Abstract
In addition to unwinding double-stranded nucleic acids, helicase activity can also unfold noncanonical structures such as G-quadruplexes. We previously characterized Pif1 helicase catalyzed unfolding of parallel G-quadruplex DNA. Here we characterized unfolding of the telomeric G-quadruplex, which can fold into antiparallel and mixed hybrid structures and found significant differences. Telomeric DNA sequences are unfolded more readily than the parallel quadruplex formed by the c-MYC promoter in K+. Furthermore, we found that under conditions in which the telomeric quadruplex is less stable, such as in Na+, Pif1 traps thermally melted quadruplexes in the absence of ATP, leading to the appearance of increased product formation under conditions in which the enzyme is preincubated with the substrate. Stable telomeric G-quadruplex structures were unfolded in a stepwise manner at a rate slower than that of duplex DNA unwinding; however, the slower dissociation from G-quadruplexes compared with duplexes allowed the helicase to traverse more nucleotides than on duplexes. Consistent with this, the rate of ATP hydrolysis on the telomeric quadruplex DNA was reduced relative to that on single-stranded DNA (ssDNA), but less quadruplex DNA was needed to saturate ATPase activity. Under single-cycle conditions, telomeric quadruplex was unfolded by Pif1, but for the c-MYC quadruplex, unfolding required multiple helicase molecules loaded onto the adjacent ssDNA. Our findings illustrate that Pif1-catalyzed unfolding of G-quadruplex DNA is highly dependent on the specific sequence and the conditions of the reaction, including both the monovalent cation and the order of addition.
Keywords: DNA helicase, G-quadruplex, DNA-protein interaction, enzyme mechanism, enzyme kinetics, pre-steady-state kinetics, DNA unwinding, Pif1, secondary structure
Introduction
G-quadruplexes (G4) 2 are four-stranded structures that can form when four guanines associate through Hoogsteen hydrogen bonds to form a planar tetrad. Multiple tetrads stack to form a G4 structure, which is stabilized by monovalent cations bound in the central channel (reviewed in Refs. 1–4). More stable G4s are formed in K+, which binds between two tetrads, than in Na+, which binds in the center of a tetrad. Li+ does not support G4 formation. Inside a mammalian cell, K+ is the predominant cation (5).
Multiple different types of G4 structures can form that are described based on the number of strands (unimolecular/intramolecular, bimolecular, and tetramolecular) and the orientation of the strands. In parallel G4s, each strand runs in the same direction, whereas in antiparallel quadruplexes, the strand directionality alternates. This results in loops that span the outside of a parallel quadruplex (Fig. S1) and simply fold back at the top and bottom of antiparallel quadruplexes. Hybrid quadruplexes have a combination of parallel and antiparallel loops.
DNA and RNA quadruplexes have been visualized in cells (6–11). The G4s most likely to form are intramolecular, although bimolecular quadruplexes have been reported such as the RNA-DNA hybrid quadruplex that forms in the mitochondria during the transition from transcription to replication (12). The promoters of many proto-oncogenes contain GDNA sequences (13), and these form predominantly parallel G4s. The telomeric repeats of eukaryotic organisms are sequences that can form quadruplex structures (14). The human telomeric sequence (hTEL) forms a mixture of two-hybrid quadruplexes in K+ (15).
Formation of quadruplexes has also been shown to cause genetic instability (16–20) and potential G4 forming sequences are associated with known breakpoints in the mitochondrial DNA (21–23). This suggests that removal of G4s is essential for proper replication to occur. Helicases are the proteins that unwind double-stranded DNA (dsDNA) and a subset of them have been shown to resolve noncanonical secondary structures in DNA including Holliday junctions (24, 25), triplexes (26, 27), and quadruplexes (28, 29). Several DNA helicases have been shown to have G4DNA resolving activity, including human BLM (30), WRN (31), and FANCJ (32, 33), and yeast Pif1 (17). Pif1 (reviewed in Refs. 34 and 35) localizes to G4DNA sequences at the end of the S phase and these sites are susceptible to double strand breaks (DSB) in the absence of Pif1 (36). Pif1 has been suggested to resolve G4DNA structures, thereby reducing subsequent formation of DSBs (37) due to unresolved G4DNA persisting through mitosis (38). In the absence of Pif1, gross chromosomal rearrangements at G4DNA sequences increase (17). In addition to its role at G4s, Pif1 is a negative regulator of telomerase both at telomeres and DSBs (39). Pif1 is also involved in Okazaki fragment processing (40), recombination-dependent telomere maintenance in the absence of telomerase (41), maintenance of mitochondrial DNA (42), and DNA repair by both homologous recombination (43) and break-induced replication (44).
Unfolding of intramolecular G4DNAs by Pif1 has been investigated by numerous groups with somewhat conflicting conclusions. Some groups have reported that Pif1 monomers repetitively and rapidly unfold G4DNA structures but do not unwind the dsDNA beyond the G4 (45, 46). Others have reported that multiple Pif1 molecules slowly unfold the G4DNA and dsDNA beyond (47, 48), and still others have reported that G4DNA stimulates unwinding of downstream dsDNA (49, 50). It is likely that these conflicting results are due to differences in G4DNA structures, substrate design, and reaction conditions. We investigated G4DNA unfolding by Pif1 under a variety of conditions with two different quadruplexes that have been previously used to characterize Pif1's activity on G4DNA. The results support the conclusion that reaction conditions and the sequence of the specific G4DNA structures contribute greatly to the experimental outcome. Experiments performed in Na+ rather than K+ can lead to spurious results. Furthermore, we illustrate that G4DNA does not stimulate unwinding of downstream dsDNA.
Results
Quadruplex structure and stability vary with sequence and salt
The quadruplex forming sequences used most frequently to study Pif1 unwinding are the hTEL sequence and a modified version of the G4DNA sequence in the NHE III1 of the human c-MYC promoter. This modified version (Table S1) contains guanine runs 2–5 of the naturally occurring sequence with guanines 14 and 23 mutated to thymidines to prevent folding of alternative structures (51). Importantly, this sequence folds into the same structure as the predominant structure formed by the WT sequence (51–53). The c-MYC and hTEL G4DNA sequences have been reported to fold into very different structures (51, 54), which could affect the ability of Pif1 to unfold these structures. G4DNA structures are also affected by the identity of the monovalent cation, which has been quite variable in in vitro experimental conditions with some studies using 50–60 mm KCl (45, 47) and others using 50–100 mm NaCl (46, 48, 50, 55–57).
The G4 formed by the human telomeric sequence folds into an antiparallel G4 in Na+ and a hybrid G4 in K+ (Fig. S1, A and B). Two different hybrid G4s form, one with a parallel loop as the 5′-loop and one with a parallel loop as the 3′-loop (Fig. 1A) (15, 58). In a mixture of 139 mm K+ and 12 mm Na+ similar to that in a cell (called physiological salt here), the telomeric sequence also forms hybrid G4 structures (Fig. S1B). The one from the human c-MYC promoter folds into a parallel quadruplex in Na+, K+, or a mixture of both salts (Fig. S1, C and D).
Different G4 structures can have very different stabilities, which could also affect the ability of a helicase to unfold the structures. The melting temperatures of the c-MYC and hTEL G4DNAs were measured (Table 1 and Fig. S2, A and B). The c-MYC G4DNA is more stable than the hTEL G4DNA and both are more stable in K+ than Na+.
Table 1.
Melting temperatures of G4DNA sequences in different salts
Another indicator of quadruplex stability is the ability of the quadruplex to remain folded in the presence of the complementary strand (59). Quadruplexes were folded in K+, Na+, or a mixture of both and mixed with the complementary strand (Fig. 1A) to determine the stability of the G4s (Fig. 1, B–D). Both hTEL and c-MYC are trapped by the complementary strand in Na+ but c-MYC is not trapped in K+ or a mixture of salts. Some trapping of hTEL occurs in K+ solutions; although not to the degree seen in Na+.
Figure 1.
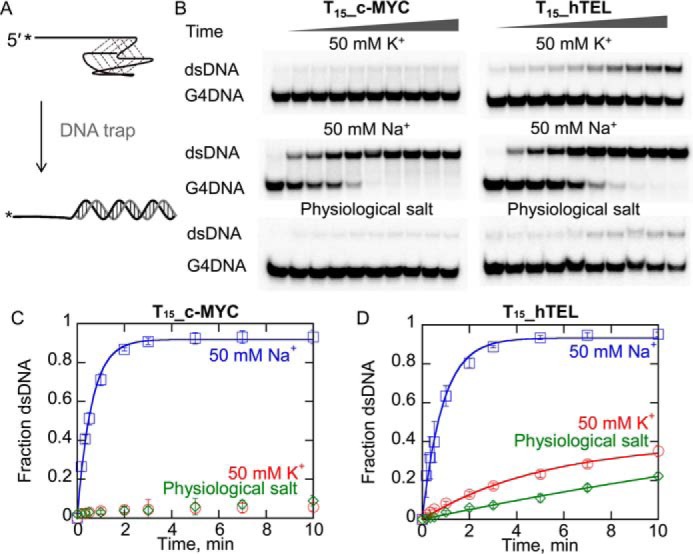
Quadruplex stability in the presence of a complementary strand is affected by both the sequence and the monovalent cation. A, schematic illustration of the trapping reaction. Folded G4DNA (2 nm) is mixed with a complementary strand (120 nm) as a DNA trap and reactions are quenched by adding an excess of unlabeled G4DNA (750 nm) to sequester the trap. Products were separated by electrophoresis (B). Melting of T15_c-MYC (C) and T15_hTEL (D) by the complementary strand was measured in 50 mm K+ (red), 50 mm Na+ (blue), and physiological salt (green). Data were fit to a single exponential to obtain rate constants for melting of 1.7 ± 0.3 s−1 for c-MYC in Na+, 0.19 ± 0.02 s−1, 1.2 ± 0.4 s−1, and 0.16 ± 0.06 s−1, for melting of hTEL in 50 mm K+, 50 mm Na+, and physiological salt, respectively. Errors bars represent the standard deviation of three independent experiments.
c-MYC in Na+ and hTEL in physiological salt have the same Tm (Table 1, Fig. S2) but c-MYC in Na+ is trapped more rapidly by the complementary strand. Assuming that similar Tm values reflect similar stability, then one way to explain the differences in trapping is that the kinetics of folding and unfolding of the two species are very different. For example, the rates of unfolding and refolding of c-MYC in Na+ could be much faster than those of hTEL in physiological salt. Thus trapping of c-MYC in Na+ would be expected to occur more rapidly than trapping of hTEL in physiological salt even though the ratio of the folding and unfolding rate constants and therefore the thermal stability is very similar. In this case, the trapping by the complementary strand reflects the kinetics of the equilibrium between the folded and unfolded states as it is easier for a complementary strand (or an enzyme) to gain access to the unfolded ssDNA when these rates are more rapid than when they are slower. Thus, even substrates with the same thermal stability (c-MYC in Na+ and hTEL in physiological salt) can have different unfolding rates in the presence of a trapping strand. These data indicate that the c-MYC G4DNA is quite different from the hTEL G4DNA in terms of the stability and that both G4DNAs are much less stable in Na+ as they can be trapped simply by the addition of a complementary strand.
Previous investigation of hTEL folding kinetics from multiple laboratories have found that folding is slow in K+ and that antiparallel intermediates persist for minutes to hours using CD (15, 60). Folding of a different version of the c-MYC sequence than utilized here in K+ has been shown to be faster than hTEL, but an antiparallel intermediate has been shown to remain for about 5 min (60). hTEL has also been shown to unfold about 75-fold more rapidly in Na+ than K+ (about 30 min half-life with 1–5 μm complementary PNA in 100 mm Na+ and about 40 h half-life in 100 mm K+) (59). However, the Balci lab (61) found that hTEL in 100 mm K+ was unfolded by 100 nm RPA in about 0.5 s using single molecule FRET. These measurements were all conducted by different approaches. Clearly, dramatic differences are found when different methods are used. PNAs may be limited due to their propensity to self-aggregate, which could slow the apparent capture of melted G4DNA. It has also been reported that unfolding of fully folded G4DNA structures to partially folded intermediates is not always detectable by FRET (15), indicating that some methods may miss some unfolding events. Based on data in Fig. 1, the half-lives of both c-MYC and hTEL G4DNA structures in 50 mm Na+ with 120 nm complementary strand are both about 0.5 min, whereas the half-life of hTEL in K+ is 15–30 min. Very little melting of the c-MYC G4DNA in K+ occurred during the time frame of the reaction suggesting that the half-life is likely at least an hour.
Telomeric G4DNA is destabilized during preincubation with the enzyme
Data in Fig. 1 indicates that a complementary strand can trap thermally unfolded structures, particularly in Na+, suggesting that a similar process could occur during preincubation with an enzyme. The high stability of c-MYC in K+ (Fig. 1C, Fig. S2) may preclude rapid unfolding of G4DNA (47). We turned our attention to the less stable G4DNA formed by the human telomeric sequence and to conditions in which G4DNA is less stable. We investigated the possibility that hTEL can be melted during the preincubation phase with the enzyme using an assay in which G4DNA unfolding is measured using a duplex reporter assay (47, 48, 55, 57). The substrate contains a ssDNA-binding site prior to the G4DNA, then a short duplex after the G4DNA (Fig. 2A). Using a similar assay, some previous reports have suggested that hTEL unfolding increases downstream duplex unwinding, which involved preincubation of the enzyme with the DNA (55). Substrates with a 3-nucleotide ssDNA gap between the G4DNA and the dsDNA were designed because dsDNA immediately adjacent to G4DNA has been shown to destabilize the G4DNA structure and a gap of at least 2 nucleotides is required for proper folding (57). Pif1 cannot load in the 3-nucleotide gap so G4DNA unfolding is required for dsDNA unwinding to occur (47). However, some previous reports have used substrates with G4 structures immediately adjacent to the dsDNA (46, 50, 55) even though destabilization of the G4 structure has been reported for such substrates (57). Additionally, we used a reporter duplex that has a 3′-ssDNA overhang because forked duplexes are preferred relative to nonforked duplexes by Pif1 (62–64). Finally, some of the articles reporting activation of Pif1 dsDNA unwinding by an upstream G4DNA utilized substrates with a fluorophore in the path of the helicase (45, 46, 50); we tested the ability of Pif1 to translocate past two common Cy3 modifications and found that Pif1 was blocked by iCy3 but unaffected by Cy3 attached through an NHS ester-amine linkage (Fig. S3).
Figure 2.
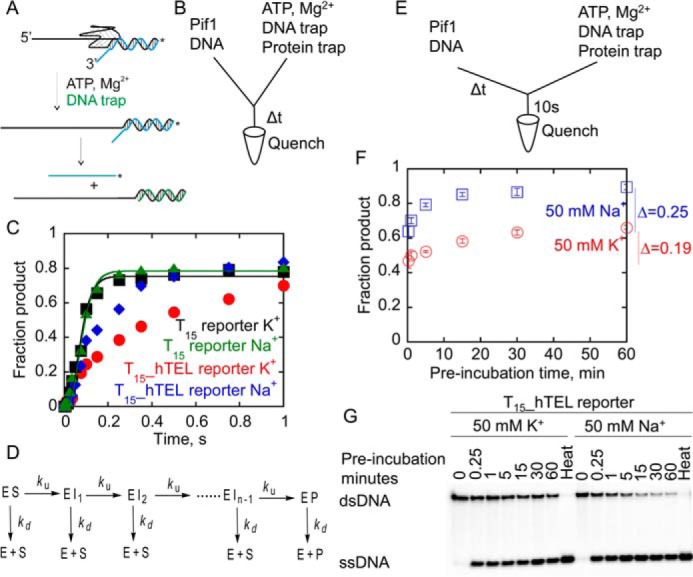
hTEL reporter substrates are partially melted during the preincubation with Pif1. A, G4DNA unfolding by Pif1 is monitored by measuring the rate of unfolding of a short reporter duplex in the presence of protein and annealing traps. After unfolding the G4DNA, Pif1 unwinds the dsDNA, which is forked due to Pif1's preference for forked duplexes. The unlabeled strand is trapped with excess complementary reporter trap, and the products are separated by electrophoresis. B, Pif1 (400 nm) was preincubated for 5 min with 2 nm radiolabeled DNA before loading into one sample port of a rapid chemical quench flow instrument. Time points were initiated by mixing with 5 mm ATP, 10 mm Mg2+, 60 nm reporter trap, and 10 μm T50 protein trap from the other sample port and quenched with 400 mm EDTA. The incubation time of the enzyme/DNA mixture increased with each successive time point in the reaction. C, unwinding of T15 reporter in K+ (black squares) and Na+ (green triangles) was fit to a 4-step sequential mechanism (D) using KinTek Explorer (85) to obtain rate constants for unwinding of 46.0 ± 3.9 and 43.2 ± 3.9 s−1, respectively. Data for unwinding the T15_hTEL reporter in K+ (red circles) and Na+ (blue diamonds) could not be fit to a simple n-step sequential mechanism. Data are the result of a single experiment due to the variable preincubation times. D, the reaction scheme describes unwinding by the helicase in a series of n sequential steps. ES, enzyme-substrate complex; EI, intermediate; EP, enzyme-product complex; E, free enzyme; S, substrate; P, ssDNA product. Each step is defined by an unwinding rate constant, ku, and a dissociation rate constant, kd. E and F, Pif1 (400 nm) was preincubated with 2 nm T15_hTEL reporter in 50 mm K+ (red) or 50 mm Na+ (blue) for varying amounts of time before mixing with 5 mm ATP, 10 mm Mg2+, 60 nm reporter trap, and 10 μm T50 protein trap for 10 s before quenching with 400 mm EDTA. Samples were separated by electrophoresis (G) and plotted to determine the quantity of G4 reporter unwound. The quantity of product formed in a 10-s reaction with ATP increased 19 ± 3% as the preincubation time increased from 15 s to 1 h in K+ (red) and 25 ± 2% as the preincubation time increased from 15 s to 1 h in Na+ (blue). Data are the average and standard deviation of three independent experiments.
The rates of hTEL unfolding in Na+ and K+ solutions when the enzyme and substrate were preincubated were measured (Fig. 2, A–C). Due to the relatively fast rates of T15_hTEL reporter melting by Pif1, unfolding rates were measured using a rapid chemical quench flow instrument. Pif1 was preincubated with the substrate in either Na+ or K+ containing buffer. Each time point was initiated by mixing this solution with ATP in the presence of annealing and protein traps (Fig. 2, B and C). As a control, the T15 duplex reporter (with no quadruplex) was unwound to ∼80% completion in both salts in a single turnover. Data were fit to a 4-step sequential mechanism (Fig. 2D) based on the known 1-bp step size of Pif1 (63) and considering that the final 8-bp of the duplex spontaneously melt at 25 °C (65). Therefore, only 4-bp of the 12-bp reporter duplex must be unwound by the enzyme.
The T15_hTEL reporter in both Na+ and K+ exhibited a lag phase followed by appearance of ssDNA product. Data for unwinding the T15_hTEL reporter in K+ and Na+ could not be fit to an n-step sequential mechanism (Fig. 2D), suggesting that a process in addition to stepping along the substrate is occurring. Because this experiment was performed using a rapid chemical quench-flow instrument, the time that the Pif1 was preincubated with the DNA substrate increased with each time point. Thus, if Pif1 can trap the thermally melted G4DNA in an ATP-independent manner during the variable preincubation period, less G4DNA will be present as the experiment proceeds. This would result in the appearance of greater product formation over time.
To directly determine the degree of ATP-independent unfolding during the preincubation, Pif1 was preincubated with hTEL reporter for increasing times, followed by initiation of the ATP-dependent phase of the reaction for 10 s as depicted in the diagram of Fig. 2E. In Na+, the quantity of product formed in 10 s increased 25% as the preincubation time increased from 15 s to 1 h, and in K+, the quantity of product formed in 10 s increased 19% as the preincubation time increased from 15 s to 1 h (Fig. 2, F and G). The quantity of product formed in 10 s increased more rapidly in Na+ than K+, which is consistent with the faster melting by a complementary strand in Na+ than K+ (Fig. 1D). This suggests that during the preincubation time, Pif1 is able to trap these partially melted intermediates, thereby providing the ssDNA overhang necessary for ATP-dependent dsDNA unwinding without the G4DNA structure (Fig. 3). Thus, unfolding of the hTEL G4DNA in an ATP-independent manner during the preincubation period could be misinterpreted as rapid, ATP-dependent unfolding.
Figure 3.
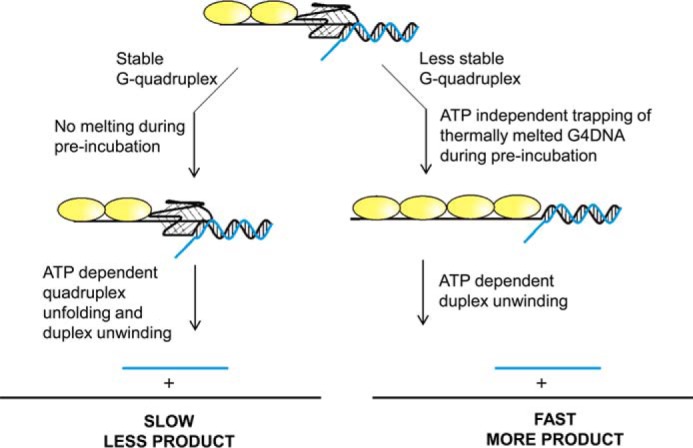
Model for ATP independent partial melting of G4DNA. When Pif1 is preincubated with less stable G4DNA structures, particularly those stabilized by Na+, ATP independent trapping of thermally melted G4DNA structures can occur during the time in which the enzyme is preincubated with the substrate, resulting in a long ssDNA overhang on a partial duplex substrate. After addition of ATP, rapid unwinding of the duplex can occur. This can lead to the appearance of increased product formation when an unstable G4DNA structure is present relative to experiments with stable G4DNA structures performed in K+ where the G4DNA structure is unaffected by preincubation with the enzyme.
Measurement of the rate of unfolding of telomeric G4DNA by Pif1
Because ATP independent melting occurs during the preincubation of enzyme with substrate, the rates of T15_hTEL reporter and T15 reporter unwinding were measured without preincubating the enzyme with the substrate (Fig. 4). The rate constants for unwinding of the T15_hTEL reporter were reduced relative to the T15 reporter. This result supports the conclusion that the G4DNA does not stimulate the activity of Pif1 for unwinding the duplex after unfolding the quadruplex. However, we note that the mechanism for unfolding G4DNA may differ from unwinding of duplex DNA, making direct comparisons difficult.
Figure 4.
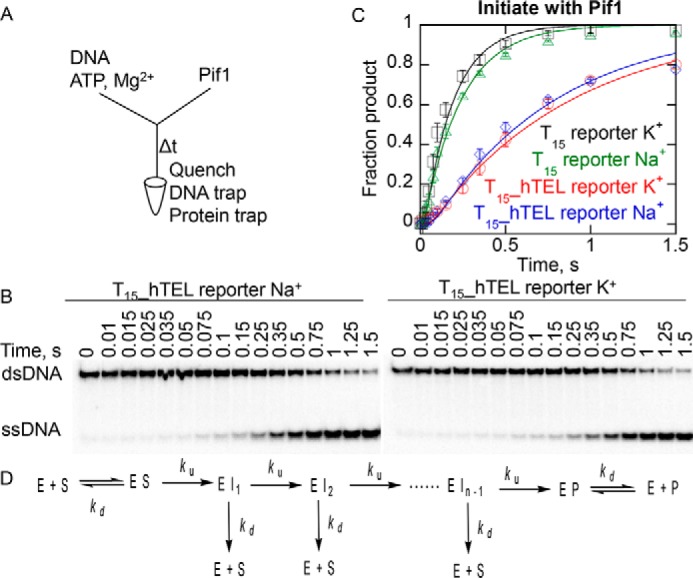
Unwinding hTEL reporter substrates and dsDNA without preincubation with Pif1. A, radiolabeled DNA (2 nm), 5 mm ATP, and 10 mm Mg2+ were loaded in one sample port of a rapid chemical quench flow instrument and time points were initiated by mixing with 400 nm Pif1 from the other sample port and quenched with 400 mm EDTA, 60 nm reporter trap, and 10 μm T50 protein trap. Samples were separated by electrophoresis (B) and data (C) was fit to a 4-step sequential mechanism, which includes binding steps (D) to obtain rate constants of 120 ± 40, 110 ± 20, 28 ± 7, and 23 ± 2 s−1 for unwinding of the T15 reporter in K+ (black squares) and Na+ (green triangles) and T15_hTEL reporter in K+ (red circles) and Na+ (blue diamonds), respectively. Data are the average and standard deviation of three independent experiments.
The T15_hTEL reporter was unwound at a similar rate in both salts and the reaction went nearly to completion in 1.5 s. These conditions were multiturnover, in which multiple molecules of Pif1 can participate in the reaction through multiple binding events. The lack of a large difference in the rates in Na+ compared with K+ indicates that the stability of the folded substrate has less influence on the rate of unfolding in multiturnover reaction conditions compared with reactions in which the enzyme and substrate are preincubated as in Fig. 2, B and C. By initiating the reaction via addition of the enzyme, the quadruplex did not have an opportunity to be unwound prior to the initiation of the reaction. This suggests that ATP independent melting before initiation of the reaction was indeed contributing to product formation in the reaction which involved preincubation of the enzyme and hTEL substrate (Fig. 2, B and C).
The rate of ATP hydrolysis by Pif1 is reduced on G4DNA relative to ssDNA
Although the rate of G4 unfolding cannot be compared directly to the rate of dsDNA unwinding on a per bp basis, the rates of ATP hydrolysis on different substrates can be compared. ATP hydrolysis provides the energy for G4DNA unfolding, dsDNA unwinding, and ssDNA translocation. Pif1 hydrolyzes 1 ATP per nucleotide translocated or bp unwound, and all three of these rates are essentially the same (63). We have previously reported the ATP hydrolysis rates on ssDNA and c-MYC G4DNA (47) and found that ATP hydrolysis was reduced on c-MYC 2–3-fold relative to ssDNA (47). The rates of ATP hydrolysis stimulated by T15 ssDNA and T15_hTEL G4DNA were measured (Fig. 5). The maximal rate of ATP hydrolysis, kcat, was reduced on hTEL G4DNA relative to ssDNA. This is consistent with what we previously observed for c-MYC G4DNA (47). The Kact, or concentration of DNA required to stimulate half of the maximal activity, was also reduced on hTEL G4DNA relative to ssDNA. This is likely due to the increased affinity for Pif1 for G4DNA relative to ssDNA (16, 45, 47). The reduced rate of ATP hydrolysis on G4DNA relative to ssDNA indicates that G4DNA does not enhance Pif1's catalytic activity.
Figure 5.
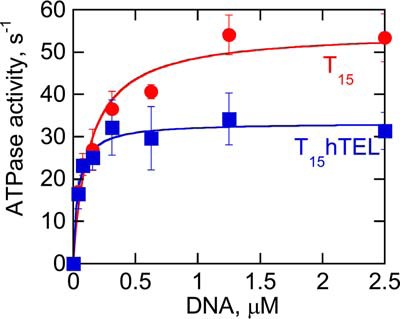
Pif1 ATP hydrolysis kinetics on ssDNA and G4DNA. The rate of ATP (5 mm) hydrolysis by 50 nm Pif1 was measured with varying quantities of T15 or T15_hTEL DNA cofactors in physiological salt and the specific activity was determined by dividing the ATP hydrolysis rate by the enzyme concentration. A plot of the ATPase activity versus the concentration of DNA was fit to a hyperbola to obtain the kcat of 55 ± 4 s−1 on T15 and 33 ± 7 s−1 on T15_hTEL. The Kact was 130 ± 9 nm on T15 and 39 ± 17 nm on T15_hTEL. Data are the average and standard deviation of three independent experiments.
Pif1 unfolds hTEL G4DNA more slowly but traverses more nucleotides relative to dsDNA
In light of the slower ATP hydrolysis rates on G4DNA relative to ssDNA, and the tight coupling of ATP hydrolysis to duplex unwinding, we anticipate unfolding of G4DNA to be slower than the ssDNA translocation and dsDNA unwinding rates because all of these processes are driven by ATP hydrolysis. To quantitatively measure the rate of hTEL G4DNA unfolding without competing processes such as association of enzyme and substrate interfering, we sought conditions in which Pif1 could be preincubated with the T15_hTEL reporter without pre-melting occurring. The degree of ATP independent unfolding of hTEL G4DNA during the preincubation was measured to determine whether Pif1 could be preincubated with T15_hTEL reporter G4DNA under conditions with limiting Pif1 concentration. The quantity of product formed in 10 s was low but remained essentially unchanged as the preincubation time increased (Fig. 6). These results indicate that subsaturating concentrations of Pif1 can be preincubated with T15_hTEL reporter in K+ without pre-melting.
Figure 6.
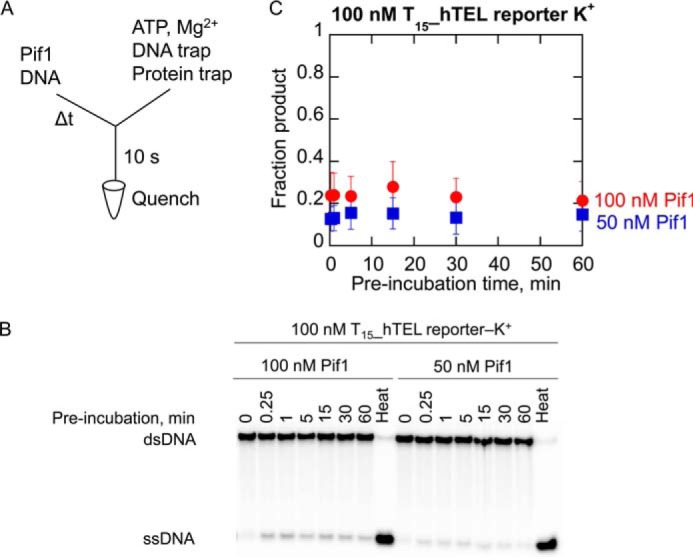
Melting does not occur during preincubation when Pif1 is not in excess with hTEL reporter substrates in K+. A, Pif1 (100 nm in red; 50 nm in blue) was preincubated with 100 nm T15_hTEL reporter in 50 mm K+ for varying amounts of time before mixing with 5 mm ATP, 10 mm Mg2+, 3 μm reporter trap, and 10 μm T50 protein trap for 10 s before quenching with 400 mm EDTA. Samples were separated by electrophoresis (B) and plotted (C) to determine the quantity of G4 reporter unwound. The quantity of product formed in a 10-s reaction with ATP was essentially unchanged as the preincubation time increased from 15 s to 1 h. Data are the average and standard deviation of three independent experiments.
These conditions were used to measure the first turnover of hTEL, cMYC, and T15 reporter substrates by Pif1 (Fig. 7). Very little product was produced with the T15_cMYC reporter, which was not surprising because very little product is produced with saturating concentrations of Pif1 with this substrate (47). The T15 reporter was rapidly unwound to about 30% completion. The T15_hTEL reporter was also unwound, over a longer time period, to about 20% completion. Data were fit using nonlinear least squares fitting (66, 67) to an n-step sequential mechanism (Fig. 2D) (68, 69) to determine the minimum number of steps (n) required for unwinding and the rate constants for unwinding (ku) and dissociation (kd). The unwinding rate was reduced on the T15_hTEL reporter relative to the T15 reporter, and the dissociation rate was also reduced, consistent with the known tight binding of Pif1 to G4DNA (16, 47). It should be noted that even though less product is formed with T15_hTEL reporter than T15 reporter, the slower dissociation from the T15_hTEL reporter allows Pif1 to traverse more nucleotides than on a duplex before dissociation.
Figure 7.
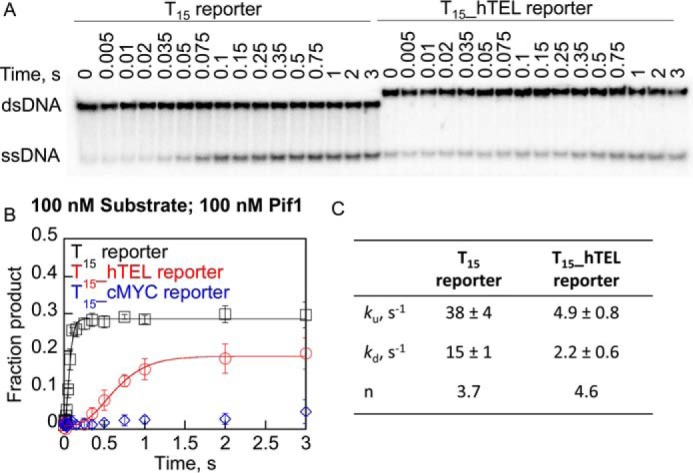
Pif1 unfolds hTEL G4DNA with slower rate but more bases are traversed compared with dsDNA. A, radiolabeled DNA (100 nm) and 100 nm Pif1 were loaded in one sample port of a rapid chemical quench flow instrument and time points were initiated by mixing with 5 mm ATP, 10 mm Mg2+, 3 μm reporter trap, and 10 μm T50 protein trap from the other sample port and quenched with 400 mm EDTA. Samples were separated by electrophoresis and plotted (B) for the T15 reporter in K+ (black squares), the T15_hTEL reporter in K+ (red circles), and T15_cMYC reporter in K+ (blue diamonds). C, data were fit using nonlinear least squares analysis to an n-step sequential mechanism (Fig. 2D) to obtain rate constants for unwinding (ku) and dissociation (kd) and the number of apparent steps (n). Data are the average and standard deviation of three independent experiments. The calculated processivity using Equation 3 is 0.72 for the T15 reporter and 0.69 for the T15_hTEL reporter.
Increasing length ssDNA overhangs result in increased G4DNA unfolding by Pif1
Although product formation with hTEL G4DNA is slower than dsDNA, Pif1 is able to unfold the substrate. However, little unfolding of c-MYC G4DNA that has a ssDNA-loading site of only 15 nucleotides occurs (Fig. 7) (47). Others have reported that longer ssDNA-loading sites resulted in unfolding of hTEL G4DNA on almost every encounter (45, 46, 50, 55). The amplitude of product formation from dsDNA unwinding by Pif1 has been shown to increase when multiple Pif1 molecules can bind to the 5′ ssDNA-loading site (63) so variations in the length of the ssDNA-binding site could result in large differences in product formation during G4DNA unfolding.
Unfolding of c-MYC G4DNA substrates in a single turnover with increasing lengths of ssDNA-loading sites was measured under excess enzyme conditions (Fig. 8). As observed previously for dsDNA unwinding (63), the quantity of product increased as the number of Pif1-binding sites increased. The quantities of product formed are reduced with c-MYC G4DNA relative to dsDNA, but increasing the number of bound Pif1 molecules increases product formation. However, the rate of c-MYC G4DNA unfolding was slow relative to dsDNA unwinding and hTEL G4DNA unfolding (Fig. 7), suggesting that Pif1 unfolds G4DNA structures at different rates that are affected by the sequence and stability of the G4DNA structure. However, in both cases, G4DNA unfolding is slower than dsDNA unwinding.
Figure 8.
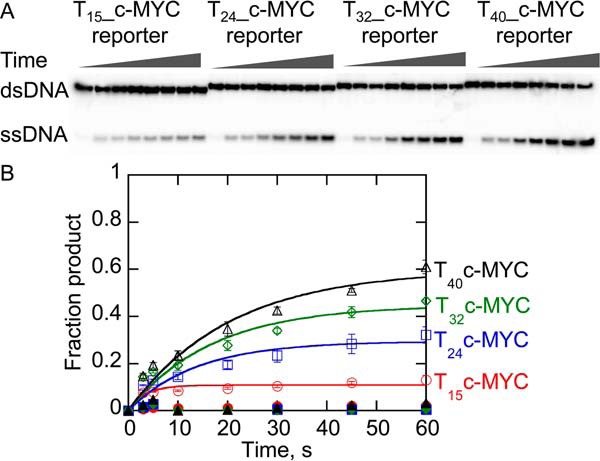
Unfolding of c-MYC G4DNA substrates with varying length ssDNA overhangs under single turnover conditions by Pif1. A, products of unfolding 2 nm G4DNA by 400 nm Pif1 were separated by electrophoresis. B, data were fit to a single exponential to obtain rate constants for unwinding of the T15_c-MYC reporter (open red circles), T24_c-MYC reporter (open blue squares), T32_c-MYC reporter (open green diamonds), and T40_c-MYC reporter (open black triangles) of 0.29 ± 0.08, 0.075 ± 0.012, 0.059 ± 0.004, and 0.049 ± 0.009 s−1, respectively. Data represent the average and standard deviation of three independent experiments. Data shown in closed symbols had the protein trap (T50) preincubated with the Pif1 to test the efficiency of the trapping strand.
Discussion
Different groups have reported different activities for Pif1 on G4DNA that are possibly substrate and condition dependent and may not reflect ATP-dependent G4DNA destabilization. It has been reported that G4DNA stimulates Pif1 unwinding of dsDNA (50, 55). We investigated this possibility using a variety of different substrates and reaction conditions and conclude that some conditions do not support stable G4DNA formation, which can lead to the appearance of increased product formation when the enzyme is preincubated with the substrate. Pif1 unfolding of substrates with G4DNA structures followed by forked duplexes is limited by the rate of G4DNA unfolding. For less stable G4DNA structures, our data shows them to be destabilized prior to addition of ATP when preincubated with enzyme, indicating that the helicase is not unfolding the G4DNA structure in an active process but simply trapping ssDNA that transiently forms during preincubation (Fig. 2). Trapping of unfolded G4DNA by Pif1 produces a substrate with a longer ssDNA overhang, which allows binding of additional molecules of Pif1 (Fig. 3). The quantity of product formed by Pif1 (63) and some other helicases (70–74) increases as the length of the ssDNA overhang and the number of bound helicase molecules increases. This gives the appearance of increased product formation but it actually reflects trapping of unfolded G4DNA before ATP is added. Therefore, statements of activation of Pif1 by G4DNA are possibly the result of this phenomenon.
The data in Fig. 2 supports the conclusion that hTEL, particularly in Na+ but also in K+, is destabilized during preincubation with excess enzyme resulting in increased product formation when enzyme and G4DNA are preincubated. However, when the reaction is started upon addition of enzyme to the G4DNA, product formation in Na+ and K+ is similar (Fig. 4). Thus, preincubation of excess Pif1 with less stable G4DNA results in ATP-independent trapping of the thermally melted G4DNA before initiation of the reaction with ATP (Fig. 3).
This ATP-independent reaction is sensitive to the stability of the G4DNA because more trapping occurs with hTEL in Na+ than K+ (Fig. 2). Similarly, hTEL in Na+ is melted faster by a complementary strand than in K+ (Fig. 1). In contrast, c-MYC in K+, which is very stable, does not appear to be melted by a complementary strand (Fig. 1) or during preincubation with the enzyme (Fig. 8) (47). However, in Na+ c-MYC is less stable and some trapping by the complementary strand is observed (Fig. 1).
Although ATP independent pre-melting is quite sensitive to the stability of the G4DNA structure, ATP-dependent melting appears to be less affected by the G4 stability. hTEL in Na+ and K+ have different structures (Fig. S1) and stabilities (Fig. S2, Table 1, and Fig. 1) but are unfolded by Pif1 at the same rate (Fig. 7), suggesting that G4 stability is not limiting the G4DNA unfolding rate. For some helicases, such as Dda and RecQ, the stability of the duplex has little to no effect on the rate of dsDNA unwinding (47, 75). For dsDNA unwinding by highly efficient helicases such as Pif1, the rate of unwinding has been suggested to be limited by the rate of ATP hydrolysis (63). Likewise, the rate of G4DNA unfolding may be limited by the rate of ATP hydrolysis on G4DNA. The interaction of the helicase with the G4DNA appears to support lower ATPase rates than are supported by purely ssDNA (Fig. 5) (47). However, the ATPase, at this lower rate, may limit the reaction. Thus, it is possible that quadruplexes, which are below a stability threshold are all unfolded at rates that are dependent on the length of the sequence as opposed to the stability. We suggest that above a certain stability threshold, G4DNA structures are too stable to be readily melted by Pif1 and the unfolding rate decreases (i.e. c-MYC unfolding). Similarly, Pif1 unwinds dsDNA at the same rate as it translocates on ssDNA but it displaces streptavidin bound to biotinylated DNA at a much slower rate (63). These observations suggest that barriers requiring minimal energy expenditure (dsDNA unwinding) do not reduce the rate of the helicase but barriers requiring more energy (streptavidin displacement or unfolding of highly stable G4DNAs) result in a reduced rate.
G4DNA unfolding rates are difficult to compare directly with dsDNA unwinding rates because it is not known how many Hoogsteen bp must be disrupted by the helicase to cause the structure to collapse. It is possible that only a few bp need to be removed from the G4DNA structure to cause complete unfolding. Telomeric G4DNA structures have previously been suggested to melt with disruption of only a few bp (76). Additionally, under multiturnover conditions, partial unfolding due to Pif1 activity could expose ssDNA, which could be bound by additional Pif1 molecules resulting in increased dsDNA unwinding (63) through formation of helicase “trains.”
The presence of a lag phase for hTEL reporter unfolding indicates that like dsDNA unwinding by Pif1 (63) and other helicases (69), G4DNA unfolding is a stepwise process, as has been suggested recently for DHX36 unfolding of G4DNA (77). For dsDNA unwinding, each physical step is generally thought to represent movement of a single nucleotide along the DNA lattice (78) although each of these physical steps may not be visible kinetically.
As would be expected due to the longer lag phase with the T15_hTEL reporter, the apparent number of steps involved in unwinding is greater on the T15_hTEL reporter than the T15 reporter. The ∼4 steps required for unwinding of the 12 bp T15 reporter duplex (Fig. 7) is consistent with the known single bp step size for unwinding of dsDNA by Pif1 (63) and the known 8 bp that spontaneously melt at 25 °C (79). Pif1 has previously been suggested to unfold hTEL in 3 steps (45) corresponding to transitions from G4 to triplex, triplex to hairpin, and hairpin to unfolded states. Within these three kinetic steps, it was suggested that Pif1 translocates along the DNA in single nucleotide steps based on the distribution of the dwell times at each state (45). Our experiments in Fig. 7 suggest Pif1 unwinds the T15_hTEL reporter in 5 apparent steps. This is less than the one step per bp that has been observed for dsDNA unwinding by Pif1 (63), indicating that in addition to multiple slow steps in the kinetic mechanism, additional faster steps may occur. Hence, the number of steps observed here is the minimum number of steps required for hTEL unfolding. After slow removal of 4–5 nucleotides from the G4 structure, the remaining nucleotides may spontaneously unfold, allowing Pif1 to rapidly translocate on the ssDNA and unwind the downstream dsDNA.
We observed unwinding of the downstream duplex after G4DNA unfolding (Figs. 2, 4, 7, and 8) and the rate of the overall reaction appears to be limited by the rate of G4DNA unfolding. However, Zhou et al. (45) observed repetitive unfolding of hTEL G4DNA without unwinding of the downstream duplex. This difference is likely due to differences in the substrate design. The duplex used here is only 12 bp and has a 3′ ssDNA overhang on the displaced strand to produce a forked duplex. The duplex used by Zhou et al. (45) was 21 bp and was not forked. Pif1 is a nonprocessive helicase, and it preferentially unwinds forked duplexes (62, 63, 80) so the difference in duplex design could have a dramatic effect on Pif1's ability to unwind the duplex.
Unfolding of c-MYC G4DNA is both slower and less efficient than unwinding of dsDNA or hTEL G4DNA (Figs. 7 and 8). Unwinding of downstream dsDNA is limited by either c-MYC or hTEL G4DNA (Figs. 4 and 7), which is consistent with the reduced rate of ATP hydrolysis on both hTEL (Fig. 5) and c-MYC (47) G4DNA as the kcat, or maximal rate of ATP hydrolysis reflects the movement of the enzyme along the DNA (81). However, due to the reduced dissociation rate on hTEL G4DNA relative to dsDNA, Pif1 progresses further on the G4DNA (Fig. 7). This is likely the result of the strong affinity of Pif1 for G4DNA (16, 47).
Binding energy is sufficient for Pif1 to melt some G4DNA structures, especially in the presence of Na+. A number of proteins readily unfold G4DNA in the absence of ATP, for example, RPA (75) and POT1 (76). Thus, capture of transient ssDNA can lead to unfolding. DHX36 helicase has been shown to disrupt G4DNA structure upon binding, in the absence of ATP (82).
Pif1 oligomerization has been investigated carefully by the Galletto lab (64, 83). They suggested that DNA induced Pif1 dimerization (83). However, they also showed that under conditions where Pif1 is not in excess of the DNA, a monomer of Pif1 can unwind dsDNA (64). The Xi lab proposed (50, 55) that G4DNA stimulates dimerization of Pif1, which results in increased unwinding of downstream duplex DNA. Preincubation of the enzyme with the substrate is thought to allow the active species to form. However, we found that preincubation can result in ATP independent unfolding of some G4DNAs. Therefore, we developed conditions that allowed preincubation of the enzyme and substrate without inadvertently unfolding the G4DNA (Fig. 6). Under these conditions, we do not observe increased duplex unwinding downstream of G4DNA, even when using hTEL, the G4DNA species used by the Xi lab (Fig. 7). With a more stable G4DNA species, c-MYC, we observe no duplex unwinding under these same conditions.
There is a general trend that more stable G4DNAs are unfolded more slowly by Pif1, but unfolding of less stable G4DNA structures appears to be limited by other factors. ATP-independent trapping of spontaneously unfolded G4DNA can occur and is limited by the kinetics of the partial unfolding and refolding. We suggest that the rate of ATP-dependent Pif1 catalyzed unfolding of hTEL G4DNA is controlled by the rate of ATP hydrolysis, rather than the stability per se. However, for highly stable G4DNAs, unfolding is limited by the stability. The c-MYC substrate can be unfolded by Pif1 in a single turnover reaction but requires multiple Pif1 molecules to be bound to the substrate. Thus, the relationships between the G4DNA structure and stability with helicase-catalyzed unfolding rates are complex and vary depending on the specific sequence and reaction conditions.
Experimental procedures
Oligonucleotides and protein
Oligonucleotides (Table S1) were purchased from Integrated DNA Technologies and gel purified as described (71). Oligonucleotides were resuspended in 10 mm Tris-Cl, 1 mm EDTA, pH 7.5, and the concentrations were determined using calculated extinction coefficients (84). For some experiments, DNA was radiolabeled as described (71). Salts were added to the indicated concentrations and G4DNA was folded by heating to 95 °C for 5 min and slowly cooling to room temperature. Duplexes were formed by adding a 1.2-fold excess of the unlabeled strand and heating as described for G4DNA. Saccharomyces cerevisiae Pif1 was purified as described (63).
CD
CD spectra of 10 μm DNA were obtained using a Jasco J-715 spectropolarimeter. Spectra in K+ were obtained in 25 mm Tris-Ac, pH 7.4, 50 mm KCl, 2 mm β-mercaptoethanol, 1 mm EDTA. Spectra in Na+ were obtained in 25 mm Tris-Ac, pH 7.4, 50 mm NaCl, 2 mm β-mercaptoethanol, 1 mm EDTA. Spectra in physiological salt were obtained in 25 mm Tris-Ac, pH 7.4, 135 mm KOAc, 4 mm KCl, 12 mm NaHCO3, 2 mm β-mercaptoethanol, 1 mm EDTA. Scan speed was 50 nm/min; data were acquired 5 times, averaged, and smoothed using the Savitzky-Golay method after subtracting the spectrum of a sample containing only buffer. Melting curves were measured using a Jasco J-1100 spectropolarimeter at 1 °C/min in the same buffers used for CD spectra. Measurements were at 268 nm for c-MYC and 290 nm for hTEL.
G4DNA unfolding by trapping
All concentrations are final, after mixing. The G4DNA substrates were radiolabeled. DNA (2 nm) in the buffers described for CD spectra was mixed with 120 nm annealing trap complementary to the G4DNA. Samples were removed at various times and quenched with 100 mm EDTA, 0.35% SDS, 750 nm unlabeled G4DNA to sequester the annealing trap, and 2 μm T50. Duplex DNA product was separated from G4DNA substrate by 20% native PAGE, visualized using a Typhoon Trio phosphorimager, and quantified using ImageQuant software. The fraction product formed was fit to a single exponential using Kaleidagraph software.
Excess enzyme reporter assay for G4DNA unfolding
All concentrations are final, after mixing. The reporter (displaced) strand of a substrate containing a ssDNA-loading site, a G4DNA sequence, and duplex reporter (Table S1) was radiolabeled. DNA (2 nm) was preincubated with 400 nm Pif1 for 5 min, unless otherwise specified, in the buffers described for CD spectra with the addition of 1 mg/ml of BSA. Reactions were initiated with the addition of 5 mm ATP, 10 mm MgCl2, and 60 nm annealing trap. T50 (10 μm) was added with the ATP as a protein trap for all single-turnover reactions. Some reactions had a different order of addition as indicated in the figure legends. Samples were removed at various times and quenched with 200 mm EDTA, 0.7% SDS, 2 μm T50, 0.1% bromphenol blue, 0.1% xylene cyanol, and 6% glycerol. Reactions in Fig. 2 were performed using a KinTek rapid chemical quench-flow instrument maintained at 25 °C. These reactions were quenched with 400 mm EDTA and loading dye (0.1% bromphenol blue, 0.1% xylene cyanol, and 6% glycerol) was added. Multiturnover reactions (Fig. 4) were quenched with 400 mm EDTA; the receiving vial contained 60 nm annealing trap and 10 μm T50. Samples were separated by 20% native PAGE, visualized using a Typhoon Trio phosphorimager, and quantified using ImageQuant software. For the c-MYC reporters in K+ (Fig. 8), the fraction of ssDNA product formed was fit to a single exponential equation using Kaleidagraph software. Data in Fig. 2 for the T15 reporter was fit to a 4-step sequential mechanism (Fig. 2D) using KinTek Explorer Software (85). Data from multiturnover reactions (Fig. 4) was fit to a 4-step sequential mechanism, which includes binding steps with the association rate constant fixed at diffusion controlled (108 m−1 s−1) (Fig. 4B) using KinTek Explorer Software (85).
Pre-steady state rapid mixing reporter assay for G4DNA unfolding
Pre-steady state reporter assays were performed with a KinTek rapid chemical quench-flow instrument maintained at 25 °C. All concentrations listed are final. The reporter (displaced) strand of a substrate containing a ssDNA-loading site, a G4DNA sequence, and duplex reporter (Table S1) was radiolabeled and mixed with an equivalent of an unlabeled complementary strand. Radiolabeled duplex was mixed with a 99-fold excess of unlabeled duplex and samples were heated in 100 mm KCl to 95 °C for 5 min before slowly cooling to room temperature. DNA (100 nm) was preincubated with 100 nm Pif1 for 5 min in K+ buffer (25 mm Tris-Ac, pH 7.4, 50 mm KCl, 2 mm β-mercaptoethanol, 1 mm EDTA, 1 mg/ml of BSA) before mixing the first time point. Reactions were initiated with the addition of 5 mm ATP, 10 mm MgCl2. Annealing trap (3 μm) complementary to the unlabeled strand was added with ATP to prevent reannealing of the duplex after unwinding. A protein trap (10 μm T50) was also added at the initiation of the reaction to prevent rebinding of the enzyme to the substrate after dissociation. The reactions were quenched with 400 mm EDTA at increasing times. Loading buffer consisting of 0.1% bromphenol blue, 0.1% xylene cyanol, and 6% glycerol was subsequently added to each sample. The duplex substrate and ssDNA product were resolved by 20% native PAGE, visualized using a Typhoon Trio phosphorimager, and quantified using ImageQuant software. Data were fit by nonlinear least squares analysis. Product formation in a series of n apparent steps (Fig. 2D) has been defined by the function fss(t) in Equation 1 (66, 67), where kobs is the sum of the unwinding and dissociation rate constants, ku and kd, respectively (Equation 2) and the processivity (P), or average number of bp unwound in a single binding event, is defined by Equation 3.
| (Eq. 1) |
| (Eq. 2) |
| (Eq. 3) |
The differential equations have been solved by Laplace transforms (66, 67) for the reaction scheme in Fig. 2D, which results in Equation 4 where s is the Laplace variable of the fraction of ssDNA formed over time.
| (Eq. 4) |
The inverse Laplace transform (L−1) is given by Equation 5.
| (Eq. 5) |
Triplicate data were globally fit to Equation 5 using the numerical integration software Scientist (Micromath) to obtain values for ku and kd for each individual data set and a single value for n.
ATP hydrolysis
ATPase activity was measured using a spectrophotometric assay in which the hydrolysis of ATP by Pif1 is coupled to NADH oxidation as described (47).
Author contributions
A. K. B. and K. D. R. conceptualization; A. K. B. and K. D. R. formal analysis; A. K. B. and M. R. B. investigation; A. K. B. writing-original draft; M. R. B. and K. D. R. writing-review and editing; K. D. R. funding acquisition.
Supplementary Material
This work was supported by Grant R35GM122601 (to K. D. R.) from the National Institutes of Health, NIGMS. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S3.
- G4
- G-quadruplex
- DSB
- DNA double-strand break
- hTEL
- human telomeric DNA
- Ac
- acetyl
- PNA
- peptide nucleic acid.
References
- 1. Hansel-Hertsch R., Di Antonio M., and Balasubramanian S. (2017) DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 18, 279–284 10.1038/nrm.2017.3 [DOI] [PubMed] [Google Scholar]
- 2. Maizels N. (2015) G4-associated human diseases. EMBO Rep. 16, 910–922 10.15252/embr.201540607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rhodes D., and Lipps H. J. (2015) G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 43, 8627–8637 10.1093/nar/gkv862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neidle S. (2016) Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 59, 5987–6011 10.1021/acs.jmedchem.5b01835 [DOI] [PubMed] [Google Scholar]
- 5. Thier S. O. (1986) Potassium physiology. Am. J. Med. 80, 3–7 10.1016/0002-9343(86)90334-7 [DOI] [PubMed] [Google Scholar]
- 6. Biffi G., Tannahill D., McCafferty J., and Balasubramanian S. (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5, 182–186 10.1038/nchem.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Biffi G., Tannahill D., Miller J., Howat W. J., and Balasubramanian S. (2014) Elevated levels of G-quadruplex formation in human stomach and liver cancer tissues. PLoS One 9, e102711 10.1371/journal.pone.0102711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Biffi G., Di Antonio M., Tannahill D., and Balasubramanian S. (2014) Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 6, 75–80 10.1038/nchem.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lam E. Y., Beraldi D., Tannahill D., and Balasubramanian S. (2013) G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 4, 1796 10.1038/ncomms2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang W. C., Tseng T. Y., Chen Y. T., Chang C. C., Wang Z. F., Wang C. L., Hsu T. N., Li P. T., Chen C. T., Lin J. J., Lou P. J., and Chang T. C. (2015) Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res. 43, 10102–10113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rodriguez R., Miller K. M., Forment J. V., Bradshaw C. R., Nikan M., Britton S., Oelschlaegel T., Xhemalce B., Balasubramanian S., and Jackson S. P. (2012) Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 8, 301–310 10.1038/nchembio.780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wanrooij P. H., Uhler J. P., Shi Y., Westerlund F., Falkenberg M., and Gustafsson C. M. (2012) A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 40, 10334–10344 10.1093/nar/gks802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eddy J., and Maizels N. (2006) Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 34, 3887–3896 10.1093/nar/gkl529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brázda V., Haroniková L., Liao J. C., and Fojta M. (2014) DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 15, 17493–17517 10.3390/ijms151017493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gray R. D., Trent J. O., and Chaires J. B. (2014) Folding and unfolding pathways of the human telomeric G-quadruplex. J. Mol. Biol. 426, 1629–1650 10.1016/j.jmb.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paeschke K., Bochman M. L., Garcia P. D., Cejka P., Friedman K. L., Kowalczykowski S. C., and Zakian V. A. (2013) Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 497, 458–462 10.1038/nature12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ribeyre C., Lopes J., Boulé J. B., Piazza A., Guédin A., Zakian V. A., Mergny J. L., and Nicolas A. (2009) The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 5, e1000475 10.1371/journal.pgen.1000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tran P. L. T., Pohl T. J., Chen C. F., Chan A., Pott S., and Zakian V. A. (2017) PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat. Commun. 8, 15025 10.1038/ncomms15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sarkies P., Reams C., Simpson L. J., and Sale J. E. (2010) Epigenetic instability due to defective replication of structured DNA. Mol. Cell 40, 703–713 10.1016/j.molcel.2010.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sarkies P., Murat P., Phillips L. G., Patel K. J., Balasubramanian S., and Sale J. E. (2012) FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 40, 1485–1498 10.1093/nar/gkr868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bharti S. K., Sommers J. A., Zhou J., Kaplan D. L., Spelbrink J. N., Mergny J. L., and Brosh R. M. Jr. (2014) DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative Twinkle helicase. J. Biol. Chem. 289, 29975–29993 10.1074/jbc.M114.567073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dong D. W., Pereira F., Barrett S. P., Kolesar J. E., Cao K., Damas J., Yatsunyk L. A., Johnson F. B., and Kaufman B. A. (2014) Association of G-quadruplex forming sequences with human mtDNA deletion breakpoints. BMC Genomics 15, 677 10.1186/1471-2164-15-677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oliveira P. H., da Silva C. L., and Cabral J. M. (2013) An appraisal of human mitochondrial DNA instability: new insights into the role of non-canonical DNA structures and sequence motifs. PLoS One 8, e59907 10.1371/journal.pone.0059907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsaneva I. R., Müller B., and West S. C. (1993) RuvA and RuvB proteins of Escherichia coli exhibit DNA helicase activity in vitro. Proc. Natl. Acad. Sci. U.S.A. 90, 1315–1319 10.1073/pnas.90.4.1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whitby M. C., Ryder L., and Lloyd R. G. (1993) Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell 75, 341–350 10.1016/0092-8674(93)80075-P [DOI] [PubMed] [Google Scholar]
- 26. Jain A., Bacolla A., Chakraborty P., Grosse F., and Vasquez K. M. (2010) Human DHX9 helicase unwinds triple-helical DNA structures. Biochemistry 49, 6992–6999 10.1021/bi100795m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo M., Hundseth K., Ding H., Vidhyasagar V., Inoue A., Nguyen C. H., Zain R., Lee J. S., and Wu Y. (2015) A distinct triplex DNA unwinding activity of ChlR1 helicase. J. Biol. Chem. 290, 5174–5189 10.1074/jbc.M114.634923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mendoza O., Bourdoncle A., Boulé J. B., Brosh R. M. Jr., and Mergny J. L. (2016) G-quadruplexes and helicases. Nucleic Acids Res. 44, 1989–2006 10.1093/nar/gkw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sauer M., and Paeschke K. (2017) G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 45, 1173–1182 10.1042/BST20170097 [DOI] [PubMed] [Google Scholar]
- 30. Sun H., Karow J. K., Hickson I. D., and Maizels N. (1998) The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 273, 27587–27592 10.1074/jbc.273.42.27587 [DOI] [PubMed] [Google Scholar]
- 31. Fry M., and Loeb L. A. (1999) Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 274, 12797–12802 10.1074/jbc.274.18.12797 [DOI] [PubMed] [Google Scholar]
- 32. Wu Y., Shin-ya K., and Brosh R. M. Jr. (2008) FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 28, 4116–4128 10.1128/MCB.02210-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. London T. B., Barber L. J., Mosedale G., Kelly G. P., Balasubramanian S., Hickson I. D., Boulton S. J., and Hiom K. (2008) FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 283, 36132–36139 10.1074/jbc.M808152200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Geronimo C. L., and Zakian V. A. (2016) Getting it done at the ends: Pif1 family DNA helicases and telomeres. DNA Repair 44, 151–158 10.1016/j.dnarep.2016.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Byrd A. K., and Raney K. D. (2017) Structure and function of Pif1 helicase. Biochem. Soc. Trans. 45, 1159–1171 10.1042/BST20170096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paeschke K., Capra J. A., and Zakian V. A. (2011) DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145, 678–691 10.1016/j.cell.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Osmundson J. S., Kumar J., Yeung R., and Smith D. J. (2017) Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes. Nat. Struct. Mol. Biol. 24, 162–170 10.1038/nsmb.3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lemmens B., van Schendel R., and Tijsterman M. (2015) Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat. Commun. 6, 8909 10.1038/ncomms9909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schulz V. P., and Zakian V. A. (1994) The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76, 145–155 10.1016/0092-8674(94)90179-1 [DOI] [PubMed] [Google Scholar]
- 40. Budd M. E., Reis C. C., Smith S., Myung K., and Campbell J. L. (2006) Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase δ. Mol. Cell. Biol. 26, 2490–2500 10.1128/MCB.26.7.2490-2500.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dewar J. M., and Lydall D. (2010) Pif1- and Exo1-dependent nucleases coordinate checkpoint activation following telomere uncapping. EMBO J. 29, 4020–4034 10.1038/emboj.2010.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Foury F., and Dyck E. V. (1985) A PIF-dependent recombinogenic signal in the mitochondrial DNA of yeast. EMBO J. 4, 3525–3530 10.1002/j.1460-2075.1985.tb04112.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wilson M. A., Kwon Y., Xu Y., Chung W. H., Chi P., Niu H., Mayle R., Chen X., Malkova A., Sung P., and Ira G. (2013) Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 502, 393–396 10.1038/nature12585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saini N., Ramakrishnan S., Elango R., Ayyar S., Zhang Y., Deem A., Ira G., Haber J. E., Lobachev K. S., and Malkova A. (2013) Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502, 389–392 10.1038/nature12584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou R., Zhang J., Bochman M. L., Zakian V. A., and Ha T. (2014) Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife 3, e02190 10.7554/eLife.02190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hou X. M., Wu W. Q., Duan X. L., Liu N. N., Li H. H., Fu J., Dou S. X., Li M., and Xi X. G. (2015) Molecular mechanism of G-quadruplex unwinding helicase: sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem. J. 466, 189–199 10.1042/BJ20140997 [DOI] [PubMed] [Google Scholar]
- 47. Byrd A. K., and Raney K. D. (2015) A parallel quadruplex DNA is bound tightly but unfolded slowly by pif1 helicase. J. Biol. Chem. 290, 6482–6494 10.1074/jbc.M114.630749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li J. R., Lu C. Y., Lin J. J., and Li H. W. (2016) Multiple Pif1 helicases are required to sequentially disrupt G-quadruplex structure and unwind duplex DNA. Biochem. Biophys. Res. Commun. 473, 1235–1239 10.1016/j.bbrc.2016.04.046 [DOI] [PubMed] [Google Scholar]
- 49. Chen W. F., Dai Y. X., Duan X. L., Liu N. N., Shi W., Li N., Li M., Dou S. X., Dong Y. H., Rety S., and Xi X. G. (2016) Crystal structures of the BsPif1 helicase reveal that a major movement of the 2B SH3 domain is required for DNA unwinding. Nucleic Acids Res. 44, 2949–2961 10.1093/nar/gkw033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang B., Wu W. Q., Liu N. N., Duan X. L., Li M., Dou S. X., Hou X. M., and Xi X. G. (2016) G-quadruplex and G-rich sequence stimulate Pif1p-catalyzed downstream duplex DNA unwinding through reducing waiting time at ss/dsDNA junction. Nucleic Acids Res. 44, 8385–8394 10.1093/nar/gkw669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ambrus A., Chen D., Dai J., Jones R. A., and Yang D. (2005) Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter: implications for G-quadruplex stabilization. Biochemistry 44, 2048–2058 10.1021/bi048242p [DOI] [PubMed] [Google Scholar]
- 52. Siddiqui-Jain A., Grand C. L., Bearss D. J., and Hurley L. H. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598 10.1073/pnas.182256799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seenisamy J., Rezler E. M., Powell T. J., Tye D., Gokhale V., Joshi C. S., Siddiqui-Jain A., and Hurley L. H. (2004) The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 126, 8702–8709 10.1021/ja040022b [DOI] [PubMed] [Google Scholar]
- 54. Ambrus A., Chen D., Dai J., Bialis T., Jones R. A., and Yang D. (2006) Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 34, 2723–2735 10.1093/nar/gkl348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Duan X. L., Liu N. N., Yang Y. T., Li H. H., Li M., Dou S. X., and Xi X. G. (2015) G-quadruplexes significantly stimulate Pif1 helicase-catalyzed duplex DNA unwinding. J. Biol. Chem. 290, 7722–7735 10.1074/jbc.M114.628008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu K. Y., Chen W. F., Rety S., Liu N. N., Wu W. Q., Dai Y. X., Li D., Ma H. Y., Dou S. X., and Xi X. G. (2018) Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase. Nucleic Acids Res. 46, 1486–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mendoza O., Gueddouda N. M., Boulé J. B., Bourdoncle A., and Mergny J. L. (2015) A fluorescence-based helicase assay: application to the screening of G-quadruplex ligands. Nucleic Acids Res. 43, e71 10.1093/nar/gkv193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Phan A. T., Luu K. N., and Patel D. J. (2006) Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 34, 5715–5719 10.1093/nar/gkl726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Green J. J., Ying L., Klenerman D., and Balasubramanian S. (2003) Kinetics of unfolding the human telomeric DNA quadruplex using a PNA trap. J. Am. Chem. Soc. 125, 3763–3767 10.1021/ja029149w [DOI] [PubMed] [Google Scholar]
- 60. Marchand A., and Gabelica V. (2016) Folding and misfolding pathways of G-quadruplex DNA. Nucleic Acids Res. 44, 10999–11012 10.1093/nar/gkw970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Qureshi M. H., Ray S., Sewell A. L., Basu S., and Balci H. (2012) Replication protein A unfolds G-quadruplex structures with varying degrees of efficiency. J. Phys. Chem. B 116, 5588–5594 10.1021/jp300546u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lahaye A., Leterme S., and Foury F. (1993) PIF1 DNA helicase from Saccharomyces cerevisiae: biochemical characterization of the enzyme. J. Biol. Chem. 268, 26155–26161 [PubMed] [Google Scholar]
- 63. Ramanagoudr-Bhojappa R., Chib S., Byrd A. K., Aarattuthodiyil S., Pandey M., Patel S. S., and Raney K. D. (2013) Yeast Pif1 helicase exhibits a one-base pair stepping mechanism for unwinding duplex DNA. J. Biol. Chem. 288, 16185–16195 10.1074/jbc.M113.470013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Singh S. P., Koc K. N., Stodola J. L., and Galletto R. (2016) A monomer of Pif1 unwinds double-stranded DNA and it is regulated by the nature of the non-translocating strand at the 3′-end. J. Mol. Biol. 428, 1053–1067 10.1016/j.jmb.2016.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Eoff R. L., and Raney K. D. (2006) Intermediates revealed in the kinetic mechanism for DNA unwinding by a monomeric helicase. Nat. Struct. Mol. Biol. 13, 242–249 10.1038/nsmb1055 [DOI] [PubMed] [Google Scholar]
- 66. Lucius A. L., Maluf N. K., Fischer C. J., and Lohman T. M. (2003) General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 85, 2224–2239 10.1016/S0006-3495(03)74648-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lucius A. L., Vindigni A., Gregorian R., Ali J. A., Taylor A. F., Smith G. R., and Lohman T. M. (2002) DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J. Mol. Biol. 324, 409–428 10.1016/S0022-2836(02)01067-7 [DOI] [PubMed] [Google Scholar]
- 68. Ali J. A., Maluf N. K., and Lohman T. M. (1999) An oligomeric form of E. coli UvrD is required for optimal helicase activity. J. Mol. Biol. 293, 815–834 10.1006/jmbi.1999.3185 [DOI] [PubMed] [Google Scholar]
- 69. Ali J. A., and Lohman T. M. (1997) Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275, 377–380 10.1126/science.275.5298.377 [DOI] [PubMed] [Google Scholar]
- 70. Levin M. K., Wang Y. H., and Patel S. S. (2004) The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 279, 26005–26012 10.1074/jbc.M403257200 [DOI] [PubMed] [Google Scholar]
- 71. Byrd A. K., and Raney K. D. (2004) Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 11, 531–538 10.1038/nsmb774 [DOI] [PubMed] [Google Scholar]
- 72. Byrd A. K., and Raney K. D. (2005) Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry 44, 12990–12997 10.1021/bi050703z [DOI] [PubMed] [Google Scholar]
- 73. Byrd A. K., and Raney K. D. (2006) Displacement of a DNA binding protein by Dda helicase. Nucleic Acids Res. 34, 3020–3029 10.1093/nar/gkl369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rad B., Forget A. L., Baskin R. J., and Kowalczykowski S. C. (2015) Single-molecule visualization of RecQ helicase reveals DNA melting, nucleation, and assembly are required for processive DNA unwinding. Proc. Natl. Acad. Sci. U.S.A. 112, E6852–E6861 10.1073/pnas.1518028112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Manosas M., Xi X. G., Bensimon D., and Croquette V. (2010) Active and passive mechanisms of helicases. Nucleic Acids Res. 38, 5518–5526 10.1093/nar/gkq273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Long X., Parks J. W., Bagshaw C. R., and Stone M. D. (2013) Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 41, 2746–2755 10.1093/nar/gks1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yangyuoru P. M., Bradburn D. A., Liu Z., Xiao T. S., and Russell R. (2018) The G-quadruplex (G4) resolvase DHX36 efficiently and specifically disrupts DNA G4s via a translocation-based helicase mechanism. J. Biol. Chem. 293, 1924–1932 10.1074/jbc.M117.815076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Velankar S. S., Soultanas P., Dillingham M. S., Subramanya H. S., and Wigley D. B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97, 75–84 10.1016/S0092-8674(00)80716-3 [DOI] [PubMed] [Google Scholar]
- 79. Eoff R. L., and Raney K. D. (2010) Kinetic mechanism for DNA unwinding by multiple molecules of Dda helicase aligned on DNA. Biochemistry 49, 4543–4553 10.1021/bi100061v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chib S., Byrd A. K., and Raney K. D. (2016) Yeast helicase Pif1 unwinds RNA:DNA hybrids with higher processivity than DNA:DNA duplexes. J. Biol. Chem. 291, 5889–5901 10.1074/jbc.M115.688648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dillingham M. S., Wigley D. B., and Webb M. R. (2000) Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry 39, 205–212 10.1021/bi992105o [DOI] [PubMed] [Google Scholar]
- 82. Chen M. C., Tippana R., Demeshkina N. A., Murat P., Balasubramanian S., Myong S., and Ferré-D'Amare A. R. (2018) Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 558, 465–469 10.1038/s41586-018-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Barranco-Medina S., and Galletto R. (2010) DNA binding induces dimerization of Saccharomyces cerevisiae Pif1. Biochemistry 49, 8445–8454 10.1021/bi100984j [DOI] [PubMed] [Google Scholar]
- 84. Carroll S. S., Benseler F., and Olsen D. B. (1996) Preparation and use of synthetic oligoribonucleotides as tools for study of viral polymerases. Methods Enzymol. 275, 365–382 10.1016/S0076-6879(96)75022-X [DOI] [PubMed] [Google Scholar]
- 85. Johnson K. A., Simpson Z. B., and Blom T. (2009) Global Kinetic Explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 387, 20–29 10.1016/j.ab.2008.12.024 [DOI] [PubMed] [Google Scholar]
- 86. Hatzakis E., Okamoto K., and Yang D. (2010) Thermodynamic stability and folding kinetics of the major G-quadruplex and its loop isomers formed in the nuclease hypersensitive element in the human c-Myc promoter: effect of loops and flanking segments on the stability of parallel-stranded intramolecular G-quadruplexes. Biochemistry 49, 9152–9160 10.1021/bi100946g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kim B. G., Evans H. M., Dubins D. N., and Chalikian T. V. (2015) Effects of salt on the stability of a G-quadruplex from the human c-MYC promoter. Biochemistry 54, 3420–3430 10.1021/acs.biochem.5b00097 [DOI] [PubMed] [Google Scholar]
- 88. Wlødarczyk A., Grzybowski P., Patkowski A., and Dobek A. (2005) Effect of ions on the polymorphism, effective charge, and stability of human telomeric DNA: photon correlation spectroscopy and circular dichroism studies. J. Phys. Chem. B 109, 3594–3605 10.1021/jp045274d [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


