Abstract
A unique chemically induced proximity method is engineered based on mutant antibody VL domain using a fluorogenic malachite green derivative as the inducer, which gives fluorescent signals upon VL domain dimerization while simultaneously inducing downstream biological effects.
Graphical Abstract
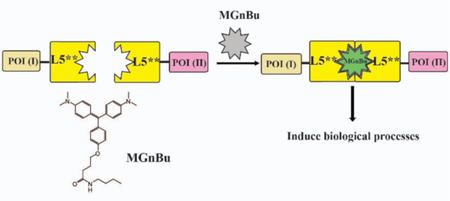
Authors are required to submit a graphic entry for the Table of Contents (TOC) that, in conjunction with the manuscript title, should give the reader a representative idea of one of the following: A key structure, reaction, equation, concept, or theorem, etc., that is discussed in the manuscript. Consult the journal’s Instructions for Authors for TOC graphic specifications.
The chemically induced proximity (CIP), or chemically induced dimerization (CID), method uses a small molecule inducer that homo- or hetero-dimerizes two unique inducer-binding protein domains fused individually to any two proteins of interest (POIs). Depending on the choice of linked POIs, a variety of downstream cellular events can be triggered temporally by the chemical inducer 1–4. Several CIP systems have been engineered that each uses a unique chemical inducer to dimerize a corresponding pair of inducer-binding proteins 5–14. These orthogonal CIP systems allow multiple biological events to be controlled simultaneously and independently in the same cell and the construction of sophisticated bio-computation systems 15. Recently, several studies have shown that these CIP inducers can be chemically modified to become activatable by artificial or endogenous cellular signals that greatly expands the utility of CIP methods in synthetic biology 16–21. One limitation of existing CIP systems is that the inducer-triggered dimerization cannot be directly monitored, therefore, the kinetics of dimerization and that between dimerization and the induced downstream effects is difficult to follow.
Fluorogen activating proteins (FAPs) derived from single chain antibodies (scFv) induce fluorescent signals upon binding from certain organic dyes (fluorogens) which are otherwise non-fluorescent when free in solution 22–33. Structural studies of the malachite green (MG-based FAP system revealed that MG forms a 1:2 ligand:protein complex with two L5** proteins, the VL domain of an antibody, which leads to the activation of intense fluorescence 34. Importantly, these L5** protein domains do not self-dimerize in the absence of dye, and association is induced in the presence of fluorogenic dye. We reason that by linking two POIs individually to each L5** protein, a new MG inducible CIP system can be built that can trigger downstream effects through MG-induced VL-POI dimerization and also self-report the dimerization by activation of MG fluorescent signals upon dimerization, produc-ing a self-reporting chemically induced proximity system (Figure 1).
Figure 1.
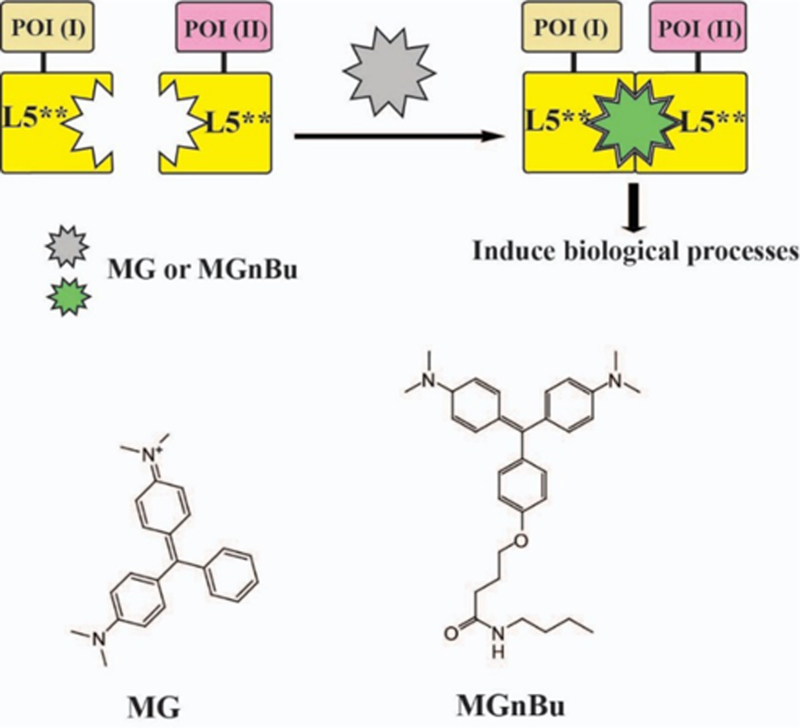
(A) MG-based chemically induced proximity method to control biological processes.
To create an MG-based fluorogenic CIP system, we first constructed an MG inducible transcriptional activation system based on L5**-fusion proteins (Figure 1). We generated DNA plasmids encoding the MG-inducible gene expression cassette including a yeast Gal4 DNA binding domain (Gal4DBD)-L5** and a herpes simplex virusVP16 transactivation domain (VP16AD)-L5** fusion proteins. A reporter construct with five copies of the upstream activation sequence (UAS) that Gal4DBD recognizes inserted upstream of a luciferase gene was used to test the transcriptional activation. In this system, luciferase expression can be turned on only when MG induces hetero-dimerization of GAL4DBD-L5** and L5**-VP16AD. Furthermore, fluorescence will be observed only when MG forms a complex with two L5** fusion proteins. Although the homo-dimerization of two identical L5** fusion proteins can also induce MG fluorescence, we expect that any activated fluorescence will report combined occurring dimerization events from both hetero- and homo-dimerizations and provide the information regarding the dimerization kinetics.
To test this system, we co-transfected HEK293T cells with the MG-inducible gene expression cassette and the UAS-luciferase reporter (Figure 2A) for 24 h and then treated cells with 500 nM MG or a more cell-permeant MG derivative, MGnBu 35 (Figure 1) for another 24 h. Under these conditions, MG and MGnBu were expected to induce protein interactions and drive reporter expression. Treated cells were then washed and analyzed under a fluorescence plate reader to detect the activated MG fluorescence (excitation 640 nm/emission 670 nm) or lysed for luciferase assays. A 70-fold (for MG) to 250-fold (for MGnBu) fluorescence increase from activated MG fluorescence was observed (Figure 2B), as well as a 3-fold increase in luciferase expression in the case of MGnBu induction (Figure 2C). This study showed that an MG-based fluorogenic CIP system can be established and the observed activated fluorescence of MGnBu is correlated with the observed induced luciferase expression. The higher induction efficiency of MGnBu as compared to MG is likely due to the superior cell permeability of MGnBu. The low induction fold in luciferase expression is likely a result of the competition between the hetero- and homo-dimerization of GAL4DBD-L5** and L5**-VP16AD. Since MGnBu gave the highest fluorescent signal upon dimerization and more effectively induced the luciferase expression, we focused on MGnBu as the inducer for the following studies.
Figure 2.
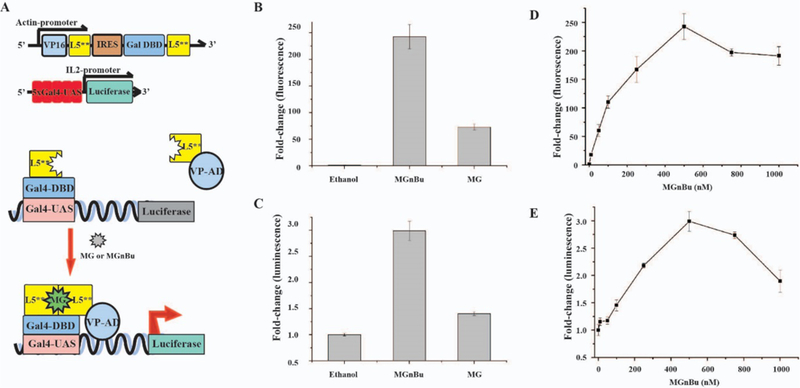
(A) Scheme and constructs of MG-induced luciferase expression system. (B) Activated MG fluorescence fold changes and (C) induced luciferase expression in HEK293T cells by MG and MGnBu. Dosage response of (D) activated MGnBu fluorescence fold change and (E) Induced luciferase expression in HEK293T cells. For (B) through (E), the cells were transfected with the constructs shown in (A) for 24 h before addition of 500 nM MG or MGnBu (B) and (C), or 0–1000 nM of MGnBu (D) and (E), for another 24 h. Cells were washed and analyzed by fluorescence plate reader in (B) and (D) or luciferase assays in (C) and (E). For all experiments, the induced fold change was calculated by comparison to the values of ethanol-treated samples. Independent experiments were repeated five or more times. Errors bars are SD (n = 5).
We next tested if there is a dosage dependence in this new MG-based CIP system as seen in some other CIP systems. We transfected HEK293T cells with the MG-inducible gene expression cassette and the UAS-luciferase reporter for 24 h, and then treated cells with MGnBu (from 0 to 1000 nM) for another 24 h. We observed that activated MGnBu fluorescence showed a dosage-dependent increase and gave the highest signal at 500 nM (Figure 2D). A similar dosage-dependent increase was observed in luciferase induction, which also reached the highest expression at 500 nM (Figure 2F). Decreases in the activated MGnBu fluorescence signal and luciferase expression were observed when cells were treated with higher than 500 nM of MGnBu. It is known that when a bi-functional ligand was used at high concentrations, the dimerization efficiency can reduce due to the saturation of the ligand binding site on each dimerizable protein by excess ligand 36. A similar effect can potentially account for the observed phenomena. Another possibility to cause the observed effects is the potential cytotoxicity of MGnBu at high concentrations. As a result, we tested the viability of cells treated with 10 to 1000 nM of MGnBu and analyzed using MTT assays. Although a gradual decrease in cell viability was detected at higher concentrations of MGnBu (Figure S1), the results are not consistent with the significant decreases observed in fluorescence signals and luciferase expression.
Next, we tested if the MG-induced CIP system can be used to control protein translocation. We constructed DNA plasmids encoding the nuclear export sequence-tagged L5** (NES-L5**) and the EYFP-L5** fusion proteins (Figure 3A). We transfected CHO cells with both constructs for 24 h and followed by treating cells with 500 nM MGnBu (or ethanol as a negative control). Cells were then collected 2 h after MGnBu addition for the analysis of MGnBu fluorescence activation and nuclear exporting of EYFP-L5**. We quantified the fold changes of activated MGnBu fluorescence due to dimerization with a fluorescence plate reader. The results showed that there were up to 1200-fold increasing after 2 h incubation (Figure 3B). Meanwhile, the subcellular distribution of EYFP-L5** proteins was monitored under a fluorescence microscope. When expressed in cells, EYFP-L5** proteins were found to freely cross the nuclear envelope and distributed throughout the cell, although a preferential nuclear localization was observed (Figure 3C). In the presence of MGnBu, the dimerization between of EYFP-L5** and NES-L5** led to the translocation of EYFP-L5** out of the nucleus (Figure 3C). To further confirm that the nuclear localization of EYFP-L5** is due to the MGnBu induced dimerization with NES-L5**, we transfected cells with plasmids encoding EYFP-ABI and NES-L5** or EYFP-ABI only, and then treated cells with or without 500 nM MGnBu. The results showed that EYFP-ABI was distributed evenly over the cell in the presence of MGnBu and no activated MGnBu fluorescence signal was observed (Figure 3D), indicating that L5** was required to activate MGnBu fluorescence. When we transfected cells with EYFP-ABI & NES-L5** (that EYFP and NES fusion proteins cannot be heter-odimerized by MGnBu) and treated cells with MGnBu, no translocation of EYFP was observed (Figure 3D), which confirmed that the observed nuclear exportation of EYFP in Figure 3C was resulted from the heterodimerization of EYFP-L5** and NES-L5**. Although no nuclear exportation of EYFP-ABI was observed, we observed the activated red fluorescence signal in the cytoplasm (Figure 3D), indicating the self-dimerization of NES-L5** occurred as expected. We quantified the percentage of cells showing nuclear exportation (defined by a lower EYFP fluorescence intensity inside the nucleus versus outside). More than 30 percent of cells showing nucleus exportation after 2 h incubation with MGnBu (Figure 3E). Also, the co-localization under the channels of EYFP and the MGnBu fluorescence was examined, which gave a Pearson’s correlation coefficient of 0.96 (Figure 3F), indicating a significantly high correlation between these two signals.
Figure 3.
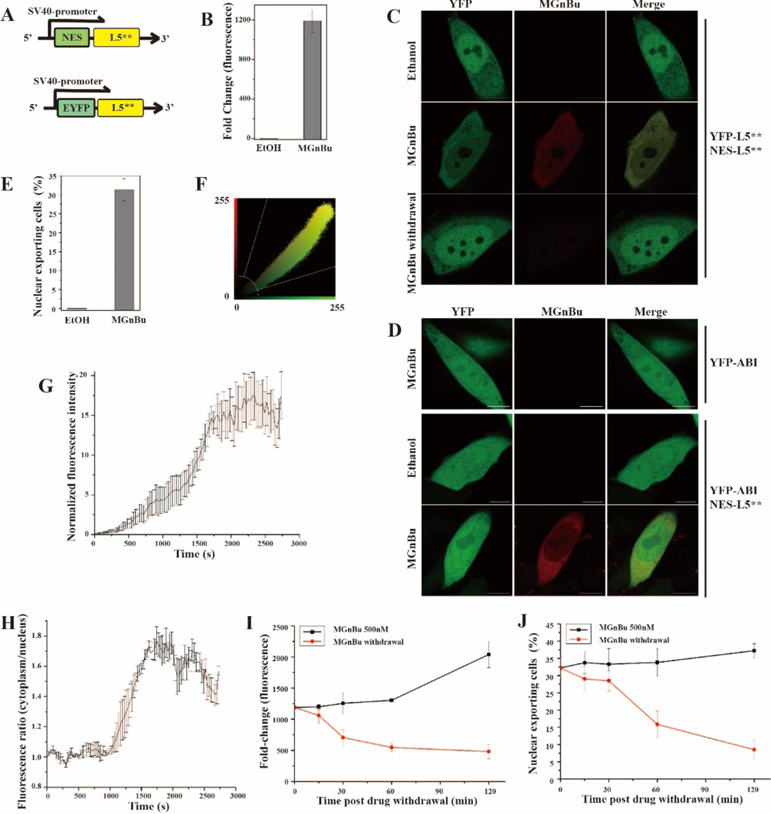
(A) DNA constructs for EYFP translocation experiments. (B) Activated MGnBu fluorescence fold change (2h). (C) Representative images of transfected CHO cells with ethanol, or with 500 nM MGnBu, or removal of MGnBu after treatment. Scale bar is 10 µm. (D) Representative images of transfected CHO cells with ethanol, or with 500 nM MGnBu. Scale bar is 10 µm. (E) The percentage of cells showing EYFP nuclear exporting in transfected CHO cells treated with MGnBu for 2 h. (F) Colocalization analysis for red (MGnBu) channel and YFP channel with 2h MGnBu treatment as shown in (C), with Pearson’s correlation coefficient r = 0.96. (G) Normalized EYFP fluorescence ratio of cytoplasm over nucleus after MGnBu treatment. The initial ratio was normalized to 1. Data is the average of four independent cells. (H) Normalized fluorescence intensity under MGnBu fluorescence channel. The initial fluorescence intensity was normalized to 1. Data is the average of four independent cells. (I) & (J) Time course (0 to 2 h) of activated MGnBu fluorescence fold change (I) and the percentage of cells showing EYFP nuclear exportation (J) after MGnBu withdrawal in transfected CHO cells originally treated with MGnBu for 4h. For (B)(E)(G)(H)(I)(J), cells were transfected with the constructs in (A) for 24 h before treating with 500 nM MGnBu for indicated time periods. In reversibility experiments, cells were washed and collected after indicated time periods for analysis by fluorescence plate reader in (B) and (H) or under a fluorescence microscope in (E) and (J). The statistical data in (E) and (J) were obtained by counting cells showing EYFP unclear exporting over the total EYFP positive cells to give the percentage of translocated cells. Cells were counted from three separate experiments, and 100–200 cells were counted for each sample. Error bars are SD (n = 3).
To compare the kinetics of MGnBu-induced protein dimerization (indicated by activated MGnBu fluorescence) and the resulting translocation of EYFP-L5*, we transfected CHO cells with both NES-L5** and EYFP-L5**constructs and treated cells with 500 nM MGnBu. We performed live-cell imaging following individual cells after adding MGnBu under a confocal microscope. We quantified in real time the MGnBu fluorescence and the EYFP fluorescence intensity inside and outside of the nucleus in each single cell. We observed a rapid increase in the fluorescence of MGnBu (indicating the dimerization of EYFP fusion proteins) within 5 min, which continued to increase during the observation time period up to 1 h (Figure 3G). The nuclear exportation of EYFP-L5** occurred and continued to increase shortly after the dimerization was induced (Figure 3H), and the kinetics of these two events are well correlated. MGnBu can also induce the homodimerization of EYFP-L5**, which may lead to the exclusion of homodimerized EYFP-L5** from the nucleus due to the increased size and gives the observed results. To examine this possibility, we transfected cells with only EYFP-L5** (without NES-L5**) and then treated cells with MGnBu for 24 h. We observed that a much lower percentage of cells showing nuclear exportation upon MGnBu addition in the absence of NES-L5** when compared to the condition with NES-L5** transfected, although a low degree of nuclear exclusion was indeed observed (Figure S2).
To investigate whether the MG-induced CIP system is reversible, after CHO cells were transfected with EYFP-L5** and NES-L5** constructs and treated with MGnBu for 2 h, cells were washed with fresh media (without MGnBu) and collected at different time points after washing for analysis. The activated MGnBu fluorescence fold change and the percentage of cells showing nuclear exportation were quantified as described above. We observed that both the activated MGnBu fluorescence and the induced EFYP-L5** translocation was readily reversible and correlated with each other (Figure 3C, 3I, 3J).
Finally, we tested whether this new MG-based CIP system is orthogonal to other existing CIP systems including abscisic acid (ABA)-based system that dimerizes ABI/PYL fusion proteins 13, gibberellin acid (GA)-based system that dimerizes GAI/GID1 fusion proteins 14, and rapamycin (Rap)-based system that dimerize FRB/FKBP fusion proteins 5. We constructed a new plasmid encoding GA-inducible transcriptional activation cassette (Figure 4A-iii) and used the MG, ABA and Rap inducible constructs that were developed in this study or previously (Figure 4A). We co-transfected CHO cells with a UAS luciferase reporter and individually with each of these inducible transcriptional activation cassettes for 24 h. MGnBu, GA-AM, ABA, Rap or ethanol (negative control) was then separately added to each transfected condition for another 24 h. Afterwards, cells were collected and analysed by a fluorescence plate reader for activated inducer fluorescence or by luciferase assays for induced luciferase expression. We observed that only cells transfected with the MG-inducible cassette and treated with MGnBu gave detectable fluorescence signals (Figure 4B). We also confirmed that these four tested CIP systems are orthogonal in that only the corresponding inducer can dimerize the matching fusion protein pairs to induce luciferase expression (Figure 4C). The MGnBu induced CIP system is the only method that allows direct monitoring the induced dimerization and also triggering downstream cellular effects.
Figure 4.
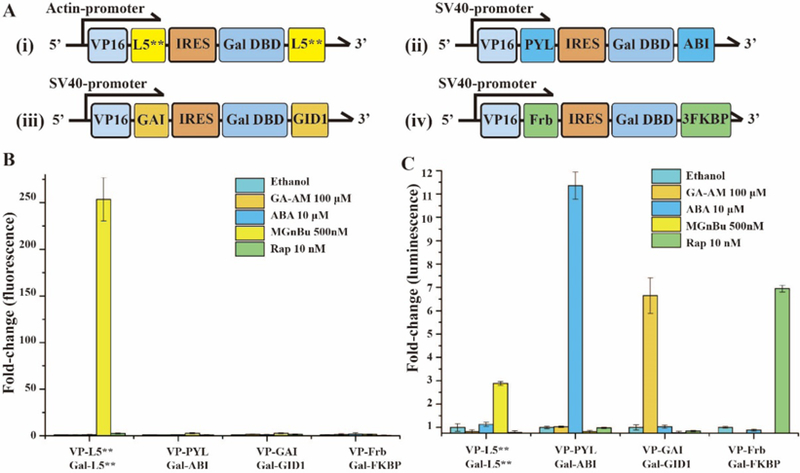
(A) DNA constructs for inducible gene expression induced by (i) MGnBu, (ii) ABA, (iii) GA, (iv) Rap. (B) Activated inducer fluorescence fold change and (C) induced luciferase expression after transfected HEK293T cells were treated by different CIP inducers for 24 h. For all experiments, the induction fold change was calculated relative to the values of non-induced (i.e. ethanol treated) samples. Independent experiments were repeated five or more times. Errors bars are SD (n = 5).
In summary, we developed a unique new fluorogenic CIP system that uses an MG derivative (MGnBu) as an inducer that is able to trigger specific downstream cellular events through the dimerization of two L5** fusion proteins and also generates fluorescence signal upon forming the ternary complex. This is the first CIP system that is able to achieve direct reporting of dimerization. Although MGnBu showed certain toxicity at higher concentrations and the induced effects are not as high as other CIP systems that dimerize two non-identical inducer-binding proteins, we expect that new MG derivatives and FAP-based MG binding proteins can be engineered based on this work to offer new and improved fluorogenic CIP systems with specific heterodimerization. In addition to the range of MG-analog binding FAP proteins, a variety of additional fluorogens with cognate fluorogen activating proteins, several of which have been found to form ternary complexes sandwiching a single ligand suggests that a range of additional CIP systems could be derived from FAP-fluorogen pairs 27, 37.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health R21 HG008776 (FSL, GZ) and R21NS092019 (MB, YW). We thank Prof. Diane S. Lidke at the University of New Mexico to provide the MG compounds.
Footnotes
The authors declare no competing financial interests.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: xx.xxxx/acs.bioconj-chem.xxxxxxx.
Experimental methods and materials for mammalian cell culture and transfection, cloning and plasmid construction, fluorescence microscopy, live cell imaging experiment for CHO cells with nuclear export experiments and its analysis, statistical analysis of cell population. Cytotoxicity of MGnBu tested using MTT assays. (Figure S1), Analysis of nuclear exclusion of dimerized EYFP-L5** (Figure S2). (PDF)
REFERENCES
- (1).Fegan A; White B; Carlson JC; Wagner CR, Chemically controlled protein assembly: techniques and applications. Chemical reviews 2010, 110 (6), 3315–36. [DOI] [PubMed] [Google Scholar]
- (2).Gestwicki JE; Marinec PS, Chemical control over protein-protein interactions: beyond inhibitors. Combinatorial chemistry & high throughput screening 2007, 10 (8), 667–75. [DOI] [PubMed] [Google Scholar]
- (3).DeRose R; Miyamoto T; Inoue T, Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Archiv : European journal of physiology 2013, 465(3), 409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Voss S; Klewer L; Wu YW, Chemically induced dimerization: reversible and spatiotemporal control of protein function in cells. Current opinion in chemical biology 2015, 28, 194–201. [DOI] [PubMed] [Google Scholar]
- (5).Spencer DM; Wandless TJ; Schreiber SL; Crabtree GR, Controlling signal transduction with synthetic ligands. Science 1993, 262 (5136), 1019–24. [DOI] [PubMed] [Google Scholar]
- (6).Ho SN; Biggar SR; Spencer DM; Schreiber SL; Crabtree GR, Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature 1996, 382 (6594), 822–6. [DOI] [PubMed] [Google Scholar]
- (7).Lin HN; Abida WM; Sauer RT; Cornish VW, Dexamethasone-methotrexate: An efficient chemical inducer of protein dimerization in vivo. Journal of the American Chemical Society 2000, 122 (17), 4247–4248. [Google Scholar]
- (8).Clemons PA; Gladstone BG; Seth A; Chao ED; Foley MA; Schreiber SL, Synthesis of calcineurin-resistant derivatives of FK506 and selection of compensatory receptors. Chem Biol 2002, 9 (1), 49–61. [DOI] [PubMed] [Google Scholar]
- (9).Bayle JH; Grimley JS; Stankunas K; Gestwicki JE; Wandless TJ; Crabtree GR, Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem Biol 2006, 13(1), 99–107. [DOI] [PubMed] [Google Scholar]
- (10).Czlapinski JL; Schelle MW; Miller LW; Laughlin ST; Kohler JJ; Cornish VW; Bertozzi CR, Conditional glycosylation in eukaryotic cells using a biocompatible chemical inducer of dimerization. Journal of the American Chemical Society 2008, 130 (40), 13186–13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Skwarczynska M; Molzan M; Ottmann C, Activation of NF-kappaB signalling by fusicoccin-induced dimerization. Proceedings of the National Academy of Sciences of the United States of America 2013, 110(5), E377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Erhart D; Zimmermann M; Jacques O; Wittwer MB; Ernst B; Constable E; Zvelebil M; Beaufils F; Wymann MP, Chemical development of intracellular protein heterodimerizers. Chem Biol 2013, 20(4), 549–57. [DOI] [PubMed] [Google Scholar]
- (13).Liang FS; Ho WQ; Crabtree GR, Engineering the ABA plant stress pathway for regulation of induced proximity. Science signaling 2011, 4 (164), rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Miyamoto T; DeRose R; Suarez A; Ueno T; Chen M; Sun TP; Wolfgang MJ; Mukherjee C; Meyers DJ; Inoue T, Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. Nature chemical biology 2012, 8 (5), 465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Miyamoto T; Razavi S; DeRose R; Inoue T, Synthesizing biomolecule-based Boolean logic gates. ACS synthetic biology 2013, 2 (2), 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zeng G; Zhang R; Xuan W; Wang W; Liang FS, Constructing de novo H2O2 signaling via induced protein proximity. ACS chemical biology 2015, 10 (6), 1404–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zeng G; Li H; Wei Y; Xuan W; Zhang R; Breden LE; Wang W; Liang FS, Engineering Iron Responses in Mammalian Cells by Signal-Induced Protein Proximity. ACS synthetic biology 2017, 6 (6), 921–927. [DOI] [PubMed] [Google Scholar]
- (18).Wright CW; Guo ZF; Liang FS, Light Control of Cellular Processes by Using Photocaged Abscisic Acid. Chembiochem : a European journal of chemical biology 2015, 16 (2), 254–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Karginov AV; Zou Y; Shirvanyants D; Kota P; Dokholyan NV; Young DD; Hahn KM; Deiters A, Light Regulation of Protein Dimerization and Kinase Activity in Living Cells Using Photocaged Rapamycin and Engineered FKBP. Journal of the American Chemical Society 2011, 133 (3), 420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Umeda N; Ueno T; Pohlmeyer C; Nagano T; Inoue T, A Photocleavable Rapamycin Conjugate for Spatiotemporal Control of Small GTPase Activity. Journal of the American Chemical Society 2011, 133 (1), 12–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Brown KA; Zou Y; Shirvanyants D; Zhang J; Samanta S; Mantravadi PK; Dokholyan NV; Deiters A, Light-cleavable rapamycin dimer as an optical trigger for protein dimerization. Chem Commun 2015, 51 (26), 5702–5705. [DOI] [PubMed] [Google Scholar]
- (22).Feldhaus MJ; Siegel RW; Opresko LK; Coleman JR; Feldhaus JM; Yeung YA; Cochran JR; Heinzelman P; Colby D; Swers J, et al. , Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nature biotechnology 2003, 21 (2), 163–70. [DOI] [PubMed] [Google Scholar]
- (23).Fisher GW; Adler SA; Fuhrman MH; Waggoner AS; Bruchez MP; Jarvik JW, Detection and quantification of beta2AR internalization in living cells using FAP-based biosensor technology. Journal of biomolecular screening 2010, 15 (6), 703–9. [DOI] [PubMed] [Google Scholar]
- (24).Szent-Gyorgyi C; Schmidt BF; Creeger Y; Fisher GW; Zakel KL; Adler S; Fitzpatrick JA; Woolford CA; Yan Q; Vasilev KV, et al. , Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nature biotechnology 2008, 26 (2), 235–40. [DOI] [PubMed] [Google Scholar]
- (25).Ozhalici-Unal H; Pow CL; Marks SA; Jesper LD; Silva GL; Shank NI; Jones EW; Burnette JM 3rd; Berget PB; Armitage BA, A rainbow of fluoromodules: a promiscuous scFv protein binds to and activates a diverse set of fluorogenic cyanine dyes. Journal of the American Chemical Society 2008, 130 (38), 12620–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Szent-Gyorgyi C; Schmidt BF; Fitzpatrick JAJ; Bruchez MP, Fluorogenic Dendrons with Multiple Donor Chromophores as Bright Genetically Targeted and Activated Probes. Journal of the American Chemical Society 2010, 132 (32), 11103–11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Zanotti KJ; Silva GL; Creeger Y; Robertson KL; Waggoner AS; Berget PB; Armitage BA, Blue fluorescent dye-protein complexes based on fluorogenic cyanine dyes and single chain antibody fragments. Organic & biomolecular chemistry 2011, 9 (4), 1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Telmer CA; Verma R; Teng H; Andreko S; Law L; Bruchez MP, Rapid, specific, no-wash, far-red fluorogen activation in subcellular compartments by targeted fluorogen activating proteins. ACS chemical biology 2015, 10 (5), 1239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Xu SN; Hu HY, Fluorogen-activating proteins: beyond classical fluorescent proteins. Acta Pharm Sin B 2018, 8 (3), 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Saurabh S; Perez AM; Comerci CJ; Shapiro L; Moerner WE, Super-resolution Imaging of Live Bacteria Cells Using a Genetically Directed, Highly Photostable Fluoromodule. Journal of the American Chemical Society 2016, 138 (33), 10398–10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ackerman DS; Vasilev KV; Schmidt BF; Cohen LB; Jarvik JW, Tethered Fluorogen Assay to Visualize Membrane Apposition in Living Cells. Bioconjugate chemistry 2017, 28 (5), 1356–1362. [DOI] [PubMed] [Google Scholar]
- (32).Boeck JM; Spencer JV, Effect of human cytomegalovirus (HCMV) US27 on CXCR4 receptor internalization measured by fluorogen-activating protein (FAP) biosensors. PloS one 2017, 12 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhang QY; Wang QH; Sun Y; Zuo LM; Fetz V; Hu HY, Superior Fluorogen-Activating Protein Probes Based on 3-Indole-Malachite Green. Org Lett 2017, 19 (17), 4496–4499. [DOI] [PubMed] [Google Scholar]
- (34).Szent-Gyorgyi C; Stanfield RL; Andreko S; Dempsey A; Ahmed M; Capek S; Waggoner AS; Wilson IA; Bruchez MP, Malachite green mediates homodimerization of antibody VL domains to form a fluorescent ternary complex with singular symmetric interfaces. Journal of molecular biology 2013, 425 (22), 4595–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Lydia A, Perkins GWF, Matharishwan Naganbabu, Brigitte F. Schmidt, Frederick Mun, Marcel P. Bruchez, High-content Surface and Total Expression siRNA Kinase Library Screen with VX-809 Treatment Reveals Synergistic Kinase Targets that Enhance F508del-CFTR Rescue. Molecular Pharmaceutics 2017. [DOI] [PMC free article] [PubMed]
- (36).Winter GE; Buckley DL; Paulk J; Roberts JM; Souza A; Dhe-Paganon S; Bradner JE, Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015, 348 (6241), 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Senutovitch N; Stanfield RL; Bhattacharyya S; Rule GS; Wilson IA; Armitage BA; Waggoner AS; Berget PB, A Variable Light Domain Fluorogen Activating Protein Homodimerizes To Activate Dimethylindole Red. Biochemistry 2012, 51 (12), 2471–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


