Abstract
Objectives:
The Sahel is a semi-arid zone stretching from the Atlantic Ocean in the west to the Red Sea in the east and from the Sahara in the north to the Sudanian Savanna in the south. Here, we investigated the genetic history of the spread of Northern African ancestry common among Berbers, the Y DNA haplogroup R1b-V88, and Chadic languages throughout the Sahel, with a focus on Chad.
Materials and Methods:
We integrated and analyzed genotype data from 751 individuals from Chad, Burkina Faso, Mali, South Sudan, and Sudan in the context of a global reference panel of 5,966 individuals.
Results:
We found that genetic diversity in Chad was broadly divided by a north-south axis. The core ancestry of Southern Chadians was Central African, most closely related to Pygmies. Southern Chadians then experienced four waves of gene flow over the last 3,000 years from West-Central Africans, Eastern Africans, West-Central Africans again, and then Arabians. In contrast, Northern Chadians did not share Central African ancestry and were not influenced by the first wave of West-Central Africans but were influenced by Northern African ancestry.
Discussion:
We found that Y DNA haplogroup R1b entered the Chadian gene pool during Baggarization. Baggara Arabs spoke Arabic, not Chadic, implying that people carrying R1b-V88 were not responsible for the spread of Chadic languages, which may have spread approximately 3,700 years ago. We found no evidence for migration of Near Eastern farmers or any ancient episodes involving Eurasian backflow.
Keywords: Baggara Arabs, Berber, Chadic, genetic anthropology, Y DNA haplogroup R1b
1 |. INTRODUCTION
The Sahel is a zone between the Sahara in the north and savanna in the south. It spans across the African continent from the Atlantic Ocean to the Red Sea, including parts of Senegal, Mauritania, Mali, Burkina Faso, Algeria, Niger, Cameroon, Chad, the Central African Republic, the Republic of the Sudan, South Sudan, Eritrea, and Ethiopia. As such, the Sahel houses substantial genetic diversity distributed among many ethno-linguistic groups. Recent population genetic studies have revealed geographically distributed ancestries on a global scale (Baker, Rotimi, & Shriner, 2017; Shriner, Tekola-Ayele, Adeyemo, & Rotimi, 2014; Tishkoff et al., 2009). In Chad, five ancestries have been identified: Western African, West-Central African, Eastern African, Northern African, and Arabian (Tishkoff et al., 2009; Triska et al., 2015). Each of these ancestries has been found to correlate with a primary language family or branch: Western African ancestry with Mande; West-Central African ancestry with Niger-Congo, Bantu and non-Bantu combined; Eastern African ancestry with Nilo-Saharan; Northern African ancestry with Berber; and Arabian ancestry with Semitic (Baker et al., 2017). In addition to the Berber and Semitic branches, the Afroasiatic language family includes Chadic, Cushitic, Egyptian, and Omotic branches. Separate ancestries have been inferred for Cushitic and Omotic speakers (Shriner et al., 2014), but the ancestry of Chadic speakers is unclear. Two possibilities are that Chadic speakers have a distinct ancestry or that they have Eastern African ancestry and experienced a language shift (Tishkoff et al., 2009).
The Y DNA haplogroup R-M207 arose 31.9 [29.8, 34.0] kiloyears ago (kya) (Urasin, 2017a), probably in Central Asia. Its descendant R1b1a2-V88 arose 17.1 [15.3, 19.0] kya (Urasin, 2017b). The presence of R1b1a2-V88 in many populations across the Sahel attests to one clear instance of Eurasian immigration, but when this haplogroup entered Africa and by whom it was carried is unclear (Bekada et al., 2013; Bučková, Černý, & Novelletto, 2013; Cruciani et al., 2010a; Cruciani et al., 2010b).
Presently, the distributions of R1b1a2-V88 and speakers of Chadic languages show partial overlap (Bučková et al., 2013; Cruciani et al., 2010a), but whether there is a causal, historical association is unknown. Based on the distribution of the mitochondrial DNA haplogroup L3f3, it has been suggested that Chadic-speaking pastoralists migrated from Northeastern or Eastern Africa, with an upper bound of the early Holocene (Černý et al., 2009). Chadic languages are classified in the Afroasiatic family but the genealogical relationships with the other branches in the Afroasiatic family are disputed. The spread of Chadic languages has been hypothesized to have proceeded either from the Northern Sahara south toward Lake Chad, with Chadic being most closely related to Berber (Ehret, 2002), or westward from the Nile Valley through Sudan, with Chadic being most closely related to Cushitic (Blench, 1999).
To shed light on these issues, we integrated genotype data from diversity projects in Chad, the Sahel, Sudan, and South Sudan with a previously curated collection of 5,966 individuals sampled from around the world (Baker et al., 2017). Using a combination of supervised clustering and decay of linkage disequilibrium, we revealed a layered history of migrations across Chad spanning almost 3,000 years. Disentangling these layers yielded insights regarding the source and spread of R1b and Chadic.
2 |. MATERIALS AND METHODS
2.1 |. Genotype and Sequence Data
We retrieved genotype data (references EGAD00010001101, EGAD00010001102, and EGAD00010001103) and sequence data (reference EGAD00001002742), both from Chad, from the European Genome-Phenome Archive at the European Bioinformatics Institute under agreement with the Wellcome Trust Sanger Institute. Due to the unknown ethnic mix of people sampled from N’Djamena, the capital city of Chad, we excluded this sample as a potential reference sample. We retrieved genotype data from the Sahel (reference EGAD00010000943) from the European Genome-Phenome Archive at the European Bioinformatics Institute under agreement with Dr. Luísa Pereira, Institute of Molecular Pathology and Immunology of the University of Porto, Portugal. We retrieved genotype data from Sudan (Dobon et al., 2015) from the website of Dr. Jaume Bertranpetit, Universitat Pompeu Fabra. For all three genotype data sets, all individuals had genotyping call rates >90%. Due to the mixed ethnic composition of each of the three genotype data sets, we did not filter markers for minor allele frequency or Hardy-Weinberg disequilibrium. In the Sahel data set, three Oromo were identical to three Somali; since we did not use any of the Oromo or Somali data, this was not a problem. All other genotype data were retrieved as previously described (Baker et al., 2017). For sample locations, sizes, genotyping platforms, and sources, see Figure 1 and Supporting Information Table S1.
Figure 1.
Geographic locations of the samples from Chad, South Sudan, Sudan, and the Sahel.
2.2 |. Projection Analysis
There are three ways to perform clustering analysis to learn about population structure in a sample. In unsupervised clustering, there are no reference data and population structure in the sample is learned de novo. The number of ancestries, ancestry-specific allele frequencies, and mixing proportions are all unknown. In semi-supervised clustering, the number of ancestries is fixed by the reference panel. Initial estimates of the ancestry-specific allele frequencies are also provided by the reference panel and then are updated, jointly considering the sample and reference data, while mixing proportions in the sample are learned. In supervised clustering, both the number of ancestries and the ancestry-specific allele frequencies are fixed by the reference panel. Only the mixing proportions in the sample are learned. With a small amount of sample data and a large amount of reference data, supervised clustering is the most efficient of the three approaches. Autosomal ancestry was analyzed using supervised clustering in ADMIXTURE version 1.3 (Alexander, Novembre, & Lange, 2009), based on a global reference panel of 21 global ancestries (Baker et al., 2017) to which we integrated Cushitic ancestry (Shriner et al., 2014). This analysis was performed by running ADMIXTURE with the option -P. To determine standard errors for the proportions of ancestral components for each individual, we reran ADMIXTURE with the addition of 200 bootstrap replicates. Accounting for both within and between individual variances, we calculated the proportions for average ancestry using inverse variance weights. We then calculated 95% confidence intervals for each ancestry and individual, zeroed out any average proportions for which the 95% confidence intervals included 0, and renormalized the remaining averages to sum to 1. ADMIXTURE makes no assumption regarding the historical process(es) that brought together previously isolated populations.
2.3 |. Decay of Linkage Disequilibrium
We used the liftover tool to convert all data sets to GRCh37/hg19 coordinates. We merged genotype data using PLINK 1.9.0-beta4.4, orienting markers to the plus strand and excluding strand-ambiguous SNPs. We then incorporated genetic distances. After using convertf in EIGENSOFT version 6.1.4 to reformat the data, we used MALDER version 1.0 to infer mixing times (Pickrell et al., 2014). MALDER is based on weighted linkage disequilibrium statistics estimated from genotype data, with the weighted statistics expected to decay exponentially as a function of genetic distance (Loh et al., 2013). MALDER assumes instantaneous admixture, possibly over multiple events, and the estimates of time refer to generations after admixture (Loh et al., 2013). If admixture occurred continuously over several generations, then the estimates of time should be interpreted as an average or midpoint but could be biased toward the recent past (Loh et al., 2013). We converted generations to years assuming 28 years/generation (Fenner, 2005; Moorjani et al., 2016).
2.4 |. Analysis of Uniparental Markers
Y DNA haplogroups were called using YFitter (Jostins et al., 2014). Mitochondrial DNA haplogroups were called using HaploFind (Vianello et al., 2013).
2.5 |. Ethics Statement
This project was excluded from IRB review by the Office of Human Subjects Research Protections, National Institutes of Health (OHSRP ID# 17-NHGRI-00282).
3 |. RESULTS
3.1 |. Supervised clustering of Sahelian populations
We performed supervised clustering individuals from four Chadian populations, eight Sahelian populations, and seven Sudanese and South Sudanese populations, and integrated these results with two additional Chadian populations and two populations of Chadic speakers (Table 1). Broad trends conform to geographic expectations: Western and West-Central African ancestries were found predominantly in the west; Eastern African, Omotic, and Cushitic ancestries were found predominantly in the east; Northern African ancestry was found predominantly in the north; and Central African ancestry was found predominantly in the south. A combination of primarily Arabian, secondarily Western Asian, and a low level of Southern Asian ancestries formed a gradient that decreased from east to west. We found evidence for Southern European ancestry only in Arabians, Copts, and Nubians from Sudan; we found no evidence for Northern European ancestry.
Table 1.
Autosomal ancestry profiles.
| Group | Country | West-Central African | Omotic | Eastern African | Northern African | Southern Asian | Western Asian | Central African | Southern European | Southern African | Western African | Arabian | Cushitic |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arab | Sudan | 0.040 | 0.080 | 0.331 | 0.106 | 0.027 | 0.055 | 0.008 | 0.018 | 0 | 0.051 | 0.284 | 0 |
| Arabs | Sudan | 0 | 0 | 0.469 | 0 | 0 | 0.036 | 0 | 0 | 0 | 0.007 | 0.488 | 0 |
| Beja | Sudan | 0 | 0.057 | 0.313 | 0.023 | 0 | 0.042 | 0 | 0 | 0 | 0 | 0.347 | 0.218 |
| Bulala | Chad | 0.175 | 0 | 0.583 | 0.003 | 0 | 0 | 0.020 | 0 | 0 | 0.211 | 0.008 | 0 |
| Copts | Sudan | 0 | 0 | 0 | 0.040 | 0.015 | 0.481 | 0 | 0.038 | 0 | 0 | 0.426 | 0 |
| Darfurian | Sudan | 0.059 | 0.012 | 0.789 | 0 | 0 | 0 | 0 | 0 | 0 | 0.141 | 0 | 0 |
| Daza | Chad | 0.126 | 0.028 | 0.457 | 0.142 | 0 | 0 | 0.008 | 0 | 0 | 0.144 | 0.079 | 0.011 |
| Gurmantche | Burkina Faso | 0.476 | 0 | 0.051 | 0 | 0 | 0 | 0 | 0 | 0 | 0.473 | 0 | 0 |
| Gurunsi | Burkina Faso | 0.453 | 0 | 0.062 | 0 | 0 | 0 | 0.017 | 0 | 0.005 | 0.462 | 0 | 0 |
| Hausa | Nigeria | 0.563 | 0 | 0.084 | 0 | 0 | 0 | 0.021 | 0 | 0.006 | 0.326 | 0 | 0 |
| Kaba | Chad | 0.448 | 0 | 0.190 | 0 | 0 | 0 | 0.052 | 0 | 0.013 | 0.296 | 0 | 0 |
| Kanembu | Chad | 0.238 | 0 | 0.370 | 0.079 | 0 | 0 | 0.016 | 0 | 0 | 0.260 | 0.037 | 0 |
| Laal | Chad | 0.328 | 0.003 | 0.340 | 0 | 0 | 0 | 0.067 | 0 | 0 | 0.262 | 0 | 0 |
| Mada | Cameroon | 0.283 | 0 | 0.370 | 0 | 0 | 0 | 0.030 | 0 | 0 | 0.317 | 0 | 0 |
| Mossi | Burkina Faso | 0.443 | 0.004 | 0.055 | 0 | 0 | 0 | 0.016 | 0 | 0 | 0.481 | 0 | 0 |
| N’Djamena | Chad | 0.273 | 0.016 | 0.429 | 0.002 | 0 | 0 | 0.041 | 0 | 0 | 0.237 | 0.001 | 0 |
| Nilotes | South Sudan | 0.058 | 0 | 0.881 | 0 | 0 | 0 | 0.047 | 0 | 0 | 0.014 | 0 | 0 |
| Nuba | Sudan | 0.035 | 0.018 | 0.845 | 0 | 0 | 0 | 0 | 0 | 0 | 0.102 | 0 | 0 |
| Nubian | Sudan | 0.044 | 0.084 | 0.308 | 0.111 | 0.034 | 0.110 | 0 | 0.032 | 0 | 0.013 | 0.263 | 0 |
| Nubians | Sudan | 0 | 0.035 | 0.521 | 0 | 0 | 0.076 | 0 | 0.020 | 0 | 0 | 0.349 | 0 |
| Sara | Chad | 0.411 | 0 | 0.226 | 0 | 0 | 0 | 0.069 | 0 | 0.003 | 0.291 | 0 | 0 |
| Songhai | Mali | 0.388 | 0.019 | 0.062 | 0 | 0 | 0 | 0 | 0 | 0 | 0.532 | 0 | 0 |
| Tubu | Chad | 0.138 | 0.031 | 0.409 | 0.177 | 0 | 0 | 0.003 | 0 | 0 | 0.167 | 0.075 | 0 |
3.2 |. Analysis of Sudanese patrilines
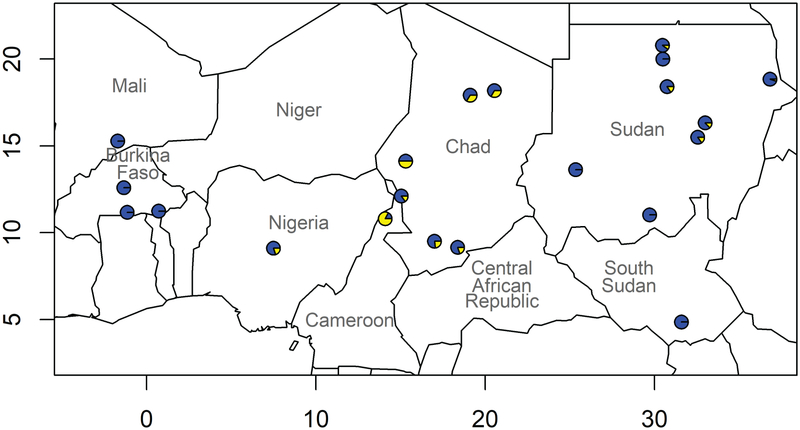
The Sudanese and South Sudanese populations can be divided into those with significant amounts of Arabian ancestry, including Arabs, Beja, Copts, and Nubians, and those without, including Darfurians, Nilotes, and Nuba (Table 1). We correlated these autosomal results with previously published Y DNA haplogroups (Hassan, Underhill, Cavalli-Sforza, & Ibrahim, 2008). Specifically, R1b was present in Arabs, Copts, Nubians, and Beja, i.e., groups with Arabian and Western Asian ancestries (Fig. 2 and Table 2). In contrast, R1b was absent in Darfurians (Fur and Masalit), Nilotes (Dinka, Nuer, and Shilluk), and Nuba, i.e., groups without Arabian or Western Asian ancestries (Fig. 2 and Table 2). These results suggest that R1b was introduced into indigenous Eastern African peoples by Arabs who had already experienced introgression with Western Asian ancestry.
Figure 2.
Distribution of Y DNA haplogroup R across the Sahel. The size of the yellow sector is proportional to the frequency of the haplogroup.
Table 2.
Y DNA haplogroups.
| Group | Country | A | B | E | J | R1b | T | Other |
|---|---|---|---|---|---|---|---|---|
| Arab | Sudan | 0 | 0 | 0.500 | 0.375 | 0.125 | 0 | 0 |
| Arabs† | Sudan | 0.029 | 0 | 0.167 | 0.471 | 0.157 | 0 | 0.098 F, 0.039 I, 0.029 K*, 0.010 R1(xb) |
| Beja† | Sudan | 0.048 | 0 | 0.524 | 0.381 | 0.048 | 0 | 0 |
| Copts† | Sudan | 0 | 0.152 | 0.212 | 0.455 | 0.152 | 0 | 0.030 K* |
| Darfurian† | Sudan | 0.250 | 0.031 | 0.656 | 0.063 | 0 | 0 | 0 |
| Daza | Chad | 0.056 | 0 | 0.111 | 0.056 | 0.333 | 0.444 | 0 |
| Gurmantche | Burkina Faso | 0 | 0 | 1.000 | 0 | 0 | 0 | 0 |
| Gurunsi | Burkina Faso | 0 | 0 | 1.000 | 0 | 0 | 0 | 0 |
| Kanembu | Chad | 0 | 0.500 | 0 | 0 | 0.500 | 0 | 0 |
| Mossi | Burkina Faso | 0 | 0 | 1.000 | 0 | 0 | 0 | 0 |
| Nilotes† | South Sudan | 0.528 | 0.302 | 0.170 | 0 | 0 | 0 | 0 |
| Nuba† | Sudan | 0.464 | 0.143 | 0.393 | 0 | 0 | 0 | 0 |
| Nubian | Sudan | 0.083 | 0 | 0.250 | 0.667 | 0 | 0 | 0 |
| Nubians† | Sudan | 0 | 0.077 | 0.231 | 0.436 | 0.103 | 0 | 0.103 F, 0.051 I |
| Songhai | Mali | 0 | 0 | 1.000 | 0 | 0 | 0 | 0 |
Adapted from (Hassan et al., 2008).
3.3 |. Dating the entry of Arabian ancestry into the Sudanese gene pool
We used the decay of linkage disequilibrium to estimate when admixture occurred between Arabians and Eastern Africans. To do this, we first combined the Sudanese Arabs and Nubians into the test sample. We next combined the Anuak, Gumuz, and Sudanese (i.e., Nilotes from South Sudan) into one reference sample reflecting predominantly Eastern African ancestry. We then tested for admixture using a series of second parental references (Table 3). We estimated that admixture occurred between Arabian ancestry and Eastern African ancestry 20 generations ago. This result was robust to both the second parental reference, marker density, and the source of the genotype data (i.e., whole genome sequencing or any of several genotyping arrays).
Table 3.
Admixture dating analysis of Sudanese Arabs.
| SNPs | Test | Reference 1 | Reference 2 | Time | SE | Z | 95% CI |
|---|---|---|---|---|---|---|---|
| 647,775 | Sudanese Arab† | Nilo-Saharan‡ | Cushitic§ | 19.7 | 1.8 | 11.0 | [16, 23] |
| 647,775 | Sudanese Arab | Nilo-Saharan | European¶ | 20.6 | 1.9 | 11.0 | [17, 24] |
| 647,775 | Sudanese Arab | Nilo-Saharan | South Asian†† | 20.5 | 2.1 | 9.9 | [16, 24] |
| 348,945 | Sudanese Arab | Nilo-Saharan | Arabian‡‡ | 20.5 | 1.6 | 12.8 | [17, 24] |
| 348,945 | Sudanese Arab | Nilo-Saharan | Cushitic | 20.4 | 1.5 | 13.3 | [17, 23] |
| 174,057 | Sudanese Arab | Nilo-Saharan | Cushitic | 19.4 | 1.5 | 12.9 | [16, 22] |
| 174,057 | Sudanese Arab | Nilo-Saharan | European | 21.8 | 1.7 | 13.1 | [19, 25] |
| 174,057 | Sudanese Arab | Nilo-Saharan | South Asian | 21.9 | 1.9 | 11.4 | [18, 26] |
| 174,057 | Sudanese Arab | Nilo-Saharan | North African§§ | 20.3 | 1.6 | 12.8 | [17, 23] |
| 70,116 | Sudanese Arab | Nilo-Saharan | Qatari | 21.5 | 1.7 | 12.4 | [18, 25] |
Arab + Nubian
Anuak + Gumuz + Sudanese [Nilotes]
Ethiopian Somali + Somali
CEU + GBR + FIN + IBS + TSI
BEB + GIH + ITU + PJL + STU
Bedouin + Druze + Palestinian
Algeria + Egypt + Libya + NMorocco + Sahrawi + SMorocco + Tunisia
3.4 |. Dating the mixing of Arabian and Western Asian ancestries
Given the results from the analysis of Sudanese patrilines, we estimated when admixture occurred between Arabian and Western Asian ancestries. To do this, we combined the Sudanese Arabs and Nubians into the test sample; we used a Bedouin sample as a reference reflecting Arabian ancestry (Li et al., 2008); and we combined Abkhazian, North Ossetian, and Armenian samples into one reference sample reflecting predominantly Western Asian ancestry (Yunusbayev et al., 2012). We detected one gene flow event that occurred 28 generations ago (Table 4).
Table 4.
Admixture dating analysis for Chad and Chadic.
| SNPs | Test | Reference 1 | Reference 2 | Time (generations ago) | SE | Z | 95% CI |
|---|---|---|---|---|---|---|---|
| 359,346 | Sudanese Arab† | Western Asian‡ | Arabian§ | 28.1 | 8.0 | 3.5 | [12, 44] |
| 348,945 | Southern Chad¶ | Arabian†† | West-Central‡‡ | 15.4 | 8.9 | 1.7 | [0, 33] |
| 647,775 | Southern Chad | Nilo-Saharan§§ | West-Central | 26.7 | 11.9 | 2.2 | [3, 50] |
| 348,945 | Southern Chad | Nilo-Saharan | Pygmy¶¶ | 36.0 | 4.9 | 7.3 | [26, 46] |
| 348,945 | Southern Chad | Pygmy | West-Central | 95.1 | 24.9 | 3.8 | [46, 144] |
| 348,945 | Northern Chad††† | Arabian | Southern Chad | 15.7 | 1.6 | 9.9 | [13, 19] |
| 348,945 | Northern Chad | Arabian | West-Central | 15.2 | 1.4 | 10.9 | [12, 18] |
| 174,057 | Northern Chad | North African‡‡‡ | Southern Chad | 15.6 | 1.6 | 9.7 | [12, 19] |
| 647,775 | Northern Chad | Nilo-Saharan | West-Central | 32.4 | 12.9 | 2.5 | [7, 58] |
| 70,116 | Chadic§§§ | Nilo-Saharan¶¶¶ | Arabian†††† | 9.6 | 4.5 | 2.1 | [1, 18] |
| 70,116 | Chadic | Nilo-Saharan | West-Central | 133.3 | 61.5 | 2.2 | [13, 254] |
Arab + Nubian
Abkhasian + North Ossetia + Armenia
Bedouin
Laal + Sara
Bedouin + Druze + Palestinian
Gurmantche + Gurunsi + Mossi + Songhai
Anuak + Gumuz + Sudanese [Nilotes]
BiakaPygmy + MbutiPygmy
Daza + Kanembu + Tubu
Algeria + Egypt + Libya + NMorocco + Sahrawi + SMorocco + Tunisia
Hausa + Mada
Anuak + Bulala + Gumuz + Sudanese [Nilotes]
Arab + Nubian + Qatari
3.5 |. Gene flow in Southern Chad
We next investigated the genetic history of Southern Chad. Based on similar ancestral profiles, we combined the Laal and Sara to increase the sample size of the admixed test sample. Notably, these samples share Central African ancestry and do not share Northern African ancestry (Table 1). We detected four waves of gene flow (Table 4). The first wave occurred 95 generations ago between the Pygmy and West-Central African references. The second wave resulted in the introduction of Eastern African ancestry 36 generations ago. The third wave resulted in a second introduction of West-Central African ancestry 27 generations ago. Finally, Arabian ancestry was introduced 15 generations ago.
3.6 |. Gene flow in Northern Chad
We then investigated the genetic history of Northern Chad. To increase the sample size of the admixed test sample, we combined the Daza, Kanembu, and Tubu samples. In contrast to the Laal and Sara, the Daza, Kanembu, and Tubu share Northern African ancestry. We detected two waves of gene flow (Table 4). The first wave occurred 32 generations ago between the Eastern African and West-Central African references. The second wave introduced Arabian ancestry and/or Northern African ancestry 15 generations ago, coinciding with the entry of Arabian ancestry into Southern Chad.
3.7 |. Gene flow in the Afroasiatic Chadic speakers
We investigated gene flow in the Hausa from Nigeria and the Mada from Cameroon. Both peoples showed four ancestries: Western African, West-Central African, Eastern African, and Central African (Table 1). Both peoples also carried R1b (82.4% in the Mada and 20.0% in the Hausa (Cruciani et al., 2010b)) and spoke Chadic. We detected two waves of gene flow (Table 4). The first wave occurred 133 generations ago between Eastern African and West-Central African references. The second wave introduced Arabian ancestry 10 generations ago.
3.8 |. Distribution of uniparental markers in Chad and the Sahel
We called the Y DNA and mitochondrial DNA haplogroups from whole genome sequences of 11 Chadians (Table 5). Among the patrilines, the Laal possessed two B1 and one R1, the Tubu possessed two E1b1b1b2 and two R1, and the Sara possessed two E1b1a7a. All 11 matrilines were of the L macro-haplogroup, with five L0a, one L1b, five L3 haplogroups. Although underpowered, these data suggest a relative excess of non-African patrilines, i.e., sex-biased gene flow. We also called Y DNA haplogroups from the Sahelian genotype data (Table 2). The frequency of R1 was 12.5% in the Sudanese Arabs, 33.3% in the Daza, and 50% in the Kanembu. The frequency of T, another patriline indicative of Eurasian immigration, was 44.4% in the Daza (Table 2).
Table 5.
Uniparental haplogroups from whole genome sequence data from Chad.
| Subject ID | Group | Y DNA haplogroup | Mitochondrial DNA haplogroup |
|---|---|---|---|
| yemcha6089639 | Laal | B1 | L1b1a10a |
| yemcha6089631 | Laal | B1 | L3f1b3 |
| yemcha6089662 | Laal | R1 | L0a1a |
| yemcha6089629 | N’Djamena | E1b1a7a | L0a1b |
| yemcha6089637 | N’Djamena | E1b1b1a1 | L3d3b |
| yemcha6089633 | Sara | E1b1a7a | L0a1a |
| yemcha6089634 | Sara | E1b1a7a | L3e3b |
| yemcha6089778 | Tubu | E1b1b1b2 | L0a1c |
| yemcha6089746 | Tubu | E1b1b1b2 | L3d4a |
| yemcha6089731 | Tubu | R1 | L0a1b1a |
| yemcha6089762 | Tubu | R1 | L3d5 |
4 |. DISCUSSION
We investigated the genetic history of Chad, in the context of the Sahel as well as in the global context. We did not find evidence of immigration of Near Eastern farmers or ancient Eurasians of any sort. We found that the Y DNA haplogroup R1b correlated with the presence of Arabian and Western Asian ancestries in the Sudanese gene pool. Using autosomal data, we found that Western Asian and Arabian ancestries mixed in the 13th century prior to entering the Sudanese gene pool. We found that Arabian ancestry migrated from east to west, having entered the Sudanese gene pool 20 generations ago, the Chadian gene pool 15 generations ago, and the Cameroonian and Nigerian gene pools 10 generations ago. Linguistic data support this path: Nigerian Arabic and Chadian Arabic are closer to Sudanese Arabic than to Arabic in Northern Africa (Owens, 1994). We also found that R1b was present in many populations other than Chadic speakers.
Historical records indicate that Arabs entered Sudan through Egypt by the 12th century (Wilson, 1888). Assuming a generation interval of 28 years, our results indicated that Arabian ancestry entered the Sudanese gene pool between the mid-1300s and mid-1500s, giving rise to both Sudanese Arabs as well as Arabized Sudanese (such as Nubians). This period follows the decline of the Nubian Kingdom of Makuria and the end of the Baqt, a peace treaty between Muslim Egyptians and Christian Nubians. We observed that this gene flow event involved a combination of Arabian and Western Asian ancestries. Since Arabs in the Arabian Peninsula most commonly carry Y DNA haplogroup J1-M267 (Abu-Amero et al., 2009; Cadenas, Zhivotovsky, Cavalli-Sforza, Underhill, & Herrera, 2008; Chiaroni et al., 2010; Mohammad, Xue, Evison, & Tyler-Smith, 2009), and since R originated in Central Asia, we hypothesize that R entered the Arabian gene pool via Western Asian ancestry. Although most studies lack the resolution to state whether R1b was specifically R1b1a2-V88, we found no evidence supporting the hypothesis of multiple entries of R1b at different historical times. Furthermore, these Arabs spoke Arabic, not Chadic, implying no causal association between R1b and Chadic. These Arabs are also known as Baggara, which is derived from Arabic for cattle-herder, because as they migrated from Sudan the environment became unsuitable for camels and they learned cattle-herding from Fulani (Braukämper, 1994).
We estimated that Arabian and Western Asian ancestries mixed in the mid-13th century. This time is well after the Muslim conquest of Egypt in the 7th century and shortly before Arabian ancestry entered the Sudanese gene pool, suggesting that this mixture event occurred in Egypt. We hypothesize that this event involved the Mamluks, who included military slaves from the Caucasus region (Hathaway, 1997; McGregor, 2006; Philipp & Haarmann, 1998). Thus, our results suggest that the entry of R1b into Africa occurred in the last millennium, not in the early to mid-Holocene (Cruciani et al., 2010a), during the Neolithic revolution (Haber et al., 2016), or with the first pastoralists (Kulichová et al., 2017).
Previously, it has been suggested that Nubians experienced admixture with incoming Eurasians prior to the early Islamic conquests in the 7th century (Hollfelder et al., 2017). Our results do not support such an old event; it is possible that this finding is a false positive resulting from small sample sizes. It has also been suggested that recent admixture of Eastern African and Arabian ancestries in Sudanese Arabs indicates that these peoples were indigenously Northeast African, but kept the language and culture of the incoming Arabs (Hollfelder et al., 2017). A simpler explanation is that the incoming Arabs received Eastern African ancestry while keeping their original language and culture.
The genetic history of the Laal and Sara in Southern Chad started with a mixture event approximately 2,700 years ago between people with Central African ancestry, proxied by Biaka and Mbuti Pygmies, and West-Central African ancestry, proxied collectively by Gurmantche, Gurunsi, Mossi, and Songhai. The frequency of B-M181 in the Laal is 54%; it is not B2-M182 but rather B1-M146. Previously, B1-M146 has been found in 1 of 44 individuals from Mali (Underhill et al., 2000) and 2 of 49 Mossi from Burkina Faso (Cruciani et al., 2002). A plausible scenario is that the indigenous Southern Chadian people had Central African ancestry and experienced admixture with the founders of the Sao civilization. The Sao civilization is thought to have begun circa 600 BC around Lake Chad and the Chari River (DeLancey & DeLancey, 2000). According to Ahmad Ibn Furtu, the grand Imam of the Bornu Empire during the reign of Mai Idris Alooma in the 16th century AD, the Sao or “others” were local settlers who did not speak Kanuri, a Nilo-Saharan language, but rather spoke Chadic (Ibn Furtu, 1576; Ibn Furtu, 1578), consistent with Chadic predating R1b in this region. This scenario supports the hypothesis that the original phylum of the Laal language, which is considered to be an isolate and contains a core vocabulary that does not appear to belong to the Afroasiatic, Niger-Congo, or Nilo-Saharan families, is a grouping from Central Africa (Blench, 2006). Next, there was an influx of Eastern African ancestry approximately 1,000 (~700 to 1,300) years ago. This period corresponds to the Kanem Empire. The Kanem Empire was founded by the nomadic Tebu-speaking people. Tebu is a Western Saharan language in the Nilo-Saharan family spoken by the two groups of Tubu people, the Teda and the Daza. A second wave of immigration proxied by West-Central Africans occurred approximately 750 (100 to 1,400) years ago. This event may be explained by the arrival of the Kanembu at the end of the 12th century AD. This hypothesis is supported by the finding that the present-day sample of Kanembu has 50% West-Central and Western African ancestry (Table 1). Finally, Arabian ancestry entered the Chadian gene pool approximately 400 years ago, five generations after having entered the Sudanese gene pool.
The genetic history of the Daza, the Kanembu, and the Tubu in Northern Chad started with a mixture event between Eastern Africans and West-Central Africans approximately 950 years ago. This finding may be explained by mixture between Nilo-Saharan-speaking Tubu and West-Central Kanembu. This event may have had the same participants as the event detected in Southern Chad approximately 750 years ago, consistent with a southward migration of the Kanembu. Then, Arabian ancestry and North African ancestry entered the gene pool approximately 400 years ago. The entry of Arabian ancestry at this time is consistent with westward migration of R1b-carrying Arabs from Sudan. We hypothesize that Y DNA haplogroup T entered Northern Chad via a southward migration of Berbers that occurred around the same time as the westward migration of Arabs.
The westward migration of R1b-carrying Arabs reached the Hausa and the Mada approximately 300 years ago, or five generations after reaching Chad. This result is supported by the dating method but not by the projection analysis, which did not detect a statistically significant presence of Arabian ancestry in the autosomes of either the Hausa or the Mada. Based on the distributions of patrilines and matrilines in the Chad sequence data, we hypothesize that gene flow was sex-biased such that males carried Arabian ancestry and females were preferentially indigenous, i.e., females carried L haplogroups (Priehodová, Abdelsawy, Heyer, & Černý, 2014). This hypothesis predicts that the association of autosomal Arabian ancestry and R1b weakened as the Arabs migrated west.
The mitochondrial haplogroups suggest multiple source populations. L0a is thought to have originated in Eastern Africa, spreading to Central Africa during the transition from the Pleistocene to Holocene (Silva et al., 2015). L1 is common in Pygmy populations in Central Africa and, more specifically, L1b is common in Central and West Africa (Harich et al., 2010). L3 is thought to originated in Eastern Africa and spread to Central Africa, with L3d and L3e subsequently spreading from Central Africa back to Eastern Africa during the Bantu Expansion (Bandelt et al., 2001; Soares et al., 2012). L3f is thought to have spread from Eastern Africa into the Sahel and Central Africa at the beginning of the Holocene (Soares et al., 2012).
Our samples included only two speakers of Chadic, the Hausa in Nigeria and the Mada in Cameroon. We did not see genetic evidence of a southward migration of Northern African ancestry past Northern Chad (Ehret, 2002). Similarly, we saw no genetic evidence of a westward migration of Cushitic ancestry (Blench, 1999). However, in both the Hausa and the Mada, we did see Eastern African ancestry. Therefore, the genetic evidence supports the hypothesis that Chadic speakers were originally from the Nile Valley and that these people reached the west approximately 3,700 years ago. Dense sampling from Egypt and Libya are necessary to further address this issue.
Taken together, our results provide a detailed genetic history of Chad. We found that Berber ancestry spread into Northern Chad but did not reach Southern Chad. We also found that the spread of R1b can be attributed to Baggara Arabs, not ancient Eurasians or Near Eastern farmers. Furthermore, the spread of Chadic languages likely predated the arrival of R1b by over 3,000 years. Finally, the genetic data support the tantalizing prospect that the Laal language can give insight into an otherwise lost Central African linguistic phylum.
Supplementary Material
ACKNOWLEDGEMENTS
This work utilized the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of the National Institutes of Health. This research was supported by the Intramural Research Program of the Center for Research on Genomics and Global Health (CRGGH). The CRGGH is supported by the National Human Genome Research Institute, the National Institute of Diabetes and Digestive and Kidney Diseases, the Center for Information Technology, and the Office of the Director at the National Institutes of Health (1ZIAHG200362).
Funding information: US National Institutes of Health, 1ZIAHG200362
Footnotes
DISCLOSURE STATEMENT
The authors declare no competing interests.
WEB RESOURCES
Sudan genotype data, http://biologiaevolutiva.org/jbertranpetit/wp-content/uploads/2015/02/SudanImmunochip.zip; UCSC liftover tool, https://genome.ucsc.edu/cgi-bin/hgLiftOver; PLINK, https://www.cog-genomics.org/plink2; genetic maps, http://mathgen.stats.ox.ac.uk/impute/ALL_1000G_phase1integrated_feb2012_impute/; EIGENSOFT, https://data.broadinstitute.org/alkesgroup/EIGENSOFT/EIG-6.1.4.tar.gz
LITERATURE CITED
- Abu-Amero KK, Hellani A, González AM, Larruga JM, Cabrera VM, & Underhill PA (2009). Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions. BMC Genetics, 10, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, & Lange K (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19, 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Rotimi CN, & Shriner D (2017). Human ancestry correlates with language and reveals that race is not an objective genomic classifier. Scientific Reports, 7, 1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Alves-Silva J, Guimarães PEM, Santos MS, Brehm A, Pereira L, … Pena SDJ (2001). Phylogeography of the human mitochondrial haplogroup L3e: a snapshot of African prehistory and Atlantic slave trade. Annals of Human Genetics, 65, 549–563. [DOI] [PubMed] [Google Scholar]
- Bekada A, Fregel R, Cabrera VM, Larruga JM, Pestano J, Benhamamouch S, & González AM (2013). Introducing the Algerian mitochondrial DNA and Y-chromosome profiles into the North African landscape. PLOS ONE, 8, e56775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blench R (1999). The westward wandering of Cushitic pastoralists: explorations in the prehistory of Central Africa In: Baroin C & Boutrais J (Eds.), Man and animal in the Lake Chad Basin: Proceedings of the Mega-Chad Network Symposium (pp. 39–80). Orléans: Research Institute for Development. [Google Scholar]
- Blench R (2006). Archaeology, Language, and the African Past. Lanham, Maryland: AltaMira Press. [Google Scholar]
- Braukämper U (1994). Notes on the origin of Baggara Arab culture with special reference to the Shuwa In: Owens J (Ed.), Arabs and Arabic in the Lake Chad Region. Koln, Germany: Rüdiger Köppe Verlag. [Google Scholar]
- Bučková J, Černý V, & Novelletto A (2013). Multiple and differentiated contributions to the male gene pool of pastoral and farmer populations of the African Sahel. American Journal of Physical Anthropology, 151, 10–21. [DOI] [PubMed] [Google Scholar]
- Cadenas AM, Zhivotovsky LA, Cavalli-Sforza LL, Underhill PA, & Herrera RJ (2008). Y-chromosome diversity characterizes the Gulf of Oman. European Journal of Human Genetics, 16, 374–386. [DOI] [PubMed] [Google Scholar]
- Černý V, Fernandes V, Costa MD, Hájek M, Mulligan CJ, & Pereira L (2009). Migration of Chadic speaking pastoralists within Africa based on population structure of Chad Basin and phylogeography of mitochondrial L3f haplogroup. BMC Evolutionary Biology, 9, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaroni J, King RJ, Myres NM, Henn BM, Ducourneau A, Mitchell MJ, … Underhill PA (2010). The emergence of Y-chromosome haplogrouap J1e among Arabic-speaking populations. European Journal of Human Genetics, 18, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, … Underhill PA (2002). A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. The American Journal of Human Genetics, 70, 1197–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Trombetta B, Sellitto D, Massaia A, Destro-Bisol G, Watson E, … Scozzari R (2010a). Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages. European Journal of Human Genetics, 18, 800–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Trombetta B, Sellitto D, Massaia A, Destro-Bisol G, Watson E, … Scozzari R (2010b). Reply to Lancaster. European Journal of Human Genetics, 18, 1186–1187. [Google Scholar]
- DeLancey MW, & DeLancey MD (2000). Historical Dictionary of the Republic of Cameroon. Lanham, Maryland: The Scarecrow Press. [Google Scholar]
- Dobon B, Hassan HY, Laayouni H, Luisi P, Ricaño-Ponce I, Zhernakova A, … Bertranpetit J (2015). The genetics of East African populations: a Nilo-Saharan component in the African genetic landscape. Scientific Reports, 5, 9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret C (2002). The Civilizations of Africa: A history to 1800. Virginia: The University Press of Virginia. [Google Scholar]
- Fenner JN (2005). Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. American Journal of Physical Anthropology, 128, 415–423. [DOI] [PubMed] [Google Scholar]
- Haber M, Mezzavilla M, Bergström A, Prado-Martinez J, Hallast P, Saif-Ali R, … Tyler-Smith, C. (2016). Chad genetic diversity reveals an African history marked by multiple Holocene Eurasian migrations. The American Journal of Human Genetics, 99, 1316–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harich N, Costa MD, Fernandes V, Kandil M, Pereira JB, Silva NM, & Pereira L (2010). The trans-Saharan slave trade - clues from interpolation analyses and high-resolution characterization of mitochondrial DNA lineages. BMC Evolutionary Biology, 10, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan HY, Underhill PA, Cavalli-Sforza LL, & Ibrahim ME (2008). Y-chromosome variation among Sudanese: restricted gene flow, concordance with language, geography, and history. American Journal of Physical Anthropology, 137, 316–323. [DOI] [PubMed] [Google Scholar]
- Hathaway J (1997). The Politics of Households in Ottoman Egypt: The Rise of the Qazdağlis. Cambridge, United Kingdom: Cambridge University Press. [Google Scholar]
- Hollfelder N, Schlebusch CM, Günther T, Babiker H, Hassan HY, & Jakobsson M (2017). Northeast African genomic variation shaped by the continuity of indigenous groups and Eurasian migrations. PLOS Genetics, 13, e1006976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibn Furtu A (1576). The Book of the Bornu Wars.
- Ibn Furtu A (1578). The Book of the Kanem Wars.
- Jostins L, Xue Y, McCarthy S, Ayub Q, Durbin R, Barrett J, & Tyler-Smith C (2014). YFitter: maximum likelihood assignment of Y chromosome haplogroups from low-coverage sequence data. arXiv, 1407.7988.
- Kulichová I, Fernandes V, Deme A, Nováčková J, Stenzl V, Novelletto A, … Černý, V. (2017). Internal diversification of non-Sub-Saharan haplogroups in Sahelian populations and the spread of pastoralism beyond the Sahara. American Journal of Physical Anthropology, 164, 424–434. [DOI] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, … Myers RM (2008). Worldwide human relationships inferred from genome-wide patterns of variation. Science, 319, 1100–1104. [DOI] [PubMed] [Google Scholar]
- Loh P-R, Lipson M, Patterson N, Moorjani P, Pickrell JK, Reich D, & Berger B (2013). Inferring admixture histories of human populations using linkage disequilibrium. Genetics, 193, 1233–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor A (2006). A Military History of Modern Egypt: from the Ottoman Conquest to the Ramadan War. Westport, Connecticut: Praeger Security International. [Google Scholar]
- Mohammad T, Xue Y, Evison M, & Tyler-Smith C (2009). Genetic structure of nomadic Bedouin from Kuwait. Heredity, 103, 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorjani P, Sankararaman S, Fu Q, Przeworski M, Patterson N, & Reich D (2016). A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proceedings of the National Academy of Sciences of the United States of America, 113, 5652–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens J (1994). Arabs and Arabic in the Lake Chad region. Köln, Germany: Rüdiger Köppe Verlag. [Google Scholar]
- Philipp T, & Haarmann U (Eds.) (1998). The Mamluks in Egyptian Politics and Society. Cambridge, United Kingdom: Cambridge University Press. [Google Scholar]
- Pickrell JK, Patterson N, Loh P-R, Lipson M, Berger B, Stoneking M, … Reich D (2014). Ancient west Eurasian ancestry in southern and eastern Africa. Proceedings of the National Academy of Sciences of the United States of America, 111, 2632–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priehodová E, Abdelsawy A, Heyer E, & Černý V (2014). Lactase persistence variants in Arabia and in the African Arabs. Human Biology, 86, 7–18. [DOI] [PubMed] [Google Scholar]
- Shriner D, Tekola-Ayele F, Adeyemo A, & Rotimi CN (2014). Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry. Scientific Reports, 4, 6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M, Alshamali F, Silva P, Carrilho C, Mandlate F, Jesus Trovoada M, … Soares P (2015). 60,000 years of interactions between Central and Eastern Africa documented by major African mitochondrial haplogroup L2. Scientific Reports, 5, 12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, Alshamali F, Pereira JB, Fernandes V, Silva NM, Afonso C, … Pereira L (2012). The Expansion of mtDNA Haplogroup L3 within and out of Africa. Molecular Biology and Evolution, 29, 915–927. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, … Williams SM (2009). The genetic structure and history of Africans and African Americans. Science, 324, 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triska P, Soares P, Patin E, Fernandes V, Cerny V, & Pereira L (2015). Extensive admixture and selective pressure across the Sahel Belt. Genome Biology and Evolution, 7, 3484–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, … Oefner, P. J. (2000). Y chromosome sequence variation and the history of human populations. Nature Genetics, 26, 358–361. [DOI] [PubMed] [Google Scholar]
- Urasin V (2017a). R YTree. https://www.yfull.com/tree/R. Accessed August 11, 2017.
- Urasin V (2017b). R-V88 YTree. https://www.yfull.com/tree/R-V88. Accessed August 11, 2017.
- Vianello D, Sevini F, Castellani G, Lomartire L, Capri M, & Franceschi C (2013). HAPLOFIND: a new method for high-throughput mtDNA haplogroup assignment. Human Mutation, 34, 1189–1194. [DOI] [PubMed] [Google Scholar]
- Wilson CW (1888). On the tribes of the Nile Valley, north of Khartúm. The Journal of the Anthropological Institute of Great Britain and Ireland, 17, 3–25. [Google Scholar]
- Yunusbayev B, Metspalu M, Järve M, Kutuev I, Rootsi S, Metspalu E, … Villems R (2012). The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Molecular Biology and Evolution, 29, 359–365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.