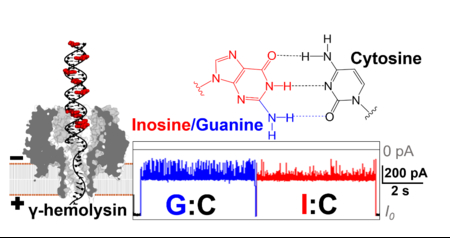
Abstract
Biological nanopores provide a unique single-molecule sensing platform to detect target molecules based on their specific electrical signatures. The γ-hemolysin (γ-HL) protein produced by Staphylococcus aureus is able to assemble into an octamer nanopore with a ~2.3 nm diameter β-barrel. Herein, we demonstrate the first application of γ-HL nanopore for DNA structural analysis. To optimize conditions for ion-channel recording, the properties of the γ-HL pore (e.g., conductance, voltage-dependent gating, ion-selectivity) were characterized at different pH, temperature, and electrolyte concentrations. The optimal condition for DNA analysis using γ-HL corresponds to 3 M KCl, pH = 5, and T = 20 ⁰C. The γ-HL protein nanopore is able to translocate dsDNA at about ~20 bp/ms, and the unique current-signature of captured dsDNA can directly distinguish guanine-to-inosine substitutions at the single-molecule level with ~99% accuracy. The slow dsDNA threading and translocation processes indicate this wild-type γ-HL channel has potential to detect other base modifications in dsDNA.
Graphical Abstract
Introduction
Over the past two decades, resistive sensing using nanopores has been demonstrated for label-free, single-molecule recognition of analytes.1–4 Charged molecules, such as DNA, are electrophoretically driven into a nanopore under an electric potential, and a volume element of the electrolyte solution is displaced by the analyte as it passes through the channel, resulting in a corresponding decrease in the nanopore conductance, i.e., a “resistive pulse”.5–7 The event frequency, duration time, and unique current modulation signature can reveal the identity and concentration of the target molecule.3, 7–8
Protein ion channels are particularly attractive for molecule sensing due to their ability to fold into well-defined nanopores that yield reproducible electrical signals. Further, site-directed mutagenesis allows modifications of protein channels with molecular precision.3, 7, 9–11 The sensitivity and specificity of molecular sensing with protein nanopores are determined by the unique interactions between the target molecule and the protein sensor.8 As a consequence of the small aperture diameters, α-hemolysin (d ~1.4 nm), MspA (d ~1.2 nm), and aerolysin (d ~1 nm), have shown potential for DNA sequencing, peptide/protein characterization, and single nucleotide substitutions in short oligonucleotides.6, 12–26 As the most thoroughly studied protein nanopore, the α-hemolysin (α-HL) nanopore has been demonstrated to discriminate DNA sequences in the β-barrel, and to analyze DNA secondary structures such as hairpins,27–29 G-quadruplexes,30–31 and i-motifs32 in the vestibule. The latch zone of α-HL also provides a sensitive region for discrimination of base pairs and their modifications in double-stranded DNA (dsDNA).33–40 However, a key feature of α-HL is that dsDNA is unable to translocate through the β-barrel, preventing the continuous analysis of base pairs and their modifications. Several other nanopores have been demonstrated to translocate dsDNA. The channel either has a constriction zone large enough to allow direct passage of dsDNA, including phi 29 connector channel (~3.3 nm constriction diameter)41–43 and ClyA (~3.3 nm constriction diameter),44–46 or is able to deform the constriction to translocate the dsDNA molecules, i.e., Fragaceatoxin C nanopore (~1.2 nm constriction diameter).47 However, discrimination of base modifications in the dsDNA has not been demonstrated using these nanopores. Since the pore geometry plays a determining role in sensing ability, sensitivity, and specificity for the DNA analyte, the search for protein nanopores with optimal geometries for sequencing base modifications on the dsDNA is ongoing.
Herein, the γ-hemolysin (γ-HL) secreted by Staphylococcus aureus, which has a larger nanocavity (vestibule) and a wider β-barrel compared with α-HL, is investigated as a potential nanopore for dsDNA analysis (Figure 1).48 However, to our knowledge, the capture of negatively charged DNA molecules in γ-HL has not been demonstrated, likely due to the cation-selective property and problematic voltage-dependent gating of the channel.49–51 Only the detection of the permethylated γ-cyclodextrin at positive potential (cis vs trans) in γ-HL has been explored.52 In the present work, we optimize the experimental conditions for γ-HL for DNA analysis by increasing the electrolyte concentration to 3 M and lowering the pH to 5.0, resulting in a continuous open-channel current, allowing the capture and characterization of DNA molecules from the vestibule side of the γ-HL nanopore.
Figure 1.
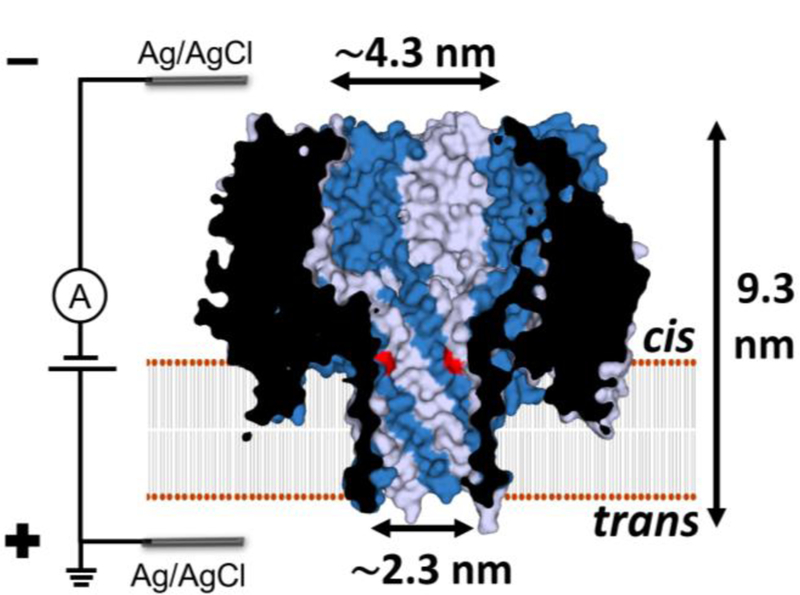
Cross section structure of the γ-HL nanopore showing the dimension of the lumen. The γ-HL pore is an octamer with four HlgB chains (gray) and four HlgC chains (blue) alternating around a central axis.48 This putative HlgC-HlgB pore structure is based on the crystal structure of the HlgA-HlgB homologue (PDB: 3B07)48 with one mutation in the β-barrel: lysine to threonine in position 108 of HlgA (highlighted in red). The surface representation, K108T mutations, and the dimensions of the protein were generated using PyMOL.
Results and Discussion
Optimize conditions for ion channel measurement using γ-HL.
Previous ion channel recordings showed that γ-HL pore is cation-selective and exhibits frequent voltage-dependent gating events at potentials more negative than −60 mV (cis vs trans).49–51 Both the cation selectivity and the gating at negative potentials impede the electrophoretic capture and subsequent analysis of DNA using γ-HL. However, previous investigations were performed at neutral pH with either 0.1 or 1 M electrolyte. The experimental conditions, including pH, electrolyte concentration, and temperature, can often affect the ion selectivity and voltage-dependent gating of the nanopore. Therefore, we first optimized the experimental conditions of γ-HL nanopore for DNA analysis.
γ-HL ion selectivity.
The ion selectivity was determined from the ion-channel reversal potential (Vr, also known as Nernst potential) using the Goldman-Hodgkin-Katz (GHK) equation.53 The reversal potential corresponds to the electrical potential at zero current with different electrolyte concentrations on the two sides of the ion channel (3 M KCl (cis) and 1 M KCl (trans)). Figures 2a-b displays I-V relationships for γ-HL at different pH. As the pH decreased from 8.0 to 4.0, the channel changed from being cation selective (, Table S2) to near non-selective (, Table S2). Therefore, negatively charged DNA is likely to be captured by the γ-HL in acidic pH. The modulation of ion selectivity by solution pH is likely due to the alteration of charges in the channel via protonation/deprotonation of ionizable residuals. Similar pH-dependent ion selectivity has been observed in other β-barrel toxins, including α-hemolysin.54
Figure 2.
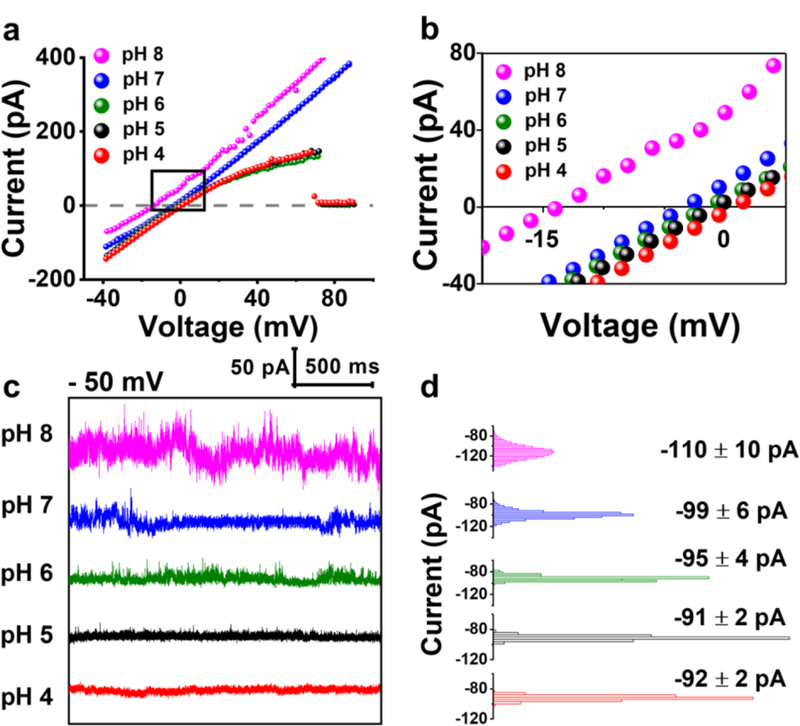
(a) Single-channel I-V curves of γ-HL at different pH in asymmetrical KCl solutions (3 M vs 1 M KCl, cis vs trans) for measurement of ion selectivity of the protein channel. Note in acid conditions (pH 4–6), the protein exhibits voltage-induced gating at positive potentials (indicated by ~0 current at > 70 mV). The sudden drop in current at pH 4 to 6 is due to voltage gating of the channel. Potential difference between the two Ag/AgCl reference electrodes has been accounted for. (b) Expanded view of the I-V curves near 0 pA. (c) I-t traces showing the open-channel current at −50 mV from pH 4 to 8 in 1 M KCl, 20 mM HOAc/KOAc or PB, at 20.0 °C. I-t traces were sampled at 50 kHz and filtered at 10 kHz using a low-pass Bessel filter. (d) All-point histograms of the open-channel current during the 2 s period (100,000 data points) at each pH as shown in (c).
Effect of pH on the open-channel current.
The γ-HL channel remains mostly open at −50 mV at pH from 8.0 to 4.0, as indicated by the non-interrupted open-channel current (Figure 2c). The histogram of the open-channel current is Gaussian-shaped showing a mean conductance of ~2.4 nS in 1 M KCl (−50 mV, 5 mM PB, pH 8.0), consistent with the previous report.49 The fluctuation of the open-channel current (measured by the standard deviation of the current) decreased from 10 to 2 pA as the pH was reduced from 8.0 to 4.0 (Figure 2 c-d). The fluctuation in the I-t traces show stepwise transitions between several similar conductance levels (Figure 2c and Figure S1), which may correspond to open-close transitions of the protein channel.55 Although the current fluctuation is significantly reduced in acidic conditions, DNA depurination process becomes significant at pH 4.0 and below.56–57 Therefore, a pH of 5.0 was selected as the optimal condition for DNA analysis using γ-HL.
Figure 4.
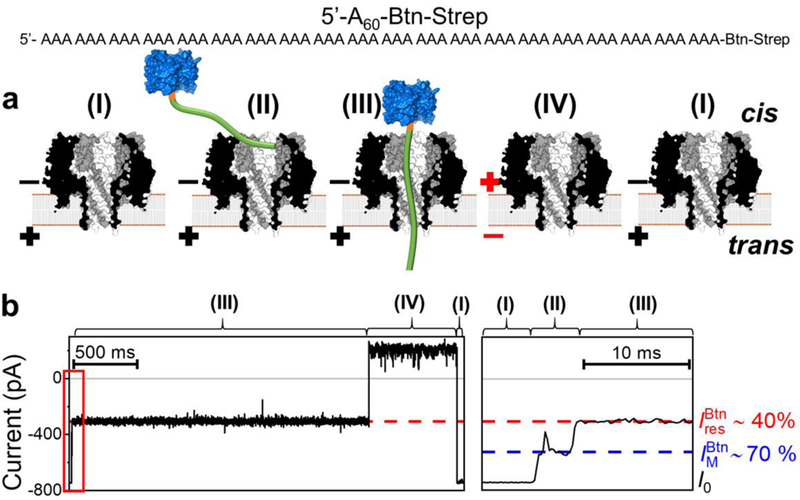
Capture and immobilization of A60-Btn-Strep in γ-HL. (a) Schematic showing: (I) open channel of γ-HL; (II) electrophoretically captured ssDNA exploring the vestibule; (III) ssDNA threading through the β-barrel; (IV) release of ssDNA by reversing the potential polarity. The ssDNA [poly(dA)60] (green) is attached to streptavidin (blue) using a biotin linker (orange). (b) Left: sample I-t trace labeled with proposed steps, (I)-(IV). Right: expanded view of the mid-level blocking current indicated in the red box. Recordings were carried out at −120 mV, in 3 M KCl, 20 mM HOAc/KOAc, pH 5.0, at 20 °C. The I-t traces were post-filtered at 2 kHz for presentation. Open-channel current (I0), mid-level blockade (), and deep current blockade correspond to (I), (II), and (III), respectively.
Dependence of gating on electrolyte concentration.
As noted above, another challenge for DNA analysis using γ-HL is the frequent voltage-dependent gating at negative potentials (< −60 mV, cis vs trans), which results from the stochastic transition between open- and close-state conformations. Here, we discuss the effect of electrolyte concentration on the protein gating. In 1 M KCl (pH 5.0), the protein channel shows frequent gating events at −80 and −120 mV (Figure 3). The reversal of gating (transition from the closed state to the open state) may occur spontaneously in tens of milliseconds, but sometimes takes longer periods (> 1 s) and requires toggling between positive and negative potentials (indicated by the arrows in Figure 3). The frequency of these long-duration closed states increases as the voltage changes from at −50 mV to −120 mV in 1 M KCl (Figure 3). However, as the electrolyte concentration increases to 3 M, the gating frequency of the channel decreases, and the long-duration closed states becomes very rare (<1 per minute, at −120 mV). The less favorable close-state conformation at high salt condition is probably due to efficient screening of charged surface residues; a similar salt-dependent gating phenomenon has been reported for the α-hemolysin nanopore.58 Overall, as the electrolyte concentration increases, the open-channel state is more favorable, which is desirable for DNA analysis.
Figure 3.
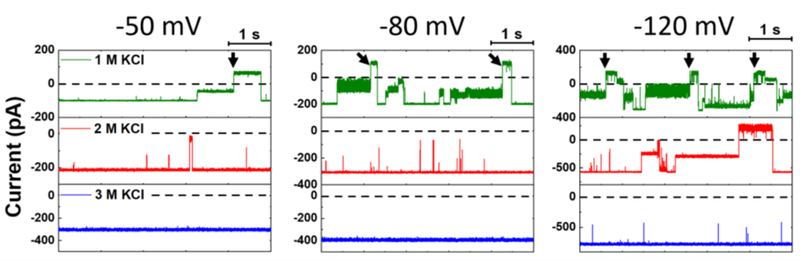
Single-channel I-t traces (5 s) obtained in 1, 2, and 3 M KCl at applied potentials of −50, −80, and −120 mV (cis vs trans). When the channel transits to a closed state and does not recover to the open state, a reversal potential (+50, +80, +120 mV) is applied as indicated by the arrow. Experiments were carried out at 20.0 °C in pH 5.0 solutions. The I-t traces were post-filtered at 2 kHz for presentation.
Effect of temperature.
The effect of temperature on current fluctuation and gating frequency of γ-HL nanopore was also investigated. Figure S2a shows the sample I-t traces for γ-HL at temperatures ranging from 10.0 to 30.0 °C, using the optimized solution condition (3 M KCl, buffered at pH 5.0). The open-channel conductance shows an approximately linear dependence on temperature (Figure S2b), reflecting the bulk temperature-dependent conductivity of the KCl buffer. The fluctuation of the open-channel current remains relatively low (1% relative deviation) between 10.0 and 25.0 °C and increases significantly at 30.0 °C (3% relative deviation). On the other hand, the protein gating events (~50% current blockade) are largely temperature independent in the range of 10 – 30 °C. Considering the above, 20.0 °C was selected as the temperature for further experiments.
In summary, we find the optimal conditions for using γ-HL are 3 M KCl, pH 5.0 and 20.0 °C. Under these conditions, the protein channel shows no cation selectivity, infrequent voltage-dependent gating, and low fluctuation in open-channel current at negative potentials (cis vs trans). These features allow for the capture and characterization of ssDNA or dsDNA molecules in the vestibule of the γ-HL nanopore.
Threading of ssDNA into the γ-HL protein channel.
We investigated the current signature and blockade level of tethered ssDNA captured by the γ-HL nanopore, Figure 4. At −120 mV (cis vs trans), 0.4 μM of streptavidin-biotinylated poly(dA)60 (hereinafter termed “A60-Btn-Strep”) was added in the cis chamber. The ssDNA portion of the A60-Btn-Strep was electrophoretically driven into the γ-HL protein channel and held for ~2 s, generating a residual current blockade of ~40% (Figure 4). The ssDNA was then released by reversing the applied potential, and the current was restored to the open-channel current. More examples out of a total of 294 individual events are shown in Figure S3.
The proposed sequential processes of the capture-release cycle of A60-Btn-Strep is illustrated in Figure 5a: (I) open channel; (II) the ssDNA portion of A60-Btn-Strep threads in the vestibule and attempts to enter the β-barrel, resulting in a shallow level blockade ; (III) ssDNA poly(dA)60 occupies the β-barrel, generating a deeper blocking current ; (IV) ssDNA is driven back to the bulk by voltage reversal, and (I) the γ-HL returns to open channel.
Figure 5.
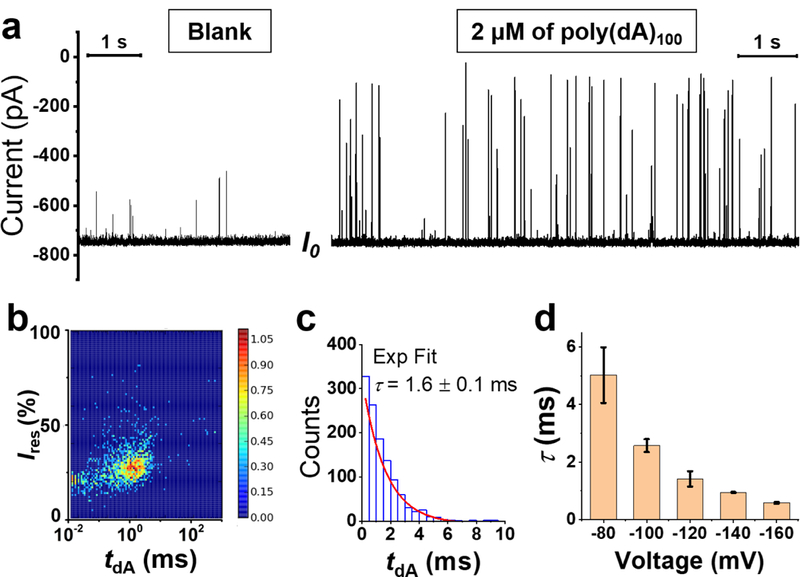
Translocation of ssDNA (poly(dA)100) through the γ-HL nanopore. (a) Left: open-channel I-t trace at −120 mV in the absence of ssDNA. Right: The addition of 2 μM ssDNA in the cis chamber results in short but deep current blockades. Expanded I-t trace is shown in Figure S9. (b) Density plot (N=1175) for residual current (Ires) vs event duration (tdA). (c) Histogram of ssDNA translocation duration (N=1039) by excluding the fast collision events (tdA <0.04 ms) and incomplete translocation events (Ires ≥40%). The distribution of tdA is well fitted by a first-order exponential kinetic with a time constant (τ) of 1.6 ms (red line). (d) Effect of voltage on the translocation time constant, τ. The error bars represent standard deviations from the fittings of 3 repeated experiments (N=800–2500 in each experiment). Recordings were carried out at 20 °C in 3 M KCl, 20 mM HOAc/KOAc, pH 5.0. I-t traces were sampled at 500 kHz and post-filtered at 50 kHz for presentation.
In contrast to a single blockade current by immobilization of ssDNA in α-HL (11 ± 2%, Figure S3, under the identical experimental conditions: in 3 M KCl, 20 mM HOAc/KOAc, pH 5.0, at −120 mV), two current blockade levels, (~70%) and (42 ± 3%), were observed as the ssDNA is immobilized by the γ-HL (Figure 4b). These two current blockade levels are voltage independent, while the duration of is voltage dependent. As the applied potential changes from −80 to −120 mV, the duration of decreased from several seconds to less than several microseconds (Figure S4–6). These results are consistent with the assignment of the shallower blockade level, , to Step II (ssDNA exploring the vestibule), and the deeper blockade level, , to Step III (ssDNA occupying the β-barrel), as the transition from Step II to Step III is favored at more negative potentials.
Translocation of single-stranded DNA in the γ-HL protein channel.
Having established that tethered ssDNA can be captured by the γ-HL protein channel, we then explored the translocation of free ssDNA. The addition of homopolymeric strand, poly(dA)100, to the cis chamber produced transient current blockades (Figure 5a). The transient current blockades caused by poly(dA)100 molecules display deep blockades (< 50 % of I0), and the frequency of the events is proportional to the concentration of the ssDNA in the cis chamber (Figure S7). At low applied potential (−60 mV), over 50% of events showed shallow current blockade (> 50% of I0) and had relatively long residence times (> 10 ms), resulting from incomplete translocation events, i.e. ssDNA enters the vestibule and then escapes from the cis side of the protein channel without threading into the narrower β-barrel (Figure S8). At potentials more negative than −120 mV, >90% of events showed successful ssDNA translocation, indicated by the deep-blockade current ≤ 40% of I0. As the applied potential increased from −60 to −160 mV, the average blockade current decreased from 42±17 % to 25±5 % of I0 (Figure S8–10). However, for the very fast events (< 0.04 ms) with ~20% residual current (Figure 5b), neither duration nor blockade level were dependent on the applied potential (Figure S10a), suggesting these events likely result from the collision between ssDNA molecules and the protein channel.
Thousands of successful translocation events in each experiment were used to generate the dwell time histogram, Figure 6c, which is well determined by an exponential decay. The time constant (τ) for translocation of poly(dA)100 is ~1.6 ms at −120 mV (~60 nt/ms) (Figure 5c). The dwell time constant (τ) decreases as the applied potential increases (Figure 5d), consistent with the voltage-driven translocation of ssDNA through the nanopore. The observed translocation speed of ssDNA in γ-HL (~2.3 nm diameter β-barrel) is slower compared to other narrower protein channels, including α-HL (500–1,000 nt/ms, ~1.4 nm diameter constriction)17, 59 and MspA (2,000–10,000 nt/ms,~1.2 nm diameter β-barrel),19 suggesting that protein-DNA interactions using γ-HL slow down the translocation of DNA. Strong protein-DNA interactions have also been proposed to explain the slow tranloscation rate (2–40 nt/ms) of ssDNA in aerolysin (d ~1 nm).21
Figure 6.
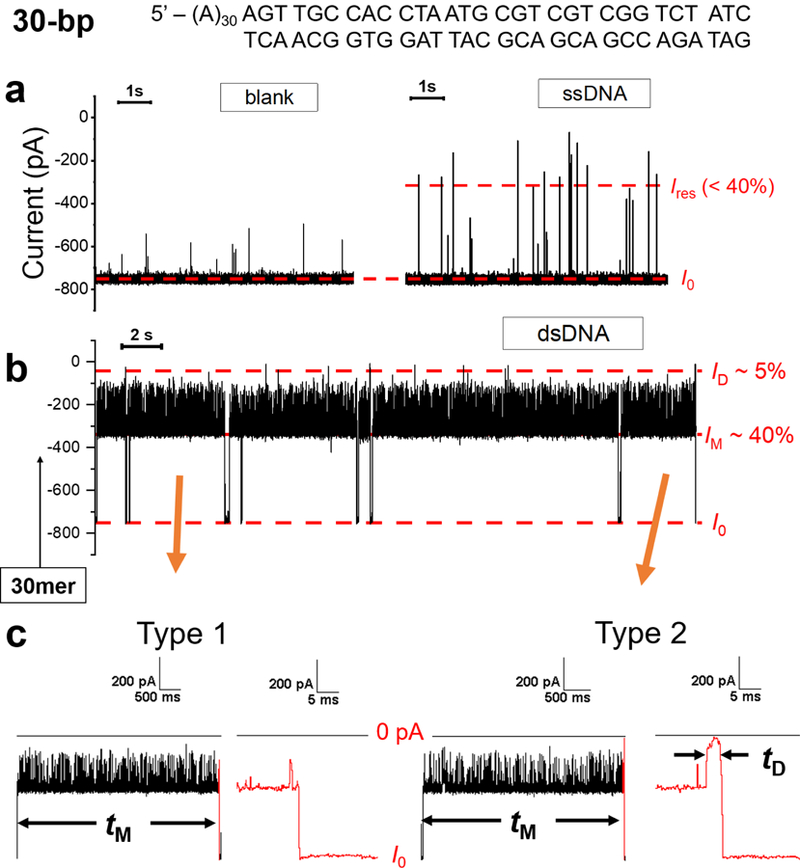
Capture and release of dsDNA using the γ-HL nanopore. (a) I-t trace in the absence of DNA (left) and in the presence of 2 μM 60-mer ssDNA in the cis chamber (right). (b) I-t traces corresponding to 30-bp dsDNA molecules with a poly(dA)30 tail (sequence shown on the top). The long blockades (> 100 ms) display modulation between a mid-level blockade (IM) of ~40% and a deeper blockade (~20%). (c) I-t traces for two types of dsDNA blockade events. Type 1 events terminate from IM to the open-channel current I0 directly. Type 2 events exhibit a deep-blockade current of ~5% at the end before returning to the open channel current. Data were filtered by a Bessel filter at 100 kHz for (a) and 10 kHz for (b)-(c). The I-t traces were post-filtered at 10 kHz (a) and 2 kHz for presentation (b-c), respectively.
Translocation double-stranded DNA in the γ-HL protein channel.
We next evaluated the ability of γ-HL to translocate dsDNA. A 60-mer ssDNA, 5’-(A)30-AGTTGCCACCTAATGCGTCGTCGGTCTATC, was first added to the cis chamber, which produced transient current blockades of ≤ 40% (Figure 6a), similar to that of the ssDNA translocation events described above (Figure 5). A 30-mer complementary strand was subsequently added, which hybridized with the 60-mer ssDNA in the chamber to form a 30-bp duplex with a poly(dA)30 tail (sequence shown in Figure 6). The conversion of ssDNA to dsDNA results in long blocking currents events (> 100 ms) in the I-t traces as shown in Figures 6b and S11. Close inspection of the I-t traces revealed that these long blocking dsDNA events can be categorized into two types (Figure 6c). Type 1 events exhibit a mid-blockade current, IM (~40% of I0), with upward current spikes, and then terminate from IM to the open-channel current. Type 2 events have a similar current pattern as Type 1 events but terminate with a deep-blockade current, ID (~5% of I0). The amplitude of IM in both Type 1 and Type 2 events are similar to that generated by immobilized A60-Btn-Strep molecules (Figure 3), suggesting that the single-stranded tail (dA)30 has threaded into the β-barrel while the duplex is occupying the vestibule to generate IM. The spikes on top of IM in both Type 1 and Type 2 events likely result from the attempts of the duplex portion of the DNA to enter the barrel. For Type 1 events, the event terminates by either diffusing back to the cis compartment, or unzipping into its single-stranded components, which will be further discussed later. For Type 2 events, the deep-blockade current ID (~5% of I0) likely results from the translocation of the duplex part of the DNA (see below). For both events, the current returns to baseline (I0) after the dsDNA has fully translocated, retracted from the nanopore, or unzipped.
To better understand the mechanisms of both Type 1 and 2 events, we investigated the effects of voltage on the fraction and the duration of the two types of event. As shown in Figure 7 a-b, the duration of both Type 1 and Type 2 events can be fitted by an exponential distribution, from which the respective time constants, τ1 and τ2 can be obtained. As the applied potential changed from −100 to −160 mV, the fraction of Type 1 events decreased from 90% to 20%, while their duration (τ1) increased from 0.6 ± 0.2 s to 4.3 ± 0.4 s (Figure 7 c-d). These observations suggest that Type 1 events likely correspond to the dsDNA sampling the vestibule of γ-HL and then escaping back to the cis compartment rather than translocating or unzipping, as the probability of DNA escaping from the cis side decreases as the applied potential increases.60 In contrast, the fraction of Type 2 events increased from 10% to 80% and their dwell times decreased as the voltage increased (Figure 7c-d), which is consistent with the duplex translocating or unzipping in the γ-HL protein channel (vide infra).
Figure 7.
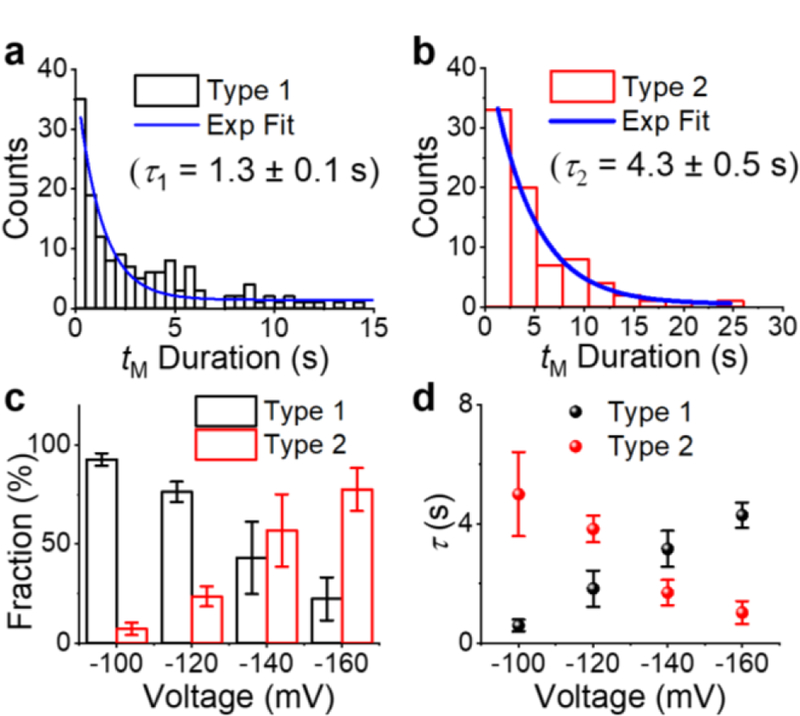
(a-b) Histograms of event duration of (a) Type 1 and (b) Type 2 events at −120 mV. Blue curves are the best exponential fits. Effect of voltage on the (c) fraction and (d) duration (τ) of Type 1 (black) and Type 2 (red) events. Error bars represent standard deviations from 3 or 4 repeated measurements (N=30–500 in each experiment). Experiments were carried out in 3 M KCl, 20 mM HOAc/KOAc, pH 5.0, at 20.0 °C.
We used the HlgA-HlgB assembled γ-HL crystal structure48 to estimate the internal diameter of the β-barrel of the nanopore to be ~2.3 nm (measured from the side chains, Figure 1). Note that the residues of the central stem domains that generate the β-barrel are homologous between HlgA and HlgC, in support of such a measurement.61–62 As the dsDNA is captured by the protein channel, the poly(dA)30 tail threads into the barrel and pulls the duplex part into the β-barrel. However, the β-barrel of γ-HL is too narrow for the duplex, with a diameter of ~2.0 nm, to freely translocate without a close alignment between the helix and channel axes. When the duplex is partly inside the barrel, it can diffuse out of the barrel via thermal motion. The intermittent partial entrance and the back diffusion of the duplex result in the millisecond deep-blocking spikes in the I-t traces (Figure 6 b-c). Since the duplex is driven into the barrel by electric force, the attempted entrance frequency is enhanced by increasing the applied potential, as shown by the increased frequency of the spikes in the I-t traces for Type 1 and 2 events (Figures S12–14).
As mentioned above, the deep-blockade current ID at the end of Type 2 events likely corresponds to either duplex translocation through γ-HL, or the capture of the DNA during unzipping. To clarify this dichotomy, nine G → I mutations were synthetically introduced into the 30-mer strand of the duplex to obtain an I:C-containing duplex (sequence shown in Figure 8). The I:C base pairs significantly decrease the stability of the duplex due to the loss of one H-bond per base pair compared G:C (Figure 8a).63 However, the overall geometry of the duplex remains unchanged with the substitutions.64 Therefore, if ID in Type 2 events corresponds to the unzipping of the duplex, the event duration would be much shorter for the less stable I:C-containing DNA duplex than the G:C-containing one.35, 65 Conversely, if the dsDNA translocates through the protein channel, the residence times for both duplexes should remain nearly the same.
Figure 8.
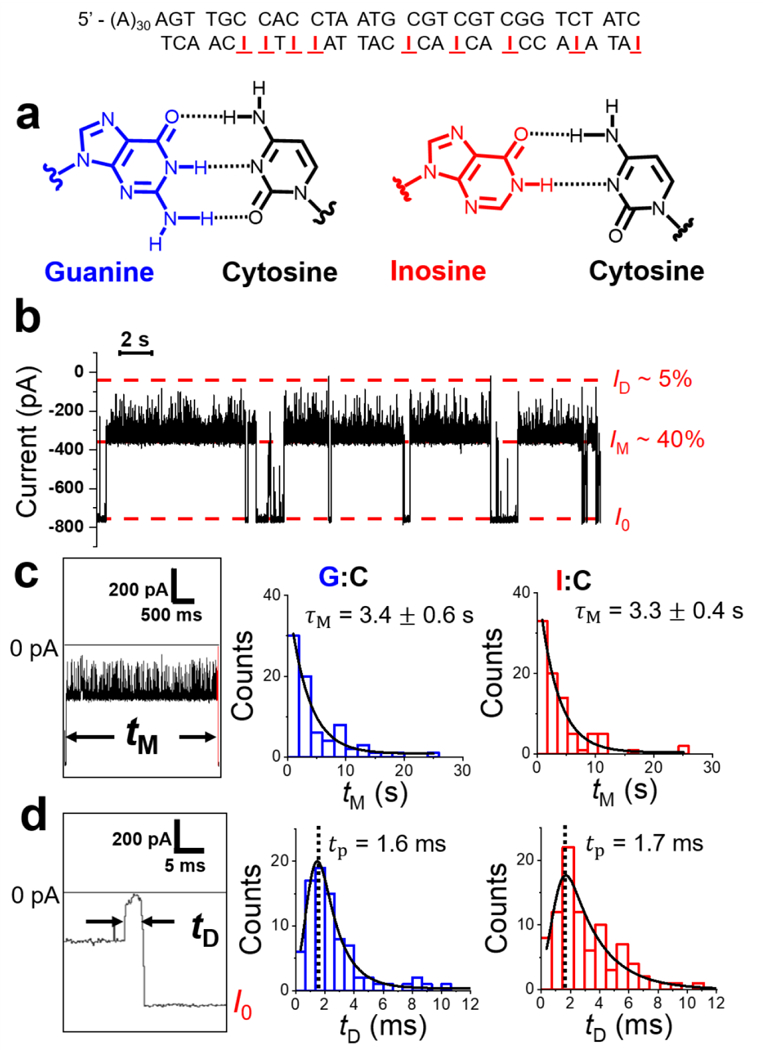
Determining the dsDNA translocation rate and mechanism through the γ-HL protein channel. (a) Structure of G:C and I:C base pairs. (b) I-t trace of I:C-containing duplexes captured by the protein channel (post filtered to 2 kHz). Extended trace is shown in Figure S15. (c-d) Histograms of mid-blockade duration (tM) and deep-blockade duration (tD) of G:C and I:C duplexes, respectively. Black curves are the best fits of exponentials (c) and exponentially modified Gaussian (d). Experiments were carried out at −120 mV in 3 M KCl, 20 mM HOAc/KOAc, pH 5.0 at 20.0 °C. M corresponds to the fitted exponential decay constant for tM distributions, while tp corresponds to the peak value of the tD distributions.
As shown in Figure 8b, the I:C-containing dsDNA generated a similar current signature as the G:C dsDNA (Figure 6b). Furthermore, the residence time of the whole events (including both Type 1 and 2 events), tM, for I:C dsDNA and G:C dsDNA were indistinguishable (~3.4 ± 0.6 s for G:C vs ~3.3 ± 0.4 s for I:C, −120 mV), suggesting that the DNA duplex did not unzip (Figure 8c). tD of type 2 events for I:C- and G:C-containing duplex were similar (Figure 8d), consistent with the translocation of the duplex through the γ-HL protein channel. The distribution of tD shows the shape of an exponentially modified Gaussian, which is predicted for free translocation of DNA through a nanopore.66 The translocation speed for the 30-bp duplex is ~20 bp/ms at −120 mV as measured from the histogram peak, tp (see Figure 8d). Furthermore, the results of voltage-dependent studies of td are consistent with dsDNA translocating through the γ-HL protein channel, as td decreases with increasing applied potential for both duplexes (Figure S16). Noteably, the translocation speed of dsDNA is much slower than that reported in most nanopores, including phi29 connector channel (850 bp/ms at −75 mV),43 and 4–8 nm synthetic pores (30,000 bp/ms at −120 mV).67–68 The slow translocation speed holds potential for direct sequencing of dsDNA using γ-HL, if distinct blockade currents can be obtained for different base pairs.
In summary, based on the current blockade levels, dsDNA residence time, and duplex translocation duration, we propose that the dsDNA is initially captured inside the vestibule of the γ-HL, and subsequently attempts to enter the β-barrel until an appropriate alignment is obtained to translocate through the protein channel (Figure 9).
Figure 9.
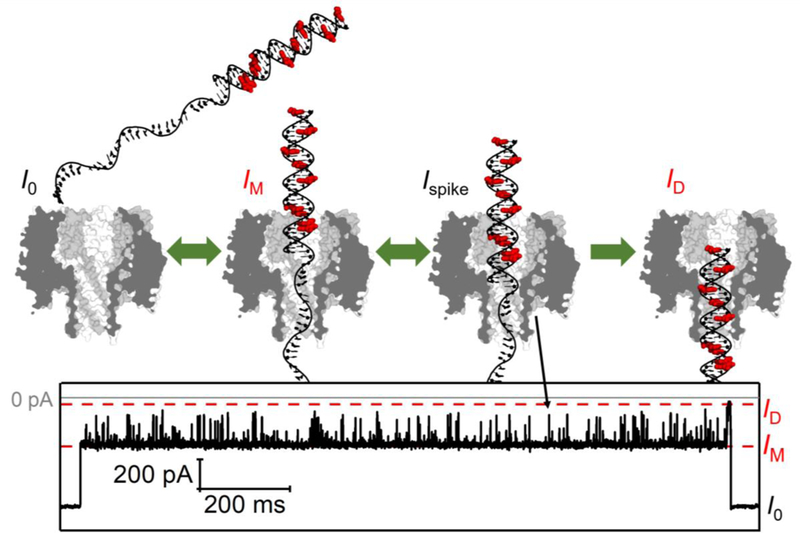
Schematic of dsDNA translocation through the γ-HL protein channel and the corresponding I-t trace. There are nine G → I substitutions on the 30-mer complementary strand, indicated in red.
Differentiation G to I substitutions in dsDNA using γ-HL protein channel.
As an initial demonstration of γ-HL to differentiate base pairs in dsDNA, the current signature of captured dsDNA was closely examined for both G:C- and I:C-containing duplexes (Figure 10). The average blockade residual currents of G:C and I:C dsDNA show ~1% difference (43.1 ± 0.5% and 44.1 ± 0.5% of I0, respectively, N=75 events for each). The shallower blockade from I:C-containing duplexes compared to the G:C-containing ones can be explained by its slightly smaller occupancy in the protein channel due to the lack of the 2-amino group on the purine in the minor groove of the duplex. Interestingly, a less blocking IM from the I:C dsDNA is observed even though five out of the nine guanine-to-inosine substitutions are located outside the cis opening of the γ-HL (Figure 9).
Figure 10.
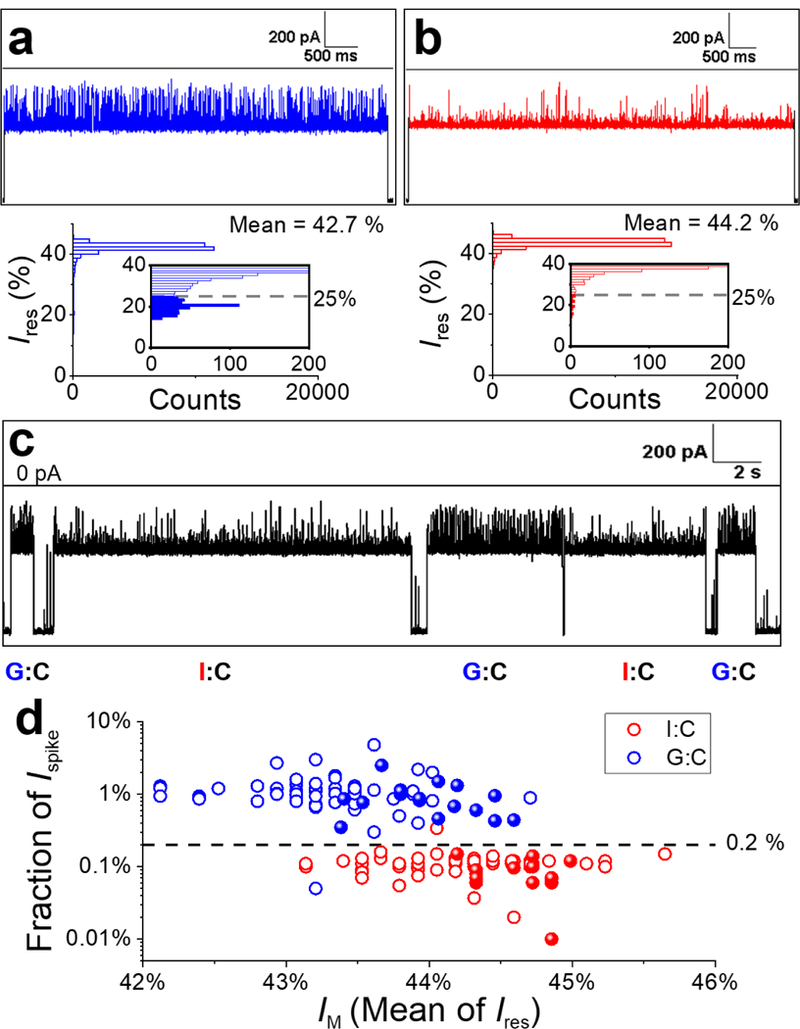
Single-molecule differentiation of G:C- and I:C-containing duplexes using the γ-HL nanopore. (a-b) Sample I-t trace corresponding to a G:C (a) or I:C (b) duplex captured by the protein channel in separate experiments and the corresponding all-point histograms of blockade current. Inset: Zoomed-in view of the histograms showing the current distributions of upward spikes for G:C and I:C duplex, respectively. The deeper blockade currents (< 25%) are plotted as filled columns in the insets. (c) Sample I-t trace for solution containing a 1:1 mixture of G:C and I:C duplexes. Extended I-t trace is shown in Figure S17. (d) Scatter plot of fraction of Ispike (percentage of counts with Ires< 25% in all-point histograms) vs mean of the blockade current (IM) for G:C (blue) and I:C (red) duplexes. Each data point represents a measurement of a single dsDNA molecule. Open circles represent data from a solution of a single component (G:C or I:C) for establishing the threshold for differentiation (N=75 for each). Solid spheres are data from the 1:1 mixture of G:C and I:C dsDNA molecules and are identified based on the threshold (N=32). Experiments were carried out at −120 mV in 3 M KCl, 20 mM HOAc/KOAc, pH 5 at 20.0 °C. Data were filtered by a Bessel filter at 10 kHz. The I-t traces (a-c) were post-filtered at 1 kHz for presentation.
The all-point current histograms generated for individual events indicate that the deep blocking current spikes can be used to differentiate the two duplexes (Figure 10 a-b). Specifically, the current spikes for the G:C duplex (Ispike, ~20% of I0) are readily observed in the expanded current amplitude histogram (inset of Figure 10a). However, the current spikes for the I:C-containing duplex are greatly reduced in the expanded current histograms (inset of Figure 10b), indicating the fraction of the counts for these spikes is much smaller (Figure 10b). Therefore, we used the fraction of Ispike in the current histograms to differentiate the I:C-containing duplex from the G:C duplex at the single-molecule level. A threshold of 25% of I0 is selected as the cutoff blockade current when counting the current spikes (Ispike) (Figure 10 a-b). The fraction of Ispike were first obtained for G:C- or I:C-containing duplexes (N=75 events for each) from two separate experiments in which the identities of the molecules were unambiguously assigned. As shown in Figure 10d, a scatter plot of the fraction of Ispike vs IM for G:C vs I:C duplexes shows two distinct clusters that can be separated by a boundary of 0.2% fraction of Ispike with ~99% accuracy.
To further demonstrate the ability to differentiate base pairs in dsDNA, γ-HL was challenged with a mixture of G:C- and I:C-containing duplexes (1:1). In fact, the differences in the frequency of Ispike for the two duplexes can be directly observed in the I-t traces (Figure 10b). To be more quantitative, the molecules in the mixture of G:C- and I:C-containing duplexes were identified by comparing the fraction of Ispike to the threshold of 0.2%, which indicates the five events in Figure 10c to be duplexes containing G:C, I:C, G:C, I:C, and G:C, respectively. Current-time traces with more examples of differentiation of I:C- and G:C-containing duplexes at the single-molecule level are shown in Figure S17. The results were also summarized as solid spheres in Figure 10d. Again, the events can be well separated based on the threshold of 0.2% fraction of Ispike, demonstrating >99% accuracy in the single-molecule identification of base pairs in dsDNA.
Dependence of the translocation time on dsDNA length.
To better estimate the translocation time of dsDNA molecules through the γ-HL nanopore, we characterized the current signatures of translocation events for different lengths of dsDNA (9-, 17-, 30-, and 60-bp). These dsDNA molecules also contained different tails for threading; the 9- and 17-bp dsDNA contained poly(dT)24 tails, while the 30- and 60-bp dsDNA contained poly(dA)30 tails (see Table S1 for the dsDNA sequences).
Figure 11 shows typical I-t traces of 9-, 17- and 60-bp dsDNA translocation events, which are very similar to that of the 30-bp dsDNA (previously shown in Figure 6). Each translocation event is initiated by a long mid-level blockade (>10 ms) with upward spikes, and the events terminate with a deep-level blockade as the duplex portion translocates through the β-barrel. The average mid-blockade currents of 9-, 17-, and 60-bp dsDNA are 44.3 ± 0.8%, 42.9 ± 0.6%, and 43.2 ± 0.9% of I0, respectively, (N = 75 events for each), which are similar to that of the 30-bp G:C-containing dsDNA (43.1 ± 0.5 %). As discussed earlier, the mid-blockade current corresponds to the 5’ ssDNA tail occupying the β-barrel and the duplex occupying the vestibule. The similar mid-blockade currents for the dsDNA molecules containing either poly(dT)24 or poly(dA)30 tails, ~43%, suggests that the ~2.3 nm wide β-barrel is too wide for ssDNA differentiation or sequencing.
Figure 11.
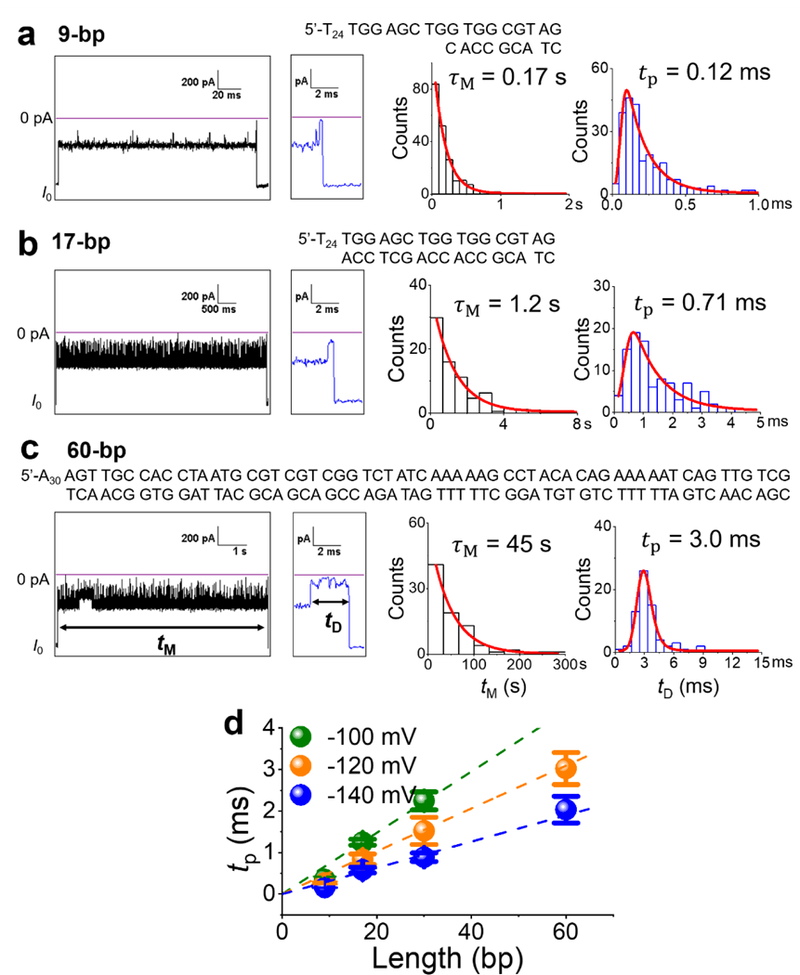
(a-c) Typical I-t traces recorded at −120 mV for the translocation of 9-, 17-, and 60-bp dsDNA, with an expanded view of the end of the event. Only successful translocation events are analyzed. Additional examples for each dsDNA are shown in Figure S18-S20. Duration histograms of and are fitted by exponential decay and exponentially modified Gaussians (red curves), respectively. The most probable translocation times are obtained from the fitted curves and plotted in (d) for applied potentials of−100, −120, and −140 mV. Note that the 60-bp dsDNA did not translocate through the protein channel at the lowest applied potential, i.e., −100 mV. The y-axis error bars were obtained from 3 or 4 experiments. The data were collected in 3 M KCl, 20 mM HOAc/KOAc, pH 5.0, using a 10-kHz Bessel filter (100-kHz Bessel filter for 9-bp data) and post-filtered at 3 kHz for presentation.
Analysis of the duration event histograms for 9-, 17-, and 60-bp dsDNA are shown in Figures 11a-11c. As previously discussed in Figure 8, the two time constants, and , correspond to the mid-blockade duration and deep-blockade translocation time, respectively. In general, dsDNA molecules with random sequences share the same geometry, with an ~2.0 nm diameter and ~0.34 nm rise per base-pair, as shown in Figure 1b. We speculate that both and of dsDNA should be shorter at the higher applied potential and also proportional to the length of the duplex, regardless of the specific DNA sequence. The dependence of and of the applied potentials were investigated on all four dsDNA (9-, 17-, 30-, and 60-bp), and the results for tp are presented in Figure 11d. Overall, the duration of both and increased approximately linearly with the length of the dsDNA duplex, at about 10 ± 3 bp/s and 19 ± 1 bp/ms, respectively, at −120 mV.
No deep blockages were observed for 60-bp dsDNA at −100 mV, indicating that these longer duplexes diffused back to the cis chamber instead of translocating (thus, no value of tp is reported for 60-bp dsDNA at −100 mV in Figure 11d). As the applied potential increased from the −120 mV to −140 mV, of the 60-bp dsDNA decreased by about one order of magnitude from 45 ± 6 s to 3.3 ± 0.6 s. Considering the length of the duplex portion of the 60-bp dsDNA (~20 nm) and the height of the protein vestibule (~4.6 nm), we propose that the longer duplex more readily diffuse away from the protein channel during the mid-level duration, which is on the order of tens of seconds at −100 and −120 mV.
Conclusions
We demonstrated the application of wild-type γ-HL for direct analysis of dsDNA after optimizing the experimental conditions (lowering the pH and increasing the electrolyte concentration) to reduce the current noise and frequency of gating. In γ-HL, 9- to 30-bp dsDNA translocates at a rate of 19 ± 1 bp/ms (−120 mV), which is much slower than that reported in other biological and solid-state nanopores.43, 67–68 Based on their unique current signature, a DNA duplex containing guanine-to-inosine substitutions can be distinguished from the unsubstituted G:C complementary duplex at the single-molecule level, indicating that this protein nanopore has considerable potential for analyzing dsDNA sequences. Mapping the sensing zone of γ-HL for dsDNA will be valuable in improving the resolution of nucleobase recognition in dsDNA. Additionally, the slow translocation rate of dsDNA in γ-HL may also allow measurement of modified bases along the duplex.
Supplementary Material
Acknowledgments
The authors thank Electronic Biosciences Inc. (San Diego, CA) for donating the ion-channel recording instruments and software utilized for recording the I−t traces. A.M.F. Consults for Electronic Biosciences Inc. (San Diego, CA). The authors gratefully acknowledge Professor Robert P. Johnson for helpful discussions on protein oligomerization methods. This work was funded by a grant from the U. S. National Institute of General Medical Sciences (R01 GM093099).
Footnotes
Notes. The authors declare no competing financial interest.
References
- 1.Bayley H; Cremer PS, Stochastic Sensors Inspired by Biology. Nature 2001, 413 (6852), 226. [DOI] [PubMed] [Google Scholar]
- 2.Bezrukov SM; Vodyanoy I; Parsegian VA, Counting Polymers Moving through a Single Ion Channel. Nature 1994, 370 (6487), 279. [DOI] [PubMed] [Google Scholar]
- 3.Howorka S; Siwy Z, Nanopore Analytics: Sensing of Single Molecules. Chem. Soc. Rev 2009, 38 (8), 2360. [DOI] [PubMed] [Google Scholar]
- 4.Venkatesan BM; Bashir R, Nanopore Sensors for Nucleic Acid Analysis. Nat. Nanotechnol 2011, 6 (10), 615. [DOI] [PubMed] [Google Scholar]
- 5.Coulter WH Means for Counting Particles Suspended in a Fluid. U.S. Patent 2,656,508, October 20, 1953. [Google Scholar]
- 6.Kasianowicz JJ; Brandin E; Branton D; Deamer DW, Characterization of Individual Polynucleotide Molecules Using a Membrane Channel. Proc. Natl. Acad. Sci. U.S.A 1996, 93 (24), 13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayley H; Martin CR, Resistive-Pulse Sensing from Microbes to Molecules. Chem. Rev 2000, 100 (7), 2575. [DOI] [PubMed] [Google Scholar]
- 8.Majd S; Yusko EC; Billeh YN; Macrae MX; Yang J; Mayer M, Applications of Biological Pores in Nanomedicine, Sensing, and Nanoelectronics. Curr. Opin. Biotechnol 2010, 21 (4), 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasianowicz JJ; Robertson JW; Chan ER; Reiner JE; Stanford VM, Nanoscopic Porous Sensors. Annu. Rev. Anal. Chem 2008, 1, 737. [DOI] [PubMed] [Google Scholar]
- 10.Wanunu M, Nanopores: A Journey Towards DNA Sequencing. Phys. Life. Rev 2012, 9 (2), 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoddart D; Heron AJ; Mikhailova E; Maglia G; Bayley H, Single-Nucleotide Discrimination in Immobilized DNA Oligonucleotides with a Biological Nanopore. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (19), 7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akeson M; Branton D; Kasianowicz JJ; Brandin E; Deamer DW, Microsecond Time-Scale Discrimination among Polycytidylic Acid, Polyadenylic Acid, and Polyuridylic Acid as Homopolymers or as Segments within Single Rna Molecules. Biophys. J 1999, 77 (6), 3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howorka S; Cheley S; Bayley H, Sequence-Specific Detection of Individual DNA Strands Using Engineered Nanopores. Nat. Biotechnol 2001, 19 (7), 636. [DOI] [PubMed] [Google Scholar]
- 14.Branton D; Deamer DW; Marziali A; Bayley H; Benner SA; Butler T; Di Ventra M; Garaj S; Hibbs A; Huang X; Jovanovich SB; Krstic PS; Lindsay S; Ling XS; Mastrangelo CH; Meller A; Oliver JS; Pershin YV; Ramsey JM; Riehn R; Soni GV; Tabard-Cossa V; Wanunu M; Wiggin M; Schloss JA, The Potential and Challenges of Nanopore Sequencing. Nat. Biotechnol 2008, 26 (10), 1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoddart D; Heron AJ; Klingelhoefer J; Mikhailova E; Maglia G; Bayley H, Nucleobase Recognition in Ssdna at the Central Constriction of the Alpha-Hemolysin Pore. Nano Lett 2010, 10 (9), 3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke J; Wu HC; Jayasinghe L; Patel A; Reid S; Bayley H, Continuous Base Identification for Single-Molecule Nanopore DNA Sequencing. Nat. Nanotechnol 2009, 4 (4), 265. [DOI] [PubMed] [Google Scholar]
- 17.Meller A; Nivon L; Brandin E; Golovchenko J; Branton D, Rapid Nanopore Discrimination between Single Polynucleotide Molecules. Proc. Natl. Acad. Sci. U.S.A 2000, 97 (3), 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laszlo AH; Derrington IM; Ross BC; Brinkerhoff H; Adey A; Nova IC; Craig JM; Langford KW; Samson JM; Daza R; Doering K; Shendure J; Gundlach JH, Decoding Long Nanopore Sequencing Reads of Natural DNA. Nat. Biotechnol 2014, 32 (8), 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler TZ; Pavlenok M; Derrington IM; Niederweis M; Gundlach JH, Single-Molecule DNA Detection with an Engineered Mspa Protein Nanopore. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (52), 20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derrington IM; Butler TZ; Collins MD; Manrao E; Pavlenok M; Niederweis M; Gundlach JH, Nanopore DNA Sequencing with Mspa. Proc. Natl. Acad. Sci. U. S. A 2010, 107 (37), 16060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao C; Ying YL; Hu ZL; Liao DF; Tian H; Long YT, Discrimination of Oligonucleotides of Different Lengths with a Wild-Type Aerolysin Nanopore. Nat. Nanotechnol 2016, 11 (8), 713. [DOI] [PubMed] [Google Scholar]
- 22.Cao C; Yu J; Wang YQ; Ying YL; Long YT, Driven Translocation of Polynucleotides through an Aerolysin Nanopore. Anal. Chem 2016, 88 (10), 5046. [DOI] [PubMed] [Google Scholar]
- 23.Movileanu L; Schmittschmitt JP; Scholtz JM; Bayley H, Interactions of Peptides with a Protein Pore. Biophys. J 2005, 89 (2), 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oukhaled G; Mathe J; Biance AL; Bacri L; Betton JM; Lairez D; Pelta J; Auvray L, Unfolding of Proteins and Long Transient Conformations Detected by Single Nanopore Recording. Phys. Rev. Lett 2007, 98 (15), 158101. [DOI] [PubMed] [Google Scholar]
- 25.Cao C; Yu J; Li MY; Wang YQ; Tian H; Long YT, Direct Readout of Single Nucleobase Variations in an Oligonucleotide. Small 2017, 13 (44). [DOI] [PubMed] [Google Scholar]
- 26.Yu J; Cao C; Long YT, Selective and Sensitive Detection of Methylcytosine by Aerolysin Nanopore under Serum Condition. Anal. Chem 2017, 89 (21), 11685. [DOI] [PubMed] [Google Scholar]
- 27.Vercoutere W; Winters-Hilt S; Olsen H; Deamer D; Haussler D; Akeson M, Rapid Discrimination among Individual DNA Hairpin Molecules at Single-Nucleotide Resolution Using an Ion Channel. Nat. Biotechnol 2001, 19 (3), 248. [DOI] [PubMed] [Google Scholar]
- 28.Vercoutere WA; Winters-Hilt S; DeGuzman VS; Deamer D; Ridino SE; Rodgers JT; Olsen HE; Marziali A; Akeson M, Discrimination among Individual Watson-Crick Base Pairs at the Termini of Single DNA Hairpin Molecules. Nucleic Acids Res 2003, 31 (4), 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding Y; Fleming AM; White HS; Burrows CJ, Internal Vs Fishhook Hairpin DNA: Unzipping Locations and Mechanisms in the Alpha-Hemolysin Nanopore. J. Phys. Chem. B 2014, 118 (45), 12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An N; Fleming AM; Middleton EG; Burrows CJ, Single-Molecule Investigation of G-Quadruplex Folds of the Human Telomere Sequence in a Protein Nanocavity. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (40), 14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An N; Fleming AM; Burrows CJ, Interactions of the Human Telomere Sequence with the Nanocavity of the Alpha-Hemolysin Ion Channel Reveal Structure-Dependent Electrical Signatures for Hybrid Folds. J. Am. Chem. Soc 2013, 135 (23), 8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Y; Fleming AM; He L; Burrows CJ, Unfolding Kinetics of the Human Telomere I-Motif under a 10 Pn Force Imposed by the Alpha-Hemolysin Nanopore Identify Transient Folded-State Lifetimes at Physiological Ph. J. Am. Chem. Soc 2015, 137 (28), 9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan CS; Riedl J; Fleming AM; Burrows CJ; White HS, Kinetics of T3-DNA Ligase-Catalyzed Phosphodiester Bond Formation Measured Using the Α-Hemolysin Nanopore. ACS Nano 2016, 10 (12), 11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson RP; Fleming AM; Perera RT; Burrows CJ; White HS, Dynamics of a DNA Mismatch Site Held in Confinement Discriminate Epigenetic Modifications of Cytosine. J. Am. Chem. Soc 2017, 139 (7), 2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin Q; Fleming AM; Burrows CJ; White HS, Unzipping Kinetics of Duplex DNA Containing Oxidized Lesions in an Alpha-Hemolysin Nanopore. J. Am. Chem. Soc 2012, 134 (26), 11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson RP; Fleming AM; Beuth LR; Burrows CJ; White HS, Base Flipping within the Alpha-Hemolysin Latch Allows Single-Molecule Identification of Mismatches in DNA. J. Am. Chem. Soc 2016, 138 (2), 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin Q; Fleming AM; Johnson RP; Ding Y; Burrows CJ; White HS, Base-Excision Repair Activity of Uracil-DNA Glycosylase Monitored Using the Latch Zone of Alpha-Hemolysin. J. Am. Chem. Soc 2013, 135 (51), 19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren H; Cheyne CG; Fleming AM; Burrows CJ; White HS, Single-Molecule Titration in a Protein Nanoreactor Reveals the Protonation/Deprotonation Mechanism of a C:C Mismatch in DNA. J. Am. Chem. Soc 2018, 140 (15), 5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng T; Fleming AM; Ding Y; White HS; Burrows CJ, Interrogation of Base Pairing of the Spiroiminodihydantoin Diastereomers Using the Alpha-Hemolysin Latch. Biochemistry 2017, 56 (11), 1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng T; Fleming AM; Ding Y; Ren H; White HS; Burrows CJ, Nanopore Analysis of the 5-Guanidinohydantoin to Iminoallantoin Isomerization in Duplex DNA. J. Org. Chem 2018, 83 (7), 3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wendell D; Jing P; Geng J; Subramaniam V; Lee TJ; Montemagno C; Guo P, Translocation of Double-Stranded DNA through Membrane-Adapted Phi29 Motor Protein Nanopores. Nat. Nanotechnol 2009, 4 (11), 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haque F; Li J; Wu HC; Liang XJ; Guo P, Solid-State and Biological Nanopore for Real-Time Sensing of Single Chemical and Sequencing of DNA. Nano Today 2013, 8 (1), 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haque F; Wang S; Stites C; Chen L; Wang C; Guo P, Single Pore Translocation of Folded, Double-Stranded, and Tetra-Stranded DNA through Channel of Bacteriophage Phi29 DNA Packaging Motor. Biomaterials 2015, 53, 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franceschini L; Soskine M; Biesemans A; Maglia G, A Nanopore Machine Promotes the Vectorial Transport of DNA across Membranes. Nat. Commun 2013, 4, 2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soskine M; Biesemans A; De Maeyer M; Maglia G, Tuning the Size and Properties of Clya Nanopores Assisted by Directed Evolution. J. Am. Chem. Soc 2013, 135 (36), 13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franceschini L; Brouns T; Willems K; Carlon E; Maglia G, DNA Translocation through Nanopores at Physiological Ionic Strengths Requires Precise Nanoscale Engineering. ACS Nano 2016, 10 (9), 8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wloka C; Mutter NL; Soskine M; Maglia G, Alpha-Helical Fragaceatoxin C Nanopore Engineered for Double-Stranded and Single-Stranded Nucleic Acid Analysis. Angew. Chem. Int. Ed Engl 2016, 55 (40), 12494. [DOI] [PubMed] [Google Scholar]
- 48.Yamashita K; Kawai Y; Tanaka Y; Hirano N; Kaneko J; Tomita N; Ohta M; Kamio Y; Yao M; Tanaka I, Crystal Structure of the Octameric Pore of Staphylococcal Gamma-Hemolysin Reveals the Beta-Barrel Pore Formation Mechanism by Two Components. Proc. Natl. Acad. Sci. U.S.A 2011, 108 (42), 17314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miles G; Cheley S; Braha O; Bayley H, The Staphylococcal Leukocidin Bicomponent Toxin Forms Large Ionic Channels. Biochemistry 2001, 40 (29), 8514. [DOI] [PubMed] [Google Scholar]
- 50.Comai M; Dalla Serra M; Coraiola M; Werner S; Colin DA; Monteil H; Prevost G; Menestrina G, Protein Engineering Modulates the Transport Properties and Ion Selectivity of the Pores Formed by Staphylococcal Gamma-Haemolysins in Lipid Membranes. Mol. Microbiol 2002, 44 (5), 1251. [DOI] [PubMed] [Google Scholar]
- 51.Menestrina G; Dalla Serra M; Comai M; Coraiola M; Viero G; Werner S; Colin DA; Monteil H; Prévost G, Ion Channels and Bacterial Infection: The Case of Β-Barrel Pore-Forming Protein Toxins of Staphylococcus Aureus. FEBS Lett 2003, 552 (1), 54. [DOI] [PubMed] [Google Scholar]
- 52.Holden MA; Jayasinghe L; Daltrop O; Mason A; Bayley H, Direct Transfer of Membrane Proteins from Bacteria to Planar Bilayers for Rapid Screening by Single-Channel Recording. Nat. Chem. Biol 2006, 2 (6), 314. [DOI] [PubMed] [Google Scholar]
- 53.Hille B Ion Channels of Excitable Membranes, 3rd ed.; Sinauer Associates, Inc.: Sunderland, MA, 2001. [Google Scholar]
- 54.Kasianowicz JJ; Bezrukov SM, Protonation Dynamics of the Alpha-Toxin Ion Channel from Spectral Analysis of Ph-Dependent Current Fluctuations. Biophys. J 1995, 69 (1), 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sigworth FJ, Open Channel Noise. I. Noise in Acetylcholine Receptor Currents Suggests Conformational Fluctuations. Biophys J 1985, 47 (5), 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hevesi L; Wolfson-Davidson E; Nagy JB; Nagy OB; Bruylants A, Contribution to the Mechanism of the Acid-Catalyzed Hydrolysis of Purine Nucleosides. J. Am. Chem. Soc 1972, 94 (13), 4715. [DOI] [PubMed] [Google Scholar]
- 57.Zoltewicz JA; Clark DF; Sharpless TW; Grahe G, Kinetics and Mechanism of the Acid-Catalyzed Hydrolysis of Some Purine Nucleosides. J. Am. Chem. Soc 1970, 92 (6), 1741. [DOI] [PubMed] [Google Scholar]
- 58.Kasianowicz JJ, Voltage-Dependent Gating Kinetics of an Ion Channel Modulated by Ionic Strength. Biophys. J 1994, 66 (2). [Google Scholar]
- 59.Butler TZ; Gundlach JH; Troll M, Ionic Current Blockades from DNA and Rna Molecules in the Alpha-Hemolysin Nanopore. Biophys. J 2007, 93 (9), 3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lathrop DK; Ervin EN; Barrall GA; Keehan MG; Kawano R; Krupka MA; White HS; Hibbs AH, Monitoring the Escape of DNA from a Nanopore Using an Alternating Current Signal. J. Am. Chem. Soc 2010, 132 (6), 1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cooney J; Kienle Z; Foster TJ; O’Toole PW, The Gamma-Hemolysin Locus of Staphylococcus Aureus Comprises Three Linked Genes, Two of Which Are Identical to the Genes for the F and S Components of Leukocidin. Infect. Immun 1993, 61 (2), 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gouaux E; Hobaugh M; Song L, Alpha-Hemolysin, Gamma-Hemolysin, and Leukocidin from Staphylococcus Aureus: Distant in Sequence but Similar in Structure. Protein Sci 1997, 6 (12), 2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krepl M; Otyepka M; Banas P; Sponer J, Effect of Guanine to Inosine Substitution on Stability of Canonical DNA and Rna Duplexes: Molecular Dynamics Thermodynamics Integration Study. J. Phys. Chem. B 2013, 117 (6), 1872. [DOI] [PubMed] [Google Scholar]
- 64.Xuan JC; Weber IT, Crystal-Structure of a B-DNA Dodecamer Containing Inosine, D(Cgciaattcgcg), at 2.4 a Resolution and Its Comparison with Other B-DNA Dodecamers. Nucleic Acids Res 1992, 20 (20), 5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schibel AE; Fleming AM; Jin Q; An N; Liu J; Blakemore CP; White HS; Burrows CJ, Sequence-Specific Single-Molecule Analysis of 8-Oxo-7,8-Dihydroguanine Lesions in DNA Based on Unzipping Kinetics of Complementary Probes in Ion Channel Recordings. J. Am. Chem. Soc 2011, 133 (37), 14778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meller A; Nivon L; Branton D, Voltage-Driven DNA Translocations through a Nanopore. Phys. Rev. Lett 2001, 86 (15), 3435. [DOI] [PubMed] [Google Scholar]
- 67.Fologea D; Uplinger J; Thomas B; McNabb DS; Li J, Slowing DNA Translocation in a Solid-State Nanopore. Nano Lett 2005, 5 (9), 1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J; Gershow M; Stein D; Brandin E; Golovchenko JA, DNA Molecules and Configurations in a Solid-State Nanopore Microscope. Nat. Mater 2003, 2 (9), 611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


