Abstract
miR-224 has recently emerged as a driver oncomiR in sporadic colorectal carcinogenesis, but its pathogenetic role is still controversial. A large phenotypical and molecularly characterized series of preinvasive and invasive colorectal lesions was investigated for miR-224 expression by qRT-PCR and in situ hybridization. The caspase-3 and caspase-7 status was also assessed and correlated to miR-224 dysregulation. miR-224 was significantly upregulated during the adenoma-carcinoma sequence and in the context of inflammatory bowel disease dysplastic lesions, whereas its expression was significantly downregulated among BRAF-mutated tumors and in the presence of a DNA mismatch repair deficiency. miR-224 targets caspase-3 and caspase-7 in colorectal cancer, and this inverse relation was already evident from the earliest phases of transformation in intestinal mucosa. The miR-224/caspases axis may represent an interesting field of study for innovative biomarkers/therapeutics for BRAF-mutated/DNA mismatch repair-deficient tumors.
Introduction
For many years, sporadic colorectal cancer (CRC) has been considered to be a homogeneous condition developing within the context of the adenoma-carcinoma phenotypic sequence driven by the accumulation in genetic alterations of CRC key genes such as APC, KRAS, and TP53 [1]. However, comprehensive genomic profiling studies pinpointed the existence of multiple phenotypic and molecular CRC cascades with specific clinical and therapeutic implications. This heterogeneity includes colon mucosa transformation through the serrated pathway, alterations in the BRAF gene, and the presence of microsatellite instability (MSI) [1], [2], [3]. Another important colon mucosa cancerization field is represented by inflammatory bowel diseases (IBD), and both Crohn's disease (CD) and ulcerative colitis (UC) patients carry a significantly increased risk of developing CRC [4].
miRs aberrant expression is common in CRC and has been associated to cancer progression and clinical outcome [5], [6]. Among the others, miR-224 has emerged as a driver oncomiR involved in colorectal carcinogenesis [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17] and has been demonstrated to be a prognostic marker for survival of patients with CRC [7], [12]. When analyzed in the context of the CRC molecular landscape, miR-224 has been shown to be significantly downregulated in BRAF mutated tumors and in presence of MSI [8], [12].
Several cancer-associated genes, such as SMAD4 [12], [14], [18], [19] and p21 [13], have been validated to be directly targeted by miR-224 in the colorectal setting. In other cancers, miR-224 has been described to be a negative regulator of RASSF8 [20], [21], [22], HOXD10 [23], [24], and caspases [25].
Here, we tested miR-224 expression and dysregulation in a large phenotypical and molecular characterized series of preinvasive and invasive colorectal lesions, and we provide evidence supporting a driver role for miR-224 in the neoplastic progression of CRC. We also demonstrated that miR-224 is targeting caspases expression in CRC.
Materials and Methods
Ethical Approvals
Investigation has been conducted in accordance to the Declaration of Helsinki and according to national and international guidelines. Paraffin-embedded tissues considered in this study were retrospectively collected and analyzed following approval of the ethical committees from the files of the 1) Ohio State University Pathology Archive; 2) Department of Pathology, University of Ferrara; and 3) Surgical Pathology Unit, University of Padua.
Tissue Samples
Original slides (4-6 μm thick) obtained from archival paraffin-embedded tissue samples (H&E) were reassessed by expert gastrointestinal pathologists. In all cases, the histology assessment of IEN and CRC was done according to the current histology criteria [26], [27], [28].
Four hundred eleven tissue samples from a total of 377 patients were tested in the study:
-
i)
Sporadic carcinogenesis. A series of 60 endoscopy biopsy samples was obtained from patients with different types of left-side sporadic colic lesions: 20 tubular adenomas with low-grade intraepithelial neoplasia (LG-IEN, formerly known as LG-dysplasia), 20 tubular adenomas with high-grade IEN (HG-IEN), and 20 well-differentiated (G1) pT1/pT2 CRCs. Another 20 normal colonic mucosa biopsy samples were obtained from patients who underwent colonoscopy for irritable bowel syndrome. No sample was obtained from cases of familial adenomatous polyposis syndrome.
-
ii)
IBD-related carcinogenesis. A total of 175 IBD-related samples (98 from UC patients and 77 from CD patients) were obtained from colectomy samples performed for histologically proven dysplastic/invasive lesions. The assessment of IBD acute inflammation versus nonactive inflammation was based on microscopic criteria, activity being defined as the presence of neutrophils or unequivocal damage of the surface and crypt epithelium typically in conjunction with neutrophils [27], [29]. For UC, a total of inactive disease (n = 50), IEN (n = 26; both low and high grade), and CRC (n = 22) specimens were considered. The 77 CD-related specimens included inactive disease (n = 50), IEN (n = 13; both low and high grade), and CRC (n = 14) specimens.
-
iii)
A further series of 156 colectomy specimens was considered according to pathological and molecular features of the tumors. In 17 stage IV cases, matched samples of normal colorectal mucosa (>3 cm far from CRC), primary CRC, and synchronous liver metastasis were selected, for a total of 51 samples. Another 105 CRC cases were retrospectively selected according their molecular profiling, as assessed for clinical purposes by Sequenom MassArray sequencing (for the purposes of the study, only the mutational status of KRAS exons 2-4, NRAS exons 2-4, and BRAF exon 15 was considered; Diatech Pharmacogenetics, Jesi, Italy) and by immunohistochemical evaluation of DNA mismatch repair (MMR) proteins (MSH2, MSH6, MLH1, PMS2; Dako, Carpinteria, CA) to assess MMR proficiency (MMRp) or deficiency (MMRd). In case of MMRd, cases were evaluated for microsatellite status (Titano kit; Diatech Pharmacogenetics), and in all the considered MMRd cases, microsatellite instability (MSI) was confirmed. These 105 CRCs were classified as: KRAS/NRAS/BRAF wild type and mismatch repair proficient (MMRp) (n = 25), KRAS exon 2G12-mutated (n = 25), KRAS exon 2G13-mutated (n = 25), BRAF exon 15 V600E-mutated and mismatch repair deficient (MMRd; which usually correspond to a MSI phenotype) (n = 20), BRAF exon 15 V600E-mutated and MMRp (n = 5), and KRAS/NRAS/BRAF wild type and MMRd (n = 5). The 25 KRAS/NRAS/BRAF wild-type and MMRp samples were selected as age- and stage-matched to the MMRd tumors series.
Mutational Profiling
Screening for mutations was performed on a Sequenom MassARRAY platform by applying the Myriapod Colon status panel (Diatech Pharmacogenetics, Jesi, Italy). Details on targeted hotspots of the commercial panel are at http://www.diatechpharmacogenetics.com. In each case, DNA was prepared after enrichment for neoplastic cellularity to at least 60% using manual microdissection of 5 consecutive 4-μm FFPE sections and purified using the QIAamp DNA FFPE Tissue Kit (Qiagen; Milan, Italy). In a random set of 20 samples, mutational profiling was validated by Sanger sequencing.
miR-224 In Situ Hybridization (ISH) and Immunohistochemical (IHC) Analyses
Fifty-five primary CRC surgical samples (25 KRAS mutated, 5 BRAF mutated, 10 MMRd) were processed using the Galileo CK3500 Arrayer (www.isenet.it), a semiautomatic and computer-assisted tissue microarray platform, as previously described [30], [31]. Three tissue cores (1 mm in diameter) were obtained from each considered lesion.
Locked nucleic acid (LNA) probes with complementarity to miR-224 were labeled with 5′-biotin and synthesized using Exiqon (Vedbaek, Denmark). Tissue sections were digested with ISH protease 1 (Ventana Medical Systems, Milan, Italy), and ISH was performed as previously described [25], [32], with minor modifications. Positive (U6; Exiqon) and negative scrambled LNA probes were used as controls (Supplementary Figure 1). Only cytoplasmic miR-224 intensity was retained for scoring purposes.
Protein/microRNA co-expression analysis was carried out as previously described with minor modifications [25]. After ISH staining, we used the Benchmark LT automated system from Leica Microsystems Bondmax (Leica, Wetzlar, Germany) according to the manufacturer's specifications to perform the immunohistochemistry for CASP3 or CASP7 (Santa Cruz Biotechnology, Inc.; Dallas, TX). The expression of CASP3 or CASP7 was cytoplasmic and nuclear. Color-relative staining intensities were evaluated by 1) semiquantitative evaluation considering a three-tier scale according to the intensity of the reaction (0 = negative; 1+ = faint/moderate staining; 2+ strong and diffuse staining) and 2) ImageJ software evaluation.
DNA mismatch repair machinery deficient tumors (MMRd) were defined in the absence of nuclear immunostaining for one of the couples MLH1/PMS2 or MSH2/MSH6 (Dako, Carpinteria, CA) in tumor cells, as assessed in the colorectal setting [33].
Reverse Transcription and Quantitative Real-Time PCR
Total RNA was extracted using the RecoverAll kit (Ambion, Austin, TX). The NCodeTM miRNA qRT-PCR method (Invitrogen, Carlsbad, CA) was applied to detect and quantify mature hsa-miR-224 according to the manufacturer's instructions using an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Normalization was performed with the small nuclear RNA U6B. All reactions were run in duplicate, including no-template controls.
Plasmid Construction, Cell Lines, and Reagents
All the human CRC-derived cell lines used in this study were purchased from the American Type Culture Collection (ATCC, Manassas, VA). RKO and HCT116 cells were regularly maintained in McCoy-5A media (Life Technologies, Carlsbad, CA) containing 10% fetal bovine serum (Life Technologies) and 1% streptomycin-penicillin (Life Technologies) at 37°C in a CO2 incubator (5% CO2, 100% H2O).
For transfection experiments, the human miR-224 precursor miRNA and the negative control were purchased from Thermo Fisher Scientific (Waltham, MA). The experiments were run in triplicates.
For Western blot analysis, antibody against CASP7 was purchased from Cell Signaling (Danvers, MA), whereas anti-vinculin antibody was from Sigma-Aldrich (Saint Louis, MO).
Array Database Meta-Analysis
The TCGA (https://cancergenome.nih.gov/) and the Oncomine (www.oncomine.org) databases and gene microarray analysis tools were explored (15 June 2016) to assess miR-224 and caspases expression in colorectal cancer samples. Oncomine algorithms, which enable multiple comparisons among different studies, were used for the statistical analysis of the differences in mRNA expression between the aforementioned comparisons.
Statistical Analysis
Differences between groups were tested by applying the unpaired/paired t test, ordinary one-way ANOVA with the Greenhouse-Geisser correction, and Fisher’s exact test, as appropriate. P values <.05 were considered significant. The statistical analysis was performed using STATA software (Stata Corporation, College Station, TX).
Results
miR-224 Is Significantly Upregulated in Sporadic and IBD-Related Colorectal Carcinogenesis
To extend previous findings on miR-224 expression in CRC [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], miR-224 expression (qRT-PCR) was investigated in 80 endoscopy biopsy samples representative of 20 normal colon mucosa, 40 sporadic tubular adenomas, and 20 early-stage CRCs. A significant miR-224 upregulation was observed in LG-IEN (P = .033), in HG-IEN (P < .001), and in CRC (P < .001) in comparison to normal mucosa, and among LG-IEN and HG-IEN lesions (P < .001), LG-IEN and CRC (P < .001) (Figure 1A). Overall, miR-224 expression increased progressively and significantly along with the adenoma-carcinoma sequence (ANOVA, P < .001).
Figure 1.
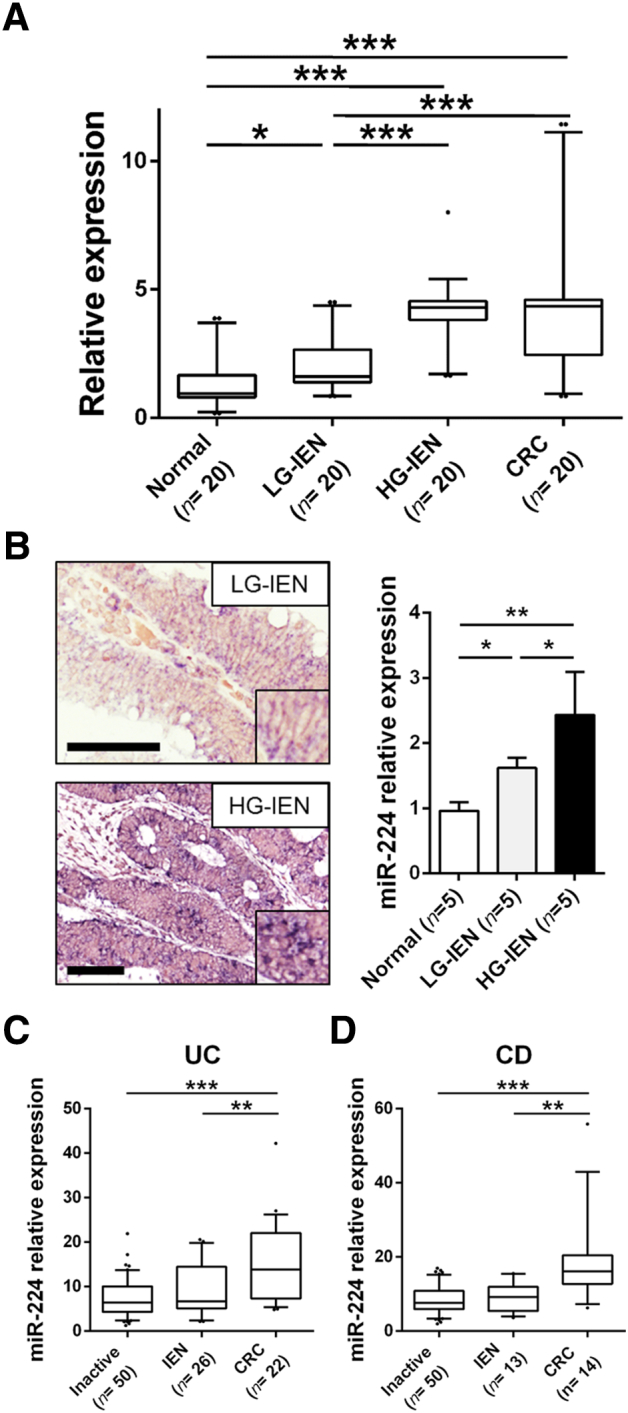
miR-224 is significantly upregulated in sporadic and IBD-related colorectal carcinogenesis. (A) A significant miR-224 upregulation was observed by qRT-PCR in LG-IEN (P = .033), in HG-IEN (P < .001), and in early-stage CRCs (P < .001) in comparison to normal colon mucosa. Also, HG-IEN lesions and CRCs showed a significantly higher miR-224 expression in comparison to LG-IEN adenomas (both P < .001). (B) Representative ISH analysis pictures showing a significant miR-224 overexpression in HG lesions in comparison to LG adenomas (ORIGINAL magnifications 20×, scale bars 100 μm) and the relative expression as assessed by image software analysis. (C-D) miR-224 expression (qRT-PCR) was also investigated in a large series of IBD-related IEN (n = 39) and CRC (n = 36) lesions. In both ulcerative colitis (UC; C) and Crohn's disease (CD; D), miR-224 levels were significantly upregulated between inactive IBD disease or IEN and full-blown CRC (UC, P < .001 and P = .006, respectively; CD, P < .001 and P = .010, respectively). Although not significant, miR-224 expression was higher in IEN samples in comparison to inactive disease.
To further confirm miR-224 overexpression according to IEN grading, miR-224 ISH analysis was performed in 5 normal colon mucosa, 5 LG-IEN, and 5 HG-IEN samples. The dysplastic samples showed a weak to moderate granular blue miR-224 cytoplasmic staining in neoplastic epithelia. Also, lymphocytes and plasma cells showed a faint cytoplasmic staining. The analysis of the relative miRNA expression as acquired by Image J software demonstrated a higher miR-224 overexpression in dysplastic lesions in comparison to normal mucosa (P = .012 for LG-IEN and P = .002 for HG-IEN) and in HG-IEN compared to LG-IEN lesions (P = .041) (Figure 1B).
We also investigated miR-224 expression in two series of endoscopy biopsy samples obtained from IBD patients. Of note, these series included a significant number of IBD-related IEN (n = 39) and CRC (n = 36) samples. In both UC and CD (Figure 1C) samples, a significant miR-224 upregulation was observed in full-blown CRC compared to the inactive IBD disease or IEN. Even if the difference was not statistically significant, miR-224 expression was higher in IEN samples in comparison to inactive disease.
miR-224 Dysregulation in Advanced CRCs Is Dependent on the Molecular Landscape of the Lesion
The role of miR-224 during CRC progression is controversial [9], [12], [15]. However, most studies have demonstrated that miR-224 overexpression promotes CRC metastasis and is an important prognostic marker in CRC patients. To further test this hypothesis, we investigated miR-224 expression by qRT-PCR in a series of 17 stage IV CRCs (Figure 2A). A significant overexpression of miR-224 was observed in both primary CRCs (P < .001) and liver metastases (P < .001) compared to matched normal colon mucosa. Metastatic samples also showed a significantly higher miR-224 expression in comparison to primary tumors (P < .001). Overall, miR-224 expression increased progressively and significantly along with CRC metastatic sequence (P < .001).
Figure 2.
miR-224 is significantly upregulated in metastatic disease and is downregulated in MMRd tumors. (A) miR-224 expression was evaluated by qRT-PCR in matched normal, primary CRC and synchronous liver metastasis obtained from a series of 17 stage IV CRCs. A significant overexpression of miR-224 was observed in primary CRCs in comparison to normal colon mucosa (P < .001) and CRC liver metastases in comparison to primary CRCs (P < .001) compared to normal colon mucosa. (B) ISH analysis further confirmed qRT-PCR data, with a significant mIR-224 overexpression in paired primary tumors and CRC liver metastasis in comparison to normal colon mucosa. Note in the metastasis picture, the miR-224 negative hepatic parenchyma in comparison to the miR-224 positive metastatic lesion (original magnifications 10× and 40×, scale bars 100 μm). On the right is the relative value as assessed by image software analysis, which confirmed qRT-PCR data. (C) No significant differences in miR-224 expression were observed among CRCs stratified according to KRAS status, as confirmed by both qRT-PCR and ISH analyses. BRAF-mutated tumors showed a lower miR-224 expression in comparison to wild-type and KRAS-mutated tumors (P = .013 and P < .001, respectively). On the bottom of the figure is the representation of the results of Sanger sequencing. (D) MMRp tumors showed a significantly higher miR-224 expression in comparison to an age- and stage-matched MMRd tumors series (P < .001). Representative images of mismatch repair protein immunohistochemical status are also shown.
miR-224 expression was also investigated by ISH in five cases of stage IV CRCs. A consistently significant overexpression was observed in paired primary tumors and CRC liver metastasis in comparison to normal colon mucosa in all the tested samples (Figure 2B). Normal colocytes showed a negative or faint cytoplasmic staining.
A retrospective series of 105 molecularly characterized CRCs was selected according to their KRAS/NRAS/BRAF status and the expression of four mismatch repair proteins to test miR-224 dysregulation according to the CRC molecular landscape. No significant differences were observed among CRCs stratified according to KRAS status (Figure 2C), whereas a significant downregulation was observed in BRAF-mutated tumors in comparison to wild-type (P = .013) and KRAS-mutated (P < .001) samples. Further supporting these observations, in two cases per group, miR-224 ISH analysis was performed and revealed a moderate to strong cytoplasmic expression in all wild-type and KRAS-mutated tested samples; the two BRAF-mutated tumors showed a moderate expression of the miR-224. Moreover, as previously observed [12], miR-224 was significantly overexpressed in a small series of MMRp tumors in comparison to an age- and stage-matched MMRd tumors group (P < .001; Figure 2D).
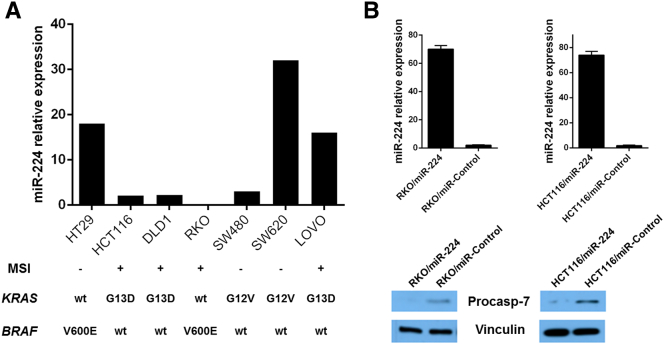
In keeping with tissue data, analysis of miR-224 expression in 7 CRC cell lines demonstrated that 3 out of 4 MSI cell lines were characterized by a low miR-224 expression (Figure 3A).
Figure 3.
miR-224 is downregulated in MSI CRC-derived cancer cell line, and its overexpression targets caspase-7 expression. (A) We tested miR-224 expression in seven CRC-derived cell lines and observed that three out of four MSI cell lines were characterized by a significantly low miR-224 expression (the molecular profiling of the single cell lines is also shown). (B) miR-224 overexpression in RKO and HCT116 corresponded to a significant reduction in procaspase-7 expression, as assessed by Western blot analysis (92.2% reduction for RKO cells and 87.9% reduction for HCT116).
Caspases 3 and 7 as miR-224 Targets in Colorectal Carcinogenesis
We have previously reported that miR-224 is implicated in lung cancer pathogenesis through targeting caspase-3 and caspase-7. Our luciferase reporter assays also confirmed miR-224 directly binding to the 3’UTR of caspase3 and caspase-7 [25]. Among the seven CRC cell lines, we selected the two characterized by the lowest miR-224 expression (i.e., RKO and HCT116) to overexpress miR-224. We transiently transduced these two cell lines with miR-224 precursor and confirmed miR-224 overexpression by qRT-PCR (Figure 3B). The overexpression resulted in a significant reduction in pro-caspase-7 expression, as assessed by Western blot analysis (92.2% reduction for RKO cells and 87.9% reduction for HCT116), whereas no significant alteration was observed in pro-caspase-3.
The correlation between miR-224 and its target proteins (i.e., caspase-3 and caspase-7) was further evaluated by miR-224 ISH and caspase immunohistochemical analyses in a series of 10 normal colon mucosae, 5 tubular adenomas with low-grade dysplasia, and 55 CRCs.
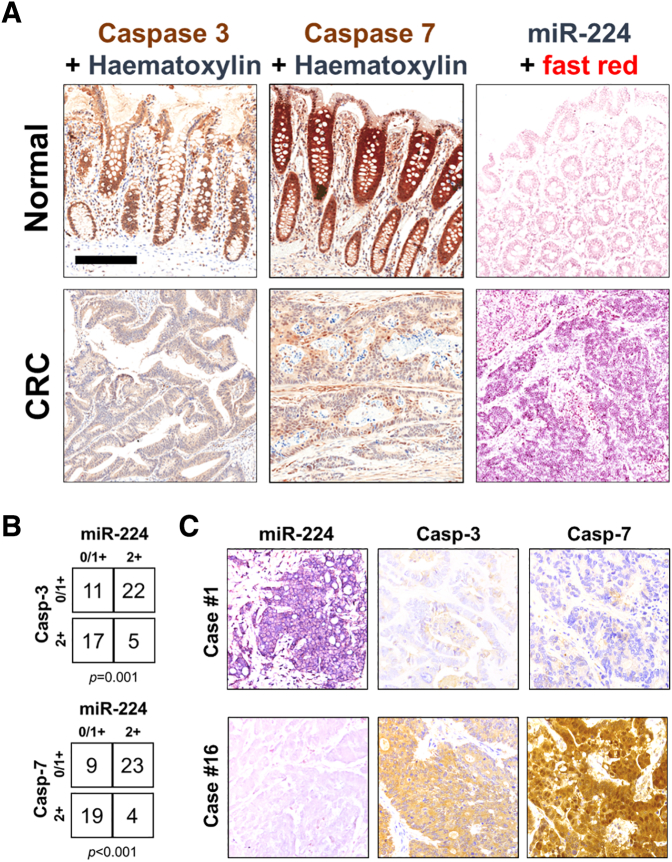
Both caspases showed a moderate/strong cytoplasmic and nuclear immunoreaction in normal crypts, with a decreased expression within gland's neck and superficial epithelial layer (Figure 4A and Supplementary Figure 2). CRCs showed a negative to moderate caspases expression. A significant inverse correlation between miR-224 and caspases expression was observed in the series of 55 CRCs (Figure 4, B and C). Of note, 7 out of the 10 MSI CRCs showed a negative to faint miR-224 expression, which was associated to a moderate caspases expression.
Figure 4.
CRCs are characterized by a significant miR-224/caspases inverse correlation. (A) Representative images of miR-224 ISH and caspase-3 and caspase-7 immunohistochemical analyses in a series of normal colic mucosa and CRC samples. (original magnifications 10×, scale bars 200 μm). (B) The analysis of 55 primary CRCs samples demonstrated a significant inverse correlation between miR-224 and caspases expression. (C) Two representative cases in which there was a significant inverse relationship between the expression of the miR-224 and of the two caspases.
This inverse relationship (i.e., lower the caspases, higher the miR-224) was also observed in the five tubular adenomas, supporting an early oncogenic role for miR-224. The dysplastic glands showed a significant downregulation in caspase expression concurrent to a miR-224 overexpression (Supplementary Figure 3).
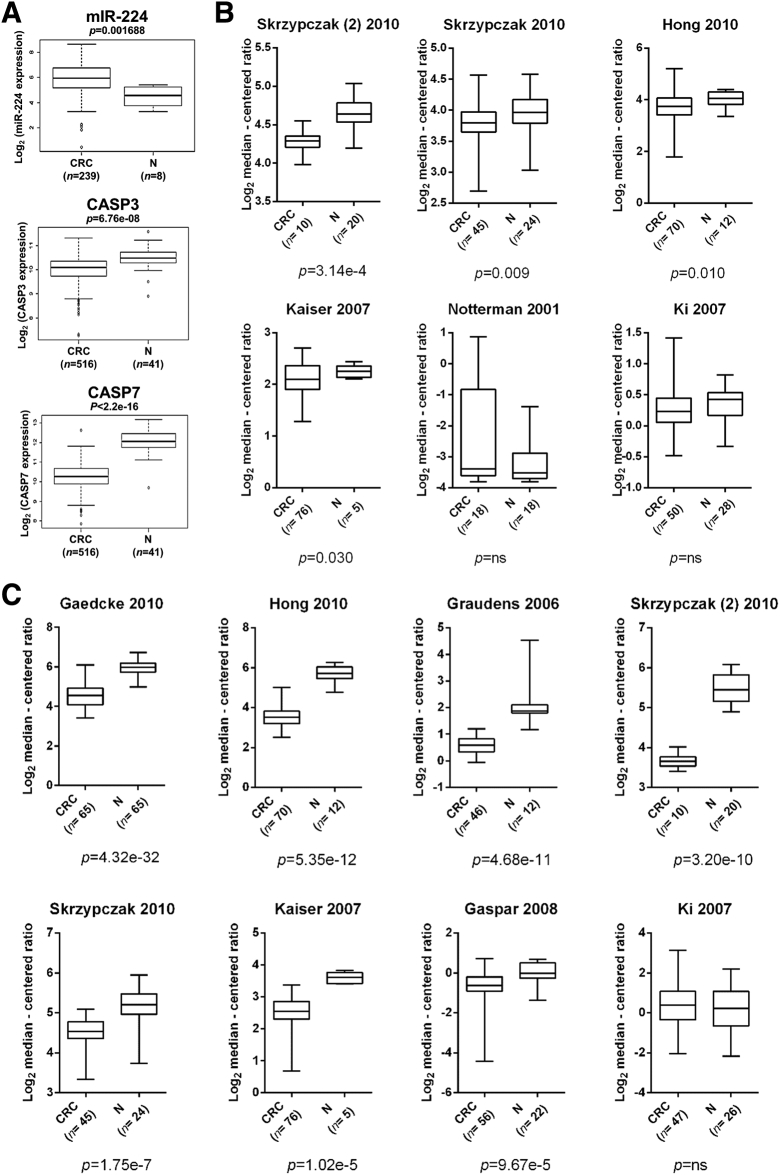
To further support these data, we explored the TCGA and the Oncomine databases (Figure 5). In the TCGA series, miR-224 resulted in being significantly upregulated in CRCs in comparison to normal samples (P = .001688), whereas both caspases were significantly downregulated (caspase-3, P = 6.67e-08; caspase-7, P < 2.2e-16) (Figure 5A). Caspase-3 expression was significantly downregulated in CRC in comparison to normal mucosa in 4 out of 6 Oncomine deposited studies (a total of 269 CRCs and 107 normal samples analyzed; P = .010; Figure 5B), whereas Caspase-7 was significantly downregulated in 7 out of 8 studies (a total of 415 CRCs and 186 normal samples analyzed; P = 1.02e-5; Figure 5C).
Figure 5.
Caspases are significantly downregulated in CRC. (A) In the TCGA CRC series, miR-224 resulted significantly upregulated in CRCs in comparison to normal samples (P = .001688), whereas both caspases were significantly downregulated (caspase-3, P = 6.67e-08; caspase-7, P < 2.2e-16). (B) Caspase-3 expression was significantly downregulated in CRC in comparison to normal mucosa in 4 out of 6 Oncomine deposited studies (a total of 269 CRCs and 107 normal samples analyzed; P = .010). (C) Caspase-7 was significantly downregulated in 7 out of 8 studies (a total of 415 CRCs and 186 normal samples analyzed; P = 1.02e-5).
Discussion
Despite recent improvements in therapy mainly due to the introduction of predictive biomarkers into clinical practice, CRC still remains a major health burden with significant morbidity and mortality worldwide [34].
Among the different classes of cancer biomarkers, miRs have emerged as novel noninvasive tools for diagnostic, predictive, and prognostic applications in tissues and biofluids [17] and as having a potential role as therapeutics [35].
miR-224 has emerged as a potential driver of CRC acting as an oncomiR; however, some contrasting data related to its expression have been reported [9], [12], [15].
Here we demonstrated an early upregulation of miR-224 in both sporadic and IBD-related carcinogenetic process. In the adenoma-carcinoma pathway, both low-grade and high-grade intraepithelial lesions (i.e., dysplasia) were characterized by miR-224 overexpression, which was demonstrated by both in vitro and in situ studies. On the other hand, in the context of IBD, dysplastic lesions did not show a significant overexpression in comparison to colon mucosa with inactive disease, which support an important oncogenic role of inflamed mucosa in tumor initiation via early dysregulation of several carcinogenetic pathways. Thus, our data support the use of miR-224 expression as an early biomarker of colon mucosa transformation in the sporadic setting; however, miR-224 has no significant diagnostic impact in discriminating dysplastic epithelia from surrounding mucosa in IBD patients.
We further demonstrated that miR-224 is also involved in the acquisition of the metastatic phenotype. It has to be underlined that we examined matched samples obtained from 17 stage IV CRC patients, and miR-224 expression was always higher in metastatic deposits in comparison to the corresponding primary tumor. In their seminal article, Yuan and colleagues showed an antimetastatic effect of miR-224 on CRC-derived cancer cell lines [15]; however, this could have been related to the molecular characteristics of the cell lines adopted for their in vivo experiments.
Another important observation gathered by the analysis of our samples is the validation of previous results published by Ling and colleagues [12] on the different miR-224 expression profiles depending on the molecular landscape of the tumors. This concordance corroborates the quality of the presented data. In our series, we did not observe any difference in miR-224 expression according to KRAS status, which is in contrast to Amankwatia and colleagues [8] but in line with the larger series of Ling [12]. Moreover, we observed a significant miR-224 downregulation among tumors characterized by a BRAFV600E mutation and by the MMRd phenotype. Because most of the BRAFV600E-mutated samples were also MMRd, it is difficult to understand the genetic driver of this difference, and larger series of samples should be investigated to clarify this point.
As previously demonstrated in lung cancer, miR-224 targets caspase-3 and caspase-7 also in CRC, and this was also observed in vivo by co-staining primary CRC samples for miR-224 and caspases. This was already evident in a series of preinvasive lesions (i.e., LG-IEN) supporting a strong pathogenetic role for caspase downregulation by miR-224 during colon mucosa carcinogenesis.
A low miR-224 coupled with high caspases expression was observed in 7/10 BRAF-mutated/MMRd samples. According to this information, data produced within the Consensus Molecular Subtype (CMS) of CRC [3] showed a significant caspase activation (Biocarta) in the CMS1 group which is characterized by the BRAF-mutated/MMRd status in comparison to the other three CMS groups. These tumors are characterized by good overall and disease-free survivals but show the worst survival in relapsing disease after adjuvant chemotherapies. Of note, a high caspase-3 activity has been significantly correlated with a higher risk of recurrence and has been preferentially found in tumors of the right side of the colon (which are more frequently BRAF-mutated/MMRd) [36]. Overall, these findings support the miR-224/caspases axis as an interesting field of study for innovative biomarkers/therapeutics for BRAF-mutated/MMRd tumors.
Further data on the correlation between BRAF status, MMR deficiency, and metastatic disease would be of paramount importance in the near future given the specific targeted option currently under investigation for these special subgroups of CRC [37], [38], [39].
Acknowledgements
All authors took part in writing the manuscript and approved the final, submitted version. We would like to thank Dr. Roberta Salmaso, Dr. Martina Grigiante, and Dr. Mariangela Balistreri for their technical assistance.
Footnotes
Disclosure of potential conflicts of interest: The authors have no competing interests to disclose.
Financial support: This work was partly supported by the grants from the Italian Association for Cancer Research (AIRC Regional grant 2008 N. 6421) to Massimo Rugge and National Natural Science Foundation of China (81672305) to Ri Cui. Nicola Valeri is supported by Cancer Research UK (grant number CEA A18052), the National Institute for Health Research (NIHR) Biomedical Research Centre (BRC) at The Royal Marsden NHS Foundation Trust and The Institute of Cancer Research (grant numbers A62, A100, A101, A159), and the European Union FP7 (grant number CIG 334261).
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2018.10.013.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Fassan M, Baffa R, Kiss A. Advanced precancerous lesions within the GI tract: The molecular background. Best Pract Res Clin Gastroenterol. 2013;27(2):159–169. doi: 10.1016/j.bpg.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saraggi D, Fassan M, Mescoli C, Scarpa M, Valeri N, Michielan A, D'Incá R, Rugge M. The molecular landscape of colitis-associated carcinogenesis. Dig Liver Dis. 2017;49(4):326–330. doi: 10.1016/j.dld.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat F. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25(4):469–483. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Angelo E, Vicentini C, Agostini M, Kiss A, Baffa R, Scarpa A, Fassan M. MicroRNAs as tools and effectors for patient treatment in gastrointestinal carcinogenesis. Curr Drug Targets. 2015;16(4):383–392. doi: 10.2174/1389450116666141210091454. [DOI] [PubMed] [Google Scholar]
- 7.Adamopoulos PG, Kontos CK, Rapti SM, Papadopoulos IN, Scorilas A. miR-224 overexpression is a strong and independent prognosticator of short-term relapse and poor overall survival in colorectal adenocarcinoma. Int J Oncol. 2015;46(2):849–859. doi: 10.3892/ijo.2014.2775. [DOI] [PubMed] [Google Scholar]
- 8.Amankwatia EB, Chakravarty P, Carey FA, Weidlich S, Steele RJ, Munro AJ, Wolf CR, Smith G. MicroRNA-224 is associated with colorectal cancer progression and response to 5-fluorouracil-based chemotherapy by KRAS-dependent and -independent mechanisms. Br J Cancer. 2015;112(9):1480–1490. doi: 10.1038/bjc.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ke TW, Hsu HL, Wu YH, Chen WT, Cheng YW, Cheng CW. MicroRNA-224 suppresses colorectal cancer cell migration by targeting Cdc42. Dis Markers. 2014;2014:617150. doi: 10.1155/2014/617150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T, Lai Q, Wang S, Cai J, Xiao Z, Deng D, He L, Jiao H, Ye Y, Liang L. MicroRNA-224 sustains wnt/beta-catenin signaling and promotes aggressive phenotype of colorectal cancer. J Exp Clin Cancer Res. 2016;35(1) doi: 10.1186/s13046-016-0287-1. [21-016-0287-1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao WT, Li TT, Wang ZG, Wang SY, He MR, Ye YP, Qi L, Cui YM, Wu P, Jiao HL. microRNA-224 promotes cell proliferation and tumor growth in human colorectal cancer by repressing PHLPP1 and PHLPP2. Clin Cancer Res. 2013;19(17):4662–4672. doi: 10.1158/1078-0432.CCR-13-0244. [DOI] [PubMed] [Google Scholar]
- 12.Ling H, Pickard K, Ivan C, Isella C, Ikuo M, Mitter R, Spizzo R, Bullock M, Braicu C, Pileczki V. The clinical and biological significance of MIR-224 expression in colorectal cancer metastasis. Gut. 2016;65(6):977–989. doi: 10.1136/gutjnl-2015-309372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olaru AV, Yamanaka S, Vazquez C, Mori Y, Cheng Y, Abraham JM, Bayless TM, Harpaz N, Selaru FM, Meltzer SJ. MicroRNA-224 negatively regulates p21 expression during late neoplastic progression in inflammatory bowel disease. Inflamm Bowel Dis. 2013;19(3):471–480. doi: 10.1097/MIB.0b013e31827e78eb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Ren J, Gao Y, Ma JZ, Toh HC, Chow P, Chung AY, Ooi LL, Lee CG. MicroRNA-224 targets SMAD family member 4 to promote cell proliferation and negatively influence patient survival. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0068744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan K, Xie K, Fox J, Zeng H, Gao H, Huang C, Wu M. Decreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology. 2013;145(4):853–864.e9. doi: 10.1053/j.gastro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. Up-regulation of miR-224 promotes cancer cell proliferation and invasion and predicts relapse of colorectal cancer. Cancer Cell Int. 2013;13(1) doi: 10.1186/1475-2867-13-104. [104-2867-13-104] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Xu A, Li J, J Fu, Wang G, Yang Y, Cui L, Sun J. Fecal miR-29a and miR-224 as the noninvasive biomarkers for colorectal cancer. Cancer Biomark. 2016;16(2):259–264. doi: 10.3233/CBM-150563. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J, Hu M, Wang F, Song M, Huang Q, Ge B. miR-224 controls human colorectal cancer cell line HCT116 proliferation by targeting Smad4. Int J Med Sci. 2017;14(10):937–942. doi: 10.7150/ijms.19565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Yang J, Di J, Cui M, Xing J, Wu F, Wu W, Yang H, Zhang C, Yao Z. Downregulated USP3 mRNA functions as a competitive endogenous RNA of SMAD4 by sponging miR-224 and promotes metastasis in colorectal cancer. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-04368-3. [4281-017-04368-3] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He C, Wang L, Zhang J, Xu H. Hypoxia-inducible microRNA-224 promotes the cell growth, migration and invasion by directly targeting RASSF8 in gastric cancer. Mol Cancer. 2017;16(1) doi: 10.1186/s12943-017-0603-1. [35-017-0603-1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Li Y, Wang FF, Lv W, Xie X, Cheng X. Over-expressed miR-224 promotes the progression of cervical cancer via targeting RASSF8. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0162378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Liu W, Zhang YP, Huang XR. The miR-224 promotes non-small cell lung cancer cell proliferation by directly targeting RASSF8. Eur Rev Med Pharmacol Sci. 2017;21(14):3223–3231. [PubMed] [Google Scholar]
- 23.Li S, Zhang J, Zhao Y, Wang F, Chen Y, Fei X. miR-224 enhances invasion and metastasis by targeting HOXD10 in non-small cell lung cancer cells. Oncol Lett. 2018;15(5):7069–7075. doi: 10.3892/ol.2018.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Ding C, Chen C, Zhang X, Xiao H, Xie F, Lei L, Chen Y, Mao B, Jiang M. miR-224 promotion of cell migration and invasion by targeting homeobox D 10 gene in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2014;29(4):835–842. doi: 10.1111/jgh.12429. [DOI] [PubMed] [Google Scholar]
- 25.Cui R, Kim T, Fassan M, Meng W, Sun H, Jeon YJ, Vicentini C, Tili E, Peng Y, Scarpa A. MicroRNA-224 is implicated in lung cancer pathogenesis through targeting caspase-3 and caspase-7. Oncotarget. 2015;6(26):21802–21815. doi: 10.18632/oncotarget.5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosman FT, Carneiro F, Hruban RH, Theise N, editors. WHO Classification of Tumours of the Digestive System. Fourth edition. IARC; France: 2014. [Google Scholar]
- 27.Cornaggia M, Leutner M, Mescoli C, Sturniolo CG, Gullotta R, Gruppo Italiano Patologi Apparato Digerente (GIPAD), Società Italiana di Anatomia Patologica e Citopatologia Diagnostica/International Academy of Pathology, Italian division (SIAPEC/IAP) Chronic idiopathic inflammatory bowel diseases: The histology report. Dig Liver Dis. 2011;43(Suppl. 4):S293–S303. doi: 10.1016/S1590-8658(11)60585-9. [DOI] [PubMed] [Google Scholar]
- 28.Lanza G, Messerini L, Gafa R, Risio M, Gruppo Italiano Patologi Apparato Digerente (GIPAD), Societa Italiana di Anatomia Patologica e Citopatologia Diagnostica/International Academy of Pathology, Italian division (SIAPEC/IAP) Colorectal tumors: the histology report. Dig Liver Dis. 2011;43(Suppl. 4):S344–S355. doi: 10.1016/S1590-8658(11)60590-2. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig K, Fassan M, Mescoli C, Pizzi M, Balistreri M, Albertoni L, Pucciarelli S, Scarpa M, Sturniolo GC, Angriman I. PDCD4/miR-21 dysregulation in inflammatory bowel disease-associated carcinogenesis. Virchows Arch. 2013;462(1):57–63. doi: 10.1007/s00428-012-1345-5. [DOI] [PubMed] [Google Scholar]
- 30.Saraggi D, Galuppini F, Fanelli GN, Remo A, Urso EDL, Bao RQ, Bacchin D, Guzzardo V, Luchini C, Braconi C. MiR-21 up-regulation in ampullary adenocarcinoma and its pre-invasive lesions. Pathol Res Pract. 2018;214(6):835–839. doi: 10.1016/j.prp.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 31.Saraggi D, Galuppini F, Remo A, Urso EDL, Bacchin D, Salmaso R, Lanza C, Bao RQ, Fanelli GN, Guzzardo V. PD-L1 overexpression in ampulla of vater carcinoma and its pre-invasive lesions. Histopathology. 2017;71(3):470–474. doi: 10.1111/his.13254. [DOI] [PubMed] [Google Scholar]
- 32.Cui R, Meng W, Sun HL, Kim T, Ye Z, Fassan M, Jeon YJ, Li B, Vicentini C, Peng Y. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc Natl Acad Sci U S A. 2015;112(31):E4288–E4297. doi: 10.1073/pnas.1502068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remo A, Fassan M, Lanza G. Immunohistochemical evaluation of mismatch repair proteins in colorectal carcinoma: The AIFEG/GIPAD proposal. Pathologica. 2016;108:104–109. [PubMed] [Google Scholar]
- 34.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 35.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonges LE, Nagelkerke JF, Ensink NG, van der Velde EA, Tollenaar RA, Fleuren GJ, van de Velde CJ, Morreau H, Kuppen PJ. Caspase-3 activity as a prognostic factor in colorectal carcinoma. Lab Invest. 2001;81(5):681–688. doi: 10.1038/labinvest.3780277. [DOI] [PubMed] [Google Scholar]
- 37.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, McDermott R, Hill A. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 38.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huijberts S, Schellens JHM, Elez E, Cuyle P, Van Cutsem E, Yaeger R, Fakih M, Montagut C, Peeters M, Desai J. BEACON CRC: safety lead-in (SLI) for the combination of binimetinib (BINI), encorafenib (ENCO), and cetuximab (CTX) in patients (pts) with BRAF-V600E mutated colorectal cancer. Ann Oncol. 2017;28(Suppl_5):v158–v208. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures