Sphagnum-dominated peatlands play an important role in maintaining atmospheric carbon dioxide levels by modifying conditions in the surrounding soil to favor the growth of Sphagnum over that of other plant species. This lowers the rate of decomposition and facilitates the accumulation of fixed carbon in the form of partially decomposed biomass. The unique environment produced by Sphagnum enriches for the growth of a diverse microbial consortia that benefit from and support the moss's growth, while also maintaining the hostile soil conditions. While a growing body of research has begun to characterize the microbial groups that colonize Sphagnum, little is currently known about the ecological factors that constrain community structure and define ecosystem function. Top-down population control by viruses is almost completely undescribed. This study provides insight into the significant viral influence on the Sphagnum microbiome and identifies new potential model systems to study virus-host interactions in the peatland ecosystem.
KEYWORDS: viruses, RNA-seq, Sphagnum, peat bogs, microbial ecology
ABSTRACT
Sphagnum-dominated peatlands play an important role in global carbon storage and represent significant sources of economic and ecological value. While recent efforts to describe microbial diversity and metabolic potential of the Sphagnum microbiome have demonstrated the importance of its microbial community, little is known about the viral constituents. We used metatranscriptomics to describe the diversity and activity of viruses infecting microbes within the Sphagnum peat bog. The vegetative portions of six Sphagnum plants were obtained from a peatland in northern Minnesota, and the total RNA was extracted and sequenced. Metatranscriptomes were assembled and contigs were screened for the presence of conserved virus marker genes. Using bacteriophage capsid protein gp23 as a marker for phage diversity, we identified 33 contigs representing undocumented phages that were active in the community at the time of sampling. Similarly, RNA-dependent RNA polymerase and the nucleocytoplasmic large DNA virus (NCLDV) major capsid protein were used as markers for single-stranded RNA (ssRNA) viruses and NCLDV, respectively. In total, 114 contigs were identified as originating from undescribed ssRNA viruses, 22 of which represent nearly complete genomes. An additional 64 contigs were identified as being from NCLDVs. Finally, 7 contigs were identified as putative virophage or polinton-like viruses. We developed co-occurrence networks with these markers in relation to the expression of potential-host housekeeping gene rpb1 to predict virus-host relationships, identifying 13 groups. Together, our approach offers new tools for the identification of virus diversity and interactions in understudied clades and suggests that viruses may play a considerable role in the ecology of the Sphagnum microbiome.
IMPORTANCE Sphagnum-dominated peatlands play an important role in maintaining atmospheric carbon dioxide levels by modifying conditions in the surrounding soil to favor the growth of Sphagnum over that of other plant species. This lowers the rate of decomposition and facilitates the accumulation of fixed carbon in the form of partially decomposed biomass. The unique environment produced by Sphagnum enriches for the growth of a diverse microbial consortia that benefit from and support the moss's growth, while also maintaining the hostile soil conditions. While a growing body of research has begun to characterize the microbial groups that colonize Sphagnum, little is currently known about the ecological factors that constrain community structure and define ecosystem function. Top-down population control by viruses is almost completely undescribed. This study provides insight into the significant viral influence on the Sphagnum microbiome and identifies new potential model systems to study virus-host interactions in the peatland ecosystem.
INTRODUCTION
Peatlands represent one of the most significant biological carbon sinks on the planet, storing an estimated 25% of terrestrial carbon in the form of partially decomposed organic matter (1–3). This accumulation of carbon is achieved through much lower rates of respiration and decomposition than those observed in soil, due in large part to the low pH, nutrient-poor, and anaerobic environments created by the dominant moss population (4, 5), of which the genus Sphagnum is the most prevalent (6, 7). As these environmental conditions appear to favor the growth of Sphagnum over that of vascular plants, primary production is dominated by the moss, which further retards decomposition due to the production of antimicrobial compounds such as sphagnic acid (8–10) and sphagnan (11, 12). Despite this, Sphagnum and other peat mosses cultivate a diverse, symbiotic microbiome that appears to diminish nutritional gaps for the moss and also contribute to the unique biogeochemical characteristics of the peatland ecosystem (13–15). In addition to their value as reservoirs of microbial diversity, the partially decomposed organic matter, known as Sphagnum peat, serves as an important economic resource for use in horticulture. As many peat bogs have begun to experience stress due to anthropogenic disturbances (16–18) and possibly climate change (19), the Sphagnum microbiome is of interest in peatland conservation and the ecosystem's services to the surrounding environment.
While there is a growing body of research characterizing the microbial groups that colonize Sphagnum (15), little is currently known about the ecological factors that define community structure and ecosystem function. Studies suggest that subtle differences in pH and available nutrients, manipulated by different Sphagnum species and strains, create distinct microbial consortia (14, 20, 21). Other observations suggest a more homogenous community (22), highlighting a need for further study. Culture-dependent experiments isolating endophytic bacteria indicate that Sphagnum cultivates symbionts with abilities that include antifungal activity (20, 23) and nitrogen fixation (14) and that these microbiomes may be passed vertically to the moss progeny (21). While there have been examinations of how environmental conditions and host-microbe symbiotic interactions shape the structure and function of microbial communities, however, the influence of virus populations on the Sphagnum microbiome remains unexplored.
Viruses are the most abundant biological entities on Earth and are central to global ecosystems, as they can drive the host evolution through predator-prey interactions and horizontal gene transfer (24). Moreover, viruses can lyse single-celled primary producers and heterotrophs, releasing nutrient elements from the biomass of prokaryotes and eukaryotic protists (25, 26). Viruses may also act as a top-down control on the composition and evenness of microbial communities, targeting hosts that reach higher cell densities, a phenomenon referred to as the “kill-the-winner” model (27).
As laboratory studies of viruses require hosts that can be grown in culture, many environmentally relevant viruses are poorly understood and their representation in reference databases is often skewed. Previous efforts to describe environmental viromes have focused primarily on the sequencing of shotgun or PCR-targeted metagenomes. While these methods have proven powerful, rapidly expanding the available reference material for bacteriophage (28, 29), it leaves the considerable diversity of RNA viruses largely untapped (30). Moreover, the common approach of selecting for viruses based on size exclusion with filters removes many of the nucleocytoplasmic large DNA viruses (NCLDVs, or commonly “giant viruses”) that are also environmentally relevant and phylogenetically informative (31, 32). Metagenomic sequencing also limits observations to virus particles: from these data, inferences on viral activity require tenuous assumptions. The advent of high-throughput RNA sequencing offers viral ecologists the opportunity to study active infections in the environment, as DNA viruses only produce transcripts inside a host. Moreover, this approach also captures fragments of RNA virus genomes. When sequencing is of sufficient depth and multiple samples are collected with spatial and temporal variability, these data present an opportunity to develop hypothetical relationships between virus and host markers (33) for subsequent in-laboratory testing.
In this study, we analyzed metatranscriptomes from the microbial community inhabiting the vegetative portion of Sphagnum fallax and S. magellanicum plants in northern Minnesota, with the goal of describing active viral infections within the Sphagnum microbiome. Using marker genes conserved within several viral taxa, we identified an active and diverse bacteriophage population, largely undescribed in previous studies. We also identified ongoing infections by a diverse consortium of “giant” viruses and potentially corresponding virophage/polinton-like viruses (here referred to as virophage), including several giant viruses closely related to the recently discovered klosneuviruses (34). Finally, a number of novel positive-sense single-stranded RNA (ssRNA) viruses, some of which assembled into nearly complete genomes, were observed. With this information in hand, we developed statistical network analyses, correlating coexpression of viral marker genes with housekeeping transcripts from potential hosts. The resulting observations propose several virus-host pairings that, moving forward, can be tested in a laboratory setting. Together, these results demonstrate new potential model systems to study virus-host interactions in the peat bog ecosystem and provide insight into the significant viral influence on the Sphagnum microbiome.
RESULTS
Identification of resident phage populations.
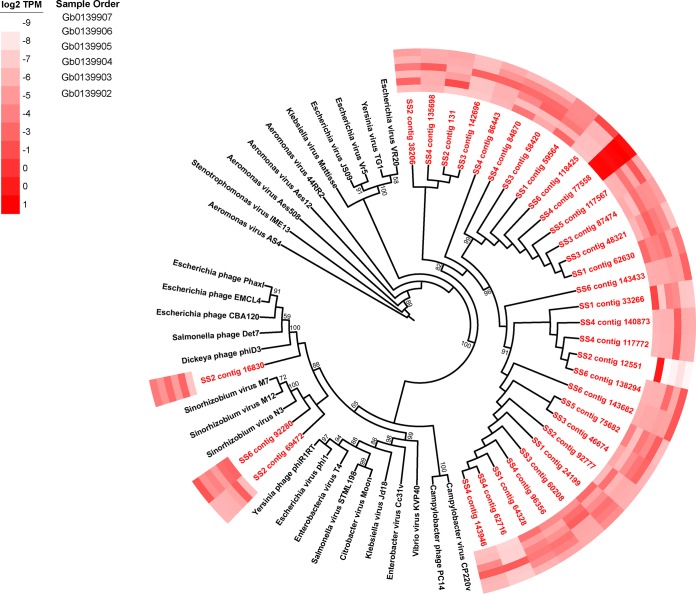
To identify active virus populations in the Sphagnum phyllosphere, we obtained S. fallax and S. magellanicum plant matter samples (three from each species) from peatland terrariums as a part of the Spruce and Peatlands Responses Under Changing Environments (SPRUCE) project for metatranscriptomic sequencing. Across all six Sphagnum phyllosphere samples, 33 contigs were identified as transcripts encoding major capsid protein (MCP; gp23) originating from bacteriophage, while only 6 contigs were identified using three other marker genes. Concurrent with this, more reads mapped to gp23 contigs than to the other marker genes combined, the most abundant of which were three ribonucleotide reductase contigs.
Of the 33 contigs, 18 were assigned to the Eucampyvirinae subfamily with Campylobacter viruses CP220 and PC18, while the rest were spread among the other Myovirus taxa, predominantly the Tevenvirinae (Fig. 1). SS4 contig 77559 was the most abundant, with consistently high expression across all samples, whereas other contigs dominated just one or two samples. Of the six contigs identified using the other three viral marker genes, one was identified as a potential gp20 homologue, originating within Myoviridae with Clostridium virus phiCD119 as the closest relative (see Fig. S1 in the supplemental material). Two contigs were identified as recA contigs, likely originating in Myovirus and Siphovirus relatives (Fig. S2), while the remaining three contigs were identified as ribonucleotide reductase transcripts (Fig. S3).
FIG 1.
Phylogenetic placement of identified phage major capsid protein contigs (red) on a Myovirus gp23 maximum likelihood reference tree (references in black). Full alignment length, 477 amino acids. Node support values (aLRT-SH statistic) of >50% are shown. Contigs are shown with their abundance (log2 transformed TPM) in a heat map surrounding the tree. The sample order on the heat map is provided in the inset.
Single-stranded RNA virus diversity and abundance.
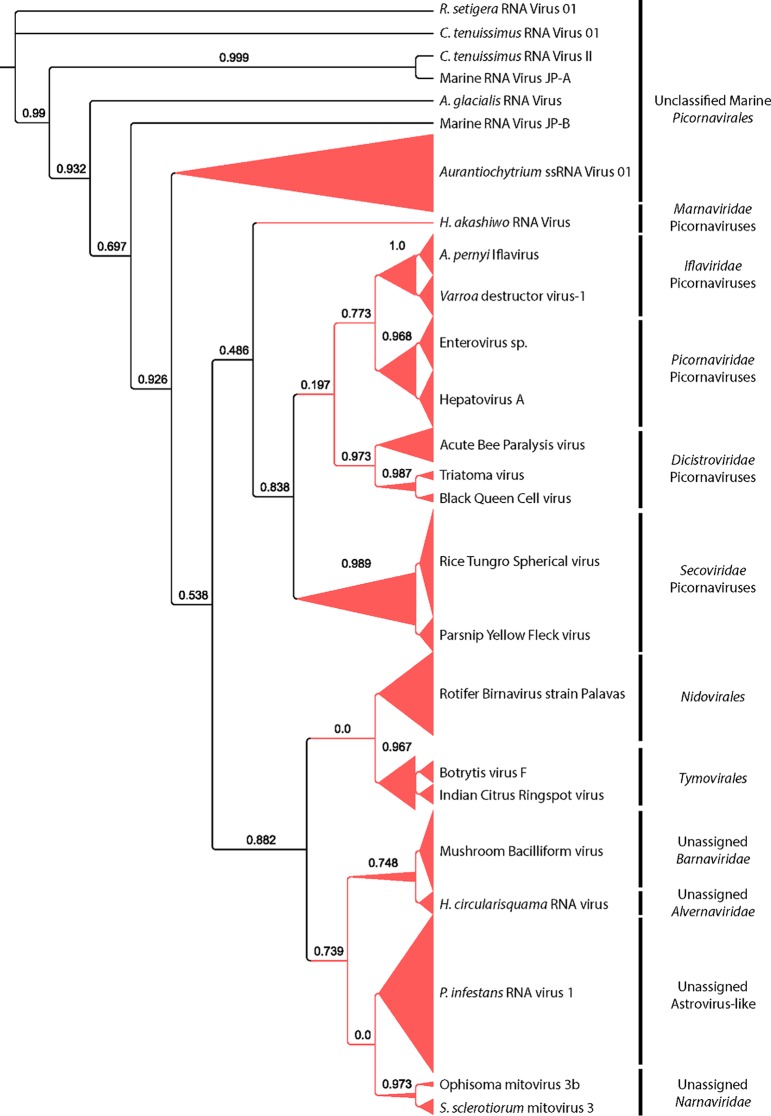
Within our samples, 114 contigs originated from RNA viruses, the majority of which belonged to the currently unassigned Barnaviridae and astrovirus-like families (Fig. 2). Additionally, a large number of picornaviruses were observed, most of which were closely related to the unclassified marine Aurantiochytrium single-stranded RNA virus and to Secoviridae plant viruses. Lastly, several contigs were closely related to the Nidovirales clade, which generally infect animal species.
FIG 2.
Phylogenetic placement of identified ssRNA virus RNA-dependent RNA polymerase contigs on a maximum likelihood reference tree. Full alignment length, 551 amino acids. Branch width represents the number of contigs placed on the reference branch. Node support values (aLRT-SH statistic) of >50% are shown. Viruses include (but are not limited to) Rhizosolenia setigera RNA virus 01, Chaetoceros tenuissimus RNA virus 01 and II, Asterionellopsis glacialis RNA virus, Heterosigma akashiwo RNA virus, Antheraea pernyi iflavirus, Heterocapsa circularisquama RNA virus, Phytophtora infestans RNA virus 1, and Sclerotinia sclerotiorum mitovirus 3.
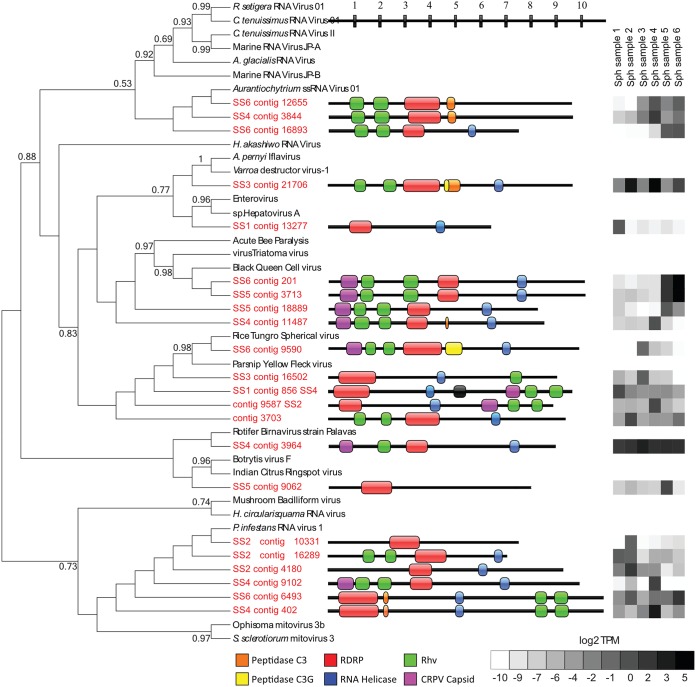
Among these, 22 contigs were found to be nearly complete ssRNA virus genomes (based on gene content and size), encoding multiple viral gene products in addition to RNA-dependent RNA polymerase (RDRP). Gene regions were identified and annotated using the NCBI conserved domain and Pfam HMM search tools, and the full-length RDRP sequence was used to construct a maximum likelihood phylogenetic tree (Fig. 3). Of the partial ssRNA genomes that were assembled, two were missing the conserved Rhv structural genes, while one was missing an RNA virus helicase gene. The majority of these contigs fall under the Picornavirales order, which also included the most complete viral genomes. As was observed with the shorter RDRP contigs described above, most of the Picornavirales contigs were most closely related to the unclassified marine species, or members of the family Secoviridae clade, whose membership includes the Parsnip yellow fleck virus. A number of partial Picornavirus genomes were also identified as members of the family Dicistroviridae. Outside the Picornavirales, most contigs clustered closely with the unassigned astrovirus-like Phytophthora infestans RNA virus. To determine the relative abundance of different RNA virus genomes in the peat bog samples, we mapped reads back to contigs and calculated transcripts per million (TPM) to account for contig length and library size. Overall, NCLDV transcripts were the most abundant (range, 4,465 to 16,887 reads mapped to MCP contigs, or 46.4 to 200.1 TPM), followed by RNA viruses (range, 13,373 to 166,337 total reads mapped to RNA virus contigs, or 7.11 to 97.5 TPM), and bacteriophage (range, 287 to 1,405 total reads mapped to gp23 contigs, or 3.4 to 5.2 TPM). The most abundant contig across all samples was SS4 contig 3964, which was most closely related to the rotifer birnavirus. All other contigs appear to be abundant prominently in one or two samples and absent or in low abundance in the others, with no patterns of abundance apparent.
FIG 3.
Phylogeny, genome architecture, and abundance of partial ssRNA virus genomes. The tree represents phylogenetic placement of RDRP gene regions from partial ssRNA virus genome contigs (red) on a maximum likelihood reference tree (references in black). Full alignment length, 551 amino acids. Node support values (aLRT-SH statistic) of >50% are shown. The center panel represents the genome architecture determined by conserved domain search and ORF prediction. The lengths of contigs and gene regions are measured in kilobases. The heat map in the right panel shows the abundance of reads mapped to partial genome contigs in log2 TPM from each of the six metatranscriptome libraries.
Giant viruses and virophage in Sphagnum microbiome.
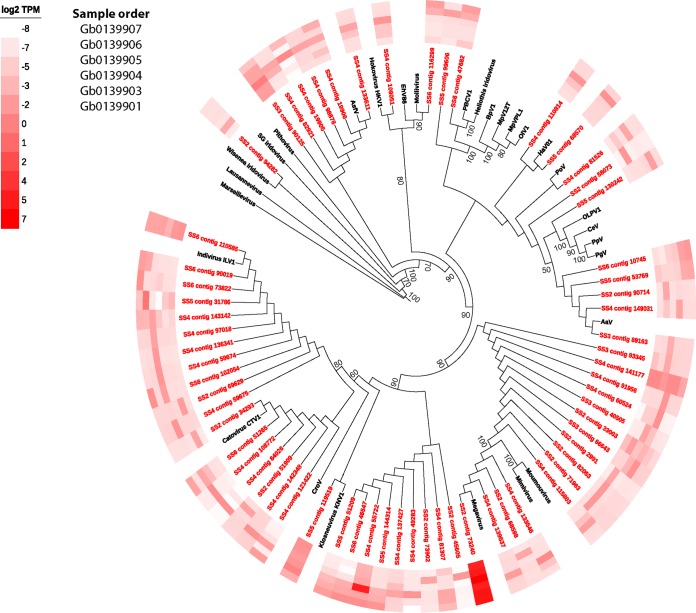
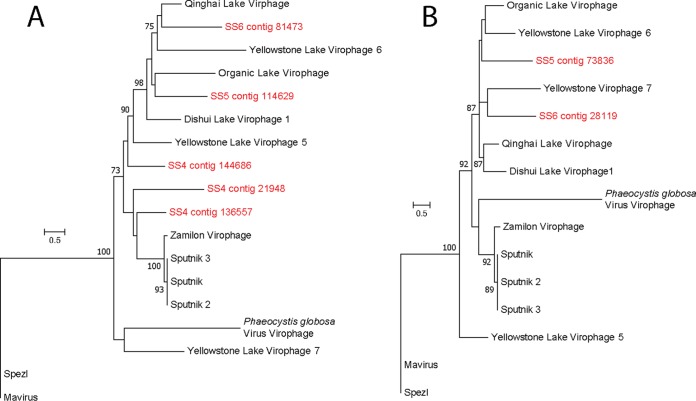
Of the 10 gene markers tested to identify nucleocytoplasmic large DNA viruses (NCLDVs), only the giant virus MCP was detected in the metatranscriptome. Sixty-four contigs were observed with homology to MCP, representing every known group of NCLDVs (Fig. 4). Out of the 64 MCP contigs, 46 were placed within the Mimiviridae taxa. Most contigs (25 contigs) closely aligned with the recently discovered klosneuviruses, with Indivirus and Catovirus representing the most diversity in these samples. The next most abundant group were the “extended Mimiviridae” (7 contigs), species with known similarity to mimiviruses but that infect eukaryotic algae. Six contigs were phylogenetically similar to the Asfarviridae, here represented by the African swine fever virus. Potential relatives of the giant virus outliers, Pandoravirus and Pithovirus, were not observed (due to methodological limitations), and the Iridoviridae were poorly represented (1 contig). Using the virophage MCP and packaging ATPase as markers, we identified 7 contigs as transcripts originating in putative virophage or polinton-like viruses, all of which were phylogenetically placed among isolates identified from freshwater ecosystems (Fig. 5).
FIG 4.
Phylogenetic placement of identified NCLDV major capsid protein contigs (red) on a maximum likelihood reference tree (references in black). Full alignment length, 477 amino acids. Node support values (aLRT-SH statistic) of >50% are shown. Contigs are shown with their abundance (log2 transformed TPM) in a heat map surrounding the tree. Abbreviations: SG iridovirus, Singapore grouper iridovirus; AsfV, African swine fever virus; EhV86, Emiliania huxleyi virus 86; PBCV1, Paramecium bursaria chlorella virus 1; BpV1, Bathycoccus prasinos virus; MpV12T, Micromonas pusilla virus 12T; MpVPL1, Micromonas pusilla virus PL1; OlV1, Ostreococcus lucimarinus virus; HaV01, Heterosigma akashiwo virus; PoV, Pyramimonas orientalis virus; OLPV1, organic lake phycodnavirus 1; CeV, Chrysochromulina ericinia virus; PpV, Phaeocystis pouchetii virus; PgV, Phaeocysis globosa virus; AaV, Aureococcus anophagefferens virus; CroV, Cafeteria roenbergensis virus.
FIG 5.
Phylogenetic placement of identified virophage major capsid protein (full alignment length, 549 amino acids) (A) and ATPase (full alignment length, 251 amino acids) (B) contigs (red) on a maximum likelihood reference tree (references in black). Node support values (aLRT-SH statistic) of >50% are shown.
As was observed with the other major viral taxa described, the majority of contigs were most abundantly expressed in one or two samples and present at very low levels in the rest. The most abundant NCLDV-MCP contig in the samples was SS2 contig 73240, most closely related to Megavirus chilensis, which was the most highly expressed giant virus contig across all samples. Four other contigs (SS6 contig 110585, SS4 contigs 55722 and 141177, and SS5 contig 119519) were highly expressed across all six samples.
Prediction of virus-host pairs.
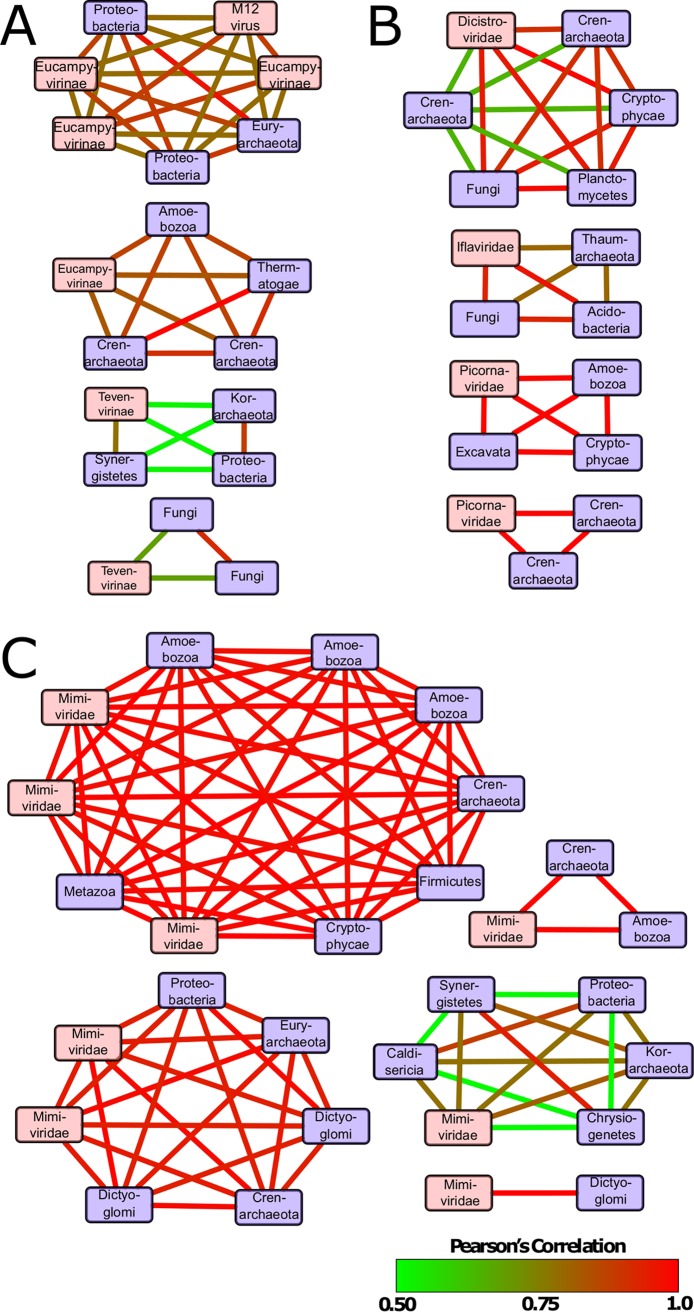
By comparing and correlating expression of virus marker genes to rpb1 expression from cellular organisms, we endeavored to predict potential virus-host groups in the Sphagnum phyllosphere. Figure 6 shows statistically robust networks containing at least one virus and one host, where co-occurrence and correlation were observed in more than one sample. A total of 13 virus-host groups were detected, spread across the major viral taxa detected in this data set. We note that no networks containing the virophage/polinton-like viruses emerged. Four relationships were predicted from bacteriophage gp23 abundance, the simplest of which was a Tevenvirinae phage-Fungi-Fungi group with moderate correlations (Fig. 6A). The other three relationships are more complicated, containing multiple potential hosts and, for the largest predicted group, multiple virus transcripts. Some of the potential hosts in these groups were identified as eukaryotic.
FIG 6.
Correlation co-occurrence network analysis of conserved viral gene and host RNA polymerase expression for bacteriophage (Gp23) (A), ssRNA viruses (RDRP) (B), and NCLDVs (NCLDV MCP) (C). Nodes in red represent virus contigs, and nodes in blue represent potential hosts. Nodes are connected by edges colored according to the Pearson correlation coefficient values between two contigs. Only relationships with contigs expressed in more than one sample are shown.
We observed four predicted RNA virus-host clusters, all of which contained multiple hosts grouped with a single virus (Fig. 6B). Many of the predicted hosts appear closely related to eukaryotic single-celled protists, including members of the Cryptophyceae, Excavata, and Amoebozoa, as well as a variety of bacterial and archaeal species. Correlation coefficients observed in these relationships are generally higher than observed in the phage-host clusters. The five predicted NCLDV-host clusters (Fig. 6C) were the most highly correlated and complex. Predicted hosts were highly varied, ranging from bacteria to fungi, although all virus members were placed within either Mimiviridae or the extended Mimivirus group. MCP contigs originating in close relatives of the recently discovered klosneuviruses are present in both the 7- and 10-member clusters, in addition to a pair of contigs most closely related to Aureococcus anophagefferens virus (AaV). An additional 15 statistically significant clusters across all three viral taxa were observed where the virus and host were present in only one sample (not shown).
DISCUSSION
Understanding the virus burden on microbial communities in ecologically rich ecosystems is an important step forward in resolving their function and predicting how they might respond to various drivers of ecosystem scale change. In the present study, we used metatranscriptomes to describe the diversity and activity of the resident virus populations in a peat moss (Sphagnum) microbiome. We identified previously undescribed virus activity from multiple taxa, most of which are poorly represented in either the literature or reference sequence databases. We used read mapping to quantify the relative abundance of active viral infections. Lastly, we compared the expression of viral transcripts to that of potential hosts, using a correlation co-occurrence network approach (33) to predict putative hosts for the observed virus populations. Together, our results suggest that the Sphagnum phyllosphere represents a significant and largely untapped source of virus diversity and activity. Viruses were highly active across all samples, with some individual viruses exhibiting abundant activity in single samples, while others were more pervasive. Given that our observations were based on RNA sequencing data, they do not represent a full accounting of the virus particles present in the community. However, metatranscriptomic data allow us to distinguish virus populations active at the time of sampling. In addition, as viruses transcribe their genes only during infection, virus and host transcripts are expected to co-occur, and it is possible that the abundance of transcripts (at least for DNA viruses) could be used to predict natural hosts of viruses observed in the ecosystem which can be tested in a laboratory or field setting. Ultimately, this study identifies from within a complex community a number of candidate virus-host model systems for future study.
Viral diversity and activity in Sphagnum plants.
As viruses lack a universal genetic marker like the bacterial 16S rRNA gene, we opted to screen metatranscriptome assemblies for genes previously demonstrated to be largely or wholly conserved among individual viral taxa. Within the expanded and diverse genetic potential of giant viruses, only a few genes are currently conserved among all members (32, 35), and these, in addition to several markers conserved among a large portion of giant viruses, were used to identify activity in the Sphagnum phyllosphere. For the 10 genes used to screen the metatranscriptomes for giant viruses, only MCP transcripts were found. This is not surprising, given the number of capsid proteins needed for viral assembly; indeed, this transcriptional pattern was previously observed in both cultures (36) and marine systems (33) by Moniruzzaman et al. It should be noted that the transcriptome sequencing (RNA-seq) data set used in those studies was poly(A) selected, enriching for eukaryotic transcripts, and thus coverage of eukaryotic virus gene expression is much higher than in the Sphagnum metatranscriptome. That we observed MCP expression in abundance suggests that a significant number of infections occurred at the time of sampling. While the magnitude of giant virus diversity in Sphagnum-dominated ecosystems is, to our knowledge, completely unexplored, the richness observed here is considerably larger than expected in comparison to better documented systems. Sixty-four distinct MCP genotypes were identified in the Sphagnum phyllosphere metatranscriptomes, which is high compared to one recent survey that identified 30 novel MCP transcripts from multiple environmental data sets (37) and another which observed 107 NCLDV sequences in 16 publicly available environmental metagenomes of comparable sequencing depth isolated from different ecosystems (38). Most of the MCP contigs identified were placed in clusters around a small number of virus relatives, highlighting the undersampled diversity of giant viruses in the literature, poor representation in reference databases, and the considerable diversity present in Sphagnum peat bogs. The significant giant virus diversity observed here implies a corresponding eukaryotic richness that is also underdescribed (39). Additionally, a series of virophage transcripts were detected, indicating a significant response to infections by giant viruses in the system. Many of these are phylogenetically grouped with the polintoviruses, transposable elements that produce virion particles that can exploit the replication machinery of actively infecting giant viruses to reproduce, often at the expense of the giant virus (40, 41). These observations suggest that while an active picoeukaryotic population may persist, mortality mechanisms beyond grazer-driven losses are at play and likely important to carbon flow in the system.
The use of RNA-seq presents a unique opportunity to capture the genomic material of RNA viruses that is lost in metagenomic sequencing. As such, RNA virus representation in sequencing databases and the literature is largely constrained to culture-based studies. All known RNA viruses require a functional RNA-dependent RNA polymerase (RDRP) to copy their genome inside the host cell, a function exclusive to viruses, making it a highly specific marker for RNA virus discovery (42, 43). Recent attempts to use metatranscriptomes to describe environmental RNA viruses have proven successful, not only in identifying marker gene fragments in data sets but also in assembling complete and near-complete genomes (33, 43). The diversity and composition of RNA virus populations in Sphagnum peatlands are largely unknown: these populations are currently limited to the small group of RNA-DNA hybrid chimeric cruciviruses (44). Here, as was observed with the giant viruses, most RNA virus contigs were placed in clusters with a single represented species, suggesting a significant degree of uncharacterized diversity. This is not entirely surprising, as RNA viruses are expected to make up as much as half of the virus particles in the Earth's oceans, and yet they are almost as poorly understood and represented in sequencing databases as giant viruses (30). Similarly, we assembled and identified 22 near-complete RNA virus genomes, where completeness was determined primarily by size and the presence of the six core genes. As there are currently only 265 sequenced genomes within the Picornavirales, most of which grouped within the Picornaviridae, this represents a sizeable addition to the known diversity of ssRNA viruses. This is especially true for the unassigned and unclassified taxa and establishes a strong foundation for future efforts to describe RNA virus populations in Sphagnum.
Description of bacteriophage populations in Sphagnum peatlands is currently limited to the ssDNA viruses of the Microviridae (45) and Caudovirales (46) observed in metagenomics data, although it appears that phages are the most abundant biological entities in the Sphagnum phyllosphere (46). Given this, and the dominance of bacteria in the Sphagnum microbiome as previously described (15), the relatively low abundance of active bacteriophage in our samples was a surprise. Marker genes to identify bacteriophage were chosen based on their conservation across phage taxa and their success in other environmental data sets. Gp20 (phage portal protein) and Gp23 (major capsid protein) have been shown previously to be highly conserved and effective for phylogenetic assignment of members of the Myoviridae (47–49). RecA is conserved across all three bacteriophage taxa and could illuminate lysogeny, and ribonucleotide reductase (RNR) has been used as an effective marker for screening novel viruses from marine sequencing data sets (50). As such, we identified 39 bacteriophage contigs using these markers, 33 of which were from Gp23. This may represent a phenomenon similar to that of MCP in the giant viruses described above, where transcripts encoding structural proteins are much more abundant than other genes and sequencing lacked the depth to detect them. For the purpose of discovering novel phage species, DNA sequencing through metagenomics may prove more successful.
Virus-host predictions.
Future study of viral dynamics in peatlands will require the establishment of model virus-host pairs for in vitro experimentation and in situ tracking. While culture-based techniques can yield model systems, it is not always clear whether the isolated organisms are environmentally relevant. In order to address this, we attempted to use statistical methods to propose virus-host pairs as potential future model systems based on their co-occurrence in samples and the correlation of their abundance. As viruses produce transcripts only when actively infecting a host, positive correlation and co-occurrence between virus and host transcripts are expected and might be used to predict host-virus relationships, provided that an appropriate transcriptional proxy for growth and activity is available (33). In this study, we used the eukaryotic RNA polymerase gene rpb1 as a marker for abundance and activity in potential hosts, as it is conserved among all eukaryotic organisms, is phylogenetically informative, and has been previously described as one of the more consistently expressed eukaryotic genes in marine systems, scaling well with the activity of the organism (51), although the stability of its expression has not been evaluated in terrestrial ecosystems. We used NCLDV MCP abundance as a proxy for giant virus production and Gp23 for phage production, as transcription is necessary for the assembly of new virus particles and transcript abundance in some appears to be closely linked to viral replication. We also used RDRP as a proxy for RNA virus production, acknowledging the caveat that we cannot distinguish between abundance of free virus particles and active infections (33).
Correlation and co-occurrence matrices, clustered into groups by similarity and tested with the SIMPROF permutation test, yielded 13 predicted groups of viruses and hosts. For ssRNA and giant viruses, several of the networks produced in the analysis included multiple bacterial and archaeal sequences picked up in the RNA polymerase screen. As we have no reason to believe that bacterial species are infected by NCLDVs or picornaviruses, it is likely that these predictions represent a confounding relationship between prokaryotes and potential eukaryotic hosts, observed in network analyses for all three viral taxa described here, where a beneficial interaction results in an indirect correlation with viral infection. Indeed, previous use of this method in marine systems showed a similar phenomenon, where an algal Mimivirus and a known host were grouped with a fungal species and another virus (33). Even after the consideration of bacterial species within the predicted groups, some remain complicated with multiple viruses and potential eukaryotic hosts, which may be explained by a broader host range among giant viruses enabled by the expansion of genetic material and increased independence from host machinery. Similar relationships were observed among RNA viruses, though these are more tenuous, as we are unable to distinguish whether sequencing reads originated transcripts or genomic material.
All together, we have identified a considerable amount of viral diversity from several major viral taxa active within a poorly understood microbial ecosystem. As they were identified from transcript sequencing data, the viruses described here likely represent only a fraction of the whole virus community, which may be elucidated through further culture-independent work. We have also used transcript abundance within a statistical framework to predict several host-virus relationships which can be sought out and tested in culture. These results establish an important and much needed foundation for future research into the microbial ecology in Sphagnum peat bogs.
MATERIALS AND METHODS
Sample collection and survey of environmental conditions.
Triplicate individual plants of Sphagnum magellanicum and Sphagnum fallax were collected on August 2015 from the SPRUCE experiment site at the S1 bog in the Marcell Experimental Forest (U.S. Forest Service, http://mnspruce.ornl.gov/). The S1 bog is an acidic and nutrient-deficient ombrotrophic Sphagnum-dominated peatland bog (surface pH ≤4.0) located approximately 40 km north of Grand Rapids, Minnesota, USA (47°30.476′N, 93°27.162′W; 418 m above mean sea level) (52–54). To characterize the Sphagnum virome, Sphagnum samples were collected as previously described (54). Only green living plants were sampled: samples focused on the capitulum plus about 2 to 3 cm of green living stem. Sphagnum stems (phyllosphere) were cleaned from unrelated plant debris and frozen immediately on dry ice. Frozen samples were shipped overnight to the Georgia Institute of Technology for RNA extraction.
RNA extraction and sequencing.
One gram of Sphagnum phyllosphere tissue was ground with a mortar and pestle under liquid nitrogen. The fine powder was transferred to 10 extraction tubes, and total RNA was isolated using the PowerPlant RNA isolation kit with DNase according to the manufacturer's protocol (MoBio Laboratories, Carlsbad, CA, USA). DNA-depleted RNA was quantified using the Qubit RNA HS assay kit (Invitrogen, Carlsbad, CA, USA), and quality was assessed on the Agilent 2100 BioAnalyzer using the Agilent RNA 6000 Pico kit (Agilent Technologies). Additionally, the absence of DNA contamination was confirmed by running a PCR using universal bacterial 16S rRNA primers 515F and 806R. Finally, RNA samples without detectible DNA contamination and exhibiting an RNA integrity number (RIN) of >6 were pooled. Extracted total environmental RNA samples were sent on dry ice to the Joint Genome Institute (JGI) facilities for metatranscriptome library construction and sequencing. All protocols employed were standard JGI protocols, and rRNA subtraction from total environmental RNA was completed using the Ribo-Zero rRNA removal kit (Illumina, San Diego, CA). rRNA-depleted environmental RNA was used to construct paired-end metatranscriptome libraries using the TruSeq kit and sequenced on the Illumina HiSeq 2000 platform at the JGI facilities using a single-end 250-bp flow cell.
RNA-seq data processing.
Raw sequences (see Table S1 in the supplemental material, sequencing stats tab) were downloaded from the Department of Energy Joint Genome Institute server and processed using CLC Genomics Workbench v.10.0.1 (Qiagen, Hilden, Germany). Reads below a 0.03 quality score cutoff were removed from subsequent analyses, and the remaining reads were trimmed of any ambiguous and low-quality 5′ bases. Samples were subjected to a subsequent in silico rRNA reduction using the SortMeRNA v.2.0 software package (55). Filtered reads were de novo assembled with cutoffs of a 300-base minimum contig length and an average coverage of 2, leaving a total of 705,526 contigs across all samples (Table S1, contig mappings tab).
Screening assemblies for marker genes.
Marker genes to identify bacteriophage were chosen based on their conservation across phage taxa and their success in other environmental data sets. Gp20 (phage portal protein) and Gp23 (major capsid protein) have been shown previously to be highly conserved and effective for phylogenetic assignment of members of the Myoviridae (47–49). RecA is conserved across all three bacteriophage taxa and could illuminate lysogeny, and ribonucleotide reductase (RNR) has been used as an effective marker for screening novel viruses from marine sequencing data sets (50). To identify contigs specific to the nucleocytoplasmic large DNA virus (NCLDV) clade, contig libraries were screened for the presence of 10 genes previously identified as core NCLDV genes as previously described (33). Briefly, contig libraries were queried against nucleocytoplasmic virus orthologous group (NCVOG) protein databases (ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/NCVOG/) for each of the following 10 marker genes in a BLASTX search with a minimum E-value cutoff of 10−3: A32 virion packaging ATPase (NCVOG0249), VLFT-like transcription factor (NCVOG0262), superfamily II helicase II (NCVOG0024), mRNA capping enzyme (NCVOG1117), D5 helicase-primase (NCVOG0023), ribonucleotide reductase small subunit (NCVOG0276), RNA polymerase large subunit (NCVOG0271), RNA polymerase small subunit (NCVOG0274), B-family DNA polymerase (NCVOG0038), and major capsid protein (NCVOG0022). The resulting hits were then queried against the NCBI RefSeq protein database (56), and only contigs with top hits to virus genes were maintained for subsequent analyses. A similar method was used to identify virophage transcripts, where the virophage major capsid protein and packaging ATPase genes were used as markers.
Contigs derived from ssRNA viruses were identified by screening the contig library for RNA-dependent RNA polymerase (RDRP), a distinctive and wholly conserved RNA virus gene and a strong phylogenetic marker (57). A BLAST database of RDRP sequences was downloaded from the Pfam database (58) under code pf00680. Contigs were aligned using BLASTX with a minimum E value of 10−4. Hits were queried against the NCBI RefSeq protein database, and only hits to viral RDRP genes were retained for downstream analyses. Contigs derived from rpb1 transcripts were similarly identified using a BLAST database of rpb1 sequences downloaded from the UniProt database under the K03006 group.
To identify RNA virus genome fragments, contig libraries were screened as described above using the following core set of genes observed in RNA viruses: genes encoding CRPV capsid (Pfam 08762), VP4 (Pfam 11492), RDRP (Pfam 00680), peptidase C3 (Pfam 00548), peptidase C3G (Pfam 12381), Rhv (Pfam 00073), and RNA helicase (Pfam 00910). BLAST databases for core RNA virus genes were constructed from reference sequences downloaded from Pfam. Query sequences were then cross-referenced to identify contigs with hits to multiple RNA virus core genes. Only contigs of >1,000 bases with at least one viral RDRP region were retained for further analysis. Open reading frames (ORFs) on these putative partial genomes were predicted using the CLC Genomics Workbench. Features on the partial genomes were predicted using the Pfam HMM domain and the NCBI Conserved Domain Database searches (59, 60). Genome architecture was visualized using the Illustrator for Biological Sequences (IBS) software package (61).
Phylogenetic analysis.
Reference sequences for viral marker genes and host Rpb1 were downloaded from the InterPro and RefSeq databases (see Table S2 in the supplemental material) (62). Reference sequences were aligned using the MUSCLE alignment algorithm (63) in the MEGA v7.0.26 software package (64). Maximum likelihood phylogenetic trees were constructed in PhyML from reference sequences and contigs containing the respective full-length genes (65) with the LG substitution model and the aLRT-SH-like likelihood method. Putative viral and Rpb1 contigs assembled from the metatranscriptomes were translated into proteins according to the reading frame of the top BLAST hit. Translated proteins were placed on the reference trees in a maximum likelihood framework in pplacer (66), and contigs were identified based on the most closely related clade in the tree. Trees with abundance data were visualized using the iToL web interface (67).
Statistical analysis.
Quality filtered and trimmed reads were stringently mapped to the selected contigs (0.97 identity fraction, 0.7 length fraction) in CLC Genomics Workbench 10.0.1. Expression values were calculated as a modification of the transcripts per million (TPM) metric. Read counts were normalized by contig length in kilobases to determine the reads per kilobase (RPK) values for every contig within each library. These RPK values were then summed and divided by 1 million, to determine the sequencing depth scaling factor for each library. The TPM for a contig was calculated by dividing its RPK value by the scaling factor for the library.
Expression values for contigs were imported into the PRIMER7 (68) statistical software package and log2 transformed. Expression values from each contig were correlated (Pearson's rho) to one another and statistically grouped by co-occurrence using group average hierarchical clustering. The SIMPROF test (69) was used to determine the statistical significance level of resulting clusters (alpha = 0.05, 1,000 permutations). Statistically significant clusters with at least one viral contig, one rpb1 contig, and less than 10 total members were visualized and annotated in Cytoscape 3.5.1 (70).
Accession number(s).
Full RNA-seq libraries have been made publicly available on the JGI website under accession number Gs0118677.
Supplementary Material
ACKNOWLEDGMENTS
Research was sponsored by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory and the Joint Directed Research and Development Program of the University of Tennessee. Support for the SPRUCE experimental site is from the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725. Support at the University of Tennessee was received from the Kenneth & Blaire Mossman Endowment to the University of Tennessee (S.W.W.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01124-18.
REFERENCES
- 1.Post WM, Emanuel WR, Zinke PJ, Stangenberger AG. 1982. Soil carbon pools and world life zones. Nature 298:156–159. doi: 10.1038/298156a0. [DOI] [Google Scholar]
- 2.Gorham E. 1991. Northern peatlands: role in the carbon cycle and probable responses to climatic warming. Ecol Appl 1:182–195. doi: 10.2307/1941811. [DOI] [PubMed] [Google Scholar]
- 3.Bridgham SD, Patrick Megonigal J, Keller JK, Bliss NB, Trettin C. 2006. The carbon balance of North American wetlands. Wetlands 26:889–916. doi: 10.1672/0277-5212(2006)26[889:TCBONA]2.0.CO;2. [DOI] [Google Scholar]
- 4.van Breemen N. 1995. How Sphagnum bogs down other plants. Trends Ecol Evol 10:270–275. doi: 10.1016/0169-5347(95)90007-1. [DOI] [PubMed] [Google Scholar]
- 5.Lamers LPM, Bobbink R, Roelofs JGM. 2000. Natural nitrogen filter fails in polluted raised bogs. Global Change Biol 6:583–586. doi: 10.1046/j.1365-2486.2000.00342.x. [DOI] [Google Scholar]
- 6.Turetsky MR. 2003. The role of bryophytes in carbon and nitrogen cycling. Bryologist 106:395–409. doi: 10.1639/05. [DOI] [Google Scholar]
- 7.Turetsky MR, Bond-Lamberty B, Euskirchen E, Talbot J, Frolking S, McGuire AD, Tuittila ES. 2012. The resilience and functional role of moss in boreal and arctic ecosystems. New Phytol 196:49–67. doi: 10.1111/j.1469-8137.2012.04254.x. [DOI] [PubMed] [Google Scholar]
- 8.Verhoeven JTA, Liefveld WM. 1997. The ecological significance of organochemical compounds in Sphagnum. Acta Bot Neerlandica 46:117–130. doi: 10.1111/plb.1997.46.2.117. [DOI] [Google Scholar]
- 9.Mellegard H, Stalheim T, Hormazabal V, Granum PE, Hardy SP. 2009. Antibacterial activity of sphagnum acid and other phenolic compounds found in Sphagnum papillosum against food-borne bacteria. Lett Appl Microbiol 49:85–90. doi: 10.1111/j.1472-765X.2009.02622.x. [DOI] [PubMed] [Google Scholar]
- 10.Freeman C, Ostle N, Kang H. 2001. An enzymic ‘latch’ on a global carbon store—a shortage of oxygen locks up carbon in peatlands by restraining a single enzyme. Nature 409:149–149. doi: 10.1038/35051650. [DOI] [PubMed] [Google Scholar]
- 11.Stalheim T, Ballance S, Christensen BE, Granum PE. 2009. Sphagnan—a pectin-like polymer isolated from Sphagnum moss can inhibit the growth of some typical food spoilage and food poisoning bacteria by lowering the pH. J Appl Microbiol 106:967–976. doi: 10.1111/j.1365-2672.2008.04057.x. [DOI] [PubMed] [Google Scholar]
- 12.Hajek T, Ballance S, Limpens J, Zijlstra M, Verhoeven JTA. 2011. Cell-wall polysaccharides play an important role in decay resistance of Sphagnum and actively depressed decomposition in vitro. Biogeochemistry 103:45–57. doi: 10.1007/s10533-010-9444-3. [DOI] [Google Scholar]
- 13.Lin X, Tfaily MM, Green SJ, Steinweg JM, Chanton P, Imvittaya A, Chanton JP, Cooper W, Schadt C, Kostka JE. 2014. Microbial metabolic potential for carbon degradation and nutrient (nitrogen and phosphorus) acquisition in an ombrotrophic peatland. Appl Environ Microbiol 80:3531–3540. doi: 10.1128/AEM.00206-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leppanen S, Rissanen A, Tiirola M. 2015. Nitrogen fixation in Sphagnum mosses is affected by moss species and water table level. Plant Soil 389:185–196. doi: 10.1007/s11104-014-2356-6. [DOI] [Google Scholar]
- 15.Kostka JE, Weston DJ, Glass JB, Lilleskov EA, Shaw AJ, Turetsky MR. 2016. The Sphagnum microbiome: new insights from an ancient plant lineage. New Phytol 211:57–64. doi: 10.1111/nph.13993. [DOI] [PubMed] [Google Scholar]
- 16.Dudova L, Hajkova P, Buchtova H, Opravilova V. 2013. Formation, succession and landscape history of Central-European summit raised bogs: a multiproxy study from the Hruby Jesenik Mountains. Holocene 23:230–242. doi: 10.1177/0959683612455540. [DOI] [Google Scholar]
- 17.Ireland AW, Clifford MJ, Booth RK. 2014. Widespread dust deposition on North American peatlands coincident with European land-clearance. Veg Hist Archaeobot 23:693–700. doi: 10.1007/s00334-014-0466-y. [DOI] [Google Scholar]
- 18.Swindles GT, Turner TE, Roe HM, Hall VA, Rea HA. 2015. Testing the cause of the Sphagnum austinii (Sull. ex Aust.) decline: multiproxy evidence from a raised bog in Northern Ireland. Rev Palaeobot Palynol 213:17–26. doi: 10.1016/j.revpalbo.2014.11.001. [DOI] [Google Scholar]
- 19.Galka M, Tobolski K, Gorska A, Lamentowicz M. 2017. Resilience of plant and testate amoeba communities after climatic and anthropogenic disturbances in a Baltic bog in Northern Poland: implications for ecological restoration. Holocene 27:130–141. doi: 10.1177/0959683616652704. [DOI] [Google Scholar]
- 20.Opelt K, Chobot V, Hadacek F, Schonmann S, Eberl L, Berg G. 2007. Investigations of the structure and function of bacterial communities associated with Sphagnum mosses. Environ Microbiol 9:2795–2809. doi: 10.1111/j.1462-2920.2007.01391.x. [DOI] [PubMed] [Google Scholar]
- 21.Bragina A, Cardinale M, Berg C, Berg G. 2013. Vertical transmission explains the specific Burkholderia pattern in Sphagnum mosses at multi-geographic scale. Front Microbiol 4:394. doi: 10.3389/fmicb.2013.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bragina A, Maier S, Berg C, Muller H, Chobot V, Hadacek F, Berg G. 2011. Similar diversity of Alphaproteobacteria and nitrogenase gene amplicons on two related Sphagnum mosses. Front Microbiol 2:275. doi: 10.3389/fmicb.2011.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opelt K, Berg G. 2004. Diversity and antagonistic potential of bacteria associated with bryophytes from nutrient-poor habitats of the Baltic Sea coast. Appl Environ Microbiol 70:6569–6579. doi: 10.1128/AEM.70.11.6569-6579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brussaard CPD, Wilhelm SW, Thingstad F, Weinbauer MG, Bratbak G, Heldal M, Kimmance SA, Middelboe M, Nagasaki K, Paul JH, Schroeder DC, Suttle CA, Vaque D, Wommack KE. 2008. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J 2:575–578. doi: 10.1038/ismej.2008.31. [DOI] [PubMed] [Google Scholar]
- 25.Jover LF, Effler TC, Buchan A, Wilhelm SW, Weitz JS. 2014. The elemental composition of virus particles: implications for marine biogeochemical cycles. Nat Rev Microbiol 12:519–528. doi: 10.1038/nrmicro3289. [DOI] [PubMed] [Google Scholar]
- 26.Wilhelm SW, Suttle CA. 1999. Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49:781–788. doi: 10.2307/1313569. [DOI] [Google Scholar]
- 27.Thingstad TF, Lignell R. 1997. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Microb Ecol 13:19–27. doi: 10.3354/ame013019. [DOI] [Google Scholar]
- 28.Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, Poulos BT, Solonenko N, Lara E, Poulain J, Pesant S, Kandels-Lewis S, Dimier C, Picheral M, Searson S, Cruaud C, Alberti A, Duarte CM, Gasol JM, Vaque D, Tara Oceans Coordinators, Bork P, Acinas SG, Wincker P, Sullivan MB. 2016. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537:689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 29.Simmonds P, Adams MJ, Benko M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, Hull R, King AMQ, Koonin EV, Krupovic M, Kuhn JH, Lefkowitz EJ, Nibert ML, Orton R, Roossinck MJ, Sabanadzovic S, Sullivan MB, Suttle CA, Tesh RB, van der Vlugt RA, Varsani A, Zerbini M. 2017. Virus taxonomy in the age of metagenomics. Nat Rev Microbiol 15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 30.Steward GF, Culley AI, Mueller JA, Wood-Charlson EM, Belcaid M, Poisson G. 2013. Are we missing half of the viruses in the ocean? ISME J 7:672–679. doi: 10.1038/ismej.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilhelm SW, Bird JT, Bonifer KS, Calfee BC, Chen T, Coy SR, Gainer PJ, Gann ER, Heatherly HT, Lee J, Liang XL, Liu J, Armes AC, Moniruzzaman M, Rice JH, Stough JMA, Tams RN, Williams EP, LeCleir GR. 2017. A student's guide to giant viruses infecting small eukaryotes: from Acanthamoeba to Zooxanthellae. Viruses 9:E46. doi: 10.3390/v9030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moniruzzaman M, Wurch LL, Alexander H, Dyhrman ST, Gobler CJ, Wilhelm SW. 2017. Virus-host relationships of marine single-celled eukaryotes resolved from metatranscriptomics. Nat Commun 8:16054. doi: 10.1038/ncomms16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulz F, Yutin N, Ivanova NN, Ortega DR, Lee TK, Vierheilig J, Daims H, Horn M, Wagner M, Jensen GJ, Kyrpides NC, Koonin EV, Woyke T. 2017. Giant viruses with an expanded complement of translation system components. Science 356:82–85. doi: 10.1126/science.aal4657. [DOI] [PubMed] [Google Scholar]
- 35.Moniruzzaman M, LeCleir GR, Brown CM, Gobler CJ, Bidle KD, Wilson WH, Wilhelm SW. 2014. Genome of brown tide virus (AaV), the little giant of the Megaviridae, elucidates NCLDV genome expansion and host-virus coevolution. Virology 466-467:60–70. doi: 10.1016/j.virol.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 36.Moniruzzaman M, Gann ER, Wilhelm SW. 2018. Infection by a giant virus (AaV) induces widespread physiological reprogramming in Aureococcus anophagefferens CCMP1984—a harmful bloom algae. Front Microbiol 9:752. doi: 10.3389/fmicb.2018.00752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilhelm SW, Coy SR, Gann ER, Moniruzzaman M, Stough JMA. 2016. Standing on the shoulders of giant viruses: five lessons learned about large viruses infecting small eukaryotes and the opportunities they create. PLoS Pathog 12:e1005752. doi: 10.1371/journal.ppat.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerepesi C, Grolmusz V. 2017. The “Giant Virus Finder” discovers an abundance of giant viruses in the Antarctic dry valleys. Arch Virol 162:1671–1676. doi: 10.1007/s00705-017-3286-4. [DOI] [PubMed] [Google Scholar]
- 39.Rusin LY. 2016. Metagenomics and biodiversity of sphagnum bogs. Mol Biol 50:645–648. doi: 10.1134/S0026893316050150. [DOI] [PubMed] [Google Scholar]
- 40.Krupovic M, Koonin EV. 2014. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci Rep 4:5347. doi: 10.1038/srep05347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krupovic M, Koonin EV. 2015. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat Rev Microbiol 13:105. doi: 10.1038/nrmicro3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomaru Y, Nagasaki K. 2007. Flow cytometric detection and enumeration of DNA and RNA viruses infecting marine eukaryotic microalgae. J Oceanogr 63:215–221. doi: 10.1007/s10872-007-0023-8. [DOI] [Google Scholar]
- 43.Miranda JA, Culley AI, Schvarcz CR, Steward GF. 2016. RNA viruses as major contributors to Antarctic virioplankton. Environ Microbiol 18:3714–3727. doi: 10.1111/1462-2920.13291. [DOI] [PubMed] [Google Scholar]
- 44.Quaiser A, Krupovic M, Dufresne A, Francez A-J, Roux S. 2016. Diversity and comparative genomics of chimeric viruses in Sphagnum-dominated peatlands. Virus Evol 2:vew025. doi: 10.1093/ve/vew025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quaiser A, Dufresne A, Ballaud F, Roux S, Zivanovic Y, Colombet J, Sime-Ngando T, Francez A-J. 2015. Diversity and comparative genomics of Microviridae in Sphagnum-dominated peatlands. Front Microbiol 6:375. doi: 10.3389/fmicb.2015.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ballaud F, Dufresne A, Francez A-J, Colombet J, Sime-Ngando T, Quaiser A. 2015. Dynamics of viral abundance and diversity in a sphagnum-dominated peatland: temporal fluctuations prevail over habitat. Front Microbiol 6:1494. doi: 10.3389/fmicb.2015.01494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorigo U, Jacquet S, Humbert JF. 2004. Cyanophage diversity, inferred from g20 gene analyses, in the largest natural lake in France, Lake Bourget. Appl Environ Microbiol 70:1017–1022. doi: 10.1128/AEM.70.2.1017-1022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roux S, Enault F, Robin A, Ravet V, Personnic S, Theil S, Colombet J, Sime-Ngando T, Debroas D. 2012. Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS One 7:e33641. doi: 10.1371/journal.pone.0033641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comeau AM, Krisch HM. 2008. The capsid of the T4 phage superfamily: the evolution, diversity, and structure of some of the most prevalent proteins in the biosphere. Mol Biol Evol 25:1321–1332. doi: 10.1093/molbev/msn080. [DOI] [PubMed] [Google Scholar]
- 50.Sakowski EG, Munsell EV, Hyatt M, Kress W, Williamson SJ, Nasko DJ, Polson SW, Wommack KE. 2014. Ribonucleotide reductases reveal novel viral diversity and predict biological and ecological features of unknown marine viruses. Proc Natl Acad Sci U S A 111:15786–15791. doi: 10.1073/pnas.1401322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alexander H, Jenkins BD, Rynearson TA, Dyhrman ST. 2015. Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc Natl Acad Sci U S A 112:E2182–E2190. doi: 10.1073/pnas.1421993112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson RM, Hopple AM, Tfaily MM, Sebestyen SD, Schadt CW, Pfeifer-Meister L, Medvedeff C, McFarlane KJ, Kostka JE, Kolton M, Kolka RK, Kluber LA, Keller JK, Guilderson TP, Griffiths NA, Chanton JP, Bridgham SD, Hanson PJ. 2016. Stability of peatland carbon to rising temperatures. Nat Commun 7:13723. doi: 10.1038/ncomms13723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanson PJ, Riggs JS, Nettles WR, Phillips JR, Krassovski MB, Hook LA, Gu L, Richardson AD, Aubrecht DM, Ricciuto DM. 2017. Attaining whole-ecosystem warming using air and deep-soil heating methods with an elevated CO2 atmosphere. Biogeosciences 14:861. doi: 10.5194/bg-14-861-2017. [DOI] [Google Scholar]
- 54.Warren MJ, Lin XJ, Gaby JC, Kretz CB, Kolton M, Morton PL, Pett-Ridge J, Weston DJ, Schadt CW, Kostka JE, Glass JB. 2017. Molybdenum-based diazotrophy in a sphagnum peatland in northern Minnesota. Appl Environ Microbiol 83:14. doi: 10.1128/AEM.01174-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kopylova E, Noe L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 56.O′Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koonin EV. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol 72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 58.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR. 2015. HMMER web server: 2015 update. Nucleic Acids Res 43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu SN, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang ZX, Yamashita RA, Zhang DC, Zheng CJ, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu WZ, Xie YB, Ma JY, Luo XT, Nie P, Zuo ZX, Lahrmann U, Zhao Q, Zheng YY, Zhao Y, Xue Y, Ren J. 2015. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31:3359–3361. doi: 10.1093/bioinformatics/btv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztanyi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang HZ, Huang XS, Letunic I, Lopez R, Lu SN, Marchler-Bauer A, Mi HY, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Xenarios I, Yeh LS, Young SY, Mitchell AL. 2017. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res 45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 66.Matsen FA, Kodner RB, Armbrust EV. 2010. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clark KR, Gorley RN. 2015. PRIMER v7: user manual/tutorial. PRIMER-E, Plymouth, United Kingdom. [Google Scholar]
- 69.Clarke KR, Somerfield PJ, Gorley RN. 2008. Testing of null hypotheses in exploratory community analyses: similarity profiles and biota-environment linkage. J Exp Mar Biol Ecol 366:56–69. doi: 10.1016/j.jembe.2008.07.009. [DOI] [Google Scholar]
- 70.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.