Abstract
The white rhinoceros (Ceratotherium simum) has a discontinuous African distribution, which is limited by the extent of sub-Saharan grasslands. The southern population (SWR) declined to its lowest number around the turn of the nineteenth century, but recovered to become the world's most numerous rhinoceros. In contrast, the northern population (NWR) was common during much of the twentieth century, declining rapidly since the 1970s, and now only two post-reproductive individuals remain. Despite this species's conservation status, it lacks a genetic assessment of its demographic history. We therefore sampled 232 individuals from extant and museum sources and analysed ten microsatellite loci and the mtDNA control region. Both marker types reliably partitioned the species into SWR and NWR, with moderate nuclear genetic diversity and only three mtDNA haplotypes for the species, including historical samples. We detected ancient interglacial demographic declines in both populations. Both populations may also have been affected by recent declines associated with the colonial expansion for the SWR, and with the much earlier Bantu migrations for the NWR. Finally, we detected post-divergence secondary contact between NWR and SWR, possibly occurring as recently as the last glacial maximum. These results suggest the species was subjected to regular periods of fragmentation and low genetic diversity, which may have been replenished upon secondary contact during glacial periods. The species's current situation thus reflects prehistoric declines that were exacerbated by anthropogenic pressure associated with the rise of late Holocene technological advancement in Africa. Importantly, secondary contact suggests a potentially positive outcome for a hybrid rescue conservation strategy, although further genome-wide data are desirable to corroborate these results.
Keywords: white rhinoceros, anthropogenic declines, demographic history, secondary contact, conservation
1. Introduction
The white rhinoceros (Ceratotherium simum) is the most common of the world's five remaining rhinoceros species. It has borne the brunt of rhinoceros losses during the global acceleration in illegal hunting, which began in 2008 because of increasing demand for horn products in southeast and east Asia. The species is an obligate grazer, thriving historically in two geographically separated grassland areas in sub-Saharan Africa, and has consequently been divided by taxonomists. The southern white rhinoceros (SWR) is endemic to southern Africa, historically occurring in much of the sub-region, south of the Zambezi river, including Namibia, Botswana, Zimbabwe and South Africa (electronic supplementary material, figure S1A,B, after [1]). The northern white rhinoceros (NWR) was endemic to a narrow belt of grassland from west of the Nile River and Albertine Rift, comprising parts of Uganda, South Sudan, the Democratic Republic of the Congo (DRC), Chad and the Central African Republic (electronic supplementary material, figure S1A,B). The recent histories of both populations are well known and independent, and contrastingly reflect events occurring in Africa and the Middle East since the eighteenth century (electronic supplementary material, figure S1C).
In southern Africa, the northwards spread of colonialism from the Cape of Good Hope resulted in the extermination of the SWR across most of the sub-region [2]. Even before the turn of the nineteenth century, the SWR had undergone a population decline so severe that only 100–200 individuals remained, restricted to around the confluence of the Black and White Umfolozi Rivers in Zululand [3]. However, in 1895 colonial authorities declared the white rhinoceros royal game and proclaimed the area the Umfolozi Junction Reserve [4]. With the dedicated conservation action of wildlife authorities in South Africa, this small population increased steadily throughout the twentieth century (electronic supplementary material, figure S1C) to become a conservation success story. The current severe poaching epidemic is threatening to undo these gains, and it is predicted that if present trends continue, the SWR population will start to decline again in 2018 [5]. Efforts to curb recent losses are ineffective with only marginal decreases in poaching rates in 2015 and 2016, with more than 1000 African rhinoceros killed every year since 2013. Such a population contraction, in the absence of gene flow from other sources, could negatively affect the genetic diversity and evolutionary potential of the SWR through genetic drift.
The demographic recovery of the SWR is all the more remarkable because the twentieth century also brought the near eradication of all other rhinoceros populations across the world. The NWR was still common throughout most of its range at the turn of the nineteenth century [6,7], and numbers were still relatively high until the 1960s [8], when demand for rhino horn, mainly on the Arabian peninsula, precipitated the penultimate poaching epidemic. Political instability and ineffective conservation measures during the ensuing period saw the rapid decline of NWR numbers in the wild (electronic supplementary material, figure S1C), with the last wild individuals extirpated in Uganda by 1980 [9], in Sudan by 1984 [8] and finally in Garamba National Park, Democratic Republic of the Congo [10], declared extinct in 2008. The NWR now survives only in captivity, and with two post-reproductive females remaining, its chances of survival look bleak. The imminent extinction of the NWR has sparked several conservation efforts to prevent the loss of what little remains of the population's genetic diversity.
The plight of the NWR has also precipitated a debate on whether the evolutionary relationship between the two populations could allow for interbreeding and genetic rescue as a conservation strategy [11], enabling the retention of at least some of the NWR's genetic diversity. The only known NWR–SWR hybrid was a female (Nasi), born in captivity in 1977. Although she survived 30 years in captivity, she never bred, and this has raised questions about the level of reproductive isolation between the two white rhinoceros populations. Although studies have revealed morphological, behavioural and genetic differences between the SWR and NWR [12–14], the evolutionary processes giving rise to this differentiation have not been discussed. Several authors have attempted to compare fossils with extant SWR and/or NWR [15–17], but with limited success due to the scarcity of well-preserved fossil material and difficulty in delimiting species/populations from fossil remains.
However, the fossil record demonstrates clearly that the prehistoric distribution of the white rhinoceros was wider than its recognized historical range. The presence of anatomically modern white rhinoceros in Pleistocene Tanzania, Ethiopia, Libya, Eritrea and Kenya [17] suggests a demographic history of population contraction and expansion. Fluctuation between cold and arid glacial periods with wet and warm interglacials would have respectively expanded and contracted the grassland biomes on which the white rhinoceros is dependent (electronic supplementary material, figure S1D). The evolutionary consequences of such climatic fluctuations, especially with regard to demographic isolation, depends on whether climatically driven range expansions allowed NWR and SWR populations to come into demographic secondary contact. The deep divergence between NWR and SWR implied by analysis of mtDNA (0.46–0.97 Ma [14]) is indicative of a prolonged period of demographic isolation between NWR and SWR maternal lineages. However, due to the maternal inheritance of mtDNA, it has a lower effective population size (Ne) than nuclear DNA and its lineages assort more quickly into monophyletic clades. Nuclear markers, especially those that evolve rapidly (such as microsatellites), would be expected to perform reliably in an analysis of demography and isolation by quantifying prehistoric levels of differentiation and gene-flow between populations [18].
Here we analysed genetic variation in the white rhinoceros with the aim of more appropriately informing conservation management. We use both nuclear microsatellites and mtDNA to determine levels of genetic variation across a sample of NWR and SWR populations, and from both wild and captive populations. To estimate the losses in genetic diversity resulting from twentieth century population declines, we also measured the genetic diversity of historical (pre-bottleneck) museum material for comparison. Additionally, we also tested the hypothesis that both populations underwent prehistoric demographic size changes, and determined whether the NWR and SWR came into secondary genetic contact after their initial Pleistocene divergence.
2. Methods
(a). Samples and loci
Samples were collected from wild (electronic supplementary material, table S1) and captive (electronic supplementary material, table S2) animals for both SWR and NWR. A total of 217 SWR samples (174 wild, 42 captive) and 15 NWR samples (8 wild, 7 captive) were obtained from extant and historical (museum) material representing the entire species range (full details are provided in electronic supplementary material 2, and permit information in electronic supplementary material, table S3). The 5′ end of the control region using primers mt15996 L (5′-TCCACCATCAGCACCCAAAGC-3′) and mt16502H (5′-TTTGATGGCCCTGAAGTAAGAACCA-3′) were used to amplify a 477 bp fragment of the control region. Samples were also amplified for 10 microsatellite loci (electronic supplementary material, table S4). The number of markers used in this study is comparable both with the number and identity of markers used in other publication on rhinoceros [19–21]. Markers were selected at random and were developed from a variety of target species (black rhinoceros, SWR and pig). For detailed molecular and quality control methods, see electronic supplementary material 2.
(b). Genetic diversity
We included four previously published mtDNA control region sequences (GenBank accessions AF187836, AF187837, AF187838 and AF187839 [22]), as well as seven mitochondrial genomes from wild individuals, three of which were from wild SWR and four from wild NWR prior to that population's extirpation [14]. Diversity was estimated for all populations separately. Since captive animals were from a variety of zoos and animal parks, we pooled all captive individuals into SWR and NWR groups. For microsatellites, we calculated the mean number of alleles, observed (HO) and unbiased expected heterozygosity (HE) using GENETIX [23]. Allelic richness (AR) was computed by resampling to correct for sample size differences among populations. Both AR and inbreeding coefficients (FIS) were calculated in FSTAT [24]. Mitochondrial DNA diversity for both control region and whole genomes was assessed for levels of polymorphism and haplotype diversity, as well as nucleotide diversity (π), in Arlequin v. 3.5 [25]. Tajima's D [26] and Fu's Fs [27] statistics were also calculated in Arlequin to determine whether sequences showed evidence for population size changes.
(c). Genetic structure
Population structure using microsatellite variation was assessed using Bayesian k-means clustering in Structure [28]. We assumed an admixture model and analysis was run ten times for k = 1–7 with each randomly started run consisting of 500 000 Markov chain Monte Carlo (MCMC) iterations, assuming correlated allele frequencies, discarding the first 100 000 iterations. The optimal k for the microsatellite data was determined as the highest value that was biologically interpretable. MtDNA structure was deduced by constructing a phylogenetic network of control region sequences. We used the median-joining method in Network v. 5.0.0.1 [29] with equal weighting on all nodes and using a correction cost algorithm.
(d). Evolutionary time frame
In order to obtain a time frame for the evolutionary history of the species, we reconstructed a species level maternal phylogeny from the seven mitochondrial genomes sequenced by Harley et al. [14]. We conducted Bayesian phylogenetic dating using BEAST v. 2.4.3 [30] as this allowed us to parametrize splits in the tree with soft-bounded priors based on known fossil information, using the same mammalian mtDNA genomes and priors described by Harley et al. [14], except that all five calibration times were used simultaneously in a single analysis. We used the Tamura–Nei model for nucleotide substitution with gamma correction, as deduced by jModelTest v. 2 [31], placing a relaxed, lognormal prior on the clock rate to account for potential differences in the molecular clock. The analysis was facilitated by a heuristic 100 million-step exploration of the likelihood surface using MCMC simulation, sampling the chain every 100 000 steps and discarding the first 10%.
(e). Ancient and recent changes in effective population size
Ancient demographic change in both SWR (n = 20, excluding zoo individuals) and NWR (n = 15) populations was inferred using MSVar v. 1.3 [32,33], with Ne being the size of a model population that has the same rate of genetic drift as the rhinoceros population of interest. Wide priors were set for all parameter estimates to allow for uncertainties in the data. Three potential scenarios were performed separately for SWR and NWR, assuming different ancestral (N1) and current (N0) effective population sizes. These were (i) a stable population (N1 = N0), (ii) a population decline (N1 > N0) and (iii) a population expansion (N1 < N0). Further details of priors and MCMC runs are given in electronic supplementary material, table S5.
The more recent demographic history of the white rhinoceros, during which humans may have driven population size changes, was investigated through Approximate Bayesian Computation (ABC) simulations [34]. This approach is unlike the likelihood calculations of the data performed by MSVar, but instead simulates a finite set of potential demographic scenarios, which are then compared to the observed data using sets of summary statistics. Demographic histories for SWR and NWR were thus tested independently by exploratory simulations of six scenarios in ABCToolbox v. 1.1 [35]: a null model, two expansion models, two bottleneck models and one model with two bottlenecks (electronic supplementary material, figure S2). Under expansion and bottleneck scenarios we tested whether the timing of the demographic event coincided with sub-Saharan Africa's two most important anthropogenic events—the migration of Iron Age, agriculturalist Niger–Congo language speakers (Bantu) into eastern and southern Africa 400–2000 years ago (ya) [36] and the expansion of colonial-era European influence into the region (present to 400 ya).
(f). Secondary contact between northern and southern white rhinoceros
We also tested for the possibility that NWR and SWR could have come into secondary genetic contact since they diverged from each other. This may have occurred during the late Pleistocene during which the grassland biome would have periodically been continuous between eastern and southern Africa (electronic supplementary material, figure S1D). We therefore built a two-population model that included uni- and bidirectional migration (electronic supplementary material, figure S3). First, we tested for migration (uni- and bidirectional) at any time during the last glacial period (LGP) of the late Pleistocene (14 000–106 000 ya, scenarios 2–4) which followed the end of the Eemian interglacial. We then subdivided the LGP to attempt to differentiate between recent migration during the last glacial maximum (LGM, 14 000–26 000 ya, scenarios 5–7) and earlier migration during the LGP (26 000–106 000 ya, scenarios 8–10). Last, we tested the null hypothesis against a model of ancient (pre-Eemian) migration (130 000–500 000 ya, scenarios 11–13). For details of model parametrization see electronic supplementary material 2.
We were concerned that individuals in our dataset could be closely related and we therefore removed all individuals with a relatedness values (r) of 0.3 or higher and reran all one- and two-population ABC simulations.
3. Results
A total of 232 white rhinoceros were genotyped at ten microsatellite loci (electronic supplementary material 3) and 419 bp of the mitochondrial control region (electronic supplementary material 4) was sequenced in 63 individuals. The level of missing data is given as the number and percentage (%) of failed genotypes for the historical and modern data (electronic supplementary material, table S6). Three loci (RHI32A, RH17B and RH17C) for the historical NWR sample showed a high proportion of missing data (greater than 50%). To determine if missing data at these loci affected the overall observed structure between populations, we reran the Structure analyses for k = 1–7 without these loci, and found that there was no change in the overall result.
(a). Genetic diversity
The effect of ascertainment bias was limited in this study as our results were consistent with previous studies using different types of markers in rhinoceros [37]. All populations were found to be in Hardy–Weinberg equilibrium and individual loci were randomly associated (in linkage equilibrium). Nuclear microsatellite genetic diversity was moderate to low, with an average of eight alleles per locus and heterozygosity ranging from 0.48 to 0.56 (electronic supplementary material, table S7). SWR (HO = 0.48) were more diverse than NWR (HO = 0.46), but both populations had lower observed than expected heterozygosity and positive, but not significant, inbreeding coefficients (FIS SWR = 0.09, FIS NWR = 0.33). The extant wild SWR population (six subpopulations) had slightly lower diversity (HO = 0.47) than captive SWR individuals (HO = 0.51). Within the captive SWR there was no difference in genetic diversity between founders and their offspring (zoo-born). In contrast, the wild (museum sampled) NWR population was more diverse than our sample of captive NWR animals, returning a higher mean number of alleles (3.4 versus 2.4) and heterozygosity (0.48 versus 0.43). Yet, despite comprising only seven individuals, the captive NWR sample had similar allelic diversity to all sampled wild SWR subpopulations from reserves in South Africa today.
Mitochondrial genetic diversity was low for both SWR and NWR (electronic supplementary material, table S8), comprising three haplotypes in total, with SWR comprising two haplotypes and NWR just one. Captive SWR contained both haplotypes but had lower nucleotide diversity than wild SWR (0.003 versus 0.005). In both populations, genetic diversity of nineteenth and twentieth century haplotypes were the same as extant levels. We amplified the control region for one ancient SWR individual (shot in South Africa in 1869) which possessed one of the two haplotypes detected in extant SWR populations. Surprisingly, our entire historical NWR sample, containing early twentieth century individuals from the three range states in which the NWR was most common, all harboured the same haplotype as NWR population in captivity.
(b). Genetic structure
Both nuclear and mitochondrial markers structured the species into two distinct populations/clades (figure 1), corresponding to SWR and NWR. For microsatellite data, k = 2 returned the highest likelihood, with no recent admixture detected between populations (figure 1a). When the data were analysed at higher, less likely, k models, substructure within SWR was revealed with the separation of mainly captive individuals (k = 4), Mthethomusha Game Reserve (k = 5) and Origstad and Nkomazi Game Reserves (k = 6, electronic supplementary material, figure S4,). However, since we could not assume that our captive sample was taken from a naturally breeding population, only wild SWR were used for subsequent ABC analyses. However, the multilocus profile of the only known SWR–NWR hybrid individual Nasi clearly shows an admixed profile. The mitochondrial control region showed two monophyletic clades, separated by 30 mutational steps (figure 1b). Haplotypes within the SWR were separated by four mutations.
Figure 1.
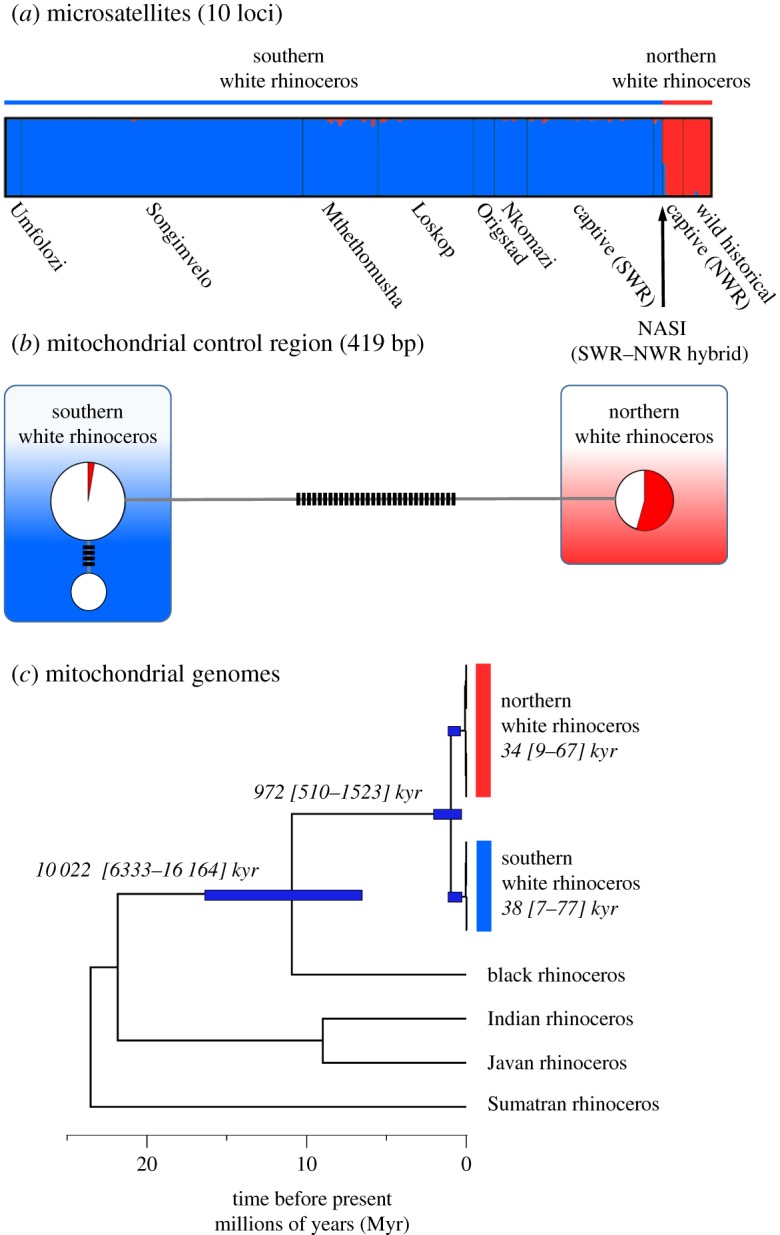
Genetic structure of the white rhinoceros showing clear separation into two populations. (a) Structuring of multilocus nuclear microsatellite profiles (k = 2). (b) Network of three mitochondrial control region haplotypes (circles) dividing the two white rhinoceros populations into distinct clades. Mutational steps are given as small black bars. (c) Bayesian-dated whole mitochondrial genome phylogeny of the extant Rhinocerotidae. (Online version in colour.)
(c). Evolutionary time frame
Using complete mitochondrial genomes (electronic supplementary material 5), which contained greater levels of within-population polymorphism relative the control region, BEAST analysis inferred a divergence time between the two white rhino lineages at approximately 0.97 million years (Myr), but with a large 95% highest posterior density (HPD) of 0.5–1.5 Myr (figure 1c), with African rhinoceros species (black and white) identified as sister taxa with a most recent common ancestor (MRCA) of 11 Myr (HPD95: 6.4–16.0 Myr). Low mitochondrial DNA diversity within each white rhinoceros population resulted in very shallow but similar MRCA times. SWR lineages coalesce to 38 000 years ago (ya, HPD95: 7400–77 400 ya) and NWR mitochondrial genomes shared a common ancestor 34 000 ya (HPD95: 8800–67 000 ya).
(d). Ancient changes in effective population size
While analyses of mtDNA neutrality indicated a history of population contraction for the species, with the majority yielding positive Fu's Fs values, these were not significant for the control region or for mitochondrial genomes (electronic supplementary material, table S8). However, microsatellite data analyses using MSVar revealed that both SWR and NWR have undergone an ancient reduction in effective population size (electronic supplementary material, table S9 and figure S5). Independent runs invoking stable, expansion and contraction models all converged to the same posterior values placing current population size (N0) consistently lower than ancestral population size (N1, electronic supplementary material, table S9 and figure S5A,C). The timing of these population contraction events could be dated to the mid-Holocene, between 3400–5800 ya for SWR and the early Holocene to late Pleistocene (7000–29 000 ya) for NWR (electronic supplementary material, table S9 and figure S5B,D).
(e). Recent demographic change
The null ABC model (scenario 1 SWR and NWR; electronic supplementary material, figure S2) of no recent change in population size could be rejected for both SWR and NWR. Instead, highest model support for both populations was for a single decline or bottleneck (table 1; BF > 3; electronic supplementary material, figure S6). For SWR, the best model selected was that of a population bottleneck during the colonial period (scenario 4 SWR; table 1) with a modal time for the beginning of the decline of 264 years (HPD90 138–394 years). In contrast, the best model for NWR was a demographic bottleneck during the time of the Bantu expansion into eastern Africa (scenario 5 NWR; table 1), occurring about 1370 ya (HPD90 518–1869 years).
Table 1.
Posterior estimates for demographic scenarios within and between northern and southern white rhinoceros based on approximate Bayesian computation. The best model among one- and two-population scenarios are indicated in italics.
| scenario | scenario description | migration prior (kyr) |
posterior mode (kyr) |
posterior NeM |
marginal density |
p-value | Bayes factor (BF) | |
|---|---|---|---|---|---|---|---|---|
| southern white rhinoceros | 1 | null model (stable population size) | — | — | — | 0.01 | 0.00 | — |
| 2 | expansion during the colonial period | — | — | — | 7.49 × 10−5 | 0.00 | 0.007 | |
| 3 | expansion during the Bantu migrations | — | — | — | 2.59 × 10−37 | 0.00 | 2.59 × 10−35 | |
| 4 | bottleneck during the colonial period | — | — | — | 2.04 | 1.00 | 204.00 | |
| 5 | bottleneck during the Bantu migrations | — | — | — | 0.12 | 0.18 | 1.2 | |
| 6 | two bottlenecks colonial period/Bantu migrations | — | — | — | 3.61 × 10−50 | 0.00 | 3.61 × 10−48 | |
| northern white rhinoceros | 1 | null model (stable population size) | — | — | — | 0.003 | 0.00 | — |
| 2 | expansion during the colonial period | — | — | — | 1.28 × 10−7 | 0.00 | 4.27 × 10−05 | |
| 3 | expansion during the Bantu migrations | — | — | — | 6.45 × 10−35 | 0.00 | 2.15 × 10−32 | |
| 4 | bottleneck during the colonial period | — | — | — | 2.68 × 10−68 | 0.00 | 8.93 × 10−66 | |
| 5 | bottleneck during the Bantu migrations | — | — | — | 1.04 | 1.00 | 346.67 | |
| 6 | two bottlenecks colonial period/Bantu migrations | — | — | — | 1.72 × 10−18 | 0.00 | 5.73 × 10−16 | |
| both populations | 1 | null model; no migration | — | — | — | 739.66 | 0.56 | — |
| 2 | unidirectional migration (S-N) | last glacial maximum (14–26) | 21 054 | 5 | 7211.49 | 0.76 | 9.75 | |
| 3 | unidirectional migration (N-S) | 18.090 | 72 | 2432.03 | 0.80 | 3.29 | ||
| 4 | bidirectional migration |
S-N: 18 112
N-S: 21 753 |
S-N: 7
N-S: 112 |
19 241.30 | 0.92 | 26.01 | ||
| 5 | unidirectional migration (S-N) | last glacial period (LGP) (14–106) |
43 729 | 7 | 4937.74 | 0.70 | 6.68 | |
| 6 | unidirectional migration (N-S) | 40 011 | 87 | 1929.91 | 0.77 | 2.61 | ||
| 7 | bidirectional migration |
S-N: 43 729
N-S: 40 940 |
S-N: 7
N-S: 95 |
10 643.00 | 0.86 | 14.39 | ||
| 8 | unidirectional migration (S-N) | early LGP (26–106) | 72 870 | 7 | 4819.14 | 0.65 | 6.52 | |
| 9 | unidirectional migration (N-S) | 49 436 | 85 | 2099.64 | 0.83 | 2.84 | ||
| 10 | bidirectional migration |
S-N: 73 678
N-S: 47 011 |
S-N: 9
N-S: 98 |
9950.02 | 0.79 | 13.45 | ||
| 11 | unidirectional migration (S-N) | pre-Eemian interglacial (130–540) | 274 690 | 6 | 2377.90 | 0.65 | 3.21 | |
| 12 | unidirectional migration (N-S) | 276 255 | 80 | 1691.94 | 0.69 | 2.29 | ||
| 13 | bidirectional migration | S-N: 357 599 N-S: 241 527 |
S-N: 7 N-S: 62 |
5124.45 | 0.91 | 6.93 |
(f). Secondary contact and gene flow
Combining both SWR and NWR data, parametrized according to the two best single-population scenarios above, we found the marginal densities for all migration models to be higher than the null model of no post-divergence migration (table 1). Among migration scenarios, bidirectional migration was more likely than any equivalent unidirectional scenario. The highest marginal densities and Bayes factors among bidirectional models were for scenarios set within the LGP (table 1). Within the LGP, recent LGM secondary contact was the most likely of all tested scenarios, but could not be significantly differentiated from later LGP migration.
All ABC simulations were also run without closely related individuals (r > 0.3). This reduced the sample size of SWR and NWR to 11 and 10 respectively. Nevertheless, all runs returned similar results to those above, with the exception that colonial and Bantu period population bottleneck scenarios could no longer be distinguished from each other for NWR. The results of these additional simulations are provided in electronic supplementary material, tables S10 and S11.
4. Discussion
We generated molecular data from a sample that included the recent recorded white rhinoceros range, with samples from extant and historical specimens back to the nineteenth century. Our sample for both marker sets was low for the NWR (electronic supplementary material, tables S1 and S2), especially among historical specimens where mtDNA amplified more readily than mtDNA. This is because available NWR material is limited to erstwhile captive populations at the Dvůr Králové and San Diego zoos and a handful of museum specimens from Europe and the United States. We observed much higher allelic variation at microsatellites than for mtDNA, likely reflecting the differences in effective population size between the two markers (approximately 4 : 1). Levels of microsatellite heterozygosity in white rhinoceros are lower than eastern, western and southern African black rhinoceros populations (HE: 0.71–0.74, [21]) but higher than the relatively unmanaged southwestern black rhinoceros of Namibia or Angola (HE: 0.42–0.49). We also found that historical levels of NWR diversity were greater than extant levels, demonstrating the negative genetic consequences of the NWR's colonial-era history of hunting and habitat destruction during the latter part of the twentieth century. However, for mtDNA, we found that even in colonial times maternal genetic diversity was already as low as it is in extant populations. Therefore, the evolutionary process which reduced maternal variation in both white rhinoceros populations is very likely to have occurred prior the time of sampling in the late 1800s.
The lower diversity of SWR individuals born in captivity relative to wild-born founders may also indicate a loss of diversity, even in the space of one or two generations. Owing to the increased erosive power of genetic drift in small populations, this effect may increase as time progresses. We therefore suggest an active management plan for captive bred individuals, where multilocus genetic profiles can be used to maintain genetic diversity. Similarly, genetic drift in isolation has differentiated some SWR populations, and more active management between reserves is encouraged to help ameliorate these effects.
(a). Structure and evolutionary time frame
Microsatellite clustering clearly differentiated the white rhinoceros into two distinct populations (figure 1), an observation already made using mtDNA [12,14] and for the nuclear amelogenin gene [12]. Both these studies, however, made use of more limited datasets comprising two and seven individuals, respectively. The present study therefore is the first to use large sample sizes and microsatellite markers, and in the case of the NWR the historical sample covered much of the population's range. For maternally inherited mtDNA, dated using five mammalian calibration points, we estimated the divergence of mtDNA lineages at just under a million years, but with wide confidence limits (±500 000 years). ABC simulations were not able to narrow this estimate. These wide limits underscore model uncertainty and the wide prior distributions on fossil calibration points. Nevertheless, these divergence estimates provide a general time frame for the initial split between NWR and SWR populations. Genomic analysis will likely be needed to date the divergence of the two white rhinoceros populations more precisely.
(b). Ancient population size changes
A coalescent analysis of prehistoric effective population size changes using our microsatellite data allowed us to infer late Pleistocene to mid-Holocene population contractions for both SWR and NWR (electronic supplementary material, figure S5). Since the effective population size inferred through coalescent simulation is a measure of the effect of genetic drift on the genealogical process, reported numbers reflect the minimum number of effective breeders required by the studied population to ameliorate the loss of further diversity through drift. The inferred time frame largely overlaps with the confidence limits for the coalescence of intra-population mtDNA lineages (figure 1c), which could potentially have occurred as recently as 6000–7000 ya and as early as 77 000 ya. Although confidence limits on the posterior distribution of these times were large, for SWR, the inferred decline suggests that this population was adversely affected by the grassland contraction that occurred after the LGM. In the NWR, we obtained a signal for both a post-LGM (7000 ya) and a pre-LGM (26 000–29 000 ya) population decline, also possibly in response to grassland contraction, highlighting the white rhinoceros' dependence on suitable grassland habitats. These prehistoric population contractions may have been partly responsible for low mtDNA genetic variation detected among colonial-era NWR and SWR samples.
(c). Recent human-associated population declines
Microsatellite analysis also allowed us to infer very recent population declines associated with human movements in Africa. We stress that although the time frames for the recent NWR and SWR bottlenecks were defined to test for an association with known human historical events, our ABC simulations do not provide a causal link between the human activity and white rhinoceros demography. NWR precolonial population decline, may be coincident with the arrival of Bantu speakers from western Africa. Recent reconstructions have inferred that the Bantu expansion proceeded first in a south-easterly direction from Cameroon, avoiding rainforest and taking advantage of a savannah corridor that started to open approximately 4000 ya (e.g. [38]), accelerating approximately 2500 ya [39] and leading to colonization of eastern Africa and the Great Lakes region around 2000 ya [36]. While the Bantu were predominantly agriculturalists, using grassland habitats on which to grow newly domesticated strains of millet and sorghum, they were also in possession of iron age smelting technology, and thus capable of hunting larger game animals, either directly, or through interactions with and spread of iron-age technology through local hunter–gatherers (e.g. [40]). It is also possible that Bantu speakers associated with people from further afield, either with Arab and south Asian traders via the eastern coast of Africa or with Romans via the Nile Valley. In either case, demand for rhinoceros products, and potentially even live animals, may have helped intensify the decline in effective population size in the NWR observed during this period.
In comparison, we recovered a clear signal for a more recent human-induced population decline in the SWR, during the occupation of southern Africa by Europeans. This population decline is historically well documented, with the SWR reaching its lowest number of approximately 100 animals over a hundred years after the median time of decline (electronic supplementary material, figure S6), although the actual time at which the SWR was at its lowest number falls well within the confidence limits of our posterior distribution. Interestingly, although both populations were reduced to low numbers by humans, current effective population size confidence limits did not overlap, showing that the SWR was reduced to significantly lower effective numbers. Although the effective numbers of NWR destroyed by humans was greater, significantly lower effective size for SWR could reflect the more efficient destruction of white rhinoceros by mechanized hunting during the colonial times.
(d). Post-divergence gene flow between SWR and NWR
We used two-population ABC analyses to demonstrate that although the NWR may have diverged from the SWR over a million years ago, both populations came into post-divergence secondary contact more recently during the LGP, and potentially even as recently as the LGM. The implications of this finding may prove central in ongoing debates about the specific status of the two white rhinoceros populations, and how best to manage their remaining genetic diversity in the future. The inferred post-divergence gene-flow was likely facilitated by savannah grassland expansions after the Eemian interglacial (115 000–130 000 ya), but has ceased completely since the Holocene when NWR and SWR populations declined as their grassland habitat diminished (electronic supplementary material, figure 1D). A potentially continuous distribution of the white rhinoceros is also supported by evidence of its occurrence east of the Nile river from the middle Pleistocene [41], the LGP [10] and as recently as the Holocene [42]. Therefore, the present-day absence of the white rhinoceros east of the Nile River can only be explained by the local extirpation of an eastern African population during the Holocene contraction, and with repopulation of Uganda, Kenya and Tanzania subsequently attenuated by the flow of the Nile. Taken together, these results suggest that the white rhinoceros has been resilient to population size contractions, which would have subjected local populations to periods of low genetic diversity during interglacial periods, but with diversity being potentially replenished during glacial periods by secondary contact.
(e). Conservation implications
The contrasting histories of the northern and southern white rhinoceros have substantial implications for their conservation. Low diversity at both mtDNA and microsatellite loci implies that maintenance of genetic diversity should be a core conservation action for the species. Although the African Rhino Specialist Group advocates a lower limit of 20 founding individuals [43] for new populations, some wild SWR populations like Mthethomusha, Origstad and Nkomazi have already differentiated from the original SWR stock due to management in isolation. Our results suggest not only a minimum number of founders for new populations, but also that microsatellite profiles should be used to select founding individuals from more than one source population. Additionally, low diversity of some populations should be ameliorated by regular and targeted translocations.
With most endangered species intensive genetic management of populations would be prohibitively expensive and/or logistically challenging. However, population genetic analysis carried out in a forensic context is increasingly being applied in large African mammals, for example in both the forest and savannah elephant to identify the origins of seized animal products [44] and to identify demographic units for conservation management (e.g. [45]). Forensic studies require large genetic reference databases, thus a large and growing number of white rhinoceros have been routinely genotyped for forensic purposes [46], and we advocate making use of this unique genetic resource to aid the management of genetic diversity. With this database, it should be possible to monitor population diversity levels in real time, and select the profiles of immigrant individuals that would maximize population genetic diversity. Since landowners in South Africa are legally obliged to genotype their rhinoceros, it would also be possible to monitor the breeding success of immigrant individuals as the calves of the next generation are added to the expanding database.
The situation for the NWR is very different, and here we show that this population is the endpoint of a long period of both prehistoric and anthropogenic decline. With only two female individuals remaining, the role of genetics is presently confined to an evaluation of the potential outcomes of hybrid rescue involving the use of SWR genomes. The recent LGP secondary contact is a key result in this context, as it increases the likelihood that hybrid rescue could be positive and that the recently reported NWR–SWR hybrid embryos may provide a viable strategy for conservation of the NWR [47]. However drawing such inference could be premature using a handful of genetic markers alone, and for this reason the resequencing of whole genomes using next-generation sequencing (NGS) could be extremely useful. NGS approaches yield data for millions of loci across the genome, providing more power to infer demographic histories and time more precisely the onset of demographic events like population bottlenecks and bouts of gene flow. From whole-genome data, it is also possible to document locally adapted regions of the white rhinoceros genome that may be a priority for genetic management in white rhinoceros occupying the northern edge of the species's historical distribution, regardless of the origin of the animals.
An additional conservation implication of this work is that managed translocation of SWR into some portion of the NWR's historical range might be a viable approach to restore the ecological functionality that this large grazing mammal previously contributed to the northern savannah ecosystem it once occupied [48], although any such introductions would need to be closely monitored for evidence of a lack of local adaptation, genetic drift and inbreeding.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank the following museums for making their collections available for this study: American Museum of Natural History, New York; National Museum of Natural History, Prague; Naturhistorisches Museum, Vienna; Powell Cotton Museum, Birchington; Royal Museum of Central Africa, Tervuren. J.R. is very grateful to Luděk Čulík, Jiří Hrubý, Jan Ždárek, Jiří Váhala (Zoo Dvůr Králové) and Petr Benda (National Museum, Prague) for enabling him to collect samples for genetic analysis. A.K. thanks Mpumalanga Parks and Tourism Agency, in particular Annelize Steyn, for making samples available to the project.
Ethics
Samples collected for this study received ethical approval from Cardiff University and were collected in accordance with the protocols/guidelines of the National Zoological Gardens of South Africa (NZG). Where relevant, animals were handled under the guidelines of the American Society of Mammologists (ASM; Animal Care and Use Committee, 2011). All required permits are listed in electronic supplementary material, table S3. All museum samples were collected in accordance with the relevant guidelines and regulations of each museum.
Data accessibility
All mitochondrial DNA sequences/genomes and microsatellite data generated in this study were uploaded to the Dryad Digital Repository at http://dx.doi.org/10.5061/dryad.6mc1606 [49].
Authors' contributions
Y.M., I.-R.M.R., J.R., A.K., C.W. and M.W.B. conceived the study. Y.M., J.R., J.S., O.A.R., R.H. and C.W. performed the fieldwork. I.-R.M.R., D.L.D. and S.S. performed the laboratory work. Y.M., I.-R.M.R., D.L.D., S.S. and M.W.B. conducted the analyses. Y.M., I.-R.M.R. and M.W.B. wrote the initial draft which was subsequently critically revised and approved by co-authors.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.Rookmaaker K, Antoine PO. 2012. New maps representing the historical and recent distribution of the African species of rhinoceros: Diceros bicornis, Ceratotherium simum and Ceratotherium cottoni. Pachyderm 52, 91–96. [Google Scholar]
- 2.Player IC, Feely JM. 1960. A preliminary report on the square-lipped rhinoceros Ceratotherium simum simum. Lammergeyer 1, 3–23. [Google Scholar]
- 3.Vaughan-Kirby F. 1920. The white rhinoceros, with special reference to its habits in Zululand. Ann. Durban Mus. 2, 223–242. [Google Scholar]
- 4.Brooks S. 2006. Human discourses, animal geographies: imagining Umfolozi's white rhinos. Curr. Writ., Durban 18, 6–27. (doi:10.13929x.2006.9678230) [Google Scholar]
- 5.Ferreira SM, Greaver C, Knight GA, Knight MH, Smit IP, Pienaar D. 2015. Disruption of rhino demography by poachers may lead to population declines in Kruger National Park, South Africa. PLoS ONE 10, e0127783 ( 10.1371/journal.pone.0127783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roosevelt T, Heller E. 1914. Life histories of African game animals, vol. 2, pp. 420–798. New York: NY: Charles Scribner's Sons. [Google Scholar]
- 7.Pitman CRS. 1931. Hobnobbing with the white rhinoceros. Asia 31, 446–451. [Google Scholar]
- 8.Emslie R, Brooks M. 1999. African rhino: status survey and conservation action plan. Gland, Switzerland: IUCN. [Google Scholar]
- 9.Edroma EL. 1982. White rhino extinct in Uganda. Oryx 16, 352–355. ( 10.1017/S0030605300017841) [DOI] [Google Scholar]
- 10.Hillman-Smith K, ma Oyisenzoo M, Smith F. 1986. A last chance to save the northern white rhino? Oryx 20, 20–26. ( 10.1017/S0030605300025862) [DOI] [Google Scholar]
- 11.Saragusty J, et al. 2016. Rewinding the process of mammalian extinction. Zoo Biol. 35, 280–292. ( 10.1002/zoo.21284) [DOI] [PubMed] [Google Scholar]
- 12.Groves CP, Fernando P, Robovský J. 2010. The sixth rhino: a taxonomic re-assessment of the critically endangered northern white rhinoceros. PLoS ONE 5, e9703 ( 10.1371/journal.pone.0009703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cinková I, Policht R. 2014. Contact calls of the northern and southern white rhinoceros allow for individual and species identification. PLoS ONE 9, e98475 ( 10.1371/journal.pone.0098475) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harley EH, de Waal M, Murray S, O'Ryan C. 2016. Comparison of whole mitochondrial genome sequences of northern and southern white rhinoceroses (Ceratotherium simum): the conservation consequences of species definitions. Conserv. Genet. 17, 1285–1291. ( 10.1007/s10592-016-0861-2) [DOI] [Google Scholar]
- 15.Groves CP. 1975. Taxonomic notes on the white rhinoceros Ceratotherium simum (Burchell, 1817). Saugetierkd. Mitt. 23, 200–212. [Google Scholar]
- 16.Geraads D. 2005. Pliocene Rhinocerotidae (Mammalia) from Hadar and Dikika (lower Awash, Ethiopia), and a revision of the origin of modern African rhinos. J. Vertebr. Paleontol. 25, 451–461. ( 10.1671/0272-4634(2005)025%5B0451:PRMFHA%5D2.0.CO;2) [DOI] [Google Scholar]
- 17.Geraads D. 2010. Rhinocerotidae. In Cenozoic mammals of Africa (eds Werdelin L, Sanders WJ), pp. 669–683. Berkeley, CA: University of California Press. [Google Scholar]
- 18.Kuhner MK, Yamato J, Felsenstein J. 1998. Maximum likelihood estimation of population growth rates based on the coalescent. Genetics 149, 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harley EH, Baumgarten I, Cunningham J, O'Ryan C. 2005. Genetic variation and population structure in remnant populations of black rhinoceros, Diceros bicornis, in Africa. Mol. Ecol. 14, 2981–2990. ( 10.1111/j.1365-294X.2005.02660.x) [DOI] [PubMed] [Google Scholar]
- 20.Kotzé A, Dalton D, du Toit R, Anderson N, Moodley Y. 2014. Genetic structure of the black rhinoceros (Diceros bicornis) in south-eastern Africa. Conserv. Genet. 15, 1479–1489. ( 10.1007/s10592-014-0632-x) [DOI] [Google Scholar]
- 21.Moodley Y, et al. 2017. Extinctions, genetic erosion and conservation options for the black rhinoceros (Diceros bicornis). Sci. Rep. 7, 41417 ( 10.1038/srep41417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown SM, Houlden BA. 2000. Conservation genetics of the black rhinoceros (Diceros bicornis). Conserv. Genet. 1, 365–370. ( 10.1023/A:1011579807460) [DOI] [Google Scholar]
- 23.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. 2004. GENETIX 4.05, Population genetics software for Windows TM. Montpellier, France: Université de Montpellier II.
- 24.Goudet J. 1995. FSTAT (version 1.2): a computer program to calculate F-statistics. J. Hered. 86, 485–486. ( 10.1093/oxfordjournals.jhered.a111627) [DOI] [Google Scholar]
- 25.Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. ( 10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 26.Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu YX. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. ( 10.1093/oxfordjournals.molbev.a026036) [DOI] [PubMed] [Google Scholar]
- 30.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 ( 10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772. ( 10.1038/nmeth.2109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beaumont M. 1999. Detecting population expansion and decline using microsatellites. Genetics 153, 2013–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Storz J, Beaumont M. 2002. Testing for genetic evidence of population expansion and contraction: an empirical analysis of microsatellite DNA variation using a hierarchical Bayesian model. Evolution 56, 154–166. ( 10.1111/j.0014-3820.2002.tb00857.x) [DOI] [PubMed] [Google Scholar]
- 34.Beaumont MA, Zhang W, Balding DJ. 2002. Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wegmann D, Excoffier L. 2010. Bayesian inference of the demographic history of chimpanzees. Mol. Biol. Evol. 27, 1425–1435. ( 10.1093/molbev/msq028) [DOI] [PubMed] [Google Scholar]
- 36.Grollemund R, Branford S, Bostoen K, Meade A, Venditti C, Pagel M. 2015. Bantu expansion shows that habitat alters the route and pace of human dispersals. Proc. Natl Acad. Sci. USA 112, 13 296–13 301. ( 10.1073/pnas.1503793112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merenlender AM, Woodruff DS, Ryder OA, Kock R, Vahala J. 1889. Allozyme variation and differentiation in African and Indian rhinoceroses. J. Hered. 80, 377–382. ( 10.1093/oxfordjournals.jhered.a110878) [DOI] [PubMed] [Google Scholar]
- 38.Vincens A, Schwartz D, Bertaux J, Elenga H, de Namur C. 1998. Late Holocene climatic changes in western equatorial Africa inferred from pollen from Lake Sinnda, southern Congo. Quat. Res. 50, 34–45. ( 10.1006/qres.1998.1979) [DOI] [Google Scholar]
- 39.Bayon G, Dennielou B, Etoubleau J, Ponzevera E, Toucanne S, Bermell S. 2012. Intensifying weathering and land use in Iron Age Central Africa. Science 335, 1219–1222. ( 10.1126/science.1215400) [DOI] [PubMed] [Google Scholar]
- 40.Patin E, et al. 2014. The impact of agricultural emergence on the genetic history of African rainforest hunter-gatherers and agriculturalists. Nat. Commun. 5, 3163 ( 10.1038/ncomms4163) [DOI] [PubMed] [Google Scholar]
- 41.Clark JD, Brown KS. 2001. The Twin Rivers Kopje, Zambia: stratigraphy, fauna, and artefact assemblages from the 1954 and 1956 excavations. J. Archaeol. Sci. 28, 305–330. ( 10.1006/jasc.2000.0563) [DOI] [Google Scholar]
- 42.Gifford DP, Issac GL, Nelson CM. 1980. Evidence for predation and pastoralism at prolonged drift: a pastoral Neolithic site in Kenya. Azania 15, 57–108. ( 10.1080/00672708009511277) [DOI] [Google Scholar]
- 43.Emslie R, Amin R, Kock R (eds). 2009. Guidelines for the in situ re-introduction and translocation of African and Asian rhinoceros. Gland, Switzerland: IUCN.
- 44.Wasser SK, Brown L, Mailand C, Mondol S, Clark W, Laurie C, Weir BS. 2015. Genetic assignment of large seizures of elephant ivory reveals Africa's major poaching hotspots. Science 349, 84–87. ( 10.1126/science.aaa2457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishida Y, Gugala NA, Georgiadis NJ, Roca AL. 2018. Evolutionary and demographic processes shaping geographic patterns of genetic diversity in a keystone species, the African forest elephant (Loxodonta cyclotis). Ecol. Evol. 8, 4919–4931. ( 10.1002/ece3.4062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harper C, et al. 2018. Robust forensic matching of confiscated horns to individual poached African rhinoceros. Curr. Biol. 28, R13–R14. ( 10.1016/j.cub.2017.11.005) [DOI] [PubMed] [Google Scholar]
- 47.Hildebrandt TB, et al. 2018. Embryos and embryonic stem cells from the white rhinoceros. Nat. Commun. 9, 2589 ( 10.1038/s41467-018-04959-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Griffiths CJ, Hansen DM, Jones CG, Zuël N, Harris S. 2011. Resurrecting extinct interactions with extant substitutes. Curr. Biol. 21, 762–765. ( 10.1016/j.cub.2011.03.042) [DOI] [PubMed] [Google Scholar]
- 49.Moodley Y, et al. 2018. Data from: Contrasting evolutionary history, anthropogenic declines and genetic contact in the northern and southern white rhinoceros (Ceratotherium simum) Dryad Digital Repository. ( 10.5061/dryad.6mc1606) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Moodley Y, et al. 2018. Data from: Contrasting evolutionary history, anthropogenic declines and genetic contact in the northern and southern white rhinoceros (Ceratotherium simum) Dryad Digital Repository. ( 10.5061/dryad.6mc1606) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All mitochondrial DNA sequences/genomes and microsatellite data generated in this study were uploaded to the Dryad Digital Repository at http://dx.doi.org/10.5061/dryad.6mc1606 [49].


