Fig. 2), which appears to be the only difference from SAGE-seq or DGETP (digital gene expression tag profiling) methods described below.
Fig. 2.
Whole transcriptome target/tag sequencing with restriction digestion. SAGE, MPSS, DGETP and TALEST share many common steps in library preparation: both anchoring (AE) and tagging enzymes (TE) are used with adaptors added in slightly different ways. In PAT-A, rSAGE and 3′ end cDNA amplification, only the anchor enzyme cut site is used for 5′ adaptor ligation, while the 3′ adaptor/linker is combined with reverse transcription. For 3′ most target/tag collection, only 3′ end cDNA amplification uses specific primers, while the remaining methods rely on magnetic bead selection
SAGE-seq
To further simplify SAGE analysis procedures, several strategies were developed to increase the number of tags sequenced per transcriptome/library using high-throughput sequencing platforms. In 2000, Brenner and colleagues [51] developed a novel sequencing approach, called the “massively parallel signature sequencing (MPSS)” method, which combined non-gel-based signature sequencing with in vitro cloning of millions of templates on separate 5 micron diameter microbeads. A 17 base sequence was generated for each transcript using enzymes DpnII and BbvI, followed by cloning and sequencing on beads (Fig. 2) [52]. The authors claimed that MPSS generated over one million signature sequences (tags), which provided enough sequence depth to identify low-expressed transcripts with high accuracy. The MPSS design led to development of several next generation sequencing (NGS) platforms, such as the Roche/454 FLX, the Illumina/Solexa Genome Analyzer, the Applied Biosystems SOLiD™ System, the Helicos Heliscope™ and Pacific Biosciences SMRT instruments [53]. Therefore, the traditional SAGE method has been easily adapted into NGS platforms, such as DGETP on the Illumina/Solexa Genome Analyzers. Without formation of di-tags and concatamers, the tags are simply “sandwiched” by the Illumina GEX Adaptors 1 and 2 (5′ and 3′ adaptors) for amplification and sequencing [54] (Fig. 2). SuperSAGE has also been successfully integrated with NGS as high-throughput SuperSAGE with Illumina Genome Analyzers and the Applied Biosystems SOLiD™ System [55].
Elongation of tag/target sizes
Although the tag length produced by SAGE-related methods has increased from 10 to 26 bp by use of different tagging enzymes, assigning them to known transcripts remains a challenging task. To overcome this problem, there are at least three methods that have been invented to collect long tags or targets and they are 3′ end cDNA amplification [56], rSAGE (reverse serial analysis of gene expression) [57] and PATs (polyA tags) using restriction digestion [58] (Fig. 2). The 3′ end cDNA amplification method uses a 2-base anchored oligo(dT) primer with a heel (like a linker sequence) for first-strand cDNA synthesis, followed by the second-strand cDNA synthesis. The cDNA products are then digested with restriction enzymes and ligated to a so-called Y-shaped adaptor, which blocks amplification of the Y–Y ligated products. Therefore, only 3′ ends can be amplified for sequencing and profiling [56]. The rSAGE method uses primers containing 64 nucleotides (30 Ts included) as linkers for reverse transcription to synthesize cDNA molecules, which are then digested with NlaIII and ligated with 5′ adaptors, but without further digestion with any tagging enzymes. Both 5′ adaptor and 3′ linker sequences are used to design primers (rSAGEF1 and rSAGER1) that amplify the long tags or targets for sequencing (Fig. 2). In the PAT method using restriction digestion, switching primers containing enzyme cut sites are used in reverse transcription along with linker [58]. The cDNA products are then digested with NlaIII or TaiI and ligated with new adaptors that have overhangs complementary to the enzyme cut sites (Fig. 2). The remaining steps for PATs are the same as those for rSAGE, but only the PATs that are between 100 and 600 bp in size are selected for sequencing [58]. The advantage of PATs over rSAGE is that the former method might recover some transcripts that do not contain native restriction cut sites for both enzymes.
Challenges and potential solutions
Tag size and data amount are two major drawbacks associated with the conventional SAGE method that have been well addressed. By exploring different tagging enzymes, tag length has been increased from 10 bp (digested with BsmFI) to 26 bp (digested with EcoP15I). In particular, rSAGE [57] and PATs using restriction digestion [58] can further extend the tag/targets sizes up to a few hundred bp in length, depending on the sequencing platforms. The long tags or targets certainly make it easier and more accurate to assign them to genes/transcripts. In terms of data amount, a conventional SAGE analysis usually results in less than 20,000 tag reads per library [59]. However, a typical SAG-seq run (DGETP) can now provide many tag reads, ranging from 3 to 30 million reads.
To our knowledge, however, a real challenge facing whole transcriptome tag/target sequencing with restriction digestion is the restriction enzymes themselves. First, none of the enzymes can make whole transcriptome analysis a reality, because a transcript may be cut by one enzyme, but lack the recognition site for another. For example, we performed a survey on a total of 36,590 X. tropicalis mRNA sequences downloaded from the GenBank database and made a in silico digestion using a total of 11 restriction enzymes (so-called four cutter enzymes), including Tsp509I (AATT), TaiI (ACGT), NlaIII (CATG), AciI (CCGC), MspI (CCGG), DpnII (GATC), HhaI (GCGC), BfaI (CTAG), MseI (TTAA), TaqI (TCGA) and Csp6I (GTAC). We found that 23 (23/36,590 = 0.06 %) mRNA sequences did not have recognition sequences for any of these enzymes. The transcriptome coverage ranged from 76 % (27,735/36,590) with MseI (TTAA) to 95 % (34,653/36,590) with DpnII (GATC). Second, when an enzyme cuts only at the polyA junction site, the traditional SAGE method collects tags with all As, while rSAGE will not collect targets for these genes/transcripts. Therefore, these transcripts will be missed in the transcriptome analysis. Third, generally speaking, SAGE and its derivatives focus on collection of tags or targets associated with 3′ most cut sites of transcripts. As such, rSAGE will certainly collect targets of various lengths. When PCR is performed to enrich the targets for sequencing, the short fragments might be favored for amplification, and thus indicate false abundance.
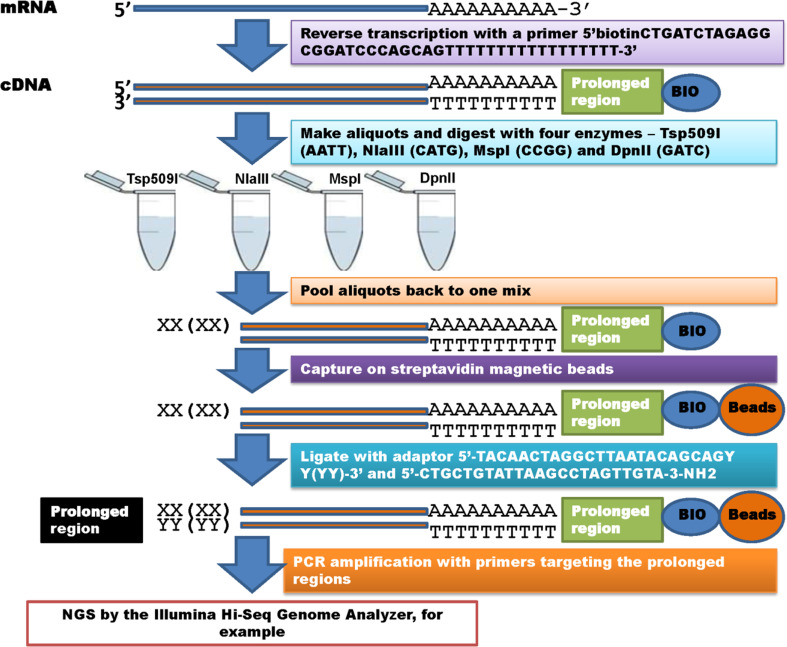
To overcome the limitations related to enzymes themselves, we propose a combined set of enzymes be used to conduct multiple rSAGE analysis (Fig. 3). Our data indicates that a combination of four enzymes: Tsp509I (AATT), NlaIII (CATG), MspI (CCGG) and DpnII (GATC) would cover 99.82 % of the X. tropicalis transcriptome. Up to date, both NlaIII and DpnII have been heavily used in tag-based RNA-seq analyses. We assume that addition of Tsp509I (AATT) and MspI (CCGG) to the enzyme combination would help produce tags for AT-rich and CG-rich mRNA sequences, respectively. As shown in Fig. 3, multiple rSAGE processes will involve (1) mRNA extraction: extract total RNA from each sample and enrich polyA+ RNA; (2) Reverse transcription: convert the mRNA to cDNA molecules; (3) Enzyme digestion: digest the cDNA with four enzymes (Tsp509I, NlaIII, MspI and DpnII) in separate aliquots; (4) Bead collection: collect fragments with polyA tails using streptavidin magnetic beads; (5) Adaptor ligation: ligate adaptors with different 3′ ends designed based on the restriction enzymes to the collected products; (6) PCR amplification: amplify the collected fragments with primers designed based on the sequences of the reverse transcription primer and the adaptors and (7) NGS: sequence fragments of sizes ranging from 150 to 450 bp with NGS platforms. Even so, we believe that this process will create novel challenges in data analysis because the use of multiple enzymes may result in one or more collected tags per transcript.
Fig. 3.
SAGE-seq with multiple enzymes. The process represents a combination of SAGE and DDRT-PCR methods as the 5′ prolonged region was adapted from the former, while the 3′ prolonged region from the latter technique, respectively. Adaptors for the 5′ prolonged regions are designed according to the enzyme cut sites: Tsp509I (AATT), NlaIII (CATG), MspI (CCGG) and DpnII (GATC)
Whole transcriptome tag/target sequencing without restriction digestion
DDRT-PCR (differential display reverse transcription polymerase chain reaction)
DDRT-PCR considered as the prototype of whole transcriptome profiling of 3′ termini, was developed by Liang and Pardee [60] to compare and measure gene expression changes between biological samples under different conditions. The DDRT-PCR method starts with reverse transcription in fractions using a set of anchored primers containing T11 to the 3′ polyadenylation (polyA) sites with either one or two additional bases to a subset of polyA+ mRNAs, followed by amplification of cDNA species from each fraction using a set of arbitrary primers (10 bp in length) and anchored primers. The resulting fragments are then electrophoretically separated for identification of differentially expressed fragments, which are excised from the gel for reamplification, cloning and sequencing for gene identification. Other methods, such as Northern blotting, RNase protection, and/or nuclear run-on are often applied to validate the differentially expressed genes [61]. However, due to low sensitivity, quantitation difficulties and false positives, this old and simple method has been replaced by other methods.
Transcriptome profiling of 3′-ends with enrichment of polyA+ RNA
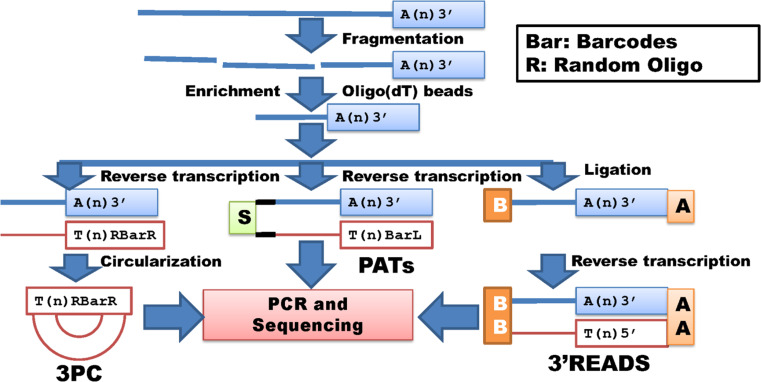
Three methods: PATs using RNA fragmentation [62], 3PC (3′Poly(A) site mapping using cDNA circulation) [63], and 3′READS (3′ region extraction and deep sequencing) [64] can be classified into this group with enrichment of polyA+ RNA (Fig. 4). A common step involved in these methods is fragmentation of total RNA or polyA+ RNA followed by purification of fragmented polyA+ RNA using the Life Technologies oligo(dT) magnetic beads in 3PC [63], the New England Biolabs oligo(dT) magnetic beads in PATs [62] or the Sigma CU5T45 coated beads in 3′READ [64]. The polyA+ containing fragments are used for reverse transcription in both 3PC and PATs, but the former method employs a circularization for formation of 5′ and 3′ adaptors, while the latter method combines reverse transcription with integration of both 5′ and 3′ adaptors (Fig. 4). In 3′READS, the enriched polyA+ fragments are first ligated to 5′ and 3′ adaptors and then reverse transcribed to cDNA. All three methods use PCR to amplify products that are then submitted for deep sequencing [62–64].
Fig. 4.
Whole transcriptome target/tag sequencing without restriction digestion (I). 3PC, PATs and 3′READS represent methods with enrichment of fragmented polyA+ RNA to conduct polyA site sequencing. These methods share common steps including size selection, PCR amplification and next generation sequencing
Transcriptome profiling of 3′-ends with enrichment of polyA+ cDNA
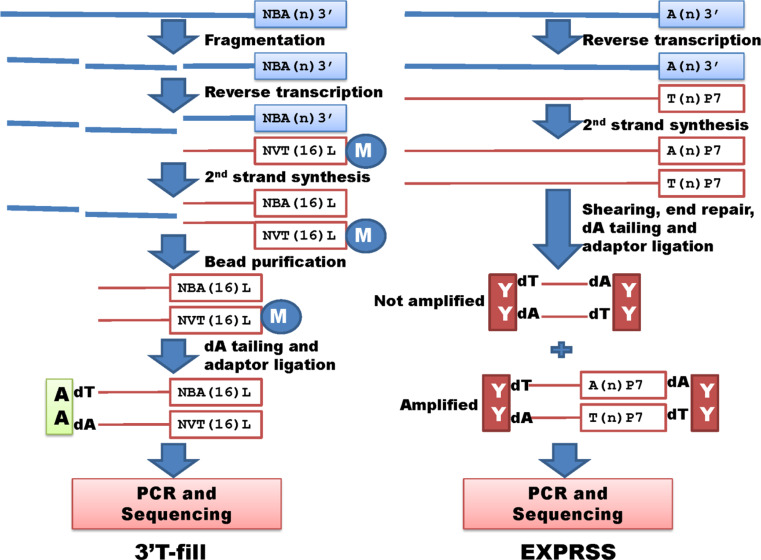
Although two methods, 3′T-fill [37, 65] and EXPRSS (Expression Profiling through Random Sheared cDNA Tag Sequencing) [42] both enrich polyA+ cDNA, the procedures are quite different (Fig. 5). In the 3′T-fill method, total RNA is fragmented and first-strand cDNA is synthesized by reverse transcription with a biotinylated oligo (dT16VN). The first-strand cDNA is treated with RNase H and second-strand cDNA synthesized using DNA polymerase I [37, 65]. In comparison, the EXPRSS method uses total RNA as a template to synthesize first-strand cDNA with oligo(dT) primers containing the P7 sequence of the Illumina flow cell. Second-strand cDNA synthesis is based on a traditional protocol [42]. In the 3′T-fill method, the double-stranded cDNA molecules are enriched for polyA+ cDNA using Dynabeads (Life Technologies), followed by dA tailing with ligation to a 5′ end adaptor. Eighteen cycles of PCR are performed and the products are then size-selected by gel electrophoresis for deep sequencing. In the EXPRSS method, however, the double-stranded cDNA products are physically sheared using Covaris AFA to a target size of 200 bp, followed by end repair, dA tailing, ligation with Y-shaped adaptors and size selection with an agarose gel. The Y-shaped adaptors were invented by Prashar and Weissman in 1996 [56], which allow amplification of only fragments derived from the 3′ end primer, and thus enrich polyA+ cDNA for sequencing [42].
Fig. 5.
Whole transcriptome target/tag sequencing without restriction digestion (II). 3′T-fill and EXPRSS represent methods with enrichment of fragmented polyA+ cDNA to conduct polyA site sequencing. These methods share common steps including size selection, PCR amplification and next generation sequencing
Transcriptome profiling of 3′-ends with custom primers containing oligo(dT) at 3′ end for sequencing
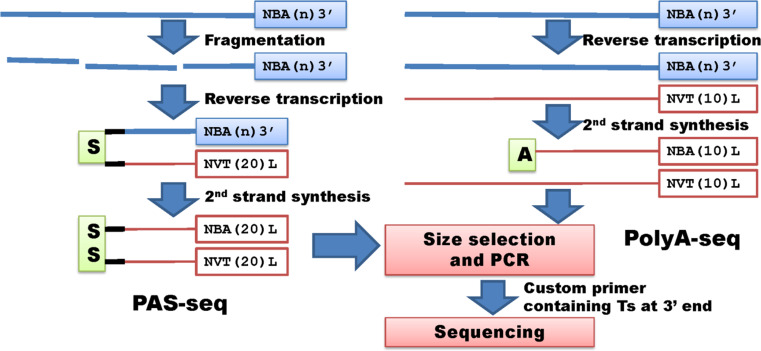
Two technologies were developed to profile 3′ ends of transcripts using oligo(dT) containing primers for sequencing (Fig. 6). PAS-seq (PolyA site sequencing) [66, 67] utilizes purified poly(A+) RNAs that are fragmented and reverse transcribed into cDNA using both an oligo(dT) primer and a switching primer. To sequence the products on the Illumina platform, both PE 1.0 and PE 2.0 primers are used for the first round of PCR using only 3 cycles. The amplified products are then size-selected (200–300 bp in size) and used for the second round of PCR with 15 cycles. The products are purified and submitted for sequencing using a custom primer containing oligo(dT20) at the 3′ end [67]. Library preparation for PolyA-seq (polyA sequencing) [68] begins with reverse transcription of unfragmented poly(A+) RNA molecules using an oligo (T10VN plus a 5′ heel of 10 bp). The first-strand cDNA is treated with RNase H and second-strand cDNA is synthesized using N7 random primers plus a 5′ heel of 10 bp. A total of 32 cycles of PCR is performed to amplify the cDNA products for sequencing using a custom primer containing oligo(dT10) at 3′ end.
Fig. 6.
Whole transcriptome target/tag sequencing without restriction digestion (III). PAS-seq and PolyA-seq represent methods with custom primers containing dTs at 3′ end to conduct polyA site sequencing. These methods share common steps including size selection, PCR amplification and next generation sequencing
Challenges and potential solutions
As described above, oligo(dT) primers/linkers are frequently used to initiate the first-strand cDNA synthesis in library preparation. It is possible that during the step when mRNA is converted to cDNA, internal polyA runs (encoded in the genome) can be primed off to produce spurious polyA sites. Here, we propose two solutions to address this issue. The first solution is to examine these polyA site reads using well-developed models for prediction. A multispecies polyadenylation site model was, for example, proposed by Ho et al. [69], which was validated using two machine learning methods: logistic regression and linear discriminant analysis. The authors found that the comparative model could reach 85–92 % sensitivity and 85–96 % specificity using data from seven species of animals and plants. Another model is the “PolyA-iEP method” [70] and its unique features include taking advantage of emerging alternative polyadenylation patterns in combination with a distance-based scoring method. The authors also concluded that their PolyA-iEP method achieves high scores of sensitivity and specificity [70]. The second option is to scan currently available mRNA sequences for internal polyA stretches of ≥5, for example, followed by collecting 300 bp of their upstream sequences. These sequences can be used as “seeds” to bait the reads derived from the internal polyA sites. When these experimental reads are “ignored” by the prediction models or there is evidence indicating they are derived from internal polyA sites, we can examine them further for clarification.
Ma and coworkers [62] compared different library preparation procedures for PATs (polyA tags). The PAT-A method relies on restriction digestion (Fig. 2); while the PAT-B method uses fragmentation (Fig. 4). However, the difference between PAT-B1 and PAT-B2 is related to application of size selection: only PAT-B2 is subject to size selection before PCR. The authors made a total of 20 libraries, including 5 for PAT-A, 11 for PAT-B1 and 4 for PAT-B2. Surprisingly, the percentage of reads mapped to the 3′UTR within each type of library varied widely, ranging from 1.8 to 44.4 % within 5 PAT-A libraries, 15.6 to 52.8 % within PAT-B1 libraries and 5.8 to 47.9 % within PAT-B2 libraries, respectively [62]. These results clearly indicate that noisy data can be overwhelmingly produced at some point during the library preparation process. We would argue here that PCR amplification of the libraries for sequencing can introduce bias. In particular, sequencing primers can amplify not only the 3′ termini of targets/tags, but can also amplify internal mRNA sequences that happen to contain nucleotides identical to the 3′ end of the primers. Our recent experience showed that direct usage of sequencing primers in PCR can amplify the noisy reads and account for up to 65 % of total number of reads (data not shown). We have modified the method and have reduced the noisy data to less than 3.5 % of the total reads.
Both PAS-seq and PolyA-seq methods use a custom primer that contains either oligo(dT20) or oligo(dT10) for sequencing [66–68]. Unfortunately, these customized oligo(dT) primers cannot be used for sequencing of other libraries. Switching primers for sequencing different libraries make it difficult to adapt to currently available sequencing platforms. The customized primer might also make it impossible to use barcodes to sequence multiple libraries in one run. In addition, both sense and antisense sequencing strategies have been used to profile 3′ ends of mRNAs. To our knowledge, antisense sequencing should provide the best option to map reads because it begins at the polyA junction sites. However, some of sense reads might not be useful if they are too short to reach the polyA sites.
Direct RNA sequencing (DRS) is a single molecule sequencing-by-synthesis method developed by Ozsolak and co-workers in 2009 [71]. This technology involves hybridization of 3′ blocked polyA RNA molecules to polyT oligonucleotides coated on sequencing surfaces, followed by dTTP fills and initiation of sequencing by synthesis. The sequencing is done on the Heliscope Sequencer [71, 72]. This method does not require cDNA synthesis and PCR amplification, thus achieving bias-free transcriptome analysis [73]. However, the service company, Helicos BioSciences Corporation filed for bankruptcy in 2012 (http://business-bankruptcies.com/cases/helicos-biosciences-corporation).
Whole transcriptome analysis with sequencing: other developments
Profiling of nonpolyadenylated RNAs
All of the methods described above use the presence of the 3′ polyA tail as an essential structure to enrich polyA+ transcripts and synthesize cDNA by using oligo-dT. However, whole transcriptomes also contain transcripts without polyA tails, or polyA− RNA molecules. The currently known polyA− transcripts include ribosomal RNAs, other small RNAs, replication-dependent histone RNAs and long non-coding RNAs [74–76]. It seems that a large amount of polyA− RNAs remain uncharacterized. To date, only two methods have been developed to profile polyA− RNAs. Wu et al. (2008) [74] actually profiled both polyA− and polyA+ in one pipeline. Like the RNA ligase mediated amplification of cDNA end approach created by Liu and Gorovsky [77], the authors ligated a common RNA adaptor to the 3′ ends of all RNA transcripts, followed by removal of 18S and 28S ribosomal RNAs using biotinylated ribosomal specific probes and removal of other small size RNAs using size selection. The common adaptor ligated to the 3′ end of RNAs was then used to synthesize cDNAs, followed by digestion with NlaIII for addition of the 5′ end adaptor. The products that contained both 5′ and 3′ end adaptors were amplified and sequenced from their 3′ ends using the Roche 454 system. The relatively long reads produced by this platform are required for this method because short reads with only As are useless in the analysis. The method reported by Yang et al. [75] and Zhang et al. [76] starts with separation of the polyA− from the polyA+ RNA. The process is relatively simple: polyA+ RNAs are removed using the oligo-dT beads. The “leftover” polyA− RNAs are then analyzed by a routine RNA-seq method.
Profiling of circular RNAs
Circular RNAs (circRNAs), also known as nonpolyadenylated RNAs are a type of long non-coding RNA (lncRNA) generated by exon backsplicing during RNA splicing or directly generated from spliced out intron fragments. Exonic circRNAs act as miRNA sponges or act as an “mRNA trap” that is involved in regulation of parent mRNA expression [78]. Exonic circRNA and intronic circRNA are two types of circular RNA that can be detected by different RNA-seq strategies. Several studies investigated thousands of exonic circRNAs by sequencing rRNA-free and RNase R treated RNAs [79–81]. RNase R treatment degrades linear RNA molecular and Y-structure RNAs so that only circular RNAs are retained in the reaction. Special algorithms based on existing transcript models or based on genomic sequences were developed to identify exonic circRNAs. Generally speaking, pair end reads of a single cDNA fragment may be assigned to different locations of annotated transcripts with different directions, indicating the existence of the backsplicing for RNA splicing [81, 82]. On the other hand, if multiple reads fail to follow genomic order or a read is split so it maps to different regions of the genome, provides additional evidence for backsplicing [78]. Zhang and colleagues added a polyA− enrichment step to reduce noisy data and directly identified abundant intronic circular RNAs [83].
Profiling of RNA methylation
RNA methylation is a common phenomenon in both eukaryotes and prokaryotes. To investigate cytosine methylation within RNAs, several methods based on RNA sequencing techniques were developed to identify methylation at nucleotide resolution of RNAs with or without chemical treatment. RNA bisulfite sequencing is quite similar to bisulfite genomic DNA sequencing methods. Basically, RNAs are treated with bisulfite to convert methylated cytosine to uracil. Schaefer and colleagues successfully investigated the RNA methylation pattern based on this method combined with deep sequencing [84]. Beside chemical treatment, other techniques are based on immunoprecipitation to detect RNA cytosine methylation. For example, Khoddami and his colleagues [85, 86] used both mammalian cytosine RNA methyltransferases (m5C-RMTs) and the cytosine analog 5-azacytidin (5-aza-C) and developed an Aza-immunoprecipitation (Aza-IP) methodology to form stable m5C-RMT-RNA linkages in cell culture. The products were then immunoprecipitated and subjected to high-throughput sequencing. Similarly, RNA methyltransferase Nsun2 was also used to develop the methylation individual-nucleotide-resolution crosslinking and immunoprecipitation(miCLIP) [87] method to detect cytosine methylation in RNA species.
Profiling of 5′-ends with enrichment of 5′G-CAP
Methods and technologies have been well developed to map transcription start sites, such as the cap analysis of gene expression (CAGE) approach using Sanger sequencing [88]. This method has also been converted to next generation sequencing platforms to form deep CAGE, PEAT [paired-end analysis of TSSs (transcription start sites)], nanoCAGE and CAGEscan methodologies [89–91]. These methods were previously reviewed by Ozsolak and Milos [92]. The first step usually involves treatment of RNAs with bacterial alkaline phosphatase (BAP) to degrade the phosphate group of RNA without 5′G-cap. Tobacco acid pyrophosphatase (TAP) is then used to hydrolyze the phosphodiester bond of the 5′ triphosphate of an mRNA molecule and generate mRNA molecules with one phosphate group at the 5′-end. After that, the BAP–TAP-treated RNAs are ligated with sequence adaptors. As a result, only RNAs with cap structures can be sequenced [93, 94]. Using this strategy, Yamashita and colleagues generated 140 million TSS tags in 12 human cell types [94]. In addition, a modified method was developed to map transcriptional start sites based on 5′ cap catching [95, 96].
Future perspectives
Alternative transcription start sites, alterative splicing isoforms, and alternative polyadenylation sites present three major transcriptional events that significantly contribute to transcript variants and transcriptome diversity. Understanding transcriptome dynamics and patterns will certainly deliver new insights into mechanisms controlling many biological events and processes, such as the cell cycle and mitosis, nuclear reprogramming and stem cell biology, organogenesis and tissue remodeling during metamorphosis and regeneration, and innate response to pathogens and environmental challenges. As discussed above, the genome research community has been developing various methods and technologies to map alternative transcription start sites, assemble alternative splicing isoforms and profile alternative polyadenylation sites. In particular, next generation sequencing platforms, such as Illumina Genome Analyzers, Ion Torrent™ Sequencers and PacBio SMRT® Sequencing technology have accelerated the development of methods that produce large amounts of data. It is now time for the genome research community to assemble whole transcriptomes and collect signature targets for each gene/transcript, and thus use known genes/transcripts to determine known transcriptomes directly in near future.
Acknowledgments
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number R21HD076845 to ZJ. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest
The authors have declared that no competing interest exists.
References
- 1.Granjeaud S, Bertucci F, Jordan BR. Expression profiling: DNA arrays in many guises. Bioessays. 1999;21(9):781–790. doi: 10.1002/(SICI)1521-1878(199909)21:9<781::AID-BIES10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 2.Altman RB, Raychaudhuri S. Whole-genome expression analysis: challenges beyond clustering. Curr Opin Struct Biol. 2001;11(3):340–347. doi: 10.1016/s0959-440x(00)00212-8. [DOI] [PubMed] [Google Scholar]
- 3.Hsiao LL, Stears RL, Hong RL, Gullans SR. Prospective use of DNA microarrays for evaluating renal function and disease. Curr Opin Nephrol Hypertens. 2000;9(3):253–258. doi: 10.1097/00041552-200005000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Celis JE, Kruhøffer M, Gromova I, Frederiksen C, Ostergaard M, Thykjaer T, Gromov P, Yu J, Pálsdóttir H, Magnusson N, Orntoft TF. Gene expression profiling: monitoring transcription and translation products using DNA microarrays and proteomics. FEBS Lett. 2000;480(1):2–16. doi: 10.1016/s0014-5793(00)01771-3. [DOI] [PubMed] [Google Scholar]
- 5.Manger ID, Relman DA. How the host ‘sees’ pathogens: global gene expression responses to infection. Curr Opin Immunol. 2000;12(2):215–218. doi: 10.1016/s0952-7915(99)00077-1. [DOI] [PubMed] [Google Scholar]
- 6.Peffers MJ, Fang Y, Cheung K, Wei TK, Clegg PD, Birch HL. Transcriptome analysis of ageing in uninjured human Achilles tendon. Arthritis Res Ther. 2015;17(1):33. doi: 10.1186/s13075-015-0544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe R, Gemma C, Rakyan VK, Holland ML. Sexually dimorphic gene expression emerges with embryonic genome activation and is dynamic throughout development. BMC Genom. 2015;16(1):295. doi: 10.1186/s12864-015-1506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4(12):e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elkon R, Drost J, van Haaften G, Jenal M, Schrier M, Oude Vrielink JA, Agami R. E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol. 2012;13:R59. doi: 10.1186/gb-2012-13-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Z, Rokhsar DS, Harland RM. Old can be new again: HAPPY whole genome sequencing, mapping and assembly. Int J Biol Sci. 2009;5(4):298–303. doi: 10.7150/ijbs.5.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodkinson BP, Grice EA. Next-generation sequencing: a review of technologies and tools for wound microbiome research. Adv Wound Care (New Rochelle) 2015;4(1):50–58. doi: 10.1089/wound.2014.0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deschamps S, Llaca V, May GD. Genotyping-by-sequencing in plants. Biology (Basel) 2012;1(3):460–483. doi: 10.3390/biology1030460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252(5013):1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 15.Quackenbush J, Liang F, Holt I, Pertea G, Upton J. The TIGR gene indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 2000;28(1):141–145. doi: 10.1093/nar/28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. The TIGR gene indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 2001;29(1):159–164. doi: 10.1093/nar/29.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang DM, Dempsey AA, Lee CY, Liew CC. Identification of differentially expressed genes in cardiac hypertrophy by analysis of expressed sequence tags. Genomics. 2000;66(1):1–14. doi: 10.1006/geno.2000.6171. [DOI] [PubMed] [Google Scholar]
- 18.Nelson PS, Han D, Rochon Y, Corthals GL, Lin B, Monson A, Nguyen V, Franza BR, Plymate SR, Aebersold R, Hood L. Comprehensive analyses of prostate gene expression: convergence of expressed sequence tag databases, transcript profiling and proteomics. Electrophoresis. 2000;21(9):1823–1831. doi: 10.1002/(SICI)1522-2683(20000501)21:9<1823::AID-ELPS1823>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Z, Zhang M, Wasem VD, Michal JJ, Zhang H, Wright RW., Jr Census of genes expressed in porcine embryos and reproductive tissues by mining an expressed sequence tag database based on human genes. Biol Reprod. 2003;69(4):1177–1182. doi: 10.1095/biolreprod.103.018127. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Z, Wu XL, Garcia MD, Griffin KB, Michal JJ, Ott TL, Gaskins CT, Wright RW., Jr Comparative gene-based in silico analysis of transcriptomes in different bovine tissues and (or) organs. Genome. 2004;47(6):1164–1172. doi: 10.1139/g04-084. [DOI] [PubMed] [Google Scholar]
- 21.Wu XL, Griffin KB, Garcia MD, Michal JJ, Xiao Q, Wright RW, Jr, Jiang Z. Census of orthologous genes and self-organizing maps of biologically relevant transcriptional patterns in chickens (Gallus gallus) Gene. 2004;340(2):213–225. doi: 10.1016/j.gene.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 22.Rodríguez-Ezpeleta N, Teijeiro S, Forget L, Burger G, Lang BF. Construction of cDNA libraries: focus on protists and fungi. Methods Mol Biol. 2009;533:33–47. doi: 10.1007/978-1-60327-136-3_3. [DOI] [PubMed] [Google Scholar]
- 23.Okayama H, Berg P. High-efficiency cloning of full-length cDNA. Mol Cell Biol. 1982;2(2):161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okubo K, Hori N, Matoba R, Niiyama T, Fukushima A, Kojima Y, Matsubara K. Large scale cDNA sequencing for analysis of quantitative and qualitative aspects of gene expression. Nat Genet. 1992;2(3):173–179. doi: 10.1038/ng1192-173. [DOI] [PubMed] [Google Scholar]
- 25.Wan KH, Yu C, George RA, Carlson JW, Hoskins RA, Svirskas R, Stapleton M, Celniker SE. High-throughput plasmid cDNA library screening. Nat Protoc. 2006;1(2):624–632. doi: 10.1038/nprot.2006.90. [DOI] [PubMed] [Google Scholar]
- 26.Matsubara K, Okubo K. cDNA analyses in the human genome project. Gene. 1993;135(1–2):265–274. doi: 10.1016/0378-1119(93)90076-f. [DOI] [PubMed] [Google Scholar]
- 27.Gautheret D, Poirot O, Lopez F, Audic S, Claverie JM. Alternate polyadenylation in human mRNAs: a large-scale analysis by EST clustering. Genome Res. 1998;8(5):524–530. doi: 10.1101/gr.8.5.524. [DOI] [PubMed] [Google Scholar]
- 28.Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11(9):1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33(1):201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, McDonald H, Varhol R, Jones S, Marra M. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45(1):81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 31.Costa V, Angelini C, De Feis I, Ciccodicola A. Uncovering the complexity of transcriptomes with RNA-seq. J Biomed Biotechnol. 2010;2010:853916. doi: 10.1155/2010/853916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilhelm BT, Landry JR. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48(3):249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 34.Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, Eisman RC, Andrews J, Kaufman T, Cherbas P, Celniker SE, Graveley BR, Lai EC. Global patterns of tissue-specific alternative polyadenylation in Drosophila . Cell Rep. 2012;1(3):277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlackow M, Marguerat S, Proudfoot NJ, Bähler J, Erban R, Gullerova M. Genome-wide analysis of poly(A) site selection in Schizosaccharomyces pombe. RNA. 2013;19(12):1617–1631. doi: 10.1261/rna.040675.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finotello F, Di Camillo B. Measuring differential gene expression with RNA-seq: challenges and strategies for data analysis. Brief Funct Genomics. 2015;14:130–142. doi: 10.1093/bfgp/elu035. [DOI] [PubMed] [Google Scholar]
- 37.Pelechano V, Wilkening S, Järvelin AI, Tekkedil MM, Steinmetz LM. Genome-wide polyadenylation site mapping. Methods Enzymol. 2012;513:271–296. doi: 10.1016/B978-0-12-391938-0.00012-4. [DOI] [PubMed] [Google Scholar]
- 38.Baker KE, Parker R. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol. 2004;16:293–299. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao L, Fang Z, Zhang K, Zhi D, Cui X. Length bias correction for RNA-seq data in gene set analyses. Bioinformatics. 2011;27(5):662–669. doi: 10.1093/bioinformatics/btr005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12(3):R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rallapalli G, Kemen EM, Robert-Seilaniantz A, Segonzac C, Etherington GJ, Sohn KH, MacLean D, Jones JD. EXPRSS: an Illumina based high-throughput expression-profiling method to reveal transcriptional dynamics. BMC Genom. 2014;15:341. doi: 10.1186/1471-2164-15-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;9:34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steijger T, Abril JF, Engström PG, Kokocinski F, Hubbard TJ, Guigó R, Harrow J, Bertone P; RGASP Consortium Assessment of transcript reconstruction methods for RNA-seq. Nat Methods. 2013;10(12):1177–1184. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharon D, Tilgner H, Grubert F, Snyder M. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 2013;31(11):1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tilgner H, Grubert F, Sharon D, Snyder MP. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc Natl Acad Sci USA. 2014;111(27):9869–9874. doi: 10.1073/pnas.1400447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270(5235):484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 48.Saha S, Sparks AB, Rago C, Akmaev V, Wang CJ, Vogelstein B, Kinzler KW, Velculescu VE. Using the transcriptome to annotate the genome. Nat Biotechnol. 2002;20(5):508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- 49.Matsumura H, Reich S, Ito A, Saitoh H, Kamoun S, Winter P, Kahl G, Reuter M, Kruger DH, Terauchi R. Gene expression analysis of plant host-pathogen interactions by SuperSAGE. Proc Natl Acad Sci USA. 2003;100(26):15718–15723. doi: 10.1073/pnas.2536670100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spinella DG, Bernardino AK, Redding AC, Koutz P, Wei Y, Pratt EK, Myers KK, Chappell G, Gerken S, McConnell SJ. Tandem arrayed ligation of expressed sequence tags (TALEST): a new method for generating global gene expression profiles. Nucleic Acids Res. 1999;27(18):e22. doi: 10.1093/nar/27.18.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, Roth R, George D, Eletr S, Albrecht G, Vermaas E, Williams SR, Moon K, Burcham T, Pallas M, DuBridge RB, Kirchner J, Fearon K, Mao J, Corcoran K. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18(6):630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 52.Reinartz J, Bruyns E, Lin JZ, Burcham T, Brenner S, Bowen B, Kramer M, Woychik R. Massively parallel signature sequencing (MPSS) as a tool for in-depth quantitative gene expression profiling in all organisms. Brief Funct Genomic Proteomic. 2002;1(1):95–104. doi: 10.1093/bfgp/1.1.95. [DOI] [PubMed] [Google Scholar]
- 53.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 54.Asmann YW, Klee EW, Thompson EA, Perez EA, Middha S, Oberg AL, Therneau TM, Smith DI, Poland GA, Wieben ED, Kocher JP. 3′ tag digital gene expression profiling of human brain and universal reference RNA using Illumina Genome Analyzer. BMC Genom. 2009;10:531. doi: 10.1186/1471-2164-10-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsumura H, Urasaki N, Yoshida K, Krüger DH, Kahl G, Terauchi R. SuperSAGE: powerful serial analysis of gene expression. Methods Mol Biol. 2012;883:1–17. doi: 10.1007/978-1-61779-839-9_1. [DOI] [PubMed] [Google Scholar]
- 56.Prashar Y, Weissman SM. Analysis of differential gene expression by display of 3′ end restriction fragments of cDNAs. Proc Natl Acad Sci USA. 1996;93(2):659–663. doi: 10.1073/pnas.93.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richards M, Tan SP, Chan WK, Bongso A. Reverse serial analysis of gene expression (SAGE) characterization of orphan SAGE tags from human embryonic stem cells identifies the presence of novel transcripts and antisense transcription of key pluripotency genes. Stem Cells. 2006;24(5):1162–1173. doi: 10.1634/stemcells.2005-0304. [DOI] [PubMed] [Google Scholar]
- 58.Wu X, Liu M, Downie B, Liang C, Ji G, Li QQ, Hunt AG. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci USA. 2011;108(30):12533–12538. doi: 10.1073/pnas.1019732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang Z, Zhou X, Michal JJ, Wu XL, Zhang L, Zhang M, Ding B, Liu B, Manoranjan VS, Neill JD, Harhay GP, Kehrli ME, Jr, Miller LC. Reactomes of porcine alveolar macrophages infected with porcine reproductive and respiratory syndrome virus. PLoS One. 2013;8(3):e59229. doi: 10.1371/journal.pone.0059229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257(5072):967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 61.Bauer D, Warthoe P, Rohde M, Strauss M. Detection and differential display of expressed genes by DDRT-PCR. PCR Methods Appl. 1994;4(2):S97–S108. doi: 10.1101/gr.4.2.s97. [DOI] [PubMed] [Google Scholar]
- 62.Ma L, Pati PK, Liu M, Li QQ, Hunt AG. High throughput characterizations of poly(A) site choice in plants. Methods. 2014;67(1):74–83. doi: 10.1016/j.ymeth.2013.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mata J. Genome-wide mapping of polyadenylation sites in fission yeast reveals widespread alternative polyadenylation. RNA Biol. 2013;10(8):1407–1414. doi: 10.4161/rna.25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods. 2013;10(2):133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilkening S, Pelechano V, Järvelin AI, Tekkedil MM, Anders S, Benes V, Steinmetz LM. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41(5):e65. doi: 10.1093/nar/gks1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-seq. RNA. 2011;17(4):761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yao C, Shi Y. Global and quantitative profiling of polyadenylated RNAs using PAS-seq. Methods Mol Biol. 2014;1125:179–185. doi: 10.1007/978-1-62703-971-0_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22(6):1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ho ES, Gunderson SI, Duffy S. A multispecies polyadenylation site model. BMC Bioinform. 2013;14(Suppl 2):S9. doi: 10.1186/1471-2105-14-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kavakiotis I, Tzanis G, Vlahavas I. Polyadenylation site prediction using PolyA-iEP method. Methods Mol Biol. 2014;1125:131–140. doi: 10.1007/978-1-62703-971-0_11. [DOI] [PubMed] [Google Scholar]
- 71.Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM. Direct RNA sequencing. Nature. 2009;461(7265):814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 72.Ozsolak F, Milos PM. Transcriptome profiling using single-molecule direct RNA sequencing. Methods Mol Biol. 2011;733:51–61. doi: 10.1007/978-1-61779-089-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ozsolak F. Quantitative polyadenylation site mapping with single-molecule direct RNA sequencing. Methods Mol Biol. 2014;1125:145–155. doi: 10.1007/978-1-62703-971-0_13. [DOI] [PubMed] [Google Scholar]
- 74.Wu Q, Kim YC, Lu J, Xuan Z, Chen J, Zheng Y, Zhou T, Zhang MQ, Wu CI, Wang SM. Poly A- transcripts expressed in HeLa cells. PLoS One. 2008;3(7):e2803. doi: 10.1371/journal.pone.0002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12(2):R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X, Yin Q, Chen L, Yang L. Gene expression profiling of non-polyadenylated RNA-seq across species. Genomics Data. 2015;2:237–241. doi: 10.1016/j.gdata.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu X, Gorovsky MA. Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE) Nucleic Acids Res. 1993;21(21):4954–4960. doi: 10.1093/nar/21.21.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 80.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 84.Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37(2):e12. doi: 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Khoddami V, Cairns BR. Transcriptome-wide target profiling of RNA cytosine methyltransferases using the mechanism-based enrichment procedure Aza-IP. Nat Protoc. 2014;9(2):337–361. doi: 10.1038/nprot.2014.014. [DOI] [PubMed] [Google Scholar]
- 86.Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31(5):458–464. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, Frye M. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4(2):255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, Fukuda S, Sasaki D, Podhajska A, Harbers M, Kawai J, Carninci P, Hayashizaki Y. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci USA. 2003;100(26):15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Valen E, Pascarella G, Chalk A, Maeda N, Kojima M, Kawazu C, Murata M, Nishiyori H, Lazarevic D, Motti D, Marstrand TT, Tang MH, Zhao X, Krogh A, Winther O, Arakawa T, Kawai J, Wells C, Daub C, Harbers M, Hayashizaki Y, Gustincich S, Sandelin A, Carninci P. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19(2):255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ni T, Corcoran DL, Rach EA, Song S, Spana EP, Gao Y, Ohler U, Zhu J. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat Methods. 2010;7(7):521–527. doi: 10.1038/nmeth.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Plessy C, Bertin N, Takahashi H, Simone R, Salimullah M, Lassmann T, Vitezic M, Severin J, Olivarius S, Lazarevic D, Hornig N, Orlando V, Bell I, Gao H, Dumais J, Kapranov P, Wang H, Davis CA, Gingeras TR, Kawai J, Daub CO, Hayashizaki Y, Gustincich S, Carninci P. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat Methods. 2010;7(7):528–534. doi: 10.1038/nmeth.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12(2):87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsuchihara K, Suzuki Y, Wakaguri H, Irie T, Tanimoto K, Hashimoto S, Matsushima K, Mizushima-Sugano J, Yamashita R, Nakai K, Bentley D, Esumi H. Sugano S (2009) Massive transcriptional start site analysis of human genes in hypoxia cells. Nucleic Acids Res. 2009;37(7):2249–2263. doi: 10.1093/nar/gkp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamashita R, Sathira NP, Kanai A, Tanimoto K, Arauchi T, Tanaka Y, Hashimoto S, Sugano S, Nakai K, Suzuki Y. Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res. 2011;21(5):775–789. doi: 10.1101/gr.110254.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, Hess WR. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 2011;108(5):2124–2129. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cortes T, Schubert OT, Rose G, Arnvig KB, Comas I, Aebersold R, Young DB. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis . Cell Rep. 2013;5(4):1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]