Abstract
Background
In systemic lupus erythematosus (SLE), immune complexes (ICs) containing self-derived nucleic acids trigger the synthesis of proinflammatory cytokines by immune cells. We asked how an interleukin (IL)-1 receptor-associated kinase 4 small molecule inhibitor (IRAK4i) affects RNA-IC-induced cytokine production compared with hydroxychloroquine (HCQ).
Methods
Plasmacytoid dendritic cells (pDCs) and natural killer (NK) cells were isolated from peripheral blood mononuclear cells (PBMCs) of healthy individuals. PBMCs from SLE patients and healthy individuals were depleted of monocytes. Cells were stimulated with RNA-containing IC (RNA-IC) in the presence or absence of IRAK4i I92 or HCQ, and cytokines were measured by immunoassay or flow cytometry. Transcriptome sequencing was performed on RNA-IC-stimulated pDCs from healthy individuals to assess the effect of IRAK4i and HCQ.
Results
In healthy individuals, RNA-IC induced interferon (IFN)-α, tumor necrosis factor (TNF)-α, IL-6, IL-8, IFN-γ, macrophage inflammatory protein (MIP)1-α, and MIP1-β production in pDC and NK cell cocultures. IFN-α production was selective for pDCs, whereas both pDCs and NK cells produced TNF-α. IRAK4i reduced the pDC and NK cell-derived cytokine production by 74–95%. HCQ interfered with cytokine production in pDCs but not in NK cells. In monocyte-depleted PBMCs, IRAK4i blocked cytokine production more efficiently than HCQ. Following RNA-IC activation of pDCs, 975 differentially expressed genes were observed (false discovery rate (FDR) < 0.05), with many connected to cytokine pathways, cell regulation, and apoptosis. IRAK4i altered the expression of a larger number of RNA-IC-induced genes than did HCQ (492 versus 65 genes).
Conclusions
The IRAK4i I92 exhibits a broader inhibitory effect than HCQ on proinflammatory pathways triggered by RNA-IC, suggesting IRAK4 inhibition as a therapeutic option in SLE.
Electronic supplementary material
The online version of this article (10.1186/s13075-018-1702-0) contains supplementary material, which is available to authorized users.
Keywords: SLE, pDC, NK, HCQ, IRAK4
Background
Systemic lupus erythematosus (SLE) is characterized by circulating immune complexes (ICs), an activation of the type I interferon (IFN) system, and production of proinflammatory cytokines and chemokines which cause an autoimmune reaction with organ inflammation [1]. The cellular and molecular mechanisms behind the ongoing inflammatory process in SLE have been partially clarified, and a number of different disease-associated pathways identified [2]. One important event is the induction of type I IFN production by plasmacytoid dendritic cells (pDCs) in response to ICs consisting of autoantibodies and apoptotic or necrotic cell-derived nucleic acids [3]. Such interferogenic ICs are internalized in pDCs via fragment crystallizable receptor IIA (FcγRIIA) and directed to the endosomes, where RNA and DNA interact with Toll-like receptor (TLR)7 and 9, respectively [4]. Activation of TLR7/9 triggers a signaling cascade, involving myeloid differentiation primary response protein 88 (MyD88), interleukin (IL)-1 receptor-associated kinase (IRAK)1, and IRAK4, that eventually leads to transcription of type I IFN genes. In addition to type I IFN production, MyD88 and IRAK4 signaling triggers the production of other proinflammatory cytokines, such as tumor necrosis factor (TNF)-α and IL-6, via activation of nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) or IFN regulatory factor (IRF) 5 [5]. Besides the pDCs, interferogenic ICs will also activate several other immune cells, such as natural killer (NK) cells, which contribute to enhanced cytokine production [6]. The final outcome in SLE is a complex inflammatory response that is difficult to bring into complete remission.
Current therapies in SLE aim to downregulate the autoimmune reaction. Treatment with antimalarials, such as hydroxychloroquine (HCQ), is considered the standard of care [7, 8]. The presumed central mechanism of action of HCQ is a reduction in the IFN-α production by inhibition of endosomal TLR signaling [9]. Studies have also shown that SLE patients treated with HCQ have a decreased type I IFN production after stimulation of pDCs with TLR ligands [10]. However, despite continuous HCQ treatment, few patients with SLE experience complete remission and flares still occur. A possible reason could be the limited number of disease-associated pathways affected by HCQ. Consequently, targeting a broader repertoire of inflammatory cytokines in SLE, yet avoiding severe infections, is needed. A potential therapeutic target in SLE is IRAK4 due to its essential role in MyD88 signaling [11]. IRAK4-deficient children are susceptible to life-threatening pyogenic infections that are reported to cease in adolescence, making IRAK4 inhibition an attractive therapeutic possibility [12].
In this study, we compared the effect of HCQ and the IRAK4 inhibitor (IRAK4i) I92 on the RNA-IC-induced cytokine production by pDCs and NK cells from healthy individuals and monocyte-depleted peripheral blood mononuclear cells (PBMCs) from SLE patients and healthy controls. Gene expression profiles of RNA-IC-stimulated pDCs treated with IRAK4i or HCQ were compared with nontreated cells to clarify the inflammatory response modulated by the drugs.
Methods
Patients and controls
All SLE patients (n = 15) fulfilled ≥ 4 of the American College of Rheumatology criteria for SLE [13]. Patients were a median 52 (range 32–81) years old with a disease duration of 15 (1–46) years (Additional file 1). Healthy age- and gender-matched controls (n = 12) were 53 (32–68) years old. The local ethics committee at Uppsala University approved the study and informed consent was obtained from all patients and controls.
Cell isolation and culture conditions
PBMCs were prepared from healthy donor buffy coats by Ficoll density-gradient centrifugation. pDCs (25 × 103/well) and NK cells (50 × 103/well) were isolated and cultivated as previously described [6, 14]. Cell purity was > 95% for pDC (blood dendritic cell antigen (BDCA)2+) and NK cells (cluster of differentiation (CD)56+) as determined by flow cytometry. Cell viability as measured by flow cytometry after 20 h was approximately 90%.
Interferon inducers
Necrotic material from U937 cell line and U1snRNP particles were prepared as previously described [3, 15]. Immunoglobulin (Ig)G was isolated from two Smith nuclear antigen (Sm) and ribonucleoprotein (RNP) antibody-containing SLE patient sera [14]. U1snRNP particles and SLE IgG were used at final concentrations of 2.5 μg/ml and 1 mg/ml, respectively.
Drugs
The small molecule drug IRAK4i I92 (ND-2158, Nimbus Discovery) [16] and HCQ (Sigma-Aldrich) were pretitrated and used at final concentrations of 10 μM and 5.8 μM (pDCs and NK cells) or 7.8 μM (monocyte-depleted PBMCs) (Additional files 2, 3 and 4). The cells were preincubated with I92 or HCQ for 30 min at 5% CO2 and 37 °C before adding IFN inducers.
Flow cytometry
pDCs and NK cells were cultivated for 5 or 9 h, with the final 4 h with brefeldin A. After gating live cells, singlets, and lymphocytes (Additional file 5), the cells were identified with anti-CD56-phycoerythrin (PE)Cy7 (NCAM 16.2, BD Biosciences) and BDCA-2-fluorescein isothiocyanate (FITC) (AC144, Miltenyi Biotech) monoclonal antibodies (mAbs). Intracellular cytokines were detected with anti-TNF-α-allophycocyanin (APC) (Mab11, BD Pharmingen) and anti-IFN-α-PE mAbs (LT27:295, Miltenyi Biotech). Isotype-matched irrelevant mAbs were used as controls. Live/dead Fixable Near-InfraRed Dead Cell Stain (Invitrogen) was used to distinguish live cells. Data were acquired with a FACS CantoII instrument and analyzed with Diva 6.1.3 software (BD Biosciences).
Immunoassays
TNF-α, IL-6, IL-8, IFN-γ, macrophage inflammatory protein (MIP)1-α, and MIP1-β were measured after 20 h by multiplex immunoassays (Milliplex Human Cytokine/Chemokine (Millipore) or Luminex Screening Assay (R&D systems)). Lower limits of quantification (LLoQ) of cytokines were: TNF-α, 3.8 pg/ml; IL-6, 5.2 pg/ml; IL-8, 1.4 pg/ml; IFN-γ, 1.6 pg/ml; MIP1-α, 54.8; and MIP1-β, 162.2 pg/ml. IFN-α was measured by dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA; LLoQ, 2 U/ml) [17].
RNA sequencing
pDCs from four healthy individuals were stimulated with RNA-IC for 6 h. RNA (RNA integrity number (RIN) ≥ 8) was extracted by the RNeasy 96 plus kit (Qiagen). RNA libraries were prepared with the TruSeq Stranded mRNA kit and sequenced by NextSeq500 (Illumina). RNAseq fastq files were processed using bcbio-nextgen (v.0.9.7) and mapped to the human genome GRCh38.79 [18]. Gene-level quantifications were generated with featureCounts software (v.1.4.4) [19] and Sailfish (version 0.9.0) [20]. Pheatmap and ggplot2 (v.2.2.1, http://ggplot2.org/) were used for visualizations [21].
Statistical analysis
Statistical analysis was performed using GraphPad Prism software 7.0. Differences were analyzed with Friedman’s test, and p values ≤ 0.05 were considered significant. For transcriptome analysis, a false discovery rate (FDR) < 0.05 was considered significant. Analyses were performed using R (version 3.3.3). Differential gene expression was assessed with DESeq2 (v.1.14.1) [22] using raw counts as input. Pathway enrichments were obtained from Pathway Studio® (Elsevier). A one-sided Mann-Whitney U test was performed to calculate the significance of the differences in distribution between the background (from the differential gene expression analysis) and the gene subnetworks (upstream regulators) or the gene sets (pathways).
Results
RNA-containing ICs induce TNF-α production more rapidly in NK cells than in pDCs
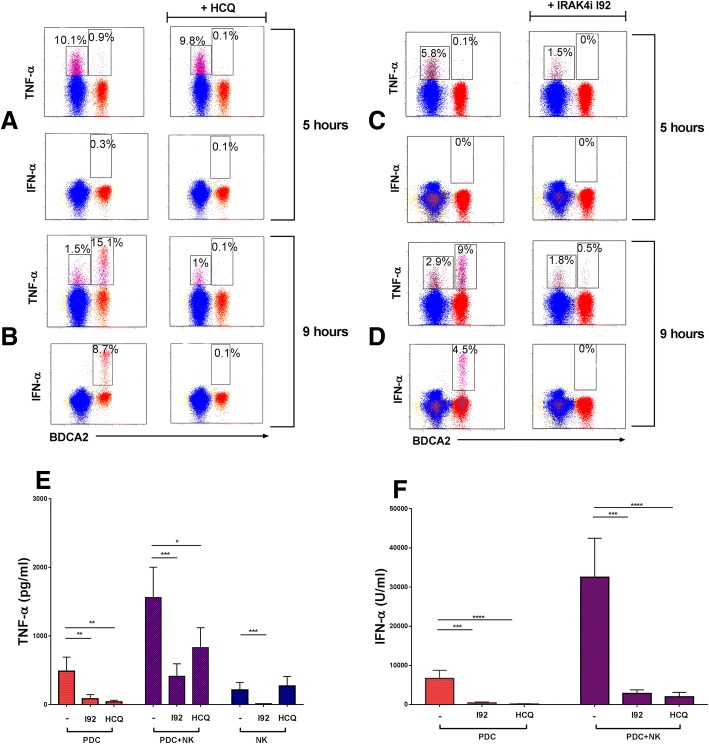
TNF-α and IFN-α are important drivers of inflammation in SLE and large amounts are produced in RNA-IC-stimulated cocultures of pDCs and NK cells [6]. However, the cellular source and quantity produced by each cell type have not been determined. Therefore, we initially analyzed the frequency of TNF-α- and IFN-α-producing pDCs and NK cells in cocultures at 5 and 9 h, due to expected differences regarding peak cytokine production by the different cell types. A minority (< 20%) of both pDCs and NK cells produced TNF-α in response to RNA-IC (Fig. 1a, b; left panels). Furthermore, the TNF-α production was prominent in NK cells at 5 h, but occurred later in pDCs when the NK cell response had decreased.
Fig. 1.
Regulatory effect of hydroxychloroquine (HCQ) and the interleukin-1 receptor associated kinase 4 inhibitor (IRAK4i) I92 on tumor necrosis factor (TNF)-α and interferon (IFN)-α production. a–d Cocultures of plasmacytoid dendritic cells (pDC) and natural killer (NK) cells from healthy donors were stimulated with RNA-containing immune complexes (RNA-IC) for 5 h (a,b) or 9 h (c,d) in the absence or presence of HCQ or I92. The frequencies of TNF-α- and IFN-α-producing NK cells (blue) and pDCs (red) were determined by flow cytometry. The dot plots represent one representative individual donor from two (HCQ) and four (IRAK4i) donors analyzed. e,f pDCs or NK cells were cultivated separately or in coculture in the presence of RNA-IC with or without I92 or HCQ. Cytokine levels were measured by immunoassays after 20 h. No IFN-α is produced by NK cells (data not shown). No cytokines were detected in the cell cultures in the absence of RNA-IC. Bars represent the mean with standard error of the mean (SEM) of nine donors from at least three independent experiments. Friedman’s test, uncorrected Dunn’s test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. BDCA blood dendritic cell antigen
Only pDCs produced IFN-α in response to RNA-IC (Fig. 1a, b; left panels), and synthesis of both TNF-α and IFN-α was most prominent at 9 h (Fig. 1b). Nearly all IFN-α-producing cells expressed TNF-α, whereas a fraction of the pDCs produced TNF-α only (Fig. 1b, left panel; Additional file 6). Almost no IFN-α-containing pDCs were detected at 5 h.
Therefore, RNA-containing ICs trigger the production of TNF-α in a fraction of pDCs and NK cells at different time points.
IRAK4i I92 inhibits TNF-α production by activated pDCs and NK cells, while HCQ affects TNF-α production by pDCs only
Next, we asked if HCQ inhibits the RNA-IC-induced TNF-α production in pDC/NK cell cocultures. As shown in Fig. 1a, b (right panels), HCQ completely blocked TNF-α production in pDCs but not in NK cells at 9 h. At 20 h, HCQ significantly reduced TNF-α production in cultures of pDCs and pDC/NK cells, but not in NK cells (Fig. 1e). In contrast, IFN-α production was completely blocked by HCQ. Subsequently, we investigated if IRAK4i I92 could inhibit the cytokine response in the cell cultures. As shown in Fig. 1c, d (right panels), intracellular IFN-α and TNF-α production by pDCs was effectively blocked by I92. The drug reduced the early (5-h) TNF-α response by approximately 75% in NK cells, whereas the inhibitory effect at a later time point (9 h) was less prominent. At 20 h, I92 reduced the TNF-α production by 70–95% in all cell cultures (Fig. 1e). In addition, the IFN-α levels in RNA-IC-stimulated pDC and pDC-NK cell cocultures were reduced by > 90% (Fig. 1f). No IFN-α was produced by NK cells. Thus, HCQ blocked TNF-α and IFN-α production by pDCs, whereas I92 reduced TNF-α production in both pDCs and NK cells, as well as the IFN-α production by pDCs.
IRAK4 inhibition reduces proinflammatory cytokine production by activated pDCs and NK cells more extensively than HCQ
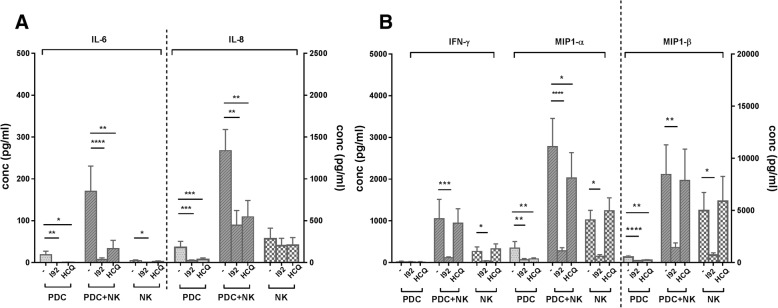
To investigate whether IRAK4 inhibition affects production of other proinflammatory cytokines, levels of IL-6, IL-8, IFN-γ, MIP1-α, and MIP1-β were measured in RNA-IC-stimulated cultures with pDCs, NK cells, or pDC/NK cell cocultures in the presence or absence of I92 or HCQ (Fig. 2). In pDC cultures and pDC/NK cocultures, both I92 and HCQ blocked IL-6, IL-8, and MIP1-α, whereas I92 also blocked IFN-γ and MIP1-β production. In NK cell cultures, I92 significantly reduced IL-6, IFN-γ, MIP1-α, and MIP1-β levels, whereas IL-8 was unaffected. HCQ did not inhibit cytokine production by NK cells. Hence, I92 showed a broader inhibitory effect than HCQ on proinflammatory cytokines produced by RNA-IC-stimulated pDCs and NK cells.
Fig. 2.
The interleukin-1 receptor associated kinase 4 inhibitor (IRAK4i) I92 displays a more prominent inhibitory effect than hydroxychloroquine (HCQ) on the production of proinflammatory cytokines in plasmacytoid dendritic cells (pDC) and natural killer (NK) cell cocultures and in NK cells alone. pDCs and NK cells from healthy donors were cultivated separately or in coculture in the presence of RNA-containing immune complexes (RNA-IC), with or without I92 or HCQ. The levels of a interleukin (IL)-6 and IL-8, and b interferon (IFN)-γ, macrophage inflammatory protein (MIP)1-α, and MIP1-β were measured by immunoassays after 20 h. Bars represent the mean with standard error of the mean (SEM) of nine donors from at least three independent experiments. Friedman’s test, uncorrected Dunn’s test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
IRAK4 inhibition blocks production of proinflammatory cytokines by PBMCs from SLE patients
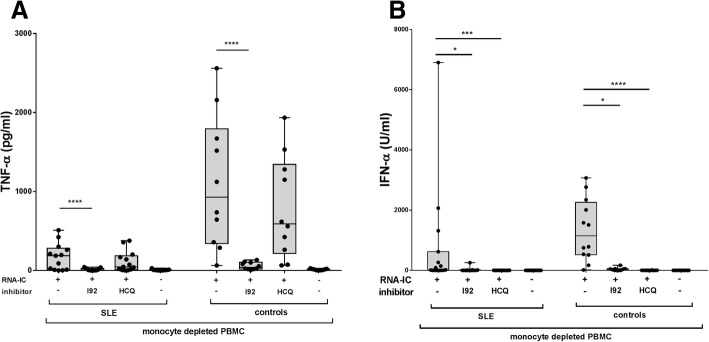
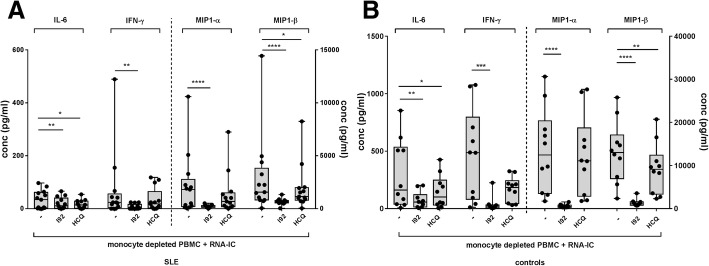
As healthy individuals and SLE patients may respond differently, we investigated if cytokine production in cells from patients with SLE also could be targeted with an IRAK4i. PBMCs from patients with SLE or healthy controls were stimulated after depletion of monocytes, due to their suppressive effect on the IFN-α response [14]. I92 inhibited TNF-α and IFN-α production by 80% and 97%, respectively, whereas HCQ interfered significantly only with IFN-α production (99%) (Fig. 3a, b). Furthermore, IL-6, IFN-γ, MIP1-α, and MIP1-β production was inhibited by I92, whereas only IL-6 and MIP1-β were significantly inhibited by HCQ (Fig. 4a, b). HCQ and I92 displayed the same inhibitory profile in monocyte-depleted PBMCs from healthy individuals. In summary, I92 reduced cytokine production in SLE patients and demonstrated a more extensive inhibitory profile on proinflammatory cytokines than did HCQ.
Fig. 3.
The interleukin-1 receptor associated kinase 4 inhibitor (IRAK4i) I92 reduces both a tumor necrosis factor (TNF)-α and b interferon (IFN)-α production by monocyte-depleted peripheral blood mononuclear cells (PBMCs) from systemic lupus erythematosus (SLE) patients and healthy controls. The cells were stimulated with RNA-containing immune complexes (RNA-IC) in the presence or absence of I92 or hydroxychloroquine (HCQ), or were mock stimulated. TNF-α and IFN-α production in cell cultures was measured after 20 h by immunoassays. Box plots show median with interquartile range, based on 10–15 donors, in at least 10 independent experiments. Friedman’s test, uncorrected Dunn’s test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Fig. 4.
The interleukin-1 receptor associated kinase 4 inhibitor (IRAK4i) I92 exhibits a broader inhibitory effect than hydroxychloroquine (HCQ) on cytokine production by monocyte-depleted peripheral blood mononuclear cells (PBMCs) from a systemic lupus erythematosus (SLE) patients and b healthy controls. Monocyte-depleted PBMCs were stimulated with RNA-containing immune complexes (RNA-IC) in the presence or absence of I92 or HCQ. Levels of interleukin (IL)-6, interferon (IFN)-γ, macrophage inflammatory protein (MIP)1-α, and MIP1-β in the cell cultures were measured after 20 h by immunoassays. Box plots show median with interquartile range, based on 10–13 donors from 10 independent experiments. Friedman’s test, uncorrected Dunn’s test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Gene expression changes in RNA-IC-stimulated pDCs are reversed by both IRAK4 inhibition and HCQ
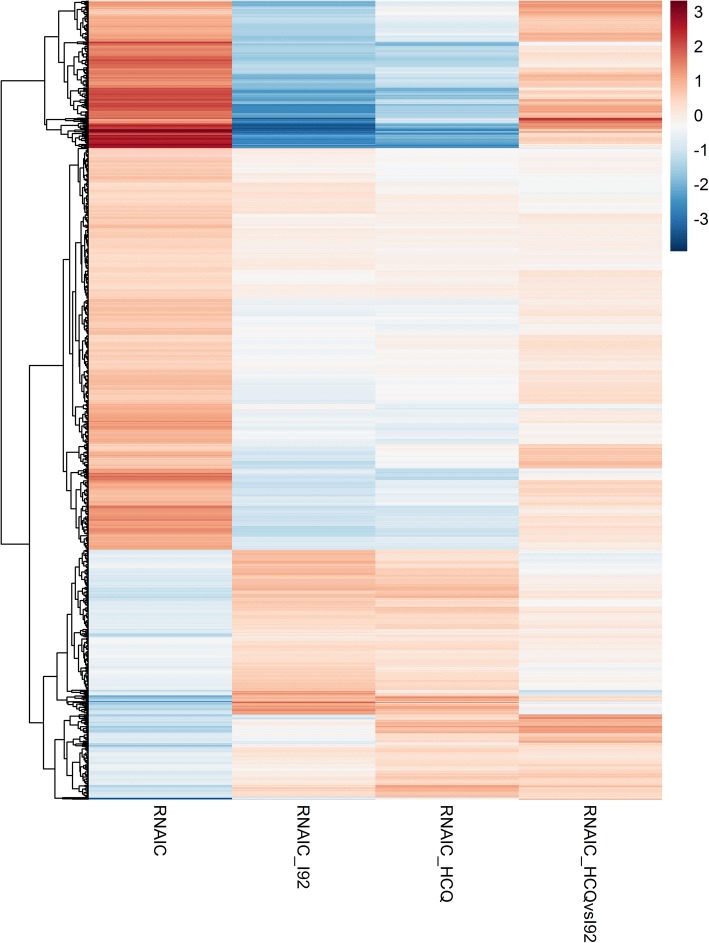
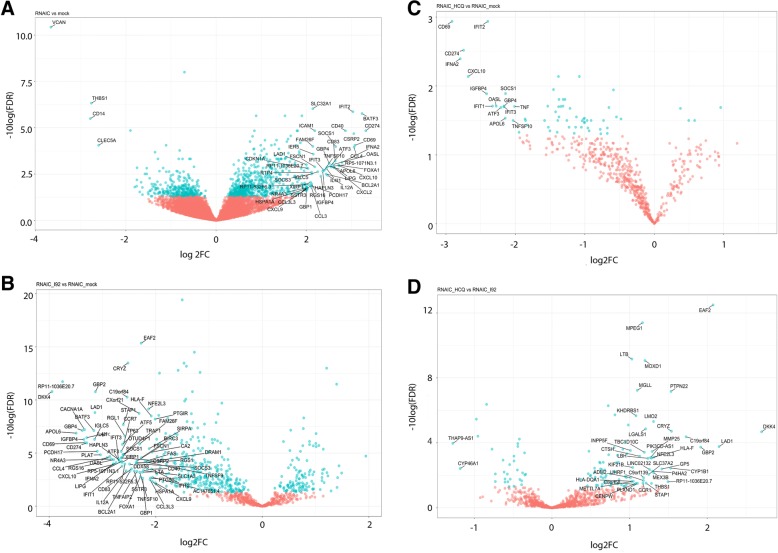
To clarify the pathways affected by I92 and HCQ, we performed transcriptome sequencing (RNA-seq) of RNA-IC-stimulated pDCs. An RNA-IC activation signature was identified consisting of 975 differentially expressed genes (DEGs) compared with unstimulated pDCs (RNA-IC-DEGs, FDR < 0.05; Fig. 5, Additional file 7). A majority of the responding genes showed an upregulation after RNA-IC stimulation (n = 670), with 48 genes increased at least fourfold (Fig. 6a, Additional file 7). Among the 48 top RNA-IC-DEGs were genes mapping to the type I IFN signaling pathway (IFNA2, IFIT1–3, GBP1, and OASL), clearance of apoptotic material (BCL2A1, CDKN1A, and TNFSF10), and chemokine genes (CXCL2, CXCL9, CXCL10, and CCL4). Several regulators of inflammation and activation (e.g., IRF1, IRF3, STAT1, STAT3, NFκB, RELA/B, and SP1) were predicted to drive the RNA-IC-DEGs (Additional file 8).
Fig. 5.
Differential effects on gene expression in RNA-containing immune complex (RNA-IC)-stimulated plasmacytoid dendritic cells (pDCs) by the interleukin-1 receptor associated kinase 4 inhibitor (IRAK4i) I92 and hydroxychloroquine (HCQ). RNA-sequencing analysis was performed on RNA-IC-stimulated pDCs from healthy donors (n = 4), cultivated for 6 h in the presence or absence of I92 or HCQ. The heat map showing the mean log2 fold change values for the 975 activation signature genes from RNA-IC versus mock-stimulated cells (left), RNA-IC I92 versus RNA-IC (middle left), RNA-IC HCQ versus RNA-IC (middle right), and the difference between the two compounds RNA-IC I92 versus RNA-IC HCQ treated cultures (right)
Fig. 6.
The effect of interleukin-1 receptor associated kinase 4 inhibitor (IRAK4i) I92 or hydroxychloroquine (HCQ) treatment on gene expression in plasmacytoid dendritic cells (pDCs) stimulated with RNA-containing immune complex (RNA-IC). RNA sequencing analysis was performed on RNA-IC-stimulated pDCs from healthy donors (n = 4), cultivated for 6 h in the presence or absence of I92 or HCQ. Volcano plots comparing false discovery rate (FDR) versus log2 fold change (FC) for genes from a RNA-IC-stimulated pDCs relative to mock-treated cells, b RNA-IC I92 relative to RNA-IC-treated cells, c RNA-IC HCQ relative to RNA-IC-treated cells, or d RNA-IC HCQ relative to RNA-IC I92-treated cells. Genes labeled in blue are statistically significantly different between the two treatments (FDR < 0.05)
I92 significantly altered the expression of almost 4000 genes (FDR < 0.05) in RNA-IC-activated pDCs, of which 492 overlapped with the RNA-IC-DEGs. In contrast, HCQ significantly altered only 73 genes, with 65 overlapping with the RNA-IC-DEGs (FDR < 0.05) (Fig. 5, Additional file 9). More RNA-IC-DEGs were strongly downregulated (log2 fold change > 2) by I92 (n = 73) than by HCQ (n = 15) (Fig. 6b, c). The expression of several top upregulated genes in the RNA-IC-DEGs was reversed by both I92 and HCQ, including IFNA2, IFIT2–3, OASL, CXCL10, CD274, TNFSF10, and APOL6.
Between HCQ- and I92-treated, RNA-IC-stimulated pDCs, 125 genes were differentially expressed. The greatest relative difference in expression was observed for DKK4, LAD1, and EAF2 (Fig. 6d), which were more strongly downregulated by I92 than by HCQ. These genes have mainly been studied in the context of tumorigenesis [23–25]. The top enriched biological function pathway for these genes was macroautophagy decline, represented by ATG14, AMBRA1, and BECN1, contributing to the regulation of autophagy, autophagosomal maturation, endocytosis, and apoptosis (Additional file 10). Several STAT-related signaling pathways were more suppressed by I92 than by HCQ (Additional file 11). The expression of cytokine genes TNF, IFNA2, CXCL8, CCL3, and CCL4 was significantly suppressed by I92, compared with only TNF and IFNA2 being suppressed by HCQ (Additional file 12). In conclusion, both I92 and HCQ reversed the effects of RNA-IC stimulation on the pDC gene expression profile, but I92 more extensively affected gene expression and modulated more cellular pathways than HCQ.
Discussion
This study demonstrates that the cytokine production by RNA-IC-stimulated pDCs and NK cells can be suppressed by HCQ and, more profoundly, by an IRAK4 inhibitor. The strong TNF-α induction by RNA-IC is interesting since TNF-α plays a critical role in several SLE disease manifestations, such as nephritis, skin lesions, and arthritis, all characterized by tissue deposition of ICs [26–28]. Increased IC formation precedes SLE flares and our findings may therefore partly explain the observed association between increased serum TNF-α levels and disease activity in SLE [29]. The difference in the TNF-α production rate between pDCs and NK cells indicates that RNA-ICs activate different induction pathways for TNF-α synthesis in these two cell types. Supporting this conjecture is the observed difference between pDCs and NK cells in response to HCQ. pDCs are mainly activated by ligation of endosomal TLRs [2] and this pathway is inhibited by HCQ [7]. TNF-α production by NK cells, on the other hand, can be induced by a number of different receptors, including TLR7 [30–32]. However, RNA-IC-induced production of cytokines and chemokines from NK cells was not dependent on endosomal TLR signaling since HCQ had no inhibitory effect. Consistent with a TLR7-independent activation of NK cells, heat-aggregated IgG was as efficient as RNA-IC in inducing TNF-α from purified NK cells, and a synthetic TLR7 agonist (DSR6434) did not induce TNF-α in NK cells (Additional file 13). Although no studies were performed regarding specific RNA-IC-responding NK cell receptors, the fact that TNF-α production was observed only in the CD56dim, CD16-expressing NK cell population (Additional file 14) suggests NK cell activation by RNA-IC via CD16. The prominent effect of the IRAK4 inhibitor I92 on the TNF-α production by NK cells implies that NFκB-mediated and/or mitogen-activated protein kinase activation was involved in the NK cell response [33]. However, we cannot exclude that other protein kinases were also affected by I92, despite the previously demonstrated high selectivity for IRAK4 by this drug [16].
When investigating the effect of HCQ and IRAK4 inhibitor I92 on other RNA-IC-induced cytokines, we noted that HCQ almost completely blocked the production of all investigated cytokines by pDCs. This was in stark contrast to the lack of effect on the cytokine response in NK cells. Conversely, HCQ markedly reduced the production of most cytokines in the pDC/NK cell cocultures. The reason for this is unclear, but an optimal cytokine production in cell cocultures depends on both cell types since RNA-IC-activated pDC and NK cells promote the function of each other [6]. Consequently, inhibition of the pDC function in pDC/NK cell cocultures will also reduce the NK cell cytokine-producing capacity. However, in pDC/NK cocultures the production of IFN-γ and MIP-1β was not affected by HCQ, suggesting a pDC-independent production by NK cells, although the exact cellular source of these cytokines was not investigated. Nevertheless, this observation indicates the need for a therapeutic agent with broader effects than HCQ to achieve better control of IC-driven inflammatory processes.
The IRAK4 inhibitor I92 blocked the NK cell production of all cytokines in healthy individuals, except for IL-8. This could imply yet another induction mechanism for IL-8 production in NK cells. In fact, IL-8 production was also remarkably high in monocyte-depleted PBMC cultures from SLE patients (Additional file 15) but, due to a shortage of patient material, the effects of I92 and HCQ on the IL-8 production could not be clarified. Studies have shown that patients with SLE have increased serum levels of IL-8 despite continuous standard treatment and being in remission [34]. An association between IL-8 gene polymorphisms and SLE further supports a role for IL-8 in SLE [35]. Additional studies are needed to determine the regulation of IL-8 production in patients with SLE, and some are now in progress. Notably, I92 inhibited all other investigated cytokines produced by RNA-IC-stimulated cells from SLE patients, whereas HCQ only reduced IL-6 and MIP1-β production significantly.
The RNA-IC activation signature in pDCs revealed an enrichment of pathways with connection to the IFN signaling system, antigen presentation, and apoptosis. This demonstrates that nucleic acid containing ICs elicit a powerful inflammatory response, but also trigger other cellular processes of importance in SLE. Both I92 and HCQ largely reversed the RNA-IC activation in pDCs, although some differences were observed. I92 increased the expression of genes involved in protein degradation and the autophagy process, in contrast to HCQ which downregulated these genes. The most strongly downregulated genes by I92 compared with HCQ were DKK4, LAD1, and EAF2, and suppression of these genes could have several effects on the SLE disease process. DKK4 is an inhibitor of the canonical Wnt signaling pathway, which has been suggested to contribute to disrupted T effector cell differentiation and the immune dysfunction in SLE [36, 37]. Ladinin 1, encoded by LAD1, modulates the EGF to ERK pathway and increased ERK activation is associated with organ damage in SLE [24, 38]. EAF2, on the other hand, is selectively upregulated in germinal center B cells and promotes their apoptosis [39]. Possibly, inhibition of EAF2 could therefore increase autoantibody production. The increased activation of the autophagy pathway by I92 might be beneficial in SLE since autophagy is reduced in SLE regulatory T cells and enhanced autophagy has been shown to improve both murine and human SLE [40]. On the other hand, activation of autophagy favors plasmablast development, enabling expansion of self-reactive B cells in SLE, as well as type I IFN production by facilitating intracellular IC transport [41, 42]. These observations merit further studies of the effects of I92 on different cell types, not least considering that IRAK4 inhibition ameliorates experimental murine lupus, suggesting a favorable effect also in human SLE [43]. Translating results of in-vitro studies of pharmaceutical compounds to potential drug effects in vivo has limitations. However, the approach to investigate drug candidates in cell cultures can be useful to determine the effects on central immune cells in the disease process [44]. Thus, we consider our system with IC-stimulated immune cells from SLE patients as one relevant model for an initial screening of potential drugs that target disease-associated pathways in SLE.
Conclusions
In conclusion, the IRAK4 inhibitor I92 reduced a number of proinflammatory cytokines triggered by RNA-IC that are involved in the immune pathogenesis of SLE equally, or more effectively, than HCQ. For the first time, we show the effects of an IRAK4 inhibitor on both transcription and protein synthesis in RNA-IC-activated pDCs, which demonstrates that IRAK4 inhibition affects many cellular pathways of importance in an autoimmune disease process.
Additional files
Table S1. Patient clinical characteristics. (PDF 36 kb)
Figure S1. Titration of IRAK4 inhibitor (I92) on cytokine production by cocultured plasmacytoid dendritic cells and NK cells. (PDF 194 kb)
Figure S2. Titration of hydroxychloroquine in cocultured plasmacytoid dendritic cells and NK cells with regard to interferon-α production. (PDF 126 kb)
Figure S3. Titration of hydroxychloroquine in cocultured plasmacytoid dendritic cells and NK cells. (PDF 221 kb)
Figure S4. Flow cytometric gating strategy of stimulated plasmacytoid dendritic cells and NK cells. (PDF 211 kb)
Figure S5. Flow cytometry showing total proportion of cytokine-producing cells in RNA-IC-stimulated pDC and NK cells. (PDF 282 kb)
Table S2. Gene list of 975 differentially expressed genes. (PDF 767 kb)
Table S3. Upstream regulators. (PDF 291 kb)
Figure S6. Overlap of differentially expressed genes in plasmacytoid dendritic cells. (PDF 135 kb)
Table S4. Enriched biological function pathways. (PDF 249 kb)
Table S5. Enriched signal processing pathways. (PDF 251 kb)
Figure S7. RNA-seq analysis of cytokine expression in plasmacytoid dendritic cells stimulated for 6 h in the presence of IRAK4 inhibitor or hydroxychloroquine. (PDF 186 kb)
Figure S8. TNF-α production in NK cell cultures and NK cell/pDC cocultures. (PDF 179 kb)
Figure S9. Flow cytometric analysis of TNF-α in NK cells. (PDF 165 kb)
Figure S10. Interleukin-8 production by stimulated blood cells from SLE patients. (PDF 104 kb)
Table S6. Gene expression in plasmacytoid dendritic cells (pDCs) from healthy donors. (XLSX 4030 kb)
Acknowledgements
We thank Lisbeth Fuxler for excellent technical assistance, Rezvan Kiani Dehkordi for collecting the patient blood samples, and Dr. Gert Weber, Ernst-Moritz-Arndt University of Greifswald, for kindly providing the U1snRNP particles.
Funding
The study was supported by The Swedish Rheumatism Association, King Gustaf V's 80-years Foundation, Agnes and Mac Rudberg’s Foundation, AstraZeneca Science for Life Research Collaboration grant (DISSECT), The Swedish Research Council, and the Swedish Society of Medicine (the Ingegerd Johansson donation). The funding bodies had no role in any aspect of study design, analysis, interpretation, or manuscript writing.
Availability of data and materials
All data analyzed during this study are included in this published article and its supplementary information files. The RNA sequencing datasets are provided as aggregated data (Additional file 16).
Abbreviations
- AMBRA1
Autophagy and beclin 1 regulator 1
- APC
Allophycocyanin
- APOL6
Apolipoprotein L6
- ATG14
Autophagy related 14
- BCL2A1
BCL2 related protein A1
- BDCA
Blood dendritic cell antigen
- BECN1
Beclin 1
- CCL
C-C motif chemokine ligand
- CD
Cluster of differentiation
- CD274
CD274 molecule
- CDKN1A
Cyclin dependent kinase inhibitor 1A
- CXCL
C-X-C motif chemokine ligand
- DEG
Differentially expressed gene
- DELFIA
Dissociation-enhanced lanthanide fluorescence immunoassay
- DKK4
Dickkopf WNT signaling pathway inhibitor 4
- EAF2
ELL associated factor 2
- FcγRIIA
Fragment crystallizable receptor IIA
- FDR
False discovery rate
- FITC
Fluorescein isothiocyanate
- HCQ
Hydroxychloroquine
- IC
Immune complex
- IFIT
Interferon induced protein with tetratricopeptide repeats
- IFN
Interferon
- IFNA2
Interferon alpha 2
- Ig
Immunoglobulin
- IL
Interleukin
- IRAK
Interleukin-1 receptor-associated kinase
- IRAK4i
Interleukin-1 receptor-associated kinase 4 inhibitor
- IRF
Interferon regulatory factor
- LAD1
Ladinin 1
- LLoQ
Lower limits of quantification
- mAb
Monoclonal antibody
- MIP
Macrophage inflammatory protein
- MyD88
Myeloid differentiation primary response protein 88
- NFκB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- NK
Natural killer
- OASL
2′-5′-Oligoadenylate synthetase like protein
- PBMC
Peripheral blood mononuclear cell
- pDC
Plasmacytoid dendritic cell
- PE
Phycoerythrin
- RELA
RELA proto-oncogene, NF-κB subunit
- RELB
RELB proto-oncogene, NF-κB subunit
- RIN
RNA integrity number
- RNA-IC
Ribonucleic acid containing immune complex
- RNP
Ribonucleoprotein
- SLE
Systemic lupus erythematosus
- Sm
Smith nuclear antigen
- snRNP
Small nuclear ribonucleoprotein
- SP1
Specificity protein 1 transcription factor
- STAT
Signal transducer and activator of transcription
- TLR
Toll-like receptor
- TNF
Tumor necrosis factor
- TNFSF10
TNF superfamily member 10
Authors’ contributions
Study design: KH, JM, MLE, LR. Laboratory data acquisition: KH, NH, LJ, OB, KT, MLE. Data analysis and interpretation: KH, NH, EI, JKS, KT, JM, MLE. Draft writing: KH, NH, EI, MLE, LR. Final revision: KH, NH, EI, LJ, OB, JKS, KT, JM, MLE, LR. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The local ethics committee of Uppsala approved the study and informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
EI, LJ, KT, and JM are employees of AstraZeneca. LR received a research grant from AstraZeneca. The remaining authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Karin Hjorton, Phone: +46-18-4714534, Email: karin.hjorton@medsci.uu.se.
Niklas Hagberg, Email: niklas.hagberg@medsci.uu.se.
Elisabeth Israelsson, Email: elisabeth.israelsson@astrazeneca.com.
Lisa Jinton, Email: lisa.jinton@astrazeneca.com.
Olof Berggren, Email: olof.berggren@medsci.uu.se.
Johanna K. Sandling, Email: johanna.sandling@medsci.uu.se
Kristofer Thörn, Email: kristofer.thorn@astrazeneca.com.
John Mo, Email: john.mo@astrazeneca.com.
Maija-Leena Eloranta, Email: maija-leena.eloranta@medsci.uu.se.
Lars Rönnblom, Email: lars.ronnblom@medsci.uu.se.
References
- 1.Bengtsson AA, Ronnblom L. Systemic lupus erythematosus: still a challenge for physicians. J Intern Med. 2017;281:52–64. doi: 10.1111/joim.12529. [DOI] [PubMed] [Google Scholar]
- 2.Tsokos GC, Lo MS, Reis PC, Sullivan KE. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol. 2016;12:716–730. doi: 10.1038/nrrheum.2016.186. [DOI] [PubMed] [Google Scholar]
- 3.Lovgren T, Eloranta ML, Bave U, Alm GV, Ronnblom L. Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum. 2004;50:1861–1872. doi: 10.1002/art.20254. [DOI] [PubMed] [Google Scholar]
- 4.Eloranta ML, Alm GV, Rönnblom L. Disease mechanisms in rheumatology—tools and pathways: plasmacytoid dendritic cells and their role in autoimmune rheumatic diseases. Arthritis Rheum. 2013;65:853–863. doi: 10.1002/art.37821. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Hagberg N, Berggren O, Leonard D, Weber G, Bryceson YT, Alm GV, Eloranta ML, Ronnblom L. IFN-alpha production by plasmacytoid dendritic cells stimulated with RNA-containing immune complexes is promoted by NK cells via MIP-1beta and LFA-1. J Immunol. 2011;186:5085–5094. doi: 10.4049/jimmunol.1003349. [DOI] [PubMed] [Google Scholar]
- 7.Wallace DJ, Gudsoorkar VS, Weisman MH, Venuturupalli SR. New insights into mechanisms of therapeutic effects of antimalarial agents in SLE. Nat Rev Rheumatol. 2012;8:522–533. doi: 10.1038/nrrheum.2012.106. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz-Irastorza G, Ramos-Casals M, Brito-Zeron P, Khamashta MA. Clinical efficacy and side effects of antimalarials in systemic lupus erythematosus: a systematic review. Ann Rheum Dis. 2010;69:20–28. doi: 10.1136/ard.2008.101766. [DOI] [PubMed] [Google Scholar]
- 9.Rainsford KD, Parke AL, Clifford-Rashotte M, Kean WF. Therapy and pharmacological properties of hydroxychloroquine and chloroquine in treatment of systemic lupus erythematosus, rheumatoid arthritis and related diseases. Inflammopharmacology. 2015;23:231–269. doi: 10.1007/s10787-015-0239-y. [DOI] [PubMed] [Google Scholar]
- 10.Sacre K, Criswell LA, McCune JM. Hydroxychloroquine is associated with impaired interferon-alpha and tumor necrosis factor-alpha production by plasmacytoid dendritic cells in systemic lupus erythematosus. Arthritis Res Ther. 2012;14:R155. doi: 10.1186/ar3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrao R, Zhou H, Shan Y, Liu Q, Li Q, Shaw DE, Li X, Wu H. IRAK4 dimerization and trans-autophosphorylation are induced by Myddosome assembly. Mol Cell. 2014;55:891–903. doi: 10.1016/j.molcel.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picard C, von Bernuth H, Ghandil P, Chrabieh M, Levy O, Arkwright PD, McDonald D, Geha RS, Takada H, Krause JC, et al. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine (Baltimore) 2010;89:403–425. doi: 10.1097/MD.0b013e3181fd8ec3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 14.Eloranta ML, Lövgren T, Finke D, Mathsson L, Rönnelid J, Kastner B, Alm GV, Rönnblom L. Regulation of the interferon-alpha production induced by RNA-containing immune complexes in plasmacytoid dendritic cells. Arthritis Rheum. 2009;60:2418–2427. doi: 10.1002/art.24686. [DOI] [PubMed] [Google Scholar]
- 15.Weber G, Trowitzsch S, Kastner B, Luhrmann R, Wahl MC. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010;29:4172–4184. doi: 10.1038/emboj.2010.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly PN, Romero DL, Yang Y, Shaffer AL, 3rd, Chaudhary D, Robinson S, Miao W, Rui L, Westlin WF, Kapeller R, et al. Selective interleukin-1 receptor-associated kinase 4 inhibitors for the treatment of autoimmune disorders and lymphoid malignancy. J Exp Med. 2015;212:2189–2201. doi: 10.1084/jem.20151074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cederblad B, Blomberg S, Vallin H, Perers A, Alm GV, Ronnblom L. Patients with systemic lupus erythematosus have reduced numbers of circulating natural interferon-alpha- producing cells. J Autoimmun. 1998;11:465–470. doi: 10.1006/jaut.1998.0215. [DOI] [PubMed] [Google Scholar]
- 18.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 20.Patro R, Mount SM, Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol. 2014;32:462–464. doi: 10.1038/nbt.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer Verlag; 2009. [Google Scholar]
- 22.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pendas-Franco N, Aguilera O, Pereira F, Gonzalez-Sancho JM, Munoz A. Vitamin D and Wnt/beta-catenin pathway in colon cancer: role and regulation of DICKKOPF genes. Anticancer Res. 2008;28:2613–2623. [PubMed] [Google Scholar]
- 24.Roth Lee, Srivastava Swati, Lindzen Moshit, Sas-Chen Aldema, Sheffer Michal, Lauriola Mattia, Enuka Yehoshua, Noronha Ashish, Mancini Maicol, Lavi Sara, Tarcic Gabi, Pines Gur, Nevo Nava, Heyman Ori, Ziv Tamar, Rueda Oscar M., Gnocchi Davide, Pikarsky Eli, Admon Arie, Caldas Carlos, Yarden Yosef. SILAC identifies LAD1 as a filamin-binding regulator of actin dynamics in response to EGF and a marker of aggressive breast tumors. Science Signaling. 2018;11(515):eaan0949. doi: 10.1126/scisignal.aan0949. [DOI] [PubMed] [Google Scholar]
- 25.Ai J, Pascal LE, Wei L, Zang Y, Zhou Y, Yu X, Gong Y, Nakajima S, Nelson JB, Levine AS, et al. EAF2 regulates DNA repair through Ku70/Ku80 in the prostate. Oncogene. 2017;36:2054–2065. doi: 10.1038/onc.2016.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Postal M, Appenzeller S. The role of tumor necrosis factor-alpha (TNF-α) in the pathogenesis of systemic lupus erythematosus. Cytokine. 2011;56:537–543. doi: 10.1016/j.cyto.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 27.Aringer M, Smolen JS. The role of tumor necrosis factor-alpha in systemic lupus erythematosus. Arthritis Res Ther. 2008;10:202. doi: 10.1186/ar2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cortes-Hernandez J, Egri N, Vilardell-Tarres M, Ordi-Ros J. Etanercept in refractory lupus arthritis: an observational study. Semin Arthritis Rheum. 2015;44:672–679. doi: 10.1016/j.semarthrit.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Studnicka-Benke A, Steiner G, Petera P, Smolen JS. Tumour necrosis factor alpha and its soluble receptors parallel clinical disease and autoimmune activity in systemic lupus erythematosus. Br J Rheumatol. 1996;35:1067–1074. doi: 10.1093/rheumatology/35.11.1067. [DOI] [PubMed] [Google Scholar]
- 30.Fauriat C, Long EO, Ljunggren HG, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115:2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu F, Maniar A, Diaz MQ, Chapoval AI, Medvedev AE. Activation of cytokine-producing and antitumor activities of natural killer cells and macrophages by engagement of toll-like and NOD-like receptors. Innate Immun. 2011;17:375–387. doi: 10.1177/1753425910372000. [DOI] [PubMed] [Google Scholar]
- 32.Adib-Conquy M, Scott-Algara D, Cavaillon JM, Souza-Fonseca-Guimaraes F. TLR-mediated activation of NK cells and their role in bacterial/viral immune responses in mammals. Immunol Cell Biol. 2014;92:256–262. doi: 10.1038/icb.2013.99. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Wesche H, Stevens T, Walker N, Yeh WC. IRAK-4 inhibitors for inflammation. Curr Top Med Chem. 2009;9:724–737. doi: 10.2174/156802609789044407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vega L, Barbado J, Almansa R, Gonzalez-Gallego R, Rico L, Jimeno A, Nocito M, Ortiz de Lejarazu R, Bermejo-Martin JF. Prolonged standard treatment for systemic lupus erythematosus fails to normalize the secretion of innate immunity-related chemokines. Eur Cytokine Netw. 2010;21:71–76. doi: 10.1684/ecn.2009.0176. [DOI] [PubMed] [Google Scholar]
- 35.Sandling J, Garnier S, Sigurdsson S, Wang C, Nordmark G, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jönsen A, et al. A candidate gene study of the type I interferon pathway implicates IKBKE and IL8 as risk loci for SLE. Eur J Hum Genet. 2011;19:479–484. doi: 10.1038/ejhg.2010.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 37.Olferiev M, Jacek E, Kirou KA, Crow MK. Novel molecular signatures in mononuclear cell populations from patients with systemic lupus erythematosus. Clin Immunol. 2016;172:34–43. doi: 10.1016/j.clim.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 38.Bloch O, Amit-Vazina M, Yona E, Molad Y, Rapoport MJ. Increased ERK and JNK activation and decreased ERK/JNK ratio are associated with long-term organ damage in patients with systemic lupus erythematosus. Rheumatology (Oxford) 2014;53:1034–1042. doi: 10.1093/rheumatology/ket482. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Takahashi Y, Fujii S, Zhou Y, Hong R, Suzuki A, Tsubata T, Hase K, Wang JY. EAF2 mediates germinal centre B-cell apoptosis to suppress excessive immune responses and prevent autoimmunity. Nat Commun. 2016;7:10836. doi: 10.1038/ncomms10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato Hiroshi, Perl Andras. Blockade of Treg Cell Differentiation and Function by the Interleukin-21-Mechanistic Target of Rapamycin Axis Via Suppression of Autophagy in Patients With Systemic Lupus Erythematosus. Arthritis & Rheumatology. 2018;70(3):427–438. doi: 10.1002/art.40380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clarke AJ, Ellinghaus U, Cortini A, Stranks A, Simon AK, Botto M, Vyse TJ. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Ann Rheum Dis. 2015;74:912–920. doi: 10.1136/annrheumdis-2013-204343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gros F, Muller S. Pharmacological regulators of autophagy and their link with modulators of lupus disease. Br J Pharmacol. 2014;171:4337–4359. doi: 10.1111/bph.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dudhgaonkar S, Ranade S, Nagar J, Subramani S, Prasad DS, Karunanithi P, Srivastava R, Venkatesh K, Selvam S, Krishnamurthy P, et al. Selective IRAK4 inhibition attenuates disease in murine lupus models and demonstrates steroid sparing activity. J Immunol. 2017;198:1308–1319. doi: 10.4049/jimmunol.1600583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy Anuradha. Early Probe and Drug Discovery in Academia: A Minireview. High-Throughput. 2018;7(1):4. doi: 10.3390/ht7010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Patient clinical characteristics. (PDF 36 kb)
Figure S1. Titration of IRAK4 inhibitor (I92) on cytokine production by cocultured plasmacytoid dendritic cells and NK cells. (PDF 194 kb)
Figure S2. Titration of hydroxychloroquine in cocultured plasmacytoid dendritic cells and NK cells with regard to interferon-α production. (PDF 126 kb)
Figure S3. Titration of hydroxychloroquine in cocultured plasmacytoid dendritic cells and NK cells. (PDF 221 kb)
Figure S4. Flow cytometric gating strategy of stimulated plasmacytoid dendritic cells and NK cells. (PDF 211 kb)
Figure S5. Flow cytometry showing total proportion of cytokine-producing cells in RNA-IC-stimulated pDC and NK cells. (PDF 282 kb)
Table S2. Gene list of 975 differentially expressed genes. (PDF 767 kb)
Table S3. Upstream regulators. (PDF 291 kb)
Figure S6. Overlap of differentially expressed genes in plasmacytoid dendritic cells. (PDF 135 kb)
Table S4. Enriched biological function pathways. (PDF 249 kb)
Table S5. Enriched signal processing pathways. (PDF 251 kb)
Figure S7. RNA-seq analysis of cytokine expression in plasmacytoid dendritic cells stimulated for 6 h in the presence of IRAK4 inhibitor or hydroxychloroquine. (PDF 186 kb)
Figure S8. TNF-α production in NK cell cultures and NK cell/pDC cocultures. (PDF 179 kb)
Figure S9. Flow cytometric analysis of TNF-α in NK cells. (PDF 165 kb)
Figure S10. Interleukin-8 production by stimulated blood cells from SLE patients. (PDF 104 kb)
Table S6. Gene expression in plasmacytoid dendritic cells (pDCs) from healthy donors. (XLSX 4030 kb)
Data Availability Statement
All data analyzed during this study are included in this published article and its supplementary information files. The RNA sequencing datasets are provided as aggregated data (Additional file 16).