Endosidin 4, which interferes with endomembrane protein trafficking, represents a chemical tool for studying processes mediated by ARF GTPase exchange factors, including their role in plant development.
Abstract
The trafficking of subcellular cargos in eukaryotic cells crucially depends on vesicle budding, a process mediated by ARF-GEFs (ADP-ribosylation factor guanine nucleotide exchange factors). In plants, ARF-GEFs play essential roles in endocytosis, vacuolar trafficking, recycling, secretion, and polar trafficking. Moreover, they are important for plant development, mainly through controlling the polar subcellular localization of PIN-FORMED transporters of the plant hormone auxin. Here, using a chemical genetics screen in Arabidopsis thaliana, we identified Endosidin 4 (ES4), an inhibitor of eukaryotic ARF-GEFs. ES4 acts similarly to and synergistically with the established ARF-GEF inhibitor Brefeldin A and has broad effects on intracellular trafficking, including endocytosis, exocytosis, and vacuolar targeting. Additionally, Arabidopsis and yeast (Saccharomyces cerevisiae) mutants defective in ARF-GEF show altered sensitivity to ES4. ES4 interferes with the activation-based membrane association of the ARF1 GTPases, but not of their mutant variants that are activated independently of ARF-GEF activity. Biochemical approaches and docking simulations confirmed that ES4 specifically targets the SEC7 domain-containing ARF-GEFs. These observations collectively identify ES4 as a chemical tool enabling the study of ARF-GEF-mediated processes, including ARF-GEF-mediated plant development.
INTRODUCTION
Structural integrity, chemical homeostasis, and, thus, the functionality of eukaryotic cells, including plant cells, depends on a complex network of intracellular membrane trafficking routes that act in concert with each other. A large number of critical components of endomembrane trafficking have been identified using various approaches; some of them are evolutionarily conserved, and some are more specific for particular model systems. Among the most prominent regulators of trafficking are ARF (ADP-ribosylation factor) GTPases that, together with their activators ARF-GEFs (ARF guanine nucleotide exchange factors), regulate the budding of trafficking vesicles (Yorimitsu et al., 2014). ARF proteins constantly switch between active (GTP-bound) and inactive (GDP-bound) states (Yorimitsu et al., 2014). The inactive ARF-GDP form localizes to the cytosol or associate loosely with membranes and become activated by the catalytic SEC7 domain of ARF-GEFs by exchanging GDP for GTP (Nielsen et al., 2008). Following activation to the GTP-bound state, ARFs bind to membranes and recruit cytosolic coat proteins Coat Protein Complex I (COPI), COPII, and clathrin to specific sites of vesicle budding at the Golgi apparatus (GA), trans-Golgi network (TGN), plasma membrane (PM), and endosomal compartments (Serafini et al., 1991; Bonifacino and Lippincott-Schwartz, 2003).
The Arabidopsis thaliana genome encodes eight large (179–200 kD) ARF-GEFs, which fall into two subfamilies: the GGG class, comprising GNOM, GNOM-LIKE1 (GNL1), and GNL2; and the BIG class, comprising five members (BIG1, BIG2, BIG3, BIG4, and BIG5) (Anders and Jürgens, 2008). The activity of ARF-GEFs and their cellular and developmental roles have been extensively studied using Brefeldin A (BFA), a fungal inhibitor of protein trafficking that specifically targets ARF-GEFs (Mossessova et al., 2003; Anders and Jürgens, 2008).
Unlike in other eukaryotes (where all large ARF-GEFs are BFA sensitive), in Arabidopsis, two of them, GNL1 and BIG3, are BFA resistant (Anders and Jürgens, 2008). Insensitivity to BFA is conveyed by defined residues in the conserved SEC7 domain of ARF-GEFs (Geldner et al., 2003; Richter et al., 2007). GNOM is the most prominent of Arabidopsis ARF-GEFs, playing important roles in many aspects of plant development. These roles have been linked to the function of GNOM in controlling the cellular, polar localization of PIN proteins, which are efflux transporters for the plant hormone auxin (Paciorek and Friml, 2006; Viaene et al., 2014; Adamowski and Friml, 2015; Nodzyński et al., 2016).
Cellular polarity of PIN proteins at the PM is crucial for directional auxin transport and contributes to the establishment of differential auxin distributions within tissues, which mediates many polarization and patterning processes in plants (Adamowski and Friml, 2015). To maintain their polar distribution, PINs undergo constant cycles of endocytosis and recycling between the PM and endosomes (Kleine-Vehn et al., 2011). PIN recycling is mediated predominantly by the BFA-sensitive GNOM, which was originally hypothesized to act at elusive recycling endosomes (Geldner et al., 2003) but rather seems to be associated with, and act at, GA under normal, undisturbed conditions (Naramoto et al., 2014). ARF-GEFs are also involved in PIN endocytosis (Teh and Moore, 2007; Naramoto et al., 2010), but this process is less BFA sensitive than PIN recycling back to the PM, thus leading to the intracellular accumulation and aggregation of PINs and other PM cargos in so-called “BFA compartments” after BFA treatment (Geldner et al., 2001). The inhibition of PIN recycling by BFA or in Arabidopsis gnom mutants results in polarity loss and, after prolonged treatment, induces transcytosis of PIN proteins from the basal to the apical side of the cell (Kleine-Vehn et al., 2008a, 2008b). ARF-GEF-mediated PIN trafficking is also involved in the dynamic rearrangement of PIN polarity in response to light and gravity, which is crucial for plant phototropism and gravitropism, respectively (Ding et al., 2011; Rakusová et al., 2016). BFA-insensitive GNL1 and BFA-sensitive GNOM also display distinct, but overlapping, functions at the GA, regulating retrograde COPI-dependent vesicle transport from the GA to the endoplasmic reticulum (ER) (Richter et al., 2007). Another ARF-GEF, BFA-VISUALIZED ENDOCYTIC TRAFFICKING DEFECTIVE1 (BEN1), mediates early endosomal trafficking; correspondingly, the ben1 mutant is defective in PIN trafficking and polarity (Tanaka et al., 2009, 2014). The BEN1 gene encodes the SEC7-domain containing protein AtMIN7/BIG5 (Tanaka et al., 2009). The functionally redundant ARF-GEFs BIG1, BIG2, BIG3, and BIG4 mediate the late secretory pathway and transport of newly synthesized and recycled proteins to the cell division plane during cytokinesis (Richter et al., 2014; Doyle et al., 2015).
The Arabidopsis ARF-GEFs substrate ARF1 is involved in retrograde trafficking from the GA to the ER and from the TGN to the endosome (Dascher and Balch, 1994; Ooi et al., 1998; Goldberg, 1999; Poon et al., 1999; Jackson and Casanova, 2000), regulating the sequence-specific vacuolar sorting route to lytic vacuoles (Pimpl et al., 2003) and dynamin-independent endocytosis (Kumari and Mayor, 2008). In plants, ARF1 localizes to the TGN and GA (Robinson et al., 2011) and is implicated in ER-to-GA and cargo-dependent GA-to-PM transport (Lee et al., 2002; Takeuchi et al., 2002). ARF1 is also involved in polar localization of PIN2 in epidermis (Xu and Scheres, 2005) and the recycling of PIN to the PM (Tanaka et al., 2014). Thus, genetic and pharmacological (specifically BFA) tools that manipulate the functions of ARFs and ARF-GEFs have been instrumental in elucidating the cellular and developmental functions of these important trafficking regulators.
In general, forward-genetic approaches in Arabidopsis have been successfully used to identify mutants defective in trafficking, whether based on developmental defects or by using subcellular markers such as PIN1-GFP to study intracellular trafficking (Tanaka et al., 2009, 2013; Feraru et al., 2010, 2012; Zwiewka and Friml, 2012). Nonetheless, classical genetic approaches for studying the complexity of dynamic vesicular trafficking are often limited by mutant lethality or gene redundancy. One approach that can be used to overcome these problems is chemical genomics: Small molecules can interfere with intracellular trafficking in a dose-dependent manner, resulting in severe phenotypes (Drakakaki et al., 2011).
Here, we characterized the compound Endosidin 4 (ES4), which interferes with endomembrane protein trafficking and directly targets SEC7 domain-containing ARF-GEFs, thus providing a tool to study the cellular and developmental roles of these crucial trafficking regulators.
RESULTS
Identification of ES4, Which Affects PIN1 Polarity in a PIN2pro:PIN1-HA pin2 Line
Chemical libraries had initially been screened using germinating tobacco (Nicotiana tabacum) pollen and subsequently on diverse Arabidopsis subcellular marker lines to select small molecules that interfere with the endomembrane trafficking system. Out of 46,418 tested chemicals, 360 were selected as inhibitors of pollen growth (Drakakaki et al., 2011; Doyle et al., 2015); further analysis in Arabidopsis helped to cluster these chemicals based on intracellular phenotypes (Drakakaki et al., 2011). One of the clusters consisted of molecules affecting the basal, polar localization of PIN1 proteins in the epidermis of PIN2pro:PIN1-GFP pin2 roots (Drakakaki et al., 2011).
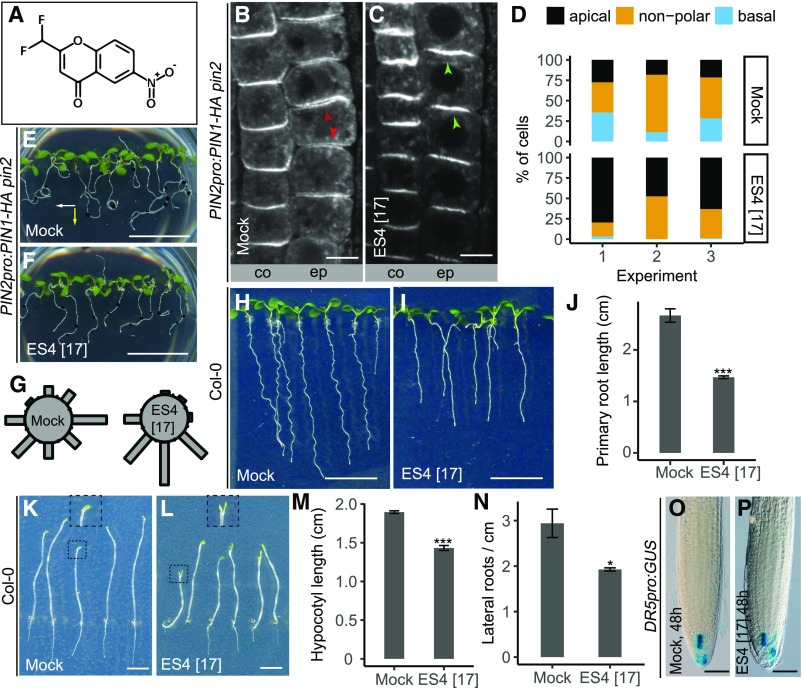
In wild-type plants, PIN2 localizes to the apical sides of epidermal cells, directing auxin from the root tip upwards. PIN2-mediated auxin transport is required for the asymmetric translocation of auxin following gravistimulation and, thus, for root growth along the gravity vector (Müller et al., 1998; Abas et al., 2006; Baster et al., 2013). In the PIN2pro:PIN1-HA pin2 line, replacement of endogenous, apically localized PIN2 by ectopic, predominantly basal localized PIN1 leads to agravitropic root growth (Wiśniewska et al., 2006). We reasoned that PIN1 apicalization would facilitate the flow of auxin in the correct direction and rescue the impaired gravity response. Of the clusters of chemicals able to apicalize epidermal PIN1 in the PIN2pro:PIN1-GFP pin2 root (Drakakaki et al., 2011), we analyzed a set of 11 molecules in terms of their effect on PIN1 localization and the gravity response. Three-day-old seedlings were grown on medium supplemented with chemicals for 2 d. The localization of PIN1 tagged with hemagglutinin (HA) was verified by immunolocalization with anti-HA antibodies and by analyzing the gravitropic responses of roots by turning a plate with vertically grown seedlings 90° (Supplemental Figures 1A and 1B). Among the tested molecules (besides ES6, which is the subject of another study), ES4 (Figure 1A) displayed the strongest effect. ES4 strongly increased the number of epidermal cells with apically localized PIN1 (Figures 1B to 1D) and partially rescued the gravitropic response (Figures 1E to 1G) of PIN2pro:PIN1-HA pin2. Obvious changes in polar localization were observed for ectopically expressed PIN1 in PIN2pro:PIN1-HA pin2, but not for the native apically localized PIN2 in wild-type root epidermis (Supplemental Figures 2A and 2B).
Figure 1.
Identification of ES4.
(A) Chemical structure of ES4.
(B) to (D) Immunolocalization of PIN1-HA in epidermal (ep) and cortex (co) cells in root elongation zones of PIN2pro:PIN1-HA pin2 line in 5-d-old seedlings treated for 48 h with mock or 17 µM ES4. Red arrowheads in (B) indicate basal or nonpolar localization, whereas green arrowheads in (C) indicate apical localization of PIN1-HA in epidermal cells. In each of the three experiments, 4 to 22 roots were analyzed per treatment, per line (in total, 34 roots for mock and 31 roots for ES4 treatments). The number of epidermal cells with apical, nonpolar, or basal PIN1-HA localization from each root has been summed up and represented as a percentage of the total number of cells (D). Bars = 10 µm.
(E) to (G) Gravitropic response of 7-d-old PIN2pro:PIN1-HA pin2 seedlings grown on medium supplemented with mock or 17 µM ES4 and gravistimulated for 48 h. The white and yellow arrows mark gravity vectors after the first and second gravistimulation, respectively. Black dots on the roots indicate the localization of the root tips at the time of plate turning. In contrast to mock (E), PIN2pro:PIN1-HA pin2 grown on 17 µM ES4 (F) shows a positive gravitropic response. Gravistimulated roots were assigned into one of the eight 45° sectors on the gravitropism diagram (G). The length of bars in the diagram represents the percentage of pooled number of seedlings from four independent experiments assigned to a respective sector. In each experiment, 7 to 16 roots per treatment were analyzed (in total, 43 roots for mock and 41 roots for ES4 treatments). Bars = 1 cm.
(H) to (J) Root length of 7-d-old Col-0 seedlings grown on medium supplemented with mock or 17 µM ES4. Values represent means ± se of three independent experiments (J), each consisting of 13 to 22 analyzed roots per treatment (in total, 48 roots per treatment). Asterisks indicate significant difference between mock and ES4 treatment (two-tailed Student’s t test, ***P < 0.001). Bars = 1 cm.
(K) to (M) Hypocotyl length of 7-d-old Col-0 seedlings grown in the dark on medium supplemented with mock or 17 µM ES4. Insets represent magnified cotyledons. Values represent means ± se of three independent experiments (M), each consisting of 20 to 30 analyzed roots per treatment. Asterisks indicate significant difference between mock and ES4 treatment (two-tailed Student’s t test, ***P < 0.001). Bars = 0.5 cm.
(N) Quantification of lateral roots density in 11-d-old Col-0 seedlings grown on medium supplemented with mock or 17 µM ES4. Values represent means ± se of three independent experiments, each consisting of at least 20 analyzed roots per treatment. Asterisks indicate significant difference between mock and ES4 treatment (two-tailed Student’s t test, *P < 0.05).
(O) and (P) GUS staining of 5-d-old DR5pro:GUS seedlings. Three-day-old seedlings were transferred to growth medium supplemented with mock (O) or 17 µM ES4 (P) and grown for 48 h. In two independent experiments, 7 to 12 roots per treatment were analyzed. Bars = 100 µm.
The weak polarity mutants have a preferentially disrupted basal polar localization, leading to the basal-to-apical switch in PIN localization, pointing to a more robust control of the apical versus basal polar proteins (Kleine-Vehn et al., 2008b). Therefore, we also examined the effect of ES4 on native basally localized PIN1 and PIN2 in stele and cortex tissue. In the cortex, dual expression of PIN2 occurs; in young cortex cells, PIN2 localizes to the basal cell side, whereas in older cells (more distant from the root tip), PIN2 polarity switches its localization to the apical cell side (Kleine-Vehn et al., 2008b). After ES4 treatment for 48 h at a concentration of 17 µM, both PIN1 and PIN2 signals became weaker; however, neither the basal localization of PIN1 in the stele nor the basal localization of PIN2 in young cortex cells was visibly affected (Supplemental Figures 2A to 2E). Accordingly, ES4 applied at a concentration of 17 µM did not affect the normal gravitropic response of gravistimulated wild-type roots (Supplemental Figures 2F to 2H). The observation that PIN1 polarization was affected only when expressed ectopically is not surprising, since it is known that the ectopic basal polar localization of PIN1 in the epidermis is more sensitive to perturbations than in the endogenous situation. This was previously indicated by analysis of the regulator of PIN polarity3 mutant, which is defective in cellulose biosynthesis. This mutant also showed partial apicalization of ectopic PIN1-HA in epidermal cells correlating with the rescue of the gravitropic response, whereas the polar localization of endogenous PIN proteins was not defective in this mutant (Feraru et al., 2011).
We also analyzed the effects of ES4 on primary root growth, hypocotyl growth, and lateral root density in the wild type. At a concentration of 17 µM, primary root length was reduced by nearly 50% (Figures 1H to 1J) and hypocotyl length of dark-grown Arabidopsis seedlings by 25% (Figures 1K to 1M). The cotyledons of treated dark-grown seedlings were straight and open, compared with the control, which had closed cotyledons in the apical hook (Figures 1K and 1L, insets). Additionally, after ES4 treatment, the lateral root density of Col-0 seedlings was reduced (Figure 1N). To test whether ES4 influences the auxin response in root tissues, we used the auxin-responsive promoter DR5pro:GUS. Treatment for 48 h with 17 µM ES4 had no visible effect compared with the control (Figures 1O and 1P).
These studies identified ES4 as a chemical with effects on the ectopic basal polar localization of PIN auxin transporters and on multiple, possibly auxin transport-regulated processes.
ES4 Broadly Affects Endocytic Trafficking and Multiple Endomembrane Compartments
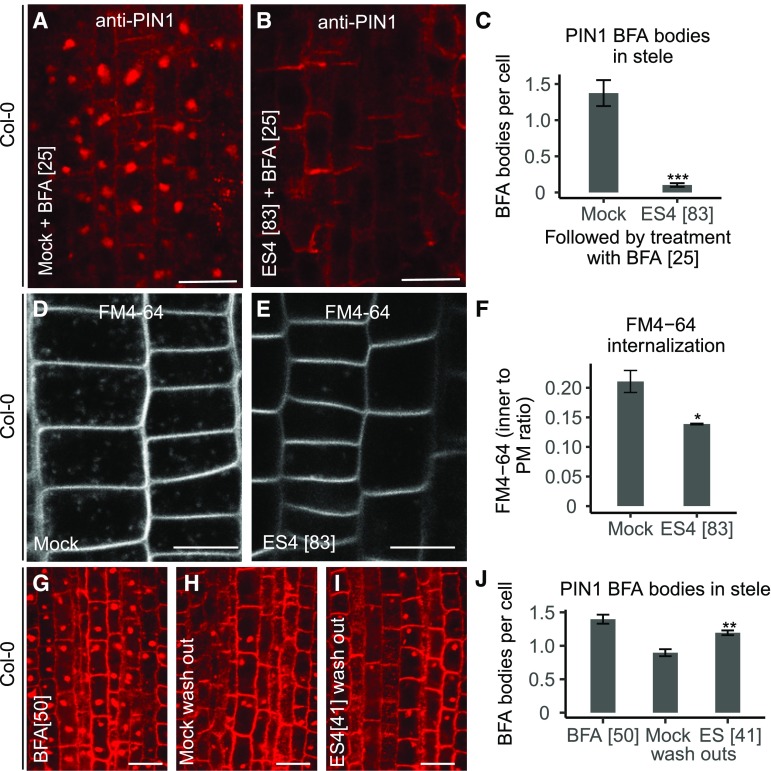
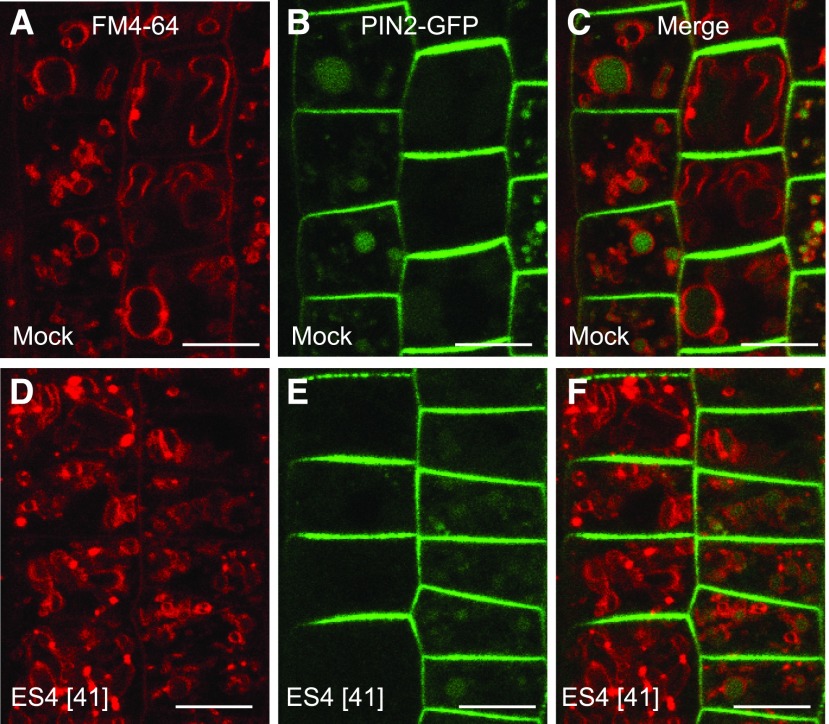
PM-localized PIN1 proteins undergo constitutive cycling between the PM and endosomes (Dhonukshe et al., 2007). In presence of the recycling inhibitor BFA, internalized PIN1 proteins accumulate in BFA-induced intracellular agglomerations called BFA bodies (Geldner et al., 2001). Compared with mock treatment, ES4 treatment reduced the number of PIN1-labeled BFA bodies depending on the concentration, up to 93% at 83 µM (Figures 2A to 2C; Supplemental Figures 3A to 3D). The observed effect was not specific to polar PIN1 but was also visible for the apolarly localized PM marker PIP2-GFP, since after treatment with 17 µM ES4, the number of PIP2-labeled BFA bodies significantly decreased (Supplemental Figures 3E to 3G). To test whether the reduced number of BFA bodies after ES4 treatment resulted from inhibited endocytosis in response to ES4, we examined the uptake of the endocytosis tracer FM4-64 (Jelínková et al., 2010). At 17 µM ES4, the internalization of FM4-64 was not affected, but the morphology of FM4-64-stained endosomes differed from that of the control (Supplemental Figures 3H to 3J). At higher concentration (83 µM), the uptake of FM4-64 was reduced, hinting at the inhibition of endocytosis by ES4 (Figures 2D to 2F). Additionally, we tested the effect of ES4 on exocytosis via BFA washout in the presence of ES4: Washing removes the inhibitory effect of BFA on exocytosis, restoring vesicle trafficking to the PM and resulting in the disappearance of BFA bodies (Geldner et al., 2001). After treatment with 25 µM BFA followed by washout with 41 µM ES4, the number of BFA bodies appeared to be slightly higher than that after washout with mock medium (Supplemental Figures 3K to 3N). Recycling of PIN1 to the PM is inhibited by 25 µM BFA, whereas 50 µM BFA also inhibits the degradation pathway to the vacuoles, resulting in the formation of enlarged BFA bodies (Kleine-Vehn et al., 2008c). Washout of 50 µM BFA with ES4 reduced the disappearance of BFA bodies that occurred after washout with mock (Figures 2G to 2J). This observation suggests that ES4, besides its effect on recycling, also affects the vacuolar trafficking pathway. Therefore, we examined the localization of FM4-64 and PIN2-GFP after treatment in the dark, as dark treatment stabilizes GFP proteins in the lytic vacuoles (Tamura et al., 2003). After FM4-64 uptake, followed by 4 h of mock treatment in the dark, the FM4-64 dye almost completely reached the tonoplast and PIN2-GFP signal was detected in the vacuoles. After ES4 treatment at 41 µM, the morphology of tonoplasts was altered compared with the control: Vacuoles appeared more fragmented, tonoplast labeling was less pronounced, and small agglomerates of the FM4-64 signal (not present in the mock treatment) were observed (Figures 3A to 3F). Additionally, the vacuolar PIN2-GFP signal was less abundant after ES4 treatment, although the ratio of intracellular PIN2-GFP signal versus the PM signal was comparable with that of the control, indicating that PIN2 was internalized from the PM but was inhibited from reaching the vacuole (Supplemental Figure 4).
Figure 2.
Effect of ES4 on Intracellular Trafficking.
(A) to (C) Immunolocalization of PIN1 (red signal) in Col-0 stele cells ([A] and [B]) and mean number of BFA bodies per cell (C). Five-day-old seedlings were pretreated for 30 min with mock or 83 µM ES4 before 25 µM BFA was added for an additional 90 min. Values represent means ± se of four independent experiments, each consisting of 4 to 17 analyzed roots per treatment (in total, 41 roots per treatment) (C). The number of BFA bodies was counted for 10 cells in the stele of each root. Asterisks indicate significant difference between mock and ES4 treatment (two-tailed Mann-Whitney U test, ***P < 0.001). Bars = 10 µm.
(D) to (F) Uptake of endocytic tracer FM4-64 (2 µM) after 10 min. Five-day-old Col-0 seedlings were pretreated for 2 h with mock (D) or 83 µM ES4 (E). FM4-64 uptake was quantified by dividing the mean gray value of the fluorescent signal inside the cell to the adjacent PM signal (F). Values represent means ± se of three independent experiments, each consisting of 5 to 11 analyzed roots per treatment. Asterisks indicate significant difference between mock and ES4 treatment (two-tailed Student’s t test, *P < 0.05). Bars = 10 µm.
(G) to (J) Immunolocalization of PIN1 (red signal) in the stele ([G] to [I]) and number of BFA bodies per cell after washout with ES4 (J). Five-day-old seedlings were treated for 2 h with 50 µM BFA (G), followed by 30 min of washout with medium complemented with mock (H) or 41 µM ES4 (I). Values represent means ± se of three independent experiments (J), each consisting of 4 to 11 analyzed roots per treatment (in total, 16 to 23 roots per treatment). The number of BFA bodies was counted for 5 to 49 cells in the stele of each root. Asterisks indicate significant difference between mock and ES4 washout (two-tailed Mann-Whitney U test, **P < 0.01). Bars = 10 µm.
Figure 3.
Effect of ES4 on Vacuolar Trafficking.
Vacuolar trafficking of PIN2-GFP. Seedlings were pretreated for 2 h with mock ([A] to [C]) or 41 µM ES4 ([D] to [F]) and stained with 8 µM FM4-64, followed by a 4-h treatment in the dark on growth medium supplemented with mock or ES4, respectively. Bars = 10 µm.
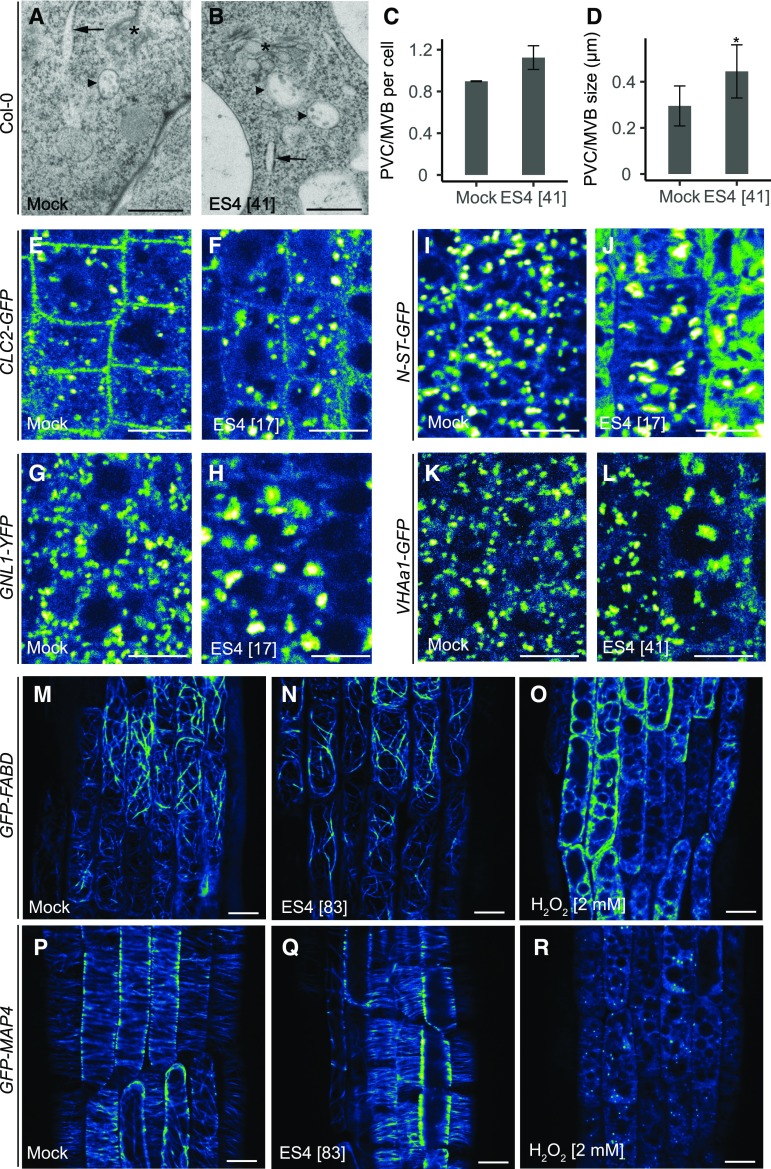
Furthermore, ultrastructural analysis by electron microscopy after ES4 treatment revealed enlarged and an (apparently) slightly increased number of prevacuolar compartment/multivesicular bodies (PVC/MVBs) per cell (Figures 4A to 4D). In Arabidopsis, MVBs that mature from the TGN/early endosome (EE) mediate transport to the lytic vacuoles (Scheuring et al., 2011). Enlarged MVBs, together with the altered FM4-64 and PIN2-GFP tonoplast staining/signal patterns, indicate that ES4 also affects the vacuolar trafficking pathway. Additionally, we observed a higher number of small vesicles in close proximity to the TGN after 2 h of 41 µM ES4 treatment compared with the mock treatment (Supplemental Figures 5A and 5B). To further examine the impact of ES4 on intracellular compartments, we tested different marker lines: BRASSINOSTEROID INSENSITIVE1 (BRI1)-GFP (PM and endosomes), CLATHRIN LIGHT CHAIN2 (CLC2)-GFP (PM and TGN), GNL1-YFP (GA and TGN/EE), GNOM-GFP, SIALYLTRANSFERASE (N-ST)-GFP (GA), and VACUOLAR H+-ATPASE SUBUNIT A1 (VHAa1)-GFP (TGN/EE) (Figures 4E to 4L; Supplemental Figures 5C to 5F). At a concentration of 17 µM ES4, intracellular signal agglomerations were observed for almost all tested marker lines except VHAa1, for which the lowest active concentration inducing agglomeration was 41 µM. In the CLC2-GFP marker line, in addition to the intracellular agglomerations, we also observed a lower signal at the PM after ES4 treatment (Figures 4E and 4F). We compared these effects of ES4 with those of BFA (treatment for 90 min at 50 µM) and found that the effects of BFA on CLC2-GFP were similar, including signal dissociation from the PM and its intracellular aggregation (Supplemental Figures 5G and 5H). This is consistent with a role of BFA-sensitive ARF-GEFs including GNOM not only in recycling, but also to a lesser extent in endocytosis, as previously indicated (Naramoto et al., 2010).
Figure 4.
Effect of ES4 on Intracellular Compartments.
(A) to (D) Transmission electron microscopy images of MVBs in 5-d-old Col-0 seedlings after 2 h mock (A) and 41 µM ES4 (B) treatment and quantification of the number (C) and size (D) of MVBs. Asterisks indicate GA, arrows point at the rough ER, and arrowheads mark the PVC/MVBs. Values represent means ± sd of MVBs number per cell in two (mock) or three (ES4 treatment) roots. In total, MVBs were counted in 39 epidermal and cortex cells for mock and 46 cells for ES4 (C). Values represent means ± sd of the PVC/MVBs size measured in 12 epidermal and cortex root cells for mock and 21 cells for ES4 (D). After ES4 treatment, the size of MVBs increased compared with the control (two-tailed Student’s t test, *P < 0.05) (D), whereas there was no significant difference in MVBs number per cell between ES4 treatment and control (two-tailed Mann-Whitney U test, P > 0.05) (C). Bars = 0.5 µm.
(E) to (L) Intracellular localization of CLC-GFP ([E] and [F]), GNL1-YFP ([G] and [H]), N-ST-GFP ([I] and [J]), and VHAa1-GFP ([K] and [L]) after mock ([E], [G], [I], and [K]) and 17 µM ([F], [H], and [J]) or 41 µM (L) ES4 treatment in 5-d-old seedlings. After 2 h of ES4 treatment, all markers displayed increased intracellular agglomerations. Bars = 10 µm.
(M) to (R) Intracellular localization of actin filament marker GFP-FABD ([M] to [O]) and microtubule marker GFP-MAP4 ([P] to [R]) after 2 h of mock, 83 µM ES4, or 2 mM H2O2 treatment. There was no visible effect of ES4 on actin filaments or microtubule markers, as it was observed after H2O2 treatments. Bars = 10 µm.
To test the possibility that the broad effects of ES4 may be due to its cytotoxicity, we examined the influence of ES4 on cytoskeleton integrity. We used the microtubule marker line GFP-MAP4 and the actin filaments marker GFP-FABD. H2O2 was used as a positive control, since it is known to have a toxic effect on all living cells (Halliwell et al., 2000) and causes depolymerization of cortical microtubules in leaf pavement cells in Arabidopsis (Yao et al., 2012). Treatment with 2 mM H2O2 affected both the microtubule and actin filament markers, whereas treatment with ES4 had no visible effect on cytoskeleton integrity (Figures 4M to 4R).
These observations show that ES4 affects many intracellular trafficking processes, including endocytosis, recycling, and vacuolar trafficking in a manner similar to the established trafficking inhibitor BFA, suggesting possible overlapping actions of these compounds.
ES4 Targets ARF-GEF-Dependent Processes
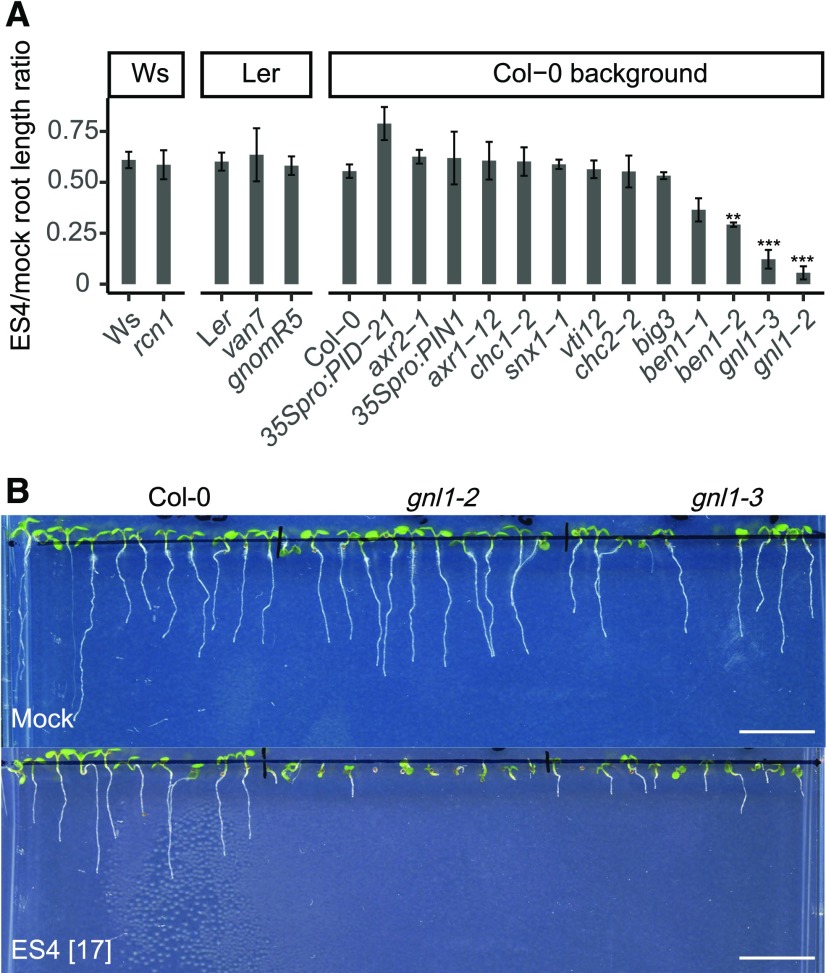
To gain further insight into the mode of action of ES4, we tested the sensitivity of different Arabidopsis trafficking mutants to ES4 by measuring their root length after growth on ES4-supplemented medium (Figure 5A). Among all tested lines, the gnl1-2, gnl1-3, and ben1-2 mutants—defective in different ARF-GEFs—displayed the highest sensitivity to ES4 (Figure 5B). Mutation in another ARF-GEF, GNOM, known for its important function in PIN protein trafficking (Geldner et al., 2003; Naramoto et al., 2014), resulted in agravitropic, short roots that made the analysis after ES4 treatment more difficult to interpret. In contrast, 35Spro:PID-21 plants overexpressing PINOID (PID) kinase, which phosphorylates PIN proteins and promotes their localization to the apical sides of cells (Michniewicz et al., 2007), appeared to show slight resistance to ES4 (Figure 5A). PID kinase-mediated apical PIN polarity has been shown to be independent from GNOM trafficking (Kleine-Vehn et al., 2009), whereas ES4 preferentially affects the basal PIN cargo without strongly affecting the apical PIN proteins, thus explaining the partial ES4 resistance of this line. Other trafficking-related or auxin-related mutants, such as roots curl in NPA (rcn1), auxin resistant (axr), 35Spro:PIN1, clathrin heavy chain (chc), sorting nexin1 (snx1), vesicle transport v-snare (vti12), and big3, did not show an observable difference in sensitivity to ES4 (Figure 5A). These observations suggest a link between ES4 activity and ARF-GEF-dependent processes.
Figure 5.
Mutants with Altered Sensitivity to ES4.
(A) Quantification of root growth sensitivity of the wild type (Ws, Ler, and Col-0), mutants (rcn1, van7, gnomR5, axr2-1, axr1-12, chc1-2, snx1-1, vti12, chc2-2, big3, ben1-1, ben1-2, gnl1-3, and gnl1-2), and overexpression lines (35Spro:PID21 and 35Spro:PIN1). Seedlings were grown for 7 d on growth medium supplemented with mock or 17 µM ES4. Ratio of root lengths grown on ES4 to those grown on mock was calculated. Values represent means ± se of three independent experiments, each consisting of 5 to 32 analyzed roots per treatment for each line. Asterisks indicate significant difference between mutant and corresponding wild type (two-tailed Student’s t test, **P < 0.01 and ***P < 0.001).
(B) Representative images of the ES4 sensitivity of gnl1 mutants manifested by a highly reduced root length compared with the control. Wild-type, gnl1-2, and gnl1-3 seedlings were grown for 7 d on growth medium supplemented with 17 µM ES4. Bars = 10 µm.
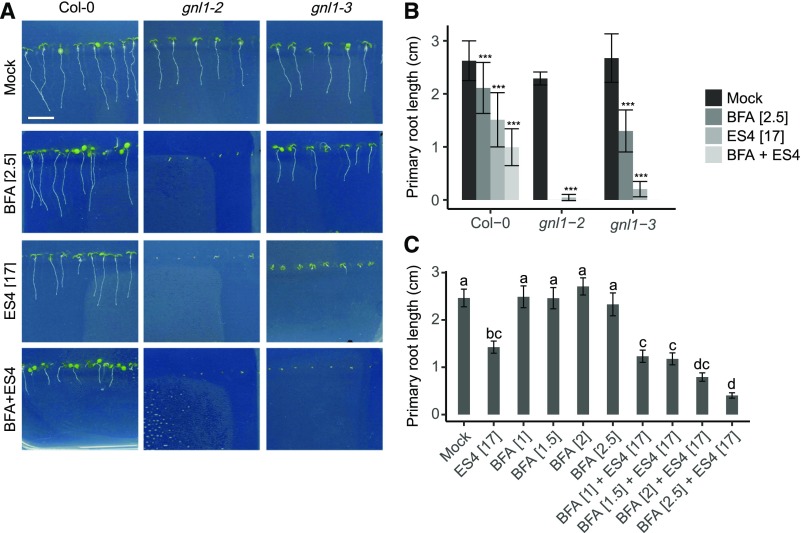
BFA is a well-characterized inhibitor of SEC7 domain-containing ARF-GEFs. Like plants under ES4 treatment, gnl1 showed increased sensitivity to BFA (Figures 6A and 6B). Moreover, the combined treatment with BFA and ES4 completely inhibited germination in gnl1 mutants compared with single treatments with BFA or ES4. In BFA-treated gnl1, the activities of both ARF-GEFs, GNOM and GNL1, are inhibited (Richter et al., 2007; Teh and Moore, 2007). GNOM can functionally substitute for GNL1 in the GA-ER-dependent pathway, but not vice versa (Richter et al., 2007). Accordingly, after BFA treatment, the BFA-sensitive protein GNOM cannot substitute for GNL1 in the gnl1 mutant, resulting in strongly reduced primary root growth. Similarly, the ben1 mutant is also hypersensitive to both BFA (Tanaka et al., 2013) and ES4 treatment (Figure 5A). Among the four BIG ARF-GEFs that play redundant roles in regulating post-GA trafficking, BIG3 is the only one that is insensitive to BFA; hence, BFA treatment of big3 but not the other big mutants results in more pronounced inhibition of plant growth (Richter et al., 2014). In contrast, the big3 mutant displayed a level of sensitivity to ES4 similar to that of the wild type (Figure 5A).
Figure 6.
Characterization of the Sensitivity of GNL1 ARF-GEF and PIN1-GFP to ES4 and BFA.
(A) and (B) Quantification of root growth sensitivity of 7-d-old Col-0, gnl1-2, and gnl1-3 seedlings grown on medium supplemented with mock, 2.5 µM BFA, 17 µM ES4, or 2.5 µM BFA plus 17 µM ES4. Three independent experiments were performed with similar results. Values represent mean ± sd of at least 13 roots analyzed per treatment from one representative experiment. Asterisks indicate significant difference between treatment and mock (two-tailed Student’s t test, ***P < 0.001). Bars = 10 µm.
(C) Root growth sensitivity of PIN1-GFP to ES4 and BFA. Seven-day-old seedlings were grown on medium supplemented with mock, 17 µM ES4, BFA (1, 1.5, 2, and 2.5 µM), and ES4 together with BFA. Values represent means ± se of 10 to 22 analyzed roots per treatment. Different letters indicate statistically significant differences between treatments (ANOVA, P < 0.05). Asterisks indicate statistically significant differences between genotypes (two-tailed Student’s t test, *P < 0.05 and ***P < 0.001).
To characterize the effects of ES4 and BFA, we tested the sensitivity of plants to a range of BFA concentrations in combination with ES4 (Figure 6C). ES4 inhibited root growth in PIN1-GFP plants. BFA in concentrations ranging from 0.5 to 2.5 µM did not visibly reduce root growth in the tested lines. However, when BFA and ES4 were combined, the root length of the wild type was shorter than that of plants grown separately on each of the chemicals, pointing to their additive or synergistic effects.
These results further suggest that ES4 acts similarly to BFA and presumably targets SEC7 domain-containing ARF-GEF-mediated processes.
ES4 Interferes with ARF-GEF-Dependent Activation of ARF1
Previous experiments suggested that ES4 might target ARF-GEF proteins. The direct role of ARF-GEFs is to activate ARF1 proteins by catalyzing the exchange from GDP to GTP. Activated, GTP-bound ARFs recruit coat proteins from the cytosol to the membranes and initiate vesicle formation and transport processes (Beck et al., 2009).
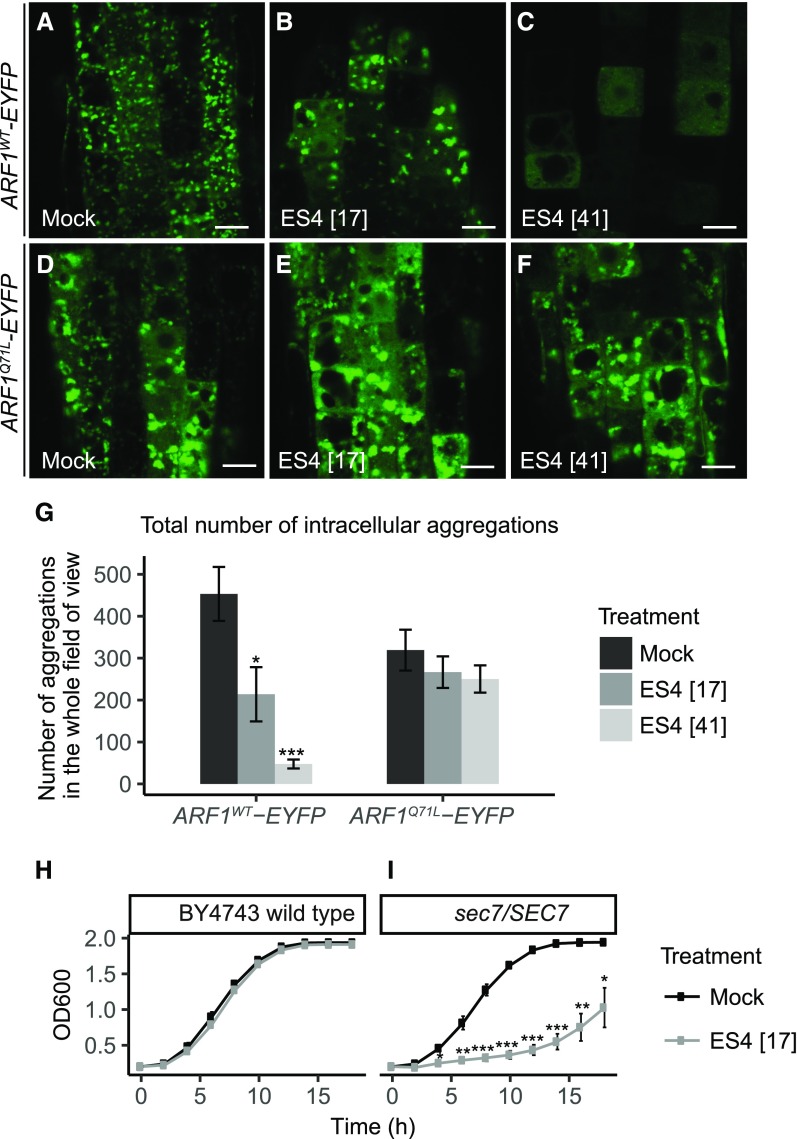
We tested the impact of ES4 on the wild type (ARF1WT), GTP-locked (ARF1Q71L), and GDP-locked (ARF1T31N) forms of ARF1 (Xu and Scheres, 2005) in transgenic Arabidopsis plants. The expression of these constructs coupled to fluorescent protein tags was driven by the Arabidopsis heat shock-inducible promoter of HEAT SHOCK PROTEIN18.2 (HSP18.2). For activation, the lines were incubated for 2 h at 37°C, followed by chemical treatment for 2 h at room temperature. ARF1WT-EYFP localized to the GA structures and endocytic organelles (Xu and Scheres, 2005) (Figure 7A). After treatment with 17 µM ES4, the EYFP label was agglomerated and partially localized to the cytosol (Figure 7B), which was also reflected by reduced number of aggregations after treatment (Figure 7G). A higher (41 µM) concentration led to an almost completely cytosolic fluorescence signal (Figure 7C) and a further reduction in aggregation number (Figure 7G). Similar cytosolic localization and no organellar labeling were observed for the inactive GDP-locked ARF1T31N form (Supplemental Figures 6A and 6B). On the contrary, the GTP-locked ARF1Q71L form was more often associated with membranes than the wild-type ARF1WT form (Figures 7A and 7D). Importantly, GTP-locked ARF1Q71L was resistant to ES4 treatment and, in contrast to ES4-treated ARF1WT, the cytosolic fluorescent signal did not increase (Figures 7E and 7F). Also, the number of agglomerations after ES4 treatment was similar to that of the mock control (Figure 7G).
Figure 7.
Effect of ES4 on ARF1-GFP Localization and the sec7 Yeast Deletion Mutant.
(A) to (G) Intracellular localization of ARF1WT-EYFP and ARF1Q71L-EYFP in 5-d-old seedlings after 2 h treatment with mock, 17 µM, or 41 µM ES4. Values represent means ± se of aggregation number in epidermal cells of 8 to 11 analyzed roots per treatment (G). Asterisks indicate significant difference between mock and ES4 treatment (two-tailed Student’s t test, *P < 0.05 and ***P < 0.001). Bars = 10 µm.
(H) and (I) Growth curves of wild-type (H) and sec7/SEC7 (I) heterozygous deletion strains of yeasts grown in YPD liquid medium at 28°C under mock and 17 µM ES4 treatment. OD600 was measured every 2 h. Values represent means ± se of three independent experiments, each consisting of three technical replicates. Asterisks indicate significant difference between mock and ES4 treatment (two-tailed Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001).
These results provide strong support for the notion that ES4 inhibits the activity of ARF-GEFs, which is required for the activation of ARF1WT and its binding to membranes. The permanently activated GTP-bound and already membrane-bound ARF1Q71L does not require ARF-GEF activity and is therefore unaffected by ES4 treatment, whereas the GDP-locked ARFT31N form shows a localization similar to that of ES4-treated ARF1WT (wild-type form), i.e., an inactive, cytosolic localization.
ES4 Targets SEC7-Containing ARF-GEF-Mediated Processes in Yeast
All results obtained from the experiments with Arabidopsis described above are consistent with the notion that ES4 acts in an inhibitory fashion on ARF-GEFs. As these ARF-GEFs are evolutionarily conserved, we tested the effect of ES4 in yeast. Many proteins in plants and yeast share some conserved sequences, enabling one to search for or confirm known targets from plants that are also found in yeast (; Klutstein et al., 2008). We therefore performed a yeast growth assay on a set of heterozygous yeast deletion strains, including strains with deletions in subunits of the COPI coatomer (a protein complex that coats membrane-bound transport vesicles) and ARF-GEFs (Supplemental Table 1). Treatment of heterozygous yeast strains has been reported to trigger drug-induced haploinsufficiency, meaning that a deletion of one gene copy in diploid cells results in increased sensitivity to the applied chemical (Giaever et al., 1999; Baetz et al., 2004). To determine the sensitivity of yeast to ES4, the wild-type diploid yeast strain (BY4743) was grown in liquid culture with a range of ES4 concentrations (Supplemental Figure 7A). At a concentration 41 µM, ES4 severely inhibited yeast growth, whereas 17 µM ES4, the concentration used for the Arabidopsis experiments, slightly reduced yeast growth and therefore was chosen for the growth assays of heterozygous deletion strains. From the tested deletion strains, the only mutant that displayed severely increased sensitivity to ES4 was a sec7/SEC7 ARF-GEF line (Figures 7H and 7I; Supplemental Figures 7B to 7H). SEC7p is the major ARF-GEF in yeasts and is required for membrane trafficking from the ER to and through the GA and from the TGN (Franzusoff et al., 1991; Deitz et al., 2000; Richardson et al., 2012). Like the other ARF-GEFs, SEC7p contains a highly conserved SEC7 domain that was first identified in this protein, from which its name was derived (Achstetter et al., 1988). A line deficient in SEC12, a GEF that activates another GTPase (Sar1) not from the ARF class, was not affected by ES4 (Supplemental Figure 7H). ES4 also had no effect on the deletion strains of the COPI subunits α-COPI (RET1), β-COPI (SEC26), β’-COPI (SEC27), γ-COPI (SEC21), δ-COPI (RET2), and ζ-COPI (RET3). Two other major ARF-GEF proteins involved in the ER-GA secretory pathway in yeast, GEA1p and GEA2p, play redundant roles in ARF1 activation at the early GA compartments to regulate COPI-mediated vesicle formation (Peyroche et al., 1996, 2001). Strains with a complete knockout of GEA1p protein showed a level of ES4 sensitivity comparable to that of the wild type (Supplemental Figures 7I and 7J), presumably due to the presence of the second redundant protein, GEA2p.
These results are in line with the observations from Arabidopsis and confirm that in yeast, the mode of action of ES4 is also related specifically to the SEC7-domain-containing ARF-GEFs.
ES4 Directly Binds to a Subset of Arabidopsis SEC7-Containing ARF-GEFs
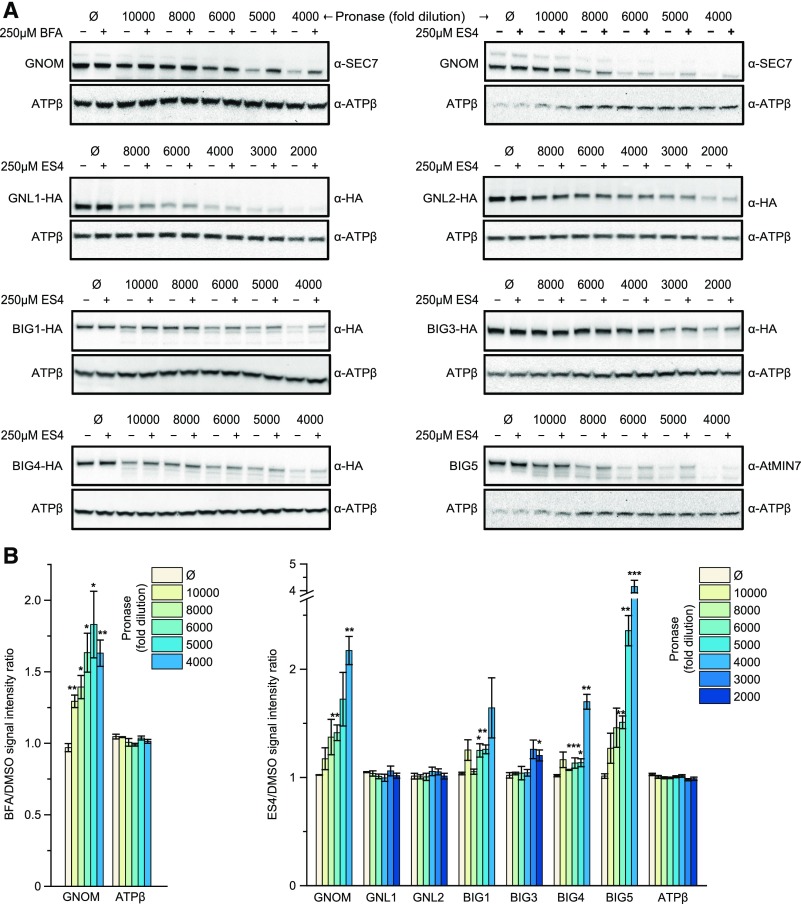
To biochemically validate ARF-GEFs as potential direct binding targets of ES4, we took advantage of the drug affinity responsive target stability (DARTS) assay (Lomenick et al., 2009) and tested the proteolytic degradation of all Arabidopsis ARF-GEFs excluding BIG2 in the presence of ES4 in lysates of Arabidopsis cell cultures. We used specific antibodies to detect GNOM and BIG5 in wild-type cell cultures and generated cell lines expressing HA-tagged ARF-GEF versions to allow for detection with anti-HA antibodies. BFA was used as a positive control, since it is a well-known noncompetitive inhibitor of BFA-sensitive ARF-GEFs (Mossessova et al., 2003; Renault et al., 2003). As expected, treatment with BFA resulted in protection from protease-induced degradation of the BFA-sensitive ARF-GEF GNOM (Figures 8A and 8B). When ES4 was tested, we found that this compound stabilized the BFA-sensitive proteins GNOM, BIG1, BIG4, and BIG5 (Figures 8A and 8B). Our DARTS experiments with the BFA-resistant BIG3 protein showed a borderline response to ES4 treatment, while GNL1 and GNL2 were equally digested by pronase in the presence or absence of ES4 (Figures 8A and 8B), thus not supporting the binding of these ARF-GEFs by ES4.
Figure 8.
DARTS-Based Validation of the Association of ES4 with Arabidopsis ARF-GEFs.
(A) DARTS analysis of the susceptibility of protein to proteolytic degradation in the presence of 250 µM BFA or 250 µM ES4. Total protein extracts from Arabidopsis PSB-D cell cultures were incubated with the respective compound and then challenged with different dilutions of protease. The ARF-GEF protein levels were detected through protein gel blot analysis. ATPβ was used as reference protein control.
(B) Quantification of the protein band intensity reveals that the presence of ES4 stabilizes all Arabidopsis ARF-GEFs with exception of GNL1 and GNL2. Values represent means ± se of representative immunoblots from three biological repeats. Asterisks indicate significant difference between BFA (or ES4) and DMSO treatments (two-tailed Student’s t test, ***P < 0.001, **P < 0.01, and *P < 0.05).
Collectively, these results indicate that ES4 selectively interacts with some, but not all, Arabidopsis ARF-GEF proteins, which is fully consistent with the observed effects of ES4 on trafficking and development, and in particular with ES4 inhibiting the activation of ARF proteins.
ES4 Target Sites Predicted by Docking Simulations
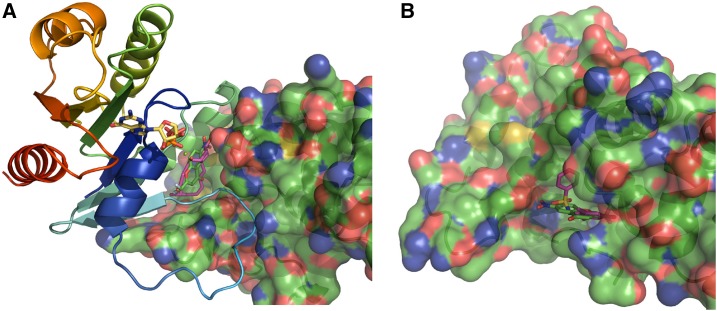
Next, we used ligand-docking simulations to obtain insights into the possible binding sites of ES4. No crystal structure of an Arabidopsis SEC7-containing ARF-GEF has thus far been resolved. Nonetheless, based on sequence comparisons, the SEC7 catalytic domain from all ARF-GEFs is highly conserved (Cox et al., 2004). We therefore attempted to predict the target site of ES4 by docking on the liganded crystal structures of Saccharomyces cerevisiae ARF1-GDP bound to the GEA1-SEC7 domain complexed with BFA (PDB entry 1RE0; 40% overall sequence identity with Arabidopsis GNOM ARF-GEF) (Mossessova et al., 2003) and the human ARNO-cytohesin SEC7 domain in complex with inhibitor N-(4-hydroxy-2,6-dimethylphenyl) benzenesulfonamide (PDB entry 4JWL; 43% overall sequence identity with GNOM) (Rouhana et al., 2013). In the ARF1-SEC7 complex (1RE0), BFA binds tightly in a cavity at the interface between ARF1 and SEC7, while residues 48 to 52 of ARF1’s long switch-1 element loop reside in a hydrophobic groove of SEC7, of which all contact-involved residues are highly conserved. In the ARNO SEC7 complex (4JWL), its cocrystallized inhibitor resides in a depression along the hydrophobic groove that recognizes the ARF’s switch-1 element loop.
With only the SEC7 domain of 1RE0 and of 4JWL, blind (search space encompassing the whole SEC7 domain) and local (search space encompassing the depression in the SEC7 domain where 4JWL’s ligand resides) redocks of 4JWL’s ligand yielded essentially the same docking pose, as it is observed in the 4JWL crystal structure in the depression along the hydrophobic groove that is recognized by an ARF switch-1 element loop. Dockings with ES4 predict a similar docking pose in the same depression, also with similar or slightly higher predicted affinities (Figure 9; Supplemental Table 2).
Figure 9.
ES4 Docking Simulations.
(A) Model of the crystal structure 1RE0 of yeast ARF1 (cartoon colored from the N terminus [blue] to the C terminus [red]) containing GDP (sticks, yellow) and a Mg ion (green sphere) with the GEA1-SEC7 domain (cartoon and transparent surface) in complex with BFA (sticks, purple) residing in a cavity at the ARF1-SEC7 interface. Shown is a position of ES4 (sticks, green) obtained by local docking in this cavity. ARF1’s switch-1 element loop (light-blue tube) occupies a hydrophobic groove of the SEC7 domain.
(B) Model of the crystal structure 4JWL of the human ARNO SEC7 domain (cartoon and transparent surface, similarly orientated as the SEC7 domain in [A]) in complex with N-(4-hydroxy-2,6-dimethylphenyl) benzenesulfonamide (sticks, purple). With blind dockings, ES4 (sticks, green) prefers to dock in the same depression where 4JWL’s ligand resides; this depression lies along SEC7’s hydrophobic groove that is recognized by an ARF switch-1 element loop.
Blind dockings using the BFA-emptied 1RE0 ARF1-SEC7 complex suggest another possible binding site. A redock of BFA yielded the same docking pose as observed in the interfacial cavity of the crystal structure. ES4 also docks in the same cavity with promising poses where the flat aromatic system of ES4 can have sandwich-type overlaps with the aromatic amino acid residues from ARF1 that are lining this cavity, i.e., Phe-51, Trp-66, and Tyr-81, and showing a 3 Å proximity of the ES4’s nitro group to Trp78’s NE1 nitrogen (Figure 9A).
To summarize, our docking simulations show that ES4 docks with similar affinities to those of 4JWL’s ligand and BFA to their respective protein regions; however, thus far, we cannot definitely state which zone is the actual ES4 target site.
ES4 Does Not Inhibit the Activation of ARF1 by SEC7 Domains from Different ARF-GEFs
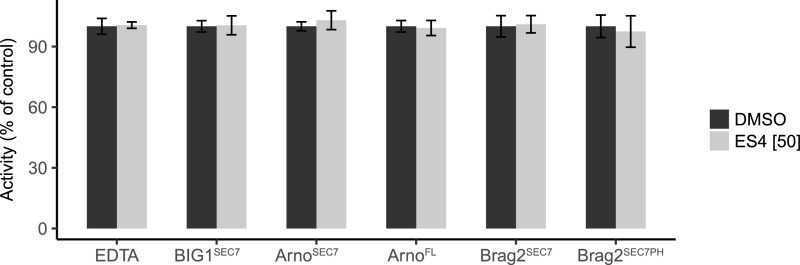
To compare the biochemical action of ES4 and BFA, we heterologously produced and purified three different SEC7 domains from human ARF-GEFs, including the SEC7 domain of the large ARF-GEF BIG1, which is sensitive to BFA and the SEC7 domains of ARNO and BRAG2, which are resistant to BFA. The human SEC7 domains share 29 to 52% sequence identity with the SEC7 domains of the eight Arabidopsis ARF-GEFs, which is in the same range as the sequence identity between the Arabidopsis BIG and GNOM/GNL groups (38–48%). Of note, BIG1, ARNO, and BRAG2 are inhibited by the small molecule AMF-26, and ARNO is also inhibited by SECIN-H3 (Benabdi et al., 2017). ES4 had no effect on nucleotide exchange on ARF1 catalyzed by any of these SEC7 domains (Figure 10; Supplemental Figure 8). It also did not inhibit full-length ARNO and a longer BRAG2, both of which carry membrane binding elements. We conclude from these experiments that ES4 does not inhibit representative human SEC7 domains with high sequence identity to Arabidopsis ARF-GEFs, suggesting that it blocks the functions of Arabidopsis and other ARF-GEF via a more complex mechanism: either not binding the SEC7 domain directly or requiring a more natural membrane environment for its inhibitory activity.
Figure 10.
Analysis of the Inhibitory Activity of 50 μM ES4 on ARF-GEF Constructs.
Nucleotide exchange rates (kobs) were determined by fluorescence kinetics of ARF1 in the presence of ARF-GEF constructs as indicated. kobs rates are expressed as a percentage of the rate obtained with DMSO. Assays were performed with 1 µM ∆17ARF1 and ARF-GEFs at either 100 nM (ARNOSEC7 or ARNOFL) or 250 nM (BRAG2SEC7, BRAG2SEC7-PH, and BIG1SEC7) or 5 mM EDTA with 50 µM ES4 or DMSO for the control. Values are the mean of triplicates ± sd.
DISCUSSION
ES4 Specifically Interferes with ARF-GEF-Dependent Trafficking
Here, we identified and characterized a chemical compound that affects endomembrane trafficking, particularly the ARF/ARF-GEF-dependent pathway. ES4 works in a dose-dependent manner, slowing down or inhibiting multiple trafficking processes including endocytosis, recycling, and trafficking to the vacuole. ES4 treatment changes the basal polarity of ectopically expressed PIN1 without visibly affecting the localization of native PIN. This effect is in accordance with previous findings (Kleine-Vehn et al., 2008b), showing that the polarity of ectopic basal PIN1 in the epidermis is easily perturbed by trafficking defects. ES4 does not specifically affect PIN trafficking, but it does interfere with vesicular routing of other PM proteins. Defects at the cellular level are mirrored by macroscopically observed phenotypes, including inhibited primary root and hypocotyl growth and a decrease in lateral root density, revealing that ES4 is an inhibitor of a broad range of trafficking processes.
A number of independent experimental approaches strongly indicate that ES4 specifically acts on ARF-GEFs: (1) gnl1 and ben1 mutants defective in different ARF-GEFs show the highest ES4 sensitivity; (2) ES4 affects the subcellular localization of ARF-GEF substrates—the ARF1 proteins; (3) the wild-type version of ARF1 is sensitive to ES4, but the GTP-locked ARF1Q71L, which does not require functional ARF-GEFs for its activation, is resistant to this compound; (4) the sec7/SEC7 yeast deletion mutant, which is defective in the major ARF-GEF-regulating subcellular trafficking in yeast (Wolf et al., 1998), shows the highest sensitivity to ES4; (5) in the DARTS assay, ES4 stabilizes (suggesting its binding) several members belonging to the two protein subfamilies of large ARF-GEFs in Arabidopsis, i.e., GNOM from the GBF/Gea subfamily and the BIG/SEC7 proteins BIG1, 3, 4, and 5; and (6) docking simulations support the binding of ES4 to the SEC7 domain of ARF-GEFs with an affinity comparable to that of the established ARF-GEF inhibitor BFA. All of these results strongly suggest that ES4, similar to BFA but with a different mechanism and/or specificity, inhibits ARF-GEF-dependent pathways, presumably by directly targeting members of the SEC7 domain ARF-GEF class.
ES4 Works in a Similar but Not the Same Manner as BFA
Vesicle trafficking in plants and other eukaryotes is regulated by ARF proteins, whose activity is controlled by ARF-GEFs. The most well known and widely used chemical, BFA, inhibits most of the large ARF-GEFs in plants, except GNL1 and BIG3. Although our biochemical and root sensitivity experiments demonstrated that ES4 affects ARF-GEFs, as does BFA, these compounds appear to have different potencies against the same targets. The most pronounced difference is the lack of BFA body-like structures after ES4 treatment. Additionally, in contrast to BFA treatment at instances where PIN1 protein localization at the PM becomes nonpolar, ES4 does not exert a similar effect. Moreover, ES4 overcomes the effects of BFA on the formation of BFA bodies. One possible explanation is that ES4 has higher affinity for ARF-GEFs, which act upstream of GNOM in the endocytic trafficking of PIN from the PM. The ARF-GEF BIG5/MIN7/BEN1 was shown to play a role in the early endocytic pathway at the level of early endosomes (Tanaka et al., 2009), whereas GNOM plays a role in recycling processes, which direct PIN1 back to the PM. In line with this, ben1 has a reduced number of BFA bodies after BFA treatment (Tanaka et al., 2009). Our DARTS assays suggest that BIG5 has the greatest affinity to ES4, consistent with a strong effect on early endocytic trafficking of PIN1, thus preventing its accumulation in more interior BFA bodies.
Moreover, the ARF-GEF BIG3 is resistant to BFA (Nielsen et al., 2006) and according to our DARTS assay has only a moderate affinity to ES4. Additionally, BIG1 and BIG4, which share redundant functions with BIG3, are sensitive to both ES4 and BFA. However, miscellaneous phenotypes were obtained after treatment of the big3 mutant with ES4 or BFA. It was reported that big3 had impaired seed germination and seedling root growth (Richter et al., 2014) after BFA treatment, whereas ES4 had no effect on root growth in big3 compared with the control. This could indicate that BIG1 and BIG4 are less sensitive to ES4 than to BFA.
The inactive GDP-bound form of ARF GTPases is cytosolic, and its conversion by ARF-GEF proteins to an active GTP-bound form induces membrane association (Nielsen et al., 2008). It was reported that BFA treatment results in agglomeration of signals from wild-type ARF1-GFP (Xu and Scheres, 2005), which is consistent with the finding that the abortive complex of ARF-GDP-SEC7-BFA is able to bind to membranes due to its intermediate level of membrane affinity between ARF-GDP and nucleotide-free ARF1-SEC7 (Béraud-Dufour et al., 1999; Mossessova et al., 2003). On the other hand, our observations show that ARF1WT-EYFP treated with ES4 localizes to the cytosol (Figure 7C). These diverse responses of ARF1WT-EYFP suggest that ARF-GEFs inhibited by either ES4 or BFA have different effects on ARF proteins. This notion is also supported by the results of a nucleotide exchange assay, where a change in tryptophan fluorescence reflects a conformational change of ARF-GDP into ARF-GTP (Benabdi et al., 2017). We show that ES4 does not inhibit GDP to GTP exchange by any of the tested SEC7-containing ARF-GEFs, unlike BFA, which specifically inhibits large Golgi ARF-GEFs such as BIG1 (Benabdi et al., 2017). Although the structural basis for the activation of large ARF-GEFs of the BIG and GBF1/GNOM families and their binding to membranes is currently not well understood, it is more than likely that large conformational rearrangements take place through their interactions with other proteins and with membranes. These conformational changes provide potential opportunities for inhibition by small molecules. In the case of ES4, our kinetic data rule out a simple competition mechanism with the ARF binding site on the SEC7 domain and favor a mode of action where ES4 affects a regulatory mechanism instead.
Years of experimentation and the discovery of its target, the SEC7 domain of ARF-GEFs (Peyroche et al., 1999), have established BFA as an important tool for dissecting endomembrane trafficking pathways. ES4 is an ARF-GEF inhibitor that targets a subset of members of this family with different specificity compared with BFA, which in turn determines the differences in the intracellular phenotypes observed in the presence of these compounds. ES4 has a clear potential to become an important tool to dissect different roles of ARF/ARF-GEF-dependent trafficking pathways, including those in polar targeting. Future work should focus on defining the precise mode of action of this inhibitor. Nonetheless, at this stage of knowledge, ES4 already enables the targeted manipulation of ARF-GEF-mediated processes, circumventing the obstacles posed by BFA-insensitive ARF-GEF versions.
ES4 Treatment Confirms a Role for ARF-GEF-Dependent Trafficking in the Basal Polarity of PIN
Two of the characterized ARF-GEF mutants, gnom and ben1, displayed defects preferentially in the basal trafficking of PIN1 proteins, indicating a role for GNOM and BEN1 in the regulation of this process (Kleine-Vehn et al., 2008a; Tanaka et al., 2009, 2013). In the partial loss-of-function gnomR5 mutant, the basal-to-apical shift of PIN1 in the stele and of PIN2 in the cortex was demonstrated, whereas in ben1, the defect was weaker, leading to less pronounced basal localization. We originally identified ES4 as a compound causing the basal-to-apical shift of ectopically expressed PIN1. The observation that ES4 interferes specifically with ARF-GEF-mediated processes confirms the important role of ARF-GEFs in trafficking of basal PIN cargos. Initially, it was suggested that GNOM localizes to endosomal compartments, presumably at its (so far elusive) recycling subdomain (Geldner et al., 2003). Nonetheless, studies have shown that GNOM predominantly localizes to the GA and that after BFA treatment, it translocates to the TGN/EE (Naramoto et al., 2014). Colocalization studies of GNL1 and GNOM revealed their close, but not completely overlapping, localization to the same GA stacks. Defects in the secretion of polysaccharides and secGFP in gnom and gnl1 mutants, respectively, hint at a role in regulating the transport of different cargos (Shevell et al., 2000; Teh and Moore, 2007). Immunogold labeling of plant coatomers localized all COPI subunits only to the GA stacks or in their close vicinity (Pimpl et al., 2000). Interestingly, GNOM and GNL1 localize to the GA in ring-like structures (Naramoto et al., 2014) similar to the ring-like distribution of COPI (Pimpl et al., 2000). This spatial correlation of the localizations of COPI and ARF-GEFs to the GA hints at a so far unclear role for GA-based, COP-mediated trafficking in the basal polar targeting of PIN proteins and possibly other cargos. Further research should clarify the so far elusive role of COPI in intracellular trafficking in plants and its possible involvement in polar trafficking. ES4 could be used as a tool to help dissect the roles of a coatomer and the GA in general polar trafficking processes in plants.
METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis thaliana were stratified for 2 d in the dark at 4°C and grown vertically at 21°C under continuous white light emitted by fluorescent lamps with an intensity of 120 µmol m−2 s−1 on 0.8% agar half-strength Murashige and Skoog medium (Duchefa) with 1% sucrose (pH 5.9). The Arabidopsis lines PIN2pro:PIN1-HA eir1-1/pin2 (Wiśniewska et al., 2006); DR5pro:GUS (Ulmasov et al., 1997); N-ST-GFP (Batoko et al., 2000); 35Spro:PID21 (Benjamins et al., 2001); BRI1pro:BRI1-GFP (Friedrichsen et al., 2000); CLC2pro:CLC2-GFP (Konopka and Bednarek, 2008); VHAa1pro:VHAa1-GFP (Dettmer et al., 2006); PIN1pro:PIN1-GFP (Benková et al., 2003); 35Spro:GFP-PIP2a (Cutler et al., 2000); PIN2pro:PIN2-GFP (Xu and Scheres, 2005); GNOMpro:GNOM-GFP (Geldner et al., 2003); GNL1pro:GNL1-YFP (Richter et al., 2007); 35Spro:GFP-MAP4 (Marc et al., 1998); 35Spro:GFP-FABD (Ketelaar et al., 2004); HSP18.2pro:ARF1WT-EYFP, HSP18.2pro:ARF1Q71L-EYFP and HSP18.2pro:ARF1T31N-ECFP (Xu and Scheres, 2005); axr2-1 (Wilson et al., 1990); ben1-1 and ben1-2 (SALK_013761) (Tanaka et al., 2009); big3 (SALK_044617) (Richter et al., 2014); rcn1 (Garbers et al., 1996); gnl1-2 and gnl1-3 (Teh and Moore, 2007); gnomR5 (Geldner et al., 2004); van7 (Koizumi et al., 2000); snx1-1 (Jaillais et al., 2006); chc1-2 (SALK_103252) and chc2-2 (SALK_028826) (Kitakura et al., 2011); and vti12 (Surpin et al., 2003) have been described previously. The Columbia (Col-0) accession was used for immunolocalization, transmission electron microscopy, FM4-64 uptake, and as the wild-type control in the seedling growth experiments, except for growth of rcn1 and gnomR5 and van7 for which the Wassilewskija (Ws) and Landsberg erecta (Ler) accessions were used, respectively.
Chemical Treatments
Stock solutions of BFA (Sigma-Aldrich), ES4 (Chembridge ID 6938485), and FM4-64 (Invitrogen) were prepared in DMSO and diluted in liquid half-strength MS (or growth) medium for treatments with the indicated concentrations and times. Equal volumes of solvents were used as mock treatments for controls. For germination and growth of seedlings on ES4 and BFA, seeds were sown directly onto ES4/BFA/ES4+BFA-supplemented growth medium. For polar localization and DR5pro:GUS experiments, 3-d-old seedlings were transferred for 48 h from solid growth medium to solid medium supplemented with ES4. For shorter treatment (4 h) of DR5pro:GUS, 5-d-old seedlings were transferred to liquid medium supplemented with ES4. After treatment, the seedlings were stained overnight at 37°C in darkness in GUS staining buffer [100 mM Na-phosphate buffer, pH 7, 0.1% Triton X-100, 10 mM EDTA, pH 8, and 2 mM of each K3FeIII(CN)6 and K4FeII(CN)6] containing X-Gluc to visualize GUS activity. X-Gluc was added at a final concentration of 1 mg/mL from a freshly prepared 10-mg/mL stock dissolved in DMSO. For BFA treatments, 5-d-old seedlings were pretreated for 30 min with ES4 before the addition of 25 µM BFA to the treatment for a further 90 min. For BFA washouts, 5-d-old seedlings were treated with 25 or 50 µM BFA for 2 h, followed by 30 min of wash treatment. For FM4-64 uptake experiments, after 2 h of treatment, 5-d-old seedlings were transferred directly to 2 µM FM4-64 in treatment medium on ice for 5 min, followed by two washes in treatment medium on ice. Endocytosis was started by removing seedlings from the ice-cold conditions. For visualization of vacuolar GFP labeling, 5-d-old seedlings were transferred directly after 2 h of treatment with 41 µM ES4 to 8 µM FM4-64 in treatment medium on ice for 5 min, followed by two washes in treatment medium on ice and transferred to 41 µM ES4-supplemented growth medium and incubated vertically in darkness for 4 h. For live imaging and transmission electron microscopy, 5-d-old seedlings were transferred for 2 h to treatment medium. For induction of HSP18.2pro:ARF1 expression, the plates were incubated at 37°C for 2 h followed by ES4 treatment for 2 h at room temperature.
Immunolocalization and Transmission Electron Microscopy of Roots
Whole-mount immunolocalization on 5-d-old seedlings of Arabidopsis was done with the InSituProrobot (Intavis) according to the described protocol (Sauer et al., 2006). Primary antibodies and final dilutions were as follows: rabbit anti-PIN1 (Paciorek et al., 2005), 1:1000; rabbit anti-PIN2 (generously provided by C. Luschnig), 1:1000; and mouse anti-HA (AbCam/HA.C5), 1:500. Secondary antibodies and final dilutions were as follows: Cy3 anti-rabbit (Sigma-Aldrich), 1:600; and Alexa488 anti-mouse (Invitrogen), 1:600. Root tips of 5-d-old Col-0 seedlings untreated and treated for 2 h with 41 μM ES4 were excised and processed as described (Tanaka et al., 2009).
Yeast Strains and Media
The diploid heterozygous and homozygous gea1 deletion mutants generated by the International Deletion Consortium (Winzeler et al., 1999) were obtained from EUROSCARF. BY4743 and BY4742 were used as the diploid wild-type control for the diploid heterozygous and gea1 homozygous deletion mutants, respectively. Yeast cells were grown on standard rich medium (yeast extract/peptone/dextrose [YPD]) with and without ES4. For analysis of growth rate and ES4 sensitivity, BY4743 cells were grown in 1 mL liquid YPD medium in 50-mL Falcon tubes for 4 h at 28°C with shaking at 200 rpm to an OD600 of ∼1. After 4 h, 2 μL of the liquid cell culture was transferred into 96-well plates containing 198 μL of liquid YPD with mock treatment or with different concentrations of ES4 (17, 21, 28, and 41 µM) at a final OD600 of ∼0.1 and grown overnight at 28°C with shaking in a H1 Microplate Reader (BioTek). The deletion mutants were grown under 17 µM ES4 and mock treatment as described for the wild type.
Quantitative and Statistical Analyses
For PIN polarity quantification in the cortex, the ratio of the cell numbers with basal PIN2 to the total number of cortical cells was calculated from the confocal laser scanning microscopy images of the root apical meristem. For polarity of PIN2pro:PIN1 in epidermal cells, the ratio of the cell numbers with basal, both basal and apical, or apical PIN localization was compared with the total number of epidermal cells from the confocal laser scanning microscopy images of the root apical meristem. For analysis of the gravitropic response, 5-d-old seedlings grown vertically in light were gravistimulated by a 90° rotation (Col-0). PIN2pro:PIN1-HA pin2 seedlings were gravistimulated twice, at the 5-d-old stage and 24 h after the first gravistimulation. The bending angle was measured with Java-based ImageJ software (http://rsb.info.nih.gov/ij/) 48 h after the first gravistimulation. All gravitropically stimulated roots were assigned to one of the eight 45° sectors on the gravitropism diagram. The length of the bars in the diagram represents the percentage of seedlings assigned to the respective sector. Wild-type root lengths and hypocotyl lengths were measured with ImageJ software. For mutant root lengths, the ratio of the root length of seedlings grown on ES4 and that of seedlings grown on mock medium was calculated with ImageJ software. For quantification of FM4-64 and PIN2-GFP internalization, the ratio of the mean pixel intensity of the internal cell fluorescence and the mean pixel intensity of the adjacent PM fluorescence was obtained by ImageJ. The size of the PVC/MVBs was calculated with ImageJ software. For the quantification of intracellular aggregations of ARF1-XFP in transgenic fluorescent marker lines, Imaris software version 9.1.2 was used utilizing a surface identification algorithm for counting the aggregations. The parameters were set using images acquired for the wild-type mock treated sample (ARF1WT-EYFP; see Figure 7). Next, the whole wild-type image set was analyzed and quantified and subsequently (using the same Imaris parameters) all images of ARF1-XFP fluorescent marker lines (ARF1Q71L-EYFP and ARF1T31N-ECFP) were analyzed. The minimal aggregation (spot) size (XY diameter) considered for detection was set at 0.7 µm. The results were presented as the number of aggregations in the whole field of view. For the yeast growth assay, the growth curves of 12 strains (including wild types) were analyzed, each grown with mock treatment or in the presence of 17 µM of ES4. OD600 of the cell density was determined every 2 h for nine time points. Three or two replications per strain per experiment for ES4 or mock treatment were done, respectively. The statistical significances of differences of data were quantified with Student’s t test, Mann-Whitney U test, or ANOVA (Supplemental Table 3).
Construct Generation and Cell Culture Transformation
The BIG1 and BIG4 fragments without stop codons were amplified by PCR with iProof high-fidelity DNA polymerase (Bio-Rad) from wild-type Col-0 genomic DNA with the following primer pairs with Gateway system-compatible attB sites: BIG1 forward 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGTCGTCGTCGCAGAAC-3′ and BIG1 reverse 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTTCATCCATCATTGCACCC-3′; BIG4 forward 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTCAACGTCACAAACC-3′ and BIG4 reverse 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTAAGCCAAAATAGGACCAAT-3′. The PCR products were then introduced into pDONR221 donor vectors (Invitrogen). The cDNA fragments of BIG3, GNL1, and GNL2 were synthesized with Gateway system-compatible attL sites (Invitrogen) and introduced into the pG9m-2 vector (Gen9 Company). A 1694-bp fragment of the RPS5A promoter was introduced into the pDONRP4-P1R donor vector. The entry clones pDONRP4-P1R-pRPS5A, pDONR221-BIG1/BIG3/BIG4/GNL1/GNL2, and pDONRP2R-P3-HA were recombined in a multisite LR reaction with pH7m34GW (Invitrogen) as the destination vector. All clones were confirmed by sequencing. The generated constructs were used for transformation of dark-grown Arabidopsis PSB-D cell suspension cultures as described previously (Van Leene et al., 2007).
DARTS Assays
The DARTS assay for validation of protein interactors of ES4 was performed as described (Lomenick et al., 2011). Arabidopsis PSB-D cell cultures were used for total protein extraction. All extraction steps were performed at 4°C. After harvesting, the cells were ground in liquid nitrogen, resuspended in total protein extraction buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% IGEPAL CA-630, and Roche cOmplete ULTRA protease inhibitor cocktail, EDTA free) at a 1:2 (w/v) ratio, and centrifuged to discard the cell debris. After determining the protein concentration with Quick Start Bradford 1× dye reagent (Bio-Rad), the cell lysate was split into LoBind tubes and incubated with the respective chemical (BFA or ES4) at 250 µM concentration for 30 min at room temperature with slow mixing (control treatments were with equal volumes of DMSO). The used concentrations were much higher than the biologically relevant doses in order to saturate the protein with ligand and ensure maximal protection from proteolysis (Lomenick et al., 2011). The treated protein extracts were further aliquoted, and each of the aliquots was mixed with Pronase (Roche) at the corresponding dilution prepared in Pronase buffer (25 mM Tris-HCl, pH 7.5. and 150 mM NaCl) to achieve the aimed ratio of total enzyme to total protein substrate. After incubation for 30 min at room temperature, the proteolytic digestion was stopped by adding the protease inhibitor cocktail (Roche) and the tubes were left on ice for 10 min. The protein samples were then mixed with 4× NuPAGE LDS sample buffer (Invitrogen), heated at 70°C for 10 min, and loaded onto NuPAGE 4 to 12% Bis-Tris protein gels (Invitrogen). The protein transfer to PVDF membranes was performed using the iBlot dry blotting system (Thermo), and protein detection was done according to standard protein gel blotting procedures. The membranes were probed with the following antibodies: rabbit α-AtMIN7/BEN1/BIG5 (1:6000) (Nomura et al., 2006), rabbit α-SEC7 (1:10,000) (Steinmann et al., 1999), rat α-HA (1:1000; Roche), and rabbit α-ATPβ (1:2000) (Agrisera) as a reference for protein loading. The secondary antibodies were ECL α-rabbit/rat IgG, horseradish peroxidase-linked whole antibody (GE Healthcare). Blots were developed with Western Lightning Plus-ECL, Enhanced Chemiluminescence Substrate (Perkin-Elmer), and imaging was done with the Bio-Rad ChemiDoc XRS+ molecular imager. Intensity of protein bands was measured with the Bio-Rad Image Lab software package, and the ratio between the compound- and mock-treated samples for each of the pronase concentrations was calculated.
Docking Simulation Settings
All water and ligands of crystal structures 1RE0 (yeast ARF1-GDP with GEA1-SEC7 domain in complex with BFA) (Mossessova et al., 2003) and 4JWL (human ARNO-SEC7 domain in complex with N-(4-hydroxy-2,6-dimethylphenyl) benzenesulfonamide) (Rouhana et al., 2013) were manually deleted from the PDB text file. The emptied structures were subjected to a local minimization with the GROMOS96 (43B1 parameter set) implementation within the Swiss-PdbViewer (Guex and Peitsch, 1997), and polar hydrogens were added. Ligands were 3D-drawn with Avogadro 1.1.1 (Hanwell et al., 2012) and minimized with the built-in MMFF94s force field (Halgren, 1999). The AutoDockTools 1.5.4 suite (Sanner, 1999) was used for pdbqt format preparation of the proteins and ligands. Dockings were performed with AutoDock-Vina 1.1.0 (Trott and Olson, 2010). For local dockings on the 1RE0 ARF1-SEC7 structure in the interfacial space where BFA resides, the grid-box size was x, y = 24 Å and z = 30 Å centered at x = 45.0, y = 11.4, and z = 48.0, and for local dockings on the 4JWL SEC7 structure where its ligand resides, the grid-box size was x, y, z = 24 Å centered at x = 0.6, y = 40.1 and z = 59.0; exhaustiveness for both was set at 32. Blind dockings were with grid-box sizes encompassing the full protein structure(s): x, y, and z = 50 Å centered at x = 47.4, y = 20.8, and z = 49.2 for the GEA1-SEC7 domain of 1RE0 (chain B only), and x, y = 50 Å and z = 60 Å centered at x = 9.2, y = 39.7, and z = 46.9 for the ARNO-SEC7 structure of 4JWL; exhaustiveness for both was set at 124. Visualization was with PyMOL (DeLano Scientific).
Image Analysis
Imaging was done on LSM 710 (Zeiss), LSM 700 (Zeiss), and Leica SP2 confocal laser scanning microscopes.
Protein Expression and Purification
N-terminally truncated bovine ∆17ARF1 (identical to the human form) was expressed, purified, and loaded with GDP prior to use as described (Zeeh et al., 2006). Human BRAG2SEC7, BRAG2SEC7-PH, ARNOSEC7, and BIG1SEC7 were produced in Escherichia coli and purified as described (Benabdi et al., 2017). Expression in E. coli and purification of full-length human ARNO (ARNOFL) will be described elsewhere. All proteins were >90% pure, as controlled by SDS-PAGE (Supplemental Figure 9A) and had high GEF activity toward ARF1.
Nucleotide Exchange Assays
Inhibition was analyzed by fluorescence kinetics as described (Benabdi et al., 2017). Briefly, nucleotide exchange kinetics were monitored by the change in tryptophan fluorescence that follows the conformational change from ARF-GDP to ARF-GTP (excitation and emission wavelengths of 292 and 340 nm, respectively). Exchange rates (kobs) were determined from monoexponential fits over the entire kinetics and expressed as a percentage of control activity. Experiments were performed with 1 μM ∆17ARF1 and ARF-GEFs at either 100 nM (ARNOSEC7 or ARNOFL) or 250 nM (BRAG2SEC7, BRAG2SEC7-PH, and BIG1SEC7) with 50 µM ES4 or DMSO for control. It should be noted that ES4 slightly reduces the fluorescence intensity (Supplemental Figure 9B), which is due to absorbance at 290 nm (Supplemental Figure 9C), explaining why kinetics measured with the compound have a lower plateau. This effect is not due to inhibition and is taken into account by fitting the entire kinetics curve rather than using initial velocities.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PIN1 (At1g73590), PIN2 (At5G57090), PIP2a (At3G53420), VHAa1 (At2g28520), CLC2 (At2G40060), N-ST (AJ243198), BEN1 (At3G43300), SYP61 (AF355754), GNL1 (At5G39500), ARF1 (At2g47170), GNOM (At1g13980), BRI1 (At4g39400), PID (At2g34650), CHC1 (At3g11130), CHC2 (At3g08530), SNX1 (At5g06140), VTI12 (At1g26670), RCN1 (At1g25490), AXR2 (At3g23050), BIG3 (At1g01960), FABD (AT4G26700), and MAP4 (M72414).
Supplemental Data
Supplemental Figure 1. Chemical screen of a set of 11 small molecules implicated in polarity changes.
Supplemental Figure 2. Characterization of the effects of ES4.
Supplemental Figure 3. Effect of ES4 on intracellular trafficking.
Supplemental Figure 4. Effect of ES4 on vacuolar trafficking.
Supplemental Figure 5. Effect of ES4 on intracellular compartments.
Supplemental Figure 6. Effect of ES4 on ARF1T31N-CFP marker.
Supplemental Figure 7. ES4 sensitivity of yeast deletion strains.
Supplemental Figure 8. Representative nucleotide exchange kinetic curves.
Supplemental Figure 9. SDS-PAGE and fluorescence intensity and absorbance of ES4.
Supplemental Table 1. Yeast deletion strains used in the growth assay.
Supplemental Table 2. Docking-calculated affinities.
Supplemental Table 3. ANOVA tables.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Gerd Jürgens, Sandra Richter, and Sheng Yang He for providing antibodies; Maciek Adamowski, Fernando Aniento, Sebastian Bednarek, Nico Callewaert, Matyás Fendrych, Elena Feraru, and Mugurel I. Feraru for helpful suggestions; Siamsa Doyle for critical reading of the manuscript and helpful comments and suggestions; and Stephanie Smith and Martine De Cock for help in editing and language corrections. We acknowledge the core facility Cellular Imaging of CEITEC supported by the Czech-BioImaging large RI project (LM2015062 funded by MEYS CR) for their support with obtaining scientific data presented in this article. Plant Sciences Core Facility of CEITEC Masaryk University is gratefully acknowledged for obtaining part of the scientific data presented in this article. We acknowledge support from the Fondation pour la Recherche Médicale and from the Institut National du Cancer (J.C.). The research leading to these results was funded by the European Research Council under the European Union’s 7th Framework Program (FP7/2007-2013)/ERC grant agreement numbers 282300 and 742985 and the Czech Science Foundation GAČR (GA18-26981S; J.F.); Ministry of Education, Youth, and Sports/MEYS of the Czech Republic under the Project CEITEC 2020 (LQ1601; T.N.); the China Science Council for a predoctoral fellowship (Q.L.); a joint research project within the framework of cooperation between the Research Foundation-Flanders and the Bulgarian Academy of Sciences (VS.025.13N; K.M. and E.R.); Vetenskapsrådet and Vinnova (Verket för Innovationssystem; S.R.), Knut och Alice Wallenbergs Stiftelse via “Shapesystem” Grant 2012.0050 (S.R.), Kempe stiftelserna (P.G.), Tryggers CTS410 (P.G.).
AUTHOR CONTRIBUTIONS
U.K., S.R., G.R.H., and J.F. conceived the project. U.K., S.R., T.N., and J.F. designed the research strategies. R.D.R. conducted transmission electron microscopy experiments. U.K. and P.G. performed root growth assays. W.N. performed the docking simulations. Q.L., K.M., and E.R. performed the DARTS assays. T.N. performed Imaris software analysis and imaging of cytoskeleton marker lines. F.P. and J.C. performed nucleotide exchange assay and protein expression and purification. U.K. performed the majority of the remaining experiments and analyzed the data. U.K., T.N., G.R.H., and J.F. edited the manuscript. U.K., T.N., and J.F. wrote the article. All authors revised the article.
Footnotes
Articles can be viewed without a subscription.
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256. [DOI] [PubMed] [Google Scholar]
- Achstetter T., Franzusoff A., Field C., Schekman R. (1988). SEC7 encodes an unusual, high molecular weight protein required for membrane traffic from the yeast Golgi apparatus. J. Biol. Chem. 263: 11711–11717. [PubMed] [Google Scholar]
- Adamowski M., Friml J. (2015). PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders N., Jürgens G. (2008). Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell. Mol. Life Sci. 65: 3433–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz K., McHardy L., Gable K., Tarling T., Rebérioux D., Bryan J., Andersen R.J., Dunn T., Hieter P., Roberge M. (2004). Yeast genome-wide drug-induced haploinsufficiency screen to determine drug mode of action. Proc. Natl. Acad. Sci. USA 101: 4525–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baster P., Robert S., Kleine-Vehn J., Vanneste S., Kania U., Grunewald W., De Rybel B., Beeckman T., Friml J. (2013). SCF(TIR1/AFB)-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J. 32: 260–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H., Zheng H.Q., Hawes C., Moore I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12: 2201–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R., Rawet M., Wieland F.T., Cassel D. (2009). The COPI system: molecular mechanisms and function. FEBS Lett. 583: 2701–2709. [DOI] [PubMed] [Google Scholar]
- Benabdi S., Peurois F., Nawrotek A., Chikireddy J., Cañeque T., Yamori T., Shiina I., Ohashi Y., Dan S., Rodriguez R., Cherfils J., Zeghouf M. (2017). Family-wide analysis of the inhibition of ARF guanine nucleotide exchange factors with small molecules: evidence of unique inhibitory profiles. Biochemistry 56: 5125–5133. [DOI] [PubMed] [Google Scholar]
- Benjamins R., Quint A., Weijers D., Hooykaas P., Offringa R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067. [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Béraud-Dufour S., Paris S., Chabre M., Antonny B. (1999). Dual interaction of ADP ribosylation factor 1 with Sec7 domain and with lipid membranes during catalysis of guanine nucleotide exchange. J. Biol. Chem. 274: 37629–37636. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Lippincott-Schwartz J. (2003). Coat proteins: shaping membrane transport. Nat. Rev. Mol. Cell Biol. 4: 409–414. [DOI] [PubMed] [Google Scholar]
- Cox R., Mason-Gamer R.J., Jackson C.L., Segev N. (2004). Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell 15: 1487–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher C., Balch W.E. (1994). Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J. Biol. Chem. 269: 1437–1448. [PubMed] [Google Scholar]
- Deitz S.B., Rambourg A., Képès F., Franzusoff A. (2000). Sec7p directs the transitions required for yeast Golgi biogenesis. Traffic 1: 172–183. [DOI] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527. [DOI] [PubMed] [Google Scholar]
- Ding Z., Galván-Ampudia C.S., Demarsy E., Łangowski Ł., Kleine-Vehn J., Fan Y., Morita M.T., Tasaka M., Fankhauser C., Offringa R., Friml J. (2011). Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 13: 447–452. [DOI] [PubMed] [Google Scholar]
- Doyle S.M., Haeger A., Vain T., Rigal A., Viotti C., Łangowska M., Ma Q., Friml J., Raikhel N.V., Hicks G.R., Robert S. (2015). An early secretory pathway mediated by GNOM-LIKE 1 and GNOM is essential for basal polarity establishment in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: E806–E815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G., et al. (2011). Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc. Natl. Acad. Sci. USA 108: 17850–17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E., Paciorek T., Feraru M.I., Zwiewka M., De Groodt R., De Rycke R., Kleine-Vehn J., Friml J. (2010). The AP-3 β adaptin mediates the biogenesis and function of lytic vacuoles in Arabidopsis. Plant Cell 22: 2812–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E., Feraru M.I., Kleine-Vehn J., Martinière A., Mouille G., Vanneste S., Vernhettes S., Runions J., Friml J. (2011). PIN polarity maintenance by the cell wall in Arabidopsis. Curr. Biol. 21: 338–343. [DOI] [PubMed] [Google Scholar]
- Feraru E., Feraru M.I., Asaoka R., Paciorek T., De Rycke R., Tanaka H., Nakano A., Friml J. (2012). BEX5/RabA1b regulates trans-Golgi network-to-plasma membrane protein trafficking in Arabidopsis. Plant Cell 24: 3074–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A., Redding K., Crosby J., Fuller R.S., Schekman R. (1991). Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J. Cell Biol. 112: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D.M., Joazeiro C.A.P., Li J., Hunter T., Chory J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbers C., DeLong A., Deruére J., Bernasconi P., Söll D. (1996). A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 15: 2115–2124. [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.-D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230. [DOI] [PubMed] [Google Scholar]
- Geldner N., Richter S., Vieten A., Marquardt S., Torres-Ruiz R.A., Mayer U., Jürgens G. (2004). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131: 389–400. [DOI] [PubMed] [Google Scholar]
- Giaever G., Shoemaker D.D., Jones T.W., Liang H., Winzeler E.A., Astromoff A., Davis R.W. (1999). Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet. 21: 278–283. [DOI] [PubMed] [Google Scholar]
- Goldberg J. (1999). Structural and functional analysis of the ARF1-ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell 96: 893–902. [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Halgren T.A. (1999). MMFF VII. Characterization of MMFF94, MMFF94s, and other widely available force fields for conformational energies and for intermolecular-interaction energies and geometries. J. Comput. Chem. 20: 730–748. [DOI] [PubMed] [Google Scholar]
- Halliwell B., Clement M.V., Ramalingam J., Long L.H. (2000). Hydrogen peroxide. Ubiquitous in cell culture and in vivo? IUBMB Life 50: 251–257. [DOI] [PubMed] [Google Scholar]
- Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. (2012). Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.L., Casanova J.E. (2000). Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 10: 60–67. [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miège C., Rollin C., Gaude T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109. [DOI] [PubMed] [Google Scholar]
- Jelínková A., Malínská K., Simon S., Kleine-Vehn J., Parezová M., Pejchar P., Kubeš M., Martinec J., Friml J., Zazímalová E., Petrásek J. (2010). Probing plant membranes with FM dyes: tracking, dragging or blocking? Plant J. 61: 883–892. [DOI] [PubMed] [Google Scholar]
- Ketelaar T., Allwood E.G., Anthony R., Voigt B., Menzel D., Hussey P.J. (2004). The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14: 145–149. [DOI] [PubMed] [Google Scholar]
- Kitakura S., Vanneste S., Robert S., Löfke C., Teichmann T., Tanaka H., Friml J. (2011). Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., et al. (2011). Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol. Syst. Biol. 7: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Sauer M., Brewer P.B., Wiśniewska J., Paciorek T., Benková E., Friml J. (2008a). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18: 526–531. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Łangowski L., Wiśniewska J., Dhonukshe P., Brewer P.B., Friml J. (2008b). Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol. Plant 1: 1056–1066. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008c). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Huang F., Naramoto S., Zhang J., Michniewicz M., Offringa R., Friml J. (2009). PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutstein M., Shaked H., Sherman A., Avivi-Ragolsky N., Shema E., Zenvirth D., Levy A.A., Simchen G. (2008). Functional conservation of the yeast and Arabidopsis RAD54-like genes. Genetics 178: 2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi K., Sugiyama M., Fukuda H. (2000). A series of novel mutants of Arabidopsis thaliana that are defective in the formation of continuous vascular network: calling the auxin signal flow canalization hypothesis into question. Development 127: 3197–3204. [DOI] [PubMed] [Google Scholar]
- Konopka C.A., Bednarek S.Y. (2008). Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant J. 53: 186–196. [DOI] [PubMed] [Google Scholar]
- Kumari S., Mayor S. (2008). ARF1 is directly involved in dynamin-independent endocytosis. Nat. Cell Biol. 10: 30–41. [DOI] [PubMed] [Google Scholar]
- Lee M.H., Min M.K., Lee Y.J., Jin J.B., Shin D.H., Kim D.H., Lee K.-H., Hwang I. (2002). ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol. 129: 1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B., et al. (2009). Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 106: 21984–21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B., Jung G., Wohlschlegel J.A., Huang J. (2011). Target identification using drug affinity responsive target stability (DARTS). Curr. Protoc. Chem. Biol. 3: 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc J., Granger C.L., Brincat J., Fisher D.D., Kao Th., McCubbin A.G., Cyr R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056. [DOI] [PubMed] [Google Scholar]
- Mossessova E., Corpina R.A., Goldberg J. (2003). Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol. Cell 12: 1403–1411. [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S., Kleine-Vehn J., Robert S., Fujimoto M., Dainobu T., Paciorek T., Ueda T., Nakano A., Van Montagu M.C.E., Fukuda H., Friml J. (2010). ADP-ribosylation factor machinery mediates endocytosis in plant cells. Proc. Natl. Acad. Sci. USA 107: 21890–21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S., Otegui M.S., Kutsuna N., de Rycke R., Dainobu T., Karampelias M., Fujimoto M., Feraru E., Miki D., Fukuda H., Nakano A., Friml J. (2014). Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell 26: 3062–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Cheung A.Y., Ueda T. (2008). The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol. 147: 1516–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Albrethsen J., Larsen F.H., Skriver K. (2006). The Arabidopsis ADP-ribosylation factor (ARF) and ARF-like (ARL) system and its regulation by BIG2, a large ARF-GEF. Plant Sci. 171: 707–717. [Google Scholar]
- Nodzyński T., Vanneste S., Zwiewka M., Pernisová M., Hejátko J., Friml J. (2016). Enquiry into the topology of plasma membrane-localized PIN auxin transport components. Mol. Plant 9: 1504–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223. [DOI] [PubMed] [Google Scholar]
- Ooi C.E., Dell’Angelica E.C., Bonifacino J.S. (1998). ADP-Ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J. Cell Biol. 142: 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T., Friml J. (2006). Auxin signaling. J. Cell Sci. 119: 1199–1202. [DOI] [PubMed] [Google Scholar]
- Paciorek T., Zazímalová E., Ruthardt N., Petrásek J., Stierhof Y.-D., Kleine-Vehn J., Morris D.A., Emans N., Jürgens G., Geldner N., Friml J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256. [DOI] [PubMed] [Google Scholar]
- Peyroche A., Paris S., Jackson C.L. (1996). Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature 384: 479–481. [DOI] [PubMed] [Google Scholar]
- Peyroche A., Antonny B., Robineau S., Acker J., Cherfils J., Jackson C.L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3: 275–285. [DOI] [PubMed] [Google Scholar]
- Peyroche A., Courbeyrette R., Rambourg A., Jackson C.L. (2001). The ARF exchange factors Gea1p and Gea2p regulate Golgi structure and function in yeast. J. Cell Sci. 114: 2241–2253. [DOI] [PubMed] [Google Scholar]
- Pimpl P., Movafeghi A., Coughlan S., Denecke J., Hillmer S., Robinson D.G. (2000). In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell 12: 2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl P., Hanton S.L., Taylor J.P., Pinto-daSilva L.L., Denecke J. (2003). The GTPase ARF1p controls the sequence-specific vacuolar sorting route to the lytic vacuole. Plant Cell 15: 1242–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon P.P., Cassel D., Spang A., Rotman M., Pick E., Singer R.A., Johnston G.C. (1999). Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 18: 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakusová H., Abbas M., Han H., Song S., Robert H.S., Friml J. (2016). Termination of shoot gravitropic responses by auxin feedback on PIN3 polarity. Curr. Biol. 26: 3026–3032. [DOI] [PubMed] [Google Scholar]
- Renault L., Guibert B., Cherfils J. (2003). Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 426: 525–530. [DOI] [PubMed] [Google Scholar]
- Richardson B.C., McDonold C.M., Fromme J.C. (2012). The Sec7 Arf-GEF is recruited to the trans-Golgi network by positive feedback. Dev. Cell 22: 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S., Geldner N., Schrader J., Wolters H., Stierhof Y.D., Rios G., Koncz C., Robinson D.G., Jürgens G. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448: 488–492. [DOI] [PubMed] [Google Scholar]
- Richter S., Kientz M., Brumm S., Nielsen M.E., Park M., Gavidia R., Krause C., Voss U., Beckmann H., Mayer U., Stierhof Y.D., Jürgens G. (2014). Delivery of endocytosed proteins to the cell-division plane requires change of pathway from recycling to secretion. eLife 3: e02131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Scheuring D., Naramoto S., Friml J. (2011). ARF1 localizes to the golgi and the trans-golgi network. Plant Cell 23: 846–849, author reply 849–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana J., Hoh F., Estaran S., Henriquet C., Boublik Y., Kerkour A., Trouillard R., Martinez J., Pugnière M., Padilla A., Chavanieu A. (2013). Fragment-based identification of a locus in the Sec7 domain of Arno for the design of protein-protein interaction inhibitors. J. Med. Chem. 56: 8497–8511. [DOI] [PubMed] [Google Scholar]
- Sanner M.F. (1999). Python: a programming language for software integration and development. J. Mol. Graph. Model. 17: 57–61. [PubMed] [Google Scholar]
- Sauer M., Paciorek T., Benková E., Friml J. (2006). Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat. Protoc. 1: 98–103. [DOI] [PubMed] [Google Scholar]
- Scheuring D., Viotti C., Krüger F., Künzl F., Sturm S., Bubeck J., Hillmer S., Frigerio L., Robinson D.G., Pimpl P., Schumacher K. (2011). Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini T., Orci L., Amherdt M., Brunner M., Kahn R.A., Rothman J.E. (1991). ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell 67: 239–253. [DOI] [PubMed] [Google Scholar]
- Shevell D.E., Kunkel T., Chua N.-H. (2000). Cell wall alterations in the arabidopsis emb30 mutant. Plant Cell 12: 2047–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann T., Geldner N., Grebe M., Mangold S., Jackson C.L., Paris S., Gälweiler L., Palme K., Jürgens G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286: 316–318. [DOI] [PubMed] [Google Scholar]
- Surpin M., Zheng H., Morita M.T., Saito C., Avila E., Blakeslee J.J., Bandyopadhyay A., Kovaleva V., Carter D., Murphy A., Tasaka M., Raikhel N. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15: 2885–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M., Ueda T., Yahara N., Nakano A. (2002). Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 31: 499–515. [DOI] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.I., Watanabe M., Nishimura M., Hara-Nishimura I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35: 545–555. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19: 391–397. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., Rakusová H., Uemura T., Feraru M.I., De Rycke R., Robert S., Kakimoto T., Friml J. (2013). Cell polarity and patterning by PIN trafficking through early endosomal compartments in Arabidopsis thaliana. PLoS Genet. 9: e1003540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Nodzyłski T., Kitakura S., Feraru M.I., Sasabe M., Ishikawa T., Kleine-Vehn J., Kakimoto T., Friml J. (2014). BEX1/ARF1A1C is required for BFA-sensitive recycling of PIN auxin transporters and auxin-mediated development in Arabidopsis. Plant Cell Physiol. 55: 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh O.K., Moore I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448: 493–496. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238. [DOI] [PubMed] [Google Scholar]
- Viaene T., et al. (2014). Directional auxin transport mechanisms in early diverging land plants. Curr. Biol. 24: 2786–2791. [DOI] [PubMed] [Google Scholar]
- Wilson A.K., Pickett F.B., Turner J.C., Estelle M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222: 377–383. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wiśniewska J., Xu J., Seifertová D., Brewer P.B., Růžička K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J. (2006). Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Wolf J., Nicks M., Deitz S., van Tuinen E., Franzusoff A. (1998). An N-end rule destabilization mutant reveals pre-Golgi requirements for Sec7p in yeast membrane traffic. Biochem. Biophys. Res. Commun. 243: 191–198. [DOI] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L.-L., Pei B.-L., Zhou Q., Li Y.-Z. (2012). NO serves as a signaling intermediate downstream of H2O2 to modulate dynamic microtubule cytoskeleton during responses to VD-toxins in Arabidopsis. Plant Signal. Behav. 7: 174–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Sato K., Takeuchi M. (2014). Molecular mechanisms of Sar/Arf GTPases in vesicular trafficking in yeast and plants. Front. Plant Sci. 5: 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeh J.-C., Zeghouf M., Grauffel C., Guibert B., Martin E., Dejaegere A., Cherfils J. (2006). Dual specificity of the interfacial inhibitor brefeldin a for arf proteins and sec7 domains. J. Biol. Chem. 281: 11805–11814. [DOI] [PubMed] [Google Scholar]
- Zwiewka M., Friml J. (2012). Fluorescence imaging-based forward genetic screens to identify trafficking regulators in plants. Front. Plant Sci. 3: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]