Abstract
Transcription factors and signaling pathways that regulate stem cells and specialized hormone-producing cells in the pituitary gland have been the subject of intense study and have yielded a mechanistic understanding of pituitary organogenesis and disease. However, the regulation of stem cell proliferation and differentiation, the heterogeneity among specialized hormone-producing cells, and the role of nonendocrine cells in the gland remain important, unanswered questions. Recent advances in single-cell RNA sequencing (scRNAseq) technologies provide new avenues to address these questions. We performed scRNAseq on ∼13,663 cells pooled from six whole pituitary glands of 7-week-old C57BL/6 male mice. We identified pituitary endocrine and stem cells in silico, as well as other support cell types such as endothelia, connective tissue, and red and white blood cells. Differential gene expression analyses identify known and novel markers of pituitary endocrine and stem cell populations. We demonstrate the value of scRNAseq by in vivo validation of a novel gonadotrope-enriched marker, Foxp2. We present novel scRNAseq data of in vivo pituitary tissue, including data from agnostic clustering algorithms that suggest the presence of a somatotrope subpopulation enriched in sterol/cholesterol synthesis genes. Additionally, we show that incomplete transcriptome annotation can cause false negatives on some scRNAseq platforms that only generate 3′ transcript end sequences, and we use in vivo data to recover reads of the pituitary transcription factor Prop1. Ultimately, scRNAseq technologies represent a significant opportunity to address long-standing questions regarding the development and function of the different populations of the pituitary gland throughout life.
The pituitary gland is a highly vascularized neuroendocrine organ made up of a heterogeneous mix of hormone lineages, stem cells, pituitary-specific support cells, and general support cells. The five hormone-producing populations in the mature anterior pituitary gland arise from a Sox2, Prop1-positive progenitor population (1) and diverge during development to become distinct cell types. Many of the signaling and molecular mechanisms governing the embryonic specification and postnatal function of pituitary cell types have been elucidated, but there continue to be gaps in understanding how particular populations are regulated. This is partly due to the population heterogeneity of pituitary gland cells and difficulty in pinpointing population-specific gene expression when performing bulk RNA sequencing or microarrays. Genetically engineered mouse strains have been developed for most of the major known pituitary cell types that can label individual populations for fluorescence-assisted cell sorting (FACS). Population-specific gene expression patterns have been described for gonadotropes using this approach, revealing differences influenced by sex, age, and physiological changes (2). Corticotropes and melanotropes have been similarly profiled (3), but most cell types have not been examined individually to reveal transcriptomes. Heterogeneity within a hormone-producing cell population has been observed with a variety of methods (4, 5), but its extent has been controversial.
Single-cell RNA sequencing (scRNAseq) is a relatively new technique that allows transcriptome analyses from single cells, and it is well suited for the analysis of multiple pituitary cell types. Several methods of scRNAseq have been developed, each with certain advantages and disadvantages (6–9). Some, including the 10x Genomics Chromium platform (10x Genomics, Pleasanton, CA) we use in the present study (10), are based on capturing single cells on a scaffold such as a gel bead, allowing for barcoded RNA library preparation from each cell separately. The barcoded libraries are then multiplexed and sequenced using next-generation technology. The major advantage of scRNAseq for pituitary cells is that a heterogeneous mix of pituitary cells may be processed for scRNAseq in bulk without the need of genetic markers, and separate populations can be reconstituted by in silico cell sorting to produce population-specific gene expression patterns. It also means that most pituitary populations can be captured simultaneously, rather than capturing marker-positive and marker-negative fractions. Gene expression changes in GnRH-treated LβT2 gonadotrope cells were recently studied by scRNAseq (11), and there have been previous studies exploring pituitary cell gene expression on the single-cell level by single-cell quantitative PCR (12).
In this study, we present the first scRNAseq cell census of in vivo pituitary cells and describe the identification of population-specific gene expression patterns of many classical pituitary cell populations from 13,663 total cells representing whole pituitary glands of six 7-week-old C57BL/6 male mice. We identified endocrine cell types, pituitary stem cells, proliferating Pou1f1 cells, and support tissues in silico based on their expression of known gene markers. Using two agnostic clustering algorithms, we provide evidence to suggest that a subpopulation of Gh-expressing cells is enriched in genes in the sterol/cholesterol biosynthesis pathway. We show that scRNAseq-based gene expression is independently reproducible in vivo, such as the enrichment of Foxp2 expression in gonadotropes. Alternatively, we show that incomplete annotation of 3′ transcript sequences on the standard mm10/Grcm38 reference transcriptome can result in false negatives because aligned sequence reads at unannotated loci appear intergenic. For example, the pituitary transcription factor Prop1 is expressed postnatally (13, 14), but it is poorly detected in our scRNAseq data unless the transcriptome is updated to include the complete 3′ untranslated region (UTR) as detected in full-length embryonic pituitary cDNA clones of Prop1 (15). Ultimately, we demonstrate that scRNAseq is a powerful technique for identifying novel transcripts that are specific to distinct endocrine and stem cell populations of the pituitary gland. This rich research resource will be valuable for identifying the molecular mechanisms that regulate cell specialization during organogenesis.
Materials and Methods
Mice
Wild-type C56BL/6 mice used for this study were approved by the University of Michigan Institutional Animal Care and Use Committee and the Animal Care and Use Office, as well as by the Southern Illinois University Animal Care and Use Office. Animals were housed in 12-hour light:12-hour dark cycles, and food and water were provided ad libitum.
Pituitary gland dispersion
The pituitary glands of six individually housed 7-week-old male C57BL/6 mice were dissected, and the tissue was dispersed as previously described (14, 16, 17), as two pools of three glands each. Briefly, the dissected tissue is placed into an enzyme mix containing 0.5% w/v collagenase type 2 (Lorne Laboratories, Reading, UK), 0.1× trypsin (Invitrogen, Carlsbad, CA), 50 μg/mL DNase I (Worthington Biochemical, Lakewood, NJ), and 2.5 μg/mL amphotericin B (Fungizone; Invitrogen) in Hanks balanced salt solution (Invitrogen) for 4 hours at 37°C. The tissue and cells were gently triturated, counted on a hemocytometer, pelleted at 200g for 5 minutes, and resuspended at ∼1000 cells per μL in 0.4% BSA/Hanks balanced salt solution.
Cell capture and library preparation on chromium platform
Single-cell 3′ library generation was performed on the 10x Genomics Chromium Controller following the manufacturer’s protocol for the v2 reagent kit (10x Genomics) (10). Cell suspensions were loaded onto a Chromium Single Cell A Chip along with reverse transcription master mix and single-cell 3′ gel beads, aiming for 10,000 cells per channel. For each pool, 17,400 cells with 71% to 82% viability were encapsulated into emulsion droplets at a concentration of 700 to 1200 cells per μL, which targets 10,000 single cells with an expected multiple cell rate of 7.6%. Following generation of single-cell gel bead-in-emulsions (GEMs), reverse transcription was performed and the resulting post-GEM reverse transcription product was cleaned up using DynaBeads MyOne silane beads (Thermo Fisher Scientific, Waltham, MA). The cDNA was amplified, SPRIselect (Beckman Coulter, Brea, CA) cleaned and quantified, and then enzymatically fragmented and size selected using SPRIselect beads to optimize the cDNA amplicon size prior to library construction. An additional round of double-sided SPRI bead cleanup was performed after end repair and A-tailing. Another single-sided cleanup was done after adapter ligation. Indexes were added during PCR amplification and a final double-sided SPRI cleanup was performed. Libraries were quantified by KAPA quantitative PCR for Illumina adapters (Roche, Pleasanton, CA) and size was determined by Agilent TapeStation D1000 tapes. Read 1 primer sequences were added to the molecules during GEM incubation. P5, P7, and sample index and read 2 primer sequences were added during library construction via end repair, A-tailing, adaptor ligation, and PCR. Libraries were generated with unique sample indices for each sample. Libraries were sequenced on a HiSeq 4000 (Illumina, San Diego, CA) using a HiSeq 4000 paired-end cluster kit with HiSeq 4000 sequencing by synthesis kit (100 cycles, reagents, loaded at 200 pM) following Illumina’s denaturing and dilution recommendations. PhiX was spiked into the load at 1%. The run configuration was 26 × 8 × 98 (115) cycles for read 1, index, and read 2, respectively. Cell Ranger Single Cell Software Suite 2.1.1 was used to perform sample demultiplexing and barcode processing at the University of Michigan Biomedical Core Facilities DNA Sequencing Core.
Bioinformatic analyses of scRNAseq data
Sequence alignment maps and transcript quantification of data discussed in this publication are available on National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (18, 19) through accession no. GSE120410 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120410). Demultiplexed fastq files generated by the University of Michigan Biomedical Core Facilities DNA Sequencing Core were analyzed with the 10x Genomics Cell Ranger 2.1.1 alignment and gene counting software, a self-contained scRNAseq pipeline developed by 10x Genomics that utilizes the RNAseq alignment program STAR (10, 20). The pipeline aligned reads to the University of California Santa Cruz (UCSC) mm10 (mGRC38) transcriptome using STAR and performed transcript counting. Principle component analysis, t-distributed stochastic neighbor embedding (tSNE), and K-means clustering were performed as previous described (10) using Cell Ranger. Manually generated clusters, tSNE plots, differential gene expression analyses, and volcano plots were produced using a combination of the Loupe software (10x Genomics), the Cell Ranger R kit (10x Genomics) within R (21), and Photoshop CS6 (Adobe). Venn diagrams were generated by the BioVenn Web application (22). Gene ontology analyses were performed using the NCBI Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (23, 24).
Custom annotation of transcriptome reference
The UCSC mm10 reference mouse transcriptome was amended by annotating chromosome 11: 50,948,572 to 50,949,192 in the gene transfer file as an artificial single exon gene on the negative strand named “Prop1L.” A custom reference was then compiled using the cellranger mkref command, and the custom reference was used for Cell Ranger/STAR alignment instead of the original UCSC mm10 transcriptome.
Histology and immunohistochemistry
Pituitary glands from C57BL/6 male mice were dissected and fixed in 4% paraformaldehyde in PBS (pH 7.2) for 24 hours. All samples were washed in PBS, dehydrated in a graded series of ethanol, embedded in paraffin, and sectioned at 5 μm. To visualize FOXP2, tissue sections were deparaffinized in xylene and rehydrated through a series of graded ethanol washes, and 1.5% peroxide in PBS was used to quench endogenous peroxidases. After epitopes were unmasked by boiling in 10 mM citric acid for 10 minutes, tissue sections were blocked with blocking reagent from the Tyramide Signal Amplification kit (PerkinElmer, Waltham, MA). Sections were incubated overnight at 4°C with a rabbit anti-FOXP2 antibody [1:1000; Abcam catalog no. ab16046; RRID: AB_2107107 (25)]. Tissue sections were incubated with anti-rabbit secondary antibody (1:100; Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 hour at room temperature. Next, sections were incubated sequentially with streptavidin–horseradish peroxidase and fluorescein isothiocyanate (FITC) from the Tyramide Signal Amplification kit (PerkinElmer). Following a 5-minute incubation with water, sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (167 nM; Molecular Probes/Thermo Fisher Scientific). Colocalization of FOXP2 with hormones was performed as described above with the addition of a blocking step (anti-rabbit IgG; 1:200; Jackson ImmunoResearch Laboratories) after incubation with primary antibody. To visualize pituitary hormones, tissue sections were incubated with antibodies against GH [1:10,000; A. F. Parlow National Hormone and Peptide Program catalog no. mGH; RRID: AB_2629219 (26)], ACTH [1:500; A. F. Parlow National Hormone and Peptide Program catalog no. hACTH; RRID: AB_2665562 (27)], TSH β subunit [1:2000; A. F. Parlow National Hormone and Peptide Program catalog no. rTSHb; RRID: AB_2665563 (28)], LH β subunit [LHB; 1:500; A. F. Parlow National Hormone and Peptide Program catalog no. r-gp-LHb; RRID: AB_2665565 (29)], FSH β [FSHB; 1:1120; A. F. Parlow National Hormone and Peptide Program catalog no. AFP-C0972881; RRID: AB_2687903 (30)], or prolactin [1:10,000; A.F. Parlow National Hormone and Peptide Program catalog no. PRL; RRID: AB_2629220 (31)] for 1 hour at room temperature, followed by the appropriate secondary antibodies anti-rabbit tetramethylrhodamine isothiocyanate [1:100; Jackson ImmunoResearch Laboratories catalog no. 711-025-152; RRID: AB_2340588 (32)] or anti-guinea pig FITC [1:100; Jackson ImmunoResearch Laboratories catalog no. 706-095-148; RRID: AB_2340453 (33)]. Digital images were captured with a Leica DM 5000B fluorescent microscope and Retiga 2000R digital camera. FITC, tetramethylrhodamine isothiocyanate, and DAPI images were merged using Adobe Photoshop CS3. In some instances, images were brightened for illustrative purposes.
Results
scRNAseq of pituitary tissue generates highly reproducible datasets
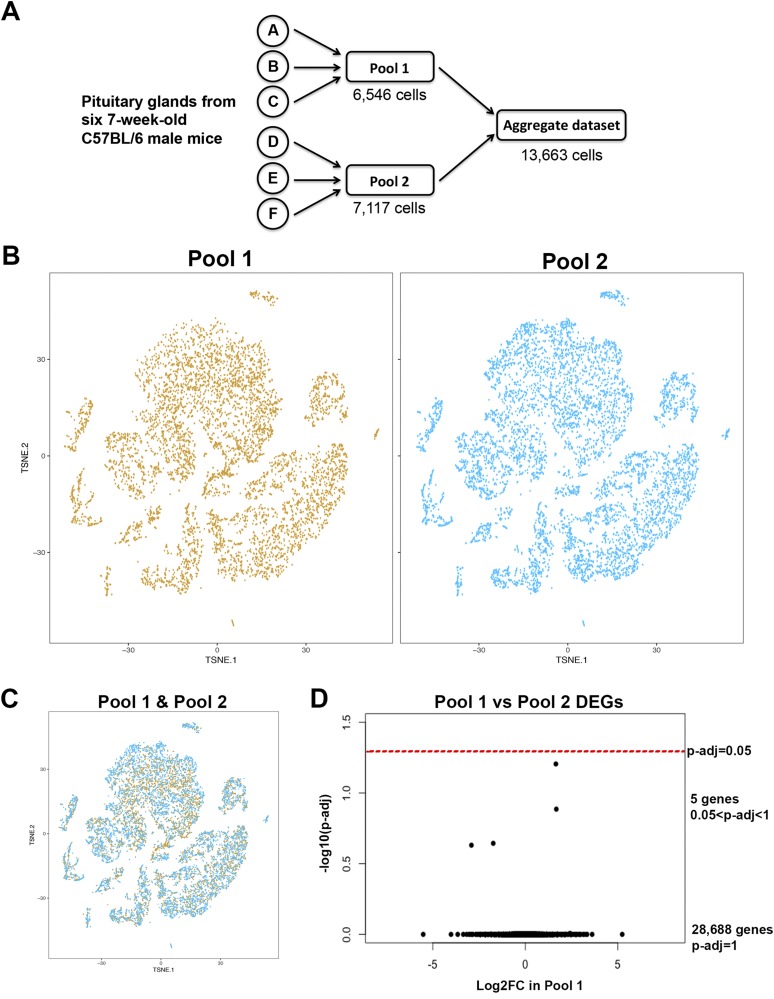
We processed pituitary cells from six individual 7-week-old C57BL/6 male mice in two pools of three each for scRNAseq (Fig. 1A). Data were evaluated from each pool separately, using 10 different parameters, to assess differences that could arise from biological variation among individual animals or technical variation in library preparation and/or sequencing (Table 1). Although pool 2 was sequenced more deeply than pool 1, there is low variation for all other parameters. Sequencing metrics for each pool showed 65% to 70% cell capture efficiency, and a high percentage of reads confidently mapped to the UCSC mm10 mGRC38 genome (Table 1, 88%). The data from each pool were normalized for read depth and aggregated into one dataset containing a total of 13,663 cells (Fig. 1A). Each pool was compared with the aggregate.
Figure 1.
Low biological and technical variability between pooled samples processed together. (A) Schematic of processing of dispersed cells from six pituitary glands for single-cell capture and RNA sequencing. (B) tSNE plots for each pool show highly similar cell signatures, and there are no clusters that are present in only one pool. (C) Merged tSNE plots for the two pools show high overlap between the clusters from each pool. (D) Volcano plot of differentially expressed genes between the two pools. No genes are significantly differentially expressed.
Table 1.
Cell Capture and Sequencing Metrics for scRNAseq of Six Whole Pituitary Glands From 7-Week-Old C57BL/6 Mouse Pituitary Gland, Processed as Two Pools of Three Each
| Pool 1 | Pool 2 | |
|---|---|---|
| Number of captured cells | 6546 | 7117 |
| Total sequence reads | 273,079,474 | 326,042,253 |
| Fraction of reads in cells | 89.30% | 91.50% |
| Total genes detected | 19,330 | 19,589 |
| Mean reads per cell | 41,716 | 45,811 |
| Median genes detected per cell | 2113 | 2213 |
| Median transcripts per cell | 10,128 | 11,035 |
| Reads mapped to genome | 89.10% | 88.10% |
| Reads mapped uniquely to genome | 87.20% | 86.20% |
| Reads mapped uniquely to transcriptome | 70.90% | 69.80% |
All 13,663 cells from the two pools were analyzed together and plotted onto the same tSNE plot, and visualized by which pool they originated from (Fig. 1B). The tSNE plots of each pool are highly similar, and all cell populations are common to both pools. There are no cell clusters that are produced by only one of the pools. Plotting the pools together shows high overlap again between the two pools (Fig. 1C). Differential gene expression analysis between pool 1 and pool 2 shows no significantly differentially expressed genes between the two samples (Fig. 1D). Therefore, there is limited biological and/or technical variability between the two pools, supporting their analyses as an aggregated dataset.
All classical pituitary endocrine cell populations are detected and cluster closely together
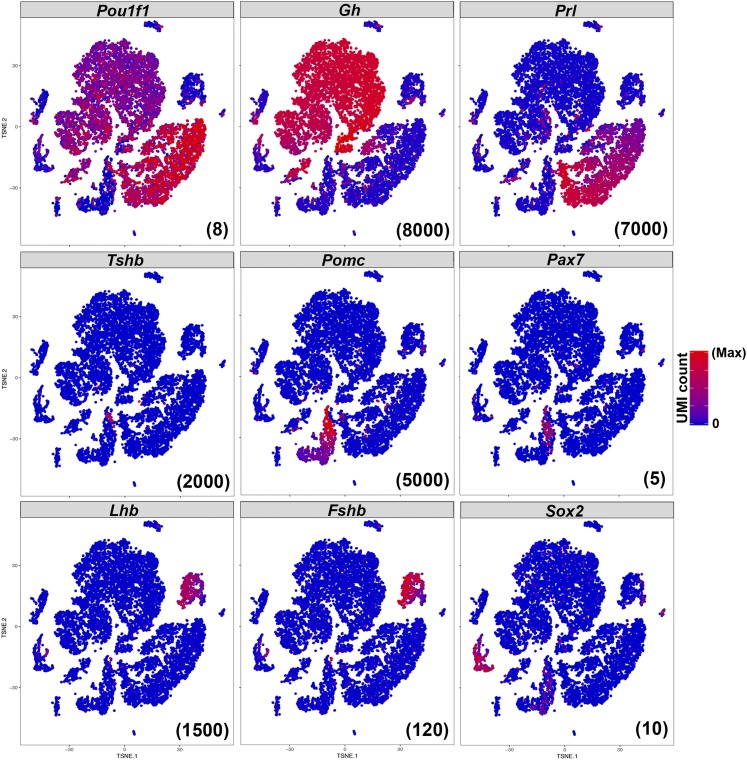
tSNE plots indicate unique molecular identifier (UMI) counts, which reflect transcript number, as each transcript has a unique UMI that is only counted once (even if it is sequenced more than once). Expression of anterior pituitary endocrine markers Pou1f1, Gh, Prl, Tshb, Pomc, Pax7, Lhb, and Fshb shows that somatotropes, lactotropes, thyrotropes, corticotropes, melanotropes, and gonadotropes, respectively, are all detected in our sample (Fig. 2). Gh-expressing cells are the most abundant cell population, representing somatotropes. Prl-expressing lactotropes are the second most abundant cell type. Cells expressing Pomc are corticotropes or melanotropes; the portion of Pomc-expressing cells that also express Pax7 distinguish them as melanotropes. Tshb-expressing cells are thyrotropes, and cells expressing the gonadotropins Lhb and Fshb represent gonadotropes. Adult pituitary stem cells are identified by their expression of Sox2 similar to the expected proportion of 3% to 5% (34) (∼2.5% in our sample). Expression of hormone transcripts are much higher than those of transcription factors.
Figure 2.
Classical pituitary endocrine and stem cell markers are all detected by scRNAseq. Expression of Pou1f1, Gh, Prl, Tshb, Pomc, Pax7, Lhb, Fshb, and Sox2 are all detectable, and cells expressing those markers cluster closely together. The hormones are much more highly expressed. The maximum UMI count plotted for each gene is indicated in parentheses at the bottom right of each panel.
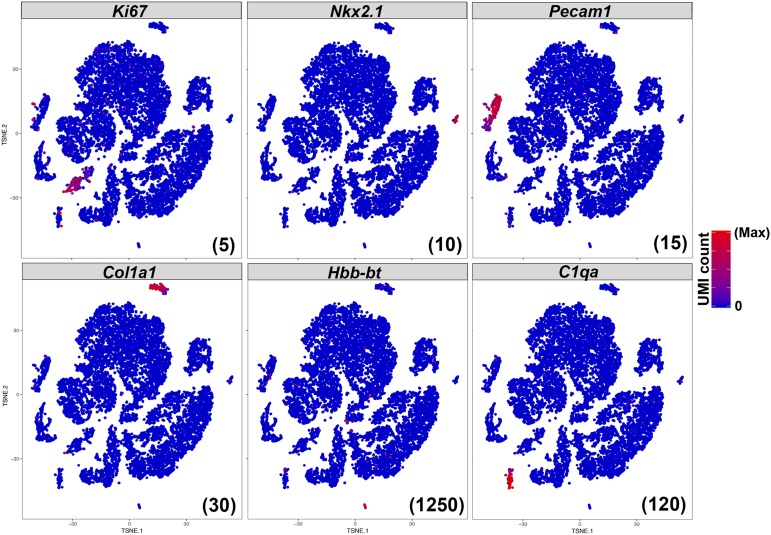
The proliferative cells in the embryonic and juvenile pituitary are Sox2-positive, but rapidly switch to being Pou1f1-positive in the first 3 weeks of life (35). Transcripts for the marker of active proliferation, Ki67, are enriched in a subpopulation of Pou1f1-positive cells and are not observed in the Sox2 stem cell population (Fig. 3), consistent with previous findings (35). Expression of platelet and endothelial cell adhesion molecule Pecam1, the neural transcription factor Nkx2.1, collagen Col1a1, adult hemoglobin β-chain Hbb-bt, and complement protein C1qa mark additional cell populations as endothelia, posterior pituitary, connective tissue, red blood cells, and white blood cells, respectively. We processed the pituitary tissue without any microdissection or perfusion, and therefore the detection of these additional cell types is expected.
Figure 3.
Identification of the proliferating Pou1f1 population and nonendocrine populations. A subset of Pou1f1-positive cells expresses the proliferative marker Ki67, whereas adult Sox2-positive cells do not. Expression of Nkx2.1, Pecam1, Col1a1, Hbb-bt, and C1qa mark distinct peripheral populations as posterior lobe axons, endothelia, connective tissue, red blood cells, and white blood cells, respectively. The maximum UMI count plotted for each gene is indicated in parentheses at the bottom right of each panel.
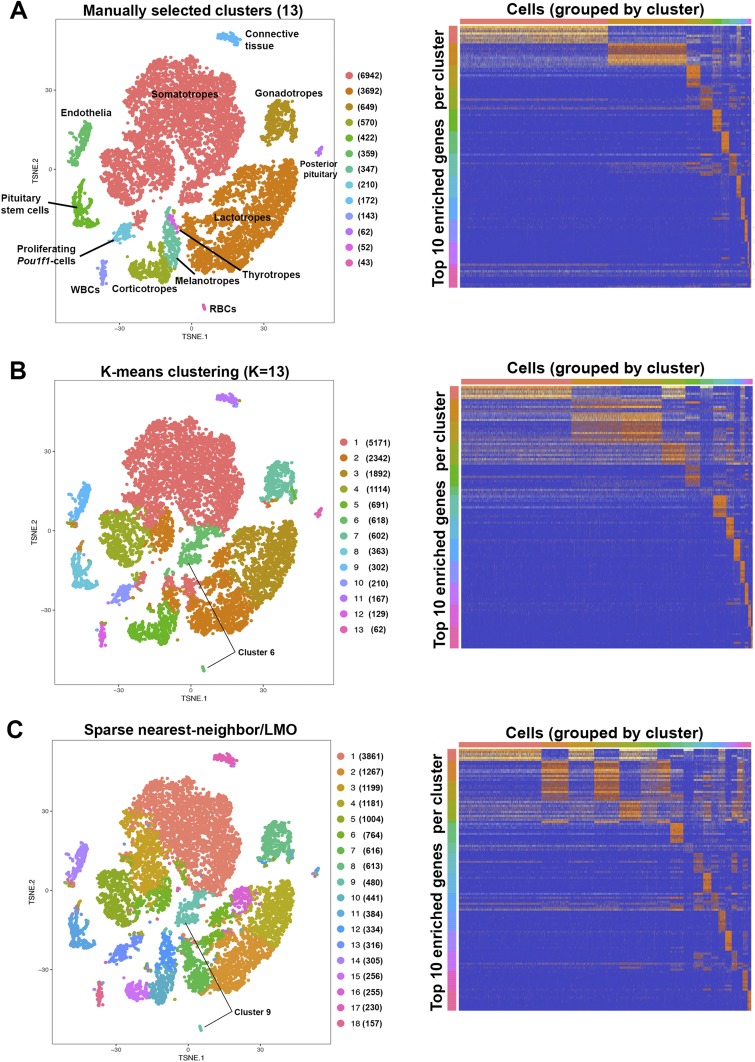
Cluster analyses of pituitary populations using biased and unbiased methods
Conventionally, an intact, adult pituitary gland is expected to contain the five anterior hormone-producing cell types, proliferating Pou1f1-cells, Sox2-expressing stem cells, intermediate lobe melanotropes, posterior lobe axon terminals and pituicytes, endothelia, surrounding connective tissue, red blood cells, and white blood cells; we therefore expect at least 13 cell populations in our dataset. We manually created cell clusters based on expression of known gene markers as shown in Figs. 2 and 3 (Fig. 4A) for each of the 13 expected cell types to yield a gene expression heat map with distinct molecular signatures. We also used two different statistical methods to generate unbiased cell clusters within our aggregated dataset. Both are based on calculating near-neighbor Euclidean distance between cells as placed on the tSNE plot. Unbiased clustering by traditional K-means clustering for 13 clusters (Fig. 4B) identified subpopulations within the somatotropes and lactotropes, but it clustered Tshb- and Pax7-expressing cells with Gh- and Prl-expressing cells in clusters 1 and 2, respectively. It also grouped hemoglobin-expressing cells together with a subset of Gh-expressing cells (which do not express hemoglobins) in cluster 6. The graph-based clustering algorithm developed by 10x Genomics, consisting of building a sparse nearest neighbor graph and Louvain modularity optimization (36), automatically distinguished 18 clusters within the dataset (Fig. 4B). This method similarly created somatotrope and lactotrope subpopulations, but clustered Tshb- and Pax7-expressing cells together in cluster 12, and clustered hemoglobin-expressing cells with a Gh subpopulation not expressing hemoglobins in cluster 9. A heat map of gene enrichment per cluster showed several clusters have similar molecular signatures.
Figure 4.
Cluster analyses of pituitary hormone-producing cells and other cell types. Numbers in parentheses beside each cluster label indicate number of cells in that cluster. (A) Manual assignment of cells to expected cell types based on gene expression of known markers. The gene expression heat map shows the distinct patterns for each cell cluster. (B) Traditional K-means clustering for 13 populations finds subpopulations within Gh- and Prl-expressing populations, but does not distinguish thyrotropes, melanotropes, or red blood cells as separate populations. (C) Cell clustering by the graph-based sparse nearest neighbor and Louvain modularity optimization method generates 18 cell clusters, identifying subpopulations within Gh- and Prl-expressing populations, but does not distinguish Tshb-, Pax7-, or hemoglobin-expressing cells as separate populations.
Enrichment of sterol/cholesterol biosynthesis genes in agnostically defined subsets of Gh-expressing cells
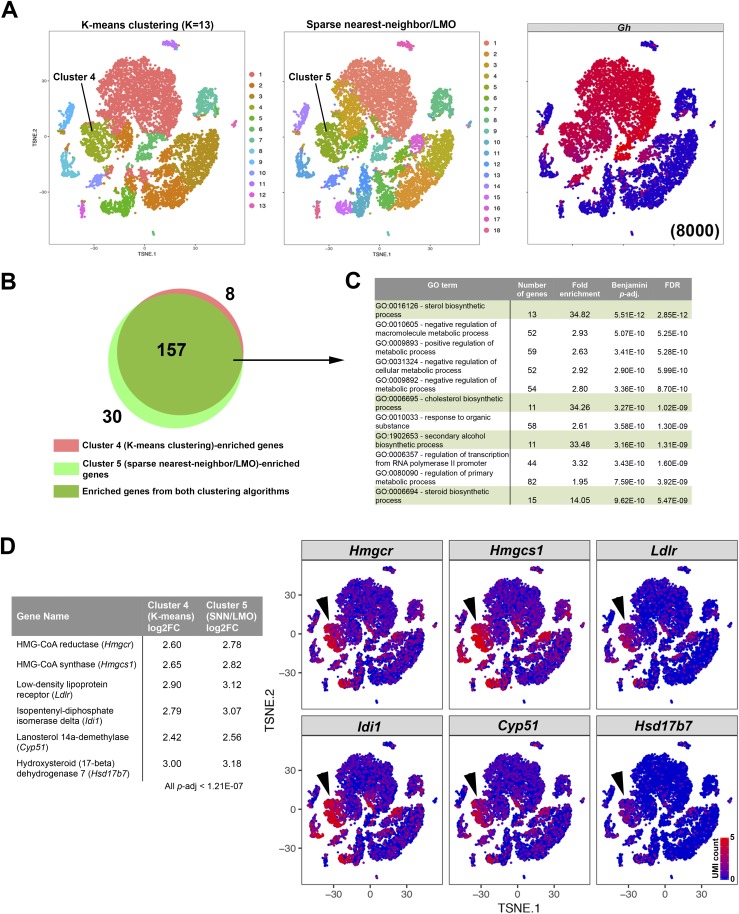
Both the K-means and sparse nearest neighbor/linear moveout (LMO) agnostic clustering algorithms identified subpopulations within the Gh-expressing populations (Fig. 4B and 4C). Cluster 4 of the K-means method and cluster 5 of the sparse nearest neighbor/LMO method represent a subset of somatotropes with similar tSNE boundaries (Fig. 5A). Genes enriched in each cluster have a high overlap, most likely because the clusters encompass many of the same cells (Fig. 5B). Using only genes that are enriched in both clustering methods, we performed gene ontology analysis with DAVID and found an enrichment of genes related to sterol and/or cholesterol biosynthesis (Fig. 5C), in particular, enzymes that metabolize upstream intermediates of cholesterol (Fig. 5D), although transcripts of these genes are also detected in other cells. Therefore, two different agnostic clustering algorithms identified a somatotrope subpopulation that is enriched in cholesterol-related genes.
Figure 5.
Sterol/cholesterol biosynthesis genes are enriched in a subpopulation of Gh-expressing cells identified by two agnostic clustering algorithms. (A) Subsets of Gh-expressing cells are clustered as a distinct population by two different clustering algorithms. (B) Venn diagram showing a large overlap between cluster-enriched genes calculated from cluster 4 of the K-means method and cluster 5 of the sparse nearest neighbor/LMO method. (C) DAVID gene ontology analysis of the 157 commonly enriched genes in this Gh-expressing subset. Pathways related with synthesis of sterols/cholesterol are highly enriched. (D) Genes coding for proteins that synthesize upstream intermediates of cholesterol are enriched in these Gh-expressing subsets (arrowheads), although some of these genes are also detected in other cells. FDR, false discovery rate; GO, gene ontology.
Independent confirmation of scRNAseq gene expression data: detection of FOXP2 in gonadotropes in vivo
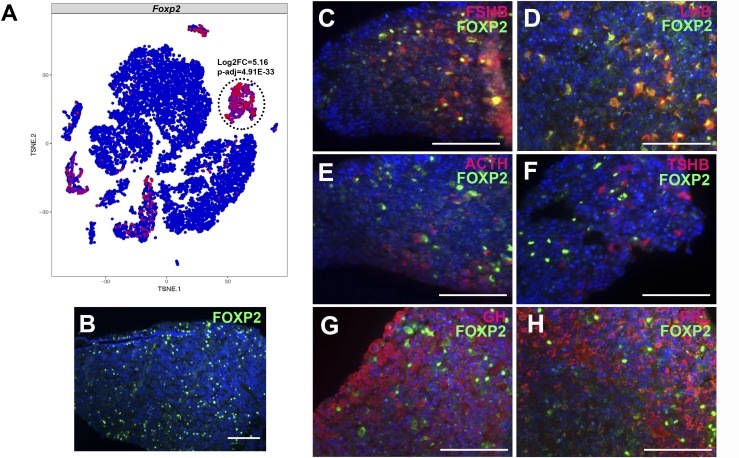
Lists of genes that are differentially expressed in the manually assigned clusters (Fig. 4A) provide an overview of genes enriched in classically defined pituitary hormone–producing cell populations, which include known and novel markers of pituitary cell types (Table 2). We sought to validate an scRNAseq observation by independent means in vivo to demonstrate the veracity of the scRNAseq technique in identifying novel population markers. Transcripts encoding the forkhead homeobox transcription factor Foxp2 were significantly enriched in the gonadotrope cluster (5.16 log2 fold change, P = 4.91 × 10−33) (Fig. 6A). Foxp2 expression has been detected in the mouse brain (37–40), but its expression has not previously been explored in the pituitary gland. Immunohistochemistry for FOXP2 in C57BL/6 8-week-old male mice confirmed its expression in a subset of anterior pituitary cells (Fig. 6B), and costaining of FOXP2 with each anterior pituitary hormone showed FOXP2 colocalization with LHB and FSHB, hormones expressed in gonadotropes (Fig. 6C–6H). Therefore, we demonstrate that FOXP2 is a novel transcription factor enriched in gonadotropes that is observed both using our scRNAseq technique and by immunohistochemistry in vivo.
Table 2.
Novel Markers of Classical Pituitary Populations
| Log2 Fold Change |
Adjusted P Value |
|
|---|---|---|
| Somatotrope cluster | ||
| Pappa2 | 3.17 | 2.49 × 10−22 |
| Gh | 3.05 | 6.16 × 10−22 |
| Ceacam10 | 2.74 | 2.57 × 10−17 |
| Ghrhr | 2.57 | 2.00 × 10−16 |
| Tcerg1l | 2.67 | 5.12 × 10−16 |
| Cabp2 | 3.00 | 7.94 × 10−16 |
| Wnt10a | 2.57 | 8.39 × 10−16 |
| RP24-294M17.1 | 2.89 | 2.29 × 10−15 |
| Mmp7 | 3.23 | 3.40 × 10−15 |
| Gm43569 | 2.95 | 5.85 × 10−15 |
| Sstr5 | 2.64 | 2.47 × 10−14 |
| Sstr2 | 2.39 | 1.02 × 10−11 |
| Rxrg | 2.10 | 1.41 × 10−11 |
| Rad18 | 1.27 | 5.68 × 10−5 |
| Lactotrope cluster | ||
| Prl | 5.10 | 3.30 × 10−69 |
| Agtr1a | 4.90 | 2.71 × 10−60 |
| Syndig1 | 5.27 | 1.29 × 10−55 |
| Angpt1 | 4.75 | 1.29 × 10−55 |
| Edil3 | 4.55 | 1.38 × 10−52 |
| 6030419C18Rik | 4.68 | 2.59 × 10−52 |
| Hepacam2 | 4.41 | 1.09 × 10−51 |
| Cilp | 4.84 | 2.31 × 10−48 |
| Akr1c14 | 5.01 | 1.80 × 10−44 |
| Gpr83 | 5.33 | 2.95 × 10−43 |
| Gonadotrope cluster | ||
| Lhb | 7.82 | 8.01 × 10−99 |
| Tgfbr3l | 7.72 | 5.60 × 10−97 |
| Fshb | 7.49 | 2.50 × 10−90 |
| Gnrhr | 7.78 | 5.79 × 10−90 |
| Cga | 6.78 | 1.30 × 10−73 |
| Spp1 | 6.91 | 6.99 × 10−71 |
| Grem1 | 7.43 | 6.08 × 10−59 |
| Nr0b1 | 6.87 | 1.14 × 10−57 |
| Speer4e | 7.55 | 5.15 × 10−57 |
| Rarres1 | 7.26 | 1.61 × 10−55 |
| Nr5a1 | 6.04 | 2.69 × 10−50 |
| Foxp2 | 5.16 | 4.91 × 10−33 |
| Corticotrope cluster | ||
| Crhr1 | 7.59 | 6.98 × 10−50 |
| Gpc5 | 8.04 | 2.27 × 10−40 |
| Tbx19 | 6.52 | 1.99 × 10−36 |
| Tnnt1 | 7.12 | 3.89 × 10−36 |
| Tnni3 | 7.41 | 6.26 × 10−28 |
| Gm15543 | 7.21 | 1.05 × 10−27 |
| Cplx3 | 7.51 | 1.05 × 10−27 |
| Egr4 | 7.47 | 6.98 × 10−24 |
| Cdh8 | 7.71 | 2.61 × 10−23 |
| Galnt9 | 6.51 | 2.61 × 10−23 |
| Pomc | 3.06 | 3.34 × 10−4 |
| Melanotrope cluster | ||
| Pomc | 7.23 | 5.80 × 10−51 |
| Oacyl | 7.16 | 3.01 × 10−38 |
| Pax7 | 7.37 | 3.12 × 10−31 |
| Esm1 | 6.11 | 1.95 × 10−21 |
| Pkib | 5.77 | 2.29 × 10−21 |
| Pcsk2 | 5.46 | 2.53 × 10−21 |
| Gulo | 6.97 | 3.38 × 10−20 |
| Megf11 | 5.86 | 5.55 × 10−18 |
| Rbfox3 | 5.77 | 2.60 × 10−17 |
| Scn2a1 | 5.30 | 2.92 × 10−15 |
| Thyrotrope cluster | ||
| Tshb | 8.42 | 1.56 × 10−6 |
| Trhr | 7.86 | 6.12 × 10−4 |
| Dio2 | 6.30 | 1.85 × 10−3 |
| Stem cell cluster | ||
| Cyp2f2 | 9.60 | 9.78 × 10−122 |
| Lcn2 | 9.39 | 1.33 × 10−106 |
| Mia | 8.72 | 2.68 × 10−103 |
| Aldh3a1 | 9.30 | 7.07 × 10−93 |
| Aldh1a2 | 8.42 | 2.50 × 10−90 |
| Cpxm2 | 8.38 | 3.43 × 10−87 |
| Rbpms | 7.77 | 1.08 × 10−84 |
| Gm266 | 8.49 | 5.21 × 10−84 |
| Cdh26 | 11.46 | 7.29 × 10−83 |
| Aqp3 | 9.08 | 1.53 × 10−81 |
| Aqp4 | 9.77 | 1.06 × 10−75 |
| Sox2 | 6.86 | 1.78 × 10−62 |
| Proliferating Pou1f1 cluster | ||
| Pbk | 7.92 | 6.98 × 10−61 |
| Spc25 | 7.83 | 6.49 × 10−60 |
| Cdca3 | 7.24 | 2.95 × 10−52 |
| Birc5 | 7.29 | 9.17 × 10−52 |
| Cenpf | 7.57 | 4.29 × 10−51 |
| Ube2c | 7.68 | 1.14 × 10−50 |
| Hmmr | 7.86 | 4.76 × 10−50 |
| Top2a | 7.56 | 2.17 × 10−49 |
| Ckap2l | 7.60 | 3.93 × 10−48 |
| Ccnb1 | 7.77 | 7.30 × 10−47 |
| Cdk1 | 6.99 | 5.61 × 10−46 |
| Ccna2 | 6.96 | 8.84 × 10−44 |
| Ccnb2 | 6.80 | 1.23 × 10−39 |
Log2 fold change values and Benjamini–Hochberg adjusted P values for the somatotrope, lactotrope, corticotrope, melanotrope, thyrotrope, Sox2 stem cell, and proliferating Pou1f1 cell clusters as labeled in Fig. 4A. Many novel candidate genes are more significantly enriched than the cell type–identifying hormones. Rows in bold type indicate novel genes of that cell type.
Figure 6.
scRNAseq detection of Foxp2 expression in gonadotropes is independently observed by immunofluorescence in vivo. (A) Foxp2 expression by scRNAseq in 7-week-old male pituitary cells. Foxp2 expression is enriched in the Lhb-expressing cluster. (B) Immunohistochemistry for FOXP2 (green) was performed in 8-week-old male mice. (C–H) Coimmunohistochemistry was performed for FOXP2 (green) and anterior pituitary hormones in red: (C) FSHB, (D) LHB, (E) ACTH, (F) TSH β subunit (TSHB), (G) GH, and (H) prolactin (PRL). Cell nuclei are labeled with DAPI (blue). Scale bars, 100 μm. (B–H) Each panel is a representative image of immunohistochemistry that was performed on samples from four different litters.
Incomplete transcriptome annotation of 3′ end sequences can cause false negatives in gene expression analyses
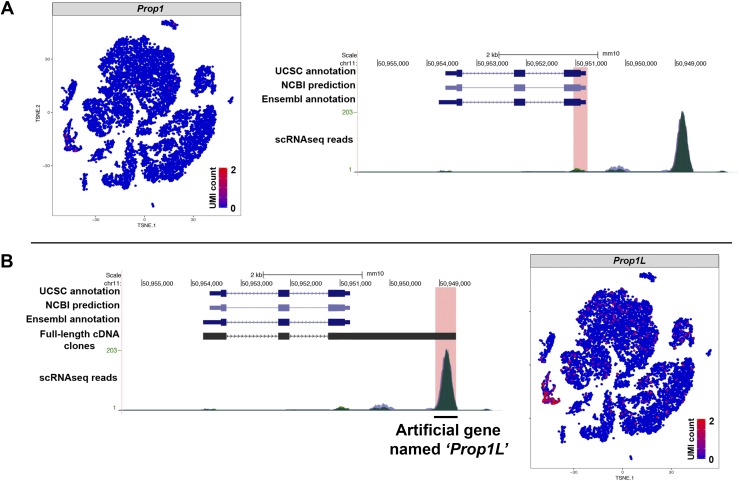
RNA libraries generated on the 10x Genomics Chromium platform produce ∼100-bp sequence reads from the 3′ end of mRNA transcripts, because intact transcripts are UMI barcoded before being fragmented prior to sequencing. This can cause a problem for genes that do not have fully annotated 3′ ends/UTRs. In this experiment, we observed that there are fewer than expected reads of the pituitary transcription factor Prop1, which led us to investigate whether incomplete annotation could have caused a false negative. Prop1 is expressed in postnatal pituitary stem cells within the marginal zone and in scattered cells throughout the anterior lobe (13, 14). Analysis of the sequence alignment map for each sample pool found a minor peak at the annotated Prop1 3′ UTR, and a much higher peak aligning to an intergenic region downstream of the annotated Prop1 gene on the same strand of chromosome 11 (Fig. 7A). These genome-aligned reads are discarded because no gene is annotated there on the transcriptome. We previously generated full-length cDNA libraries from the embryonic pituitary gland using a 5′ end m7G cap-trap and poly-A selection technique that identified bona fide 5′ ends and polyadenylation sites previously unknown for many of the 12,000 genes represented in the cDNA libraries (15). The 5′ and 3′ end sequencing of several full-length Prop1 cDNAs present in these libraries consistently revealed an extensive Prop1 3′ UTR (Fig. 7B). The 5′ end begins at the same coordinates as the annotation. Sanger sequencing confirmed that in vivo Prop1 transcripts possess a longer 3′ UTR than what is shown on the mm10 transcriptome reference. The extended 3′ UTR ends at mGRC38 chromosome 11: 50,948,669 on the negative strand, 1.5 kb after the annotated end of the 3′ UTR at chromosome 11: 50,950,206. The chromosomal position of the major alignment peak in the scRNAseq matches the position of the extended 3′ UTR of Prop1. Custom amendment of the mm10 transcriptome to annotate chromosome 11: 50,948,572 to 50,949,192 on the negative strand as an artificial gene named Prop1L allows assignment of those reads separately from Prop1. We find that Prop1L is enriched in the Sox2 stem cell cluster. Therefore, incomplete 3′ end sequence transcriptome annotation can cause false negatives that can be corrected with custom amendments.
Figure 7.
Incomplete annotation of 3′ UTRs in the reference transcriptome can cause false negatives on gene expression analyses. (A) Prop1 is not detected by scRNAseq despite previous descriptions of its postnatal expression. Analyses of the sequence alignment map for each pool showed a mapping peak in an apparently intergenic region 2.5 kb downstream of Prop1 on the negative strand of mouse chromosome 11 (chr11). (B) This peak coincides with the end of full-length Prop1 cDNAs derived from embryonic mouse pituitaries that were confirmed by Sanger sequencing. Custom annotation of that genomic location as the artificial gene Prop1L recovers those reads, which are enriched in Sox2-expressing stem cells.
Discussion
scRNAseq is a novel technique that allows for in silico sorting of tissues that contain heterogeneous cell populations, such as the multiple interdigitated hormone and support populations of the postnatal pituitary gland. Transgenic reporter mice have been generated to fluorescently mark four of the five anterior endocrine populations: Gh-eGFP (41), Prl-dsRed/Prl-mRFP (42), Pomc-eGFP (43), and Lhβ-Cerulean (3) mice. Tshb-Cre transgenic mice (44) can be crossed to a reporter strain such as the Rosa26LSL-GFP mice to produce fluorescently marked thyrotropes, similar to the approach taken with Gnrhr-iCre mice to purify gonadotropes and analyze gene expression (2, 45). Pituitary glands from these mice can be dispersed and the cells undergo FACS to isolate purified pituitary hormone cell populations for RNAseq or other genomic and epigenetic analyses. However, these approaches can be limited by the fidelity of the transgenic reporter mice and the FACS capture efficiency. Our results demonstrate that scRNAseq of dispersed pituitary cells can identify all pituitary endocrine populations in silico without the need of any genetic marker or FACS. Another advantage of this technique is that multiple populations can be captured simultaneously, without the need to produce double- or triple-transgenic mice, especially when at least two strains are tagged with the same fluorescent marker and would be uninformative if used together (Gh-eGFP and Pomc-eGFP). Furthermore, although it is presumed that transgenic insertions of fluorescent markers do not have negative effects, there is some evidence to suggest that expression of fluorescent proteins can affect endocrine physiology, because total pituitary GH content is reduced by ∼50% to 67% in Gh-eGFP mice (41).
The statistical algorithms we used in the present study distinguished somatotrope and lactotrope subpopulations, but they both did not distinguish Tshb- and Pax7-expressing cells as separate populations, despite the general consensus that they are distinct cell types. A subpopulation of Gh-expressing somatotropes is enriched in sterol/cholesterol-biosynthesis genes. The role of a cholesterol-synthesizing somatotrope subpopulation is not clear, and there is no generally known interaction between sterols and GH. In humans who undertake GH replacement therapy, there is some evidence to suggest that recombinant GH reduces plasma cholesterol (46–48), but the relationship between this observation and the actual synthesis of sterols from some somatotropes is again unclear. Further studies may investigate whether this somatotrope subpopulation has a biological role in vivo.
Manually assigning cells on a region of the tSNE plot based on expression of known cell-type markers does not use a statistical algorithm but allows us to generate the cell populations that are expected based on expression of known genes. In this study, we provide gene enrichment lists for traditional pituitary cell types as a community resource for potential regulators of different pituitary populations. We identified novel genes that are more highly expressed and more significantly enriched than classically accepted characteristic markers of most pituitary cell types. As examples, transcripts for the metalloproteinase gene Pappa2 are greatly enriched in somatotropes, angiotensin receptor Agtr1a in lactotropes, TGF receptor Tgfbr3l in gonadotropes, a glypican of the heparin sulfate proteoglycan family Gpc5 in corticotropes, and an O-acyltransferase Oacyl in melanotropes (Table 2). We established the population specificity of this technique by showing the enriched expression of Foxp2 in gonadotropes by scRNAseq and immunohistochemistry. Some of the most enriched genes are presented in Table 2. Rad18 is an E3 ubiquitin-protein ligase that is highly enriched in somatotropes. Its expression in somatotropes is novel, and Rad18 knockout/knockdown mice have variably decreased [(49) and EUCOMM/IMPC data (http://www.mousephenotype.org/data/genes/MGI:1890476)] or normal (50) body weights. Neither Gh expression nor the nature of the decreased body weight was investigated in the affected Rad18-mutant mice. Cdh8 is an adherens junction cell adhesion molecule that was previously detected, using sorted Gh-eGFP cells, in the GFP-negative fraction, although at the time no antibody was available to determine its localization (51). Here, we find that Cdh8 expression is enriched in the corticotropes. Aquaporins 3 and 4 (Aqp3 and Aqp4), which are transmembrane water channel proteins, are both highly enriched in the Sox2 stem cell population, but little is known about their role there. AQP4 and AQP5 are expressed in the rat pituitary gland (52, 53), an aquaporin 3 homolog AQP3-h3BL in the tree frog is involved in the formation of secretory granules in gonadotropes (54), and Aqp1 knockout mice show reduced ACTH levels (55).
We demonstrate that the scRNAseq analysis accurately predicts the cell-specific expression of Foxp2 in gonadotropes in vivo. We verified the in silico observation with immunohistochemistry of the transcription factor Foxp2 in gonadotropes. FOXP2 mutations or deletions in humans are associated with severe speech impairments and developmental verbal dyspraxia (56–59), but no pituitary defects have been reported. Foxp2 expression has been detected in the mouse brain (37–40), esophagus, and lungs (60, 61) in mice. Our study is one of the first to demonstrate Foxp2 expression in the pituitary gland, and its enrichment in adult gonadotropes is suggestive of a role there. Further studies may investigate its expression in female adult mice, at different embryonic time points, or pituitary phenotypes of Foxp2-null mice (62, 63).
The 10x Genomics Chromium platform generates transcript sequences only from the 3′ end due to the fragmentation of cDNAs after barcoding. The preparation process allows quantification of transcript counts from UMIs because full-length transcripts are kept intact through the initial library preparation. However, cDNA fragments are later enzymatically fragmented to retain only the last ∼100 bp, so that there is no gene body coverage from this protocol (splicing events that do not occur at the end of the transcript are also not observed). Approximately 20% of sequencing reads map uniquely to the genome, but not within the UCSC mm10 transcriptome. This may be due, in part, to genome-aligned reads that are not annotated as transcripts or exons in the reference transcriptome, which causes them to appear intergenic and therefore uncounted, causing a false negative. In our experiment, this occurred for the known pituitary factor Prop1. We demonstrate the utility of using sequence information from our full-length pituitary cDNA libraries to properly annotate 3′ UTR sequences of Prop1, improving the accuracy of identifying transcripts in pituitary stem cells and a low proportion of endocrine cells. This highlights the value of curated analysis of in vivo 3′ transcript ends to rectify false negatives in both bulk and scRNAseq analyses and recover expression data for genes of particular interest.
Although a practical consideration of the scRNAseq technique is the high cost of cell capture and library preparation and high-throughput sequencing, we demonstrate in the present study that scRNAseq of pituitary cells can be highly reproducible. Our scRNAseq data are aggregated from two pools of three animals each, representing a total of six pituitary glands. The two pools have no significantly differentially expressed genes, indicating remarkably low biological and technical variability.
In this scRNAseq study, we identified novel markers enriched in classically defined pituitary populations. Using a transcription factor as an example, we validate these in silico data with in vivo immunohistochemistry to confirm that FOXP2 expression is enriched in gonadotropes, demonstrating the ability of the scRNAseq to find biologically significant and valid candidates. Additionally, we can separate proliferating Pou1f1-expressing cells from those that are quiescent. We provide evidence to suggest a somatotrope subpopulation enriched in sterol synthesis genes. We also present data to show that false negatives can occur from incomplete 3′ transcript annotation and one method to resolve it. Ultimately, recent advances in scRNAseq technologies provide an exciting opportunity to investigate gene expression patterns of individual pituitary populations at virtually any stage of life.
Acknowledgments
We thank Dr. Robert Lyons, Judith Opp, and other members of the University of Michigan’s DNA Sequencing Core Facility for help in experimental planning, single-cell capture and library preparation, and Illumina sequencing.
Financial Support: This work was supported by the National Institutes of Health through National Human Genome Research Institute Grant T32HG000040 (to A.Z.D.), National Institute of General Medical Sciences Grant T32GM007544 (to A.Z.D.), Eunice Kennedy Shriver National Institute of Child Health and Human Development Grants R15HD078885 (to B.S.E.), R01HD34283 (to S.A.C.), and R01HD30428 (to S.A.C.), as well as by University of Michigan Reproductive Science Program Grant U060475 (to S.A.C.).
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- DAPI
4′,6-diamidino-2-phenylindole
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- FACS
fluorescence-assisted cell sorting
- FITC
fluorescein isothiocyanate
- FSHB
FSH β
- GEM
gel bead-in-emulsion
- LHB
LH β subunit
- LMO
linear moveout
- NCBI
National Center for Biotechnology Information
- scRNAseq
single-cell RNA sequencing
- tSNE
t-distributed stochastic neighbor embedding
- UCSC
University of California Santa Cruz
- UMI
unique molecular identifier
- UTR
untranslated region
References
- 1. Davis SW, Keisler JL, Pérez-Millán MI, Schade V, Camper SA. All hormone-producing cell types of the pituitary intermediate and anterior lobes derive from Prop1-expressing progenitors. Endocrinology. 2016;157(4):1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Qiao S, Nordström K, Muijs L, Gasparoni G, Tierling S, Krause E, Walter J, Boehm U. Molecular plasticity of male and female murine gonadotropes revealed by mRNA sequencing. Endocrinology. 2016;157(3):1082–1093. [DOI] [PubMed] [Google Scholar]
- 3. Budry L, Lafont C, El Yandouzi T, Chauvet N, Conéjero G, Drouin J, Mollard P. Related pituitary cell lineages develop into interdigitated 3D cell networks. Proc Natl Acad Sci USA. 2011;108(30):12515–12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Childs GV, Unabia G, Wu P. Differential expression of growth hormone messenger ribonucleic acid by somatotropes and gonadotropes in male and cycling female rats. Endocrinology. 2000;141(4):1560–1570. [DOI] [PubMed] [Google Scholar]
- 5. Frawley LS, Boockfor FR, Hoeffler JP. Identification by plaque assays of a pituitary cell type that secretes both growth hormone and prolactin. Endocrinology. 1985;116(2):734–737. [DOI] [PubMed] [Google Scholar]
- 6. Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35–45. [DOI] [PubMed] [Google Scholar]
- 7. Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W. Comparative analysis of single-cell RNA sequencing methods. Mol Cell. 2017;65(4):631–643.e4. [DOI] [PubMed]
- 8. Baran-Gale J, Chandra T, Kirschner K. Experimental design for single-cell RNA sequencing. Brief Funct Genomics. 2018;17(4):233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Svensson V, Natarajan KN, Ly LH, Miragaia RJ, Labalette C, Macaulay IC, Cvejic A, Teichmann SA. Power analysis of single-cell RNA-sequencing experiments. Nat Methods. 2017;14(4):381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ruf-Zamojski F, Fribourg M, Ge Y, Nair V, Pincas H, Zaslavsky E, Nudelman G, Tuminello SJ, Watanabe H, Turgeon JL, Sealfon SC. Regulatory architecture of the LβT2 gonadotrope cell underlying the response to gonadotropin-releasing hormone. Front Endocrinol (Lausanne). 2018;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hodne K, Haug TM, Weltzien FA. Single-cell qPCR on dispersed primary pituitary cells—an optimized protocol. BMC Mol Biol. 2010;11(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garcia-Lavandeira M, Quereda V, Flores I, Saez C, Diaz-Rodriguez E, Japon MA, Ryan AK, Blasco MA, Dieguez C, Malumbres M, Alvarez CV. A GRFa2/Prop1/Stem (GPS) cell niche in the pituitary. PLoS One. 2009;4(3):e4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pérez Millán MI, Brinkmeier ML, Mortensen AH, Camper SA. PROP1 triggers epithelial-mesenchymal transition-like process in pituitary stem cells. eLife. 2016;5:e14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brinkmeier ML, Davis SW, Carninci P, MacDonald JW, Kawai J, Ghosh D, Hayashizaki Y, Lyons RH, Camper SA. Discovery of transcriptional regulators and signaling pathways in the developing pituitary gland by bioinformatic and genomic approaches. Genomics. 2009;93(5):449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andoniadou CL, Matsushima D, Mousavy Gharavy SN, Signore M, Mackintosh AI, Schaeffer M, Gaston-Massuet C, Mollard P, Jacques TS, Le Tissier P, Dattani MT, Pevny LH, Martinez-Barbera JP. Sox2+ stem/progenitor cells in the adult mouse pituitary support organ homeostasis and have tumor-inducing potential. Cell Stem Cell. 2013;13(4):433–445. [DOI] [PubMed] [Google Scholar]
- 17. Gaston-Massuet C, Andoniadou CL, Signore M, Jayakody SA, Charolidi N, Kyeyune R, Vernay B, Jacques TS, Taketo MM, Le Tissier P, Dattani MT, Martinez-Barbera JP. Increased Wingless (Wnt) signaling in pituitary progenitor/stem cells gives rise to pituitary tumors in mice and humans. Proc Natl Acad Sci USA. 2011;108(28):11482–11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. R Core Team R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 22. Hulsen T, de Vlieg J, Alkema W. BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9(1):488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 24. Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. RRID: AB_2107107.
- 26. RRID: AB_2629219.
- 27. RRID: AB_2665562.
- 28. RRID: AB_2665563.
- 29. RRID: AB_2665565.
- 30. RRID: AB_2687903.
- 31. RRID: AB_2629220.
- 32. RRID: AB_2340588.
- 33. RRID: AB_2340453.
- 34. Fauquier T, Rizzoti K, Dattani M, Lovell-Badge R, Robinson IC. SOX2-expressing progenitor cells generate all of the major cell types in the adult mouse pituitary gland. Proc Natl Acad Sci USA. 2008;105(8):2907–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu X, Tollkuhn J, Taylor H, Rosenfeld MG. Notch-dependent pituitary SOX2+ stem cells exhibit a timed functional extinction in regulation of the postnatal gland. Stem Cell Reports. 2015;5(6):1196–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech. 2008;2008:P1008.
- 37. Lai CS, Gerrelli D, Monaco AP, Fisher SE, Copp AJ. FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain. 2003;126(Pt 11):2455–2462. [DOI] [PubMed] [Google Scholar]
- 38. Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 2003;460(2):266–279. [DOI] [PubMed] [Google Scholar]
- 39. Takahashi K, Liu FC, Hirokawa K, Takahashi H. Expression of Foxp2, a gene involved in speech and language, in the developing and adult striatum. J Neurosci Res. 2003;73(1):61–72. [DOI] [PubMed] [Google Scholar]
- 40. Fujita E, Tanabe Y, Shiota A, Ueda M, Suwa K, Momoi MY, Momoi T. Ultrasonic vocalization impairment of Foxp2 (R552H) knockin mice related to speech-language disorder and abnormality of Purkinje cells. Proc Natl Acad Sci USA. 2008;105(8):3117–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Magoulas C, McGuinness L, Balthasar N, Carmignac DF, Sesay AK, Mathers KE, Christian H, Candeil L, Bonnefont X, Mollard P, Robinson IC. A secreted fluorescent reporter targeted to pituitary growth hormone cells in transgenic mice. Endocrinology. 2000;141(12):4681–4689. [DOI] [PubMed] [Google Scholar]
- 42. He Z, Fernandez-Fuente M, Strom M, Cheung L, Robinson IC, Le Tissier P. Continuous on-line monitoring of secretion from rodent pituitary endocrine cells using fluorescent protein surrogate markers. J Neuroendocrinol. 2011;23(3):197–207. [DOI] [PubMed] [Google Scholar]
- 43. Lavoie PL, Budry L, Balsalobre A, Drouin J. Developmental dependence on NurRE and EboxNeuro for expression of pituitary proopiomelanocortin. Mol Endocrinol. 2008;22(7):1647–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castinetti F, Brinkmeier ML, Gordon DF, Vella KR, Kerr JM, Mortensen AH, Hollenberg A, Brue T, Ridgway EC, Camper SA. PITX2 and PITX1 regulate thyrotroph function and response to hypothyroidism. Mol Endocrinol. 2011;25(11):1950–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wen S, Schwarz JR, Niculescu D, Dinu C, Bauer CK, Hirdes W, Boehm U. Functional characterization of genetically labeled gonadotropes. Endocrinology. 2008;149(6):2701–2711. [DOI] [PubMed] [Google Scholar]
- 46. Leese GP, Wallymahmed M, VanHeyningen C, Tames F, Wieringa G, MacFarlane IA. HDL-cholesterol reductions associated with adult growth hormone replacement. Clin Endocrinol (Oxf). 1998;49(5):673–677. [DOI] [PubMed] [Google Scholar]
- 47. Tonstad S, Sundt E, Ose L, Hagve TA, Fruchart JC, Bard JM, Edén S. The effect of growth hormone on low-density lipoprotein cholesterol and lipoprotein(a) levels in familial hypercholesterolemia. Metabolism. 1996;45(11):1415–1421. [DOI] [PubMed] [Google Scholar]
- 48. O'Neal DN, Hew FL, Best JD, Alford F. The effect of 24 months recombinant human growth hormone (rh-GH) on LDL cholesterol, triglyceride-rich lipoproteins and apo [a] in hypopituitary adults previously treated with conventional replacement therapy. Growth Horm IGF Res. 1999;9(3):165–173. [DOI] [PubMed]
- 49. Inagaki A, Sleddens-Linkels E, Wassenaar E, Ooms M, van Cappellen WA, Hoeijmakers JH, Seibler J, Vogt TF, Shin MK, Grootegoed JA, Baarends WM. Meiotic functions of RAD18. J Cell Sci. 2011;124(Pt 16):2837–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun J, Yomogida K, Sakao S, Yamamoto H, Yoshida K, Watanabe K, Morita T, Araki K, Yamamura K, Tateishi S. Rad18 is required for long-term maintenance of spermatogenesis in mouse testes. Mech Dev. 2009;126(3–4):173–183. [DOI] [PubMed] [Google Scholar]
- 51. Chauvet N, El-Yandouzi T, Mathieu MN, Schlernitzauer A, Galibert E, Lafont C, Le Tissier P, Robinson IC, Mollard P, Coutry N. Characterization of adherens junction protein expression and localization in pituitary cell networks. J Endocrinol. 2009;202(3):375–387. [DOI] [PubMed] [Google Scholar]
- 52. Kuwahara S, Maeda S, Tanaka K, Hayakawa T, Seki M. Expression of aquaporin water channels in the rat pituitary gland. J Vet Med Sci. 2007;69(11):1175–1178. [DOI] [PubMed] [Google Scholar]
- 53. Matsuzaki T, Inahata Y, Sawai N, Yang CY, Kobayashi M, Takata K, Ozawa H. Immunohistochemical localization of the water channels AQP4 and AQP5 in the rat pituitary gland. Acta Histochem Cytochem. 2011;44(6):259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sato M, Nakakura T, Ogushi Y, Akabane G, Kurabuchi S, Suzuki M, Tanaka S. Expression of a mammalian aquaporin 3 homolog in the anterior pituitary gonadotrophs of the tree frog, Hyla japonica. Cell Tissue Res. 2011;343(3):595–603. [DOI] [PubMed] [Google Scholar]
- 55. Arnaoutova I, Cawley NX, Patel N, Kim T, Rathod T, Loh YP. Aquaporin 1 is important for maintaining secretory granule biogenesis in endocrine cells. Mol Endocrinol. 2008;22(8):1924–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lai CS, Fisher SE, Hurst JA, Levy ER, Hodgson S, Fox M, Jeremiah S, Povey S, Jamison DC, Green ED, Vargha-Khadem F, Monaco AP. The SPCH1 region on human 7q31: genomic characterization of the critical interval and localization of translocations associated with speech and language disorder. Am J Hum Genet. 2000;67(2):357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Feuk L, Kalervo A, Lipsanen-Nyman M, Skaug J, Nakabayashi K, Finucane B, Hartung D, Innes M, Kerem B, Nowaczyk MJ, Rivlin J, Roberts W, Senman L, Summers A, Szatmari P, Wong V, Vincent JB, Zeesman S, Osborne LR, Cardy JO, Kere J, Scherer SW, Hannula-Jouppi K. Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am J Hum Genet. 2006;79(5):965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rice GM, Raca G, Jakielski KJ, Laffin JJ, Iyama-Kurtycz CM, Hartley SL, Sprague RE, Heintzelman AT, Shriberg LD. Phenotype of FOXP2 haploinsufficiency in a mother and son. Am J Med Genet A. 2012;158A(1):174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Žilina O, Reimand T, Zjablovskaja P, Männik K, Männamaa M, Traat A, Puusepp-Benazzouz H, Kurg A, Ounap K. Maternally and paternally inherited deletion of 7q31 involving the FOXP2 gene in two families. Am J Med Genet A. 2012;158A(1):254–256. [DOI] [PubMed] [Google Scholar]
- 60. Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D, Morrisey EE. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development. 2007;134(10):1991–2000. [DOI] [PubMed] [Google Scholar]
- 61. Shu W, Yang H, Zhang L, Lu MM, Morrisey EE. Characterization of a new subfamily of winged-helix/forkhead (Fox) genes that are expressed in the lung and act as transcriptional repressors. J Biol Chem. 2001;276(29):27488–27497. [DOI] [PubMed] [Google Scholar]
- 62. French CA, Groszer M, Preece C, Coupe AM, Rajewsky K, Fisher SE. Generation of mice with a conditional Foxp2 null allele. Genesis. 2007;45(7):440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shu W, Cho JY, Jiang Y, Zhang M, Weisz D, Elder GA, Schmeidler J, De Gasperi R, Sosa MA, Rabidou D, Santucci AC, Perl D, Morrisey E, Buxbaum JD. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc Natl Acad Sci USA. 2005;102(27):9643–9648. [DOI] [PMC free article] [PubMed] [Google Scholar]