Abstract
Planarian behavior, physiology, and pattern control offer profound lessons for regenerative medicine, evolutionary biology, morphogenetic engineering, robotics, and unconventional computation. Despite recent advances in the molecular genetics of stem cell differentiation, this model organism’s remarkable anatomical homeostasis provokes us with truly fundamental puzzles about the origin of large-scale shape and its relationship to the genome. In this review article, we first highlight several deep mysteries about planarian regeneration in the context of the current paradigm in this field. We then review recent progress in understanding of the physiological control of an endogenous, bioelectric pattern memory that guides regeneration, and how modulating this memory can permanently alter the flatworm’s target morphology. Finally, we focus on computational approaches that complement reductive pathway analysis with synthetic, systems-level understanding of morphological decision-making. We analyze existing models of planarian pattern control and highlighting recent successes and remaining knowledge gaps in this interdisciplinary frontier field.
Keywords: planaria, Dugesia japonica, regeneration, patterning, morphostasis
Graphical abstract
1. Introduction
1.1 A primer on planarians’ functional features
Planarian flatworms are free-living bilaterian organisms with a complex set of organ systems and cell types [1, 2]. Planaria possess rich behavioral repertoires that include sensing of a wide range of environmental cues ranging from chemicals [3] to gravity [4], to weak gamma radiation [5]. Though planaria have a true brain [6, 7], the real wonder of this remarkable model system is revealed most clearly when one considers the robust decision-making capabilities of its individual somatic pieces [8]. Amputated fragments of planaria regenerate a complete worm, growing precisely what is missing – no more and no less – with events at the wound edge coordinated tightly with body-wide remodeling of the intact tissue to ensure that a perfectly proportioned animal results within about a week after it is cut along any plane [9]. Every piece of a planarian is able to grow and remodel towards its species-specific large-scale pattern, stopping precisely when that specific anatomical pattern (the target morphology) is achieved. This is most obvious in the head-tail polarity of their primary axis: like a bar magnet cut into pieces (Fig. 1A), each piece determines head/tail identity at the wound based on an invariant axial polarity, regenerating into a worm with one head and one tail.
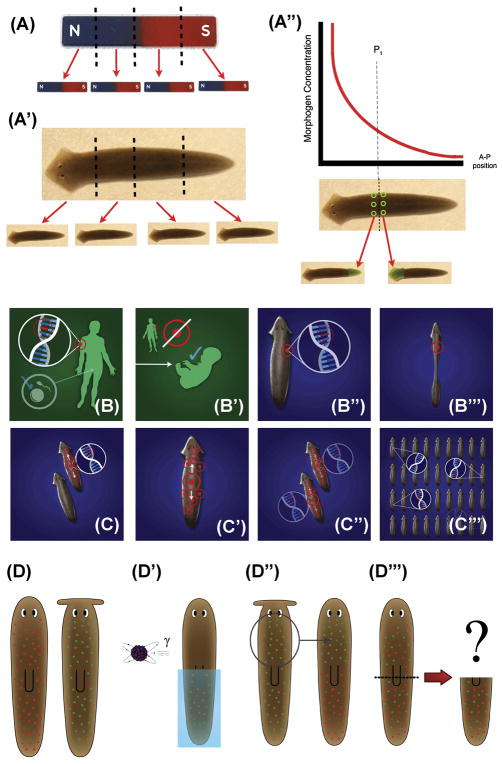
Figure 1. Planarian regeneration: fundamental puzzles of pattern control.
(A) Bar magnets illustrate a basic property of polarity re-scaling: having a North and South pole, a magnet can be cut into pieces and each piece reorganizes its polarity to likewise have a North and South pole by orientating small magnetic domains into large-scale axial patterning.
(A′) Planarian’s anterior-posterior (AP) axis likewise re-scales: every piece cut from a planarian (D. japonica shown here) regenerates a head and tail at the correct end.
(A″) Common models of axial patterning postulate a chemical gradient that indicates positional information for cells along the AP axis. The ability of cells on either side of a bisection to develop distinct anatomical fates (head vs. tail), even though they began as adjacent neighbors with identical positional information (green circles), suggests the need for long-range communication across the fragment so that decisions at the wound could be made based on the rest of the fragment.
(B, C) For most metazoans, sexual reproduction results in Weissmann’s barrier – somatic mutations (B) do not persist into the next generation (B′). However, planaria such as D. japonica largely reproduce through fission; thus, mutations in neoblasts anywhere in the worm body (B″) that do not kill the neoblast persist into the next generation upon fission (B‴). Within that individual (C), the progeny of the neoblast inherit the mutation(s), which spread throughout the body (C′) and are propagated to both offspring of the next fission event (C″). This gives rise to a fundamental puzzle about the relationship between genetics and patterning: over hundreds of millions of years, planarian lineages have thus accumulated diverse mutations in their bodies; despite the resulting very messy genomes, planaria regenerate with 100% anatomical fidelity and offer no genetic lines of patterning mutants.
(D) A thought experiment involving two different species of flatworms with distinct head shapes, illustrates knowledge gaps in the field. If one of the worms is irradiated so that half of its neoblasts are killed (D′), and neoblasts from another worm type are injected into this host (D″), what kind of head shape will this hybrid worm regenerate after amputation (D‴)? Existing models do not make a prediction as to whether a dominant shape, a combination shape, or never-ending remodeling will result.
Panels B–C‴ were made by Jeremy Guay of Peregrine Creative. Panels D-D‴ are used with permission from [8].
Planarians’ unparalleled pattern homeostasis – the ability to adjust cellular activity to a large-scale anatomical specification [10, 11] – is an on-going process, not an injury-specific one. Starving animals will shrink, and well-fed animals will grow, repeating this process ad infinitum while maintaining correct body proportions [12]. Because of this capability, planaria have conquered aging. While single cells senesce and die within relatively short time spans (~1 month) [13–18], the animal regenerates tissues continuously; planaria like D. japonica appear to be immortal at the level of the individual. Planaria are the champions of life-long functional health and damage repair. However, unlike simpler forms of life such as bacteria or Hydra, planaria combine these abilities with significant cognitive potential – they can learn in instrumental and classical conditioning contexts [19–24]. In keeping with their incredible somatic plasticity, they can transfer information between the body and the brain [25–28], and reprogram cancerous tissue when regeneration is activated [29, 30].
Planaria serve as a proof-of-principle – showing what is possible for biological systems to achieve. Therefore, truly understanding planaria would crack open most of the pressing problems in the fields of aging, regenerative medicine, cancer, and primitive cognition. Beyond biology, they also serve as a design challenge for fields such as morphogenetic engineering [31, 32], unconventional computation [33–35], and soft robotics [36, 37], which look to the biological world for clues as to designing robust, parallel, fault-tolerant systems. The planarian’s ability to integrate patterning and functional control is a superb example of morphological computation [38] and distributed decision-making [39, 40]. The dynamic adaptability of planaria dwarfs engineers’ efforts on this front – no human artifacts even come close to their capabilities yet.
1.2 Fundamental knowledge gaps
“No paradox, no progress.”
- Niels Bohr
As befits a model system with fundamental lessons to teach, planaria starkly reveal the large gaps in current knowledge. While much excellent work has drilled deeply into the molecular genetics of stem cell regulation [41, 42] and specific pathways necessary for regeneration [43, 44], we are still largely missing an understanding of the dynamics that are sufficient to build an organism with the observed remodeling and repair functions. Here, we argue that some of the most fascinating aspects of this field concern not only the mechanisms involved at the cellular and molecular levels, but also the algorithms that harness individual cell behaviors toward specific large-scale anatomical outcomes. We begin by highlighting a few of the mysteries that still confront us despite the recent advances in planarian molecular biology.
Establishing correct anterior-posterior (AP) polarity involves making sure every fragment cut from a planarian ends up with exactly one head and one tail, at the appropriate locations (setting aside for now the issue of orienting and scaling them correctly, giving them the right shape, and stopping when they are done). Planaria axially pattern each fragment no matter where it came from in the parent structure (Fig. 1A′) – the challenging nature of this process becomes clear when one considers a single bisected worm (Fig. 1A″). One wound site will make a head while the other will make a tail; the cells in those two wound sites were adjacent neighbors (before an arbitrary cut separated them) and yet go on to make radically different anatomical structures. A fundamental challenge is to understand the decision-making process that occurs at each wound site – what are the cells measuring, and from how far away, to determine anatomical identity of the new tissue to be made? This is difficult for simple gradient models to explain (as the wound cells had identical positional information before the cut, Fig. 1A″, green circles); head-inhibits-head models of this phenomenon run afoul of the fact that a brain has no trouble building another brain right next to it, if the head is amputated longitudinally, as well as being unable to explain axial patterning in mid-fragments with open wounds on both sides.
The second puzzle concerns the conspicuous lack of stable lines of planarian patterning mutants. Most other model organisms – Drosophila, C. elegans, chick, mouse, zebrafish, and humans – all offer stably transmitted lines with characteristic and striking anatomical deviations from wild-type (patterning mutants) that can be studied to forge a link between genetics and anatomy (central to understanding the genotype-phenotype relationship in evolution). It is rarely mentioned in our field that, despite almost 120 years of every conceivable experiment including irradiation, with the exception of a physiologically-induced double-headed line described below, there are no planarian mutant lines with patterns (in shape, number, or placement of specific organs) that differ from the standard species-specific target morphology. Why?
The answer might be linked to the third puzzle: their remarkable anatomical stability in the face of a highly variable genome. Weissmann’s Barrier (Fig. 1B,B′) does not apply to species like D. japonica which reproduce primarily asexually by fission and regeneration. In this case, any mutation that does not kill a neoblast is propagated to the next generation and can expand into a clonal line (Fig. 1B″,B‴). This will be especially true of the inevitable dominant mutations, which increase relative cellular fitness and could become something akin to cancer stem cells. This somatic inheritance (Fig. 1C–C‴) is predicted to generate extremely divergent genomes over time [45]; indeed not only are some planarian species mixoploid (not every cell has the same chromosome number) [46], but they also accumulate immense amount of change: up to 74% in protein-coding genes [47]. Indeed, large numbers of mutations have been found both outside and within gene-coding regions, including many amino acid substitutions and non-synonymous SNPs. Furthermore, an analysis of the genome of the planaria Schmidtea mediterranea revealed that many essential genes are missing from the genome, including components of many core pathways ranging from cell division, to DNA repair and metabolism [48]. And yet, they regenerate under control conditions with 100% anatomical fidelity, making perfect planaria each time despite their messy genomes. How is it possible for the genome to accumulate somatic mutations over hundreds of millions of years and yet maintain perfect anatomical fidelity? This paradox is pointing to a fundamental lack of knowledge of “where pattern comes from” and what part genetics plays in the answer. We are currently investigating this issue by raising generations of planaria in heavy doses of mutagens, to fully probe the stability of their pattern homeostasis under genetic change.
The fourth puzzle to consider is a thought experiment. Suppose a hybrid was made between planaria with two distinct head shapes: removing half of the neoblasts of one species of planaria, and populating it with 50% of the neoblasts from another species (Fig. 1D). After the neoblasts get a chance to accustom to their new home, the head is amputated (Fig. 1D′,D″). As the neoblasts start rebuilding the head, what shape will form (Fig. 1D‴)? Is one dominant over the other, or will it be an intermediate shape, or will the remodeling never cease, as each set of neoblasts is never given the “stop” signal derived from having a complete, normally-shaped head appropriate to its species? The value of this Gedankenexperiment is not so much only in the answer, but in the fact that we currently have no models that make a prediction about this basic scenario. The combined communities’ best quantitative models, including extensive RNAseq profiling, do not constrain the outcome of such experiments – they are silent about shape and what determines it, and it is not yet clear how much of the necessary information is generated by the soma and how much by the neoblasts. Thus, our field lacks theoretical frameworks for thinking about large-scale states (e.g., head shape) as targets and stop conditions for pattern homeostasis loops regulating cell behavior. This is a necessity if we are to understand how cells know what to build and when to stop.
1.3 Perspective
We highlight these puzzles as examples of questions that emphasize major knowledge gaps, and thus opportunities, in this field. We argue that significant new conceptual and technological approaches must be added to the highly successful mainstream efforts to understand specific gene products and cell-level phenotypes. Here, we use planarian regeneration as a lens through which to view several broad issues of biological computation, pattern control, and the genotype-phenotype relation. Advances in biochemical and genetic controls of regeneration have been expertly reviewed elsewhere [44, 49–51]. Focusing on synthesis and understanding global morphological decision-making in this model system, we review recent developments in this field that have the potential to drive progress on fundamental aspects of the origin and control of growth and form. These include (1) endogenous physiological signals that underlie pattern control: bioelectric and neurotransmitter-mediated signaling mechanisms in numerous cell types, which enable long-range coordination and morphological decision-making by cell collectives [40]; and (2) advances in computational modeling and automated model discovery, which are helping to understand the algorithms by which planarian shape is controlled.
2. Physiological Controls of Patterning
Regeneration of significant injury requires rebuilding structures that are properly coordinated in position, orientation and size with the large-scale anatomy of the remaining body, which implies that cells need non-local information to make patterning decisions (Fig. 1A′). The ability to re-create the same structure time and again can be understood as a “pattern memory” [39, 52], while the ability to reach the same correct pattern from different initial starting conditions (location and extent of damage) implies a robust goal-directed process. One way to think about the remarkable decision-making properties of planarian tissues is to consider the algorithms and molecular mechanisms exploited by brains – our best example of biological systems that implement memory, distributed processing, decision-making, and flexible goal-achieving cell networks. Interestingly, planaria were one of the earliest model systems in which data suggested roles for ion- and voltage-mediated processes in guiding regeneration [53–57]. Since then, advances in developmental bioelectricity (reviewed in [58, 59]) have driven hypotheses about the role of ion channels in guiding cell behavior, and more broadly, about the relationships between the activity of multi-cellular electric circuits and developmental morphospace [60, 61]. This perspective made several specific and counter-intuitive predictions, driving new experiments that complement the biochemical/genetic research programs. Recent work has tested some of these predictions, uncovering novel biology in the planarian model system.
2.1 Prediction 1: Ion channels and voltage gradients are involved in planarian patterning
Neurons compute and transmit information long-distance by virtue of electrical signaling. It is now well-recognized that even neural computations can involve graded (not spiking) potentials [62–66], which are more similar to the slower, steady non-neural bioelectric events operating in regeneration and development (Fig. 2A). Could non-neural somatic tissues be performing information-processing tasks by exploiting ion flows, albeit in different ways and on different timescales than brains? While developmental biologists are not yet used to thinking of non-neural tissues making decisions, the emerging field of primitive cognition [67, 68] and the recent data on the phylogeny of ion channel and neurotransmitter signals [69–71] have highlighted the fact that brains did not invent their tricks de novo – the basic machinery of bioelectrical computation (Fig. 2A′) was present very early on in evolution and is ubiquitous in animal, plant, and fungal bodies. Even before multicellularity, cells were using ion channels and neurotransmitters to process information and communicate; from bacteria, fungi, to the earliest metazoans, cells exploited ion currents to regulate individual and group behaviors (data in vertebrate as well as invertebrate systems are reviewed in [59, 72–74]).
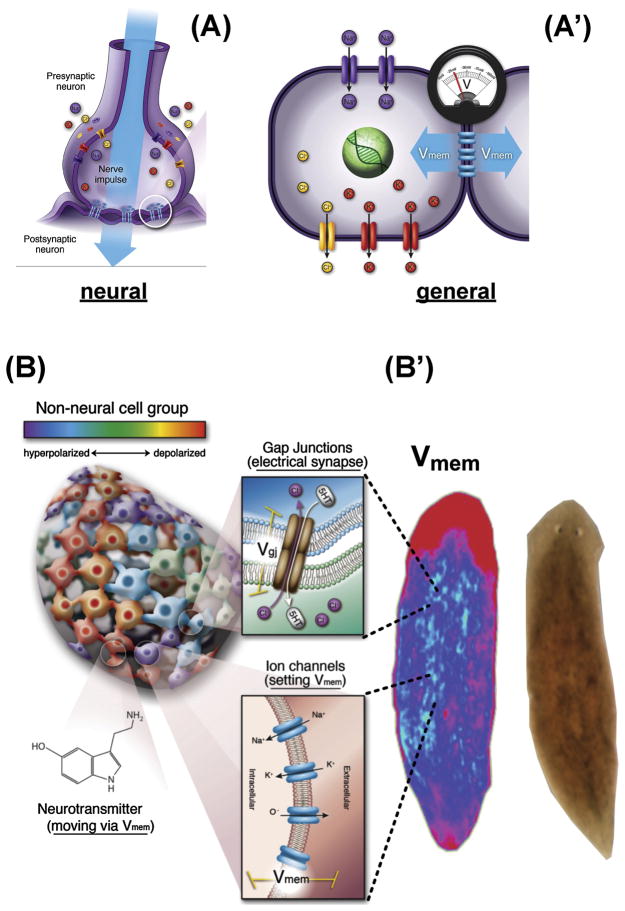
Figure 2. Bioelectric signaling among somatic tissues.
(A) Neurons implement memory and distributed decision-making by virtue of electrical potentials (Vmem) set by ion channels, which are propagated to neighboring cells via electrochemical synapses known as gap junctions.
(A′) The same machinery is present in most cells, where ion channels and pumps set Vmem, and gap junctions allow its propagation to some neighboring cells.
(B) Tissues sustain physiological compartments, whose borders and patterns of small molecule connectivity that are driven by the complex gating of ion channels and gap junctions. As in the central nervous system, neurotransmitters are among the key small molecule morphogens moved across tissues by bioelectric properties.
(B′) These dynamics result in spatio-temporal distributions of resting potential across anatomical distances (shown here in a planarian) – bioelectrical prepatterns that underlie subsequent gene expression and other cell behaviors during regeneration and development.
Panels A, A′ were created by Jeremy Guay of Peregrine Creative.
Research into bioelectric aspects of patterning has a rich classical history [75, 76]. The ideal fit of bioelectric circuits to the control of regeneration did not escape prescient early workers such as T. H. Morgan, who postulated electrical polarity to underlie regenerative axial polarity [77, 78], and C. M. Child, who focused on physiological gradients in pattern control [79]. Subsequent work used applied electric fields and biochemical analysis to manipulate planarian head/tail polarity [55, 56, 80, 81] and suggested the electrophoretic movement of morphogens during this process [82, 83] – a scheme that applies also to vertebrate left-right axial patterning [84].
Recent work (summarized in Tables 1–3) has moved beyond applied electric fields to probe the regenerative involvement of proteins and signaling molecules most often associated with the nervous system. Bioelectric signals (both slow and rapid) occur among all cell types, not just neurons, and are driven by several main classes of components (Fig. 2B): a) ion channel and pump proteins that set resting potential (Vmem), b) gap junction proteins (such as Connexins or Innexins) which form cytosolic connections to share a cell’s electrical and chemical state with neighboring cells, c) neurotransmitters and other small signaling molecules that move (by electrophoresis or voltage-powered transporters) across cell groups, and d) transduction machinery that converts changes in resting potential to downstream processes such as transcriptional changes (see [58] for review). Advances in imaging endogenous anatomical gradients of resting potential across tissues in vivo [85–87], and the development of techniques for specific modification of endogenous bioelectric aspects of morphogenesis [88, 89] have uncovered roles for bioelectric prepatterns (Fig. 2B′) in morphogenesis across phyla, from the alignment of the left-right axis [90] (sea urchin, frog, chick), control of developing organ size and shape [91–93] (Drosophila, frog, and zebrafish), induction of appendage regeneration [94] (frog), and craniofacial patterning [95, 96] (frog, mouse, and human).
Table 1.
Cell-level properties/behaviors controlled by bioelectric events
Table 3.
Ion channels and pumps proteins implicated in patterning by genetic screens
| Protein | Morphogenetic role or LOF phenotype | Species | References |
|---|---|---|---|
| TRH1 K+ transporter | Root hair patterning | Arabidopsis | [249] |
| Kir2.1 potassium channel | Wing patterning | Drosophila | [250] |
| Kir7.1 K+ channel | Craniofacial patterning, lung development | Mouse | [251] |
| NHE2 Na+/H+ exchanger | Epithelial patterning | Drosophila | [252] |
| V-ATPase proton pump | Wing hair patterning, Pigmentation and brain patterning Craniofacial patterning |
Drosophila Medaka, Human |
[253, 254] [255] |
| HCN1, Kv3.1 K+ channels | Forebrain patterning | Mouse | [256, 257] |
| KCNC1 K+ channel | Growth deficits | Mouse | [256] |
| TWIK-1 K+ channel (KCNK1) | Cardiac (atrial) size | Mouse | [258] |
| KCNJ6 K+channel | Keppen-Lubinsky syndrome –craniofacial and brain | Human | [96] |
| KCNH1 (hEAG1) K+ channel and ATP6V1B2 V- ATPase proton pump | Zimmermman-Laband and Temple-Baraitser syndrome –craniofacial and brain defects, dysplasia/aplasia of nails of thumb and great toe. | Human | [259, 260] |
| GLRa4 chloride channel | Craniofacial anomalies | Human | [261] |
| KCNA1 potassium channel | Megencephaly | Mouse | [262] |
| NCX-9 (Na+/Ca++) exchanger | Neural patterning | C. Elegans | [263] |
| GLRa4 chloride channel | Craniofacial anomalies | Human | [261] |
| KCNJ8 K+ | Cantu syndrome – face, heart, skeleton, and brain defects | Human | [264–266] |
| NALCN (Na+ leak channel) | Freeman-Sheldon syndrome – limbs, face, brain | Human | [267] |
| CFTR chloride channel | Bilateral absence of vas deferens | Human | [268, 269] |
| KCNK9, TASK3 K+ channels | Birk-Barel Dysmorphism Syndrome – craniofacial defects | Human | [270, 271] |
| Kir6.2 K+ channel | Craniofacial defects | Human | [272] |
| KCNQ1 K+ channel (via epigenetic regulation) | Hypertrophy of tongue, liver, spleen, pancreas, kidneys, adrenals, genitalia – Beckwith- Wiedemann syndrome; craniofacial and limb defects | Human, Mouse | [273–275] |
| KCNQ1 K+ channel | Jervell and Lange-Nielsen syndrome - inner ear and limb | Human, mouse | [276–278] |
| Kir2.1 K+ channel (KNCJ2) | Andersen-Tawil syndrome – craniofacial, limb, ribs | Human, mouse | [250, 279, 280] |
| GABA-A receptor (chloride channel) | Angelman Syndrome -craniofacial (e.g., cleft palate) and hand patterning | Human, mouse | [281–283] |
| TMEM16A chloride channel | Tracheal morphogenesis | Mouse | [284] |
| Girk2 K+ channel | Cerebellar development defects | Mouse | [285–288] |
| KCNH2 K+ channel | Cardiac, craniofacial patterning defects | Mouse | [289] |
| KCNQ1 K+ channel | Abnormalities of rectum, pancreas, and stomach | Mouse | [290] |
| NaV1.2 | Muscle and nerve repair defects | Xenopus | [94] |
| Kir6.1 K+ channel | Eye patterning defects | Xenopus | [248] |
| V-ATPase ion pump | Left-right asymmetry defects, muscle and nerve repair | Xenopus, chick, zebrafish | [237, 291] |
| H,K-ATPase ion pump | Left-right asymmetry defects | Xenopus, sea urchin | [90, 292, 293] |
| Kir7.1 K+ channel | Melanosome development defects | Zebrafish | [294] |
| Kv channels | Fin size regulation, heart size regulation | Zebrafish, mouse | [92, 295] |
| NaV 1.5, Na+/K+-ATPase | Cardiac morphogenesis | Zebrafish | [296, 297] |
Bioelectric pathways are most efficiently probed by misexpressing dominant channels, including optogenetic actuators [95, 97–99]; however, the expression of exogenous DNA is not yet possible in planaria (it is not yet known if this is related to the lack of genetic mutant lines in planaria). Instead, loss-of-function via RNAi or small molecule inhibitors (which also offer the benefit of mass spectrometry-verified wash-out experiments) can be used to make predictable changes in resting potential patterns and downstream outcomes. Targeting native bioelectric components in planaria has revealed a dependence of the head-tail decision on voltage gradients driven by the H+/K+-ATPase [100]; alteration of the normal bioelectric gradient in regenerative fragments [100] (using RNAi or drug inhibitors) can produce double-head or no-head heteromorphoses (Fig. 3A–A″) in D. japonica. Similar work in S. mediterranea identified a role for bioelectric signaling in size control and rescaling of the head [101], a function that appears conserved in brain [91], eye [102], and tail [92]. In addition to proteins that generate electrical gradients, gap junctions (a.k.a. electrical synapses) are crucial to the function of networks because they are highly controllable valves [103], which cells can use to regulate the spatial propagation of electrochemical signaling through tissues [104]. It is thus no surprise that both RNAi-based and pharmacological targeting of innexin-based gap junctions revealed roles in the control of stem cell dynamics [105] and head/tail polarity [106].
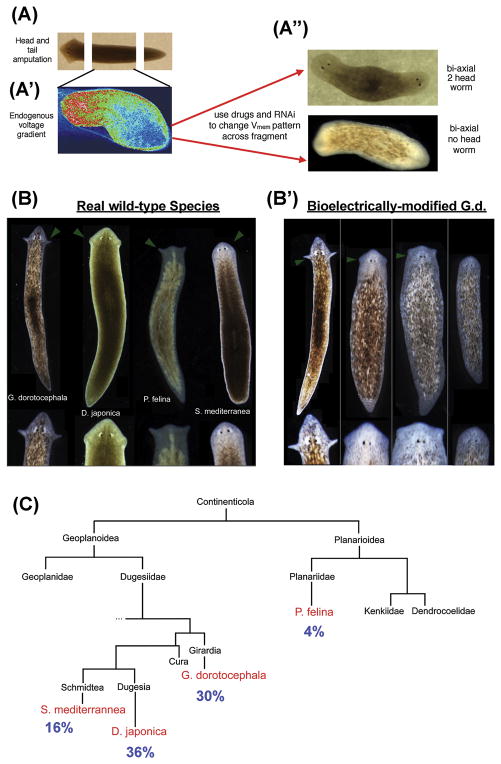
Figure 3. Bioelectrically-mediated changes of patterning in planaria.
(A) D. japonica mid-fragments exhibit bioelectric gradients, with anterior ends’ cellular Vmem depolarized compared to those of posterior cells. This pattern can be detected [85] via voltage-sensitive fluorescent dyes (A′), and modified with ion channel drugs, which alter the endogenous bioelectrical gradient toward bi- or no-head heteromorphoses respectively (A″), demonstrating that the bioelectric pattern is instructive for large-scale anatomical polarity along the AP axis [100, 132].
(B) G. dorotocephala planaria exhibit a characteristically distinct head shape, compared to other species. (B′) When fragments of G. dorotocephala were briefly treated with a gap junction blocker [124], they regenerated heads whose shapes matched those of other extant species of planaria.
(C) The appearance of these head shapes in a single cohort of worms treated together was stochastic, appearing at frequencies proportional to the evolutionary distance between the species.
Panel A′ was courtesy of Taisaku Nogi. Panels B–C used with permission from [124].
2.2 Prediction 2: Neurotransmitters are involved in planarian patterning control
A key component to the function of neural circuits are neurotransmitters – small signaling molecules that move as a result of bioelectric dynamics. These are evolutionarily ancient [107], long predating the appearance of nervous systems, and are known to be utilized as a kind of morphogen in development, for example in left-right axial polarity [108, 109] and early blastomere dynamics [110]. Are neurotransmitters important for planarian regenerative control [111]? The Marchant lab’s elegant use of a combination of pharmacology and molecular genetics identified important functional roles for serotonin, voltage-gated calcium channels, and other neurotransmitter machinery in determining anterior-posterior polarity in planarian regeneration [112–118]. Unlike RNA or protein, neurotransmitters are too small to effectively tag, hindering efforts to observe their movement through tissues during regeneration. Recent development of fluorescent sensors of neurotransmitters such as [119, 120] and light-based techniques for precise spatio-temporal control of neurotransmitter signaling [121, 122] will revolutionize this area, as soon as misexpression of exogenous proteins is available in planaria.
2.3 Prediction 3: Anatomical outcome and genetic default can diverge
Another feature of bioelectric pathways is that they confer a degree of robustness and plasticity that enable outcomes diverging from genetic default. For example, in Xenopus, metastatic melanoma can be initiated on a background of a perfectly normal genome (no carcinogens, no oncogenic mutagenesis) [123], tumors induced by KRAS mutation can be normalized [97], and brain defects induced by dominant Notch mutations can be circumvented [91], all by the appropriate modulation of native bioelectrical communication among cells.
Large-scale anatomical pattern (the species-specific target morphology) is thought to be determined by the genomic sequence and its chromatin modifications. Recent data, however, suggest that bioelectric networks also store key components of this information, as another epigenetic layer of pattern control [52]. This is consistent with Waddington’s original sense of the word, which included much more than chromatin modifications. G. dorotocephala planarians regenerate their specific head morphology with 100% fidelity under normal conditions. However (Fig. 3B,B′), when their heads were amputated, and the fragments exposed to a reagent that alters the bioelectrical connectivity among the cells [124], they regenerated heads that closely resembled those of several other species (as confirmed by quantitative morphometrics). Moreover, it was not only external head shapes that were converted, but also the shape of the brain and distribution of neoblasts in the head became similar to that observed in the heads of these other extant species [124].
One notable aspect is that the choice of heads was stochastic (Fig. 3C) – the different types of heads appeared in the same cohort of animals treated identically and raised in the same dish, but in frequencies proportional to the evolutionary distance between the species they resembled [124]. These data showed that a wild-type animal exposed briefly to a non-mutagenic modulator of gap junctions can generate heads belonging to species ~150 million years distant. This drastic effect of a transient change to bioelectric circuit dynamics, suggests these as a novel form of epigenetics, and one with convenient master-regulator properties that could readily have been exploited by evolution (e.g., via mutations in ion channel coding or promoter regions) as part of the exploration of a morphospace. Another notable aspect is that the change was not permanent: several weeks after completion of head construction, remodeling suddenly began, and converted the heads back to a normal G. dorotocephala shape. Unlike the example of permanent patterning change described below, this head-shape switch most resembles short-term memory without consolidation. Work is currently on-going to formulate and test quantitative dynamical systems models which reveal why head shape changes represent shallower attractor states than head-tail decisions in the physico-chemical state space of the circuits driving regeneration in planaria (thus, easier for the system to escape from) [61, 125].
2.4 Prediction 4: Pattern memory can be over-written
The dominant paradigm for regeneration is that of emergence, with cells behaving according to specific rules, and the combination of a large number of these individual activities somehow resulting in the same complex body being created from diverse starting conditions. A complementary view is that a specific pattern memory (the organism’s target morphology) is encoded, at least in very general terms, by some physical mechanism in tissue that is read out and elaborated during regeneration and regulative embryogenesis (and serves as reference for the stopping point for new growth and remodeling). Neuroscientists are comfortable with cellular networks that guide flexible activity to achieve stored goal states. Could a more ancient version of this system be more widely utilized for pattern control in biology? One of the major benefits emerging from cybernetics and control theory over the last six decades is a solid grounding for teleological-seeming processes in rigorous engineering principles. Systems that implement specific goal states can be modeled as homeostatic processes that do not require any anthropomorphisms, and are routinely constructed (from thermostats to self-driving cars). Thus, one way to model regeneration is as a kind of pattern homeostasis – a TOTE (Test-Operate-Test-Exit) loop [39, 126]. Planaria tissues can be envisioned as executing a continuous error minimization, striving to reduce the difference between the current morphology and the species’ target morphology (a kind of least-action model, as is often used to understand the role of other physical forces in morphogenesis [127–129]). A key aspect of any homeostatic process is that it has to store a setpoint – planarian tissue would have to represent (encode) some amount of information about the bodyplan they must regenerate to (and stop when the current anatomy matches this pattern memory). Thus, one prediction of this highly speculative viewpoint is that it should be possible to over-write the setpoint and permanently change the shape to which the animals regenerate in the future.
Precisely this was discovered when D. japonica animals were treated with 1-octanol (Fig. 4A) – an experiment motivated by the fact that gap junctions are a key component of memory in the brain and also an ideal candidate for the long-range communication between the wound site and remote tissues. The result was double-headed bipolar heteromorphoses [106], which, remarkably, continued to regenerate as double-headed in perpetuity with no further treatments (Fig. 4A′). These animals, which lose all trace of the gap junction-blocking reagent in a few days after initial treatment, are permanently converted to regenerating a different target morphology by a transient physiological perturbation. This phenomenon was first described as “trophic memory” in Bubenik’s work on deer antler injuries and pattern changes in subsequent years of regeneration [130]; planaria provide the first molecularly-tractable model system in which this fascinating aspect of regeneration biology can be studied [131]. The permanent double-head state can be re-set back to a wild-type single-head target morphology by a different transient modulation of the bioelectric circuit using the ion pump blocker SCH28080 (Fig. 4B).
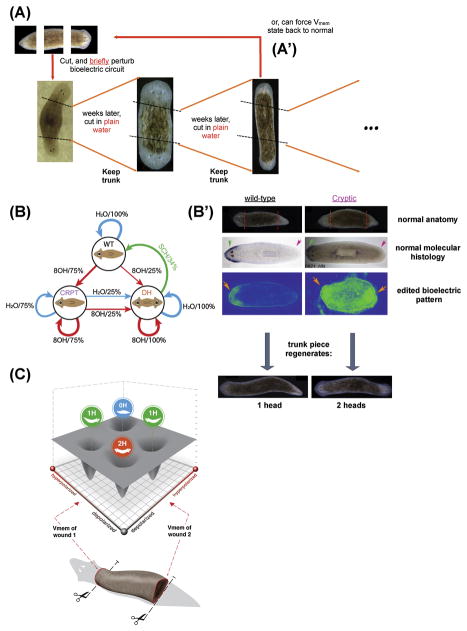
Figure 4. Permanent change to planarian target morphology: resetting bioelectrical pattern memories.
(A) Exposing planarian fragments to the gap junction blocker 1-octanol for three days results in a permanent re-setting of the target morphology [193]. Trunk fragments of such worms continue to regenerate as double-headed animals in perpetuity, in plain water. 1-octanol is washed out of the worm tissues within 2–3 days (as shown by mass spectrometry), this demonstrates transient physiological changes becoming consolidated as long-term pattern memory, without genomic editing. The animals’ target morphology can be re-set back to normal by altering the bioelectric circuit back to a wild-type distribution, using ion pump drugs such as SCH28080 (A′).
(B) Animals that did not become double-headed after an initial exposure to 1-octanol are not wild-type because when cut in plain water they give rise to the same percentage of double-headed worms in each generation. Here shown as a state transition diagram with double-headed worms always regenerating as double-headed (a terminal state) while cryptics continue to generate double-headed worms at the same ratio (each arrow is labeled with the reagent applied at regeneration and the percentage of outcomes).
(B′) Cryptic worms are identical to wild-type worms in their anatomy, expression of head and tail marker, and stem cell distribution [132]. However, a uniform depolarization of endogenous bioelectrical gradient reveals the difference between normal and cryptic animals. This altered bioelectric distribution is the key functional component of the re-writable pattern memory mediating the regenerative control.
(C) The same body can contain at least two diverse bioelectric patterns guiding future growth: wild-type (permanent single1-headed) or cryptic (destabilized, stochastic). One way to understand stable, discrete anatomical outcomes emerging from bioelectric circuit activity is as stable attractors in a morphospace defined by the voltage states of the two ends of the body.
Panel C made by Jeremy Guay of Peregrine Creative.
A biophysical model of the planarian pattern memory has been analyzed [132], explaining how patterning in regenerates can be templated off of stable biophysical properties of fragments despite wild-type genomic sequence. While it is very possible that the permanent change of pattern memory also involves chromatin modification machinery [133], the field of epigenetics does not yet offer an explanation of how large-scale anatomical patterning outcomes would result from specific chromatin states in individual cells. At the same time, the long-range organizing properties of electric fields arising from ion channel activity provide a natural medium in which to understand these questions of how large-scale order arises [125, 134–136]. The interaction of bioelectrics and chromatin modification machinery [137, 138] remains therefore an important area for future work.
As with any intervention (molecular-genetic or pharmacological), the effects of gap junction inhibiting reagents are not 100% penetrant in a cohort of planaria treated together. However, an interesting phenomenon was discovered when the unaffected “escapees”, which looked like normal one-headed planaria, were re-cut weeks after the initial treatment without any further interventions. It was found [132] that these animals were in fact not wild-type: they differed from normal planaria in that when cut, in plain water, they generated double-headed worms in the same proportion as the original treated cohort (Fig. 4B). The same is true for subsequent generations from these “cryptic” worms. The cryptic worms were analyzed and shown to have normal anatomy, histology, stem cell distribution, and expression of several head and tail markers (Fig. 4B′). What makes these worms, bearing apparently normal hardware, regenerate in a stochastic manner in plain water? It is that they bear an aberrant pattern memory: voltage-sensitive fluorescent dyes reveal that their tissues are uniformly more depolarized than true wild-type worms (Fig. 4B′).
Numerous examples in engineering (e.g., flip flops) exist of how the same electrical circuit can be bi-stable, able to store several discrete possible patterns of gene expression or ion flow [139]. It is not yet known whether these concepts will directly translate to understanding planarian pattern control; however, the cryptic worm data described above provide a simple illustration of how an anatomically-normal body can support distinct bodyplan encodings that are latent, but recalled if the animal is challenged to regenerate in the future. In this, they fulfill the basic properties that define memory (whether neural, electronic, or molecular): encoding discrete large-scale outcomes, which are long-term stable, and yet sufficiently labile to be able to be rewritten by appropriate stimuli. A dynamical systems perspective on such discrete, stable, yet potentially labile end-states is to view them as attractors in the state space of the relevant circuits, which demarcate regions of morphospace corresponding to different bodyplan layouts (Fig. 4C).
2.5 Summary: Physiological controls of growth and form
The planarian model system offers experimentally-tractable examples of rewriting the anatomical setpoint for regeneration without genomic editing: permanent changes of the pattern to which animals regenerate following future injury, driven by transient alterations of physiological state (Fig. 4A,B-B′). Currently, our ability to control biological patterns is limited, and the full range of possible patterns is unknown. Future work must focus on a better understanding of the interplay of transcriptional, chromatin-based, and physiological layers to explain stochasticity in large-scale anatomical outcomes and long-term stability of target morphology. The key challenge is to convert pathway and physiological circuit information at the level of single cells into an understanding of stable large-scale anatomical attractor states, and to achieve a systems level understanding of shape homeostasis and regeneration (Fig. 4A). AP polarity can be explained using chemical positional information gradients coupled with directed transport [125, 140]; it remains to be seen whether the full complexity of planarian pattern homeostasis (including shape, cell number and proportion, and patterning along DV/LR axes) will require connectionist or other neural-like computational models [40, 141]. Just how much global information is encoded in physiological circuits, to what resolution a target morphology might be represented in tissue, the size of the smallest unit that processes bioelectric states (single cells, or cell groups), and how much predictive control can be gained over patterning in planaria, are open questions that will require not only technique development but conceptual advances that may need to borrow from neuroscience, control theory, and cybernetics.
3. Computational Approaches to an Integrative Understanding
The mysteries of planarian regeneration have been with us for ~120 years [142], and one of the most challenging aspects has been the discovery of specific models that exhibit the desired patterning properties matching the huge base of functional knowledge in this model system. As with many difficult problems in science, this field is ripe for assistance from the revolution in information technology. Alongside new recent databases of transcriptomic, phylogenetic, and biochemical resources (such as PlanNET and PlanMine) [143, 144], two new directions are emerging: simulation environments for interrogating the complex dynamics of patterning models, and machine learning tools for helping to derive models with desired properties.
3.1 Current state of the art in understanding regenerative dynamics: gradients and beyond
Next, we summarize the current state of the art in computational understanding of planarian regeneration, focusing primarily on anterior-posterior axial polarity.
The mechanisms underlying body-plan control in planaria have been explored from a variety of different perspectives, from biochemical gradients to neural network dynamics [141, 145, 146]. Models exploring gradients of positional information (mediated by gene expression and secondary messenger gradients) have shown particular promise [147–149]. The concept of morphogen gradients underlying control of body plan in planaria regeneration in a concentration-dependent manner is an old idea which was first proposed at the turn of the century (reviewed in [10]). With the advent of molecular genetics and RNAi-mediated loss-of-function in planaria, recent experiments have revealed the existence of gene expression and signaling gradients along the anterior-posterior axis, which have been found to be crucial for anterior-posterior body-plan control [10, 44, 150]. Most distinctly, canonical Wnt/β-Catenin signaling has been strongly implicated in posterior development [151, 152], via the inhibition of signaling pathways such as extracellular receptor kinase (ERK) that are associated with head development [153–155]. Graded Hedgehog (Hh), fibroblast growth factor receptor like (FGFR), and Notum signaling have also been observed [140, 149, 156–158]. RNAi knockdown of these factors results in dramatic alteration of the regenerated planarian body plan, including doubled-headed (RNAi of Wnts), missing tails (RNAi of Hh), and double-tailed (e.g. RNAi of Notum) heteromorphoses.
A remarkable feature of morphogen gradients in planaria is that their polarity in the original organism is spontaneously regenerated with amputation [159]. For example, Notum is detectable at strong concentrations at the anterior of a whole planaria in homeostasis (and the opposite is true of Wnt1, which is expressed most strongly at the posterior). When the animal is cut into fragments, each fragment will spontaneously reform a concentration gradient so that Notum is expressed strongly at the fragment end oriented towards the previous head location, even if the fragments are left adjacent to one another (and vice-versa for Wnt1) [159]. This observation of spontaneously reforming polarized Wnt and Notum gradients provides strong substantiation for the concept of morphogen gradients underlying anatomical polarity control.
Several models have been proposed to account for the spontaneous emergence of morphogen gradients underlying planaria body plan control [21]. Meinhardt and Gierer proposed a mathematical reaction-diffusion model consisting of a self-activating substance acting over a short range in combination with a long-range acting inhibitor, which was used to describe the formation of an emergent β-Catenin gradient along the anterior-posterior axis [146, 160, 161]. Working with a different underlying mathematical premise, Stuckeman et al. proposed two mutually antagonistic signaling circuits — one for the anterior and one instructive for the posterior – which together could function as a molecular switch to control anterior-posterior polarity [140]; mathematical modeling in a spatialized context would be an important step for future work. Mutually repressive signals function as a distinctly bimodal system capable of efficiently switching between one of two states [139], in this case, between head and tail signaling modalities. In Stuckeman’s conception, posterior development would be comprised of Wnt/β-Catenin signaling, which is proposed to repress, and to be in turn repressed by, a second unknown signaling modality crucial for anterior development [140].
While reaction-diffusion models are valuable in contextualizing body plan control in regeneration in terms of an experimentally tractable output, and show promise in explaining positional information generation and control in planaria, these models suffer from a fatal flaw: they are highly dependent on size scale, meaning the type of pattern resulting from the mechanism is dependent on the size of the organism [145]. For a particular model capable of forming a gradient on a particular organism size, when the organism is cut into pieces a gradient may no longer form in these smaller pieces; or, if the organism grows in size, the simple gradient changes to a more complex pattern such as a collection of spots or stripes, which would no longer map to a clean anatomical outcome such as one head and one tail [145]. The scale-dependence of many reaction-diffusion models was addressed by Werner et al., who developed, and evaluated in a one-dimensional model, an elegant regulatory circuit that is able to rescale a monopolar concentration gradient virtually independent of scale.
An alternative solution to the scale-dependence of traditional reaction-diffusion schemes is to consider polar transport of gene products and/or secondary messengers [125]. Endogenous bioelectricity, which comprises very strong electric fields active across cell membranes (~1.0x106 V/m) and between gap junction-coupled cells (~1.0x104 V/m), with weaker fields (~1.0 V/m) in the global environment [162, 163], offers a tractable mechanism through which electrically charged substances may be subjected to directional transport. Passage of small, charged signaling molecules such as ATP4−, or neurotransmitters such as 5HT+, across gap junctions in transmembrane potential (Vmem) gradients is a particularly promising mechanism that has previously been implicated in the establishment of developmental left-right asymmetry [164] and neural pathfinding [165] in vertebrate models.
A simple model of bioelectricity-induced polar transport in gap junction connected cells was recently reported, and computationally analyzed in a physiologically realistic tissue context. It exhibited effective self-assembly and reassembly of highly polarized concentration gradients in whole organisms and cut fragments of highly diverse sizes for a simple model where an anion moves between cells in a Vmem gradient that is most depolarized at the anterior [125]. Interestingly, Lange and Steel experimentally detected a highly negatively charged proteinaceous substance moving in the posterior direction along the anterior-posterior axis of planaria, which was furthermore found to inhibit head formation [82, 83]. These observations are also consistent with early reports from Marsh and Beams, who were able to switch the anterior-posterior axis of regenerating planaria fragments using applied electric fields [56, 166]. Thus, the electrophoretic movement of signaling molecules in endogenous electric fields provides a straightforward explanation for the regenerating polarized morphogen gradients of planaria in a manner that is essentially independent of size scale.
3.2 Advances in modeling and simulation: testing available models
A mature understanding of patterning requires algorithmic models, which make each step in the process explicit, require clarification of the mechanisms sufficient for pattern control, enable testable predictions, and can be inverted to infer specific interventions. As in other areas of developmental biology, recent progress has allowed planarian research to move beyond arrow diagrams of pathways to generative models that highlight the spatio-temporal dynamics that must be implemented to explain regeneration [167]. This is essential to bridge the well-recognized gulf between the growing deluge of transcriptomic and genomic data and the anatomical outcomes reported in the functional literature that we seek to understand and control. Simulation modules for planaria include ones that focus on biochemical diffusible factors [168], and the BETSE modeling environment [134], in which a very rich set of dynamics (including biochemical signaling as well as bioelectric/physiological processes), in a bio-realistic spatialized virtual tissue context, can be explicitly modeled [125].
The fields of genetics and cell biology have been revolutionized by bioinformatics - computational tools that help scientists deal with high-volume molecular/genetic data. Planarian regeneration was the domain of some of the first efforts at a new bioinformatics of shape – software for extracting control principles of multicellular anatomical control from functional published data. One aspect of this effort is formalizing results of functional regenerative experiments and capturing the relationships between interventions and anatomical outcomes (Fig. 5A–B′). PlanForm formalized both the possible functional experiments in the planarian model system (including gene targeting, surgical cut/paste manipulations, etc.), and possible planarian body configurations as outcomes of such experiments (via graph representations) [169, 170]. The database currently contains most of the papers in the planarian research field, matching published experiments to their patterning outcomes. This expert system not only allows new workers in the field to rapidly determine what has been done and what the outcomes result from specific manipulations, but also forms the body of knowledge against which models of regeneration can be formally tested, to determine how well they recapitulate the known dynamics of planarian repair. This database is a flexible, general knowledge system to which newly published papers’ results can be continuously added resulting in a standardized resource akin to UniProt for molecular data.
Figure 5. Bioinformatics of shape, applied to planaria.
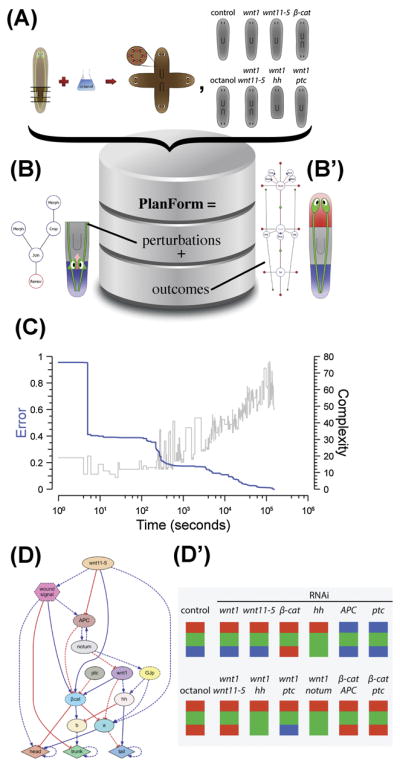
(A, B) The continued development of knowledge in this field will require computational tools going beyond bioinformatics of genes and proteins, to assist in development and analysis of models. One effort, PlanForm [169], comprises over 1,000 experiments from the literature, matching the functional manipulations performed (e.g., specific cuts, joins, RNAi, bioelectric change; see B) and the resulting anatomical outcomes represented by a graph notation (B′).
(C) One recent application of artificial intelligence to discovery of regulator pathways [168] used evolutionary selection over a population of biochemical models. Here shown as the progressive reduction of error in the predictions of top candidate models at each generation.
(D) This process uncovered a gene regulatory network whose patterning properties matched observed data on canonical pathways (a sample is shown in D′).
3.3 Tools for model discovery
Environments are coming on-line for quantitative simulations of available models, whose predictions can be tested against prior data. Anterior-posterior patterning and re-scaling have been solved by models created in the traditional way [125, 140, 145, 146, 171]. However, future work will also have to deal with the full range of remarkable capabilities of planarian regeneration. Arbitrary cuts and punctures require a worm to restore precisely-patterned and intricate shapes such as intestinal branches, brains, pharynxes, and numerous other tissues/organ systems in the right number, shape, placement, and orientation (stopping precisely when the right shape is complete). A bio-realistic model combining molecular genetics and biophysics to quantitatively explain the full spectrum of planarian regenerative capabilities is likely to be incredibly complex. Moreover, the rapidly increasing dataset on perturbations and outcomes in this (and other systems) makes it increasingly more difficult for scientists to come up with models that exhibit the correct large-scale patterning behavior from a specification of cellular pathways and include all available results. Thus, it is likely that this field, as many others, is poised to benefit from machine learning approaches – using techniques from artificial intelligence to assist human scientists in model discovery (a branch of the nascent Robot Scientist field [172]).
A recent example of model inference from functional data [168] used evolutionary search algorithms (progressive rounds of evaluation and proportional mutation of candidate models, Fig. 5C) to rapidly uncover a small biochemical network whose behavior in a biochemical simulator matched key regeneration experiments on anterior-posterior polarity (Fig. 5D,D′). Surprisingly, this first example of a non-human-derived model in regenerative biology resulted in a fully-specified model that is simple enough for human scientists to understand (unlike neural network approaches, the result is not simply a black box that gets the answers correct but provides limited insight into the dynamics involved) (Fig 5D). The use of evolutionary principles to identify models with desired functional properties (the process by which real planaria arose) is a powerful strategy for future work, as the search can be re-run as new data appear in the database, and more powerful simulators come on-line. The latter is important as a major limitation of this work is that the machine learning-derived models did not include physiological components and have not yet been tested in spatially-realistic simulators to probe all of their patterning properties.
Such machine-learning approaches can uncover networks with unknown components. Fortunately, computational techniques are now available to provide putative identities for unknown elements, that can be tested in model validation [173]. This strategy was used to identify one of the novel elements in the planarian network as the HNF4 factor, which subsequent functional testing via RNAi confirmed [174].
An important aspect of this effort is being able to invert the models: using them to predict functional interventions that will have a specific desired outcome. This is an essential component of leveraging model systems such as planaria for progress in regenerative medicine. An example of this is the recent automated discovery of a bioelectric and serotonergic signaling network explaining stochastic conversion of normal pigment cells to a melanoma phenotype by bioelectrical disregulation [175]. While the molecular signaling components at the cell-level were known [123], the system-level dynamics were unclear. For any specific disruption of bioelectric signaling among somatic cells, some percentage of the animals in a cohort converted normal melanocytes to a melanoma-like behavior. What was completely unclear was how all of the cells within a certain animal coordinated their decision to react in an all-or-none manner, why animals treated in the same dish exhibited distinct outcomes, and how the number of affected animals in any specific perturbation could be predicted. This problem has important parallels understanding the stochastic outcomes of planaria in which bioelectrical systems have been altered [132]. A computational search analyzed the network as a dynamical system, identified the main drivers that lock the system into one of several global attractor states, and proposed an intervention consisting of two drugs and one specific protein misexpression, that would break the concordance [176]. Testing confirmed the prediction, producing the first partially-hyperpigmented animals [177]. Future developments in the field of planarian regeneration will likewise make use of dynamical systems analysis and computational model inference to not only identify explanatory models but also identify specific interventions to drive desired morphological outcomes in the context of increasingly-complex regulatory networks. Applicability of other frameworks, such as P-systems [178], agent-based dynamics of target morphology [179–181] and connectionist models [141] remains to be investigated.
4. Conclusion
Planaria reflect many of the fundamental mysteries facing us in the new century of interdisciplinary biology. As a model amenable to molecular-genetic, developmental, regenerative, and behavioral research, this remarkable model species is at the intersection of not only evolutionary biology and biomedicine but also synthetic bioengineering and information science. We argue that one of the main benefits of this model is to facilitate a focus not only on the mechanisms that control regeneration, but also on the algorithms and information-processing mechanisms implemented in planarian tissues. Some of the most exciting advances in the biosciences revolve around morphogenetic engineering [31, 32], morphological computation [34, 182, 183], and cellular perception/decision-making [184–186]. Planaria represent a proof of principle of a remarkable “computational medium” – a material that actively fulfills a complex design spec while itself being drastically remodeled, and a system in which the control circuitry and the body it controls are one and the same. Planaria are an ideal lens through which to develop new techniques, data, and conceptual approaches to advance the intersection of these fields, with numerous applications for the biomedicine of anatomical control and the understanding of the relationship between genome and anatomy.
Given that the genome directly encodes proteins, not anatomical structures, how do tissues store information about the pattern to which they must regenerate? By regenerating from pieces (no obligate Weismann’s barrier), planaria are helping to reveal new perspectives on the question of where anatomical pattern is specified [45, 61, 187]. Planaria with a normal histological configuration and genome can permanently store (and regenerate to) one of several target morphologies [132]. The recent work on bioelectrical re-specification of pattern memory, producing permanent lines of double-headed or stochastically-destabilized (cryptic) planaria, may have important implications for the evolution of bodyplans [52, 61]. Future work will determine to what extent evolution exploits the plasticity of physiological software in concert with classical genetic change, in the implementation of bodies and their repair circuits [188]. Here, we have argued that the dynamics of regenerating planaria offer an ideal system in which to quantitatively integrate the perspectives of molecular-genetics, dynamical systems theory, biophysical self-organizing processes, and computation. Recent approaches provide rich fodder for this effort, including advances in the mechanisms of bioelectrical pattern control and biorealistic modeling that facilitates machine learning approaches to model discovery and extraction of systems-level insights from molecular mechanisms.
Importantly, future efforts must begin to expand from AP polarity and head number, to understanding of actual shape (of species-specific heads, and overall planarian anatomy in three dimensions). At the moment, our understanding of planarian shape is insufficient to derive planaria-specific morphologies from genomic or any other data. A focus on shape is essential, not only because of the demonstrated multi-stability of the planarian regenerative outcome but because sometimes molecular marker expression and anatomy diverge; for example tail markers can be expressed in tissues that have the overall shape of heads [151], challenging the community to be explicit of what criteria are considered the gold standard by which “identity” of a structure can be determined.
Specific directions for future research provide a fertile ground for young new scientists entering this field, linking planarian regeneration to profound directions facing biology at large. The understanding of variability is one; how can clonal animals, raised in the same container and exposed to the same reagent/stimuli in the same dish, exhibit such different responses, as observed in the stochastic outcomes of bioelectric modulation [132] and in the behavioral responses in memory and drug addiction research [24, 25, 189]? Robustness is another; despite the huge variability in cell number, damage type, and genetics, planaria reliably exhibit unfailing anatomical homeostasis. This kind of goal-directed process, able to harness individual cell behaviors toward the anatomical needs of the host organism, poses a fascinating design challenge not only for biologists but also for roboticists and engineers seeking to improve on today’s brittle technology.
It is clear that biologists will have to expand not only the toolkits (bringing (opto)genetics to planaria) but also the conceptual apparatus, if we hope to understand what regenerating planaria are telling us about biology. It remains to be seen which type of paradigm for understanding pattern memory and its elaboration during regeneration will be the most effective; connectionist and Least Action/Active Inference ideas from neuroscience and physics [190–192], are possible candidates. Importantly however, the explosion of molecular, genetic, physiological, and functional data in this field also provide a context for learning to extract wisdom and actionable intelligence from large volumes of data. New efforts in the bioinformatics of shape, with experimental testing in the planarian model, will facilitate the contributions of artificial intelligence to assist human researchers in cracking the secrets of planaria and exploiting them for unprecedented advances in biomedicine.
Table 2.
Experimental data implicating endogenous bioelectric signal roles in morphogenesis
| Developmental Role | Species / model system | References |
|---|---|---|
| Cellular polarization (anatomical asymmetry of cell or epithelium) | Alga Fucus, yeast | [230, 231] |
| Migration of neurons and positional information | Chick, Amphibia | [232, 233] |
| Patterning in gastrulation, neurulation, and organogenesis | Chick, axolotl, frog | [90, 232, 234–237] |
| Directional transport of maternal components into the oocyte | Moth, Drosophila | [238] |
| Growth control and size determination | segmented worms | [239] |
| Neural differentiation | Xenopus embryo | [225, 240] |
| Polarity during regeneration | Planaria, plants, and annelids | [55, 56, 80, 81, 100, 241–243] |
| Induction of limb and spinal cord regeneration | Amphibia | [244–246] |
| Control of gene expression and anatomy in craniofacial patterning | Xenopus embryo | [247] |
| Induction of eye development | Xenopus embryo | [248] |
Acknowledgments
We thank the Joshua LaPalme, other members of the Levin lab, Emili Salò, and many members of the planarian community for helpful discussions, and Joshua Finkelstein for comments on the draft. This paper is dedicated to the memory of C. M. Child, G. Marsh, and H. W. Beams – original pioneers in the physiology of planarian regeneration. This work was supported by an Allen Discovery Center award from The Paul G. Allen Frontiers Group (12171). The authors gratefully acknowledge support from the National Institutes of Health (AR055993, AR061988), the G. Harold and Leila Y. Mathers Charitable Foundation (TFU141), National Science Foundation award #CBET-0939511, the W. M. KECK Foundation (5903), and the Templeton World Charity Foundation (TWCF0089/AB55).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gentile L, Cebria F, Bartscherer K. The planarian flatworm: an in vivo model for stem cell biology and nervous system regeneration. Disease models & mechanisms. 2011;4(1):12–9. doi: 10.1242/dmm.006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddien PW, Sanchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–57. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 3.Mason PR. Chemo-klino-kinesis in planarian food location. Animal Behaviour. 1975;23(2):460–9. doi: 10.1016/0003-3472(75)90095-0. [DOI] [PubMed] [Google Scholar]
- 4.Adell T, Salo E, van Loon JJ, Auletta G. Planarians sense simulated microgravity and hypergravity. Biomed Res Int. 2014;2014:679672. doi: 10.1155/2014/679672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown F, Park Y. Seasonal variations in sign and strength of gamma-taxis in planarians. Nature. 1964;202:469–471. doi: 10.1038/202469a0. [DOI] [PubMed] [Google Scholar]
- 6.Sarnat HB, Netsky MG. The brain of the planarian as the ancestor of the human brain. Can J Neurol Sci. 1985;12(4):296–302. doi: 10.1017/s031716710003537x. [DOI] [PubMed] [Google Scholar]
- 7.Pagán OR. The first brain : the neuroscience of planarians. 2014 [Google Scholar]
- 8.Lobo D, Beane WS, Levin M. Modeling planarian regeneration: a primer for reverse-engineering the worm. PLoS computational biology. 2012;8(4):e1002481. doi: 10.1371/journal.pcbi.1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheiman IM, Kreshchenko ND. Regeneration of Planarians: Experimental Object. Russ J Dev Biol+ 2015;46(1):1–9. [PubMed] [Google Scholar]
- 10.Adell T, Cebria F, Salo E. Gradients in planarian regeneration and homeostasis. Cold Spring Harbor perspectives in biology. 2010;2(1):a000505. doi: 10.1101/cshperspect.a000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salo E, Abril JF, Adell T, Cebria F, Eckelt K, Fernandez-Taboada E, Handberg-Thorsager M, Iglesias M, Molina MD, Rodriguez-Esteban G. Planarian regeneration: achievements and future directions after 20 years of research. Int J Dev Biol. 2009;53(8–10):1317–27. doi: 10.1387/ijdb.072414es. [DOI] [PubMed] [Google Scholar]
- 12.Oviedo NJ, Newmark PA, Sanchez Alvarado A. Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev Dyn. 2003;226(2):326–33. doi: 10.1002/dvdy.10228. [DOI] [PubMed] [Google Scholar]
- 13.Bardeen CR, Baetjer FH. The inhibitive action of the Roentgen rays on regeneration in planarians. Journal of Experimental Zoology. 1904;1(1):191–195. [Google Scholar]
- 14.Salvetti A, Rossi L, Bonuccelli L, Lena A, Pugliesi C, Rainaldi G, Evangelista M, Gremigni V. Adult stem cell plasticity: neoblast repopulation in non-lethally irradiated planarians. Dev Biol. 2009;328(2):305–14. doi: 10.1016/j.ydbio.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 15.Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8(5):635–49. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner DE, Ho JJ, Reddien PW. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell stem cell. 2012;10(3):299–311. doi: 10.1016/j.stem.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332(6031):811–6. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubois F. Contribution a l’etude de la migration des cellules de regeneration chez les Planaires dulcicoles. Bull Biol Fr Belg. 1949;83:213–83. [Google Scholar]
- 19.Hicks C, Sorocco D, Levin M. Automated analysis of behavior: A computer-controlled system for drug screening and the investigation of learning. J Neurobiol. 2006;66(9):977–90. doi: 10.1002/neu.20290. [DOI] [PubMed] [Google Scholar]
- 20.Cherkashin AN, Sheiman IM. Conditioning in planarians and RNA content. J Biol Psychol. 1967;9(1):5–11. [Google Scholar]
- 21.Westerman RA. Somatic Inheritance of Habituation of Responses to Light in Planarians. Science. 1963;140(3567):676–677. doi: 10.1126/science.140.3567.676. [DOI] [PubMed] [Google Scholar]
- 22.Thompson R, McConnell JV. Classical conditioning in the planarian Dugesia dorotocephala. Journal of comparative and physiological psychology. 1955;48:65–68. doi: 10.1037/h0041147. [DOI] [PubMed] [Google Scholar]
- 23.Humpheries B, McConnell JV. Factors affecting maze learning in planarians. Worm Runner’s Digest. 1964;6:52–59. [Google Scholar]
- 24.McConnell JV. The Worm Runner’s Digest. Ann Arbor: Michigan; 1965. A manual of psychological experimentation on planarians; p. 111. [Google Scholar]
- 25.Shomrat T, Levin M. An automated training paradigm reveals long-term memory in planarians and its persistence through head regeneration. The Journal of experimental biology. 2013;216(Pt 20):3799–810. doi: 10.1242/jeb.087809. [DOI] [PubMed] [Google Scholar]
- 26.Corning WC. Retention of a position discrimination after regeneration in planarians. Psychanomic Science. 1966;5:17–18. [Google Scholar]
- 27.McConnell JV. Memory transfer through cannibalism in planarians. Journal of Neuropsychiatry. 1962;3:42–48. [Google Scholar]
- 28.La A, Jacobson M, James V. Research on learning in the planarian. Carolina Tips. 1962;XXV(7):25–27. [Google Scholar]
- 29.Oviedo NJ, Beane WS. Regeneration: The origin of cancer or a possible cure? Semin Cell Dev Biol. 2009;20(5):557–64. doi: 10.1016/j.semcdb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seilern-Aspang F, Kratochwill L. Relation between regeneration and tumor growth, Regeneration in animals and related problems. North-Holland Publishing Company; Amsterdam: 1965. pp. 452–73. [Google Scholar]
- 31.Doursat R, Sanchez C. Growing fine-grained multicellular robots. Soft Robotics. 2014;1(2):110–121. [Google Scholar]
- 32.Doursat R, Sayama H, Michel O. A review of morphogenetic engineering. Nat Comput. 2013;12(4):517–535. [Google Scholar]
- 33.Katz E. Biocomputing - tools, aims, perspectives. Curr Opin Biotechnol. 2015;34:202–8. doi: 10.1016/j.copbio.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Fuchslin RM, Dzyakanchuk A, Flumini D, Hauser H, Hunt KJ, Luchsinger RH, Reller B, Scheidegger S, Walker R. Morphological computation and morphological control: steps toward a formal theory and applications. Artificial life. 2013;19(1):9–34. doi: 10.1162/ARTL_a_00079. [DOI] [PubMed] [Google Scholar]
- 35.Hauser H, Ijspeert AJ, Fuchslin RM, Pfeifer R, Maass W. Towards a theoretical foundation for morphological computation with compliant bodies. Biological cybernetics. 2012 doi: 10.1007/s00422-012-0471-0. [DOI] [PubMed] [Google Scholar]
- 36.Kano T, Sato E, Ono T, Aonuma H, Matsuzaka Y, Ishiguro A. A brittle star-like robot capable of immediately adapting to unexpected physical damage. Roy Soc Open Sci. 2017;4(12) doi: 10.1098/rsos.171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bongard J, Zykov V, Lipson H. Resilient machines through continuous self-modeling. Science. 2006;314(5802):1118–21. doi: 10.1126/science.1133687. [DOI] [PubMed] [Google Scholar]
- 38.Pfeifer R, Gomez G. Morphological Computation - Connecting Brain, Body, and Environment. Creating Brain-Like Intelligence: From Basic Principles to Complex Intelligent Systems. 2009;5436:66–83. [Google Scholar]
- 39.Pezzulo G, Levin M. Re-membering the body: applications of computational neuroscience to the top-down control of regeneration of limbs and other complex organs. Integr Biol (Camb) 2015;7(12):1487–517. doi: 10.1039/c5ib00221d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friston K, Levin M, Sengupta B, Pezzulo G. Knowing one’s place: a free-energy approach to pattern regulation. J R Soc Interface. 2015;12(105) doi: 10.1098/rsif.2014.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangel M, Bonsall MB, Aboobaker A. Feedback control in planarian stem cell systems. BMC Syst Biol. 2016;10(1):17. doi: 10.1186/s12918-016-0261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang B, An Y, Agata K, Umesono Y. Two distinct roles of the yorkie/yap gene during homeostasis in the planarian Dugesia japonica. Dev Growth Differ. 2015;57(3):209–17. doi: 10.1111/dgd.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill EM, Petersen CP. Wnt/Notum spatial feedback inhibition controls neoblast differentiation to regulate reversible growth of the planarian brain. Development. 2015 doi: 10.1242/dev.123612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Owlarn S, Bartscherer K. Go ahead, grow a head! A planarian’s guide to anterior regeneration. Regeneration (Oxf) 2016;3(3):139–55. doi: 10.1002/reg2.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuhof M, Levin M, Rechavi O. Vertically- and horizontally-transmitted memories - the fading boundaries between regeneration and inheritance in planaria. Biol Open. 2016;5(9):1177–88. doi: 10.1242/bio.020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo L, Accorsi A, He S, Guerrero-Hernández C, Sivagnanam S, McKinney S, Gibson M, Sánchez Alvarado A. An adaptable chromosome preparation methodology for use in invertebrate research organisms. BMC Biology. 2018;16(1):25. doi: 10.1186/s12915-018-0497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimura O, Hosoda K, Kawaguchi E, Yazawa S, Hayashi T, Inoue T, Umesono Y, Agata K. Unusually Large Number of Mutations in Asexually Reproducing Clonal Planarian Dugesia japonica. PLoS One. 2015;10(11):e0143525. doi: 10.1371/journal.pone.0143525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grohme MA, Schloissnig S, Rozanski A, Pippel M, Young GR, Winkler S, Brandl H, Henry I, Dahl A, Powell S, Hiller M, Myers E, Rink JC. The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature. 2018;554(7690):56–61. doi: 10.1038/nature25473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross KG, Currie KW, Pearson BJ, Zayas RM. Nervous system development and regeneration in freshwater planarians. Wiley Interdiscip Rev Dev Biol. 2017 doi: 10.1002/wdev.266. [DOI] [PubMed] [Google Scholar]
- 50.Barghouth PG, Thiruvalluvan M, Oviedo NJ. Bioelectrical regulation of cell cycle and the planarian model system. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbamem.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robb SM, Sanchez Alvarado A. Histone modifications and regeneration in the planarian Schmidtea mediterranea. Current topics in developmental biology. 2014;108:71–93. doi: 10.1016/B978-0-12-391498-9.00004-8. [DOI] [PubMed] [Google Scholar]
- 52.Fields C, Levin M. Multiscale memory and bioelectric error correction in the cytoplasm–cytoskeleton-membrane system. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2017:e1410. doi: 10.1002/wsbm.1410. n/a. [DOI] [PubMed] [Google Scholar]
- 53.Robertson JA. Galvanotropic reactions of Polycelis nigra in relation to inherent electrical polarity. Brit J Exp Biol. 1927;5:66–88. [Google Scholar]
- 54.Hyman L. Studies on the correlation between metabolic gradients, electrical gradients, and galvanotaxis II: Galvanotaxis of the brown hydra and some non-fissioning planarians. Physiol Zool. 1932;5(2):185–190. [Google Scholar]
- 55.Marsh G, Beams HW. Electrical Control of Growth Polarity in Regenerating Dugesia- Tigrina. Federation Proceedings. 1947;6(1):163–164. [PubMed] [Google Scholar]
- 56.Marsh G, Beams HW. Electrical control of morphogenesis in regenerating dugesia tigrina. I. Relation of axial polarity to field strength. J Cell Comp Physiol. 1952;39(2):191–213. doi: 10.1002/jcp.1030390203. [DOI] [PubMed] [Google Scholar]
- 57.Bonaventure N. Galvanotropisme De Regenerats Monstrueux De Planaires - Monstres Bifides Et Heteromorphoses. Comptes Rendus Des Seances De La Societe De Biologie Et De Ses Filiales. 1957;151(3):598–602. [PubMed] [Google Scholar]
- 58.Levin M, Pezzulo G, Finkelstein JM. Endogenous Bioelectric Signaling Networks: Exploiting Voltage Gradients for Control of Growth and Form. Annual Review of Biomedical Engineering. 2017;19:353–387. doi: 10.1146/annurev-bioeng-071114-040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bates E. Ion Channels in Development and Cancer. Annu Rev Cell Dev Biol. 2015;31:231–47. doi: 10.1146/annurev-cellbio-100814-125338. [DOI] [PubMed] [Google Scholar]
- 60.Rasskin-Gutman D, Izpisua-Belmonte JC. Theoretical morphology of developmental asymmetries. Bioessays. 2004;26(4):405–12. doi: 10.1002/bies.10410. [DOI] [PubMed] [Google Scholar]
- 61.Sullivan KG, Emmons-Bell M, Levin M. Physiological inputs regulate species-specific anatomy during embryogenesis and regeneration. Commun Integr Biol. 2016;9(4):e1192733. doi: 10.1080/19420889.2016.1192733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wildman MH, Cannone AJ. Action potentials in a ‘non-spiking’ neurone: graded responses and spikes in the afferent P fibre of the crab thoracic-coxal muscle receptor organ. Brain Res. 1990;509(2):339–42. doi: 10.1016/0006-8993(90)90562-p. [DOI] [PubMed] [Google Scholar]
- 63.Min R, Santello M, Nevian T. The computational power of astrocyte mediated synaptic plasticity. Front Comput Neurosci. 2012;6:93. doi: 10.3389/fncom.2012.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Angarita-Jaimes N, Dewhirst OP, Simpson DM, Kondoh Y, Allen R, Newland PL. The dynamics of analogue signalling in local networks controlling limb movement. The European journal of neuroscience. 2012;36(9):3269–82. doi: 10.1111/j.1460-9568.2012.08236.x. [DOI] [PubMed] [Google Scholar]
- 65.Marin-Burgin A, Szczupak L. Processing of sensory signals by a non-spiking neuron in the leech. J Comp Physiol (A) 2000;186(10):989–97. doi: 10.1007/s003590000152. [DOI] [PubMed] [Google Scholar]
- 66.Victor JD. Temporal aspects of neural coding in the retina and lateral geniculate. Network. 1999;10(4):R1–66. [PubMed] [Google Scholar]
- 67.Lyon P. The biogenic approach to cognition. Cogn Process. 2006;7(1):11–29. doi: 10.1007/s10339-005-0016-8. [DOI] [PubMed] [Google Scholar]
- 68.Baluska F, Levin M. On Having No Head: Cognition throughout Biological Systems. Front Psychol. 2016;7:902. doi: 10.3389/fpsyg.2016.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liebeskind BJ, Hillis DM, Zakon HH. Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci U S A. 2011;108(22):9154–9. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harris KD, Weiss M, Zahavi A. Why are neurotransmitters neurotoxic? An evolutionary perspective. F1000Res. 2014;3:179. doi: 10.12688/f1000research.4828.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Levin M, Buznikov GA, Lauder JM. Of minds and embryos: left-right asymmetry and the serotonergic controls of pre-neural morphogenesis. Dev Neurosci. 2006;28(3):171–85. doi: 10.1159/000091915. [DOI] [PubMed] [Google Scholar]
- 72.Pitcairn E, McLaughlin KA. Bioelectric signaling coordinates patterning decisions during embryogenesis. Trends in Developmental Biology. 2016;9:1–9. [Google Scholar]
- 73.Levin M, Martyniuk CJ. The bioelectric code: An ancient computational medium for dynamic control of growth and form. Biosystems. 2017 doi: 10.1016/j.biosystems.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McLaughlin KA, Levin M. Bioelectric signaling in regeneration: Mechanisms of ionic controls of growth and form. Dev Biol. 2018;433(2):177–189. doi: 10.1016/j.ydbio.2017.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pullar CE. The physiology of bioelectricity in development, tissue regeneration, and cancer. CRC Press; Boca Raton: 2011. [Google Scholar]
- 76.Lund E. Bioelectric fields and growth. Univ. of Texas Press; Austin: 1947. [Google Scholar]
- 77.Morgan TH, Dimon AC. An examination of the problem of physiological ‘polarity’ and electrical polarity in the earthworm. Journal of Experimental Zoology I. 1904 [Google Scholar]
- 78.Morgan TH. Regeneration in planarians. Bryn Mawr. Pa; 1901. [Google Scholar]
- 79.Child CM. The problem of pattern in organisms II. The physiological gradients. American Naturalist. 1924;58:322–336. [Google Scholar]
- 80.Marsh G, Beams HW. Electrical Control of Growth Axis in a Regenerating Annelid. Anatomical Record. 1950;108(3):512–512. [Google Scholar]
- 81.Marsh G, Beams HW. Electrical Control of Axial Polarity in a Regenerating Annelid. Anatomical Record. 1949;105(3):513–514. [Google Scholar]
- 82.Lange CS, Steele VE. The mechanism of anterior-posterior polarity control in planarians. Differentiation. 1978;11(1):1–12. doi: 10.1111/j.1432-0436.1978.tb00965.x. [DOI] [PubMed] [Google Scholar]
- 83.Steele VE, Lange CS. Characterization of an organ-specific differentiator substance in the planarian Dugesia etrusca. J Embryol Exp Morphol. 1977;37(1):159–72. [PubMed] [Google Scholar]
- 84.Levin M. Is the early left-right axis like a plant, a kidney, or a neuron? The integration of physiological signals in embryonic asymmetry. Birth Defects Res C Embryo Today. 2006;78(3):191–223. doi: 10.1002/bdrc.20078. [DOI] [PubMed] [Google Scholar]
- 85.Oviedo NJ, Nicolas CL, Adams DS, Levin M. Live Imaging of Planarian Membrane Potential Using DiBAC4(3) Cold Spring Harb Protoc. 2008;(11) doi: 10.1101/pdb.prot5055. 2008 pdb.prot5055- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adams DS, Levin M. Measuring resting membrane potential using the fluorescent voltage reporters DiBAC4(3) and CC2-DMPE. Cold Spring Harbor protocols. 2012;2012(4):459–64. doi: 10.1101/pdb.prot067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adams DS, Levin M. General principles for measuring resting membrane potential and ion concentration using fluorescent bioelectricity reporters. Cold Spring Harbor protocols. 2012;2012(4):385–97. doi: 10.1101/pdb.top067710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adams DS. A new tool for tissue engineers: ions as regulators of morphogenesis during development and regeneration. Tissue engineering. 2008;14(9):1461–8. doi: 10.1089/ten.tea.2008.0080. [DOI] [PubMed] [Google Scholar]
- 89.Adams DS, Levin M. Endogenous voltage gradients as mediators of cell-cell communication: strategies for investigating bioelectrical signals during pattern formation. Cell Tissue Res. 2013;352(1):95–122. doi: 10.1007/s00441-012-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Levin M, Thorlin T, Robinson KR, Nogi T, Mercola M. Asymmetries in H+/K+-ATPase and cell membrane potentials comprise a very early step in left-right patterning. Cell. 2002;111(1):77–89. doi: 10.1016/s0092-8674(02)00939-x. [DOI] [PubMed] [Google Scholar]
- 91.Pai VP, Lemire JM, Pare JF, Lin G, Chen Y, Levin M. Endogenous Gradients of Resting Potential Instructively Pattern Embryonic Neural Tissue via Notch Signaling and Regulation of Proliferation. The Journal of Neuroscience. 2015;35(10):4366–85. doi: 10.1523/JNEUROSCI.1877-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Perathoner S, Daane JM, Henrion U, Seebohm G, Higdon CW, Johnson SL, Nusslein-Volhard C, Harris MP. Bioelectric signaling regulates size in zebrafish fins. PLoS genetics. 2014;10(1):e1004080. doi: 10.1371/journal.pgen.1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dahal GR, Pradhan SJ, Bates EA. Inwardly rectifying potassium channels influence Drosophila wing morphogenesis by regulating Dpp release. Development. 2017;144(15):2771–2783. doi: 10.1242/dev.146647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tseng AS, Beane WS, Lemire JM, Masi A, Levin M. Induction of vertebrate regeneration by a transient sodium current. J Neurosci. 2010;30(39):13192–200. doi: 10.1523/JNEUROSCI.3315-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Adams DS, Uzel SG, Akagi J, Wlodkowic D, Andreeva V, Yelick PC, Devitt-Lee A, Pare JF, Levin M. Bioelectric signalling via potassium channels: a mechanism for craniofacial dysmorphogenesis in KCNJ2-associated Andersen-Tawil Syndrome. J Physiol. 2016;594(12):3245–3270. doi: 10.1113/JP271930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Masotti A, Uva P, Davis-Keppen L, Basel-Vanagaite L, Cohen L, Pisaneschi E, Celluzzi A, Bencivenga P, Fang M, Tian M, Xu X, Cappa M, Dallapiccola B. Keppen-Lubinsky Syndrome Is Caused by Mutations in the Inwardly Rectifying K(+) Channel Encoded by KCNJ6. Am J Hum Genet. 2015;96(2):295–300. doi: 10.1016/j.ajhg.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chernet BT, Adams DS, Lobikin M, Levin M. Use of genetically encoded, light-gated ion translocators to control tumorigenesis. Oncotarget. 2016;7(15):19575–88. doi: 10.18632/oncotarget.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Adams DS, Lemire JM, Kramer RH, Levin M. Optogenetics in Developmental Biology: using light to control ion flux-dependent signals in Xenopus embryos. The International journal of developmental biology. 2014;58:851–861. doi: 10.1387/ijdb.140207ml. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Adams DS, Tseng AS, Levin M. Light-activation of the Archaerhodopsin H(+)-pump reverses age-dependent loss of vertebrate regeneration: sparking system-level controls in vivo. Biology open. 2013;2(3):306–13. doi: 10.1242/bio.20133665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beane WS, Morokuma J, Adams DS, Levin M. A Chemical genetics approach reveals H,K-ATPase-mediated membrane voltage is required for planarian head regeneration. Chemistry & Biology. 2011;18(1):77–89. doi: 10.1016/j.chembiol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beane WS, Morokuma J, Lemire JM, Levin M. Bioelectric signaling regulates head and organ size during planarian regeneration. Development. 2013;140(2):313–22. doi: 10.1242/dev.086900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nuckels RJ, Ng A, Darland T, Gross JM. The vacuolar-ATPase complex regulates retinoblast proliferation and survival, photoreceptor morphogenesis, and pigmentation in the zebrafish eye. Invest Ophthalmol Vis Sci. 2009;50(2):893–905. doi: 10.1167/iovs.08-2743. [DOI] [PubMed] [Google Scholar]
- 103.Palacios-Prado N, Bukauskas FF. Heterotypic gap junction channels as voltage-sensitive valves for intercellular signaling. Proc Natl Acad Sci U S A. 2009;106(35):14855–60. doi: 10.1073/pnas.0901923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mathews J, Levin M. Gap junctional signaling in pattern regulation: Physiological network connectivity instructs growth and form. Developmental neurobiology. 2017;77(5):643–673. doi: 10.1002/dneu.22405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oviedo NJ, Levin M. smedinx-11 is a planarian stem cell gap junction gene required for regeneration and homeostasis. Development. 2007;134(17):3121–31. doi: 10.1242/dev.006635. [DOI] [PubMed] [Google Scholar]
- 106.Oviedo NJ, Morokuma J, Walentek P, Kema IP, Gu MB, Ahn JM, Hwang JS, Gojobori T, Levin M. Long-range neural and gap junction protein-mediated cues control polarity during planarian regeneration. Dev Biol. 2010;339(1):188–99. doi: 10.1016/j.ydbio.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Buznikov GA, Peterson RE, Nikitina LA, Bezuglov VV, Lauder JM. The Pre-nervous Serotonergic System of Developing Sea Urchin Embryos and Larvae: Pharmacologic and Immunocytochemical Evidence. Neurochem Res. 2005;30(6–7):825–37. doi: 10.1007/s11064-005-6876-6. [DOI] [PubMed] [Google Scholar]
- 108.Fukumoto T, Blakely R, Levin M. Serotonin transporter function is an early step in left-right patterning in chick and frog embryos. Dev Neurosci. 2005;27(6):349–63. doi: 10.1159/000088451. [DOI] [PubMed] [Google Scholar]
- 109.Fukumoto T, Kema I, Nazarenko D, Levin M. Serotonin is a novel very early signaling mechanism in left-right asymmetry. Developmental Biology. 2003;259:490a. [Google Scholar]
- 110.Buznikov G, Shmukler Y, Lauder J. From oocyte to neuron: do neurotransmitters function in the same way throughout development? Cell Molec Neurobiol. 1996;16(5):537–59. doi: 10.1007/BF02152056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Villar D, Schaeffer DJ. Morphogenetic action of neurotransmitters on regenerating planarians--a review. Biomed Environ Sci. 1993;6(4):327–47. [PubMed] [Google Scholar]
- 112.Chan JD, Zhang D, Liu X, Zarowiecki M, Berriman M, Marchant JS. Utilizing the planarian voltage-gated ion channel transcriptome to resolve a role for a Ca2+ channel in neuromuscular function and regeneration. Biochim Biophys Acta. 2017;1864(6):1036–1045. doi: 10.1016/j.bbamcr.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chan JD, Zhang D, Liu X, Zarowiecki MZ, Berriman M, Marchant JS. Dataset for a Dugesia japonica de novo transcriptome assembly, utilized for defining the voltage-gated like ion channel superfamily. Data Brief. 2016;9:1044–1047. doi: 10.1016/j.dib.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chan JD, McCorvy JD, Acharya S, Johns ME, Day TA, Roth BL, Marchant JS. A Miniaturized Screen of a Schistosoma mansoni Serotonergic G Protein-Coupled Receptor Identifies Novel Classes of Parasite-Selective Inhibitors. PLoS Pathog. 2016;12(5):e1005651. doi: 10.1371/journal.ppat.1005651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chan JD, Grab T, Marchant JS. Kinetic profiling an abundantly expressed planarian serotonergic GPCR identifies bromocriptine as a perdurant antagonist. Int J Parasitol Drugs Drug Resist. 2016;6(3):356–363. doi: 10.1016/j.ijpddr.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chan JD, Agbedanu PN, Zamanian M, Gruba SM, Haynes CL, Day TA, Marchant JS. ‘Death and Axes’: Unexpected Ca(2+) Entry Phenologs Predict New Anti-schistosomal Agents. PLoS Pathog. 2014;10(2):e1003942. doi: 10.1371/journal.ppat.1003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang D, Chan JD, Nogi T, Marchant JS. Opposing roles of voltage-gated Ca2+ channels in neuronal control of regenerative patterning. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(44):15983–95. doi: 10.1523/JNEUROSCI.3029-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nogi T, Zhang D, Chan JD, Marchant JS. A novel biological activity of praziquantel requiring voltage-operated ca channel Beta subunits: subversion of flatworm regenerative polarity. PLoS neglected tropical diseases. 2009;3(6):e464. doi: 10.1371/journal.pntd.0000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nguyen QT, Schroeder LF, Mank M, Muller A, Taylor P, Griesbeck O, Kleinfeld D. An in vivo biosensor for neurotransmitter release and in situ receptor activity. Nat Neurosci. 2010;13(1):127–32. doi: 10.1038/nn.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Balaconis MK, Clark HA. Biodegradable optode-based nanosensors for in vivo monitoring. Anal Chem. 2012;84(13):5787–93. doi: 10.1021/ac301137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cabrera R, Filevich O, Garcia-Acosta B, Athilingam J, Bender KJ, Poskanzer KE, Etchenique R. A Visible-Light-Sensitive Caged Serotonin. ACS Chem Neurosci. 2017;8(5):1036–1042. doi: 10.1021/acschemneuro.7b00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rea AC, Vandenberg LN, Ball RE, Snouffer AA, Hudson AG, Zhu Y, McLain DE, Johnston LL, Lauderdale JD, Levin M, Dore TM. Light-activated serotonin for exploring its action in biological systems. Chemistry & biology. 2013;20(12):1536–46. doi: 10.1016/j.chembiol.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Blackiston D, Adams DS, Lemire JM, Lobikin M, Levin M. Transmembrane potential of GlyCl-expressing instructor cells induces a neoplastic-like conversion of melanocytes via a serotonergic pathway. Disease models & mechanisms. 2011;4(1):67–85. doi: 10.1242/dmm.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Emmons-Bell M, Durant F, Hammelman J, Bessonov N, Volpert V, Morokuma J, Pinet K, Adams DS, Pietak A, Lobo D, Levin M. Gap Junctional Blockade Stochastically Induces Different Species-Specific Head Anatomies in Genetically Wild-Type Girardia dorotocephala Flatworms. Int J Mol Sci. 2015;16(11):27865–96. doi: 10.3390/ijms161126065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pietak A, Levin M. Bioelectric gene and reaction networks: computational modelling of genetic, biochemical and bioelectrical dynamics in pattern regulation. J R Soc Interface. 2017;14(134) doi: 10.1098/rsif.2017.0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pezzulo G, Levin M. Top-down models in biology: explanation and control of complex living systems above the molecular level. J R Soc Interface. 2016;13(124) doi: 10.1098/rsif.2016.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thompson DAW, Whyte LL. On growth and form. The University Press; Cambridge Eng: 1942. A new ed. [Google Scholar]
- 128.Lau K, Tao H, Liu H, Wen J, Sturgeon K, Sorfazlian N, Lazic S, Burrows JT, Wong MD, Li D, Deimling S, Ciruna B, Scott I, Simmons C, Henkelman RM, Williams T, Hadjantonakis AK, Fernandez-Gonzalez R, Sun Y, Hopyan S. Anisotropic stress orients remodelling of mammalian limb bud ectoderm. Nat Cell Biol. 2015;17(5):569–79. doi: 10.1038/ncb3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Munjal A, Philippe JM, Munro E, Lecuit T. A self-organized biomechanical network drives shape changes during tissue morphogenesis. Nature. 2015;524(7565):351–5. doi: 10.1038/nature14603. [DOI] [PubMed] [Google Scholar]
- 130.Bubenik AB, Pavlansky R. Trophic responses to trauma in growing antlers. J Exp Zool. 1965;159(3):289–302. doi: 10.1002/jez.1401590302. [DOI] [PubMed] [Google Scholar]
- 131.Lobo D, Solano M, Bubenik GA, Levin M. A linear-encoding model explains the variability of the target morphology in regeneration. Journal of the Royal Society, Interface / the Royal Society. 2014;11(92):20130918. doi: 10.1098/rsif.2013.0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Durant F, Morokuma J, Fields C, Williams K, Adams DS, Levin M. Long-Term, Stochastic Editing of Regenerative Anatomy via Targeting Endogenous Bioelectric Gradients. Biophys J. 2017;112(10):2231–2243. doi: 10.1016/j.bpj.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hubert A, Henderson JM, Ross KG, Cowles MW, Torres J, Zayas RM. Epigenetic regulation of planarian stem cells by the SET1/MLL family of histone methyltransferases. Epigenetics. 2013;8(1):79–91. doi: 10.4161/epi.23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pietak A, Levin M. Exploring Instructive Physiological Signaling with the Bioelectric Tissue Simulation Engine (BETSE) Frontiers in Bioengineering and Biotechnology. 2016;4 doi: 10.3389/fbioe.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Burr HS, Northrop FSC. The electro-dynamic theory of life. Quarterly Review of Biology. 1935;10(3):322–333. [Google Scholar]
- 136.Cervera J, Meseguer S, Mafe S. The interplay between genetic and bioelectrical signaling permits a spatial regionalisation of membrane potentials in model multicellular ensembles. Sci Rep. 2016;6:35201. doi: 10.1038/srep35201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tseng AS, Levin M. Transducing bioelectric signals into epigenetic pathways during tadpole tail regeneration. Anatomical record. 2012;295(10):1541–51. doi: 10.1002/ar.22495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ryland KE, Hawkins AG, Weisenberger DJ, Punj V, Borinstein SC, Laird PW, Martens JR, Lawlor ER. Promoter Methylation Analysis Reveals That KCNA5 Ion Channel Silencing Supports Ewing Sarcoma Cell Proliferation. Mol Cancer Res. 2016;14(1):26–34. doi: 10.1158/1541-7786.MCR-15-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cherry JL, Adler FR. How to make a biological switch. J Theor Biol. 2000;203(2):117–33. doi: 10.1006/jtbi.2000.1068. [DOI] [PubMed] [Google Scholar]
- 140.Stuckemann T, Cleland JP, Werner S, Thi-Kim Vu H, Bayersdorf R, Liu SY, Friedrich B, Julicher F, Rink JC. Antagonistic Self-Organizing Patterning Systems Control Maintenance and Regeneration of the Anteroposterior Axis in Planarians. Dev Cell. 2017;40(3):248–263e4. doi: 10.1016/j.devcel.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 141.De A, Chakravarthy VS, Levin M. A computational model of planarian regeneration. International Journal of Parallel, Emergent and Distributed Systems. 2016:1–17. [Google Scholar]
- 142.Morgan TH. Experimental studies of the regeneration of Planaria maculata. Arch Entwicklungsmech Org. 1898;7:364–397. [Google Scholar]
- 143.Castillo-Lara S, Abril JF. PlanNET: Homology-based predicted interactome for multiple planarian transcriptomes. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Brandl H, Moon H, Vila-Farre M, Liu SY, Henry I, Rink JC. PlanMine--a mineable resource of planarian biology and biodiversity. Nucleic Acids Res. 2016;44(D1):D764–73. doi: 10.1093/nar/gkv1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Werner S, Stückemann T, Beirán Amigo M, Rink JC, Jülicher F, Friedrich BM. Scaling and Regeneration of Self-Organized Patterns. Physical Review Letters. 2015;114(13):138101. doi: 10.1103/PhysRevLett.114.138101. [DOI] [PubMed] [Google Scholar]
- 146.Meinhardt H. Beta-catenin and axis formation in planarians. Bioessays. 2009;31(1):5–9. doi: 10.1002/bies.080193. [DOI] [PubMed] [Google Scholar]
- 147.Lander AD. Morpheus unbound: reimagining the morphogen gradient. Cell. 2007;128(2):245–56. doi: 10.1016/j.cell.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 148.Witchley JN, Mayer M, Wagner DE, Owen JH, Reddien PW. Muscle cells provide instructions for planarian regeneration. Cell Rep. 2013;4(4):633–41. doi: 10.1016/j.celrep.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Scimone ML, Cote LE, Rogers T, Reddien PW. Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. Elife. 2016;5 doi: 10.7554/eLife.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Almuedo-Castillo M, Sureda-Gomez M, Adell T. Wnt signaling in planarians: new answers to old questions. Int J Dev Biol. 2012;56(1–3):53–65. doi: 10.1387/ijdb.113451ma. [DOI] [PubMed] [Google Scholar]
- 151.Gurley KA, Rink JC, Sanchez Alvarado A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319(5861):323–7. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sureda-Gomez M, Pascual-Carreras E, Adell T. Posterior Wnts Have Distinct Roles in Specification and Patterning of the Planarian Posterior Region. Int J Mol Sci. 2015;16(11):26543–54. doi: 10.3390/ijms161125970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Agata K, Tasaki J, Nakajima E, Umesono Y. Recent identification of an ERK signal gradient governing planarian regeneration. Zoology (Jena) 2014;117(3):161–2. doi: 10.1016/j.zool.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 154.Umesono Y, Tasaki J, Nishimura Y, Hrouda M, Kawaguchi E, Yazawa S, Nishimura O, Hosoda K, Inoue T, Agata K. The molecular logic for planarian regeneration along the anterior-posterior axis. Nature. 2013;500(7460):73–6. doi: 10.1038/nature12359. [DOI] [PubMed] [Google Scholar]
- 155.Tasaki J, Shibata N, Nishimura O, Itomi K, Tabata Y, Son F, Suzuki N, Araki R, Abe M, Agata K, Umesono Y. ERK signaling controls blastema cell differentiation during planarian regeneration. Development. 2011;138(12):2417–27. doi: 10.1242/dev.060764. [DOI] [PubMed] [Google Scholar]
- 156.Lander R, Petersen CP. Wnt, Ptk7, and FGFRL expression gradients control trunk positional identity in planarian regeneration. Elife. 2016;5 doi: 10.7554/eLife.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang IE, Lapan SW, Scimone ML, Clandinin TR, Reddien PW. Hedgehog signaling regulates gene expression in planarian glia. Elife. 2016;5 doi: 10.7554/eLife.16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yazawa S, Umesono Y, Hayashi T, Tarui H, Agata K. Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc Natl Acad Sci U S A. 2009;106(52):22329–34. doi: 10.1073/pnas.0907464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Petersen CP, Reddien PW. Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science. 2011;332(6031):852–5. doi: 10.1126/science.1202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Meinhardt H. Models for the generation and interpretation of gradients. Cold Spring Harbor perspectives in biology. 2009;1(4):a001362. doi: 10.1101/cshperspect.a001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Meinhardt H, Gierer A. Pattern formation by local self-activation and lateral inhibition. Bioessays. 2000;22(8):753–60. doi: 10.1002/1521-1878(200008)22:8<753::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 162.Nuccitelli R. Endogenous ionic currents and DC electric fields in multicellular animal tissues. Bioelectromagnetics Suppl. 1992;1:147–57. doi: 10.1002/bem.2250130714. [DOI] [PubMed] [Google Scholar]
- 163.Nuccitelli R. Endogenous electric fields in embryos during development regeneration and wound healing. Radiat Prot Dosimetry. 2003;106(4):375–83. doi: 10.1093/oxfordjournals.rpd.a006375. [DOI] [PubMed] [Google Scholar]
- 164.Zhang Y, Levin M. Particle tracking model of electrophoretic morphogen movement reveals stochastic dynamics of embryonic gradient. Dev Dyn. 2009;238(8):1923–35. doi: 10.1002/dvdy.22016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Blackiston DJ, Anderson GM, Rahman N, Bieck C, Levin M. A novel method for inducing nerve growth via modulation of host resting potential: gap junction-mediated and serotonergic signaling mechanisms. Neurotherapeutics. 2015;12(1):170–84. doi: 10.1007/s13311-014-0317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dimmitt J, Marsh G. Electrical Control of Morphogenesis in Regenerating Dugesia- Tigrina. 2. Potential Gradient Vs Current Density as Control Factors. J Cell Comp Physiol. 1952;40(1):11–23. doi: 10.1002/jcp.1030400103. [DOI] [PubMed] [Google Scholar]
- 167.Ghosh S. Application of Computational Methods in Planaria Research: A Current Update. J Integr Bioinform. 2017 doi: 10.1515/jib-2017-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lobo D, Levin M. Inferring Regulatory Networks from Experimental Morphological Phenotypes: A Computational Method Reverse-Engineers Planarian Regeneration. PLoS computational biology. 2015;11(6):e1004295. doi: 10.1371/journal.pcbi.1004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lobo D, Malone TJ, Levin M. Planform: an application and database of graph-encoded planarian regenerative experiments. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lobo D, Malone TJ, Levin M. Towards a bioinformatics of patterning: a computational approach to understanding regulative morphogenesis. Biology Open. 2013;2(2):156–69. doi: 10.1242/bio.20123400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Meinhardt H. Models of biological pattern formation: from elementary steps to the organization of embryonic axes. Current topics in developmental biology. 2008;81:1–63. doi: 10.1016/S0070-2153(07)81001-5. [DOI] [PubMed] [Google Scholar]
- 172.Sparkes A, Aubrey W, Byrne E, Clare A, Khan MN, Liakata M, Markham M, Rowland J, Soldatova LN, Whelan KE, Young M, King RD. Towards Robot Scientists for autonomous scientific discovery. Autom Exp. 2010;2:1. doi: 10.1186/1759-4499-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lobo D, Hammelman J, Levin M. MoCha: Molecular Characterization of Unknown Pathways. J Comput Biol. 2016;23(4):291–7. doi: 10.1089/cmb.2015.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lobo D, Morokuma J, Levin M. Computational discovery and in vivo validation of hnf4 as a regulatory gene in planarian regeneration. Bioinformatics. 2016;32(17):2681–5. doi: 10.1093/bioinformatics/btw299. [DOI] [PubMed] [Google Scholar]
- 175.Lobikin M, Chernet B, Lobo D, Levin M. Resting potential, oncogene-induced tumorigenesis, and metastasis: the bioelectric basis of cancer in vivo. Physical biolog. 2012;9(6):065002. doi: 10.1088/1478-3975/9/6/065002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lobikin M, Lobo D, Blackiston DJ, Martyniuk CJ, Tkachenko E, Levin M. Serotonergic regulation of melanocyte conversion: A bioelectrically regulated network for stochastic all-or-none hyperpigmentation. Sci Signal. 2015;8(397):ra99. doi: 10.1126/scisignal.aac6609. [DOI] [PubMed] [Google Scholar]
- 177.Lobo D, Lobikin M, Levin M. Discovering novel phenotypes with automatically inferred dynamic models: a partial melanocyte conversion in Xenopus. Sci Rep. 2017;7:41339. doi: 10.1038/srep41339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.García-Quismondo M, Levin M, Lobo D. Modeling regenerative processes with membrane computing. Inf Sci. 2017;381:229–249. [Google Scholar]
- 179.Bessonov N, Levin M, Morozova N, Reinberg N, Tosenberger A, Volpert V. Target morphology and cell memory: a model of regenerative pattern formation. Neural Regen Res. 2015;10(12):1901–5. doi: 10.4103/1673-5374.165216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bessonov N, Levin M, Morozova N, Reinberg N, Tosenberger A, Volpert V. On a Model of Pattern Regeneration Based on Cell Memory. PloS one. 2015;10(2):e0118091. doi: 10.1371/journal.pone.0118091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tosenberger A, Bessonov N, Levin M, Reinberg N, Volpert V, Morozova N. A Conceptual Model of Morphogenesis and Regeneration. Acta biotheoretica. 2015;63(3):283–94. doi: 10.1007/s10441-015-9249-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pfeifer R, Iida F, Lungarella M. Cognition from the bottom up: on biological inspiration, body morphology, and soft materials. Trends Cogn Sci. 2014;18(8):404–413. doi: 10.1016/j.tics.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 183.Zahedi K, Ay N. Quantifying Morphological Computation. Entropy-Switz. 2013;15(5):1887. [Google Scholar]
- 184.Wilson MZ, Ravindran PT, Lim WA, Toettcher JE. Tracing Information Flow from Erk to Target Gene Induction Reveals Mechanisms of Dynamic and Combinatorial Control. Mol Cell. 2017;67(5):757–769e5. doi: 10.1016/j.molcel.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bugaj LJ, O’Donoghue GP, Lim WA. Interrogating cellular perception and decision making with optogenetic tools. J Cell Biol. 2017;216(1):25–28. doi: 10.1083/jcb.201612094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mitchell A, Lim W. Cellular perception and misperception: Internal models for decision-making shaped by evolutionary experience. Bioessays. 2016;38(9):845–9. doi: 10.1002/bies.201600090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Levin M. Endogenous bioelectrical networks store non-genetic patterning information during development and regeneration. The Journal of Physiology. 2014;592(11):2295–2305. doi: 10.1113/jphysiol.2014.271940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Noble D. A theory of biological relativity: no privileged level of causation. Interface Focus. 2012;2(1):55–64. doi: 10.1098/rsfs.2011.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pagan OR, Baker D, Deats S, Montgomery E, Tenaglia M, Randolph C, Kotturu D, Tallarida C, Bach D, Wilk G, Rawls S, Raffa RB. Planarians in pharmacology: parthenolide is a specific behavioral antagonist of cocaine in the planarian Girardia tigrina. The International journal of developmental biology. 2012;56(1–3):193–6. doi: 10.1387/ijdb.113486op. [DOI] [PubMed] [Google Scholar]
- 190.Mcculloch WS, Pitts W. A Logical Calculus of the Ideas Immanent in Nervous Activity. Bulletin of Mathematical Biology. 1990;52(1–2):99–115. [PubMed] [Google Scholar]
- 191.Friston K, Ao P. Free energy, value, and attractors. Comput Math Methods Med. 2012;2012:937860. doi: 10.1155/2012/937860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Friston KJ, Daunizeau J, Kilner J, Kiebel SJ. Action and behavior: a free-energy formulation. Biological cybernetics. 2010;102(3):227–60. doi: 10.1007/s00422-010-0364-z. [DOI] [PubMed] [Google Scholar]
- 193.Nogi T, Levin M. Characterization of innexin gene expression and functional roles of gap-junctional communication in planarian regeneration. Dev Biol. 2005;287(2):314–35. doi: 10.1016/j.ydbio.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 194.Binggeli R, Weinstein R. Membrane potentials and sodium channels: hypotheses for growth regulation and cancer formation based on changes in sodium channels and gap junctions. J Theor Biol. 1986;123:377–401. doi: 10.1016/s0022-5193(86)80209-0. [DOI] [PubMed] [Google Scholar]
- 195.MacFarlane SN, Sontheimer H. Changes in ion channel expression accompany cell cycle progression of spinal cord astrocytes. Glia. 2000;30(1):39–48. doi: 10.1002/(sici)1098-1136(200003)30:1<39::aid-glia5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 196.Rouzaire-Dubois B, Gerard V, Dubois JM. Involvement of K+ channels in the quercetin-induced inhibition of neuroblastoma cell growth. Pflugers Arch. 1993;423(3–4):202–5. doi: 10.1007/BF00374395. [DOI] [PubMed] [Google Scholar]
- 197.Wonderlin WF, Strobl JS. Potassium channels, proliferation and G1 progression. J Membr Biol. 1996;154(2):91–107. doi: 10.1007/s002329900135. [DOI] [PubMed] [Google Scholar]
- 198.Cone CD. The role of the surface electrical transmembrane potential in normal and malignant mitogenesis. Ann NY Acad Sci. 1974;238:420–35. doi: 10.1111/j.1749-6632.1974.tb26808.x. [DOI] [PubMed] [Google Scholar]
- 199.Arcangeli A, Carla M, Bene M, Becchetti A, Wanke E, Olivotto M. Polar/apolar compounds induce leukemia cell differentiation by modulating cell-surface potential. PNAS. 1993;90:5858–5862. doi: 10.1073/pnas.90.12.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Liebau S, Propper C, Bockers T, Lehmann-Horn F, Storch A, Grissmer S, Wittekindt OH. Selective blockage of Kv1.3 and Kv3.1 channels increases neural progenitor cell proliferation. J Neurochem. 2006;99(2):426–37. doi: 10.1111/j.1471-4159.2006.03967.x. [DOI] [PubMed] [Google Scholar]
- 201.Morokuma J, Blackiston D, Adams DS, Seebohm G, Trimmer B, Levin M. Modulation of potassium channel function confers a hyperproliferative invasive phenotype on embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105(43):16608–13. doi: 10.1073/pnas.0808328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cone CD, Cone CM. Induction of mitosis in mature neurons in central nervous system by sustained depolarization. Science. 1976;192(4235):155–8. doi: 10.1126/science.56781. [DOI] [PubMed] [Google Scholar]
- 203.Cone CD., Jr Variation of the transmembrane potential level as a basic mechanism of mitosis control. Oncology. 1970;24(6):438–70. doi: 10.1159/000224545. [DOI] [PubMed] [Google Scholar]
- 204.Cone CD. Unified theory on the basic mechanism of normal mitotic control and oncogenesis. J Theor Biol. 1971;30(1):151–81. doi: 10.1016/0022-5193(71)90042-7. [DOI] [PubMed] [Google Scholar]
- 205.Cone CD, Tongier M. Control of somatic cell mitosis by simulated changes in the transmembrane potential level. Oncology. 1971;25(2):168–82. doi: 10.1159/000224567. [DOI] [PubMed] [Google Scholar]
- 206.Cone CD, Tongier M. Contact inhibition of division: involvement of the electrical transmembrane potential. J Cell Physiol. 1973;82(3):373–86. doi: 10.1002/jcp.1040820307. [DOI] [PubMed] [Google Scholar]
- 207.Stillwell EF, Cone CM, Cone CD. Stimulation of DNA synthesis in CNS neurones by sustained depolarisation. Nat New Biol. 1973;246(152):110–1. doi: 10.1038/newbio246110a0. [DOI] [PubMed] [Google Scholar]
- 208.Wang L, Zhou P, Craig RW, Lu L. Protection from cell death by mcl-1 is mediated by membrane hyperpolarization induced by K(+) channel activation. J Membr Biol. 1999;172(2):113–20. doi: 10.1007/s002329900589. [DOI] [PubMed] [Google Scholar]
- 209.Lang F, Foller M, Lang KS, Lang PA, Ritter M, Gulbins E, Vereninov A, Huber SM. Ion channels in cell proliferation and apoptotic cell death. J Membr Biol. 2005;205(3):147–57. doi: 10.1007/s00232-005-0780-5. [DOI] [PubMed] [Google Scholar]
- 210.Miki T, Iwanaga T, Nagashima K, Ihara Y, Seino S. Roles of ATP-sensitive K+ channels in cell survival and differentiation in the endocrine pancreas. Diabetes. 2001;50(Suppl 1):S48–51. doi: 10.2337/diabetes.50.2007.s48. [DOI] [PubMed] [Google Scholar]
- 211.Lauritzen I, Zanzouri M, Honore E, Duprat F, Ehrengruber MU, Lazdunski M, Patel AJ. K+-dependent cerebellar granule neuron apoptosis. Role of task leak K+ channels. J Biol Chem. 2003;278(34):32068–76. doi: 10.1074/jbc.M302631200. [DOI] [PubMed] [Google Scholar]
- 212.Shen YA, Lin CH, Chi WH, Wang CY, Hsieh YT, Wei YH, Chen YJ. Resveratrol Impedes the Stemness, Epithelial-Mesenchymal Transition, and Metabolic Reprogramming of Cancer Stem Cells in Nasopharyngeal Carcinoma through p53 Activation. Evidence-based complementary and alternative medicine : eCAM. 2013;2013:590393. doi: 10.1155/2013/590393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Anderson JD. GALVANOTAXIS OF SLIME MOLD. J Gen Physiol. 1951;35(1):1–16. doi: 10.1085/jgp.35.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hyman L, Bellamy A. Studies on the correlation between metabolic gradients, electrical gradients, and galvanotaxis I. Biological Bulletin. 1922;XLIII(5):313–347. [Google Scholar]
- 215.Stump RF, Robinson KR. Xenopus neural crest cell migration in an applied electrical field. J Cell Biol. 1983;97(4):1226–1233. doi: 10.1083/jcb.97.4.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhao M, McCaig CD, Agius-Fernandez A, Forrester JV, Araki-Sasaki K. Human corneal epithelial cells reorient and migrate cathodally in a small applied electric field. Curr Eye Res. 1997;16(10):973–84. doi: 10.1076/ceyr.16.10.973.9014. [DOI] [PubMed] [Google Scholar]
- 217.Pullar CE, Isseroff RR. Cyclic AMP mediates keratinocyte directional migration in an electric field. J Cell Sci. 2005;118(Pt 9):2023–34. doi: 10.1242/jcs.02330. [DOI] [PubMed] [Google Scholar]
- 218.Schwab A. Function and spatial distribution of ion channels and transporters in cell migration. Am J Physiol Renal Physiol. 2001;280(5):F739–47. doi: 10.1152/ajprenal.2001.280.5.F739. [DOI] [PubMed] [Google Scholar]
- 219.Schwab A, Gabriel K, Finsterwalder F, Folprecht G, Greger R, Kramer A, Oberleithner H. Polarized ion transport during migration of transformed Madin-Darby canine kidney cells. Pflugers Arch. 1995;430(5):802–7. doi: 10.1007/BF00386179. [DOI] [PubMed] [Google Scholar]
- 220.Yan X, Han J, Zhang Z, Wang J, Cheng Q, Gao K, Ni Y, Wang Y. Lung cancer A549 cells migrate directionally in DC electric fields with polarized and activated EGFRs. Bioelectromagnetics. 2009;30(1):29–35. doi: 10.1002/bem.20436. [DOI] [PubMed] [Google Scholar]
- 221.Fraser SP, Diss JK, Chioni AM, Mycielska ME, Pan H, Yamaci RF, Pani F, Siwy Z, Krasowska M, Grzywna Z, Brackenbury WJ, Theodorou D, Koyuturk M, Kaya H, Battaloglu E, De Bella MT, Slade MJ, Tolhurst R, Palmieri C, Jiang J, Latchman DS, Coombes RC, Djamgoz MB. Voltage-gated sodium channel expression and potentiation of human breast cancer metastasis. Clin Cancer Res. 2005;11(15):5381–9. doi: 10.1158/1078-0432.CCR-05-0327. [DOI] [PubMed] [Google Scholar]
- 222.McCaig CD, Rajnicek AM, Song B, Zhao M. Controlling cell behavior electrically: current views and future potential. Physiol Rev. 2005;85(3):943–78. doi: 10.1152/physrev.00020.2004. [DOI] [PubMed] [Google Scholar]
- 223.Hinard V, Belin D, Konig S, Bader CR, Bernheim L. Initiation of human myoblast differentiation via dephosphorylation of Kir2.1 K+ channels at tyrosine 242. Development. 2008;135(5):859–67. doi: 10.1242/dev.011387. [DOI] [PubMed] [Google Scholar]
- 224.Konig S, Beguet A, Bader CR, Bernheim L. The calcineurin pathway links hyperpolarization (Kir2.1)-induced Ca2+ signals to human myoblast differentiation and fusion. Development. 2006;133(16):3107–14. doi: 10.1242/dev.02479. [DOI] [PubMed] [Google Scholar]
- 225.Lange C, Prenninger S, Knuckles P, Taylor V, Levin M, Calegari F. The H(+) vacuolar ATPase maintains neural stem cells in the developing mouse cortex. Stem cells and development. 2011;20(5):843–50. doi: 10.1089/scd.2010.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sundelacruz S, Levin M, Kaplan DL. Membrane potential controls adipogenic and osteogenic differentiation of mesenchymal stem cells. PLoS One. 2008;3(11):e3737. doi: 10.1371/journal.pone.0003737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Barth LG, Barth LJ. Ionic regulation of embryonic induction and cell differentiation in Rana pipiens. Dev Biol. 1974;39(1):1–22. doi: 10.1016/s0012-1606(74)80004-7. [DOI] [PubMed] [Google Scholar]
- 228.Barth LJ, Barth LG. Effect of the potassium ion on induction of notochord from gastrula ectoderm of Rana pipiens. Biol Bull. 1974;146(3):313–25. doi: 10.2307/1540407. [DOI] [PubMed] [Google Scholar]
- 229.Harrington DB, Becker RO. Electrical stimulation of RNA and protein synthesis in the frog erythrocyte. Exp Cell Res. 1973;76(1):95–8. doi: 10.1016/0014-4827(73)90423-0. [DOI] [PubMed] [Google Scholar]
- 230.Jaffe L. Developmental currents, voltages, and gradients. In: Subtelny S, editor. Developmental Order: its origin and regulation. Alan R Liss; New York: 1982. pp. 183–215. [Google Scholar]
- 231.Minc N, Chang F. Electrical control of cell polarization in the fission yeast Schizosaccharomyces pombe. Curr Biol. 2010;20(8):710–6. doi: 10.1016/j.cub.2010.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Shi R, Borgens RB. Three-dimensional gradients of voltage during development of the nervous system as invisible coordinates for the establishment of embryonic pattern. Dev Dyn. 1995;202(2):101–14. doi: 10.1002/aja.1002020202. [DOI] [PubMed] [Google Scholar]
- 233.Pan L, Borgens RB. Perpendicular organization of sympathetic neurons within a required physiological voltage. Exp Neurol. 2010;222(1):161–4. doi: 10.1016/j.expneurol.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 234.Stern C. Experimental reversal of polarity in chick embryo epiblast sheets in vitro. Exp Cell Res. 1982;140:468–471. doi: 10.1016/0014-4827(82)90143-4. [DOI] [PubMed] [Google Scholar]
- 235.Hotary KB, Robinson KR. Evidence of a role for endogenous electrical fields in chick embryo development. Development. 1992;114(4):985–96. doi: 10.1242/dev.114.4.985. [DOI] [PubMed] [Google Scholar]
- 236.Borgens RB, Shi R. Uncoupling histogenesis from morphogenesis in the vertebrate embryo by collapse of the transneural tube potential. Developmental Dynamics. 1995;203(4):456–67. doi: 10.1002/aja.1002030408. [DOI] [PubMed] [Google Scholar]
- 237.Adams DS, Robinson KR, Fukumoto T, Yuan S, Albertson RC, Yelick P, Kuo L, McSweeney M, Levin M. Early, H+-V-ATPase-dependent proton flux is necessary for consistent left-right patterning of non-mammalian vertebrates. Development. 2006;133:1657–1671. doi: 10.1242/dev.02341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Woodruff RI. Calmodulin transit via gap junctions is reduced in the absence of an electric field. J Insect Physiol. 2005;51(8):843–52. doi: 10.1016/j.jinsphys.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 239.Kurtz I, Schrank AR. Bioelectrical properties of intact and regenerating earthworms Eisenia foetida. Physiol Zool. 1955;28:322–330. [Google Scholar]
- 240.Uzman JA, Patil S, Uzgare AR, Sater AK. The role of intracellular alkalinization in the establishment of anterior neural fate in Xenopus. Developmental Biology. 1998;193(1):10–20. doi: 10.1006/dbio.1997.8782. [DOI] [PubMed] [Google Scholar]
- 241.Bentrup F, Sandan T, Jaffe L. Induction of Polarity in Fucus Eggs by Potassium Ion Gradients. Protoplasma. 1967;64(3):254. [Google Scholar]
- 242.Novák B, Bentrup FW. An electrophysiological study of regeneration in Acetabularia Mediterranea. Planta. 1972;108:227–244. doi: 10.1007/BF00384111. [DOI] [PubMed] [Google Scholar]
- 243.Novak B, Sirnoval C. Inhibition of regeneration of Acetabularia mediterranea enucleated posterior stalk segments by electrical isolation. Plant Science Letters. 1975;5:183–188. [Google Scholar]
- 244.Borgens RB. The role of natural and applied electric fields in neuronal regeneration and development. Progress in Clinical & Biological Research. 1986;210:239–50. [PubMed] [Google Scholar]
- 245.Borgens RB, Blight AR, Murphy DJ. Axonal regeneration in spinal cord injury: a perspective and new technique. Journal of Comparative Neurology. 1986;250(2):157–67. doi: 10.1002/cne.902500203. [DOI] [PubMed] [Google Scholar]
- 246.Borgens RB, Blight AR, McGinnis ME. Functional recovery after spinal cord hemisection in guinea pigs: the effects of applied electric fields. J Comp Neurol. 1990;296(4):634–53. doi: 10.1002/cne.902960409. [DOI] [PubMed] [Google Scholar]
- 247.Vandenberg LN, Morrie RD, Adams DS. V-ATPase-dependent ectodermal voltage and pH regionalization are required for craniofacial morphogenesis. Dev Dyn. 2011;240(8):1889–904. doi: 10.1002/dvdy.22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Pai VP, Aw S, Shomrat T, Lemire JM, Levin M. Transmembrane voltage potential controls embryonic eye patterning in Xenopus laevis. Development. 2012;139(2):313–23. doi: 10.1242/dev.073759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Rigas S, Debrosses G, Haralampidis K, Vicente-Agullo F, Feldmann KA, Grabov A, Dolan L, Hatzopoulos P. TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. The Plant cell. 2001;13(1):139–51. doi: 10.1105/tpc.13.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Dahal GR, Rawson J, Gassaway B, Kwok B, Tong Y, Ptacek LJ, Bates E. An inwardly rectifying K+ channel is required for patterning. Development. 2012;139(19):3653–64. doi: 10.1242/dev.078592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Villanueva S, Burgos J, Lopez-Cayuqueo KI, Lai KM, Valenzuela DM, Cid LP, Sepulveda FV. Cleft Palate, Moderate Lung Developmental Retardation and Early Postnatal Lethality in Mice Deficient in the Kir7.1 Inwardly Rectifying K+ Channel. PloS one. 2015;10(9):e0139284. doi: 10.1371/journal.pone.0139284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Simons M, Gault WJ, Gotthardt D, Rohatgi R, Klein TJ, Shao Y, Lee HJ, Wu AL, Fang Y, Satlin LM, Dow JT, Chen J, Zheng J, Boutros M, Mlodzik M. Electrochemical cues regulate assembly of the Frizzled/Dishevelled complex at the plasma membrane during planar epithelial polarization. Nat Cell Biol. 2009;11(3):286–94. doi: 10.1038/ncb1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Hermle T, Saltukoglu D, Grunewald J, Walz G, Simons M. Regulation of Frizzled-dependent planar polarity signaling by a V-ATPase subunit. Curr Biol. 2010;20(14):1269–76. doi: 10.1016/j.cub.2010.05.057. [DOI] [PubMed] [Google Scholar]
- 254.Muller C, Maeso I, Wittbrodt J, Martinez-Morales JR. The medaka mutation tintachina sheds light on the evolution of V-ATPase B subunits in vertebrates. Sci Rep. 2013;3:3217. doi: 10.1038/srep03217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Borthwick KJ, Kandemir N, Topaloglu R, Kornak U, Bakkaloglu A, Yordam N, Ozen S, Mocan H, Shah GN, Sly WS, Karet FE. A phenocopy of CAII deficiency: a novel genetic explanation for inherited infantile osteopetrosis with distal renal tubular acidosis. Journal of medical genetics. 2003;40(2):115–21. doi: 10.1136/jmg.40.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Zheng J, Trudeau MC. Handbook of ion channels. CRC Press; Boca Raton: 2015. [Google Scholar]
- 257.Duque A, Gazula VR, Kaczmarek LK. Expression of Kv1.3 potassium channels regulates density of cortical interneurons. Developmental neurobiology. 2013;73(11):841–55. doi: 10.1002/dneu.22105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Christensen AH, Chatelain FC, Huttner IG, Olesen MS, Soka M, Feliciangeli S, Horvat C, Santiago CF, Vandenberg JI, Schmitt N, Olesen SP, Lesage F, Fatkin D. The two-pore domain potassium channel, TWIK-1, has a role in the regulation of heart rate and atrial size. J Mol Cell Cardiol. 2016 doi: 10.1016/j.yjmcc.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 259.Kortum F, Caputo V, Bauer CK, Stella L, Ciolfi A, Alawi M, Bocchinfuso G, Flex E, Paolacci S, Dentici ML, Grammatico P, Korenke GC, Leuzzi V, Mowat D, Nair LD, Nguyen TT, Thierry P, White SM, Dallapiccola B, Pizzuti A, Campeau PM, Tartaglia M, Kutsche K. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat Genet. 2015 doi: 10.1038/ng.3282. [DOI] [PubMed] [Google Scholar]
- 260.Simons C, Rash LD, Crawford J, Ma L, Cristofori-Armstrong B, Miller D, Ru K, Baillie GJ, Alanay Y, Jacquinet A, Debray FG, Verloes A, Shen J, Yesil G, Guler S, Yuksel A, Cleary JG, Grimmond SM, McGaughran J, King GF, Gabbett MT, Taft RJ. Mutations in the voltage-gated potassium channel gene KCNH1 cause Temple-Baraitser syndrome and epilepsy. Nat Genet. 2015;47(1):73–7. doi: 10.1038/ng.3153. [DOI] [PubMed] [Google Scholar]
- 261.Labonne JD, Graves TD, Shen Y, Jones JR, Kong IK, Layman LC, Kim HG. A microdeletion at Xq22.2 implicates a glycine receptor GLRA4 involved in intellectual disability, behavioral problems and craniofacial anomalies. BMC Neurol. 2016;16:132. doi: 10.1186/s12883-016-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Petersson S, Persson AS, Johansen JE, Ingvar M, Nilsson J, Klement G, Arhem P, Schalling M, Lavebratt C. Truncation of the Shaker-like voltage-gated potassium channel, Kv1.1, causes megencephaly. Eur J Neurosci. 2003;18(12):3231–40. doi: 10.1111/j.1460-9568.2003.03044.x. [DOI] [PubMed] [Google Scholar]
- 263.Sharma V, Roy S, Sekler I, O’Halloran DM. The NCLX-type Na+/Ca2+ Exchanger NCX-9 Is Required for Patterning of Neural Circuits in Caenorhabditis elegans. J Biol Chem. 2017;292(13):5364–5377. doi: 10.1074/jbc.M116.758953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hiraki Y, Miyatake S, Hayashidani M, Nishimura Y, Matsuura H, Kamada M, Kawagoe T, Yunoki K, Okamoto N, Yofune H, Nakashima M, Tsurusaki Y, Satisu H, Murakami A, Miyake N, Nishimura G, Matsumoto N. Aortic aneurysm and craniosynostosis in a family with Cantu syndrome. American journal of medical genetics. Part A. 2014;164A(1):231–6. doi: 10.1002/ajmg.a.36228. [DOI] [PubMed] [Google Scholar]
- 265.Cooper PE, Reutter H, Woelfle J, Engels H, Grange DK, van Haaften G, van Bon BW, Hoischen A, Nichols CG. Cantu syndrome resulting from activating mutation in the KCNJ8 gene. Hum Mutat. 2014;35(7):809–13. doi: 10.1002/humu.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Brownstein CA, Towne MC, Luquette LJ, Harris DJ, Marinakis NS, Meinecke P, Kutsche K, Campeau PM, Yu TW, Margulies DM, Agrawal PB, Beggs AH. Mutation of KCNJ8 in a patient with Cantu syndrome with unique vascular abnormalities - support for the role of K(ATP) channels in this condition. European journal of medical genetics. 2013;56(12):678–82. doi: 10.1016/j.ejmg.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Chong JX, McMillin MJ, Shively KM, Beck AE, Marvin CT, Armenteros JR, Buckingham KJ, Nkinsi NT, Boyle EA, Berry MN, Bocian M, Foulds N, Uzielli ML, Haldeman-Englert C, Hennekam RC, Kaplan P, Kline AD, Mercer CL, Nowaczyk MJ, Klein Wassink-Ruiter JS, McPherson EW, Moreno RA, Scheuerle AE, Shashi V, Stevens CA, Carey JC, Monteil A, Lory P, Tabor HK, Smith JD, Shendure J, Nickerson DA, Bamshad MJ. De novo mutations in NALCN cause a syndrome characterized by congenital contractures of the limbs and face, hypotonia and developmental delay. Am J Hum Genet. 2015;96(3):462–73. doi: 10.1016/j.ajhg.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Uzun S, Gokce S, Wagner K. Cystic fibrosis transmembrane conductance regulator gene mutations in infertile males with congenital bilateral absence of the vas deferens. The Tohoku journal of experimental medicine. 2005;207(4):279–85. doi: 10.1620/tjem.207.279. [DOI] [PubMed] [Google Scholar]
- 269.Wilschanski M, Dupuis A, Ellis L, Jarvi K, Zielenski J, Tullis E, Martin S, Corey M, Tsui LC, Durie P. Mutations in the cystic fibrosis transmembrane regulator gene and in vivo transepithelial potentials. Am J Respir Crit Care Med. 2006;174(7):787–94. doi: 10.1164/rccm.200509-1377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Veale EL, Hassan M, Walsh Y, Al-Moubarak E, Mathie A. Recovery of current through mutated TASK3 potassium channels underlying Birk Barel syndrome. Mol Pharmacol. 2014;85(3):397–407. doi: 10.1124/mol.113.090530. [DOI] [PubMed] [Google Scholar]
- 271.Barel O, Shalev SA, Ofir R, Cohen A, Zlotogora J, Shorer Z, Mazor G, Finer G, Khateeb S, Zilberberg N, Birk OS. Maternally inherited Birk Barel mental retardation dysmorphism syndrome caused by a mutation in the genomically imprinted potassium channel KCNK9. Am J Hum Genet. 2008;83(2):193–9. doi: 10.1016/j.ajhg.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, Howard N, Srinivasan S, Silva JM, Molnes J, Edghill EL, Frayling TM, Temple IK, Mackay D, Shield JP, Sumnik Z, van Rhijn A, Wales JK, Clark P, Gorman S, Aisenberg J, Ellard S, Njolstad PR, Ashcroft FM, Hattersley AT. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. The New England journal of medicine. 2004;350(18):1838–49. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 273.Lee MP, Ravenel JD, Hu RJ, Lustig LR, Tomaselli G, Berger RD, Brandenburg SA, Litzi TJ, Bunton TE, Limb C, Francis H, Gorelikow M, Gu H, Washington K, Argani P, Goldenring JR, Coffey RJ, Feinberg AP. Targeted disruption of the Kvlqt1 gene causes deafness and gastric hyperplasia in mice. J Clin Invest. 2000;106(12):1447–55. doi: 10.1172/JCI10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Weksberg R, Nishikawa J, Caluseriu O, Fei YL, Shuman C, Wei C, Steele L, Cameron J, Smith A, Ambus I, Li M, Ray PN, Sadowski P, Squire J. Tumor development in the Beckwith-Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum Mol Genet. 2001;10(26):2989–3000. doi: 10.1093/hmg/10.26.2989. [DOI] [PubMed] [Google Scholar]
- 275.Moore ES, Ward RE, Escobar LF, Carlin ME. Heterogeneity in Wiedemann-Beckwith syndrome: anthropometric evidence. Am J Med Genet. 2000;90(4):283–90. [PubMed] [Google Scholar]
- 276.Rivas A, Francis HW. Inner ear abnormalities in a Kcnq1 (Kvlqt1) knockout mouse: a model of Jervell and Lange-Nielsen syndrome. Otol Neurotol. 2005;26(3):415–24. doi: 10.1097/01.mao.0000169764.00798.84. [DOI] [PubMed] [Google Scholar]
- 277.Casimiro MC, Knollmann BC, Yamoah EN, Nie L, Vary JC, Jr, Sirenko SG, Greene AE, Grinberg A, Huang SP, Ebert SN, Pfeifer K. Targeted point mutagenesis of mouse Kcnq1: phenotypic analysis of mice with point mutations that cause Romano-Ward syndrome in humans. Genomics. 2004;84(3):555–64. doi: 10.1016/j.ygeno.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 278.Chouabe C, Neyroud N, Guicheney P, Lazdunski M, Romey G, Barhanin J. Properties of KvLQT1 K+ channel mutations in Romano-Ward and Jervell and Lange-Nielsen inherited cardiac arrhythmias. Embo J. 1997;16(17):5472–9. doi: 10.1093/emboj/16.17.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Bendahhou S, Donaldson MR, Plaster NM, Tristani-Firouzi M, Fu YH, Ptacek LJ. Defective potassium channel Kir2.1 trafficking underlies Andersen-Tawil syndrome. J Biol Chem. 2003;278(51):51779–85. doi: 10.1074/jbc.M310278200. [DOI] [PubMed] [Google Scholar]
- 280.Yoon G, Oberoi S, Tristani-Firouzi M, Etheridge SP, Quitania L, Kramer JH, Miller BL, Fu YH, Ptacek LJ. Andersen-Tawil syndrome: prospective cohort analysis and expansion of the phenotype. Am J Med Genet A. 2006;140(4):312–21. doi: 10.1002/ajmg.a.31092. [DOI] [PubMed] [Google Scholar]
- 281.Culiat CT, Stubbs LJ, Woychik RP, Russell LB, Johnson DK, Rinchik EM. Deficiency of the beta 3 subunit of the type A gamma-aminobutyric acid receptor causes cleft palate in mice. Nat Genet. 1995;11(3):344–6. doi: 10.1038/ng1195-344. [DOI] [PubMed] [Google Scholar]
- 282.Wee EL, Zimmerman EF. GABA uptake in embryonic palate mesenchymal cells of two mouse strains. Neurochem Res. 1985;10(12):1673–88. doi: 10.1007/BF00988609. [DOI] [PubMed] [Google Scholar]
- 283.Homanics GE, DeLorey TM, Firestone LL, Quinlan JJ, Handforth A, Harrison NL, Krasowski MD, Rick CE, Korpi ER, Makela R, Brilliant MH, Hagiwara N, Ferguson C, Snyder K, Olsen RW. Mice devoid of gamma-aminobutyrate type A receptor beta3 subunit have epilepsy, cleft palate, and hypersensitive behavior. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(8):4143–8. doi: 10.1073/pnas.94.8.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Rock JR, Futtner CR, Harfe BD. The transmembrane protein TMEM16A is required for normal development of the murine trachea. Dev Biol. 2008;321(1):141–9. doi: 10.1016/j.ydbio.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 285.Rakic P, Sidman RL. Sequence of developmental abnormalities leading to granule cell deficit in cerebellar cortex of weaver mutant mice. The Journal of comparative neurology. 1973;152(2):103–32. doi: 10.1002/cne.901520202. [DOI] [PubMed] [Google Scholar]
- 286.Rakic P, Sidman RL. Weaver mutant mouse cerebellum: defective neuronal migration secondary to abnormality of Bergmann glia. Proceedings of the National Academy of Sciences of the United States of America. 1973;70(1):240–4. doi: 10.1073/pnas.70.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Hatten ME, Liem RK, Mason CA. Weaver mouse cerebellar granule neurons fail to migrate on wild-type astroglial processes in vitro. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1986;6(9):2676–83. doi: 10.1523/JNEUROSCI.06-09-02676.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Patil N, Cox DR, Bhat D, Faham M, Myers RM, Peterson AS. A potassium channel mutation in weaver mice implicates membrane excitability in granule cell differentiation. Nat Genet. 1995;11(2):126–9. doi: 10.1038/ng1095-126. [DOI] [PubMed] [Google Scholar]
- 289.Teng GQ, Zhao X, Lees-Miller JP, Quinn FR, Li P, Rancourt DE, London B, Cross JC, Duff HJ. Homozygous missense N629D hERG (KCNH2) potassium channel mutation causes developmental defects in the right ventricle and its outflow tract and embryonic lethality. Circ Res. 2008;103(12):1483–91. doi: 10.1161/CIRCRESAHA.108.177055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Than BL, Goos JA, Sarver AL, O’Sullivan MG, Rod A, Starr TK, Fijneman RJ, Meijer GA, Zhao L, Zhang Y, Largaespada DA, Scott PM, Cormier RT. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene. 2013 doi: 10.1038/onc.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Adams DS, Masi A, Levin M. H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration. Development. 2007;134(7):1323–35. doi: 10.1242/dev.02812. [DOI] [PubMed] [Google Scholar]
- 292.Monteiro J, Aires R, Becker JD, Jacinto A, Certal AC, Rodriguez-Leon J. V-ATPase Proton Pumping Activity Is Required for Adult Zebrafish Appendage Regeneration. PloS one. 2014;9(3):e92594. doi: 10.1371/journal.pone.0092594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Duboc V, Rottinger E, Lapraz F, Besnardeau L, Lepage T. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev Cell. 2005;9(1):147–58. doi: 10.1016/j.devcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 294.Iwashita M, Watanabe M, Ishii M, Chen T, Johnson SL, Kurachi Y, Okada N, Kondo S. Pigment Pattern in jaguar/obelix Zebrafish Is Caused by a Kir7.1 Mutation: Implications for the Regulation of Melanosome Movement. PLoS Genet. 2006;2(11):e197. doi: 10.1371/journal.pgen.0020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Tur J, Chapalamadugu KC, Padawer T, Badole SL, Kilfoil PJ, 2nd, Bhatnagar A, Tipparaju SM. Deletion of Kvbeta1.1 subunit leads to electrical and haemodynamic changes causing cardiac hypertrophy in female murine hearts. Exp Physiol. 2016;101(4):494–508. doi: 10.1113/EP085405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Chopra SS, Stroud DM, Watanabe H, Bennett JS, Burns CG, Wells KS, Yang T, Zhong TP, Roden DM. Voltage-gated sodium channels are required for heart development in zebrafish. Circ Res. 2010;106(8):1342–50. doi: 10.1161/CIRCRESAHA.109.213132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Shu X, Cheng K, Patel N, Chen F, Joseph E, Tsai HJ, Chen JN. Na,K-ATPase is essential for embryonic heart development in the zebrafish. Development. 2003;130(25):6165–73. doi: 10.1242/dev.00844. [DOI] [PubMed] [Google Scholar]