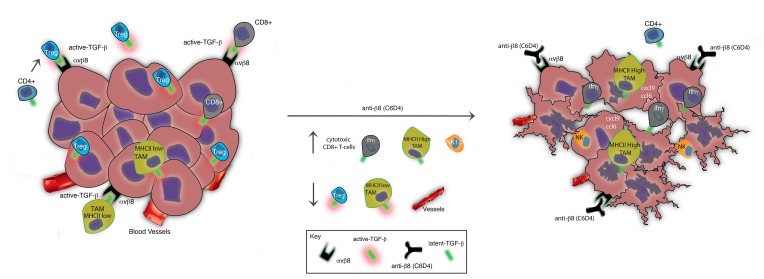
Abstract
TGF-β is a promising immunotherapeutic target. It is expressed ubiquitously in a latent form that must be activated to function. Determination of where and how latent TGF-β (L-TGF-β) is activated in the tumor microenvironment could facilitate cell- and mechanism-specific approaches to immunotherapeutically target TGF-β. Binding of L-TGF-β to integrin αvβ8 results in activation of TGF-β. We engineered and used αvβ8 antibodies optimized for blocking or detection, which — respectively — inhibit tumor growth in syngeneic tumor models or sensitively and specifically detect β8 in human tumors. Inhibition of αvβ8 potentiates cytotoxic T cell responses and recruitment of immune cells to tumor centers — effects that are independent of PD-1/PD-L1. β8 is expressed on the cell surface at high levels by tumor cells, not immune cells, while the reverse is true of L-TGF-β, suggesting that tumor cell αvβ8 serves as a platform for activating cell-surface L-TGF-β presented by immune cells. Transcriptome analysis of tumor-associated lymphoid cells reveals macrophages as a key cell type responsive to β8 inhibition with major increases in chemokine and tumor-eliminating genes. High β8 expression in tumor cells is seen in 20%–80% of various cancers, which rarely coincides with high PD-L1 expression. These data suggest tumor cell αvβ8 is a PD-1/PD-L1–independent immunotherapeutic target.
Keywords: Immunology
Keywords: Cancer immunotherapy, Integrins, Lung cancer, Macrophages
Tumor cell expression of the integrin avb8 regulates TGF-b activation by immune cells and maintains an immunosuppressive microenvironment.
Introduction
The majority of cancer patients receiving existing immunotherapies to the programed death receptor, PD-1, or its ligands PD-L1/2 do not respond, demonstrating a need for an expanded immunotherapeutic armamentarium (1). In many solid tumors, the immunosuppressive TGF-β pathway is associated with poor prognosis (2). In mice, TGF-β and PD-1/PD-L1 pathways can be experimentally coopted to favor tumor elimination (3, 4). Recent studies suggest that increased TGF-β activity in tumor stroma prevents penetration of tumor by cytotoxic CD8+ T cells and may prevent response to PD-L1 inhibitors (4, 5). Alternatively, TGF-β has been suggested to directly upregulate PD-L1 on tumor cells (6). A recent study demonstrates that TGF-β blockade can render a previously resistant colon cancer lesion susceptible to PD-1/PD-L1 blockade (4). Therefore, the function of TGF-β in antitumor immunity and interactions between the TGF-β and PD-1/PD-L1 pathways require further elucidation to understand the potential therapeutic benefit of targeting the TGF-β pathway.
TGF-β is expressed in a latent form (L-TGF-β) by most cell types and must be activated in order to exert its function in immune regulation, cell growth, vasculogenesis, matrix remodeling, and invasion (7). TGF-β is expressed in 3 partially functionally redundant isoforms (TGF-β1, -2, -3), which utilize the same receptors and signaling apparatus, making TGF-β a challenging therapeutic target, both for potential toxicities and for patient selection and stratification (8). Latency is conferred by noncovalent association of mature TGF-β with the TGF-β propeptide, latency associated peptide (LAP), which in TGF-β1 and TGF-β3 contains the Arg-Gly-Asp (RGD) integrin recognition motif (8). The αvβ8 integrin binds to and supports the activation of TGF-β (9). The amino acid sequence immediately c-terminal to the RGD in TGF-β1 and TGF-β3 contributes to high binding affinity and accounts for the specificity of the integrin αvβ8 for the LAPs of TGF-β1 and TGF-β3 (9, 10). TGF-β2, lacking the RGD sequence, is activated by non–integrin-mediated mechanisms. Most of the vascular, stromal, and immune effects relevant to cancer have been attributed to TGF-β1 (11–13).
Mice that lack integrin-mediated TGF-β activation recapitulate the developmental vascular pathology and lethal autoimmunity seen in tgfb1-deficient mice (14, 15). Animals deficient in either itgav or itgb8 show developmental vascular pathology due to defects in vessel differentiation similar to mice deficient in tgfb1, demonstrating that αvβ8 is an essential mechanism of TGF-β activation during development (13, 16). In postnatal mice, αvβ8-mediated TGF-β activation plays a major role in immune tolerance (14). However, β8 protein expression in vivo has mainly been demonstrated in epithelial, neural, and stromal cells and not in immune cells (17–20). The role of αvβ8 in the tumor immune response has not been reported.
αvβ8-mediated TGF-β activation is thought to mainly exert its functional effects through autocrine or paracrine stimulation (11–13, 21). However, whether the crucial source of L-TGF-β is αvβ8-expressing cells or adjacent cells, or whether it is freely diffusible remains unknown. For immune regulation, there is mounting evidence that it is not freely diffusible L-TGF-β1, but rather L-TGF-β1 expressed as a cell-surface complex via adaptor proteins such as GARP (lrrc32) that is the functionally important L-TGF-β1 biorepository in vivo (22–25). Thus, it is crucial to determine which cell types express αvβ8 and L-TGF-β in the tumor microenvironment to gain a fundamental understanding into the roles and function of TGF-β–mediated immune suppression in the neoplastic process. Here, we seek to address the role and cell type–specific mechanisms whereby αvβ8-mediated TGF-β activation influences tumorigenesis, as well as the relationship of αvβ8-mediated TGF-β activation with the PD-1/PD-L1/2 pathway.
Results
Simultaneous inhibition of αvβ8 and PD-1/PD-L1 pathways dramatically inhibit MC38 colon carcinoma growth.
To determine the role of αvβ8-mediated TGF-β activation in tumorigenesis, we used the murine MC38 colon carcinoma cancer cell line. MC38 cells are syngeneic to C57BL/6 and are TGF-β responsive, since blocking TGF-β alone partially blocks tumor growth (26). However, when used in combination with PD-1/PD-L1 blockade, TGF-β inhibition dramatically inhibits MC38 tumor growth (3, 5). MC38 cells express cell-surface αvβ8, as determined using a newly developed highly specific engineered blocking monoclonal antibody to the ligand-binding region of αvβ8 (C6D4), which reacts with both human and mouse β8 (Figure 1A, Supplemental Figure 1, Supplemental Methods; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.122591DS1). MC38 cells express PD-L1 but do not express PD-L2 (Figure 1B, Supplemental Figure 2, A–D). Treatment with C6D4 has no effect on PD-L1/2 levels either in vitro or from ex vivo isolated tumor cells (Supplemental Figure 2). MC38 cells support TGF-β activation that is mostly dependent on αvβ8, since C6D4 inhibits TGF-β activation almost as well as 1D11, a pan–TGF-β inhibitory antibody (Figure 1C). MC38 cells efficiently form s.c. tumors. The growth of established MC38 tumors 7 days after the initiation of antibody treatment is inhibited by treatment with C6D4 (Figure 1, D–F). To confirm the effects of αvβ8 on tumor growth using another naturally αvβ8-expressing tumor cell line, we used prostatic carcinoma TRAMP-C2 cells, which are syngenic to C57BL/6, express relatively high levels of β8 on the cell surface, and support αvβ8-mediated TGF-β activation (Supplemental Figure 3). Established TRAMP-C2 tumor growth is significantly inhibited by C6D4 (Supplemental Figure 3).
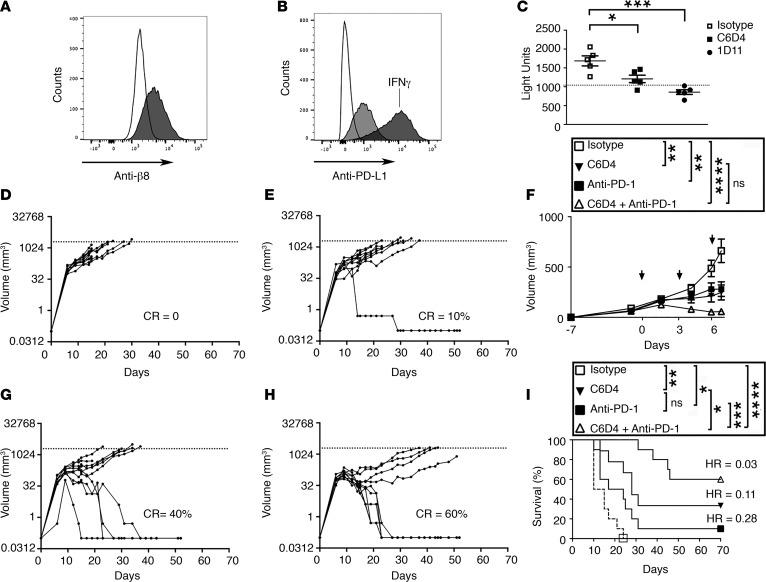
Figure 1. MC38 colon carcinoma tumor growth in vivo is reduced by anti-αvβ8 and eliminated in the majority of mice by combination with anti–PD-1.
(A) MC38 cells express cell-surface β8. Shown is a representative histogram overlay of anti-β8 (C6D4) stained cells (gray histogram) compared with isotype control (unfilled histogram) (a representative experiment of 4 is shown). For engineering and characterization of C6D4, see Supplemental Methods and Supplemental Figure 1. (B) MC38 cells express cell-surface PD-L1. Cells were stained with anti–PD-L1 (light gray). As a positive control cells were stimulated with IFN-γ (dark gray) (10 ng/ml) for 24 hours (hr). Shown is a representative histogram overlay of PD-L1 expression of IFN-γ–stimulated or nonstimulated compared with control (a representative experiment of 4 is shown). (C) MC38 cells support αvβ8-mediated TGF-β activation. MC38 cells were cocultured with TGF-β reporter cells in the presence or absence of C6D4, isotype control, or pan–TGF-β neutralizing antibody (1D11). Dotted line indicates basal level of TGF-β activation in MC38 cells. (D–I) MC38 colon carcinoma cells (5 × 105) were s.c. implanted into the flank of C57BL/6 mice. After tumors became palpable (7 days after inoculation), isotype control (anti-SV5 and 2A3), anti-β8 (C6D4), anti–PD-1 (RMP1-14), or both in combination were injected on days 0, 3, 6 (10 mg/kg i.p.), and RMP1-14 was added alone on day 9 (10 mg/kg i.p.). Spider plots of tumor growth of each treatment arm from mice treated with isotype control (D) SV5 and 2A3 (mouse and rat IgG2a, respectively) (n = 10) and neutralizing antibodies to (E) αvβ8 (C6D4) (n = 10), (G) PD-1 (RMP1-14) (n = 9), or (H) αvβ8 and PD-1 (C6D4 and RMP1-14) (n = 10). (F) Average tumor volumes from D, E, G, and H 15 days after tumor cell injection and 7 days after antibody administration is shown. (I) Kaplan-Meier survival plots. In legends F and I, ANOVA with Tukey’s post-hoc test of day 7 volume, or day 70 survival data, respectively, is shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. In D, E, G, H, complete response percentages (CR) and, in I, hazard ratios (Mantel-Haenszel) are shown. Arrows in F indicate antibody injection days.
Therapeutic treatment of established MC38 tumors with anti–PD-1 has a similar tumor inhibitory effect as C6D4 (Figure 1, D–G), but the two in combination produce a dramatic growth inhibitory effect (Figure 1, F and H). Survival is significantly improved by C6D4, or anti-PD-1, which can be further significantly improved by using both in combination (Figure 1I). In the combined treatment group, 60% of tumors show complete response 70 days after treatment initiation (Figure 1I).
Expression of αvβ8 by tumor cells potentiates in vivo lung tumor growth.
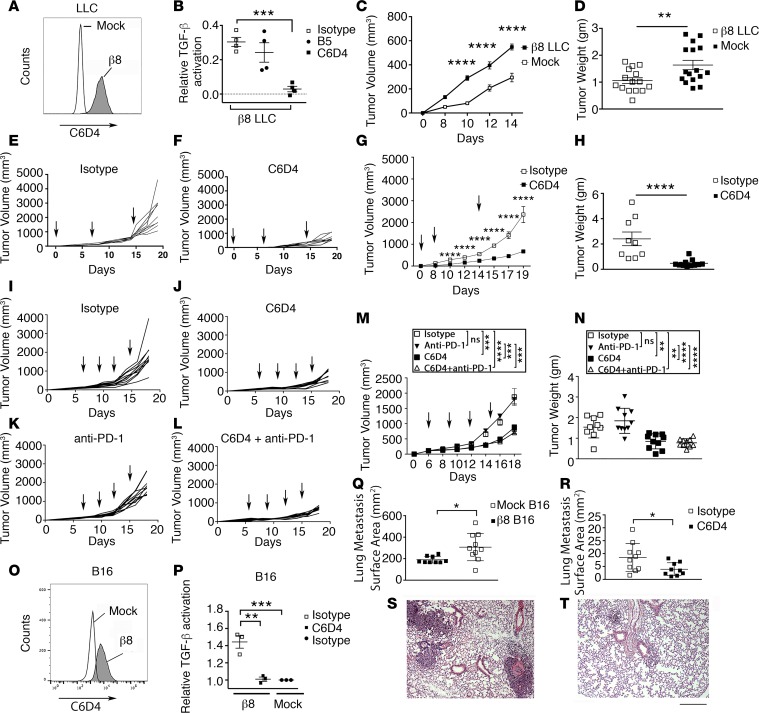
To understand the role of αvβ8 expressed by tumor cells, independent of the PD-1/PD-L1 pathway, we used the murine Lewis Lung Carcinoma (LLC) cell line, which is known to be PD-1/PD-L1 nonresponsive and is an established model cell line for tumorigenicity assays (27). LLC cells do not express detectable αvβ8 on their cell surface (Figure 2A), and C6D4 treatment does not significantly affect tumor growth of WT LLC cells (Supplemental Figure 4), indicating that host cells expressing αvβ8 do not significantly impact primary LLC growth. Mouse β8-expressing transfected LLC cells were created by stable transfection with a β8 cDNA expression vector (Figure 2A). Expression of β8 on LLC cells results in TGF-β activation, which can be efficiently blocked by C6D4 (Figure 2B). β8 expression increases the growth of LLC cell tumors compared with WT LLC cells (Figure 2, C and D). Prophylactic (Figure 2, E–H) or therapeutic (Figure 2, I, J, M, and N) dosing of C6D4 dramatically inhibits β8 LLC tumor growth (Figure 2, E–J, M, and N).
Figure 2. Expression of β8 increases in vivo tumor growth.
(A) LLC cells were transfected with itgb8. Stable pools were established and sorted for uniform expression and C6D4 (filled histogram, 1 μg/ml), and stained cells were compared with mock LLC (unfilled histogram). (B) β8-expressing LLC cells were tested for their ability to support TGF-β activation in the presence of C6D4 (filled boxes) or isotype controls SV5 (open boxes) or B5 (closed circles), n = 4. (C) Tumor growth of s.c. injected β8 LLC cells compared with mock LLC cells. Shown is a representative experiment (n = 14–16, repeated 3 times). (D) Tumor weight from individual mice bearing mock or β8 LLC tumors harvested at day 14. Open boxes, β8 LLC; filled boxes, mock LLC. (E and F) Spider plots of tumor cell growth in individual mice followed until day 19 after injection with β8 LLC cells. Mice were treated with isotype control (E) or C6D4 (F). Arrows indicate treatments (7 mg/kg i.p.). n = at least 9/group from a representative experiment of 3. (G) Average tumor volumes and (H) weights from tumors harvested at day 19 in E and F. Open boxes, isotype control; filled boxes, C6D4. (I–N) Established β8 LLC tumors were treated with isotype control (I), C6D4 (J), anti–PD-1 (RMP1-14) (K), or C6D4 + RMP1-14 (L). Arrows indicate injection time points. (M) Average tumor volumes and (N) weights from tumors harvested at day 19 in I–L. n = 10/ group. (O) B16 melanoma cells, which normally do not express αvβ8, were stably transfected with itgb8, sorted for uniform expression, and stained with C6D4 (1 μg/ml) and compared with mock-transfected B16 cells. Shown is a representative experiment of 4. (P) β8 B16 cells were tested for their ability to support TGF-β activation in the presence of isotype (filled bar) or C6D4 (open bar) (10 μg/ml) or were compared with mock-transfected B16 cells. n = 3. (Q) β8 or mock-transfected B16 cells were injected (i.v.), and 14 days later, lungs were morphometrically assessed for metastatic burden. n = 9–10/group. Shown is a representative experiment repeated twice. (R–T) β8 B16 cells were injected (i.v.), and mice were treated with isotype control or C6D4 (7 mg/kg i.p.) at day 0 and 7 and assessed for metastasis at day 14. In Q, open boxes, β8 B16; filled boxes, mock-transfected B16. In R, open boxes, isotype; filled boxes, C6D4. Shown are representative micrographs of lungs taken at 40× magnification from isotype (S) or C6D4-treated mice (T). Scale bar: 200 μm. Significance was determined by unpaired Student’s t test or ANOVA for multiple comparisons followed by Tukey’s post-hoc test to find where the difference lay. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
β8 LLC cells express PD-L1, not PD-L2, and treatment with C6D4 has no effect on PD-L1/2 levels either in cultured cells or ex vivo (Supplemental Figure 2). Anti–PD-1 has no therapeutic benefit alone and does not potentiate the effect of C6D4 on β8 LLC growth (Figure 2, K, L, M, and N). Thus, αvβ8 surface expression increases the tumorigenicity of β8 LLC lung cancer cells, and C6D4 can effectively inhibit β8 LLC growth independently of PD-1/PD-L1/2.
To determine whether β8 expression on tumor cells influences metastasis, we employed the B16 murine melanoma cell line, which rapidly colonizes the lung (27). B16 cells do not normally express αvβ8 on their cell surface but, when stably transfected with a β8 cDNA expression vector, express high levels (Figure 2O) and can significantly activate TGF-β relative to mock-transfected B16 cells (Figure 2P). Expression of β8 on B16 cells significantly enhances in vivo tumor growth of lung metastasis as determined by increased lung tumor surface area (Figure 2Q). Treatment of mice injected with β8 B16 cells with C6D4 significantly decreases lung tumor surface area compared with isotype treatment (Figure 2, R–T). Therefore, β8 expression not only affects primary tumor growth, but also affects growth of lung metastases.
β8 inhibition decreases tumor vascular growth and promotes tumor cell apoptosis in mice.
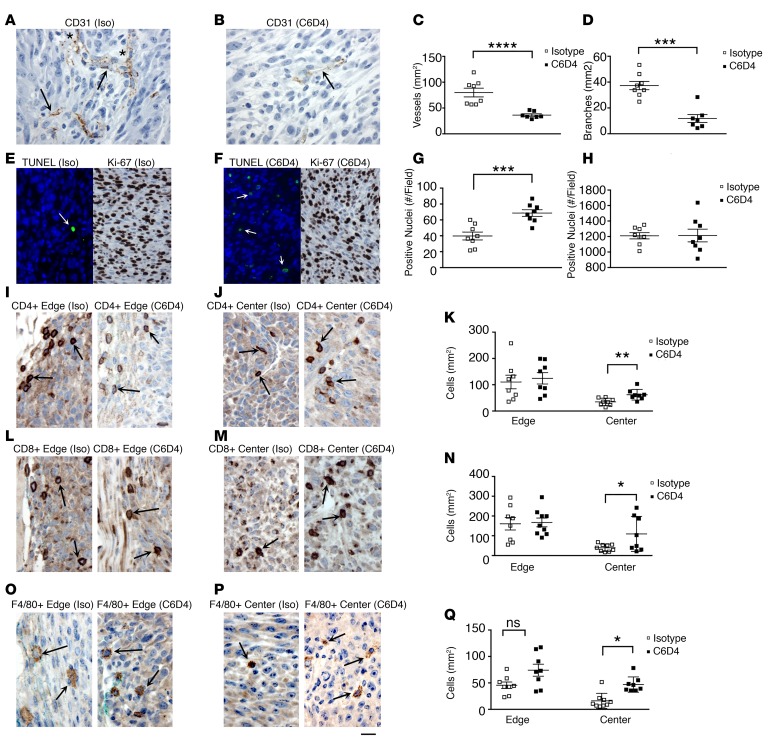
To address the mechanism of β8-induced tumor growth, we assessed vessel growth and morphology, apoptosis, proliferation, and tumor immune infiltration in established MC38 tumors from mice treated with isotype or C6D4. We first assessed MC38 tumors for vessel growth and morphology, since tumor cell growth is dependent on angiogenesis and is influenced by TGF-β (28). Expression of β8 on MC38 cells is associated with increased angiogenesis and vessel differentiation, since treatment with C6D4 significantly decreases vessel density and branching compared with isotype-treated tumors (Figure 3, A–D). To address the role of cell survival and proliferation in increasing β8-dependent tumor cell growth, we assessed apoptosis and cell proliferation of isotype- and C6D4-treated MC38 tumors. Established MC38 tumors from mice treated with isotype control antibodies have low levels of apoptotic cells, which are significantly increased by C6D4, while no treatment-specific differences in cell proliferation are seen (Figure 3, E–H).
Figure 3. β8 expression increases tumor vessel growth, promotes tumor cell survival and immune exclusion.
(A–R) Established MC38 tumors were therapeutically treated with isotype (Iso) or C6D4 (10 mg/kg i.p. on days 0, 3, and 6) and harvested when tumors reached endpoint volume (2,000 mm3). Tumors were immunohistochemically assessed for vascular growth (A–D), apoptosis and proliferation (E–H), and immune infiltration (I–R). Representative 400× fields from tumor centers are shown for CD31+ vascular staining of (A) isotype-treated (Iso-treated) (A) or C6D4-treated (B) MC38 tumors. Arrows indicate representative CD31+ vessels, and in A, asterisks indicate representative branch points; (C) vascular density and (D) vessel branching assessed morphometrically from five 200× fields. Open boxes, isotype; filled boxes, C6D4. (E and F) Representative 400× fields from MC38 tumor centers from mice treated with isotype (E) or C6D4 (F) stained by TUNEL (green fluorescence, TUNEL+ (arrows) and blue, DAPI; left panels) or Ki-67 (right panels) and assessed for apoptosis (G) or Ki-67 positivity as a proliferation marker (H). (I, J, L, M, P, Q) Representative 400× fields from MC38 tumor edge (I, L, O) or center (J, M, P), as indicated above each photomicrograph, from mice treated with Iso (left panel) or C6D4 (right panel), as indicated above each figure, stained with anti-CD4 (I and J), anti-CD8 (L and M), or anti-F4/80 (O and P). Representative positively stained immune cells are indicated by arrows. Scale bar: 25 μm. Quantification from individual tumors for CD4+ T cells (K), CD8+ T cells (N), or F4/80 + macrophages (Q) from the tumor edge or center are shown. n = 8, in each antibody treatment group. Open boxes, isotype control; filled boxes, C6D4. Significance determined by unpaired Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The β8-dependent effects on vessel growth and morphology are confirmed in β8-expressing LLC tumors, since they display increased vessel density and branching compared with mock-transfected tumors (Supplemental Figure 5). C6D4 significantly decreases vessel density and branching compared with isotype control of β8-expressing LLC tumors (Supplemental Figure 5) to a similar level as seen in mock-transfected LLC tumors. Similar to MC38 tumors, β8 LLC tumors from mice treated with isotype control antibodies have low levels of apoptotic cells, which are significantly increased by C6D4 (Supplemental Figure 5). Apoptosis was increased and cell proliferation unchanged by inhibiting β8 expressed on LLC cells. β8-mediated TGF-β activation does not impact cell growth of LLC cells, and in vitro and β8-LLC cells cultured on L- TGF-β1 coated substrates show no differences in growth compared with mock transfected cells (mock, 0.48 ± 0.28 (A450); β8, 0.37 ± 0.29 (A450); n = 3; P = 0.25). In vivo, cell proliferation is not different between isotype- and C6D4-treated β8 LLC tumors (Supplemental Figure 5). These data suggest a prosurvival role for αvβ8-mediated TGF-β activation specifically in the tumor microenvironment.
In both MC38 and β8-LLC tumors, vessel density, and branch point numbers, apoptotic cell number and proliferation rates are not significantly correlated with tumor weight, while tumor volume and weight are highly correlated, indicating that C6D4-dependent differences are not due to tumor size (Supplemental Figure 6 and 7).
β8 inhibition increases the infiltration of cytotoxic T cells and proinflammatory tumor-associated macrophages to the tumor center in mice.
Increased penetration of immune cells to the tumor center from the tumor periphery is a hallmark of an effective tumor immune response and has been reported to be inhibited by TGF-β (4, 29). Movement of CD4+ and CD8+ T cells, and F4/80+ macrophages into MC38 and β8 LLC tumor centers, as assessed by IHC, is significantly increased by C6D4 compared with isotype controls, while the numbers of CD4+, CD8+, and F4/80+ cells at the tumor periphery remain unchanged, indicating that αvβ8 expression by tumor cells suppresses T cell and macrophage infiltration into the tumor center (Figure 3, I–Q, Supplemental Figure 5). The most abundant tumor-infiltrating immune cell types as assessed by IHC in MC38 and β8 LLC tumors are CD8+ and CD4+ T cells, respectively (Figure 3, I–Q, Supplemental Figure 5). Immune cell counts in tumor centers are not significantly correlated with tumor weight, indicating that C6D4-dependent differences are independent of tumor size (Supplemental Figure 7).
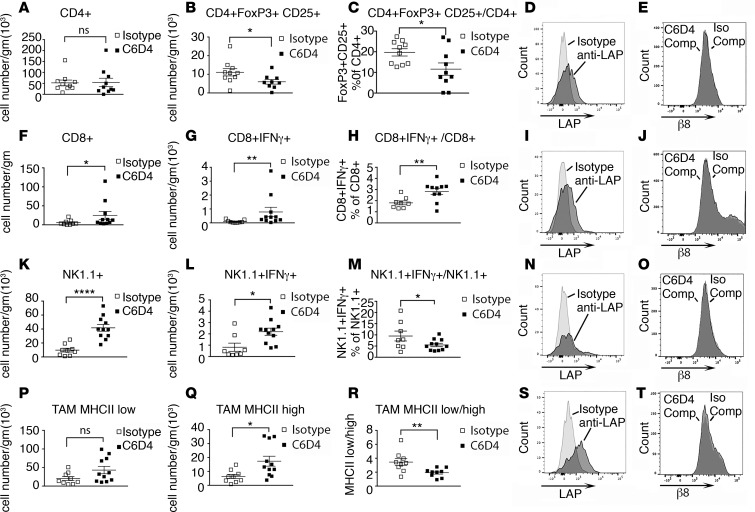
To address whether changes in immune cell compartments were consistent with a tumor-eliminating immune cell phenotype, we performed detailed immune cell analysis of β8 LLC tumors using multicolor cell staining. Immunosuppressive CD4+ Tregs (30), granulocytic (g) and monocytic (m) myeloid–derived suppressor cells (MDSC) (31), and tumor-associated macrophages (TAM) expressing low surface levels of MHC class II (32) favor tumor progression; IFN-γ–secreting CD4+, CD8+ T cells and NK cells, and TAM expressing high surface levels of MHC class II favor tumor elimination (32–34). Immune cell phenotyping of tumor-infiltrating lymphoid cells from β8 LLC tumors treated with C6D4 or isotype control reveal C6D4-dependent decreases in numbers and percentages of Treg, without significant effects on total CD4+ T cell number (Figure 4, A–C) or numbers or percentages of CD4+IFN-γ–secreting cells (Supplemental Figure 8). Total numbers of CD8+ and number and percentage of CD8+IFN-γ+ T cells are significantly increased by C6D4 treatment (Figure 4, F–H). Total numbers of NK1.1+ and NK1.1+IFN-γ–secreting cells are significantly increased by C6D4 treatment, while percentages of NK1.1+IFN-γ–secreting cells significantly decreased, indicating that recruitment of non–IFN-γ–secreting NK1.1+ cells exceeds that of NK1.1+IFN-γ–secreting cells (Figure 4, K–M). The numbers of NK1.1+ T-like cells and numbers and percentages NK1.1+ T-like IFN-γ–secreting cells are not significantly changed by C6D4 (Supplemental Figure 8). Numbers of TAM expressing high levels of MHCII are significantly increased, and the ratio of MHCIIlo to MHCIIhi TAM is significantly decreased by C6D4; the numbers of MHCIIlo TAM are not significantly changed by C6D4 (Figure 4, P and R). Numbers and percentages of g-MDSCand m-MDSC are not significantly changed by C6D4 (Supplemental Figure 8). The most abundant immune cell types in β8 LLC tumors, as determined using flow cytometry, are CD4+ T cells, consistent with immunohistochemical staining data (Figure 4, Supplemental Figure 5). In summary, these data show that inhibition of αvβ8-mediated TGF-β activation by C6D4 changes the tumor immune cell profile to favor tumor elimination by reducing immunosuppressive Treg differentiation, increasing recruitment and differentiation of tumor-eliminating IFN-γ–secreting CD8+ T cells, increasing recruitment of NK1.1+ cells and NK1.1+ cells secreting IFN-γ, and increasing numbers of tumor-eliminating MHCIIhi TAM relative to tumor-promoting MHCIIlo TAM.
Figure 4. The antibody C6D4 decreases CD4+ Tregs, increases CD8+ and NK1.1+ IFN-γ–secreting cells, and alters differentiation of TAM.
Tumor-infiltrating lymphoid cells from β8-LLC tumors harvested at day 14 after injection from mice treated with isotype or C6D4 (7 mg/kg) on days 0 and 7 underwent multicolor cell staining and analysis. (A–C, F–H, K–M, and P–R) Tumor-associated lymphoid cells isolated from tumors from mice treated with isotype (open boxes) or C6D4 (closed boxes) are stained to identify CD4+ T cells (A–E), CD8+ T cells (F-J), NK1.1+TCRβ– cells (K–O), or tumor-associated macrophages (TAM) (P–T). (A) CD4+ cells, (B) CD4+FoxP3+CD25+ Tregs (gating strategy Supplemental Figure 9), (C) percentages of Tregs of CD4+ cells. (D) Histogram overlays of CD4+ T cells, from isotype-treated mice, stained with isotype (light gray) or for latency-associated peptide of TGF-β1 (LAP-β1) to detect cell-surface L-TGF-β1 (dark gray). (E) Histogram overlays of CD4+ T cells, from isotype-treated mice, stained with C6D4-PE with a 100-fold excess of unlabeled C6D4 (C6D4-Comp, light gray) or B5, a human-specific β8 antibody, as an isotype control (Iso-Comp, dark gray). (F–H) CD8+ cells total (F), IFN-γ–secreting (G), or percentages (H) of CD8+ IFN-γ–secreting T cells of all CD8+ T cells. (I) CD8+ cells stained as in D (gating strategy shown in Supplemental Figure 10). (J) All TCR-β+ cells stained, analyzed, and labeled as in E. (K–O) NK1.1+ cells analyzed and labeled as F–J (gating strategy shown in Supplemental Figure 10). (P–T) CD45+, CD11bhi, Ly6g–, Ly6c–, F4/80+, MHCII high, or MHCII low TAM (gating strategy shown in Supplemental Figure 11) were analyzed and the numbers of MHCII low TAM (P), numbers of MHCII high TAM (Q), and the ratios of MHCII low/MHCII high TAM determined (R). (S) Histogram overlays of CD45+, CD11bhi, F4/80+, TCR-β– cells, from isotype-treated mice, stained with isotype (light gray) or LAP-β1(dark gray). (T) Histogram overlays of CD45+, CD11bhi, F4/80+, TCRβ– macrophages stained and labeled as in E. n = 9–11/group. Shown are representative experiments of at least 3. *P < 0.05, **P < 0.01, ****P < 0.0001, by Student’s t test.
L-TGF-β, but not αvβ8, is expressed on the surface of tumor-associated T cells and macrophages.
To address the cell types expressing cell-surface L-TGF-β that could be interacting with αvβ8 on tumor cells, we isolated and stained β8 LLC tumor-infiltrating CD4+, CD8+ T cells, NK cells, and macrophages for cell-surface L-TGF-β1 using an antibody that detects the LAP of TGF-β1. CD4+, CD8+ T cells, Tregs, and macrophages — but not NK cells — express detectable levels of cell-surface L-TGF-β (Figure 4, D, I, N, and S, and Supplemental Figure 12). β8 LLC, MC38, or TRAMP-C2 tumor cells do not express cell-surface L-TGF-β (Supplemental Figures 2 and 3).
To determine whether β8 protein is expressed by tumor immune cells, we isolated and stained immune cells from β8 LLC tumors using fluorescently conjugated C6D4. Staining with fluorescently conjugated C6D4, including several controls for specificity, revealed no β8 staining in T cells, Tregs, NK cells, DCs, or macrophages (Figure 4, E, J, O, and T, and Supplemental Figures 12 and 13). These results suggest that tumor cell αvβ8 can modulate immune cell TGF-β function by activation of L-TGF-β on the immune cell surface, and this is likely to be the primary mode of αvβ8-mediated L-TGF-β activation in the tumor environment.
αvβ8 inhibition leads to suppression of macrophage chemokine gene expression.
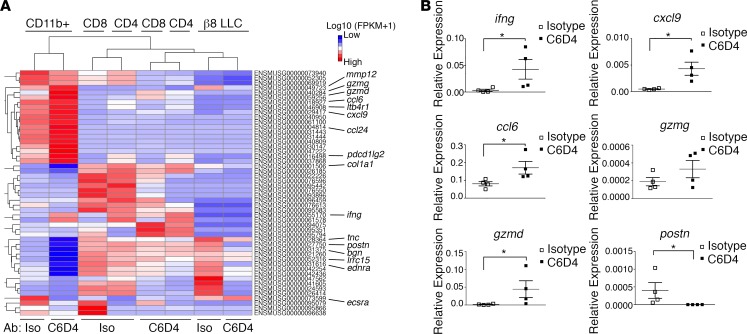
To identify which L-TGF-β–expressing immune cell types are impacted by β8 expression on tumor cells, we performed transcriptome analysis of isolated CD11b+ and T cells from β8 LLC tumors treated with isotype vs. C6D4. Expression values across all samples revealed a total of 39.6 × 106 to 42.4 × 106 uniquely mapped reads representing a range of 88.1%–90.3% of total reads. The highest number of significantly differentially expressed (DE) genes between isotype- and C6D4-treated paired samples (Padj < 0.005) was found in macrophages (18 increased, 12 decreased), with fewer in CD4+ T cells (2 increased, 10 decreased), CD8+ T cells (2 increased, 7 decreased), and β8 LLC cells (0 increased, 5 decreased). Functional annotation of the macrophage DE genes increased by C6D4 include those involved in inflammation and chemotaxis (ifng, cxcl9, ccl6, ccl24, ltb4r1, chil3), response to IFN-γ (ccl6, ccl24), cytolysis and proteolysis (gzmd, gzme, mmp12), and immune suppression (pdcd1lg2); those decreased by C6D4 include genes involved in vasculogenesis (bgn, ecscr, ednra) and TGF-β–responsive extracellular-matrix protein production (postn, tnc, col1a1, lrrc15) (Figure 5A, Supplemental Tables 1 and 2). Functional annotation of DE genes (Padj < 0.005) from CD4+ and CD8+ T cells and β8 LLC cells did not reveal any statistically significant enrichment in gene ontology classes induced by C6D4, and no relevant gene ontology classes decreased by C6D4 (Supplemental Table 2).
Figure 5. Differential gene expression by tumor cells, CD4+ T cells, CD8+ T cells, and CD11b+ cells reveals a role for β8 in suppression of a tumor-eliminating macrophage signature.
(A) Heatmap of hierarchical clustering of differentially expressed (DE) genes determined by RNAseq across pooled CD11b+, CD4+, CD8+ T cells, and β8 LLC tumor cell samples isolated from mice (n = 10 in each group) treated with isotype or C6D4 (7 mg/kg i.p.) on days 5 or 12 after β8 LLC tumor cell injection. Tumors were harvested on day 15 after tumor cell injection. Hierarchical clustering analysis was carried out with the log10 (FPKM+1) of union of DE genes. DE was defined by q < 0.005 and |log2 (fold change)| > 1. Red represents highly expressed genes. Blue represents genes with low expression. Color gradient from red to blue reflects log10 (FPKM+1) values from large to small. ENSEMBL IDs are shown, and select gene names are indicated. The majority (60%) of DE genes were found in macrophages (18 up; 12 down), which drives the clustering of T cells to be defined by antibody treatment rather than CD4+ or CD8+ lineage. (B) Validation of key macrophage DE genes from RNA isolated from pooled macrophage samples (n = 4 pools) that had sorted for purity (CD45+, F4/80, CD11b+) and assessed by qPCR for ifng, cxcl9, ccl6, gzmg, gzmd, and postn. Shown is expression (2-Δct) relative to hprt. *P < 0.05 by unpaired Mann-Whitney U test.
Representative significantly DE genes identified by RNA sequencing (RNAseq) were validated using quantitative PCR (qPCR) using RNA isolated from purified TAM. Expression of the TGF-β–induced extracellular matrix gene postn was decreased, and IFN-γ signature chemokines (ifng, cxcl9, ccl6) and the cytolytic gene (gzmd) were significantly increased by C6D4 (Figure 5B). Overall, gene expression patterns reveal that macrophages are the main cell class influenced by C6D4, with major increases in chemokines and cytotoxic genes and decreases in extracellular matrix genes.
Transcriptomic analysis of human cancer specimens and cell lines reveals that β8 is commonly expressed in human carcinoma cells.
Public human RNAseq databases were evaluated to confirm that the tumor cell is the main compartment where ITGB8 is expressed. ITGB8 transcripts are significantly enriched in carcinoma, glioma, and melanoma cell lines relative to hematogenous- and lymphoid-derived malignant lines (Supplemental Figure 14A). This general pattern of ITGB8 expression is seen in public RNAseq datasets of normal and malignant human tissue types. Enhanced expression is seen in several carcinomas — in particular, ovarian and pancreatic carcinomas with little or no expression in lymphoid or mesenchymal tissues (Supplemental Figure 14B).
β8 protein is highly expressed in tumor cells of human epithelial malignancies.
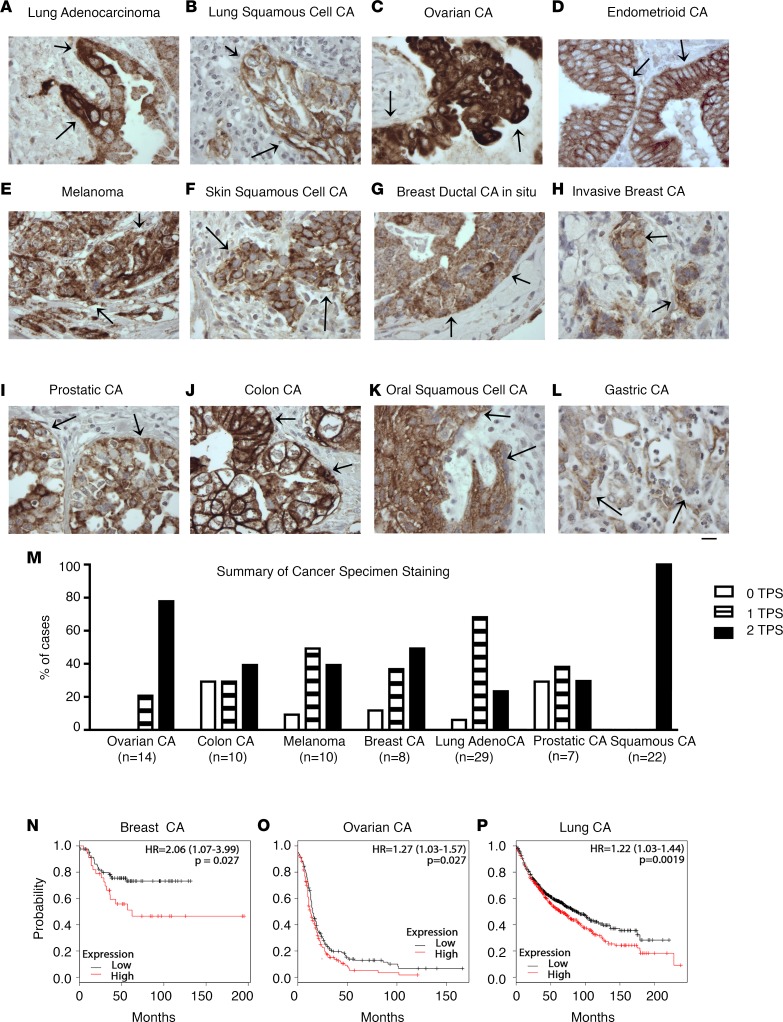
To confirm the high tumor cell expression of β8 in human cancers, we engineered and developed a highly human-specific and sensitive antibody, F9, to stain β8 in archival formalin-fixed paraffin embedded (FFPE) human tissues (Supplemental Figures 15–19 and Supplemental Methods). Using F9, β8 immunostaining is detected in tumor cells of various carcinomas including lung, ovarian, endometrial, melanoma, breast, prostate, colon, skin, and stomach (Figure 6, A–L). No convincing β8 staining is noted in stromal or immune cell compartments. The absence of tumor stromal and immune cell staining is confirmed in transgenic mice engineered to express only human ITGB8 and not mouse itgb8 (Supplemental Figure 18). Taken together, these data demonstrate that tumor cells are the major β8-expressing cell type in human cancers. High β8 expression (i.e., greater than 50% tumor cells showing membrane staining [>50% tumor proportion score (TPS)]) is seen in a substantial proportion of cancers — in particular, high-grade serous ovarian carcinoma and squamous cell carcinoma (Figure 6M).
Figure 6. β8 expression in human carcinoma and melanoma specimens.
A highly specific and sensitive anti-β8 antibody suitable for staining formalin-fixed paraffin-embedded tissues (FFPE) was engineered, optimized, and extensively characterized (Supplemental Methods and Supplemental Figures 15–19). Representative F9 immunostaining results of archival FFPE tissues of (A) lung adenocarcinoma; (B) lung squamous cell carcinoma; (C) ovarian carcinoma; (D) endometrioid adenocarcinoma (n = 3, all positive); (E) melanoma; (F) skin (n = 3, all positive); (G) breast ductal carcinoma in situ, high grade (n = 3, all positive); (H) breast invasive carcinoma; (I) prostatic adenocarcinoma; (J) colonic adenocarcinoma; (K) oral squamous cell carcinoma; and (L) gastric adenocarcinoma (n = 3, all positive). All images taken at 400×. Scale bar: 25 μm. Arrows indicate representative positive staining of tumor cell islands. (M) Summary of staining data from cohorts of tumor samples depicted in A–L, where n > 3. High-grade serous ovarian (n = 14), colon (n = 10), melanoma (n = 10), breast (n = 8), lung (n = 29), prostate (n = 7), and mucosal squamous cell carcinoma (lung, oropharyngeal) (n = 22) were stained with F9, and staining was expressed as tumor proportion score (TPS), which represents the percentage of tumor cells with membrane staining compared with all tumor cells. 0 = TPS <1% (open bars); 1 = TPS 1%–49% (striped bars); 2 = TPS 50% or greater (filled bars). (N–P) High ITGB8 mRNA expression is correlated with decreased survival in subsets of breast, ovarian, and lung carcinomas. Kaplan-Meier curves of high vs. low ITGB8 mRNA expression in (N) triple-negative basal-type breast carcinoma (n = 186), (O) stage 3 or 4 serous ovarian carcinoma (n = 1,001), or (P) non–small cell lung carcinoma (n = 1,926). Kaplan-Meier plots were generated using KM plotter (http://kmplot.com), a web application supporting biomarker assessment for multiple carcinoma types and subtypes (35). HR, hazard ratio; significance found with logrank tests.
High β8 expression is associated with decreased survival in triple-negative basal-type breast cancer, advanced stage serous ovarian cancer, and non–small cell lung carcinoma (NSCLC).
As a first step to understand the clinical relevance of high β8 expression, we evaluated a public microarray expression database (35) to reveal that cancers with reported high β8 expression are associated with decreased survival: triple-negative basal-type breast cancers (36), advanced stage serous ovarian carcinomas (37) and NSCLC (Figure 6, N–P). Taken together, these data suggest that increased expression of β8 in human cancer cells plays a role in the progression of epithelial malignancies.
High β8 and PD-L1 expression are rarely seen concurrently in NSCLC.
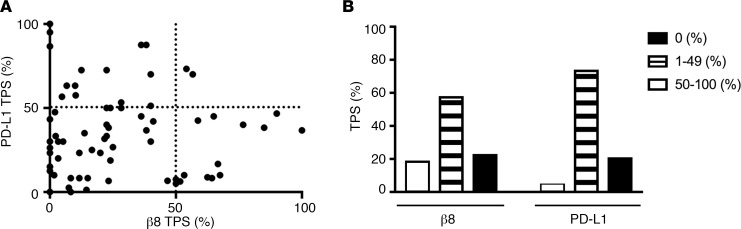
PD-L1 immunohistochemical staining is currently recommended as part of biomarker testing for all advanced NSCLC (38). Approximately, 30% of advanced NSCLC patients have high PD-L1 staining (>50% of tumor cells) and have significantly improved response rates to PD-1 inhibitors compared with tradition chemotherapy (39). We assessed the relationship of β8 and PD-L1 expression on tumor cells with F9 using an extended cohort of 73 archival lung NSCLC cases. Strong tumor cell β8 staining is seen in 21.9% of cases, while strong PD-L1 tumor cell staining (≥50% TPS) is seen on 28.8% of cases. Strong β8 and PD-L1 immunostaining (≥50% TPS) are concurrently seen in only 2.7% of cases, suggesting that high β8 and PD-L1 occurring together is rare. The majority of cases (60.3%) show weak staining (1%–49% tumor proportion score) for β8 and PD-L1. Negative staining for β8 and PD-L1 are seen in 17.8% and 4.1% of cases, respectively (Figure 7). These results demonstrate that high β8 and PD-L1 expression are rarely seen together on NSCLC tumor cells.
Figure 7. High expression of the integrin β8 subunit and PD-L1 are rarely seen concurrently in non–small cell lung carcinoma (NSCLC).
(A and B) Seventy-three archival lung adenocarcinoma cases were stained for the β8 integrin subunit and PD-L1 with antibody clone F9 for β8 and antibody clone E1N3 for PD-L1. Expression was determined by tumor proportion score (TPS) for both β8 and PD-L1 and plotted on an (A) xy graph divided into TPS quadrants 0%–49% or ≥50% (P = 0.7194 by McNemar’s test; Pearson r = –0.0394; P = 0.7403), or shown by (B) TPS as 0% (open bars), 1%–49% (striped bars), or 50%–100% (filled bars) in column format.
Discussion
We have demonstrated that αvβ8 expression by tumor cells drives tumor growth in vivo, and antibody inhibition can significantly reduce tumor growth and improve survival. In an in vivo model using a PD-1–responsive cell line, inhibition of αvβ8 and PD-1 in combination achieve a 60% complete response rate. β8 expression by tumor cells is important, since β8 expression increased the tumorigenicity of LLC and B16 cell lines. Furthermore, inhibiting β8 expressed on LLC cells decreases tumor growth independently of PD-1 or PD-L1/2, since β8 LLC tumors are completely refractory to PD-1 inhibition alone or in combination with β8 inhibition. Anti-αvβ8 increases effective antitumor immunity coincident with increased IFN-γ secretion by CD8+ T cells, increased tumor cell apoptosis, and decreased angiogenesis. β8 protein is not detected on immune cells, while the reverse is true for cell-surface TGF-β1, suggesting that tumor cell αvβ8 serves as a platform for TGF-β1 activation on tumor-infiltrating immune cells. Transcriptome profiling identifies macrophages as a key immune target in β8-mediated tumor promotion, since β8 inhibition increases a macrophage proinflammatory tumor-eliminating gene expression signature, while decreasing expression of macrophage angiogenesis and TGF-β–responsive extracellular matrix genes. These findings may be relevant to human cancers, since αvβ8 is highly expressed by tumor cells in a variety of human carcinomas. In subsets of ovarian, breast, and lung cancers, high β8 expression is associated with decreased survival. Finally, high protein expression of the integrin β8 subunit and PD-L1 are rarely seen concurrently in human NSCLC. These results raise the hypothesis that patients with high β8 expression in tumor cells may benefit from αvβ8 inhibition with or without concurrent anti–PD-1/PD-L1 therapy.
The widespread immunohistochemical detection of the β8 protein in malignant cells of a variety of human carcinomas and melanomas and lack of β8 detection in tumor stroma or immune cells is consistent with the hypothesis that β8 expression on tumor cells is the main driver of β8-dependent protumorigenic effects. This does not exclude the possibility that immune or stromal cells with β8 expression below the level of antibody detection also play a role in αvβ8 function. A recent study suggests that low levels of αvβ8 protein are expressed on the surface of human intestinal DCs and Tregs, which is concordant with reports of β8 mRNA in both cell types (40–42). However, we are unable to detect specific β8 expression on freshly isolated macrophages, DCs, T cells, Tregs, or NK cells from tumors using cell-surface staining. These results indicate that β8 is abundantly expressed by epithelial cells and neural derivatives, and not by immune cells. The fact that antibody inhibition of αvβ8 only impacted the growth of β8 LLC and not the non–β8-expressing mock LLC cells reinforces the relative importance of β8 expression on tumor cells rather than host immune cells.
L-TGF-β is secreted mainly as a protein complex bound to immune, fibroblast, or endothelial cell surfaces or incorporated into the extracellular matrix by TGF-β binding proteins (e.g., LTBP1-4, GARP) or other proteins (8, 23, 43, 44). Attempts to immunohistochemically localize TGF-β isoforms to specific cellular compartments within tumors have been largely inconclusive due to antibody specificity (45). Therefore, staining of cells isolated from disaggregated tumors is more reliable as a method to understand what cells present cell-surface L-TGF-β. In our models, tumor cells themselves do not express cell-surface L-TGF-β — rather, TAM and CD4+ and CD8+ T cells express cell-surface L-TGF-β. L-TGF-β has been previously reported on the surface of DCs, B cells, and various T cells— in particular, CD4+ Tregs (46–51); however, to our knowledge, L-TGF-β has not been shown on the surface of macrophages.
We hypothesize that αvβ8-expressing tumor cells serve as a platform for TGF-β activation when contacted by TGF-β–presenting TAM. As such, tumor cells could direct TGF-β–mediated activation at the tumor-stromal interface through direct contact with macrophages, an abundant immune cell type in tumors (52). In vivo, inhibition of β8 increased macrophage expression of mRNA for cxcl9, ccl6, and ccl24. Increased expression of these molecules has been shown to promote chemotaxis and clonal expansion of immune cells expressing their cognate receptors (53–55). Consistent with this, blocking tumor cell β8 leads to redistribution of tumor-infiltrating lymphoid cells into the tumor center, and it increased IFN-γ–producing intratumoral NK1.1+ and CD8+ T cells and decreased MHClo TAM, hallmarks of effective antitumor immunity (56–58). Ultimately, treatment with β8 antibodies increases tumor cell apoptosis, which likely results from enhanced antitumor immunity (59) coupled with hypoxia due to decreased angiogenesis (60). Therefore, αvβ8-mediated TGF-β activation is likely to support a complex interaction network between tumor cells and immune cells within the tumor environment.
A tumor stromal TGF-β signature is associated with nonresponse to PD-1 inhibitors in patients with urothelial carcinoma, which is associated with exclusion of T cells from the tumor center (5). In a colon carcinoma model, TGF-β is associated with decreased PD-1 expression by T cells (4). How the TGF-β and PD-1/PD-L1 pathways interact is an area of active investigation. We did not find any evidence that blocking αvβ8 impacts tumor cell-surface PD-L1/2 expression, nor does it enhance responsiveness to PD-1 inhibition, in PD-1 nonresponsive β8 LLC tumors. Thus, while interactions between TGF-β and PD-1/PD-L1 pathways may exist, our data suggest that αvβ8-mediated TGF-β activation and the PD-1/PD-L1 pathways are independent pathways of tumor immune evasion.
TGF-β can be targeted directly through inhibiting interactions of mature TGF-β with its receptor, or through inhibition of TGF-β signaling. However, such global approaches perturb the essential homeostatic functions of TGF-β and have been associated with toxicities in preclinical studies (61, 62). Targeting the L-TGF-β activation mechanism provides improved cell type and context specificity, mitigating risk of toxicity, a goal that targeting the L-TGF-β binding protein GARP or TGF-β activating integrins, such as αvβ8, potentially both accomplish (49, 63). However, antibodies targeting GARP have reduced efficacy in vivo compared with anti-β8, as might be expected because of the restricted expression distribution of GARP and the mechanism of action of targeting an adaptor protein rather than directly perturbing ligand-receptor interactions (40). All therapeutic approaches targeting the ubiquitously expressed TGF-β ligand or the L-TGF-β complex are complicated by the challenge of selecting and stratifying patients for clinical trials. Our study addresses this challenge by laying the groundwork for an intriguing therapeutic approach: targeting tumors with high expression of β8 with neutralizing αvβ8 antibodies either alone or in combination with PD-1/PD-L1 inhibitors, depending on the PD-L1 staining status.
Methods
Cell lines and reagents.
LL/2 (LLC1) (ATCC, CRL-1642), B16-F10 (ATCC, CRL-6475), TRAMP-C2 (gift from Larry Fong, UCSF, San Francisco, California, USA), (CHO-K1 [ATCC, CCL-61], MC38 (UCSF Preclinical Core), TMLC (gift from John Munger, NYU Medical Center, New York, New York, USA), and CHOlec3.2.8.1 (gift from Pamela Stanley, Albert Einstein College of Medicine, New York, New York, USA) were used throughout experimentation. CHOlec3.2.8.1 cells were stably transfected with human αv and β8 expression constructs, as previously described (64). Mouse tracheal epithelial cells were isolated from mouse tracheal epithelium and grown using conditional reprogramming (65). L-TGF-β1 was produced as described (66). Isotype control anti-SV5 (67) hybridomas were grown and purified using FPLC, as previously described (17). Antibodies were tested for endotoxin to confirm levels <0.2 EU/μg as determined by LAL method, (Genscript). Anti–mouse PD-1 (RMP1-14) and rat IgG2a isotype control (clone 2A3) were purchased (Bio X cell). Clone 1D11 (ATCC) is a pan–TGF-β isoform monoclonal antibody that cross-reacts with TGF-β1, -2, and -3 of human, mink, and mouse origin. Development and characterization of antibodies C6D4 and F9 are described in Supplemental Methods and Supplemental Figures 1 and 15–19. Recombinant integrins (αvβ1, αvβ3, αvβ5, αvβ6, αvβ8) were from R&D Systems.
Isolation and staining of tumor and immune cells.
Single cell suspensions were prepared from tumors by mincing and digesting with Collagenase NB8 (1 mg/ml; Crescent Chemical Co. Inc.), 0.1% hyaluronidase and 30 μg/ml DNAse I (MilliporeSigma) in RPMI 1640 for 90 minutes in a shaking 25°C incubator. After passing through a 70-μM nylon cell strainer (BD Biosciences), live tumor cells were negatively selected by magnetic beads (030-110-187, Miltenyi Biotec) or infiltrating lymphoid cells were enriched by density gradient centrifugation in Percoll (GE Heathcare) and harvested at the 40%/80% interface. For RNAseq, macrophages were positively selected by CD11b+, and CD4+ and CD8+ T cells were selected by negative magnetic bead separation (Miltenyi Biotech). For surface staining analysis or sorting, Fc receptors were blocked with 10 μg/ml (2.4G2, BD Biosciences), followed by fluorochrome-labeled antibodies: CD45 (30.F11), CD25 (PC61), LAP-β1 (TW716B4), PD-L1 (B7-H1), and PD-L2 (TY75) (all from BioLegend); Ly6c (AL21), F4/80 (BM8), Gr1 (RB6-8C5), CD11b (M1/70), TCRβ (H57-597), CD19 (ID3), and NK1.1 (PK136) (all from eBiosciences); MHCII (M5/114.14.2), CD11c (N418), CD4 (GK1.5), B220 (RA3-6B2), and CD206 (MMR) (all from BioLegend); and CD8α (5H10; Invitrogen). Cell-surface cytokine capture assay for IFN-γ (Miltenyi Biotec) was performed as described (68). Briefly, cells were washed once with 2 ml of cold RPMI buffer to remove any exogenous IFN-γ prior to binding to the IFN-γ catch reagent (Miltenyi Biotec). Cells were mixed briefly and incubated on ice for 5 minutes, prior to warming (37°C) and incubation for a 45-minute cytokine secretion window with the IFN-γ capture reagent, followed by washing and detection using the kit-supplied IFN-γ detection antibody. Flow cytometry acquisition was performed on a LSR II Flow Cytometer (BD Biosciences) and cell sorting by FACSAria (BD Biosciences) available through the Liver Center Flow Cytometry, or the Core Immunology Laboratory/CFAR Immunology Cores, respectively, at the UCSF. Gating for myeloid populations (g-MDSC, m-MDSC, TAM) was performed essentially as described (69, 70), with the exception that we did not use CD206, as it does not discriminate between MHCIIhi and MHCIIlo macrophage populations (70). All gating strategies were performed as shown (Supplemental Figures 9–13).
In silico gene expression analysis.
The EMBL-EBI expression database (https://www.ebi.ac.uk/gxa/home) was used to access curated RNAseq data from 675 commonly used human cell lines (71). Data were expressed as transcripts/million (TPM). KM plotter (http://kmplot.com) — a web application for meta-analysis–based biomarker assessment of publically available gene expression data (GEO [https://www.ncbi.nlm.nih.gov/geo/], EGA [https://www.ebi.ac.uk/ega/home], and TCGA [https://tcga-data.nci.nih.gov/docs/publications/tcga/?]) for breast, ovarian, lung, gastric, and liver cancer (35) — was used to generate Kaplan-Meier survival statistics for patients with triple-negative basal-type breast carcinomas (n = 186), serous ovarian carcinomas (n = 1,001), and NSCLC (n = 1,926). Cutoffs were determined by the auto-select setting and optimal probe selection by Jetset (72).
Mice.
C57BL/6 mice expressing GFP inserted into FoxP3 downstream of the endogenous Foxp3 STOP codon (B6.Cg-Foxp3tm2Tch/J) and WT C57BL/6 mice (all female except for TRAMP-C2 experiments using male), 8–10 weeks of age, were purchased (The Jackson Laboratory). Mice expressing the human bacterial artificial chromosome ITGB8 and not mouse itgb8 (itgb8–/–) have been described (73) and are congenic to C57BL/6 (Jax Speed Congenic Service). For the B16 lung colonization model, 2 × 105 cells were injected via tail vein and lungs of WT C57BL/6 mice and harvested at day 18–19. For the MC38, TRAMP-C2, and LLC models, cells (MC38, 5 × 105; TRAMP-C2, 10 × 106; LLC, 1 × 106) were injected in the flank of C57BL/6 mice and harvested when tumor reached tumor volumes of 1,000–2,000 mm3. Tumor volumes were calculated using the formula (length × width2)/2, which was validated in MC38 and LLC models where calculated volumes show a high degree of correlation with tumor weight (Supplemental Figure 6). Isotype or C6D4 (7 mg/kg i.p.) was given on days 7, 10, 13, or 16 for the MC38 model, or on days 17, 22, and 27 after tumor cell injection for the TRAMP-C2 model. Isotype, C6D4, RMP1-14, or a combination of C6D4 and RMP1-14 were given on days 0 and then once weekly for preventive dosing and once or twice weekly for therapeutic dosing on days 6, 9, 12, 15 for the LLC model (10 mg/kg i.p.). For β8 LLC RNAseq experiments, isotype or C6D4 was therapeutically administered on days 5 and 12, and tumors were resected on day 18. For qPCR experiments, isotype or C6D4 was therapeutically administered on days 6, 10, and 12, and tumors were resected on day 18.
TGF-β bioassay and adhesion assays.
TGF-β bioassays were performed exactly as previously described (9). L-TGF-β was used at 0.5 μg/ml to coat a 96-well ELISA plate. CHOlec3.2.8.1 cells stably transfected with αvβ8 were allowed to bind to the protein-coated wells for 30 minutes at room temperature. Unbound cells were washed off with PBS. The mAb C6D4 was added at the indicated concentration. Results were presented as stained cells detected after staining with crystal violet (OD590) with the values from cells adhering to BSA-coated wells subtracted.
Cell proliferation assay.
Wells of a 96-well were coated with L-TGF-β1 (2.5 μg/ml) blocked with 5% BSA and were washed. Cells (5 × 105) were added to coated wells, and after 24 hours, cell number was estimated calorimetrically (EZQuant, Alstem).
DNA constructs and transfection.
Mouse full-length itgb8 cDNA was generated from a truncated itgb8 construct (itgb8 tr pcDNA3.1, gift from Louis Reichardt, UCSF, San Francisco, California, USA) by inserting a PCR fragment generated using 5′-GTACTGATCCCAGAAGCATTGG-3′, 5′-CTCTGCGGCCGCTCATTAGAAGTTGCACCTGAAGGCC-3′. ΔSDL was amplified using β8 pcDNAneo as a template by splice overlap extension PCR with the mutagenic primer pair: 5-CTTTCGTCTTGGATTTGGCTCATACGTAGATAAAGGATACATCCATGTGCTGTCTTTGACAGAGAAC-3′, 5′-GTTCTCTGTCAAAGACAGCACATGGATGTATCCTTTATCTACGTATGAGCCAAATCCAAGACGAAAG-3′. cDNAs were inserting into pcDNA3.1 and pcDNA6. Transfections of B16 and LLC cells were performed using the Amaxa nucleofector system (Lonza).
RNAseq and qPCR.
Total RNA was isolated using kits according to the manufacturer’s instructions (Qiagen). The quality of the input RNA was confirmed by agarose gel electrophoresis and confirmed using the Agilent 2100 Bioanalyzer system. mRNA was purified from total RNA using poly-T oligo-magnetic beads, was fragmented, and was then used for first-strand cDNA synthesis with random hexamer primers and M-MuLV Reverse Transcriptase (RNase H). Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Double-stranded cDNA (ds-cDNA) was purified using AMPure XP beads (Beckman Coulter). The remaining overhangs of the purified ds-cDNA were converted into blunt ends using exonuclease/polymerase. After adenylation of 3′ ends of DNA fragments, adaptors with hairpin loop structure were ligated (NEBNext UltraTM RNA Library Prep Kit). cDNA fragments of 150–200 bp in length were purified using the AMPure XP system. The final library was achieved by PCR amplification and purification of PCR products by AMPure XP beads. Library quality was assessed by Qubit2.0, the insert size verified by Agilent 2100, and qPCR used to quantify the library effective concentration (>2nM). The cDNA library was sequenced using an Illumina HiSeqTM 2000 platform. Clean reads were obtained by removing reads from the raw-seq reads containing low-quality reads and/or adaptor sequences and mapped to the mouse genome using STAR. The gene expression level was then calculated using the reads per kilo bases per million reads (RPKM) method. For pairwise comparisons, read count was adjusted by trimmed mean of M values (TMM), and then, significance of differential expression was determined using the EdgeR package, with significance criterion set to q < 0.005 (Padj) and |log2 (fold change)| > 1. Differential expression analysis was performed using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Gene Ontology (GO) was used to determine significant overrepresentation of GO terms of DE genes. Data is available in the sequence read archive (accession no. SRP149751; https://www.ncbi.nlm.nih.gov/sra). qPCR was performed as previously described, with the exception that KiCqStart SYBR Green Primers (MilliporeSigma) were used (19).
Statistics.
All data are reported as means ± SEM. Comparisons between 2 different groups were determined using 2-tailed Student’s t test. One-way ANOVA was used for multiple comparisons and Tukey’s or Bonferroni’s post-hoc tests used to test for statistical significance. Significance was defined as P < 0.05. All statistical analyses were performed using the software package Prism 7.0b (GraphPad Software).
Study approval.
Archival surgical pathology material was obtained with full approval of the UCSF IRB in full accordance with Declaration of Helsinki principles. Written informed consent was received from participants prior to inclusion in the study. All animal studies have been approved by the UCSF IACUC.
Author contributions
NT, RIS, and SLN established and developed methodology and performed and analyzed in vivo and gene analysis experiments; AC, AJB, SI, JL, HY, MH, RM, MDL, JDM, FC, JLB, and SLN participated in antibody development, production, and characterization; AC, MGC, SI, and YC participated in structural biology experiments; AE, SI, and NT developed IHC staining protocols and performed IHC; RIS, SI, JP, RP, JMJ, and JLB performed flow cytometry experiments; SLN conceived, designed, and oversaw all experimentation and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported, in part, by grants the NIH (U54HL119893, R01HL113032 to SLN, R01HL134183 to SLN and YC, S10OD020054 to YC, and P41CA196276 to JM) and from the UC-CAI UCSF Catlyst2 (AB2664) and University of California Office of the President Tobacco-Related Disease Research Program to SLN. This work was also supported by R01DK093646 (JLB), the UCSF Liver Center - P30DK026743 (JLB, JP, and RP), and the Ibrahim El-Hefni Technical Training Foundation (JLB). JP was supported by NIH 1K01DK099405. JMJ was supported by NIH T32 AI 007334-27 and NIH F31 DK112607. YC is an investigator of the Howard Hughes Medical Institute. We thank Larry Fong (UCSF, San Francisco, California, USA) for the TRAMP-C2 cell line and helpful suggestions.
Version 1. 10/18/2018
Electronic publication
Footnotes
AJB’s present address is: Department of Molecular and Cell Biology, University of Washington, Seattle, Washington, USA.
Conflict of interest: JL, JM, JLB, AC, NT, and SLN are inventors on the international patent application number PCT/US17/54306, assigned to The Regents of the University of California.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: JCI Insight. 2018;3(20):e122591. https://doi.org/10.1172/jci.insight.122591.
Contributor Information
Naoki Takasaka, Email: ntakasak@hotmail.com.
Robert I. Seed, Email: robert.seed@ucsf.edu.
Anthony Cormier, Email: anthony.cormier@ucsf.edu.
Jianlong Lou, Email: LouJ@anesthesia.ucsf.edu.
Ahmed Elattma, Email: ahmedelattma@berkeley.edu.
Saburo Ito, Email: sabu-s55@jikei.ac.jp.
Haruhiko Yanagisawa, Email: haruhikov@mac.com.
Mitsuo Hashimoto, Email: mitsuoha-georgetown@live.jp.
Royce Ma, Email: ma.royce@gmail.com.
Jean Publicover, Email: Jean.publicover@ucsf.edu.
Rashaun Potts, Email: Rashaun.Potts@ucsf.edu.
Jillian M. Jespersen, Email: Jillian.Jespersen@ucsf.edu.
Fraser Conrad, Email: fraser.conrad@ucsf.edu.
Yifan Cheng, Email: yifan.cheng@ucsf.edu.
References
- 1.Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer. 2018;118(1):9–16. doi: 10.1038/bjc.2017.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neuzillet C, et al. Targeting the TGFβ pathway for cancer therapy. Pharmacol Ther. 2015;147:22–31. doi: 10.1016/j.pharmthera.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Lan Y, et al. Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci Transl Med. 2018;10(424):eaan5488. doi: 10.1126/scitranslmed.aan5488. [DOI] [PubMed] [Google Scholar]
- 4.Tauriello DVF, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538–543. doi: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- 5.Mariathasan S, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David JM, Dominguez C, McCampbell KK, Gulley JL, Schlom J, Palena C. A novel bifunctional anti-PD-L1/TGF-β Trap fusion protein (M7824) efficiently reverts mesenchymalization of human lung cancer cells. Oncoimmunology. 2017;6(10):e1349589. doi: 10.1080/2162402X.2017.1349589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickup M, Novitskiy S, Moses HL. The roles of TGFβ in the tumour microenvironment. Nat Rev Cancer. 2013;13(11):788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116(Pt 2):217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 9.Mu D, et al. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J Cell Biol. 2002;157(3):493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozawa A, Sato Y, Imabayashi T, Uemura T, Takagi J, Sekiguchi K. Molecular Basis of the Ligand Binding Specificity of αvβ8 Integrin. J Biol Chem. 2016;291(22):11551–11565. doi: 10.1074/jbc.M116.719138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold TD, et al. Excessive vascular sprouting underlies cerebral hemorrhage in mice lacking αVβ8-TGFβ signaling in the brain. Development. 2014;141(23):4489–4499. doi: 10.1242/dev.107193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenton TM, et al. Inflammatory cues enhance TGFβ activation by distinct subsets of human intestinal dendritic cells via integrin αvβ8. Mucosal Immunol. 2017;10(3):624–634. doi: 10.1038/mi.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarty JH, et al. Defective associations between blood vessels and brain parenchyma lead to cerebral hemorrhage in mice lacking alphav integrins. Mol Cell Biol. 2002;22(21):7667–7677. doi: 10.1128/MCB.22.21.7667-7677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aluwihare P, et al. Mice that lack activity of alphavbeta6- and alphavbeta8-integrins reproduce the abnormalities of Tgfb1- and Tgfb3-null mice. J Cell Sci. 2009;122(Pt 2):227–232. doi: 10.1242/jcs.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, et al. Absence of integrin-mediated TGFbeta1 activation in vivo recapitulates the phenotype of TGFbeta1-null mice. J Cell Biol. 2007;176(6):787–793. doi: 10.1083/jcb.200611044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, Motejlek K, Wang D, Zang K, Schmidt A, Reichardt LF. beta8 integrins are required for vascular morphogenesis in mouse embryos. Development. 2002;129(12):2891–2903. doi: 10.1242/dev.129.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Araya J, et al. Squamous metaplasia amplifies pathologic epithelial-mesenchymal interactions in COPD patients. J Clin Invest. 2007;117(11):3551–3562. doi: 10.1172/JCI32526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cambier S, et al. A role for the integrin alphavbeta8 in the negative regulation of epithelial cell growth. Cancer Res. 2000;60(24):7084–7093. [PubMed] [Google Scholar]
- 19.Kitamura H, et al. Mouse and human lung fibroblasts regulate dendritic cell trafficking, airway inflammation, and fibrosis through integrin αvβ8-mediated activation of TGF-β. J Clin Invest. 2011;121(7):2863–2875. doi: 10.1172/JCI45589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimura SL, Boylen KP, Einheber S, Milner TA, Ramos DM, Pytela R. Synaptic and glial localization of the integrin alphavbeta8 in mouse and rat brain. Brain Res. 1998;791(1-2):271–282. doi: 10.1016/S0006-8993(98)00118-8. [DOI] [PubMed] [Google Scholar]
- 21.Cambier S, et al. Integrin alpha(v)beta8-mediated activation of transforming growth factor-beta by perivascular astrocytes: an angiogenic control switch. Am J Pathol. 2005;166(6):1883–1894. doi: 10.1016/S0002-9440(10)62497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longenecker G, et al. Endocrine expression of the active form of TGF-beta1 in the TGF-beta1 null mice fails to ameliorate lethal phenotype. Cytokine. 2002;18(1):43–50. doi: 10.1006/cyto.2002.1025. [DOI] [PubMed] [Google Scholar]
- 23.Rachidi S, et al. Platelets subvert T cell immunity against cancer via GARP-TGFβ axis. Sci Immunol. 2017;2(11):eaai7911. doi: 10.1126/sciimmunol.aai7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, Shevach EM. GARP (LRRC32) is essential for the surface expression of latent TGF-beta on platelets and activated FOXP3+ regulatory T cells. Proc Natl Acad Sci USA. 2009;106(32):13445–13450. doi: 10.1073/pnas.0901944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshinaga K, et al. Perturbation of transforming growth factor (TGF)-beta1 association with latent TGF-beta binding protein yields inflammation and tumors. Proc Natl Acad Sci USA. 2008;105(48):18758–18763. doi: 10.1073/pnas.0805411105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knudson KM, Hicks KC, Luo X, Chen JQ, Schlom J, Gameiro SR. M7824, a novel bifunctional anti-PD-L1/TGFβ Trap fusion protein, promotes anti-tumor efficacy as monotherapy and in combination with vaccine. Oncoimmunology. 2018;7(5):e1426519. doi: 10.1080/2162402X.2018.1426519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin H, et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest. 2018;128(4):1708. doi: 10.1172/JCI120803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Meeteren LA, Goumans MJ, ten Dijke P. TGF-β receptor signaling pathways in angiogenesis; emerging targets for anti-angiogenesis therapy. Curr Pharm Biotechnol. 2011;12(12):2108–2120. doi: 10.2174/138920111798808338. [DOI] [PubMed] [Google Scholar]
- 29.Galon J, et al. Cancer classification using the Immunoscore: a worldwide task force. J Transl Med. 2012;10:205. doi: 10.1186/1479-5876-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Facciabene A, Motz GT, Coukos G. T-regulatory cells: key players in tumor immune escape and angiogenesis. Cancer Res. 2012;72(9):2162–2171. doi: 10.1158/0008-5472.CAN-11-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umansky V, Blattner C, Gebhardt C, Utikal J. The Role of Myeloid-Derived Suppressor Cells (MDSC) in Cancer Progression. Vaccines (Basel) 2016;4(4):E36. doi: 10.3390/vaccines4040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B, Li Q, Qin L, Zhao S, Wang J, Chen X. Transition of tumor-associated macrophages from MHC class II(hi) to MHC class II(low) mediates tumor progression in mice. BMC Immunol. 2011;12:43. doi: 10.1186/1471-2172-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dyck L, Lynch L. New Job for NK Cells: Architects of the Tumor Microenvironment. Immunity. 2018;48(1):9–11. doi: 10.1016/j.immuni.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Swann JB, Smyth MJ. Immune surveillance of tumors. J Clin Invest. 2007;117(5):1137–1146. doi: 10.1172/JCI31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Culhane AC, Quackenbush J. Confounding effects in “A six-gene signature predicting breast cancer lung metastasis”. Cancer Res. 2009;69(18):7480–7485. doi: 10.1158/0008-5472.CAN-08-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coscia F, et al. Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nat Commun. 2016;7:12645. doi: 10.1038/ncomms12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riely GL. What, When, and How of Biomarker Testing in Non-Small Cell Lung Cancer. J Natl Compr Canc Netw. 2017;15(5S):686–688. doi: 10.6004/jnccn.2017.0073. [DOI] [PubMed] [Google Scholar]
- 39.Reck M, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 40.Stockis J, et al. Blocking immunosuppression by human Tregs in vivo with antibodies targeting integrin αVβ8. Proc Natl Acad Sci USA. 2017;114(47):E10161–E10168. doi: 10.1073/pnas.1710680114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Travis MA, et al. Loss of integrin alpha(v)beta8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 2007;449(7160):361–365. doi: 10.1038/nature06110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Worthington JJ, et al. Integrin αvβ8-Mediated TGF-β Activation by Effector Regulatory T Cells Is Essential for Suppression of T-Cell-Mediated Inflammation. Immunity. 2015;42(5):903–915. doi: 10.1016/j.immuni.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuende J, et al. Monoclonal antibodies against GARP/TGF-β1 complexes inhibit the immunosuppressive activity of human regulatory T cells in vivo. Sci Transl Med. 2015;7(284):284ra56. doi: 10.1126/scitranslmed.aaa1983. [DOI] [PubMed] [Google Scholar]
- 44.Rifkin DB. Latent transforming growth factor-beta (TGF-beta) binding proteins: orchestrators of TGF-beta availability. J Biol Chem. 2005;280(9):7409–7412. doi: 10.1074/jbc.R400029200. [DOI] [PubMed] [Google Scholar]
- 45.Riemenschneider MJ, et al. TGF-ß isoforms in cancer: Immunohistochemical expression and Smad-pathway-activity-analysis in thirteen major tumor types with a critical appraisal of antibody specificity and immunohistochemistry assay validity. Oncotarget. 2015;6(29):26770–26781. doi: 10.18632/oncotarget.5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen ML, Yan BS, Kozoriz D, Weiner HL. Novel CD8+ Treg suppress EAE by TGF-beta- and IFN-gamma-dependent mechanisms. Eur J Immunol. 2009;39(12):3423–3435. doi: 10.1002/eji.200939441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gandhi R, Anderson DE, Weiner HL. Cutting Edge: Immature human dendritic cells express latency-associated peptide and inhibit T cell activation in a TGF-beta-dependent manner. J Immunol. 2007;178(7):4017–4021. doi: 10.4049/jimmunol.178.7.4017. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura K, Kitani A, Strober W. Cell contact-dependent immunosuppression by CD4(+)CD25(+) regulatory T cells is mediated by cell surface-bound transforming growth factor beta. J Exp Med. 2001;194(5):629–644. doi: 10.1084/jem.194.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oida T, Weiner HL. TGF-β induces surface LAP expression on murine CD4 T cells independent of Foxp3 induction. PLoS One. 2010;5(11):e15523. doi: 10.1371/journal.pone.0015523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rezende RM, et al. Identification and characterization of latency-associated peptide-expressing γδ T cells. Nat Commun. 2015;6:8726. doi: 10.1038/ncomms9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Mammary-tumor-educated B cells acquire LAP/TGF-β and PD-L1 expression and suppress anti-tumor immune responses. Int Immunol. 2016;28(9):423–433. doi: 10.1093/intimm/dxw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41(1):49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coelho AL, Schaller MA, Benjamim CF, Orlofsky AZ, Hogaboam CM, Kunkel SL. The chemokine CCL6 promotes innate immunity via immune cell activation and recruitment. J Immunol. 2007;179(8):5474–5482. doi: 10.4049/jimmunol.179.8.5474. [DOI] [PubMed] [Google Scholar]
- 54.Jin L, et al. CCL24 contributes to HCC malignancy via RhoB- VEGFA-VEGFR2 angiogenesis pathway and indicates poor prognosis. Oncotarget. 2017;8(3):5135–5148. doi: 10.18632/oncotarget.14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peperzak V, et al. CD8+ T cells produce the chemokine CXCL10 in response to CD27/CD70 costimulation to promote generation of the CD8+ effector T cell pool. J Immunol. 2013;191(6):3025–3036. doi: 10.4049/jimmunol.1202222. [DOI] [PubMed] [Google Scholar]
- 56.Wendel M, Galani IE, Suri-Payer E, Cerwenka A. Natural killer cell accumulation in tumors is dependent on IFN-gamma and CXCR3 ligands. Cancer Res. 2008;68(20):8437–8445. doi: 10.1158/0008-5472.CAN-08-1440. [DOI] [PubMed] [Google Scholar]
- 57.Kurachi M, et al. Chemokine receptor CXCR3 facilitates CD8(+) T cell differentiation into short-lived effector cells leading to memory degeneration. J Exp Med. 2011;208(8):1605–1620. doi: 10.1084/jem.20102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oghumu S, et al. CXCR3 deficiency enhances tumor progression by promoting macrophage M2 polarization in a murine breast cancer model. Immunology. 2014;143(1):109–119. doi: 10.1111/imm.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akbay EA, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3(12):1355–1363. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Selvakumaran M, Yao KS, Feldman MD, O’Dwyer PJ. Antitumor effect of the angiogenesis inhibitor bevacizumab is dependent on susceptibility of tumors to hypoxia-induced apoptosis. Biochem Pharmacol. 2008;75(3):627–638. doi: 10.1016/j.bcp.2007.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderton MJ, et al. Induction of heart valve lesions by small-molecule ALK5 inhibitors. Toxicol Pathol. 2011;39(6):916–924. doi: 10.1177/0192623311416259. [DOI] [PubMed] [Google Scholar]
- 62.Vitsky A, et al. Homeostatic role of transforming growth factor-beta in the oral cavity and esophagus of mice and its expression by mast cells in these tissues. Am J Pathol. 2009;174(6):2137–2149. doi: 10.2353/ajpath.2009.080723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishimura SL. Integrin-mediated transforming growth factor-beta activation, a potential therapeutic target in fibrogenic disorders. Am J Pathol. 2009;175(4):1362–1370. doi: 10.2353/ajpath.2009.090393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nishimura SL, Sheppard D, Pytela R. Integrin alpha v beta 8. Interaction with vitronectin and functional divergence of the beta 8 cytoplasmic domain. J Biol Chem. 1994;269(46):28708–28715. [PubMed] [Google Scholar]
- 65.Liu X, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180(2):599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi M, et al. Latent TGF-β structure and activation. Nature. 2011;474(7351):343–349. doi: 10.1038/nature10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Razai A, et al. Molecular evolution of antibody affinity for sensitive detection of botulinum neurotoxin type A. J Mol Biol. 2005;351(1):158–169. doi: 10.1016/j.jmb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 68.Hashimoto M, et al. TGF-β-Dependent Dendritic Cell Chemokinesis in Murine Models of Airway Disease. J Immunol. 2015;195(3):1182–1190. doi: 10.4049/jimmunol.1500348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cassetta L, et al. Isolation of Mouse and Human Tumor-Associated Macrophages. Adv Exp Med Biol. 2016;899:211–229. doi: 10.1007/978-3-319-26666-4_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su X, et al. Antagonizing Integrin β3 Increases Immunosuppression in Cancer. Cancer Res. 2016;76(12):3484–3495. doi: 10.1158/0008-5472.CAN-15-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klijn C, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33(3):306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 72.Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics. 2011;12:474. doi: 10.1186/1471-2105-12-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minagawa S, et al. Selective targeting of TGF-β activation to treat fibroinflammatory airway disease. Sci Transl Med. 2014;6(241):241ra79. doi: 10.1126/scitranslmed.3008074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.