Most Frankia-actinorhiza plant symbioses are capable of high rates of nitrogen fixation comparable to those found on legumes. Yet, our understanding of the ecology and distribution of Frankia spp. is still very limited. Several studies have focused on the distribution patterns of Frankia spp., demonstrating a combination of host and pedoclimatic parameters in their biogeography. However, very few have considered the in planta sporulation form of the strain, although it is a unique feature among all symbiotic plant-associated microbes. Compared with Sp− Frankia strains, Sp+ strains would be obligate symbionts that are highly dependent on the presence of a compatible host species and with lower efficiency in nitrogen fixation. Understanding the biogeographical drivers of Sp+ Frankia strains might help elucidate the ecological role of in planta sporulation and the extent to which this trait mediates host-partner interactions in the alder-Frankia-ECM fungal symbiosis.
KEYWORDS: Frankia, actinorhizal symbiosis, in planta sporulation, Alnus, host specificity, ectomycorrhizae
ABSTRACT
The Alnus genus forms symbiosis with the actinobacteria Frankia spp. and ectomycorrhizal fungi. Two types of Frankia lineages can be distinguished based on their ability to sporulate in planta. Spore-positive (Sp+) strains are predominant on Alnus incana and Alnus viridis in highlands, while spore-negative (Sp−) strains are mainly associated with Alnus glutinosa in lowlands. Here, we investigated whether the Sp+ predominance in nodules is due to host selection of certain Frankia genotypes from soil communities or the result of the ecological history of the alder stand soil, as well as the effect of the sporulation genotype on the ectomycorrhizal (ECM) communities. Trapping experiments were conducted using A. glutinosa, A. incana, and A. viridis plantlets on 6 soils, differing in the alder species and the frequency of Sp+ nodules in the field. Higher diversity of Frankia spp. and variation in Sp+ frequencies were observed in the trapping than in the fields. Both indigenous and trapping species shape Frankia community structure in trapped nodules. Nodulation impediments were observed under several trapping conditions in Sp+ soils, supporting a narrower host range of Sp+ Frankia species. A. incana and A. viridis were able to associate equally with compatible Sp+ and Sp− strains in the greenhouse. Additionally, no host shift was observed for Alnus-specific ECM, and the sporulation genotype of Frankia spp. defined the ECM communities on the host roots. The symbiotic association is likely determined by the host range, the soil history, and the type of in planta Frankia species. These results provide an insight into the biogeographical drivers of alder symbionts in the Holarctic region.
IMPORTANCE Most Frankia-actinorhiza plant symbioses are capable of high rates of nitrogen fixation comparable to those found on legumes. Yet, our understanding of the ecology and distribution of Frankia spp. is still very limited. Several studies have focused on the distribution patterns of Frankia spp., demonstrating a combination of host and pedoclimatic parameters in their biogeography. However, very few have considered the in planta sporulation form of the strain, although it is a unique feature among all symbiotic plant-associated microbes. Compared with Sp− Frankia strains, Sp+ strains would be obligate symbionts that are highly dependent on the presence of a compatible host species and with lower efficiency in nitrogen fixation. Understanding the biogeographical drivers of Sp+ Frankia strains might help elucidate the ecological role of in planta sporulation and the extent to which this trait mediates host-partner interactions in the alder-Frankia-ECM fungal symbiosis.
INTRODUCTION
The role of the host species in the biogeography of symbiotic microorganisms has been intensively studied and has often confirmed codispersal patterns (1, 2), particularly for associations with a high level of specialization between the host and its symbionts. The impact of the symbiont's life history traits has been investigated far less, except for species of mycorrhizal fungi with different mechanisms of spore dispersion (3). Alders (Alnus spp., Betulaceae family) are involved in tripartite symbioses with ectomycorrhizal (ECM) fungi and nitrogen-fixing actinobacteria belonging to the Frankia genus. According to their subgenus or species, alders are associated with specific Frankia subgroups (4–6) and a few specific ECM fungi (7–10). This specificity has been confirmed on a worldwide scale, as the biogeography of ECM fungi and Frankia is mainly explained by their host biogeography and by abiotic factors, such as elevation or organic matter content (11–13). The life history traits of Frankia strains could also explain their high specificity and biogeography. Similar to what is seen with ECM fungi, spore dispersion probably differs among Frankia strains. On the one hand, some saprophytic strains only sporulate when cultured in vitro and probably in soils but never in the nodules (Sp− strains). On the other hand, uncultured strains are supposedly obligate symbionts and sporulate profusely within the host root nodules (Sp+ strains). These two sporulation phenotypes are, with few exceptions, genetically and phylogenetically distinct, yet they can occur at the same site or colonize the same host species (6, 14). The production of abundant asexual spores, which have high potential for propagation and for resistance to unfavorable conditions, may play an important role in the microbe's fitness and biogeography. Thus, the huge number of spores produced by Sp+ Frankia strains may enhance both their survival and dispersion capacity and could therefore explain their high infectivity and competitiveness on host plant roots compared to Sp− strains (15). In addition, spore production may compete with nitrogen fixation by using part of the plant's investment (photosynthesis products and energy) in the nodules and by occupying the infected cells with spores instead of diazovesicles (the specialized cells where nitrogen fixation occurs), thus limiting the benefits for the plant (14). Overall, these studies have led to the assumption that the ability to sporulate in planta is an important life trait in Frankia spp. (15, 16).
The abundance of Sp+ strains varies among Alnus species and stands. Indeed, some sites are exclusively nodulated by Sp+ or Sp− strains, while at other sites, both types cooccur in various proportions (17). Recent studies on European alder species demonstrated that Sp+ strains are dominant on Alnus viridis (or Alnus alnobetula) and Alnus incana strains and are far less abundant on Alnus glutinosa (13). Moreover, Sp+ strains from high-altitude alder stands were genetically very close to strains from boreal stands (high latitude) for a given Alnus taxon, suggesting that climate could be a driving factor in their distribution. However, since these three Alnus species are staggered according to altitude in subalpine, montane, and lowland areas (for A. viridis, A. incana, and A. glutinosa, respectively), the question remains whether the distribution of Sp+ strains reflects high dependency on the host (specificity) or selection by the environmental conditions prevalent at the different elevations of its host.
The phylogenetic clustering of Sp+ strains isolated from different host species (13) and their limited host range deduced from cross-inoculation experiments (15–18) suggest a stronger specificity of Sp+ strains to their host plant than that of Sp− strains. Compatible interactions between Alnus species and their specific Frankia strains may also affect their interaction with ECM fungi. Indeed, root nodule formation often precedes specific ECM association, and several studies have demonstrated that nodulation efficiency affects ECM diversity (10, 19, 20). As a consequence, and given that Sp+ and Sp− Frankia strains differ in their infectivities (15), nitrogen fixation activities (16), and secondary metabolite profiles (21), in planta sporulation of Frankia spp. may lead to marked differences in the ECM community structures of alders.
To clarify the ecological role of the Frankia in planta sporulation phenotype and to explore its consequences on host-symbiont interactions, we decided to study (i) whether the dominance of the Sp+ phenotype in A. incana and A. viridis stands in highlands can be explained by a preferential selection by the host (A. incana and A. viridis specificity) or by the environmental conditions, (ii) whether soil ecological history (Frankia genotype and presence over time of a given Alnus species) shapes soil symbiont communities, and (iii) whether the Frankia sporulation phenotype could affect ECM communities associated with alder roots. For this purpose, a full factorial plant-trapping experiment was performed with three Alnus species, used as traps, on six soils that differed in regards the alder species present (A. glutinosa, A. incana, or A. viridis) and the frequency of the Sp+ Frankia phenotype (high versus low) in the field. Frankia spp. and ECM fungi from seedling roots were identified, and community assemblages were compared under the different conditions. Furthermore, trapped Frankia spp. and ECM fungi were compared with the pool of Frankia spp. and ECM fungi sequenced from each native soil.
RESULTS
Trapping plant nodulation and Frankia Sp+ frequencies.
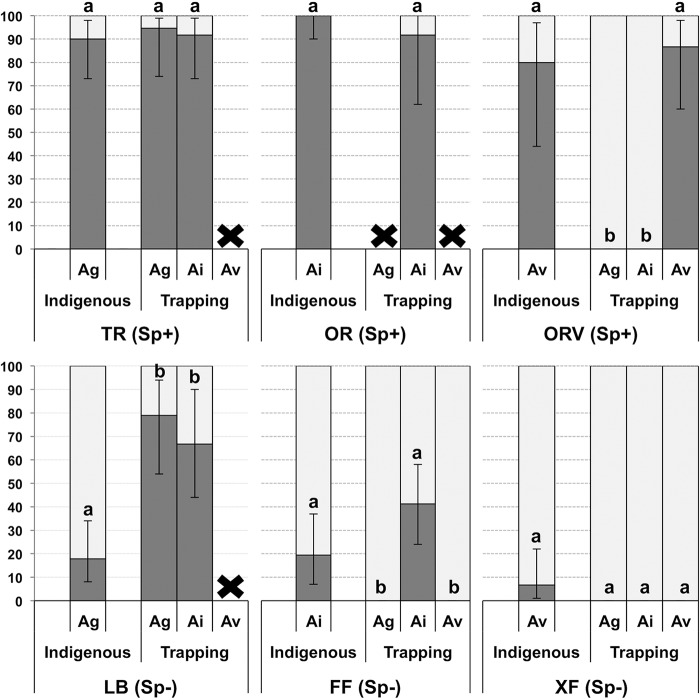
The number of nodules obtained on the different treatments ranged from 0 (no nodule trapped, e.g., A. viridis on Le Blanchet [LB] soil) to 405 nodules (i.e., A. viridis on Croix-de-Fer [XF] soil). A total of 168 field nodules and 236 trapping nodules were successfully phenotyped. The three Sp+ sites (Le Tremblay [TR], Ornon site 1 [OR], and Ornon site 2 [ORV]) and the three Sp− sites (LB, Fond-de-France [FF], and XF) harbored more than 80% and less than 20% of the Sp+ field nodules, respectively (Fig. 1). In trapping assays, the three Sp+ sites produced high Sp+ nodule frequencies when the same species present in the field was used as a trapping species, with TR, OR, and ORV harboring 94.7, 91.7, and 86.7% of the Sp+ nodules on A. glutinosa, A. incana, and A. viridis plantlets, respectively (Fig. 1). The three Sp− sites harbored different Sp+ frequencies in the trapping experiment from those in the field (Fig. 1). Higher Sp+ frequencies were observed on LB soil when trapped with A. glutinosa and on FF soil when trapped with A. incana than those in the field (78.9% and 41.2% compared with 17.9% and 19.4%, respectively) (Fig. 1). For XF soil, the rare Sp+ nodules present in the field were not recovered from any A. viridis plantlets (6.7% versus 0% of Sp+ nodules) (Fig. 1).
FIG 1.
Histograms of spore-positive percentages among indigenous host species nodules compared to trapping plant species nodules. Percentages were calculated using the estimation of the binomial law. Error bars correspond to 95% confidence intervals. Black crosses indicate an absence of nodules on the plantlets. Panels correspond to the combinations of indigenous host species (Ag, A. glutinosa; Ai, A. incana; Av, A. viridis) and of spore-positive (Sp+) or spore-negative (Sp−) soils. Lowercase letters indicate results obtained by the binomial law test.
When an alder species other than the indigenous one was used for trapping assays, contrasting results were obtained. A. glutinosa plantlets trapped only Sp− strains from FF (A. incana Sp− soil), XF (A. viridis Sp− soil), and ORV (A. viridis Sp+ soil), with average nodule numbers of 8.3, 5.9, and 2.1 per plant, respectively (Fig. 1 and Table 1). On OR (A. incana Sp+ soil), A. glutinosa plantlets formed only prenodules whose small size and early stage of development precluded any phenotype diagnostics. A. incana plantlets produced an average of 3.0 and 10.8 nodules per plant, of which 91.7 and 66.7% were Sp+ nodules, respectively, on TR and LB soils both coming from A. glutinosa alder stands in the field. Exclusively Sp− strains were trapped on ORV and XF soils, both corresponding to A. viridis alder stands in the field, forming 3.5 and 11.3 nodules per plant, respectively (Table 1). A. viridis plantlets formed an average of 6.2 nodules per plant on FF soil, all of them being Sp−. On the three other non-A. viridis soils (TR, LB, and OR), A. viridis plantlets did not form nodules, and most individuals died after a few weeks.
TABLE 1.
Nodules per trapping plant for each plant-trapping assay
| Predominant phenotype | Alder stand (indigenous species) | Trapping species (avg ± SD) |
||
|---|---|---|---|---|
| A. glutinosa | A. incana | A. viridis | ||
| Sp+ | TR (A. glutinosa) | 3.2 ± 0.4 | 3.0 ± 0.5 | 0 |
| OR (A. incana) | 0 | 3.7 ± 1.2 | 0 | |
| ORV (A. viridis) | 2.1 ± 0.3 | 2.1 ± 0.3 | 3.5 ± 0.9 | |
| Sp− | LB (A. glutinosa) | 10.9 ± 1.0 | 10.8 ± 2.4 | 0 |
| FF (A. incana) | 8.3 ± 0.8 | 9.4 ± 1.8 | 6.2 ± 1.4 | |
| XF (A. viridis) | 5.9 ± 0.8 | 5.3 ± 0.6 | 11.3 ± 1.4 | |
aSpore-positive (Sp+) and spore-negative (Sp−) are the predominant types of Frankia strains observed in the field nodules from the 6 alder stand soils used for the trapping. Average values and standard deviations of the nodule numbers per plant are reported for each of the three trapping species (Alnus glutinosa, Alnus incana, and Alnus viridis).
Frankia genetic diversity in nodules.
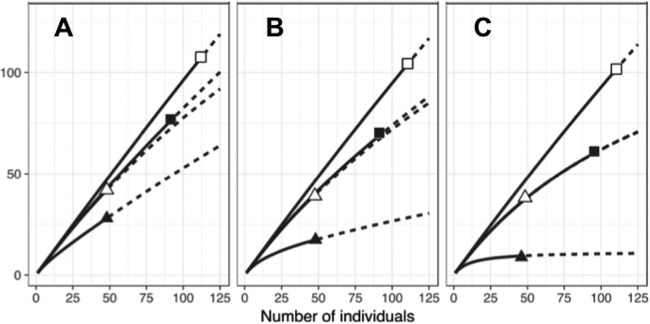
DNA extractions and pgk gene fragment (695 bp) amplifications were successfully conducted for 99 and 207 nodules from the field and the greenhouse trapping plants, respectively (i.e., about 16 per field site and 15 per trapping condition). None of the rarefaction curves obtained from pgk sequences of field and trapped nodules reached saturation (Fig. 2).
FIG 2.
Accumulation curves of Frankia richness (A), diversity (Shannon index) (B), and equitability (Simpson index) (C) estimated from pgk sequences at unique sequence threshold. Spore-positive (Sp+; filled symbols) and spore-negative (Sp−; open symbols) Frankia strains from indigenous host plant (squares) or trapping plants (triangles). Continuous line, interpolation; dashed line, extrapolation.
The richness of Frankia strains was higher in the trapped nodules than in field nodules (85.9 against 68 operational taxonomic units [OTUs], respectively; data not shown), regardless of the Sp+ or Sp− phenotype. Frankia Sp+ strains always harbored a lower level of richness than Frankia Sp− strains in both field nodules and trapped nodules (28 versus 42 OTU in field nodules, respectively, and 39.8 versus 45.9 OTU in trapped nodules, respectively).
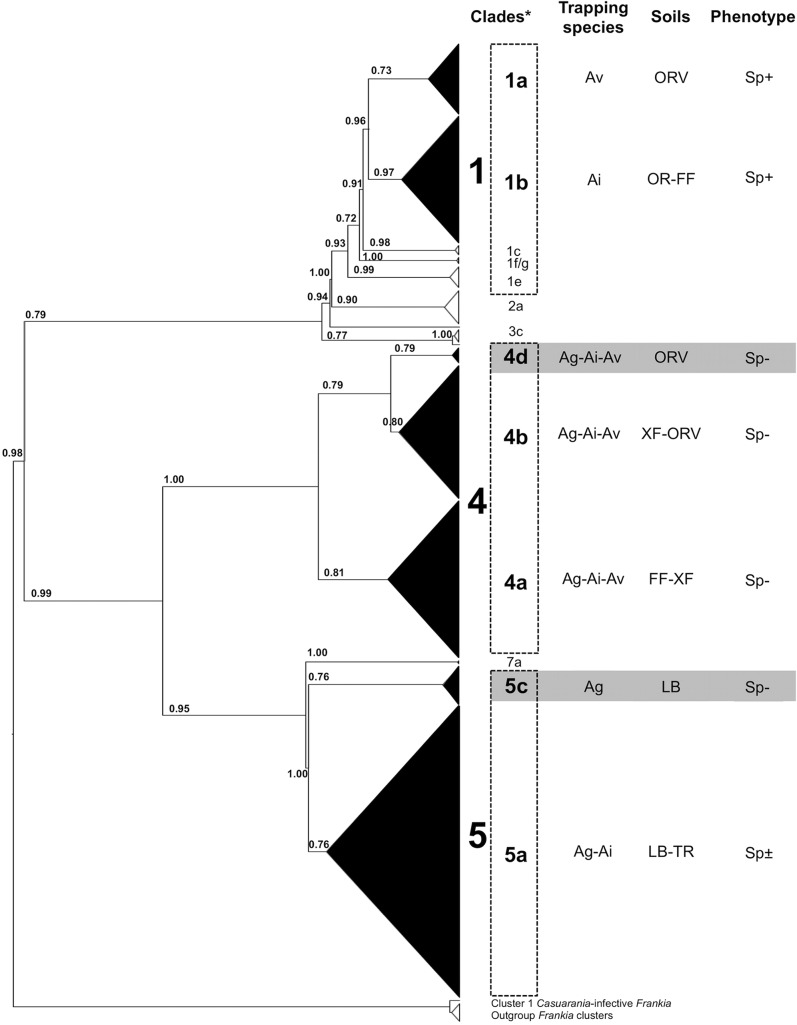
The phylogenetic analysis revealed that all pgk sequences of the trapped Frankia strains could be grouped into three previously described clades (1, 4, and 5) (Fig. 3). Three subclades contained narrow-range strains (1a, 1b, and 5c trapped only by A. viridis, A. incana, and A. glutinosa, respectively), and two subclades contained large-range strains (4a, 4b, and 4c trapped by the three alder species used). Subclade 5a contained middle-range strains (trapped by A. glutinosa and A. incana but not A. viridis). All Sp+ strains trapped on A. viridis Sp+ soil (ORV) belonged to clade 1a, while all Sp+ strains trapped on both A. incana soils OR and FF were grouped into clade 1b. Clade 4 contained all Sp− Frankia strains trapped with the three trapping species on A. incana and A. viridis soils (FF, XF, and ORV), creating three subclades (labeled a, b, and d). Subclades 4d and 4b contained all Sp− strains trapped on ORV soil, subclades 4b and 4a contained all Sp− strains trapped on XF soil, and subclade 4a contained all Sp− strains trapped on FF soil. Sp+ and Sp− strains trapped with A. glutinosa and A. incana host species from both A. glutinosa soils, TR and LB, were grouped in clade 5. No strain of this clade 5 was trapped by the A. viridis host species.
FIG 3.
Positions of trapped Frankia strains in the pgk phylogenetic tree of Alnus-infective (cluster 1) Frankia species. The phylogeny was estimated by maximum likelihood (PhyML). Statistical support of the nodes was estimated by aLRT SH-like method. Clades were annotated according to Pozzi et al. (6), and gray zones correspond to the 2 new subclades identified in the present study. Subclades in black are those that contain Frankia strains from our trapping assays. Clades 1, 4, and 5 correspond to OTUs 1, 4, and 5 (at the 0.05 threshold) in the present study.
Frankia community structure in soils and nodules resulting from trapping.
From the 1,310,346 reads obtained in the nifH run, 222,678 soil reads and 336,655 nodule reads were considered to be of good quality, with an average of 12,371 and 11,222 reads per sample, respectively. Sequences clustered at the 97% threshold into 163,020 soil reads and 199,432 nodule reads. For each sample, the first 250 Frankia OTUs at a 0.03 dissimilarity threshold, each containing a minimum of 200 sequences, were delineated. On average, 10,720 and 266 Frankia sequences per sample were obtained for the nodules and the soils, respectively. Frankia OTU compositions were compared between soils and trapped nodules. For all the sites studied, the most highly represented Frankia OTU in trapping nodules was not the most abundant in the soil (Data S1 in the supplemental material). The dominant OTUs in nodules represent between 3.9 and 18.5% relative abundance, depending on the soil. The predominant OTUs in each soil grouped together in a single clade, while predominant OTUs in nodules formed two distinct clades (data not shown).
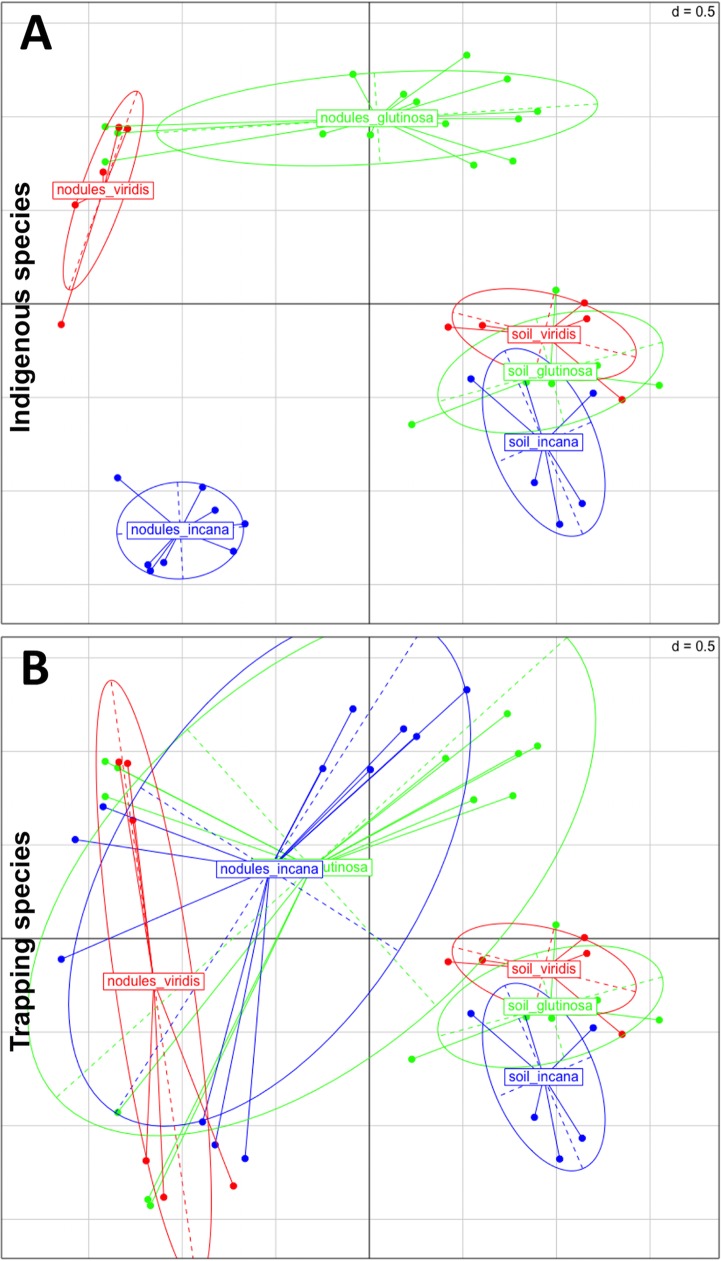
To compare Frankia OTU compositions in the different soils and in trapped nodules, nonmetric multidimensional scaling (NMDS) was used. Frankia OTU compositions in soils were always significantly different from those in trapped nodules (Fig. 4A and B, F = 14.1, R2 = 0.23, P = 0.001). There was no difference between indigenous soil communities, but the indigenous host species had a strong effect on OTU composition in trapped nodules (Fig. 4A, F = 9.72, R2 = 0.41, P = 0.001). The trap plant species alone had no statistically significant structuring effect (Fig. 4B, F = 1.61, R2 = 0.10, P = 0.091). However, there was an interaction between the indigenous species and the trap plant species that explained the Frankia OTU compositions in trapped nodules (F = 7.88, R2 = 0.71, P = 0.001).
FIG 4.
Effect of the indigenous species (A) and the trapping species (B) on Frankia community structure in soils and in the trapped nodules, illustrated by nonmetric multidimensional scaling (NMDS). NMDS were performed on Frankia OTU matrices computed from nifH sequences. Colors refer to the alder species, both on the field and in the trapping experiment. P values of permutational multivariate analysis of variance (adonis function) are given for both factors tested (A and B). Ellipses are graphical overviews.
Fungal and ectomycorrhizal diversity in soils and on plantlet roots.
After removing singletons and putative chimeras, 302,972 sequences could be assigned to fungi, of which 30,503 sequences (circa 10%) could be attributed to ECM taxa and 17,114 sequences could be assigned to 10 specific ECM species (Data S2). Species accumulation curves revealed that plantlets always associated with fewer fungi than did the soil samples. Considering ECM fungi, all trapping conditions led to a successful growth of ECM fungi on alder roots, and each alder species trapped more fungal and ECM OTUs than on its own soil (Data S3). The six specific ECM OTUs detected were associated with host species according to their known specificity pattern. No host shifts were detected. However, the identification of three Alnicola OTUs and 65 Tomentella spp. was not precise enough to investigate their specificity patterns and detect possible host shifts (Data S2).
In the soil, fungal and ECM species richness was not shaped by the same parameters. The sporulation phenotype present in the field had an effect on the number of different fungal species but not on that of ECM (Data S4). Instead, the variations in ECM species richness could be explained by the ecoregion (Data S4). The species richness measured on plantlets followed a different pattern, as the number of both fungal and ECM species was determined first by the indigenous host and, to a lesser extent for the ECM community only, by the trapping host and the ecoregion (Data S4). The soil community structure of the fungal and ECM community was shaped primarily by the indigenous host and then by the ecoregion and the sporulation phenotype present in the field. On plantlets, the fungal and ECM communities were shaped first by the indigenous alder species and then by the ecoregion, the trapping sporulation phenotype, the field sporulation phenotype and, to a lesser extent, the trapping host (Data S4). To summarize, species richness and community structures of all fungi and ECM fungi were mostly shaped by the indigenous host but also partly by the sporulation phenotype and its interaction with the indigenous host (ecoregion).
DISCUSSION
Frankia diversity and distribution.
All trapped strain sequences were grouped into the previously described clades, sometimes constituting new subclades (4d and 5c) that had never been sampled during previous field studies at the same soil collection sites (6, 13). The seven subclades identified differed in their host specificities (i.e., narrow, middle, or large range) and the sporulation phenotypes of the strains. No other clear indication of phylogenetic clustering (trapping species or ecoregion) emerged from our results. The predominance of Sp+ Frankia strains in high-altitude zones, associated with A. incana and A. viridis, was thought to be a result of host species selection and/or climatic factors associated with the habitat (13). Indeed, in the field, very few A. viridis stands had a low Sp+ frequency (16, 22), suggesting that alpine habitats may promote Sp+ strains over Sp− strains. Conversely, in the trapping experience, we showed that all the strains trapped from alpine soils belonged to either clade 1 (all Sp+ strains) or 4 (all Sp− strains) and none to clade 5, no matter the trapping species that was used. Under greenhouse conditions, A. incana and A. viridis form nodules with the strains present, whatever their sporulation phenotype, but with the sole condition that they belong to clade 1 or 4. Thus, they did not actively select Sp+ strains over Sp− strains.
The trapping assays revealed a greater richness of Frankia species than previously described in the field, suggesting that greenhouse conditions could affect biomass and function of microbial populations. Both soil preparation (i.e., sieving and mixing) (23–25) and plant age and phenology (26) could influence the recruitment of soil microbial communities in the rhizosphere (27, 28). Young plants often exude substantially higher quantities of organic substances than do mature individuals, in particular phenolic compounds known to play a defensive role in plant-microorganism interactions (29, 30). For instance, age-related resistance (ARR) has been linked with the production of defense compounds (31, 32) that might affect alder symbiotic interactions.
Soils collected from A. glutinosa and A. incana Sp− alder stands (LB and FF, respectively) revealed higher Sp+ frequencies in trapping trials than with the field when their original alder species was used for the trapping. This result could be explained either by the underestimation of the proportion of Sp+ strains in the field or by the experimental conditions. Although the Sp+ strain proportions in the field have been confirmed by various research studies (data not shown), none of the available tools permit the identification of Frankia Sp+ and Sp− strains directly from soil communities. Thus, the proportion of infective Sp+ propagules in the soil sampling spots is not known and might be higher than the proportion determined from the nodules. There are currently no available data to explain a difference in the sensitivities of the young plantlets toward Sp+ symbionts, although recent studies on the production of defensin-like peptides by Alnus spp. (33) and the detection of plant defense compounds differentially produced in Sp+ and Sp− nodules (21) are both promising lines of research.
This increased proportion of Sp+ strains on trapping plants compared to those under field conditions was not observed in A. viridis Sp− soil (XF). Based on crushed-nodule inoculations, Sp+ Frankia strains have been described as being about 100 to 2,000 times more infective than Sp− strains (13, 34–36). As discussed above, the low Sp+ frequency observed on nodules (XF soil) would be due to a low abundance in soil rather than host filtering. Therefore, the low probability of an encounter between the roots and Sp+ propagules might explain the finding that Sp− rather than Sp+ nodules are trapped in the greenhouse.
It is worth noting that whatever the trapping species used, Sp− soils always induce a higher number of nodules than do Sp+ soils, independently of the proportion of Sp+/Sp− strains obtained. The most likely hypotheses could be differences in rhizospheric Frankia abundance (discussed below) or differences in plant development between Sp+ and Sp− soils. This last hypothesis is supported by significant differences of plantlet root lengths when grown on Sp+ and Sp− soils (15.8 and 17.8 cm, respectively, P = 0.01; data not shown).
Compatibility patterns between soils and trapping species.
Different patterns of compatibility were found between soils and trapping plants depending on the alder species and the proportion of Sp+ nodules present in the field (Fig. 5). Frankia strains isolated from alders were long thought to belong to a unique host specificity group, i.e., a group of strains sharing the same compatible hosts, in this case plant species that belong to the Alnus genus and Myricaceae (37–40). While this Alnus-Myricaceae specificity group concept was confirmed for most Alnus-cultured strains, cross-inoculation experiments, using crushed nodules as inocula, suggested that Sp+ strains had a narrower host range than that of Sp− strains (15–18, 41). However, the use of crushed nodules may have three major side effects. First, the presence of plant secondary metabolites could prevent root-Frankia recognition and association (5). Second, the inoculum concentration used is generally much higher than the natural Frankia concentration in the field (42, 43). Finally, due to the dominance of one strain in the nodules, only a few strains were tested, and this was not representative of the diversity of Frankia communities in soils (44), thus giving a simplistic version of the Alnus-Frankia compatibility patterns compared with the trapping experiments.
FIG 5.
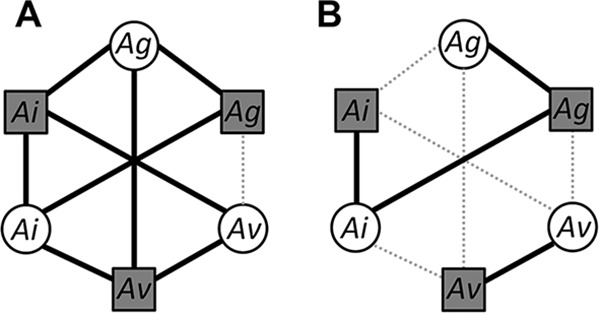
Compatibility patterns between the plant-trapping species and the spore-negative (A) and spore-positive (B) soils used in the assays. The compatibility between the 3 trapping species (white circles) Alnus glutinosa (Ag), Alnus incana (Ai), and Alnus viridis (Av), and the 6 alder stand soils (gray squares) differing in the indigenous host species (A. glutinosa, A. incana, or A. viridis) is symbolized by a continuous line (presence of compatible strains and formation of mature nodules). The incompatibility is symbolized by a dotted gray line (impeded prenodules and no compatible strain trapped).
Our results mirrored the narrower host range of Sp+ strains previously described, since most incompatibility patterns concerned Sp+ soils (Fig. 5). Indeed, Sp+ A. incana soils and Sp+ A. viridis soils never led to compatible associations when alder species other than the indigenous field species (A. incana and A. viridis, respectively) were used for trapping (Fig. 5B), since nodules were either absent or very small (impeded prenodules) and not functional (absence of diazovesicles). Moreover, our results corroborate the lack of compatibility between A. incana Sp+ strains and A. glutinosa hosts, which were previously described using crushed-nodule inocula (15, 16). This incompatibility is even more marked in our soil trapping experiment (no nodules formed) compared to the crushed-nodule inoculations (less infectivity). The high Frankia density in crushed-nodule inoculations could force the occurrence of some symbiotic associations never observed in the field, thus indicating an artificially broad host range (15). This result suggests that although A. incana Sp+ strains are genetically capable (symbiotic signalization is not lacking) of infecting A. glutinosa roots, they actually display low infectivity on this host species.
Equally, several results suggest very narrow specificity patterns between the A. viridis host and its associated Frankia strains, and especially for the Sp+ strains, as follows: (i) the absence of nodulation of A. viridis plantlets on A. glutinosa soils, suggesting a total incompatibility between A. viridis and clade 5 strains, whatever their sporulation phenotype (Fig. 3); (ii) the absence of nodulation of A. viridis plantlets on A. incana Sp+ soil, although strains from subclade 1b (all A. incana Sp+ strains) and subclade 1a (A. viridis Sp+ strains) are genetically close (Fig. 3); and (iii) the strict specificity of A. viridis Sp+ strains from subclade 1a that were exclusively trapped by A. viridis plantlets. This result is consistent with findings from previous studies (6, 13) where Sp+ strains from clade 1a were described as a monophyletic group considered to be highly specific to their host.
The colonization of a new environment by nonindigenous nitrogen-fixing plants (as shown with a cross-trap plant experiment) often leads to the establishment of novel associations through the recruitment of cosmopolitan strains, implying the presence of compatible low-specificity strains in the soil (45–47). In light of our results, the cosmopolitan status of Frankia strains in alder stand soils mainly depends on their Sp+ or Sp− identity. In contrast to earlier hypotheses (48, 49), our findings suggest that specificity within a particular set of mutualists may result in a failure of some alder species to colonize new habitats.
Impact of alder stand soil history and Frankia sporulation phenotype on symbiont diversity.
The next-generation sequencing (NGS) approach, targeting both bacterial and fungal symbionts, was used to determine the extent to which soil communities are shaped by the host species in the field, to explain the symbiotic associations observed on the trapping plants, and to test the potential impact of the in planta sporulation of Frankia spp. on root ECM fungi.
Although soil Frankia communities were the same among the three alder species in the field, differences in Frankia communities in trapped nodules were observed according to the field host species. These data indicate that, in agreement with findings from a previous study (50), host species in the field determined the nodule-forming populations of Frankia species. Nodule formation has been shown not to be a function of Frankia population abundance (51). Similarly, in this study, the sequences of trapped Frankia strains harbored relatively low abundances compared to those of soil-dwelling Frankia strains, possibly indicating that our metabarcoding approach did not allow for the detection of differences among soil Frankia communities beneath the different alder species.
The use of NGS on alder roots revealed mostly ECM fungi; however, sequences were too short to allow a deeper investigation of fungal specificity. By searching for single nucleotide polymorphisms, we did identify some of the Alnus symbionts, and, interestingly, all the associations observed in the experiment were congruent with the current information about Alnus specificity (8). Here, more ECM fungi were observed on plantlets than in their own soil, probably because, in addition to specialist ECM fungi, general ones were also recruited. This pattern is often observed on Alnus spp. in new environments, as recently shown in the case of invasion in New Zealand (48) and in an isolation case in Corsica (52). In these studies, generalist fungi never dominate Alnus roots, and we observed the same pattern in our study, as Tomentella and Alnicola represented the most abundant genera for the three hosts.
In the context of the numerous studies and reviews on Alnus-ECM specificity, our results highlight the importance of the indigenous host and, therefore, the soil history on specificity, and we have also shown that the three hosts can grow and develop ECM on other soils. Interestingly, A. incana grew well on both its own soil and on A. glutinosa ones, a characteristic that might be correlated with its ability to grow in a wide altitudinal range. As for Frankia spp., the indigenous host always had a stronger effect than the trapping host on the fungal and ECM community and diversity. This effect is probably due to the following two major factors: the soil chemistry, when it is particularly distinct between the sites and the hosts (9), and the spore bank.
We investigated the influence of the Frankia sporulation phenotype, both predicted from the ecoregion (the soil and the indigenous host present) and as observed on the trapping plantlets. We detected a significant effect of the sporulation phenotype (both predicted and observed) on the ECM community structure but not on ECM species diversity, suggesting that the abundance may change but the identity of the fungi does not. This effect of the sporulation phenotype was often stronger than the trapping host effect, suggesting that the specificity could result from a strong interaction between the host and its diverse symbionts. Several clues suggest an intimate relationship between Frankia spp. and ECM fungi (10), and plant investment to nodules and mycorrhizae might depend on their cooccurrence on the host. Here, we did not measure plant investment, but we observed that ECM communities are partly determined by Frankia spp. and their phenotype of in planta sporulation, even on young plantlets. The underlying mechanism still needs to be deciphered. As the profuse sporulation of Frankia spp. within the nodules would require sizable amounts of carbon (spore coat layers), nitrogen (DNA bases for genome copy), and possibly phosphorus (DNA bases and replication enzymes) from the host plant, we hypothesize that the sporulation phenotype of Frankia spp. may modify the levels of these elements in the host plant to such an extent that it might mediate host-partner interactions, and even partner-partner interactions, through the host, particularly when these partners are also involved in the C:N:P economy of the symbiotic system.
MATERIALS AND METHODS
Sites and sampling.
Six well-developed alder stands about 100 years old were selected in the French alpine region to include three Alnus species (A. glutinosa, A. incana, and A. viridis) and the two Frankia sporulation phenotypes (Sp+ and Sp−), resulting in 6 distinct ecoregions. These sites included two lowland A. glutinosa stands, Le Blanchet (LB) and Le Tremblay (TR); two montane A. incana stands, Fond-de-France (FF) and Ornon site 1 (OR); and two subalpine A. viridis stands, Croix-de-Fer (XF) and Ornon site 2 (ORV). LB, FF, and XF alder stands had been previously identified as Sp− sites (sites harboring predominantly Sp− nodules), while TR, OR, and ORV were identified as Sp+ sites (13) (Table 2). For each site, soil samples were collected in mid-autumn at 3 different points (0 to 10 cm depth without the upper organic layer), 50 m apart, sieved at 4 mm, and mixed to form a composite soil sample. Three subsamples of 0.5 g from each composite sample were frozen for subsequent genomic analyses. About 25 root nodules were also collected per site from at least 5 alders.
TABLE 2.
Factorial plan and conditions
| Sporulation phenotype | Soils (indigenous host)a |
||
|---|---|---|---|
| A. glutinosa | A. incana | A. viridis | |
| Sp+ | TR | OR | ORV |
| Sp− | LB | FF | XF |
aFor each condition, these 3 same trapping plant species were used.
Plant growth and trapping experiments.
Seeds from the three Alnus species (A. glutinosa from Grand Lemps, Rhône-Alpes, and A. incana and A. viridis from Vanoise National Park, France) were sterilized for 30 s in absolute ethanol and then rehydrated, for 48 h at 4°C, under magnetic stirring in sterile water. On each of the six soils, seed germination, plantlet precultivation for 3 weeks, and trapping experiences were conducted. The greenhouse conditions were 16 h of daylight at 10,000 lx, 22°C, and 60% humidity and 8 h of dark at 18°C and 75% humidity. For each soil, 36 plantlets were used for each of the 3 host species (A. glutinosa, A. incana, and A. viridis). Plants were watered twice a week with sterile deionized water, and plant positions within the climatic chamber were randomized weekly. After 4 to 6 months of growth, depending on the species growth rate, plants were harvested. Nodules collected from roots were counted and used for Frankia phenotypic and genotypic analyses. Circa 50 root tips per seedling were sampled randomly and used for ECM fungal genotypic characterization.
From each field and trapping nodule, two adjacent lobes were sampled, with one lobe to determine the sporulation phenotype and the other one to genotype the Frankia strains. About of 25 field nodules per site and 15 nodules per trapping condition were phenotyped and genotyped.
Sporulation phenotype determination of nodules.
The sporulation phenotype was determined by microscopic observation of hand-cut sections of nodule lobes stained with lactophenol blue (Réactifs RAL, Martillac, France), as previously described (13). The lactophenol blue stain discriminates between spores from hyphae (refringent and nonstained) and those from diazovesicles (stained deep blue). Nodules were considered to have the Sp+ phenotype when more than one sporangium was observed out of 50 infected plant cells, and the others were Sp−. The proportion of Sp+ strains was estimated for each trapping condition.
DNA extraction from nodules and pgk gene sequencing.
Genotyping consisted of targeting a partial sequence (695 bp) of the housekeeping gene pgk coding for the phosphoglycerate kinase. Total nodular DNA was extracted from each nodule lobe individually, using the method previously described (13). For each lobe, amplification was performed using the specific primers and PCR protocol previously described (13). PCR products were single-strand sequenced by Biofidal-DTAMB (Villeurbanne, France) using the Sanger method with the same set of primers. pgk sequences were checked, trimmed, and manually corrected using 4Peaks version 1.7.2 (53). Sequences were confirmed to belong to Frankia using BLASTN searches against the National Center for Biotechnology Information (NCBI) databases (54) and aligned using the Muscle version 3.8.31 package (55) in SeaView version 4.4.2. The distance matrices were calculated using the DNADIST program. OTU matrices were computed from the alignment in mothur version 1.31.2 (56), with the furthest neighbor clustering and using 0.0049 as the maximum pairwise distance between sequences from an identical OTU. Accumulation curves of Frankia richness were computed from the matrices using the iNEXT package in R version 3.0.1 (57), at a 95% confidence interval.
Phylogenetic analyses were performed from our aligned nucleotide sequences and pooled with sequences previously submitted (13), using the maximum likelihood method in the software PhyML (53) with a general time reversible (GTR) + G4 model and the NNI + SPR option for topology exploration. Topologies were rooted with Frankia pgk sequences that did not belong to the Alnus infective cluster 1 (outgroup). The branch robustness of maximum likelihood (ML) trees was estimated by the approximate likelihood ratio test (aLRT) using the nonparametric Shimodaira-Hasegawa-like branch test (58), implemented in SeaView version 4.4.2. Clades were delineated on the basis of the strain habitat (host plant and/or site) and the in planta sporulation phenotype, as previously proposed (13), and using the same numbering (6).
nifH metabarcoding analyses from soils and trapped nodules.
A metabarcoding approach was used to assess the nifH bacterial community structure from soils and nodules. Soil DNA was extracted from defrosted soil samples using the PowerSoil DNA kit (Qiagen), according to the manufacturer's instructions. Three plants of each trapping condition were randomly selected, and about 8 nodules per plant were sampled for the same DNA extraction protocol previously described (59). Soil and nodule nucleic acid solutions were amplified using IGK3/DVV primers (60), and the PCR conditions were the same as previously described (42). Soil and nodule barcoded amplicons were pooled and used as the template for a single run of Illumina MiSeq sequencing, using the paired-end sequencing technology (2 × 250 bp) at the Genotoul GIS facility, Toulouse, France.
nifH reads were processed using the open-source software mothur (version 1.38.0) (56), following the method previously described (42). Once the reads had been processed and the OTUs had been delimited at a 97% dissimilarity threshold, the first 250 most abundant OTUs were screened against the nucleotide database of the National Center for Biotechnology Information (NCBI), using the BLASTn tool to identify Frankia OTUs.
Global and ectomycorrhizal fungal diversities in soils and on plantlet roots.
Root apices were harvested with sterile tweezers and stored in cetyltrimethylammonium bromide (CTAB) buffer until DNA extraction. DNA extraction was performed as previously described (9). The internal transcribed spacer 1 (ITS1) was chosen as a barcode and amplified, both on root apex extracts and on soil DNA, as previously described (42). All ITS1 amplicons were pooled and sequenced on a single run of Illumina MiSeq sequencing. Sequence analysis, according to Schwob et al. (42), enabled us to attribute sequences to seedlings or to soil samples, to group sequences in OTUs at a 97% threshold, and to assign sequences to fungal taxa based on a comparison with GenBank sequences using ecotag in the OBITools package (61). To determine the specificity of the ECM symbionts, sequences belonging to Alnicola, Alpova, Lactarius, Melanogaster, Paxillus, and Russula spp. were aligned with the previously published sequences of specific species associated with alders (8, 9). The alignments, handled with MAFFT (62), were restricted to the ITS1 region. These included reference sequences and all sequences attributed to a given genus. These alignments allowed sequences to be grouped based on their shared single nucleotide polymorphism, and these groups could be identified to the species level. Sequence assignation enabled us to subset ECM OTUs for further analysis. Accumulation curves of ECM OTU richness were computed from the matrices using an iNEXT package in R version 3.0.1 at a 95% confidence interval.
Community and statistical analyses.
Ninety-five percent confidence intervals of Sp+ frequencies were estimated using the normal approximation of the binomial distribution with the function binom.test implemented in the R software (63). Comparisons of Sp+ frequencies on trapping plants, versus those in the field, were performed using the same function. Fungal and ECM OTU richness was computed for each seedling and soil sample using the ade4 package (64). Differences between ecoregions (indigenous host × dominant Frankia phenotype) and the effect of trapping host or indigenous host were tested on plantlet samples using analysis of variance (ANOVA). The Bray-Curtis distance matrices between communities, sampled on seedlings or in the soil, were computed for each gene using the vegan package (65). Bray-Curtis matrices were used to perform nonmetric multidimensional scaling (NMDS) with the metaMDS function available in the vegan package (65). The effect of the indigenous host species on fungal, ectomycorrhizal, and Frankia communities in soils was tested through a permutational multivariate analysis of variance (PERMANOVA), using the adonis function in the vegan package. Likewise, the effect of the trapping host species, the dominant spore phenotype, and the indigenous host species were tested on fungal, ECM, and Frankia communities on trapping plantlets. The order in the models was permuted to determine which factor could best explain the variations, and, finally, the model with the least residuals was chosen.
Data availability.
All Frankia pgk sequences were previously deposited with EMBL (https://www.ebi.ac.uk/ena) under accession numbers LT599837 to LT600328. Fastq files were deposited with EMBL under BioProject PRJEB26577 and accession number ERS2462111 for bacteria and under BioProject PRJEB18608 and accession number ERS1473494 for fungi.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Elise Lacroix from the Greenhouses and Climatic Chambers Platform for nursing our alder plantlets throughout the study. We also thank Florian Vautrin, Guillaume Laliberté, Lucile Daniel, Claire Bellehumeur, and Samuel Jean for their precise technical assistance.
G. Schwob was granted a doctoral fellowship by the Ministry of Higher Education and Research (France). The sequencing was funded by a regional project ARC and the French Laboratory of Excellence project “TULIP” (grants ANR-10-LABX-41 and ANR-11-IDEX-0002-02).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01737-18.
REFERENCES
- 1.Higgins LM, Kennedy PG. 2012. Symbiotic Frankia bacteria in Alnus forests in Mexico and the United States of America: is geographic location a good predictor of assemblage structure? Botany 90:423–431. doi: 10.1139/b2012-006. [DOI] [Google Scholar]
- 2.Hughes Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Adams Krumins J, Kuske CR. 2006. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 3.Douhan GW, Vincenot L, Gryta H, Selosse M-A. 2011. Population genetics of ectomycorrhizal fungi: from current knowledge to emerging directions. Fungal Biol 115:569–597. doi: 10.1016/j.funbio.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Huguet V, Batzli JM, Zimpfer JF, Normand P, Dawson JO, Fernandez MP. 2001. Diversity and specificity of Frankia strains in nodules of sympatric Myrica gale, Alnus incana, and Shepherdia canadensis determined by rrsGene polymorphism. Appl Environ Microbiol 67:2116–2122. doi: 10.1128/AEM.67.5.2116-2122.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torrey JG. 1990. Cross-inoculation groups within Frankia and host-endosymbiont associations, p 83–106. In Schwintzer CR, Tjepkema JD (ed), The biology of Frankia and actinorhizal plants. Academic Press, San Diego, CA. [Google Scholar]
- 6.Pozzi AC, Bautista-Guerrero HH, Abby SS, Herrera-Belaroussi A, Abrouk D, Normand P, Menu F, Fernandez MP. 2018. Robust Frankia phylogeny, species delineation and intraspecies diversity based on MLSA and a simplified protocol adapted to intensive field sampling. Syst Appl Microbiol 41:311–323. doi: 10.1016/j.syapm.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Molina R. 1981. Ectomycorrhizal specificity in the genus Alnus. Can J Bot 59:325–334. doi: 10.1139/b81-045. [DOI] [Google Scholar]
- 8.Rochet J, Moreau P-A, Manzi S, Gardes M. 2011. Comparative phylogenies and host specialization in the alder ectomycorrhizal fungi Alnicola, Alpova and Lactarius (Basidiomycota) in Europe. BMC Evol Biol 11:40. doi: 10.1186/1471-2148-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy M, Rochet J, Manzi S, Jargeat P, Gryta H, Moreau PA, Gardes M. 2013. What determines Alnus-associated ectomycorrhizal community diversity and specificity? A comparison of host and habitat effects at a regional scale. New Phytol 198:1228–1238. doi: 10.1111/nph.12212. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy PG, Walker JKM, Bogar LM. 2015. Interspecific mycorrhizal networks and non-networking hosts: exploring the ecology of the host genus Alnus. 224:227–254. [Google Scholar]
- 11.Põlme S, Bahram M, Koljalg U, Tedersoo L. 2014. Global biogeography of Alnus-associated Frankia actinobacteria. New Phytol 204:979–988. doi: 10.1111/nph.12962. [DOI] [PubMed] [Google Scholar]
- 12.Põlme S, Bahram M, Yamanaka T, Nara K, Dai YC, Grebenc T, Kraigher H, Toivonen M, Wang PH, Matsuda Y. 2013. Biogeography of ectomycorrhizal fungi associated with alders (Alnus spp.) in relation to biotic and abiotic variables at the global scale. New Phytol 198:1239–1249. doi: 10.1111/nph.12170. [DOI] [PubMed] [Google Scholar]
- 13.Pozzi AC, Bautista-Guerrero HH, Nouioui I, Cotin-Galvan L, Pepin R, Fournier P, Menu F, Fernandez MP, Herrera-Belaroussi A. 2015. In-planta sporulation phenotype: a major life history trait to understand the evolution of Alnus-infective Frankia strains. Environ Microbiol 17:3125–3138. doi: 10.1111/1462-2920.12644. [DOI] [PubMed] [Google Scholar]
- 14.Torrey JG. 1987. Endophyte sporulation in root nodules of actinorhizal plants. Physiologia Plantarum 70:279–288. doi: 10.1111/j.1399-3054.1987.tb06145.x. [DOI] [Google Scholar]
- 15.Cotin-Galvan L, Pozzi AC, Schwob G, Fournier P, Fernandez PM, Herrera-Belaroussi A. 2015. In-planta sporulation capacity enhances infectivity and rhizospheric competitiveness of Frankia strains. Microbes Environ 31:11–18. doi: 10.1264/jsme2.ME15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurdali F, Domenach A-M, De La Paz Fernandez M, Capellano A, Moiroud A. 1988. Compatibility of Frankiae spore positive and spore negative inocula with Alnus glutinosa and Alnus incana. Soil Sci Plant Nutr 34:451–459. doi: 10.1080/00380768.1988.10415700. [DOI] [Google Scholar]
- 17.Schwintzer CR. 1990. Spore-positive and spore-negative nodules, p 177–193. In Schwintzer CR, Tjepkema JD (ed), The biology of Frankia and actinorhizal plants. Academic Press, San Diego, CA. [Google Scholar]
- 18.Markham JH. 2008. Variability of nitrogen-fixing Frankia on Alnus species. Botany 86:501–510. doi: 10.1139/B08-023. [DOI] [Google Scholar]
- 19.Miller SL, Koo C, Molina R. 1992. Early colonization of red alder and Douglas fir by ectomycorrhizal fungi and Frankia in soils from the Oregon coast range. Mycorrhiza 2:53–61. doi: 10.1007/BF00203250. [DOI] [Google Scholar]
- 20.Yamanaka T, Li C-Y, Bormann BT, Okabe H. 2003. Tripartite associations in an alder: effects of Frankia and Alpova diplophloeus on the growth, nitrogen fixation and mineral acquisition of Alnus tenuifolia. Plant Soil 254:179–186. doi: 10.1023/A:1024938712822. [DOI] [Google Scholar]
- 21.Hay A-E, Boubakri H, Buonomo A, Rey M, Meiffren G, Cotin-Galvan L, Comte G, Herrera-Belaroussi A. 2017. Control of endophytic Frankia sporulation by Alnus nodule metabolites. Mol Plant Microbe Interact 30:205–214. doi: 10.1094/MPMI-11-16-0235-R. [DOI] [PubMed] [Google Scholar]
- 22.Daniere C, Capellano A, Moiroud A. 1986. Dynamique de l'azote dans un peuplement naturel d'Alnus incana (L.) Moench. Act Oecol (Montrouge) 7:165–175. [Google Scholar]
- 23.Jasper D, Abbott L, Robson A. 1991. The effect of soil disturbance on vesicular—arbuscular mycorrhizal fungi in soils from different vegetation types. New Phytol 118:471–476. doi: 10.1111/j.1469-8137.1991.tb00029.x. [DOI] [Google Scholar]
- 24.Petersen SO, Klug MJ. 1994. Effects of sieving, storage, and incubation temperature on the phospholipid fatty acid profile of a soil microbial community. Appl Environ Microbiol 60:2421–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zabinski CA, Gannon JE. 1997. Effects of recreational impacts on soil microbial communities. Environ Manage 21:233–238. doi: 10.1007/s002679900022. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy PG, Weber MG, Bluhm AA. 2010. Frankia bacteria in Alnus rubra forests: genetic diversity and determinants of assemblage structure. Plant Soil 335:479–492. doi: 10.1007/s11104-010-0436-9. [DOI] [Google Scholar]
- 27.Micallef SA, Shiaris MP, Colón-Carmona A. 2009. Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J Exp Bot 60:1729–1742. doi: 10.1093/jxb/erp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore BD, Andrew RL, Kulheim C, Foley WJ. 2014. Explaining intraspecific diversity in plant secondary metabolites in an ecological context. New Phytol 201:733–750. doi: 10.1111/nph.12526. [DOI] [PubMed] [Google Scholar]
- 29.Achakzai AKK, Achakzai P, Masood A, Kayani SA, Tareen RB. 2009. Response of plant parts and age on the distribution of secondary metabolites on plants found in Quetta. Pak J Bot 41:2129–2135. [Google Scholar]
- 30.Gransee A, Wittenmayer L. 2000. Qualitative and quantitative analysis of water-soluble root exudates in relation to plant species and development. J Plant Nutr Soil Sci 163:381–385. doi:. [DOI] [Google Scholar]
- 31.Carella P, Wilson DC, Cameron RK. 2015. Some things get better with age: differences in salicylic acid accumulation and defense signaling in young and mature Arabidopsis. Front Plant Sci 5:775. doi: 10.3389/fpls.2014.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kus JV, Zaton K, Sarkar R, Cameron RK. 2002. Age-related resistance in Arabidopsis is a developmentally regulated defense response to Pseudomonas syringae. Plant Cell 14:479–490. doi: 10.1105/tpc.010481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carro L, Pujic P, Alloisio N, Fournier P, Boubakri H, Hay AE, Poly F, François P, Hocher V, Mergaert P, Balmand S, Rey M, Heddi A, Normand P. 2015. Alnus peptides modify membrane porosity and induce the release of nitrogen-rich metabolites from nitrogen-fixing Frankia. ISME J 9:1723–1733. doi: 10.1038/ismej.2014.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Dijk C, Simmer A, Weber A. 1988. Host range differentiation of spore-positive and spore-negative strain types of Frankia in stands of Alnus glutinosa and Alnus incana in Finland. Physiol Plant 72:349–358. doi: 10.1111/j.1399-3054.1988.tb05844.x. [DOI] [Google Scholar]
- 35.Houwers A, Akkermans A. 1981. Influence of inoculation on yield of Alnus glutinosa in the Netherlands. Plant Soil 61:189–202. doi: 10.1007/BF02277374. [DOI] [Google Scholar]
- 36.Quispel A. 1990. Discoveries, discussions, and trends in research on actinorhizal root nodule symbioses before 1978, p 15–33. In Schwintzer CR, Tjepkema JD (ed). The biology of Frankia and actinorhizal plants. Academic Press, San Diego, CA. [Google Scholar]
- 37.Baker DD. 1987. Relationships among pure cultured strains of Frankia based on host specificity. Physiol Plant 70:245–248. doi: 10.1111/j.1399-3054.1987.tb06139.x. [DOI] [Google Scholar]
- 38.Normand P, Orso S, Cournoyer B, Jeannin P, Chapelon C, Dawson J, Evtushenko L, Misra AK. 1996. Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int J Syst Bacteriol 46:1–9. doi: 10.1099/00207713-46-1-1. [DOI] [PubMed] [Google Scholar]
- 39.Dawson J. 2007. Ecology of actinorhizal plants, p 199–234. In Pawlowski K, Newton W (ed), Nitrogen-fixing actinorhizal symbioses. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 40.Chanway CP, Turkington R, Holl F. 1991. Ecological implications of specificity between plants and rhizosphere micro-organisms. Adv Ecol Res 21:121–169. [Google Scholar]
- 41.Weber A, Nurmiaho-Lassila EL, Sundman V. 1987. Features of the intrageneric Alnus-Frankia specificity. Physiol Plant 70:289–296. doi: 10.1111/j.1399-3054.1987.tb06146.x. [DOI] [Google Scholar]
- 42.Schwob G, Roy M, Manzi S, Pommier T, Fernandez M. 2017. Green alder (Alnus viridis) encroachment shapes microbial communities in subalpine soils and impacts its bacterial or fungal symbionts differently. Environ Microbiol 19:3235–3250. doi: 10.1111/1462-2920.13818. [DOI] [PubMed] [Google Scholar]
- 43.Samant SS, Dawson JO, Hahn D. 2015. Growth responses of indigenous Frankia populations to edaphic factors in actinorhizal rhizospheres. Syst Appl Microbiol 38:501–505. doi: 10.1016/j.syapm.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Mirza BS, Welsh A, Rasul G, Rieder JP, Paschke MW, Hahn D. 2009. Variation in Frankia populations of the Elaeagnus host infection group in nodules of six host plant species after inoculation with soil. Microb Ecol 58:384–393. doi: 10.1007/s00248-009-9513-0. [DOI] [PubMed] [Google Scholar]
- 45.Moora M, Berger S, Davison J, Öpik M, Bommarco R, Bruelheide H, Kühn I, Kunin WE, Metsis M, Rortais A. 2011. Alien plants associate with widespread generalist arbuscular mycorrhizal fungal taxa: evidence from a continental-scale study using massively parallel 454 sequencing. J Biogeogr 38:1305–1317. doi: 10.1111/j.1365-2699.2011.02478.x. [DOI] [Google Scholar]
- 46.Lafay B, Burdon JJ. 2006. Molecular diversity of rhizobia nodulating the invasive legume Cytisus scoparius in Australia. J Appl Microbiol 100:1228–1238. doi: 10.1111/j.1365-2672.2006.02902.x. [DOI] [PubMed] [Google Scholar]
- 47.Parker MA, Malek W, Parker IM. 2006. Growth of an invasive legume is symbiont limited in newly occupied habitats. Divers Distrib 12:563–571. doi: 10.1111/j.1366-9516.2006.00255.x. [DOI] [Google Scholar]
- 48.Bogar LM, Dickie IA, Kennedy PG. 2015. Testing the co-invasion hypothesis: ectomycorrhizal fungal communities on Alnus glutinosa and Salix fragilis in New Zealand. Divers Distrib 21:268–278. doi: 10.1111/ddi.12304. [DOI] [Google Scholar]
- 49.Pringle A, Bever JD, Gardes M, Parrent JL, Rillig MC, Klironomos JN. 2009. Mycorrhizal symbioses and plant invasions. Annu Rev Ecol Evol Syst 40:699–715. doi: 10.1146/annurev.ecolsys.39.110707.173454. [DOI] [Google Scholar]
- 50.Anderson MD, Ruess RW, Myrold DD, Taylor DL. 2009. Host species and habitat affect nodulation by specific Frankia genotypes in two species of Alnus in interior Alaska. Oecologia 160:619–630. doi: 10.1007/s00442-009-1330-0. [DOI] [PubMed] [Google Scholar]
- 51.Ben Tekaya S, Guerra T, Rodriguez D, Dawson JO, Hahn D. 2018. Frankia diversity in host plant root nodules is independent of abundance or relative diversity of Frankia populations in corresponding rhizosphere soils. Appl Environ Microbiol 84:e02248-17. doi: 10.1128/AEM.02248-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pozzi AC, Roy M, Nagati M, Schwob G, Manzi S, Gardes M, Moreau P-A, Fernandez MP. 2018. Patterns of diversity, endemism and specialization in the root symbiont communities of alder species on the island of Corsica. New Phytol 219:336–349. doi: 10.1111/nph.14996. [DOI] [PubMed] [Google Scholar]
- 53.Griekspoor A, Groothuis T. 2005. 4Peaks: a program that helps molecular biologists to visualize and edit their DNA sequence files v1. 7.
- 54.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 55.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsieh T, Ma K, Chao A. 2016. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol Evol 7:1451–1456. doi: 10.1111/2041-210X.12613. [DOI] [Google Scholar]
- 58.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 59.Rouvier C, Prin Y, Reddell P, Normand P, Simonet P. 1996. Genetic diversity among Frankia strains nodulating members of the family Casuarinaceae in Australia revealed by PCR and restriction fragment length polymorphism analysis with crushed root nodules. Appl Environ Microbiol 62:979–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaby JC, Buckley DH. 2012. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS One 7:e42149. doi: 10.1371/journal.pone.0042149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boyer F, Mercier C, Bonin A, Le Bras Y, Taberlet P, Coissac E. 2016. obitools: a unix-inspired software package for DNA metabarcoding. Mol Ecol Resour 16:176–182. doi: 10.1111/1755-0998.12428. [DOI] [PubMed] [Google Scholar]
- 62.Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 63.R Core Team. 2012. Package ‘ade4’: analysis of ecological data: exploratory and Euclidean methods in environmental sciences. R package; version 15-1. [Google Scholar]
- 64.Chessel D, Dufour A-B, Dray S. 2009. Package ‘ade4’: analysis of ecological data: exploratory and Euclidean methods in environmental sciences. R package version 15-1. [Google Scholar]
- 65.Oksanen J, Blanchet FG, Kindt R, Legendre P, O'Hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2011. vegan: community ecology package; R package version 1.17-8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All Frankia pgk sequences were previously deposited with EMBL (https://www.ebi.ac.uk/ena) under accession numbers LT599837 to LT600328. Fastq files were deposited with EMBL under BioProject PRJEB26577 and accession number ERS2462111 for bacteria and under BioProject PRJEB18608 and accession number ERS1473494 for fungi.