Abstract
Background:
Digital Imaging and Communications in Medicine (DICOM®) is the standard for the representation, storage, and communication of medical images and related information. A DICOM file format and communication protocol for pathology have been defined; however, adoption by vendors and in the field is pending. Here, we implemented the essential aspects of the standard and assessed its capabilities and limitations in a multisite, multivendor healthcare network.
Methods:
We selected relevant DICOM attributes, developed a program that extracts pixel data and pixel-related metadata, integrated patient and specimen-related metadata, populated and encoded DICOM attributes, and stored DICOM files. We generated the files using image data from four vendor-specific image file formats and clinical metadata from two departments with different laboratory information systems. We validated the generated DICOM files using recognized DICOM validation tools and measured encoding, storage, and access efficiency for three image compression methods. Finally, we evaluated storing, querying, and retrieving data over the web using existing DICOM archive software.
Results:
Whole slide image data can be encoded together with relevant patient and specimen-related metadata as DICOM objects. These objects can be accessed efficiently from files or through RESTful web services using existing software implementations. Performance measurements show that the choice of image compression method has a major impact on data access efficiency. For lossy compression, JPEG achieves the fastest compression/decompression rates. For lossless compression, JPEG-LS significantly outperforms JPEG 2000 with respect to data encoding and decoding speed.
Conclusion:
Implementation of DICOM allows efficient access to image data as well as associated metadata. By leveraging a wealth of existing infrastructure solutions, the use of DICOM facilitates enterprise integration and data exchange for digital pathology.
Keywords: Computational pathology, DICOMweb, image compression, slide scanning, whole slide imaging
INTRODUCTION
Tissue-based diagnostics relies heavily on optical (bright field) microscopy. Digital pathology promises advances over this 150-year-old technology.[1] First, scanned slides may be navigated seamlessly on a display at multiple magnifications for diagnosis and research (“virtual microscopy”).[2] Second, different pathologists may remotely review the slides simultaneously in real time (“telepathology”).[3] Third, images may be analyzed by computer algorithms and the resulting quantitative biomarkers can be integrated with clinical data (“computational pathology”).[4,5] Despite these potential benefits, widespread adoption of digital pathology in a clinical setting has not yet been achieved. Regulatory hurdles in the United States can, in part, be held historically accountable;[6] however, the US Federal Drug Administration recently cleared the first whole slide imaging system for marketing[6] based on a recent multisite clinical trial (NCT02699970) that demonstrated noninferiority to optical microscopy across a range of use cases.[7] It is now clear that digitized slides provide an acceptable level of clinical performance when compared with conventional light microscopy.
Whole slide imaging applications extend well beyond interactive viewing on screen. In particular, computer vision and machine learning techniques hold great promise to unlock the potential of digital pathology by extending human capabilities with decision-support tools and automating laborious mechanical tasks.[8,9,10,11,12,13,14,15] These tools require machine-readable data in a standardized format–not only for the image pixel data but also for their annotations and associated descriptive metadata. In the current digital pathology landscape, whole slide imaging systems store image data in proprietary file formats. While these systems allow interactive viewing, the proprietary nature of data formats and interfaces create vendor lock-in and impede data access.[16] Open-source and commercial software solutions to read proprietary formats have been developed.[17,18,19] However, these solutions primarily provide access to image pixel data whereas crucial metadata related to the clinical context (and the acquisition process) remain largely inaccessible. Furthermore, proliferation of competing vendor solutions increases the number of proprietary formats, which represents a barrier for interoperability and maintainability.[20] In other words, there is a compelling need for data standardization in digital pathology to facilitate the clinical integration and to support the computational development streams.[21,22]
Digital Imaging and Communications in Medicine (DICOM) is the globally accepted standard for communication and management of a wide range of medical images and related information.[21,22,23] DICOM further supports encoding, storage, and exchange of image annotations as well as quantitative measurements derived from images.[24] While the standard has been extended to support digital pathology,[25] it has seen little adoption in pathology practice. Specifically, the DICOM standard comprises an extensive set of documents that specify various technical aspects of digital pathology. DICOM addresses primarily information technology experts who have the necessary technical expertise to implement it. In contrast, practicing pathologists without a solid computer science background may not instantly appreciate the value of DICOM data models and communication protocols for their everyday work. This disconnect has resulted in an apparent lack of prioritization of interoperability,[20] and vendors lack a compelling return on investment for building DICOM turn-key solutions.
There are several misperceptions among pathologists about the scope, applicability, and suitability of the DICOM standard. For example, DICOM is often perceived as being only an open file format for storage of image pixel data whereas the metadata integration, communication, and data exchange aspects are often disregarded. Recently, the need to achieve interoperability of whole slide imaging between different systems has been emphasized by the Digital Pathology Association[26] as well as DICOM Working Group 26.[27] These groups (in which many vendors participate) recently met for the first time to evaluate the image data exchange using DICOM representations and protocols.[28] Intellectual property obstacles, which previously hindered implementation by vendors, have been resolved.[28,29] Vendors now generally embrace the standard and agree on implementation details for improved interoperability.[28] These large-scale efforts need to be supplemented by pilot implementations in pathology departments for various practical reasons: (1) the evaluation of capabilities and limitations requires first-hand experience at the user level (especially by content experts in pathology); (2) the demonstration of compatibility with existing clinical systems (e.g., pathology laboratory information systems [LIS], enterprise-wide Picture Archiving and Communication Systems) requires local, laboratory-based proofs of principle; and (3) reliance on external advice and guidelines cannot replace local stakeholder involvement and active definition of resource requirements. Importantly, adoption of the DICOM standard represents an opportunity for pathology to leverage established enterprise medical imaging infrastructure and software solutions. Ultimately, a common data standard will enable convergence between radiology and pathology for multidisciplinary integrated diagnostics.[4,30,31]
Based on the compelling need for data standardization and interoperability in digital pathology, we initiated a cross-departmental prospective quality improvement project to implement the DICOM standard for digital pathology and outline resource requirements for implementation. The solutions presented here empower pathologists to gain an appreciation of and enable the assessment of the appropriateness of the DICOM standard for pathology practice. In addition, we demonstrate that existing software solutions developed for radiology can be reused for pathology through conformance with the DICOM standard.
METHODS
Study sites, ethics approval
Two pathology laboratories and a clinical data science center within the authors’ tertiary healthcare network served as the study sites. The project is part of a prospective and ongoing interdepartmental clinical quality-improvement initiative (institutional checklist, Human Research Committee, version May 25, 2012). Use of deidentified patient samples was performed under institutional review board approval “Feasibility Assessment of a Standard File Format for Digital Pathology” (IRB: 2018P000082); research was performed in accordance with the Declaration of Helsinki.
Study objectives
The primary goal was to assess whether DICOM is a practical format for digital pathology. We generated DICOM files from available pixel and metadata. For pixel data, we used proprietary image files from four different slide-scanning systems (Aperio CS2, Leica Biosystems, Buffalo Grove, IL, USA; Hamamatsu NanoZoomer S60, Hamamatsu Photonics, Boston, MA, USA; Motic EasyScan Pro6, Motic Microscopy, Richmond, British Columbia, Canada; Philips IntelliSite Ultra Fast Scanner, Eindhoven, The Netherlands). For metadata, we extracted the relevant information from two LISs (CoPathPlus™ 6.1MR1, PowerPath 10.0.1.10, Sunquest Information Systems, Tucson, AZ, USA). Secondary goals were: assessment of performance, evaluation of compatibility with existing DICOM software, assessment of pixel compression, pixel load times, evaluation of querying and retrieving data, and tracking of development efforts.
Selection and encoding of DICOM attributes
DICOM defines application-specific representations of images and their metadata in information object definitions (IODs), which are composed of a set of attributes grouped in modules. DICOM defines some attributes as mandatory, those always required to be present or present under specified conditions, and others as optional, which may be included or omitted at the implementer's discretion. For our main routine surgical pathology use case, we chose to implement 114 attributes (93 required or conditionally required, 21 optional) of the Whole Slide Microscopy Image IOD, defined in DICOM PS3.3 IODs.[32] The attributes we selected are listed in Supplementary Table 1 (2.6MB, tif) . The rules common to all applications for encoding, transmitting, and storing these attributes are defined in other parts of DICOM.[33,34,35,36] DICOM also defines an extensive set of controlled terminology for various applications by reference, when possible, to external lexicons, such as the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT).[37] The coded values to use for attributes of the Specimen module, which includes both identifying and preparation descriptive attributes, are defined by DICOM in templates and context groups (value sets) in PS3.16.[38] Templates and context groups represent generic mechanisms to describe the information in coded form and are cataloged by their corresponding template identifiers (TIDs) and context group identifiers (CIDs), respectively. We used the standard template TID 8001 (Specimen Preparation) to code items of the Specimen Preparation Step Content Item Sequence (FHIR) attribute and a variety of standard context groups to code different concepts contained by items. To code the Preparation Step Type concept, we used the coded values from context group CID 8111 (Specimen Preparation Procedure) and to code specific preparation steps, we used the following standard context groups: CID 8109 (Specimen Collection Procedure) for the collection step, CID 8110 (Specimen Sampling Procedure) for the sampling step, CID 8112 (Specimen Stains) for the staining step, and CID 8113 (Specimen Preparation Steps) for the processing step. Fixatives and embedding media can be applicable to one or more of the above steps, and we accounted for this variability by including values from CID 8114 (Specimen Fixatives) and CID 8115 (Specimen Embedding Media) as appropriate.
Selected DICOM attributes for digital pathology
Generation of DICOM files
To create DICOM files, we defined a program that consists of the following steps: (1) extract pixel data and pixel-related metadata from proprietary file formats of different whole slide imaging vendors (see above); (2) obtain patient-and specimen-related identifying and descriptive metadata from the LIS (see above); (3) populate DICOM attributes with the obtained data; (4) encode attributes as DICOM data elements; and (5) store DICOM data sets in files on disk. We implemented this program in Python, using the pydicom Python package (version 1.0.2).[39]
Pixel data and pixel-related metadata were extracted from proprietary image file formats using the open slide-python Python package (version 1.1.1),[18,40,41] which depends on the OpenSlide C library (version 3.4.1).[40,41] The OpenSlide library enables accessing regions of pixel data through a uniform active programming interface (API) that abstracts the specific details and encoding issues of the proprietary pixel data organization and representation. Pixel data were extracted through this interface from the original lossy JPEG compressed images and re-encoded in compressed form to enable comparison of different compression methods. Specifically, we compared JPEG (lossy), JPEG-LS (lossless), and JPEG 2000 (lossless) compression methods. JPEG and JPEG 2000 compressions were performed using the Pillow Python package (version 5.1.0)[42] together with the libjpeg-turbo C library for JPEG (version 1.5.3)[43] and the openjpeg C library for JPEG 2000 (version 2.1.2).[44] The C libraries were installed from source with default compiler flags. A lossy quality factor of 95 was used for JPEG. JPEG-LS compression was performed using the CharPyLS Python package[45] interfacing with the CharLS C++ library.[46]
Patient-and specimen-related metadata were extracted from the LISs. Patient-related information is captured in discrete fields, and its extraction was straightforward. However, extraction of specimen-related information proved difficult due to the variability in capturing this data. For example, anatomic location and certain details of the sampling, fixation, embedding, and staining process are not tracked explicitly and had to be inferred from other data fields or even standard operating procedures stored outside the LIS. For this study, we focused on formalin-fixed, paraffin-embedded, and hematoxylin and eosin -stained surgical pathology specimens. We obtained the values required to populate the corresponding DICOM attributes in the Specimen module by retrieving relevant data fields from the LIS in the form of comma-separated value files (.csv), using a proprietary database API to perform queries using slide identifiers. It was not possible to retrieve these values using a standard API -such as by Health Level Seven (HL7) version two queries or messages- since this is not a functionality available in generic LIS systems. We created dictionaries to map composite LIS data fields to discrete HL7 key-value pairs and to map these values to their corresponding SNOMED CT codes (when such coding was required by DICOM). For input to our DICOM conversion program, the mapped patient-and specimen-related metadata key-value pairs were then stored in JavaScript Object Notation (JSON) documents, structured according to the JSON template of the HL7 Fast Healthcare Interoperability Resources Specimen resource.[47] To speed up DICOM file generation through parallelization, the conversion program was installed and executed on an IBM Platform LSF Linux cluster comprised of HPE blade servers with Intel Xeon processors and RedHat Enterprise Linux operating system, Panasas PanFS parallel network attached file system and 40 Gb/s network.
Validation of DICOM files
It is important to note that the DICOM standard does not specify any testing or validation procedures to assess conformance to the standard. However, software tools have been developed for the validation of DICOM files. We used the dciodvfy tool from the dicom3tools package[48] to validate the generated files in an automated manner. The validation tool checks whether metadata are encoded in conformity to the standard; however, it does not attempt to decode and interpret the pixel data. To ensure that the generated DICOM files fully conform with the latest 2018b edition of the DICOM standard and that compressed pixel data can be decoded and displayed correctly, we further performed an in-depth manual review using a variety of DICOM software libraries including pydicom (version 1.0.2),[39,49] GDCM (version 2.8.6),[50,51] and DCMTK (version v3.6.3).[52]
Performance measurements
To assess the performance of our conversion program, we evaluated the conversion times (in minutes per gigabyte) and relative file sizes by submitting separate jobs for each combination of the compression method and image/metadata file pair to the cluster. We tracked total elapsed execution times using the GNU “time” command[53] and measured file sizes using the Unix “du” command. To assess the performance of DICOM as a file format for pixel data access, we considered the reading of individual frame items (within a DICOM series) as a typical use case; for example, the steps necessary to automate pixel data retrieval for building machine-learning applications. Specifically, we emulated pixel data access using a custom-built Python command line program that: (1) reads and decodes metadata of each DICOM image instance of a given series stored in PS3.10 files on disc, (2) identifies the image instance that contains the pixel data frame item for a given tile position within the image pyramid based on the corresponding DICOM metadata attributes, (3) determines the position of the frame item within the Pixel Data element (using the basic offset table item), (4) reads the frame pixel content into memory, and (5) decompresses the pixel data. To assess pixel data access efficiency, we measured the access times across three DICOM series from different vendor-specific file formats and compared performance between compression algorithms using the pydicom Python package (version 1.0.2)[39,49] as well as the Python interface of the GDCM C++ library (version 2.8.6).[50]
Network storage and retrieval of DICOM data
To enable interchange of images over both local and wide-area networks, DICOM provides various protocols and services for communication. Conventionally, DICOM has defined its own services, messages, and protocols that form the backbone of radiology departments worldwide;[33,54,55] however, the standard has recently been extended with a family of Hypertext Transfer Protocol (HTTP)-based resources and transactions, particularly to facilitate the access by browsers and mobile devices. The family of RESTful resources[56] and transactions specified in DICOM PS3.18, collectively referred to as DICOMweb™, includes storage (STOW-RS), query (QIDO-RS) and retrieval (WADO-RS).[57] We used the open-source DICOM archive DCM4CHEE,[58,59] which exposes DICOMweb RESTful services, as the origin server.[59] To assess appropriate web-based network functionality for storage, query, and retrieval, we implemented DICOMweb user agent (client) interfaces in Python[60,61] and JavaScript.[62]
Effort tracking and data analysis
To estimate resources for DICOM implementation we prospectively tracked project efforts of personnel using Jira.[63] For visualization of results, we used the Pandas Python package (version 0.22.0)[64] and the Plotly Python package (version 2.5.1).[65] For repeated measures, we provide averages ± standard deviation; statistical significance was defined as P ≤ 0.01.
RESULTS
In the following, we first present the DICOM data models and describe how the standard represents multi-scale, tiled whole slide image pixel data together with related clinical metadata, including detailed descriptions of pathology laboratory workflows. Then, we report the generation of DICOM files from existing vendor files and assess encoding, storage and access performance using different lossy and lossless compression methods. We demonstrate the query as well as retrieval of image data over the web using DICOM RESTful services (DICOMweb) and report our implementation efforts.
DICOM enables modeling of image pixel data together with clinical metadata
Clinical diagnosis by light microscopy requires integration of image and clinical metadata. At the very least, a pathologist needs a slide label and a patient identifier to uniquely assign a diagnosis to a given patient (currently captured in the LIS) and part-type (typically referring to an anatomical region). We consider the integration of additional imaging-derived (e.g., annotations of the slide findings) and imaging-related information (e.g., tumor stage, biopsy sampling approach) invaluable. This level of integration will also allow for reuse of the clinical data for training and testing of machine learning applications, as well as automated data queries. To account for various use cases, the DICOM standard provides the syntax and semantics to describe both pixel and metadata by defining two data models: (1) the Model of the Real World and (2) the Information Model.
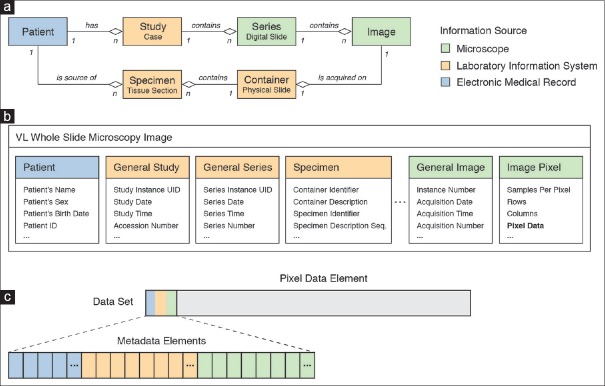
The Model of the Real World defines real-world entities and their relationships [Figure 1a]. The relevant entities for digital pathology are patient, study, series, image, container, and specimen. Importantly, the DICOM model follows the nomenclature of laboratory specimens in HL7,[66] which differs slightly from the terminology commonly used in anatomical pathology practice. For example, a DICOM specimen represents any distinct tissue unit that may be subject to processing in the pathology laboratory (and not necessarily just the surgical specimen). In other words, according to the DICOM nomenclature, parts, blocks, and tissue sections may all represent DICOM specimens (of different types). Likewise, a DICOM container refers to any physical object that holds a specimen such as a jar, a syringe, a cassette, or a glass slide. Specifically, the DICOM container that is relevant in the context of digital pathology is most often the glass slide, which holds the imaged tissue section specimen. A DICOM series typically comprises all the digital images of a tissue section specimen mounted on a single physical glass slide container. A slide tray (paper folder, or multiple folders) with numerous physical slides in the real world would typically map to a DICOM study when digitized. In general, a DICOM study is equivalent to one case with one accession number. It is particularly important to mention the slide as a container because it generally also holds a label (with or without a barcode) for identification of the imaged tissue section specimen [Figure 1a]. There is a great variation between sites (laboratories) as to what information this label includes, ranging from identifiers of the patient, study, specimen, and/or container through human or machine-readable descriptive data such as the patient's name or the stain. In current pathology workflows, different sites and systems variously track studies, containers and/or specimens, depending on local standard operating procedures and naming-conventions. While the DICOM standard contains an informative, nonexhaustive description of various use cases that extend beyond current anatomic pathology practice,[67] the priority for the purpose of managing digital images in the clinic is to track the slide container and link it to other information entities in the Model of the Real World (the patient, study, series, image, or specimen) as necessary.
Figure 1.
Representation of digital pathology information in DICOM. (a) Extension of the DICOM model of the real world for specimens. Diagram showing different DICOM information entities and their hierarchical relationships. Entities are color-coded based on information source. Information about Patient entities (shown in blue) is obtained from the electronic medical record, information about Study, Container, and Specimen entities (shown in orange) is obtained from the laboratory information system, and information about Series and Image entities is provided by the microscope/slide scanning system (shown in green). (b) DICOM information model. The VL Whole Slide Microscopy Image composite information object definition class provides attributes to describe real-world information entities. Related attributes are grouped into modules (visualized as boxes). The Pixel Data attribute, which represents the actual image pixel values, is highlighted in bold. (c) DICOM data set. Attributes of an instance of the VL Whole Slide Microscopy Image class are encoded as data elements. The pixel data element is generally the last element in the dataset (shown in grey). The elements preceding the pixel data element encode metadata that are required for interpretation of the image. These meta data elements are often collectively (but inaccurately) referred to as the “header” (blue, orange, green)
The DICOM Information Model provides a set of IODs to describe the properties of entities of the Model of the Real World. The model uses object-oriented semantics to define IODs (classes) and attributes for a description of real-world entities in the context of distinct imaging modalities. Importantly, IODs do not map to individual entities one to one, but an IOD includes a variety of attributes that hold information about different entities. Specifically, the Visible Light Whole Slide Microscopy Image IOD provides attributes to describe the actual image as well as the related patient, study, series, image, specimen, and container entities [Figure 1b]. For storage and exchange, IODs are instantiated by populating attributes with actual values and are serialized into data sets by encoding each attribute as a separate data element [Figure 1c]. Each data element is identified in the data set by a tag and data elements are ordered in the data set by their tag's numerical value. The encoded dataset represents data in “unnormalized form” in database parlance, which in practice means that all information (including information about the patient, study, series, specimen, and container) is encoded at the level of the image. While this introduces redundancy (the complete information is entirely repeated in every image instance), it assures that each image can still be identified and interpreted safely in an unambiguous and self-describing manner, even when completely separated from any management system.
DICOM allows encoding of multiscale, tiled whole slide images
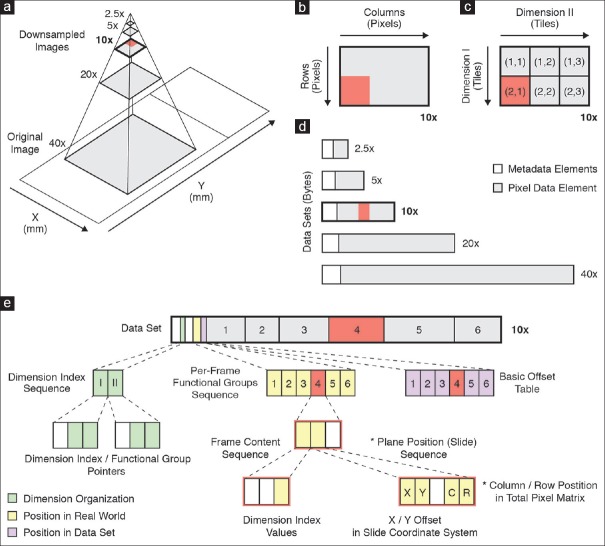
A digital slide is typically represented as a multiresolution image pyramid. The scanned image, for example, acquired at 0.25 or 0.5 μm resolution (corresponding to ×40 or ×20 objectives), forms the base (highest resolution) level of the pyramid. Other magnifications (higher levels of the pyramid) are derived computationally by successive down-sampling [Figure 2a]. In DICOM, unlike most proprietary formats, each resolution level is represented as a separate instance of the VL Whole Slide Microscopy Image IOD and therefore encoded as a separate DICOM data set and stored in a separate DICOM file. The two-dimensional array of pixel values that represents the image is referred to as the total pixel matrix [Figure 2b]. For improved access performance and to circumvent limitations on total frame size, each matrix is tiled into smaller, continuous, rectangular, equally sized pixel regions along the row and column dimension [Figure 2c]. Each tile is compressed and encoded as a separate frame [Figure 2d], each of which is encapsulated as one or more fragments (frame items) within the pixel data element. For example, a digital slide with five magnification levels may be represented in DICOM as a series consisting of five image instances. DICOM provides two options for organizing and encoding the tiled pixel matrix: Full and sparse. Which method is used is indicated by the value of the Dimension Organization Type attribute.
Figure 2.
Encoding of whole slide imaging information in DICOM data set. (a) Schematic of multiresolution whole slide image pyramid. The pyramid base level represents the original image that was acquired by the microscope/slide scanning system. Higher pyramid level images (=lower power) are derived through successive downsampling. The frame of reference for images is the slide coordinate system in millimeter units, where the origin is defined as the lower left corner of the upright standing slide. (b) Total pixel matrix. An image is defined as a continuous, rectangular area of pixels. Shown is the image at ×10 resolution. Note that it is rotated 90° (counterclockwise) relative to the slide coordinate system. (c) Tiled image grid. The pixel matrix may be tiled into smaller rectangular, equally sized regions. Tiles are organized such that the first dimension spans the matrix rows and the second dimension the matrix columns, respectively. (d) Image pyramid encoded as series of VL Whole Slide Microscopy Image instances. Each image instance (=pyramid layer =downsampled magnification) is encoded as a separate DICOM data set. (e) DICOM data set of a multiframe image instance with encapsulated Pixel Data element. Each tile is compressed and encoded as a separate Frame item. Frames are implicitly numbered, based on the order of encoding. The dimensional organization of frames in the real world is described by the dimension index sequence attribute (green), which contains a dimension index pointer and a functional group pointer attribute for each dimension. The values of these attributes point to other attributes in the per-frame functional groups sequence attribute (yellow), which encode the actual values for each frame. The frame content sequence attribute hereby describes the relative position of each frame in the tiled image grid whereas the plane position (slide) sequence attribute describes the absolute position of each frame in the slide coordinate system as well as the total pixel matrix. The byte offset to individual frame items in the pixel data element (grey box labeled 1-6) is specified by the basic offset table item (purple), which is itself part of (the first item of) the encapsulated pixel data element
In the case of full organization, a frame must exist for every tile of the rectangular total pixel matrix; the order in which the tiles are encoded in the pixel data element is predictable. Specifically, for two-dimensional images, the order is such that frames are encoded first along the row direction and then along the column direction of the pixel matrix. Since the position of frames is defined implicitly, the explicit tile coordinates may be omitted. A recipient may need to re-compute them based on column and row dimensions of the tiles and the total pixel matrix. In the case of sparse organization, tile coordinates and position are required to be explicitly recorded for each tile, not all tiles need to be present, and the frame items can be encoded in the pixel data element in any order. The position of each frame item is encoded in the Per-frame Functional Groups Sequence attribute [Figure 2e]. The full organization was recently added to DICOM as a result of preliminary implementation experience that suggested improvement was needed for the most common use case due to the size of the Per-frame Functional Groups Sequence.[68] We chose to implement the full encoding scheme because it improves access performance by reducing overhead when transmitting metadata over the network as well as loading and parsing it into memory.[28] The DICOM standard thereby enables encoding of multiscale, tiled whole-slide images in a standardized fashion.
DICOM enables encoding of pathology laboratory workflow metadata
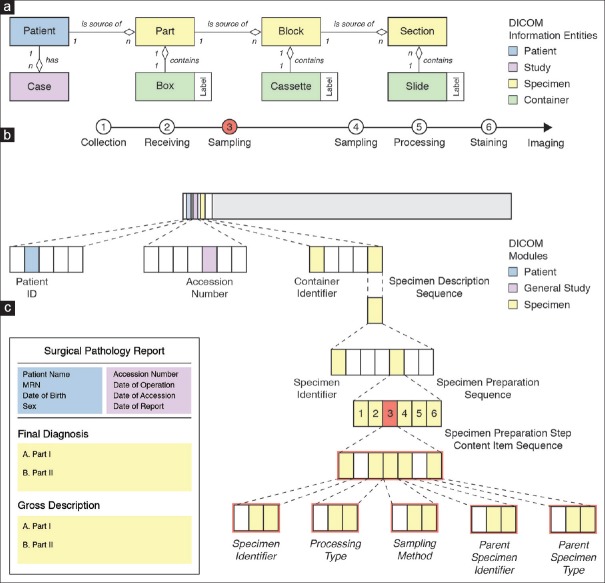
Documentation of clinical metadata in pathology is complicated by the fact that the imaging subject is not the patient itself, but tissues obtained from the patient. Before digitization, tissues undergo several preparation steps including one or more sampling steps [Figure 3a]. At first glance, this may appear trivial to pathologists because they receive, sample, subsample, process, and stain hundreds of specimens per day. However, capturing and describing this information digitally in a standardized format is by no means trivial, regardless of whether optical or digital microscopy is used, because workflows differ greatly between laboratories and use cases. Here, we applied the terms part, block, and section to refer to different steps in a typical surgical pathology workflow [Figure 3a]. It is important to note, however, that DICOM does not provide designated attributes for these concepts. Instead, it provides a generic mechanism that enables encoding a variety of different laboratory workflows in a general and flexible manner. For example, based on the DICOM Specimen Preparation template, we defined a set of five preparation steps that apply to the vast majority of our routine surgical pathology use cases: Collection (e.g., surgical procedure), receiving, sampling (e.g., grossing or sectioning), processing (e.g., decalcification), and staining (e.g., trichrome stain) [Figure 3a]. The fixative (e.g., formalin) and embedding medium (e.g., paraffin wax) can be specified as part of the appropriate preparation step. This allows, for example, distinguishing specimens received fresh for frozen sectioning (original frozen section) from subsequent sections after fixation and from those received in formalin. Attributes for a description of patient and study (in a typical workflow, assigned during accessioning of the case) are part of the Patient General Study and Patient Study modules. Attributes for description of containers (e.g., glass slides) as well as specimens (e.g., tissue sections) are part of the Specimen module [Figure 3b]. DICOM requires that preparation steps be coded using a defined coding scheme, such as SNOMED CT, to permit the standardized interpretation of specimen-related information beyond the confines of one department and one LIS. Thereby, DICOM enables the encoding of relevant metadata together with the pixel data.
Figure 3.
Encoding of specimen information in DICOM data set. (a) DICOM model of the real world for pathology laboratory workflow. Top: Hierarchical relationships between the patient (blue), study =case (purple), specimen (yellow), and container (green) information entities. Bottom: Sequence of specimen preparation steps (1-6) for a typical surgical pathology encounter. (b) DICOM data set of image instance with encoded laboratory information. Patient and Study entities are described by attributes of the Patient module (blue) and general study and patient study modules (purple), respectively. Specimen and container entities are described by attributes of the Specimen module (yellow). The container used for imaging (glass slide) is described at the root dataset level whereas imaged specimens (tissue sections) are described by the nested specimen description sequence attribute. Laboratory procedures that were performed to prepare a specimen for image acquisition are described by the specimen preparation sequence attribute, which includes a specimen preparation step content item sequence attribute for each performed procedure. The description of a content item is based on the Specimen Preparation template, which provides concepts to encode a procedure as name-value pairs using a specified coding scheme, such as SNOMED CT. Mandatory concepts are the specimen identifier, which describes the identifier of the processed specimen, and processing type, which describes the kind of procedure that was performed. Additional concepts may be included depending on the value of the Processing Type concept. In case of a sampling preparation step (highlighted in red , compare sequence step 3 in a), Sampling method, Parent Specimen Identifier and Parent Specimen Type concepts are included. (c) Schematic of a current pathology report. Top: Identifiable information about patient (blue) and accession (purple), which map to attributes of the patient and general study module. Bottom: Final diagnosis and gross description for each specimen of type “part.” Identifiable information about specimen of type “section” is often not included in the report (i.e., final diagnoses generally lack complete slide-level annotations). SNOMED CT: Systematized Nomenclature of Medicine Clinical Terms
The level of detail described by DICOM attributes of the Specimen module is generally not reflected in the pathology report: Specifically, a surgical pathology report contains sufficient information to uniquely identify the patient (name, sex, date of birth, medical record number, etc.) and the study (accession number, accession date, etc.) and it provides a gross (and sometimes a histological) description of the parts [Figure 3c]. When viewing the images to render a diagnosis and to create the report, DICOM metadata are crucial to the display software for visualizing the images correctly and providing related information to the pathologist who needs to interpret the image in the appropriate clinical context. The identifiers for the glass slide (container) and the tissue section (specimen) are generally not explicitly provided in current pathology reports, which makes it difficult to map the free text description in the report to the corresponding digital image and the spatial position within the image. In the future, there is considerable opportunity for improvement of such reports, particularly when they are encoded as synoptic (structured) or multimedia reports, at which time inclusion of links and hyperlinks to individual slides, images, and annotations become feasible in an integrated digital imaging environment. The level of detail in the DICOM attributes is also critically important for machine learning workflows, whether it be for the simple task of selecting the appropriate images to route for digital processing, or for more complex parameterization of algorithm behavior. Regardless of the downstream use case, implementing DICOM effectively requires integrating relevant data across different information sources into one data model [Figure 3c].
DICOM data sets can be generated from existing vendor files
Only very recently have commercially available whole slide imaging systems started to produce images encoded according to the DICOM standard.[28,69] In the interim, to support the installed based of scanners – only capable of producing proprietary format images, commercial and open-source implementers have begun to develop conversion tools.[70,71] To evaluate the capabilities and limitations of DICOM for the representation of digital pathology whole slide images, we implemented a process to generate DICOM PS3.10 files based on existing vendor-specific file formats and validated their integrity and standard conformance using established automated validation tools as well as manual expert review as described in the Methods section.
DICOM provides efficient lossy and lossless compression methods
Whole slide imaging data sets are generally relatively large (GB range for individual slides) compared to other medical imaging applications and are therefore usually stored in compressed form. DICOM supports three image compression schemes relevant for visible light microscopy images: JPEG, JPEG-LS, and JPEG 2000.[16] Most systems routinely apply a baseline lossy JPEG compression algorithm, before storing the images in their proprietary format. This allows for reasonable file sizes suitable for rapid access balanced against sufficient image quality for most current diagnostic tasks. Some systems can be configured to allow the user to select alternative compression schemes, including lossy forms of JPEG 2000 and other proprietary formats.[28]
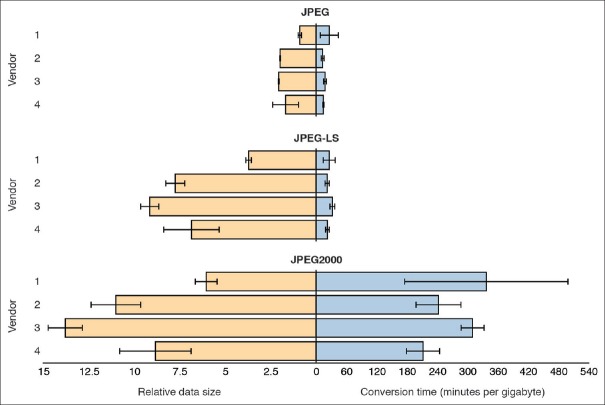
We measured the influence of all three compression schemes supported by DICOM on overall relative data size [Figure 4, left x-axis] and encoding times for VL Whole Slide Microscopy Image instances [Figure 4, right x-axis] generated from original lossy compressed vendor images. Recompressing previously lossy compressed images potentially biases performance measurements; repeated lossy compression also degrades image quality and should not be used in production. Recompression can be avoided by copying the binary pixel data “blobs” directly from original files into DICOM data sets without decoding and re-encoding. However, the recompression approach was necessary for our comparison of different methods since original uncompressed (or lossless compressed images) were not available. The resulting data size after DICOM conversion depends on many parameters including tile size, number of resolution levels, the compression method and in the case of lossy compression, the quality factor or target bitrate. When the same compression method, tile size and number of resolution levels are chosen for DICOM encoding or when the JPEG blocks are simply moved from the TIFF container to the DICOM datasets, the resulting data size will be almost identical [Figure 4, left axis, top]. The conversion time is primarily a factor of the data size and the compression method. As expected, the lossy baseline JPEG method has the overall smallest data size and lossless JPEG-LS and JPEG 2000 compression results in larger files [Figure 4, left x-axis]. Notably, the JPEG-LS method compresses images more efficiently when compared to JPEG 2000 (6.85 ± 2.17 versus 9.90 ± 3.14 min/GB; P < 0.000001, t-test). The baseline JPEG method also has the shortest encoding times [Figure 4, right x-axis]. Interestingly, JPEG-LS encoding is almost as fast as baseline JPEG (37.03 ± 19.86 min/GB versus 26.58 ± 19.20 min/GB; P > 0.01, t-test) and orders of magnitude faster than JPEG 2000 (37.03 ± 19.86 min/GB versus 405.56 ± 239.52 min/GB; P < 10−17, t-test) [Figure 4]. The observed performance gains with JPEG-LS over JPEG 2000 are not surprising given the low complexity of the underlying algorithm and are consistent with the performance of the same schemes on digital photographs.[72]
Figure 4.
Measurements for encoding and storing DICOM data sets in PS3.10 files. Relative file sizes and encoding times for different vendor-specific formats and image compression methods. A set of 16 vendor-specific whole slide image files (4 files per vendor) were converted to DICOM files using lossy (JPEG with quality set to 95) and lossless (JPEG-LS and JPEG 2000) image compression methods. Left: Size of generated DICOM files with respect to the size of the corresponding original image files. The lossy JPEG size is shown to exemplify the difference between lossy and lossless schemes. Right: Program execution times for reading original files, encoding the contained information as DICOM data sets and writing generated data sets to DICOM files on disk. Execution times were normalized with respect to original file sizes for comparison between digital slides. Shown are averages (bars) and standard deviation (error bars) of measurements
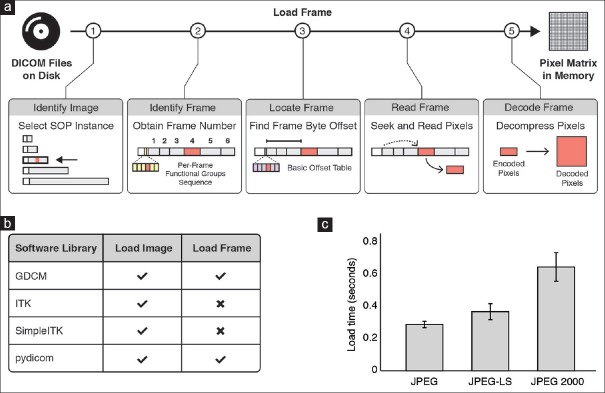
DICOM enables efficient frame-level data access from local files
Most digital pathology applications require efficient access to subsets of pixel data without having to load the entire data set into memory. We evaluated existing, actively maintained software libraries for retrieving individual frames from the DICOM PS3.10 files that we previously generated. As a use case, we considered a machine-learning program that requires the loading of specific frames from files on disk into memory [Figure 5a]. Specifically, in contrast to reading all pixel data into memory, our program reads and interprets the image metadata to locate and to load only relevant subsets of the pixel data. Since many machine-learning algorithms are often orchestrated using the Python programming language, we explored software libraries that provide Python interfaces for reading of metadata as well as pixel data of VL Whole Slide Microscopy Image SOP instances stored in DICOM files. We identified the four libraries, which support loading of JPEG, JPEG-LS, and JPEG 2000 compressed multiframe DICOM images in encapsulated format: GDCM,[50] ITK,[73] SimpleITK,[74] and pydicom[39,49] [Figure 5b]. However, only two of the four libraries [Figure 5b] expose interfaces for fetching individual frames without loading the entire pixel data element into memory. We measured the time to retrieve an individual frame for a given position in the slide coordinate system and resolution level using the pydicom library and compared retrieval times between compression methods [Figure 5c]. Our results show that JPEG (average: 0.29 ± 0.02s; P < 0.0001, t-test) and JPEG-LS (average: 0.37 ± 0.05s; P < 0.0002, t-test) allow significantly faster pixel data access when compared to JPEG 2000 (average: 0.65 ± 0.09s). We consider these observations good evidence that DICOM enables efficient retrieval of pixel data from files (e.g., for machine learning applications).
Figure 5.
Retrieving pixel data from DICOM data sets stored in PS3.10 files on disk. (a) Sequence of steps required for loading an individual frame of a multiframe DICOM image from a file on disk into a pixel matrix in memory. First, the file that contains the frame of interest is identified by reading and interpreting metadata (“DICOM header”) of each file. Second, the relative position of the frame within the image is identified using the available metadata. Third, the absolute position of the frame is determined within the encapsulated pixel data element. Fourth, the binary content of the frame item is read into memory. Lastly, the pixel data are decompressed. (b) Software library support for reading images from DICOM files. Selected software libraries available for the Python programming language and their level of support for reading multiframe DICOM images. Only a subset of libraries provides the functionality (check icon) for selectively loading an individual frame into memory. (c) Frame access efficiency for different image compression methods. Shown is the average time (±standard deviation) it takes to identify, read, and decompress an individual frame of a multi-frame DICOM image using an implementation of the algorithm (shown in a) in the pydicom software library
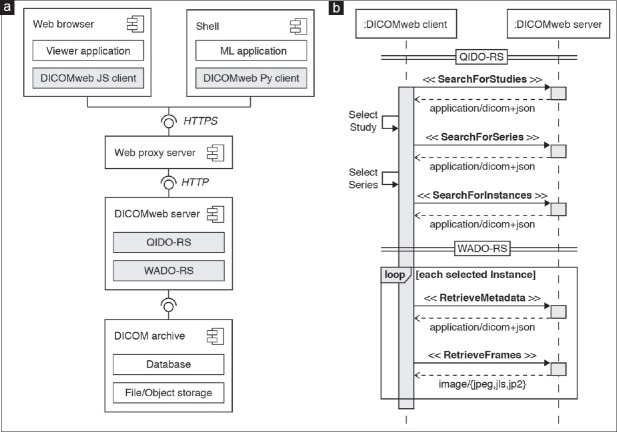
DICOMweb facilitates remote frame-level data access
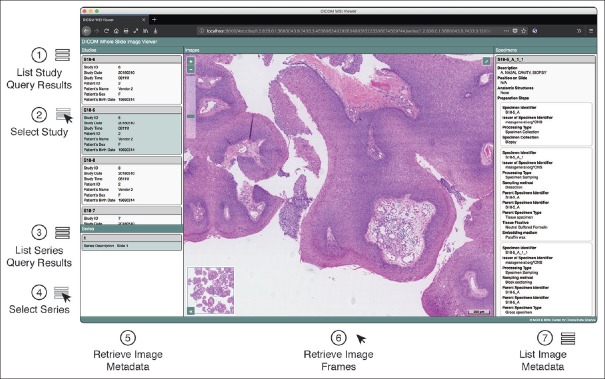
Working directly with files stored on local disk is impracticable in a clinical setting and DICOM enables exchange of image data between devices over network. In the past, accessing data in DICOM format over network required specialized client programs that implemented the DICOM networking protocol.[54] Recently, RESTful web services have been made available for storage, query, and retrieval over HTTP protocol.[75] We decided to use DICOM PS3.18 RESTful web services (DICOMweb™) and investigated the compatibility of VL Whole Slide Microscopy Image instances with existing DICOM archives [Figure 6a]. A key feature of DICOMweb is that it allows efficient access to subsets of server-side DICOM data sets from thin clients, such as smartphones, and tablet computers. Specifically, clients can request representations of DICOM objects in web-friendly formats, such as JSON and JPEG from a DICOMweb server using the HTTP protocol. Functionally, this service-oriented architecture allows clients to first search for objects in the archive and retrieve only relevant subsets of the data [Figure 6b]. The approach is particularly useful for whole slide imaging because it avoids the unnecessary transfer of large amounts of data over the network when only a small fraction of the pixel data is required, as is usually the case for virtual microscopy. We created DICOMweb client implementations in JavaScript[76] and Python[59] to facilitate the programmatic access to remote DICOM objects for visualization and machine learning applications. For visualization, we built a browser-based viewer for interactive display of whole slide images in DICOM format,[77] which uses our JavaScript DICOMweb client implementation to search and retrieve DICOM objects from an archive over the web [Figure 7]. The viewer provides users an intuitive graphical interface for selection of available DICOM studies and series, interactive multiscale viewing of pixel data, and inspection of specimen-related metadata. By allowing pathologists to review the images next to relevant clinical information in the same user interface, our implementation demonstrates a key feature of DICOM, the ability to interpret pixel data and metadata in context. For machine learning, we established a computational workflow that leverages our Python DICOMweb client implementation to dynamically retrieve frames from a DICOMweb server to feed the pixel data into a machine learning-based image analysis algorithm.
Figure 6.
Querying and retrieving DICOM data sets from an archive through DICOMweb RESTful services. (a) Component diagram of DICOMweb client and server components as well as their respective interfaces. Client and server communicate over network using the HTTP. The client may be an interactive viewer application running in a web browser or a machine learning (ML) program running in a Unix shell. The server provides RESTful services for query and retrieval of DICOM objects: Query based on ID for DICOM Objects (QIDO-RS) and Web Access to DICOM Objects (WADO-RS). (b) Sequence diagram of DICOMweb client-server interactions for query and retrieval of DICOM objects. The client searches for studies (cases) or series (digital slides) resources via a SearchForStudies or SearchForSeries request, respectively. The client may provide query parameters to filter DICOM objects based on given attribute values. The server responds with resource representations for each matched object in JSON format according to the DICOM JSON model. The client retrieves image metadata resources from the server for each image (resolution level) belonging to the matched study or series through a RetrieveMetadata request. The server responds with metadata resource representations in DICOM JSON format for each image instance, which provide the client the necessary information to interpret the images and identify relevant frames (e.g., based on their position in the total pixel matrix or the slide coordinate system). The client requests a subset of frames from the server through a RetrieveFrames request. The server responds with a message containing the requested frames in the requested image format (e.g., JPEG, JPEG-LS, JPEG2000). JSON: JavaScript Object Notation, HTTP: Hypertext Transfer Protocol, library
Figure 7.
Interactive DICOMweb viewer for whole slide images and related metadata. Screenshot of browser-based graphical user interface for display of DICOM VL Whole Slide Microscopy Image instances. The viewer represents a single-page application that uses the DICOMweb JavaScript client implementation to query and retrieve DICOM objects from an archive using RESTful web services. The viewer searches for studies using QIDO-RS and displays the list of matched study resources (1). Each list item shows relevant metadata (e.g., patient name, medical record number, accession number). Users can select a study by clicking on a list item (2), which triggers a search for all series that belong to the selected study (3). When the user selects a series, i.e., digital slide (4), the viewer retrieves the metadata for all images that belong to the series using WADO-RS and interprets the metadata to determine the image pyramid structure (5). The viewer then automatically retrieves the frames for a navigated resolution image using WADO-RS and displays them to the user (6). When the user navigates through the image pyramid, the viewer automatically retrieves the frames for the given slide coordinate position using WADO-RS. In addition, the viewer displays the image-level metadata (7). Standardized metadata enable interactive query and retrieval of images for visualization, human interpretation and data validation
DICOM standard can be implemented with reasonable efforts
To obtain a real-world estimate of the time, skills, and budget necessary to implement DICOM for digital pathology, we prospectively tracked our implementation efforts. At time of submission of the manuscript, our efforts encompassed 120 h of documentation review, 273 h of software/algorithm development (including code review, writing documentation and developing tests), and 86 h of consultation with content experts. These numbers will equate to 0.8 full-time equivalents (FTE) for a 120-day period or 0.4 FTE over 1 year if one assumes that person can focus 100% on the DICOM implementation. With respect to the required computational expertise, in our case, the lead computational physician-scientist (MDH) had 4 years of in-depth coding experience in image applications. A project budget estimate accounting for the personnel salary USD $95,687 (expressed as fraction of average salary from publicly available resources for the FTE fraction described), plus computational resources (USD $28,434) amounts to roughly $125,000. Since we had direct access to DICOM experts and received support from the open-source DICOM community, the above estimates may differ in other settings. We provide these data as a reasonable point of reference for similar efforts.
DISCUSSION
We implemented a pilot of the DICOM standard for whole slide imaging for digital pathology in a multisite, multivendor healthcare network setting. Since a reference implementation is currently lacking, we generated valid DICOM files by combining pixel data from vendor-specific file formats with clinical metadata from LISs. We used the generated data sets to test storage as well as access performance and evaluated overall practicability, placing an emphasis on compatibility with existing software libraries, and available archives.
The complexity of the DICOM standard is initially daunting and its adoption poses several challenges. A common misperception among pathologists is that DICOM is merely yet another file format, and there is uncertainty about what benefits and drawbacks adoption of the standard would entail. We hope we have convinced the reader that, although the standard does include a file format, its scope is much broader than the typical proprietary file format and includes standardized communication of image data and a rich and extensible model describing related identifying and descriptive information between devices and over network. Another misperception is that the standard is static and unchangeable. DICOM has proven to be a living standard that has been continuously adapting to technological change. Most notably, DICOMweb services and the DICOM JSON model have significantly improved data accessibility over the web. One can now understand DICOM as a set of URI templates for RESTful API endpoints and standardized JSON schemas for the corresponding resources. Crucially, these are defined in a manner consistent with the use of DICOM for all other imaging specialties. Our results demonstrate that once implemented, DICOM enables data access and exchange in a practical and vendor-neutral manner. We further show that, by relying on DICOM, one can leverage available medical imaging infrastructure and software systems to store, search, and retrieve whole slide imaging data efficiently.
Despite the obvious advantages of DICOM with respect to interoperability and enterprise integration,[16,21,22] challenges remain. In the following, we discuss some key open questions that need to be addressed.
First, large whole slide imaging data sets complicate storage and network transmission. DICOM was originally designed for radiology image data sets more than a quarter of a century ago.[78] At the time, data sets were orders of magnitude smaller than whole slide images in pathology. The total length of the compressed Pixel Data element itself is theoretically unlimited. The number of frames (tiles) is somewhat arbitrarily limited to 4 gigabytes; however, this is generally more than sufficient. An optional Basic Offset Table allows the sender to encode an index of the physical byte offsets to the positions of individual frames within the Pixel Data element. In other words, the receiver does not have to build its own index of the frames. However, currently, only 32-bit integers are used in the Basic Offset Table, which means that images larger than ~4 gigabytes have to omit it. There is no need to include the Basic Offset Table for a server since it can easily build its own index to satisfy frame-level retrieval requests; however, it is a significant convenience when attempting to retrieve selected frames from a PS3.10 file stored on disk. Given the typical size of pixel data in pathology, this size limit can be easily exceeded, in particular when lossless rather than lossy compression methods are applied [Figure 4, left axis]. A simple extension of the DICOM standard is in progress that addresses this Basic Offset Table weakness and will allow for an Extended Offset Table with 64-bit pointers as well as length information.[79] That said, the information will remain optional and recipients will need to be prepared to rebuild their own index if necessary.
Concatenations are a DICOM feature that provides a workaround for size-related issues. Specifically, concatenations allow splitting a multiframe DICOM data set into several smaller parts. For example, with a pixel size threshold at 100MB, the number of concatenation parts in our series ranged from 11 to 46. Concatenations offer several other advantages, such as storing large DICOM objects over the web. Unfortunately, existing DICOM archives don’t interpret concatenations they receive as a single image instance for the purpose of query and frame-level retrieval. Since each concatenation part still represents a valid DICOM object, archives can handle storage and retrieval of instances that are parts of concatenations; however, this places the burden on client implementations, which need to understand concatenations to retrieve the appropriate frames. A new generation of archive implementations that understand and reassemble concatenations would be highly desirable.
Second, vendors currently do not store files in DICOM format. Converting images from existing vendor proprietary file formats introduces overhead, requires constant maintenance to track variations as well as generate updates for each new model/vendor, and important information about the image acquisition process may be lost. For example, the orientation of images and the absolute position of tiles relative to the slide-based coordinate system are generally not specified in proprietary file formats and need to be assumed. Obviously whole slide scanning systems should store data directly in DICOM format and do so efficiently such that DICOM generation does not become a rate-limiting and error-prone step. We embrace ongoing efforts by whole slide scanner vendors to implement DICOM,[69] and we anticipate that adoption of DICOM will advance digital pathology by providing standardized access to pixel data together with clinical metadata.
Third, choosing the optimal image compression method represents a challenge. Our comparison of the three image compression methods currently supported by DICOM highlights the importance of the choice of compression algorithm for data encoding and decoding performance. Currently, vendors store images in lossy compressed format, sometimes using proprietary compression schemes. Lossless compression would maximize image quality and circumvent potential issues related to recompression; however, its benefits may not justify the considerably larger data size. Our performance measurements indicate that JPEG-LS is considerably faster than JPEG 2000 for lossless compression and decompression, at speeds comparable to lossy, baseline JPEG. Since potential consequences of lossy image compression for machine learning applications are yet unexplored, JPEG-LS may present a potential solution for storing and accessing images efficiently without losing information. However, there is limited support for JPEG-LS in existing enterprise-level or radiology DICOM archives and the appropriate color space transformation to use for lossless schemes like JPEG-LS remains an open question.
Fourth, we emphasize the importance of integrating imaging and information systems. Digital pathology involves more than digitizing slides. It requires the integration of image pixel data with clinical metadata to be useful in pathology practice. Current anatomic pathology LISs are generally designed around graphical user interfaces and optimized toward reporting and billing rather than digital pathology workflows. As a result, these systems often do not expose an application programming interface (API) for programmatic communication of image-related information between systems. Creation of custom middleware represents a solution;[80,81] however, integration with imaging systems remains challenging because relevant details may not even be tracked by the information system in structured form. In particular, information about specimen preparation steps is generally not readily available. Interpretation of acquired images in clinical context demands substantially different approaches to tracking specimen-related information, which are less case-centric and place more emphasis on the imaged tissue sections. There are various alternative standards routinely used for communicating information from departmental information systems; to DICOM acquisition devices and clinical laboratory devices, specifically DICOM Modality Worklist (MWL) and various HL7 version 2 messages. Existing profiles for anatomical pathology and laboratory workflows[82] are under investigation by the Integrating the Healthcare Enterprise (IHE)[83] Pathology and Laboratory Medicine (PaLM) group[84] in collaboration with DICOM Working Group 26,[27] for whom standardization of the LIS to scanner interface is a high priority. However, mapping proprietary LIS internal data structures to standard HL7 specimen representations or the DICOM Specimen Preparation template is nontrivial and currently may require customization of the HL7 message triggers, segments, and fields (including private segments) with the vendor, or use of third-party middleware solutions for extraction and transformation of the data. The IHE PaLM group has recognized that the underlying HL7 V2 messages and Domain Analysis Model (DAM) for histopathology and slide specimen description have gaps and need mapping to the model that was defined in DICOM Supplements 122 and 145, and are in the process of submitting a Project Scope Statement from the Orders and Observations Work Group to fill these gaps.[85] The earlier effort by the predecessor of IHE PaLM, IHE Anatomical Pathology, which defined a DICOM MWL-based Anatomical Pathology Workflow profile for communication between anatomic pathology AP-LIS and scanners, has not been adopted. We hope that the next generation of LIS will implement any new HL7 V2 messages and future IHE PaLM technical framework(s) to provide standard-compliant interfaces for data query and retrieval. In the interim, it would be desirable if LIS would use customized HL7 messages and fields to allow automated query and retrieval of specimen information, rather than relying on completely proprietary mechanisms.
Fifth, the operational challenges of incorporating DICOM for digital pathology should not be underestimated. One major difference between implementing whole slide imaging and other technologies into routine practice is the need for expert IT involvement (to establish the necessary IT infrastructure, storage, and personnel). Regardless of the selected storage format, any implementation of digital pathology requires storage of vast amounts of data.[16] If the same compression scheme and parameters are used, a DICOM file containing a multiframe image with encapsulated pixel data element is similar in total size to a TIFF or proprietary format file. The DICOM file serves as a container for compressed pixel data blobs (frame items) and the additional metadata is minimal in size by comparison. Some optimizations may be lost (such as factoring out JPEG tables and only sending them once, which TIFF allows for but DICOM does not); however, these have minimal effect. There are exceptions though, for example, significant size expansion may occur when images are lossy compressed with a proprietary scheme by the scanner vendor and can only be recompressed without further loss using a standard lossless scheme. Accordingly, scanner vendors should be strongly discouraged from using proprietary schemes and only use standard schemes supported by DICOM such as JPEG, JPEG-LS and JPEG 2000. Thus, there is normally no storage size penalty for using DICOM.
The storage requirements remain enormous. For example, the lead authors’ clinical facilities are characterized by ~185,000 cases/year with a slide output of 3800–5800/day, which equates to approximately 1,420,000 slides per year. Assuming lossy compressed data storage (e.g., 1–3 GB per slide), we will require ~4.3 petabytes storage space per year. We have established IT divisions and a center for clinical data science with extensive experience in enterprise medical imaging (20+ years); however, digital pathology images are on average 10 times the size of radiology images and will require significantly more resources for image management. One should not overestimate the impact of this factor, however, since radiology too has been faced with periodic challenges involving nontrivial leaps in data volume, such as thin slice CT and breast tomosynthesis; one just needs to be suitably prepared. By relying on DICOM as a data format for storage and exchange of whole slide images, healthcare enterprises can leverage a wealth of already existing medical imaging infrastructure representing billions of dollars of investments and build on the extensive DICOM expertise of IT specialists in hospitals and companies around the world.
Finally, clearly defining the “added clinical value” remains as the key challenge of implementing digital pathology into routine clinical practice. The core vision of digital pathology is that availability of pixel and metadata can unlock the full potential of integrated histopathological data analytics. We foresee that decision-support tools and machine-learning applications will facilitate adoption; however, these solutions require interoperability of systems and effective data access. Once completely realized, the advantages of digital pathology will be clear and fundamentally change how pathologists work. Researchers, practitioners, and vendors are now tasked to collectively navigate the transition from theoretical availability towards a meaningful return on investment. An enterprise-level communication standard (i.e., DICOM) is an essential component of streamlining data workflows to ultimately improve patient care.
Financial support and sponsorship
This work was funded in part by the NIH (RO1 CA225655) to J.K.L, NIH (U24 CA180918, P41 EB015898) to A.Y.F., NIH (P41 EB015902, U24 CA180918, U24 CA199460) to S.P, and the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health or any other organization.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The authors would like to thank A. Smith, N. Jones, B. R. Friedman, L. Chen, and P. H. Norwood for expert technical assistance. We thank C. Ayala, J. Floyd, S. Shafieha, and H. I. Paik for help with LIS access and E. Ziegler for technical discussions. We also thank N. Tenenholtz and R. W. Larson for project management as well as K. Burke and J. Reyes for administrative support. We further thank Enterprise Research Infrastructure and Services at Partners Healthcare for their support and for the provision of the ERISOne cluster. A special thanks is extended to J. Gilbertson and J. Kalpathy-Cramer for advice and for being awesome.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2018/9/1/37/244882
REFERENCES
- 1.Schultz M. Rudolf Virchow. Emerg Infect Dis. 2008;14:1480–1. [Google Scholar]
- 2.Weinstein RS, Graham AR, Richter LC, Barker GP, Krupinski EA, Lopez AM, et al. Overview of telepathology, virtual microscopy, and whole slide imaging: Prospects for the future. Hum Pathol. 2009;40:1057–69. doi: 10.1016/j.humpath.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Pantanowitz L, Dickinson K, Evans AJ, Hassell LA, Henricks WH, Lennerz JK, et al. American telemedicine association clinical guidelines for telepathology. J Pathol Inform. 2014;5:39. doi: 10.4103/2153-3539.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louis DN, Feldman M, Carter AB, Dighe AS, Pfeifer JD, Bry L, et al. Computational pathology: A path ahead. Arch Pathol Lab Med. 2016;140:41–50. doi: 10.5858/arpa.2015-0093-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis DN, Gerber GK, Baron JM, Bry L, Dighe AS, Getz G, et al. Computational pathology: An emerging definition. Arch Pathol Lab Med. 2014;138:1133–8. doi: 10.5858/arpa.2014-0034-ED. [DOI] [PubMed] [Google Scholar]
- 6.Abels E, Pantanowitz L. Current state of the regulatory trajectory for whole slide imaging devices in the USA. J Pathol Inform. 2017;8:23. doi: 10.4103/jpi.jpi_11_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukhopadhyay S, Feldman MD, Abels E, Ashfaq R, Beltaifa S, Cacciabeve NG, et al. Whole slide imaging versus microscopy for primary diagnosis in surgical pathology: A multicenter blinded randomized noninferiority study of 1992 cases (Pivotal study) Am J Surg Pathol. 2018;42:39–52. doi: 10.1097/PAS.0000000000000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gheisari S, Catchpoole DR, Charlton A, Kennedy PJ. Convolutional deep belief network with feature encoding for classification of neuroblastoma histological images. J Pathol Inform. 2018;9:17. doi: 10.4103/jpi.jpi_73_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golden JA. Deep learning algorithms for detection of lymph node metastases from breast cancer: Helping artificial intelligence be seen. JAMA. 2017;318:2184–6. doi: 10.1001/jama.2017.14580. [DOI] [PubMed] [Google Scholar]
- 11.Granter SR, Beck AH, Papke DJ., Jr Straw men, deep learning, and the future of the human microscopist: Response to “Artificial intelligence and the pathologist: Future frenemies?“. Arch Pathol Lab Med. 2017;141:624. doi: 10.5858/arpa.2017-0023-ED. [DOI] [PubMed] [Google Scholar]
- 12.Litjens G, Bandi P, Ehteshami Bejnordi B, Geessink O, Balkenhol M, Bult P, et al. 1399 H&E-stained sentinel lymph node sections of breast cancer patients: The CAMELYON dataset. Gigascience. 2018;7 doi: 10.1093/gigascience/giy065. doi: 101093/gigascience/giy065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moskowitz CS. Using free-response receiver operating characteristic curves to assess the accuracy of machine diagnosis of cancer. JAMA. 2017;318:2250–1. doi: 10.1001/jama.2017.18686. [DOI] [PubMed] [Google Scholar]
- 14.Sharma G, Carter A. Artificial intelligence and the pathologist: Future frenemies? Arch Pathol Lab Med. 2017;141:622–3. doi: 10.5858/arpa.2016-0593-ED. [DOI] [PubMed] [Google Scholar]
- 15.van Smeden M, Van Calster B, Groenwold RH. Machine learning compared with pathologist assessment. JAMA. 2018;319:1725–6. doi: 10.1001/jama.2018.1466. [DOI] [PubMed] [Google Scholar]
- 16.Clunie DA, Dennison DK, Cram D, Persons KR, Bronkalla MD, Primo HR, et al. Technical challenges of enterprise imaging: HIMSS-SIIM collaborative white paper. J Digit Imaging. 2016;29:583–614. doi: 10.1007/s10278-016-9899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Rojo M, Sanchez A, Bueno G, De Mena D. Standardization of pathology whole slide images according to DICOM 145 supplement and storage in PACs. Diagn Pathol. 2016;8:175. [Google Scholar]
- 18.Goode A, Gilbert B, Harkes J, Jukic D, Satyanarayanan M. OpenSlide: A vendor-neutral software foundation for digital pathology. J Pathol Inform. 2013;4:27. doi: 10.4103/2153-3539.119005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marques Godinho T, Lebre R, Silva LB, Costa C. An efficient architecture to support digital pathology in standard medical imaging repositories. J Biomed Inform. 2017;71:190–7. doi: 10.1016/j.jbi.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Balis UJ. Digital imaging standards and system interoperability. Clin Lab Med. 1997;17:315–22. [PubMed] [Google Scholar]
- 21.Roth CJ, Lannum LM, Joseph CL. Enterprise imaging governance: HIMSS-SIIM collaborative white paper. J Digit Imaging. 2016;29:539–46. doi: 10.1007/s10278-016-9883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth CJ, Lannum LM, Persons KR. A foundation for enterprise imaging: HIMSS-SIIM collaborative white paper. J Digit Imaging. 2016;29:530–8. doi: 10.1007/s10278-016-9882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larobina M, Murino L. Medical image file formats. J Digit Imaging. 2014;27:200–6. doi: 10.1007/s10278-013-9657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedorov A, Clunie D, Ulrich E, Bauer C, Wahle A, Brown B, et al. DICOM for quantitative imaging biomarker development: A standards based approach to sharing clinical data and structured PET/CT analysis results in head and neck cancer research. PeerJ. 2016;4:e2057. doi: 10.7717/peerj.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh R, Chubb L, Pantanowitz L, Parwani A. Standardization in digital pathology: Supplement 145 of the DICOM standards. J Pathol Inform. 2011;2:23. doi: 10.4103/2153-3539.80719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Digital Pathology Association (DPA) 2018. [Last accessed on 2018 Jun 12]. Available from: https://www.digitalpathologyassociation.org/
- 27.DICOM Working Group. [Last accessed on 2018 Sep 10]. Available from: https://www.dicomstandard.org/wgs/wg-26/
- 28.Clunie D, Hosseinzadeh D, Wintell M, De Mena D, Lajara N, Garcia-Rojo M, et al. Digital imaging and communications in medicine whole slide imaging connectathon at digital pathology association pathology visions 2017. J Pathol Inform. 2018;9:6. doi: 10.4103/jpi.jpi_1_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cucoranu IC, Parwani AV, Vepa S, Weinstein RS, Pantanowitz L. Digital pathology: A systematic evaluation of the patent landscape. J Pathol Inform. 2014;5:16. doi: 10.4103/2153-3539.133112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundström CF, Gilmore HL, Ros PR. Integrated diagnostics: The computational revolution catalyzing cross-disciplinary practices in radiology, pathology, and genomics. Radiology. 2017;285:12–5. doi: 10.1148/radiol.2017170062. [DOI] [PubMed] [Google Scholar]
- 31.Sorace J, Aberle DR, Elimam D, Lawvere S, Tawfik O, Wallace WD, et al. Integrating pathology and radiology disciplines: An emerging opportunity? BMC Med. 2012;10:100. doi: 10.1186/1741-7015-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DICOM Standard Website for DICOMweb. 2018. [Last accessed on 2018 Sep 10]. Available from: http://www.dicomstandard.org .
- 33.National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM) Standard PS3.4-Service Class Specifications. Report No.: PS3.4. Rosslyn, VA: National Electrical Manufacturers Association (NEMA); 2018. [Last accessed on 2018 Jun 23]. Available from: http://www.dicom.nema.org/medical/dicom/current/output/pdf/part 04.pdf . [Google Scholar]
- 34.National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM) Standard PS3.5 - Data Structures and Encoding. Report No.: PS3.5. Rosslyn, VA: National Electrical Manufacturers Association (NEMA); 2018. [Last accessed on 2018 Jun 23]. Available from: http://www.dicom.nema.org/medical/dicom/current/output/pdf/part 05.pdf . [Google Scholar]
- 35.National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM) Standard PS3.6 - Data Dictionary. Report No.: PS3.6. Rosslyn, VA: National Electrical Manufacturers Association (NEMA); 2018. [Last accessed on 2018 Jun 23]. Available from: http://www.dicom.nema.org/medical/dicom/current/output/pdf/part 06. pdf . [Google Scholar]
- 36.National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM) Standard PS3.10 - Media Storage and File Format for Media Interchange. Report No.: PS3.10. Rosslyn, VA: National Electrical Manufacturers Association (NEMA); 2018. [Last accessed on 2018 Jun 23]. Available from: http://www.dicom.nema.org/medical/dicom/current/output/pdf/part 10.pdf . [Google Scholar]
- 37.National Health Institute website for the SCOMED CT United States edition. [Last accessed on 2018 Jun 11]. Available from: http://www.nlm.nih.gov/healthit/snomedct/us_edition.html .
- 38.National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM) Standard PS3.16 - Content Mapping Resource. Report No.: PS3.16. Rosslyn, VA: National Electrical Manufacturers Association (NEMA); 2018. [Last accessed on 2018 Jun 23]. Available from: http://www.dicom.nema.org/medical/dicom/current/output/pdf/part 16.pdf . [Google Scholar]
- 39.Mason D. Pydicom: An open source DICOM library. Med Phys. 2011;38:3493. [Google Scholar]
- 40.Github Website for the Git Repository of the Openslide Library. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.github.com/openslide .
- 41.PyPi Website for the Openslide-Python Python Package. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.openslide.org/
- 42.PyPi Website for the Pillow Python Package. 2018. [last accessed on 2018 Jun 11]. Available from: https://www.pypi.org/project/Pillow/
- 43.Github Website for Git Repository of the Libjpeg-Turbo Library. 2018. [Last accessed on 2018Jun 11]. Available from: https://www.github.com/libjpeg-turbo/libjpeg-turbo .
- 44.Openjpeg Library. 2018. [Last accessed on 2018 Jun 11]. Available from: http://www.openjpeg.org/
- 45.Github Website for the Git Repository of the CharPyLS Python Package. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.github.com/Who8MyLunch/CharPyLS .
- 46.Github Website for the Git Repository of the CharLS library. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.github.com/team-charls/charls .
- 47.Fast Healthcare Interoperability Resources (FHIR) - 10.6 Resource Specimen – Content. 2018. [Last accessed on 2018 Sep 10]. Available from: https://wwwhl7org/fhir/specimen.html#json .
- 48.Dicom3tools, Snapshot 20180401144051. 2018. [Last accessed on 2018 Jun 11]. Available from: http://www.dclunie.com/dicom3tools/workinprogress/
- 49.PyPi Website for the Pydicom Python Package. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.pypi.org/project/pydicom/
- 50.Malaterre M. GDCM Reference Manual. Manual. 2008. Available from: https://www.sourceforge.net/projects/gdcm/files/gdcm%202.x/ Last accessed on 2018 Jun 11.
- 51.Sourceforge Website for Git Repository of the Grassroots DICOM Library. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.sourceforge.net/projects/gdcm/
- 52.DCMTK - DICOM Toolkit. 2018. [Last accessed on 2018 Jun 26]. Available from: https://www.dcmtk.org/dcmtk.php.en .
- 53.GNU Website for Time Command. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.gnu.org/software/time/
- 54.National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM) Standard PS3.8 - Network Communication Support for Message Exchange. Report No.: PS3.8. Rosslyn, VA: National Electrical Manufacturers Association (NEMA); 2018. [Last accessed on 2018 Jun 23]. Available from: http://www.dicom.nema.org/medical/dicom/current/output/pdf/part 08.pdf . [Google Scholar]
- 55.National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM) Standard PS3.7 - Message Exchange. Report No.: PS3.7. Rosslyn, VA: National Electrical Manufacturers Association (NEMA); 2018. [Last accessed on 2018 Jun 23]. Available from: http://www.dicom.nema.org/medical/dicom/current/output/pdf/part 07. pdf . [Google Scholar]
- 56.Fielding RT. Architectural Styles and the Design of Network-based Software Architectures. PhD Thesis, UC Irvine; 2000. [Google Scholar]
- 57.National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM) Standard PS3.18 - Web Services. Report No.: PS3.18. Rosslyn, VA: National Electrical Manufacturers Association (NEMA); 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.dicomstandard.org/dicomweb/ [Google Scholar]
- 58.Github Website for the Git Repository of the DCM4CHEE Archive. Miscellaneous. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.dcm4che.org/
- 59.Readthedocs Website for the Conformance Statement of the DCM4CHEE Archive. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.github.com/dcm4che/dcm4che/blob/master/README.md .
- 60.Github Website for the Git Repository of the Dicomweb-Client Python Package. 2018. [Last accessed on 2018 Jun 1]. Available from: https://www.github.com/clindatsci/dicomweb.client .
- 61.Herrmann MD. DICOMweb Client Documentation Release 0.3.0. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.media.readthedocs.org/pdf/dicomweb.client/stable/dicomweb.client.pdf .
- 62.Github Website for the Git Repository of the Dicomweb-Client JavaScript Package. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.github.com/DICOMcloud/DICOMweb-js .
- 63.Jira Software (Development Tool used by Agile Teams); Atlassian Inc. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.atlassian.com/software/jira .
- 64.PyPi website for the Pandas Python Package. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.pandas.pydata.org/
- 65.PyPi Website for the Plotly Python Package. 2018. [Last accessed on 2018 Jun 11]. Available from: https://www.pypi.org/project/plotly/
- 66.2018. [Last accessed on 2018 Jun 23]. Available from: http://www.medical.nema.org/medical/dicom/final/sup122_ft2.pdf .
- 67.National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM) Standard PS3.17 - Explanatory Information. Annex NN Specimen Identification and Management. Report No.: PS3.17. Rosslyn, VA: National Electrical Manufacturers Association (NEMA); 2018. [Last accessed on 2018 Jun 23]. Available from: http://www. dicom.nema.org/medical/dicom/current/output/pdf/part 17.pdf . [Google Scholar]
- 68.DICOM Correction Proposal 1713. More Compact Use of Per-Frame Functional Group Macros in Non-Sparse VL Whole Slide Microscopy Image IOD. 2018. Mar 25, [Last accessed on 2018 Jun 22]. Available from: http://www.medical.nema.org/medical/dicom/final/cp1713_ft_WSIPerFrameFunctionalGroupMacro.pdf .
- 69.Hosseinzadeh D. DICOM Digital Pathology Connectathon Proposal Version 6; A Multi-Vendor Demonstration of DICOM Workflow for Pathology. 2018. [Last accessed on 2018 Jun 11]. Available from: http://www.medical.nema.org/medical/dicom/DICOMWSI/Connectathon/PV2017/documents/DICOM%20Digital%20Pathology%20Connectathon%20Proposal.pdf .
- 70.Jodogne S. The orthanc ecosystem for medical imaging. J Digit Imaging. 2018;31:341–52. doi: 10.1007/s10278-018-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jodogne S, Lenaerts E, Marquet L, Erpicum C, Graimers R, Gillet P, et al. Open Implementation of DICOM for Whole-Slide Microscopic Imaging. 2017. [Last accessed on 2018 Sept 10]. Available from: https://www.semanticscholar.org/paper/Open-Implementation-of-DICOM-for-Whole-Slide-Jodogne-Lenaerts/5f1dc26f21f80b2218574fd13ad867484f476ce0 .
- 72.Rhatushnyak A. Lossless Photo Compression Benchmark. 2013. [Last accessed on 2018 Jun 24]. Available from: http://www.imagecompression.info/gralic/LPCB.html .
- 73.McCormick M, Liu X, Jomier J, Marion C, Ibanez L. ITK: Enabling reproducible research and open science. Front Neuroinform. 2014;8:13. doi: 10.3389/fninf.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lowekamp BC, Chen DT, Ibáñez L, Blezek D. The design of simpleITK. Front Neuroinform. 2013;7:45. doi: 10.3389/fninf.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Genereaux BW, Dennison DK, Ho K, Horn R, Silver EL, O’Donnell K, et al. DICOMweb™: Background and application of the web standard for medical imaging. J Digit Imaging. 2018;31:321–326. doi: 10.1007/s10278-018-0073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Github Website: DICOMweb Client Side JavaScript Implementation. 2018. [Last accessed on 2018 Aug 14]. Available from: https://www.github.com/dcmjs-org/dicomweb-client .
- 77.Web-Based Viewer for DICOM Visible Light Whole Slide Microscopy Images. 2018. [Last accessed 2018 Sep 10]. Available from: http://www.github.com/dcmjs-org/dicom-microscopy-viewer .
- 78.Bidgood WD, Jr, Horii SC. Introduction to the ACR-NEMA DICOM standard. Radiographics. 1992;12:345–55. doi: 10.1148/radiographics.12.2.1561424. [DOI] [PubMed] [Google Scholar]
- 79.Clunie D, Busbridge R. Correction Number CP-1818: Large Compressed Images may Have More Frames than Fit in the Basic Offset Table. 2018. [Last accessed on 2018 Jun 24]. Available from: http://www.medical.nema.org/medical/dicom/cp/cp1818_01_whenoffsettabletoosmall.pdf .
- 80.Guo H, Birsa J, Farahani N, Hartman DJ, Piccoli A, O’Leary M, et al. Digital pathology and anatomic pathology laboratory information system integration to support digital pathology sign-out. J Pathol Inform. 2016;7:23. doi: 10.4103/2153-3539.181767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Isaacs M, Lennerz JK, Yates S, Clermont W, Rossi J, Pfeifer JD, et al. Implementation of whole slide imaging in surgical pathology: A value added approach. J Pathol Inform. 2011;2:39. doi: 10.4103/2153-3539.84232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anatomic Pathology Workflow. 2016. [Last accessed on 2018 Jun 24]. Available from: https://www.wiki.ihe.net/index.php/Anatomic_Pathology_Workflow .
- 83.Daniel C, Rojo MG, Klossa J, Della Mea V, Booker D, Beckwith BA, et al. Standardizing the use of whole slide images in digital pathology. Comput Med Imaging Graph. 2011;35:496–505. doi: 10.1016/j.compmedimag.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 84.IHE Pathology and Laboratory Medicine (PaLM) 2018. [Last accessed on 2018 Jun 24]. Available from: https://www.wiki.ihe.net/index.php/Pathology_and_Laboratory_Medicine_(PaLM))
- 85.IHE Pathology and Laboratory Medicine (PaLM). HL7 Project Scope Statement. IHE Digital Pathology Workflow Metadata Requirements (e.g. DICOM) to Specimen DAM Mapping and HL7 Product Family Use. [Last accessed on 2018 Aug 05]. Available from: http://wiki.hl7.org/images/1/17/HL7_CG_20180904.pdf .
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selected DICOM attributes for digital pathology