Abstract
Viroids were described 47 years ago as the smallest RNA molecules capable of infecting plants and autonomously self-replicating without an encoded protein. Work on viroids initiated the development of a number of innovative methods. Novel chromatographic and gelelectrophoretic methods were developed for the purification and characterization of viroids; these methods were later used in molecular biology, gene technology and in prion research. Theoretical and experimental studies of RNA folding demonstrated the general biological importance of metastable structures, and nuclear magnetic resonance spectroscopy of viroid RNA showed the partially covalent nature of hydrogen bonds in biological macromolecules. RNA biochemistry and molecular biology profited from viroid research, such as in the detection of RNA as template of DNA-dependent polymerases and in mechanisms of gene silencing. Viroids, the first circular RNA detected in nature, are important for studies on the much wider spectrum of circular RNAs and other non-coding RNAs.
INTRODUCTION
Viroids are single-stranded, covalently closed circular RNA molecules. As a distinct class of pathogens, they are clearly distinguished from viruses by their small size (∼250–400 nt), do not encode any protein and lack a capsid. The phenomenon of a small infectious RNA was described first by Diener (1), Singh & Clark (2), Sänger (3) and Semancik & Weathers (4). Viroids are infectious for a variety of higher plants including potato, tomato, avocado, ornamental plants and coconut palm, and can cause disease leading to significant agricultural damage. By contrast, viroids have not been observed as infectious agents in animals or humans. Despite some structural similarity, the hepatitis delta virus (HDV) is about 5 times larger and encodes two proteins from one open reading frame (5). Viroids, as isolated RNA molecules with the complete biology of a virus, received a lot of scientific attraction (6–8).
More than a decade after the discovery of the circularity of viroid RNA (9), cellular circular RNAs (circRNA) were observed as genomic transcripts with potential functions in gene regulation (10). Thus, viroids were not only molecules of enormous biological interest on their own, but stimulated the developments of new experimental and conceptual approaches. Novel methods of nucleic acid purification and gel electrophoresis developed during viroid research were later applied to nucleic acid research in general. Combining the experimental and theoretical approaches, the concept of metastable structures was developed and could be verified in vivo, when the replication cycle of viroids was studied in detail. The application of the improved electrophoretic methods showed that nucleic acids are not essential for prion infectivity, confirming that prions are indeed infectious proteins. Analysis of a viroid fragment by nuclear magnetic resonance (NMR) spectroscopy demonstrated directly that hydrogen bonds in RNA have partially covalent character. Predictions from evolution theory about the relationship between error rate and genome size were verified with viroids.
In the following we will describe developments in RNA technology. Furthermore basic insights into RNA biology originating from viroid research will be presented.
VIROIDS—THE FIRST CIRCULAR RNA DISCOVERED IN NATURE
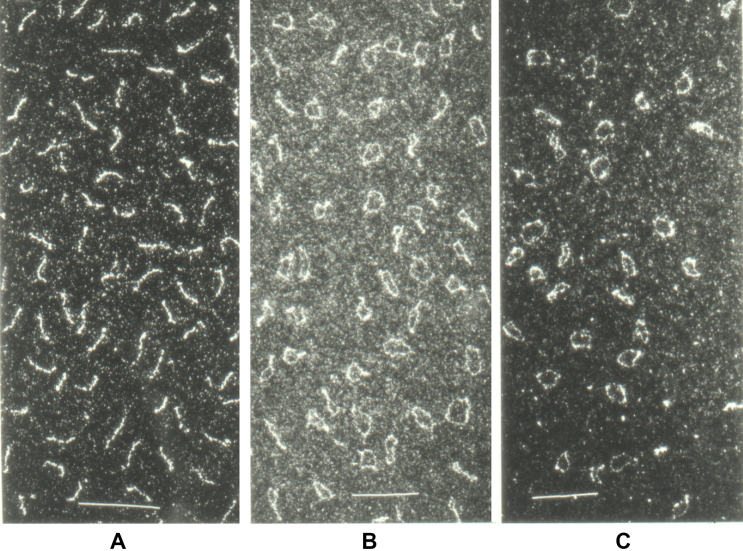
Electron micrographs of highly purified viroid RNA showed rod-like structures under native conditions (Figure 1A); in contrast, under fully denaturing conditions (Figure 1C), >80% of viroid RNAs were visible as circular single-stranded molecules (9,11). Accessible 5′ or 3′ ends of viroid RNA could not be detected by enzymatic and chemical modification reactions (9). Potato spindle tuber viroid (PSTVd) was the first pathogen of a eukaryotic host for which the sequence was established (12). Without reverse transcription at hand at the time, highly purified viroid RNA was enzymatically cleaved into fragments, which were sequenced by nucleolytic degradation and separation by either polyacrylamide gel electrophoresis (PAGE) (13) or two-dimensional chromatography (14). The full sequence was assembled by hand from the sequences of overlapping fragments. The sequence of PSTVd unequivocally demonstrated that it consists of a single-stranded, covalently closed, circular RNA molecule. The circularity of viroid RNA is essential for the replication cycle; it increases thermodynamic stability and supports the switching between different structural and functional states (see below). CircRNAs are now recognized as a large class of RNA which are non-coding and non-infectious, have a prolonged half-life, function in the regulation of gene expression or are of unknown function (15,16).
Figure 1.
Electron micrographs of viroid RNA. (A) PSTVd RNA spread onto water at room temperature showing rod-shaped structures. (B) PSTVd RNA denatured in 3 M urea, 0.1 M Tris–HCl, 0.1 mM ethylenediaminetetraacetic acid, pH 7 and spread onto water at 40 °C; the micrograph shows various structures ranging from rods and tennis rackets to single-stranded circles. (C) PSTVd RNA denatured as in (B), spread onto water at 70 °C showing mostly single-stranded circles. Calibration bars: 100 nm. For details see references (9,11).
Circular RNA as basis for autonomous replication
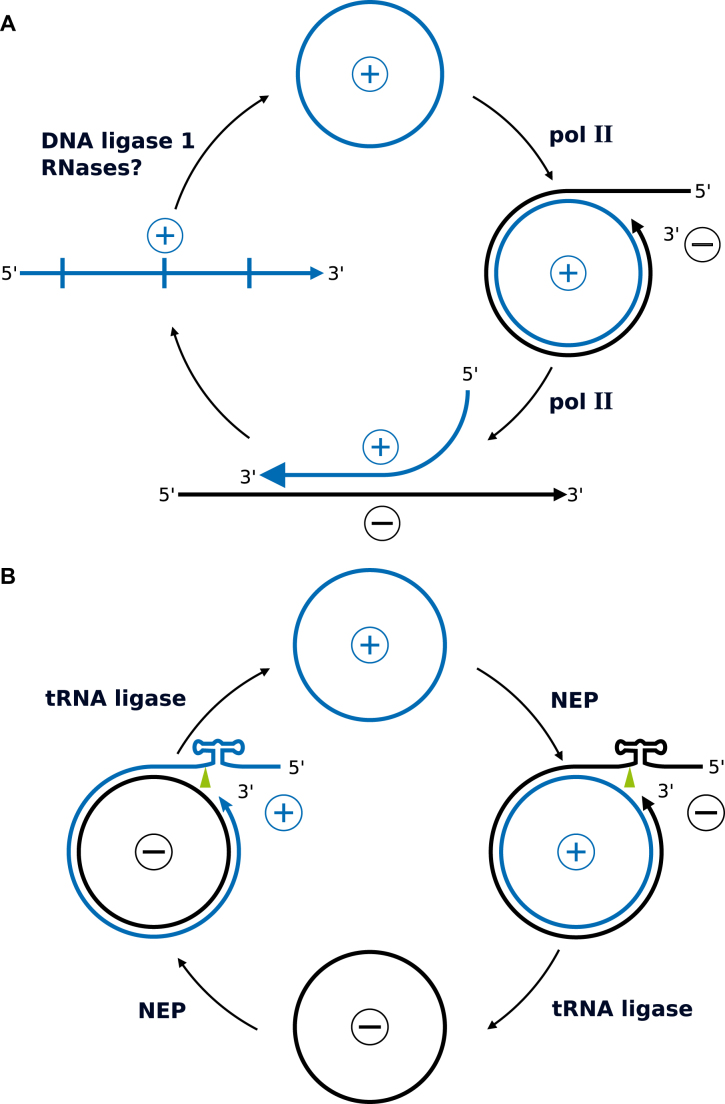
Viroids are replicated by two different rolling-circle mechanisms (Figure 2), depending on the viroid family: a symmetric cycle with (+)- and (−)-circles in case of Avsunviroidae members (with type species Avocado sunblotch viroid) or an asymmetric cycle with only (+)-circles in case of Pospiviroidae members (with type species Potato spindle tuber viroid) (17–19). As non-coding RNAs, viroids have to rely on host enzymes in most steps of their replication cycle. Very surprisingly, DNA-dependent RNA polymerases were discovered to be responsible for viroid transcription from RNA templates (20,21), in contrast to the expectation of replication by an RNA-directed RNA polymerase of the host (see below). DNA ligase I (22) and transfer RNA (tRNA) ligase (23) support the circularization of specific viroid monomers from Pospiviroidae and Avsunviroidae members, respectively. The unprecedented finding of cellular DNA-dependent enzymes accepting viroid RNA as template is considered to be due to the particular secondary structure of viroids and other unknown factors.
Figure 2.
Replication cycle of viroids. (A) Members of Pospiviroidae replicate in an asymmetric cycle. DNA-dependent RNA polymerase (polII) transcribes mature circular RNA into oligomeric (−) intermediates and these into oligomeric (+) intermediates. The (+) intermediates are enzymatically cleaved into monomers and ligated with DNA ligase 1 into mature circles. (B) Members of Avsunviroidae replicate in a symmetric cycle. Circular RNA is transcribed into oligomeric intermediates by nuclear-encoded polymerase (NEP). The intermediates are cleaved into monomeric units by the internal hammerhead ribozyme and ligated by tRNA ligase into circles. Molecules are shown here without structure, but the presence of thermodynamically stable as well as metastable structures (Figure 11) is critical for each step of the different replication cycles.
The first step in the asymmetric cycle of Pospiviroidae members is the synthesis of a linear oligomeric (−)-stranded intermediate by DNA-dependent RNA polymerase II (polII) in the nucleoplasm using the circular (+)-stranded RNA as template. The oligomeric (−)-strand is then transcribed by polII into (+)-stranded oligomers, enzymatically processed to monomers, circularized, and ‘stored’ in the nucleolus (24) as demonstrated by confocal laser scanning microscopy (25), an early application of 3D microscopy.
The first step in the symmetric cycle of Avsunviroidae members is synthesis of a linear oligomeric (−)-stranded intermediate by nuclear-encoded polymerase in the chloroplast (21). The oligomeric (−)-strand is cleaved to monomers by the internal hammerhead ribozyme (HHrz; see below) and circularized (23,26,27). Subsequently, these steps are repeated from (−)-circles to yield (+)-circles (28).
Characterization of the DNA-like features of viroids by ultracentrifugation with fluorescence detection
The specific binding of viroid RNA to polII and the flexibility or stiffness of the rod-like structure of viroids were studied by analytic ultracentrifugation. The shortage of purified viroid material was overcome by establishing fluorescence detection optics (29), which is more sensitive than the commercially available ultraviolet (UV) absorption optics by one to two orders of magnitude. A laser beam with the aperture of ∼0.8° scans the sample cell in radial direction, excites the fluorescent dye ethidium bromide intercalated into the double-stranded regions of the nucleic acids, and absorbed totally in a light trap outside the cell. The fluorescence emission is collected with an aperture of ∼50° and recorded. In the sedimentation profile, the boundaries of free viroid and viroid-polymerase complexes were clearly separated, with two polymerase molecules bound per viroid confirming electron microscopic analysis (30).
The stiffness of viroids was compared to that of double-stranded RNA (dsRNA) and double-stranded DNA (dsDNA). It was observed that viroids are less stiff than dsRNA but similar to dsDNA (31). In biophysical terms, the persistence length of dsRNA is ∼1200 Å, that of dsDNA ∼600 Å, and that of viroids 300 Å, confirming the rod-like structure of viroids in solution. The concept of fluorescence optics originally developed for viroid studies was later applied by researchers in other areas, for example in studies on protein–protein interaction (32–35).
Gene silencing
It was not obvious for some time whether DNA-dependent RNA polymerase II was the only enzyme involved in PSTVd replication. In Sänger’s group, the RNA-directed RNA polymerase from tomato (LeRDR1) was purified to homogeneity, its gene isolated and sequenced, and its enzymatic properties characterized (36–38). RDR1 expression is enhanced after viroid (or virus) infection, is not responsible for viroid (or virus) replication, but is involved in control of virus accumulation (39). The isolation of RDR1 was facilitated by the non-coding nature of viroid in contrast to viruses, which code for and express their own RNA-dependent RNA polymerases in large amounts.
The first association of RNA silencing with DNA methylation and suppression of transcription was found with viroids: members of Pospiviroidae are able to trigger RNA-directed DNA methylation of homologous sequences, a process discovered in viroid-infected tobacco plants containing genome-integrated PSTVd complementary DNA (cDNA) copies (40). Furthermore, viroids are able to induce post-transcriptional gene silencing via viroid-derived small RNAs (41–43).
Hammerhead ribozymes—first found in pathogens
While the research groups of Cech (44) and Altman (45) described ribozymes in a cellular context as self-splicing introns and RNase P, respectively, HHrz were first detected in exogenous, infectious RNAs, the satellite RNA of tobacco ringspot virus (46) and avocado sunblotch viroid (26). In both cases the HHrz cleaves replication intermediates in cis. The catalytic motifs were later shown to be ubiquitously present in the genomes of organisms from all branches of the tree of life (47–49). A minimal HHrz consists of only 13 highly conserved nucleotides in a junction loop from which three helical regions consisting of any base pair composition emerge. These observations led to the design and development of trans-acting HHrz (50,51). In addition to the minimal motif, natural HHrz encompass tertiary interactions that lower the requirement for divalent cations to those present in vivo (52–54). Minimal trans-acting HHrz, however, are only catalytically active at Mg2 + concentrations that are higher than those available in cells but they can also be designed to include tertiary interactions and thus to act at lower Mg2 + (55,56). HHrz with trans-activity are now considered as drugs to induce post-transcriptional gene silencing (57–60).
Viroid error rates—confirmation of a theory in nature
The cellular DNA-dependent polymerases introduce many mutations into viroid progeny sequences, presumably due to the non-physiological nature of the RNA templates (61,62). Natural selection acts on existing genetic variants to optimize fitness, termed ‘survival of the fittest’. Quasispecies theory predicts that slower replicators will be favored over faster replicators if mutations of the slow replicator give rise to progeny that is similar fit as the parent and mutations of the fast replicator dramatically decrease its fitness. This effect is termed ‘survival of the flattest’ (63,64) and was demonstrated with two different viroids: the more slowly replicating viroid—with a lower influence of mutations on the replication rate—outcompeted a faster replicating viroid, if the mutation rate was raised by UV radiation (65).
The error rates of chrysanthemum chlorotic mottle viroid (66) and eggplant latent viroid (67), two members of Avsunviroidae, were found to be in the range of 1/1000 to 1/400, while the error rate of PSTVd seems to be lower by a factor of ∼5 (67,68). The high error rate observed fits well into the theory of the viral quasispecies developed by Eigen (69) and of the inverse correlation between genome size and replication fidelity (70,71).
VIROID PURIFICATION—THE BASIS OF PLASMID-PURIFICATION KITS
In the early days of viroid research, purification of viroid RNA was a tedious procedure (1,7,72). PAGE under denaturing conditions was the method of choice. Identification of the viroid band in the gel had to be carried out by eluting the RNA from many gel slices, followed by testing the content of each slice for infectivity. Figure 3 shows testing the infectivity of different gel slices in tomato plants. Only micrograms of pure viroid RNA could be eluted, which, however, have sufficed for the identification of viroid circles by electron microscopy, early biophysical characterization and extended work of sequence analysis.
Figure 3.
An early assay method for viroid. A crude RNA extract from a PSTVd-infected tomato plant was separated by denaturing PAGE (cf. lane ‘M’ of Figure 5B). After electrophoresis the gel under the slot of application was cut horizontally into about 30 slices, the RNA eluted and a series of tomato plants inoculated. The photograph shows a ‘tomatogram’, in which the front row of tomato plants was inoculated with a higher concentration of RNA than the back row. Clearly, the highest concentration of PSTVd RNA was in gel slices 19 and 20. The photograph is courtesy of Heinz-Ludwig Sänger(†), Martinsried.
In order to obtain larger amounts of viroid RNA for biophysical studies, a chromatographic procedure was developed (73,74), in which size exclusion and anionic exchange principles were combined. In Figure 4 the structure of silica modifications with an anionic exchange group is shown. After optimization of the silica gel’s pore size and of the elution conditions the chromatographic fractionation of RNA crude extract from viroid-infected plants was achieved (Figure 5). Milligram amounts of highly purified viroid RNA were obtained as required for example for NMR experiments (described below). This chromatographic method also separated supercoiled plasmids from crude bacterial extracts, restriction fragments and oligonucleotides (75–77).
Figure 4.
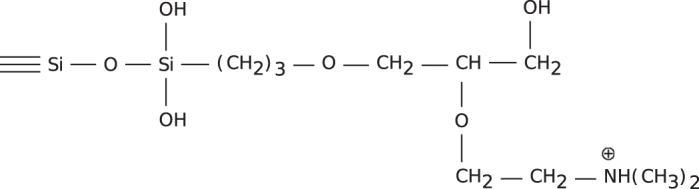
Unit structure of silica modifications with an anionic exchange group. A silica gel (10 μm, 500 Å pore size) was reacted with (1,2-epoxy-3-propylpropoxy)-trimethoxysilane and condensed with N,N-dimethylaminoethanol to yield DMA-500. From (74).
Figure 5.
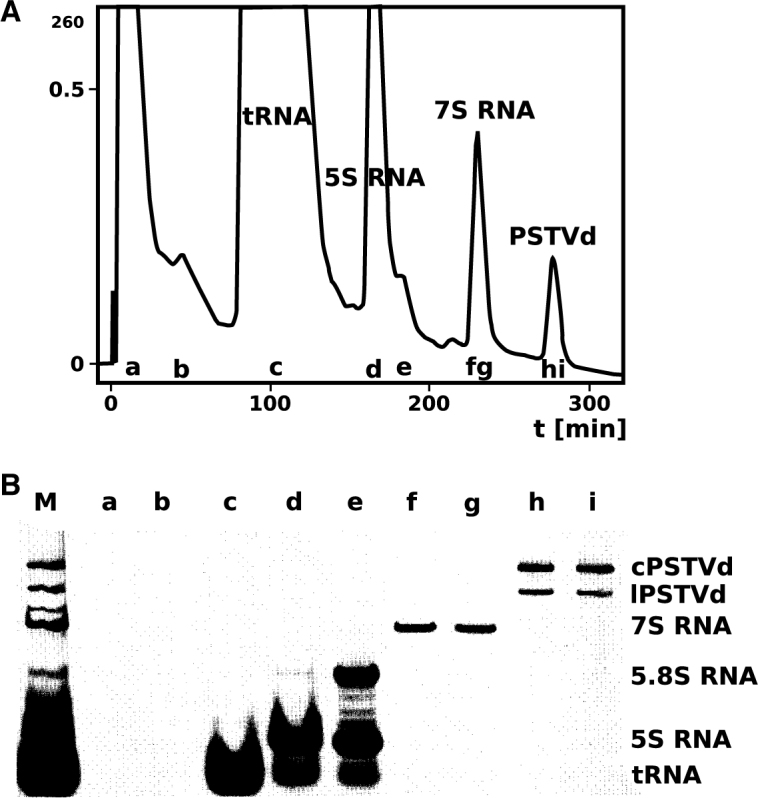
Chromatography of crude RNA extract from PSTVd-infected tomato plants. (A) Preparative chromatographic fractionation of 30 mg RNA crude extract from viroid-infected plants on a DMA-500 column. (B) Gel electrophoretic analysis (5% PAGE, denaturing conditions) of fractions as in (A); the non-chromatographed sample is shown in slot M. Modified from (74).
A limitation of this method became obvious when the columns were used for plasmid purification in microbiology laboratories: the columns had a slight ‘memory effect’; i. e. only minute amounts of one plasmid were eluted when the columns were used in the next run with another plasmid. This observation led to the development of commercially available kits (Qiagen, Hilden, Germany) for single use in plasmid preparation with complete exclusion of cross-contamination. Very soon most researchers realized that the kits now available from several companies were not only safe but also easy and fast to use and less expensive.
OPTIMAL AND SUBOPTIMAL STRUCTURES—A GENERAL PROBLEM OF RNA STRUCTURE RESEARCH
Without knowledge of viroid sequences Langowski et al. (78) concluded from optical melting curves of several different viroid RNAs and simulations based on partition functions that the high cooperativity but relatively low temperature midpoints of denaturation point toward a rod-like secondary structure composed of short helices and loops but no or only a low number of junctions. The high cooperativity contradicted a highly branched tRNA- or messenger RNA-like structure, while the low denaturation temperature contradicted a dsRNA-like structure. This view was supported by results from analytical ultracentrifugation experiments that indicated a highly extended structure similar to that of dsDNA but contradicted a more globular shape like that of tRNA (9).
Gross and his research group (12) proposed a secondary structure of PSTVd based on manually maximizing the number of base pairs taking into consideration the nuclease and bisulfite accessibility data. ‘Tinoco plots’ (79), a 2D graphic representation of all possible base pairs and their neighbors were applied. Thermodynamically optimal structures at temperatures relevant for interpretation of equilibrium or kinetic denaturation curves were identified (80,81). A plot for the first HDV sequence with close to 1700 nts (82) could only be laid out on several tables (Steger, unpublished).
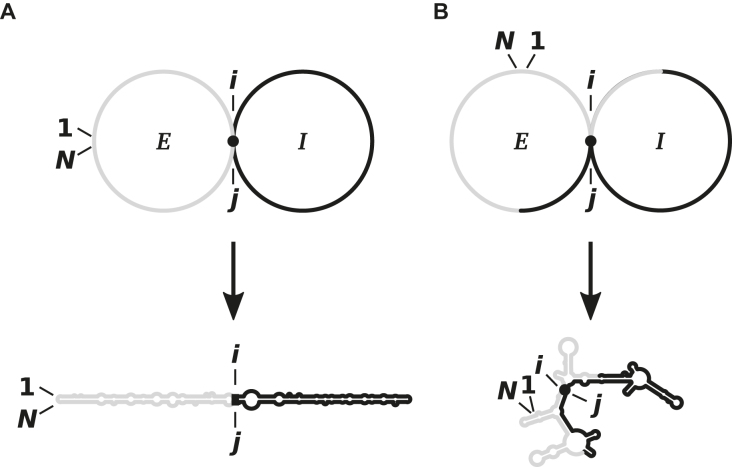
The RNA folding program of M. Zuker called mfold (83) was modified to allow the calculation of thermodynamically optimal as well as suboptimal secondary structures for circRNAs. This enhancement was based on the argument that each basepair at position i, j divides a structure in two parts, the so-called ‘included fragment’ with nucleotide range i < j and and the ‘excluded fragment’ with range j ≤ N < 1 ≤ i (Figure 6). A doubling of memory space then permitted to find the optimal structure of both parts in relation to the basepair i, j. If the given basepair does not belong to the structure of minimum free energy, a suboptimal structure is found (84). At that time we were unaware of the general importance of suboptimal structures and circularity in RNA structure research and published the advanced algorithm as an appendix to (84). A detailed description of the same algorithm can be found in (85).
Figure 6.
Two examples to the algorithm for prediction of suboptimal structures. The nucleotides are numbered from 1 to N. Sequence parts I and E belong to the ‘included fragment’ with nucleotide indices from i to j and to the ‘excluded fragment’ with j ≤ N < 1 ≤ i, respectively. Doubling of memory space (84) in comparison to that used in (83) permitted the algorithm to find for any given basepair i, j the optimal structures in direction of I and E. (A) If the basepair i, j belongs to a structure with minimum free energy, the optimal structure is found. (B) If the basepair i, j is not part of a structure with minimum free energy, a suboptimal structure is found.
Since their introduction, these programs were enhanced (86–88), included better and more parameters (89), and enabled the prediction of partition functions (86,88,90,91) or even all suboptimal structures (92,93). On the experimental side, structural mapping with ‘selective 2′-hydroxyl acylation analyzed by primer extension’ superseded previously developed techniques (94–97), while combinations of mutational studies and predictions gave insight into the 3D structure of many viroid loops and their functions (98–100).
NOVEL GEL ELECTROPHORETIC METHODS FOR VIROIDS, OTHER RNAS AND PRIONS
Since the 1970’s, PAGE has become a well established analytical and preparative method. Circular and linear viroids under native and fully denaturing conditions migrate as narrow, well defined bands. In particular, the circularity of viroid RNA leads to extraordinary low electrophoretic mobility under denaturing conditions, whereas the rod-like native structure behaves similarly to a dsRNA of comparable length. The extraordinary PAGE properties of viroids prompted for other applications (see below).
Two-dimensional and return gel electrophoresis for diagnostics of circular RNA
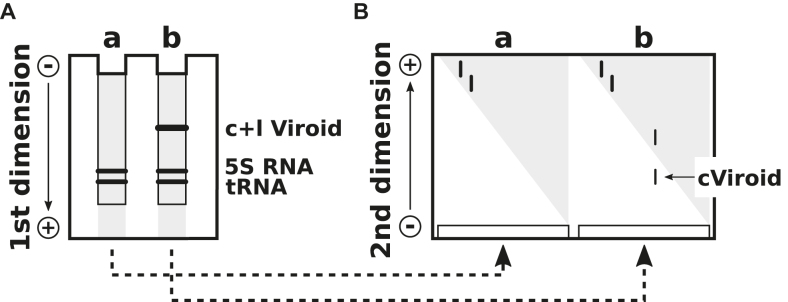
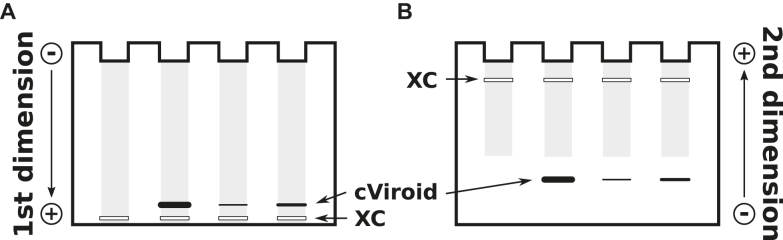
Denaturing conditions can be established by high concentrations of urea or of other denaturants or by high temperature. In 2D gel electrophoresis, native conditions are established in the first and fully denaturing conditions in the second dimension (Figure 7). All RNAs and DNAs from a crude extract run on or faster than a diagonal-like front (Figure 7B); because the sample is a crude extract, isolated bands are not visible. Only the circular viroid RNA is well separated as a clear band behind the front (101). In a modification of the same principle (Figure 8), all nucleic acids running faster than the viroid RNA and in front of the xylene cyanol marker have left the gel in the first dimension. The denaturing conditions were established by changing the running buffer, raising the temperature and reversing the direction of gel electrophoretic migration. The circRNAs appear to be well separated behind all other nucleic acids (102).
Figure 7.
Two-dimensional gel electrophoresis. (A) The first electrophoresis is a conventional PAGE with native conditions. Circular (c) and linear (l) viroid RNA can not be visualized by standard staining techniques due to the high background of other nucleic acids (gray). After electrophoresis the lanes a and b are cut from the gel; these are polymerized at the bottom of a new gel matrix. (B) Due to the denaturing conditions used for the second dimension, the circular viroid, which migrates very slowly, is well separated from other nucleic acids. Modified from (101).
Figure 8.
Return gel electrophoresis. (A) The first electrophoresis is a conventional PAGE with high-salt conditions (89 mM Tris, 89 mM boric acid, pH 8.3) and low temperature (20 °C). If the dye xylene cyanol (XC), present in the samples, is nearly running out of the gel matrix, the buffer is exchanged to low-salt conditions (10 mM Tris, 10 mM boric acid, pH 8.3, 8 M urea), the temperature is raised to 60°C and the direction of the electric field is inverted. (B) During the second electrophoresis step all nucleic acids are denatured; then the circular viroid migrates more slowly than all other, larger nucleic acids. Modified from (102).
Both methods have been applied by several research groups for diagnostic purposes (e. g. (103–105)); thus, the use of radioactively labeled or fluorescent oligonucleotides for hybridization could be completely avoided. Viroid-like, circular satellite RNAs, so-called virusoids, were diagnosed by these procedures as well (101,102). Not only 3′-5′ covalently closed circles but also 2′-5′ circles as in lariat structures were analyzed by 2D gel electrophoresis (106,107). The advantage of return gel electrophoresis is due to the ability of analyzing more samples in a single gel and to the lack or requiring repolymerization of a second gel.
Temperature gradient gel electrophoresis for conformational analysis of nucleic acids and proteins
Denaturing conditions were applied also as a continuous gradient in one and the same gel but in the second dimension, either as denaturant gradient or as temperature gradient gel electrophoresis (TGGE). Thus, continuous structural transition curves can be visualized. Temperature gradients have the advantage over denaturant gradients that handling is easier and the transition curves correlate better with optical melting and theoretical transition curves.
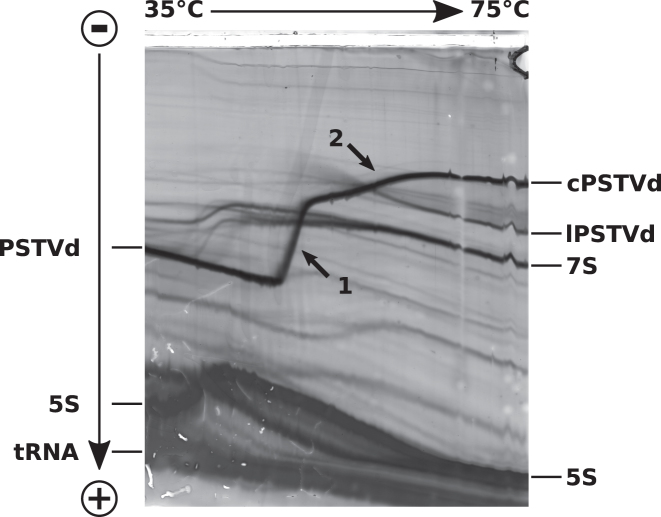
Figure 9 shows the TGGE analysis of a crude RNA extract from tomato plants infected with PSTVd. The rod-shaped secondary structure of circular PSTVd (cPSTVd) has a relatively high electrophoretic mobility at low temperature; it denatures completely in a highly cooperative main transition (see arrow 1 in Figure 9) into a mostly single-stranded structure with at least two extra-stable hairpins I and II (see below); these hairpins denature above the main transition (see arrow 2 in Figure 9). The branched hairpin-containing structure and especially the open circle occupy a larger hydrodynamic volume, and thus migrate much slower than the rod-like structure.
Figure 9.
Temperature gradient gel electrophoresis. TGGE analysis of a crude RNA extract from tomato plants infected with PSTVd. A linear temperature gradient is applied perpendicular to the electric field. The sample is loaded into the broad central slot (top). RNAs were visualized by silver staining. The rod-like structure of circular PSTVd (cPSTVd; 359 nt) migrates relatively fast at low temperature; it denatures in a highly cooperative transition (see arrow 1) into a mostly single-stranded structure with at least two extrastable hairpins I and II; these hairpins denature above the main transition (see arrow 2). Linear PSTVd (lPSTVd) molecules migrate proportional to their length after full denaturation and faster than fully denatured cPSTVd. The temperature of the main transition of lPSTVd depends on the site of linearization; lPSTVd is not visible at low temperatures due the low concentration of individual lPSTVd molecules with different 5′- and 3′-ends. The different 7 S RNAs (∼305 nt) are separated at low temperature due to their different thermodynamic stability but co-migrate at high temperature due to their nearly identical length. 5S: 5 S ribosomal RNA (120 nt).
The advantage of TGGE over optical melting curves are also obvious from Figure 9 (108): co-existing structures and transition curves of different RNAs are demonstrated as well-separated transition curves, mono- and bimolecular transitions are differentiated, mixture of mutants even with single nucleotide mutations are shown as separated transition curves, and even in a crude extract the transition curve of a specific RNA can be analyzed, if a radioactive or a fluorescent probe is used for staining (108,109).
The TGGE method subsequently was applied to the analysis of protein denaturation and of protein–nucleic acid interactions (110,111). Screening for thermostable or thermolabile mutations was an obvious and simple application (112).
Analysis of prion samples by return refocusing gel electrophoresis
Based on the experience with viroids it was challenging and exciting to identify or rule out nucleic acids of unknown nature or unknown origin in infectious prion material. The problem was: do such samples contain sufficient nucleic acid to explain the infectivity? In 1982, Prusiner (113) had already claimed the absence of a nucleic acid component based on several aspects of experimental evidence. Since direct evidence by an infectious prion protein gene was obtained only more than 20 years later (114,115), the search for a potential prion-specific nucleic acid was an urgent problem in the late 1980s and early 1990s. The unknown but potentially essential nucleic acid could be a priori DNA or RNA, circular or linear, single- or double-stranded, chemically modified and possibly heterogeneous in size. The only generally applicable and sensitive detection method was silver staining (116), but silver staining shows limited sensitivity with heterogeneous nucleic acids and detects also proteins (116,117).
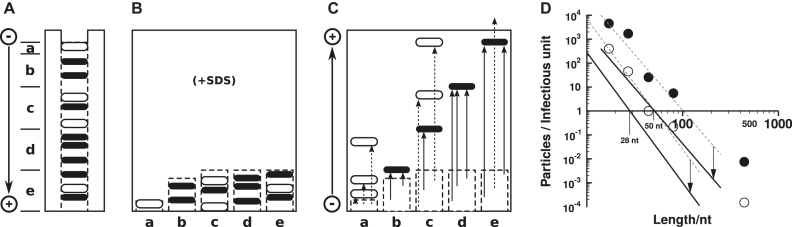
When comparable amounts of control nucleic acids (3 × 1010 molecules) and highly purified but still infectious prion units (1.2 × 1010) were analyzed by 9 or 15% PAGE, respectively, control nucleic acids (viroids and even decameric deoxyoligonucleotides) were detectable but no nucleic acid band in the prion containing sample, except fast migrating bands, possibly peptides. If, however, the potential prion nucleic acid would be heterogeneous in length, it would not be detected. Return-gel electrophoresis was applied in order to separate proteins from nucleic acids and to detect heterogeneous nucleic acids with high sensitivity (Figure 10) (118,119). Electrophoresis in both directions were carried out for the same running time in 8 M urea, i. e. nucleic acid denaturing conditions, but the buffer in the second direction contained 1% sodium dodecyl sulphate (SDS). The presence of SDS did not alter the electrophoretic mobility of nucleic acids significantly, but transformed the proteins into the well-known peptide-SDS complexes (120) raising their electrophoretic mobility. The gel lane of the first direction was cut into slices, containing for example RNA fragments of 10–20, 21–40, 41–70 nt in length, and polymerized at the bottom of a new gel matrix containing SDS. During the second electrophoresis in the reverse direction all nucleic acids from one slice form one band at their starting position, whereas all proteins migrate faster leaving the gel matrix or at least are well separated from the nucleic acid band. From the nucleic acid bands the total amount of nucleic acids in this slice, e. g. 21–40 nt, could be estimated. The method, termed ‘return refocusing gel electrophoresis’, permits the calculation of the number of potential nucleic acid molecules (118,119,121). Calculating the ratio of nucleic acid molecules per infectious unit, we concluded from the first analysis that <1 nucleic acid molecule longer than 75 nt was present in one infectious unit, and consequently could not be essential for infectivity. After the improvements of the method and also the preparation procedures and including the clearance correction (118), it was concluded that no nucleic acid longer than 25 nt was present as essential component.
Figure 10.
Return refocusing gel electrophoresis. (A) The first electrophoresis is a conventional PAGE to separate macromolecules of a prion sample. Nucleic acids (black bars) and proteins (hollow bars) migrate according to their charge and size. After electrophoresis the gel is cut into slices marked a–e. (B) The slices are polymerized at the bottom of a new gel matrix containing 1% SDS. (C) In a second electrophoresis in the reverse direction, the nucleic acids migrate back the same distance as in the first run (arrows with continuous lines) thus refocusing into single bands per slice. Due to the presence of SDS, proteins migrate differently from the first run (arrows with dotted lines) and thus are not refocused. (D) Particle-to-infectivity ratio (P/I) are plotted as a function of the average length of residual nucleic acid. The white and the black circles are from two independent preparations (cf. (118)). The dashed lines are linear regression lines of the experimental values leading to P/I = 1 at ∼50 and 100 nt average length of the nucleic acid. Due to the clearance effect in the animal brain only 4% of the injected infectious particles are present after 20 h and induce further infectivity; consequently, only 4% of the inoculum could be essential for infectivity (for further details see (118)). When the data were corrected for the clearance effect (vertical arrows and continuous lines), 28 and 50 nt average length, respectively, of nucleic acid molecules were present in one infectious unit. Modified from (119) and (118).
In summary, viroids and prions, infectious nucleic acids and infectious proteins, respectively, are both infectious molecules. The methodological developments to characterize infectious viroid RNAs helped to prove the absence of nucleic acid in prions and thereby to confirm the unprecedented phenomenon of an infectious protein (118).
METASTABLE STRUCTURES IN VIROIDS AS A COMMON PRINCIPLE IN RNA BIOLOGY
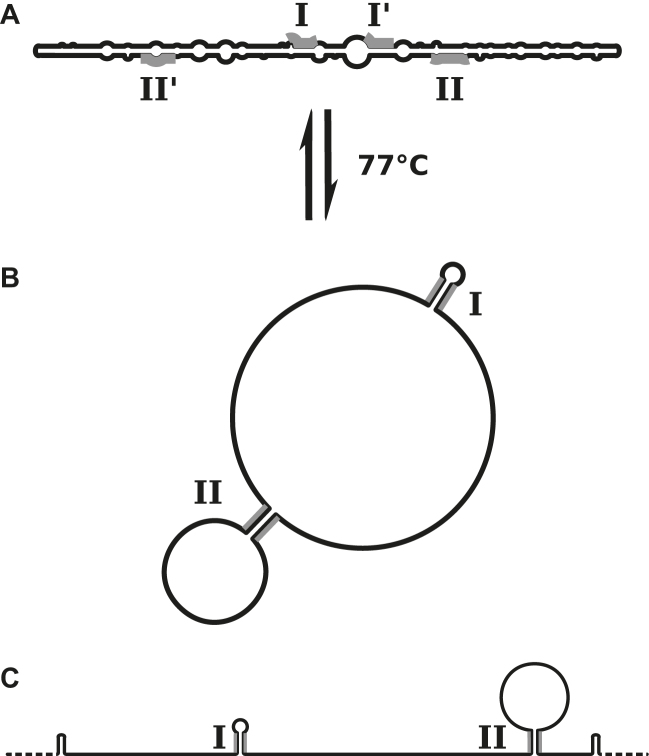
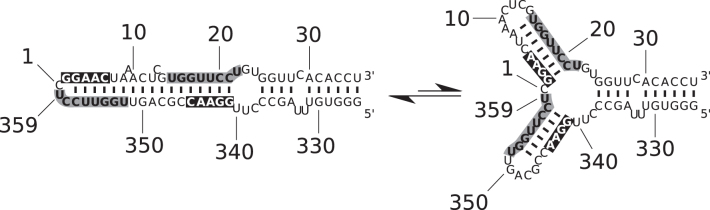
Calculation had shown that the rod-like structure of viroid RNA is unstable at higher temperatures (>70 °C, 1 M NaCl) when the rod-like structure is completely disrupted and two stable hairpins are newly formed (Figure 11) (11,80,122). The stable hairpins I and II (HPI and HPII) in PSTVd are statistically highly significant, i. e. would not be present in random sequences, and can be identified at similar positions in most members of Pospiviroidae. The condition of high temperature for the presence of HPI and HPII, however, is irrelevant for any in vivo function in plants. Consequently, at low temperature the stable hairpins may have only a function as thermodynamically metastable structures: ‘metastable’ meaning that the structure is formed under particular conditions and the transition into the most stable structure is prevented or very slow (123). Metastable structures may be kinetically preferred: RNA is synthesized at a rate of ∼10 nt/s, and hairpin helices with small loops are formed in the microsecond to milisecond range in newly synthesized RNA segments of 20–40 nt, but their rearrangements into the thermodynamically preferred global structure involving pairing of bases quite distant in the full-length RNA strand, may occur at a much slower rate. In summary, stable hairpins are formed fast but may be disrupted in a slow process in favor of basepairs characterizing the most stable structure.
Figure 11.
Thermodynamically and kinetically optimal structures of PSTVd. (A) The rod-like native structure of PSTVd is thermodynamically stable up to temperatures of 75 °C in 1 M ionic strength. (B) In the main transition the rod-like structure denatures with high cooperativity in a structure with two hairpins (HPI and II, which denature at even higher temperature (cf. Figure 9). (C) Structures containing HPII and further hairpins are thermodynamically metastable, but kinetically favored during and after synthesis of replication intermediates in ionic strength conditions of plant cells.
Metastable structures in vivo
To verify the theoretical concept of metastable structures in vivo, we introduced several single- (124) or double-point mutations (62) into the HPII helix. The mutation sites were selected to perturb the native structure only marginally. The mutations in the central part of the HPII helix reverted to wild-type sequence, pointing to the importance of HPII, while mutations, either shortening or elongating the helix by a base pair, were stably maintained after infection of tomato plants. In detail, spontaneous reversions were generated during (−)-strand synthesis, G:U pairs were tolerated in HPII helix of the (−)-strand and a diminished stability of the hairpin structure was compensated for by additional mutations outside the HPII helix destabilizing the rod-like structure (62,124).
A similar result was obtained in a synthetic chimeric recombinant in which the right half of the rod-like structure of citrus exocortis viroid was replaced by the corresponding half of chrysanthemum stunt viroid. This chimera was non-viable, probably due to a mismatch in the HPII helix. Restoration of the base pairing, and possibly a few additional mutations, in progeny occurring from plants infected with an Agrobacterium strain carrying the recombinant sequence led to viability (125).
While the appearance and rate of HPII-favoring mutations can be interpreted in accordance with HPII-containing structures during replication, structure-specific hybridization with a site-specific oligonucleotide also demonstrated the presence of HPII-containing structures in multimeric replication intermediates of PSTVd in vivo (126).
Metastable structures in vitro
To quantify the appearance or preference of metastable structures during PSTVd replication, we measured in vitro transcription of PSTVd cDNA inserted into a plasmid downstream from a T7 promoter in dependence on rate and duration of synthesis. Under optimized laboratory conditions, the synthesis rate by T7-polymerase is much higher than that expected for polII using DNA or viroid RNA as templates. The rate of synthesis may be lowered by a decrease in concentrations of nucleoside triphosphates. The resulting structure distribution of the PSTVd transcripts was analyzed by TGGE (109). Metastable structures were generated preferentially at low transcription rates similar to those observed in vivo. At higher transcription rates, longer products were synthesized before metastable structures of short segments could be formed and more rod-like structures of higher thermodynamic stability were recorded (109).
Metastable structures in silico
To predict the preferred metastable structures of PSTVd, either generated during synthesis or after rapid cooling from fully or partially denatured states, we developed a method, based on ‘simulated annealing’, that describes the kinetically controlled folding process of any RNA (127). Using kinetic data, the process of RNA structure formation and/or structural rearrangements was simulated taking into account the probabilities for opening and closing of randomly selected single base-pairs. Depending on RNA elongation rates, the process of ‘sequential folding’ during transcription could be described. From the kinetically controlled folding of circular as well as of linear PSTVd the importance of HPI and HPII for the folding pathway was obvious and supported experimental data from kinetics (80,109) and from the HPII mutagenesis studies (62,124).
Unexpected biological features of viroid RNA such as biological functionality of the structure of minimum free energy, of metastable structures and of co-transcriptional or sequential folding of RNA were found. The principles of structure formation emerging from folding kinetics are now widely accepted (128–131). The most intriguing examples are riboswitches (132–134), which may switch between two mutually exclusive, functional structures due to presence/absence of a ligand, thermosensors (135–137), which regulate gene expression in dependence on environmental temperature and pseudoknots, which might lead to frameshifting depending on their thermodynamic and kinetic characteristics (138,139). Computational methods based on different algorithms were developed for prediction of the kinetic behavior of RNA folding and design of functional RNA molecules (140–149).
EVIDENCE FOR THE PARTIALLY COVALENT NATURE OF HYDROGEN BONDS AND ESTABLISHMENT OF THE BASE PAIRING NETWORK
The left terminal domain of PSTVd, termed TL and consisting of the terminal hairpin loop and three stem regions separated by two internal loops, was probed in a study combining theoretical modeling with nuclease digestion and chemical reactivity (150). The presence of 2-fold complementary sequence repeats in the TL domain allows for the formation of two possible secondary structures: an elongated-rod and a bifurcated form (Figure 12). The problem to differentiate between these conformations was addressed in collaboration with the NMR group of Grzesiek (151). Milligram amounts of the 69 nt PSTVd-specific TL segment were prepared by T7-polymerase transcription with C13 and N15 labeled nucleotides and purified over Nucleogen 500 columns (Macherey-Nagel, Düren, Germany; Figure 4). While the elongated structure was clearly confirmed, an additional and unexpected finding resulted from the quantitative interpretation of NMR results (152,153). It was observed that nuclear magnetic polarization is transmitted across the hydrogen bond between the donor D and acceptor A nuclei in Watson Crick A·U and G·C base pairs. This two-bond H-bond scalar coupling (h2JDA) follows the same electron-mediated polarization mechanism as observed for their covalent counterparts (154), thereby suggesting that H-bonds possess some covalent character. This has been studied extensively by theoretical studies (155). Backbone H-bonds in proteins have been detected by h3JDA-couplings (153,156) and H-bond couplings for non-canonical base pairs have been observed in both RNA and DNA molecules (157–163). Hydrogen-bond couplings provide a direct approach to establish unambiguous hydrogen-bond networks in secondary and tertiary structures, and also allow the study of H-bond cooperativity (164). Whereas before only stacked nucleotides in one strand could be identified through π-electron polarization, H-bond scalar couplings link nucleotides on opposing strands, thereby providing unique structural information.
Figure 12.
Structural alternatives of PSTVd’s left-terminal domain. The gray-boxed and black-boxed nucleotides highlight two complementary repeat units, which principally give rise to the two alternative structures: rod-like (left) and branched (right). The rod-like structure is thermodynamically highly favored according to analyses of sequence covariation, energy predictions, TGGE, UV melting experiments and NMR (151).
In conclusion, a study designed to answer detailed questions of viroid structure led to discoveries that are important for chemistry in general, and provided impetus to further characterize physical aspects of H-bond (154).
ORIGIN OF VIROIDS
The high error rate of viroid transcription, their circular nature, and the presence of HHrz in members of Avsunviroidae led to many speculations on the evolutionary history of viroids. The first documentation of a viroid-infected fruit (etrog, Citrus medica L.) is depicted on a mosaic from the 6th century CE (165), while corresponding diseases were described close to 100 years ago (166,167). The circularity of viroids reminded of introns (168), and the HHrz might even stem from the pre-cellular RNA world (169,170). As Diener (171) speculated, the ‘small size and circularity [of viroids] would have enhanced probability of their survival in error-prone, primitive self-replicating RNA systems and assured complete replication without the need for initiation or termination signals’. However, circRNAs are now recognized as a large RNA class with prolonged half-life (in comparison to linearized RNAs), may act as miRNA sponges and be involved in gene expression and splicing (172–175). The HHrz motif is found in many genomes, quite often as tandem repeats, but mostly with unknown functions. More specifically, however, eukaryotic Penelope-like retroelements encode HHrz motifs (176,177) and may thus be related to viroids, at least those of the Avsunviroidae family.
CONCLUSIONS AND OUTLOOK
This article describes methodological improvements achieved in viroid research as potential paradigm for developments in other areas of basic research. At the onset viroid research was a biologically very attractive subject for structure-function studies. Viroids were an unprecedented phenomenon of a medium-sized, capsid-free RNA molecule with the complex biology of a virus. Improvements of RNA technology developed for the study of viroid biology were applied to other nucleic acid research projects. The characterization of metastable viroid structures or the classification of hydrogen bonding as a partially covalent bond significantly contributed to knowledge in basic molecular biology.
Definitely, viroid research is not at its end but its further contributions to RNA biology or basic biochemistry cannot be foreseen. We consider several research areas of future interest. Particular efforts will be directed toward the agricultural impact of viroids (178–181). The host range of viroids is not as restricted as originally assumed (182–184) but particular environmental conditions can induce viroid transmission to new crop plants, as shown for Matricaria chamomilla (185) and recently for hop (186). Careful monitoring may help to avoid such transmissions in the future. Locally restricted viroid infections, such as dwarfing of citrus trees for easier harvesting and less water consumption (187,188), will require exploration.
Further investigation of viroid structure will be of interest. Structural details of a viroid part were elucidated by NMR (151), but the complete viroid molecule resisted structural analysis at atomic resolution by x-ray. The improvements in electron cryomicroscopy and atomic force microscopy will bring further insights.
The functional relevance of circular RNA in viroids is different from that of cellular circRNAs, with rolling circle replication differing from back-splicing. However, parallels will be found when the secondary or even tertiary structure of circRNA is characterized. Stability of the RNA in the cellular medium, protection by proteins and segments of the circRNA, which have to be accessible for their function, might be studied in future and further insight might be expected from comparison with viroid features.
ACKNOWLEDGEMENTS
We thank U. Desselberger (Cambridge) for productive discussion. We thank all former members of the collaborating laboratories of H. L. Sänger (†) (Martinsried), H. J. Gross (Würzburg), G. Klotz and A. K. Kleinschmidt(†) (Ulm) and D. Riesner (Düsseldorf), called the ‘German team’ (8).
FUNDING
This survey was not funded, but original work by the authors was mostly funded for many years by the German Research Foundation and the Alexander von Humboldt-Foundation.
Conflict of interest statement. D. R. states that he is a co-founder of Qiagen N. V. and was a board member up to 2014. Since then D. R. has no influence on the business of Qiagen. G. S. has no conflicts of interest to declare.
REFERENCES
- 1. Diener T. Potato spindle tuber ‘virus’ IV. A replicating, low molecular weight RNA. Virology. 1971; 45:411–428. [DOI] [PubMed] [Google Scholar]
- 2. Singh R., Clark M.. Infectious low-molecular weight ribonucleic acid from tomato. Biochem. Biophys. Res. Commun. 1971; 44:1077–1083. [DOI] [PubMed] [Google Scholar]
- 3. Sänger H. An infectious and replicating RNA of low molecular weight: the agent of the exocortis disease of citrus. Adv. Biosc. 1972; 8:103–116. [Google Scholar]
- 4. Semancik J., Weathers L.. Exocortis disease: evidence for a new species of ‘infectious’ low molecular weight RNA in plants. Nature New Biol. 1972; 237:242–244. [DOI] [PubMed] [Google Scholar]
- 5. Flores R., Owens R., Taylor J.. Pathogenesis by subviral agents: viroids and hepatitis delta virus. Curr. Opin. Virol. 2016; 17:87–94. [DOI] [PubMed] [Google Scholar]
- 6. Hadidi A, Randles J, Flores R, Palukaitis P. Viroids and satellites. 2017; San Diego: Academic Press. [Google Scholar]
- 7. Diener T. Discovering viroids—a personal perspective. Nat. Rev. Microbiol. 2003; 1:75–80. [DOI] [PubMed] [Google Scholar]
- 8. Singh R. The discovery and eradication of potato spindle tuber viroid in Canada. Virusdisease. 2014; 25:415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sänger H., Klotz G., Riesner D., Gross H., Kleinschmidt A.. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 1976; 73:3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilusz J. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip. Rev. RNA. 2018; 9:e1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riesner D., Henco K., Rokohl U., Klotz G., Kleinschmidt A., Domdey H., Jank P., Gross H., Sänger H.. Structure and structure formation of viroids. J. Mol. Biol. 1979; 133:85–115. [DOI] [PubMed] [Google Scholar]
- 12. Gross H., Domdey H., Lossow C., Jank P., Raba M., Alberty H., Sänger H.. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature. 1978; 273:203–208. [DOI] [PubMed] [Google Scholar]
- 13. Maxam A., Gilbert W.. A new method for sequencing DNA. Proc. Natl. Acad. Sci. U.S.A. 1977; 74:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Domdey H., Jank P., Sänger L., Gross H.. Studies on the primary and secondary structure of potato spindle tuber viroid: products of digestion with ribonuclease A and ribonuclease T1, and modification with bisulfite. Nucleic Acids Res. 1978; 5:1221–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H.. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015; 365:141–148. [DOI] [PubMed] [Google Scholar]
- 16. Lei K., Bai H., Wei Z., Xie C., Wang J., Li J., Chen Q.. The mechanism and function of circular RNAs in human diseases. Exp. Cell Res. 2018; 368:147–158. [DOI] [PubMed] [Google Scholar]
- 17. Branch A., Robertson H.. A replication cycle for viroids and other small infectious RNA’s. Science. 1984; 223:450–455. [DOI] [PubMed] [Google Scholar]
- 18. Branch A., Benenfeld B., Robertson H.. Evidence for a rolling circle in the replication of potato spindle tuber viroid. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:9128–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flores R., Gas M., Molina-Serrano D., Nohales M., Carbonell A., Gago S., De la Peña M., Darós J.. Viroid replication: rolling-circles, enzymes and ribozymes. Viruses. 2009; 1:317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mühlbach H., Sänger H.. Viroid replication is inhibited by α-amanitin. Nature. 1979; 278:185–188. [DOI] [PubMed] [Google Scholar]
- 21. Navarro J., Vera A., Flores R.. A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology. 2000; 268:218–225. [DOI] [PubMed] [Google Scholar]
- 22. Nohales M.-Á., Flores R., Darós J.. Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:13805–13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nohales M.-Á., Molina-Serrano D., Flores R., Darós J.. Involvement of the chloroplastic isoform of tRNA ligase in the replication of viroids belonging to the family Avsunviroidae. J. Virol. 2012; 86:8269–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qi Y., Ding B.. Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell. 2003; 15:2566–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harders J., Lukács N., Robert-Nicoud M., Jovin T., Riesner D.. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 1989; 8:3941–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hutchins C., Rathjen P., Forster A., Symons R.. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986; 14:3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Daròs J., Marcos J., Hernández C., Flores R.. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:12813–12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Flores R., Daròs J., Hernández C.. Avsunviroidae family: viroids containing hammerhead ribozymes. Adv. Virus Res. 2000; 55:271–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmidt B., Rappold W., Rosenbaum V., Fischer R., Riesner D.. A fluorescence detection system for the analytical ultracentrifuge and its application to proteins, nucleic acids, and viruses. Colloid Polym. Sci. 1990; 268:45–54. [Google Scholar]
- 30. Goodman T., Nagel L., Rappold W., Klotz G., Riesner D.. Viroid replication: equilibrium association constant and comparative activity measurements for the viroid-polymerase interactions. Nucleic Acids Res. 1984; 12:6231–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kapahnke R., Rappold W., Desselberger U., Riesner D.. The stiffness of dsRNA: hydrodynamic studies on fluorescence-labelled RNA segments of bovine rotavirus. Nucleic Acids Res. 1986; 14:3215–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. MacGregor I., Anderson A., Laue T.. Fluorescence detection for the XLI analytical ultracentrifuge. Biophys. Chem. 2004; 108:165–185. [DOI] [PubMed] [Google Scholar]
- 33. Zhao H., Casillas E., Shroff H., Patterson G., Schuck P.. Tools for the quantitative analysis of sedimentation boundaries detected by fluorescence optical analytical ultracentrifugation. PLoS One. 2013; 8:e77245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chaturvedi S., Ma J., Zhao H., Schuck P.. Use of fluorescence-detected sedimentation velocity to study high-affinity protein interactions. Nat. Protoc. 2017; 12:1777–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wright R., Hayes D., Stafford W., Sherwood P., Correia J.. Characterization of therapeutic antibodies in the presence of human serum proteins by AU-FDS analytical ultracentrifugation. Anal. Biochem. 2018; 550:72–83. [DOI] [PubMed] [Google Scholar]
- 36. Schiebel W., Pélissier T., Riedel L., Thalmeir S., Schiebel R., Kempe D., Lottspeich F., Sänger H., Wassenegger M.. Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell. 1998; 10:2087–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schiebel W., Haas B., Marinković S., Klanner A., Sänger H.. RNA-directed RNA polymerase from tomato leaves. I. Purification and physical properties. J. Biol. Chem. 1993; 268:11851–11857. [PubMed] [Google Scholar]
- 38. Schiebel W., Haas B., Marinković S., Klanner A., Sänger H.. RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J. Biol. Chem. 1993; 268:11858–11867. [PubMed] [Google Scholar]
- 39. Wassenegger M., Krczal G.. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006; 11:142–151. [DOI] [PubMed] [Google Scholar]
- 40. Wassenegger M., Heimes S., Riedel L., Sänger H.. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994; 76:567–576. [DOI] [PubMed] [Google Scholar]
- 41. Navarro B., Gisel A., Rodio M., Delgado S., Flores R., Di Serio F.. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012; 70:991–1003. [DOI] [PubMed] [Google Scholar]
- 42. Hammann C., Steger G.. Viroid-specific small RNA in plant disease. RNA Biol. 2012; 9:809–819. [DOI] [PubMed] [Google Scholar]
- 43. Dadami E., Dalakouras A., Wassenegger M.. Hadidi A, Randles J, Flores R, Palukaitis P. Viroids and RNA silencing. Viroids and satellites. 2017; San Diego: Academic Press; 115–124. [Google Scholar]
- 44. Cech T., Zaug A., Grabowski P.. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981; 27:487–496. [DOI] [PubMed] [Google Scholar]
- 45. Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S.. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983; 35:849–857. [DOI] [PubMed] [Google Scholar]
- 46. Prody G., Bakos J., Buzayan J., Schneider I., Bruening G.. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986; 231:1577–1580. [DOI] [PubMed] [Google Scholar]
- 47. De la Peña M., García-Robles I.. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA. 2010; 16:1943–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Seehafer C., Kalweit A., Steger G., Gräf S., Hammann C.. From alpaca to zebrafish: hammerhead ribozymes wherever you look. RNA. 2011; 17:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. De la Peña M., García-Robles I., Cervera A.. The hammerhead ribozyme: a long history for a short RNA. Molecules. 2017; 22:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Uhlenbeck O. A small catalytic oligoribonucleotide. Nature. 1987; 328:596–600. [DOI] [PubMed] [Google Scholar]
- 51. Citti L., Rainaldi G.. Synthetic hammerhead ribozymes as therapeutic tools to control disease genes. Curr. Gene Ther. 2005; 5:11–24. [DOI] [PubMed] [Google Scholar]
- 52. De la Peña M., Gago S., Flores R.. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003; 22:5561–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Khvorova A., Lescoute A., Westhof E., Jayasena S.. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003; 10:708–712. [DOI] [PubMed] [Google Scholar]
- 54. Przybilski R., Gräf S., Lescoute-Phillips A., Nellen W., Westhof E., Steger G., Hammann C.. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell. 2005; 17:1877–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Burke D., Greathouse S.. Low-magnesium, trans-cleavage activity by type III, tertiary stabilized hammerhead ribozymes with stem 1 discontinuities. BMC Biochem. 2005; 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fedoruk-Wyszomirska A., Szymańki M., Wyszko E., Barciszewska M., Barciszewski J.. Highly active low magnesium hammerhead ribozyme. J. Biochem. 2009; 145:451–459. [DOI] [PubMed] [Google Scholar]
- 57. Mulhbacher J., St-Pierre P., Lafontaine D.. Therapeutic applications of ribozymes and riboswitches. Curr. Opin. Pharmacol. 2010; 10:551–556. [DOI] [PubMed] [Google Scholar]
- 58. Wieland M., Ausländer D., Fussenegger M.. Engineering of ribozyme-based riboswitches for mammalian cells. Methods. 2012; 56:351–357. [DOI] [PubMed] [Google Scholar]
- 59. Sullivan J., Yau E., Taggart R., Butler M., Kolniak T.. Relieving bottlenecks in RNA drug discovery for retinal diseases. Adv. Exp. Med. Biol. 2012; 723:145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li C., Chen Y.. Targeting long non-coding RNAs in cancers: progress and prospects. Int. J. Biochem. Cell Biol. 2013; 45:1895–1910. [DOI] [PubMed] [Google Scholar]
- 61. Lehmann E., Brueckner F., Cramer P.. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007; 450:445–449. [DOI] [PubMed] [Google Scholar]
- 62. Qu F., Heinrich C., Loss P., Steger G., Tien P., Riesner D.. Multiple pathways of reversion in viroids for conservation of structural elements. EMBO J. 1993; 12:2129–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wilke C. Quasispecies theory in the context of population genetics. BMC Evol. Biol. 2005; 5:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lauring A., Andino R.. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010; 6:e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Codoñer F., Darós J., Solé R., Elena S.. The fittest versus the flattest: experimental confirmation of the quasispecies effect with subviral pathogens. PLoS Pathog. 2006; 2:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gago S., Elena S., Flores R., Sanjuán R.. Extremely high mutation rate of a hammerhead viroid. Science. 2009; 323:1308. [DOI] [PubMed] [Google Scholar]
- 67. López-Carrasco A., Ballesteros C., Sentandreu V., Delgado S., Gago-Zachert S., Flores R., Sanjuán R.. Different rates of spontaneous mutation of chloroplastic and nuclear viroids as determined by high-fidelity ultra-deep sequencing. PLoS Pathog. 2017; 13:e1006547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brass J., Owens R., Matoušek J., Steger G.. Viroid quasispecies revealed by deep sequencing. RNA Biol. 2016; 14:317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971; 58:465–523. [DOI] [PubMed] [Google Scholar]
- 70. Drake J. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:7160–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sanjuán R., Nebot M., Chirico N., Mansky L., Belshaw R.. Viral mutation rates. J. Virol. 2010; 84:9733–9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Palukaitis P., Symons R.. Purification and characterization of the circular and linear forms of chrysanthemum stunt viroid. J. Gen. Virol. 1980; 46:477–489. [Google Scholar]
- 73. Colpan M., Schumacher J., Brüggemann W., Sänger H., Riesner D.. Large-scale purification of viroid RNA using Cs2SO4 gradient centrifugation and high-performance liquid chromatography. Anal. Biochem. 1983; 131:257–265. [DOI] [PubMed] [Google Scholar]
- 74. Colpan M., Riesner D.. High-performance liquid chromatography of high-molecular-weight nucleic acids on the macroporous ion exchanger, Nucleogen. J. Chromat. A. 1984; 296:339–353. [Google Scholar]
- 75. Hecker R., Colpan M., Riesner D.. High-performance liquid chromatography of DNA restriction fragments. J. Chromatogr. 1985; 326:251–261. [DOI] [PubMed] [Google Scholar]
- 76. Colpan M., Riesner D.. Neuberger A, van Deenen L. High performance liquid chromatography of nucleic acids. Modern Physical Methods in Biochemistry. 1985; 11:Amsterdam: Elsevier; 85–105. [Google Scholar]
- 77. Hecker R., Riesner D.. Chromatographic separation of DNA restriction fragments. J. Chromatogr. B Biomed. Sci. Appl. 1987; 418:97–114. [DOI] [PubMed] [Google Scholar]
- 78. Langowski J., Henco K., Riesner D., Sänger H.. Common structural features of different viroids: serial arrangement of double helical sections and internal loops. Nucleic Acids Res. 1978; 5:1589–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tinoco I. Jr, Uhlenbeck O., Levine M.. Estimation of secondary structure in ribonucleic acids. Nature. 1971; 230:362–367. [DOI] [PubMed] [Google Scholar]
- 80. Henco K., Sänger H., Riesner D.. Fine structure melting of viroids as studied by kinetic methods. Nucleic Acids Res. 1979; 6:3041–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Randles J., Steger G., Riesner D.. Structural transitions in viroid-like RNAs associated with cadang-cadang disease, velvet tobacco mottle virus, and Solanum nodiflorum mottle virus. Nucleic Acids Res. 1982; 10:5569–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang K., Choo Q., Weiner A., Ou J., Najarian R., Thayer R., Mullenbach G., Denniston K., Gerin J., Houghton M.. Structure, sequence and expression of the hepatitis delta (δ) viral genome. Nature. 1986; 323:508–514. [DOI] [PubMed] [Google Scholar]
- 83. Zuker M., Stiegler P.. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981; 9:133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Steger G., Hofmann H., Förtsch J., Gross H., Randles J., Sänger H., Riesner D.. Conformational transitions in viroids and virusoids: Comparison of results from energy minimization algorithm and from experimental data. J. Biomol. Struct. Dyn. 1984; 2:543–571. [DOI] [PubMed] [Google Scholar]
- 85. Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989; 244:48–52. [DOI] [PubMed] [Google Scholar]
- 86. Markham N., Zuker M.. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008; 453:3–31. [DOI] [PubMed] [Google Scholar]
- 87. Reuter J., Mathews D.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lorenz R., Bernhart S., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P., Hofacker I.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Andronescu M., Condon A., Turner D., Mathews D.. The determination of RNA folding nearest neighbor parameters. Methods Mol. Biol. 2014; 1097:45–70. [DOI] [PubMed] [Google Scholar]
- 90. McCaskill J. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990; 29:1105–1119. [DOI] [PubMed] [Google Scholar]
- 91. Gruber A., Lorenz R., Bernhart S., Neuböck R., Hofacker I.. The Vienna RNA websuite. Nucleic Acids Res. 2008; 36:W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wuchty S., Fontana W., Hofacker I., Schuster P.. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers. 1999; 49:145–165. [DOI] [PubMed] [Google Scholar]
- 93. Stone J., Bleckley S., Lavelle S., Schroeder S.. A parallel implementation of the Wuchty algorithm with additional experimental filters to more thoroughly explore RNA conformational space. PLoS One. 2015; 10:e0117217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Giguère T., Adkar-Purushothama C., Bolduc F., Perreault J.. Elucidation of the structures of all members of the Avsunviroidae family. Mol. Plant Pathol. 2014; 15:767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Steger G., Perreault J.-P.. Structure and associated biological functions of viroids. Adv. Virus Res. 2016; 94:141–172. [DOI] [PubMed] [Google Scholar]
- 96. López-Carrasco A., Flores R.. Dissecting the secondary structure of the circular RNA of a nuclear viroid in vivo: a ‘naked’ rod-like conformation similar but not identical to that observed in vitro. RNA Biol. 2017; 14:1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Giguère T., Perreault J.. Classification of the Pospiviroidae based on their structural hallmarks. PLoS ONE. 2017; 12:e0182536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhong X., Leontis N., Qian S., Itaya A., Qi Y., Boris-Lawrie K., Ding B.. Tertiary structural and functional analyses of a viroid RNA motif by isostericity matrix and mutagenesis reveal its essential role in replication. J. Virol. 2006; 80:8566–8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Steger G. Modelling the three-dimensional structure of the right-terminal domain of pospiviroids. Sci Rep. 2017; 7:711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wang Y., Zirbel C., Leontis N., Ding B.. RNA 3-dimensional structural motifs as a critical constraint of viroid RNA evolution. PLoS Pathog. 2018; 14:e1006801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Schumacher J., Randles J., Riesner D.. A two-dimensional electrophoretic technique for the detection of circular viroids and virusoids. Anal. Biochem. 1983; 135:288–295. [DOI] [PubMed] [Google Scholar]
- 102. Schumacher J., Meyer N., Riesner D., Weidemann H.. Diagnostic procedure for detection of viroids and viruses with circular RNAs by ‘return’-gel electrophoresis. J. Phytopath. 1986; 115:332–343. [Google Scholar]
- 103. Flores R., Duran-Vila N., Pallas V., Semancik J.. Detection of viroid and viroid-like RNAs from grapevine. J. Gen. Virol. 1985; 66:2095–2102. [Google Scholar]
- 104. Owens R. Identification of viroids by gel electrophoresis. Curr. Protoc. Microbiol. 2008; 10:doi:10.1002/9780471729259.mc16g01s10. [DOI] [PubMed] [Google Scholar]
- 105. Gucek T., Trdan S., Jakse J., Javornik B., Matousek J., Radisek S.. Diagnostic techniques for viroids. Plant Pathol. 2017; 66:339–358. [Google Scholar]
- 106. Tabak H., Van der Horst G., Smit J., Winter A., Mul Y., Groot Koerkamp M.. Discrimination between RNA circles, interlocked RNA circles and lariats using two-dimensional polyacrylamide gel electrophoresis. Nucleic Acids Res. 1988; 16:6597–6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Jeck W., Sharpless N.. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014; 32:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Rosenbaum V., Riesner D.. Temperature-gradient gel electrophoresis. Thermodynamic analysis of nucleic acids and proteins in purified form and in cellular extracts. Biophys. Chem. 1987; 26:235–246. [DOI] [PubMed] [Google Scholar]
- 109. Repsilber D., Wiese U., Rachen M., Schröder A., Riesner D., Steger G.. Formation of metastable RNA structures by sequential folding during transcription: Time-resolved structural analysis of potato spindle tuber viroid (−)-stranded RNA by temperature-gradient gel electrophoresis. RNA. 1999; 5:574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Riesner D., Steger G., Zimmat R., Owens R., Wagenhöfer M., Hillen W., Vollbach S., Henco K.. Temperature-gradient gel electrophoresis: Analysis of conformational transitions, sequence variations, and protein-nucleic acid interactions. Electrophoresis. 1989; 10:377–389. [DOI] [PubMed] [Google Scholar]
- 111. Klaff P., Mundt S., Steger G.. Complex formation of the spinach chloroplast psbA mRNA 5′ untranslated region with proteins is dependent on the RNA structure. RNA. 1997; 3:1468–1479. [PMC free article] [PubMed] [Google Scholar]
- 112. Sättler A., Riesner D.. Temperature-gradient gel electrophoresis for analysis and screening of thermostable proteases. Electrophoresis. 1993; 14:782–788. [DOI] [PubMed] [Google Scholar]
- 113. Prusiner S. Novel proteinaceous infectious particles cause scrapie. Science. 1982; 216:136–144. [DOI] [PubMed] [Google Scholar]
- 114. Legname G., Baskakov I., Nguyen H., Riesner D., Cohen F., DeArmond S., Prusiner S.. Synthetic mammalian prions. Science. 2004; 305:673–676. [DOI] [PubMed] [Google Scholar]
- 115. Colby D., Zhang Q., Wang S., Groth D., Legname G., Riesner D., Prusiner S.. Prion detection by an amyloid seeding assay. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:20914–20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Whitton J., Hundley F., O’Donnell B., Desselberger U.. Silver staining of nucleic acids. Applications in virus research and in diagnostic virology. J. Virol. Methods. 1983; 7:185–198. [DOI] [PubMed] [Google Scholar]
- 117. Rabilloud T., Vuillard L., Gilly C., Lawrence J.. Silver-staining of proteins in polyacrylamide gels: a general overview. Cell. Mol. Biol. (Noisy-le-grand). 1994; 40:57–75. [PubMed] [Google Scholar]
- 118. Safar J., Kellings K., Serban A., Groth D., Cleaver J., Prusiner S., Riesner D.. Search for a prion-specific nucleic acid. J. Virol. 2005; 79:10796–10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Meyer N., Rosenbaum V., Schmidt B., Gilles K., Mirenda C., Groth D., Prusiner S., Riesner D.. Search for a putative scrapie genome in purified prion fractions reveals a paucity of nucleic acids. J. Gen. Virol. 1991; 72:37–49. [DOI] [PubMed] [Google Scholar]
- 120. Reynolds J., Tanford C.. The gross conformation of protein-sodium dodecyl sulfate complexes. J. Biol. Chem. 1970; 245:5161–5165. [PubMed] [Google Scholar]
- 121. Kellings K., Meyer N., Mirenda C., Prusiner S., Riesner D.. Further analysis of nucleic acids in purified scrapie prion preparations by improved return refocusing gel electrophoresis. J. Gen. Virol. 1992; 73:1025–1029. [DOI] [PubMed] [Google Scholar]
- 122. Henco K., Riesner D., Sänger H.. Conformation of viroids. Nucleic Acids Res. 1977; 4:177–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Riesner D., Baumstark T., Qu F., Klahn T., Loss P., Rosenbaum V., Schmitz M., Steger G.. Lilley D, Heumann H, Suck D. Physical basis and biological examples of metastable RNA structures. Structural Tools for the Analysis of Protein-Nucleic Acids Complexes. 1992; Basel: Advances in Life Sciences, Birkhäuser Verlag AG; 401–435. [Google Scholar]
- 124. Loss P., Schmitz M., Steger G., Riesner D.. Formation of a thermodynamically metastable structure containing hairpin II is critical for infectivity of potato spindle tuber viroid RNA. EMBO J. 1991; 10:719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Candresse T., Góra-Sochacka A., Zagórski W.. Restoration of secondary hairpin II is associated with restoration of infectivity of a non-viable recombinant viroid. Virus Res. 2001; 75:29–34. [DOI] [PubMed] [Google Scholar]
- 126. Schröder A., Riesner D.. Detection and analysis of hairpin II, an essential metastable structural element in viroid replication intermediates. Nucleic Acids Res. 2002; 30:3349–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Schmitz M., Steger G.. Description of RNA folding by ‘simulated annealing’. J. Mol. Biol. 1996; 255:254–266. [DOI] [PubMed] [Google Scholar]
- 128. Xayaphoummine A., Viasnoff V., Harlepp S., Isambert H.. Encoding folding paths of RNA switches. Nucleic Acids Res. 2006; 35:614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Isambert H. The jerky and knotty dynamics of RNA. Methods. 2009; 49:189–196. [DOI] [PubMed] [Google Scholar]
- 130. Wong T., Pan T.. RNA folding during transcription: protocols and studies. Meth. Enzymol. 2009; 468:167–193. [DOI] [PubMed] [Google Scholar]
- 131. Lai D., Proctor J., Meyer I.. On the importance of cotranscriptional RNA structure formation. RNA. 2013; 19:1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Breaker R. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012; 4:a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Serganov A., Nudler E.. A decade of riboswitches. Cell. 2013; 152:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Gong S., Wang Y., Wang Z., Zhang W.. Co-transcriptional folding and regulation mechanisms of riboswitches. Molecules. 2017; 22:1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Schumann W. Thermosensor systems in eubacteria. Adv. Exp. Med. Biol. 2012; 739:1–16. [DOI] [PubMed] [Google Scholar]
- 136. Krajewski S., Narberhaus F.. Temperature-driven differential gene expression by RNA thermosensors. Biochim. Biophys. Acta. 2014; 1839:978–988. [DOI] [PubMed] [Google Scholar]
- 137. Quereda J., Cossart P.. Regulating bacterial virulence with RNA. Annu. Rev. Microbiol. 2017; 71:263–280. [DOI] [PubMed] [Google Scholar]
- 138. Green L., Kim C., Bustamante C., Tinoco I.. Characterization of the mechanical unfolding of RNA pseudoknots. J. Mol. Biol. 2007; 375:511–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Giedroc D., Cornish P.. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2008; 139:193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Isambert H., Siggia E.. Modeling RNA folding paths with pseudoknots: application to hepatitis delta virus ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6515–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Shcherbakova I., Mitra S., Laederach A., Brenowitz M.. Energy barriers, pathways and dynamics during folding of large, multidomain RNAs. Curr. Opin. Chem. Biol. 2008; 12:655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Cao H., Xie H., Zhang W., Wang K., Li W., Liu C.. Dynamic extended folding: modeling the RNA secondary structures during co-transcriptional folding. J. Theor. Biol. 2009; 261:93–99. [DOI] [PubMed] [Google Scholar]
- 143. Zhu J., Steif A., Proctor J., Meyer I.. Transient RNA structure features are evolutionarily conserved and can be computationally predicted. Nucleic Acids Res. 2013; 41:6273–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Senter E., Dotu I., Clote P.. RNA folding pathways and kinetics using 2D energy landscapes. J. Math. Biol. 2014; 70:173–196. [DOI] [PubMed] [Google Scholar]
- 145. Mann M., Kucharík M., Flamm C., Wolfinger M.. Memory-efficient RNA energy landscape exploration. Bioinformatics. 2014; 30:2584–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Hofacker I. Energy-directed RNA structure prediction. Methods Mol. Biol. 2014; 1097:71–84. [DOI] [PubMed] [Google Scholar]
- 147. Kucharík M., Hofacker I., Stadler P., Qin J.. Pseudoknots in RNA folding landscapes. Bioinformatics. 2015; 32:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Sun L., Zhang D., Chen S.. Theory and modeling of RNA structure and interactions with metal ions and small molecules. Annu. Rev. Biophys. 2017; 46:227–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Meyer I. In silico methods for co-transcriptional RNA secondary structure prediction and for investigating alternative RNA structure expression. Methods. 2017; 120:3–16. [DOI] [PubMed] [Google Scholar]
- 150. Gast F., Kempe D., Spieker R., Sänger H.. Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicons provides evidence for central non-canonical base-pairs, large A-rich loops, and a terminal branch. J. Mol. Biol. 1996; 262:652–670. [DOI] [PubMed] [Google Scholar]
- 151. Dingley A., Steger G., Esters B., Riesner D., Grzesiek S.. Structural characterization of the 69 nucleotide potato spindle tuber viroid left-terminal domain by NMR and thermodynamic analysis. J. Mol. Biol. 2003; 334:751–767. [DOI] [PubMed] [Google Scholar]
- 152. Dingley A., Grzesiek S.. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998; 120:8293–8297. [Google Scholar]
- 153. Grzesiek S., Cordier F., Dingley A.. Scalar couplings across hydrogen bonds. Meth. Enzymol. 2001; 338:111–133. [DOI] [PubMed] [Google Scholar]
- 154. Grzesiek S., Cordier F., Jaravine V., Barfield M.. Insights into biomolecular hydrogen bonds from hydrogen bond scalar couplings. Prog. Nucl. Magn. Reson. Spectrosc. 2004; 45:275–300. [Google Scholar]
- 155. Barfield M. Structural dependencies of interresidue scalar coupling h3JNC' and donor 1H chemical shifts in the hydrogen bonding regions of proteins. J. Am. Chem. Soc. 2002; 124:4158–4168. [DOI] [PubMed] [Google Scholar]
- 156. Cornilescu G., Hu J.-S., Bax A.. Identification of the hydrogen bonding network in a protein by scalar couplings. J. Am. Chem. Soc. 1999; 121:2949–2950. [Google Scholar]
- 157. Dingley A., Masse J., Peterson R., Barfield M., Feigon J., Grzesiek S.. Internucleotide scalar couplings across hydrogen bonds in Watson-Crick and Hoogsteen base pairs of a DNA triplex. J. Am. Chem. Soc. 1999; 121:6019–6027. [Google Scholar]
- 158. Hennig M., Geierstanger B.. Direct detection of a histidine-histidine side chain hydrogen bond important for folding of apomyoglobin. J. Am. Chem. Soc. 1999; 121:5123–5126. [Google Scholar]
- 159. Liu A., Majumdar A., Jiang F., Chernichenko N., Skripkin E., Patel D.. NMR detection of intermolecular N-H⋅⋅⋅N hydrogen bonds in the Human T Cell Leukemia Virus-1 Rex peptide-RNA aptamer complex. J. Am. Chem. Soc. 2000; 122:11226–11227. [Google Scholar]
- 160. Dingley A., Masse J., Feigon J., Grzesiek S.. Characterization of the hydrogen bond network in guanosine quartets by internucleotide 3hJNC' and 2hJNN scalar couplings. J. Biomol. NMR. 2000; 16:279–289. [DOI] [PubMed] [Google Scholar]
- 161. Wöhnert J., Dingley A., Stoldt M., Görlach M., Grzesiek S., Brown L.. Direct identification of NH⋅⋅⋅N hydrogen bonds in non-canonical base pairs of RNA by NMR spectroscopy. Nucleic Acids Res. 1999; 27:3104–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Majumdar A., Kettani A., Skripkin E.. Observation and measurement of internucleotide 2JNN coupling constants between 15N nuclei with widely separated chemical shifts. J. Biomol. NMR. 1999; 14:67–70. [DOI] [PubMed] [Google Scholar]
- 163. Majumdar A., Kettani A., Skripkin E., Patel D.. Observation of internucleotide NH⋅⋅⋅N hydrogen bonds in the absence of directly detectable protons. J. Biomol. NMR. 1999; 15:207–211. [DOI] [PubMed] [Google Scholar]
- 164. Salvador P., Kobko N., Wieczorek R., Dannenberg J.. Calculation of trans-hydrogen-bond 13C-15N three-bond and other scalar J-couplings in cooperative peptide models. A density functional theory study. J. Am. Chem. Soc. 2004; 126:14190–14197. [DOI] [PubMed] [Google Scholar]
- 165. Bar-Joseph M. A contribution to the natural history of viroids. International Organization of Citrus Virologists Conference Proceedings. 1996; 13:226–229. https://escholarship.org/uc/item/5ww463md. [Google Scholar]
- 166. Martin W. Spindle Tuber, a new potato trouble. Hints to potato growers. N.Y. State Potato Assoc. 1922; 3:8. [Google Scholar]
- 167. Schultz E., Folsom D.. Transmission, variation and control of certain degeneration diseases of Irish potatoes. J. Agric. Res. 1923; 25:43–118. [Google Scholar]
- 168. Diener T. Are viroids escaped introns?. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:5014–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Flores R., Gago-Zachert S., Serra P., Sanjuán R., Elena S.. Viroids: survivors from the RNA world?. Annu. Rev. Microbiol. 2014; 68:395–414. [DOI] [PubMed] [Google Scholar]
- 170. Diener T. Viroids: ‘living fossils’ of primordial RNAs?. Biol. Direct. 2016; 11:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Diener T. Circular RNAs: relics of precellular evolution. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:9370–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S., Gregersen L., Munschauer M. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338. [DOI] [PubMed] [Google Scholar]
- 173. Lasda E., Parker R.. Circular RNAs: diversity of form and function. RNA. 2014; 20:1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Han C., Seebacher N., Hornicek F., Kan Q., Duan Z.. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget. 2017; 8:64622–64637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Huang S., Yang B., Chen B., Bliim N., Ueberham U., Arendt T., Janitz M.. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017; 109:401–407. [DOI] [PubMed] [Google Scholar]
- 176. Cervera A., De la Peña M.. Eukaryotic Penelope-like retroelements encode hammerhead ribozyme motifs. Mol. Biol. Evol. 2014; 31:2941–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Lünse C., Weinberg Z., Breaker R.. Numerous small hammerhead ribozyme variants associated with Penelope-like retrotransposons cleave RNA as dimers. RNA Biol. 2017; 14:1499–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Hammond R. Hadidi A, Randles J, Flores R, Palukaitis P. Economic significance of viroids in vegetable and field crops. Viroids and satellites. 2017; San Diego: Academic Press; 5–14. [Google Scholar]
- 179. Hadidi A., Vidalakis G., Sano T.. Hadidi A, Randles J, Flores R, Palukaitis P. Economic significance of fruit tree and grapevine viroids. Viroids and satellites. 2017; San Diego: Academic Press; 15–21. [Google Scholar]
- 180. Verhoeven J., Hammond R., Stancanelli G.. Hadidi A, Randles J, Flores R, Palukaitis P. Economic significance of viroids in ornamental crops. Viroids and satellites. 2017; San Diego: Academic Press; 27–38. [Google Scholar]
- 181. Rodriguez M., Vadamalai G., Randles J.. Hadidi A, Randles J, Flores R, Palukaitis P. Economic significance of palm tree viroids. Viroids and satellites. 2017; San Diego: Academic Press; 27–38. [Google Scholar]
- 182. Verhoeven J., Jansen C., Roenhorst J.. First report of pospiviroids infecting ornamentals in the Netherlands: Citrus exocortis viroid in Verbena sp., Potato spindle tuber viroid in Brugmansia suaveolens and Solanum jasminoides, and Tomato apical stunt viroid in Cestrum sp. Plant Pathol. 2007; 57:399. [Google Scholar]
- 183. Matoušek J., Orctová L., Ptáček J., Patzak J., Dědič P., Steger G., Riesner D.. Experimental transmission of pospiviroid populations to weed species characteristic of potato and hop fields. J. Virol. 2007; 81:11891–11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Verhoeven J., Hüner L., Marn M., Plesko I., Roenhorst J.. Mechanical transmission of Potato spindle tuber viroid between plants of Brugmansia suaveoles, Solanum jasminoides and potatoes and tomatoes. Eur. J. Plant Pathol. 2010; 128:417–421. [Google Scholar]
- 185. Matoušek J., et al. Biological and molecular analysis of the pathogenic variant C3 of potato spindle tuber viroid (PSTVd) evolved during adaptation to chamomilla (Matricaria chamomilla). Biol. Chem. 2012; 393:605–615. [DOI] [PubMed] [Google Scholar]
- 186. Jaksé J., Radišek S., Pokorn T., Matoušek J., Javornik B.. Deep-sequencing revealed Citrus bark cracking viroid (CBCVd) as a highly aggressive pathogen on hop. Plant Pathol. 2015; 64:831–842. [Google Scholar]
- 187. Semancik J., Rakowski A., Bash J., Gumpf D.. Application of selected viroids for dwarfing and enhancement of production of ‘Valencia’ orange. J. Hort. Sci. 1997; 72:563–570. [Google Scholar]
- 188. G. Vidalakis G., Pagliaccia D., Bash J., Afunian M., Semancik J.. Citrus dwarfing viroid: effects on tree size and scion performance specific to Poncirus trifoliata rootstock for high-density planting. Ann. Appl. Biol. 2011; 158:204–217. [Google Scholar]