Abstract
The contribution of basal cellular processes to the regulation of tissue homeostasis has just started to be appreciated. However, our knowledge of the modulation of ribosome biogenesis activity in situ within specific lineages remains very limited. This is largely due to the lack of assays that enable quantitation of ribosome biogenesis in small numbers of cells in vivo. We used a technique, named Flow-FISH, combining cell surface antibody staining and flow cytometry with intracellular ribosomal RNA (rRNA) FISH, to measure the levels of pre-rRNAs of hematopoietic cells in vivo. Here, we show that Flow-FISH reports and quantifies ribosome biogenesis activity in hematopoietic cell populations, thereby providing original data on this fundamental process notably in rare populations such as hematopoietic stem and progenitor cells. We unravel variations in pre-rRNA levels between different hematopoietic progenitor compartments and during erythroid differentiation. In particular, our data indicate that, contrary to what may be anticipated from their quiescent state, hematopoietic stem cells have significant ribosome biogenesis activity. Moreover, variations in pre-rRNA levels do not correlate with proliferation rates, suggesting that cell type-specific mechanisms might regulate ribosome biogenesis in hematopoietic stem cells and progenitors. Our study contributes to a better understanding of the cellular physiology of the hematopoietic system in vivo in unperturbed situations.
Keywords: hematopoietic stem cell, ribosome biogenesis, rRNA maturation
INTRODUCTION
Continuous blood cells production over the lifetime of the organism requires the coordinated activity of hematopoietic stem cells (HSCs) and progenitor cells (Orkin and Zon 2008). Tight regulation of renewal, proliferation, and differentiation of the different populations is key during both steady-state and stress hematopoiesis and involves the complex interactions between intrinsic and extrinsic factors. Because of their ubiquitous nature, direct contribution of basal cellular processes to the regulation of tissue homeostasis (Buszczak et al. 2014) has been largely underappreciated for a long time.
Ribosome biogenesis is a fundamental and universal cellular process tightly coupled to cell growth and proliferation (Lempiäinen and Shore 2009). Ribosome biogenesis occurs in the nucleolus of eukaryotic cells starting with transcription by RNA polymerase I of the 47S pre-rRNA (precursor of the 5.8S, 18S, and 28S mature rRNAs), which then undergoes a series of cleavages and modifications while being assembled into ribosomal pre-particles upon hierarchical addition of ribosomal proteins. Large and small ribosomal pre-particles are then exported to the cytoplasm to terminate their maturation and assemble into functional ribosomes.
Ribosome biogenesis is highly connected to diseases. In humans, disorders of ribosome dysfunction are called ribosomopathies and correspond to a collection of inherited or somatically acquired human syndromes associated with haploinsufficiency in genes encoding ribosomal proteins or mutations in ribosome biogenesis factors. They represent a set of clinically distinct diseases presenting with tissue-specific developmental defects (bone marrow [BM] failure, skeletal abnormalities, asplenia, …) and increased risk of cancer (Narla and Ebert 2010). The fact that mutations of ribosomal proteins or ribosome biogenesis factors cause tissue-specific diseases in humans is puzzling (McCann and Baserga 2013; Mills and Green 2017; Sulima and De Keersmaecker 2018) and several mechanisms have been put forward to explain the physiopathology of these diseases. In particular, a shortage in ribosome supply may affect translation efficiency but not equally for all mRNA species depending on their initiation rates (Mills and Green 2017).
The existence of differences in ribosome biogenesis between stem cells and their progenies is supported by several studies (Brombin et al. 2015). For example, ribosomal small subunit processing factors are down-regulated during mouse embryonic stem (ES) cell differentiation and knockdown of the corresponding genes in undifferentiated ES cells impairs pluripotency maintenance (You et al. 2015). Similarly, genes coding for ribosomal proteins and ribosome biogenesis factors were identified in screens for genes regulating maintenance or differentiation of drosophila germ stem cells (Fichelson et al. 2009; Zhang et al. 2014; Sanchez et al. 2015; Yu et al. 2016), drosophila neuroblasts (Neumüller et al. 2011), and ES cells (Fortier et al. 2015). In the hematopoietic tissue, we recently identified previously unsuspected differences in 60S ribosomal subunit production between stem cells and committed progenitors (Le Bouteiller et al. 2013). Contrasting with these data, our knowledge on the variation of ribosome biogenesis activity in vivo within specific lineages remains very poor.
Because extracellular growth and stress signals impinge upon the production of ribosomal components and biogenesis factors, studies on ribosome biogenesis activity in cultured cells cannot be informative on the in vivo situation. In addition, the paucity of HSCs has largely impeded direct measurements of ribosome biogenesis activity using traditional methods. Recently, we developed a technique combining rRNA FISH with flow cytometry to reveal unbalanced 40S and 60S subunit biosynthesis in situ in hematopoietic cells following disruption of Notchless (Le Bouteiller et al. 2013). Here, we used this approach to quantify ribosome biogenesis activity within the hematopoietic lineage of the adult mouse with special emphasis on the rare populations of HSCs and immature progenitors.
RESULTS
HSCs display low translational activity
We first monitored ribosome activity in the immature cell populations of the BM using the RiboPuromycylation Method (RPM) (David et al. 2012; Seedhom et al. 2016). This method is based on puromycin incorporation into the A site of elongating ribosome and specific and covalent association with nascent peptidic chains. Eight- to 10-wk-old C57Bl/6 mice were injected with a single dose of puromycin and killed 10 min after injection. Immediately after, BM cells were harvested and placed at 4°C in emetine-containing medium in order to freeze ribosome elongation and block the eventual release of puromycylated nascent chains from ribosomes. BM cells were first stained using cell surface antibodies and then processed for intracellular puromycin immunodetection before flow cytometry analysis. Because labeling is limited to a short period of time, incorporation of puromycin is likely limited to one round of translation, and puromycin immunodetection is therefore a good proxy of translation rate.
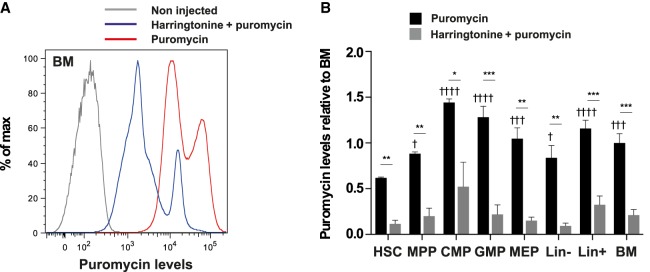
Total BM cells from injected mice showed a marked increase in puromycin staining compared to untreated mice (Fig. 1A). When mice were treated with harringtonine, an inhibitor of the initial steps of translation, 15 min prior to puromycin administration, staining was significantly reduced (Fig. 1A,B) confirming that RPM indeed measured translation activity in BM cells. The wide distribution of fluorescence intensity suggested the existence of differences in translation rate between BM cells. Interestingly, HSC (Lin– Sca1+ cKit+ CD34–) exhibited lower puromycin incorporation compared to other immature progenitors including multipotent progenitors (MPP; Lin– Sca1+ cKit+ CD34+), common myeloid progenitors (CMP; Lin– Sca1– cKit+ CD34+ FCγR-II/IIIhi), granulocyte and macrophage progenitors (GMP; Lin– Sca1– cKit+ CD34low FCγR-II/IIIlow), and megakaryocyte and erythrocyte progenitors (MEP; Lin– Sca1– cKit+ CD34– FCγR-II/IIIlow) (Fig. 1B). Consistent with a previous study (Signer et al. 2014), our data suggest that HSCs have a reduced protein synthesis rate compared to committed progenitors.
FIGURE 1.
Low translation activity in HSCs. Mice were injected with 2 mg puromycin only for 10 min or with 20 µg harringtonine for 15 min, then 2 mg puromycin for 10 min, and noninjected mice were used as controls. (A) FACS profile of puromycin levels in whole BM cells of control mice (gray), mice injected with puromycin only (red), or harringtonine and puromycin (blue). (B) Quantification of puromycin levels in the indicated populations, relative to puromycin levels in the unsorted BM cells of mice injected with puromycin alone (HSC: Lin– Sca1+ cKit+ CD34–; MPP: Lin– Sca1+ cKit+ CD34+; CMP: Lin– Sca1+ cKit+ CD34hi FCγRII/IIIhi; GMP: Lin– Sca1+ cKit+ CD34low FCγRII/IIIlo; MEP: Lin– Sca1+ cKit+ CD34low FCγRII/IIIlo). For comparisons of puromycin levels between HSCs and other populations in mice injected with puromycin only, statistical significance was calculated using one-way ANOVA with Dunnett's correction for multiple comparisons: (†) P < 0.05, (†††) P < 0.001, and (††††) P < 0.0001. For comparisons of each population between mice injected with puromycin ± harringtonine, statistical significance was calculated using unpaired two-tailed Student's t-tests: (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. n = 3 mice for each sample group; results representative of two independent experiments.
Flow-FISH allows quantification of pre-rRNA levels in the hematopoietic tissue
To monitor ribosome biogenesis activity of hematopoietic cells, we used a combination of cell surface antibody staining and flow cytometry with intracellular RNA FISH using rRNA probes. This method, named Flow-FISH, allows quantifying rRNAs levels at the single cell level in the different hematopoietic cell populations (Le Bouteiller et al. 2013). We used its1 and its2 FISH probes hybridizing to nucleolar rRNA precursors but not to the mature rRNA species found in cytoplasmic ribosome and specific for precursors of the small and the large subunit, respectively.
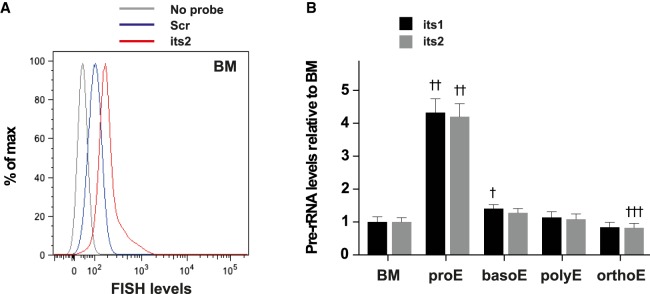
First, we used Flow-FISH to determine pre-rRNA levels during erythroid differentiation. Indeed, terminal differentiation of erythroid progenitors is characterized by a gradual decrease of the cell volume and ribosomes content (Dolznig et al. 1995). A decrease in ribosome production is therefore expected during this process although this has not been documented in vivo so far. BM cells were collected from femurs and tibias of 6- to 12-wk-old, wild-type C57Bl/6J mice, and stained using fluorescently labeled antibodies against CD71 and Ter119 allowing to separate four different steps of erythroid maturation: CD71+ Ter119low pro-erythroblasts (proE), CD71+ Ter119+ basophilic erythroblasts (basoE), CD71low Ter119+ late basophilic and polychromatophilic erythroblasts (polyE), and CD71− Ter119+ orthochromatophilic erythroblasts (orthoE) (Socolovsky et al. 2001) (for gating strategy, see Supplemental Fig. S1). Cells were then fixed and hybridized with either its1 or its2 fluorescently labeled probes. We found that CD71+ Ter119low pro-erythroblasts expressed high levels of its1 and its2 (Fig. 2, 4.3-fold and 4.2-fold increase compared to whole BM cell). Strikingly, pre-rRNA levels dropped dramatically at the transition from pro- to basophilic erythrobasts and remained at similar levels in more mature cells (Fig. 2). We conclude from these experiments that pre-RNAs levels are tightly regulated during BM erythroid maturation.
FIGURE 2.
Flow-FISH allows quantification of pre-rRNA levels during erythroid differentiation. BM cells were stained using Flow-FISH with probes directed against its1 or its2 or a scramble control probe (scr). (A) FACS profile of FISH levels in the whole BM population of samples stained with either no probe (gray) or the scr (blue) and its2 (red) probes. (B) Quantification of its1 and its2 levels in cells of the erythroid lineage, relative to its1 and its2 levels in the BM population: pro-erythroblasts (proE, CD71+ Ter119low), basophilic erythroblasts (basoE, CD71+ Ter119+), late basophilic and polychromatophilic erythroblasts (polyE, CD71low Ter119+), and orthochromatophilic erythroblasts (orthoE, CD71− Ter119+). For comparisons of pre-rRNA levels between BM and erythroid populations of samples stained with the same probe, statistical significance was calculated using one-way ANOVA with Dunnett's correction for multiple comparisons: (†) P < 0.05, (††) P < 0.01, (†††) P < 0.001. n = 3 mice; representative results of four independent experiments.
Immature hematopoietic populations display robust pre-rRNA levels
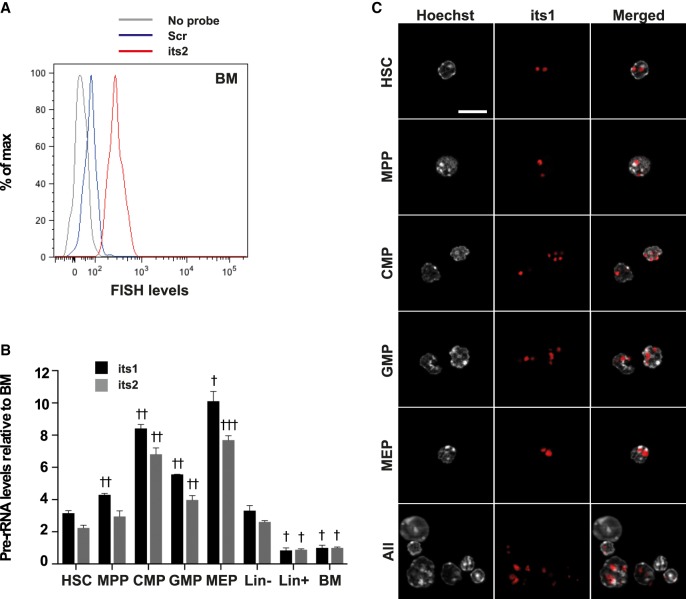
We then used Flow-FISH to measure pre-rRNA levels in different populations of immature hematopoietic cells of the BM (Fig. 3A). Because the FCγR-II/III staining did not resist to the FlowFISH procedure, a different gating strategy was used to define CMP, GMP, and MEP populations (Supplemental Fig. S1D). Immature Lin– cells, which represent around 10% of whole BM cells, displayed higher levels of its1 and its2 than more mature Lin+ cells (Fig. 3B). Among Lin– cells, MEPs showed the highest levels of its1 and its2 and HSCs the lowest (Fig. 3B). HSCs are in a vast majority quiescent, with very low division rate and low mitochondrial metabolic activity (Nakamura-Ishizu et al. 2014; Chandel et al. 2016) and therefore are expected to require less ribosome production than more active cells. However, we found that pre-rRNA levels in HSCs were only slightly less than that of actively dividing MPPs (1.4-fold and 1.3-fold reduction for its1 and its2, respectively) and higher than Lin+ progenitors and differentiated cells of the BM (3.8-fold and 2.5-fold increase for its1 and its2, respectively).
FIGURE 3.
Flow-FISH uncovers variations in pre-rRNA levels of immature hematopoietic populations. BM cells were stained using Flow-FISH with probes directed against its1 or its2 or a scramble control probe (scr). (A) FACS profile of FISH levels in the HSC population of samples stained with either no probe (gray) or the scr (blue) and its2 (red) probes. (B) Quantification of its1 and its2 levels in the indicated populations, relative to its1 and its2 levels in the BM population (HSC: Lin– Sca1+ cKit+ CD34–; MPP: Lin– Sca1+ cKit+ CD34+; CMP: Lin– Sca1+ cKit+ CD34hi; GMP: Lin– Sca1+ cKit+ CD34lo; MEP: Lin– Sca1+ cKit+ CD34lo). Statistical significance of the difference in pre-rRNA levels between HSCs and other populations of samples stained with the same probe was calculated using one-way ANOVA with Dunnett's correction for multiple comparisons: (†) P < 0.05, (††) P < 0.01, and (†††) P < 0.001. n = 3 mice; representative results of four independent experiments. (C) Lin–-depleted BM cells were stained using Flow-FISH with the its1 probe and sorted to obtain HSC, MPP, CMP, GMP, and MEP populations. Nuclei were then stained using Hoechst. Bar, 10 μm.
These relatively high levels of pre-rRNA levels suggest that ribosomes are actively produced in HSCs. However, they could also correspond to a pool of stored pre-ribosomal particles that could be rapidly mobilized to produce new ribosomes upon HSCs activation. To discriminate between these two possibilities, we first examined the subcellular distribution of pre-rRNAs in HSCs and progenitors. We performed Flow-FISH staining on Lin– BM cells and then FACS-sorted the different immature BM populations before imaging. As shown in Figure 3C, its1 staining in HSCs appeared as a few discrete spots located in DNA sparse regions of the nucleus likely corresponding to nucleoli. Importantly, the subcellular distribution of its1 staining was identical in HSCs (Lin– Sca1+ cKit+ CD34–), MPPs (Lin– Sca1+ cKit+ CD34+), CMPs (Lin– Sca1– cKit+ CD34hi), GMPs (Lin– Sca1– cKit+ CD34lo), and MEPs (Lin– Sca1– cKit+ CD34–) (Fig. 3C). Identical results were obtained with the its2 probe (not shown). These data argue against the idea that Flow-FISH would measure the levels of different types of pre-rRNAs in HSCs (stored pre-ribosomal particles) versus other cells (pre-ribosomal particles neosynthesis).
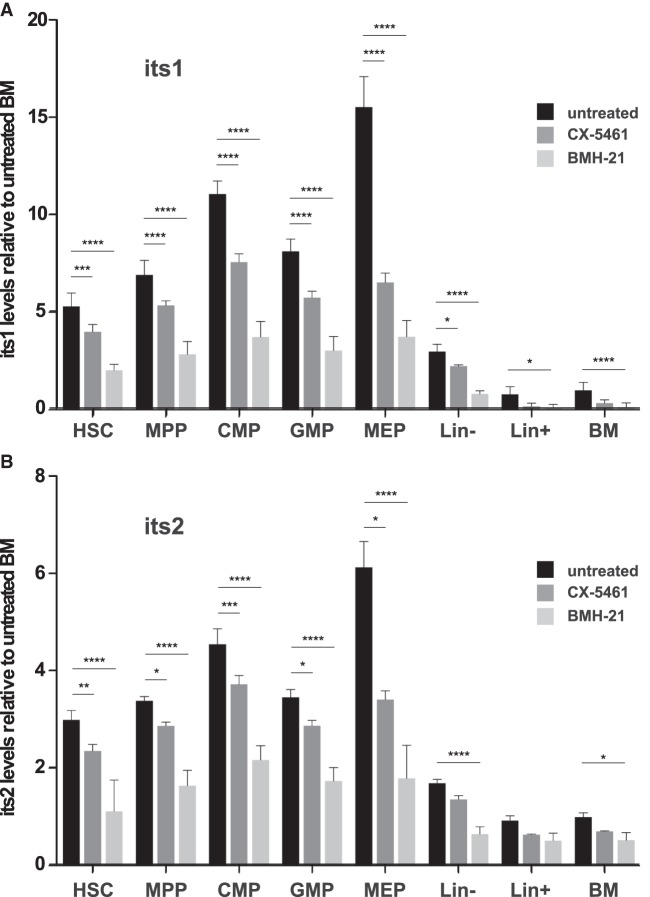
We next examined the effects of the inhibition of rRNA transcription on the levels of pre-rRNAs in hematopoietic cells. BM cells were harvested and cultured for 3 h in the presence of 10 µM CX-5461, a selective inhibitor of RNA polymerase I-driven rRNA transcription (Bywater et al. 2012; Quin et al. 2014), prior to Flow-FISH staining. CX-5461 treatment resulted in a significant decrease of its1 and its2 levels in HSCs, MPPs, CMPs, GMPs, and MEPs (Fig. 4). Because only a modest reduction in its1 and its2 levels was observed, we analyzed in more details the effects of CX-5461 on pre-rRNAs processing using mouse ES cells. We found that although CX-5461 efficiently inhibited rRNA transcription, as visualized by the drastic reduction in the nucleolar incorporation of the nucleotide analog 5-ethynyl-uridine (EU), it also caused a delay in pre-rRNA processing as suggested by the retention of previously labeled nucleolar RNAs (Supplemental Fig. S2). Such inhibitory effects of CX-5461 on pre-rRNA processing likely explain its limited impact on its1 and its2 levels in hematopoietic cells. Recently, BMH-21 was reported as another selective inhibitor of RNA polymerase I (Peltonen et al. 2014). Importantly, BMH-21 was shown to repress rRNA synthesis without affecting its processing (Peltonen et al. 2014). We thus repeated the experiment using BMH-21 and obtained similar but more pronounced reduction of its1 and its2 levels in HSCs, MPPs, CMPs, GMPs, and MEPs (Fig. 4). We conclude from these experiments that de novo rDNA transcription largely contributes to pre-rRNA levels measured by Flow-FISH in HSCs and other BM cell populations.
FIGURE 4.
RNA Pol I inhibitors lead to significant decrease of pre-rRNA levels detected by Flow-FISH. BM cells were incubated or not in the presence of 10 µM CX-5461 or 2.5 µM BMH-21 for 3 h, and were then stained using Flow-FISH with probes directed against its1 or its2 or a nontargeting probe (scr). Quantification of its1 (A) and its2 (B) levels in the indicated populations, relative to levels in the BM population (HSC: Lin– Sca1+ cKit+ CD34–; MPP: Lin– Sca1+ cKit+ CD34+; CMP: Lin– Sca1+ cKit+ CD34hi; GMP: Lin– Sca1+ cKit+ CD34lo; MEP: Lin– Sca1+ cKit+ CD34lo). Statistical significance of the differences between treated and untreated cells was calculated using unpaired two-tailed Student's t-tests: (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001. n = 3 mice for each sample group.
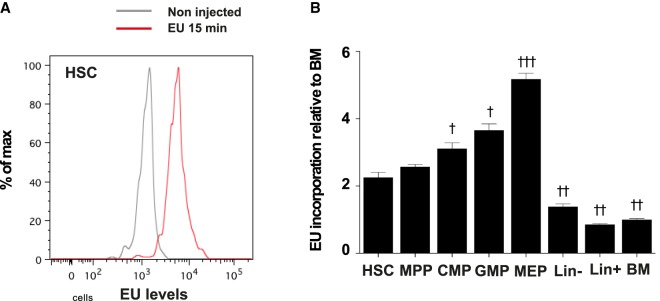
Finally, we measured transcription in HSCs (Lin– Sca1+ cKit+ CD34–), MPPs (Lin– Sca1+ cKit+ CD34+), CMPs (Lin– Sca1– cKit+ CD34hi FCγR-II/IIIhi), GMPs (Lin– Sca1– cKit+ CD34low FCγR-II/IIIlo), and MEPs (Lin– Sca1– cKit+ CD34– FCγR-II/IIIlo) by labeling nascent RNAs with a pulse of uridine analog. Because rRNA synthesis represents the majority of transcription, levels of labeled nascent RNAs after a short pulse should reflect the levels of rRNA transcription. We incubated BM cells for 15 min in the presence of 1 mM 5-ethynyl-uridine (EU) and measured the levels of incorporated EU in the different hematopoietic subsets using a fluorescently labeled azide (Jao and Salic 2008). BM cells showed a clear EU staining (Fig. 5). Similarly to pre-rRNA levels, we found that EU incorporation was higher in Lin– cells compared to Lin+ progenitors and differentiated cells and that among immature cell populations MEPs showed the highest levels of EU staining and HSCs the lowest (Fig. 5B). However, levels of nascent RNA in HSCs was not massively reduced compared to MPPs (1.1-fold reduction) and higher than in Lin+ cells (2.6-fold increase) suggesting that rRNA transcription is effective is HSCs.
FIGURE 5.
RNA transcription in BM cells. BM cells were incubated or not in the presence of 1 mM EU for 15 min. (A) FACS profile of EU levels in HSC population of samples incubated without EU (gray) or with EU for 15 min (red). (B) Quantification of EU levels in the indicated populations, relative to EU levels in the BM population (HSC: Lin– Sca1+ cKit+ CD34–; MPP: Lin– Sca1+ cKit+ CD34+; CMP: Lin– Sca1+ cKit+ CD34hi FCγRII/IIIhi; GMP: Lin– Sca1+ cKit+ CD34low FCγRII/IIIlo; MEP: Lin– Sca1+ cKit+ CD34low FCγRII/IIIlo). Statistical significance of the difference in EU levels between HSCs and other populations was calculated using one-way ANOVA with Dunnett's correction for multiple comparisons: (†) P < 0.05, (††) P < 0.01, and (†††) P < 0.001. n = 4 mice; results representative of two independent experiments.
Altogether, these experiments demonstrate that ribosome biogenesis is active in HSCs.
DISCUSSION
Here, we take advantage of the Flow-FISH method for measuring the levels of pre-rRNAs in hematopoietic cells in vivo. We show that Flow-FISH reports and quantifies ribosome biogenesis activity in hematopoietic cell populations, thereby providing original data on this fundamental process notably in rare populations such as HSCs. We unravel variations in pre-rRNA levels between different hematopoietic progenitor compartments and during erythroid differentiation. Thus, our study contributes to a better knowledge of the cellular physiology of the hematopoietic system in vivo in unperturbed situations.
An important parameter known to modulate ribosome biogenesis activity is the proliferation rate (Ruggero and Pandolfi 2003). Using Flow-FISH, we found that oligopotent myeloid progenitors, CMPs, MEPs, and GMPs, which are highly proliferative cells, had higher levels of pre-rRNAs compared to HSCs and MPPs, which are non- and slowly proliferative cell populations, respectively (Passegué et al. 2005; Busch et al. 2015). However, among oligopotent myeloid progenitors, pre-rRNA levels do not match with the proliferation index. Indeed, GMPs have the highest proliferation index but the lowest pre-rRNA levels, while MEPs have the lowest proliferation index but the highest pre-rRNA levels. On the same line, our data show that pre-rRNA levels are only slightly higher in MPPs compared to HSCs, although their proliferation status is completely different. Therefore, our data demonstrate that pre-rRNA levels are not simply a reflection of proliferation and suggest that cell type-specific mechanisms might regulate ribosome biogenesis in hematopoietic stem cells and progenitors. Consistent with our conclusion is the recent observation that ANGIOGENIN, a niche-specific factor, stimulates rRNA transcription in MEPs, CMPs, and GMPs but not in HSCs and MPPs (Goncalves et al. 2016).
Among Lin– undifferentiated cells, MEPs showed the highest activity of pre-rRNA levels. Although classically defined as bipotent progenitors, the Lin– Sca1– cKit+ CD34– FCγR-II/IIIlow population of the BM was recently shown to be composed almost exclusively of cells having erythroid differentiation potency (Paul et al. 2015). We also detected high levels of pre-rRNAs further down along the erythroid differentiation program, in proerythroblasts. Contrary to pre-rRNA levels, global protein translation is not increased in MEPs compared to CMPs and GMPs (this study and Signer et al. 2014) indicating that high ribosome biogenesis activity in these progenitors is not driven by a particularly high need in translation. Importantly, a critical role for ribosome in erythroid development was uncovered more than a decade ago by the demonstration that mutations in ribosomal genes cause erythroid defects in patients suffering from Diamond-Blackfan anemia and the 5q syndrome (Narla and Ebert 2010). Our observation of a particularly high level of ribosome biogenesis activity in erythroid progenitors will eventually help to understand why developing erythrocytes are highly sensitive to suboptimal levels of ribosomal proteins.
We also documented a sharp diminution in pre-rRNA levels at the transition between pro-erythroblasts and basophilic erythroblasts. This is contrasting with the progressive modulation of other parameters (diminution of cell size, chromatin change, or hemoglobin accumulation) observed during erythroid differentiation. Interestingly, the polycomb group protein Bmi1 has been shown to positively regulate ribosome biogenesis in proerythroblasts and therefore is likely involved in the regulation of ribosome biogenesis activity during normal erythroid maturation (Gao et al. 2015). However, Bmi1 expression is turned off in late erythroblasts (Gao et al. 2015) after the sharp down-regulation of pre-rRNA levels, indicating that additional regulators of ribosome biogenesis during erythroid differentiation remain to be discovered. Interestingly, recent studies suggest that down-regulation of rRNA transcription may actually trigger differentiation (Hayashi et al. 2014; Zhang et al. 2014). In the future, it will be important to address whether the sharp down-regulation of pre-rRNA levels is actually the cause or the consequence of proerythroblast differentiation into basophilic erythroblast.
An unexpected result uncovered by Flow-FISH is the existence of sustained ribosome biogenesis in HSCs. Because a vast majority of HSCs are quiescent and have a low metabolic activity, it was expected to find much lower pre-rRNA levels in HSCs compared to other hematopoietic cells. This is not the case. The reasons why ribosome activity in HSCs is similar to that of MPPs and higher than that of Lin+ progenitors and differentiated cells is currently unclear. Because of ribosome degradation, a certain level of ribosome biogenesis is required for renewing ribosomes and maintaining protein translation above a threshold compatible with HSCs homeostasis. Protein translation is lower in HSCs compared to other hematopoietic cells (this study and Signer et al. 2014) and variation in the rate of translation is associated with defects in HSCs function (Signer et al. 2014). Recently, it was shown that reduced translation is not due to a reduction in the number of ribosomes in HSCs but involves 4E-BP-mediated translation inhibition (Signer et al. 2016). Cell type-specific ANGIOGENIN-mediated production of tRNA-derived stress-induced small RNAs was also recently shown to contribute to the reduced levels of global protein synthesis in HSCs and MPPS (Goncalves et al. 2016). Low protein translation rate in HSCs seems therefore to be primarily regulated at the levels of translation inhibition rather than through reduced ribosome supply.
Active ribosome biogenesis in HSCs may also play important roles beyond de novo ribosome production. Indeed, while its assembly and structure are dictated by the rate of rRNA transcription and ribosome biogenesis, the nucleolus also regulates other cellular processes (Boulon et al. 2010; Stępiński 2016). In particular, the activity of several factors involved in cell cycle regulation, differentiation, or stress responses has been shown to be regulated through nucleolar sequestration (Korgaonkar et al. 2005; Kuroda et al. 2011; Sasaki et al. 2011). Interestingly, Runx1-deficient phenotypic HSCs have decreased ribosome content and reduced cell size, yet they maintain long-term repopulation capacity (Cai et al. 2015) indicating that HSCs self-renewal does not critically rely on a precise level of ribosome biogenesis activity. Importantly, RUNX1 directly regulates the transcription of ribosome component and of ribosome biogenesis factors, and in the absence of Runx1, HSCs show a lower unfolded protein response and decreased p53 protein levels conferring on them an attenuated apoptotic response following endoplasmic reticulum or genotoxic stresses (Cai et al. 2015). These observations raise the possibility that decreased ribosome biogenesis may participate to the development of multiple hematopoietic malignancies that are associated with RUNX1 mutations (Mangan and Speck 2011). They might also explain why ribosome biogenesis is not lower in HSCs since this would attenuate their responses to stress and thus increase the risk of stem cell dysfunction and transformation.
In summary, in vivo Flow-FISH allows us to quantify the activity of ribosome biogenesis at the single cell level within a given lineage, the adult hematopoietic system, composed of cells with very different proliferation, growth, and metabolic profiles. It has uncovered higher than anticipated ribosome production in adult BM HSCs and sharp down-regulation of ribosome biogenesis during erythroid differentiation. Previously, we showed that Flow-FISH could reveal unbalanced ribosomal subunit processing caused by mutation in a factor involved in pre-60s ribosomal particle processing (Le Bouteiller et al. 2013). Adapted to humans, this approach should prove useful to identify dysregulation of ribosome biogenesis in patients suffering from hematologic disorders.
MATERIALS AND METHODS
Mouse husbandry
All experiments were performed on 8- to 12-wk-old C57Bl/6J mice of either sex obtained from Charles River and/or bred in our animal facility. All experiments were conducted according to the French and European regulations on care and protection of laboratory animals (EC Directive 86/609, French Law 2001-486 issued on June 6, 2001) and were approved by the Institut Pasteur ethics committee (n° 2014-0053).
Flow cytometry analysis
Mice were euthanized and femurs and tibias were collected and kept on ice in medium containing serum. Bones were split at one end and spun down shortly in eppendorf tubes at 4°C to recover whole BM. Single cell suspensions of BM cells were stained using different combinations of antibodies. All antibody stainings were performed in HBSS medium containing 2% fetal calf serum (FCS). For Flow-FISH analyses on the erythroid lineage, cells were stained using PeCy7-conjugated anti-Ter119 and biotin-conjugated anti-CD71 antibodies. For analyses on immature hematopoietic populations, cells were stained using biotin-conjugated lineage (B220, Nk1.1, Gr-1, Ter119, CD3, CD11c, Mac-1), PeCy7- or BV510-conjugated anti-Sca1, APC- or PeCy7-conjugated anti-cKit, and FITC- or eFluor660-conjugated anti-CD34 antibodies. For RPM and EU staining analyses, cells were also stained using PE-conjugated anti-FCγRII/III antibody. Biotin-conjugated stainings were revealed using Pacific Blue-conjugated streptavidin. The list of antibody clones is available in Supplemental Table S1. LSR Fortessa (BD) analyzer was used for analysis; AutoMACS Pro (Miltenyi) separator was used for depletion of Lin+ cells; MoFlo Astrios (Beckman Coulter) cell sorter was used for cell sorting. All cytometry data were analyzed using FlowJo (Treestar) software.
RiboPuromycylation
For RPM analyses, mice were injected intra-peritoneally with 2 mg of puromycin (Sigma P8833) 10 min prior to euthanasia. For some mice, 20 µg of harringtonine (Santa-Cruz sc-204771) was injected 15 min prior to puromycin. Puromycin and harringtonine solutions were warmed at 37°C prior to injections. BM cells were collected in cold HBSS medium containing 2% FCS and 208 µM emetine (Sigma E2375) to block protein elongation and kept on ice. Cells were then stained using surface marker antibodies as described above. After surface staining, cells were permeabilized and fixed as follows: Cells were rinsed in cold phosphate-buffered saline (PBS), then incubated for 2 min in permeabilization buffer consisting of “polysome buffer” (50 mM Tris-HCl pH 7.5; 5 mM MgCl2; 25 mM KCl) complemented with EDTA-free protease inhibitors (Roche 11836170001) and 0.015% digitonin (Sigma D141). Cells were then quickly rinsed with polysome buffer complemented with protease inhibitors, and then fixed for 15 min at room temperature (RT) in 1× PBS containing 3% PFA. Cells were then rinsed with PBS and incubated in staining buffer (0.05% saponin, 10 mM Glycine, 5% FCS, 1× PBS final) for 15 min. Puromycin detection was then performed by staining with Alexa647-conjugated anti-puromycin antibody in staining buffer for 1 h (see Supplemental Table S1 and Seedhom et al. 2016). Finally, cells were rinsed in PBS with bovine serum albumin (BSA) and kept at 4°C until analysis. For quantification of puromycin levels, mean fluorescence intensity (MFI) was measured for each mouse in each cell population analyzed and was then normalized to the average MFI of the BM population of mice injected with puromycin only.
Pre-rRNA fluorescent in situ hybridization
For Flow-FISH stainings, BM cells were collected from femurs and tibia and stained with surface marker antibodies as described above. Of note, some cell surface staining such as FCγRII/III poorly resisted the further treatments required for RNA hybridization. Consequently, fluorochrome-conjugated antibodies should be tested before being used for Flow-FISH experiments. Cells stained with surface marker antibodies were fixed for 30 min at RT in 1× PBS containing 4% PFA and rinsed twice in PBS. Cell were then dehydrated as follows: Cells were first resuspended in 1 volume of PBS, and 1 volume of 70% ethanol was added drop-by-drop; cells were then centrifuged, resuspended in 70% ethanol and left at 4°C overnight. The next day, cells were rehydrated for 5 min in 2× SSC (saline sodium citrate) buffer containing 10% formamide and 0.25 mg/mL BSA. Cells of each analyzed mouse were then incubated for 3 h at 37°C in FISH-staining buffer prepared as indicated in Supplemental Table S2 with DNA probes directed against its1 (5′-tagacacggaagagccggacgggaaaga-3′) and its2 (5′-ccagcgcaagacccaaacacacacaga-3′) or a “scramble” probe (scr; 5′-cggaatagtgcgtcacgaactcgctata-3′). After incubation, cells were washed twice in 2× SSC buffer containing 10% formamide and 0.25 mg/mL BSA, for 30 min at 37°C. Finally, cells were rinsed three times in PBS-containing 0.25 mg/mL BSA and kept at 4°C until analysis. For quantification of its1 and its2 levels, mean fluorescence intensity (MFI) was measured for each mouse in each cell population of samples stained with its1, its2, or scr probes and MFI of the scr was subtracted from the MFI of its1 and its2. The obtained “corrected” MFI (cMFI) of each cell population was then normalized to the average cMFI of unsorted BM cells. For fluorescent microscopy analyses on sorted cells stained with Flow-FISH, cell nuclei were stained using Hoechst prior to analysis.
EU staining
For EU staining experiments, BM cells were collected from tibias and femurs in OptiMEM medium containing 10% FCS, and were incubated for 15 min or 1 h in the presence of 1 mM 5-ethynyl-uridine (EU, Molecular Probes E10345) or without EU, at 37°C and 5% CO2. Cells were then rinsed in cold PBS and stained with surface marker antibodies as described above, and fixed and permeabilized 15 min using the Foxp3 Staining Buffer set (eBioscience 00-5523-00). Detection of EU was then performed using the Click-iT Plus Alexa Fluor 488 picolyl azide toolkit (Molecular Probes C10641). Cells were then kept in PBS with BSA at 4°C until analysis. For quantification of EU levels, MFI was measured for each mouse in each cell population of samples incubated without EU or in the presence of EU for 15 min, and MFI of the cells incubated without EU was subtracted from the MFI of EU-incubated samples. The obtained “corrected” MFI (cMFI) of each cell population was then normalized to the average cMFI of the unsorted BM cells of samples incubated with EU for 15 min.
RNA Pol I inhibition
For treatments with RNA Pol I inhibitors, BM cells were collected from tibias and femurs in OptiMEM medium containing 10% FCS. They were then incubated for 3 h in the presence of either 10 µM CX-5461 (Axon 2173) or 2.5 µM BMH-21 (Sigma SML1183) at 37°C and 5% C02. Cells were then rinsed in cold HBSS containing 2% FCS and analyzed by Flow-FISH as described above.
Cell culture
Wild-type CK35 ES cells (Kress et al. 1998) were cultured in DMEM medium complemented with 15% FCS, 100 µM β-mercaptoethanol, and 103 units/mL leukemia inhibitory factor. Cells were seeded on Matrigel-coated coverslips, at a density allowing near-confluence at the moment of analysis. Cells were then incubated 15 min in the presence of 1 mM EU, followed by up to 2 h incubation in the presence or absence of 10 µM CX-5461 for EU pulse-chase analysis. As controls of the efficiency of CX-5461 treatment, cells were incubated for 2 h in the presence or absence of 10 µM CX-5461, followed by 15 min with both 1 mM EU and 10 µM CX-5461. Cells were then fixed in 4% PFA in PBS for 15 min at RT and permeabilized 15 min in 0.5% Triton in PBS. EU detection was then performed using the Click-iT RNA Alexa Fluor 488 Imaging Kit (Molecular Probes C10329), and nuclei were stained with Hoechst.
Microscopy
All fluorescence microscopy analyses were performed using Apotome.2 (Zeiss) microscope and the Zen blue (Zeiss) software.
Statistical analysis
All bar graphs show mean ± SEM of normalized levels of the indicated variable; results presented are representative of at least two independent experiments. Statistical analyses were performed using Prism 6 (GraphPad). For comparisons between different populations within the same sample group, one-way ANOVA with Dunnett's correction for multiple comparisons was used, and significance was indicated as follows: (†) P < 0.05, (††) P < 0.01, (†††) P < 0.001, and (††††) P < 0.0001. For comparisons of the same population between different sample groups, unpaired two-tailed Student's t-tests were used, and significance was indicated as follows: (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, and (****) P < 0.0001.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
Imaging and cell sorting analyses were performed at the Imagopole of Institut Pasteur. We are grateful to P.-H. Commere for his assistance with FACS analysis and sorting. We thank F. Morlé, L. Da Costa, and all members of the laboratory for technical advice and helpful discussions. This work was supported by the Institut Pasteur, the Centre National de la Recherche Scientifique, and the Agence Nationale de la Recherche (ANR-10-LABX-73-01 REVIVE). L.J. was supported by the DIM biotherapies Ile-de-France, the Société Française d'Hématologie, and the REVIVE Labex. M.L.B. was supported by the Ligue Nationale Contre le Cancer.
Author contributions: L.J., M.L.B.: conception and design, acquisition of data, analysis and interpretation of data, drafting or revising the article; A.R., S.V.-P., S.C.: acquisition of data; A.D.: conception and design, provision of reagents; A.C.: conception and design, analysis and interpretation of data; M.C.-T.: conception and design, analysis and interpretation of data, drafting or revising the article.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.067843.118.
REFERENCES
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. 2010. The nucleolus under stress. Mol Cell 40: 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombin A, Joly JS, Jamen F. 2015. New tricks for an old dog: ribosome biogenesis contributes to stem cell homeostasis. Curr Opin Genet Dev 34: 61–70. [DOI] [PubMed] [Google Scholar]
- Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Höfer T, Rodewald HR. 2015. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518: 542–546. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Signer RAJ, Morrison SJ. 2014. Cellular differences in protein synthesis regulate tissue homeostasis. Cell 159: 242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, et al. 2012. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Gao L, Teng L, Ge J, Oo ZM, Kumar AR, Gilliland DG, Mason PJ, Tan K, Speck NA. 2015. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell 17: 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS, Jasper H, Ho TT, Passegué E. 2016. Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nat Cell Biol 18: 823–832. [DOI] [PubMed] [Google Scholar]
- David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, Bennink JR, Yewdell JW. 2012. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol 197: 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolznig H, Bartunek P, Nasmyth K, Müllner EW, Beug H. 1995. Terminal differentiation of normal chicken erythroid progenitors: shortening of G1 correlates with loss of D-cyclin/cdk4 expression and altered cell size control. Cell Growth Differ 6: 1341–1352. [PubMed] [Google Scholar]
- Fichelson P, Moch C, Ivanovitch K, Martin C, Sidor CM, Lepesant JA, Bellaiche Y, Huynh JR. 2009. Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat Cell Biol 11: 685–693. [DOI] [PubMed] [Google Scholar]
- Fortier S, MacRae T, Bilodeau M, Sargeant T, Sauvageau G. 2015. Haploinsufficiency screen highlights two distinct groups of ribosomal protein genes essential for embryonic stem cell fate. Proc Natl Acad Sci 112: 2127–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Chen S, Kobayashi M, Yu H, Zhang Y, Wan Y, Young SK, Soltis A, Yu M, Vemula S, et al. 2015. Bmi1 promotes erythroid development through regulating ribosome biogenesis. Stem Cells 33: 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves KA, Silberstein L, Li S, Severe N, Hu MG, Yang H, Scadden DT, Hu GF. 2016. Angiogenin promotes hematopoietic regeneration by dichotomously regulating quiescence of stem and progenitor cells. Cell 166: 894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Kuroda T, Kishimoto H, Wang C, Iwama A, Kimura K. 2014. Downregulation of rRNA transcription triggers cell differentiation. PLoS One 9: e98586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CY, Salic A. 2008. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci 105: 15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE. 2005. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol 25: 1258–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress C, Vandormael-Pournin S, Baldacci PA, Cohen-Tannoudji M, Babinet C. 1998. Nonpermissiveness for mouse embryonic stem (ES) cell derivation circumvented by a single backcross to 129/Sv strain: establishment of ES cell lines bearing the Omd conditional lethal mutation. Mamm Genome 9: 998–1001. [DOI] [PubMed] [Google Scholar]
- Kuroda T, Murayama A, Katagiri N, Ohta YM, Fujita E, Masumoto H, Ema M, Takahashi S, Kimura K, Yanagisawa J. 2011. RNA content in the nucleolus alters p53 acetylation via MYBBP1A. EMBO J 30: 1054–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bouteiller M, Souilhol C, Beck-Cormier S, Stedman A, Burlen-Defranoux O, Vandormael-Pournin S, Bernex F, Cumano A, Cohen-Tannoudji M. 2013. Notchless-dependent ribosome synthesis is required for the maintenance of adult hematopoietic stem cells. J Exp Med 210: 2351–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiäinen H, Shore D. 2009. Growth control and ribosome biogenesis. Curr Opin Cell Biol 21: 855–863. [DOI] [PubMed] [Google Scholar]
- Mangan JK, Speck NA. 2011. RUNX1 mutations in clonal myeloid disorders: from conventional cytogenetics to next generation sequencing, a story 40 years in the making. Crit Rev Oncog 16: 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann KL, Baserga SJ. 2013. Genetics. Mysterious ribosomopathies. Science 341: 849–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Green R. 2017. Ribosomopathies: there's strength in numbers. Science 358: eaan2755. [DOI] [PubMed] [Google Scholar]
- Nakamura-Ishizu A, Takizawa H, Suda T. 2014. The analysis, roles and regulation of quiescence in hematopoietic stem cells. Development 141: 4656–4666. [DOI] [PubMed] [Google Scholar]
- Narla A, Ebert BL. 2010. Ribosomopathies: human disorders of ribosome dysfunction. Blood 115: 3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller RA, Richter C, Fischer A, Novatchkova M, Neumüller KG, Knoblich JA. 2011. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 8: 580–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. 2008. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132: 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegué E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. 2005. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med 202: 1599–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren-Shaul H, Winter D, Lara-Astiaso D, Gury M, Weiner A, et al. 2015. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 163: 1663–1677. [DOI] [PubMed] [Google Scholar]
- Peltonen K, Colis L, Liu H, Trivedi R, Moubarek MS, Moore HM, Bai B, Rudek MA, Bieberich CJ, Laiho M. 2014. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell 25: 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quin JE, Devlin JR, Cameron D, Hannan KM, Pearson RB, Hannan RD. 2014. Targeting the nucleolus for cancer intervention. Biochim Biophys Acta 1842: 802–816. [DOI] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP. 2003. Does the ribosome translate cancer? Nat Rev Cancer 3: 179–192. [DOI] [PubMed] [Google Scholar]
- Sanchez CG, Teixeira FK, Czech B, Preall JB, Zamparini AL, Seifert JRK, Malone CD, Hannon GJ, Lehmann R. 2015. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell 18: 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Kawahara K, Nishio M, Mimori K, Kogo R, Hamada K, Itoh B, Wang J, Komatsu Y, Yang YR, et al. 2011. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat Med 17: 944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedhom MO, Hickman HD, Wei J, David A, Yewdell JW. 2016. Protein translation activity: a new measure of host immune cell activation. J Immunol 197: 1498–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RAJ, Magee JA, Salic A, Morrison SJ. 2014. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RAJ, Qi L, Zhao Z, Thompson D, Sigova AA, Fan ZP, DeMartino GN, Young RA, Sonenberg N, Morrison SJ. 2016. The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev 30: 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. 2001. Ineffective erythropoiesis in Stat5a−/−5b−/− mice due to decreased survival of early erythroblasts. Blood 98: 3261–3273. [DOI] [PubMed] [Google Scholar]
- Stępiński D. 2016. Nucleolus-derived mediators in oncogenic stress response and activation of p53-dependent pathways. Histochem Cell Biol 146: 119–139. [DOI] [PubMed] [Google Scholar]
- Sulima SO, De Keersmaecker K. 2018. Bloody mysteries of ribosomes. HemaSphere 2: e95 10.1097/HS9.0000000000000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You KT, Park J, Kim VN. 2015. Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev 29: 2004–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Lan X, Chen X, Yu C, Xu Y, Liu Y, Xu L, Fan HY, Tong C. 2016. Protein synthesis and degradation are essential to regulate germline stem cell homeostasis in Drosophila testes. Development 143: 2930–2945. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Shalaby NA, Buszczak M. 2014. Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science 343: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.