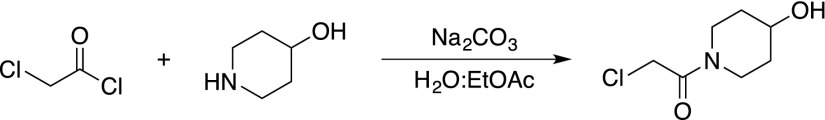A chemical genetic approach uncovered the small molecule Secdin, which impairs multiple endomembrane trafficking routes by targeting the Arabidopsis ARF GTPase exchange factors.
Abstract
Small GTP-binding proteins from the ADP-ribosylation factor (ARF) family are important regulators of vesicle formation and cellular trafficking in all eukaryotes. ARF activation is accomplished by a protein family of guanine nucleotide exchange factors (GEFs) that contain a conserved catalytic Sec7 domain. Here, we identified and characterized Secdin, a small-molecule inhibitor of Arabidopsis thaliana ARF-GEFs. Secdin application caused aberrant retention of plasma membrane (PM) proteins in late endosomal compartments, enhanced vacuolar degradation, impaired protein recycling, and delayed secretion and endocytosis. Combined treatments with Secdin and the known ARF-GEF inhibitor Brefeldin A (BFA) prevented the BFA-induced PM stabilization of the ARF-GEF GNOM, impaired its translocation from the Golgi to the trans-Golgi network/early endosomes, and led to the formation of hybrid endomembrane compartments reminiscent of those in ARF-GEF-deficient mutants. Drug affinity-responsive target stability assays revealed that Secdin, unlike BFA, targeted all examined Arabidopsis ARF-GEFs, but that the interaction was probably not mediated by the Sec7 domain because Secdin did not interfere with the Sec7 domain-mediated ARF activation. These results show that Secdin and BFA affect their protein targets through distinct mechanisms, in turn showing the usefulness of Secdin in studies in which ARF-GEF-dependent endomembrane transport cannot be manipulated with BFA.
INTRODUCTION
The small GTP-binding proteins from the ADP-ribosylation factor (ARF) family are major regulators of vesicle biogenesis and intracellular trafficking in all eukaryotes, including plants (Donaldson and Jackson, 2011; Yorimitsu et al., 2014). Like other small GTPases, the ARF proteins are controlled by a GTP-binding and GTP hydrolysis cycle that activates and inactivates them, respectively. ARF activation is facilitated by the ARF guanine-nucleotide exchange factors (ARF-GEFs), whereas ARF inactivation through GTP hydrolysis is accomplished by the ARF GTPase-activating proteins (ARF-GAPs) (Donaldson and Jackson, 2011). A distinctive feature of plant ARFs, ARF-GEFs, and ARF-GAPs is their high level of diversification, which is probably associated with the increased degree of developmental plasticity that plants have developed to cope with adverse environmental conditions (Vernoud et al., 2003; Yorimitsu et al., 2014). The ARF-GEFs contain a catalytic Sec7 domain and, according to their additional protein domains, they are classified into different families, including large ARF-GEFs as well as proteins of lower molecular weight, such as BRAGs, EFA6, and cytohesins (Jackson and Casanova, 2000; Wright et al., 2014). In contrast to animals, all plant Sec7 domain-containing proteins fall into the family of large ARF-GEFs, which are divided into two subfamilies: GBF/Gea and BIG/Sec7 (Bui et al., 2009). The Arabidopsis thaliana genome encodes three members of the GBF/Gea subfamily (GNOM, GNOM-LIKE1 [GNL1], and GNL2) and five BIG/Sec7-related proteins (BIG1 to BIG5). Besides the catalytic Sec7 domain, the large ARF-GEFs share several other conserved protein motifs that have regulatory functions as protein-protein interaction platforms and define their endomembrane localization (Mouratou et al., 2005; Bui et al., 2009; Wright et al., 2014; Nawrotek et al., 2016). Much experimental evidence indicates that members of the two protein subfamilies of Arabidopsis ARF-GEFs have overlapping functions and can complement each other. For instance, GNOM and GNL1 play redundant roles in trafficking between the endoplasmic reticulum and the Golgi apparatus, with GNOM also having additional plant-specific functions in plasma membrane (PM) protein recycling and endocytosis (Geldner et al., 2003; Richter et al., 2007; Naramoto et al., 2010), whereas BIG1 to BIG4 are redundant late secretion regulators from the trans-Golgi network/early endosome (TGN/EE) (Richter et al., 2014).
As classical genetic approaches are often inapplicable when dealing with gene redundancy, the use of small-molecule inhibitors is an alternative for probing protein functions and discovering novel regulatory mechanisms of intracellular vesicle transport (Norambuena and Tejos, 2017). The pharmacological toolbox for interference with ARF-GEF protein functions in yeast and animals comprises both natural and synthetic compounds identified through numerous chemical screens (Benabdi et al., 2017). Despite the large number of available molecules that have been meticulously analyzed for their impact on intracellular protein transport, most of them have not been characterized in terms of molecular mode of action. Furthermore, the phenotypes induced by each drug most often can be explained by the unequal efficiency of the compounds toward different members of the ARF-GEF family (Benabdi et al., 2017). By contrast, in plants, the most commonly used ARF-GEF inhibitor thus far is the fungal toxin Brefeldin A (BFA). BFA is an uncompetitive inhibitor that acts through reversible binding to the interface of the catalytic Sec7 domain in complex with ARF1-GDP (Mossessova et al., 2003; Renault et al., 2003). Overall, the number of available small-molecule modulators of the endomembrane system in plant cells is still limited (Mishev et al., 2013; Norambuena and Tejos, 2017). Only recently, attempts have been successful in defining the mode of action of several chemical intracellular trafficking modifiers discovered through plant-oriented chemical genetic screens. Small molecules from the Endosidin (ES) series were proven to interfere with endocytosis and with different steps along the exocytic pathway from the endoplasmic reticulum to the PM by perturbing known protein regulators of endomembrane transport (Doyle et al., 2015; Dejonghe et al., 2016; Zhang et al., 2016; Li et al., 2017). Here, we report on the discovery of a small molecule that alters both secretory and endocytic vesicle-trafficking routes in plant cells. Genetic and biochemical evidence in Arabidopsis revealed that both BFA-sensitive and BFA-insensitive ARF-GEFs are the molecular targets of this chemical inhibitor.
RESULTS
Identification of a Small-Molecule Inhibitor of Endomembrane Trafficking
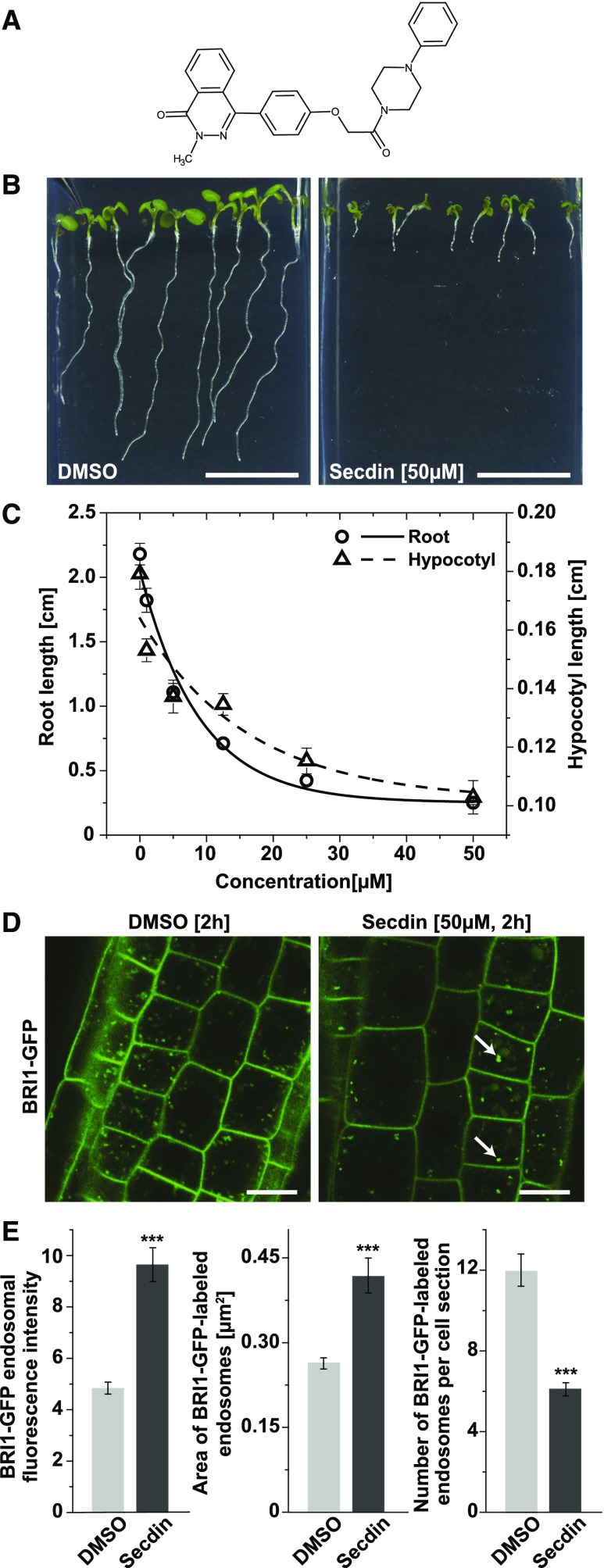
To identify small-molecule inhibitors of the endomembrane trafficking of the brassinosteroid (BR) receptor BR-INSENSITIVE1 (BRI1) (Friedrichsen et al., 2000), we screened 10,000 compounds (DIVERSet ChemBridge library) for hypocotyl growth inhibition in the light and for a localization change of GFP-tagged BRI1 in root epidermal cells of Arabidopsis (Codreanu et al., 2012). The primary screen resulted in the identification of 196 hypocotyl growth inhibitors, of which 10 structurally different compounds caused mislocalization of BRI1. One bioactive molecule, 2‐methyl‐4‐{4‐[2‐oxo‐2‐(4‐phenylpiperazin‐1‐yl)ethoxy]phenyl}‐1,2‐dihydrophthalazin‐1‐one, also designated Secdin for Sec7 domain-containing ARF-GEF inhibitor (Figure 1A), was further characterized. Secdin (50 µM) did not affect seed germination, but it substantially impaired both hypocotyl and root growth in young Arabidopsis seedlings at the half-maximal inhibitory concentration (IC50) of 4 µM (Figures 1B and 1C). Application of Secdin at the initial screening concentration (50 µM, 2 h) to Arabidopsis roots expressing BRI1-GFP (Friedrichsen et al., 2000) in the light led to the visible accumulation of a GFP signal in the vacuole and the rapid formation of abnormal GFP-positive intracellular compartments, termed “Secdin bodies” (Figure 1D). The latter were characterized by their stronger BRI1-GFP fluorescence intensity and larger size than those of the GFP-positive endosomes in mock-treated root cells (Figure 1E). Notably, the aberrant intracellular localization of BRI1-GFP was reverted to the control upon washout of the compound (Supplemental Figure 1A). The defects in endomembrane trafficking induced by Secdin were not restricted to BRI1 because several other PM proteins, including the fluorescently tagged auxin transporters PIN-FORMED1 (PIN1) (Wiśniewska et al., 2006) and PIN2 (Xu and Scheres, 2005), as well as the boron transporter BOR1 (Takano et al., 2010), accumulated in enlarged intracellular compartments in the root epidermis in the presence of the compound (50 µM, 1.5 h) (Supplemental Figure 1B). Interestingly, other tested PM proteins, such as PM INTRINSIC PROTEIN2A (PIP2A) (Cutler et al., 2000) and BR-SIGNALING KINASE1 (BSK1) (Tang et al., 2008), were insensitive to Secdin, presumably because of differences in their turnover rate and/or trafficking routes (Supplemental Figure 1B). Treatment with Secdin (50 µM, 1 h) did not induce ATP depletion or metabolic changes in cell suspension cultures of Arabidopsis, and no acidification was observed in Secdin-treated seedlings (Supplemental Figures 2A and 2B). Furthermore, Secdin (50 µM, 1.5 h) neither disrupted the cortical microtubules (Supplemental Figure 2C) nor the actin filaments (Supplemental Figure 2D), thus ruling out cytotoxic effects for the treatment dose and time used throughout this study.
Figure 1.
The Plant Growth Inhibitor Secdin Affects the Intracellular Localization of the BR Receptor BRI1.
(A) Chemical structure of Secdin.
(B) Growth phenotype of 6-d-old wild-type (Col-0) seedlings germinated and grown in the presence of Secdin (50 µM).
(C) Dose-response analysis of primary root and hypocotyl growth of 6-d-old seedlings germinated on different Secdin concentrations (IC50 = 4 µM). Values are means (n = 20 seedlings for DMSO and 18 for Secdin). Error bars indicate se.
(D) Mislocalization of BRI1-GFP in epidermal cells of root tips upon treatment with Secdin. Four-day-old BRI1-GFP seedlings were treated with either DMSO (0.5%) or Secdin (50 µM) for 2 h and imaged. White arrows mark Secdin bodies.
(E) Quantification of the BRI1-GFP fluorescence intensity as well as size and number of BRI1-GFP-positive endomembrane compartments in epidermal cells of root tips upon treatment with Secdin (50 µM, 2 h). Values are means of analyzed cells (n = 83 and n = 72 for DMSO and Secdin, respectively). Error bars indicate se. ***P < 0.001 (Student’s t test) relative to the DMSO control.
Bars = 1 cm in (B) and 10 µm in (D).
Secdin Retains PM Proteins in the Late Endosomal Compartments and Facilitates Their Vacuolar Degradation
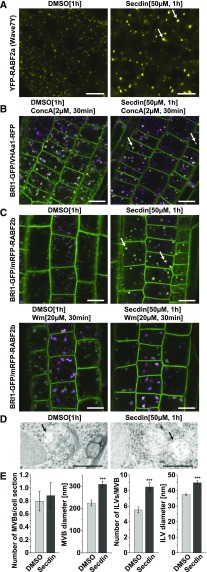
To characterize the compartments that accumulated PM proteins in the presence of Secdin, we explored the intracellular localization of a set of fluorescently tagged protein markers that labeled different endomembrane compartments in plant cells. Secdin (50 µM, 1.5 h) did not change the localization of Golgi-resident sialyltransferase (ST) fused to monomeric RFP (mRFP) (Wee et al., 1998), the TGN/EE marker vacuolar ATPase subunit a1 (VHAa1)-RFP (Dettmer et al., 2006), or the syntaxin 61 (SYP61)-CFP (Robert et al., 2008) (Supplemental Figure 1C). However, the distribution of two multivesicular body (MVB)/late endosome (LE)-localized RAB GTPases (i.e., RABF2a and RABF2b) (Ueda et al., 2004; Geldner et al., 2009) was sensitive to Secdin (Figure 2A; Supplemental Figure 1C), suggesting that the Secdin bodies are related to MVB/LEs. Unlike mock-treated plants, in the presence of Secdin, the vacuoles decorated by SYP22-YFP (Robert et al., 2008) were characterized by a rounded shape (Supplemental Figure 1C). Next, we probed different endomembrane compartments for their sensitivity to Secdin in combination with drugs known to selectively inhibit different vesicular trafficking routes in plant cells. To this end, seedlings coexpressing BRI1-GFP with various endomembrane markers were first treated with Secdin (50 µM, 0.5 h) to induce the formation of Secdin bodies and then challenged with specific inhibitors of TGN/EEs or MVB/LEs in the presence of Secdin for 0.5 h. Application of Concanamycin A (ConcA) (2 µM, 0.5 h), a specific vacuolar H+-ATPase inhibitor that blocks endocytic transport to the tonoplast from the TGN/EEs (Dettmer et al., 2006; Viotti et al., 2010), after pretreatment with Secdin had no effect on Secdin bodies, which remained spatially separated from ConcA-affected organelles colabeled by BRI1-GFP and the TGN/EE marker VHAa1-RFP (Figure 2B). In contrast, the phosphatidylinositol-3-kinase inhibitor Wortmannin (Wm), which causes swelling of MVB/LEs (Wang et al., 2009; Viotti et al., 2010) (20 µM, 0.5 h), provoked swelling of Secdin bodies colabeled with mRFP-RABF2b and BRI1-GFP, similarly to the control without Secdin (Figure 2C). Consistent with the results of confocal imaging, transmission electron microscopy (TEM) of high-pressure frozen/freeze-substituted roots showed that the Secdin treatment (50 µM, 1 h) largely modified the MVB/LE morphology (Figure 2D). Whereas the average number of MVB/LEs per cell section remained unchanged, quantitative analysis indicated a substantial increase in the average MVB/LE size, as well as in number and dimension of the intraluminal vesicles (ILVs) (Figure 2E).
Figure 2.
Secdin Affects the Morphology of MVBs in the Root Epidermal Cells of Arabidopsis.
(A) Secdin-induced accumulation of the MVB-resident RABF2a GTPase (also known as Rha1) in enlarged endomembrane compartments (white arrows).
(B) Treatment with the V-ATPase inhibitor ConcA, in the presence of Secdin, altering the morphology of the VHAa1-RFP-labeled TGN/EEs, but not affecting the BRI1-GFP-positive Secdin compartments (white arrows).
(C) Treatment with the phosphoinositide 3-kinase inhibitor Wm, in the presence of Secdin, enlarging the RABF2b GTPase (also known as ARA7)- and BRI1-GFP-positive MVBs.
(D) Ultrastructural changes in MVBs in root epidermal cells of wild-type (Col-0) seedlings treated with DMSO (0.5%) or Secdin (50 µM) (black arrows) as revealed by TEM.
(E) Quantification of MVB-related structural parameters derived from TEM micrographs. Values are means of n = 23 (DMSO) and n = 34 (Secdin) cells analyzed for MVB number and diameter; n = 37 (DMSO) and n = 75 (Secdin) MVBs analyzed for number of ILVs; n = 104 (DMSO) and n = 120 (Secdin) ILVs analyzed for their diameter. Error bars indicate se. ***P < 0.001 (Student’s t test) relative to the DMSO control.
Bars = 10 µm (A) to (C) and 1 µm in (D).
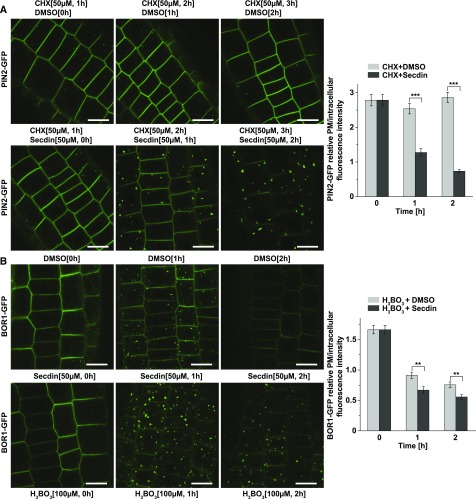
The formation of Secdin bodies was more pronounced when Secdin (50 µM, 1–2 h) was coapplied with the protein synthesis inhibitor cycloheximide (CHX) to PIN2-GFP-expressing seedlings, coinciding with a significant reduction in PM fluorescence (Figure 3A). Similarly, upon induction with boric acid and in the presence of Secdin (50 µM, 1–2 h), BOR1-GFP became depleted more rapidly from the PM than the control, but remained longer in the Secdin bodies prior to degradation into the vacuole (Figure 3B). The prominent accumulation of different PM proteins in Secdin bodies prompted us to test whether Secdin affected their vacuolar degradation. Indeed, prolonged treatment with Secdin (50 µM, 6 h) in the dark, which allows stabilization of the GFP fluorescence inside the vacuole (Tamura et al., 2003), led to stronger BRI1-GFP and PIN2-GFP signals in the vacuole than those in the mock-treated controls (Supplemental Figures 3A and 3B). In agreement, protein gel blot analysis revealed a slight decrease in total BRI1 protein content in wild-type seedlings in the presence of Secdin (50 µM, 5 h) (Supplemental Figure 3C). Altogether, our data suggest that Secdin retained the PM proteins in the MVB/LEs and that longer Secdin treatments facilitated their vacuolar degradation.
Figure 3.
Secdin Retains PM Proteins in the Late Endosomal Compartments.
(A) Treatment with Secdin in the presence of CHX induced a rapid decrease in the PM pool of PIN2-GFP with a concomitant increase in the intracellular protein fraction. At least 35 cells per treatment were analyzed for quantification of the PM/intracellular PIN2-GFP fluorescence intensity ratio.
(B) Time course of BOR1-GFP internalization under high-boron conditions in the presence of Secdin. BOR1-GFP-expressing seedlings were grown on low-boron (0.3 µM H3BO3) solid medium and then incubated with 100 µM H3BO3 in liquid medium to induce internalization in the presence of either DMSO (0.5%) or Secdin (50 µM). At least 40 cells per treatment were analyzed to assess the PM/intracellular BOR1-GFP fluorescence intensity ratio. Error bars indicate se. ***P < 0.001 and ** P < 0.01 (Student’s t test) relative to the DMSO control. Bars = 10 µm.
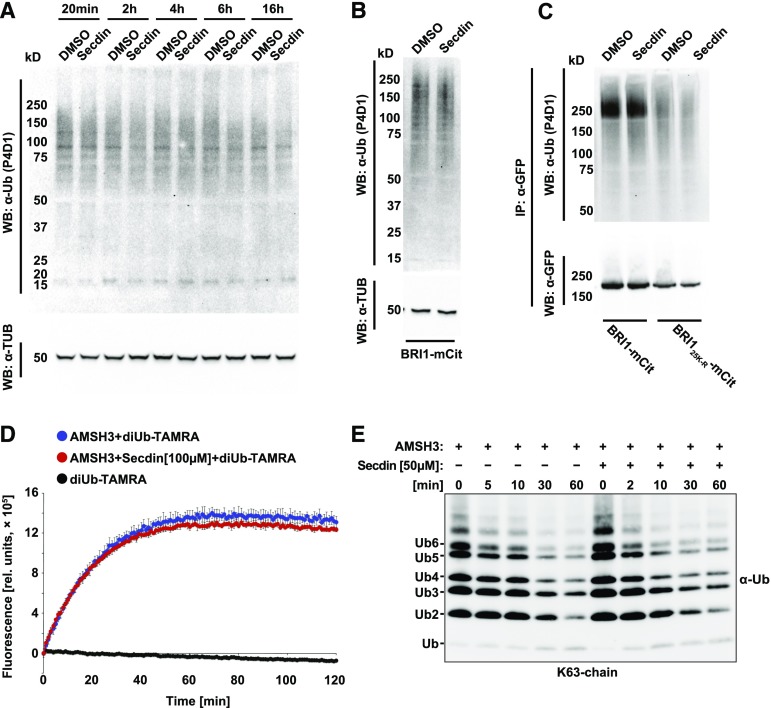
A key molecular determinant for PM protein sorting for vacuolar degradation is ubiquitination (Korbei and Luschnig, 2013). Given that a large fraction of BRI1-GFP and other PM proteins were detected in the MVB/LEs and vacuoles of Secdin-incubated seedlings, we tested whether the compound affected the ubiquitination/deubiquitination machinery. Treatment with Secdin for different periods of time (50 µM, up to 16 h) affected neither the ubiquitination pattern of total protein extracts from compound-treated seedlings nor the ubiquitination of the BRI1 receptor (Figures 4A to 4C). Similarly, Secdin (50 or 100 µM) did not affect the enzymatic activity of the deubiquitinating enzyme AMSH3 (Isono et al., 2010) (Figures 4D and 4E), indicating that the mode of action of Secdin is not directly related to the ubiquitin machinery. However, in the presence of Secdin (50 µM, 4–6 h) in the dark, the ubiquitination- and endocytosis-deficient mutant proteins BRI125K-R (Martins et al., 2015) and PIN212K-R (Leitner et al., 2012) were never observed in either Secdin bodies or vacuoles (Supplemental Figures 4A and 4B), suggesting that the PM protein accumulation in MVB/LEs and vacuoles caused by Secdin depends on ubiquitination signals that enable cargo internalization.
Figure 4.
Untargeted Ubiquitin Machinery by Secdin.
(A) Total protein ubiquitination pattern of wild-type (Col-0) seedlings treated with Secdin (50 µM) at different times and subjected to protein gel blot (Western blot [WB]) analysis with an α-Ub P4D1 antibody. Tubulin (TUB) (α-TUB antibody) was used as a loading control.
(B) Total microsomal protein ubiquitination pattern of DMSO- or Secdin-treated BRI1-mCitrine (mCit)-expressing seedlings detected with an α-Ub P4D1 antibody. Tubulin was used as a loading control.
(C) Protein gel blot analysis of the ubiquitination pattern of immunoprecipitated (IP) BRI1-mCit and BRI125K-R-mCit after microsomal protein isolation. Detection with an α-GFP antibody recognizing the mCit tag was done to estimate the BRI1-mCit protein loading. All chemical treatments ([B] and [C]) were done for 5 h in the presence of MG132 (50 µM) to enrich for ubiquitinated proteins.
(D) Unaffected AMSH3 DUB activity by Secdin. Fluorescence-based DUB assay with diubiquitin TAMRA (diUb-TAMRA) was performed with (red) or without (blue) preincubation of AMSH3 with Secdin (100 µM). DiUb-TAMRA alone was used as a negative control (black).
(E) Protein gel blot detection of AMSH3-DUB activity with K63-linked polyubiquitin (Ub2-7) chains. For the DUB assay, AMSH3 was incubated with the polyubiquitin chains with or without 10-min preincubations with 50 µM Secdin. The assay was terminated at the indicated time points and the reaction mixture was subjected to protein gel blot analysis with an α-Ub P4D1 antibody.
Secdin Impairs Secretion and Endocytosis
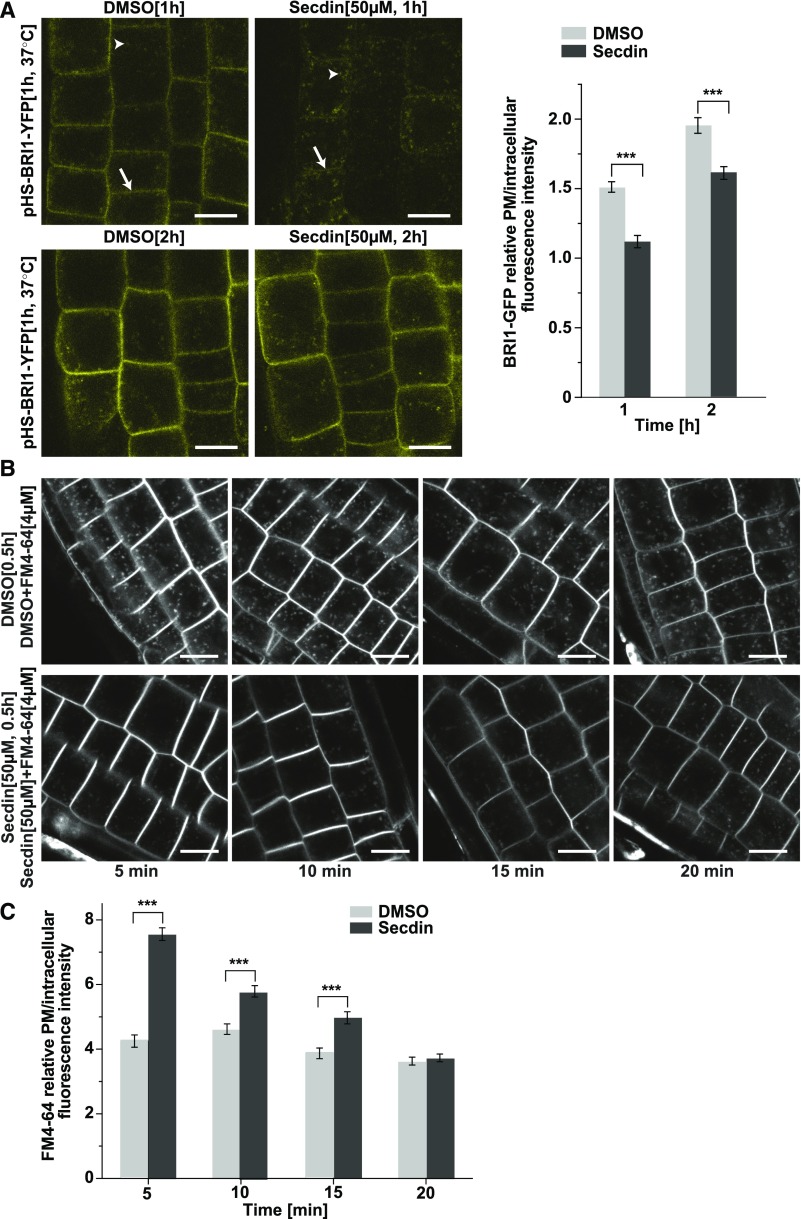
Protein abundance at the PM results from the coordinated contributions of exocytosis of de novo-synthesized proteins and their internalization by endocytosis followed by either recycling or degradation (Peer, 2011). We checked whether the targeting of PM proteins to the degradation pathway after treatment with Secdin was related to changes in either secretion or endocytosis. First, we evaluated the effect of Secdin on the secretory flux to the PM by using heat shock-inducible BRI1-YFP-expressing Arabidopsis plants (Geldner et al., 2007). Chemical treatments (Secdin, 50 µM, 1–2 h) were performed immediately after heat shock (1 h at 37°C) in the presence of Actinomycin D (20 µg/mL) (Narsai et al., 2007) to ensure the lack of transcription after the heat shock. Whereas the BRI1-YFP fluorescence signal was clearly discernible at the PM in control seedlings 1 h after the heat shock, in Secdin-treated samples, BRI1-YFP mainly accumulated intracellularly (Figure 5A). However, the abundance of BRI1-YFP in the PM after a 2-h exposure to Secdin was still lower but comparable to that in control seedlings (Figure 5A), indicating that Secdin delays but does not block the secretory pathway to the PM.
Figure 5.
Impaired Secretion and Endocytosis by Secdin.
(A) De novo BRI1-YFP protein synthesis upon heat shock (HS) induction of the BRI1-YFP gene expression for 1 h at 37°C, followed by incubation for 1 or 2 h at room temperature in liquid medium supplemented with either DMSO (0.5%) or Secdin (50 µM). Treatments were done in the presence of Actinomycin D (20 µg/mL) to ensure the absence of any transcription after the heat shock. White arrows indicate the PM fluorescent signal; arrowheads point to BRI1-YFP-positive endomembrane compartments. At least 20 (1 h) or 28 (2 h) cells per treatment were analyzed for quantification of the PM/intracellular BRI1-YFP fluorescence intensity ratio.
(B) Time-course changes in the endocytic uptake of the styryl dye FM4-64 (4 µM) in the presence of DMSO (0.5%) or Secdin (50 µM), following preincubation with the Secdin (50 µM) for 30 min. Experiments were done with wild type (Col-0) seedlings.
(C) Quantification of the PM/intracellular FM4-64 fluorescence intensity. At least 16 cells were analyzed for each treatment and time point. Error bars in (A) and (C) indicate se. ***P < 0.001 (Student’s t test) relative to the DMSO control. Bars = 10 µm.
Next, we explored the endocytosis rate by assessing the uptake of the endocytic tracer N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide (FM4-64) at different time intervals in the presence of Secdin (50 µM) (Figures 5B and 5C). At the early time points (5–15 min), FM4-64 internalization was delayed compared with mock-treated seedlings. Yet, FM4-64 uptake did not differ between Secdin-treated and control seedlings at 20 min. Together, these results demonstrate that Secdin delays both the protein secretion to the PM and the endocytosis.
Secdin Interferes with the BFA Impact
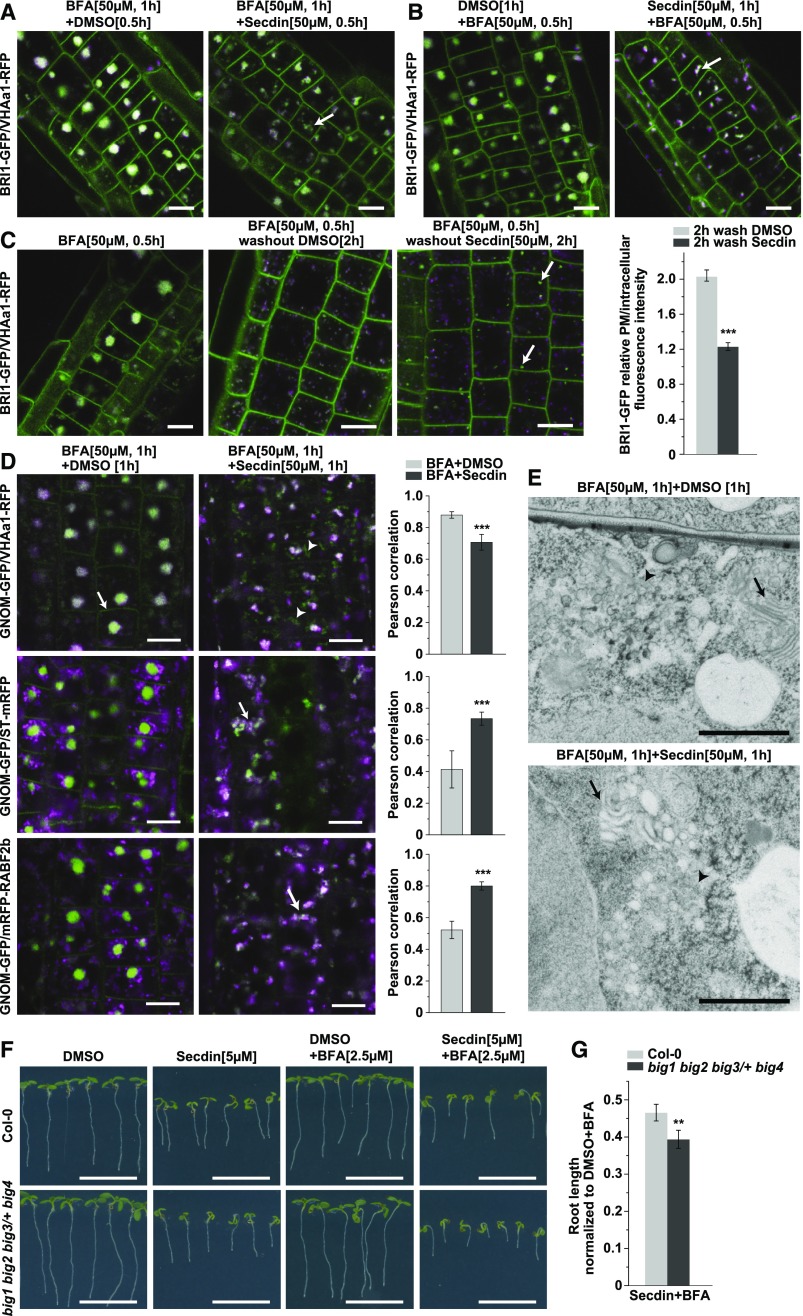
Recycling of BRI1 and PIN proteins depends on the function of the ARF-GEF GNOM, which is sensitive to the fungal toxin BFA (Geldner et al., 2003, 2007). We tested whether Secdin affected the vesicle trafficking processes mediated by BFA-sensitive ARF-GEFs by analyzing the combined effect of the two compounds on the endomembrane network. Coapplication of BFA (50 µM) and Secdin (50 µM) to BRI1-GFP/VHAa1-RFP-expressing seedlings for 0.5 h after pretreatment with either BFA (50 µM, 0.5 h) or Secdin (50 µM, 0.5 h) led to the agglomeration of BRI1-GFP and VHAa1-RFP (Figures 6A and 6B). However, these agglomerates differed from typical BFA bodies. Furthermore, BFA washout experiments in the presence of Secdin (50 µM, 2 h) led to the disappearance of agglomerates, like the control, but the released protein cargo was mostly redirected to degradation rather than recycled back to the PM, with a significant reduction in PM fluorescence and the appearance of Secdin bodies as a consequence (Figure 6C). To gain insight into the nature of the combined effects of BFA and Secdin, we examined the localization of GNOM-GFP together with different endomembrane markers (Figure 6D). In root cells, BFA treatment is known to induce both the translocation of GNOM from the Golgi to the TGN/EE membranes situated in the core of BFA bodies (Naramoto et al., 2014) and an increase in the GNOM pool at the PM (Naramoto et al., 2010). Application of Secdin and BFA together substantially reduced the PM-resident GNOM-GFP pool and led only to the partial colocalization of GNOM-GFP and VHAa1-RFP-labeled endomembranes (Figure 6D). GNOM-GFP and the Golgi-resident ST-mRFP nearly overlapped, as did GNOM-GFP and mRFP-RABF2b, which labels a TGN/EE subpopulation that would eventually mature into MVB/LEs (Scheuring et al., 2011) (Figure 6D). Hence, Secdin interfered with the BFA-induced translocation of GNOM to the TGN/EEs and led to the formation of hybrid structures of Golgi-TGN/EE origin.
Figure 6.
Secdin Interferes with the Effects of BFA.
(A) and (B) Secdin treatment (50 µM) in the presence of BFA (50 µM; [A]) or BFA treatment (50 µM) in the presence of Secdin (50 µM; [B]) altering the BFA body morphology (arrows indicate a BFA-Secdin endomembrane compartment).
(C) BFA washout for 2 h in the presence of either Secdin (50 µM) or DMSO (0.5%). Note the disappearance of the BFA bodies in both treatments but the formation of BRI1-GFP-positive Secdin compartments (arrows) and the reduced BRI1-GFP PM fluorescence intensity in the presence of Secdin. The PM/intracellular BRI1-GFP fluorescence intensity ratio was calculated for 38 cells in each treatment.
(D) Incubation for 1 h with BFA (50 µM) and Secdin (50 µM) inducing the formation of GNOM-GFP-positive compartments that were not labeled by VHAa1-RFP (arrowheads). Note the GNOM-GFP fluorescence at the PM induced by the BFA treatment (arrow) and its absence in the presence of Secdin. Unlike the DMSO controls, the combined chemical treatment led to a complete overlap of GNOM-GFP with the Golgi marker ST-mRFP (arrow) and the late endosomal marker mRFP-RABF2b (also known as ARA7, arrow). The colocalization between the GFP and RFP markers in the cytoplasm was assessed by calculating the Pearson’s correlation coefficient (rp). The mean values from at least 40 cells per treatment and genotype are presented.
(E) TEM analysis of Arabidopsis root tips treated with BFA (50 µM) and Secdin (50 µM) for 1 h. Note the lack of clear distinction between the TGN-derived vesicles (arrowheads) and the Golgi cisternae (arrows) that became rounded and dilated in the presence of the two chemicals.
(F) Growth phenotype of 7-d-old Arabidopsis big1 big2 big3/+ big4 mutant seedlings and the wild type (Col-0) germinated on solid medium without sucrose supplemented with Secdin (5 µM) alone or in combination with BFA (2.5 µM).
(G) Quantification of the primary root length of seedlings grown under the conditions in (F).
Values (n = 85 seedlings per treatment and genotype) are means from two biological replicates, i.e., independent samplings of plants. Error bars in (C), (D), and (G) indicate se. ***P < 0.001 and **P < 0.01 (Student’s t test). Bars = 10 µm in (A) to (D), 1 µm in (E), and 1 cm in (F).
We then analyzed the ultrastructure of BFA-Secdin-induced compartments via TEM. In root epidermal cells, BFA caused the typical and expected morphological changes, i.e., the aggregation of TGN/EE-derived vesicles into large clusters surrounded by Golgi stacks (Figure 6E). However, upon the combined treatment with BFA and Secdin, the membranous clusters were elongated and less compact. Unlike the BFA-treated control, the agglomerates contained dilated Golgi stacks mixed with TGN membranes (Figure 6E), and no recognizable MVBs could be detected. Such phenotypes are reminiscent of those observed in the BFA-treated ben1-1 mutant deficient in the ARF-GEF BIG5 (Tanaka et al., 2009). Collectively, these results indicate that the Secdin-induced intracellular changes might be related to the perturbation of ARF-GEF protein functions. Indeed, Arabidopsis mutants deficient in BIG1, BIG2, and BIG4, but still expressing the BFA-resistant BIG3 protein, were hypersensitive to the combined treatment with Secdin and BFA compared with wild-type seedlings (Figures 6F and 6G).
Secdin Targets All Examined Arabidopsis ARF-GEFs
To identify the molecular targets of Secdin, we utilized an affinity purification method with biotinylated Secdin. The easily distinguishable intracellular phenotype caused by Secdin allowed us to quickly evaluate the biological activity of 21 Secdin derivatives and closely related analogs (Supplemental Figure 5A), among which Secdin17 and Secdin21 were identified as the active analogs with the lowest molecular weights. Interestingly, modifications at the piperidine ring of Secdin17 that conserved its hydrophobic nature (Secdin13, Secdin14, Secdin20, and Secdin21) preserved its activity, whereas increased hydrophilicity at this site (Secdin1, Secdin12, and Secdin18) markedly diminished or abolished this activity (Supplemental Figure 5A). Replacement of the piperidine ring with a smaller (but hydrophobic) pyrrolidine ring (Secdin3) also resulted in a complete loss of activity. Having identified the minimal active scaffold required for Secdin activity, we also introduced additional groups on the bioactive scaffold to allow us to attach an affinity purification label, such as biotin. In addition to the piperidine ring, the central functionalized phenol ring was also found to be a suitable position for further substitution while preserving activity (Secdin22), which resulted in the design and preparation of the biotin conjugate Secdin24 (Supplemental Figure 5B and Supplemental Data Set 1), further utilized for affinity purification. Total protein extracts prepared from Arabidopsis PSB-D wild-type cell cultures were treated with Secdin24 and with free biotin as a negative control, and bound proteins were analyzed by mass spectrometry (Supplemental Data Set 2). We could not detect statistically significant enrichment of proteins interacting with Secdin24 compared with the biotin control, possibly the reason for the wide differences in protein ranking in the two independent pull-down experiments. Nonetheless, the presence of the ARF-GEF BIG5 (AT3G43300) in both experiments (Supplemental Data Set 2) suggests that Secdin24 may interact with BIG5 in vivo.
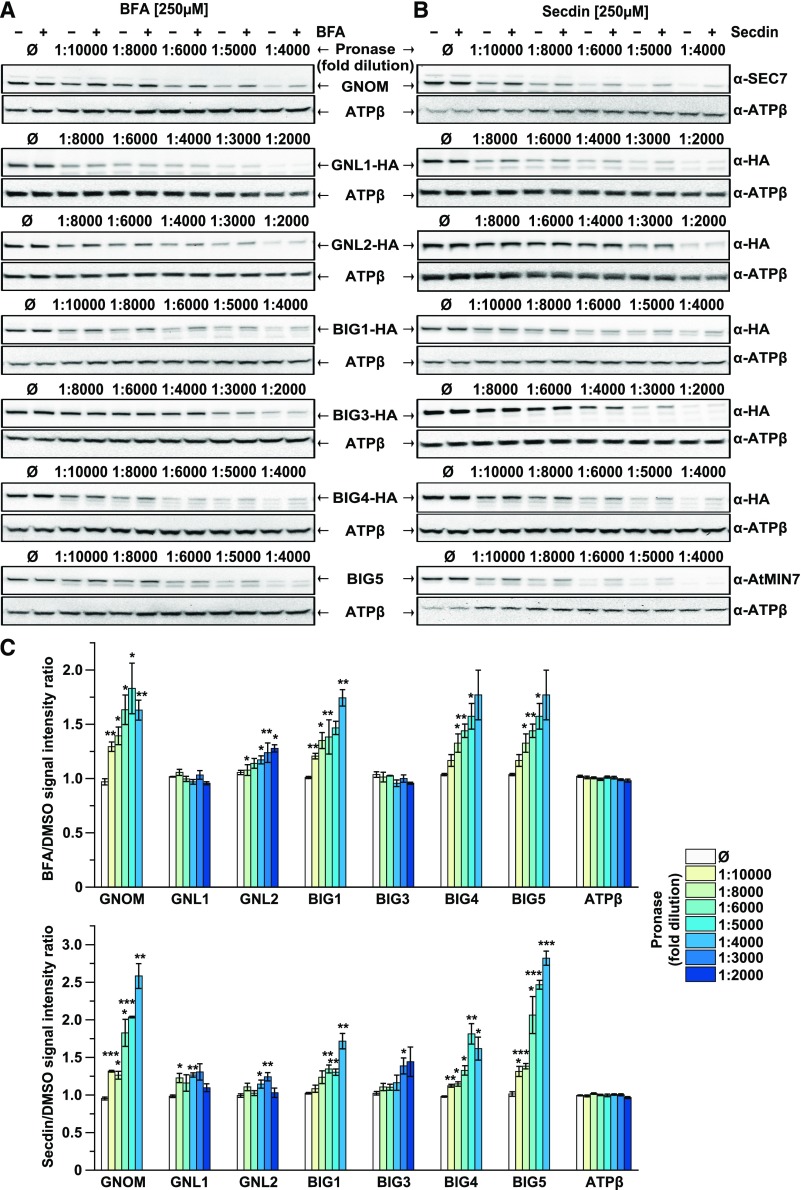
To validate whether the Arabidopsis ARF-GEFs were potential protein targets of Secdin, we took advantage of the drug affinity-responsive target stability (DARTS) assay (Lomenick et al., 2009) and checked the proteolytic degradation of all Arabidopsis ARF-GEFs, excluding BIG2 due to the low expression levels of the gene construct, in the presence of Secdin in lysates of Arabidopsis cell cultures. We used specific antibodies to detect GNOM and BIG5 in wild-type cell cultures and generated cell lines expressing hemagglutinin (HA)-tagged ARF-GEF versions to allow protein detection with anti-HA antibodies. BFA was used as a positive control (Mossessova et al., 2003; Renault et al., 2003) (Figures 7A and 7C). As expected, treatment with this compound protected all BFA-sensitive ARF-GEFs, but not BFA-resistant GNL1 (Richter et al., 2007; Teh and Moore, 2007) and BIG3 (Nielsen et al., 2006; Richter et al., 2014), from pronase-induced degradation. In contrast, Secdin protected all examined ARF-GEFs from degradation (Figures 7B and 7C). In addition, the biologically inactive Secdin analog, Secdin15 (Supplemental Figure 5A), had no effect on the stability of the two control ARF-GEFs, i.e., GNOM and BIG5 (Supplemental Figure 6A). Interestingly, neither BFA nor Secdin affected ARF1 (Supplemental Figure 6B). The lack of selectivity in Secdin binding compared with BFA allowed us to hypothesize that Secdin and BFA have different binding sites in the ARF-GEFs. Indeed, the engineered BFA-resistant version BIG4R-YFP (Richter et al., 2014) was digested equally by pronase in the presence or absence of BFA (Supplemental Figure 6C). Conversely, Secdin induced stabilization of both the BFA-sensitive and BFA-resistant versions of BIG4-YFP (Supplemental Figure 6C). Overall, the DARTS experiments suggest that all tested Arabidopsis ARF-GEFs are Secdin “protein” targets and that the binding site and binding mechanism of Secdin differ from those of BFA.
Figure 7.
Secdin Targets Both BFA-Sensitive and BFA-Resistant Arabidopsis ARF-GEFs.
(A) and (B) DARTS analysis of the protein susceptibility to proteolytic degradation in the presence of BFA and Secdin, respectively. Total protein extracts from Arabidopsis PSB-D cell cultures were incubated with BFA (250 µM) or Secdin (250 µM) and then challenged with different pronase dilutions. The ARF-GEF protein levels were detected by protein gel blot (WB) analysis. ATP synthase subunit β (ATPβ) was used as a control.
(C) Quantification of the protein band intensity relative to the DMSO control. The analysis was done with representative immunoblots from at least three biological repeats, i.e., independent sampling of cell cultures. Error bars indicate se. ***P < 0.001, **P < 0.01, and *P < 0.05 (Student’s t test).
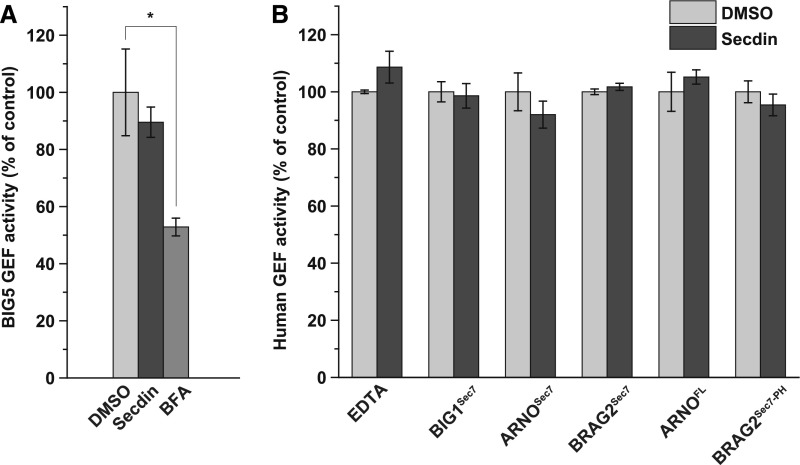
BFA is an uncompetitive inhibitor of the catalytic Sec7 domain of BFA-sensitive ARF-GEFs in yeast, mammals, and plants (Geldner et al., 2003; Mossessova et al., 2003; Renault et al., 2003). Hence, using a fluorescence kinetics-based assay with purified ARF GTPases and ARF-GEFs (Benabdi et al., 2017), we tested whether Secdin could interfere with the activity of selected plant and human Sec7 domains (Figures 8A and 8B; Supplemental Figure 7), including the Sec7 domains of the BFA-sensitive Arabidopsis and the human large ARF-GEFs, BIG5 and BIG1, respectively, and the Sec7 domains of the BFA-resistant human small ARF-GEFs, BRAG2 and cytohesin-2/ARNO. The Sec7 domains of the human ARF-GEFs had a 29 to 52% sequence identity with the Sec7 domains of the eight Arabidopsis ARF-GEFs, corresponding to the sequence identity between the Arabidopsis BIG and GNOM/GNL groups (38–48%) (Mouratou et al., 2005). Through similar assays, the human protein constructs used in our study had already been demonstrated to be effectively impaired by known ARF-GEF inhibitors (Benabdi et al., 2017). Like the human BIG1 Sec7 (Zeeh et al., 2006; Benabdi et al., 2017), we found that BFA strongly inhibited the purified Arabidopsis BIG5 Sec7 domain (Figure 8A; Supplemental Figure 7). In contrast, Secdin had no significant effect on the activity of any of the tested plant and human Sec7 domains toward the ARF1 substrate (Figures 8A and 8B; Supplemental Figure 7). Also, Secdin neither bound to (Supplemental Figure 6D) nor inhibited the activity of the full-length ARNO (Figure 8B; Supplemental Figure 7), and it had no impact on the longer BRAG2 protein construct carrying a membrane binding pleckstrin homology (PH) domain (Figure 8B; Supplemental Figure 7). Nevertheless, in human cell lines, Secdin treatment phenocopied the effects of known ARF-GEF inhibitors in terms of Golgi disassembly (Zeeh et al., 2006; Ohashi et al., 2012) (Supplemental Figures 8A and 8B). Altogether, we conclude that Secdin impairs the function of large ARF-GEFs via a mechanism more complex than direct targeting of the Sec7 domain.
Figure 8.
Effect of Secdin on ARF1 Activation.
(A) Inhibitory efficiency of BFA (50 µM) and Secdin (50 µM) on purified Arabidopsis BIG5 Sec7 protein construct.
(B) Spontaneous (EDTA) and human GEF-stimulated ARF1 activation in the presence of Secdin. Recombinant human protein constructs comprising the Sec7 domain of BIG1 (BIGSec7), ARNO (ARNOSec7), and BRAG2 (BRAG2Sec7), the Sec7 domain of BRAG2 followed by phospholipid binding PH domain (BRAG2Sec7-PH), as well as the full-length ARNO protein (ARNOFL) were assayed. The chemical impact was analyzed in vitro by monitoring the change in tryptophan fluorescence that follows the conformational alteration from ARF-GDP to ARF-GTP.
All experiments in (A) and (B) were done with N-terminally truncated bovine ∆17ARF1 (identical to the human form). The spontaneous GDP/GTP exchange in Δ17ARF1 in the absence of a GEF is examined in (B) by addition of EDTA, which displaces the bound Mg2+ ion. Values are means from three independent experiments. Error bars indicate sd. *P < 0.05 (Student’s t test).
DISCUSSION
Secdin Conditionally Hinders Multiple ARF-GEF-Dependent Trafficking Routes
Given the versatile roles of plant ARF-GEFs (Yorimitsu et al., 2014), their overlapping functions are often difficult to dissect in the context of a highly dynamic and complex endomembrane trafficking system. The concomitant genetic and pharmacological interference with both BFA-sensitive and BFA-resistant members of this protein family has turned out to be decisive for their investigation (Richter et al., 2014). We identified one small molecule, Secdin, that impaired the intracellular transport of PM-localized proteins without directly affecting general aspects of the plant cell homeostasis. In our protein pull-down experiments, we identified BIG5 as an interactor of biotinylated Secdin. Furthermore, Secdin modified the BFA body morphology by inducing the formation of a hybrid Golgi-TGN/EE compartment similar to the structures observed in the BFA-treated ben1 mutant, which carries a mutation in the ARF-GEF BIG5 (Tanaka et al., 2009). Additional validation assays via DARTS revealed that Secdin protected all examined Arabidopsis ARF-GEFs from proteolytic digestion, whereas, as expected, BFA protected only BFA-sensitive ARF-GEFs. This differential sensitivity of plant ARF-GEFs to BFA and Secdin seemingly determines the variations in the induced intracellular phenotypes. Both inhibitors dramatically affected PM protein recycling, but, as judged by the BFA washout experiments in the presence of Secdin, the internalized cargos were destined for degradation rather than recycling. Whereas BFA does not affect FM4-64 uptake (Dettmer et al., 2006), Secdin application phenocopied the endocytic defects previously observed in the BFA-treated gnl1 mutant (Naramoto et al., 2010). Additionally, at high concentrations, BFA blocked trafficking to the vacuole, whereas Secdin promoted it. Altogether, our results suggest that, like BFA, Secdin might target ARF-GEFs in Arabidopsis, but with a different impact on the ARF-GEF-assisted vesicle transport. It would be interesting to explore whether Secdin can be used as a tool to study the functions of PM proteins whose ARF-GEF-dependent endocytic trafficking cannot be manipulated with BFA. For example, the boron-induced transport of BOR1 to the vacuole is delayed by Secdin at the MVB/LE level, but is not responsive to BFA unless the gene encoding the BFA-insensitive BIG3 is knocked out (Richter et al., 2014).
As a consequence of the interference of Secdin with multiple trafficking routes, its application ultimately increased the PM protein retention into the MVB/LE compartments and enhanced protein degradation. Similar intracellular phenotypes have been reported for two small molecules derived from the ES series (Drakakaki et al., 2011). ES2 targets the EXO70 subunit of the exocyst complex and impairs exocytic transport to the PM (Zhang et al., 2016), whereas the inhibitory effect of ES16 on the exocytosis of apical, lateral, and nonpolar PM proteins is apparently determined by a disrupted RABA GTPase function (Li et al., 2017). In both cases, the inhibition of the anterograde pathway to the PM is accompanied by a concomitant increase in PM protein flux for vacuolar degradation. Regarding Secdin, the enhanced degradation of PM proteins was unrelated to impaired functions of the ubiquitination/deubiquitination machineries. The missorting of recycling cargos to the MVB/LEs is probably not restricted to proteins that can be ubiquitinated. For instance, treatment with the antitumor antibiotic geldanamycin triggers the accumulation of both the ubiquitinated EGFR receptor ErbB2 and the nonubiquitinated transferrin receptor into modified MVBs (Cortese et al., 2013). In this case, the cargo-independent effect of the drug on vesicle trafficking is attributed to the formation of aberrant endomembranes with mixed features of recycling endosomes and multivesicular compartments that are impaired in constitutive protein recycling back to the PM (Cortese et al., 2013). It remains to be tested whether Secdin can influence the trafficking of nonubiquitinated proteins in plant cells.
The Secdin Mode of Action
BFA inhibits GDP-to-GTP exchange by targeting a transient intermediate complex of the GEF Sec7 domain with the ARF GTPase substrate in its GDP-bound form (Mossessova et al., 2003; Renault et al., 2003). Several other Sec7 domain inhibitors have been predicted to bind to the interfacial site between ARF1 and the Sec7 domain. For example, molecular docking studies with Golgicide A, a potent GBF1 inhibitor in mammalian cells, have revealed that it binds within the same GBF1-ARF1 interfacial region that is accommodated by BFA (Sáenz et al., 2009). Importantly, the Golgicide A-binding region is larger than that of BFA and comprises GBF1-specific amino acid residues that, in turn, determine the drug selectivity toward this particular ARF-GEF member and the lack of effect on BIG1 and BIG2 (Sáenz et al., 2009). As suggested by molecular modeling, AMF-26, an inhibitor of the Golgi system, also targets the ARF1-GBF1 Sec7 domain surface to which BFA binds (Ohashi et al., 2012). Another ARF-GEF inhibitor, SecinH3, was identified as a cytohesin antagonist via an aptamer displacement screen, in which BFA-insensitive Sec7 domain-containing proteins were found to be involved in insulin signaling, because this inhibitor could induce insulin resistance (Hafner et al., 2006). Although the inhibitory mechanism of SecinH3 is unknown, this small molecule bound specifically to the Sec7 domain (Hafner et al., 2006). Interestingly, and in contrast to all known ARF-GEF inhibitors, our in vitro fluorescence kinetics-based assay with purified human ARF GTPase and selected human and Arabidopsis Sec7 domain-containing constructs revealed that Secdin had no inhibitory effect on the activation of ARF1 by the catalytic Sec7 domain. However, Secdin induced disruption of the Golgi in human cells, as do BFA and AMF-26 (Ohashi et al., 2012).
Besides interfering with the function of the Sec7 domain, multiple other regulatory mechanisms of ARF-GEF activity and membrane recruitment have been described (Wright et al., 2014; Nawrotek et al., 2016) and might potentially be hampered by Secdin. Growing evidence hints at the regulatory roles of the other conserved protein domains in addition to Sec7. For instance, the N-terminal dimerization and cyclophilin binding (DCB) domain has been shown to be essential for homotypic GNOM dimerization as well as for heterotypic interactions with other domains, with implications for the membrane association of the protein (Grebe et al., 2000; Anders et al., 2008). One hypothesis is that Secdin might interfere with the recruitment of the ARF-GEFs to the membrane, as implied by the reduced PM residency of GNOM in the presence of BFA. Moreover, the conserved domains are recognized by diverse interacting protein partners that determine specificity, recruitment to target membranes, and activation of large ARF-GEFs. In yeast and metazoan cells, the DCB domain is involved in the direct binding to small GTPases (Wright et al., 2014). Other experimentally validated direct interactors are subunits of the exocyst and TRAPPII tethering complexes, as well as the γ-subunit of the COPI coat complex, of which the ARF1-dependent recruitment to the target membranes is necessary for vesicle budding (Wright et al., 2014). It remains to be assessed whether Secdin exerts its effect by perturbing interactions with other regulatory proteins involved in the recruitment of ARF-GEFs to membranes.
METHODS
Plant Materials and Growth Conditions
The following mutant and transgenic Arabidopsis thaliana lines and crosses have been described previously: pBRI1-BRI1-GFP (Friedrichsen et al., 2000); pBRI1-BRI1-GFP/p35S-mRFP-RABF2b and pBRI1-BRI1-GFP/pVHAa1-VHAa1-RFP (Irani et al., 2012); pBRI1-BRI1-mCitrine/bri1 and pBRI1-BRI125K-R-mCitrine/bri1 (Martins et al., 2015); pHS-BRI1-YFP (Geldner et al., 2007); pBOR1-BOR1-GFP/bor1 (Takano et al., 2010); pUBQ10-BIG4-YFP/big3, pUBQ10-BIG4R-YFP/big3, and big1 big2 big3/+ big4 (Richter et al., 2014); pGNOM-GNOM-GFP/pVHAa1-VHAa1-mRFP, pGNOM-GNOM-GFP/p35S-mRFP-RABF2b, and pGNOM-GNOM-GFP/p35S-ST-mRFP (Naramoto et al., 2014); p35S-GFP-MAP4 (Marc et al., 1998); p35S-GFP-fABD2 (Sheahan et al., 2004); pPIN2-PIN2-GFP (Xu and Scheres, 2005); pPIN2-PIN1-GFP3 (Wiśniewska et al., 2006); pPIN2-PIN2-VEN/eir1-4 and pPIN2-pin212K-R-VEN/eir1-4 (Leitner et al., 2012); p35S-GFP-PIP2a (Cutler et al., 2000); p35S-BSK1-YFP (Tang et al., 2008); p35S-ST-mRFP (Wee et al., 1998); pVHAa1-VHAa1-RFP (Dettmer et al., 2006); pSYP61-SYP61-CFP and pSYP22-SYP22-YFP (Robert et al., 2008); p35S-mRFP-RABF2b (Ueda et al., 2004); and pUBQ10-YFP-RABF2a (Geldner et al., 2009). Arabidopsis accession Col-0 seeds were surface sterilized, stratified for 2 d at 4°C in the dark, and germinated vertically at 22°C under a 16-h-light/8-h-dark cycle (white light emitted by fluorescent lamps with an intensity of 120 µmol m−2 s−1) on 0.8% (w/v) agar plates containing half-strength Murashige and Skoog (1/2MS) medium (pH 5.6) supplemented with 1% (w/v) sucrose unless otherwise specified. BOR1-GFP seedlings were grown on MGRL medium (Fujiwara et al., 1992).
Construct Generation and Cell Culture Transformation
The BIG1 and BIG4 gene fragments without stop codons were amplified by PCR with iProof High-fidelity DNA Polymerase (Bio-Rad) from wild-type Col-0 genomic DNA with the following primer pairs with Gateway system-compatible attB sites: BIG1 forward 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGTCGTCGTCGCAGAAC-3′ and BIG1 reverse 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTTCATCCATCATTGCACCC-3′; BIG4 forward 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTCAACGTCACAAACC-3′ and BIG4 reverse 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTAAGCCAAAATAGGACCAAT-3′. The PCR products were then introduced into pDONR221 donor vectors (Invitrogen). The cDNA fragments of BIG3, GNL1, and GNL2 were synthesized with Gateway system-compatible attL sites (Invitrogen) and introduced into the pG9m-2 vector (Gen9 Company). A 1694-bp fragment of the RPS5A promoter was introduced into the pDONRP4-P1R donor vector. The entry clones pDONRP4-P1R-pRPS5A, pDONR221-BIG1/BIG3/BIG4/GNL1/GNL2, and pDONRP2R-P3-HA were recombined in a multisite LR reaction with pH7m34GW (Invitrogen) as the destination vector. All clones were confirmed by sequencing. The generated constructs were used to transform dark-grown Arabidopsis PSB-D cell suspension cultures as described previously (Van Leene et al., 2007).
Chemical Treatments
Stock solutions of BFA (50 mM), ConcA (2 mM), Wm (20 mM), Actinomycin D (1 mg/mL) (all from Sigma-Aldrich), BCECF (1 mg/mL; Invitrogen), CHX (50 mM; Calbiochem), and carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (MG132) (10 mM; Calbiochem) were prepared in DMSO. FM4-64 was obtained from Invitrogen and stored as 2 mM stock solution in deionized water. Secdin was acquired through ChemBridge (https://www.chembridge.com/; ID 7750598). Stock solutions of Secdin, its analogs, and the biotinylated Secdin derivative (10 mM for most of the assays, except for the DARTS experiments and ATP measurements) were prepared in DMSO, and aliquots were stored at −20°C. All chemical treatments were performed at room temperature in growth medium and compared with control samples incubated with equal volumes of solvent.
Synthesis of Secdin Analogs and Derivatives
A subset of the Secdin analogs was acquired from commercial chemical libraries, such as ChemBridge (Secdin1 to Secdin10), Vitas-M (Secdin12), and InterBioScreen (Secdin13 and Secdin14). Additional Secdin analogs (Secdin15 to Secdin22) were designed and synthesized for screening purposes in this study.
Secdin-Phthalazinone Fragment (Secdin16)
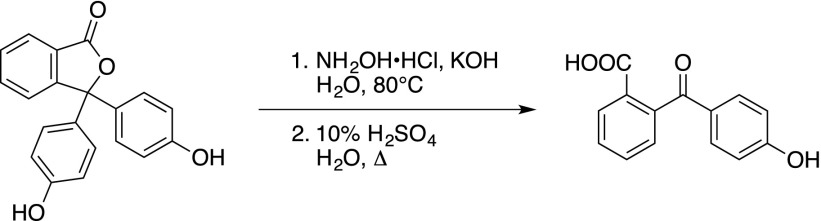
Phenolphthalein (7 g, 22 mmol, 1.0 equiv.) was dissolved in an aqueous potassium hydroxide solution (70 mL, 2 mM). Hydroxylamine hydrochloride (1.7 g, 24 mmol, 1.09 equiv.) was added to the purple solution and the mixture was heated to 80°C. Consumption of phenolphthalein was checked by taking a sample from the reaction mixture that was acidified with acetic acid, until a yellow precipitate was formed. The solid was filtered and potassium hydroxide was added to the filtrate. A color change to purple indicated the presence of unreacted phenolphthalein. After 1 h, the reaction was finished, as the color did not change anymore in the test. The reaction was cooled to room temperature and ethanol (14 mL) was added. The resulting mixture was acidified until a yellow precipitate formed, which was filtered and dried in a vacuum oven at 50°C. The dry solid was dissolved in warm sulfuric acid solution (70 mL, 10%), and the mixture was heated to reflux for 3 h. After cooling to room temperature, a precipitate formed that was filtered and recrystallized from water to yield 2-(4-hydroxybenzoyl)benzoic acid as an off-white crystalline solid (5.2 g, 20 mmol, 91%) (Hardcastle et al., 2006).
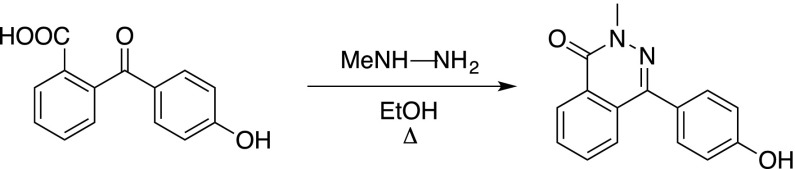
2-(4-Hydroxybenzoyl)benzoic acid (2 g, 7.7 mmol, 1.0 equiv.) was dissolved in ethanol (10 mL). Methyl hydrazine (1.4 mL, 27 mmol, 3.5 equiv.) was added and the solution was refluxed for 5 h, whereupon a yellow precipitate formed. The mixture was cooled and the formed solid was filtered, washed with cold ethanol, and dried in vacuo to yield the fragment Secdin16 as a white solid (837 mg, 3.3 mmol, 43%). The product had experimental characteristics similar to those described in the literature (Hardcastle et al., 2006).
Piperidine Fragment Synthesis
4-Hydroxypiperidine (2 g, 20 mmol) was dissolved in a mixture of ethyl acetate (300 mL) and a saturated aqueous solution of sodium carbonate (150 mL). Chloroacetylchloride (2.4 mL) was added and the mixture was stirred at room temperature. After 4 h, the organic phase was separated, dried on sodium sulfate, filtered and concentrated in vacuo to yield 2-
chloro-1-(4-hydroxypiperidin-1-yl)ethan-1-one as a pale-yellow oil that solidified upon storage at −20°C (2.9 g, 16 mmol, 80%). The product was used without further purification and had experimental characteristics similar to those reported in the literature (Wessig and Möllnitz, 2008).
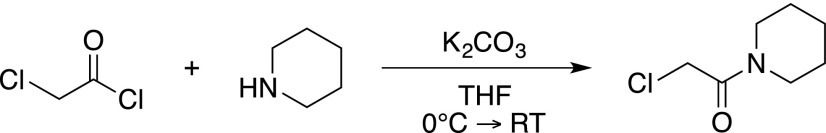
Piperidine (1.24 g, 15 mmol) and potassium carbonate (2.07 g, 15 mmol) were dissolved in tetrahydrofuran (50 mL) and the mixture was cooled to 0°C. Chloroacetyl chloride (1.3 mL) was added dropwise, after which the mixture was slowly heated to room temperature. Water (50 mL) was added carefully and the aqueous phase was extracted with ethyl acetate (3 × 250 mL). The combined organic phases were washed with a saturated sodium bicarbonate solution (75 mL), an aqueous hydrochloric acid solution (1 M, 75 mL), and a saturated sodium chloride solution (75 mL). The organic phase was dried on sodium sulfate, filtered, and concentrated in vacuo to yield 2-chloro-1-(piperidin-1-yl)ethan-1-one as a pale-yellow oil (2.1 g, 13 mmol, 87%), which was used without further purification and had experimental characteristics similar to those reported in the literature (Joshi et al., 2013).
Biotin Label Synthesis
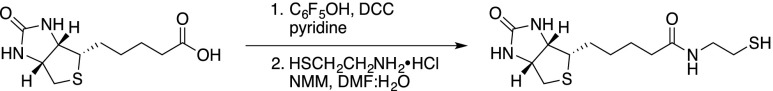
Biotin (1 g, 4.1 mmol) and pentafluorophenol (773 mg, 4.2 mmol) were dissolved in pyridine (45 mL). The clear solution was cooled to 0°C, whereafter a solution of dicyclohexylcarbodiimide (871 mg, 4.22 mmol) in pyridine (5 mL) was added and the mixture was allowed to heat to room temperature. After overnight stirring, the reaction mixture was filtered and the filtrate was concentrated in vacuo. The resulting white solid (1.6 g) was dissolved in a mixture of water and dimethylformamide (210 mL, 1:6 v/v) together with N-methylmorpholine (946 µL, 8.6 mmol) and cysteamine hydrochloride (443 mg, 3.9 mmol). The resulting mixture was stirred overnight and subsequently concentrated in vacuo. The residue was purified by flash chromatography (silica, methanol:dichloromethane 1:7, v/v) to yield N-(2-mercaptoethyl)-5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamide as a white solid (571 mg, 2 mmol, 49%) that showed an H-NMR spectrum similar to that described in the literature (Roling et al., 2013).
Secdin Analogs
In general, the phthalazinone-phenol building block was reacted with α-chloro amides to give Secdin analogs according to the procedure given for Secdin17.
Secdin17
Secdin16 (250 mg, 0.99 mmol, 1.0 equiv.) and 2-chloro-1-(piperidin-1-yl)ethan-1-one (176 mg, 1.1 mmol, 1.1 equiv.) were dissolved in dry dimethylformamide (10 mL). Potassium carbonate (274 mg, 1.98 mmol, 2.0 equiv.) was added and the mixture was heated to 60°C. When judged complete by thin-layer chromatography, water (50 mL) was added, whereupon a white precipitate formed. The solid was filtered and recrystallized from 2-propanol to yield Secdin17 as a white crystalline solid (324.4 mg, 0.86 mmol, 86%).
Formula: C22H23N3O3; Mw: 377.44 g/mol; 1H-NMR (400 MHz, CDCl3): δ 8.52 (1H, d(br), J = 7.4 Hz), 7.8–7.7 (3H, band), 7.55–7.47 (2H, m), 7.13–7.06 (2H, m), 4.76 (2H, s), 3.90 (3H, s), 3.58 (2H, dd, J = 5.6, 4.8 Hz), 3.54 (2H, dd, J = 6.0, 4.8 Hz), 1.69–1.52 (6H, band); 13C-NMR (100 MHz, CDCl3): δ 165.9 (C), 159.4 (C), 158.8 (C), 146.6 (C), 132.7 (CH), 131.4 (CH), 130.8 (CH), 129.4 (C), 128.4 (C), 128.2 (C), 127.0 (CH), 126.7 (CH), 114.9 (CH), 67.8 (CH2), 46.5 (CH2), 43.3 (CH2), 39.6 (CH3), 26.5 (CH2), 25.6 (CH2), 24.5 (CH2); HRMS (m/z): [M+H]+ calcd. for C22H24N3O2+, 378.1812; found, 378.1829.
Secdin18
Secdin16 (250 mg, 0.99 mmol) and 2-chloro-1-(4-hydroxypiperidin-1-yl)ethan-1-one (194 mg, 1.09 mmol) were dissolved in dry dimethylformamide (10 mL). Anhydrous potassium carbonate (274 mg, 1.98 mmol) was added and the suspension was stirred at 60°C. After 6 h, water (40 mL) was added and the aqueous phase extracted with ethyl acetate (3 × 400 mL). The organic phase was dried on sodium sulfate, filtered, and concentrated in vacuo to yield a colorless oil that solidified overnight upon storage at −20°C. The resulting solid was recrystallized from isopropanol to yield Secdin18 as a white crystalline solid (310 mg, 0.79 mmol, 80%).
Formula: C22H23N3O4; Mw: 393.44 g/mol; 1H-NMR (400 MHz, CDCl3): δ 8.52 (1H, d(br), J = 7.8 Hz), 7.81–7.7 (3H, band), 7.6–7.5 (2H, m), 7.13–7.06 (2H, m), 4.78 (2H, s), 4.05 (1H, ddd, J = 12.8, 6.3, 4.4Hz), 3.98 (1H, tt, J = 3.9, 3.9 Hz), 3.9 (3H, s), 3.87 (1H, ddd, J = 13.2, 6.0, 3.8 Hz), 3.34 (2H, tdd, J = 14.0, 8.9, 3.7 Hz), 2.0–1.85 (2H, m), 1.65–1.46 (2H, m); 13C-NMR (100 MHz, CDCl3): δ 166.0 (C), 159.4 (C), 158.7 (C), 146.5 (C), 132.7 (CH), 131.4 (CH), 129.3 (C), 128.7 (C), 128.1 (C), 127.0 (CH), 126.7 (CH), 114.8 (CH), 67.8 (CH2), 66.8 (CH), 42.5 (CH2), 39.5 (CH2), 39.4 (CH3), 34.5 (CH2), 33.8 (CH2); HRMS (m/z): [M+H]+ calcd. for C22H24N3O4+, 394.1761; found, 394.1779.
Synthesis of the Biotinylated Derivative Secdin24
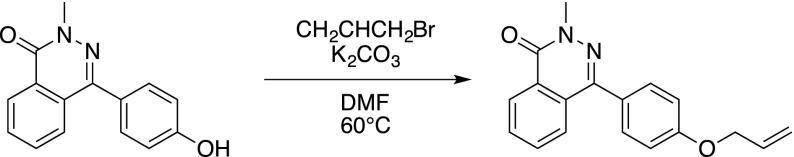
Secdin16 (0.5 g, 1.98 mmol) was dissolved in dry dimethylformamide (20 mL). Potassium carbonate (1.3 g, 9.9 mmol) was added and the mixture was stirred for 5 min at room temperature. Allyl bromide (850 µL, 9.9 mmol) was added and the reaction was heated to 60°C until judged completed based on thin-layer chromatography. After cooling to room temperature, water (20 mL) was added and the aqueous phase was extracted with ethyl acetate (3 × 200 mL). The combined organic layers were washed with an aqueous sodium hydroxide solution (15%, 60 mL) and a saturated sodium chloride solution (60 mL). The organic phase was dried on sodium sulfate, filtered, and concentrated in vacuo. The resulting yellow solid was recrystallized from 2-propanol to yield allylated Secdin16 as an off-white crystalline solid (408.6 mg, 1.4 mmol, 71%).
Formula: C18H16N2O2; Mw: 292.34 g/mol; 1H-NMR (400 MHz, (CD3)2CO): δ 8.44–8.37 (m, 1H), 7.90–7.82 (band, 2H), 7.81–7.74 (m, 1H), 7.59–7.54 (m, 2H), 7.17–7.10 (m, 2H), 6.13 (1H, ddt, J =17.4, 10.6, 5.3 Hz), 5.47 (1H, ddt, 3J = 17.3, 1.5 Hz, 4J = 1.5 Hz), 5.29 (1H, ddt, 3J = 10.6, 1.5 Hz, 4J = 1.5 Hz), 4.69 (1H, dt, 3J = 5.3 Hz, 4J = 1.5 Hz); 13C-NMR (100 MHz, (CD3)2CO): δ 160.2 (C), 159.3 (C), 146.7 (C), 134.5 (CH), 133.6 (CH), 132.1 (CH), 131.7 (CH), 130.2 (C), 129.0 (C), 128.8 (C), 127.5 (CH), 127.4 (CH), 117.6 (CH2), 115.5 (CH), 69.4 (CH2), 39.4 (CH3); HRMS (m/z): [M+H]+ calcd. for C18H17N2O2+, 293.1285; found, 293.1292.
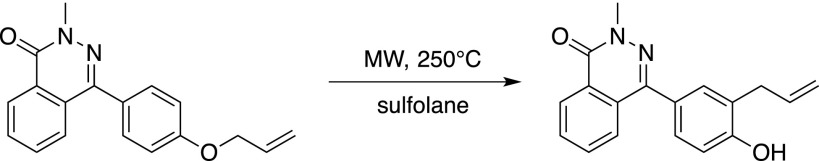
A microwave vial was charged with allylated Secdin16 (250 mg, 0.86 mmol) and sulfolane (5 mL). The mixture was heated in a microwave apparatus (75W) to 250°C for 15 min. After cooling to room temperature, the mixture was diluted with water (5 mL), whereupon a white precipitate formed. The solid was filtered and recrystallized from a water:ethanol mixture (1:1, v/v) to yield the allylated phenol derivative as an off-white crystalline solid (195 mg, 0.67 mmol, 78%).
Formula: C18H16N2O2; Mw: 292.34 g/mol; 1H-NMR (400 MHz, (CD3)2SO): δ 9.80 (1H, s(br)), 9.37–8.30 (1H, m), 7.91–7.83 (2H, band), 7.74–7.68 (1H, m), 7.30–7.24 (2H, band), 7.00–6.94 (1H, m), 6.00 (1H, ddt, J = 17.0, 10.0, 6.7 Hz), 5.08 (1H, ddt, 3J = 17.0, 2.1 Hz, 4J = 1.5 Hz), 5.20 (1H, ddt, 3J = 10.0, 2.1 Hz, 4J = 1.2 Hz), 3.76 (3H, s), 3.37 (2H, d(br), J = 6.7 Hz); 13C-NMR (100 MHz, (CD3)2SO): δ 158.2 (C), 155.8 (C), 155.7 (C), 146.1 (C), 136.8 (CH), 133.2 (CH), 131.6 (CH), 130.7 (CH), 128.9 (C), 128.4 (CH), 126.7 (CH), 126.2 (CH), 125.4 (C), 115.7 (CH2), 114.8 (CH), 39.0 (CH3), 33.7 (CH2) ; HRMS (m/z): [M+H]+ calcd. for C18H17N2O2+, 293.1285; found, 293.1286.
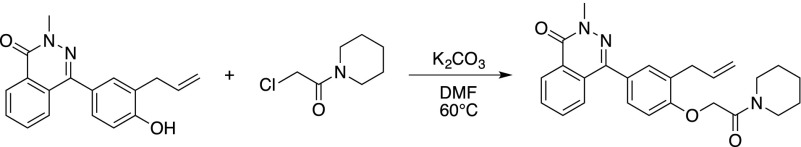
The allylated phenol derivative (150 mg, 0.51 mmol) and yield 2-chloro-1-(piperidin-1-yl)ethan-1-one (91 mg, 0.56 mmol) were dissolved in dimethylformamide (5 mL). Potassium carbonate (142 mg, 1.03 mmol) was added. The mixture was stirred for 5 min and subsequently heated to 60°C. After 4 h, water (20 mL) was added and the aqueous layer was extracted with ethyl acetate (3 × 200 mL). The combined organic phases were dried on sodium sulfate, filtered, and concentrated in vacuo to yield a dark-yellow oil that solidified upon storage at −20°C. The off-white solid was recrystallized from 2-propanol to give the Secdin17 analog as a white crystalline solid (129.8 mg, 0.31 mmol, 61%).
Formula: C25H27N3O3; Mw: 417.51 g/mol; 1H-NMR (400 MHz, CDCl3): δ 8.55–8.50 (1H, m), 7.80–7.70 (3H, band), 7.40–7.36 (2H, band), 7.04 (1H, dm(br), J = 8.6 Hz), 6.03 (1H, ddt, J = 16.9, 10.1, 6.7 Hz), 5.11 (1H, ddt, 3J = 16.9, 1.6 Hz, 4J = 1.6 Hz), 5.05 (1H, m), 4.78 (2H, s), 3.91 (3H, s), 3.63–3.48 (6H, band) 1.72–1.53 (4H, m(br)); 13C-NMR (100 MHz, CDCl3): δ 166.1 (C), 159.4 (C), 156.6 (C), 146.8 (C), 136.3 (C), 132.6 (C), 131.3 (C), 129.5 (C), 128.7 (CH), 128.2 (CH), 128.1 (CH), 127.0 (CH), 126.8 (CH), 116.2 (CH2), 111.5 (CH2), 68.2 (CH2), 46.5 (CH2), 43.3 (CH2), 39.6 (CH), 34.2 (CH2), 26.7 (CH2), 25.6 (CH2), 24.5 (CH2); HRMS (m/z): [M+H]+ calcd. for C25H28N3O3+, 418.2125; found, 418.2133.
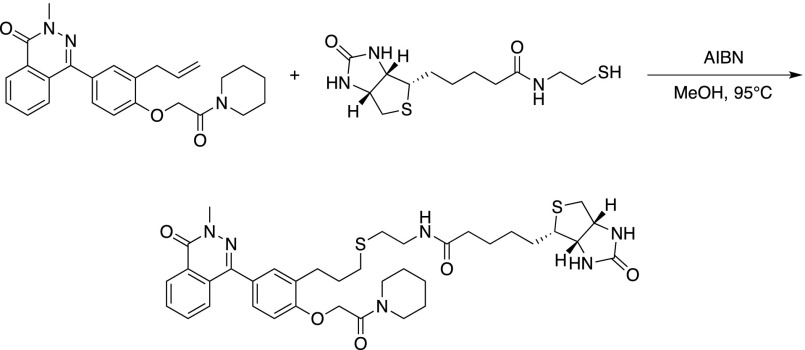
A pressure tube was charged with the Secdin17 analog (165 mg, 0.40 mmol), N-(2-mercaptoethyl)-5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamide (240 mg, 0.79 mmol), and methanol (0.2 mL). A catalytic amount of azoisobisbutyronitrile was added and the mixture was heated to 90°C. After 1.5 h, the reaction mixture was concentrated in vacuo and the residue was purified by flash chromatography to yield biotinylated Secdin17 as a white solid (29 mg, 0.04 mmol, 10%).
Formula: C37H47N5O6S2; Mw: 721.93 g/mol; 1H-NMR (400 MHz,): δ 8.58–8.47 (1H, m), 7.85–7.70 (3H, band), 7.45–7.34 (2H, band), 7.00–6.94 (1H, m), 7.80 (2H, s), 4.50 (1H, dd, J = 7.5, 5.2 Hz), 4.32 (1H, dd, J = 7.5, 4.6 Hz), 3.60 (2H, dd(br), J = 5.5, 5.5 Hz), 3.52 (2H, dd(br), J = 5.5, 5.5 Hz), 3.44–3.36 (2H, m), 3.14 (1H, td, J = 7.3, 4.4 Hz), 2.90 (1H, dd, J = 1.0, 5.0 Hz), 2.84 (2H, t, J = 7.3 Hz), 2.70 (1H, d, J = 13.0 Hz), 2.67 (2H, t, J = 6.6 Hz), 2.60 (2H, t, J = 6.9 Hz), 2.21 (2H, td, J = 7.2, 5.7 Hz), 1.97 (2H, q5, J = 7.14 Hz), 1.92–1.35 (18H, band + H2O).
Confocal Microscopy and Image Analysis
For the imaging experiments, 4-d-old Arabidopsis seedlings grown vertically on plates with 1/2MS solid medium were transferred for incubation into 1/2MS liquid medium supplemented with chemicals at the respective concentration. Whole seedlings were then mounted in compound-containing medium and imaged on an inverted confocal laser scanning microscope (Olympus FluoView 1000) equipped with a 60× water immersion lens (NA1.2) at digital zoom 4. Images of the epidermal cell layer were taken at the transition between the meristematic and elongation zones of the primary root, i.e., ∼10 cells above the quiescent center. The excitation wavelength was 458 nm for CFP, 488 nm for GFP and FM4-64, 515 nm for YFP, and 559 nm for RFP. Emission was detected between 475 and 540 nm for CFP, 500 to 530 nm for GFP, 530 to 600 nm for YFP, and 600 to 700 nm for RFP and FM4-64. For quantification, images were converted to 8-bit with ImageJ (https://imagej.nih.gov/ij/). For each analyzed cell, regions of interest (ROIs) were selected to comprise the PM and the intracellular area. Histograms listing all fluorescence intensity values per ROI were generated and the averages of the 100 most intense pixels were used to calculate the PM-to-cytoplasm fluorescence intensity ratio. To characterize the intracellular BRI1-GFP agglomerates upon Secdin treatment, cytoplasmic area-containing ROIs were selected. Identical minimum and maximum threshold values were set for mock- and Secdin-treated samples, followed by segmentation with the watershed algorithm and noise reduction (radius of 2 pixels) with the “remove outliers” filter in ImageJ to automatically delineate the endomembrane compartments of interest. After adjusting the scale (in µm), the “analyze particles” command was applied to obtain the average size, fluorescence signal intensity (i.e., integrated density), and number of intracellular compartments per cellular cross section. To quantify the colocalization of GFP- and RFP-tagged marker lines, ROIs comprising the cytoplasmic area were selected, and the PSC colocalization plug-in for ImageJ (French et al., 2008) was used with a threshold level of 10 to obtain the Pearson correlation coefficient.
For the imaging experiments with human subcellular markers, HeLa cells were plated on glass cover slips, cultured for 48 h, and treated with the respective concentration of Secdin or BFA in complete medium (Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, both from Life Technologies). Cells were then processed for immunofluorescence as described previously (Sannerud et al., 2011). The following antibodies were used: mouse α-GM130 (1:50; BD Transduction Laboratories) and rabbit α-EEA1 (1:250; Sigma-Aldrich). Images were captured on a Nikon 1AR confocal system connected to an inverse microscope (Ti-2000; Nikon) using an oil-immersion Plan-Apochromat 60× A/1.40 NA objective. Images were collected using Nikon imaging software.
TEM
Five-day-old wild-type (Col-0) Arabidopsis seedlings grown on 1/2MS solid medium in square plates were overlaid with 1/2MS liquid medium supplemented with Secdin or a combination of Secdin and BFA at the respective concentration. After 1 h of incubation with the chemicals, the root tips were excised, immersed in either 20% (w/v) BSA or 0.1 M sucrose, and subsequently frozen in a high-pressure freezer (EM PACT; Leica Microsystems or a Baltec HPM 010; Technotrade). Freeze substitution was performed in an EM AFS (Leica Microsystems). Over a period of 4 d, root tips were freeze-substituted in dry acetone containing 0.1% (w/v) uranyl acetate, with or without 1% (w/v) OsO4, and 0.2% glutaraldehyde as follows: –90°C for 26 h, 2°C increase per h for 15 h, –60°C for 16 h, 2°C increase per h for 15 h, and –30°C for 8 h. Samples were then infiltrated with Lowicryl HM20 at −60°C and polymerized under UV light or slowly warmed up to 4°C, rinsed three times with acetone for 20 min each time, infiltrated stepwise over 3 d at 4°C in Spurr’s resin, and embedded in capsules. Polymerization was done at 70°C for 16 h. Ultrathin sections were made with an ultramicrotome (Leica EM UC6) and collected on Formvar-coated copper mesh grids. Sections were poststained in an automatic contrasting instrument (Leica EM AC20) for 40 min in uranyl acetate at 20°C and for 10 min in lead stain at 20°C. Grids were viewed with a JEM 1010 transmission electron microscope (JEOL) operating at 80 kV. The MVB structural features were determined by manual counting of the number of MVBs and ILVs per cellular cross section, whereas the MVB and ILV diameters were estimated with ImageJ.
Root Growth Analysis
Wild type (Col-0) or big1 big2 big3/+ big4 mutant seeds were sown in rows in square Petri dishes containing 1/2MS solid medium supplemented with Secdin or combined Secdin and BFA, stratified for 2 d at 4°C in the dark, and germinated vertically in the light for 6 d (Figure 1) or 7 d (Figure 6). The plant phenotypes were then recorded with a flatbed scanner (Epson Perfection V850 Pro), and primary root growth was measured by tracing the roots with the Segmented line tool in ImageJ. The half-maximal inhibitory concentration (IC50) was determined after applying the nonlinear curve fit (DoseResp) function in Origin (https://www.originlab.com/).
ATP Content Determination
Wild-type PSB-D Arabidopsis cell cultures were used and maintained as described before (Van Leene et al., 2007). Three days after subculturing, the cell suspension was diluted 100 times and mixed thoroughly before distribution of 95 μL in 96-well plates. Subsequently, 5 μL of a 1/50 dilution of the Secdin stock solution (1000×) in MS with Minimal Organics medium was added to the cells (final dilution of 1000×) with a Freedom EVO robot (Tecan). ATP levels were detected by the addition of 80 μL of the ATPlite 1step Luminescence Assay System (Perkin-Elmer) after incubation of the cells in the presence of Secdin for the indicated time. Fluorescein diacetate stock solution (2% [w/v] fluorescein diacetate in acetone) was diluted 100 times in target medium and 5 μL was added to 95 μL cell culture. Measurements were done with an EnVision 2104 Multilabel Reader (Perkin-Elmer) with the Wallac EnVision manager software package. ATPlite luminescence was detected via ultrasensitive luminescence technology. Fluorescence was detected with an excitation at 485 nm (band width 14 nm) and emission at 535 nm (band width 25 nm).
Affinity Purification and LC-MS/MS Analysis of Protein Binders of Biotinylated Secdin
Wild-type Arabidopsis PSB-D cell cultures (Van Leene et al., 2007) were used for total protein extraction. All extraction steps were performed at 4°C. After harvest, the cells were ground in liquid nitrogen, resuspended in total protein extraction buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl; 0.1% [v/v] IGEPAL CA-630, and Roche cOmplete ULTRA protease inhibitor cocktail, EDTA free) in a 1:2 (w/v) ratio, and centrifuged at 15,000g to discard the cell debris. The protein extracts were incubated for 2 h with equilibrated in extraction buffer Streptavidin Sepharose High Performance beads (GE Healthcare) with slow mixing and then briefly centrifuged at 250g to discard the beads and remove endogenous biotinylated proteins. In the meantime, Dynabeads (Dynabeads MyOne Streptavidin T1; Invitrogen) equilibrated and resuspended in the extraction buffer were pretreated either with a biotinylated Secdin derivative or biotin at a 50 µM concentration for 1 h with slow mixing. After thorough removal of the supernatant, the pretreated Dynabeads were mixed with the protein extracts and incubated for 3 h or overnight at 4°C with slow mixing. The Dynabeads were then washed three times with extraction buffer without protease inhibitors to discard unbound proteins and mixed with 1× NuPAGE LDS sample buffer (Invitrogen). The mixture was heated at 95°C for 10 min and the Dynabeads were discarded. The supernatant containing the protein binders of biotinylated Secdin was loaded onto NuPAGE 4-12% Bis-Tris protein gels (Invitrogen) and run for 7 min at 200 V. Gels were stained with SYPRO Ruby protein gel stain (Invitrogen) according to the manufacturer’s rapid staining protocol. Stained gel regions were cut into small pieces and transferred to Eppendorf LoBind microcentrifuge tubes. The gel slices were washed twice with HPLC-grade water, treated consecutively with 50% (v/v) acetonitrile (AcNi) in 50 mM ammonium bicarbonate (ABC; pH 8.0) and 100% AcNi, and dried in a SpeedVac. Cysteine was reduced with 10 mM DTT in 50 mM ABC for 1 h at 56°C, followed by alkylation with 55 mM iodoacetamide in 50 mM ABC for 50 min in the dark at room temperature. The gel pieces were washed with 50 mM ABC, treated consecutively with 50% (v/v) AcNi in 50 mM ABC and 100% AcNi, and dried in a SpeedVac. Trypsin digestion was done with Trypsin Gold-mass spectrometry grade (Promega) with overnight incubation at 37°C. The peptides were extracted from the gel by sonication followed by two 1-h incubations with 50% (v/v) AcNi in 5% (v/v) trifluoroacetic acid. The pooled extracts from each sample were dried in a SpeedVac. The peptide pellets were resuspended in 0.1% (v/v) formic acid and purified as already described (Wendrich et al., 2017). The purified samples were analyzed by nanoscale liquid chromatography-tandem mass spectrometry (nLC-MS/MS) with a Proxeon EASY nLC and a LTQ-Orbitrap XL mass spectrometer as previously described (Lu et al., 2011; Wendrich et al., 2017). LC-MS data analysis (false discovery rates were set to 0.01 on peptide and protein levels) and additional result filtering (minimally two peptides are necessary for protein identification of which at least one is unique and at least one is unmodified) were performed as described previously (Smaczniak et al., 2012; Wendrich et al., 2017). To analyze the abundance of proteins, their normalized label-free quantification (LFQ) intensities were compared (Cox et al., 2014). The nLC-MS/MS results with total protein extracts were obtained from two independent biological repeats, i.e., using two independent suspensions of PSB-D cell cultures. The chemical treatments were done in triplicate within each of the repeats.
DARTS Assays
The protein interactors of Secdin were validated biochemically as described (Lomenick et al., 2011). Arabidopsis PSB-D cell cultures or 4- to 5-d-old seedlings from tagged Arabidopsis lines were used for total protein extraction according to the protocol for affinity purification. Protein extraction from HeLa cell cultures was performed with M-PER Mammalian Protein Extraction Reagent (Thermo) supplemented with cOmplete ULTRA protease inhibitor cocktail (Roche). After determining the protein concentration with the Quick Start Bradford 1× Dye Reagent (Bio-Rad), the cell lysate was split into LoBind tubes and incubated with the respective chemical (BFA, Secdin, or its inactive analog) at a 250 µM concentration for 30 min at room temperature with slow mixing (control treatments were with equal volumes of DMSO). The concentrations used were much higher than the biologically relevant doses to saturate the protein with ligand and to ensure maximal protection from proteolysis (Lomenick et al., 2011). The treated protein extracts were further aliquoted, and each of the aliquots was mixed with Pronase (Roche) at the corresponding dilution prepared in pronase buffer (25 mM Tris-HCl, pH 7.5, and 150 mM NaCl) to achieve the aimed ratio of total enzyme to total protein substrate. After incubation for 30 min at room temperature, the proteolytic digestion was stopped by adding a protease inhibitor cocktail (Roche), and the tubes were incubated on ice for 10 min. The protein samples were then mixed with 4× NuPAGE LDS sample buffer (Invitrogen), heated at 70°C for 10 min, and loaded onto NuPAGE 4-12% Bis-Tris protein gels (Invitrogen). The protein transfer to PVDF membranes was performed using the iBlot dry blotting system (Thermo) and the protein detection was done according to standard protein gel blotting procedures. The membranes were probed with the following antibodies: rabbit α-AtMIN7/BEN1/BIG5 (1:6000) (Nomura et al., 2006), rabbit α-Sec7 (1:10000) (Steinmann et al., 1999), rat α-HA (1:1000; Roche), rabbit α-ARF1 (1:2000; Agrisera), α-GFP HRP-coupled (1:5000; Miltenyi Biotec), mouse α-ARNO (cytohesin-2, 1:200; Santa Cruz Biotechnology), rabbit α-ATPβ (1:2000; Agrisera; reference for plant protein loading), and mouse α-ATPβ (1:1000; Abcam; reference for human protein loading). The secondary antibodies were ECL α-rabbit/rat/mouse IgG, horseradish peroxidase-linked whole antibody (GE Healthcare). Blots were developed with Western Lightning Plus-ECL, Enhanced Chemiluminescence Substrate (Perkin-Elmer), and imaged with the Bio-Rad ChemiDoc XRS+ molecular imager. Intensity of protein bands was measured with the Bio-Rad Image Lab software package. The ratio between the compound- and mock-treated samples for each of the pronase concentrations was calculated.
BRI1 Protein Levels, Immunoprecipitation, and Ubiquitination Analysis
Wild-type (Col-0) Arabidopsis seedlings grown for 4 d on solid 1/2MS medium were transferred for 5 h to liquid 1/2MS medium supplemented with Secdin (50 µM). Total proteins were extracted as described for the affinity purification of Secdin interactors, but with a modified extraction buffer containing 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% (w/v) SDS, and cOmplete ULTRA protease inhibitor cocktail (Roche). The protein gel blot analysis of BRI1 protein levels was done with rabbit α-BRI1 (1:2000; kind gift of Michael Hothorn). Protein extracts from wild type (Col-0) seedlings prepared as described above were also used to determine the total protein ubiquitination pattern upon Secdin treatment. To dissect the extent of the BRI1 ubiquitination, pBRI1-BRI1-mCitrine/bri1 and pBRI1-BRI125K-R-mCitrine/bri1 seedlings grown for 4 or 5 d on solid 1/2MS medium were transferred for 5 h to liquid 1/2MS medium supplemented with Secdin in combination with the proteasome inhibitor MG132. Microsomal protein enrichment was performed in MCF extraction buffer as described (Abas and Luschnig, 2010). The microsomal protein pellets were resuspended in the SDS-containing extraction buffer described above and incubated with GFP-Trap_A beads (ChromoTek) for BRI1-mCitrine immunoprecipitation. After several washes of the beads with the extraction buffer without protease inhibitors, the immunoprecipitated proteins were eluted in the SDS sample buffer by heating at 95°C for 10 min and analyzed by protein gel blots according to standard protocols. The ubiquitination pattern of the total membrane-associated protein fraction was assessed on microsomal homogenates prior to the immunoprecipitation step. The membranes were probed with mouse α-ubiquitin antibody (clone P4D1, 1:2500; Millipore), stripped, and reprobed with either mouse α-GFP (JL-8, 1:1000; Clontech) or mouse α-tubulin (1:10,000; Sigma-Aldrich). The secondary antibodies were the same as for the DARTS assays. The densitometric analyses of the signal intensities on the blots from three biological repeats were done in ImageJ.
Deubiquitination Activity Assays
Deubiquitination (DUB) assays were performed as described previously (Kalinowska et al., 2016) with slight modifications. For the immunoblot-based assay, 250 ng of K63-linked polyubiquitin chains (Ub2-7; Enzo Life Sciences) was incubated with 2 pmol of purified AMSH3 preincubated with either DMSO or 50 µM Secdin for 10 min. The fluorescent-based assay was performed by incubating 0.4 μM diubiquitin (K63-linked) FRET TAMRA Position 3 (R&D Systems) with 50 nM AMSH3 with or without preincubation with 100 µM Secdin. Fluorescence of diubiquitin-TAMRA was measured every minute for 120 min with a Synergy 2 Multi-Mode Microplate Reader (BioTek).
Protein Expression and Purification
N-terminally truncated bovine ∆17ARF1 (identical to the human form) was expressed, purified, and loaded with GDP prior to use as described (Zeeh et al., 2006). Human BRAG2Sec7 and BRAG2Sec7-PH, ARNOSec7, and BIG1Sec7 were produced in Eschercihia coli and purified as described (Benabdi et al., 2017). Expression in E. coli and purification of full-length human ARNO (ARNOFL) will be described elsewhere. The sequence coding for the Arabidopsis BIG5 Sec7 domain (residues 572–759) was cloned into the pET-15b vector between the NdeI and BamHI restriction sites and transformed into competent E. coli BL21 (DE3) pLysS cells. Transformed cells were cultured in Luria-Bertani medium supplemented with carbenicillin (100 µg/mL) at 37°C. Expression was induced by the addition of 1 mM IPTG for 4 h at 28°C. Cells were then harvested by centrifugation at 6000g, resuspended in lysis buffer (10 mM Tris-HCl, pH 8.3, 500 mM NaCl, and 5 mM DTT) supplemented with protease inhibitors (Roche), and lysed by sonication. The lysate was filtered and applied to a Ni-NTA HP 5-mL column (GE Healthcare) equilibrated with equilibration buffer (10 mM sodium phosphate buffer and 300 mM NaCl, pH 7.4). The column was subsequently washed with equilibration buffer and the bound protein was eluted with elution buffer (10 mM NaH2PO4, 300 mM NaCl, and 300 mM imidazole, pH 7.4). The eluted protein was concentrated and injected on a size-exclusion chromatography column SD75 16/600 (GE Healthcare) equilibrated on HBS (20 mM HEPES, pH 7.4, and 150 mM NaCl) for further polishing. Protein concentration was measured with a Nanodrop 1000 (Thermo). All proteins were more than 90% pure, as confirmed by SDS-PAGE, and had a high GEF activity toward ARF1.
Nucleotide Exchange Assays
Inhibition of representative Sec7 domains was analyzed by fluorescence kinetics as described (Benabdi et al., 2017). Briefly, nucleotide exchange kinetics were monitored by the change in tryptophan fluorescence that follows the conformational change from ARF-GDP to ARF-GTP (excitation and emission wavelengths of 292 and 340 nm, respectively). Exchange rates (kobs) were determined from monoexponential fits over the entire kinetics and expressed as a percentage of control activity. Experiments were performed with 1 μM ∆17ARF1 and ARF-GEFs at either 100 nM (ARNOSec7 and ARNOFL) or 250 nM (BRAG2sec7, BRAG2Sec7-PH, BIG1Sec7, and Arabidopsis BIG5Sec7), with 50 µM Secdin, 50 µM BFA, or DMSO as control treatment. Purified BIG5Sec7 was highly active toward ∆17ARF1, with a nucleotide-exchange efficiency similar to that of human BIG1Sec7 under equivalent conditions (kobs values of 0.017 s−1 and 0.013 s−1 for BIG1Sec7 and BIG5Sec7, respectively). It should be noted that Secdin absorbs light at 290 nm, which determines the differences in the plateau of the kinetics traces with and without the compound. This decrease was taken into account by fitting the entire kinetics curve. The fluorescence emission spectra of Secdin with and without ARF (excitation wavelength 290 nm) were also controlled. The emission spectra of ARF in the presence of Secdin matched well the addition of the separate spectra, revealing no obvious direct interference of Secdin with ARF. All experiments were done in triplicate.
Statistical Analysis
Unless otherwise specified, P values were calculated with a two-tailed Student’s t test with Excel software.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: GNOM (At1g13980), GNL1 (At5g39500), GNL2 (At5g19610), BIG1 (At4g38200), BIG2 (At3g60860), BIG3 (At1g01960), BIG4 (At4g35380), BIG5/BEN1 (At3g43300), ARF1 (At2g47170), BRI1 (At4g39400), PIN2 (At5g57090), BOR1 (At2g47160), VHAa1 (At2g28520), RABF2a (At5g45130), RABF2b (At4g19640), PIP2A (At3g53420), BSK1 (At2g17090), SYP61 (At1g28490), SYP22 (At5g46860), and AMSH3 (At4g16144).
Supplemental Data
Supplemental Figure 1. Subcellular localization of different plasma membrane proteins and endomembrane markers in Arabidopsis root epidermal cells treated with Secdin.
Supplemental Figure 2. Lack of cytotoxic effects of Secdin.
Supplemental Figure 3. Effects of Secdin on the vacuolar degradation pathway.
Supplemental Figure 4. The effect of Secdin on plasma membrane protein degradation is dependent on protein ubiquitination.
Supplemental Figure 5. Structure-activity relationship analysis of Secdin analogs.
Supplemental Figure 6. DARTS analysis for validation of the putative Secdin protein targets in Arabidopsis and human cells.
Supplemental Figure 7. Representative nucleotide exchange kinetics curves used to estimate the exchange rates (kobs) of spontaneous (EDTA) and GEF-stimulated ARF1 activation after small-molecule treatment.
Supplemental Figure 8. Golgi apparatus disruption in Secdin-treated human cell cultures.
Supplemental Data Set 1. NMR spectra of the synthesized Secdin-related compounds.
Supplemental Data Set 2. Affinity purification of protein interactors of biotin-tagged Secdin derivative (Secdin24) in Arabidopsis PSB-D cell cultures.
Acknowledgments
We thank V. Vassileva, D. Van Damme, S. Vanneste, N. Callewaert, F. Coppens, A. Staes, and K. Gevaert for useful discussions; G. Jürgens, S. Richter, S. Y. He, S. Naramoto, J. Friml, M. Hothorn, K. Kasai, T. Fujiwara, C. Luschnig, Z. Wang, and H. Tanaka for sharing published materials; I. Vanhoutte, E. Mylle, T. De Saedeleer, B. Vanderstraeten, and B. Canher for technical assistance; and M. De Cock for help in preparing the manuscript. This work was supported by the Agency for Innovation by Science and Technology for postdoctoral (K.M.) and predoctoral (W.D.) fellowships; the China Scholarship Council for a predoctoral fellowship (Q.L.); a joint research project within the framework of cooperation between the Research Foundation-Flanders and the Bulgarian Academy of Sciences (VS.025.13N; K.M. and E.R.); the Belgian Science Policy Office for a postdoctoral fellowship (I.S.); the German Science Foundation for Grant IS 221/4-1 (E.I.); a grant from the Fondation pour la Recherche Médicale (J.C.); a predoctoral grant from the Ecole Normale Supérieure Paris-Saclay (F.P.); the Research Foundation-Flanders for an SBO project S006617N (W.A. and M.B.); and the Hercules Foundation for Grant AKUL/09/037, 13/39 (W.A.).
AUTHOR CONTRIBUTIONS
K.M, J.W., and E.R. conceived the study and designed the experiments. B.D., J.H., A.M., and J.W. synthesized compounds and consulted with chemistry. K.M., Q.L., W.D., I.S., W.N., R.D.R., K.G., K.K., L.S.L.N., S.D.M., and A.D. performed the experiments. K.M., Q.L., V.S., D.A., M.S.O., E.I., S.N.S., and E.R. analyzed the data. S.M. and G.V. contributed materials. S.B. and S.D.V. did the mass spectrometry. M.B. and W.A. carried out the imaging in human cells. F.P. and J.C. contributed ARF-GEF activity assays. K.M. and E.R. wrote the article. All authors commented on the results and the manuscript.
References
- Abas L., Luschnig C. (2010). Maximum yields of microsomal-type membranes from small amounts of plant material without requiring ultracentrifugation. Anal. Biochem. 401: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders N., Nielsen M., Keicher J., Stierhof Y.-D., Furutani M., Tasaka M., Skriver K., Jürgens G. (2008). Membrane association of the Arabidopsis ARF exchange factor GNOM involves interaction of conserved domains. Plant Cell 20: 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdi S., Peurois F., Nawrotek A., Chikireddy J., Cañeque T., Yamori T., Shiina I., Ohashi Y., Dan S., Rodriguez R., Cherfils J., Zeghouf M. (2017). Family-wide analysis of the inhibition of ARF guanine nucleotide exchange factors with small molecules: evidence of unique inhibitory profiles. Biochemistry 56: 5125–5133. [DOI] [PubMed] [Google Scholar]
- Bui Q.T., Golinelli-Cohen M.-P., Jackson C.L. (2009). Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease. Mol. Genet. Genomics 282: 329–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codreanu M.C., Audenaert D., Nguyen L., Beeckman T., Russinova E. (2012). Small-molecule dissection of brassinosteroid signaling. Methods Mol. Biol. 876: 95–106. [DOI] [PubMed] [Google Scholar]
- Cortese K., Howes M.T., Lundmark R., Tagliatti E., Bagnato P., Petrelli A., Bono M., McMahon H.T., Parton R.G., Tacchetti C. (2013). The HSP90 inhibitor geldanamycin perturbs endosomal structure and drives recycling ErbB2 and transferrin to modified MVBs/lysosomal compartments. Mol. Biol. Cell 24: 129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13: 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejonghe W., et al. (2016). Mitochondrial uncouplers inhibit clathrin-mediated endocytosis largely through cytoplasmic acidification. Nat. Commun. 7: 11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.-D., Schumacher K. (2006). Vacuolar H+ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J.G., Jackson C.L. (2011). ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12: 362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.M., Haeger A., Vain T., Rigal A., Viotti C., Łangowska M., Ma Q., Friml J., Raikhel N.V., Hicks G.R., Robert S. (2015). An early secretory pathway mediated by GNOM-LIKE 1 and GNOM is essential for basal polarity establishment in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: E806–E815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G., et al. (2011). Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc. Natl. Acad. Sci. USA 108: 17850–17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French A.P., Mills S., Swarup R., Bennett M.J., Pridmore T.P. (2008). Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3: 619–628. [DOI] [PubMed] [Google Scholar]
- Friedrichsen D.M., Joazeiro C.A.P., Li J., Hunter T., Chory J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Hirai M.Y., Chino M., Komeda Y., Naito S. (1992). Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 99: 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Hyman D.L., Wang X., Schumacher K., Chory J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21: 1598–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.-D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230. [DOI] [PubMed] [Google Scholar]
- Grebe M., Gadea J., Steinmann T., Kientz M., Rahfeld J.-U., Salchert K., Koncz C., Jürgens G. (2000). A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12: 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M., Schmitz A., Grüne I., Srivatsan S.G., Paul B., Kolanus W., Quast T., Kremmer E., Bauer I., Famulok M. (2006). Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature 444: 941–944. [DOI] [PubMed] [Google Scholar]
- Hardcastle I.R., et al. (2006). Small-molecule inhibitors of the MDM2-p53 protein-protein interaction based on an isoindolinone scaffold. J. Med. Chem. 49: 6209–6221. [DOI] [PubMed] [Google Scholar]
- Irani N.G., et al. (2012). Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8: 583–589. [DOI] [PubMed] [Google Scholar]
- Isono E., Katsiarimpa A., Müller I.K., Anzenberger F., Stierhof Y.-D., Geldner N., Chory J., Schwechheimer C. (2010). The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell 22: 1826–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.L., Casanova J.E. (2000). Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 10: 60–67. [DOI] [PubMed] [Google Scholar]
- Joshi M.C., Wicht K.J., Taylor D., Hunter R., Smith P.J., Egan T.J. (2013). In vitro antimalarial activity, β-haematin inhibition and structure-activity relationships in a series of quinoline triazoles. Eur. J. Med. Chem. 69: 338–347. [DOI] [PubMed] [Google Scholar]
- Kalinowska K., Nagel M.K., Isono E. (2016). Measuring the enzyme activity of Arabidopsis deubiquitylating enzymes. Methods Mol. Biol. 1450: 35–44. [DOI] [PubMed] [Google Scholar]
- Korbei B., Luschnig C. (2013). Plasma membrane protein ubiquitylation and degradation as determinants of positional growth in plants. J. Integr. Plant Biol. 55: 809–823. [DOI] [PubMed] [Google Scholar]
- Leitner J., Petrášek J., Tomanov K., Retzer K., Pařezová M., Korbei B., Bachmair A., Zažímalová E., Luschnig C. (2012). Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc. Natl. Acad. Sci. USA 109: 8322–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Rodriguez-Furlan C., Wang J., van de Ven W., Gao T., Raikhel N.V., Hicks G.R. (2017). Different endomembrane trafficking pathways establish apical and basal polarities. Plant Cell 29: 90–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B., Jung G., Wohlschlegel J.A., Huang J. (2011). Target identification using drug affinity responsive target stability (DARTS). Curr. Protoc. Chem. Biol. 3: 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B., et al. (2009). Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 106: 21984–21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Boeren S., de Vries S.C., van Valenberg H.J.F., Vervoort J., Hettinga K. (2011). Filter-aided sample preparation with dimethyl labeling to identify and quantify milk fat globule membrane proteins. J. Proteomics 75: 34–43. [DOI] [PubMed] [Google Scholar]
- Marc J., Granger C.L., Brincat J., Fisher D.D., Kao T.-h., McCubbin A.G., Cyr R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins S., Dohmann E.M.N., Cayrel A., Johnson A., Fischer W., Pojer F., Satiat-Jeunemaître B., Jaillais Y., Chory J., Geldner N., Vert G. (2015). Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat. Commun. 6: 6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishev K., Dejonghe W., Russinova E. (2013). Small molecules for dissecting endomembrane trafficking: a cross-systems view. Chem. Biol. 20: 475–486. [DOI] [PubMed] [Google Scholar]
- Mossessova E., Corpina R.A., Goldberg J. (2003). Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol. Cell 12: 1403–1411. [DOI] [PubMed] [Google Scholar]
- Mouratou B., Biou V., Joubert A., Cohen J., Shields D.J., Geldner N., Jürgens G., Melançon P., Cherfils J. (2005). The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics 6: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S., Kleine-Vehn J., Robert S., Fujimoto M., Dainobu T., Paciorek T., Ueda T., Nakano A., Van Montagu M.C.E., Fukuda H., Friml J. (2010). ADP-ribosylation factor machinery mediates endocytosis in plant cells. Proc. Natl. Acad. Sci. USA 107: 21890–21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S., Otegui M.S., Kutsuna N., de Rycke R., Dainobu T., Karampelias M., Fujimoto M., Feraru E., Miki D., Fukuda H., Nakano A., Friml J. (2014). Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell 26: 3062–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R., Howell K.A., Millar A.H., O’Toole N., Small I., Whelan J. (2007). Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19: 3418–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrotek A., Zeghouf M., Cherfils J. (2016). Allosteric regulation of Arf GTPases and their GEFs at the membrane interface. Small GTPases 7: 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Albrethsen J., Larsen F.H., Skriver K. (2006). The Arabidopsis ADP-ribosylation factor (ARF) and ARF-like (ARL) system and its regulation by BIG2, a large ARF-GEF. Plant Sci. 171: 707–717. [Google Scholar]
- Nomura K., DebRoy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223. [DOI] [PubMed] [Google Scholar]
- Norambuena L., Tejos R. (2017). Chemical genetic dissection of membrane trafficking. Annu. Rev. Plant Biol. 68: 197–224. [DOI] [PubMed] [Google Scholar]
- Ohashi Y., Iijima H., Yamaotsu N., Yamazaki K., Sato S., Okamura M., Sugimoto K., Dan S., Hirono S., Yamori T. (2012). AMF-26, a novel inhibitor of the Golgi system, targeting ADP-ribosylation factor 1 (Arf1) with potential for cancer therapy. J. Biol. Chem. 287: 3885–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer W.A. (2011). Plasma membrane protein trafficking. Plant Cell Monogr. 19: 31–56. [Google Scholar]
- Renault L., Guibert B., Cherfils J. (2003). Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 426: 525–530. [DOI] [PubMed] [Google Scholar]
- Richter S., Geldner N., Schrader J., Wolters H., Stierhof Y.-D., Rios G., Koncz C., Robinson D.G., Jürgens G. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448: 488–492. [DOI] [PubMed] [Google Scholar]
- Richter S., Kientz M., Brumm S., Nielsen M.E., Park M., Gavidia R., Krause C., Voss U., Beckmann H., Mayer U., Stierhof Y.-D., Jürgens G. (2014). Delivery of endocytosed proteins to the cell-division plane requires change of pathway from recycling to secretion. eLife 3: e02131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S., Chary S.N., Drakakaki G., Li S., Yang Z., Raikhel N.V., Hicks G.R. (2008). Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. USA 105: 8464–8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roling O., Wendeln C., Kauscher U., Seelheim P., Galla H.-J., Ravoo B.J. (2013). Layer-by-layer deposition of vesicles mediated by supramolecular interactions. Langmuir 29: 10174–10182. [DOI] [PubMed] [Google Scholar]
- Sáenz J.B., Sun W.J., Chang J.W., Li J., Bursulaya B., Gray N.S., Haslam D.B. (2009). Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat. Chem. Biol. 5: 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R., Declerck I., Peric A., Raemaekers T., Menendez G., Zhou L., Veerle B., Coen K., Munck S., De Strooper B., Schiavo G., Annaert W. (2011). ADP ribosylation factor 6 (ARF6) controls amyloid precursor protein (APP) processing by mediating the endosomal sorting of BACE1. Proc. Natl. Acad. Sci. USA 108: E559–E568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring D., Viotti C., Krüger F., Künzl F., Sturm S., Bubeck J., Hillmer S., Frigerio L., Robinson D.G., Pimpl P., Schumacher K. (2011). Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan M.B., Staiger C.J., Rose R.J., McCurdy D.W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136: 3968–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C., Li N., Boeren S., America T., van Dongen W., Goerdayal S.S., de Vries S., Angenent G.C., Kaufmann K. (2012). Proteomics-based identification of low-abundance signaling and regulatory protein complexes in native plant tissues. Nat. Protoc. 7: 2144–2158. [DOI] [PubMed] [Google Scholar]
- Steinmann T., Geldner N., Grebe M., Mangold S., Jackson C.L., Paris S., Gälweiler L., Palme K., Jürgens G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286: 316–318. [DOI] [PubMed] [Google Scholar]
- Takano J., Tanaka M., Toyoda A., Miwa K., Kasai K., Fuji K., Onouchi H., Naito S., Fujiwara T. (2010). Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc. Natl. Acad. Sci. USA 107: 5220–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.-i., Watanabe M., Nishimura M., Hara-Nishimura I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35: 545–555. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19: 391–397. [DOI] [PubMed] [Google Scholar]
- Tang W., Kim T.-W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.-Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh O.-K., Moore I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448: 493–496. [DOI] [PubMed] [Google Scholar]
- Ueda T., Uemura T., Sato M.H., Nakano A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40: 783–789. [DOI] [PubMed] [Google Scholar]
- Van Leene J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238. [DOI] [PubMed] [Google Scholar]
- Vernoud V., Horton A.C., Yang Z., Nielsen E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131: 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C., et al. (2010). Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cai Y., Miao Y., Lam S.K., Jiang L. (2009). Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 60: 3075–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee E.G.-T., Sherrier D.J., Prime T.A., Dupree P. (1998). Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell 10: 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich J.R., Boeren S., Möller B.K., Weijers D., De Rybel B. (2017). In vivo identification of plant protein complexes using IP-MS/MS. Methods Mol. Biol. 1497: 147–158. [DOI] [PubMed] [Google Scholar]
- Wessig P., Möllnitz K. (2008). Nanoscale molecular rods with a new building block for solubility enhancement. J. Org. Chem. 73: 4452–4457. [DOI] [PubMed] [Google Scholar]
- Wiśniewska J., Xu J., Seifertová D., Brewer P.B., Růžička K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J. (2006). Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Wright J., Kahn R.A., Sztul E. (2014). Regulating the large Sec7 ARF guanine nucleotide exchange factors: the when, where and how of activation. Cell. Mol. Life Sci. 71: 3419–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Sato K., Takeuchi M. (2014). Molecular mechanisms of Sar/Arf GTPases in vesicular trafficking in yeast and plants. Front. Plant Sci. 5: 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeh J.-C., Zeghouf M., Grauffel C., Guibert B., Martin E., Dejaegere A., Cherfils J. (2006). Dual specificity of the interfacial inhibitor Brefeldin A for Arf proteins and Sec7 domains. J. Biol. Chem. 281: 11805–11814. [DOI] [PubMed] [Google Scholar]
- Zhang C., et al. (2016). Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc. Natl. Acad. Sci. USA 113: E41–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]