Key Points
Question
Are specific single-nucleotide polymorphisms (SNPs) associated with increased risk of warfarin-associated bleeding among patients of African descent?
Findings
In this case-control genome-wide association study involving patients of African descent taking warfarin with an international normalized ratio of less than 4, four SNPs in linkage disequilibrium on chromosome 6 were associated with an increased risk of major bleeding in a discovery cohort of 215 patients (odds ratio [OR], 8.3) and a replication cohort of 188 patients (OR, 8.2), and reached genome-wide significance when the cohorts were combined using meta-analysis (OR, 8.3).
Meaning
Four single-nucleotide polymorphisms in linkage disequilibrium on chromosome 6 were associated with an increased risk of major bleeding among patients of African descent taking warfarin; however, validation of these findings in an independent prospective cohort is required.
Abstract
Importance
Major warfarin-related bleeding occurs more frequently in African Americans than in other populations. Identification of potential genetic factors related to this adverse event may help identify at-risk patients.
Objective
To identify genetic factors associated with warfarin-related bleeding in patients of African descent at an international normalized ratio (INR) of less than 4.
Design, Setting, and Participants
A case-control genome-wide association study involving patients of African descent taking warfarin was conducted in a discovery cohort (University of Chicago [2009-2011] and the University of Illinois at Chicago [2002-2011]), and associations were confirmed in a replication cohort (University of Chicago [2015-2016]). Potential population stratification was examined in the discovery cohort by principal component analysis. Odds ratios (ORs) and 95% CIs were computed for bleeding risk by logistic regression analysis. Summary statistics from the discovery and the replication cohorts were analyzed with a fixed effects meta-analysis. The potential influence of single-nucleotide polymorphisms (SNPs) on gene expression was studied by luciferase expression assays.
Exposures
Single-nucleotide polymorphisms associated with warfarin-related bleeding.
Main Outcomes and Measures
Major bleeding—defined as bleeding requiring hospitalization, causing a decrease in hemoglobin level of more than 2 g/dL, requiring blood transfusion, or any combination of the 3—while taking warfarin at an INR of less than 4.
Results
The discovery cohort consisted of 31 cases (mean age, 60.1 years [SD, 14.9 years], 26 women [83.9%]) and 184 warfarin-treated controls (mean age, 57.1 years [SD, 15.7 years]) with no documented bleeding. The replication cohort consisted of 40 cases (mean age, 55.6 years [SD, 17.3 years], 27 women [67.5%]), and 148 warfarin-treated controls (mean age, 55.4 years [SD, 17.1 years]; 98 women [66.2%]) with no documented bleeding. In the discovery cohort, 4 SNPs in linkage disequilibrium on chromosome 6 (rs115112393, rs16871327, rs78132896, and rs114504854) were associated with warfarin-related bleeding but did not reach genome-wide significance. The SNP rs78132896 occurred in 11 cases (35.5%) and 9 controls (4.9%) in the discovery cohort (OR, 8.31; 95% CI, 3.2-21.5; P < 6.21 × 10−8), and the association was confirmed in the replication cohort (the SNP was present in 14 cases [35.0%] and 7 controls [4.8%]; OR, 8.24; 95% CI, 3.1-25.3, P = 5.64 × 10−5). Genome-wide significance of this SNP was achieved when the cohorts were combined via meta-analysis (OR, 8.27; 95% CI, 4.18-16.38; P = 2.05 × 10−11). These SNPs are found only in people of African descent. In vitro luciferase expression assays demonstrated that rs16871327 (enhancer SNP) and rs78132896 (promoter SNP) risk alleles together increased EPHA7 gene (Entrez Gene 2045) transcription by a mean of 14.95 (SD, 1.7) compared with wild-type alleles (mean, 9.56 [SD, 0.84]; difference, 5.39; 95% CI, 4.1-6.6; P < .001).
Conclusions and Relevance
In this preliminary study involving patients of African descent taking warfarin, 4 single-nucleotide polymorphisms in linkage disequilibrium on chromosome 6 were associated with an increased risk of major bleeding at INR of less than 4. Validation of these findings in an independent prospective cohort is required.
This case-control genome-wide association study identifies single nucleotide polymorphisms (SNPs) associated with major bleeding in warfarin-treated patients of African descent.
Introduction
Warfarin is an effective prophylactic therapy against thromboembolic complications of vascular diseases.1 However, balancing the benefit of anticoagulation therapy with warfarin against the inherent risk of bleeding from the drug is challenging. The risk of major bleeding from warfarin increases by 0.3% to 0.5% per year, and the risk of intracranial hemorrhage by 0.2% per year.2 However, these values are based on studies conducted among predominantly white patients and do not account for differences in responsiveness to warfarin across ethnic groups.3 Patients of African descent have a significantly higher rate of thromboembolic diseases like venous thromboembolism and stroke4,5 and a higher risk of major bleeding from warfarin than do white patients.6 Although single-nucleotide polymorphisms (SNPs) associated with bleeding have been identified in populations of European ancestry,7,8,9,10,11 the genetic basis for hemorrhagic complications in patients of African descent has not been well studied. Identifying these variants may help physicians make safer choices in anticoagulation therapy for this understudied patient population.
The objective of this study was to identify genetic factors associated with warfarin-related bleeding among patients of African descent with an international normalized ratio (INR) of less than 4, a range in which most physicians do not consider warfarin reversal.
Methods
Participants
All participants provided written informed consent as part of the University of Chicago and the University of Illinois, Chicago institutional review board–approved protocols. All study participants were self-reported African Americans based on an open-ended question and were 18 years or older currently taking warfarin. The discovery cohort was a subset of the International Warfarin Pharmacogenetics Consortium African American genome-wide association study (GWAS) cohort12 recruited at the University of Chicago (2009-2011) and the University of Illinois at Chicago (2002-2011). The replication cohort was obtained through an algorithm-driven query of the University of Chicago’s Clinical Research Data Warehouse and its DNA biobank, which identified patients who routinely received care at the University of Chicago Hospitals, identified as African American, were taking warfarin, and had International Classification of Diseases, Ninth Revision (ICD-9) codes to identify whether they had had a suspected bleeding event. All patients identified via the query (2015-2016) were then classified as cases or controls by further manual curation to validate whether bleeding occurred while they were taking warfarin at the prespecified INR threshold or whether the patient continued taking warfarin for at least 1 year with no event (eMethods in the Supplement).
Determination of Bleeding Phenotype in the Discovery and Replication Cohort
The goal of this study was to identify genetic variation associated with bleeding risk; hence, a phenotype that may have less influence from environmental causes of warfarin-related bleeding was chosen. At INR thresholds of more than 4, nongenomic factors (eg, drug-drug interactions, organ dysfunction, unintentional overdose) may have a greater association with bleeding risk than genomic factors. Thus, cases were limited to those who bled at INRs of less than 4 to provide a potential enrichment for people with a genetic predisposition to bleed. Additionally, the bleeding definition outlined below mirrors the adverse event definition in several warfarin pharmacogenomic clinical trials that report INRs of more than 4 as an adverse drug reaction. Patients with major bleeding occurring at an INR of less than 4 were considered as cases, whereas the controls were patients taking warfarin with no documented bleeding events for at least 1 year. If patients died from a fatal warfarin-related bleed within 1 year of warfarin initiation, they were retained as cases in the analysis. Major bleeding was defined as bleeding requiring hospitalization, causing a decrease in hemoglobin level of more than 2 g/dL, or requiring blood transfusion.13 Therefore, any symptom of warfarin-associated bleeding requiring hospitalization, in any area or organ (unlike the International Society on Thrombosis and Haemostasis definition14), was considered as a major bleed, to provide broader clinical applicability.
Genotyping
The discovery cohort was genotyped with the Illumina 610 Quad BeadChip (Illumina) at the RIKEN Center for Genomic Medicine, Yokohama, Japan.12 Quality control measures such as principal component analysis for potential population stratification, thresholds for SNP and sample call rates, minor allele frequency thresholds, identity by descent exclusion to ensure only unrelated individuals were retained in the analysis and Hardy-Weinberg equilibrium were used. This was followed by postimputation info-metric thresholds, used to remove poorly imputed SNPs with low imputation certainty (detailed in eMethods). A total of 8 152 232 SNPs passed quality control filters and were used for analysis. The replication cohort was genotyped by Sanger sequencing. Because CYP2C9 (Entrez Gene 1559) star variants are associated with increased risk of warfarin-associated bleeding,15 the replication cohort was genotyped for CYP2C9* variants via TaqMan genotyping. Potential functional effects of the significant variants were predicted by the Genome Wide Annotation of Variants (GWAVA),16 a tool that predicts the influence of noncoding genetic variants based on a wide range of annotations of noncoding elements.
Luciferase Reporter Assay
The potential influence of these SNPs on gene expression was studied by an in vitro functional assay. The pGL4.10[luc2] no-promoter vector (Promega Corp) containing a 6–kilobase (kb) region upstream of the EPHA7 gene (Entrez Gene 2045) was constructed, and the haplotype pGL4.10[luc2] vector containing SNPs prioritized by their GWAVA scores was generated by mutagenesis (GenScript). Human umbilical vein endolthelial cells (HUVEC) (Lonza) were transfected with 0.2 μg/well of empty vector, wild-type or mutant vector, and 0.06 μg/well of Renilla-luciferase pGL4.74[hRluc2/TK] plasmid (Promega) using the Cytofect-HUVEC Transfection Kit (Cell Applications) in 24-well plates in triplicates. After 16 hours, a dual-luciferase assay was performed according to the manufacturer’s protocols (Promega). All luciferase luminescence values were normalized to both Renilla luminescence (to control for cell-plating and transfection efficiency) and empty vector.
Bleeding Risk Prediction
To determine if the addition of an associated SNP (rs78132896) from the identified haplotype was associated with changes in bleeding risk prediction over known clinical factors, the replication cohort was scored based on the HAS-BLED (hypertension, abnormal renal and liver function, stroke, bleeding, labile INR, elderly, and drugs or alcohol) bleeding risk prediction scheme.13 A calculated HAS-BLED score ranges from 0 through 9, based on 9 parameters. This score assigns 1 point for each of the following risk factors: uncontrolled hypertension (>160 mm Hg systolic), abnormal renal function (dialysis, transplant, creatinine >200 mg/dL [to convert creatinine from μmol/L to mg/dL, divide by 88.4]), liver dysfunction (cirrhosis or bilirubin >2 × normal or aspartate aminotransferase, alanine aminotransferase, or alkaline phosphatase >3 × normal), stroke, prior major bleeding, older age (>65 years), medication usage predisposing to bleeding (antiplatelet agents, nonsteroidal anti-inflammatory drugs), and alcohol use (≥8 drinks/week). A high score (≥3) is indicative of increased risk of major bleeding. The discovery cohort could not be scored because sufficient clinical details at warfarin initiation were not available. Because the aim of this analysis was to evaluate the predictive accuracy of the HAS-BLED scoring system in determining bleeding risk prior to anticoagulation therapy, labile INR was scored at 0, as done previously in studies predicting bleeding risk with anticoagulant initiation.17,18
Statistical Analysis
To identify SNPs associated with warfarin-related bleeding, a case-control GWAS was conducted in the discovery cohort and significant SNPs with the lowest P values were studied in the replication cohort. Summary statistics from the discovery and replication cohorts were analyzed in a fixed-effect meta-analysis using METAL version 2011,19 with the assumption that the genetic effect size would be the same across the 2 cohorts of patients of African descent with the same phenotypic outcome. A 2-sided P value of 1.62 × 10−8 was considered the significance threshold for the discovery cohort20 and meta-analysis. In the replication cohort, a 2-sided P value <.05 was considered significant. Association analysis was conducted by logistic regression using an additive model. Heterogeneity of the associations across the 2 cohorts was assessed by the Cochran Q statistic. Principal component 1 (P = .016) and principal component 2 (P = .015), which reflected the genetic variation due to ancestry, showed an association with the bleeding phenotype and were thus included as covariates in the genetic analysis. Genetic analysis was conducted using SNPTEST version 2.5.2 for the discovery cohort and PLINK version 1.9 for the replication cohort. The frequency of the identified SNPs was compared among populations using data obtained from the 1000 Genomes project. Previously identified SNPs associated with bleeding disorders were tested for association with warfarin-related bleeding in the discovery cohort. The gene-region plot of the top SNP associations was generated with LocusZoom version 0.4.8.21 Differences in baseline characteristics between cases and controls were assessed by a χ2 test or Fisher exact test for categorical variables and a t test for continuous parameters. The mean INR at which bleeding occurred was determined. The predictive accuracy of the HAS-BLED score was determined by the receiver operating characteristic (ROC) curve. To test if combining the associated SNP (rs78132896) with the HAS-BLED score improved bleeding risk prediction, an additive genetic risk model was used to include the SNP into the HAS-BLED scheme. The SNP was coded as 0, 1, or 2 based on copies of the risk allele. A generalized linear model with a binomial distribution and a logit link function was generated, using bleeding outcome as the dependent variable and SNP and HAS-BLED score as the predictor variables. Missing data were handled by the listwise deletion method. Statistical analyses were performed using R version 3.4.2 statistical software. Additional information regarding the methods can be found in the eMethods section of the Supplement.
Results
The discovery cohort comprised 31 cases with major bleeding occurring at an INR of less than 4, and 184 controls with no documented history of any warfarin-related bleeding, whereas the replication cohort consisted of 40 cases and 161 controls. However, 13 controls from the replication cohort were removed from the cohort because no genotypes were created for these samples due to failure of the assay. In the discovery cohort, the mean (SD) age was 60.1 years (14.9 years) in cases and 57.1 years (15.7 years) in controls. There were 26 women (83.9%) among the cases and 128 women (69.6%) among the controls. In the replication cohort, the mean (SD) age was 55.6 years (17.3 years) in cases and 55.4 years (17.1 years) in controls. There were 27 women (67.5%) among cases and 98 women (66.2%) among controls. Mean age, weight, warfarin dosing, or sex did not differ significantly between cases and controls (Table 1). The principal component analysis showed that the discovery cohort clustered between the HapMap northern and western Europe (CEU) and Yoruba in Ibadan (YRI) samples (eFigure 1 in the Supplement). Among those with major bleeding events, 10 patients (32%) from the discovery cohort and 14 patients (35%) from the replication cohort had more than 1 major bleeding event. Gastrointestinal bleeding was the most common bleeding site in both the discovery (62%) and replication (55%) cohorts (eTable 1 in the Supplement). Abnormal renal function was significantly associated with bleeding in both the discovery (P < .001) and replication (P = .006) cohorts; therefore, all subsequent analyses were adjusted for abnormal renal function defined as dialysis, transplant, or creatinine concentrations of more than 2.6 mg/dL.
Table 1. Demographic and Clinical Characteristics of the Study Groups.
| Variable | Discovery Cohort | Replication Cohort | ||
|---|---|---|---|---|
| Cases (n = 31) |
Controls (n = 184) |
Cases (n = 40) |
Controlsa (n = 148) |
|
| Age, mean (SD), y | 60.1 (14.9) | 57.1 (15.7) | 55.6 (17.3) | 55.4 (17.1) |
| Weight, mean (SD), kg | 88.35 (22.4) | 91.89 (27.8) | 93.5 (13.6) | 96.27 (21.3) |
| Warfarin maintenance dose, mg/wk | 40.89 (20.4) | 46.07 (19.1) | 41.5 (16.2) | 44.6 (16.5) |
| INR at the time of bleed, mean (SD) | 2.67 (0.87) | 2.27 (0.83) | ||
| Sex, No. (%) | ||||
| Women | 26 (83.9) | 128 (69.6) | 27 (67.5) | 98 (66.2) |
| Men | 5 (16.1) | 56 (30.4) | 13 (32.5) | 50 (33.8) |
| Abnormal renal function, No. (%)b | 10 (32.3) | 27 (14.7) | 20 (50) | 40 (27) |
| HAS-BLED risk factors, No. (%)c | ||||
| Systolic blood pressure, >160 mm Hg uncontrolled | 8 (20) | 5 (3.4) | ||
| Abnormal renal functionb | 20 (50) | 40 (27) | ||
| Abnormal liver functiond | 5 (12.5) | 11 (7.4) | ||
| Stroke | 7 (17.5) | 37 (25) | ||
| Prior major bleeding | 27 (67.5) | 59 (40) | ||
| Age > 65 y | 18 (45) | 71 (48) | ||
| Antiplatelet agents or NSAIDs | 29 (72.5) | 93 (62.8) | ||
| 8 Alcoholic drinks/wk | 5 (12.5) | 30 (20.3) | ||
Abbreviations: HAS-BLED, hypertension, abnormal renal or liver function, stroke, bleeding history, labile international normalized ratio (INR), elderly, antiplatelet agents or nonsteroidal anti-inflammatory drugs (NSAIDs), or alcohol use.
Thirteen controls from the replication cohort failed genotyping, leading to decrease in controls to 148.
Abnormal renal function, defined as presence of chronic dialysis, renal transplantation, or serum creatinine ≥200 μmol/L (to convert creatinine from μmol/L to mg/dL, divide by 88.4).
Clinical details for HAS-BLED scheme were available only for the replication cohort.
Cirrhosis or bilirubin at more than 2 × normal or aspartate aminotransferase, alanine aminotransferase, or alkaline phosphatase more than 3 × normal.
Genetic Variants and Warfarin-Related Bleeding Outcome
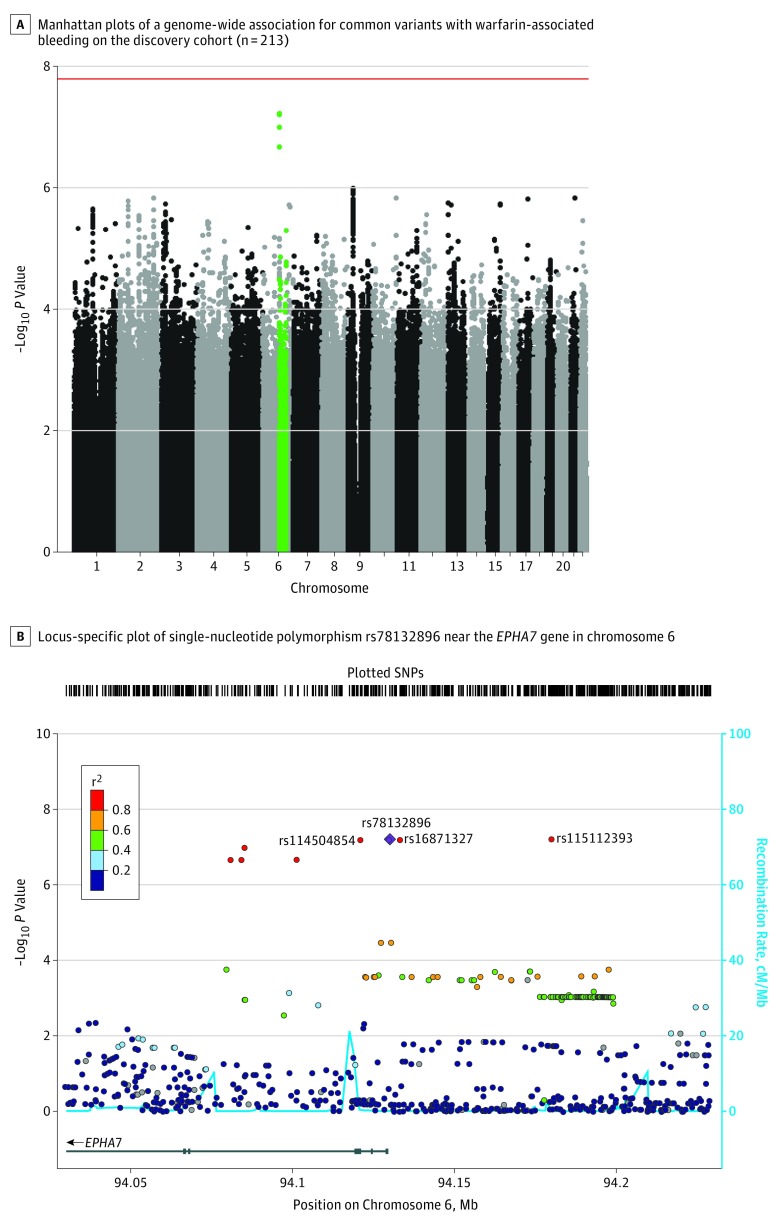
In the discovery cohort, 4 SNPs in linkage disequilibrium on chromosome 6 (rs115112393, rs16871327, rs78132896, and rs114504854) were associated with warfarin-related bleeding but did not reach genome-wide significance (Figure, A). Because the allele of 1 polymorphism in a haplotype can predict the allele of adjacent nongenotyped polymorphisms, the replication cohort was genotyped for only the rs78132896 SNP, which occurred in 11 cases (35.5%) and 9 controls (4.9%) in the discovery cohort (odds ratio [OR], 8.31; 95% CI, 3.2-21.5; P < 6.21 × 10−8), and the association was confirmed in the replication cohort (the SNP was present in 14 cases [35.0%] and 7 controls [4.8%]; OR, 8.24; 95% CI, 3.1-25.3, P = 5.64 × 10−5; Table 2 and Table 3). Genome-wide significance of this SNP was achieved when the cohorts were combined via meta-analysis (OR, 8.27; 95% CI, 4.18-16.38; P = 2.05 × 10−11), with no significant heterogeneity between cohorts detected (Cochran Q statistic P = .44). For the replication cohort, the association of the rs78132896 SNP with bleeding risk was analyzed after adjusting for the HAS-BLED score (OR, 9.38; 95% CI, 3.4-26.1; P = 1.81 × 10−5). The associated haplotype is found only in people of African ancestry (eTable 4 in the Supplement). No significant associations were observed between warfarin-related bleeding events and CYP2C9*2 (rs1799853 dbSNP; P = .40), CYP2C9*3 (rs1057910 dbSNP; P = .47), CYP2C9*5 (rs28371686 dbSNP; P = .47), CYP2C9*8 (rs7900194 dbSNP; P = .69), CYP2C9*11 (rs28371685 dbSNP; P = .12), or presence of any CYP2C9* variants (P = .50) in the replication cohort. The previously identified rs12777823 (dbSNP),12 associated with warfarin dosing among patients of African descent, showed no association with bleeding in the discovery cohort (P = .44). Previously identified SNPs associated with bleeding disorders like polycythemia vera,10 Von Willebrand disease,8,11 and Bernard-Soulier syndrome9 did not show any significant association with bleeding in the discovery cohort (eTable 5 in the Supplement).
Figure. Single-Nucleotide Polymorphisms Associated With Bleeding Risk in the African American Discovery Cohort.
A, Single-nucleotide polymorphisms (SNPs) are plotted on the x-axis according to their positions on each chromosome against association with warfarin-associated bleeding on the y-axis (−log10 P value). The red line shows genome-wide significant threshold of P = 1.62 x 10−8. Green points toward the top of graph indicate the near-significant association found in chromosome 6.
B, The colors of the circles denote linkage disequilibrium (r2) between rs78132896 (purple diamond) and nearby SNPs (based on pairwise r2 values from the 1000 Genomes Project African population). The 3 near-significant SNPs in high linkage disequilibrium with rs78132896 are shown. The blue line and right y-axis show the estimated recombination rate (obtained from HapMap). The x-axis represents the genomic position in chromosome 6 and the left y-axis represents the –log10 P of association with warfarin-related bleeding in the discovery cohort (n = 215). Genes at this locus are indicated in the lower panel of the plot. Chromosomal positions are based on hg19 genome build.
Table 2. Single-Nucleotide Polymorphisms on Chromosome 6 Associated With Major Bleeding Risk in the Discovery and Replication Cohort.
| Single-Nucleotide Polymorphism | Minor Allele | Minor Allele Frequency, % | Discovery Cohort (n = 215) |
Replication Cohort (n = 188)a |
Meta-analysis (n = 403) |
|||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | |||
| rs115112393 | G | 5 | 8.35 (3.2-21.5)b | 5.9 × 10−8 | ND | ND | ND | ND |
| rs16871327 | T | 5 | 8.31 (3.2-21.5)b | 6.21 × 10−8 | ND | ND | ND | ND |
| rs78132896c | C | 5 | 8.31 (3.2-21.5)b | 6.21 × 10−8 | 8.24 (3.1-25.3)d | 5.64 × 10−5 | 8.27 (4.18-16.38) | 2.05 × 10−11 |
| 9.38 (3.4-26.1)e | 1.81 × 10−5 | |||||||
| rs114504854 | A | 5 | 8.30 (3.2-21.5)b | 6.23 × 10−8 | ND | ND | ND | ND |
Abbreviations: ND, not determined; OR, odds ratio.
Thriteen controls from the replication cohort failed genotyping, leading to decrease in sample size to 188.
Adjusted for abnormal renal function, principal component 1, principal component 2.
Meta-analysis P value of the discovery and replication cohorts for rs78132896.
Adjusted for abnormal renal function.
Adjusted for HAS-BLED (HAS-BLED: hypertension, abnormal renal or liver function, stroke, bleeding history, labile international normalized ratio, elderly, antiplatelet agents, nonsteroidal anti-inflammatory drugs or alcohol use).
Table 3. Genotypes for rs78132896 T > C in the Discovery and Replication Cohorts.
| No. of Patients | rs78132896 Genotypes No. (%) | ||
|---|---|---|---|
| Wild-Type (TT) | Heterozygous (TC) | ||
| Discovery cohort | |||
| Cases | 31 | 20 (64.5) | 11 (35.5) |
| Controls | 184 | 175 (95.1) | 9 (4.9) |
| Replication cohort | |||
| Cases | 40 | 26 (65) | 14 (35) |
| Controls | 148 | 141 (95.2) | 7 (4.8) |
Homozygous CC individuals for the rs78132896 were not captured due to the small sample size.
Functional In Vitro Assay of the Variants
The rs78132896 SNP is a promoter regulatory–region variant, upstream of the EPHA7 gene (Figure, B). Both the rs78132896 and rs16871327 SNPs were prioritized by their highest GWAVA scores (eTable 2 in the Supplement) to construct a 6-kb haplotype vector. Luciferase reporter assays showed that the rs16871327 and rs78132896 risk alleles together increased EPHA7 gene transcription (measured as normalized luciferase activity) by a mean (SD) of 14.95 (1.7) compared with a mean 9.56 (0.84) of wild-type alleles (difference, 5.39; 95% CI, 4.1-6.6, P < .001).
Bleeding Risk Prediction
The association between individual components of the HAS-BLED scheme and bleeding risk in the replication cohort is shown in Table 1. The mean (SD) HAS-BLED score was 3.53 (1.26) in cases and 2.64 (1.53) in controls (P = .001). Among all the HAS-BLED covariates, only uncontrolled hypertension (P = .001), abnormal renal function (P = .006), and prior major bleeding (P = .002) were associated with major bleeding. A HAS-BLED score of 3 or higher was associated with increased risk of major bleeding (OR, 3.01; 95% CI, 1.34-6.76; P = .008). A HAS-BLED classification with a threshold of 3 (eTable 3 in the Supplement) had a sensitivity of 77.5% (95% CI, 69%-89%) and specificity of 46.6% (95% CI, 38%-0.55%).
The rs78132896 SNP (as a tag for the entire haplotype), had a C statistic of 0.65 (95% CI, 0.58-0.73) (eFigure 5 in the Supplement). Inclusion of the rs78132896 SNP to the HAS-BLED scheme (with a threshold of 3) increased the C statistic significantly: for major bleeding, the HAS-BLED plus the SNP C statistic was 0.78 (95% CI, 0.70-0.85) and the HAS-BLED alone C statistic was 0.67 (95% CI, 0.59-0.74; C statistic difference, 0.11; 95% CI, 0.10-0.12; P = .003). Reclassification analyses showed the inclusion of the rs78132896 SNP correctly reclassified 12.5% of major bleeding events, but incorrectly reclassified 0.7% of no bleeding events, leading to an net reclassification improvement of 11.8% (P = .02), thereby increasing sensitivity to 90% (95% CI, 75%-97%) and decreasing specificity to 45.9% (95% CI, 38%-54%). The integrated discrimination index (IDI) between HAS-BLED score alone vs HAS-BLED score plus SNP was also statistically significant (IDI, 13.6%; P = .<001) (eFigure 4 in the Supplement). Because the gastrointestinal tract was the most common site of bleeding, the predictive performance of the HAS-BLED scheme was also tested for gastrointestinal bleeding in the replication cohort (eFigure 3 and eFigure 4 in the Supplement).
Discussion
This study identified a novel association of a haplotype in patients of African descent with significantly increased risk of major bleeding while taking warfarin. The associated SNPs are relatively common in among persons of African descent, as evident by the minor allele frequency of 7% in persons of African descent from the 1000 Genomes Project.22 The haplotype is found exclusively in populations with African ancestry and therefore may represent a population-specific genetic risk factor that exposes African Americans to a higher risk of bleeding during warfarin therapy.
Single-nucleotide polymorphism rs78132896 is located 791 bp upstream of the ephrin receptor 7 (EPHA7) within the promoter region. The EPHA7 gene is expressed in peripheral lymphocytes and vascular endothelial cells, with increased expression during inflammation.23,24 Based on ChIP-sequencing data from the Encyclopedia of DNA Elements (ENCODE) consortium,25 the rs78132896 SNP is located in a binding site for the Kruppel-associated box–associated protein-1 (Entrez Gene 10155) (transcriptional repressor) and chromodomain helicase DNA binding protein 1 (Entrez Gene 9557), which associates with specific chromatin regions and mediate transcriptional control.26,27 Likewise, the rs16871327 (pairwise-linkage disequilibrium rs78132896 r2 = 1) SNP is located in a CTCF transcriptional binding site. Data from the Genotype-Tissue Expression (GTEx) project was accessed to determine the potential effect of the SNPs on EPHA7 expression. However, these significant SNPs were found in less than 1% of the GTEx cohort; hence, this cohort is underpowered to find an association with gene expression. The GTEx project may not be an appropriate data set to uncover the gene expression effects of this particular finding, given the low number of African Americans with both genotype and gene expression data and the rarity of the SNPs.
A luciferase assay showed increased activity of the haplotype construct compared with the wild-type in HUVEC, suggesting that the rs78132896 and rs16871327 SNPs are regulatory variants that together affect the transcriptional regulation of EPHA7 leading to its increased expression. Ephrin receptor–ephrin (ligand) interactions support thrombus growth and stability by regulating integrin signaling in activated platelets and thus may play an important role in hemostatic plug formation.28,29 The process also involves activation of leukocytes and endothelial cells, increased leukocyte adhesion and chemotaxis. The EPHA7 protein has been shown to promote cell repulsion and block ephrin receptor–ephrin interactions.23 Based on these findings, we hypothesize that the increased expression of the EPHA7 gene can lead to bleeding in patients who are taking warfarin and carrying the risk variant by inhibiting ephrin receptor–ephrin interaction. Additional functional work is needed to establish this mechanism as well as validation of the tissue-specific effect. Because the EPHA7 gene does not appear to exert effects directly within warfarin’s known mechanism of action, this haplotype might also have potential implications for bleeding risk with direct oral anticoagulants. Additional studies may further clarify the mechanism underlying the functional effects associated with this haplotype and the role of the EPHA7 gene in the adverse event profile of oral anticoagulants.
The HAS-BLED performance in our cohort was similar to that in other populations, as reflected by the C statistic in white patients (C statistic, 0.65)30 and Asians (C statistic, 0.64).31 Including the rs78132896 SNP in the HAS-BLED scheme correctly reclassified 12.5% of cases and significantly improved bleeding risk prediction. Although evaluation of the risk of bleeding is challenging, this study suggests that considering clinical and genetic risk factors together may lead to better bleeding risk prediction. However, improvement in bleeding risk prediction by SNP inclusion to the HAS-BLED scheme is a preliminary finding and needs to be validated prospectively.
The CYP2C9 gene variants have been associated with bleeding risk from warfarin,15 but these bleeding events occur primarily during the initial phase of therapy and can be predicted by the INR responses.32 This study showed no association of CYP2C9 variants with bleeding. Moreover, the mean (SD) INR at which bleeding occurred was 2.67 (0.87) in the discovery cohort and 2.27 (0.83) in the replication cohort; values that are least predictive of bleeding outcome, and a level at which physicians do not consider warfarin reversal.33 Therefore the use of 1 of the haplotype SNPs may help guide physicians’ drug choice and therapeutic monitoring of high-risk patients.
Limitations
This study has several limitations. First, the small sample size did not allow for identification of a genome-wide significant signal in the discovery cohort. However, the 4 SNPs in linkage disequilibrium did achieve the prespecified genome-wide significance threshold on meta-analysis of the combined cohorts. Second, the discovery cohort could not be scored for the HAS-BLED schema due to the lack of sufficient clinical details, and therefore improvement in bleeding risk prediction by the addition of the associated SNP to HAS-BLED schema in the replication cohort should be considered as a preliminary finding.
Third, both the study cohorts were recruited at the same clinical location, and it is possible that unmeasured confounding might have influenced the results of this study. Although the medical record number of the patients recruited at the University of Chicago for the discovery and replication cohorts were matched to prevent patient overlap, patient relatedness between the 2 cohorts could not be tested by identity by descent, due to lack of genome-wide data for the replication cohort. Fourth, the C statistic CIs of the models (HAS-BLED score plus SNP vs HAS-BLED score alone) overlap. However, the CIs may overlap substantially and yet yield a statistically significant result.34 Fifth, 13 controls from the replication cohort were not able to be genotyped. Given that the individuals removed were similar in both demographics and clinical attributes to the remaining controls, the removal of these individuals should have had little effect on the results.
Conclusions
In this preliminary study involving patients of African descent taking warfarin, 4 single-nucleotide polymorphisms in linkage disequilibrium on chromosome 6 were associated with an increased risk of major bleeding at INR of less than 4. Validation of these findings in an independent prospective cohort is required.
eMethods. Determination of bleeding phenotype, quality control and SNP imputation, HAS-BLED scoring for bleeding risk prediction, and statistical analyses
eFigure 1. Principal component analysis of the discovery cohort and three HapMap populations
eFigure 2. Quantile-quantile (QQ) plot of the discovery cohort
eFigure 3. Receiver operating curve (ROC) for HAS-BLED+SNP vs HAS-BLED for Major bleeding and Gastro-intestinal bleeding in the replication cohort
eFigure 4. Predictive accuracy of HAS-BLED and HAS-BLED plus SNP for detection of major bleeding and gastrointestinal (GI) bleeding in the replication cohort
eFigure 5. ROC curves for rs78132896 single nucleotide polymorphism (SNP) for major and gastrointestinal bleeding in the replication cohort
eTable1. Warfarin-related bleeding events by sites in the discovery and the replication cohort cases.
eTable 2. Genome Wide Annotation of Variants (GWAVA) scores for the significant SNPs
eTable 3. Distribution of HAS-BLED scores for major bleeding outcome in the replication cohort
eTable 4. Allele Frequency Distribution of the haplotype (rs115112393, rs16871327, rs78132896, rs114504854) in populations from the 1000 Genomes Project Phase 3 (EUR- European, ASN- Asian, AFR- African, ASW- Americans of African Ancestry in SW USA)
eTable 5. Risk alleles of bleeding disorders and their significance in the Discovery Cohort
eTable 6. Performance of ATRIA and HAS-BLED models for detection of major bleeding in the replication cohort
References
- 1.Adam SS, McDuffie JR, Ortel TL, Nagi A, Williams JW Jr. Comparative Effectiveness of Warfarin and Newer Oral Anticoagulants for the Long-Term Prevention and Treatment of Arterial and Venous Thromboembolism. Washington, DC: Quality Enhancement Research Initiative; 2012. [PubMed] [Google Scholar]
- 2.Schulman S, Beyth RJ, Kearon C, Levine MN. Hemorrhagic complications of anticoagulant and thrombolytic treatment: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines (8th Edition). Chest. 2008;133(6 suppl):257S-298S. [DOI] [PubMed] [Google Scholar]
- 3.Dang MT, Hambleton J, Kayser SR. The influence of ethnicity on warfarin dosage requirement. Ann Pharmacother. 2005;39(6):1008-1012. doi: 10.1345/aph.1E566 [DOI] [PubMed] [Google Scholar]
- 4.Pandey DK, Gorelick PB. Epidemiology of stroke in African Americans and Hispanic Americans. Med Clin North Am. 2005;89(4):739-752, vii. doi: 10.1016/j.mcna.2005.02.005 [DOI] [PubMed] [Google Scholar]
- 5.White RH, Keenan CR. Effects of race and ethnicity on the incidence of venous thromboembolism. Thromb Res. 2009;123(suppl 4):S11-S17. doi: 10.1016/S0049-3848(09)70136-7 [DOI] [PubMed] [Google Scholar]
- 6.Limdi NA, Brown TM, Shendre A, Liu N, Hill CE, Beasley TM. Quality of anticoagulation control and hemorrhage risk among African American and European American warfarin users. Pharmacogenet Genomics. 2017;27(10):347-355. doi: 10.1097/FPC.0000000000000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wadelius M, Chen LY, Lindh JD, et al. . The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113(4):784-792. doi: 10.1182/blood-2008-04-149070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campos M, Sun W, Yu F, et al. . Genetic determinants of plasma von Willebrand factor antigen levels: a target gene SNP and haplotype analysis of ARIC cohort. Blood. 2011;117(19):5224-5230. doi: 10.1182/blood-2010-08-300152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noris P, Perrotta S, Bottega R, et al. . Clinical and laboratory features of 103 patients from 42 Italian families with inherited thrombocytopenia derived from the monoallelic Ala156Val mutation of GPIBΑ (Bolzano mutation). Haematologica. 2012;97(1):82-88. doi: 10.3324/haematol.2011.050682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Percy MJ, McMullin MF. The V617F JAK2 mutation and the myeloproliferative disorders. Hematol Oncol. 2005;23(3-4):91-93. doi: 10.1002/hon.761 [DOI] [PubMed] [Google Scholar]
- 11.van Loon J, Dehghan A, Weihong T, et al. . Genome-wide association studies identify genetic loci for low von Willebrand factor levels. Eur J Hum Genet. 2016;24(7):1035-1040. doi: 10.1038/ejhg.2015.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perera MA, Cavallari LH, Limdi NA, et al. . Genetic variants associated with warfarin dose in African-American individuals: a genome-wide association study. Lancet. 2013;382(9894):790-796. doi: 10.1016/S0140-6736(13)60681-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pisters R, Lane DA, Nieuwlaat R, de Vos CB, Crijns HJ, Lip GY. A novel user-friendly score (HAS-BLED) to assess 1-year risk of major bleeding in patients with atrial fibrillation: the Euro Heart Survey. Chest. 2010;138(5):1093-1100. doi: 10.1378/chest.10-0134 [DOI] [PubMed] [Google Scholar]
- 14.Schulman S, Kearon C; Subcommittee on Control of Anticoagulation of the Scientific and Standardization Committee of the International Society on Thrombosis and Haemostasis . Definition of major bleeding in clinical investigations of antihemostatic medicinal products in non-surgical patients. J Thromb Haemost. 2005;3(4):692-694. doi: 10.1111/j.1538-7836.2005.01204.x [DOI] [PubMed] [Google Scholar]
- 15.Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnet systematic review and meta-analysis. Genet Med. 2005;7(2):97-104. doi: 10.1097/01.GIM.0000153664.65759.CF [DOI] [PubMed] [Google Scholar]
- 16.Ritchie GR, Dunham I, Zeggini E, Flicek P. Functional annotation of noncoding sequence variants. Nat Methods. 2014;11(3):294-296. doi: 10.1038/nmeth.2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kooiman J, van Hagen N, Iglesias Del Sol A, et al. . The HAS-BLED score identifies patients with acute venous thromboembolism at high risk of major bleeding complications during the first six months of anticoagulant treatment. PLoS One. 2015;10(4):e0122520. doi: 10.1371/journal.pone.0122520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen PB, Larsen TB, Lip GYH. Recalibration of the HAS-BLED score: should hemorrhagic stroke account for one or two points? Chest. 2016;149(2):311-314. doi: 10.1378/chest.15-1509 [DOI] [PubMed] [Google Scholar]
- 19.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190-2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li MX, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant P-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131(5):747-756. doi: 10.1007/s00439-011-1118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruim RJ, Welch RP, Sanna S, et al. . LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336-2337. doi: 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abecasis GR, Altshuler D, Auton A, et al. ; 1000 Genomes Project Consortium . A map of human genome variation from population-scale sequencing [published correction appears in Nature. 2011;473(7348):544]. Nature. 2010;467(7319):1061-1073. doi: 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawson DW, Hong JS, Shen RR, et al. . Global DNA methylation profiling reveals silencing of a secreted form of EPHA7 in mouse and human germinal center B-cell lymphomas. Oncogene. 2007;26(29):4243-4252. doi: 10.1038/sj.onc.1210211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Y, Lan W, Wu W, et al. . Increased expression of EPHA7 in inflamed human dental pulp. J Endod. 2013;39(2):223-227. doi: 10.1016/j.joen.2012.11.020 [DOI] [PubMed] [Google Scholar]
- 25.Thurman RE, Rynes E, Humbert R, et al. . The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75-82. doi: 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Persson J, Ekwall K. CHD1 remodelers maintain open chromatin and regulate the epigenetics of differentiation. Exp Cell Res. 2010;316(8):1316-1323. doi: 10.1016/j.yexcr.2010.02.029 [DOI] [PubMed] [Google Scholar]
- 27.Bunch H, Calderwood SK. TRIM28 as a novel transcriptional elongation factor. BMC Mol Biol. 2015;16:14. doi: 10.1186/s12867-015-0040-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prévost N, Woulfe DS, Jiang H, et al. . Eph kinases and ephrins support thrombus growth and stability by regulating integrin outside-in signaling in platelets. Proc Natl Acad Sci U S A. 2005;102(28):9820-9825. doi: 10.1073/pnas.0404065102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prevost N, Woulfe D, Tanaka T, Brass LF. Interactions between Eph kinases and ephrins provide a mechanism to support platelet aggregation once cell-to-cell contact has occurred. Proc Natl Acad Sci U S A. 2002;99(14):9219-9224. doi: 10.1073/pnas.142053899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Apostolakis S, Lane DA, Guo Y, Buller H, Lip GY. Performance of the HEMORR(2)HAGES, ATRIA, and HAS-BLED bleeding risk-prediction scores in patients with atrial fibrillation undergoing anticoagulation: the AMADEUS (evaluating the use of SR34006 compared to warfarin or acenocoumarol in patients with atrial fibrillation) study. J Am Coll Cardiol. 2012;60(9):861-867. doi: 10.1016/j.jacc.2012.06.019 [DOI] [PubMed] [Google Scholar]
- 31.Suzuki M, Matsue Y, Nakamura R, Matsumura A, Hashimoto Y. Improvement of HAS-BLED bleeding score predictive capability by changing the definition of renal dysfunction in Japanese atrial fibrillation patients on anticoagulation therapy. J Cardiol. 2014;64(6):482-487. doi: 10.1016/j.jjcc.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 32.Li C, Schwarz UI, Ritchie MD, Roden DM, Stein CM, Kurnik D. Relative contribution of CYP2C9 and VKORC1 genotypes and early INR response to the prediction of warfarin sensitivity during initiation of therapy. Blood. 2009;113(17):3925-3930. doi: 10.1182/blood-2008-09-176859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia DA, Crowther MA. Reversal of warfarin: case-based practice recommendations. Circulation. 2012;125(23):2944-2947. doi: 10.1161/CIRCULATIONAHA.111.081489 [DOI] [PubMed] [Google Scholar]
- 34.Greenland S, Senn SJ, Rothman KJ, et al. . Statistical tests, P values, confidence intervals, and power: a guide to misinterpretations. Eur J Epidemiol. 2016;31(4):337-350. doi: 10.1007/s10654-016-0149-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Determination of bleeding phenotype, quality control and SNP imputation, HAS-BLED scoring for bleeding risk prediction, and statistical analyses
eFigure 1. Principal component analysis of the discovery cohort and three HapMap populations
eFigure 2. Quantile-quantile (QQ) plot of the discovery cohort
eFigure 3. Receiver operating curve (ROC) for HAS-BLED+SNP vs HAS-BLED for Major bleeding and Gastro-intestinal bleeding in the replication cohort
eFigure 4. Predictive accuracy of HAS-BLED and HAS-BLED plus SNP for detection of major bleeding and gastrointestinal (GI) bleeding in the replication cohort
eFigure 5. ROC curves for rs78132896 single nucleotide polymorphism (SNP) for major and gastrointestinal bleeding in the replication cohort
eTable1. Warfarin-related bleeding events by sites in the discovery and the replication cohort cases.
eTable 2. Genome Wide Annotation of Variants (GWAVA) scores for the significant SNPs
eTable 3. Distribution of HAS-BLED scores for major bleeding outcome in the replication cohort
eTable 4. Allele Frequency Distribution of the haplotype (rs115112393, rs16871327, rs78132896, rs114504854) in populations from the 1000 Genomes Project Phase 3 (EUR- European, ASN- Asian, AFR- African, ASW- Americans of African Ancestry in SW USA)
eTable 5. Risk alleles of bleeding disorders and their significance in the Discovery Cohort
eTable 6. Performance of ATRIA and HAS-BLED models for detection of major bleeding in the replication cohort