Abstract
Malignant Peripheral Nerve Sheath Tumors (MPNSTs) are highly resistant sarcomas that occur in up to 13% of individuals with Neurofibromatosis Type 1 (NF1). Genomic analysis of longitudinally collected tumor samples in a case of MPNST disease progression revealed early hemizygous microdeletions in NF1 and TP53, with progressive amplifications of MET, HGF, and EGFR. To examine the role of MET in MPNST progression, we developed mice with enhanced MET expression and Nf1 ablation (Nf1fl/KO;lox-stop-loxMETtg/+;Plp-creERTtg/+; referred to as NF1-MET). NF1-MET mice express a robust MPNST phenotype in the absence of additional mutations. A comparison of NF1-MET MPNSTs with MPNSTs derived from Nf1KO/+;p53R172H;Plp-creERTtg/+ (NF1-P53) and Nf1KO/+;Plp-creERTtg/+ (NF1) mice revealed unique Met, Ras, and PI3K signaling patterns. NF1-MET MPNSTs were uniformly sensitive to the highly selective MET inhibitor, capmatinib, whereas a heterogeneous response to MET inhibition was observed in NF1-P53 and NF1 MPNSTs. Combination therapy of capmatinib and the MEK inhibitor trametinib resulted in reduced response variability, enhanced suppression of tumor growth, and suppressed RAS/ERK and PI3K/AKT signaling. These results highlight the influence of concurrent genomic alterations on RAS effector signaling and therapy response to tyrosine kinase inhibitors. Moreover, these findings expand our current understanding of the role of MET signaling in MPNST progression and identify a potential therapeutic niche for NF1-related MPNSTs.
Keywords: malignant peripheral nerve sheath tumor (MPNST), NF1, MET, MEK, capmatinib, trametinib
Introduction
Neurofibromatosis type 1 (NF1) is caused by germline mutations in the NF1 gene and is the most common single-gene disorder affecting ~1 in 3,000 live births (1). Approximately 8–13% of individuals with NF1 will develop malignant tumors, most commonly Malignant Peripheral Nerve Sheath Tumors (MPNSTs). NF1-related MPNSTs are highly aggressive sarcomas that frequently metastasize and have five-year survival rates ranging from 20–50% (2,3). The mainstay of treatment is surgical resection when possible with consideration of chemotherapy and radiation therapy in select cases. Even though chemotherapy may initially stabilize disease, early responses are typically followed by a rapid evolution of chemoresistance and metastasis (4).
The NF1 gene encodes neurofibromin, a GTPase-activating protein that regulates RAS (including HRAS, NRAS, and KRAS) and loss of NF1 leads to deregulated RAS signaling. The RAS signaling node activates multiple kinase effector cascades, including the RAF–MEK–ERK pathway. NF1-related MPNSTs have been shown to arise from NF1-null myelinating Schwann cells where neurofibromin deficiency results in RAS deregulation (5,6). Plexiform neurofibromas are the benign precursor of NF1-related MPNSTs and are formed by a recruited admixture of NF1 haploinsufficient cells (fibroblasts, mast cells, and perineurial cells) following an initial NF1-loss-of-heterozygosity event in a peripheral nerve Schwann cell (7–9).
Several studies implicate oncogenic MET signal activation in NF1-related MPNST disease progression. The oncogene MET encodes a receptor tyrosine kinase that is involved in the progression and metastasis of most solid human cancers (10). The pleiotropic effects of MET activation are mediated through a variety of effector pathways including dominant regulators of cellular proliferation and survival (i.e. RAS/ERK, PI3K/AKT/mTOR) and cellular motility (STAT3, Rho kinases). MPNSTs often develop from MET-overexpressing plexiform neurofibromas (11). High-resolution array-CGH recently identified MET and HGF gene amplifications in a significant proportion (~30%) of NF1-related MPNSTs (12). MET phosphorylation (Tyr1234/35) was demonstrated in at least half of MPNSTs, and was recently proposed as a biomarker for MET-activated MPNSTs (13). In this same study, inhibition of VEGFR2/ MET/RET (cabozantinib) mitigated tumor growth in an MPNST xenograft model. Because genomic alterations in HGF and MET occur on a backdrop of putative RAS deregulation from germline loss of NF1 in the stroma, and NF1 loss-of-heterozygosity events in the MPNST cell of origin, it is possible that RAS and MET crosstalk contributes to therapy resistance. Towards this end, MET has already been implicated in resistance to RAS pathway inhibition in several cancers, including melanoma and colorectal cancer (14,15). Although we suspect that RAS/ERK and MET/HGF cooperate to promote cancer progression and therapy resistance, the mechanisms that mediate these complex signaling interactions are not well understood. Recent studies support the concept that cooperative RAS-MET signaling is influenced by genomic alterations within the MET and RAS signaling pathways. Currently, it is unclear 1) whether MET activation is sufficient for malignant transformation of NF1-deficient Schwann cells to into MPNSTs, and 2) how the RAS and MET signaling pathways interact in NF1-deficient MPNSTs. Here, we performed a longitudinal genomic analysis to identify key genetic events underlying the transformation of a human plexiform neurofibroma to a MPNST. Our results indicate that MET and HGF copy number gains were present at the time of MPNST diagnosis and increased during disease progression and treatment. In addition to MET and HGF copy number variations, the NF1-related MPNST exhibited highly complex genomic structural variations that evolved over time leading to additional genomic gains and losses that appear to be non-random and adaptive for MPNST progression (16,17).
In order to further interrogate the role of MET signaling in MPNST progression and therapy response, we developed and characterized a unique mouse model of MET activation in p53 wild type, Nf1-null myelinating cells. Our hypothesis is that MET activation is sufficient to drive malignant tumorigenesis when combined with putative NF1 loss of function, and that MET-activated MPNSTs will respond to targeted MET inhibition. Our findings build upon prior work examining NF1 loss of heterozygosity (LOH) in myelinating cells (18,19) and present a complementary model of MPNST to established models combining Nf1 loss with p53 or Ink4a/Arf (20,21). Our results demonstrate that highly selective MET inhibition is effective against MPNSTs bearing a “MET addicted” signature whereby MET inhibition also mitigates downstream RAS-ERK and PI3K-AKT activation. Conversely, we show reduced effectiveness of MET inhibitor monotherapy in Met-amplified, p53 deficient (“NF1-P53”) NF1-related MPNSTs. These findings are attributable in part to signaling adaptations that occur early during therapy that may be affected by loss of P53 function. Cre-negative animals were used for subsequent breeding. Plp-creERT (B6.Cg-Tg(Plp1-cre/ERT)3Pop/J) signaling and therapy resistance in NF1-related MPNSTs, we also show that the combination of capmatinib (INCB28060; Novartis; MET inhibitor) and trametinib (Novartis; GSK1120212; MEK inhibitor) results in enhanced therapy effectiveness and reduced tumor response variation compared to monotherapy. Combined capmatinib and trametinib treatment was the most effective treatment in decreasing ERK, AKT, and MET activity in tumorgrafts. These results expand our current understanding of the role of RAS-MET signaling in MPNST disease progression and identify a potential therapeutic niche for NF1-related MPNSTs. Moreover, these findings highlight the influence of co-occurring genomic alterations on RAS effector signaling and therapy response to tyrosine kinase inhibitors.
Materials and Methods
Whole genome exome sequencing and TITAN analysis
The plexiform neurofibroma from which the MPNST originated was large, extending across the chest wall (Supplemental Figure 1A). We sampled a grossly distinct region that was 5cm adjacent to the visible origin of the MPNST. The specimens were analyzed by a board-certified pathologist (V.K.) to confirm diagnosis, percent content and viability. Extracted DNA samples for both human and mouse were prepared using Agilent SureSelect library prep with the Agilent SureSelect Human All Exon V5 and Agilent SureSelectXT2 exome capture systems, respectfully. Both human and mouse exome samples were sequenced with 2X 100bp reads on the Illumina HiSeq 2500 from the Michigan State University Research Technology Support Facility (MSU-RTSF) with a total mean read coverage of 50–90X and 15–20X, respectively. Read quality was assessed using FASTQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) and trimmed using seqtk (https://github.com/lh3/seqtk). After trimming, reads were mapped to the hg19 genome using BWA-MEM v 0.7.12 (http://arxiv.org/abs/1303.3997) and duplicates were removed with Picard MarkDuplicates (http://broadinstitute.github.io/picard/). Variant calling using the deduplicated BAMs was completed using GATK Best Practices with suggested default values (22–24). Somatic variants were identified using MUTECT2 (25) using default settings. Copy number alterations (CNA) of MPNST samples were assessed using TITAN (26) for pre, post, and recurrent samples compared to blood exome. Default ploidy values were set to two and converge parameters were set to default values. Segment calls generated by TITAN were used for both genome wide and inferred gene specific copy number alternation analyses.
Development of NF1-MET, NF1-P53, and NF1 mice
Nf1fl/KO;lox-stop-loxMETtg/+;Plp-creERTtg/+ mice and controls were created to examine the combination of conditional Nf1 loss of heterozygosity (27) with overexpression of a chimeric Met transgene (28,29). These two genetic events were targeted to occur in myelinating cells immediately after birth using a tamoxifen-inducible Plp-creERT transgene (18) in mice with a global, constitutive mutation of the other allele of Nf1. Nf1fl/+ mice were obtained from the NCI Frederick repository. Nf1KO/+ mice were created by breeding Nf1fl/+ mice to CMV-Cre mice (30). Only second generation or later Nf1KO/+; Cre-negative animals were used for subsequent breeding. Plp-creERT (B6.Cg-Tg(Plp1-cre/ERT)3Pop/J) and CMV-Cre(BALB/c-Tg(CMV-cre)1Cgn/J) transgenic mice were obtained from The Jackson Laboratory. Nf1fl/KO;lox-stop-loxMETtg/+;Plp-creERTtg/+ mice were produced by crossing Nf1fl/fl;Plp-creERTtg/+ mice with Nf1KO/+;lox-stop-loxMETtg/+ mice. Lactating dams were dosed with 100 μl of 20mg/ml tamoxifen (Sigma) in corn oil (Acros Organics) by oral gavage b.i.d. on postnatal days 1–5, based on similar previous studies (19). Of note, crosses using Nf1fl/fl;Plp-creERTtg/tg mice did not yield pups at expected Mendelian ratios and were not used for this study. Nf1KO/+ mice were created by breeding NF1fl/+ mice to mice carrying a CMV-Cre transgene. Only second generation or later Nf1KO/+; Cre-negative animals were used for subsequent breeding. Nf1KO/+;TP53KO/+ mice were created by breeding Nf1KO/+ mice to 129S4-Trp53tm2Tyj/Nci mice (functionally TP53KO/+) (31). All Nf1KO/+;Trp53KO/+ mice produced were bred to wildtype animals to confirm cis-conformation of Nf1 and Trp53 as performed in Vogel et al. (32). Mice producing at Nf1KO/+;Trp53KO/+ offspring when bred to wild-types were used as founders. All animal experimentation in this study was approved by the Van Andel Institute’s Internal Animal Care and Use Committee (IACUC). Details on mouse genotyping procedures are in Supplementary Data.
Mouse Genotyping
Tail biopsy DNA was prepared at the time of weaning using 25mM NaOH and 100μM EDTA at 95°C for 20 minutes. Additional DNA was extracted from tail biopsy, tumors, and sciatic nerve tissue at necropsy when possible using DirectPCR reagent (Viagen) according to the manufacturer’s instructions. Primers used were as follows: Met transgene (amplicon size = 320bp) 5’ CCT ACA GCT CCT GGG CAA CG 3’ and 5’CCA TTC GCC ATT CAG GCT GCG 3’; Met transgene recombined allele (420bp) 5’ CCT ACA GCT CCT GGG CAA CG 3’ and 5’ TGG CTT TGC TGC TGC AGT CCC 3’; Nf1 wildtype (480bp) 5’ CTT CAG ACT GAT TGT TGT ACC TGA 3’, 5’ ACC TCT CTA GCC TCA GGA ATG A 3’, Nf1 flox (350bp) 5’ CTT CAG ACT GAT TGT TGT ACC TGA 3’, 5’ TGA TTC CCA CTT TGT GGT TCT AAG 3’; Nf1 recombined (280bp) 5’ CTT CAG ACT GAT TGT TGT ACC TGA 3’, 5’ CAT CTG CTG CTC TTA GAG GAA CA 3’; and Plp-CreERT transgene (300bp) 5’ CAT GTT TAG CTG GCC CAA ATG TTG CTG 3’, 5’ CGA CCA TGC CCA AGA AGA AGA GGA AGG 3’.
Development of murine MPNST tumorgrafts and drug treatment
Immediately following euthanasia of tumor bearing mice, 15–25 mg portions of each tumor were transplanted into the flank of athymic nude mice using a 10 gauge trochar. Mice were examined weekly and euthanized when the tumor size exceeded 1500 mm3. For treatment studies, bulk tumor pieces were transplanted subcutaneously into athymic nude female mice and tumor growth was evaluated twice weekly. When tumor volume reached approximately 150 mm3, mice were randomized into treatment groups and dosed for 3 weeks or until mice reached euthanasia criteria. Respective doses across all treatment combinations were capmatinib (3, 10, 30 mg/kg twice b.i.d. oral gavage) and trametinib (1 mg/kg q.d. via oral gavage). Specific combination studies were performed with capmatinib (30 mg/kg) and trametinib (1 mg/kg) obtained from Novartis. The tumors were measured twice weekly using a caliper and the tumor volumes were calculated as length × width × depth. A minimum of 3 tumors from each GEMM were assessed. Representative experiments are shown.
Fluorescent In Situ Hybridization
Tumor touch preps were prepared by imprinting slightly thawed tissues onto glass slides and air-drying. The slides were fixed in methanol/acetic acid (3:1) for 20 min, equilibrated in 2× saline/sodium citrate (SSC) at 60°C for 45 min, digested with 0.005% pepsin at 37°C for 10 min, and washed with 1× PBS for 5 min. Slides were placed in 1% formaldehyde for 10 min at room temperature, 1× PBS for 5 min, and dehydrated in an ethanol series (70%, 85%, 95%) for 2 min each. Slides were denatured in 70% formamide/4× SSC at 73°C for 5 min, washed in a cold ethanol series (70%, 85%, 95%) for 2 min each, and air dried. Probes were denatured at 73°C for 5 min, added to the slide, and hybridized overnight at 37°C. Post-hybridization washes were with 2× SSC at 73°C for 2.5 min, cooled in 4× SSC/0.1% Tween 20, and rinsed in H2O. Slides were air dried and then counterstained with anti-fade DAPI. BAC clones RP23–416H6 and RP23–73G15 (located within A2 of chromosome 6) were labeled with Orange-dUTP (Abbot Molecular), BAC clone RP23–125O3 (located within A2 of chromosome 5) was labeled with Red-dUTP (Abbot Molecular), and the Met transgene targeting construct (containing 5.3kb of the endogenous Rosa26 gene and 12kb of the combined Rosa26-targeted mouse/human Met transgene locus) was labeled with Green-dUTP (Abbot Molecular) using nick translation. Clone RP23–151I21 (located within A1 of chromosome 8) and clone RP23–23D5 (located within B3 of chromosome 16) were labeled with either Spectrum Green or Spectrum Orange depending on the comparison. Image acquisition was performed with a CCD camera (VDS, voss Kuhler Gmbh.) mounted on an Olympus BX51 epifluorescence microscope using FISHView software version 2.1 (Applied Spectral Imaging). Hybridization signals were scored for at least 200 nuclei per slide.
Histopathology
Mouse tissues were fixed in 10% neutral buffered formalin for 72 hours (Thermo) and decalcified, when necessary, in Immunocal (Decal Chemical Corp) for 24–72 hours prior to paraffin embedding and sectioning for histology and immunohistochemistry. Immunohistochemistry was performed on formalin-fixed paraffin embedded samples using a citrate-based antigen retrieval system (Vector Labs). Samples were stained for MyoD (1:200, Dako Cytomation) and S100 (1:500, Dako Cytomation). Tumors were analyzed by a sarcoma pathologist (D.C.) who was blinded to the genotype of the animals. MPNST tumors demonstrated fascicular growth patterns of spindle cells that were MyoD-negative. RMS tumors were spindle shaped, MyoD-positive tumors that contained more rounded cells. Immunohistochemistry for Ki-67 (Spring Biosciences), phosphorylated MET (Cell Signaling, D26), and phosphorylated MAPK (Cell Signaling, 9101) were performed using a Ventana autostainer and images were obtained with an Aperio Digital Imaging system (Leica) and an Eclipse 55i microscope (Nikon). For the NF1-MET, NF1-P53, and NF1 models, all derived cell lines, primary tumors and tumorgrafts were characterized for consistency of the MPNST immunophenotype (IHC) and genetic alterations (FISH), and throughout serial passaging. With respect to the in vitro and in vivo experiments, one primary GEMM tumor per model was analyzed.
Western Blotting
Whole-cell lysates were collected in a RIPA buffer containing protease/phosphatase inhibitor cocktail (Cell Signaling Technology). Mouse derived cell lines were grown to 90% confluency overnight by seeding plates with 50,000 – 75,000 cells. Cells were then serum starved for 24 hours and then treated with capmatinib for 2 hours followed by 100ng/mL of HGF for 15 minutes. Cells were then washed with PBS and harvested in RIPA buffer plus protease inhibitor cocktail (Roche). Lysates (20 μg) were resolved on a 4%−20% TGX SDS-PAGE gel (Bio-Rad) and transferred to a PVDF membrane (Invitrogen). After blocking for 1 hour with 5% dry milk in TBST buffer (20 mmol/L Tris-HCl pH 7.4, 150 mmol/L NaCl, 0.1% Tween-20), blots were probed overnight at 4°C with the following primary antibodies from Cell Signaling Technology: Met (25H2; #3127), pMET (D26; #3077), AKT (#9272), pAKT (S473; #9271), MAPK (#9102), pMAPK (Thr202/Tyr204; #9101), and β-tubulin (#2146). Blots were reacted with peroxidase- conjugated antibody for 1 hour (Cell Signaling Technology) and visualized using ECL detection (Amersham).
Statistical Methods
Kaplan-Meier curves were used to display tumor-free survival and, after verifying the proportional hazards assumption, Cox regression with false discovery rate adjusted contrasts were used to test for differences in tumor incidence rates between mouse lines. Three mice euthanized due to hindlimb paralysis had evidence of small paraspinal neoplasms at necropsy and were therefore included as tumor events. Genotypes with no tumor events are not shown. Logistic regression with false discovery rate adjusted contrasts were used to test for differences in the frequencies at which the different mouse lines were euthanized or died early due to tumor burden. Linear mixed-effects models, with random slopes and intercepts, and false discovery rate adjusted contrasts were used to estimate and compare tumor growth rates between different capmatinib dosages within each line and between the different mono and combo therapies. Fiellers’s theorem with Bonferroni adjusted significance level was used to estimate the confidence intervals for the percent reduction in tumor growth rates for mice treated with capmatinib, trametinib, or both; relative to vehicle. All analyses were conducted using R v3.2.2 (https://cran.r-project.org/) with an assumed level of significance of α = 0.05.
Study Approval
Tissue samples were collected in accordance with US Common Rule after IRB approval at both Spectrum Health and the Van Andel Institute. Written informed consent was obtained from study participants prior to inclusion in the study.
Results
Longitudinal genomic analysis of MPNST progression confirms evolving structural and sequence variations
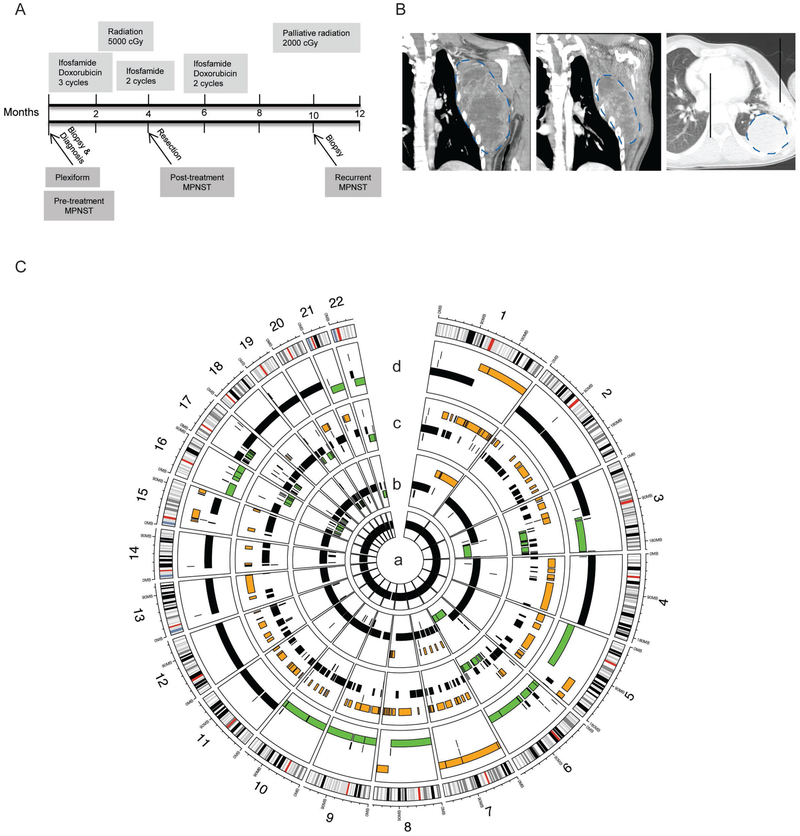
To investigate the genomic alterations that occur during MPNST progression, we followed the disease course of an NF1-affected adolescent male who developed a high grade, metastatic MPNST arising from a chest wall plexiform neurofibroma. Exome sequencing was performed on biopsies procured throughout disease progression: benign plexiform neurofibroma → MPNST pretreatment → MPNST posttreatment → MPNST metastasis (Figure 1A, Supplemental Figure 1A-C). The patient initially presented at 15 years of age with a rapidly enlarging mass in his left axilla that measured 11 × 9 × 17cm by MRI (Figure 1B, left panel). Biopsies were performed at diagnosis which verified the presence of a high grade MPNST arising within a plexiform neurofibroma (AJCC stage 3, T2bG3N0M0). The patient received neoadjuvant chemotherapy consisting of 3 cycles of ifosfamide and doxorubicin. To enhance the resectability of the tumor following neoadjuvant chemotherapy, the patient received 5000 cGy of preoperative radiation concurrently with two further cycles of ifosfamide. The combined radiation and chemotherapy reduced the size of the tumor enough to permit wide resection with chest wall reconstruction four months after diagnosis (Figure 1B, middle panel). Following resection, the patient completed two additional cycles of chemotherapy with ifosfamide and doxorubicin. Surveillance imaging obtained four months after the end of therapy revealed recurrent disease in both lungs (Figure 1B, right panel) and CT-guided biopsy confirmed the diagnosis of metastatic MPNST. The recurrence was not amenable to resection and the patient succumbed to disease two years from the date of original diagnosis.
Figure 1: Comprehensive, longitudinal genomic analysis demonstrates targetable genetic changes early in disease progression.
(A) Timeline with treatments (upper) and biopsy samples (arrows, lower; n=1 sample per time point) from an MPNST and associated plexiform neurofibroma in an adolescent male with Neurofibromatosis type 1. (B) The MPNST (indicated by blue dotted line) progression was imaged by magnetic resonance imaging at diagnosis (left panel) and post- treatment (middle panel). The recurrence was imaged by computerized axial tomography (CAT) (right panel). (C) Summary of genomic copy number changes across the four tumor samples. The plexiform neurofibroma (inner ring, a) was genomically normal. Increased copy number gains (orange), and deletions (green) throughout the progression of the pre-treatment MPNST (ring b), post-treatment MPNST (ring c) and recurrent MPNST (ring d).
To evaluate the genomic alterations that occurred during MPNST progression in this case, tumor DNA was analyzed using a shotgun whole exome sequencing approach. Copy number alterations were inferred using TITAN with the patient’s peripheral blood DNA serving as a normal control. The genomic alterations that were identified in each stage of MPNST progression are summarized in a Circos plot (Figure 1C). No copy number alterations were found in the plexiform neurofibroma (ring a, Figure 1C), but the pre-treatment MPNST demonstrated chromosomal amplifications (orange regions) on chromosomes 1, 7, and 8 (ring b, Figure 1C). Additional structural alterations and site-specific amplifications accumulated throughout the course of treatment (ring c, Figure 1C). Regional amplifications on chromosome (chr) 5, chr15, a potential whole chromosomal amplification of chr7, and deletion of chr16 (ring d, Figure 1C) developed in the metastatic lesion, specifically. Several deletions (green) were also identified in the MPNST, including a region of chr17q that contains TP53 and NF1. Notably, the deleted regions of chr17q as well as regions on chr3, chr6, and chr16 progressively expanded throughout disease progression.
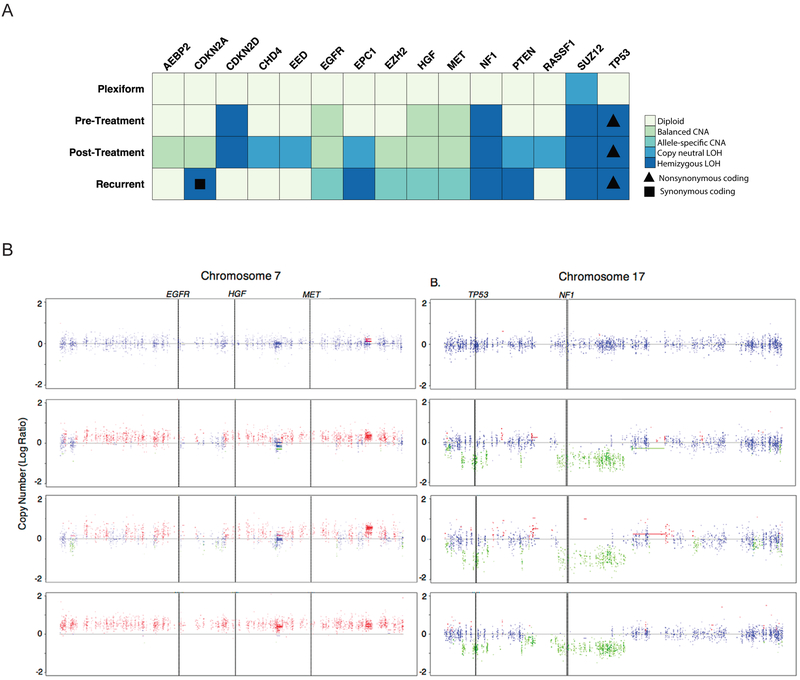
Focused analysis of known MPNST-related loci AEBP2, CDKN2A, CDKN2D, CHD4, EED, EGFR, EPC1, EZH2, HGF, MET, NF1, PTEN, RASSF1, SUZ12, and TP53 was performed (Figure 2A). The SUZ12 locus was found to be altered in the plexiform neurofibroma, within a genomic segment that while diploid, is representative of a near loss of heterozygosity event on chromosome 17. Somatic mutations in SUZ12 were not observed, but a single germline intron variant was identified. Therefore, the near loss of heterozygosity is representative of a potential genomic structural change within the SUZ12 region of chromosome 17. The observed structural alterations appear to be nonrandom and favor gain of oncogenic receptor tyrosine kinases and loss of TP53. Balanced copy number gains in MET, HGF, EGFR were noted in the pre-treatment sample and preceded other sites of additional oncogene amplification or tumor suppressor loss apart from the TP53, SUZ12, and CDKN2D alterations which were also present. The number of amplified MET, HGF, and EGFR loci was increased in the recurrent specimen (Figure 2B). More specifically, the copy numbers of MET, HGF, and EGFR progressed from a balanced conformation relative to ploidy state in the pre-treatment MPNST, to an imbalanced amplification above regional ploidy state in the recurrent MPNST (Figure 2A-B). MET copy number gain was confirmed in MPNST specimens using quantitative copy number PCR (Supplemental Figure 1D). Loss of NF1 and TP53 was also observed in the pre-, post- and recurrent-MPNST samples (Figure 2A-B). Interestingly, the non-synonymous coding alteration in the TP53 gene, NC_000017.11:g.7673223G>C, was found to be present in combination with TP53 hemizygosity in all stages of the MPNST. A similar phenomenon was observed for a synonymous coding mutation at the hemizygous CDKN2A locus in the metastatic tumor. Copy neutral loss of heterozygosity events were observed in the post-treatment sample in the genes CHD4, EED, EPC1, PTEN, and RASSF1. These observations demonstrate that copy number alterations in MET, HGF, and EGFR expanded from the time of diagnosis to metastasis. Whether these copy number alterations were due to DNA damaging therapy or a defined growth advantage is unclear.
Figure 2: Gain of MET, HGF, EGFR and loss of NF1 and TP53 copy number observed during MPNST progression.
(A) Detailed assessment of copy number changes in known MPNST-related genes with overlaid sequence alteration data identified allele-specific copy number amplifications (CNA), balanced CNA, copy neutral LOH, and hemizygous LOH. Triangles indicate a non-synonymous coding change in TP53 (NC_000017.11:g.7673223G>C). (B) TITAN copy number analysis of variants found between blood and MPNST samples for each stage of progression was performed on A) Chromosome 7 and B) Chromosome 17. Blue = Neutral, Green = Hemizygous loss of heterozygosity, Red = Gain. (n=1 sample per time point).
Additionally, our genomic analysis revealed a substantial amount of structural variation in multiple chromosomes in the post-treatment and recurrent samples (Figure 1C, ring c and d). Plexiform neurofibroma had very few regions of allelic imbalance and contained one predominant clonal cluster, whereas the pre-, post-treatment, and recurrent tumor samples contained large chromosomal regions of allele imbalance and additional subclonal clusters, corresponding to the genomic aberrations (Supplemental Figure 2). This effect is not attributable to sample necrosis as our specimens passed quality control measures for tissue viability (>99%) and DNA purity (>90%). Although some of the structural rearrangements we observed likely resulted from DNA damaging chemotherapy and radiation treatment, the underlying clonal makeup changed over time (Supplemental Figure 2A-D). For example, we observed a CDKN2D deletion in the pre-treatment and post-treatment specimens, but not in the recurrent tumor (Figure 2A). This suggests that the pre- and post-treatment tumor samples contained a majority cell population harboring a CDKN2D DLOH event (chr19), but a small subpopulation was diploid at the CDKN2D locus that later emerged in the recurrent tumor (Supplemental Figure 2C-D, Figure 2A). With our exome sequencing approach, it was not possible to characterize subpopulations in a detailed manner; however our global clonality assessment indicates probable subclonal evolution over the course of disease progression.
Met amplification and NF1 loss in peripheral nerve myelinating cells is sufficient to induce MPNST formation
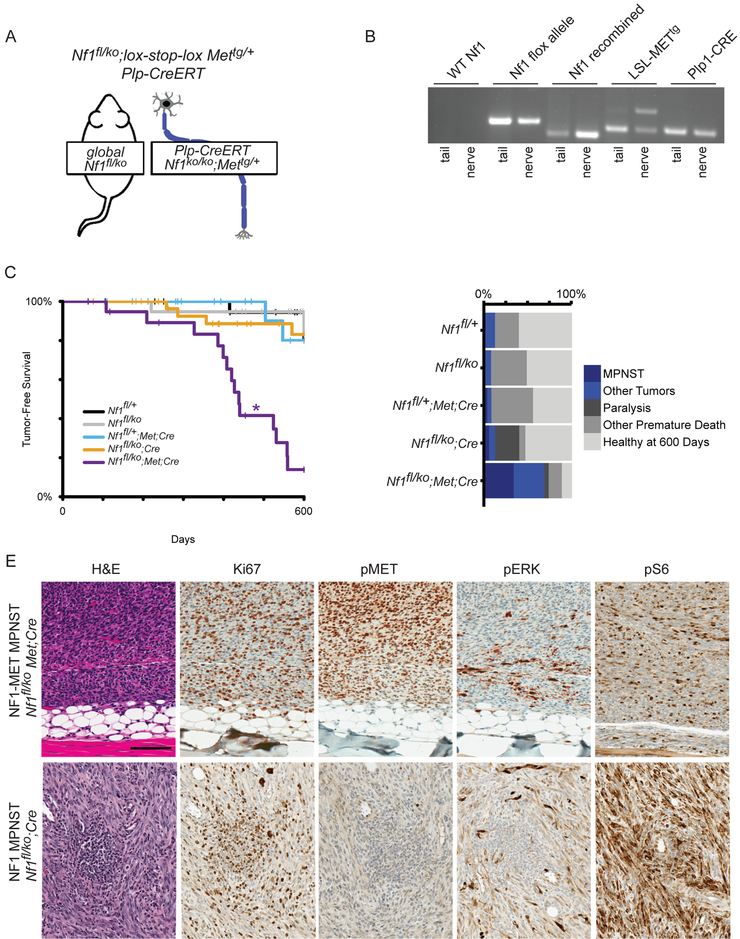
To interrogate the role of MET activation in NF1-related MPNSTs, we developed a mouse model that reflects MET amplification in the context of NF1 deficiency. In this Cre-inducible mouse model (Figure 3A), a chimeric Met transgene that was previously shown to activate Met signaling above physiologic thresholds in neuronal tissue (28,29) was combined with Nf1 deficiency in myelinating Schwann cells (18). These two genetic events were induced immediately after birth using a tamoxifen-inducible Plp-creERT transgene (33) in mice with a global, constitutive loss of the Nf1 allele. Tamoxifen induction at postnatal days 1–5 was selected to best replicate plexiform neurofibromagenesis based on previous studies of Nf1fl/ko;Plp-creERTtg/+ mice (18,19). Allele-specific PCR confirmed recombination of the loxp-flanked Nf1 gene in peripheral nerves and the presence of the recombined Met transgene (Figure 3B). For brevity we refer to the Nf1fl/ko;lox-stop-loxMettg/+;Plp-creERTtg/+ mice as “NF1-MET” mice throughout the manuscript. NF1-MET mice and controls (Nf1fl/ko;Cre, Nf1fl/+;Met;Cre, and wild-type) were aged for up to 600 days to evaluate their tumor phenotypes. The tumor free survival of NF1-MET mice was significantly decreased relative to the other four mouse lines (all p < 0.001, Figure 3C). The odds of NF1-MET mice being euthanized due to tumor burden was 14.58 times higher than Nf1fl/ko;Cre mice (p =0.0008, 95% CI [3.51, 60.59]) and 23.33 times higher than Nf1fl/ko (p= 0.0008, 95% CI [4.1, 132.9]).
Figure 3: MET activation drives malignant tumorigenesis in a mouse model of NF1-related MPNST.
(A) Schematic of mouse genetics. Mice have a global loss of one Nf1 allele, and loss of the second copy of Nf1 plus activation of a MET overexpressing transgene in myelinating cells following tamoxifen-induction of Plp1-Cre/ERT activity at postnatal day 1–5. (B) Allele-specific PCR for a mouse with the combined Nf1 loss and MET overexpression genotype shows absence of an unaltered Nf1 allele (wildtype Nf1) and presence of the functional but loxp flanked Nf1 allele (Nf1 flox); the recombined, loss-of-function Nf1 ko allele (Nf1 recombined); the inactive lox-stop-lox MET transgene (MET tg/rec, lower band) and the recombined tgMET transgene (MET tg/rec, upper band); and the Plp1-Cre/ERT transgene(Plp1-Cre). (C) Tumor-free interval data for all models plotted as Kaplan-Meier curves. Nf1fl/ko;Met;Cre mice developed tumors significantly sooner than all other mouse lines (indicated by *). (D) Cause of death in the 600 day observation period is plotted by frequency. Blue indicates death or euthanasia related to tumor burden, greys indicate healthy after 600 days or death due to other causes. E) H&E staining confirmed MPNST histology (representative images shown of 6 samples assessed in triplicate). Ki67 verified high rates of proliferation. High pERK and pS6 were observed by immunohistochemistry, however high pMET was only present in the NF1-MET MPNST.
In total, 70% of NF1-MET mice developed neoplasms, 50% of which were MPNSTs; whereas in the 13% of control mice that developed neoplasms, none were MPNSTs (Figure 3C). MPNSTs derived from NF1-MET demonstrated the characteristic spindle cell morphology with a fascicular growth pattern (Figure 3D-E). Tumors were classified as MPNSTs according to the established Genetically Engineered Mouse (GEM) nerve-sheath tumor classification: Grade III, S100+/MyoD− with nuclear atypia, high mitotic rate, and focal necrosis or hemorrhage (Figure 3E) (34). Paralysis or pseudo-paralysis of the hind-limbs, occurring in 8 of 29 NF1-MET mice, was another major cause of premature euthanasia (Figure 3D, dark grey bars). Tumor initiation occurred earlier in NF1-MET mice than the other four mouse lines (p < 0.0001), with a median tumor-free survival time of 438 days (95% CI [408, 559]). Tumors from mice with activated Met transgene expression demonstrated a high rate of cellular proliferation (Ki67) and greater amounts of active phospho-MET (pMET) by immunohistochemistry (Figure 3E).
In order to evaluate MET signaling in other genomic contexts, we isolated MPNSTs from both Nf1ko/+;p53LSL-R172Hand Nf1ko/+;Plp-creERTtg/+ mice. Nf1ko/+;p53LSL-R172H mice (referred to as “NF1-P53”) were derived by crossing the Nf1KO/+ and p53LSL-R172H mice (31). These mice did not require crossing with a Cre recombinase for tumor induction, since they are essentially a p53ko/+ model as the lox-stop-lox (LSL) cassette prevents expression of the p53R172H mutant. MPNST tumors from NF1-P53 mice have LOH of the wildtype p53 allele and we confirmed that p53LSL-R172H is in cis with Nf1 on Chr11. As a control for NF1 deficiency alone, we aged Nf1ko/+;Plp-creERTtg/+ mice and isolated an MPNST. For simplicity these mouse lines are referred to as “NF1” throughout the manuscript. Immunostaining of the NF1 MPNST showed less activated MET and ERK compared to the NF1-MET MPNST (Figure 3E).
Fluorescence in situ hybridization (FISH) was used to evaluate the Rosa26-targeted Met transgene, the endogenous mouse Met gene, and the endogenous mouse Hgf gene. Confirmation of the transgene was performed with a R26-MET probe in NF1-MET mice. In normal spleen from a NF1-MET mouse, we observed 2–4 copies of the Met transgene and in MPNSTs we observed 2–8 Met transgene copies (Supplemental Figure 3 and Supplemental Table 1). We also performed FISH analysis to determine whether endogenous Met or Hgf were amplified in the MPNST tumors from the various mouse lines. We did not observe any additional endogenous mouse Met copy number gains in NF1-MET tumors (Supplemental Figure 4A, Supplemental Table 2). Conversely, the NF1-P53 MPNST had a gain of 2–6 Met copies in 64% of tumor cells and the NF1 MPNST had a single Hgf copy number gain in 13% of MPNST cells (Supplemental Figure 3B-C, Supplemental Table 2). To ensure the robustness of these models, each founder and the derived tumors were sequenced to verify both induced and spontaneous mutations. Additional genomic characterization of these NF1-MET mouse tumors, as well as tumors from control MPNSTs in NF1 and NF1-P53 mice was performed using 15–20× whole exome sequencing and variants were cross-referenced against a list of genes with demonstrated roles in MPNST disease progression. No additional functional variants were detected in the mouse models apart from the engineered genomic modifications. In summary, we observed endogenous Met or Hgf copy number gains in the NF1-P53 and NF1 tumors; however the NF1-MET MPNSTs had 4–10 copies of Met (transgene and endogenous) and represent a novel model of Met-amplified NF1-related MPNSTs.
NF1-MET MPNST tumorgrafts are exquisitely sensitive to MET inhibition
Since MPNSTs often develop from MET-overexpressing plexiform neurofibromas and commonly exhibit MET and HGF gene amplifications (11,35), we evaluated the effect of MET inhibition with the kinase inhibitor capmatinib. Capmatinib (INC280) is an oral, highly selective and potent MET inhibitor that is well tolerated and has shown clinical activity in advanced solid tumors (36,37). Phase I and II clinical trials are ongoing to investigate the efficacy of capmatinib in cancers including melanoma, HCC, NSCLC, glioblastoma, colorectal cancer, and papillary renal cancer. Tumorgrafts and cell lines were established in immunocompromised mice from primary MPNSTs in the NF1-MET, NF1, and NF1-P53 models. Allele-specific PCR confirmed maintenance of the recombined NF1 gene and Met transgene. Because of the variability in Met alleles in these models, we evaluated three different concentrations of capmatinib (3, 10, 30 mg/kg) in each MPNST tumorgraft model (Supplemental Figure 5). Capmatinib significantly inhibited growth in the NF1-MET tumorgrafts even at the low dose of 3 mg/kg (p < 0.0001), suggesting these tumors are highly MET-dependent. Increasing the dose to 30 mg/kg produced a significant reduction and decreased tumor growth rate by 55 mm3 (p < 0.0001; Supplemental Figure 5A-C). Capmatinib treatment of NF1-P53 and NF1 MPNSTs showed variable responses among tumorgrafts harvested from the original GEMM’s. In separate experiments, we observed therapy responses to 30mg/kg capmatinib that ranged from a small subgroup of tumorgrafts (Supplemental 5B) to a statistically significant effect among an entire tumorgraft cohort (Figure 4). These results indicate that significant intertumoral heterogeneity exists within the NF1-P53 and NF1 tumorgrafts that promote resistant to MET inhibition. In comparison, NF1-related MPNSTs with high MET expression and activity (i.e. NF1-MET), significant efficacy of MET inhibition was consistently observed (Supplemental Figure 5C).
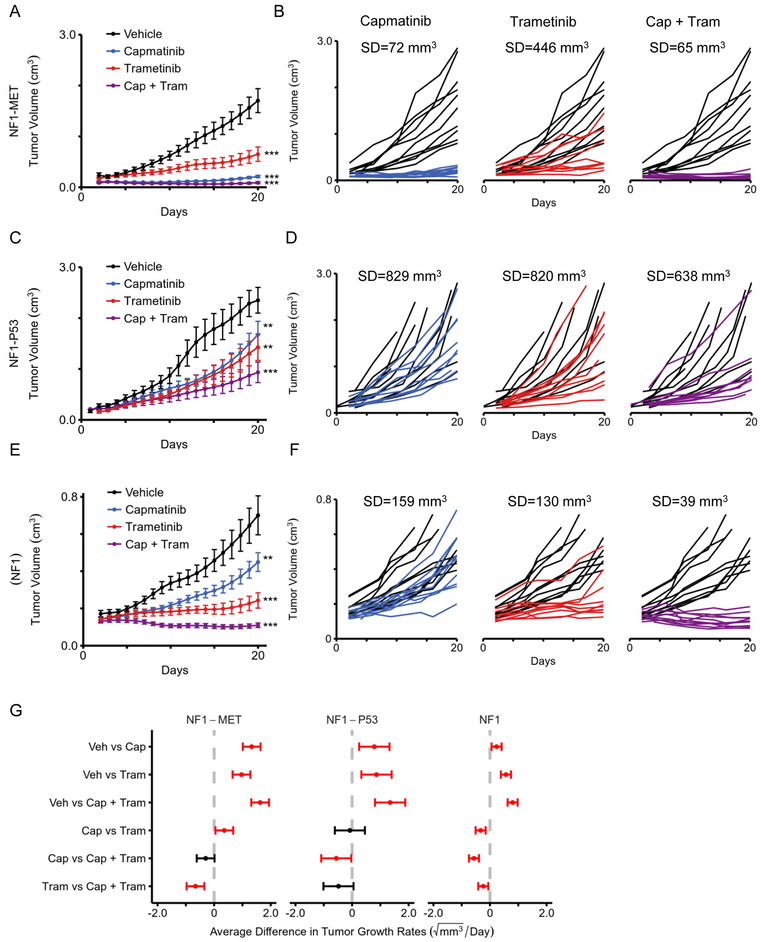
Figure 4: Combined MET and MEK inhibition significantly decreases tumor growth and response variability.
Tumor growth of (A) NF1-MET, (C) NF1-P53, and (E) NF1 tumorgrafts are plotted as means with standard errors. Tumor volume was imputed using last observation carried forward, until animal was euthanized. Curves terminate once >50% of mice have been euthanized in the respective treatment group. Individual tumor growth for (B) NF1-MET, (D) NF1-P53, and (F) NF1 tumorgrafts plotted by treatment (colored lines) compared to vehicle (black lines). (G) 95% confidence intervals for the pairwise differences between the growth rates of the select treatments, estimated and tested using linear mixed-effects models with random slopes and intercepts, and false discovery rate adjusted contrasts. Statistically significant differences (p-value < 0.05) between compared therapies are highlighted in red. * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001.
Evaluating the efficacy of MET or MEK inhibition in MPNST models of MET amplification and P53 loss
Our results and other TKI studies suggest targeted inhibition of multiple kinase nodes may be required to minimize response variability and abrogate bypass mechanisms of resistance (38–40). Since RAS deregulation is a hallmark of NF1-related tumors, we also evaluated the efficacy of MEK inhibition in our models (21). The recent clinical success of MEK inhibition (i.e. selumetinib) in NF1-related plexiform neurofibromas highlights the therapeutic potential of targeting MEK in NF1-related peripheral nerve tumors such as MPNSTs (41). We used the MEK inhibitor trametinib (Mekinist) which is a reversible, highly selective, allosteric inhibitor of MEK1 and MEK2, and is FDA approved for metastatic melanoma. One of the challenges of targeting the MEK/ERK pathway is achieving high level MEK inhibition without systemic toxicity. Trametinib has a strong pharmacokinetic profile with exceptional potency and specificity, oral bioavailability, and long half-life with a shallow Cmax (peak concentration) to Ctrough (trough concentration) profile (42). To evaluate the efficacy of MET and/or MEK inhibition in our distinct MPNST models, we measured the effect of monotherapy and combination therapy on tumor growth in the NF1-related MPNST tumorgrafts (Figure 4). We verified our previous results and again observed exquisite sensitivity to single agent capmatinib in NF1-MET (P53 wild type) tumors as well as significant tumor growth inhibition with trametinib (Figure 4A-B; p < 1.0e−16 for both capmatinib and trametinib treatment); however, pairwise comparison revealed that capmatinib was significantly more effective than trametinib in the NF1-MET tumors (p < 0.001; Figure 4G). Examining the individual growth curves for each treatment vs. vehicle control groups allowed us to assess the variability in treatment response. Capmatinib treatment was associated with minimal response variability (SD=72 mm3) while trametinib treatment response variability (446 mm3) was substantially higher (Figure 4B-C).
In NF1-P53 tumors, we observed a more aggressive growth rate than NF1-MET and NF1 tumors (Figure 4C-D). Both capmatinib and trametinib significantly inhibited NF1-P53 tumor growth (p <0.01; Figure 4G) in this experiment, although with both single agent treatments there was an aggressive growth trend after 21 days of treatment (Figure 4C). In NF1-P53 tumors, substantial variability was observed in response to capmatinib (Figure 4D; SD=829 mm3) or trametinib treatment (Figure 4D; SD=820 mm3). This was in stark contrast to the homogenous response to capmatinib in the NF1-MET tumors (Figure 4B; SD=72 mm3). We also evaluated the efficacy of MET and MEK inhibition in NF1 (P53 wildtype) tumors which had significantly slower growth rates compared to NF1-MET and NF1-P53 tumors and were suppressed with capmatinib (p < 0.004) and trametinib (p < 1.0e-16) treatment (Figure 4E-F). These results indicate that NF1-related MPNSTs containing MET amplification are highly sensitive to MET inhibitors, yet clinical response may be further improved with the addition of a MEK inhibitor.
Combination MET and MEK inhibition is more effective than single agent regardless of P53 status
Due to the diverse bypass mechanisms of resistance to kinase inhibition, achieving a durable clinical response will likely require inhibition of multiple critical kinase signaling nodes. To compare monotherapy and combination therapy, we compared the efficacy of combined MET and MEK inhibition. In NF1-MET tumors, pairwise treatment comparisons showed that combined MET and MEK inhibition was significantly more effective than monotherapy with trametinib (Figure 4G, p=0.000005). Combination therapy in NF1-MET tumors resulted in an overall growth rate reduction of 107% compared to 89% (capmatinib) and 58% (trametinib) (Supplemental Figure 6A). Even though combination therapy of capmatinib + trametinib (65 mm3) minimized the variability in response compared to capmatinib alone (72 mm3) (Figure 4B), there was no significant improvement in tumor reduction with combination therapy in the NF1-MET tumors when compared to capmatinib (Figure 4G, p=0.06). These results indicate that NF1-related MPNSTs containing MET amplification are highly sensitive to MET inhibitors, yet clinical response may be further improved with the addition of a MEK inhibitor.
In NF1-P53 tumors, combined capmatinib + trametinib treatment significantly inhibited tumor growth (p < 0.000002; Figure 4C,G) and caused a 60% growth reduction compared to monotherapy with trametinib (39%) or capmatinib (35%) (Supplemental Figure 6B). Analysis of the individual growth curves for NF1-P53 tumors revealed drastic improvement in response variability with combination therapy compared to single agent alone (Figure 4D,G). Capmatinib + trametinib reduced the variability in response to 638 mm3, yet there was one ‘non-responder’ that impacted this variability (Figure 4D). In pairwise comparisons, capmatinib + trametinib treatment was significantly better than capmatinib alone (p=0.05) but not trametinib (p=0.08) (Figure 4G). In the NF1 tumors, capmatinib + trametinib showed statistically significant improvement in tumor suppression vs single agent therapy with either capmatinib (p < 1.0e−16) or trametinib (p=0.004; Figure 4G, Supplemental Figure 6C). These results indicate that NF1-related MPNSTs without Met amplification or p53 loss may also be responsive to combination MET and MEK inhibition. Overall these findings confirm that NF1-related MPNSTs with P53 deficiency are less responsive to therapeutic approaches targeting single kinases and the best therapeutic response is achieved by inhibiting both MET and MEK signaling.
Combination MET and MEK inhibition prevents adaptive ERK and AKT response
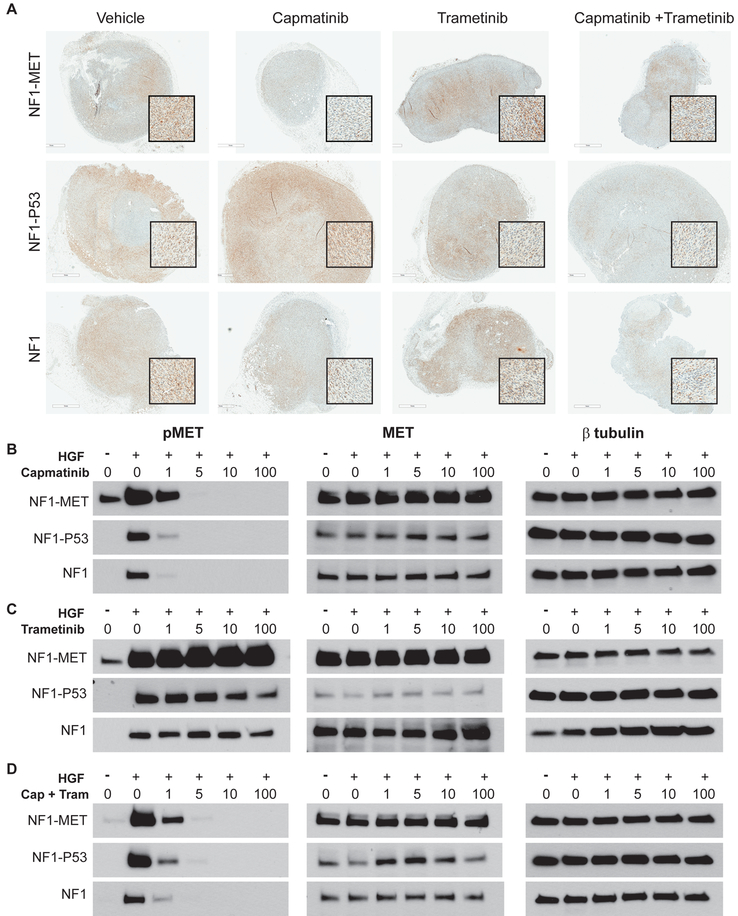
Differential patterns of MET, RAS, and AKT signaling were observed in response to monotherapy vs. combination kinase inhibition in both the in vivo and in vitro MPNST models. We examined the effect of MET and MEK inhibition on MET expression and activation (Figure 5). As expected, capmatinib resulted in complete inhibition of MET activation in NF1-MET tumors after 2 days of treatment (Figure 5A). NF1-MET cells produced extremely high levels of MET compared to NF1-P53 and NF1 cells, yet 5 nM capmatinib completely inhibited pMET in NF1-MET MPNST cells (Figure 5B). Despite the presence of minor Met amplification in NF1-P53 cells, there was no change in pMET after 2 days of capmatinib treatment in the NF1-P53 tumors (Figure 5A). Interestingly, MET was strongly activated with trametinib treatment in the NF1-MET and NF1 tumorgrafts (Figure 5A, trametinib column) and in the NF1-MET, NF1-P53, and NF1 cells with HGF treatment (Figure 5C). In both the tumorgrafts (Figure 5A) and cells (Figure 5D), dual MET and MEK inhibition effectively abrogated pMET levels in all three MPNST models. The activation of MET following trametinib treatment was unanticipated, but suggests that feedback RTK activation occurs following RAS blockade in NF1-deficient cells.
Figure 5: Effects of MET and MEK inhibition on MET activation in NF1 MPNST Models.
(A) Immunostaining of pMET (Y1234/1235) in MPNST models that were treated with kinase inhibitors for 2 days (areas of strongest signal are shown in the magnified inset boxes). (B-D) Western Blot analysis of MPNST cells treated with TKIs for 2 hours and +/− HGF for pMET (left panels), total MET (middle panels), and β-tubulin control (right panels). *The total MET exposure for the NF1-MET cells was half the time of the NF1-P53 and NF1 cells.
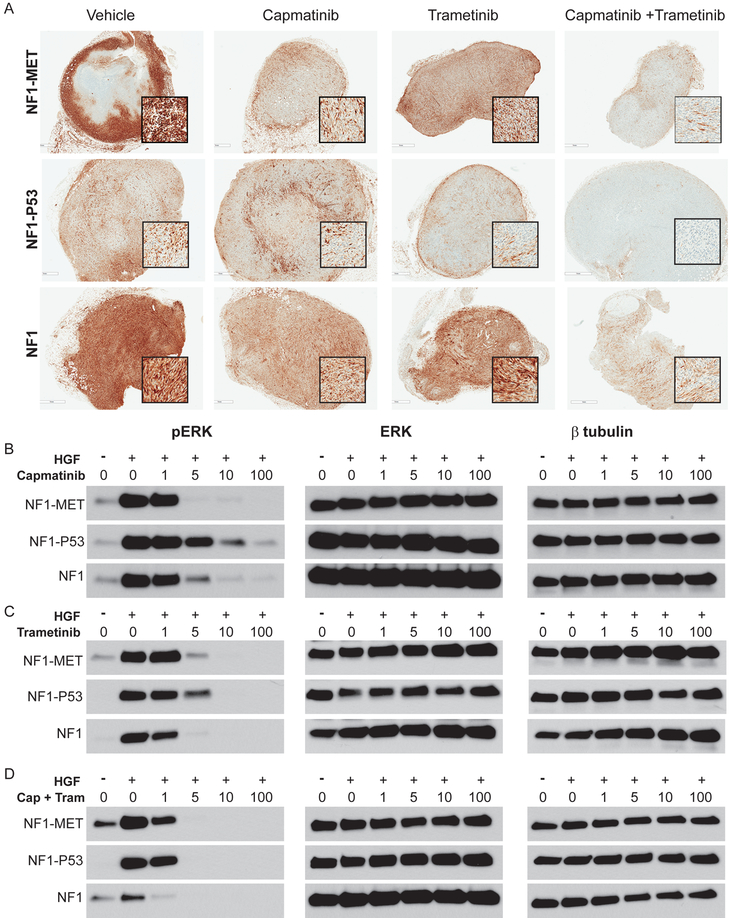
To see how MET and MEK inhibition impact RAS/MEK/ERK signaling, we examined ERK activity expression in the MPNST tumorgrafts and cells. Since NF1 deficiency results in RAS deregulation, high ERK activation was anticipated in all of the NF1-related MPNST tumors (Figure 6A). In NF1-MET cells, ERK activation was significantly inhibited by 5 nM capmatinib, whereas NF1 and especially NF1-P53 cells showed a marginal decrease in pERK in response to capmatinib (Figure 6B). Trametinib was much more effective at inhibiting ERK activation and demonstrated reduction of pERK at 10nM in NF1-P53 and NF1-MET cells and at 5nM in NF1 cells (Figure 6C). Combined capmatinib + trametinib treatment resulted in a greater than 90% reduction in pERK in all of the MPNST models (Figure 6A, right panels). In the MPNST cells, combined capmatinib and trametinib treatment mirrored the tumorgraft results and was the most effective treatment in decreasing pERK (Figure 6A,D). These findings validate the expected inhibition of RAS/ERK signaling with MEK TKIs and emphasize the importance of combined kinase inhibition to eradicate RAS signaling.
Figure 6: Responses in ERK signaling following MET and MEK inhibition in NF1 MPNST Models.
(A) Immunostaining of pERK (T202/Y204) in MPNST models that were treated with kinase inhibitors for 2 days (areas of strongest signal are shown in the magnified inset boxes). (B-D) Western Blot analysis of MPNST cells treated with TKIs for 2 hours and +/− HGF for pERK (left panels), total ERK (middle panels), and β-tubulin control (right panels).
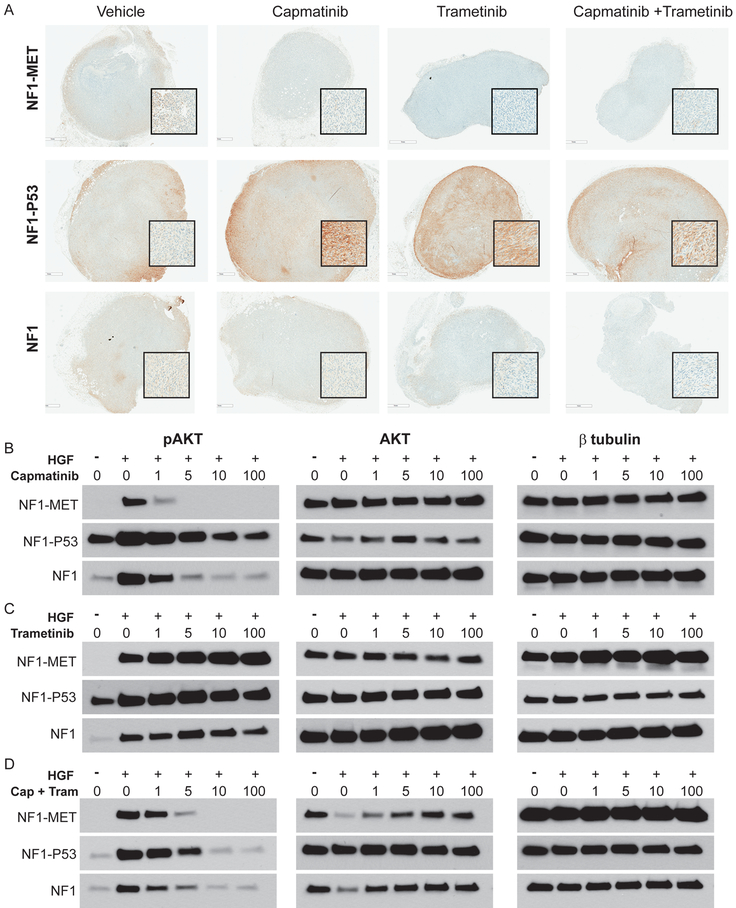
Another major signaling pathway that is activated by MET and known to interact with RAF is the PI3K/AKT pathway. We observed high pAKT at the invasive edges of the NF1-P53 tumors, yet minimal pAKT was present in NF1-MET or NF1 tumors (Figure 7A). Interestingly, increased pAKT was observed in NF1-P53 tumors after capmatinib or trametinib treatment and persisted at the tumor periphery even with combined capmatinib and trametinib treatment (Figure 7A). Analysis of AKT activity in the MPNST cell lines verified our in vivo observations. Even though AKT expression is comparable across the MPNST lines, pAKT is significantly higher in the NF1-P53 cells at basal conditions (Figures 7B-D). As expected, MET activation with HGF treatment stimulates pAKT in NF1-MET and NF1 cells, and further augmented AKT activation in NF1-P53 cells. Capmatinib treatment resulted in strong inhibition of AKT in NF1-MET and NF1 cells, but was ineffective in diminishing pAKT in the NF1-P53 cells (Figure 7B). Trametinib treatment resulted in a significant increase in pAKT in all three of the MPNST cells (Figure 7C). Combined MET and MEK inhibition was the most effective treatment for abrogating AKT activation in the NF1-P53 cells (Figure 7A,D). This analysis reveals that NF1-P53 tumors have basal AKT activity that is rapidly increased upon MET or MEK inhibition. This may indicate a distinctive mechanism of resistance that is readily activated upon kinase inhibition in P53-deficient MPNSTs.
Figure 7: Responses in AKT signaling following MET and MEK inhibition of NF1-MET, NF1-P53, and NF1 MPNSTs.
(A) Immunostaining of pAKT (S473) in MPNST models that were treated with kinase inhibitors for 2 days (areas of strongest signal are shown in the magnified inset boxes). (B-D) Western Blot analysis of MPNST cells treated with TKIs for 2 hours and +/− HGF for pAKT (left panels), total AKT (middle panels), and β-tubulin control (right panels).
Discussion
The lack of effective therapies for MPNSTs remains a significant issue for individuals affected by NF1 (2,3). By examining the evolution of genomic alterations in the case of a single, NF1-related human MPNST, we ascertained the degree of genomic instability that occurs over the course of MPNST treatment and identified targetable drivers of disease progression in MPNSTs. Intertumoral genomic heterogeneity has been assessed in NF1-related MPNSTs (12,20,27,43,44). Somatic and structural variants that appear early and accumulate throughout disease progression represent de facto driver mechanisms and in the case of NF1-related MPNSTs, could indicate fundamentally important signaling pathways in Schwann cell dedifferentiation (45). In our analysis, we observed evolution of subclonal clusters that corresponded with sampling timepoints. Additionally, we identified hemizygous microdeletions in the NF1 and TP53 loci in the pre-treatment specimen, with concomitantly observed amplifications of MET, HGF, and EGFR. It is not surprising that a TP53 variant was present in the pre-treatment MPNST given the prevalence of P53 mutations in clinical samples (46), and the driver effects of P53 in the context of NF1 deficiency (47); however, the early concomitant presence of MET, HGF, and EGFR amplifications, and the site-specific expansion of these loci over time points to an adaptive mechanism for both malignant transformation and clonal selection. The relationship between P53 haploinsufficiency and EGFR amplification has been previously demonstrated in human MPNSTs (48); however the timing and the relationship between MET/HGF and P53 genomic status has not been evaluated. Direct cooperation between MET and RAS has been shown to promote tumor resistance in other cancers. For example, KRAS and MET amplification mediates acquired resistance to MET inhibitors in MET-addicted gastric and lung cancer cells (49). In an esophagogastric cancer patient treated with a MET inhibitor, a KRAS mutation was discovered as a novel cause of acquired resistance (50). Whereas in KRAS-addicted cancer cells, KRAS mediated MET expression via increased MET translation and promoted “KRas addiction” in anchorage-independent conditions (51). Given the pleiotropic effects of MET activation (10) and its emerging role in therapy resistance (52,53), these data provide strong rationale for further exploration of MET-RAS and RAS-MET signal interactions in NF1-related MPNSTs. It should be noted that these findings were based on dependent observations in a single subject. More work is needed to determine the generalizability of these findings.
Even though therapeutically targeting RTKs has been successful in other cancers, the failure of EGFR inhibition (erlotinib) in MPNSTs indicates that other RTK pathways should be evaluated (54). To interrogate the role of MET activation in NF1-related MPNSTs, we developed a novel mouse model that reflects MET activation in the context of NF1-deficient Schwann cells. In addition to plexiform neurofibroma formation, NF1-MET mice express a robust MPNST phenotype in the absence of additional spontaneous or induced mutations. An interesting observation is that MPSNT tumorgrafts derived from NF1-P53 and NF1 mice exhibit spontaneous Met and Hgf copy number gains. Despite the presence of Met and Hgf amplifications respectively, there was no evidence of constitutive MET activation in the NF1-P53 or NF1 models, whereas the NF1-MET tumors were strongly activated without HGF stimulation. MET activation was further induced with HGF treatment in all of MPNST tumorgraft cell lines. Collectively, these data confirm that the degree of MET activation is dependent on the genomic context and that MET activation is sufficient for malignant transformation in NF1-deficient Schwann cells to MPNSTs in the setting of germline NF1 haploinsufficiency. We did not specifically assess the contribution of the tumor microenvironment to the rate of malignant transformation, but each of the models did maintain an Nf1 haploinsufficient background at baseline.
RAS-MET signal interactions are emerging as important mechanisms of therapy resistance and disease progression. NF1-related MPNSTs are no exception as they exhibit both MET and HGF amplifications, and deregulated RAS signaling owing to loss of NF1-mediated tumor suppression. The mitigated therapy response observed in other RAS-deregulated cancers following use of single agent kinase inhibitors (i.e. EGFR inhibition in non–small cell lung cancer) teaches us valuable lessons about kinome adaptation. As in KRAS mutant lung cancer, it is clear that NF1-deficient MPNSTs also rely on functionally redundant kinome signaling networks, not just the driver effects of a single therapeutic target (39,40,55). We tested the efficacy of single agent and combined inhibition of MET and MEK in MPNST models with variable levels of MET amplification and P53 deficiency. We discovered that NF1-MET tumors are “MET-addicted” and extremely sensitive to capmatinib treatment. Even though NF1-MET tumors were highly sensitive to MET inhibition, response heterogeneity was decreased with combined MET and MEK inhibition. Trametinib and capmatinib significantly reduced tumor growth as single agent therapy in both NF1-P53 and NF1 tumors. Combined capmatinib + trametinib treatment was more effective than trametinib or capmatinib alone in some of the models, however in all tumorgrafts combined MET and MEK inhibition reduced the response variability. Interestingly, we observed a differential pattern of kinase signaling between the P53-intact and P53-deficient models, and in response to MET and MEK inhibition. In all of the MPNST models, combined MET and MEK inhibition was the most effective treatment for decreasing ERK activity. The fact that NF1-P53 tumors have a lower basal ERK activity and a variable response to MEK inhibition suggests that P53-deficient tumors may not be exclusively dependent on RAS-ERK signaling. This idea is supported by high basal AKT activity in NF1-P53 tumors that was sustained our rapidly increased upon MET or MEK inhibition. Given that we observed increase AKT activation in response to trametinib in all of our MPNST tumor models, PI3K/AKT pathway may be a robust compensation mechanism in RAS-deregulated MPNSTs. As with ERK, combined MET and MEK inhibition was the most effective treatment for decreasing compensatory activation of AKT. Taken together, these data confirm that distinct but convergent compensation mechanisms drive resistance to capmatinib and trametinib therapy.
In conclusion, this work addresses important gaps in knowledge regarding the sufficiency of MET activation for malignant transformation of NF1-deficient peripheral nerve cells into MPNSTs, and the important role of MET activation in reinforcing RAS/ERK and PI3K/AKT signaling (56). These data represent the first demonstration of capmatinib treatment response in NF1-related MPNST preclinical models and support the concept that MET inhibition is a viable treatment option for the subset of MPNSTs bearing a MET-addicted signature. Currently, it is difficult to define the size or makeup of the “MET-addicted” clinical subset in the neurofibromatosis population due to the lack of suitable biomarkers, however MET Y1234/35 phosphorylation has been proposed as a candidate immunohistochemical marker to define MET-activated MPNSTs, and predict MET inhibitor response (13). More recent data demonstrates that by combining MET phosphospecies with baseline MET receptor expression levels, better accuracy can be achieved. It is important to note that pY1234/1235 antibodies did accurately predict response to combination therapy in our models (57,58). Our data also indicates that P53-deficient MPNSTs may have a unique signaling pattern that relies on MET and AKT signaling. Importantly, combined MET and MEK inhibitors were able to abrogate RAS/ERK and PI3K/AKT-based kinome adaptations that were observed with monotherapy. Based on the data presented, it is possible that MEK targeted therapy strategies can inadvertently activate MET resulting in broader TKI resistance. Overall, these results highlight potential therapeutic strategies treating genomically diverse MPNSTs with single agent tyrosine kinase inhibition strategies, as well as highlight the diverse mechanisms that promote MPNST progression and therapy resistance.
Supplementary Material
Acknowledgements
We would like to thank the MSU RTSF Genomics Core for genomic analysis of the MPNST models, Patrick Dischinger for technical assistance with this study, Amanda Schuiling for assistance with statistical analysis, and Anderson Peck from the VARI Small Animal Imaging Facility for his imaging support. We would like to thank Bryn Eagleson and the VARI Vivarium for their continuous dedication.
Grant Support
Funding for this research was made possible by the Neurofibromatosis Therapeutic Acceleration Program (NTAP), The Johns Hopkins University (JHU), and the Van Andel Institute. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NTAP and JHU.
Footnotes
Conflict of interest
The authors have declared that no conflict of interest exists.
References
- 1.Evans DG, Howard E, Giblin C, Clancy T, Spencer H, Huson SM, et al. Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am J Med Genet A 2010;152A(2):327–32. [DOI] [PubMed] [Google Scholar]
- 2.Porter DE, Prasad V, Foster L, Dall GF, Birch R, Grimer RJ. Survival in Malignant Peripheral Nerve Sheath Tumours: A Comparison between Sporadic and Neurofibromatosis Type 1-Associated Tumours. Sarcoma 2009;2009:756395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson KL, Al Sannaa GA, Kivlin CM, Ingram DR, Landers SM, Roland CL, et al. Patterns of recurrence and survival in sporadic, neurofibromatosis Type 1-associated, and radiation-associated malignant peripheral nerve sheath tumors. J Neurosurg 2017;126(1):319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peacock JD, Cherba D, Kampfschulte K, Smith MK, Monks NR, Webb CP, et al. Molecular-guided therapy predictions reveal drug resistance phenotypes and treatment alternatives in malignant peripheral nerve sheath tumors. J Transl Med 2013;11:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel AV, Eaves D, Jessen WJ, Rizvi TA, Ecsedy JA, Qian MG, et al. Ras-driven transcriptome analysis identifies aurora kinase A as a potential malignant peripheral nerve sheath tumor therapeutic target. Clin Cancer Res 2012;18(18):5020–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joseph NM, Mosher JT, Buchstaller J, Snider P, McKeever PE, Lim M, et al. The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer Cell 2008;13(2):129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le LQ, Parada LF. Tumor microenvironment and neurofibromatosis type I: connecting the GAPs. Oncogene 2007;26(32):4609–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varin J, Poulain L, Hivelin M, Nusbaum P, Hubas A, Laurendeau I, et al. Dual mTORC1/2 inhibition induces anti-proliferative effect in NF1-associated plexiform neurofibroma and malignant peripheral nerve sheath tumor cells. Oncotarget 2016;7(24):35753–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masliah-Planchon J, Pasmant E, Luscan A, Laurendeau I, Ortonne N, Hivelin M, et al. MicroRNAome profiling in benign and malignant neurofibromatosis type 1-associated nerve sheath tumors: evidences of PTEN pathway alterations in early NF1 tumorigenesis. BMC Genomics 2013;14:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer 2012;12(2):89–103. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda T, Ichimura E, Shinozaki T, Sano T, Kashiwabara K, Oyama T, et al. Coexpression of HGF and c-Met/HGF receptor in human bone and soft tissue tumors. Pathol Int 1998;48(10):757–62. [DOI] [PubMed] [Google Scholar]
- 12.Mantripragada KK, Spurlock G, Kluwe L, Chuzhanova N, Ferner RE, Frayling IM, et al. High-resolution DNA copy number profiling of malignant peripheral nerve sheath tumors using targeted microarray-based comparative genomic hybridization. Clin Cancer Res 2008;14(4):1015–24. [DOI] [PubMed] [Google Scholar]
- 13.Torres KE, Zhu QS, Bill K, Lopez G, Ghadimi MP, Xie X, et al. Activated MET is a molecular prognosticator and potential therapeutic target for malignant peripheral nerve sheath tumors. Clin Cancer Res 2011;17(12):3943–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour microenvironment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012;487(7408):500–04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oddo D, Siravegna G, Gloghini A, Vernieri C, Mussolin B, Morano F, et al. Emergence of MET hyper-amplification at progression to MET and BRAF inhibition in colorectal cancer. Br J Cancer 2017;117(3):347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Upadhyaya M, Spurlock G, Majounie E, Griffiths S, Forrester N, Baser M, et al. The heterogeneous nature of germline mutations in NF1 patients with malignant peripheral serve sheath tumours (MPNSTs). Human Mutation 2006;27(7):716–16. [DOI] [PubMed] [Google Scholar]
- 17.Miller SJ, Rangwala F, Williams J, Ackerman P, Kong S, Jegga AG, et al. Large-Scale Molecular Comparison of Human Schwann Cells to Malignant Peripheral Nerve Sheath Tumor Cell Lines and Tissues. Cancer Research 2006;66(5):2584–91. [DOI] [PubMed] [Google Scholar]
- 18.Mayes DA, Rizvi TA, Cancelas JA, Kolasinski NT, Ciraolo GM, Stemmer-Rachamimov AO, et al. Perinatal or adult Nf1 inactivation using tamoxifen-inducible PlpCre each cause neurofibroma formation. Cancer Res 2011;71(13):4675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le LQ, Liu C, Shipman T, Chen Z, Suter U, Parada LF. Susceptible stages in Schwann cells for NF1-associated plexiform neurofibroma development. Cancer Res 2011;71(13):4686–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cichowski K, Shih TS, Schmitt E, Santiago S, Reilly K, McLaughlin ME, et al. Mouse models of tumor development in neurofibromatosis type 1. Science 1999;286(5447):2172–6. [DOI] [PubMed] [Google Scholar]
- 21.Dodd RD, Mito JK, Eward WC, Chitalia R, Sachdeva M, Ma Y, et al. NF1 deletion generates multiple subtypes of soft-tissue sarcoma that respond to MEK inhibition. Mol Cancer Ther 2013;12(9):1906–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43(5):491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11 10 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31(3):213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ha G, Roth A, Khattra J, Ho J, Yap D, Prentice LM, et al. TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res 2014;24(11):1881–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller SJ, Rangwala F, Williams J, Ackerman P, Kong S, Jegga AG, et al. Large-scale molecular comparison of human schwann cells to malignant peripheral nerve sheath tumor cell lines and tissues. Cancer Res 2006;66(5):2584–91. [DOI] [PubMed] [Google Scholar]
- 28.Fan Y, Richelme S, Avazeri E, Audebert S, Helmbacher F, Dono R, et al. Tissue-Specific Gain of RTK Signalling Uncovers Selective Cell Vulnerability during Embryogenesis. PLoS Genet 2015;11(9):e1005533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Genestine M, Caricati E, Fico A, Richelme S, Hassani H, Sunyach C, et al. Enhanced neuronal Met signalling levels in ALS mice delay disease onset. Cell Death Dis 2011;2:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 1995;23(24):5080–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004;119(6):847–60. [DOI] [PubMed] [Google Scholar]
- 32.Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science 1999;286(5447):2176–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leone DP, Genoud S, Atanasoski S, Grausenburger R, Berger P, Metzger D, et al. Tamoxifen-inducible glia-specific Cre mice for somatic mutagenesis in oligodendrocytes and Schwann cells. Mol Cell Neurosci 2003;22(4):430–40. [DOI] [PubMed] [Google Scholar]
- 34.Stemmer-Rachamimov AO, Louis DN, Nielsen GP, Antonescu CR, Borowsky AD, Bronson RT, et al. Comparative pathology of nerve sheath tumors in mouse models and humans. Cancer Res 2004;64(10):3718–24. [DOI] [PubMed] [Google Scholar]
- 35.Mantripragada KK, Spurlock G, Kluwe L, Chuzhanova N, Ferner RE, Frayling IM, et al. High-Resolution DNA Copy Number Profiling of Malignant Peripheral Nerve Sheath Tumors Using Targeted Microarray-Based Comparative Genomic Hybridization. Clinical Cancer Research 2008;14(4):1015–24. [DOI] [PubMed] [Google Scholar]
- 36.Ma B, Bang YJ, Lim WT, Nam DH, Su WC, Schellens JHM, et al. P1.04Phase I dose escalation and expansion study to evaluate safety and efficacy of INC280 in patients with advanced MET-dependent solid tumors. Annals of Oncology 2015;26(suppl_2):ii17–ii17. [Google Scholar]
- 37.Bang Y-J, Su W-C, Nam D-H, Lim W-T, Bauer TM, Brana I, et al. Phase I study of the safety and efficacy of INC280 in patients with advanced MET-dependent solid tumors. Journal of Clinical Oncology 2014;32(15_suppl):2520–20. [Google Scholar]
- 38.Linklater ES, Tovar EA, Essenburg CJ, Turner L, Madaj Z, Winn ME, et al. Targeting MET and EGFR crosstalk signaling in triple-negative breast cancers. Oncotarget 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 2012;487(7408):505–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med 2013;19(11):1389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dombi E, Baldwin A, Marcus LJ, Fisher MJ, Weiss B, Kim A, et al. Activity of Selumetinib in Neurofibromatosis Type 1-Related Plexiform Neurofibromas. N Engl J Med 2016;375(26):2550–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res 2011;17(5):989–1000. [DOI] [PubMed] [Google Scholar]
- 43.Spurlock G, Knight SJ, Thomas N, Kiehl TR, Guha A, Upadhyaya M. Molecular evolution of a neurofibroma to malignant peripheral nerve sheath tumor (MPNST) in an NF1 patient: correlation between histopathological, clinical and molecular findings. J Cancer Res Clin Oncol 2010;136(12):1869–80. [DOI] [PubMed] [Google Scholar]
- 44.Castellsague J, Gel B, Fernandez-Rodriguez J, Llatjos R, Blanco I, Benavente Y, et al. Comprehensive establishment and characterization of orthoxenograft mouse models of malignant peripheral nerve sheath tumors for personalized medicine. EMBO Mol Med 2015;7(5):608–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahrmann EP, Watson AL, Keng VW, Choi K, Moriarity BS, Beckmann DA, et al. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat Genet 2013;45(7):756–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirbe AC, Dahiya S, Miller CA, Li T, Fulton RS, Zhang X, et al. Whole Exome Sequencing Reveals the Order of Genetic Changes during Malignant Transformation and Metastasis in a Single Patient with NF1-plexiform Neurofibroma. Clin Cancer Res 2015;21(18):4201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brekke HR, Kolberg M, Skotheim RI, Hall KS, Bjerkehagen B, Risberg B, et al. Identification of p53 as a strong predictor of survival for patients with malignant peripheral nerve sheath tumors. Neuro Oncol 2009;11(5):514–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rahrmann EP, Moriarity BS, Otto GM, Watson AL, Choi K, Collins MH, et al. Trp53 haploinsufficiency modifies EGFR-driven peripheral nerve sheath tumorigenesis. Am J Pathol 2014;184(7):2082–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cepero V, Sierra JR, Corso S, Ghiso E, Casorzo L, Perera T, et al. MET and KRAS gene amplification mediates acquired resistance to MET tyrosine kinase inhibitors. Cancer Res 2010;70(19):7580–90. [DOI] [PubMed] [Google Scholar]
- 50.Kwak EL, Ahronian LG, Siravegna G, Mussolin B, Godfrey JT, Clark JW, et al. Molecular Heterogeneity and Receptor Coamplification Drive Resistance to Targeted Therapy in MET-Amplified Esophagogastric Cancer. Cancer Discov 2015;5(12):1271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujita-Sato S, Galeas J, Truitt M, Pitt C, Urisman A, Bandyopadhyay S, et al. Enhanced MET Translation and Signaling Sustains K-Ras-Driven Proliferation under Anchorage-Independent Growth Conditions. Cancer Res 2015;75(14):2851–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen CT, Kim H, Liska D, Gao S, Christensen JG, Weiser MR. MET activation mediates resistance to lapatinib inhibition of HER2-amplified gastric cancer cells. Mol Cancer Ther 2012;11(3):660–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010;17(1):77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albritton KH, Rankin C, Coffin CM, Ratner N, Budd GT, Schuetze SM, et al. Phase II study of erlotinib in metastatic or unresectable malignant peripheral nerve sheath tumors (MPNST). Journal of Clinical Oncology 2006;24(18_suppl):9518–18. [Google Scholar]
- 55.Wagner JP, Wolf-Yadlin A, Sevecka M, Grenier JK, Root DE, Lauffenburger DA, et al. Receptor tyrosine kinases fall into distinct classes based on their inferred signaling networks. Sci Signal 2013;6(284):ra58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peacock JD, Pridgeon MG, Tovar EA, Essenburg CJ, Bowman M, Madaj Z, et al. Genomic MET amplification occurs early in NF1-related malignant peripheral nerve sheath tumor (MPNST) progression and is a potent therapeutic target. submitted 2017. [Google Scholar]
- 57.Huang F, Ma Z, Pollan S, Yuan X, Swartwood S, Gertych A, et al. Quantitative imaging for development of companion diagnostics to drugs targeting HGF/MET. J Pathol Clin Res 2016;2(4):210–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srivastava AK, Hollingshead MG, Weiner J, Navas T, Evrard YA, Khin SA, et al. Pharmacodynamic Response of the MET/HGF Receptor to Small-Molecule Tyrosine Kinase Inhibitors Examined with Validated, Fit-for-Clinic Immunoassays. Clin Cancer Res 2016;22(14):3683–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.