Chromate is a highly toxic oxyanion. Extensive industrial use and inadequate waste management has caused the toxic pollution of several field sites. Understanding the chromate resistance mechanisms that enable organisms to thrive under these conditions is fundamental to develop (micro)biological strategies and applications aiming at bioremediation of contaminated soils or waters. Potential detoxifying microorganisms are often not sufficient in their resistance characteristics to effectively perform, e.g., chromate reduction or biosorption. In this study, we describe the manifold strategies of L. chromiiresistens to establish an extremely high level of chromate resistance. The multitude of mechanisms conferring it make this organism suitable for consideration as a new model organism to study chromate resistance.
KEYWORDS: Leucobacter, chromate resistance, heavy-metal resistance, heavy metals
ABSTRACT
Chromate is one of the major anthropogenic contaminants on Earth. Leucobacter chromiiresistens is a highly chromate-resistant strain, tolerating chromate concentrations in LB medium of up to 400 mM. In response to chromate stress, L. chromiiresistens forms biofilms, which are held together via extracellular DNA. Inhibition of biofilm formation leads to drastically decreased chromate tolerance. Moreover, chromate is reduced intracellularly to the less-toxic Cr(III). The oxidation status and localization of chromium in cell aggregates were analyzed by energy-dispersive X-ray spectroscopy coupled to scanning transmission electron microscopy and X-ray absorption spectroscopy measurements. Most of the heavy metal is localized as Cr(III) at the cytoplasmic membrane. As a new cellular response to chromate stress, we observed an increased production of the carotenoid lutein. Carotenoid production could increase membrane stability and reduce the concentration of reactive oxygen species. Bioinformatic analysis of the L. chromiiresistens genome revealed several gene clusters that could enable heavy-metal resistance. The extreme chromate tolerance and the unique set of resistance factors suggest the use of L. chromiiresistens as a new model organism to study microbial chromate resistance.
IMPORTANCE Chromate is a highly toxic oxyanion. Extensive industrial use and inadequate waste management has caused the toxic pollution of several field sites. Understanding the chromate resistance mechanisms that enable organisms to thrive under these conditions is fundamental to develop (micro)biological strategies and applications aiming at bioremediation of contaminated soils or waters. Potential detoxifying microorganisms are often not sufficient in their resistance characteristics to effectively perform, e.g., chromate reduction or biosorption. In this study, we describe the manifold strategies of L. chromiiresistens to establish an extremely high level of chromate resistance. The multitude of mechanisms conferring it make this organism suitable for consideration as a new model organism to study chromate resistance.
INTRODUCTION
Hexavalent chromium is highly toxic and unfortunately is used intensely in different industrial branches, including, but not limited to, steel manufacturing, wood treatment, and leather tanning. Its industrial use is accompanied by rising anthropogenic chromium pollution in the last few decades (1). Contaminated groundwater and field sites are the consequences of inadequate waste management and emphasize the need for efficient decontamination and remediation.
Chromate is cancerogenic and mutagenic (2, 3). These effects are at least to a large extent caused by the formation of reactive oxygen species as side products of the process of intracellular chromate reduction. Chromate is unspecifically reduced by a variety of enzymes and can, due to its high redox potential, abiotically interact with many intracellular reductants. This generates Cr(V) and Cr(IV) as reactive side products that can interact with oxygen, leading to the formation of singlet oxygen, superoxide anions, hydroxyl radicals, and hydrogen peroxide (4). Moreover, the reactive chromium anions can form adducts with proteins and DNA, which inhibit the reactivity and lead, together with the reactive oxygen species, to mutagenic effects (3, 5). The stable end product of the reduction chain is Cr(III), which is insoluble and far less bioavailable than Cr(VI) (6). Nevertheless, Cr(III) also binds unspecifically to DNA and proteins and can lead to mutagenic effects.
Many microbes have developed an astonishing robustness toward heavy metals, like chromium. The resistance strategies known to date are complex, and usually several mechanisms are used simultaneously to achieve higher levels of resilience. Chromate is imported via anion transporters, and sulfate importers especially seem to represent a major import pathway due to the structural similarity of chromate and sulfate. Consequently, the reduced expression of transporters employed for uptake of the metal is one strategy to develop chromate tolerance (6). Moreover, the increased expression of chromate exporters has been observed as a response to chromate stress, and the best studied model transporter is the membrane potential-dependent chromate exporter ChrA (6, 7). It has been emphasized before that the reduction process of Cr(VI) to Cr(III) is accompanied by the formation of reactive oxygen species (3, 4). Therefore, a frequently observed resistance mechanism is the controlled reduction of chromate (8), since the unspecific reduction of chromate with enzymes catalyzing the transfer of only one electron can be highly detrimental, as the enhanced formation of Cr(V) leads to a higher concentration of reactive oxygen species. On the contrary, two electron-transferring enzymes, like ChrR from Pseudomonas putida, lead to reduced formation of Cr(V) and hence a decreased formation of reactive oxygen species (9–11). Interestingly, microorganisms seem to respond to chromate stress also by the formation of biofilms. The formation of a matrix consisting of different polymers and proteins around the cells might limit the diffusion of the toxic element into the cell and could also act as an unspecific biosorbent (12, 13). Of note, the reduction of chromate to insoluble Cr(III), as well as the negatively charged surface of microbial cells acting as a biosorbent, could be used for the detoxification of chromate contaminations.
Leucobacter chromiiresistens was chosen for a detailed analysis of biological chromium detoxification pathways, with the hypothesis that the very high chromate tolerance of the organism might be due to new resistance determinants (14). We observed extracellular DNA-based biofilm formation and chromate reduction as responses to chromate exposure. Moreover, the organism produced increasing amounts of carotenoids as a response to chromate stress, a potential resistance mechanism that was not reported before. In a bioinformatics approach, we compared the genomes of chromate-resistant Leucobacter strains and identified common Leucobacter but also L. chromiiresistens-specific genes encoding proteins that could result in higher heavy-metal tolerance.
RESULTS
Growth of L. chromiiresistens under chromate stress.
Growth experiments in K2CrO4-complemented LB medium were conducted to determine the maximum chromate concentration that still allows for growth of L. chromiiresistens. Growth within the chromate concentration range of 0 to 10 mM was determined via optical density at 600 nm (OD600) measurements, while growth under high K2CrO4 concentrations (up to 500 mM) was assessed by wet-mass determination after 24 h of growth. Growth in LB medium supplemented with chromate was accompanied by the formation of cell flocks. Hence, it was not possible to use OD measurements to assess growth at chromate concentrations higher than 10 mM. Although the wet cell mass after 24 h was, if at all, positively affected by the addition of 10 mM chromate, the doubling time increased from 80.5 min with 0 mM chromate to 127 min and 151 min with 5 and 10 mM chromate in the medium, respectively. Overall, L. chromiiresistens was capable of sustaining growth at chromate concentrations of up to 400 mM in the medium (Fig. 1).
FIG 1.
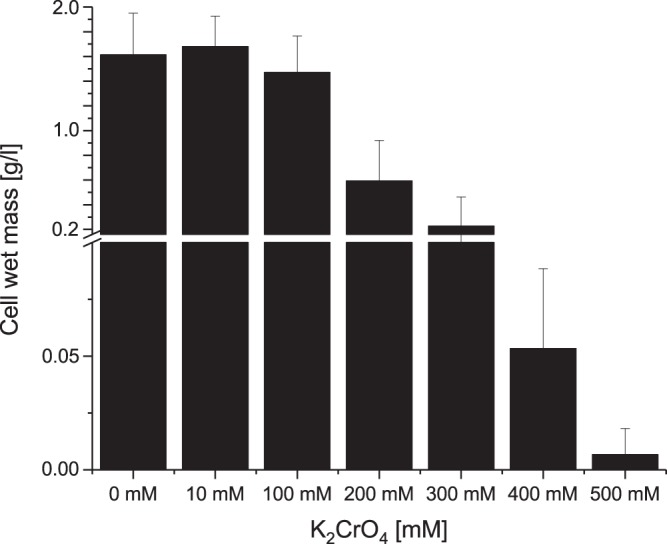
Wet-mass determination of L. chromiiresistens cells grown in K2CrO4-supplemented LB medium. Cells can grow in medium supplemented with chromate in concentrations of up to 400 mM. No growth was detectable in medium supplemented with 500 mM chromate. Error bars represent the standard deviation (SD) of the results from three independent replicates.
Importance of aggregate formation for growth under chromate stress.
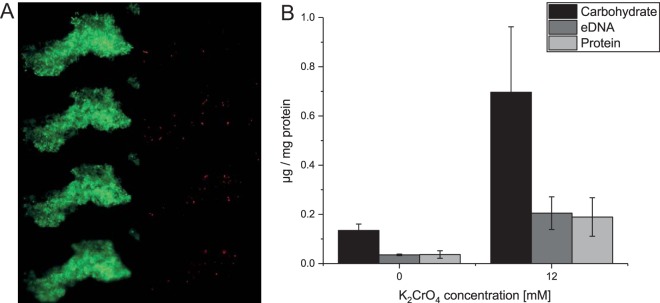
During growth under chromate stress, cells of L. chromiiresistens exhibited clearly visible aggregate formation (Fig. 2A and 3A). To investigate the nature of these aggregates, experiments were carried out to further characterize the cellular status in the cell clusters and the components of the biofilm matrix. A LIVE/DEAD stain indicated that the cells within the aggregates were mainly viable, and only a few cells with damaged membrane potentials could be detected (Fig. 2A). Component analysis of the extracellular polymeric matrix generated by the L. chromiiresistens cells revealed that it consisted of extracellular DNA (eDNA), carbohydrates, and proteins, common components of biofilms and cellular aggregates (15). Enhanced production of extracellular polymeric substances (EPS) was observed with ascending chromate concentrations during growth. Cells grown with 12 mM K2CrO4 produced significantly greater amounts of all EPS components than did cells grown in the absence of chromate (Fig. 2B). A detailed analysis of the carbohydrate compounds revealed that only glucose and mannose were present in the EPS fraction. The carbohydrate fraction of cells grown without chromate consisted of 78.6% mannose and 21.4% glucose, while fractions of chromate-grown cells were a higher mannose content of 86.7% and, consequently, a lower glucose concentration of 13.3%.
FIG 2.
Chromate stress-induced aggregate formation of L. chromiiresistens. (A) LIVE/DEAD stain of a representative aggregate. Green, 4′,6-diamidino-2-phenylindole (DAPI) stain (all cells); red, propidium iodide stain (cells with collapsed membrane potential). Shown are 4 different Z-levels of the aggregate. (B) EPS components of cellular aggregates of L. chromiiresistens cells grown without (0 mM) and with (12 mM) K2CrO4. The quantity of each of the tested components increased as a response to growth under chromate stress. All values were normalized to the initial amount of biomass (quantified as total cell protein) that was used for EPS isolation. Error bars represent SD of the results from three independent replicates.
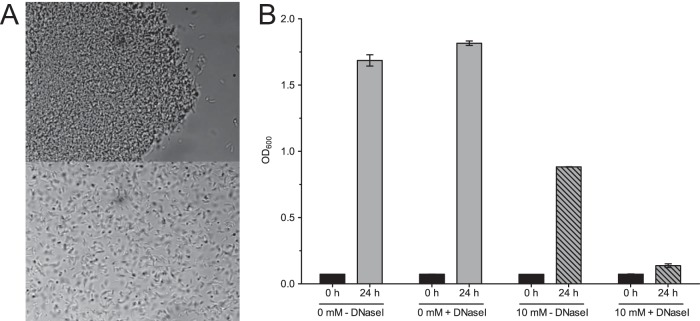
FIG 3.
Key role of eDNA. (A) Phase-contrast micrograph (×1,000) prior to (top) and 10 min after (bottom) the addition of DNase I. The addition of DNase I leads to a rapid disintegration of cellular aggregates. (B) Growth of L. chromiiresistens in the presence and absence of DNase I. The graph shows the OD values at the start of the experiment (0 h) and after 24 h. The addition of DNase I (+DNase I) did not affect the final OD achieved without chromate supplementation, while it severely impacted growth in the presence of 10 mM chromate. Error bars represent SD of the results from three independent replicates.
Since eDNA is described as a structural component in biofilms (16), experiments aimed at elucidating the importance of eDNA for the structural integrity of L. chromiiresistens aggregates were conducted. The addition of DNase I to aggregated cells of chromate-grown L. chromiiresistens led to disintegration of the biofilm structures (Fig. 3A). The importance of aggregate formation to withstand chromate stress was demonstrated through growth experiments with the addition of DNase. L. chromiiresistens grown under chromate stress (10 mM K2CrO4) was not able to sustain growth if DNase I was added to the medium, and the formation of aggregates was consequently inhibited (Fig. 3B). Of note, the addition of DNase I had no impact on the growth of L. chromiiresistens in a chromate-free medium, as well as for Escherichia coli cultures used as control (data not shown).
Sequencing of the eDNA via 454 technology revealed that the sequence reads mapped almost equally throughout the whole genome. A representative illustration of the mapping reads is given for the 3.27-Mbp scaffold 00001 (GenBank accession no. NZ_JH370377.1) (see Fig. S1 and Table S1 in the supplemental material). Based on this mapping, the origin of extracellular DNA is likely cell lysis based, and no specific DNA secretion seems to contribute to eDNA-based aggregate formation.
Localization and redox status of chromium within the L. chromiiresistens cells.
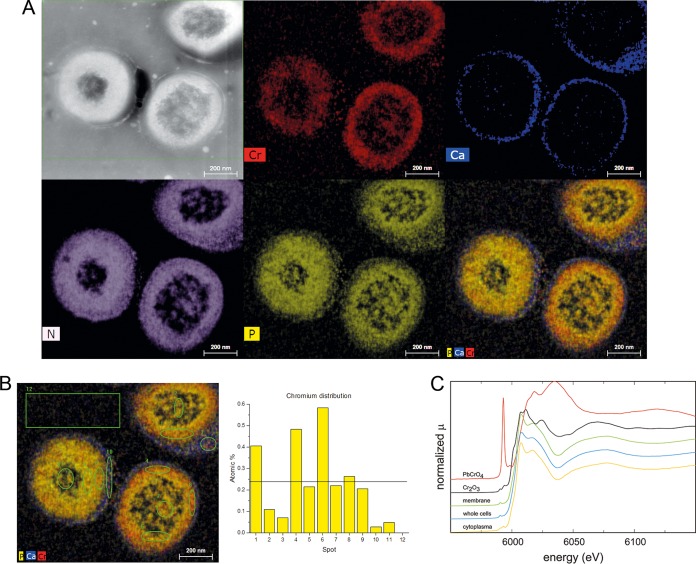
Energy-dispersive X-ray spectroscopy (EDS) was carried out to determine chromium distribution in L. chromiiresistens cells and cell aggregates. The cells were grown in 100 mM K2CrO4 and subjected to fixation and scanning transmission electron microscopy (STEM)-EDS analyses. Of note, the cells were fixed with LR white resin (Agar Scientific, Essex, UK) to reduce dehydration artifacts (Fig. 4). Chromium seems to be localized in the membrane area, as it appears below the cell wall, which is characterized by its high Ca2+ content. In contrast, only minor signals were observed at the cell center and between the cells, with the area between the cells being where the localization of EPS material was expected. Chromium signals appeared as a rather sharp zone in contrast to nitrogen signals (Fig. 4A). Hence, the analysis did not reveal a possible function of the EPS material as a specific or nonspecific biosorbent. In contrast, it probably revealed the intracellular accumulation of chromium. This is also corroborated by the quantitative EDS-based elemental analysis that displayed chromium in close proximity to the outer rim of the protoplast (Fig. 4B, spots 1, 4, and 6). Detailed EDS profiles showed a clear distinction between Ca and Cr signals and revealed a noticeable cooccurrence of Cr signals together with N, P, and O (Fig. S2).
FIG 4.
Chromium distribution in L. chromiiresistens extracellular matrix. (A) EDS images of chromate-grown cells. STEM overview (HAADF) and element distribution. Cr, chromium; Ca, calcium; N, nitrogen; P, phosphorus; PCaCr, superimposition of P, Ca, and Cr signals. (B) Detailed element analysis of the chromium dispersion. The spots are indicated on the left, and the corresponding atomic percentages of chromium are shown on the right. The black line indicates the mean chromium content of all analyzed spots (1 to 11). (C) Determination of chromium oxidation status via XANES. The predominant chromium species bound to cells was identified as Cr3+.
Since a large amount of chromium was detected via EDS-STEM, further experiments were conducted to identify the nature of the bound chromium species. Using X-ray absorption near-edge structure (XANES) spectroscopy, it was possible to determine that the chromium in chromate-grown cells occurred almost exclusively as Cr(III) (Fig. 4C). Cr(III) signals dominated in samples from whole cells as well as in the membrane and cytoplasmic fraction.
Based on the findings from EDS and XANES analysis, extended X-ray absorption fine structure (EXAFS) analysis was performed to identify the molecular environment and binding details of the chromium-containing area. The best fit was acquired for a molecular complex in which Cr(III) is bound to phosphate and guanine, a configuration that could suggest interaction with DNA (Fig. S3).
Since all conducted analyses identified the chromium species associated with the cells to be Cr(III), the question was raised whether a chromate reductase activity could be identified within the cells. Previous reports by Morais and colleagues revealed that L. chromiireducens and Leucobacter aridicollis reduce chromate during growth, and the authors suggested that chromate reduction is a general feature of Leucobacter strains. Nevertheless, the authors did not try to identify the localization of the chromate reductase (8, 17). Cells of L. chromiiresistens that were grown in LB medium were fractionated, and membrane and cytoplasmic pools were analyzed for a putative Cr(VI) reduction activity. While membrane fractions did not show any chromate reductase activity, cytoplasmic fractions reduced chromate with an activity of 7.3 nmol/min · mg protein. Surprisingly, similar rates were detectable in the presence of NADH as well as NADPH. Assays conducted with cell suspensions (OD, 8) that were grown in the presence of 5 mM chromate corroborated the identification of a chromate reductase activity, as a reduction rate of 1.9 μmol/liter · min was observed. Though, the chromate reductase activity seems to be nonspecific because it was not induced by the presence of chromate in the medium, as the chromate reduction rate of cells that were not pregrown in chromate containing medium was 2 μmol/liter · min. Higher overall reduction can be achieved by increasing cell density, as suspensions of chromate-pregrown cells (5 mM) with an OD of 12 showed a reduction rate of 3.5 μmol/liter · min.
Although the membrane fraction did not contain detectable chromate reductase activity, it was possible to macroscopically observe the color of the membranes changing from slightly yellow to bright orange as a consequence of chromate concentration in the medium during growth (Fig. S4). The putative pigment was extracted from membranes via ether extraction and spectrophotometric analyses revealed absorption maxima at 418 nm, 445 nm, and 475 nm, almost perfectly matching the absorption maxima of α-carotene (lutein) (18). Similar absorption maxima were observed for a pigment in L. chromiireducens subsp. solipictus (413 nm, 436 nm, and 466 nm, respectively) and were thought to be light induced (19). Using the absorption coefficient for lutein, we could quantify the carotenoid concentration per gram of cellular wet mass, which increased ∼4-fold from 1.46 μM to 5.4 μM at 0 mM and 6 mM chromate, respectively.
Bioinformatic analysis.
Chromate resistance is a well-known feature within the Leucobacter genus. At least 10 strains (e.g., L. chromiireducens, L. luti, L. alluvii, L. aridicollis, L. salsicius, and L. chironomi) were described to be resistant to moderate K2CrO4 concentrations of 3 to 20 mM (8, 17, 20, 21). In comparison, L. chromiiresistens shows a considerably higher level of chromate resistance and is the only organism known so far that tolerates these extreme Cr concentrations. Based on a comparison of the already-available genome sequences of L. chromiiresistens JG31, L. salsicius M1-8T, L. komagatae, L. celer subsp. astrifaciens, L. musarum subsp. musarum, and L. chironomi DSM 19883, we tried to identify genetic elements that most likely (i) enable these species to resist chromate stress and (ii) particularly allow L. chromiiresistens to thrive at extreme chromate concentrations. The obtained results indicate a number of genes that potentially confer chromate or at least heavy-metal resistance and that are common in the tested organisms (i.e., arsenate reductase; lead, cadmium, zinc, and mercury transporting ATPase; copper resistance proteins; and arsenical resistance protein). An overview of possible relevant features is given in Table S2. The NCBI automated annotation pipeline or RAST annotation, revealed the presence of the chrA gene encoding a chromate transporter that confers chromate resistance to E. coli and Pseudomonas spp. (22, 23). Of all available Leucobacter species genomes at the time of writing, chrA is present in the genomes of L. chromiiresistens JG 31, L. chironomi DSM 19883, L. musarum subsp. musarum CBX152, L. salsicius M1-8T, L. celer subsp. astrifaciens CBX151. Additionally, genes potentially involved in mercury, copper, and cadmium resistance were identified only in the genome of L. chromiiresistens (Table S2).
DISCUSSION
The aim of this study was to identify possible resistance strategies and mechanisms enabling L. chromiiresistens to withstand extremely high chromate concentrations of up to 400 mM. To the best of our knowledge, L. chromiiresistens is the organism with the highest chromate tolerance described so far. Other extremely chromate-tolerant organisms can cope with concentrations of only up to 20 mM (L. celer), 40 mM (Pseudomonas corrugata), 60 mM (Lysinibacillus fusiformis ZC1), 92 mM (Bacillus sp. strain ev3), and 200 mM (Arthrobacter sp. strain FB24) (24–28).
Based on the obtained results, we hypothesize that the extreme chromate tolerance is a consequence of biofilm formation-based reduced import, chromate export via ChrA, chromate reduction, and the detoxification of reactive oxygen species via enhanced carotenoid production.
The biofilm-based formation of a diffusion barrier is a mechanism that does not seem to be chromate specific, as a similar mechanism enables, for instance, Pseudomonas aeruginosa to thrive under immense detergent stress (29). Accordingly, it was shown that biofilm production by E. coli cells hampers the penetration of metal ions into deeper cell layers (1). Recently, a comparable behavior of chromate stress-induced aggregation of P. aeruginosa cells was also reported (30). The fundamental importance of aggregate formation for L. chromiiresistens was demonstrated by the severe growth deficiency of cells that could not aggregate due to the addition of DNase (Fig. 3B). The biofilm-based decreased chromate uptake might be assisted by the active export of chromate, which is corroborated by the findings of from EDS and EXAFS analyses. Several heavy-metal transport systems that could catalyze the export have been identified in the genome of the organism. These systems include the well-characterized ChrA chromate exporter.
Besides potentially reduced import and increased export, a chromate reductase activity was detected in the cytoplasm of the organism. This activity could be the result of an unspecific activity of one or more oxidoreductases that use NADH and/or NADPH as a cofactor. Moreover, several genes encoding heavy-metal reductases have been identified in the genome of the organism. So far, it is not clear whether these reductases play a role under chromate stress and consequently show cross-reactivity with chromate. The cytoplasmic localization of the chromate reductase in L. chromiiresistens is in line with results obtained for other Gram-positive organisms (Leucobacter sp. strain KCH4, Bacillus firmus, Bacillus subtilis, and Bacillus sphaericus AND303) (31–34). These studies also showed the chromate-reducing properties of cell extracts and likewise no discrimination between NADH and NADPH.
Chromate reduction is typically accompanied with the production of reactive oxygen species. The genome of L. chromiiresistens harbors several genes involved in oxidative stress responses (i.e., superoxide dismutases), which could help lower the concentration of reactive oxygen species (ROS) (3).
It seems likely that the observed increased production of membrane-bound carotene (Fig. S4) is a cellular response to the oxidative stress accompanying intracellular Cr(VI) reduction. All mechanisms involved in Cr(VI) reduction, whether enzymatic or not, lead to the generation of ROS (6, 11). Thus, the observed increased production of membrane-bound carotenes offers a reasonable strategy for the cells to reduce the concentration of ROS. Interestingly, Gruszecki and Strzałka showed that carotenoids increase the stability of lipid bilayer membranes and reduce their permeability for small ions and molecular oxygen (35). Along this line, it was reported that chromate reduces the stability of biological membranes and increases their permeability (36). Moreover, chromate was also detected within the cytoplasmic membrane of fungal spheroplasts, and a reduction of Cr(VI) to Cr(III) was detectable within the membranes. Hence, high concentrations of carotenoids in the cytoplasmic membrane might be a way to stabilize the chromate-destabilized membranes, to reduce Cr(VI) directly, and thereby omit the detour via ROS, and to reduce the unspecific import of the metal. Furthermore, a reduced diffusion of oxygen as a result of biofilm formation and increased membrane stability could also be a way to decrease the kinetics of ROS production. In this study, a correlation of carotene content and chromate concentration was observed. Possibly, this factor of higher carotene production is a major reason why L. chromiiresistens is more tolerant to chromate than all other so-far-characterized strains.
Based on the findings reported here, it seems reasonable to consider L. chromiiresistens a new model organism to study chromate tolerance and as a target organism for further research on its use in bioremediation. Recently, L. chromiiresistens was used in an experimental approach showing the distribution of a chromate-stressed coculture, with E. coli in a microfluidic chip. In their work, the authors showed that L. chromiiresistens-facilitated chromate reduction detoxified the rear end of a microfluidic chip and thus allowed growth of E. coli, a very convincing example of an application for potential bioremediation processes (37).
MATERIALS AND METHODS
Strain cultivation.
All experiments were conducted with L. chromiiresistens type strain JG31 (DSM 22788) and Escherichia coli (14, 38). If not otherwise stated, lysogeny broth (LB) was used as growth medium. Growth was carried out under oxic conditions at 37°C and pH 7.
Determination of cell mass.
To determine the wet cell mass of L. chromiiresistens, cells were grown aerobically in 500 ml LB medium containing 0, 10, 100, 200, 300, 400, 500, or 1,000 mM K2CrO4 at 37°C for 48 h. A volume of 5 ml of these cultures was harvested (10 min, 9,000 × g) at the starting point (T0) of the experiment and after 48 h. The cell pellets were weighted after 48 h, and the value at T0 was subtracted.
Preparation of cellular fractions.
LB-grown cells were harvested (9,000 × g) in exponential-growth phase, washed twice, and resuspended in 20 mM HEPES (pH 7.5). Cell disruption was achieved via three subsequent passages through a French press cell (Aminco; SLM Instruments, USA). Residual cells were spun down by low-spin centrifugation (9,000 × g), and the cell lysate was separated by ultracentrifugation (205,000 × g, 75 min). The cytoplasmic fraction was quickly separated from the pellet comprising the cell membranes. The membranes were resuspended in 20 mM HEPES (pH 7.5) containing 10% (wt/vol) glycerol.
Cr(VI) reduction assays.
The decrease in the Cr(VI) concentration in cell suspensions was determined by the diphenylcarbazide (DPC) method (39). Briefly, a 1-ml sample was mixed with 255 μl of a 2 M Na2CO3 solution to stop further chromate reduction. The cells were spun down, and the supernatant was transferred into a new tube containing 160 mg of pestled activated carbon-Al2O3 (1:1 [wt/wt]). The sample was mixed by vortexing. Insoluble components were separated by centrifugation (9,000 × g, 5 min). The supernatant was filtered (0.22 μm), and 200 μl of this supernatant was mixed with 200 μl detection reagent (1:1 mixture of 1% 1,5-diphenylcabazide in acetone and 2 M H2SO4 [vol/vol]). The mixture was incubated for 15 min at room temperature (RT). The absorption values at λ = 540 nm were used to determine Cr(VI) concentrations by comparison to chromate standard solutions (K2CrO4).
Cell suspension assay.
Cells were grown overnight in the presence or absence of 10 mM chromate in LB medium at 37°C. Thereafter, the cells were harvested, washed (20 mM HEPES [pH 7.5]), and resuspended in LB to an OD600 of 10. The assay was started by the addition of K2CrO4 at a final concentration of 500 μM. Samples were taken every 30 min for 4 h, and Cr(VI) concentrations were determined via the DPC method described above.
NADH/NADPH assay.
To determine whether the chromate reductase is localized in the cytoplasm or in the membrane and which cofactor, NADH or NADPH, is needed for the reaction, the membrane and cytoplasmic fraction were tested with either one of the considered electron donors. The assay was conducted in 10 mM HEPES (pH 7.5) containing 500 μM K2CrO4. The reaction was started by the addition of NADH or NADPH to a final concentration of 200 μM. The assay was incubated at 37°C, and at certain time points, a 1-ml sample was removed and the Cr(VI) concentration was determined. The assay was conducted with cell extracts from cells that were either pregrown in LB with 10 mM K2CrO4 or without Cr(VI) complementation. Protein concentrations were determined by the Bradford method using bovine serum albumin as a standard (40).
Extraction of extracellular polymeric substances.
EPS were extracted using a protocol adapted from Evans and Linker (41). Five hundred milliliters of an overnight culture grown with (12 mM) and without K2CrO4 was harvested. The cells were resuspended in 25 volumes of extraction buffer (0.9% NaCl in 10 mM EDTA). The cell suspension was mixed for 20 min in an MM 400 swing mill at 11 Hz (Retsch GmbH, Haan, Germany). Subsequently, the cell suspension was centrifuged for 45 min at 4°C and 25,000 × g. The supernatant was centrifuged again for another 2 h at 4°C and 25,000 × g. The supernatant was collected, and 3 volumes of 95% ethanol was added slowly while the mixture was continuously stirred. The precipitated EPS was collected by centrifugation for 30 min at 4°C and 3,000 × g. The precipitate was washed twice with 95% ethanol and once with 100% ethanol. Each washing step was followed by a centrifugation at 4°C and 15,000 × g for 15 min. The pellet was dried overnight (O/N) at room temperature and finally resuspended in 1 ml double-distilled water (ddH2O).
Determination of EPS.
Glycosyl composition analysis was performed by combined gas chromatography-mass spectrometry (GC/MS) of the per-O-trimethylsilyl (TMS) derivates of the monosaccharide methyl glycosides produced from the sample by acidic methanolysis. Methyl glycosides were first prepared from dry samples by methanolysis in 1 M HCl in methanol at 80°C (18 to 22 h), followed by re-N-acetylation with pyridine and acetic anhydride in methanol. The samples were then per-O-trimethylsilyated by treatment with Tri-Sil at 80°C for 30 min (42, 43). GC/MS analysis of the TMS methyl glycosides was performed on an HP 6890 GC interfaced to a 5975b mass selective detector (MSD), using an All Tech EC-1 fused silica capillary column (30 m by 0.25 mm inner diameter [i.d.]). Identification of carbohydrate components was carried out at the Complex Carbohydrate Research Center (CCRC, Georgia, USA).
The protocol of Dubois et al. was used to determine the amount of carbohydrates in the EPS (44). Briefly, 200 μl of each sample was mixed with 5 μl of 80% phenol and 500 μl of 90% H2SO4 and incubated for 10 min at room temperature. Thereafter, the sample was shaken on a rotary shaker for another 30 min at 30°C and 100 rpm. Standard solutions were prepared containing a mixture of carbohydrates corresponding to the composition of the L. chromiiresistens EPS (80% mannose, 20% glucose, as determined by analysis at the CCRC). Absorption was measured at a wavelength of λ = 490 nm.
Extracellular DNA (eDNA) was quantified using Quant-iT double-stranded DNA (dsDNA) high-sensitivity kit (Fisher Scientific, Schwerte, Germany), according to the manufacturer's instructions. Proteins were quantified using the Roti-Nanoquant kit (Carl Roth, Karlsruhe, Germany).
STEM-EDS.
The transmission electron microscopy (TEM) investigation and X-ray energy dispersive spectrometry (EDS) were performed with an FEI Tecnai Osiris microscope (Thermo Fisher Scientific, Inc.) equipped with a ChemiSTEM EDS microanalyzer (200-kV extreme field emission gun [X-FEG] Super-X EDS with 4- by 30-mm2 windowless silicon drift detector [SDD] diodes). The element distribution in samples was obtained by EDS in bright-field (BF) and high-angle annular field (HAADF) scanning TEM modes using the quantitative analysis ESPRIT (Bruker) software. Hypermaps with a complete spectrum stored for each mapped point were acquired and processed to obtain the concentrations of elements in the selected areas. Quantitative EDS analysis was carried out using the Cliff-Lorimer standardless method with thickness correction. The background was calculated based on the sample composition. Elemental concentrations in atomic % were derived from deconvoluted line intensities within a 95% confidence level.
XANES/EXAFS spectrum collection.
The micro-X-ray absorption spectroscopy (μ-XAS) data were collected at the beamline of the Synchrotron Radiation Laboratory for Environmental Studies (SUL-X) at the synchrotron radiation source at ANKA in Karlsruhe. Cultures of L. chromiiresistens were grown with 10 mM K2CrO4. Preparation of cellular fractions was conducted as described above using 500 mM phosphate-buffered saline (PBS) (pH 7.5) as a buffer. After preparation, the fractions (membrane and cytoplasm) and whole cells were lyophilized. The lyophilized samples were compression molded (tablets) and analyzed in this mode. For this work, the samples were mounted onto a Kapton tape and inserted into the beam; the tape was oriented at an angle of 45° to the beam. A silicon (111) crystal pair with a fixed-beam exit was used as a monochromator. The X-ray beam was aligned to an intermediate focus and then collimated by slits located at the distance of the intermediate focus to about 100 by 100 μm and subsequently focused with a Kirkpatrick-Baez mirror pair to about 50 by 50 μm at the sample position.
The μ-XAS spectra at the Cr K edge were measured in fluorescence mode in energy steps of 5 eV in the region from −150 to −50 eV relative to the absorption edge, of 2 eV in the region from −50 eV to −20 eV, of 0.5 eV from −20 eV to +20 eV, and with a k step of 0.05 from +20 eV to +400 eV (about k = 10). The intensity of the primary beam was measured by an ionization chamber. Fluorescence intensities were collected with a 7-element Si(Li) solid-state detector with the energy window set to the Cr K-α line. Data were dead time corrected, summed up for all seven channels, and divided by the input intensity, which was measured in an ionization chamber prior to the sample analysis. The collected data were processed by the Athena and Artemis software suite (45).
Bioinformatic analysis.
Extracellular DNA was extracted as described above and sequenced using GS FLX Titanium 454 technology (Roche). Mapping of the obtained reads was performed against the draft genome sequence of L. chromiiresistens (GenBank accession number AGCW00000000) using CLC Genomics Workbench (version 10.0.1; Qiagen, Hilden, Germany), according to the standard mapping parameters of the program.
Potential resistance factors were identified based on RAST annotation done earlier (46). The respective gene sequences from the L. chromiiresistens genome were then used in a BLAST search against all Leucobacter genomes available at the time of writing. Threshold values for identification were identity scores of ≥70% and E values of ≤10−9 by using the Blast2GO suite (47).
Isolation of carotenoids.
Cells of L. chromiiresistens were grown without and with 2, 4, and 6 mM K2CrO4 for 24 h at 37°C, and cellular fractions were prepared as described above. Colored membrane fractions of L. chromiiresistens were subjected to ether extraction. Therefore, membrane fractions were resuspended in 1 ml HEPES buffer, followed by the addition of 1 ml diethyl ether. The sample was shaken for 10 min, and the ether fraction containing the putative carotenoid was collected.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the Department of Energy-funded (DE-FG09-93ER-20097) Center for Plant and Microbial Complex Carbohydrates and by the state of Baden-Wuerttemberg.
We declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02208-18.
REFERENCES
- 1.Harrison JJ, Ceri H, Turner RJ. 2007. Multimetal resistance and tolerance in microbial biofilms. Nat Rev Microbiol 5:928–938. doi: 10.1038/nrmicro1774. [DOI] [PubMed] [Google Scholar]
- 2.Witmer CM, Park H-S, Shupack SI. 1989. Mutagenicity and disposition of chromium. Sci Total Environ 86:131–148. doi: 10.1016/0048-9697(89)90200-3. [DOI] [PubMed] [Google Scholar]
- 3.Holmes AL, Wise SS, Wise JP. 2008. Carcinogenicity of hexavalent chromium. Indian J Med Res 128:353–372. [PubMed] [Google Scholar]
- 4.Ackerley DF, Barak Y, Lynch SV, Curtin J, Matin A. 2006. Effect of chromate stress on Escherichia coli K-12. J Bacteriol 188:3371–3381. doi: 10.1128/JB.188.9.3371-3381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voitkun V, Zhitkovich A, Costa M. 1998. Cr(III)-mediated crosslinks of glutathione or amino acids to the DNA phosphate backbone are mutagenic in human cells. Nucleic Acids Res 26:2024–2030. doi: 10.1093/nar/26.8.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramírez-Díaz MI, Díaz-Pérez C, Vargas E, Riveros-Rosas H, Campos-García J, Cervantes C. 2008. Mechanisms of bacterial resistance to chromium compounds. Biometals 21:321–332. doi: 10.1007/s10534-007-9121-8. [DOI] [PubMed] [Google Scholar]
- 7.Díaz-Pérez C, Cervantes C, Campos-García J, Julián-Sánchez A, Riveros-Rosas H. 2007. Phylogenetic analysis of the chromate ion transporter (CHR) superfamily. FEBS J 274:6215–6227. doi: 10.1111/j.1742-4658.2007.06141.x. [DOI] [PubMed] [Google Scholar]
- 8.Morais PV, Paulo C, Francisco R, Branco R, Paula Chung A, da Costa MS. 2006. Leucobacter luti sp. nov., and Leucobacter alluvii sp. nov., two new species of the genus Leucobacter isolated under chromium stress. Syst Appl Microbiol 29:414–421. doi: 10.1016/j.syapm.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez CF, Aekerley DF, Lynch SV, Matin A. 2005. ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J Biol Chem 280:22590–22595. doi: 10.1074/jbc.M501654200. [DOI] [PubMed] [Google Scholar]
- 10.Viradia SH, Vala AK. 2013. In silico protein structure modeling and conservation analysis of ChrR, a class-I chromate reducing flavoenzyme from Pseudomonas putida. Protein Pept Lett 20:1049–1053. doi: 10.2174/0929866511320090011. [DOI] [PubMed] [Google Scholar]
- 11.Ackerley DF, Gonzalez CF, Keyhan M, Blake R Jr, Matin A. 2004. Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environ Microbiol 6:851–860. doi: 10.1111/j.1462-2920.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 12.Sethuraman P, Balasubramanian N. 2010. Removal of Cr(VI) from aqueous solution using Bacillus subtilis, Pseudomonas aeruginosa and Enterobacter cloacae. Int J Eng Sci Technol 2:1811–1825. [Google Scholar]
- 13.Thatheyus AJ, Ramya D. 2016. Biosorption of chromium using bacteria: an overview. Sci Int 4:74–79. doi: 10.17311/sciintl.2016.74.79. [DOI] [Google Scholar]
- 14.Sturm G, Jacobs J, Sproer C, Schumann P, Gescher J. 2011. Leucobacter chromiiresistens sp. nov., a chromate-resistant strain. Int J Syst Evol Microbiol 61:956–960. doi: 10.1099/ijs.0.022780-0. [DOI] [PubMed] [Google Scholar]
- 15.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 16.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 17.Morais PV, Francisco R, Branco R, Chung AP, da Costa MS. 2004. Leucobacter chromiireducens sp. nov, and Leucobacter aridicollis sp. nov., two new species isolated from a chromium contaminated environment. Syst Appl Microbiol 27:646–652. doi: 10.1078/0723202042369983. [DOI] [PubMed] [Google Scholar]
- 18.Hager A. 1970. Ausbildung von Maxima im Absorptionsspektrum von Carotinoiden im Bereich um 370 nm; Folgen für die Interpretation bestimmter Wirkungsspektren. Planta 91:38–53. doi: 10.1007/BF00390164. [DOI] [PubMed] [Google Scholar]
- 19.Muir RE, Tan MW. 2007. Leucobacter chromiireducens subsp. solipictus subsp. nov., a pigmented bacterium isolated from the nematode Caenorhabditis elegans, and emended description of L. chromiireducens. Int J Syst Evol Microbiol 57:2770–2776. [DOI] [PubMed] [Google Scholar]
- 20.Halpern M, Shakéd T, Pukall R, Schumann P. 2009. Leucobacter chironomi sp. nov., a chromate-resistant bacterium isolated from a chironomid egg mass. Int J Syst Evol Microbiol 59:665–670. doi: 10.1099/ijs.0.004663-0. [DOI] [PubMed] [Google Scholar]
- 21.Yun JH, Roh SW, Kim MS, Jung MJ, Park EJ, Shin KS, Nam YD, Bae JW. 2011. Leucobacter salsicius sp. nov., from a salt-fermented food. Int J Syst Evol Microbiol 61:502–506. doi: 10.1099/ijs.0.021360-0. [DOI] [PubMed] [Google Scholar]
- 22.Rivera SL, Vargas E, Ramírez-Díaz MI, Campos-García J, Cervantes C. 2008. Genes related to chromate resistance by Pseudomonas aeruginosa PAO1. Antonie Van Leeuwenhoek 94:299–305. doi: 10.1007/s10482-008-9247-x. [DOI] [PubMed] [Google Scholar]
- 23.Pimentel BE, Moreno-Sánchez R, Cervantes C. 2002. Efflux of chromate by Pseudomonas aeruginosa cells expressing the ChrA protein. FEMS Microbiol Lett 212:249–254. doi: 10.1111/j.1574-6968.2002.tb11274.x. [DOI] [PubMed] [Google Scholar]
- 24.Viti C, Decorosi F, Tatti E, Giovannetti L. 2007. Characterization of chromate-resistant and -reducing bacteria by traditional means and by a high-throughput phenomic technique for bioremediation purposes. Biotechnol Prog 23:553–559. doi: 10.1021/bp0603098. [DOI] [PubMed] [Google Scholar]
- 25.Rehman A, Zahoor A, Muneer B, Hasnain S. 2008. Chromium tolerance and reduction potential of a Bacillus sp. ev3 isolated from metal contaminated wastewater. Bull Environ Contam Toxicol 81:25–29. doi: 10.1007/s00128-008-9442-5. [DOI] [PubMed] [Google Scholar]
- 26.Shin NR, Kim MS, Jung MJ, Roh SW, Nam Do Y, Park EJ, Bae JW. 2011. Leucobacter celer sp. nov., isolated from Korean fermented seafood. Int J Syst Evol Microbiol 61:2353–2357. doi: 10.1099/ijs.0.026211-0. [DOI] [PubMed] [Google Scholar]
- 27.He M, Li X, Liu H, Miller SJ, Wang G, Rensing C. 2011. Characterization and genomic analysis of a highly chromate resistant and reducing bacterial strain Lysinibacillus fusiformis ZC1. J Hazard Mater 185:682–688. doi: 10.1016/j.jhazmat.2010.09.072. [DOI] [PubMed] [Google Scholar]
- 28.Henne KL, Nakatsu CH, Thompson DK, Konopka AE. 2009. High-level chromate resistance in Arthrobacter sp. strain FB24 requires previously uncharacterized accessory genes. BMC Microbiol 9:199. doi: 10.1186/1471-2180-9-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klebensberger J, Rui O, Fritz E, Schink B, Philipp B. 2006. Cell aggregation of Pseudomonas aeruginosa strain PAO1 as an energy-dependent stress response during growth with sodium dodecyl sulfate. Arch Microbiol 185:417–427. doi: 10.1007/s00203-006-0111-y. [DOI] [PubMed] [Google Scholar]
- 30.Kang C, Wu P, Li Y, Ruan B, Zhu N, Dang Z. 2014. Estimates of heavy metal tolerance and chromium (VI) reducing ability of Pseudomonas aeruginosa CCTCC AB93066: chromium (VI) toxicity and environmental parameters optimization. World J Microbiol Biotechnol 30:2733–2746. doi: 10.1007/s11274-014-1697-x. [DOI] [PubMed] [Google Scholar]
- 31.Pal A, Dutta S, Paul AK. 2005. Reduction of hexavalent chromium by cell-free extract of Bacillus sphaericus and 303 isolated from serpentine soil. Curr Microbiol 51:327–330. doi: 10.1007/s00284-005-0048-4. [DOI] [PubMed] [Google Scholar]
- 32.Garbisu C, Alkorta I, Llama MJ, Serra JL. 1998. Aerobic chromate reduction by Bacillus subtilis. Biodegradation 9:133–141. doi: 10.1023/A:1008358816529. [DOI] [PubMed] [Google Scholar]
- 33.Sau GB, Chatterjee S, Mukherjee SK. 2010. Chromate reduction by cell-free extract of Bacillus firmus KUCr1. Pol J Microbiol 59:185–190. [PubMed] [Google Scholar]
- 34.Sarangi A, Krishnan C. 2016. Detoxification of hexavalent chromium by Leucobacter sp. uses a reductase with specificity for dihydrolipoamide. J Basic Microbiol 56:175–183. doi: 10.1002/jobm.201500285. [DOI] [PubMed] [Google Scholar]
- 35.Gruszecki WI, Strzałka K. 2005. Carotenoids as modulators of lipid membrane physical properties. Biochim Biophys Acta 1740:108–115. doi: 10.1016/j.bbadis.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 36.Farkas N, Pesti M, Belagyi J. 2003. Effects of hexavalent chromium on the plasma membranes of sensitive and tolerant mutants of Schizosaccharomyces pombe. An EPR study. Biochim Biophys Acta 1611:217–222. doi: 10.1016/S0005-2736(03)00055-5. [DOI] [PubMed] [Google Scholar]
- 37.Hansen SH, Kabbeck T, Radtke CP, Krause S, Krolitzki E, Peschke T, Gasmi J, Rabe KS, Wagner M, Horn H, Hubbuch J, Gescher J, Niemeyer CM. 2017. Machine-assisted cultivation and analysis of biofilms. bioRxiv doi: 10.1101/210583. [DOI] [PMC free article] [PubMed]
- 38.Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 39.Sandell EB, Coogan CK, Ham NS, Stuart SN. 1959. Diphenylcarbazide method, p 392–397. In Sandell EB. (ed), Colorimetric determination of trace metals, 3rd ed Interscience Publishers, Inc., New York, NY. [Google Scholar]
- 40.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 41.Evans LR, Linker A. 1973. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol 116:915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merkle RK, Poppe I. 1994. Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol 230:1–15. doi: 10.1016/0076-6879(94)30003-8. [DOI] [PubMed] [Google Scholar]
- 43.York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P. 1986. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol 118:3–40. doi: 10.1016/0076-6879(86)18062-1. [DOI] [Google Scholar]
- 44.Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. 1951. A colorimetric method for the determination of sugars. Nature 168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 45.Ravel B, Newville M. 2005. ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT. J Synchrotron Radiat 12:537–541. doi: 10.1107/S0909049505012719. [DOI] [PubMed] [Google Scholar]
- 46.Sturm G, Buchta K, Kurz T, Rensing SA, Gescher J. 2012. Draft genome sequence of Leucobacter chromiiresistens, an extremely chromium-tolerant strain. J Bacteriol 194:540–541. doi: 10.1128/JB.06413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.