Abstract
Individuals with germline mutations in the tumor suppressor gene phosphatase and tensin homolog (PTEN), irrespective of clinical presentation, are diagnosed with PTEN hamartoma tumor syndrome (PHTS). PHTS confers a high risk of breast, thyroid, and other cancers or autism spectrum disorder (ASD) with macrocephaly. It remains unclear why mutations in one gene can lead to seemingly disparate phenotypes. Thus, we sought to identify differences in ASD vs. cancer-associated germline PTEN missense mutations by investigating putative structural effects induced by each mutation. We utilized a theoretical computational approach combining in silico structural analysis and molecular dynamics (MD) to interrogate 17 selected mutations from our patient population: six mutations were observed in patients with ASD (only), six mutations in patients with PHTS-associated cancer (only), four mutations shared across both phenotypes, and one mutation with both ASD and cancer. We demonstrate structural stability changes where all six cancer-associated mutations showed a global decrease in structural stability and increased dynamics across the domain interface with a proclivity to unfold, mediating a closed (inactive) active site. In contrast, five of the six ASD-associated mutations showed localized destabilization that contribute to the partial opening of the active site. Our results lend insight into distinctive structural effects of germline PTEN mutations associated with PTEN-ASD vs. those associated with PTEN-cancer, potentially aiding in identification of the shared and separate molecular features that contribute to autism or cancer, thus, providing a deeper understanding of genotype-phenotype relationships for germline PTEN mutations.
Keywords: PTEN, Cancer, Autism, Molecular Dynamics Simulations, Protein Structure Stability
1. Introduction
Phosphatase and Tensin homolog deleted on chromosome ten (PTEN) is a tumor suppressor gene frequently mutated somatically in tumors and in the germline of patients. Irrespective of clinical features, individuals with germline PTEN mutations are diagnosed as having PTEN hamartoma tumor syndrome [PHTS] (Eng, 2000, 2003; Marsh, Kum, Lunetta, Bennett, Gorlin et al., 1999). The clinical presentation of PHTS is broad and includes distinct subsets of patients with a predisposition to cancer or to Autism Spectrum Disorder (ASD). PTEN functions as a dual-specificity lipid and protein phosphatase that downregulates the PI3K/AKT/mTOR signaling pathway (Georgescu, 2010; Myers, Stolarov, Eng, Li, Wang et al., 1997; Steck, Pershouse, Jasser, Yung, Lin et al., 1997). Its heavily studied lipid phosphatase activity is most relevant to this inhibition, whereby PTEN catalyzes the removal of the 3’ phosphate of the inositol ring in the bulky phosphatidylinositol (3,4,5)-triphosphate (PIP3) lipid head group. This conversion of PIP3 to PI(4,5)P2 is key to restraining downstream anti-apoptotic and growth stimulatory effects that otherwise lead to unrestricted cell growth and tumorigenesis (Georgescu, 2010; Lee, Yang, Georgescu, Di Cristofano, Maehama et al., 1999; Li, Yen, Liaw, Podsypanina, Bose et al., 1997; Maehama & Dixon, 1998, 1999; Myers et al., 1997; Rodriguez-Escudero, Oliver, Andres-Pons, Molina, Cid et al., 2011; Steck et al., 1997; Tamura, Gu, Tran, & Yamada, 1999; Wu, Senechal, Neshat, Whang, & Sawyers, 1998).
PTEN dysfunction, however, does more than predisposition to cancers. Germline PTEN mutations are present in 10%−20% of children with ASD accompanied by macrocephaly (Butler, Dasouki, Zhou, Talebizadeh, Brown et al., 2005; McBride, Varga, Pastore, Prior, Manickam et al., 2010; Tan, Mester, Peterson, Yang, Chen et al., 2011). ASD is a relatively common, highly heritable neurodevelopmental disorder that is characterized by social impairment and restricted, repetitive behavior (Frazier, Youngstrom, Speer, Embacher, Law et al., 2012; Freitag, 2007). It remains unclear why mutations in a single gene such as PTEN can result in diverse phenotypes such as ASD and/or cancer, two seemingly unrelated disease states. Studies on a large patient cohort accrued at our center reveal an enrichment of germline PTEN mutations of the missense type in patients with ASD with macrocephaly compared to PHTS patients without ASD, however, an absolute genotype-phenotype correlation still remains unidentifiable (Frazier, Embacher, Tilot, Koenig, Mester et al., 2015; He, Arrotta, Radhakrishnan, Wang, Romigh et al., 2013; Leslie & Longy, 2016). Studies by Spinelli (2015) and Leslie (2016) have shown that mutations that lead to only partial loss of function of PTEN enzymatic activity seem to be preferentially associated with ASD, whereas catalytically inactive mutants lead to a more aggressive phenotype. However, the differential impact of ASD vs. cancer-associated PTEN mutations on its structure and thereby its function, remains unknown in spite of well-established literature on residues critical for PTEN phosphatase function.
Previous studies show that missense PTEN mutations in active site residues, D92, C124, and R130 result in severe reduction of phospholipid phosphatase activity of PTEN (Eng, 2003; Myers et al., 1997; Xiao, Yeong Chit Chia, Gajewski, Sio Seng Lio, Mulhern et al., 2007). These active site residues in the phosphatase domain of PTEN are known to play a key role in dephosphorylating PI(3,4,5)P3 into PI(4,5)P2 (Chia, Gajewski, Xiao, Zhu, & Cheng, 2010). Compared to other protein tyrosine phosphatases, PTEN has a wider active site pocket due to its TI loop, in order to accommodate its large PI(3,4,5)P3 substrate (Lee et al., 1999). The phosphatase domain associates with the C2 domain across an extensive inter-domain interface that is adjacent to the active site and consists of conserved residues frequently mutated in cancer (Lee et al., 1999). Structural elements within the inter-domain interface are crucial for maintaining protein stability and orientation of the catalytic active site (Das, Dixon, & Cho, 2003; Georgescu, Kirsch, Kaloudis, Yang, Pavletich et al., 2000; Lee et al., 1999). In fact, PTEN has recently been recognized as a superdomain protein, indicating that it is critical for both the phosphatase and C2 domain to exist in tandem across species (Haynie & Xue, 2015). These studies indicate the importance and far reaching consequences of point mutations in apparently restricted single residues on the PTEN structure.
Over the years, various computational methods have been utilized for predicting the impact of mutations on protein structure and function (Capriotti, Fariselli, Rossi, & Casadio, 2008; Kumar, Doss, Sneha, Tayubi, Siva et al., 2017; Masso & Vaisman, 2008; Smith & Briggs, 2016). More recently, in silico computational studies using a molecular dynamics approach have successfully identified various structural perturbations that occur as a result of the mutation to assess structural instability (Ali, Sneha, Priyadharshini Christy, Zayed, & George Priya Doss, 2017; Sutthibutpong, Rattanarojpong, & Khunrae, 2017), protein dynamics (Mohajer, Parvizpour, Razmara, Khoshkhooy Yazdi, & Shamsir, 2017; Yang, Sang, Tao, Fu, Zhang et al., 2014), protein unfolding (Anwer, Sonani, Madamwar, Singh, Khan et al., 2015; Tompa & Kadhirvel, 2017), and impact on inter-domain interactions (Krishnamoorthy, Gajendrarao, Olivotto, & Yacoub, 2017). Therefore, acquiring an in-depth understanding of the impact of missense mutations on the structure of PTEN, and by extension, its function, is imperative to elucidating the molecular mechanisms underlying development of PTEN mutation-associated ASD and cancer. To investigate this common genetic etiology of two rather disparate phenotypes, we sought to study the differences in the structural impact of germline PTEN missense mutations associated with ASD vs. those associated with cancer. Using combined in silico prediction methods and structural analysis of 17 different germline PTEN missense mutations selected from a series of 138 prospectively accrued patients with carefully annotated and documented phenotypes, we performed in-depth analyses of structural, conformational and dynamic changes, and on long-range perturbations in the PTEN structure conferred by ASD- or cancer-associated mutations. Using these computational structural biology and molecular dynamics simulation analysis tools, we present here an extensive analysis of the differential structural instability conferred by germline PTEN mutations specifically associated with ASD vs. cancer in our patient population.
2. Computational Methods
2.1. Selecting ASD-associated and cancer-associated mutations for structural analysis
This prospective study was performed within the framework of the “Molecular Mechanisms Involved in Cancer Predisposition” study (Cleveland Clinic Institutional Review Board approved IRB# 8458) and conducted with informed consent and in accordance with the World Medical Association Declaration of Helsinki. Germline PTEN missense mutations from a series of 138 prospectively accrued patients were compiled with a number of descriptive variables, which included clinical phenotypes, pathogenicity predictors, other genotype information, and structural stability predictors. The dataset was further evaluated by an in-house random forest algorithm (Khalilia, Chakraborty, & Popescu, 2011) to identify variables that contribute most significantly to any differentiation among the assortment of phenotypes observed in the cohort, specifically ASD or cancer (Table S1). These differentiating variables were further condensed with a principal component analysis [PCA] (Zhang & Castello, 2017) that aided in the visualization of specific mutations that may contribute outsized effects toward their associated phenotype [i.e. ASD vs. cancer] (Table S2).
2.2. Structural model preparation and simulations
The wild-type (WT) crystal structure of human PTEN protein (PDB ID: 1D5R) was obtained from the RCSB Protein Data Bank (PDB) (Rose, Prlic, Altunkaya, Bi, Bradley et al., 2017). The tartrate (TLA) molecule was removed and all calculations were conducted on apo PTEN. The PTEN mutant structures were constructed in silico by side-chain replacement utilizing Visual Molecular Dynamics (VMD) Mutator Plugin 1.3 (Humphrey, Dalke, & Schulten, 1996) from the WT PTEN (PDB ID 1D5R) crystal structure file. Seventeen mutant structure models were generated for each of the mutations (p.L23F, p.D24G, p.Y65C, p.Y68H, p.D92A, p.I101T, p.I122S, p.R130G, p.R130Q, p.M134R, p.C136R, p.Y155C, p.S170I, p.R173C, p.M205V, p.L220V, and p.L345V).
All-atom molecular dynamics (MD) simulations were performed using GROMACS 4.6.3 (Van Der Spoel, Lindahl, Hess, Groenhof, Mark et al., 2005) with GROMOS53a6 force field (Oostenbrink, Villa, Mark, & van Gunsteren, 2004) on (apo) WT PTEN (PDB ID: 1D5R) and each PTEN mutant structure. Each system was solvated inside a cubic box of simple point charge (SPC) water (Berendsen, Postma, van Gunsteren, & Hermans, 1981) with at least 10 Å of water between the protein and the edges of the box. All simulations were performed in explicit solvent with chloride (Cl-) counter ions added to obtain neutrality of the system. Periodic Boundary Conditions (PBC) and a 2 fs time step were employed for each simulation. Each system contained roughly 75,800 atoms. The Particle Mesh Ewald (PME) method (Darden, York, & Pedersen, 1993) was used to treat long-range electrostatic interactions and a cutoff of 9 Å was used for non-bonded interactions. Each system was subjected to a step-wise energy minimization using the steepest descent method and a series of five overall minimization steps was performed to remove steric clashes and minimize the forces as a result of the mutation that was introduced to the WT structure. The five energy minimization steps included: step 1, all protein atoms were restrained; step 2, protein heavy atoms were restrained; step 3, all protein atoms were restrained again; step 4, atoms of the protein main chain were restrained; and step 5, unrestrained energy minimization. Total minimization was carried out until convergence where the maximum atomic force was <1000 kJ/mol-nm. The minimized structures were then slowly heated from 0 to 300K over 100 ps and equilibrated for an additional 250 ps. The systems were heated linearly by increasing the temperature through velocity rescaling every 10 ps. The production simulations were carried out at constant pressure (1 atm), temperature (300 K), and particle number (NPT, isobaric-isothermal ensemble). Bond lengths were constrained using linear constraint solver (LINCS) algorithm (Hess, Bekker, Berendsen, & Fraaije, 1997) with the van der Waals forces maintained at 1.4 nm. The Berendsen (Berendsen, Postma, Vangunsteren, Dinola, & Haak, 1984) weak coupling method was employed to maintain constant temperature with a temperature coupling relaxation time of 0.1 ps, a pressure coupling constant of 0.5 ps, and a compressibility of 4.5 • 10-5. The total simulation time for each model was 200 ns with coordinates saved every 1 ps.
2.3. All-atom normal mode analysis and principal component analysis
All-atom normal mode analysis (NMA) was conducted on representative MD trajectories of each system by performing principal component analysis (PCA). Normalizing each eigenvalue of the covariance matrix to its total sum yields the percentage of all movements attributable to the corresponding eigenvector. The largest eigenvalues correspond to the principal component modes that best explain the molecular motions sampled by the all-atom system trajectories. The trajectories and principal component projections were analyzed using Visual Molecular Dynamics (VMD) version 1.9.3 (Humphrey et al., 1996).
2.4. Structural stability and free energy calculations
To explore structural stability changes induced by each germline PTEN mutation, we used Elastic Network Contact Model (ENCoM) (Frappier, Chartier, & Najmanovich, 2015; Frappier & Najmanovich, 2014) that utilizes a novel mixed coarse-grained normal mode method to account for the type and extent of pairwise atomic interaction allowing for the calculation of vibrational entropy differences as a result of the mutations (Frappier et al., 2015; Frappier & Najmanovich, 2014). ENCoM uses a potential function with four terms: (1) covalent bond stretching, (2) angle bending, (3) dihedral torsion, and (4) non-bonded interaction while taking into consideration the nature and possible effects that the orientation of the side-chains have on dynamics within the context of normal mode analysis. The WT PTEN structure file (PDB ID: 1D5R) was utilized and the residue name and mutation site were provided as input.
3. Results and Discussion
3.1. Mapping all 138 ASD- vs. cancer-associated mutations on PTEN structure
In order to test if the primary sequence or three-dimensional location of specific mutations would be associated with a particular phenotype, we mapped PTEN mutations from our cohort of 138 PHTS patients on the PTEN protein structure (data not shown). Mapping of PTEN mutations to its structure did not reveal a distinct ASD- or cancer-associated three-dimensional pattern of distribution across the PTEN molecule, thus, suggesting a complex relationship between the two phenotypes.
3.2. Germline PTEN missense mutations associated with ASD vs. cancer
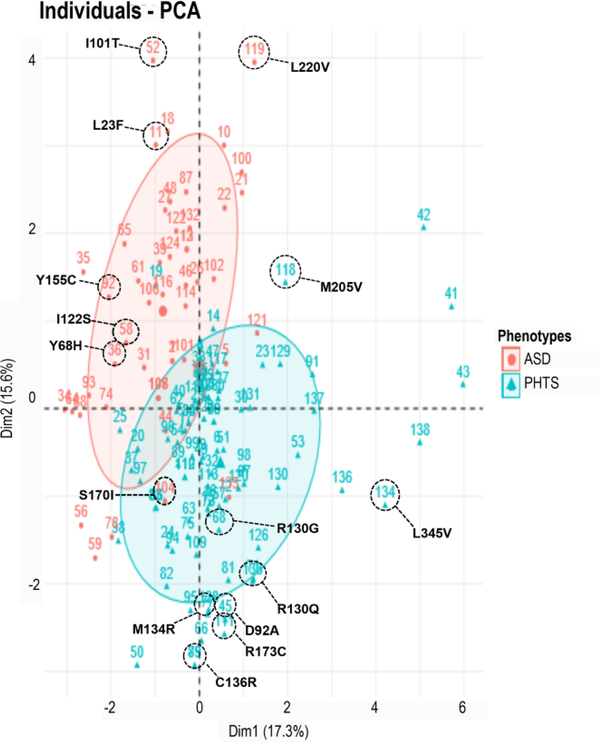
Because simple mapping of all our 138 germline missense mutations was uninformative in bifurcating cancer predisposition and autism, we applied a random forest machine learning approach (Khalilia et al., 2011) to these 138 ASD- and cancer-associated germline PTEN missense mutations based on known phenotype and other differentiating variables in each group. The strong contributing variables identified by the random forest classifier were analyzed by a principal component analysis [PCA] (Zhang & Castello, 2017), which segregated the cohort into two distinct yet overlapping phenotypic groups, of which we selected the outliers in opposing direction in order to assess mutants likely to have an outsized contribution to phenotype. Based on the PCA plot (Figure 1, Tables 1, S1, and S2), we selected for structural analyses 17 different mutations showing the most distant segregation across the PCA axes and therefore suggestive of maximal association with either ASD or cancer. These 17 mutations were identified and selected for structural analysis (Table 1).
Figure 1.
Principal component analysis (PCA) multivariate plot segregates ASD- vs. cancer-associated mutations. PCA performed on patient data using predictive genotypic and phenotypic variables including clinical phenotypes, pathogenicity predictors, other genotype information, and structural stability predictors. PCA of the first two axes explains 15.6% and 17.3% of the variation (32.9% total). Projection along the two axes is represented by a red ellipse (ASD) in the upper left quadrant and a blue ellipse (cancer) in the lower right quadrant. Red circle points correspond to ASD phenotype, whereas blue triangle points designate PHTS cancer-associated phenotype.
Table 1.
ASD- vs. cancer-associated germline PTEN mutations
| ASD (only) | Domain | Cancer (only) | Domain | ASD or Cancer (shared) | Domain | ASD and Cancer | Domain |
|---|---|---|---|---|---|---|---|
| p.L23F | Phosphatase | p.D24G | Phosphatase | p.R130Q | Phosphatase | p.S170I | Phosphatase |
| p.Y65C | p.D92A | p.C136R | |||||
| p.Y68H | p.R130G | p.Y155C | |||||
| p.I101T | p.M134R | p.R173C | |||||
| p.I122S | p.M205V | C2 | |||||
| p.L220V | C2 | p.L345V |
Based on the crystal structure of PTEN, mutations that were not part of the structure were excluded from analysis. Of the selected PTEN mutations, six mutations were observed in patients with ASD (only), six mutations in patients with PHTS cancer (only), four mutations were found to be associated with both phenotypes in different individual patients (ASD or cancer), and one mutation corresponded to an individual with coexisting phenotypes of both ASD and cancer (Table 1). Analyses on putative structural effects were conducted for individual mutations classified under the aforementioned four phenotypic categories.
In order to test if the primary sequence or three-dimensional location of specific mutations would be associated with a particular phenotype, we mapped PTEN mutations from our cohort of 138 PHTS patients on the PTEN protein structure (data not shown). Mapping of PTEN mutations to its structure did not reveal a distinct ASD- or cancer-associated three-dimensional pattern of distribution across the PTEN molecule, thus suggesting a complex relationship between the two phenotypes.
3.3. Mapping the most phenotypically divergent ASD- vs. cancer-associated mutations on PTEN structure
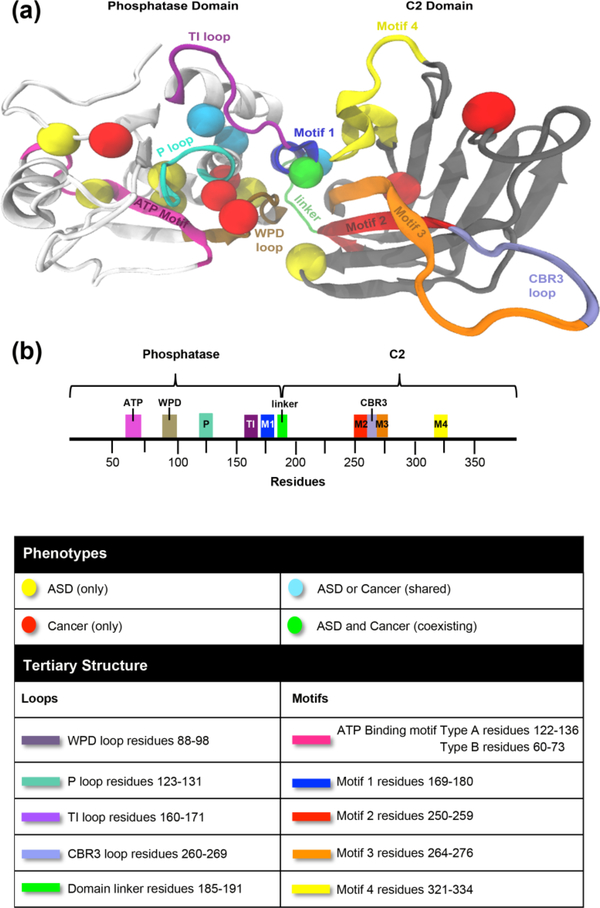
Given that the 17 mutations selected from the resulting random forest classifier and subsequent PCA segregated distinctly, we mapped them to the three-dimensional crystal structure of PTEN to obtain a more simplified picture of mutation location and associated phenotype of ASD or cancer. Mapping revealed that the majority of the mutations (irrespective of ASD or cancer association) clustered in the phosphatase domain within the two ATP-binding motifs (type A, residues 122–136 and type B, residues 60–73) and the three active site loops (TI-, P-, and WPD-), particularly the highly conserved P loop signature motif (H123CKAGKGR130) [Figures 2 and S1]. Our findings corroborate the importance of the P loop in preserving the phosphatase activity for the physiological function of PTEN. Previous studies using in vivo- and in vitro-based methods have also elucidated concentration of germline PTEN mutations in the phosphatase domain (Han, Kato, Kato, Suzuki, Shibata et al., 2000; Lee et al., 1999; Marsh, Coulon, Lunetta, Rocca-Serra, Dahia et al., 1998; Mester & Eng, 2013; Rodriguez-Escudero et al., 2011). Our computational structure-based approach, however, offers an advantage in both identifying potential spatial relationships and interaction networks between residues not previously considered in PTEN genotype-phenotype correlation studies. Moreover, our unique structural perspective provides atomic insight useful for identifying mutation clusters within the phosphatase domain that may affect the structural scaffold of the C2 domain and thus the overall function of PTEN.
Figure 2.
Mapping of ASD- vs. cancer-associated mutations on PTEN three-dimensional structure. (a) Individual spheres correspond to 17 germline missense PTEN mutations represented by ASD only (yellow), cancer only (red), mutations shared across both phenotypes (cyan), one mutation with coexisting ASD and cancer (green). Mutations occur in both the phosphatase domain (white) and C2 domain (grey) of WT PTEN structure, (b) Domain structure of PTEN protein.
We show five out of six mutations strongly associated with ASD (p.L23F, p.Y65C, p.Y68H, p.I101T, and p.I122S) were located within the ATP-binding motif type B (residues 60–73) or near the active site pocket with only one mutation (p.L220V) located within the C2 domain. Similarly, four out of the six cancer (only) mutations (p.D24G, p.D92A, p.R130G, and p.M134R) mapped to the active site, though p.D92A, p.R130G, and p.M134R mutations located specifically between the catalytic P loop (overlaps ATP-binding motif type A) and WPD loop as well as motifs 2 (residues 250–259) and 3 (residues 264–276) within the inter-domain interface unlike the ASD associated mutations which were located within the ATP-binding motif type B. Two out of six cancer-associated mutations (p.M205V and p.L345V) located within the C2 domain. Three of the four shared mutations between ASD and cancer, (p.R130Q, p.C136R, and p.Y155C) mapped to the ATP-binding motif type A within the active site, with the remaining mutation (p.R173C) located on motif 1 (pα6 helix, residues 169–180) within the inter-domain interface. Interestingly, the only mutation associated with coexisting cancer and autism phenotypes within the same patient (p.S170I) showed the same inter-domain localization as the p.R173C mutation associated with both ASD and cancer in different individuals. Mutations in S170 and R173, located within motif 1 of the inter-domain region, are among the most frequently mutated PTEN residues in cancer (Lee et al., 1999). In fact, the variant p.S170R has also been shown to ablate phosphatase activity in vivo against poly(Glu-pTyr) (Myers et al., 1997). These residues have been shown to participate in a hydrogen-bond network within the inter-domain interface and are second only to the P loop in conservation across species, (Lee et al., 1999) suggesting that the integrity of the phosphatase activity is likely influenced by the inter-domain interface and linker (residues 185–191) that bridges the two functional domains of PTEN (Haynie & Xue, 2015).
In addition to the obvious importance of the phosphatase domain structure for PTEN function, our mapping studies also highlight the impact of the inter-domain region on the PTEN structure. A notable observation is that the cancer-associated mutations, mutations shared across both ASD and cancer phenotypes, as well as the one mutation associated with both coexisting ASD and cancer phenotypes, are located in highly conserved parts of the PTEN structure (P loop as well as inter-domain region). Cancer-associated mutations p.D92A, p.R130G, and p.M134R are clustered within the active site between the catalytic P loop (residues 123–131) and WPD loop (residues 88–98) directly adjacent to the inter-domain interface. In fact, missense mutations in these three residues have been shown to significantly disrupt catalytic activity and protein function (Chia et al., 2010; Lee et al., 1999; Rodriguez-Escudero et al., 2011). Interestingly, previous studies indicate that two (p.Y155C and p.R173C) of the four mutations shared across both phenotypes result in inactivation of PTEN (Gicquel, Vabres, Bonneau, Mercie, Handiri et al., 2003; Han et al., 2000; Maehama, Taylor, & Dixon, 2001; Rodriguez-Escudero et al., 2011; Waite & Eng, 2002) and also converge within the inter-domain region. Evolutionarily, the loss of phosphatase activity by mutation but preservation of the phosphatase-C2 domain across species suggests that domain coupling may have a more significant impact on function than phosphoryl group removal or binding (Haynie & Xue, 2015; Sakata, Matsuda, Kawanabe, & Okamura, 2017).
Mapping of ASD- and cancer-associated germline PTEN mutations on the PTEN structure revealed that the vast majority of the ASD-associated mutations are located within ATP-binding motif type B and around the highly conserved P loop within the active site. In contrast, the cancer-associated mutations tend to occupy the ATP-binding motif type A in between the catalytic P and WPD loops in the active site as well as near motifs 2 and 3 within the interdomain interface, conferring a potential hotspot for cancer-associated mutations and further emphasizing that integrity of the inter-domain interface is critical to the overall function of PTEN.
3.4. Structural stability changes in ASD- vs. cancer-associated germline PTEN mutations
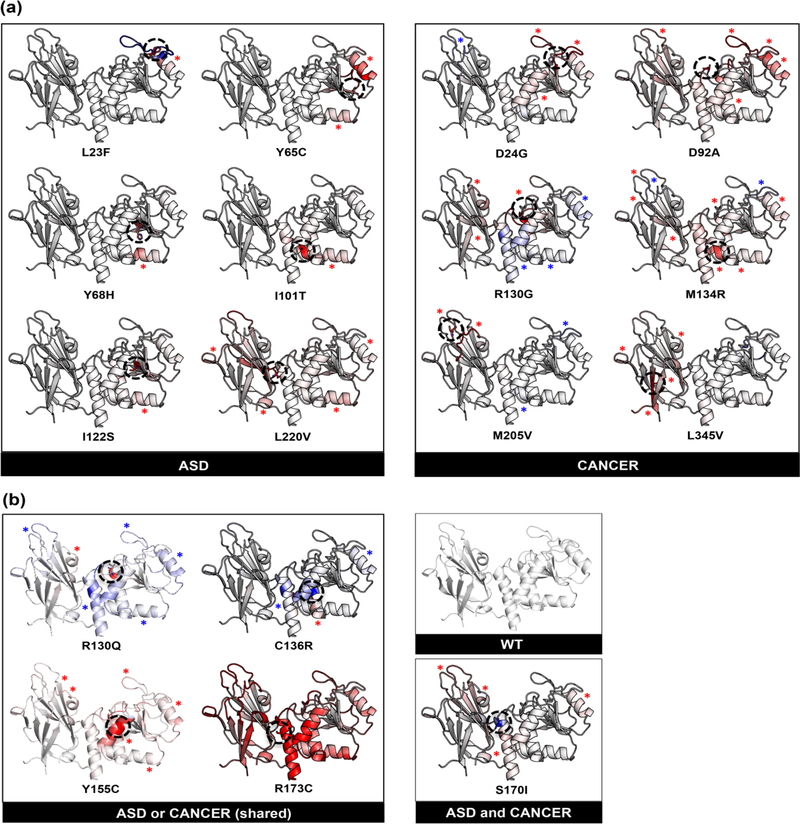
Pathogenic missense mutations tend to disrupt protein folding and structure stability typically resulting in loss-of-function. Moreover, a missense mutation that affects the three-dimensional protein structure and alters the structural stability may cause significant inter-residue perturbations or completely abolish protein function. To computationally assess the functional consequences of missense mutations, we addressed the effects of mutations on structural stability and folding free energy [ΔΔG] (Stefl, Nishi, Petukh, Panchenko, & Alexov, 2013) by grouping them into two distinct categories: (a) destabilizing and (b) stabilizing effects. We utilized ENCoM (Frappier et al., 2015; Frappier & Najmanovich, 2014) to calculate the change in the mutant protein’s structural thermostability (ΔΔG) compared to WT PTEN, as it accounts for side-chain and long-range interactions offering a realistic representation of intra-molecular interactions (Figures 3(a) and (b)). We demonstrate here that five of the six ASD-associated mutations (p.L23F, p.Y65C, p.Y68H, p.I101T, and p.I122S) show localized destabilization relative to the surrounding region of the mutation, whereas one mutation p.L220V exhibited global destabilization across both domains. In contrast, both the active site and inter-domain region were most affected by cancer-associated mutations (p.D24G, p.D92A, p.R130G, p.M134R, p.M205V, and p.L345V) as demonstrated by decreased structural stability and increased dynamics across the inter-domain interface of PTEN (Figure 3(a)). Similarly, the p.S170I mutation associated with coexisting ASD and cancer phenotypes showed similar structural stability effects to cancer (only) mutations.
Figure 3.
Effect of ASD- vs. cancer-associated mutations on structural stability and dynamics. (a) Structural stability changes induced by each ASD (only) and cancer (only) mutation. (b) Structural stability changes induced by each mutation shared across both phenotypes versus one mutation with coexisting phenotypes with respect to WT PTEN. Structural stability was determined by calculating the Gibbs free energy (ΔΔG) for each mutation with respect to WT PTEN. Blue indicates a more rigid or stabilized region (ΔΔG<0.5 kcal/mol), while red indicates a destabilized region (ΔΔG>0.5 kcal/mol). Black dashed circle represents the residue mutation. Blue and red asterisks correspond to stabilized and destabilized regions, respectively, within individual mutant PTEN structures.
Interestingly, the mutations shared across both ASD and cancer phenotypes (p.R130Q, p.C136R, p.Y155C, and p.R173C) indicate two distinct groups: stabilized (rigid) and destabilized (highly flexible), with p.R130Q and p.C136R exhibiting a more rigid inter-domain region, whereas p.Y155C and p.R173C exhibit a greater, more flexible inter-domain region (Figure 3(b)). Additionally, analysis of phenotype data from our patient cohort revealed that the p.R130Q and p.C136R mutations with the rigid inter-domain region trend towards a higher frequency of occurrence in patients with cancer – more specifically, breast, thyroid, endometrial, ovarian, cervical, and esophageal cancers (Table S4). There are two possible explanations for these results. First, mutations in the inter-domain region result in the destruction of a critical hydrogenbond network, thus impairing the structural stability within the inter-domain interface contributing to a more severe phenotype (Lee et al., 1999). Second, mutations in protein interface sites with low dynamic flexibility are more likely to be disease-associated due to the fact that these sites play a critical role in modulating inter-domain dynamics (Butler, Gerek, Kumar, & Ozkan, 2015). Furthermore, stabilizing mutations in active site residues (p.R130Q and p.C136R) can reduce enzymatic activities and may therefore be deleterious (Khan & Vihinen, 2010; Maxwell, Risinger, Gumbs, Shaw, Bentley et al., 1998). In fact, studies from our lab (Waite & Eng, 2002) and others (Han et al., 2000; Maehama et al., 2001; Rodriguez-Escudero et al., 2011) indicate that mutations within the active site pocket lead to a loss of PTEN phosphatase activity.
Results presented here reveal a distinct difference in structural stability in ASD- vs. cancer-associated mutations. We show that cancer-associated mutations demonstrate considerable changes in global stability as demonstrated by decreased structural stability within the active site and increased dynamics across the domain interface of PTEN that may result in a loss of substrate interaction, complete loss of activity, and a more severe PHTS cancer-associated phenotype. These structural findings are further validated by in vitro studies which postulate a decrease in thermostability with cancer-associated PTEN mutations that lead to cooperative unfolding of the two domains of PTEN (Johnston & Raines, 2015). Our findings on global structural instability with cancer-associated mutations are also in agreement with studies by Spinelli et al. (2015) on effects of allelic mutations in vitro that show inactive mutants lead to a more aggressive phenotype. More importantly, in our current study, we demonstrate that in contrast to cancer-associated PTEN mutations that induce global instability, ASD-associated mutations show local structural destabilization relative to the surrounding region of the mutation that may lead to impaired substrate interaction resulting in loss of function.
3.5. Effects of mutations on global dynamics fingerprint and long-range perturbations
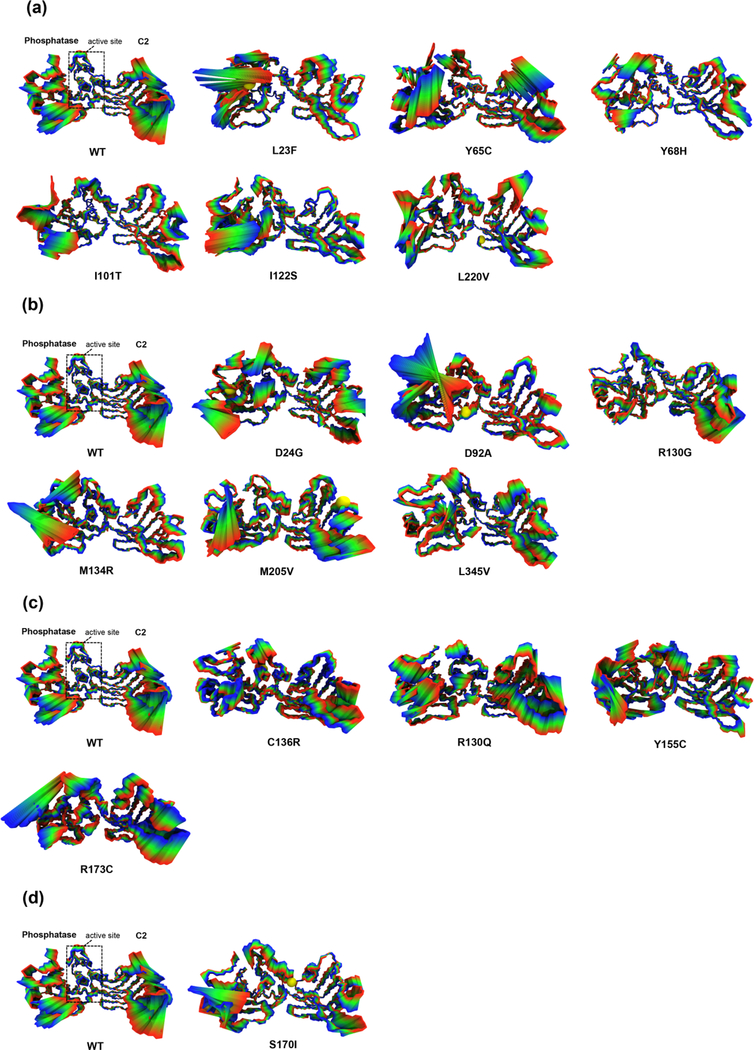
To further assess the effects on global dynamics and long-range perturbations induced by each of the mutations on the protein structure, we employed an all-atom normal mode analysis by performing principal component analysis on molecular dynamics (MD) trajectory ensembles of the WT and mutant structural models. The first combined eigenvector projection of the WT and mutant PTEN models collectively account for more than ~35% of the total motion and demonstrate a suitable approach to analyze protein structural dynamics and conformational differences (Figures 4(a)-(d)). The ASD-associated mutations (p.L23F, p.Y65C, p.Y68H, p.I101T, p.I122S, and p.L220V) reveal increased local dynamics at ATP-binding motif type B (residues 60–73) and dramatic conformational changes in motif 3 (residues 264–276) and motif 4 (residues 321–334) within the inter-domain interface that render a more rigid, compact active site and CBR3 loop (residues 260–269) compared to the WT structure of PTEN (Figure 4(a)). Three ASD-associated mutations (p.Y68H, p.I101T, and p.I122S) reveal a decrease in dynamics in motif 4 within the inter-domain interface further indicating the significance of interresidue communication across domains. Further inspection of five mutations (p. L23F, pY65C, p.Y68H, p.I101T, and p.I122S) demonstrates the impact of long-range perturbations originating in the N-terminal region of the phosphatase domain that affects the C2 domain.
Figure 4.
Local and global protein dynamics fingerprint of ASD- vs. cancer-associated mutations. The projections of the displacement and resulting long-range perturbations described by the first principle component on the structure represented by 50 frames of the entire MDS ensemble trajectory are shown for (a) ASD (only), (b) cancer (only), (c) mutations shared across both phenotypes, and (d) one mutation with coexisting phenotypes in comparison to WT PTEN. The structural segments illustrate concerted motions that represent the local and global conformational dynamic variations. The WT and mutant structures are represented in the licorice model and colored from blue to red representing the difference in minimum to maximum conformational states, respectively. A yellow sphere indicates the mutated residue.
In contrast to the ASD-associated mutations, five of the six cancer-associated mutations (p.D24G, p.D92A, p.M134R, p.M205V, and p.L345V) revealed an increase in global dynamics within the ATP-binding motif type A and TI loop, which alter the global intrinsic movement of PTEN (Figure 4(b)). The p.D92A and p.M134R mutations had a more rigid C2 domain, whereas the phosphatase domain had a substantial increase in dynamics. The p.D24G, p.M205V, and p.L345V mutations demonstrated an increase in overall global dynamics across both domains with similar dynamics within the active site and inter-domain region compared to the WT PTEN structure. In contrast, the remaining p.R130G mutation had a considerable decrease in dynamics in the N-terminal region as well as throughout the phosphatase domain including the inter-domain interface; however, the C2 domain had similar intrinsic dynamics compared to the WT PTEN structure.
Interestingly, mutations (p.C136R, p.R130Q, p.Y155C, and R173C) shared across both ASD and cancer, but more predisposed towards cancer, demonstrated an increase in overall global dynamics as well as ATP-binding motif type A and TI loop similar to cancer-associated mutations (Figure 4(c)). In addition, the p.S170I mutation associated with both coexisting ASD and cancer phenotypes exhibits a rigid C2 domain and decreased dynamics across the interdomain region and the CBR3 loop as seen in the cancer-associated mutations (Figure 4(d)). In fact, missense mutations at residue positions S170 and R173 disrupt critical hydrogen bond interactions with W274 and have been previously reported in multiple patients with cancer, (Lee et al., 1999; Marsh, Dahia, Zheng, Liaw, Parsons et al., 1997) further demonstrating variants at these positions to be more predisposed to cancer.
Overall, each of the mutation motions captures the induced effects in the conformational dynamics and thus reveals unique long-range perturbations and inter-domain communication effects within the PTEN protein. Most importantly, ASD-associated mutations exhibited increased local dynamics in ATP-binding motif B and considerable conformational changes that render a more rigid, compact active site and CBR3 loop that may possibly impair the ability for these regions to be flexible enough for substrate interaction and membrane association, respectively. In contrast, the majority of the cancer-associated mutations demonstrated an increase in overall global dynamics within the ATP-binding motif type A and TI loop across the phosphatase domain; however, p.D92A and p.M134R had a more rigid C2 domain and p.R130G had an overall decrease in dynamics throughout the phosphatase domain – all of which may result in impaired substrate interaction and thus a loss of enzymatic function. Our results illustrate mutation-driven displacements in the two ATP-binding sites and active site pocket that elicit long-range inter-residue signal propagation that may play a key role in modulating the catalytic function of PTEN.
3.6. Effects of mutations on structure and conformational dynamics
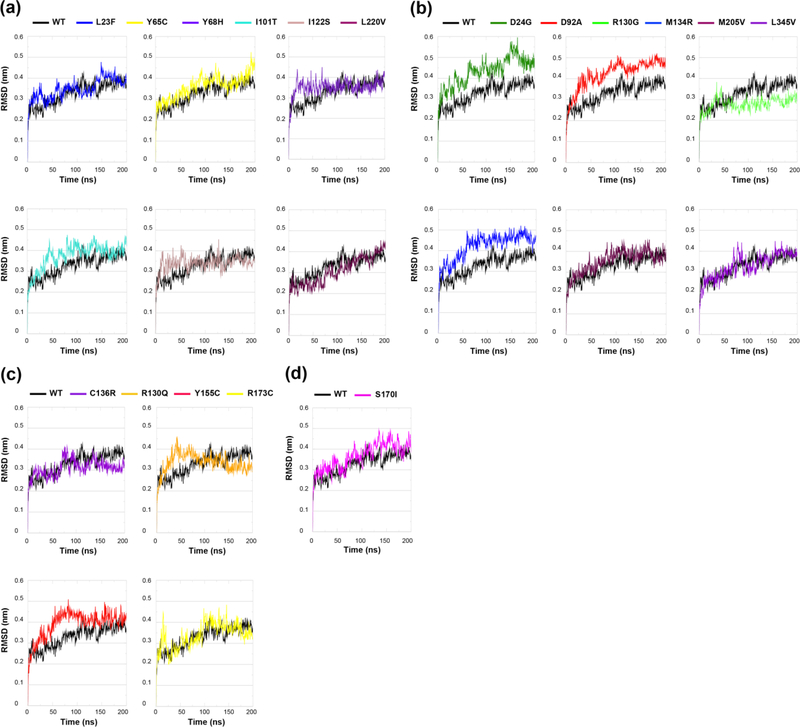
Missense mutations can affect the flexibility of the entire molecule or just a small region, can shift the equilibrium between different conformations, or can affect the entire conformational dynamics and stability of the molecule (Stefl et al., 2013). We analyzed the effects of ASD- vs. cancer-associated germline PTEN mutations where changes in conformational stability were assessed computationally via the measurement of backbone root mean square deviation (RMSD) over the course of each 200 ns MD simulation (Figures 5(a)-(d)). All six ASD-associated mutations equilibrated fairly quickly and remained fairly stable for the entire duration of the simulations, with two mutations (p.Y68H and p.I101T) initially demonstrated a slightly higher RMSD value (0.30–0.40 nm) yet stabilized within a reasonable range compared to WT PTEN (Figure 5(a)). Of the six cancer-associated mutations, three mutations (p.D24G, p.D92A, and p.M134R) had the most backbone deviation compared to WT PTEN and undergo conformational shifts at varying time points that cause relatively high spikes in RMSD (Figure 5(b)). Mutations p.M205V and p.L345V behave very similarly to the WT PTEN structure, though p.L345V demonstrates a slight initial spike in RMSD, but stabilizes immediately thereafter. Lastly, cancer-associated mutation p.R130G had a lower RMSD overall, but fluctuates often indicating that the protein structure has a packed conformation and struggles to stabilize.
Figure 5.
Comparison of backbone RMSD calculated from molecular dynamics (MD) simulation trajectory ensembles in ASD- vs. cancer-associated mutations. Time course of backbone root mean square deviations (RMSD) of (a) ASD (only), (b) cancer (only), (c) mutations shared across both phenotypes, and (d) one mutation with coexisting phenotypes in comparison to WT PTEN. Solid black line is the WT PTEN structure with the colored line representing the structure associated with each individual mutation.
Mutations shared across both ASD and cancer phenotypes (p.C136R, p.R130Q, p.Y155C, and p.R173C) demonstrated drastic changes in RMSD prior to finding a stable conformation, similar to cancer-associated mutations (Figure 5(c)). The p.R173C mutation and the p.S170I mutation associated with both coexisting ASD and cancer phenotypes had the most backbone deviation with an initial high RMSD (Figure 5(d)). Overall, the cancer (only) mutations, the mutations shared across both ASD and cancer phenotypes, as well as the one mutation associated with both coexisting ASD and cancer phenotypes demonstrated the greatest backbone deviations compared to ASD (only) mutations and in relation to WT PTEN. These results show that the conformational variability of the cancer-associated mutations is a direct result of alterations in both internal stability and structural dynamics within the mutant protein.
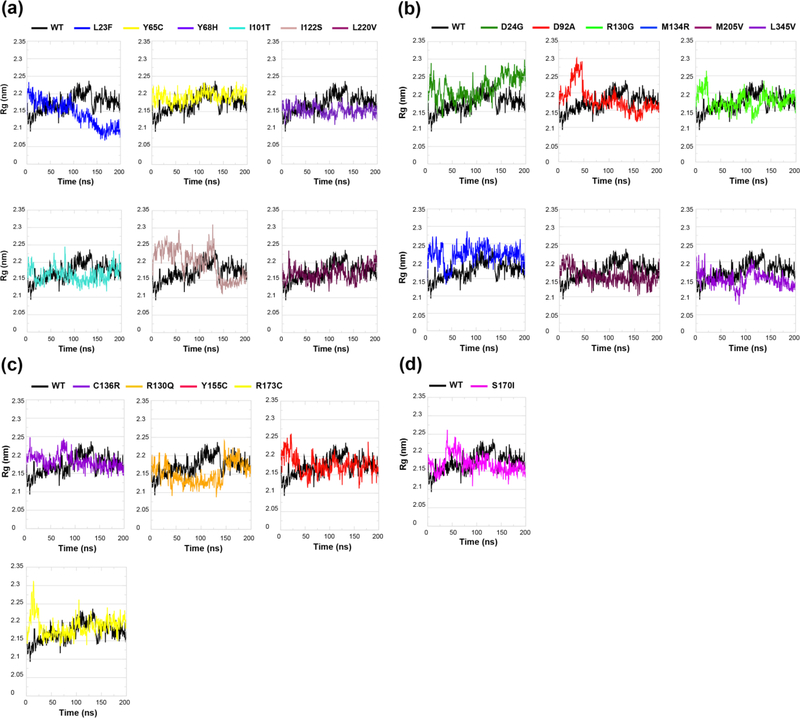
To further determine the effects of each mutation on PTEN folding/unfolding behavior or compactness, we measured the radius of gyration (Rg) of each mutant PTEN structure (Figures 6(a)-(d) and Table 2). A protein structure with a low Rg is more compactly folded, as in a spherical/globular protein, while a protein with a high Rg is more unfolded, as in a disordered protein. MD simulations reveal three ASD-associated mutations (p.L23F, p.Y68H, and p.I101T) had lower Rg values compared to WT PTEN indicating a more folded compact conformation (Figure 6(a)). The p.I122S mutation demonstrated drastic fluctuations in the Rg value throughout the trajectory indicating that it is more inclined to unfold, showing the characteristic of an overall disordered protein structure. The remaining two mutations (p.Y65C and p.L220V) had similar Rg values compared to WT PTEN indicating a fairly stable folded structure. Of the cancer-associated mutations, two mutations (p.D24G and p.M134R) trend toward high Rg values and more unfolded, disordered protein conformations compared to WT PTEN. Moreover, mutations p.D92A and p.R130G initially had higher Rg values indicating a period of unfolding at the start of each simulation (Figure 6(b)). A closer look at their simulation trajectories revealed an unfolding of the N-terminal region at two separate time points. Interestingly, p.M205V and p.L345V mutations had slightly lower Rg values suggesting an overall more compactly folded conformation throughout the duration of each simulation.
Figure 6.
Structural folding/unfolding behavior of PTEN in ASD- vs. cancer-associated mutations. Time course of backbone radius of gyration (Rg) of (a) ASD (only), (b) cancer (only), (c) mutations shared across both phenotypes, and (d) one mutation with coexisting phenotypes in comparison to WT PTEN. Solid black line is the WT PTEN structure with the colored line representing the structure associated with each individual mutation.
Table 2.
Comparison between average geometric properties of ASD- vs. cancer-associated mutations
| Phenotype | Genotype | RMSD (nm) average | Rg (nm) maximum |
|---|---|---|---|
| WT PTEN | - | 0.257 | 2.23 |
| ASD (only) | p.L23F | 0.292 | 2.23 |
| p.Y65C | 0.247 | 2.23 | |
| p.Y68H | 0.219 | 2.18 | |
| p.I101T | 0.266 | 2.24 | |
| p.I122S | 0.278 | 2.31 | |
| p.L220V | 0.277 | 2.21 | |
| Cancer (only) | p.D24G | 0.270 | 2.30 |
| p.D92A | 0.324 | 2.29 | |
| p.R130G | 0.255 | 2.26 | |
| p.M134R | 0.303 | 2.28 | |
| p.M205V | 0.237 | 2.22 | |
| p.L345V | 0.300 | 2.22 | |
| ASD or Cancer (shared) | p.R130Q | 0.291 | 2.20 |
| p.C136R | 0.255 | 2.24 | |
| p.Y155C | 0.256 | 2.26 | |
| p.R173C | 0.302 | 2.31 | |
| ASD and Cancer (coexisting) | p.S170I | 0.266 | 2.26 |
Next, we compared germline mutations shared across both ASD and cancer phenotypes as well as one mutation (p.S170I) with coexisting phenotypes (Figures 6(c)-(d)). Though mutations p.C136R, p.Y155C, p.R173C, and p.S170I initially had much higher Rg values at the start of each simulation, the Rg values later decrease indicating they adopt slightly more compact structures. Mutation p.R130Q exhibits very low Rg values throughout the entire simulation suggesting an overall more compactly folded conformation. Overall, we find that that half of the ASD-associated mutations had more compactly folded structures as demonstrated by their low Rg values, compared to the trend of high Rg values for all but two of the cancer-associated mutations with less stable, more unfolded protein structures. Interestingly, three of the four mutations shared across both ASD and cancer as well as the p.S170I mutation with coexisting phenotypes exhibited high Rg values similar to the cancer-associated mutations, albeit for a short duration, indicating these mutations have a proclivity to unfold thus destabilizing the mutant PTEN structures.
3.7. Conformational changes in catalytic active site residues
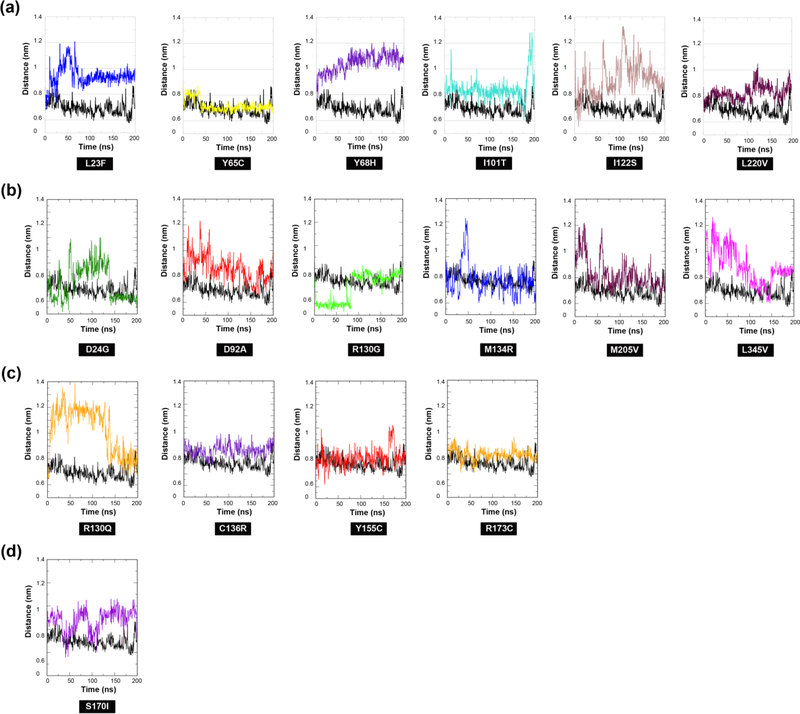
We next examined mutation-induced changes within the active site region of PTEN (Figures 7(a)-(d) and S2(a)-(b)). More specifically, we quantified the distance distribution between backbone Cα atoms of catalytic residues D92 and R130. The positioning of these two residues is essential for their interaction with the PIP3 substrate and the overall function of PTEN (Chia et al., 2010). Five of the six ASD-associated mutations (p.L23F, p.Y68H, p.I101T, p.I122S, and p.L220V) reveal a loss of interaction between D92 and R130 throughout the duration of the simulation as a result of subsequent conformational changes within the active site. A closer inspection of each of the MD simulation trajectories revealed considerable changes involving all three of the P-, WPD-, and TI- active site loops resulting in a more open active-site conformation (Figure S2(a)). Similarly, all six cancer-associated mutations (p.D24G, p.D92A, p.R130G, p.M134R, p.M205V, and p.L345V) exhibited some loss of interaction between D92 and R130, although steric hindrance of both residues dominated throughout the entire duration of MD simulations (Figure 7(b)). Interestingly, mutations p.D24G, p.D92A, and p.L345V exhibited a loss of interaction between D92 and R130 for approximately 40% of the entire simulation. Further inspection of the trajectories for these mutations revealed a dramatic unfolding of the Nterminal region and initial opening of the active site loops that subsequently settle into a closed conformation (Figure S2(b)).
Figure 7.
Distance distribution of catalytic active site loops in ASD- vs. cancer-associated mutations. The distance distribution between backbone Cα atoms of catalytic residues D92 and R130 illustrates occurrence of loss of interaction and/or steric hindrance between the two functionally crucial residues in (a) ASD (only), (b) cancer (only), (c) mutations shared across both phenotypes, and (d) one mutation with coexisting phenotypes in comparison to WT PTEN.
Of the four mutations shared across both ASD and cancer phenotypes, one mutation (p.Y155C) showed some initial steric interaction between D92 and R130 residues; however, for approximately 75% of the entire simulation there is normal interaction, which may allow for some catalytic function to occur (Figure 7(c)). Mutation p.R130Q exhibited a dramatic loss of interaction between residues D92 and R130 throughout the entire simulation. Evaluation of the trajectory revealed a considerable conformation change in the P loop and WPD loop that extends outside the active site (Figure S2(c)). Such drastic changes in the P loop where the R130 mutation resides would alter its intrinsic movement, thus affecting its role in catalytic function. The remaining two mutations (p.C136R and p.R173C) in addition to mutation p.S170I associated with both coexisting ASD and cancer phenotypes, demonstrated considerable loss of interaction between residues D92 and R130 (Figures 7(c)-(d)). However, there is obvious steric hindrance that occurs and further inspection indicates this mutation induces large conformational changes in the WPD- and TI- loops within the active site that may significantly alter the functional role of these residues (Figures S2(c)-(d)). Overall, the majority of the ASD-associated mutations demonstrated a loss of interaction between key catalytic residues D92 and R130 and considerable conformational changes in the loops involving the active site resulting in a more open active-site conformation. While cancer-associated mutations also demonstrated some loss of interaction between residues D92 and R130, similar to ASD-associated mutations, they are set apart by the dominance of steric hindrance throughout the entire simulations as well as dramatic unfolding of the N-terminal region that lead to a more closed active site. These mutations demonstrate distinct opening and closing of active site loops making this region highly unstable. Although the majority of the mutations shared across both phenotypes as well as the one mutation (p.S170I) with coexisting phenotypes reveal steric interaction between residues D92 and R130, it is of a short duration. Of these mutations, p.C136R, p.R173C, and p.S170I revealed a more closed active site similar to cancer-associated mutations.
Evidence from early research suggests that ASD-associated mutations have less severe effects on PTEN structure compared to cancer-associated mutations, which is further indicative of the possibility that these mutations partially retain their function resulting in a mild phenotype (Leslie & Longy, 2016). Moreover, mutations that lead to only partial loss of function of PTEN enzymatic activity seem to be preferentially associated with ASD.(Leslie & Longy, 2016; Spinelli, Black, Berg, Eickholt, & Leslie, 2015) We show here using in silico modeling that the conformational dynamics of ASD-associated mutations lead to a more open active site conformation. Conversely, both the cancer-associated mutations as well as mutations shared across both ASD and cancer phenotypes have significantly altered inter-domain regions and active-site loop conformational dynamics that can lead to a more closed (inactive) active site, which underscores that inactive mutations are linked to more severe PHTS phenotypes (Figures 7(b)-(c) and S2(b)-(c)). Though changes in structural stability do not necessarily equate to altered protein stability or function, given that the cancer-associated mutations mapped to two highly conserved regions within the PTEN structure (Figures 2 and S1), this suggests that these mutations may in fact inactivate the PTEN protein, further demonstrating the inter-domain region plays a key role in the function of PTEN, thus solidifying the importance of PTEN as a superdomain protein.
4. Conclusion
Our combined in silico prediction methods with MD simulation provide the first comprehensive in silico analysis on altered structural stability perturbations of germline PTEN mutations associated with ASD and cancer, potentially aiding in identification of specific mutations that contribute to each phenotype. We show that in comparison to ASD-associated mutations, cancer-associated mutations demonstrated the most damage to PTEN structural features followed by the mutations shared across both phenotypes. Using in-depth analysis on structural stability as well as intrinsic and global dynamics, our results demonstrate distinctive structural and dynamic differences in active site loop conformations and functional distortions of the interdomain region between ASD- and cancer-associated mutations. We show that ASD-associated mutations exhibit “local” destabilization and altered dynamics relative to the surrounding region of the mutation that render a more rigid, compact active site and CBR3 loop; whereas cancer-associated mutations reveal a decrease in “global” structural stability across both domains and increased dynamics across the domain interface of PTEN. Most notably, was the identification of the inter-domain disruption seen in cancer-associated mutations, mutations shared across both phenotypes as well as the one mutation with coexisting phenotypes that lead to the most dramatic effect on structural stability and long-range perturbations within the protein structure. Analysis of the eigenvector projections derived from principal component analysis of the molecular dynamic ensembles shows that the consequences of cancer-associated mutations extend to distant regions of the structure with dramatic effects in long-range inter-residue communication across the entire protein structure. A thorough examination of the large conformational displacements show that altered local dynamics, contribute to the partial opening of the active site as seen in ASD-associated mutations (Figures 7(a) and S2(a)). Conversely, altered global dynamics mediate a closed active site conformation as seen in six cancer-associated mutations (Figures 7(b) and S2(b)). Using structural metrics we find that ASD (only) mutations have a proclivity to adopt a more compact folded protein structure (Figures 5(a) and 6(a)) and cancer (only) as well as mutations shared across both phenotypes demonstrate that the structure is more inclined to unfold (Figures 5(b) and 6(b)).
Overall, the understanding of the effects of specific mutations on PTEN structure acquired through our present study can be utilized to inform future structural studies aimed at generating more robust predictive random forest classifiers and PCA models. Moreover, the results from our study will aid in a better clinical-molecular classification of the resulting phenotypes and potentially allow for translation into new diagnostic and therapeutic approaches. A limitation to our study that should be considered is that the ASD only patient cohort (mean age, 17) is much younger in comparison to the cancer only (mean age, 60) as well as the ASD or cancer (shared) [mean age, 44] cohorts (Table S3). Due to the young age range in ASD only (age range, 10–31) and ASD and cancer (shared) (age range, 8–80) cohorts in the present study, these patients have not had sufficient follow-up for late onset cancer development. However, our three-dimensional modeling of germline PTEN missense mutations in ASD vs. cancer demonstrates distinct structural differences between the two disparate phenotypes suggesting that the phenotypes are “true”. However, it would be interesting to see if specific ASD-associated mutations have subtle structural/dynamic similarities to the cancer-associated ones, and if so, those may be the ones associated with cancer development in the future. We will follow our cohort with interest to confirm or refute the present findings in distinguishing the risk of cancer in patients with ASD. The ability to accurately predict PTEN-ASD can spare patients the need to undergo high-risk cancer surveillance that is now performed in all patients with PHTS. Further computational and structural analyses will be conducted to investigate the influence of ASD- vs. cancer-associated germline PTEN mutations on functional distortions within the inter-domain region, salient inter-residue communication pathways, in facilitating membrane binding, and in substrate binding interaction.
Supplementary Material
Acknowledgements
We thank all the patients and their families whose participation was crucial to making possible this study. This study was funded, in part, by the National Cancer Institute P01CA124570, NINDS U54NS092090, the Breast Cancer Research Foundation, and a grant allocation of computing time from the Ohio Supercomputing Center (PCCF0020) [all to C.E.]. I.N.S. is funded, in part, by the Ambrose Monell Cancer Genomic Medicine Fellowship. C.E. is an American Cancer Society Clinical Research Professor and the Sondra J. and Stephen R. Hardis Endowed Chair of Cancer Genomic Medicine at the Cleveland Clinic.
List of Abbreviations:
- ASD
Autism spectrum disorder
- ATP
Adenosine triphosphate
- ENCoM
Elastic network contact model
- GROMACS
Groningen machine for chemical simulations
- LINCS
Linear constraint solver
- MD
Molecular dynamics
- NMA
Normal mode analysis
- PCA
Principal component analysis
- PDB
Protein data bank
- PHTS
PTEN hamartoma tumor syndrome
- PIP3
Phosphatidylinositol-3,4,5-triphosphate
- PIP2
Phosphatidylinositol-4,5-biphosphate
- PME
Particle mesh Ewald
- poly(Glu-pTyr)
Polymers of glutamate and tyrosine, 1:1 ratio
- PTEN
Phosphatase and tensin homolog deleted on chromosome ten
- Rg
Radius of gyration
- RMSD
Root mean square deviation
- WT
Wild-type
- VMD
Visual Molecular Dynamics
Footnotes
Disclosure statement
The authors declare no relevant conflict of interest.
References
- Ali SK, Sneha P, Priyadharshini Christy J, Zayed H, & George Priya Doss C (2017). Molecular dynamics-based analyses of the structural instability and secondary structure of the fibrinogen gamma chain protein with the D356V mutation. Journal of Biomolecular Structure and Dynmics, 35(12), 2714–2724. doi: 10.1080/07391102.2016.1229634. [DOI] [PubMed] [Google Scholar]
- Anwer K, Sonani R, Madamwar D, Singh P, Khan F, Bisetty K, … Hassan MI (2015). Role of N-terminal residues on folding and stability of C-phycoerythrin: simulation and urea-induced denaturation studies. J Biomol Struct Dyn, 33(1), 121–133. doi: 10.1080/07391102.2013.855144. [DOI] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, & Haak JR (1984). Molecular-Dynamics with Coupling to an External Bath. Journal of Chemical Physics, 81(8), 3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- Berendsen HJCP, Postma JPM, van Gunsteren WF, & Hermans J (1981). Interaction Models for Water in Relation to Protein Hydration, In: Pullman B (eds) “Intermolecular Forces” The Jerusalem Symposia on Quantum Chemistry and Biochemistry (Vol. 14): Springer, Dordrecht. [Google Scholar]
- Butler BM, Gerek ZN, Kumar S, & Ozkan SB (2015). Conformational dynamics of nonsynonymous variants at protein interfaces reveals disease association. Proteins, 83(3), 428–435. doi: 10.1002/prot.24748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, … Eng C (2005). Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. Journal of Medical Genetics, 42(4), 318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, Fariselli P, Rossi I, & Casadio R (2008). A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics, 9 Suppl 2, S6. doi: 10.1186/1471-2105-9-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia JY, Gajewski JE, Xiao Y, Zhu HJ, & Cheng HC (2010). Unique biochemical properties of the protein tyrosine phosphatase activity of PTEN-demonstration of different active site structural requirements for phosphopeptide and phospholipid phosphatase activities of PTEN. Biochimica et Biophysica Acta, 1804(9), 1785–1795. doi: 10.1016/j.bbapap.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Darden T, York D, & Pedersen L (1993). Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. Journal of Chemical Physics, 98(12), 10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- Das S, Dixon JE, & Cho W (2003). Membrane-binding and activation mechanism of PTEN. Proceedings of the National Academy of Sciences U S A, 100(13), 7491–7496. doi: 10.1073/pnas.0932835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C (2000). Will the real Cowden syndrome please stand up: revised diagnostic criteria. Journal of Medical Genetics, 37(11), 828–830. doi: 10.1136/jmg.37.11.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C (2003). PTEN: one gene, many syndromes. Human Mutation, 22(3), 183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- Frappier V, Chartier M, & Najmanovich RJ (2015). ENCoM server: exploring protein conformational space and the effect of mutations on protein function and stability. Nucleic Acids Research, 43(W1), W395–400. doi: 10.1093/nar/gkv343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier V, & Najmanovich RJ (2014). A coarse-grained elastic network atom contact model and its use in the simulation of protein dynamics and the prediction of the effect of mutations. PLoS Computional Biology, 10(4), e1003569. doi: 10.1371/journal.pcbi.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier TW, Embacher R, Tilot AK, Koenig K, Mester J, & Eng C (2015). Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Molecular Psychiatry, 20(9), 1132–1138. doi: 10.1038/mp.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier TW, Youngstrom EA, Speer L, Embacher R, Law P, Constantino J, … Eng C (2012). Validation of proposed DSM-5 criteria for autism spectrum disorder. Journal of the American Academy of Child and Adolescent Psychiatry, 51(1), 28–40 e23. doi: 10.1016/j.jaac.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag CM (2007). The genetics of autistic disorders and its clinical relevance: a review of the literature. Molecular Psychiatry, 12(1), 2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- Georgescu MM (2010). PTEN Tumor Suppressor Network in PI3K-Akt Pathway Control. Genes Cancer, 1(12), 1170–1177. doi: 10.1177/1947601911407325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu MM, Kirsch KH, Kaloudis P, Yang H, Pavletich NP, & Hanafusa H (2000). Stabilization and productive positioning roles of the C2 domain of PTEN tumor suppressor. Cancer Research, 60(24), 7033–7038. [PubMed] [Google Scholar]
- Gicquel JJ, Vabres P, Bonneau D, Mercie M, Handiri L, & Dighiero P (2003). Retinal angioma in a patient with Cowden disease. American Journal of Ophthalmology, 135(3), 400–402. doi: 10.1016/S0002-9394(02)01963-3. [DOI] [PubMed] [Google Scholar]
- Han SY, Kato H, Kato S, Suzuki T, Shibata H, Ishii S, … Ishioka C (2000). Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Research, 60(12), 3147–3151. [PubMed] [Google Scholar]
- Haynie DT, & Xue B (2015). Superdomains in the protein structure hierarchy: The case of PTP-C2. Protein Science, 24(5), 874–882. doi: 10.1002/pro.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Arrotta N, Radhakrishnan D, Wang Y, Romigh T, & Eng C (2013). Cowden syndrome-related mutations in PTEN associate with enhanced proteasome activity. Cancer Research, 73(10), 3029–3040. doi: 10.1158/0008-5472.CAN-12-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, & Fraaije JGEM (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472. doi: . [DOI] [Google Scholar]
- Humphrey W, Dalke A, & Schulten K (1996). VMD: visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38, 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Johnston SB, & Raines RT (2015). Conformational stability and catalytic activity of PTEN variants linked to cancers and autism spectrum disorders. Biochemistry, 54(7), 1576–1582. doi: 10.1021/acs.biochem.5b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalilia M, Chakraborty S, & Popescu M (2011). Predicting disease risks from highly imbalanced data using random forest. BMC Medical Informatics Decision Making, 11, 51. doi: 10.1186/1472-6947-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, & Vihinen M (2010). Performance of protein stability predictors. Human Mutation, 31(6), 675–684. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy N, Gajendrarao P, Olivotto I, & Yacoub M (2017). Impact of diseasecausing mutations on inter-domain interactions in cMyBP-C: a steered molecular dynamics study. Journal of Biomolecular Structure and Dynamics, 35(9), 1916–1922. doi: 10.1080/07391102.2016.1199329. [DOI] [PubMed] [Google Scholar]
- Kumar DT, Doss CGP, Sneha P, Tayubi IA, Siva R, Chakraborty C, & Magesh R (2017). Influence of V54M mutation in giant muscle protein titin: a computational screening and molecular dynamics approach. Journal of Biomolecular Structure and Dynamics, 917–928. doi: 10.1080/07391102.2016.1166456. [DOI] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, … Pavletich NP (1999). Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell, 99(3), 323–334. doi: 10.1016/S0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Leslie NR, & Longy M (2016). Inherited PTEN mutations and the prediction of phenotype. Seminars in Cell & Developmental Biology, 52, 30–38. doi: 10.1016/j.semcdb.2016.01.030. [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, … Parsons R (1997). PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science, 275(5308), 1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Maehama T, & Dixon JE (1998). The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. Journal of Biological Chemistry, 273(22), 13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Maehama T, & Dixon JE (1999). PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends in Cell Biology, 9(4), 125–128. doi: 10.1016/S0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- Maehama T, Taylor GS, & Dixon JE (2001). PTEN and myotubularin: novel phosphoinositide phosphatases. Annual Review of Biochemistry, 70, 247–279. doi: 10.1146/annurev.biochem.70.1.247. [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Coulon V, Lunetta KL, Rocca-Serra P, Dahia PL, Zheng Z, … et al. (1998). Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Human Molecular Genetics, 7(3), 507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Dahia PL, Zheng Z, Liaw D, Parsons R, Gorlin RJ, & Eng C (1997). Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nature Genetics, 16(4), 333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Kum JB, Lunetta KL, Bennett MJ, Gorlin RJ, Ahmed SF, … et al. (1999). PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Human Molecular Genetics, 8(8), 1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- Masso M, & Vaisman II. (2008). Accurate prediction of stability changes in protein mutants by combining machine learning with structure based computational mutagenesis. Bioinformatics, 24(18), 2002–2009. doi: 10.1093/bioinformatics/btn353. [DOI] [PubMed] [Google Scholar]
- Maxwell GL, Risinger JI, Gumbs C, Shaw H, Bentley RC, Barrett JC, … Futreal PA (1998). Mutation of the PTEN tumor suppressor gene in endometrial hyperplasias. Cancer Research, 58(12), 2500–2503. [PubMed] [Google Scholar]
- McBride KL, Varga EA, Pastore MT, Prior TW, Manickam K, Atkin JF, & Herman GE (2010). Confirmation Study of PTEN Mutations Among Individuals with Autism or Developmental Delays/Mental Retardation and Macrocephaly. Autism Research, 3(3), 137–141. doi: 10.1002/aur.132. [DOI] [PubMed] [Google Scholar]
- Mester J, & Eng C (2013). When overgrowth bumps into cancer: the PTEN-opathies. American Journal of Medical Genetics C Seminars in Medical Genetics, 163C(2), 114–121. doi: 10.1002/ajmg.c.31364. [DOI] [PubMed] [Google Scholar]
- Mohajer FS, Parvizpour S, Razmara J, Khoshkhooy Yazdi M, & Shamsir MS (2017). Structural, functional and molecular dynamics analysis of the native and mutated actin to study its effect on congenital myopathy. Journal of Biomolecular Structure and Dynamics, 35(7), 1608–1614. doi: 10.1080/07391102.2016.1190299. [DOI] [PubMed] [Google Scholar]
- Myers MP, Stolarov JP, Eng C, Li J, Wang SI, Wigler MH, … Tonks NK (1997). P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proceedings of the National Academy of Sciences U S A, 94(17), 9052–9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenbrink C, Villa A, Mark AE, & van Gunsteren WF (2004). A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. Journal of Computational Chemistry, 25(13), 1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Escudero I, Oliver MD, Andres-Pons A, Molina M, Cid VJ, & Pulido R (2011). A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Human Molecular Genetics, 20(21), 4132–4142. doi: 10.1093/hmg/ddr337. [DOI] [PubMed] [Google Scholar]
- Rose PW, Prlic A, Altunkaya A, Bi C, Bradley AR, Christie CH, … Burley SK (2017). The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Research, 45(D1), D271–D281. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata S, Matsuda M, Kawanabe A, & Okamura Y (2017). Domain-to-domain coupling in voltage-sensing phosphatase. Biophysics and Physicobiology, 14, 85–97. doi: 10.2142/biophysico.14.0_85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IN, & Briggs JM (2016). Structural mutation analysis of PTEN and its genotype-phenotype correlations in endometriosis and cancer. Proteins, 84(11), 1625–1643. doi: 10.1002/prot.25105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli L, Black FM, Berg JN, Eickholt BJ, & Leslie NR (2015). Functionally distinct groups of inherited PTEN mutations in autism and tumour syndromes. Journal of Medical Genetics, 52(2), 128–134. doi: 10.1136/jmedgenet-2014-102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, … Tavtigian SV (1997). Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nature Genetics, 15(4), 356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- Stefl S, Nishi H, Petukh M, Panchenko AR, & Alexov E (2013). Molecular mechanisms of disease-causing missense mutations. Journal of Molecular Biology, 425(21), 3919–3936. doi: 10.1016/j.jmb.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutthibutpong T, Rattanarojpong T, & Khunrae P (2017). Effects of helix and fingertip mutations on the thermostability of xyn11A investigated by molecular dynamics simulations and enzyme activity assays. Journal of Biomolecular Structure and Dynamics. doi: 10.1080/07391102.2017.1404934. [DOI] [PubMed] [Google Scholar]
- Tamura M, Gu J, Tran H, & Yamada KM (1999). PTEN gene and integrin signaling in cancer. Journal of the National Cancer Institute, 91(21), 1820–1828. doi: 10.1093/jnci/91.21.1820. [DOI] [PubMed] [Google Scholar]
- Tan MH, Mester J, Peterson C, Yang Y, Chen JL, Rybicki LA, … Eng C (2011). A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. American Journal of Human Genetics, 88(1), 42–56. doi: 10.1016/j.ajhg.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa DR, & Kadhirvel S (2017). Molecular dynamics of a far positioned SOD1 mutant V14M reveals pathogenic misfolding behavior. Journal of Biomolecular Structure and Dynamics. doi: 10.1080/07391102.2017.1407675. [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, & Berendsen HJ (2005). GROMACS: fast, flexible, and free. Journal of Computational Chemistry, 26(16), 1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Waite KA, & Eng C (2002). Protean PTEN: form and function. American Journal of Human Genetics, 70(4), 829–844. doi: 10.1086/340026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Senechal K, Neshat MS, Whang YE, & Sawyers CL (1998). The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proceedings of the National Academy of Sciences U S A, 95(26), 15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Yeong Chit Chia J, Gajewski JE, Sio Seng Lio D, Mulhern TD, Zhu HJ, … Cheng HC (2007). PTEN catalysis of phospholipid dephosphorylation reaction follows a two-step mechanism in which the conserved aspartate-92 does not function as the general acid--mechanistic analysis of a familial Cowden disease-associated PTEN mutation. Cell Signal, 19(7), 1434–1445. doi: 10.1016/j.cellsig.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Yang LQ, Sang P, Tao Y, Fu YX, Zhang KQ, Xie YH, & Liu SQ (2014). Protein dynamics and motions in relation to their functions: several case studies and the underlying mechanisms. Journal of Biomolecular Structure and Dynamics, 32(3), 372–393. doi: 10.1080/07391102.2013.770372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZH, & Castello A (2017). Principal components analysis in clinical studies. Annals of Translational Medicine, 5(17). doi: ARTN 351, 10.21037/atm.2017.07.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.