Abstract
European hake (Merluccius merluccius) is one of the most economically important fish for the Mediterranean Sea. It is an important predator of deep upper shelf slope communities currently characterized by growth overexploitation: the understanding of hake's diet might support next generation management tools. However, all current European hake diet studies depend on the morphological identification of prey remains in stomach content, with consequent limitations. In this study, we set up a metabarcoding approach based on cytochrome oxidase I PCR amplification and Miseq Illumina paired‐end sequencing of M. merluccius stomach content remains and compared the results to classic morphological analyses. A total of 95 stomach contents of M. merluccius sampled in the North‐Central Adriatic Sea were analyzed with both the metabarcoding and morphological approaches. Metabarcoding clearly outperformed the morphological method in the taxonomic identification of prey describing more complex trophic relationships even when considering the morphological identification of 200 stomach contents. Statistical analysis of diet composition revealed a weak differentiation among the hake's size classes, confirming an opportunistic feeding behavior. All the analyses performed showed the presence of a core of shared prey among the size classes and a cloud of size‐specific prey. Our study highlights the exceptional potential of metabarcoding as an approach to provide unprecedented taxonomic resolution in the diet of M. merluccius and potentially of other marine predators, due to the broad‐spectrum of detection of the primers used. A thorough description of these complex trophic relationships is fundamental for the implementation of an ecosystem approach to fisheries.
Keywords: Adriatic Sea, European hake, feeding habits, Merluccius merluccius, metabarcoding
1. INTRODUCTION
Quantitative assessment of food habits is an important aspect of fisheries management as the knowledge of both predator and prey resources can help guide management efforts aimed at increasing fish production. Accurate description of fish diet and feeding habits in aquatic environments (Chipps & Garvey, 2007) in fact provides the basis for a more comprehensive understanding of dynamics of target species by including their trophic interactions (e.g., Angelini et al., 2016; Punt, Ortiz, Aydin, Hunt, & Wiese, 2016), a basic requirement for ecosystem‐based fishery management (Möllmann et al., 2014; Pikitch et al., 2004; Zhou et al., 2010). For instance, the reconstruction of trophic links between marine fishes allows including food interactions into assessments (Punt et al., 2016) or more generally may serve as a basis for setting a balanced exploitation across trophic levels (Garcia, Rice, & Charles, 2014; Garcia et al., 2015), thereby preventing the fishing‐induced trophic level decline (Shackell, Frank, Fisher, Petrie, & Leggett, 2010). Overall, approaches based on food webs can provide a fisheries management advice based on broader and more realistic context than single species approaches (see Link, 2002; Mackinson, Deas, Beveridge, & Casey, 2009; Walters, Christensen, Martell, & Kitchell, 2005). Moreover, the study of feeding habits is necessary and useful to understand mechanisms and processes which structure and influence fish assemblages (Carlucci et al., 2018; Eriksson et al., 2011; Kotrschal & Thomson, 1986).
Nevertheless marine food webs can be extraordinarily complex with a multitude of species connected by a tangled web of predator–prey interactions. In fact omnivory is widespread in the marine environment as species have large spectrum of prey and can have large bathymetric ranges (Carpentieri, Colloca, Cardinale, Belluscio, & Ardizzone, 2005; Polunin & Pinnegar, 2002), and because typically many species undergo to important changes in feeding habits and preferences during ontogenetic growth (Belgrano, 2005). Therefore, although necessary, describing these food web structures is particularly challenging owing to data limitations. The metabarcoding approach can contribute to overcome these limitations by a better identification of trophic links.
Current studies on feeding habits (including stable isotopes), in fact, mainly relied on the morphological identification of prey remains in stomach content: this method is labor‐intensive, time expensive and depends heavily upon the skills of the taxonomist identifying semi‐digested fragments. Moreover, it precludes the identification of foods that leave no hard remains or lack diagnostic taxonomic features; thus, the contribution of some prey to the diet composition might be underestimated or neglected (Baker, Buckland, & Sheaves, 2014; Buckland, Baker, Loneragan, & Sheaves, 2017).
Conversely, recent DNA‐based approaches represent a powerful means in dietary analysis (Kress, García‐Robledo, Uriarte, & Erickson, 2015). Taxon detection from bulk samples can be achieved using PCR amplification followed by massive parallel sequencing of homologous gene regions. These short genomic DNA regions are used like unique species tag (barcodes) for specimen identification (Hebert, Cywinska, Ball, & deWaard, 2003). The obtained sequences are then compared to reference barcodes in public databases to determine similarity for taxonomic identification. This so‐called metabarcoding approach (Taberlet, Coissac, Hajibabaei, & Rieseberg, 2012) has proved to be an effective tool for characterizing the diet of predators (Deagle, Chiaradia, McInnes, & Jarman, 2010; Deagle, Kirkwood, & Jarman, 2009; Murray et al., 2011; Shehzad et al., 2012) and herbivores (Soininen et al., 2009; Valentini et al., 2009) through analysis of their feces or gut content. However, at present, metabarcoding applications in marine food webs are still limited (Albaina, Aguirre, Abad, Santos, & Estonba, 2016; Berry et al., 2015; Leray, Meyer, & Mills, 2015; Leray et al., 2013).
We tested the metabarcoding approach using an important predator species and one of the most economically important demersal fisheries resource for the Mediterranean which is the European hake (Merluccius merluccius, Linnaeus 1758, Figure 1). The European hake is a nektobenthic predator of communities inhabiting the Mediterranean shelf and upper slope, showing a very wide depth range (20–1,000 m) throughout the Mediterranean Sea and the Northeastern Atlantic (Carpentieri et al., 2005). Merluccius merluccius is a predator species of high commercial interest for the Mediterranean fisheries with 20,345 t of catch in 2014 (8,735 t by Italian fisheries, source FAO Regional capture fisheries statistics) and represents one of the main resources for Mediterranean trawl fisheries (summing up to 1.6% of total Mediterranean and Black Sea average catches in the 2000–2013 period; FAO, 2016). According to recent assessments (STECF, 2015), this species is heavily overfished in all northern Mediterranean countries. It is expected that the dynamics of this voracious predator affect other species in the ecosystem through predation control of its prey. In order to know implication of fishing management measures, therefore, it is important to have a good understanding of its food preferences.
Figure 1.

European hake picture. The European hake is a nektobenthic predator of communities inhabiting the Mediterranean shelf and upper slope (image courtesy Stefano Guerrieri).
Although the feeding habits of European hake have been described widely in the Mediterranean (Bozzano, Sardà, & Ríos, 2005; Carpentieri, Colloca, & Ardizzone, 2008; Carpentieri et al., 2005; Cartes, Rey, Lloris, & De Sola, 2004; Froglia, 1973; Papaconstantinou & Caragitsou, 1987; Sartor, Carlini, & De Ranieri, 2003; Stagioni, Montanini, & Vallisneri, 2011; Ungaro, Mannini, & Vrgoč, 2003), they show important differences justified by the opportunistic behavior of this predator. According to these works based on the morphological identification of prey remains in stomachs, adult hakes feed mainly on fish (anchovies, pilchard and gadoid species) and squids whereas the juveniles (<160 mm) feed mainly on crustaceans (preferentially euphausiids and amphipods) in the Mediterranean.
In this study, we set up a metabarcoding approach based on cytochrome oxidase I (COI) PCR amplification of stomach content remains of five size classes of M. merluccius of the Adriatic Sea (Mediterranean Sea). The North‐Central Adriatic Sea is the largest shelf area of the Mediterranean where maximum depth ranges between 75 and 100 m, with the exception of the Pomo/Jabuka Pit (200–260 m). Within the Mediterranean, the Adriatic basin represent an ideal study area because European hake spends its entire life cycle, including the spawning season, in the basin and in the Pomo/Jabuka Pit are located the nursery areas of this species (FAO resource, http://www.faoadriamed.org/italy/html/Species/MerlucciusMerluccius.html#C).
We compared the efficiency of this DNA‐based method to the classical morphological analysis to quantify dietary richness, diet composition, and potential overlap among the size classes. Moreover, using a mock positive control, we evaluated metabarcoding efficiency in species identification and the possible range of OTUs (Operational Taxonomic Units) number for each individual sample.
2. MATERIALS AND METHODS
2.1. Sampling strategy
European hake specimens were collected between 32 and 143 m depth along the coast of the Adriatic Sea (Northeast Mediterranean) from the Gulf of Trieste to Pomo/Jabuka pit (Figure 2 and Supporting Information Table S1) within the framework of International Bottom Trawl Survey in the Mediterranean (MEDITS) cruises during the campaign of the year 2014. Nineteen individuals each of the 5 size class for a total of 95 individuals were selected for the metabarcoding and morphological analyses. The five size classes (size class 1 = TL 120–149 mm, size class 2 = TL 150–199 mm, size class 3 = TL 200–249 mm, size class 4 = TL 250–299 mm, size class 5 = TL ≥300 mm) were defined on the basis of previous results, keeping in mind size distribution by bathymetric and geographical strata, abundance and feeding habits (Stagioni et al., 2011). The stomachs were dissected and preserved in 95% ethanol at −20°C.
Figure 2.
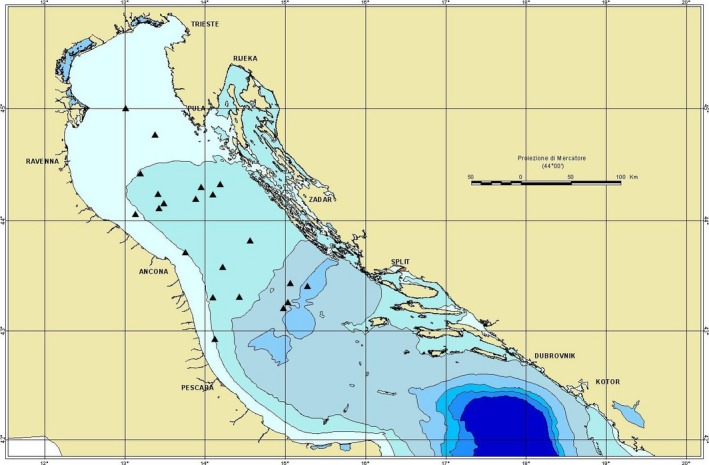
Map of the sampling hauls in the Adriatic Sea. Further details can be found in Supporting Information Table S1. This map was created using ArcViewGIS version 3.2a (https://geonet.esri.com/thread/36365). Image courtesy of Chiara Manfredi. The Adriatic cartography used is freely available at http://www.faoadriamed.org/html/adr_inf_centre.html#cart
Although the direct comparison between metabarcoding and morphological approach was carried out on the 95 stomachs, results from morphological analysis carried on additional 105 hake's stomachs collected in the same area and same campaign were also used for highlighting outperformances of the molecular approach.
2.2. DNA molecular analysis
Merluccius merluccius prey were identified to the lowest possible taxonomic level, counted, and weighed to the nearest 0.1 mg after removal of surface water by blotting paper. After morphological identification, all the content of 95 stomachs was homogenized and 100 μl of the homogenate were used for the molecular analysis. Moreover, some common prey of European hake was collected (Supporting Information Table S2) and approximately 25 mg of muscle was dissected and used for single‐species DNA extraction. These samples were used to set up PCR conditions in order to amplify at least the most common prey of M. merluccius. Furthermore, the obtained DNA was quantified with Qubit fluorometer (dsDNA HS Assay kit; Invitrogen, Supporting Information Table S2), diluted 1:100 and 1 μl of each was used to create a mock positive control (mixture of known species) to be amplified through PCR, sequenced in the same Illumina Miseq lane of the stomach samples and analyzed using the same approach as the stomach contents. All DNAs were extracted using a commercial kit (DNeasy Blood & Tissue Kit, QIAGEN). Samples were processed in small batches representing five size classes of M. merluccius (19 specimens each) with an extraction blank to monitor for potential cross‐contamination in a separated room designated to conduct molecular diet analyses. To avoid cross‐contamination, dissection tools were flame sterilized between individuals and lab surfaces were decontaminated with bleach. The primer pair selected for DNA amplification (mlCOIintF and jgHCO2198, Leray et al., 2013) was analyzed using the ecoPCR software, an in silico PCR program that allows imperfect matches between each barcode primer and its binding site to mimic in vitro PCR (Ficetola et al., 2010). The ecoPCR analysis was performed to evaluate the amplification efficiency on the two major taxonomic groups representing European hake prey (invertebrate and vertebrate subphylum, EMBL database 124 release). PCR amplification was performed in two replicates in a total volume of 25 μl with 0.75 μl of 10 μM of each forward and reverse primers, 0.2 μl of AmpliTaq Gold® DNA Polymerase (ThermoFisher) 5 U/μl, 2 μl of 25 mM Mg2+, 0.5 μl of 10 mM dNTP, 1 mg/ml BSA and 3 μl of genomic DNA. We used a “touchdown” PCR profile (Leray et al., 2013 modified) to minimize the probability of nonspecific amplifications. We carried out 16 initial cycles: denaturation for 10 s at 95°C, annealing for 30 s at 62°C (−1°C per cycle) and extension for 60 s at 72°C, followed by 25 cycles at 46°C annealing temperature (−0.2°C per cycle). All PCRs included no‐template controls, and the products were checked on 1.5% agarose gels. For DNA amplification and library preparation, 20 tagged primers were used (primers Leray et al., 2013 modified). All the tagged amplicons (313‐bp plus tag) were purified with Sera‐Mag SpeedBeads (GE Healthcare Life Sciences) purification protocol (Rohland & Reich, 2012), quantified with Qubit fluorometer and pooled in equimolar concentration. Illumina MiSeq sequencing (2 × 250) was performed by Fasteris SA (Fasteris SA, 1228 Plan‐les‐Ouates, Switzerland) following Metafast protocol (PCR‐free protocol for library preparation) and approximately 17 × 106 paired‐end sequences were obtained.
2.3. Bioinformatic and statistic methods
Sequence demultiplexing, quality control, PCR, and sequencing error filtering were performed using OBITools software (Boyer et al., 2016; http://metabarcoding.org/obitools/doc/welcome.html). The illuminapairedend command was used to perform a micro‐assembly of paired‐end reads. Sequences with Illumina fastq quality scores <30 across the head, tail, or total length of the sequence were discarded. We used ngsfilter command to assign the reads to each sample through barcode identification (14 × 106 sequences). Only the sequences longer than 100 bp were retained and dereplicated using obiuniq command. We further filtered the sequences and those with count <10 were discarded; moreover, the obiclean command were used to detect the potential PCR errors selecting only sequences with the “head” status and abundance higher than 0.05%.
Two different approaches have been used to evaluate the M. merluccius diet composition from the metabarcoding data: (a) sequence occurrence (i.e., presence/absence), (b) OTUs (Operational Taxonomic Units) Relative Abundance (ORA), the proportion of unique OTUs in a sample divided by the final number of OTUs (after bioinformatic processing) in that sample. We used ORA data to evaluate if inferences based on relative abundance differed from those obtained using occurrence data and to provide a proxy of the relative amount of marine organisms in M. merluccius diet. Most of our inferences, however, were based on occurrence data because of the semi‐quantitative nature of metabarcoding analysis (Pompanon et al., 2012; Thomas, Jarman, Haman, Trites, & Deagle, 2014).
For taxonomic assignments, we performed BLASTn (Zhang, Schwartz, Wagner, & Miller, 2000) searches of OTU representative sequences against full GenBank database (November 2015). We used BLAST algorithm optimized for very similar sequences (megablast) on the nucleotide collection (nr/nt) that includes all GenBank + EMBL + DDBJ + PDB sequences restricting the search to sequences with >95% of similarity. Moreover, we accepted a species level match when similarity to the reference barcode was ≥97%. Sample‐based Mao Tau rarefaction curves and nonparametric species richness estimators were computed in EstimateS (Colwell, 2006). Inter‐size class variability was measured using Bray‐Curtis dissimilarities (Oksanen et al., 2016), which range from 0 (complete overlap) to 1 (complete nonoverlap), to compute pairwise community distance matrices and examine differences in beta diversity. Patterns of sample dissimilarity were visualized using PCoA. A non‐parametric analysis of similarity (R‐vegan function anosim; 1,000 Monte Carlo permutations) was used to test the null hypothesis of no difference in species composition among samples. Moreover, to refine this analysis, we performed a permutational (per)MANOVA test that can accommodate both categorical and continuous predictor variables (R‐vegan function adonis, 1,000 permutations). All these analyses were carried out using the vegan package (Oksanen et al., 2016) in R (R Development Core Team, 2015). To further explore relative occurrence data, we applied generalized linear models (GLM) using the mvabund R package. Many commonly used multivariate analyses (e.g., PERMANOVA, ANOSIM, CCA, RDA, etc.), are indeed “distance‐based analyses.” This means the first step of the analysis is to calculate a measure of similarity between each pair of samples, thus converting a multivariate dataset into a univariate one. Their statistical power is very low, except for variables with high variance. GLM do not suffer for this weakness, thus was used to the multivariate hypothesis of whether species composition varied across the size classes using the mvabund package (Wang, Naumann, Wright, & Warton, 2012) in R considering sample sizes as offset (family: negative binomial) and significance was evaluated with an anova test (manyglm; resampling method “montecarlo,” number of bootstrap: 10,000) correcting p‐values for multiple comparisons (p.adjust method).
The food web representation was performed using Gephi software (https://gephi.org), which also contains routines for calculation of basic network indices such as degree (number of links per node) and other measures of centrality used to better represent the web of links (Cherven, 2013).
Indicator species analysis was performed to determine which OTU had significantly different frequency among M. merluccius size‐classes. The analysis was performed using the “signassoc” function in the “indicspecies” R package (Cáceres & Legendre, 2009) on both occurrence and relative abundance (ORA) data. We used mode = 1 (group‐based) and reported p‐values after Sidak's correction for multiple testing. Moreover, the function multipatt was used for determining lists of species that are associated to particular groups of sites (or combinations of those). Prey‐specific abundance (PSA), a function of the percentage of prey items in only those stomachs in which the prey occurs, was calculated according to the following formula
where P i is the prey‐specific abundance of prey i, S i the stomach content (number) comprising prey i and S ti the total stomach content in only those predators with prey i in their stomach (Amundsen, Gabler, & Staldvik, 1996). The PSA index was computed on both ORA data and morphological data considering only prey detected at the species level in both the analyses (namely Alpheus glaber, Engraulis encrasicolus and Solenocera membranacea).
To evaluate if prey abundance in the diet of M. merluccius can be correlated to the abundance of the prey species in the North‐Central Adriatic sampling area, we plotted the number of hake stomachs containing E. encrasicolus vs the abundance of E. encrasicolus estimated during MEDITS 2014 survey for the same hauls.
Illumina DNA sequences obtained during the current study were deposited in the ENA's Sequence Read Archive (http://www.ebi.ac.uk/ena) under the accession number PRJEB22703.
Merluccius merluccius is a commercial species; therefore, neither special permits nor ethics approval were required for their collection, stomach dissection was performed post mortem.
3. RESULTS
The two primers selected for the metabarcoding analysis of stomach contents showed a high coverage of taxa: 80,000 species were amplified and all the potential prey families among taxa were represented with >3,000 species by the in silico ecoPCR assay (Ficetola et al., 2010). Moreover, the in vitro PCR assays performed on tissue DNA of the most common M. merluccius prey (see Section 2 and Supporting Information Table S2) allowed us to set up thermal conditions in order to obtain a good amplification efficiency of all the prey species targeted (Supporting Information Figure S1).
The results obtained from the identification of species in the positive control allowed us to identify all the species present in the mock sample with the exception of Lophogaster typicus which is absent in the GeneBank; moreover, for both Alloteuthis and Sepietta species, we could assign the sequences only at the genus level (Supporting Information Figure S2) because of a lack of differentiation among species within the DNA fragment used. Interestingly we were able to detect also single species with a very low amplification success (Supporting Information Figure S1). Moreover, the number of OTUs assigned to each individual ranged from 1 to 30 with most of the species ranging between 1 and 5 OTUs per individual.
The similarity search analysis of stomach content DNA sequences carried out against the GenBank nucleotide collection (nr/nt), detected 34 prey at the species level, in spite of the only eight species detected by the morphological identification and all the other items were classified at higher taxonomic rank (e.g., Teleostei, Table 1). Moreover, when considering a total of 200 stomachs analyzed using the morphological method, items classified at the species level raised to 11 and 5 were classified only at the genus level while all other items were classified at higher taxonomic rank.
Table 1.
Species identified in M. merluccius stomach contents with both the metabarcoding and morphological approaches
| Identified species | Number of stomachs | |
|---|---|---|
| Molecular results | Morphological results | |
| Allotheutis sp. | 0 | 1 |
| Alpheus glaber | 14 | 7 |
| Anisakis pegreffii | 1 | 0 |
| Arnoglossus sp. | 1 | 0 |
| Chlorotocus crassicornis | 1 | 0 |
| Citharus linguatula | 1 | 0 |
| Decapoda | 0 | 14 |
| Eledone moschata | 1 | 0 |
| Engraulis encrasicolus | 44 | 8 |
| Gaidropsarus mediterraneus | 1 | 0 |
| Gobius niger | 0 | 3 |
| Holothuria forskali | 1 | 0 |
| Illex coindetii | 1 | 0 |
| Lesueurigobius friesii | 14 | 0 |
| Liocarcinus depurator | 1 | 0 |
| Melicertus kerathurus | 1 | 0 |
| Merlangius merlangus | 1 | 0 |
| Microchirus variegatus | 1 | 0 |
| Mullus barbatus | 4 | 0 |
| Mullus surmuletus | 2 | 0 |
| Pagellus acarne | 2 | 0 |
| Pagellus sp. | 0 | 1 |
| Philocheras bispinosus | 4 | 0 |
| Pleurobranchaea meckeli | 1 | 0 |
| Processa modica | 1 | 0 |
| Processa nouveli | 26 | 0 |
| Processa sp. | 0 | 5 |
| Raja miraletus | 2 | 0 |
| Rissoides desmaresti | 0 | 1 |
| Sardina pilchardus | 5 | 0 |
| Scomber colias | 1 | 0 |
| Scophthalmus maximus | 2 | 0 |
| Scorpaena notata | 1 | 0 |
| Sepia officinalis | 1 | 0 |
| Serranus hepatus | 1 | 0 |
| Solenocera membranacea | 24 | 2 |
| Spicara maena | 3 | 0 |
| Trachurus mediterraneus | 2 | 0 |
| Trachurus trachurus | 2 | 0 |
| Teleostei | 0 | 19 |
| Upogebia deltaura | 1 | 0 |
The adequacy of stomach sample sizes (Figure 3a, left) was assessed by generating accumulation curves (with 1,000 random iterations) of species recorded per stomach sample. Because none of the accumulation curves reached a stable plateau, the nonparametric Incidence‐based Coverage Estimator (Foggo, Attrill, Frost, & Rowden, 2003) was used to estimate total dietary richness. The identified species accounted for approximately 53% of the theoretical plateau, that is, the richness estimated at the upper limit of sampling effort. The same analysis performed using the morphological identification results (Figure 3a, right) highlighted an even stronger underestimation of the species richness for all the size classes, showing values of richness ten times lower than the values obtained by using the metabarcoding approach. Only when using a higher number of stomachs for the morphological analysis (a total of 200, Supporting Information Figure S3a), the species richness increased up to 12 only for the larger size, but overall the values were still not comparable to metabarcoding accumulation curve. The ranking order by occurrence obtained with the metabarcoding data of the first 10 species (Figure 3b, left) highlighted that they constitute from 92% (smaller size class) to 70% (larger size class) of the total species identified in the M. merluccius size classes. In particular, one teleost (E. encrasicolus) and 3 decapods (Processa nouveli holthuisi, S. membranacea, and A. glaber) are recurrent in all size classes and constitute >50% of the species identified with the metabarcoding approach. Other species (e.g., Lesueurigobius friesii and Philocheras bispinosus) showed higher frequency or were restricted to only one class (Total Length, TL 120–149 mm). The frequency of other items (out of the 10 most recurrent) is increasing with size indicating an increase in the spectrum of prey. The morphological results (Figure 3b, right) allowed the identification of a very limited spectrum of prey as expected by the accumulation curves and showed an overall lower richness of prey species compared to the metabarcoding approach. The morphological analysis performed using 200 samples showed a higher number of prey detected in comparison with the same analysis performed on 95 stomachs; however, we could appreciate this improvement only when considering also prey identified at the genus level (Supporting Information Figure S3a, i.e., species identified as spp.). Notably, the morphological analyses highlighted a higher prey diversity for the larger size class as for the metabarcoding results.
Figure 3.
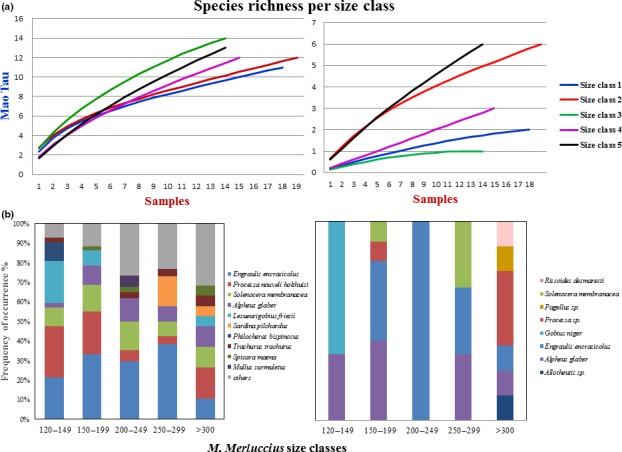
Comparison of dietary richness among M. merluccius size classes. (a) Sample‐based species richness curves for each size class for the metabarcoding data (left) and morphological data (right); (b) Main prey of M. merluccius by size classes as identified by metabarcoding approach (left) and morphological data (right). Frequency occurrence data of species are reported. The 10 most recurrent items across all classes are showed
The OTUs Relative Abundance (ORA) of the three main dietary taxa (Crustacea, Teleostei, Mollusca) across M. merluccius size classes are compared in Figure 4. In general, this analysis highlighted a slight preference for crustaceans for the size classes 2 and 3 (TL 150–199 mm and 200–249, Figure 4a), confirmed the higher abundance of molluscs in the diet of the largest size class (Figure 4b), and a greater mean ORA of teleosts for the smaller size class (TL 120–149 mm, Figure 4c). The Kruskall–Wallis test (Kruskal & Wallis, 1952) did not find any significant comparisons among the ORA of the five size classes for teleosts and crustaceans (p‐value > 0.05), while the number of molluscs families was too low to consider the test reliable.
Figure 4.
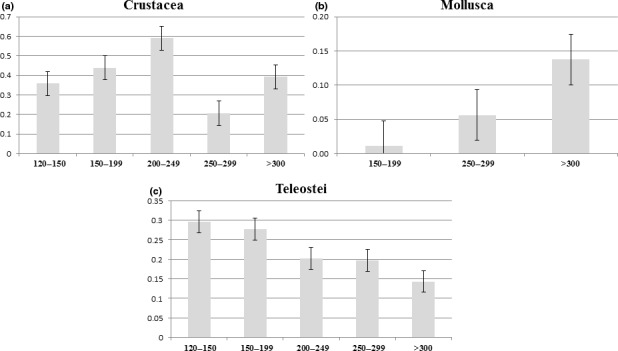
Comparison of mean ORA for each size class. Mean ORA (±SD) per sample for (a) Crustacea, (b) Mollusca and (c) Teleostei. No significant comparison was revealed after Kruskall‐Wallis test (Kruskal & Wallis, 1952) (p‐value = >0.05)
Bray‐Curtis dietary dissimilarities ranged between 0.351(size class TL 150–199 vs. TL 200–249) and 0.616 (size class TL 120–149 vs. TL 250–299) for relative occurrence data and 0.31 (size class TL 150–199 vs. TL 200–249) and 0.70 (size class TL 120–149 vs. TL ≥300) for ORA data. The Wilcoxon signed‐rank test (Wilcoxon, 1945) was not significant suggesting that both indices provided consistent measures with regard to dietary niche partitioning (Table 2). In general, the dissimilarities showed intermediate values suggesting a partial dietary overlap among the five size classes of M. merluccius.
Table 2.
Inter‐size class variability measured as Bray‐Curtis dissimilarities calculated using occurrence‐ (below diagonal) and ORA (above diagonal)‐based data
| M. merluccius classes | Class = 120–149 | Class = 150–199 | Class = 200–249 | Class = 250–299 | Class ≥ 300 |
|---|---|---|---|---|---|
| Class = 120–149 | 0.359 | 0.411 | 0.49 | 0.702 | |
| Class = 150–199 | 0.404 | 0.312 | 0.38 | 0.624 | |
| Class = 200–249 | 0.586 | 0.351 | 0.438 | 0.623 | |
| Class = 250–299 | 0.616 | 0.456 | 0.392 | 0.642 | |
| Class ≥ 300 | 0.613 | 0.483 | 0.498 | 0.517 |
The principal component analysis (PCoA) plot, based on the Bray‐Curtis distances computed on the relative occurrence data (Figure 5), showed a significant (permutest p‐value = 0.017) partial clustering among the size classes and a moderate differentiation of size classes 2 and 5 (p‐value = 0.001) and classes 2 and 4 (p‐value = 0.027). This result suggested a general homogeneity of variance within the size classes also supported by the anosim analysis (R 2 = 0.22, p‐value = 0.00099). The low although significant value of the determination coefficient suggested a lack of discrimination between groups. Similarly the permutational (per)MANOVA analysis (R 2 = 0.19, p‐value = 0.00099) showed that only 19% of variance was explained by the tested groupings.
Figure 5.
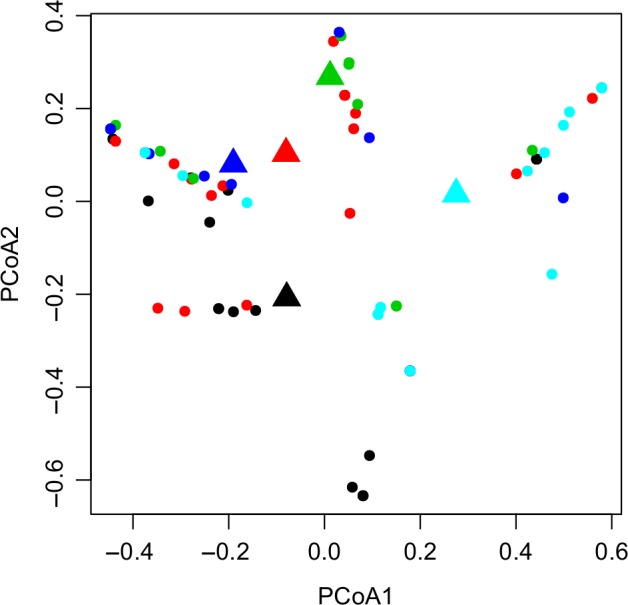
Principal Component Analysis. PCoA of relative occurrence‐based Bray‐Curtis dissimilarity of samples from all size classes (permutest p‐value = 0.017). Triangles depict the centroids of the distributions. Black color: size class 1, red: size class 2, green: size class 3, blue: size class 4, light blue: size class 5
The application of GLM to test the multivariate hypothesis of whether species composition varied across classes resulted strongly significant (Likelihood Ratio Test = 180.5 p‐value = <2e−16) highlighting the presence of an effect of groups on species composition.
The food web network (Figure 6) allowed us to identify the number of prey shared by all the size classes and highlighted a high number of size‐specific prey. In spite of the similarities identified for the relative occurrences of the highest taxonomic class (Crustacea, Mollusca, Teleostei), the Bray‐Curtis dissimilarities showed differentiations among size classes and the GLM analysis clearly detected an effect of groups. In particular, the web of trophic interactions derived from metabarcoding approach showed that prey species shared by all size classes are E. encrasicolus, P. nouveli holthuisi, S. membranacea, A. glaber (Figure 6, but also Figure 3b). Other prey species, such as Sardina pilchardus and L. friesii, for example, tend to be preferred by large and small M. merluccius individuals, respectively. Moreover, the network highlighted the presence of clouds of size‐specific prey species which were detected only in one size class, displaying an extremely high complexity of the trophic interaction of European hake in the North‐Central Adriatic.
Figure 6.
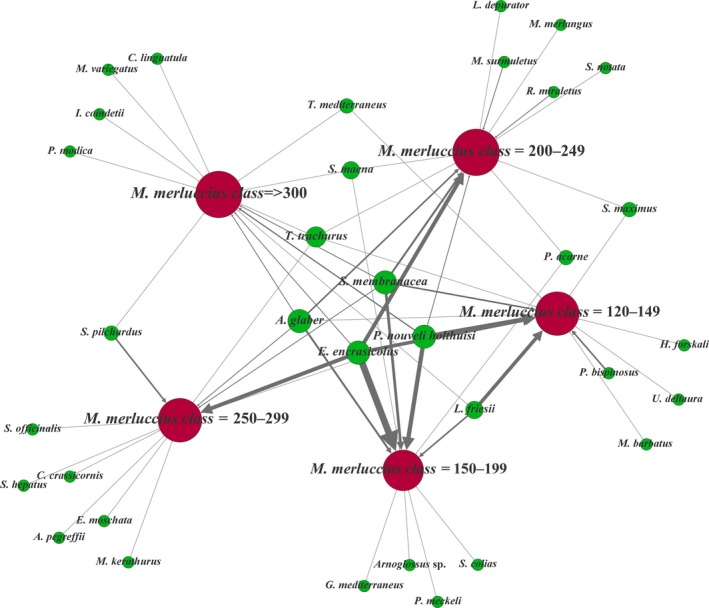
The food web related to the predator M. merluccius. Brown nodes indicate the predator divided into five size classes and green nodes represent the prey. The size of nodes is proportional to number of links connected (degree), and the size of links is proportional to the number of times the link prey–predator was found in the samples. Species are distributed according to their linkage with predator size classes: the prey species common to all size classes are in the middle
Both the multipatt and the signassoc functions performed on ORA data for identifying indicator species revealed a lower number of significant associations in comparison with sequence relative occurrence data (Supporting Information Tables S3 and S4) indicating that the relative abundance of OTUs provided the more conservative measure for our indicator‐species analysis. The multipatt function identified two species (L. friesii and P. bispinosus) significantly associated to the size class 1 (TL 120–149 mm) with high specificity and low sensitivity (Supporting Information Table S4) corroborating the food web network result (Figure 6). Moreover, E. encrasicolus showed a significant association with high specificity and sensitivity but this association concerned four hake size classes out of five suggesting that this prey is ubiquitous in the diet composition at least for the habitat and geographical area under study. The signassoc function confirmed this result also after correcting for multiple testing and highlighted the higher frequency of L. friesii and P. bispinosus in the size class 1 (Supporting Information Table S3).
The diet of M. merluccius in the North‐Central Adriatic did not show any evidence for specialization using prey‐specific abundance index (Amundsen et al., 1996; PSA < 0.5; Figure 7) computed both on metabarcoding and morphological data, and the highest value of PSA is obtained for A. glaber (0.37). This analysis showed a good agreement of PSA values obtained with the two methods of taxonomic identification and the relationship of PSA and frequency of occurrence suggested a broad niche width and low specialization for M. merluccius.
Figure 7.
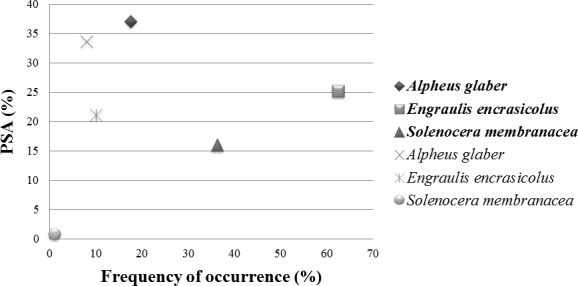
Feeding strategy diagram. Prey‐specific abundance (PSA) plotted against frequency of occurrence of prey. Only the species identified with both the metabarcoding and morphological analyses were considered. In bold the species found with the metabarcoding approach
Finally we found a good correlation between the number of stomachs containing E. encrasicolus and the estimated abundance of this species in the same hauls obtained using MEDITS 2014 survey data (Figure 8), confirming the opportunistic feeding strategy of M. merluccius.
Figure 8.
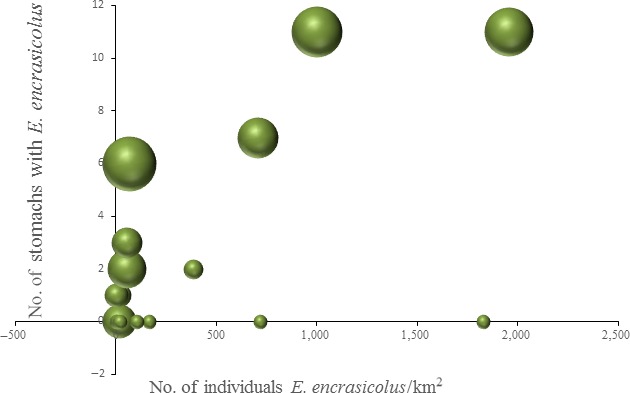
Prey–predator functional relationship. Number of M. merluccius stomachs containing E. encrasicolus in relation to the abundance of E. encrasicolus estimated for the same hauls (data from MEDITS 2014 survey). The bubbles size is proportional to the number of M. merluccius stomach data (specimen, e.g., number of individuals) available per haul
4. DISCUSSION
In this study, we developed a metabarcoding method based on COI PCR amplification to evaluate its efficiency for the analysis of European hake diet and to increase our knowledge about its feeding habits and trophic relationships. The comparison of the molecular and morphological results clearly showed that the metabarcoding approach consistently detected a wider spectrum of prey species than classical approach, providing a thorough description of M. merluccius diet and trophic links.
The wide range of habitat and the different biocenosis sampled allowed us to characterize M. merluccius overall feeding strategy in the North‐Central Adriatic, that revealed to be very diverse across sizes and sites. Unfortunately our sample did not include the Adriatic nursery area of Pomo/Jabuka pit, were the youngest individuals (TL < 120 mm, size class 0) usually live during the juvenile phase, preventing the detection of the characteristic ontogenetic shift in diet (Carpentieri et al., 2005). Juvenile hakes (TL < 120 mm) are almost restricted to this area as a consequence of their limited mobility and usually their diet is quite different from adults because of their smaller size and the different environment inhabited (between 100 and 200 m depth). Metabarcoding results in terms of richness, however, highlighted that the rarefaction curve did not reach the theoretical plateau, suggesting the need of a higher number of individuals to be sampled in future analyses. Nevertheless, these preliminary results provide a general description of the trophic preferences of this voracious predator within the North‐Central Adriatic basin and, particularly, highlighted the higher efficiency of the DNA‐based method in detecting prey compared to the classical morphological approach. For instance, metabarcoding outperformed the morphological method in identifying prey not only on the same sample size, but even when using a number of stomachs as high as 200. Furthermore, our metabarcoding results on 95 individuals allowed the identification of 34 species in the diet getting closer to a previous extensive study on European hake diet (Carpentieri et al., 2005) which allowed the identification of 46 prey species on the basis of the morphological analysis of a very high number of hake stomachs (2761). These considerations highlight the much higher efficiency of the metabarcoding approach, especially because we used a very conservative approach for the assignment of OTUs considering that only OTUs with a similarity ≥97% were assigned with confidence to a species and rejecting sequences assigned only at the genus level.
Unexpectedly our analysis highlighted significant statistical differences also among the larger size classes (>120 mm) suggesting a general opportunistic feeding behavior and the presence of some kind of differentiation when considering the single prey species as highlighted by the food network analysis. Overall our metabarcoding results describe a diet based mainly on crustaceans and teleosts with a slightly higher abundance of molluscs detected in the M. merluccius of the larger size. The low number of stomachs containing molluscs, notably cephalopods, can be attributed to the low depth of the sampling sites (<150 m), and the sub‐area of recruitment (North‐Central Adriatic) as these factors can affect the variation of the abundance of these species (Krstulovic Sifner et al., 2005). Despite an overall homogeneous composition of M. merluccius diet, there were some indicator species that were distinctive in the size class formed by the youngest individuals (TL 120–149 mm), namely L. friesii and P. bispinosus. Moreover, a clear expansion of the spectrum of prey was visible when the size of individuals increases, suggesting a reduced selectivity of the largest M. merluccius. On the other side, the largest size showed a clear decrease of teleosts in diet and a sensible increase in crustaceans and molluscs that is coherent with other results based on larger samples (Carpentieri et al., 2005) but is not clearly highlighted by our morphological data.
Interestingly, measures of dietary dissimilarities obtained with both relative occurrence and ORA data were comparable suggesting a good performance of relative abundances in describing sample diversity. Indeed our mock sample showed that each individual can be represented by a limited number of OTUs and, with the exception of few species showing more than ten OTUs per individuals (Supporting Information Figure S2), all the others were represented by a very low number of OTUs. The identification of a cloud of OTUs for each individual can be associated with both the presence of pseudogenes/intra‐individual polymorphisms that are intrinsic features of the biological complexity of genomes, and sequencing artifacts that are a well‐known limit of high‐throughput sequencing and that can be overcome using rigorous methods of analysis. The PSA analysis showed a good concordance of values obtained with both the metabarcoding and morphological methods, suggesting that, despite the semi‐quantitative nature of metabarcoding analysis of stomach content, this technique can indeed describe faithfully the diet of European hake.
In addition, the high frequency of occurrence detected for anchovy can be related to a high number of anchovies present in the area during the campaign or a high species abundance in the sampled area.
Remarkably the plot of the number of stomachs containing E. encrasicolus and the abundance obtained from MEDITS 2014 survey corroborated this evidence, showing a good concordance between the presence in the diet and the abundance of this species in the sampling area. The relationship found resemble the typical prey–predator functional response (Holling, 1959) that is largely applied in trophic ecology suggesting also the potential semi‐quantitative use of the metabarcoding results in dynamic trophic models (see, e.g., Angelini et al., 2016). Our analyses were able to describe in detail the diet of European hake, and the comparison with the classical method showed that the diet detail gained with metabarcoding approach was impossible to reproduce with the morphological data obtained from the same samples. The metabarcoding approach presented here is thus very promising for a faithful description of the food network, which is a crucial task in the context of fisheries management. There is evidence, in fact, that the increase/decrease in key predators that are often targets for exploitation can have strong effects on prey and on the whole ecosystem (Baum & Worm, 2009; Heithaus, Frid, Wirsing, & Worm, 2008; Worm & Myers, 2003). The predatory effects can propagate down to the primary producers of the food web in the so called “trophic cascade,” with possible impact on species that has fundamental role in maintaining the ecosystem functionality (e.g., Myers, Baum, Shepherd, Powers, & Peterson, 2007). Several works (Mackinson et al., 2009; Stäbler et al., 2016; Walters et al., 2005) highlighted that effects of fisheries management can propagate through the food web with possible important unexpected feedbacks and thus optimal management requires a better disentanglement of trophic interactions, especially in the case of mixed fisheries. Furthermore, the removal of keystone predators causes a loss of species diversity at trophic levels lower in the food web (Paine, 1966); therefore, the knowledge of the food chain of predators of commercial interest is decisive for a sustainable management of fisheries.
Although DNA molecular data are unable, so far, to provide information about volumes or weights of ingested prey, here we showed that the metabarcoding approach can provide a new complementary basis to morphological and stable isotope approaches for further improvement of actual knowledge on feeding preferences.
In conclusion, although still preliminary our study highlights the exceptional potential of metabarcoding as an approach to provide unprecedented taxonomic resolution in the diet of M. merluccius. These data represent an important basis to reconstruct marine food webs and provide crucial insights for a sustainable management of this precious fishery resource.
CONFLICT OF INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
G.R., S.L., C.P. conceived the project and designed research; G.R. and M.S. performed research; G.R. and S.L. analyzed data; G.R. and S.L. wrote the paper; C.P. provided laboratory space, instrumentation and funding; S.L provided funding. All authors reviewed and approved the final manuscript.
DATA ACCESSIBILITY
Illumina DNA sequences obtained during the current study were deposited in the ENA's Sequence Read Archive (http://www.ebi.ac.uk/ena) under the accession number PRJEB22703.
Supporting information
ACKNOWLEDGMENTS
The authors thank the Italian Flagship Project RITMARE – The Italian Research for the Sea (http://www.ritmare.it) coordinated by the Italian National Research Council and funded by the Italian Ministry of Education, University and Research within the National Research Program 2011–2013 for supporting this work carried out within the action “SP2‐WP2‐AZ3—Communities and food web modeling.” This work was also supported by the MEDITS (International Bottom Trawl Survey in the Mediterranean) project.
Riccioni G, Stagioni M, Piccinetti C, Libralato S. A metabarcoding approach for the feeding habits of European hake in the Adriatic Sea. Ecol Evol. 2018;8:10435–10447. 10.1002/ece3.4500
REFERENCES
- Albaina, A. , Aguirre, M. , Abad, D. , Santos, M. , & Estonba, A. (2016). 18S rRNA V9 metabarcoding for diet characterization: A critical evaluation with two sympatric zooplanktivorous fish species. Ecology and Evolution, 6(6), 1809–1824. 10.1002/ece3.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen, P. , Gabler, H. , & Staldvik, F. J. (1996). A new approach to graphical analysis of feeding strategy from stomach contents data—modification of the Costello (1990) method. Journal of Fish Biology, 48(4), 607–614. [Google Scholar]
- Angelini, S. , Hillary, R. , Morello, E. B. , Plagányi, É. E. , Martinelli, M. , Manfredi, C. , … Santojanni, A. (2016). An ecosystem model of intermediate complexity to test management options for fisheries: A case study. Ecological Modelling, 319, 218–232. https://doi.org/doi\10.1016/j.ecolmodel.2015.07.031 [Google Scholar]
- Baker, R. , Buckland, A. , & Sheaves, M. (2014). Fish gut content analysis: Robust measures of diet composition. Fish and Fisheries, 15(1), 170–177. 10.1111/faf.12026 [DOI] [Google Scholar]
- Baum, J. K. , & Worm, B. (2009). Cascading top‐down effects of changing oceanic predator abundances. Journal of Animal Ecology, 78(4), 699–714. 10.1111/j.1365-2656.2009.01531.x [DOI] [PubMed] [Google Scholar]
- Belgrano, A. (2005). Aquatic food webs: An ecosystem approach. Oxford, UK: Oxford University Press; 10.1093/acprof:oso/9780198564836.001.0001 [DOI] [Google Scholar]
- Berry, O. , Bulman, C. , Bunce, M. , Coghlan, M. , Murray, D. C. , & Ward, R. D. (2015). Comparison of morphological and DNA metabarcoding analyses of diets in exploited marine fishes. Marine Ecology Progress Series, 540, 167–181. 10.3354/meps11524 [DOI] [Google Scholar]
- Boyer, F. , Mercier, C. , Bonin, A. , Le Bras, Y. , Taberlet, P. , & Coissac, E. (2016). obitools: A unix‐inspired software package for DNA metabarcoding. Molecular Ecology Resources, 16(1), 176–182. 10.1111/1755-0998.12428 [DOI] [PubMed] [Google Scholar]
- Bozzano, A. , Sardà, F. , & Ríos, J. (2005). Vertical distribution and feeding patterns of the juvenile European hake, Merluccius merluccius in the NW Mediterranean. Fisheries Research, 73(1), 29–36. 10.1016/j.fishres.2005.01.006 [DOI] [Google Scholar]
- Buckland, A. , Baker, R. , Loneragan, N. , & Sheaves, M. (2017). Standardising fish stomach content analysis: The importance of prey condition. Fisheries Research, 196, 126–140. 10.1016/j.fishres.2017.08.003 [DOI] [Google Scholar]
- Cáceres, M. De , & Legendre, P. (2009). Associations between species and groups of sites: Indices and statistical inference. Ecology, 90(12), 3566–3574. 10.1890/08-1823.1 [DOI] [PubMed] [Google Scholar]
- Carlucci, R. , Bandelj, V. , Ricci, P. , Capezzuto, F. , Sion, L. , Maiorano, P. , … Libralato, S. (2018). Exploring spatio‐temporal changes in the demersal and benthopelagic assemblages of the north‐western Ionian Sea (central Mediterranean Sea). Marine Ecology Progress Series, 598, 1–19. 10.3354/meps12613 [DOI] [Google Scholar]
- Carpentieri, P. , Colloca, F. , & Ardizzone, G. (2008). Daily ration and feeding activity of juvenile hake in the central Mediterranean Sea. Journal of the Marine Biological Association of the United Kingdom, 88(7), 1493–1501. 10.1017/S0025315408001859 [DOI] [Google Scholar]
- Carpentieri, P. , Colloca, F. , Cardinale, M. , Belluscio, A. , & Ardizzone, G. D. (2005). Feeding habits of European hake (Merluccius merluccius) in the central Mediterranean Sea. Fishery Bulletin, 103(2), 411–416. [Google Scholar]
- Cartes, J. E. , Rey, J. , Lloris, D. , & De Sola, L. G. (2004). Influence of environmental variables on the feeding and diet of European hake (Merluccius merluccius) on the Mediterranean Iberian coasts. Journal of the Marine Biological Association of the United Kingdom, 84(4), 831–835. 10.1017/S0025315404010021h [DOI] [Google Scholar]
- Cherven, K. (2013). Network graph analysis and visualization with Gephi. Birmingham, UK: Packt Publishing Ltd. [Google Scholar]
- Chipps, S. R. , & Garvey, J. E. (2007). Assessment of food habits and feeding patterns In Guy C. S. & Brown M. L. (Eds.), Analysis and interpretation of freshwater fisheries data, chapter: Assessment of diets and feeding patterns (pp. 473–514). Bethesda, MD: American Fisheries Society. [Google Scholar]
- Colwell, R. K. (2006). ESTIMATES, Version 8.0: statistical estimation of species richness and shared species from samples (Software and User’s Guide). Freeware for Windows and Mac OS.
- Deagle, B. E. , Chiaradia, A. , McInnes, J. , & Jarman, S. N. (2010). Pyrosequencing faecal DNA to determine diet of little penguins: Is what goes in what comes out? Conservation Genetics, 11(5), 2039–2048. 10.1007/s10592-010-0096-6 [DOI] [Google Scholar]
- Deagle, B. E. , Kirkwood, R. , & Jarman, S. N. (2009). Analysis of Australian fur seal diet by pyrosequencing prey DNA in faeces. Molecular Ecology, 18(9), 2022–2038. 10.1111/j.1365-294X.2009.04158.x [DOI] [PubMed] [Google Scholar]
- Eriksson, B. K. , Sieben, K. , Eklöf, J. , Ljunggren, L. , Olsson, J. , Casini, M. , & Bergström, U. (2011). Effects of altered offshore food webs on coastal ecosystems emphasize the need for cross‐ecosystem management. Ambio, 40(7), 786–797. 10.1007/s13280-011-0158-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2016). The state of the Mediterranean and Black Sea fisheries 2016. General Fisheries Commission for the Mediterranean, 10.1163/156853010X510807 [DOI] [Google Scholar]
- Ficetola, G. F. , Coissac, E. , Zundel, S. , Riaz, T. , Shehzad, W. , Bessière, J. , … Pompanon, F. (2010). An in silico approach for the evaluation of DNA barcodes. BMC Genomics, 11, 434 10.1186/1471-2164-11-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foggo, A. , Attrill, M. J. , Frost, M. T. , & Rowden, A. A. (2003). Estimating marine species richness: An evaluation of six extrapolative techniques. Marine Ecology Progress Series, 248, 15–26. 10.3354/meps248015 [DOI] [Google Scholar]
- Froglia, C. (1973). Osservazioni sull'alimentazione del merluzzo (Merluccius merluccius L.) del Medio Adriatico In Nardò S. (Ed.), Atti V Congresso Nazionale della Societá Italiana di Biologia Marina (Vol. 5, pp. 327–341). [Google Scholar]
- Garcia, S. M. , Bianchi, G. , Charles, A. , Kolding, J. , Rice, J. , Rochet, M.‐J. , … Van Zwieten, P. A. M. (2015). Balanced harvest in the real world. Scientific, policy and operational issues in an ecosystem approach to fisheries. Gland, Switzerland: IUCN. [Google Scholar]
- Garcia, S. M. , Rice, J. , & Charles, A. (2014). Governance of marine fisheries and biodiversity conservation: Interaction and co‐evolution. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- Hebert, P. D. N. , Cywinska, A. , Ball, S. L. , & deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society of London Series B: Biological Sciences, 270(1512), 313–321. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithaus, M. R. , Frid, A. , Wirsing, A. J. , & Worm, B. (2008). Predicting ecological consequences of marine top predator declines. Trends in Ecology & Evolution, 23(4), 202–210. https://doi.org/doi\10.1016/j.tree.2008.01.003 [DOI] [PubMed] [Google Scholar]
- Holling, C. S. (1959). The components of predation as revealed by a study of small‐mammal predation of the European Pine Sawfly1. The Canadian Entomologist, 91(5), 293–320. 10.4039/Ent91293-5 [DOI] [Google Scholar]
- Kotrschal, K. , & Thomson, D. A. (1986). Feeding patterns in eastern tropical Pacific blennioid fishes (Teleostei: Tripterygiidae, Labrisomidae, Chaenopsidae, Blenniidae). Oecologia, 70(3), 367–378. 10.1007/BF00379499 [DOI] [PubMed] [Google Scholar]
- Kress, W. J. , García‐Robledo, C. , Uriarte, M. , & Erickson, D. L. (2015). DNA barcodes for ecology, evolution, and conservation. Trends in Ecology & Evolution, 30(1), 25–35. https://doi.org/doi\10.1016/j.tree.2014.10.008 [DOI] [PubMed] [Google Scholar]
- Krstulovic Sifner, S. , Lefkaditou, E. , Ungaro, N. , Ceriola, L. , Osmani, K. , Kavadas, S. , & Vrgoca, N. (2005). Composition and distribution of the cephalopod fauna in the eastern Adriatic and eastern Ionian Sea. Israel Journal of Zoology, 51(4), 315–330. 10.1560/4LT4-K01W-C9GF-7YK3 [DOI] [Google Scholar]
- Kruskal, W. H. , & Wallis, W. A. (1952). Use of ranks in one‐criterion variance analysis. Journal of the American Statistical Association, 47(260), 583–621. 10.1080/01621459.1952.10483441 [DOI] [Google Scholar]
- Leray, M. , Meyer, C. P. , & Mills, S. C. (2015). Metabarcoding dietary analysis of coral dwelling predatory fish demonstrates the minor contribution of coral mutualists to their highly partitioned, generalist diet. PeerJ, 3, e1047 10.7717/peerj.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leray, M. , Yang, J. Y. , Meyer, C. P. , Mills, S. C. , Agudelo, N. , Ranwez, V. , … Machida, R. J. (2013). A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Frontiers in Zoology, 10(1), 34 10.1186/1742-9994-10-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, J. S. (2002). What does ecosystem‐based fisheries management mean. Fisheries, 27(4), 18–21. [Google Scholar]
- Mackinson, S. , Deas, B. , Beveridge, D. , & Casey, J. (2009). Mixed‐fishery or ecosystem conundrum? Multispecies considerations inform thinking on long‐term management of North Sea demersal stocks. Canadian Journal of Fisheries and Aquatic Sciences, 66(7), 1107–1129. 10.1139/F09-057 [DOI] [Google Scholar]
- Möllmann, C. , Lindegren, M. , Blenckner, T. , Bergström, L. , Casini, M. , Diekmann, R. , … Gårdmark, A. (2014). Implementing ecosystem‐based fisheries management: From single‐species to integrated ecosystem assessment and advice for Baltic Sea fish stocks. ICES Journal of Marine Science, 71(5), 1187–1197. 10.1093/icesjms/fst123 [DOI] [Google Scholar]
- Murray, D. C. , Bunce, M. , Cannell, B. L. , Oliver, R. , Houston, J. , White, N. E. , … Haile, J. (2011). DNA‐based faecal dietary analysis: A comparison of qPCR and high throughput sequencing approaches. PLoS ONE, 6(10), e25776 10.1371/journal.pone.0025776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, R. A. , Baum, J. K. , Shepherd, T. D. , Powers, S. P. , & Peterson, C. H. (2007). Cascading effects of the loss of apex predatory sharks from a Coastal Ocean. Science, 315(5820), 1846–1850. 10.1126/science.1138657 [DOI] [PubMed] [Google Scholar]
- Oksanen, J. , Blanchet, F. G. , Kindt, R. , Legendre, P. , Minchin, P. R. , O’hara, R. B. , & Wagner, H. (2016). vegan: community ecology package R package version 2.0–7. 2013. URL http://CRAN.R-project.org/package=vegan.
- Paine, R. T. (1966). Food web complexity and species diversity. The American Naturalist, 100(910), 65–75. 10.1086/282400 [DOI] [Google Scholar]
- Papaconstantinou, C. , & Caragitsou, E. (1987). The food of hake (Merluccius merluccius) in Greek Seas. Vie et Milieu, 37(2), 77–83. [Google Scholar]
- Pikitch, E. , Santora, C. , Babcock, E. A. , Bakun, A. , Bonfil, R. , Conover, D. O. , … Heneman, B. (2004). Ecosystem‐based fishery management. Science, 305(5682), 346–347. 10.1126/science.1098222 [DOI] [PubMed] [Google Scholar]
- Polunin, N. V. C. , & Pinnegar, J. K. (2002). Trophic ecology and the structure of marine food webs In Reynolds D. J., & Hart P. J. B. (Eds.), Handbook of fish biology and fisheries (pp. 301–320). Hoboken, NJ: Blackwell Publishing Ltd; 10.1002/9780470693803 [DOI] [Google Scholar]
- Pompanon, F. , Deagle, B. E. , Symondson, W. O. C. , Brown, D. S. , Jarman, S. N. , & Taberlet, P. (2012). Who is eating what: Diet assessment using next generation sequencing. Molecular Ecology, 21(8), 1931–1950. 10.1111/j.1365-294X.2011.05403.x [DOI] [PubMed] [Google Scholar]
- Punt, A. E. , Ortiz, I. , Aydin, K. Y. , Hunt, G. L. Jr , & Wiese, F. K. (2016). End‐to‐end modeling as part of an integrated research program in the Bering Sea. Deep Sea Research Part II: Topical Studies in Oceanography, 134, 413–423. https://doi.org/doi\10.1016/j.dsr2.2015.04.018 [Google Scholar]
- R Core Team (2015). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; Retrieved from https://www.r-project.org/ [Google Scholar]
- Rohland, N. , & Reich, D. (2012). Cost‐effective, high‐throughput DNA sequencing libraries for multiplexed target capture. Genome Research, 22(5), 939–946. 10.1101/gr.128124.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor, P. , Carlini, F. , & De Ranieri, S. (2003). Diet of young European hake (Merluccius merluccius) in the Northern Tyrrhenian Sea. Genova, Italy: Società italiana di biologia marina. [Google Scholar]
- Shackell, N. L. , Frank, K. T. , Fisher, J. A. D. , Petrie, B. , & Leggett, W. C. (2010). Decline in top predator body size and changing climate alter trophic structure in an oceanic ecosystem. Proceedings of the Royal Society of London B: Biological Sciences, 277, 1353–1360. 10.1098/rspb.2009.1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad, W. , McCarthy, T. M. , Pompanon, F. , Purevjav, L. , Coissac, E. , Riaz, T. , & Taberlet, P. (2012). Prey preference of snow leopard (Panthera uncia) in South Gobi, Mongolia. PLoS ONE, 7(2), e32104 10.1371/journal.pone.0032104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soininen, E. M. , Valentini, A. , Coissac, E. , Miquel, C. , Gielly, L. , Brochmann, C. , … Yoccoz, N. G. (2009). Analysing diet of small herbivores: The efficiency of DNA barcoding coupled with high‐throughput pyrosequencing for deciphering the composition of complex plant mixtures. Frontiers in Zoology, 6(1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stäbler, M. , Kempf, A. , Mackinson, S. , Poos, J. J. , Garcia, C. , & Temming, A. (2016). Combining efforts to make maximum sustainable yields and good environmental status match in a food‐web model of the southern North Sea. Ecological Modelling, 331, 17–30. 10.1016/j.ecolmodel.2016.01.020 [DOI] [Google Scholar]
- Stagioni, M. , Montanini, S. , & Vallisneri, M. (2011). Feeding Habits of European Hake, Merluccius Merluccius (Actinopterygii: Gadiformes: Merlucciidae), from the Northeastern Mediterranean Sea. Acta Ichthyologica Et Piscatoria, 41(4), 277–284. 10.3750/AIP2011.41.4.03 [DOI] [Google Scholar]
- STECF (2015). Scientific, Technical and Economic Committee for Fisheries (STECF) – Mediterranean Assessments part 1 (STECF 15–18), Vol. 1 STECF. [Google Scholar]
- Taberlet, P. , Coissac, E. , Hajibabaei, M. , & Rieseberg, L. H. (2012). Environmental DNA. Molecular Ecology, 21, 1789–1793. 10.1111/j.1365-294X.2012.05542.x [DOI] [PubMed] [Google Scholar]
- Thomas, A. C. , Jarman, S. N. , Haman, K. H. , Trites, A. W. , & Deagle, B. E. (2014). Improving accuracy of DNA diet estimates using food tissue control materials and an evaluation of proxies for digestion bias. Molecular Ecology, 23(15), 3706–3718. 10.1111/mec.12523 [DOI] [PubMed] [Google Scholar]
- Ungaro, N. , Mannini, P. , & Vrgoč, N. (2003). The biology and stock assessment of Merluccius merluccius in the Adriatic Sea: An historical review by geographical subareas. Acta Adriatica, 44(1), 9–20. [Google Scholar]
- Valentini, A. , Miquel, C. , Nawaz, M. A. , Bellemain, E. V. A. , Coissac, E. , Pompanon, F. , … Wincker, P. (2009). New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: The trnL approach. Molecular Ecology Resources, 9(1), 51–60. 10.1111/j.1755-0998.2008.02352.x [DOI] [PubMed] [Google Scholar]
- Walters, C. J. , Christensen, V. , Martell, S. J. , & Kitchell, J. F. (2005). Possible ecosystem impacts of applying MSY policies from single‐species assessment. ICES Journal of Marine Science, 62(3), 558–568. 10.1016/j.icesjms.2004.12.005 [DOI] [Google Scholar]
- Wang, Y. , Naumann, U. , Wright, S. T. , & Warton, D. I. (2012). Mvabund – An R package for model‐based analysis of multivariate abundance data. Methods in Ecology and Evolution, 3(3), 471–474. 10.1111/j.2041-210X.2012.00190.x [DOI] [Google Scholar]
- Wilcoxon, F. (1945). Individual comparisons by ranking methods. Biometrics Bulletin, 1(6), 80–83. 10.2307/3001968 [DOI] [Google Scholar]
- Worm, B. , & Myers, R. A. (2003). Meta‐analysis of cod‐shrimp interactions reveals top‐down control in oceanic food webs. Ecology, 84(1), 162–173. 10.1890/0012-9658(2003)084[0162:MAOCSI]2.0.CO;2 [DOI] [Google Scholar]
- Zhang, Z. , Schwartz, S. , Wagner, L. , & Miller, W. (2000). A greedy algorithm for aligning DNA sequences. Journal of Computational Biology, 7(1–2), 203–214. 10.1089/10665270050081478 [DOI] [PubMed] [Google Scholar]
- Zhou, S. , Smith, A. D. M. , Punt, A. E. , Richardson, A. J. , Gibbs, M. , Fulton, E. , … Sainsbury, K. (2010). Ecosystem‐based fisheries management requires a change to the selective fishing philosophy. Proceedings of the National Academy of Sciences of the United States of America, 107(21), 9485–9489. 10.1073/pnas.0912771107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Illumina DNA sequences obtained during the current study were deposited in the ENA's Sequence Read Archive (http://www.ebi.ac.uk/ena) under the accession number PRJEB22703.


