Turnip mosaic virus replication vesicles bypass the Golgi and take an alternative, nonconventional pathway involving VTI11 in order to reach prevaculolar compartments/multivesicular bodies.
Abstract
Infection of plant cells by RNA viruses leads to the generation of organelle-like subcellular structures that contain the viral replication complex. During Turnip mosaic virus (TuMV) infection of Nicotiana benthamiana, the viral membrane protein 6K2 plays a key role in the release of motile replication vesicles from the host endoplasmic reticulum (ER). Here, we demonstrate that 6K2 contains a GxxxG motif within its predicted transmembrane domain that is vital for TuMV infection. Replacement of the Gly with Val within this motif inhibited virus production, and this was due to a relocation of the viral protein to the Golgi apparatus and the plasma membrane. This indicated that passage of 6K2 through the Golgi apparatus is a dead-end avenue for virus infection. Impairing the fusion of transport vesicles between the ER and the Golgi apparatus by overexpression of the SNARE Sec22 protein resulted in enhanced intercellular virus movement. Likewise, expression of nonfunctional, Golgi-located synaptotagmin during infection enhanced TuMV intercellular movement. 6K2 copurified with VTI11, a prevacuolar compartment SNARE protein. An Arabidopsis thaliana vti11 mutant was completely resistant to TuMV infection. We conclude that TuMV replication vesicles bypass the Golgi apparatus and take an unconventional pathway that may involve prevacuolar compartments/multivesicular bodies for virus infection.
INTRODUCTION
Plant positive-sense (+) RNA viruses remodel the endomembrane system of infected cells to produce viral factories (Laliberté and Sanfaçon, 2010; Laliberté and Zheng, 2014; Romero-Brey and Bartenschlager, 2014; Harak and Lohmann, 2015). Viral factories may take the form of spherules, which result from the invagination of the outer membrane of organelles such as the endoplasmic reticulum (ER) (Brome mosaic virus [BMV]), peroxisomes (Tomato bushy stunt virus [TBSV]), or chloroplasts (Turnip yellow mosaic virus). In plant cells infected by other RNA viruses, viral factories can be associated with vesicles that are released from the ER (Laliberté and Zheng, 2014). Spherules and vesicles contain viral replication complexes (VRCs) that are made of the viral RNA-dependent RNA polymerase (vRdRp), replication-associated viral and host proteins, and viral RNA (vRNA) as well as its associated double-stranded replicative form (Cotton et al., 2009). Besides being involved in vRNA replication, vesicles also can be involved in the movement of a vRNA-protein complex using myosin motors (Harries et al., 2009; Amari et al., 2011, 2014; Agbeci et al., 2013) to reach plasmodesmata (Grangeon et al., 2013; Tilsner et al., 2013). Then, the vRNA-protein complex enters the plasmodesmata, leading to the infection of neighboring healthy cells (Kawakami et al., 2004; Grangeon et al., 2013).
Plant (+) RNA virus genomes encode at least one membrane-associated protein that triggers the formation of spherules or vesicles (Laliberté and Zheng, 2014). These viral proteins are part of the VRCs and associate with membranes through transmembrane domains (TMDs) and/or amphipathic helices (Zhang et al., 2005; Liu et al., 2009). Host membrane proteins may further mediate the association of VRCs with cellular membranes (Nishikiori et al., 2011). Ectopic expression of these membrane-associated proteins very often produces spherules or vesicles similar to those produced during infection (Schwartz et al., 2002; Beauchemin et al., 2007). These membrane-associated viral proteins contain functional domains for direct or indirect interactions with vRNA and viral replication proteins as well as host factors that lead to the assembly of the VRCs (Kovalev et al., 2012; Diaz et al., 2015). For example, the C-terminal half of the viral protein p27 of Red clover necrotic mosaic virus (RCNMV) interacts with the p88 polymerase and further recruits the viral RNA 2 template for viral RNA replication (Mine et al., 2010; Iwakawa et al., 2011). Also, the N-terminal 16 amino acid residues of Carnation italian ringspot virus viral protein p36 mediate its interaction with host protein Vps23 (Richardson et al., 2014).
Interaction studies between membrane-associated viral proteins and host factors have emphasized the importance of protein trafficking components in the biogenesis of spherules or vesicles (Patarroyo et al., 2013; Laliberté and Zheng, 2014; Hyodo and Okuno, 2016). Endosomal sorting complexes required for transport (ESCRT) factors, which are normally required for the formation of multivesicular bodies (MVBs) (Schmidt and Teis, 2012), are hijacked by TBSV and BMV and appear to facilitate membrane curvature and VRC assembly (Barajas et al., 2009, 2014; Diaz et al., 2015). Host factors of the early secretory pathway, such as COPII Sec24A (Sec24 homolog A) and COPI ARF1, were identified as being involved in the formation of the replication vesicles of Turnip mosaic virus (TuMV) and RCNMV, respectively (Hyodo et al., 2013; Wei et al., 2013; Jiang et al., 2015). Finally, soluble N-ethylmaleimide sensitive factor attachment protein receptors (SNAREs) and synaptotagmins (SYTs), such as the ER SNARE Syp71 (Wei et al., 2013) and SYTA (Uchiyama et al., 2014), which normally work in concert to regulate fusion of transport vesicles with target membranes (Rizo et al., 2006), were shown to be important for TuMV infection. SYTA knockdown (KD) mutant also impaired Turnip vein clearing virus and Cabbage leaf curl virus movement and infection (Lewis and Lazarowitz, 2010; Uchiyama et al., 2014; Levy et al., 2015). However, it is not known if the conventional secretory pathway is required for the viral vesicles to reach plasmodesmata or whether a different, unconventional route is used.
TuMV is a (+) RNA virus in the family of Potyviridae (Mayo, 1995). Its 9.8-kb genome encodes at least 11 viral proteins, and among them are the membrane-associated P3 and 6K2 proteins (Restrepo-Hartwig and Carrington, 1994; Eiamtanasate et al., 2007; Jiang et al., 2015). 6K2 is a 6-kDa protein and, according to secondary structure predictions, is characterized by the presence of a 23-amino acid α-helix TMD, as well as a 19- and an 11-amino acid N- and C-terminal tail, respectively (Jiang et al., 2015). 6K2 is involved in the formation of vesicles (Beauchemin et al., 2007; Cotton et al., 2009; Grangeon et al., 2012) that contain vRNA and viral proteins such as the vRdRp, the cytoplasmic inclusion helicase, and the viral protein linked to the genome fused to the viral proteinase (VPg-Pro) (Cotton et al., 2009; Grangeon et al., 2012; Wan et al., 2015a), as well as host components such as translation factors (Beauchemin et al., 2007; Beauchemin and Laliberté, 2007; Thivierge et al., 2008; Huang et al., 2010). These vesicles are motile and thus support the vRNA intracellular and intercellular movement as well as systemic movement, which leads to the infection of the whole plant (Cotton et al., 2009; Grangeon et al., 2013; Wan et al., 2015b). The presence of TuMV replication complexes in vascular elements thus challenges existing conceptions of virus long-distance transport (Folimonova and Tilsner, 2018).
The molecular determinants contained in 6K2 that direct vesicle production and trafficking are largely unknown. Recently, we found both that the N-terminal cytoplasmic tail of 6K2 is required for the ER export of 6K2 and that it interacts with the COPII subunit Sec24A (Jiang et al., 2015). We report here that the predicted TMD of 6K2 contains a GxxxG motif (x being any amino acid) that is important for the production of replication vesicles. Mutating the Gly residues of this motif blocked the production of replication vesicles and delocalized 6K2 to Golgi bodies and the plasma membrane (PM), which prevented virus production. We also demonstrate that 6K2 vesicles bypass the Golgi apparatus and use a nonconventional pathway for effective virus infection. Indeed, impairing ER-Golgi trafficking did not inhibit 6K2 vesicle production and, on the contrary, enhanced the effect on virus cell-to-cell movement and whole-plant systemic infection. Finally, we found that this Golgi bypass pathway involved VTI11 (VESICLE TRANSPORT V-SNARE11) a prevacuolar compartment (PVC) SNARE protein.
RESULTS
Identification of a GxxxG Motif within the Transmembrane Domain of 6K2
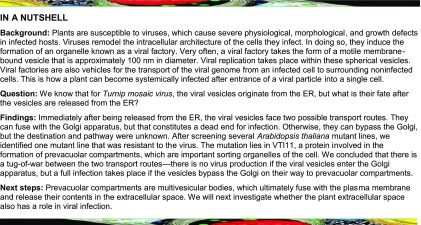
We previously demonstrated that the N-terminal tail of 6K2 is involved in ER exit by interacting with the COPII subunit Sec24A in Nicotiana benthamiana (Jiang et al., 2015). To further define the role of the 6K2 TMD in replication vesicle biogenesis, we compared the amino acid sequence of predicted TMDs of different potyviruses using the Clustal W2 online server (Larkin et al., 2007). Gly residues are found at position 30, 33, 34, and 35 of the TuMV 6K2, and this amino acid is frequently found at equivalent positions among potyviral 6K2 proteins (Figure 1A). In particular, Gly-34 is present in all the aligned sequences, and Gly-35 is present in all except for the Potato virus A 6K2 where it is replaced by an alanine residue. Gly-30 is present in four out of the seven aligned sequences or is replaced by comparable small amino acid residues (e.g., Ala or Ser) for Potato virus Y and Tobacco etch virus. Helical wheel projection of the predicted 6K2 TMD of TuMV indicates that Gly-30 and Gly-34 are located on the same side of the predicted α-helix (Figure 1A). This amino acid organization is characteristic of the GxxxG motif, which mediates protein interactions between membrane proteins (Teese and Langosch, 2015).
Figure 1.
Mutation of the GxxxG Motif in 6K2 Alters Its Localization.
(A) Sequence alignment of the predicted TMDs of several potyviral 6K2 proteins. The conserved Gly residues are shown (light blue) and the corresponding amino acid position in TuMV is indicated below. PPV, Plum pox virus; PVA, Potato virus A; PVY, Potato virus Y; SMV, Soybean mosaic virus; LMV, Lettuce mosaic virus; TEV, Tobacco etch virus. To the right is a helical wheel projection of the predicted 6K2 TMD of TuMV. The Gly residues located at positions 30 and 34 are indicated.
(B) Fluorescence imaging of N. benthamiana cells expressing GFP fusions of wild-type 6K2 and the indicated 6K2 mutants at 3 dpi. These images are three-dimensional renderings stacks of 40 1-μm-thick slices that overlap by 0.5 μm.
(C) Membrane association of wild-type 6K2 and 6K2GV protein. S3, total protein fraction; S30, soluble protein fraction; P30, membrane protein fraction. Immunoblotting was performed with anti-GFP antibodies.
(D) Confocal images of N. benthamiana epidermal cells expressing 6K2:mCherry (top middle panel) or 6K2GV:mCherry (bottom middle panel) with the cis-Golgi marker ERD2:GFP (left panels). Their coexpression is shown in the right panels (merge). Arrows in right top panel indicate 6K2:mCherry colocalizing with the cis Golgi marker.
(E) Colocalization of ERD2:GFP and 6K2:mCherry (6K2) or 6K2GV:mCherry (6K2GV). Statistical significance was calculated using the Pearson’s correlation coefficient r values of ImageJ and a t test analysis was done (*P < 0.05). Values indicate means ± sd and n = 18 cells.
(F) Fluorescence images of cells expressing 6K2GV:mCherry or 6K2:mCherry merged with the bright field image are shown.
(G) The PM marker AHA2:GFP was expressed with either 6K2GV:mCherry or mCherry. Asterisks indicate the interior of the cells. All confocal images are single optical images (1 μm thick). Three independent biological repeats were performed for (B), (D), and (F), and two for (C) and (G). Bars = 20 μm in (B), (F), and (G) and 10 μm in (D).
We performed site-directed mutagenesis to substitute each Gly with a Val to test the importance of these residues in the formation of the 6K2-induced vesicles. The functionality of 6K2 fluorescent protein fusions has been well documented by our group and others. For instance, 6K2:mCherry fusion behaves similarly as 6K2:GFP (Cotton et al., 2009), and 6K2:GFP, like 6K2, induces the formation of ER-derived vesicles and the formation of chloroplast invaginations (Wei et al., 2013). Finally, 6K2:mCherry colocalizes with vRNA and replication viral proteins (Wan et al., 2015b). We generated the following mutants 6K2G30V, 6K2G33V, 6K2G34V, 6K2G35V, and 6K2G30V-G33V-G34V-G35V (this last mutant is designated as 6K2GV), and each was fused to the N-terminal end of GFP or mCherry. Fusion proteins were produced by Agrobacterium tumefaciens-mediated transient protein expression in N. benthamiana leaves. Expression of wild-type 6K2:GFP induced the formation of punctae and aggregates of various sizes, ranging from 0.5 to 4.0 μm in diameter (Figure 1B). The expression pattern for 6K2G33V and 6K2G35V was similar to that of wild-type 6K2 protein. However, expression of 6K2G30V, 6K2G34V, and 6K2GV generated punctae of ∼2.0 μm in diameter that were dispersed and evenly distributed. In all cases, faint retention in the ER of the 6K2 protein was observed. A cellular fractionation experiment indicated that 6K2GV, like wild-type 6K2, was predominantly found in the 30,000g membrane-associated pellet (P30 fraction) (Figure 1C), thereby confirming that 6K2GV was still a membrane protein.
Mutation of the GxxxG Motif Causes 6K2 to Localize in the Golgi Apparatus
We then investigated the cellular distribution of the 6K2GV mutant protein. The diameter and homogenous size of the punctate structures induced by 6K2G30V, 6K2G34V, and 6K2GV were reminiscent of Golgi bodies (Figure 1B). We consequently expressed 6K2:mCherry or 6K2GV:mCherry with ERD2:GFP, a cis-Golgi marker that also faintly labels the ER (Saint-Jore et al., 2002). Only a portion of the wild-type 6K2 punctae overlapped with ERD2:GFP, while all 6K2GV punctae localized with this marker (Figure 1D). Twenty cells were analyzed and we calculated that 51% ± 6% of 6K2 punctae localized with the cis-Golgi marker, while the value rose up to 96% ± 4% for 6K2GV (Figure 1E). Furthermore, a strong continuous signal for 6K2GV:mCherry was visible at the cell periphery, but this was not the case for wild-type 6K2:mCherry (Figure 1F), indicating that 6K2 may have reached the PM because the GxxxG motif was mutated. The PM marker AHA2:GFP (Chen et al., 2011) was expressed with 6K2GV:mCherry or the cytoplasmic protein mCherry. The signal for 6K2GV:mCherry was on the apoplastic side, whereas mCherry was located on the cytoplasmic side of the PM marker (Figure 1G). Recalling the membranous nature of the mutated viral protein (Figure 1C, lower panel), 6K2GV is likely PM localized, but we cannot exclude the possibility that the viral protein has also been secreted into the apoplast. The lack of PM labeling by 6K2GV:GFP (Figure 1B) can be explained by the pH sensitivity of GFP, which loses its fluorescence in the acidic environment of the apoplast (Doherty et al., 2010), but this is not the case for mCherry (Ivanov and Harrison, 2014). These results indicate that mutating the GxxxG motif has modified 6K2 from a vesicle-forming protein to a default membrane protein that now traffics through the conventional secretory pathway.
Differential Sensitivity of 6K2 and 6K2GV to Inhibitors of ER-Golgi Trafficking
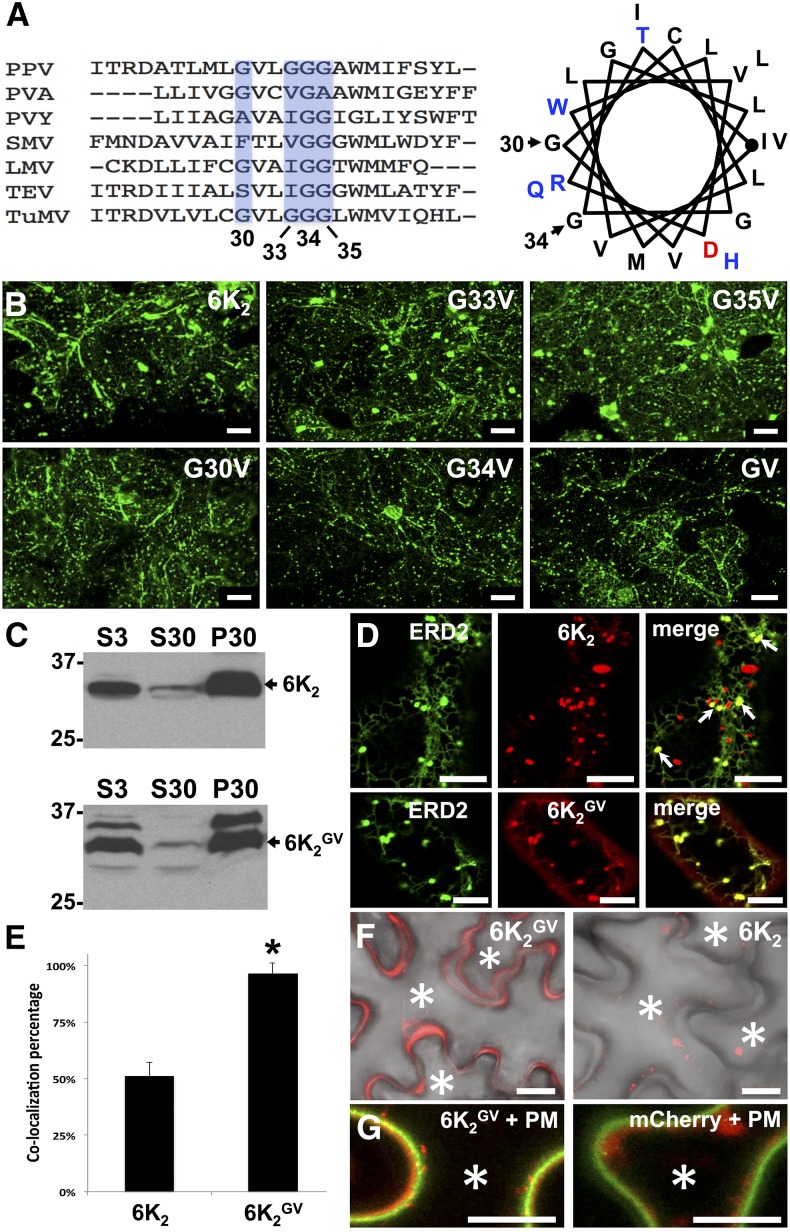
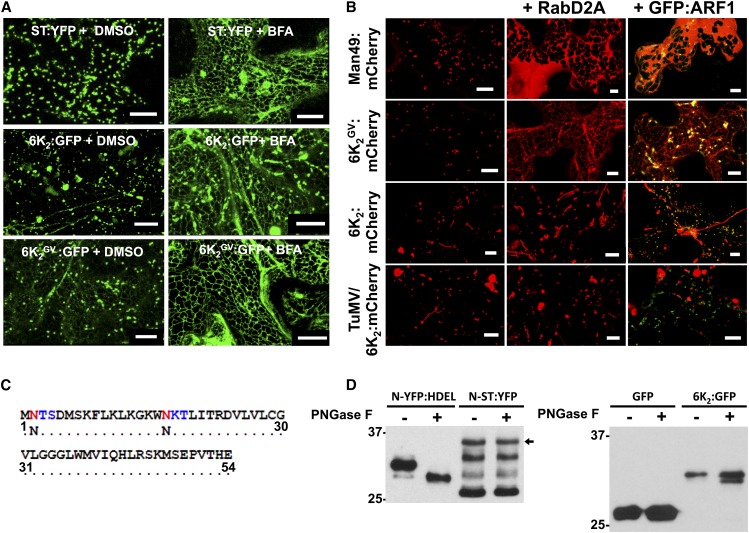
Brefeldin A (BFA) is a fungal toxin that blocks the entrance of proteins into the Golgi-dependent trafficking secretory pathway by impairing COPI function. Hence, BFA is widely used to assess if the trafficking of the protein under observation follows the conventional Golgi-dependent secretory pathway (Nebenführ et al., 2002). Leaf tissues expressing the trans-Golgi marker ST:YFP (Zheng et al., 2005), 6K2:GFP, or 6K2GV:GFP were treated with 20 μg/mL BFA 24 h prior to confocal observation. As expected, the addition of BFA induced the collapse of the Golgi apparatus, as evidenced by the retention of ST:YFP in the ER (Figure 2A). However, addition of BFA did not block the formation of 6K2-induced punctae, although some ER labeling was observed. In contrast, BFA prevented 6K2GV from reaching the Golgi apparatus, resulting in its complete ER retention. This experiment clearly shows that 6K2 and 6K2GV have a differential sensitivity to BFA, confirming that the intracellular trafficking of 6K2 is relocated toward the ER-Golgi-PM conventional secretory pathway when the GxxxG motif is mutated.
Figure 2.
Differential Sensitivity of 6K2 and 6K2GV to Inhibitors of ER-Golgi Trafficking.
(A) Confocal microscopy observation of N. benthamiana epidermal leaf cellsexpressing the trans-Golgi markers ST:YFP, 6K2:GFP, and 6K2GV:GFP were treated with DMSO (left panels) or with 20 μg/mL BFA (right panels) 24 h prior to confocal observation. Images are three-dimensional rendering stacks of 40 1-μm-thick slices that overlap by 0.5 μm.
(B) Confocal microscopy observation of N. benthamiana epidermal leaf cellexpressing the Golgi markers Man49:mCherry, 6K2GV:mCherry, and 6K2:mCherry ectopically or during TuMV infection alone (left panels) or in cells expressing Rab-D2A N123I (middle panels) or GFP:ARF1 NI (right panels). Images are three-dimensional renderings of 40 1-μm-thick slices that overlap by 0.5 μm.
(C) 6K2 N-glycosylation sites on the Asn residues in positions 2 and 17 (red) predicted by in silico analysis, forming N-X-S and N-X-T motifs (X-S/T in blue), respectively.
(D) Immunoblot performed with anti-GFP serum and protein extracts from N. benthamiana leaves expressing N-YFP:HDEL, N-ST:YFP, GFP, or 6K2:GFP that were treated with or without PNGase F. Black arrow indicates the position of N-ST:YFP. Two independent biological replicates were performed for (A) and (B), and three for (D). Bars = 20 μm in (A) and 10 μm in (B).
To support the notion that 6K2GV transits through the Golgi apparatus, we studied two dominant-negative mutant GTPases known to impair the secretory pathway. The first mutant protein, a GFP fusion with the ADP-ribosylation factor 1 mutated in its GTP/GDP exchanging site (GFP:ARF1 NI), blocks the COPI machinery and leads to the reabsorbance of Golgi membrane proteins into the ER (Stefano et al., 2006). The second mutant protein used was Rab-D2A N123I, which is a dominant-negative mutant of Rab-D2A that inhibits trafficking between the ER and Golgi (Zheng et al., 2005). Expression of GFP:ARF1 NI and Rab-D2A N123I lead to a clear retention of the Golgi marker Man49:mCherry (Saint-Jore-Dupas et al., 2006) in the ER (Figure 2B). Similarly, 6K2GV:mCherry remained in the ER when either of the two dominant negative mutants was expressed. On the contrary, the localization pattern of 6K2:mCherry (e.g., production of heterogeneous sized punctae) produced either ectopically or during infection was not affected by the presence of the protein trafficking inhibitors. These experiments confirmed that 6K2GV enters the Golgi apparatus and follows the conventional secretory pathway, whereas this is not the case for most of the wild-type 6K2.
Proteins can undergo co- and posttranslational modification, such as N-glycosylation. In silico glycosylation prediction indicates the presence of two putative N-glycosylation sites in 6K2 (Figure 2C). N-glycosylation initially takes place in the ER but can further be modified in the Golgi apparatus. Normally, N-glycans are sensitive to removal by peptide-N-glycosidase F (PNGase F), but addition in the Golgi apparatus of fucose α (1-3)-linked to the glycan core blocks removal of the glycan by PNGase F (Tretter et al., 1991). Consequently, if a population of 6K2 effectively does not enter the Golgi apparatus, this population would be expected to accumulate as a PNGase F-sensitive form. To test this hypothesis, we expressed 6K2:GFP in N. benthamiana leaves and incubated the cell-free extract with PNGase F. As controls, we expressed N-YFP:HDEL and N-ST:YFP, which are a N-glycosylable form of the ER marker YFP:HDEL and the Golgi marker ST:YFP, respectively (Batoko et al., 2000). Immunoblot analysis showed that N-YFP:HDEL and N-ST:YFP were PNGase F-sensitive and PNGase F-resistant, respectively (Figure 2D). On the other hand, 6K2 was partially sensitive to PNGase F treatment. The deglycosylation assay demonstrates that 6K2 is glycosylated in the ER and that a subset of 6K2 enters the Golgi apparatus (upper resistant band) and that another subset bypasses this organelle (lower sensitive band). No N-glycosylation site was predicted to be found in GFP and lack of GFP modification indicates that it is the 6K2 moiety of the fusion protein that was glycosylated.
The GxxxG Motif Is Required for Virus Replication
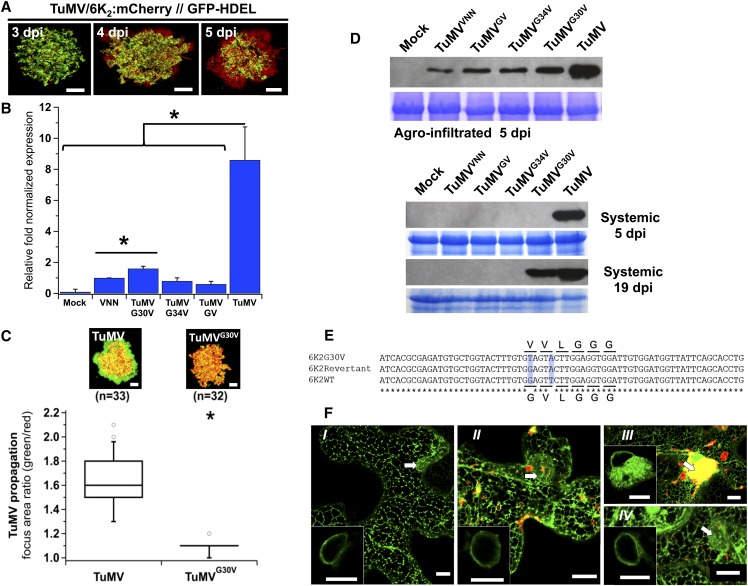
We next evaluated the impact of mutating the GxxxG motif of 6K2 on virus production. Mutations coding for 6K2G30V, 6K2G34V, and 6K2GV were introduced into the infectious clone pCambiaTuMV (Jiang et al., 2015), and the resulting modifications were identified as pCambiaTuMVG30V, pCambiaTuMVG34V, and pCambiaTuMVGV, respectively. pCambiaTuMVVNN, in which the vRdRp core motif GDD was changed to VNN, was used as a replication-defective virus control (Li and Carrington, 1995). The above viral constructs were used to inoculate plants by Agrobacterium-mediated infiltration. Viral replication was assayed 3.25 d postinfiltration (dpi) prior to any virus intercellular movement (Figure 3A) . The intercellular viral movement was evaluated by using the dual tagged TuMV infectious clone (TuMV/6K2:mCherry//GFP-HDEL) that enables to follow virus intercellular movement with the newly infected area labeled with red fluorescence only (Agbeci et al., 2013). Replication was evaluated by RT-qPCR (Figure 3B). A high quantity of vRNA was observed following infiltration with pCambiaTuMV, whereas a lesser amount was obtained with the replication-deficient pCambiaTuMVVNN. The signal from pCambiaTuMVVNN resulted from transcription of the TuMVVNN gene cassette without any vRNA amplification step. The amount of vRNA following agroinoculation with pCambiaTuMVGV, pCambiaTuMVG34V, and pCambiaTuMVG30V was close to the level obtained with pCambiaTuMVVNN, indicating that the mutations severely affected virus replication. However, comparison between pCambiaTuMVVNN and pCambiaTuMVG30V conditions highlighted that a slight, but significant, level of replication was taking place for TuMVG30V.
Figure 3.
The GxxxG Motif Is Important for Virus Replication.
(A) Time course of TuMV intercellular movement in N. benthamiana after agroinfiltration of pCambiaTuMV/6K2:mCherry//GFP-HDEL.
(B) Viral replication evaluated by RT-qPCR from N. benthamiana leaves agroinfiltrated with pCambia (Mock) and the pCambiaTuMV mutants indicated at 3.25 dpi. Values are graphically presented on a histogram. Statistical differences were determined by t test analyses (*P < 0.05). Error bars depict the sd calculated from values obtained in the three biological replicates.
(C) TuMV/6K2:GFP (TuMV) or TuMVG30V/6K2G30V:GFP (TuMVG30V) was coagroinfiltrated with mCherry-HDEL and expression of GFP and mCherry fusions is shown at 5 dpi. Upper panels are representative images of infection foci corresponding to the median value of each treatment. Data sets for TuMV and TuMVG30V treatments are graphically presented below as whisker and box plots. Statistical significant differences (*P < 0.05). n, number of infection foci analyzed.
(D) Total proteins from pCambia- (Mock), pCambiaTuMVVNN-, pCambiaTuMVGV-, pCambiaTuMVG34V-, pCambiaTuMVG30V-, and pCambiaTuMV-agroinfiltrated leaf tissues at 5 dpi, agroinfiltrated leaf tissues, and upper nonagroinfiltrated (systemic) leaf tissues at 5 and 19 dpi were analyzed by immunoblot with a rabbit serum against TuMV CP. Coomassie blue staining (bottom panel of agroinfiltrated 5 dpi, systemic 5 dpi and 19 dpi) shows equal protein loading.
(E) An alignment ofnucleotide sequence of 6K2, 6K2G30V, and its related revertant is shown. Asterisks indicate the conserved nucleotides and the differences are shadowed in light blue. The amino acid residues encoded by the underlined nucleotides are shown.
(F) Confocal microscopy observation of N. benthamiana epidermal leaf cells expressing the ER marker GFP:HDEL alone (I) or with pCambiaTuMVVNN/6K2:mCherry (II), pCambiaTuMV/6K2:mCherry (III), or pCambiaTuMVG30V/6K2G30V:mCherry (IV). Images are three-dimensional renderings of 40 1-μm-thick slices that overlap by 0.5 μm. Arrows denote the position of the nucleus and bottom left inserts are single optical slices of the area denoted by the arrow showing the ER surrounding the nucleus.
Three independent biological repeats were performed for (A) to (C) and (F), and two for (D) with the exception that reversion of TuMVG30V was noted once. Bars = 20 μm in (A) and (C) and 10 μm in (F).
TuMV intercellular movement was followed by infiltrating leaves with pCambiaTuMV/6K2:GFP or pCambiaTuMVG30V/6K2G30V:GFP. In the first construct, the 6K2:GFP coding sequence was inserted between the P1 and HC-Pro cistron of TuMV, thus allowing the release of 6K2:GFP upon production of the viral polyprotein (Cotton et al., 2009; Grangeon et al., 2012). pCambiaTuMVG30V/6K2G30V:GFP contains G30V mutation in the 6K2 cistron and also produces 6K2G30V:GFP from the same position on the viral polyprotein as for pCambiaTuMV/6K2:GFP. Each vector was expressed along with the vector expressing mCherry:HDEL in order to distinguished primary infection foci from secondary infection foci. Primary infected cells express both mCherry and GFP, while secondary foci express GFP only. After 5 dpi, confocal microscopy observations on TuMV infection foci showed that a green-only fluorescence signal was found encircling an area of both red and green fluorescence, indicative of virus movement (Figure 3C). However, only slight green-only fluorescence was measured for leaves infiltrated with pCambiaTuMVG30V/6K2G30V:GFP, suggesting that this mutant is capable of slow movement. Movement was quantified and normalized for each focus of infection by measuring the surface area of green fluorescence divided by the surface area of red fluorescence. The ratios of the focus areas were graphically represented using whisker and box plots and statistically analyzed using the nonparametric Wilcoxon-Mann-Whitney two-sample rank test. Statistical comparison of both TuMVG30V/6K2G30V:GFP and pCambiaTuMV/6K2:GFP infection area ratio distributions indicated that intercellular movement was greatly diminished for the mutant virus, but not totally abolished.
Systemic virus accumulation was determined at 5 dpi by immunoblotting leaf tissue extracts with a rabbit serum raised against the TuMV capsid protein (CP) (McClintock et al., 1998). All infiltrated leaves showed accumulation of CP, to varying levels (Figure 3D), but the viral protein was detected in upper noninoculated leaf tissues only in pCambiaTuMV-infiltrated plants. However, CP was detected in the pCambiaTuMVG30V upper noninoculated leaf tissues at 19 dpi. mRNA from pCambiaTuMVG30V-systemically infected leaf tissues was isolated and reverse-transcribed. DNA sequencing of the 6K2 coding region indicated that virus production was the result of a reversion of the G-to-T mutation back to a G, thereby reinstating the Gly residue at position 30 of 6K2 (Figure 3E). This reversion indicates that some viral replication is taking place as suggested in Figure 3B, albeit at a very low level.
TuMV infection induces the amalgamation of the ER, Golgi and chloroplasts into a large perinuclear, globular structure—a hallmark for normal TuMV replication/infection at the cellular level (Grangeon et al., 2012; Wan et al., 2015a). To qualitatively evaluate TuMV replication based on the formation of ER amalgamates, we thus expressed the ER marker GFP:HDEL alone (Figure 3F, I) or concomitantly with agroinfiltrated pCambiaTuMVVNN/6K2:mCherry (Figure 3F, II), pCambiaTuMV/6K2:mCherry (Figure 3F, III), or pCambiaTuMVG30V/6K2G30V:mCherry (Figure 3F, IV). Formation of perinuclear ER amalgamates was evaluated by confocal microscopy 5 d later. The expression pattern for GFP:HDEL, as shown in Figure 3F, panel I, bottom left insert, is characteristic of the ER surrounding the nucleus (Beauchemin et al., 2007). Although agro-infiltration of pCambiaTuMVVNN/6K2:mCherry led to the production of 6K2:mCherry punctae, no ER amalgamation near the nucleus (Figure 3F, panel II, bottom left inset) was ever noted following the observation of 20 samples. The punctae observed in Figure 3F, panel II, are due to the expression of the gene cassette expression without any vRNA amplification. On the other hand, pCambiaTuMV/6K2:mCherry always induced the formation of large perinuclear structures containing 6K2:mCherry and GFP:HDEL (n = 20) (Figure 3F, panel III, left inset), confirming that formation of ER amalgamate can be used as a qualitative indicator of TuMV replication. Finally, expression of pCambiaTuMVG30V/6K2G30V:mCherry produced red punctae, but replication of TuMVG30V was not sufficient to induce amalgamate formation (n = 20) (Figure 3F, IV, left inset). In conclusion, the low level of cellular replication and systemic infection that we observed in the mutants highlights the importance of the GxxxG motif for TuMV infection. It also demonstrates that, when 6K2 transits through the Golgi apparatus, it becomes a dead end for the virus.
Disrupting ER-Golgi Homeostasis Enhances Intercellular Movement of TuMV
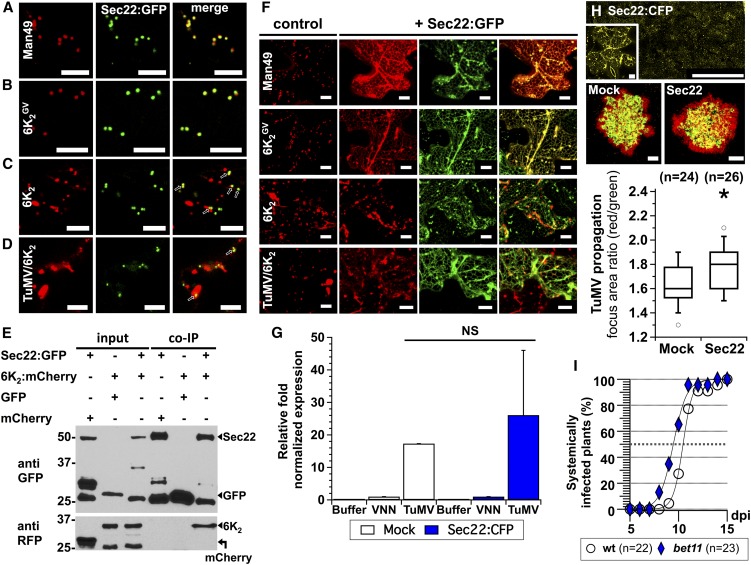
We next investigated the impact of impaired fusion of transport vesicles with the Golgi apparatus on TuMV intercellular movement. Impairment of fusion was achieved by transiently overexpressing the Sec22 SNARE protein, which induced the collapse of Golgi membrane proteins into the ER (Chatre et al., 2005). A link between Sec22 and 6K2 was first demonstrated by confocal microscopy localization experiments with fluorescent-tagged proteins. N. benthamiana leaves were agroinfiltrated to transiently coexpress Sec22:GFP with Man49:mCherry, 6K2GV:mCherry, 6K2:mCherry, and TuMV/6K2:mCherry, respectively. Man49:mCherry (Figure 4A) and 6K2GV:mCherry (Figure 4B) completely colocalized with Sec22:GFP, whereas only a small proportion of 6K2:mCherry (Figure 4C, arrows) and 6K2:mCherry punctae produced during infection did (Figure 4D, arrows). In addition to these localization experiments, we tested if 6K2 could be purified along with Sec22 by performing coimmunopurification (co-IP) experiments. We performed a series of co-IP experiments using anti-GFP beads from total protein extracts containing multiple combinations. First, control co-IPs consisting of Sec22:GFP with mCherry or GFP with 6K2:mCherry confirmed that mCherry did not bind to the beads, nor did GFP interact with 6K2:mCherry (Figure 4E). The interaction test for Sec22:GFP with 6K2:mCherry revealed that Sec22 copurified with 6K2. Hence, the above experiments indicated that Sec22 is able to interact with vesicles that contain 6K2.
Figure 4.
Impairing the Fusion of Transport Vesicles with the Golgi Apparatus Enhances TuMV Intercellular Movement.
(A) to (D) Confocal microscopy observation of N. benthamiana epidermal leaf cells coexpressing Sec22:GFP (middle panels) along with the Golgi marker Man49:mCherry ([A], left panel, 6K2GV:mCherry ([B], left panel), 6K2:mCherry ([C], left panel), or 6K2:mCherry during TuMV infection ([D], left panel). Coexpression is shown in the right panels (merge), and colocalization is indicated with arrows in (C) and (D). Confocal images are single optical images 1 μm thick.
(E) Immunoblots of immunopurifications performed from different protein expression combinations (upper table and indicated by arrows) of mCherry with Sec22:GFP, 6K2:mCherry with GFP, or 6K2:mCherry with Sec22:GFP in N. benthamiana leaves. Intermediate bands are degradation byproducts or nonspecific proteins recognized by the polyclonal GFP antiserum.
(F) Confocal microscopy of N. benthamiana epidermal leaf cells expressing the Golgi marker Man49:mCherry, 6K2GV:mCherry, or 6K2:mCherry ectopically or 6K2:mCherry during TuMV infection, alone (control panels) or in cells overexpressing Sec22 (+Sec22:GFP, left panels show Man49:mCherry, 6K2GV:mCherry or 6K2:mCherry ectopically or 6K2:mCherry during TuMV expression in red, middle panels show Sec22:GFP overexpression in green, and right panels show merged signals). Images are three-dimensional renderings of 40 1-μm-thick slices that overlap by 0.5 μm.
(G) Viral replication was evaluated by RT-qPCR at 3.25 dpi from N. benthamiana agroinfiltrated with pCambia (Buffer), pCambiaTuMVVNN (VNN), or pCambiaTuMV (TuMV) 1 d after infiltration with pCambia (Mock, white) or pCambiaSec22:CFP (blue). Error bars show the sd calculated from values obtained in the three biological replicates.
(H) Sec22:CFP was transiently expressed one day before TuMV/6K2:mCherry//GFP-HDEL inoculation, and expression of GFP and mCherry fusions was acquired at 5 dpi. Panels are representative images of infection foci corresponding to the median value of each treatment. Data sets for Mock and Sec22-overexpressed treatments are graphically presented as whisker and box plots. Statistical significance was tested with the nonparametric Wilcoxon-Mann-Whitney two sample rank test (*P < 0.05). n represents the number of infection foci analyzed.
(I) Time course of TuMV systemic infection of wild-type Arabidopsis plants (circles) and bet11 KO mutant plants (diamonds) after pCambiaTuMV/6K2:GFP agroinfiltration. Infection is scored by appearance of systemic GFP fluorescence.
Two independent biological repeats were performed for (A) to (D), and three for (E) to (I). Bars = 10 μm in (A) to (D) and (F), 1 mm in (H), and 10 μm in (H) inset.
To ascertain that Sec22 overexpression has a negative effect on ER-Golgi equilibrium in our experimental conditions, we expressed Sec22:GFP with Man49:mCherry, a Golgi marker known to travel from the ER to the Golgi (Saint-Jore-Dupas et al., 2006), 6K2GV:mCherry, 6K2:mCherry, and 6K2:mCherry during infection, respectively. Sec22 overexpression impairs its own transport through the early ER-to-Golgi secretory pathway. Hence, cells overexpressing Sec22:GFP were recognizable by the appearance of fluorescence in the ER network (Figure 4F). Under these overexpressing conditions, Man49:mCherry was retained in the ER. Similarly, 6K2GV:mCherry was redistributed to the ER, but retention in the ER neither happened in leaves coagroinfiltrated with 6K2:mCherry nor with 6K2:mCherry produced during infection. This confirmed that overexpression of Sec22 impairs transport vesicle fusion with the Golgi apparatus without affecting 6K2 replication vesicles.
We next tested TuMV intercellular movement under Sec22 overexpression conditions. TuMV intercellular movement was followed using a dual expression cassette for TuMV tagged with 6K2:mCherry and for GFP-HDEL, both under the control of the CaMV 35S promoter and within the left and right border sequences of the T-DNA (Agbeci et al., 2013). We thus expressed Sec22:CFP as in Figure 4F 1 d prior to pCambiaTuMV/6K2:mCherry//GFP-HDEL agroinoculation. Five days after virus inoculation, confocal microscopy observations on TuMV infection foci showed that a red-only fluorescence signal was found encircling an area of both red and green fluorescence, indicative of virus movement (Figure 4H). However, the focus surface area of red-only fluorescence was greater for leaves overexpressing Sec22:CFP than those that did not. Movement was quantified and normalized for each focus of infection by measuring the surface area of red fluorescence divided by the surface area of green fluorescence. The ratios of the focus areas were graphically represented using whisker and box plots and statistically analyzed using the nonparametric Wilcoxon-Mann-Whitney two-sample rank test. We found that, under Sec22 overexpression conditions, TuMV intercellular movement was significantly faster than in control conditions. TuMV replication was evaluated by RT-qPCR under Sec22:CFP-overexpression conditions (Figure 4G). We did not see any appreciable difference in replication when compared with normal conditions.
To confirm the above observation, we tried to isolate Sec22 homozygous knockout (KO) Arabidopsis thaliana plants by screening two different T-DNA insertion lines, but we were not successful. As an alternative, we challenged a KO line homozygous for the Golgi SNARE BET11 (Chatre et al., 2005) with TuMV infection. Wild-type and bet11 KO mutant plants were agroinoculated with pCambiaTuMV/6K2:GFP, and every day the number of systemically infected plants was scored by GFP fluorescence observation. Three experiments were conducted and we consistently found that the number of TuMV-infected plants over time was higher for the Golgi SNARE KO plants (Figure 4I). Indeed, the time by which 50% of plants were infected (I50) was reached earlier for bet11 (I50 = 9.5 dpi) than for wild-type plants (I50 = 10.5 dpi). Thus, these results indicate that impairing fusion of transport vesicles with the Golgi apparatus enhanced the susceptibility of Arabidopsis to TuMV infection.
ER- and Golgi-Localized Synaptotagmins Act Antagonistically to Modulate TuMV Movement
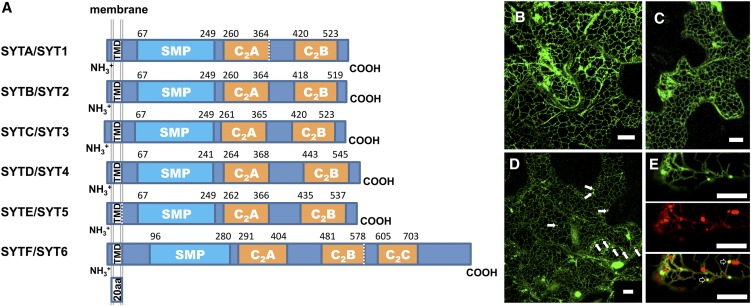
In animal cells, SYTs function in calcium-regulated membrane fusion (Rizo et al., 2006). Plant SYTs have not been studied as thoroughly, and there is evidence that they are involved in this process. For instance, SYT1/A is involved in calcium-dependent membrane resealing in Arabidopsis (Yamazaki et al., 2008) and is essential for the formation of ER-plasma membrane contact sites in plant cells (Levy et al., 2015). Hence, we decided to study their potential involvement in TuMV vesicle movement and to see if their action validates the observation that impairing transport vesicle fusion using nonfunctional Golgi SNAREs increases virus movement. The Arabidopsis genome encodes at least six different SYT-like proteins (Craxton, 2004). Arabidopsis SYTs have an N-terminal TMD, a synaptotagmin-like mitochondrial lipid binding (SMP) linker domain, and two calcium binding domains in tandem (C2) at the C-terminal end that are called C2A and C2B (Figure 5A). Mammals have a special category of SYTs that have additional C2 domains and are defined as extended synaptotagmin-like (E-SYT) proteins (Min et al., 2007). Because of its domain similarity, we considered that the sixth SYT-like protein, SYTF, might be an E-SYT. To better characterize Arabidopsis SYTs, we analyzed their subcellular distributions. We cloned the cDNA of SYTA, SYTB, SYTD, SYTE, SYTF, but not SYTC (mRNA was not detectable) and expressed the encoded proteins as GFP fusions in N. benthamiana by agroinfiltration. ER localization of SYTA has been reported by Lewis and Lazarowitz (2010). SYTE:GFP (Figure 5B) and SYTD:GFP (Figure 5C) were also located in the ER. In addition to its ER localization, SYTF expression revealed the presence of several punctate structures (Figure 5D, arrows). These punctae colocalized with the Golgi marker Man49-mCherry (Figure 5E, arrows). This indicated that SYTF was located both in the ER and the Golgi apparatus. We could not observe any expression for SYTB:GFP, although it was reported to be located in the Golgi apparatus (Zhang et al., 2011; Wang et al., 2015).
Figure 5.
Cellular Distribution of Arabidopsis Synaptotagmins.
(A) Schematic scaled domain representation of SYTA/SYT1, SYTB/SYT2, SYTC/SYT3, SYTD/SYT4, SYTE/SYT5, and SYTF/SYT6 showing their predicted TMD, SMP, and calcium binding domains (C2).
(B) to (D) Confocal images of N. benthamiana epidermal cells transiently expressing pCambiaSYTE:GFP (B), pCambiaSYTD:GFP (C), and pCambiaSYTF:GFP (D). These images are three-dimensional renderings stacks of 40 1-μm-thick slices that overlap by 0.5 μm. Punctate structures observed during SYTF expression are denoted by white arrows.
(E) Expression of SYTF:GFP (upper panel), Man49-mCherry Golgi marker (middle panel), and their coexpression (lower panel) is shown. Colocalization of SYTF punctae with Man49-mCherry is denoted by empty white arrows in the lower panel.
Three independent biological repeats were performed for (B) to (E). Bars = 10 μm in (B) to (E).
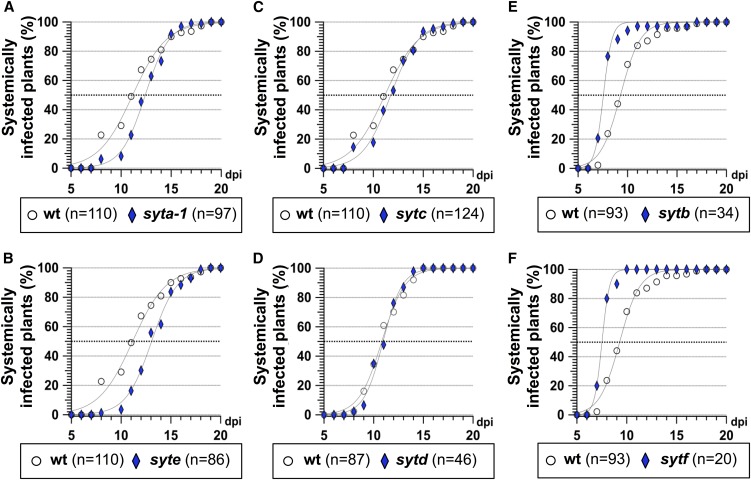
We next challenged homozygous KD/KO Arabidopsis for all SYTs to TuMV infection. syta-1 KD plants were obtained from Lewis and Lazarowitz (2010), while sytb, sytc, sytd, syte, and sytf KO mutants were obtained from TAIR. Homozygous T-DNA insertion was verified by PCR. Arabidopsis wild-type and the SYTs T-DNA insertion KD or KO plants were agroinoculated with pCambiaTuMV/6K2:GFP, and the number of systemically infected plants was scored by GFP fluorescence observation every day. Compared with the wild type (I50 = 11 dpi), TuMV infection was delayed in syta-1 KD mutant (I50 = 12 dpi), as previously reported (Uchiyama et al., 2014), and in syte KO plants (I50 = 13 dpi) (Figures 6A and 6B). No effect on the rate of TuMV infection was observed in sytc and sytd KO plants (Figures 6C and 6D). In contrast, we found that the rate of TuMV infection was faster in sytb and sytf KO plants (I50 = 8 dpi in both cases), compared with wild-type plants (I50 = 9 dpi) (Figures 6E and 6F). These results indicate that SYTs located in the ER have either a positive effect (e.g., SYTA and E) or no effect (e.g., SYTC and D), whereas those found in the Golgi (e.g., SYTB and SYTF) have a negative effect on TuMV infection.
Figure 6.
Infection Time Course of SYT KO Mutant Plants on TuMV Infection.
Time course of TuMV systemic infection of wild-type Arabidopsis plants (circles) and SYT KO mutant plants (diamonds) after pCambiaTuMV/6K2:GFP agroinfiltration. Infection is scored by appearance of GFP systemic fluorescence. The tested SYT mutant are syta-1 (A), syte (B), sytc (C), sytd (D), sytb (E), and sytf (F). n represents the number of plants agroinoculated. Three independent biological repeats were performed for each experiment.
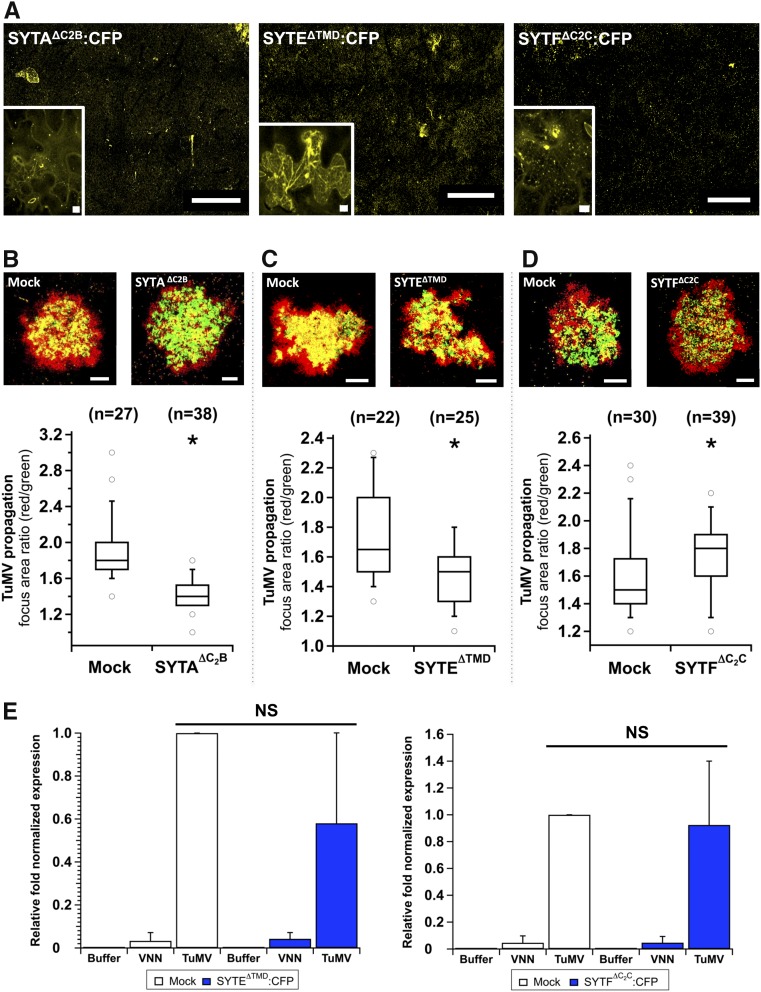
The above observations were confirmed by expressing truncated forms of selected SYTs in infected cells. Removing the TMD or C2B domain from SYTA was shown to impair its function and localization in N. benthamiana protoplasts and leaf tissues (Lewis and Lazarowitz, 2010). Similarly, by deleting the TMD, C2B domain or C2C domain, we developed truncated forms of SYTA (SYTAΔC2B), SYTE (SYTEΔTMD), and SYTF (SYTFΔC2C) that were tagged with CFP as indicated in Figure 5A (white dashed lines). The cellular distribution of the truncated SYTs fused with CFP was determined by confocal microscopy. SYTAΔC2B:CFP was mislocalized to membrane patches similar to what has been previously described (Lewis and Lazarowitz, 2010) (Figure 7A). SYTEΔTMD:CFP distributed throughout the cytosol and SYTFΔC2C:CFP was mislocalized to punctate structures. These truncated forms were expressed 1 d before pCambiaTuMV/6K2:mCherry//GFP-HDEL agroinfiltration in order to competitively impair wild-type SYT function. Confocal microscopy observations for each tested SYT showed overall less red fluorescence when SYTAΔC2B:CFP (Figure 7B) or SYTEΔTMD:CFP (Figure 7C) were expressed in comparison to mock conditions, indicating that virus intercellular movement was delayed. In contrast, confocal microscopy observations showed more red fluorescence when SYTFΔC2C:CFP was produced (Figure 7D), indicating that virus intercellular movement was faster. The surface area for red and green fluorescence of each focus of infection was quantified as mentioned above and the ratios showed that the difference in viral cell-to-cell movement between the tested conditions were statistically significant. Also, a quantitative estimate of TuMV replication was performed by RT-qPCR under SYTEΔTMD:CFP and SYTFΔC2C:CFP expression conditions and showed that viral RNA production was not significantly affected (Figure7E). Consequently, impairing ER-Golgi homeostasis by overexpressing the Golgi SNARE Sec22 or nonfunctional Golgi-located SYTF has a positive effect on TuMV infection.
Figure 7.
Impact of Truncated SYT Mutants on TuMV Intercellular Movement.
(A) Expression of SYTAΔC2B:CFP, SYTEΔTMD:CFP, and SYTFΔC2C:CFP in N. benthamiana observed by confocal microscopy with the 10× objective (main panels) and 63× objective (insets) during intercellular TuMV movement.
(B) to (D) pCambiaSYTAΔC2B:CFP (B), pCambiaSYTEΔTMD:CFP (C), and pCambiaSYTFΔC2C:CFP (D) were agroinfiltrated into N. benthamiana leaves 1 d before TuMV/6K2:mCherry//GFP-HDEL agroinoculation. Expression of GFP and mCherry fusions was acquired at 5 dpi by confocal microscopy. Panels are representative images of infection foci corresponding to the median value for the indicated treatments. Data sets for mock and SYT truncated mutants are graphically presented as whisker and box plots. Statistical significance was tested with the nonparametric Wilcoxon-Mann-Whitney two-sample rank test (*P < 0.05). n represents the number of infection foci analyzed.
(E) Viral replication was evaluated by RT-qPCR at 3.25 dpi from N. benthamiana leaves agroinfiltrated with pCambia (Buffer), pCambiaTuMVVNN (VNN), or pCambiaTuMV (TuMV) 1 d after pCambia (Mock), SYTEΔTMD:CFP (left histogram), or pCambiaSYTFΔC2C:CFP (right histogram) infiltration. Error bars show the sd calculated from values obtained in the three biological replicates. Three independent biological repeats were performed for each experiment. Bars = 1 mm in (A) main panels and (B) to (D) and 10 μm (A) insets.
The Prevacuolar Compartment SNARE VTI11 Is Essential for TuMV Infection
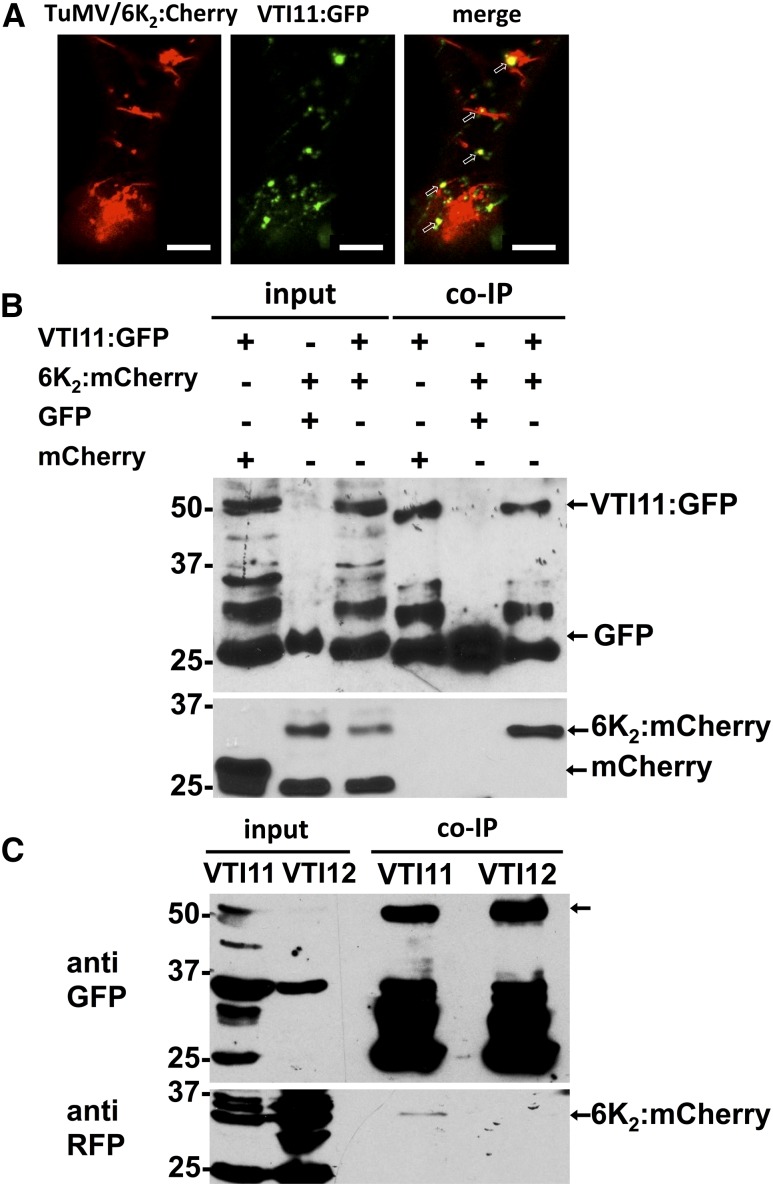
The above results suggest that TuMV replication factories bypass the Golgi apparatus during productive infections. Possible destinations following this bypass could be PVCs/MVBs, endosomes, the trans-Golgi network (TGN), and/or the central vacuole. SNAREs are distributed in different compartments and they shuttle between them (Uemura et al., 2004). This led us to perform colocalization experiments between selected GFP-tagged SNAREs associated with different compartments and 6K2:mCherry produced during infection in order to get a glimpse of 6K2 vesicle destination. Among the 25 SNAREs tested (Supplemental Table 1), we found that 6K2:mCherry partially colocalized (calculated 6% ± 2% colocalization) with VTI11:GFP during infection (Figure 8A). VTI11 is known to be located in the PVC, shuttling from the TGN to the PVC and to the vacuole, and also formed a SNARE complex with other PVC SNAREs (Zheng et al., 1999; Sanderfoot et al., 2001; Uemura et al., 2004; Ebine et al., 2008). To verify a potential association between 6K2 and VTI11, we performed different co-IPs using anti-GFP beads from total protein extracts containing a variety of combinations (Figure 8B). First, control co-IPs consisting of VTI11:GFP with mCherry or GFP with 6K2:mCherry showed that neither mCherry was binding to the beads, nor was GFP pulling down 6K2:mCherry. VTI11:GFP copurified with 6K2:mCherry. We verified that overexpression of 6K2 did not cause false interactions observed by co-IP experiments. Based on copurification with 6K2:mCherry, the vast majority of SNAREs tested did not show interaction with this fusion protein (Supplemental Figure 1). Because VTI11 interacted, but VTI12 did not (Figure 8C), this excludes nonspecific binding to SNAREs by 6K2.
Figure 8.
Interaction of the PVC SNARE VTI11 with 6K2.
(A) Confocal microscopy of N. benthamiana epidermal leaf cells expressing VTI11:GFP (middle panel) and 6K2:mCherry produced during TuMV infection (left panel). Colocalization of VTI11 with 6K2 is indicated by arrows in the merged image (right panel). Confocal images are single optical images of 1 μm thick.
(B) N. benthamiana leaves expressing combinations (upper table and indicated by arrows) of mCherry with VTI11:GFP, 6K2:mCherry with GFP, or 6K2:mCherry with VTI11:GFP were harvested 3 d after agroinfiltration. The cleared lysates (input) were used for co-IP and detection as described in Methods.
(C) N. benthamiana leaves expressing 6K2:mCherry with VTI11:GFP or VTI12:GFP were harvested 3 d after agroinfiltration. The cleared lysates were coimmunopurified as described in Methods.
Three independent biological replicates were performed for (A), four for (B), and two for (C). Bars = 10 μm in (A).
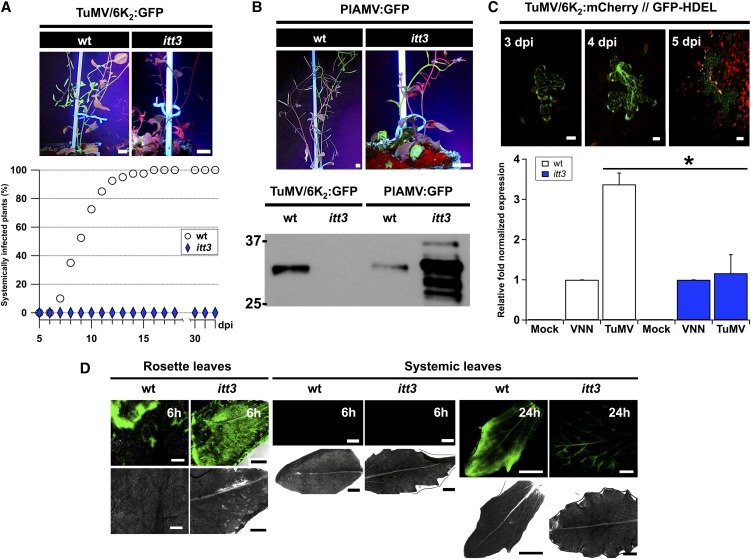
We then tested whether VTI11 was involved in TuMV infection. A homozygous VTI11 loss-of-function Arabidopsis mutant line, named itt3 for impaired tonoplast trafficking 3, was generated by EMS mutagenesis (Zheng et al., 2014). zig-1 is another VTI11-nonfunctional mutant plant that was isolated by Kato et al. (2002). Importantly, an allelism test between itt3 and zig-1 mutants did not show complementation (Zheng et al., 2014), supporting the idea that VTI11 function is specifically disrupted in the itt3 mutant line. This line was obtained from TAIR. Genomic DNA analysis revealed that the itt3 phenotype is the consequence of an early stop codon in the VTI11 mRNA, just before its SNARE coil-coiled domain and its TMD (Zheng et al., 2014).
To verify whether the itt3 mutant was still producing the VTI11 mRNA and contained the indicated point substitution, we extracted total mRNA from wild-type and itt3 mutant plants, performed an RT-PCR with primers specific for VTI11 coding region and cloned the resulting cDNA. We sequenced three independent amplified fragments and confirmed the presence of the C>T substitution, which introduces a premature stop codon. We agroinoculated Col-0 wild-type and itt3 mutant Arabidopsis plants with pCambiaTuMV/6K2:GFP. Infection kinetics showed that itt3 mutant plants were completely resistant to TuMV infection, even 4 weeks after inoculation (Figure 9A). itt3 plants showed a reduced rosette size and a characteristic zigzag stem due to a loss of polarity. The lack of susceptibility to TuMV may then be a secondary effect due to a pleiotropic effect of the mutation, resulting in abnormal development.
Figure 9.
The PVC SNARE VTI11 Is Essential for TuMV Infection.
(A) Wild-type (wt) or itt3 plants under UV light 25 d after agroinoculation with TuMV/6K2:GFP (left panel). Time course of TuMV systemic infection of wild-type Arabidopsis plants (circled) and itt3 loss-of-function mutant plants (diamonds) after pCambiaTuMV/6K2:GFP agroinoculation (right panel). Infection was scored by appearance of systemic GFP fluorescence.
(B) Wild-type or itt3 plants under UV light 25 d after agroinoculation with PlAMV:GFP (left panel). Total proteins extracts from TuMV/6K2:GFP- or PlAMV:GFP-inoculated wild-type or itt3 rosettes were analyzed by immunoblot with anti-GFP antibodies (right panel).
(C) Time course of TuMV intercellular movement after agroinoculation of pCambiaTuMV/6K2:mCherry//GFP-HDEL into wild-type Arabidopsis plants. Expression of GFP and mCherry fusions was acquired at 3, 4, and 5 dpi by confocal microscopy (upper panels). The graph below shows viral replication as evaluated by RT-qPCR from Arabidopsis wild-type and itt3 leaves that were agroinfiltrated with pCambia (Mock), pCambiaTuMVVNN (VNN), or pCambiaTuMV (TuMV) at 4 dpi. Statistical differences between samples were determined by t test analyses (*P < 0.05). Error bars show the sd calculated from values obtained in the three biological replicates.
(D) CFDA dye transport in wild-type Arabidopsis and itt3 plants observed by confocal microscopy of inoculated rosette leaves and of systemically infected leaves 6 and 24 h postinoculation. Upper panels, fluorescence; lower panels, bright-field images.
Three independent biological replicates were performed for (A) to (C) and two for (D). Bars = 10 mm in (A) and (B), 20 μm in (C), and 1 mm in (D).
To exclude this possibility, we infected itt3 plants with Plantago asiatica mosaic virus (PlAMV), a virus of the Potexvirus group. A PlAMV infectious clone expressing GFP fused to the capsid viral protein (Yamaji et al., 2012) was agroinoculated into wild-type and itt3 Arabidopsis plants. PlAMV infected primary infiltrated leaves and moved systemically in both wild-type and itt3 mutant plants (Figure 9B). Total protein extracts from wild-type and itt3 rosettes inoculated either with TuMV/6K2:GFP or with PlAMV:GFP were made and immunoblots were probed with an anti-GFP serum. Although 6K2:GFP was only produced in wild-type plants, the GFP fusion from PlAMV:GFP accumulated in both wild-type and itt3 mutant plants (Figure 9B). Viral replication in wild-type and itt3 plants was measured by RT-qPCR at 4 dpi, a time prior to any viral cell-to-cell movement of virus (Figure 9C). Also, there was very little, if any, TuMV replication in itt3 plants. We verified that itt3 symplasmic and phloem transport were unaltered by infiltrating one lower leaf with carboxyfluorescein diacetate (CFDA) and observing the dye movement in upper leaves. The itt3 plants were still capable of symplasmic and phloem transport capability, although less than in wild-type plants (Figure 9D). However, this apparent physiological impairment was not sufficient to prevent itt3 plants from being systemically infected by PlAMV.
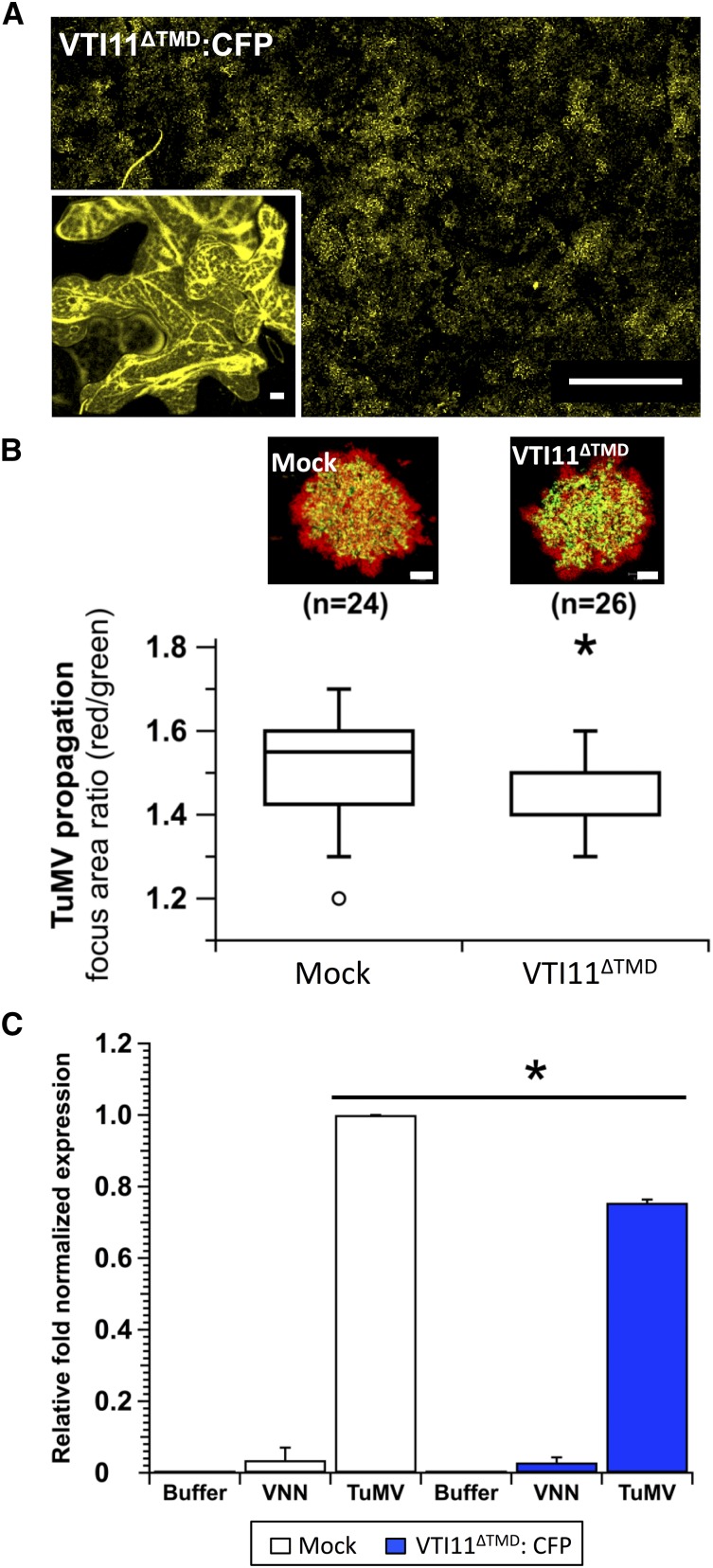
To test whether lack of infection was an indirect consequence of the altered phenotype of itt3 plants, we expressed a nonfunctional form of VTI11 and assayed its impact on TuMV movement and replication. We made a truncated form of VTI11 without its TMD and tagged with CFP (VTI11ΔTMD:CFP) and expressed it in N. benthamiana prior to pCambia/TuMV-6K2:mCherry//GFP-HDEL agroinfiltration. The expression pattern we observed is typical of a soluble protein (Figure 10A). Five days following inoculation, confocal microscopy observations of TuMV infection foci showed overall less red fluorescence when VTI11ΔTMD:CFP was expressed in comparison to mock conditions (Figure 10B), indicating that virus movement was delayed. TuMV intercellular movement was quantified as above and the data were graphically represented using whisker and box plots and statistically analyzed as described earlier. We found that TuMV intercellular movement was significantly decreased compared with mock infiltrated plants. Also, an RT-qPCR estimate of TuMV replication under the same VTI11ΔTMD:CFP expression conditions, and this showed decreased viral replication (Figure 10C). These results suggest then that one destination route for TuMV replication factory Golgi bypass involves VTI11-tagged PVCs.
Figure 10.
Impact of Truncated VTI11 Mutant on TuMV Intercellular Movement.
(A) Expression of VTI11ΔTMD:CFP in N. benthamiana as observed by confocal microscopy with the 10× objective (bigger panel) and 63× objective (inset) during intercellular TuMV movement. Bars = 10 μm in insets and 1 mm in main panels.
(B) VTI11ΔTMD:CFP was transiently expressed 1 d before TuMV/6K2:mCherry//GFP-HDEL agroinoculation. Expression of GFP and mCherry fusions was acquired at 5 dpi by confocal microscopy. Panels are representative images of infection foci corresponding to the median value for the indicated treatments. Data sets for mock and SYT truncated mutants treatments are graphically presented as whisker and box plots. Statistical significance was tested with the nonparametric Wilcoxon-Mann-Whitney two-sample rank test (*P < 0.05). n represents the number of infection foci analyzed.
(C) Viral replication was evaluated by RT-qPCR at 3.25 dpi from N. benthamiana agroinfiltrated with pCambia (Buffer), pCambiaTuMVVNN (VNN), or pCambiaTuMV (TuMV) 1 d after pCambia (mock) or pCambiaVTI11ΔTMD:CFP infiltration. Error bars show the sd calculated from values obtained in the three biological replicates. Three independent biological repeats were performed for (A) to (C).
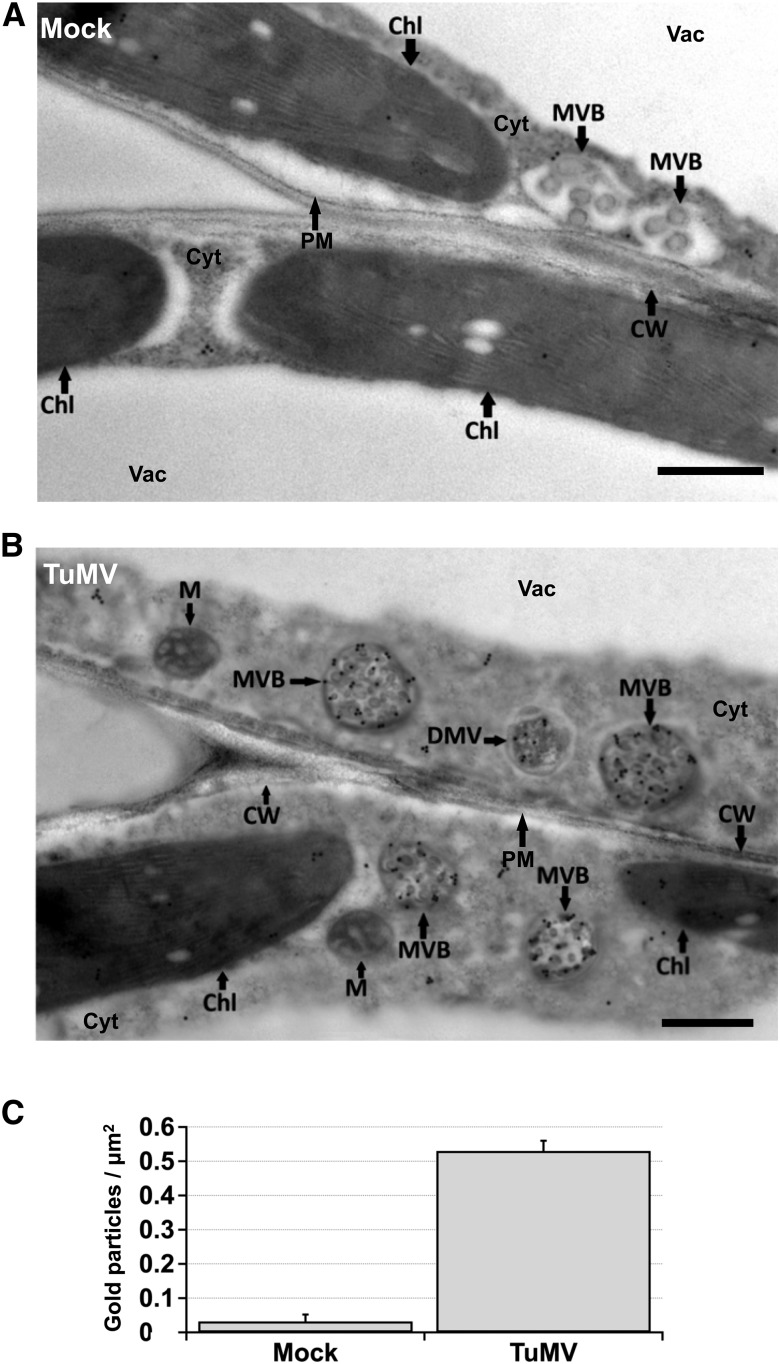
Viral RNA Is Associated with Multivesicular Bodies
As additional evidence that TuMV replication factories associate with PVCs, we performed transmission electron microscopy analyses on immunogold-labeled leaf sections using the anti-dsRNA monoclonal antibody J2. This antibody is used to immunolabel ultrastructures containing viral RNA. The validity of using the anti-dsRNA J2 monoclonal antibody for labeling viral RNA was demonstrated previously (Cotton et al., 2009; Wan et al., 2015b). We found that the RNA signal was always associated with 6K2 vesicles during infection, and there was no background noise in mock conditions or unrelated to viral factories during infection. Few dsRNA-specific gold particles were observed in mock-infected samples (Figure 11A), while a significant amount localized on MVBs from TuMV-infected leaves (Figure 11B). The number of gold particles per square micrometer found in MVBs is shown in Figure 11C, which confirms that MVBs from TuMV-infected samples were associated to significantly more gold particles compared with mock-infected samples. PVCs appear as MVBs under the transmission electron microscope (Jia et al., 2013), thus supporting the association of TuMV with these extra Golgi structures.
Figure 11.
Viral RNA is Associated with Multivesicular Bodies.
(A) and (B) Immunogold labeling was performed on mock- and TuMV-infected N. benthamiana leaf tissue cross sections using an anti-dsRNA specific antibody. Transmission electron microscopy representative images show the presence of gold particles in the mock-infected condition (A) and in TuMV-infected cells (B). Three independent biological replicates were performed. Bars = 500 nm.
(C) The density of gold particles in MVBs from mock-infected versus TuMV-infected cells is shown. Two different labeling experiments were considered for quantification, and 200 gold particles were counted for each experiment. Error bars show the sd calculated from values obtained in two biological replicates. Chl, chloroplast; CI, cytoplasmic inclusion body; Cyt, cytosol; CW, cell wall; DMV, double-membrane vesicle; M, mitochondria; Vac, vacuole.
DISCUSSION
The fate of viral vesicles immediately after their production after 6K2 exits from the ER is not known. In this study, we identified a GxxxG motif in the predicted TMD of the 6K2 viral protein of TuMV (Figure 1) that modulates the intracellular trafficking of the viral vesicles. Normally, wild-type 6K2 induces the formation of vesicles and aggregates that are only partially localized with Golgi bodies (Figure 1). However, when the Gly residues of the GxxxG motif were replaced with Val residues, the 6K2GV mutant lost its capacity to form vesicles and the protein was redirected to the Golgi apparatus and the PM (Figure 1). It has thus been converted into a default membrane protein that was trafficked through the conventional secretory pathway. One consequence of this rerouting was that virus replication and viral systemic movement were inhibited (Figure 3). 6K2 transit through the Golgi apparatus is thus a dead-end avenue for virus production.
The idea that the GxxxG motif prevents, albeit not completely, the viral protein from entering the conventional ER-Golgi secretory pathway is supported by the differential sensitivity of wild-type 6K2 and 6K2GV to BFA treatment and to dominant-negative GTPase mutant expression and the deglycosylation assay (Figure 2). These treatments had no effect on the production of wild-type 6K2 vesicles, but barred 6K2GV from leaving the ER. These observations thus indicate that the GxxxG motif somehow modulates the intracellular trafficking of 6K2. Although a proportion of 6K2 does traffic to the Golgi apparatus, there are still a sizable number of 6K2 vesicles that bypass the Golgi through this unconventional route because of this motif. This is in contrast to other TuMV/potyvirus proteins, which do not take such an unconventional pathway. For instance, P3N-PIPO and the cylindrical inclusion viral proteins from TuMV may reach plasmodesmata through the secretory pathway, where they are thought to anchor the VRC to this structure (Wei et al., 2010; Movahed et al., 2017).
TMDs are major determinants of protein-protein interactions (Langosch and Arkin, 2009). The GxxxG motif has been implicated in homo- and heterotypic interactions for more than 20 different proteins (Teese and Langosch, 2015). Structural studies indicate that the GxxxG motif induces membrane helix interactions because the short side chain of glycine, a hydrogen atom, maximizes interfacial interactions. Nevertheless, the sequence context is also important in providing interaction specificity. Interaction candidates would be host factors involved in protein sorting somewhere downstream of COPII vesicle formation and prior to Golgi fusion (e.g., tethers, GTPases, etc.). However, little is known concerning what these proteins might be and how they would function in preventing proteins from entering the Golgi apparatus (Tan et al., 2013).
The observations revealing that 6K2-induced replication vesicles are unaffected by the use of different ER to Golgi deregulating treatments (Figure 2) in addition to their partial colocalization with Golgi markers, indicates that the conventional secretory pathway and an unconventional trafficking route can be used for viral replication complex formation. Indeed, we observed an enhanced viral cell-to-cell movement when transport vesicle fusion with the Golgi apparatus was impaired by overexpression of the Golgi SNARE Sec22 (Figure 4). This increase in viral movement when ER-Golgi homeostasis was impaired was corroborated by the reproducible increase in TuMV susceptibility of the Golgi SNARE bet11 KO Arabidopsis plants (Figure 4). Observations that further support these two pathways were that SYTs localized in the Golgi apparatus (SYTB and SYTF) have an antiviral effect, while SYTs present in the ER (SYTA and SYTE) have a proviral function (Figures 6and 7). The proviral role of ER SYTs was previously reported in a study where SYTA KD plants showed delayed systemic infection by Turnip vein clearing virus, Cabbage leaf curl virus, and TuMV (Lewis and Lazarowitz, 2010; Uchiyama et al., 2014; Levy et al., 2015). Enhanced virus movement and increased virus susceptibility when ER-Golgi homeostasis is perturbed can be explained by the fact that less 6K2 vesicles fuse with the Golgi apparatus, with a concomitant increase in the number of 6K2 vesicles entering the unconventional trafficking pathway for infection.
The next question that we addressed in this investigation was what might be the destination(s) of the 6K2 vesicles once they enter this unconventional pathway. Plant cells contain multiple compartments that intercommunicate through transport vesicle fusions, and SNAREs are key players in this communication process. Our screening of SNAREs located in different compartments revealed that 6K2 vesicles appeared to contain VTI11 (Figure 8). The biological significance of this 6K2-VTI11 complex on infection was supported by the observation that loss-of-function Arabidopsis plants for this SNARE were completely resistant to TuMV, but not to an unrelated potexvirus (Figure 9). Although we cannot exclude an indirect effect due to the modified physiology of the mutant plant, expression of a truncated form of VTI11 further supports a proviral function of this SNARE protein (Figure 10). VTI11 transits from the TGN to PVCs and to the central vacuole and interacts with other PVC SNAREs (Zheng et al., 1999; Sanderfoot et al., 2001; Uemura et al., 2004; Ebine et al., 2008).
PVCs, or MVBs, are organelles that play important roles in mediating protein trafficking to vacuoles (Cui et al., 2016). The involvement of MVBs in viral infection has been proposed to explain the nonlytic spread of animal viruses (Bird and Kirkegaard, 2015). MVBs are the precursors of extracellular vesicles also known as exosomes (Alenquer and Amorim, 2015) and exosome-mediated transmissions of human viruses have been reported (Feng et al., 2013; Ramakrishnaiah et al., 2013; Longatti et al., 2015; reviewed in Anderson et al., 2016). It is also important to note that the release of Rice dwarf virus from insect vector cells involves exosomes derived from MVBs (Wei et al., 2009). We observed by transmission electron microscopy that many MVBs produced during the infection process of TuMV were decorated with a significant number of gold particles that were coupled to an anti-dsRNA specific antibody (Figure 11). These observations support the idea that TuMV replication vesicles transit through PVCs. Information on the explicit involvement of PVCs/MVBs has not been reported for plant viruses. However, importance of the proteins from the ESCRT family for BMV and TBSV infection provides some indication for the involvement of PVCs/MVBs in plant virus infections. Furthermore, Rutter and Innes (2017) isolated extracellular vesicles from Arabidopsis infected by Pseudomonas syringae and, interestingly, one protein that was identified as part of these extracellular vesicles was VTI11. Xylem vessels are dead tissues and are considered to represent the extracellular space of a plant. Xylem vessels originate from programmed cell death events of protoxylem cells, and direct continuity between the apoplast and xylem vessel has been suggested (Ligat et al., 2011). TuMV replication vesicles have been observed in xylem vessels (Wan et al., 2015b). The question now to answer is whether the 6K2 vesicles found in the xylem have any link with the apoplastic VTI11-associated extracellular vesicles.
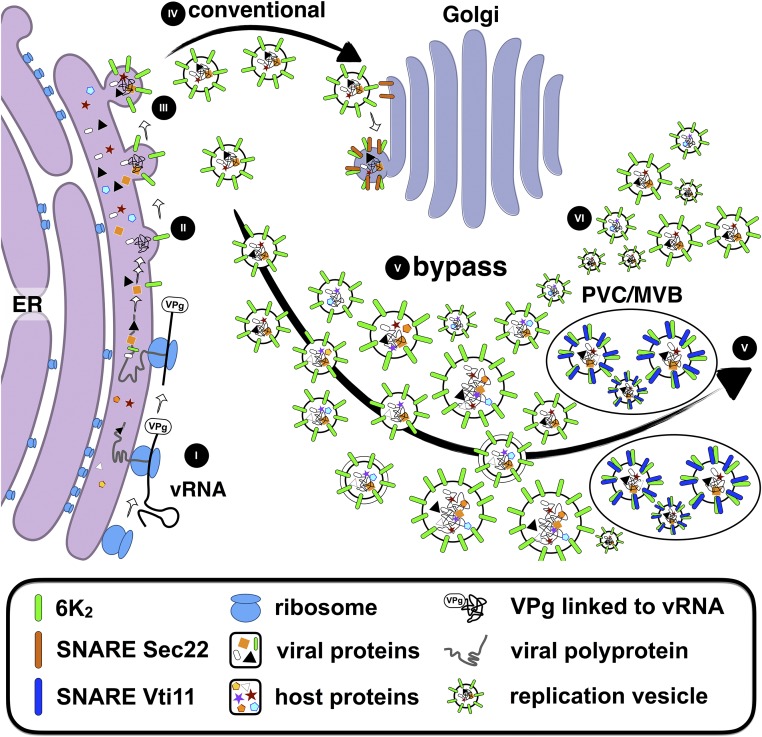
We summarize our observations and illustrate our proposed model of a tug-of-war that takes place between the conventional secretory pathway and an unconventional Golgi bypass trafficking pathway for TuMV infection (Figure 12). Early in the infection process, vRNA is translated on ER-bound ribosomes (I) and the synthetized viral proteins, along with co-opted host proteins, are assembled as proto VRCs (II). Mediated by 6K2 binding to COPII subunit Sec24A, these proto VRCs exit the ER in a COPII dependent-manner as replication vesicles (III). At this step, replication vesicles are presented with two choices. In the first case, replication vesicles may fuse with the Golgi apparatus, but this constitutes a dead end for virus infection (IV). Alternatively, replication vesicles may bypass the Golgi apparatus and enter at least a PVC SNARE VTI11-dependent unconventional pathway (V) without excluding other Golgi-bypassing potential routes (VI). Ultimately, following that unconventional pathway will result into the infection of the plant.
Figure 12.
A TuMV tug-of-war Model Bypassing the Golgi during Infection
Early during infection, vRNA is translated on ER-bound ribosomes (I) and the synthesized viral proteins, along with co-opted host proteins, are used to make proto VRCs (II). VRCs exit the ER in a COPII-dependent manner as replication vesicles (III) and face two possible fates: entering the conventional secretory dead-end pathway by going through the Golgi apparatus (IV) or bypassing it and taking an alternative non conventional pathway for reaching PVC/MVB that involves VTI11.
METHODS
Molecular Cloning
The coding sequence of GFP flanked by XbaI and BamHI restriction sites was amplified by PCR using the template pGreenPABP:GFP (Beauchemin and Laliberté, 2007). The fragment was digested with XbaI and BamHI, and it was then used to replace the mCherry coding fragment in pCambia6K2:mCherry (Jiang et al., 2015) for the construction of plasmid pCambia6K2:GFP. Mutagenesis was performed using the QuickChange II XL site-directed mutagenesis kit (Agilent), according to the manufacturer’s instructions. pCambia6K2:GFP was used as template for the production of pCambia6K2G30V:GFP, pCambia6K2G33V:GFP, pCambia6K2G34V:GFP, pCambia6K2G35V:GFP, pCambia6K2G33V-G34V-G35V:GFP, and pCambia6K2G30V-G33V-G34V-G35V:GFP. The GFP DNA fragment of pCambia6K2G30V-G33V-G34V-G35V:GFP was replaced with the coding sequence of mCherry for the construction of pCambia6K2G30V-G33V-G34V-G35V:mCherry. pCambiaTuMV (Cotton et al., 2009) was used as the template for the production of pCambiaTuMVG30V, pCambiaTuMVG34V, and pCambiaTuMVG33V-G34V-G35V. SNAREs used in this study are listed in Supplemental Table 1. Coding sequences were amplified with specific primers (Supplemental Table 2) by RT-PCR (iScript select cDNA synthesis kit; Bio-Rad) from Arabidopsis thaliana ecotype Col-0 total mRNA extraction (RNeasy Plant Mini Kit; Qiagen) and cloned in a similar manner as described above. The markers GFP:HDEL, ERD2:GFP, ST:YFP, and AHA2-GFP were described previously (Saint-Jore et al., 2002; Zheng et al., 2005; Chen et al., 2011; Grangeon et al., 2012). The mCherry-coding fragment was removed with BamHI and EcoRI from pCambia6K2:mCherry (Jiang et al., 2015) and replaced by CFP coding sequence to generate the plasmid pCambia/6K2:CFP. 6K2 was subsequently replaced by the SYTs and SNARE-truncated coding sequences forms. All constructs were verified by sequencing.
Plant Material, Genotyping, and Infectivity Assays
Nicotiana benthamiana and Arabidopsis plants were grown at 25°C under a photoperiod of 16 h light/8 h dark. Control Arabidopsis ecotype Col-0, SYTs T-DNA KO mutants sytb, sytc, sytd, syte, and sytf, BET11 SNARE T-DNA KO bet11, and VTI11 SNARE EMS KO itt3 used in this study were obtained through TAIR (https://www.arabidopsis.org/) from the ABRC. syta-1 knockdown mutant was obtained from the Lazarowtiz laboratory (Lewis and Lazarowitz, 2010). Genomic DNA was extracted with Plant DNAzol (Invitrogen) reagent following the manufacturer’s instructions, and homozygous T-DNA insertion was verified by PCR. For itt3 EMS mutants, total mRNAs were extracted as indicated by the manufacturer’s instructions (RNeasy Plant Mini Kit), RT-PCR was performed with VTI11 (At5g39510) specific primers (Supplemental Table 2) (iScript select cDNA synthesis kit), and the resulting cDNA amplification was cloned into pCambiaGFP and the homozygous mutation was confirmed by sequencing. Infectivity assays were performed on Arabidopsis wild-type and mutant plants by agroinfiltration with either pCambiaTuMV/6K2:GFP (Grangeon et al., 2012) or PlAMV:GFP infectious clones (Yamaji et al., 2012) and daily counting the number of systemically infected plants by observing GFP fluorescence under UV light.
RT-qPCR
RT-qPCR reactions were performed following to MIQE (minimum information for publication of quantitative real-time PCR experiments) guidelines (Bustin et al., 2009). Total mRNA was extracted as indicated by the manufacturer instructions (RNeasy Plant Mini Kit). After genomic DNA removal and RNA quality and purity verifications, total mRNA extracts were processed for RT as indicated by the manufacturer (iScript reverse transcription supermix for RT-qPCR) for each condition. qPCR were performed with the SYBR Green PCR master mix kit (Applied Biosystems), on CFX96 real-time PCR system thermal cycler (Bio-Rad), and data analysis was done with CFX manager software (Bio-Rad). qPCR was done in the same conditions for cDNA samples, with no-RT samples and no template control for all primers. For each pair of primers (Supplemental Table 1), specificity was validated (melt curve, gel) from pooled cDNA sample. Serial dilutions of pooled cDNA samples were done to generate standard curves, determine primer efficiency (between 90% and 110%), and select the optimal working dilution. Three reference genes, such as L23, PP2A, and F-BOX (Liu et al., 2012)were selected for N. benthamiana and three other such as ubiquitin (Cosson et al., 2012), transducin, and actin 11 (transducin and actin 11 were provided by V. Schurdi-Levraud) were selected for Arabidopsis to verify their stability for each experiment. Reference gene stability was analyzed and validated using the geNorm method (Vandesompele et al., 2002) in qbase+ software (Biogazelle). Consequently, the two most stable references genes in our conditions were PP2A and F-BOX for N. benthamiana experiments as well as ubiquitin and transducin for Arabidopsis. Normalization (ΔΔCq) was done respectively with the two validated reference genes to give a more robust qPCR data estimate of virus replication either in N. benthamiana or Arabidopsis. Values were graphically represented on histograms relative to TuMVVNN control normalized value.
Protein Domain and Glycosylation Site Prediction
Prediction of SYTs and SNAREs transmembrane domains was performed with the following online software: HMMTOP (http://www.enzim.hu/hmmtop), PHOBIUS (http://phobius.sbc.su.se/), SOSUI (http://harrier.nagahama-i-bio.ac.jp/sosui/sosui_submit.html), TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/), and TMpred (http://embnet.vital-it.ch/software/TMPRED_form.html). Complement of information for SYTs were (Craxton, 2004; Min et al., 2007; Levy et al., 2015). Prediction of SMP and C2 domains were performed with Pfam online software (http://pfam.xfam.org/). The information was used to create SYT and SNARE truncated forms of the proteins. 6K2 and GFP primary sequences were subjected to in silico N-glycosylation sites prediction with NetNGlyc 1.0 online software (http://www.cbs.dtu.dk/services/NetNGlyc/), and O-glycosylation sites were predicted with NetOGlyc 4.0 (Steentoft et al., 2013) online software (http://www.cbs.dtu.dk/services/NetOGlyc/). 6K2 presented only potential N-glycosylation modifications and was subjected to further analyses (see Total Protein Extraction, Cellular Fractionation, Deglycosylation Assay, and Immunoblot Analysis section below).
Transient Protein Expression, Inhibitor Treatments, and Co-IP
Agrobacterium tumefaciens-mediated transient protein expression was followed essentially as described by Cotton et al. (2009). Four-week-old N. benthamiana plants were used for agroinfiltration and were kept in the growth chamber until analysis. The OD600 was adjusted to 0.03 for GFP:HDEL, to 0.1 for ERD2:GFP, ST:YFP, and AHA2:GFP, to 0.3 for wild-type 6K2, mutated 6K2 fluorescent protein fusions, as well as for wild-type and mutated TuMV infectious clones. Except for the experiments in Figure 2B, the OD600 was adjusted to 0.01 to 0.015 not only for both secretory pathway inhibitors GFP-ARF1 NI and RAB-D2A N123I, but also for Man49:mCherry, 6K2GV:mCherry, 6K2:mCherry, and pCambiaTuMV/6K2:mCherry. Coexpression experiments with the secretory pathway inhibitors were done in the presence of P19 (OD600 = 0.01). OD600 of SNAREs and SYTs during expression experiments was adjusted to 0.1 in the presence of P19 (OD600 = 0.1). For RT-qPCR, infectious clones were infiltrated at an OD600 adjusted to OD600 = 0.01, pCambia (Mock) at OD600 = 0.01 or 0. 1 and dominant-negative SNAREs and SYTs at OD600 = 0. 1 with P19 (OD600 = 0. 2). For all coexpression combinations, an equal volume of Agrobacterium suspensions was mixed thoroughly prior to agroinfiltration. BFA (Millipore Sigma) was used at a final concentration of 20 μg/mL 24 h prior to confocal observations. Coimmunopurification experiments were performed with anti-GFP-Trap beads (ChromoTek) as described (Jiang et al., 2015). Briefly, N. benthamiana leaves expressing combinations of different proteins were harvested and total proteins were extracted. The cleared lysates (input) were immunopurified using anti-GFP-Trap beads, and this was followed by immunoblot analysis of input and immunopurified (co-IP) fractions using antibodies against GFP (see Total Protein Extraction, Cellular Fractionation, Deglycosylation Assay, and Immunoblot Analysis section below).
Confocal Laser Scanning Microscopy
The agroinfiltrated leaf tissues were placed between slides and cover slides. These samples were observed using 10×, 20×, or 63× oil immersion objectives on a LSM780 confocal microscope (Zeiss). GFP and YFP were excited at 488 nm, and the emission was captured between 500 and 535 nm. The mCherry signal was excited at 561 nm, and the emission was captured from 580 to 640 nm. For colocalization assay, different fluorescent signals were collected simultaneously. Image processing was performed with ZEN 2011 software and ImageJ (National Institutes of Health). For 3D reconstructions, settings and image treatment were identical for all panels.
TuMV Intercellular Movement Assays under Dominant-Negative Protein Expression
Four-week-old N. benthamiana plants were agroinfiltrated with either pCambiaSec22:CFP, pCambiaVTI11ΔTMD:CFP, or pCambiaSYTAΔC2B:CFP, or pCambiaSYTEΔTMD:CFP or pCambiaSYTFΔC2C:CFP (OD600 = 0.1) with P19 (OD600 = 0.2) 1 d prior to TuMV/6K2:mCherry//GFP-HDEL (Agbeci et al., 2013) agroinfiltration (OD600 = 0.1). For all coexpression combinations with P19, an equal volume of Agrobacterium suspensions was mixed thoroughly prior to agroinfiltration. Infection foci were acquired at 4 to 5 dpi by confocal microscopy with a 10× objective. Their corresponding confocal images were formed of juxtaposed tiles (6 × 6 × 10 tiles stacks of 40-µm-thick confocal images that overlap by 45 µm). Image processing was done with ZEN 2011 software. Primary and secondary infection area foci were calculated with ImageJ imaging software.
Graphical Representation and Statistical Analysis
All data sets were graphically represented, processed, and statistical analyzed with Igor Pro software (Wavemetrics) (except when indicated). Values obtained during infectivity kinetics were plotted and curve fitting was done. Focus area ratio distributions were graphically represented on whisker and box plots showing data sets from the 25th (lower box limit) to 75th percentile (upper box limit) with the 50th percentile (median value, middle horizontal line). The whiskers represented the 10th and 90th percentiles and the empty circles indicate the outliers. All data sets distributions were analyzed for statistical significance between treatments using the nonparametric Wilcoxon-Mann-Whitney two-sample rank test (*P value < 0.05). RT-qPCR data were statistically analyzed by performing t tests analyses (*P value < 0.05). Colocalization quantifications were calculated using Pearson’s correlation coefficient r values of ImageJ, as well as statistically analyzed by performing a t test analysis (*P value < 0.05), and graphically represented on a bar chart.
Total Protein Extraction, Cellular Fractionation, Deglycosylation Assay, and Immunoblot Analysis
Total proteins extractions were performed as described (Batoko et al., 2000). The cellular fractionation experiment was performed as described (Thivierge et al., 2008). Deglycosylation assays were performed with the PNGase F glycosidase under denaturing conditions (New England Biolabs). Briefly, proteins extracts were performed as described above in the co-IP section. Then, 40 μL of each protein extract was further processed essentially according to the manufacturer’s protocol with the following slight modification. Reaction volumes were scaled up, the denaturing step was set to 75°C for 10 min, and the digestion was performed during 3 h at 37°C. For immunoblotting analyses, protein samples were separated on 12% SDS-PAGE and transferred to a nitrocellulose membrane (Bio-Rad), and rabbit antisera were used at the following dilutions either with anti-CP at 1:2500 or anti-RFP (Thermo Fisher Scientific; R-10367) at 1:10000 or anti-GFP (Thermo Fisher Scientific; A-6455) at 1:10,000. The secondary antibody was goat anti-rabbit IgG coupled to horseradish peroxidase.
Immunogold Labeling
Immunogold labeling was performed as described (Wan et al., 2015a) with some adjustments indicated hereafter. Pieces of mock and TuMV-infected leaves were cut and fixed, washed, postfixed in reduced osmium tetroxide (Kopek et al., 2007), rinsed, and dehydrated in a graded alcohol series. Samples were rinsed in 100% alcohol an additional time for 20 min. Samples were then infiltrated with increasing concentrations of LR white resin (Electron Microscopy Science) (50, 75, and 100%) mixed with acetone for a minimum of 8 h for each step at 4°C on a rotator and embedded in pure LR white resin. Sections (90–100 nm) were successively incubated in a Dulbecco’s phosphate glycine buffer saline (DPBS; 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 6.5 mM Na2HPO4, 1 mM CaCl2, and 0.5 mM MgCl2, pH 7.4) in blocking solution containing a mouse monoclonal anti-dsRNA antibody J2 (stock solution is 1 mg/mL; English and Scientific Consulting; catalog no. 10010200; dilution 1:40) diluted in DPBS-BCO solution (2% [w/v] BSA, 2% [w/v] casein, and 0.5% [w/v] ovalbumin in DPBS), washed, and incubated again in blocking buffer. Finally, sections were incubated in a goat anti-mouse secondary antibody, conjugated to 10-nm gold particles (Millipore Sigma), and diluted in DPBS-BCO (dilution 1:20) solution. After washing with DPBS, grids were stained with uranyl acetate and Reynolds lead citrate. Quantification of the distribution of the gold particles per μm2 and relative labeling distribution was performed on mock-infected and TuMV-infected sections according to Lucocq et al. (2004). Two different labeling experiments were considered and 100 gold particles were counted per experiment.
CFDA Treatment
CFDA physiological assay was performed essentially as described (Wan et al., 2015b). Two rosette leaves per Arabidopsis wild-type and itt3 plant were gently abraded over a small area (∼2 × 3 mm2) of the adaxial surface. Five microliters of 60 μg/mL of a CFDA suspension was deposited on the abraded area. The leaf surface was covered with a plastic film, and fluorescence on leaves was acquired by confocal microscopy with the 10× objective under the 488-nm excitation laser.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database or the Arabidopsis Genome Initiative database under the following accession numbers: SYTA (At2g20990.1), SYTB (At1g20080), SYTD (At5g11100), SYTE (At1g05500), SYTF (At3g18370), and SNARE proteins Sec22 (At1g11890), MEMB11 (At2g36900), SYP31 (At5g05760), GOS11(At1g15880), GOS12 (At2g45200), BET11 (At3g58170), BET12 (At4g14455), VAMP714 (At5g22360), SYP71 (At3g09740), VAMP711 (At4g32150), VAMP713 (At5g11150), VAMP721 (At1g04750), VAMP722 (At2g33120), VAMP724 (At4g15780), VAMP725 (At2g32670), VAMP727 (At3g54300), SYP61 (At1g28490), SYP41 (At5g26980), SYP42 (At4g02195), VTI11 (At5g39510), VTI12 (At1g26670), SNAP30 (At1g13890), SNAP33 (At5g61210), Ykt61 (At5g58060), and SYP52 (At1g79590). Reference genes selected for RT-qPCR experiments were ubiquitin (At2g36060), transducin (At2g26060), actin 11 (At3g12110.1) for Arabidopsis and L23 (TC19271), PP2A (TC21939), and F-BOX (Niben.v0.3.Ctg24993647) for N. benthamiana. Mutants used in this were sytb (SALK_108104C), sytc (SALK_124835C), sytd (SAIL_359_H05), syte (SALK_036961C), sytf (SALK_033634C), BET11 SNARE T-DNA KO bet11 (SALK_150636C), VTI11 SNARE EMS KO itt3 (CS68724), and syta-1 (Sail 775A08).
Supplemental Data
Supplemental Figure 1. Interaction of Golgi and Post-Golgi SNAREs with 6K2.
Supplemental Table 1. Association of 6K2 with SNARE Proteins.
Supplemental Table 2. Primers Used in This Study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by grants from the Natural Science and Engineering Research Council of Canada and from Le Fonds Québécois de Recherche sur la Nature et les Technologies to H.Z. and J.-F.L. We thank F. Prieto Bruckner and V. Schurdi-Levraud for their advice and technical support for RT-qPCR experiments. We thank J. Tremblay for his technical expertise assistance with confocal microscopy and S.G. Lazarowitz (Cornell University) for the Arabidopsis syta-1 seeds. We thank P. Moffett (Université de Sherbrooke) for valuable advice and for critically reading the manuscript.
AUTHOR CONTRIBUTIONS
D.G.C., J.J., and J.-F.L. designed the research. D.G.C., J.J., and N.M. performed the research. D.G.C., J.J., H.Z., and J.-F.L. analyzed data. H.G. and Y.Y. contributed new analytic tools. D.G.C., J.J., H.Z., and J.-F.L. wrote the article.
References
- Agbeci M., Grangeon R., Nelson R.S., Zheng H., Laliberté J.F. (2013). Contribution of host intracellular transport machineries to intercellular movement of turnip mosaic virus. PLoS Pathog. 9: e1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenquer M., Amorim M.J. (2015). Exosome biogenesis, regulation, and function in viral infection. Viruses 7: 5066–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amari K., Lerich A., Schmitt-Keichinger C., Dolja V.V., Ritzenthaler C. (2011). Tubule-guided cell-to-cell movement of a plant virus requires class XI myosin motors. PLoS Pathog. 7: e1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amari K., Di Donato M., Dolja V.V., Heinlein M. (2014). Myosins VIII and XI play distinct roles in reproduction and transport of tobacco mosaic virus. PLoS Pathog. 10: e1004448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M.R., Kashanchi F., Jacobson S. (2016). Exosomes in viral disease. Neurotherapeutics 13: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D., Jiang Y., Nagy P.D. (2009). A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 5: e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D., Martín I.F., Pogany J., Risco C., Nagy P.D. (2014). Noncanonical role for the host Vps4 AAA+ ATPase ESCRT protein in the formation of Tomato bushy stunt virus replicase. PLoS Pathog. 10: e1004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H., Zheng H.Q., Hawes C., Moore I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin C., Laliberté J.-F. (2007). The poly(A) binding protein is internalized in virus-induced vesicles or redistributed to the nucleolus during turnip mosaic virus infection. J. Virol. 81: 10905–10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin C., Boutet N., Laliberté J.F. (2007). Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in Planta. J. Virol. 81: 775–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird S.W., Kirkegaard K. (2015). Escape of non-enveloped virus from intact cells. Virology 479-480: 444–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55: 611–622. [DOI] [PubMed] [Google Scholar]
- Chatre L., Brandizzi F., Hocquellet A., Hawes C., Moreau P. (2005). Sec22 and Memb11 are v-SNAREs of the anterograde endoplasmic reticulum-Golgi pathway in tobacco leaf epidermal cells. Plant Physiol. 139: 1244–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Stefano G., Brandizzi F., Zheng H. (2011). Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J. Cell Sci. 124: 2241–2252. [DOI] [PubMed] [Google Scholar]
- Cosson P., Schurdi-Levraud V., Le Q.H., Sicard O., Caballero M., Roux F., Le Gall O., Candresse T., Revers F. (2012). The RTM resistance to potyviruses in Arabidopsis thaliana: natural variation of the RTM genes and evidence for the implication of additional genes. PLoS One 7: e39169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton S., Grangeon R., Thivierge K., Mathieu I., Ide C., Wei T., Wang A., Laliberté J.-F. (2009). Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 83: 10460–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craxton M. (2004). Synaptotagmin gene content of the sequenced genomes. BMC Genomics 5: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Shen J., Gao C., Zhuang X., Wang J., Jiang L. (2016). Biogenesis of plant prevacuolar multivesicular bodies. Mol. Plant 9: 774–786. [DOI] [PubMed] [Google Scholar]
- Diaz A., Zhang J., Ollwerther A., Wang X., Ahlquist P. (2015). Host ESCRT proteins are required for bromovirus RNA replication compartment assembly and function. PLoS Pathog. 11: e1004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty G.P., Bailey K., Lewis P.J. (2010). Stage-specific fluorescence intensity of GFP and mCherry during sporulation in Bacillus subtilis. BMC Res. Notes 3: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., Okatani Y., Uemura T., Goh T., Shoda K., Niihama M., Morita M.T., Spitzer C., Otegui M.S., Nakano A., Ueda T. (2008). A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamtanasate S., Juricek M., Yap Y.-K. (2007). C-terminal hydrophobic region leads PRSV P3 protein to endoplasmic reticulum. Virus Genes 35: 611–617. [DOI] [PubMed] [Google Scholar]
- Feng Z., Hensley L., McKnight K.L., Hu F., Madden V., Ping L., Jeong S.H., Walker C., Lanford R.E., Lemon S.M. (2013). A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496: 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folimonova S.Y., Tilsner J. (2018). Hitchhikers, highway tolls and roadworks: the interactions of plant viruses with the phloem. Curr. Opin. Plant Biol. 43: 82–88. [DOI] [PubMed] [Google Scholar]
- Grangeon R., Agbeci M., Chen J., Grondin G., Zheng H., Laliberté J.F. (2012). Impact on the endoplasmic reticulum and Golgi apparatus of turnip mosaic virus infection. J. Virol. 86: 9255–9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeon R., Jiang J., Wan J., Agbeci M., Zheng H., Laliberté J.F. (2013). 6K2-induced vesicles can move cell to cell during turnip mosaic virus infection. Front. Microbiol. 4: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harak C., Lohmann V. (2015). Ultrastructure of the replication sites of positive-strand RNA viruses. Virology 479-480: 418–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries P.A., Park J.W., Sasaki N., Ballard K.D., Maule A.J., Nelson R.S. (2009). Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proc. Natl. Acad. Sci. USA 106: 17594–17599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.S., Wei T., Laliberté J.F., Wang A. (2010). A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol. 152: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyodo K., Okuno T. (2016). Pathogenesis mediated by proviral host factors involved in translation and replication of plant positive-strand RNA viruses. Curr. Opin. Virol. 17: 11–18. [DOI] [PubMed] [Google Scholar]
- Hyodo K., Mine A., Taniguchi T., Kaido M., Mise K., Taniguchi H., Okuno T. (2013). ADP ribosylation factor 1 plays an essential role in the replication of a plant RNA virus. J. Virol. 87: 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S., Harrison M.J. (2014). A set of fluorescent protein-based markers expressed from constitutive and arbuscular mycorrhiza-inducible promoters to label organelles, membranes and cytoskeletal elements in Medicago truncatula. Plant J. 80: 1151–1163. [DOI] [PubMed] [Google Scholar]
- Iwakawa H.O., Mine A., Hyodo K., An M., Kaido M., Mise K., Okuno T. (2011). Template recognition mechanisms by replicase proteins differ between bipartite positive-strand genomic RNAs of a plant virus. J. Virol. 85: 497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia T., Gao C., Cui Y., Wang J., Ding Y., Cai Y., Ueda T., Nakano A., Jiang L. (2013). ARA7(Q69L) expression in transgenic Arabidopsis cells induces the formation of enlarged multivesicular bodies. J. Exp. Bot. 64: 2817–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Patarroyo C., Garcia Cabanillas D., Zheng H., Laliberté J.F. (2015). The vesicle-forming 6K2 protein of Turnip mosaic virus interacts with the COPII coatomer Sec24a for viral systemic infection. J. Virol. 89: 6695–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Morita M.T., Fukaki H., Yamauchi Y., Uehara M., Niihama M., Tasaka M. (2002). SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami S., Watanabe Y., Beachy R.N. (2004). Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. USA 101: 6291–6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopek B.G., Perkins G., Miller D.J., Ellisman M.H., Ahlquist P. (2007). Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5: e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev N., Pogany J., Nagy P.D. (2012). A Co-Opted DEAD-Box RNA helicase enhances tombusvirus plus-strand synthesis. PLoS Pathog. 8: e1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliberté J.F., Sanfaçon H. (2010). Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 48: 69–91. [DOI] [PubMed] [Google Scholar]
- Laliberté J.F., Zheng H. (2014). Viral manipulation of plant host membranes. Annu. Rev. Virol. 1: 237–259. [DOI] [PubMed] [Google Scholar]
- Langosch D., Arkin I.T. (2009). Interaction and conformational dynamics of membrane-spanning protein helices. Protein Sci. 18: 1343–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Levy A., Zheng J.Y., Lazarowitz S.G. (2015). Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr. Biol. 25: 2018–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.D., Lazarowitz S.G. (2010). Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. USA 107: 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.H., Carrington J.C. (1995). Complementation of tobacco etch potyvirus mutants by active RNA polymerase expressed in transgenic cells. Proc. Natl. Acad. Sci. USA 92: 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligat L., Lauber E., Albenne C., San Clemente H., Valot B., Zivy M., Pont-Lezica R., Arlat M., Jamet E. (2011). Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 11: 1798–1813. [DOI] [PubMed] [Google Scholar]
- Liu D., Shi L., Han C., Yu J., Li D., Zhang Y. (2012). Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One 7: e46451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Westler W.M., den Boon J.A., Wang X., Diaz A., Steinberg H.A., Ahlquist P. (2009). An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 5: e1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A., Boyd B., Chisari F.V. (2015). Virion-independent transfer of replication-competent hepatitis C virus RNA between permissive cells. J. Virol. 89: 2956–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq J.M., Habermann A., Watt S., Backer J.M., Mayhew T.M., Griffiths G. (2004). A rapid method for assessing the distribution of gold labeling on thin sections. J. Histochem. Cytochem. 52: 991–1000. [DOI] [PubMed] [Google Scholar]
- Mayo M.A. (1995). The Potyviridae. New Phytol. 131: 289–290. [Google Scholar]
- McClintock K., Lamarre A., Parsons V., Laliberté J.F., Fortin M.G. (1998). Identification of a 37 kDa plant protein that interacts with the turnip mosaic potyvirus capsid protein using anti-idiotypic-antibodies. Plant Mol. Biol. 37: 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S.W., Chang W.P., Südhof T.C. (2007). E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc. Natl. Acad. Sci. USA 104: 3823–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine A., Hyodo K., Takeda A., Kaido M., Mise K., Okuno T. (2010). Interactions between p27 and p88 replicase proteins of Red clover necrotic mosaic virus play an essential role in viral RNA replication and suppression of RNA silencing via the 480-kDa viral replicase complex assembly. Virology 407: 213–224. [DOI] [PubMed] [Google Scholar]
- Movahed N., Patarroyo C., Sun J., Vali H., Laliberté J.F., Zheng H. (2017). Cylindrical inclusion protein of Turnip mosaic virus serves as a docking point for the intercellular movement of viral replication vesicles. Plant Physiol. 175: 1732–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A., Ritzenthaler C., Robinson D.G. (2002). Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol. 130: 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikiori M., Mori M., Dohi K., Okamura H., Katoh E., Naito S., Meshi T., Ishikawa M. (2011). A host small GTP-binding protein ARL8 plays crucial roles in tobamovirus RNA replication. PLoS Pathog. 7: e1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patarroyo C., Laliberté J.-F., Zheng H. (2013). Hijack it, change it: how do plant viruses utilize the host secretory pathway for efficient viral replication and spread? Front. Plant Sci. 3: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnaiah V., et al. (2013). Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc. Natl. Acad. Sci. USA 110: 13109–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo-Hartwig M.A., Carrington J.C. (1994). The tobacco etch potyvirus 6-kilodalton protein is membrane associated and involved in viral replication. J. Virol. 68: 2388–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson L.G., Clendening E.A., Sheen H., Gidda S.K., White K.A., Mullen R.T. (2014). A unique N-terminal sequence in the Carnation Italian ringspot virus p36 replicase-associated protein interacts with the host cell ESCRT-I component Vps23. J. Virol. 88: 6329–6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J., Chen X., Araç D. (2006). Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 16: 339–350. [DOI] [PubMed] [Google Scholar]
- Romero-Brey I., Bartenschlager R. (2014). Membranous replication factories induced by plus-strand RNA viruses. Viruses 6: 2826–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter B.D., Innes R.W. (2017). Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173: 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore C.M., Evins J., Batoko H., Brandizzi F., Moore I., Hawes C. (2002). Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29: 661–678. [DOI] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C., Nebenführ A., Boulaflous A., Follet-Gueye M.L., Plasson C., Hawes C., Driouich A., Faye L., Gomord V. (2006). Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18: 3182–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A.A., Kovaleva V., Bassham D.C., Raikhel N.V. (2001). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12: 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O., Teis D. (2012). The ESCRT machinery. Curr. Biol. 22: R116–R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M., Chen J., Janda M., Sullivan M., den Boon J., Ahlquist P. (2002). A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9: 505–514. [DOI] [PubMed] [Google Scholar]
- Steentoft C., et al. (2013). Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32: 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano G., Renna L., Chatre L., Hanton S.L., Moreau P., Hawes C., Brandizzi F. (2006). In tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum and of ER export sites depends on active COPI machinery. Plant J. 46: 95–110. [DOI] [PubMed] [Google Scholar]
- Tan D., Cai Y., Wang J., Zhang J., Menon S., Chou H.T., Ferro-Novick S., Reinisch K.M., Walz T. (2013). The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. USA 110: 19432–19437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teese M.G., Langosch D. (2015). Role of GxxxG motifs in transmembrane domain interactions. Biochemistry 54: 5125–5135. [DOI] [PubMed] [Google Scholar]
- Thivierge K., Cotton S., Dufresne P.J., Mathieu I., Beauchemin C., Ide C., Fortin M.G., Laliberté J.-F. (2008). Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 377: 216–225. [DOI] [PubMed] [Google Scholar]
- Tilsner J., Linnik O., Louveaux M., Roberts I.M., Chapman S.N., Oparka K.J. (2013). Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 201: 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter V., Altmann F., März L. (1991). Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached alpha 1----3 to the asparagine-linked N-acetylglucosamine residue. Eur. J. Biochem. 199: 647–652. [DOI] [PubMed] [Google Scholar]
- Uchiyama A., Shimada-Beltran H., Levy A., Zheng J.Y., Javia P.A., Lazarowitz S.G. (2014). The Arabidopsis synaptotagmin SYTA regulates the cell-to-cell movement of diverse plant viruses. Front. Plant Sci. 5: 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R.L., Nakano A., Takeyasu K., Sato M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29: 49–65. [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Basu K., Mui J., Vali H., Zheng H., Laliberté J.F. (2015a). Ultrastructural characterization of Turnip mosaic virus-induced cellular rearrangements reveals membrane-bound viral particles accumulating in vacuoles. J. Virol. 89: 12441–12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Cabanillas D.G., Zheng H., Laliberté J.F. (2015b). Turnip mosaic virus moves systemically through both phloem and xylem as membrane-associated complexes. Plant Physiol. 167: 1374–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., et al. (2015). Arabidopsis Synaptotagmin 2 participates in pollen germination and tube growth and is delivered to plasma membrane via conventional secretion. Mol. Plant 8: 1737–1750. [DOI] [PubMed] [Google Scholar]
- Wei T., Hibino H., Omura T. (2009). Release of rice dwarf virus from insect vector cells involves secretory exosomes derived from multivesicular bodies. Commun. Integr. Biol. 2: 324–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Zhang C., Hong J., Xiong R., Kasschau K.D., Zhou X., Carrington J.C., Wang A. (2010). Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 6: e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Zhang C., Hou X., Sanfaçon H., Wang A. (2013). The SNARE protein Syp71 is essential for turnip mosaic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. PLoS Pathog. 9: e1003378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji Y., et al. (2012). Lectin-mediated resistance impairs plant virus infection at the cellular level. Plant Cell 24: 778–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., Kawamura Y., Minami A., Uemura M. (2008). Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. Plant Cell 20: 3389–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang L., Gao B., Fan H., Jin J., Botella M.A., Jiang L., Lin J. (2011). Golgi apparatus-localized synaptotagmin 2 is required for unconventional secretion in Arabidopsis. PLoS One 6: e26477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.C., Zhang G., Yang L., Chisholm J., Sanfaçon H. (2005). Evidence that insertion of Tomato ring spot nepovirus NTB-VPg protein in endoplasmic reticulum membranes is directed by two domains: a C-terminal transmembrane helix and an N-terminal amphipathic helix. J. Virol. 79: 11752–11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., von Mollard G.F., Kovaleva V., Stevens T.H., Raikhel N.V. (1999). The plant vesicle-associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment. Mol. Biol. Cell 10: 2251–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Camacho L., Wee E., Batoko H., Legen J., Leaver C.J., Malhó R., Hussey P.J., Moore I. (2005). A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell 17: 2020–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Han S.W., Rodriguez-Welsh M.F., Rojas-Pierce M. (2014). Homotypic vacuole fusion requires VTI11 and is regulated by phosphoinositides. Mol. Plant 7: 1026–1040. [DOI] [PubMed] [Google Scholar]