Significance
In the past decade, the number of publicly available plant genomes and transcriptomes has steadily increased. Inspired by this genetic resource, we developed a genome-mining approach for the rapid discovery of plant ribosomal peptides from genome-sequenced plants. Herein, we introduce the hypotensive lyciumins as a class of branched cyclic ribosomal peptides in plants and show that they are widely distributed in crop and forage plants. Our results suggest that lyciumin biosynthesis is coupled to plant-specific BURP domains in their precursor peptides and that lyciumin peptide libraries can be generated in planta. This discovery sets the stage for gene-guided discovery of peptide chemistry in the plant kingdom and therapeutic and agrochemical applications of lyciumins.
Keywords: lyciumins, BURP domain, ribosomal peptides, natural products, plant metabolism
Abstract
The plant kingdom contains vastly untapped natural product chemistry, which has been traditionally explored through the activity-guided approach. Here, we describe a gene-guided approach to discover and engineer a class of plant ribosomal peptides, the branched cyclic lyciumins. Initially isolated from the Chinese wolfberry Lycium barbarum, lyciumins are protease-inhibiting peptides featuring an N-terminal pyroglutamate and a macrocyclic bond between a tryptophan-indole nitrogen and a glycine α-carbon. We report the identification of a lyciumin precursor gene from L. barbarum, which encodes a BURP domain and repetitive lyciumin precursor peptide motifs. Genome mining enabled by this initial finding revealed rich lyciumin genotypes and chemotypes widespread in flowering plants. We establish a biosynthetic framework of lyciumins and demonstrate the feasibility of producing diverse natural and unnatural lyciumins in transgenic tobacco. With rapidly expanding plant genome resources, our approach will complement bioactivity-guided approaches to unlock and engineer hidden plant peptide chemistry for pharmaceutical and agrochemical applications.
Plants have been an important source of traditional medicines in many cultures for millennia. Underlying the historic use of medicinal plants are bioactive natural products produced in these organisms for signaling and defense (1). Most plant natural products with potential pharmacological applications were discovered based on their bioactivity in bioprospecting studies inspired by traditional herbal medicines (2). However, bioactivity-guided discovery of new plant natural products faces a major bottleneck in rediscovery of known structures after purification via bioassay-guided fractionation (3). Subsequent drug development of a target plant natural product is also hindered by low isolation yields from the source plant and by structural complexity of the target compounds, where large-scale total synthesis of these compounds via organic chemistry is often infeasible (4).
Over the last two decades, advances in genome sequencing, analytical chemistry, and synthetic biology allowed researchers to address these problems in microbial and fungal natural product chemistry (5). An increasing resource of microbial and fungal genomes enabled the development of powerful computational discovery pipelines for new natural product chemistry, using gene-guided discovery approaches such as genome mining, in which a predicted genotype is connected to a chemotype by applying biosynthetic knowledge (5–7). Furthermore, mass spectrometry-based metabolomics accelerated discovery of new chemotypes from microbes and fungi in the context of growing genomic information (6, 8), while heterologous expression and metabolic engineering of biosynthetic gene clusters enabled source organism-independent production and diversification of natural products (9). More recently, analogous gene-guided approaches have been developed to characterize new plant chemistry (10–12), taking advantage of the increasing number of plant genomes (13) and transcriptomes (14), a growing biosynthetic knowledge of plant specialized metabolism (15), and the development of heterologous expression systems for plant biosynthetic pathways (16). However, there are unique challenges associated with genome mining for plant natural products because genotype prediction is complicated by knowledge gaps in plant natural product biochemistry as well as complete absence or only partial presence of clustering of plant natural product biosynthetic genes.
One compound class affords the opportunity to circumvent these challenges in plant genome mining: the ribosomally synthesized and posttranslationally modified peptides (RiPPs). RiPPs are a rapidly growing class of natural products, as the discovery of RiPP precursor genes and their corresponding biosynthetic pathways has been greatly enabled by whole-genome sequencing (17). Most RiPPs discovered to date are from bacteria and fungi, whereas a few examples were also found in plants. The two biosynthetically defined families of plant RiPPs are cyclotides and orbitides, which are “head-to-tail” cyclic peptides with or without disulfide bonds, respectively (17–21). The cyclic plant peptides with disulfide bonds are further grouped into cyclotides, which have three disulfide bonds, and PawS-derived peptides, which have one disulfide bond (18–20). Cyclotides and orbitides are biosynthesized from ribosomally derived precursor peptides. During cyclotide biogenesis, disulfide bonds are formed in the endoplasmic reticulum of the plant cell by protein disulfide isomerases (19, 22). The modified cyclotide precursor peptide is then cleaved proteolytically N-terminal of the cyclotide core peptide, and, finally, the C terminus of the core peptide is cleaved and cyclized with the N terminus by an asparagine-specific endopeptidase in the plant vacuole (19, 23, 24). Similarly, orbitides are derived from precursor peptides by endoproteolytic cleavage N-terminal of the orbitide core peptide followed by subsequent C-terminal proteolysis and cyclization catalyzed by serine proteases (19, 25, 26). Beyond head-to-tail cyclic peptides, the phytochemical repertoire of cyclic peptides suggests that the plant kingdom contains a largely undiscovered diversity of branched cyclic peptide chemistry with tremendous pharmacological potential and unknown biosynthetic mechanisms (27). Because the peptide sequence of RiPPs is directly encoded in the genome as the core peptide motif within a precursor gene (17), we hypothesized that identification of precursor genes specific to a chemically defined RiPP class would readily yield structural information that aids subsequent structure-guided chemotyping (e.g., by targeted metabolomics) (28). In addition, we expected that a precursor gene-guided genome-mining approach for plant RiPPs would not require knowledge of other biosynthetic genes encoding posttranslationally modifying enzymes or proteases to successfully connect a peptide genotype with a corresponding analyte. It is unknown whether genes encoding precursor peptides, proteases, and posttranslationally modifying enzymes involved in plant RiPP pathways are colocalized in plant genomes.
In this study, we tested this genome-mining approach on the lyciumin peptides, a candidate class of branched cyclic plant RiPPs. Lyciumins were originally isolated as inhibitors of the angiotensin-converting enzyme (ACE) and renin, from the roots of Chinese wolfberry Lycium barbarum (Solanaceae, Fig. 1A), a Chinese herbal medicine used for treating hypertension (29–31). Lyciumins contain a characteristic N-terminal pyroglutamate and an unusual macrocyclic linkage between a C-terminal tryptophan-indole nitrogen and a glycine α-carbon (Fig. 1B). Lyciumins were also previously isolated from the seeds of the medicinal plant Celosia argentea (Amaranthaceae), suggesting that the biosynthetic machineries supporting the production of lyciumin peptides may be widespread in plants (32).
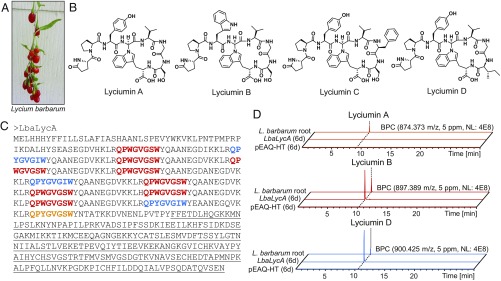
Fig. 1.
Lyciumin chemotypes and genotype in Lycium barbarum. (A) L. barbarum fruits. (B) Lyciumin structures from L. barbarum root extracts. Stereochemistry of glycine α-carbon in lyciumin B, C, and D was inferred from lyciumin A structural analysis (31). (C) Candidate precursor peptide LbaLycA of lyciumin A, B, and D chemotypes from L. barbarum root transcriptome. Putative core peptides are bold, and BURP domain is underlined. (D) Detection of lyciumin A, B, and D in peptide extracts of Nicotiana benthamiana leaves after transient expression of LbaLycA via infiltration with Agrobacterium tumefaciens LBA4404 containing pEAQ-HT-LbaLycA. L. barbarum root, peptide extract of Lycium barbarum roots; LbaLycA (6 d), peptide extract of Nicotiana benthamiana infiltrated with Agrobacterium tumefaciens LBA4404 containing pEAQ-HT-LbaLycA (6 d); pEAQ-HT (6 d), peptide extract of Nicotiana benthamiana infiltrated with Agrobacterium tumefaciens LBA4404 containing pEAQ-HT (6 d).
Results
Lyciumins Are Plant RiPPs.
Because plant genomes do not encode nonribosomal peptide synthetases (NRPSs), we hypothesized that lyciumin peptides are RiPPs. To identify the lyciumin precursor gene, we generated a de novo transcriptome of the root tissue from L. barbarum, from which the presence of lyciumin A, B, and D was confirmed based on liquid chromatography–mass spectrometry (LC-MS) and NMR analyses (SI Appendix, Figs. S1–S7 and Tables S1–S3). Tblastn search using the predicted core peptide sequences of lyciumin A (QPYGVGSW), lyciumin B (QPWGVGSW), and lyciumin D (QPYGVGIW) as queries yielded three partial transcripts of candidate lyciumin precursor genes. The full-length candidate lyciumin precursor gene was cloned guided by these partial transcripts (Fig. 1C and SI Appendix, Fig. S8). The identified lyciumin precursor protein from L. barbarum, LbaLycA, consists of an N-terminal signal peptide indicative of processing through the secretory pathway (SI Appendix, Fig. S9) (33), an N-terminal domain with 12 repeats with each including a core peptide for lyciumin A, B, or D, and a C-terminal BURP domain (Pfam 03181) (34). BURP domain proteins are terrestrial plant-specific proteins, which are often associated with abiotic stress responses and exhibit diverse temporal and spatial expression patterns in plants (35–39).
To test whether LbaLycA is the precursor gene for the identified lyciumin peptides, we expressed LbaLycA heterologously in the leaf tissue of Nicotiana benthamiana via Agrobacterium-mediated transient expression. LC-MS analysis of a peptide extract of the N. benthamiana leaves 6 d after Agrobacterium infiltration showed mass signals for lyciumin A, B, and D, identical to those detected in L. barbarum root extracts (Fig. 1D), while no lyciumin mass signals appeared in the empty vector control. This result confirmed that lyciumins are RiPPs derived from the precursor gene LbaLycA. In addition, the successful heterologous reconstitution of lyciumin biosynthesis in N. benthamiana by sole expression of the precursor gene LbaLycA suggests that N. benthamiana must contain enzymes necessary to process the precursor peptide to yield lyciumins.
Genome Mining Reveals Lyciumins in Amaranth, Legume, and Nightshade Plants.
With the first precursor gene for lyciumin biosynthesis identified, we set out to search for lyciumin genotypes and chemotypes in other plants with sequenced genomes, using precursor gene-guided genome mining. The genome-mining workflow begins with a tblastn search of plant genomes for homologous genes encoding BURP domain proteins (Pfam 03181) (Fig. 2A). A candidate lyciumin precursor was identified by one or multiple candidate core peptide sequences of the motif QP(X)5W in its N-terminal half, where X can be any amino acid. If a putative lyciumin precursor gene was identified from a plant genome, the corresponding lyciumin structures were then predicted based on the core peptide sequences. Subsequently, we searched for these predicted lyciumin chemotypes in the LC-MS–based metabolomics dataset of peptide extracts prepared from the target plant host, by querying both peptide parent masses in MS data and predicted peptide fragment masses in MS/MS data [e.g., the pyroglutamate-proline-b ion ([M+H]+, 209.09207 m/z) or amino acid-iminium ions]. Finally, MS/MS data analysis of lyciumins enabled the characterization of a planar structure, to verify the connection between a candidate lyciumin mass spectrum with a lyciumin genotype (SI Appendix, Fig. S10).
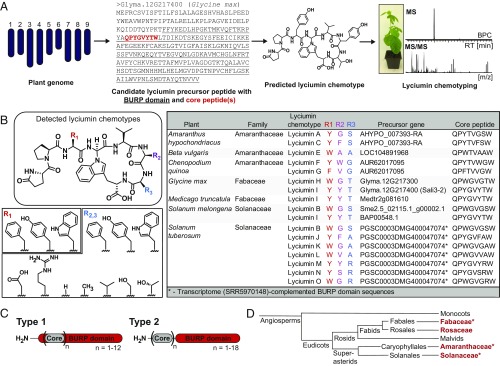
Fig. 2.
Genome mining of lyciumins in plants. (A) Precursor gene-guided genome mining of lyciumins. (B) Structural diversity of lyciumins characterized by genome mining from Amaranthaceae, Fabaceae, and Solanaceae plants. The stereochemistry of glycine α-carbon is inferred from lyciumin A and lyciumin I structure elucidation. (C) Types of lyciumin precursors based on primary structure analysis (Dataset S1). Core, core peptide. (D) Phylogenetic relationship of plant families with predicted and characterized (noted by asterisks) lyciumin chemotypes (both highlighted in red).
Genome mining revealed that 21 of 116 analyzed plant genomes harbor candidate lyciumin precursor genes (SI Appendix, Table S4). The putative lyciumin-producing plants fall into the Amaranthaceae, Fabaceae, Rosaceae, and Solanaceae families. Bioinformatic analysis of identified BURP domain proteins yielded 71 distinct core peptide sequences with 60 unique to the host species (SI Appendix, Table S5 and Dataset S1), indicating an untapped diversity of lyciumin chemotypes. Subsequently, we selected 10 plant species with candidate lyciumin genotypes and analyzed their peptide extracts for predicted lyciumin chemotypes by LC-MS. Predicted lyciumin peptides could be detected and verified by MS, MS/MS, and select NMR analysis for seven plant species, including economically important crop and forage plants such as Amaranthus hypochondriacus (amaranth), Beta vulgaris (beet), Chenopodium quinoa (quinoa), Glycine max (soybean), Solanum melongena (eggplant), and Medicago truncatula (barrelclover) (Fig. 2B and SI Appendix, Figs. S11–S23 and Tables S6 and S7). No lyciumin peptides could be detected in peptide extracts of Solanum lycopersicum Heinz 1702 (tomato), Capsicum annuum (pepper), and Trifolium pratense (red clover). Characterized lyciumin precursor genes are differentially expressed in plant tissues and developmental stages with generally the highest expression in roots and embryo-developing tissues (SI Appendix, Fig. S24). Accordingly, characterized lyciumin concentrations are generally the highest in roots and seeds, while some lyciumins are detected in the whole plant, such as in soy, quinoa, and amaranth (SI Appendix, Fig. S25).
In Solanum tuberosum (potato), several lyciumin peptides could be characterized in a tuber-sprout extract by LC-MS analysis. However, none of the detected peptides matched the predicted core peptide sequences from the genome-derived lyciumin precursor. Close examination of the corresponding genomic locus showed that the 5′-region of the lyciumin precursor gene PGSC0003DMG400047074 was incomplete (SI Appendix, Fig. S26). We therefore assembled a de novo transcriptome of the Russett potato tuber [National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), SRR5970148] to recover the missing core peptide sequences of the predicted lyciumin precursor. Blast search in the transcriptome using LbaLycA as query yielded 11 additional core peptide sequences from candidate lyciumin precursor transcripts, including all core peptide sequences that matched detected potato lyciumin peptides (SI Appendix, Fig. S26). To verify this result, one precursor peptide gene of a detected potato lyciumin, StuBURP, was cloned and transiently expressed in N. benthamiana leaves, which resulted in the detection of its predicted product, lyciumin J (SI Appendix, Fig. S26). This example of characterizing potato lyciumins illustrates that genome mining of plant RiPPs can be complicated by sequencing gaps present in the current genome assemblies and that de novo transcriptomic datasets can aid the discovery of RiPP precursor genes. Moreover, as the precursor genes typically contain repetitive core peptide motifs, de novo transcriptome assembly programs such as Trinity (40) and rnaSPAdes (41) sometimes misassemble the precursor genes, which may need to be manually corrected (SI Appendix, Figs. S8 and S26). Overall, our precursor gene-guided approach led to the rapid discovery of lyciumin peptide chemotypes and genotypes from multiple plant families by genome mining in an automatable fashion.
The Sequence Rules of Naturally Occurring Lyciumins.
All lyciumin chemotypes feature an aromatic amino acid (Phe, Tyr, Trp) at the third position and a valine at the fifth position. The sixth and seventh residues vary within 10 different amino acids including aromatic, polar, and charged residues (Fig. 2B). The cyclization site is a glycine at the fourth position of all detected lyciumins, except for the identified Amaranthaceae lyciumin precursor peptides, which contain a threonine at the fourth residue of the core peptides (Fig. 2B and SI Appendix, Tables S4 and S5). In peptide extracts of the examined Amaranthaceae plants, several lyciumin derivatives were detected, which show a mass shift at the fourth residue corresponding to C2H3O or a putative dehydrothreonine, supporting a biosynthetic route via a threonine cyclization site (SI Appendix, Fig. S27). Predicted and characterized lyciumin genotypes can be divided into two types based on primary structure. Type 1 lyciumin precursors have core peptides within the BURP domain (e.g., in Fabaceae), while type 2 lyciumin precursors contain core peptides N-terminally of the BURP domain (e.g., in Amaranthaceae and Solanaceae) (Fig. 2C and Dataset S1). Based on location of core peptides and core peptide cyclization sites, lyciumin precursors are distinct in all three lyciumin-producing plant families (Fig. 2D) and phylogenetic analysis of BURP domains from lyciumin precursors also forms plant family-specific clades (SI Appendix, Fig. S28).
The Lyciumin Biosynthetic Pathway Is Promiscuous in Substrate Preference.
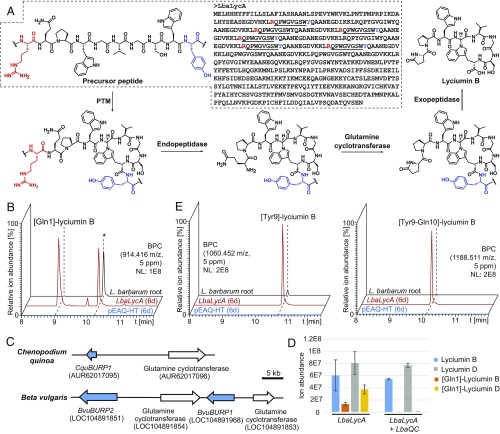
To enable heterologous production and diversification of lyciumin peptides, we next investigated the lyciumin biosynthetic pathway in planta. We establish a biosynthetic proposal for lyciumins based on several lines of evidence. Following the general dogma of RiPP biosynthesis, lyciumin biosynthesis begins with translation of a precursor peptide gene such as LbaLycA by the ribosome (Fig. 3A) (17). The precursor peptide is then cyclized between the tryptophan and glycine in each core peptide, which is supported by no detection of the linear core peptides in the LbaLycA heterologous expression experiments in N. benthamiana or in L. barbarum root extracts. Cyclization of the tryptophan-indole nitrogen to an unactivated α-carbon suggests a radical-oxidative cyclization mechanism (42), although candidate lyciumin cyclases have yet to be identified. In the next step, the modified LbaLycA is cleaved by an endopeptidase, N-terminally of the core peptide. This is supported by the detection of lyciumin derivatives with an N-terminal glutamine in leaf extracts of N. benthamiana transiently expressing LbaLycA, and in L. barbarum root extracts (Fig. 3B and SI Appendix, Fig. S29). Subsequently, core peptides are N-terminally protected by pyroglutamate formation, which can be catalyzed by a glutamine cyclotransferase (QC). Indeed, we identified QC-encoding genes next to the lyciumin precursor genes in the genomes of Chenopodium quinoa and Beta vulgaris (Fig. 3C and SI Appendix, Table S8). Furthermore, coexpression of LbaLycA with a L. barbarum homolog of these QCs in N. benthamiana resulted in the loss of mass signals of N-terminally unprotected lyciumins, confirming their enzymatic role in forming the N-terminal pyroglutamate moiety in lyciumins (Fig. 3D and SI Appendix, Fig. S30). In the final step, lyciumins are produced by C-terminal exoproteolytic maturation. This step is supported by the detection of multiple C-terminally extended lyciumin derivatives in leaf extracts of N. benthamiana transiently expressing LbaLycA, and in L. barbarum root extracts (Fig. 3E and SI Appendix, Fig. S31).
Fig. 3.
Investigation of lyciumin biosynthesis in Lycium barbarum. (A) Proposed biosynthetic pathway of lyciumin B in L. barbarum. (B) Detection of [Gln1]-lyciumin B mass signals in L. barbarum root extract and Nicotiana benthamiana leaf extracts after heterologous expression of lyciumin precursor LbaLycA for 6 d. Asterisk denotes ion source product of lyciumin B. (C) Genomic colocalization of lyciumin precursor genes and glutamine cyclotransferase genes for putative N-terminal lyciumin protection in Chenopodium quinoa and Beta vulgaris. (D) Detection of abolished mass signals for [Gln1]-lyciumin species in N. benthamiana leaf extracts after heterologous expression of lyciumin precursor LbaLycA and glutamine cyclotransferase LbaQC from Lycium barbarum root transcriptome (n = 3; error bars indicate ±1σ). (E) Detection of [Tyr9]-lyciumin B and [Tyr9-Gln10]-lyciumin B mass signals in L. barbarum root extract and N. benthamiana leaf extracts after heterologous expression of lyciumin precursor LbaLycA for 6 d. BPC, base peak chromatogram; LbaLycA (6 d), peptide extract of Nicotiana benthamiana infiltrated with Agrobacterium tumefaciens LBA4404 containing pEAQ-HT-LbaLycA (6 d); pEAQ-HT (6 d), peptide extract of Nicotiana benthamiana infiltrated with Agrobacterium tumefaciens LBA4404 containing pEAQ-HT (6 d); PTM, posttranslational modification.
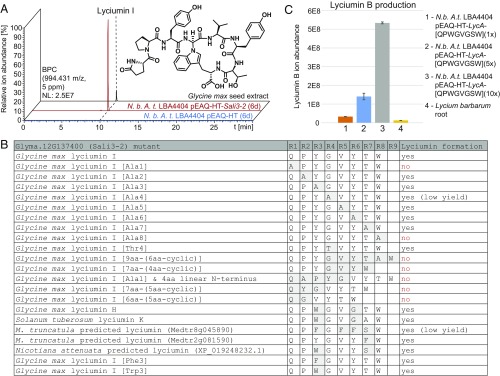
Despite the lack of a characterized lyciumin cyclase, we investigated the promiscuity of the lyciumin biosynthetic pathway in N. benthamiana. To generate lyciumin core peptide mutants, we characterized a lyciumin precursor gene from Glycine max that contains only one core peptide (QPYGVYTW) in the N-terminal domain. Heterologous expression of this precursor gene, previously named as Sali3-2 (Glyma.12G217400) (43, 44), in N. benthamiana resulted in the formation of its predicted lyciumin product, lyciumin I (Figs. 2B and 4A and SI Appendix, Figs. S16–S21 and Table S7). Using this monocore peptide precursor, a series of alanine-scanning mutagenesis experiments was performed through its core peptide region to identify mutable positions. Based on MS analysis, mutations of all residues, but the N-terminal glutamine and the C-terminal tryptophan of the Sali3-2 core peptide, to alanine resulted in lyciumin formation, indicating core peptide promiscuity of the lyciumin pathway (Fig. 4B and SI Appendix, Figs. S32–S37).
Fig. 4.
Diversification and scaled production of lyciumins in planta. (A) Heterologous expression of lyciumin I precursor Sali3-2 from Glycine max in Nicotiana benthamiana via infiltration with Agrobacterium tumefaciens LBA4404 containing pEAQ-HT-Sali3-2. (B) Investigation of lyciumin formation after Sali3-2 core peptide mutagenesis and heterologous expression in N. benthamiana (6 d). aa, amino acid; A.t., Agrobacterium tumefaciens; N.b., Nicotiana benthamiana. (C) Lyciumin B ion abundance in peptide extracts of N. benthamiana leaves 6 d after infiltration with A. tumefaciens LBA4404 pEAQ-HT-LycA-1×-QPWGVGSW, pEAQ-HT-LycA-5×-QPWGVGSW or pEAQ-HT-LycA-10×-QPWGVGSW, respectively, and of Lycium barbarum root. n = 3; error bars indicate ±1σ.
Next, we tested whether the length of the linear N-terminal branch or the size of the macrocycle of lyciumins could be modified in the N. benthamiana heterologous expression system. No lyciumin production was observed for a 4-, 2-, or 1-aa-long linear N-terminal branch, suggesting a conserved N-terminal branch length of 3 aa (Fig. 4B). Similarly, no lyciumin formation occurred from core peptides with 4- or 6-aa-long C-termini, indicating a conserved C-terminal lyciumin macrocycle of 5 aa (Fig. 4B). Finally, we tested whether the cyclization residues could be altered. Based on Amaranthaceae lyciumin precursors, mutation of Sali3-2 core peptide from glycine to threonine at the fourth position would result in lyciumin I production. When this mutant precursor peptide gene was expressed in N. benthamiana, we detected the production of a putative dehydrothreonine derivative as observed for Amaranthaceae lyciumin chemotypes with the corresponding [Thr4]-core peptides (Figs. 2B and 4B and SI Appendix, Fig. S38) in addition to lyciumin I.
Taken together, these structure–function relationship studies varying precursor core peptide sequence in heterologously reconstituted lyciumin pathway in N. benthamiana suggest restriction in peptide length but promiscuity in peptide sequence, presenting opportunity for branched cyclic RiPP diversification. We were able to produce known lyciumins such as lyciumin H and K (Figs. 2B and 4B and SI Appendix, Figs. S39 and S40), “unnatural” lyciumins such as lyciumin-[QPFGVYTW] and lyciumin-[QPWGVYTW] (Fig. 4B and SI Appendix, Figs. S41 and S42), and predicted lyciumins with up to four mutations from genome-derived BURP domain sequences such as lyciumin-[QPFGFFSW] and lyciumin-[QPYGVYFW] from M. truncatula (Fig. 4B and SI Appendix, Figs. S43 and S44) and lyciumin-[QPYGVYSW] from Nicotiana attenuata (Fig. 4B and SI Appendix, Fig. S45) via the Sali3-2-based heterologous expression system in N. benthamiana. Finally, we tested whether lyciumin yields of heterologous production in tobacco can exceed source plant extraction. Expression of an engineered LbaLycA-based lyciumin precursor with 5 or 10 core peptide repeats of lyciumin B (QPWGVGSW) in N. benthaminana leaves yielded 10 times or 40 times more lyciumin B, respectively, than peptide extraction of L. barbarum roots (Fig. 4C). These results highlight the utility of this expression platform for unlocking cryptic peptide chemotypes from diverse plant species and producing peptide libraries based on existing, predicted, or unknown lyciumin chemical space.
Discussion
The genomic resources for plants are expanding rapidly. Following the recent completion of the 1,000 plant transcriptome project (1KP), a new initiative for sequencing 10,000 plant genomes was recently announced (45), which forecasts an exponential increase in plant genomic resources within the next decade. As was the case for microbial and fungal natural product chemistry, gene-guided approaches for plant natural product discovery are now urgently needed in plant systems. This study illustrates genome mining as an automatable strategy to characterize hidden plant cyclic peptide chemistry and biochemistry by examining a class of branched cyclic RiPPs, the lyciumins. We leveraged the biosynthetic promiscuity of the identified RiPP pathway to decouple the isolation process of these peptides from their source plants by heterologous production in N. benthamiana. This approach also allows diversification of this class of branched cyclic peptides for future pharmacological and agrochemical development. Overall, our approach combining genome mining with synthetic biology for discovery, heterologous production, and diversification of peptides exemplifies how gene-guided discovery can complement bioactivity-guided approaches to overcome issues of rediscovery, production scale, and structural diversification.
Branched cyclic plant RiPPs, such as BURP domain-derived lyciumins, are particularly suitable for automated plant genome mining to identify new natural product chemistry (12). The advantage of RiPP genome mining is that no complete pathways, for example, gene clusters (46), are required to connect a predicted RiPP genotype with its chemotype. The known phytochemical space of branched cyclic peptides such as celogentins and cyclopeptide alkaloids suggests more unusual and probably unknown chemistry to be discovered from plants (27). These RiPP chemotypes can be characterized by precursor gene-guided genome mining or by querying BURP-domain proteins for repeats indicative of nonlyciumin core peptides. Moreover, plant transcriptomes can enable peptide discovery with optimized de novo assembly of repetitive sequences in less-studied nonmodel plants. The identification of unannotated precursor peptides in plant genomes and transcriptomes indicates the necessity to improve transcriptome and genome assembly for repetitive sequences, such as BURP domain genes.
The lyciumin pathway represents an example of branched cyclic RiPP biosynthesis in plants, which shares several common features with the head-to-tail plant cyclic peptide biosynthetic pathways. First, similarly to some cyclotide and orbitide precursors, lyciumin precursors can have multiple repeats of the core peptide sequences (17, 21, 47). One possible explanation for this repetitive structure is internal gene duplication of the core peptide sequences driven by strong selection to increase the yield and diversity of these bioactive peptides. Second, fusion of the RiPP core peptides, and thus RiPP biosynthesis, with other protein domains was also found to be the case for some cyclotides, for example, fusion with a seed-storage protein proalbumin (20). Finally, lyciumin biosynthesis may partially utilize general enzymatic machinery for protein processing. Cyclotides are processed by asparaginyl-endopeptidases in heterologous expression system in N. benthamiana, a nonnative cyclotide producer (23). Similarly, lyciumins can be produced in N. benthamiana through precursor peptide processing by nonspecific proteases and glutamine cyclotransferases. Whether the cyclase is specific to lyciumin biosynthesis remains to be determined. However, successful reconstitution of lyciumin production in N. benthamiana, a plant species that neither produces lyciumins endogenously nor has a lyciumin precursor in its genome, suggests that the cyclase responsible for posttranslationally modifying the lyciumin precursor proteins could be an enzyme with a specific cross-linking activity for plant proteins recruited for RiPP biosynthesis in the lyciumin pathway. It is also possible that the N. benthamiana genome may harbor cryptic lyciumin precursor genes, which are missing from the current genome assembly.
Features that distinguish lyciumin biosynthesis from head-to-tail cyclic RiPPs are N-terminal protection by pyroglutamate formation and protease-independent cyclization, both of which likely contribute to lyciumins’ in vivo stability and drug-like properties. The BURP-domain protein and lyciumin I precursor Sali3-2 has been shown to bind soft-transition metals and is targeted to plant vacuoles (43). Therefore, lyciumin cyclization may occur in vacuoles upon metal binding of the precursor peptide BURP domain and subsequent posttranslational processing of the N-terminal domain by a radical cyclase. While radical SAM enzymes are known to catalyze similar macrocyclic linkages in microbial RiPP biosynthesis (48, 49), they have not been shown to participate in plant-specialized metabolism. We therefore hypothesize that enzymes such as Fe(II)- and 2-oxoglutarate-dependent oxygenases (Fe/2OGs), cytochromes P450, laccases, or peroxidases are more likely to serve as lyciumin cyclase candidates (15, 50–52). The restriction of the lyciumin macrocycle to 5 aa suggests that the core peptide motifs likely fold into relatively confined 3D structures to facilitate radical-chemistry–based cyclization. Like cyclotides and orbitides, lyciumins likely emerged and diversified independently in multiple plant families (47). In particular, identical lyciumins are derived in Amaranthaceae, Fabaceae, and Solanaceae from precursor genes with different core peptide sequences, for example, QPYGVGSW and QPYTVGSW for lyciumin A in Lycium barbarum (Solanaceae) and Amaranthus hypochondriacus (Amaranthaceae), or different core peptide locations within the precursor gene, for example, QPYGVYTW for lyciumin I within the BURP domain in Glycine max (Fabaceae, type 1 lyciumin precursor) or N-terminally of the BURP domain in Solanum melongena (Solanaceae, type 2 lyciumin precursor), suggesting parallel evolution (Fig. 2D and SI Appendix, Fig. S28). More sampling of species representing all lineages of land plants, for example, by transcriptome mining, will enable a better picture of cyclic peptide evolution in terrestrial plants.
The fusion of lyciumin core peptides with BURP domains on a protein level indicates a connection of abiotic stress responses via heavy metal-binding BURP domains in plant vacuoles and lyciumin peptide signaling to alleviate stresses, such as drought and acidic soil (43). For example, lyciumin I precursor gene Sali3-2 is highly expressed during Al3+ stress in soybean roots as a result of acidic soil (44). Similarly, in amaranth, the precursor gene for lyciumin A and C is highly expressed during drought conditions (SI Appendix, Fig. S24). Lyciumins may be a specialized metabolic mechanism to alleviate biotic or abiotic stress in source plants, for example, by defending against pathogens and herbivores or binding harmful heavy metals within and around plant tissues. The physiological role of diverse lyciumins in their native plant hosts and their potential bioactivities for other utilities should be investigated in future research.
The lyciumin pathway presents a platform to engineer branched cyclic peptide chemistry for diverse agrochemical and pharmaceutical applications. For example, lyciumins can be produced and optimized in planta by endogenous precursor mutagenesis or precursor gene expression in crop plants to potentially improve crop fitness to specific biotic and abiotic stresses. Lyciumins are also inhibitors of pharmacologically relevant proteases such as ACE and renin, and therefore can be adapted for developing inhibitors of proteases and other protein targets. This study establishes a blueprint for genome mining of branched cyclic RiPPs in plants through the identification of pathway-specific precursor peptides and illustrates that hidden chemistry, biochemistry, and biology of numerous peptide natural products await discovery in the plant kingdom in the postgenomic era.
Materials and Methods
Materials and Instruments.
All chemicals were purchased from Sigma-Aldrich, unless otherwise specified. Oligonucleotide primers and synthetic genes were purchased as gBlocks from Integrated DNA Technologies. Solvents for LC–high-resolution MS were Optima LC-MS grade (Fisher Scientific) or LiChrosolv LC-MS grade (Millipore). High-resolution MS analysis was performed on a Thermo ESI-Q-Exactive Orbitrap MS coupled to a Thermo Ultimate 3000 UHPLC system. Low-resolution MS analysis was done on a Thermo ESI-QQQ MS coupled to a Thermo Ultimate 3000 UHPLC system. NMR analysis was performed on a Bruker Avance II 600-MHz NMR spectrometer equipped with a High Sensitivity Prodigy Cryoprobe. Preparative HPLC was performed on a Shimadzu LC-20AP liquid chromatograph equipped with a SPD-20A UV/VIS detector and a FRC-10A fraction collector.
Plant Material.
Lycium barbarum was purchased as 3-y-old plants for extraction and cultivation. Amaranthus hypochondriacus seeds for cultivation were purchased from Strictly Medicinal Seeds. Amaranth grain for extraction was Arrowhead Mills amaranth. Chenopodium quinoa seeds for cultivation were purchased from Earthcare Seeds. Quinoa for extraction was Trader Joe’s Tricolor Quinoa. Beta vulgaris seeds (Detroit Dark Red cultivar) for cultivation and extraction were purchased from David’s Garden Seeds. Glycine max seeds (Chiba green soybean) for cultivation and extraction were purchased from High Mowing Organic Seeds. Seeds of wild-type Medicago truncatula for cultivation were a gift from Prof. Dong Wang, University of Massachusetts, Amherst, MA. Capsicum annuum seeds (Jalapeno Early) for cultivation and extraction were purchased from EdenBrothers. Solanum lycopersicum seeds (cultivar Heinz 1706-BG) for cultivation were provided by the Tomato Genetics Resource Center, University of California, Davis, CA. Solanum melongena seeds for cultivation were purchased from Seedz. Solanum tuberosum tubers for cultivation (Russett or Red potato) were purchased from Trader Joe’s. Trifolium pratense seeds were purchased from OutsidePride.com. Nicotiana benthamiana seeds for cultivation were a gift from the S. L. Lindquist laboratory, Whitehead Institute, Massachusetts Institute of Technology, Cambridge, MA.
Plant Cultivation.
Lycium barbarum was grown from 3-y-old live roots in MiracleGro potting soil as a potted plant in full sun with occasional application of organic fertilizer. Lycium barbarum seeds from fruits of the 3-y-old plant were grown in Sun Gro Propagation Mix soil with added vermiculite (Whittemore) and added fertilizer in a greenhouse with a 16-h light/8-h dark cycle for 6 mo. Amaranthus hypochondriacus, Chenopodium quinoa, Beta vulgaris, Glycine max, Medicago truncatula, Capsicum annuum, Solanum lycopersicum, Solanum melongena, and Trifolium pratense were grown from seeds in Sun Gro Propagation Mix soil with added vermiculite (Whittemore) and added fertilizer in a greenhouse with a 16-h light/8-h dark cycle for 6 mo. Nicotiana benthamiana was grown from seeds in Sun Gro Propagation Mix soil with added vermiculite (Whittemore) and added fertilizer in a greenhouse with a 16-h light/8-h dark cycle for 3 mo. Solanum tuberosum tubers were sprouted under natural light for 3 wk.
Transcriptomic Analysis of Lycium barbarum and Identification of Candidate Precursor Gene LbaLycA.
Lycium barbarum roots were removed from a 3-y-old plant and washed with sterile water, and total RNA was extracted with the QIAGEN RNeasy Plant Mini kit. RNA quality was assessed by Agilent Bioanalyzer. A strand-specific mRNA library was prepared (TruSeq Stranded Total RNA with Ribo Zero Library Preparation Kit; Illumina) and sequenced with a HiSeq 2000 Illumina sequencer in HISEQRAPID mode (100 × 100). Illumina sequence raw files were combined and assembled by the Trinity package (40). Gene expression was estimated by mapping raw sequencing reads to the assembled transcriptomes using RSEM (53). The Lycium barbarum root transcriptome was analyzed for lyciumin precursors by searching predicted core peptide sequences for known lyciumins A (QPYGVGSW), lyciumin B (QPWGVGSW), lyciumin C (QPYGVFSW), and lyciumin D (QPYGVGIW) by blastp algorithm on an internal Blast server (54). To clone and sequence a lyciumin precursor gene from Lycium barbarum, cDNA was prepared from root total RNA with SuperScript III First-Strand Synthesis System (Invitrogen). Transcripts with lyciumin core peptide sequences were used to design cloning primers (LbaLycA-pEAQ-AgeI, AGACCGGTATGGAGTTGCATCACCATTAC; LbaLycA-pEAQ-XhoI, AGCTCGAGTTAGTTTTCAGACACTTGAGTTGCG) for amplification of precursor gene LbaLycA with Phusion High-Fidelity DNA polymerase (New England Biolabs) and directional cloning with restriction enzymes AgeI and XhoI (New England Biolabs) and T4 DNA ligase (New England Biolabs) into pEAQ-HT (16), which was linearized by restriction enzymes AgeI and XhoI. Cloned LbaLycA was sequenced by Sanger sequencing from pEAQ-HT-LbaLycA.
Heterologous Expression of Lyciumin Precursor Genes in Nicotiana benthamiana.
Agrobacterium tumefaciens LBA4404 was transformed with pEAQ-HT-LbaLycA, other pEAQ-HT constructs with lyciumin precursor genes (pEAQ-HT-StuBURP, pEAQ-HT-Sali3-2, pEAQ-HT-Sali-3-2-mutants) or pEAQ-HT-LbaQC by electroporation (2.5 kV), plated on YM agar [0.4 g of yeast extract, 10 g of mannitol, 0.1 g of sodium chloride, 0.2 g of magnesium sulfate (heptahydrate), 0.5 g of potassium phosphate (dibasic, trihydrate), 15 g of agar, and 1 L of Milli-Q Millipore water, adjusted to pH 7] with 100 µg/mL rifampicin, 50 µg/mL kanamycin, and 100 µg/mL streptomycin, and incubated for 2 d at 30 °C. A 5-mL starter culture of YM medium with 100 µg/mL rifampicin, 50 µg/mL kanamycin, and 100 µg/mL streptomycin was inoculated with a clone of Agrobacterium tumefaciens LBA4404 pEAQ-HT-LbaLycA and incubated for 24–36 h at 30 °C on a shaker at 225 rpm. Subsequently, the starter culture was used to inoculate a 50-mL culture of YM medium with 100 µg/mL rifampicin, 50 µg/mL kanamycin, and 100 µg/mL streptomycin, which was incubated for 24 h at 30 °C on a shaker at 225 rpm. The cells from the 50-mL culture were centrifuged for 30 min at 3,000 × g, the YM medium was discarded, and the cells were resuspended in MMA medium [10 mM MES KOH buffer (pH 5.6), 10 mM magnesium chloride, 100 µM acetosyringone] to give a final optical density of 0.8. The Agrobacterium suspension was infiltrated into the bottom of leaves of Nicotiana benthamiana plants (6 wk old). N. benthamiana plants were placed in the shade 2 h before infiltration. After infiltration, N. benthamiana plants were grown as described above for 6 d. Subsequently, infiltrated leaves were collected and subjected to chemotyping.
Chemotyping of Lyciumin Peptides from Plant Material.
For peptide chemotyping, 0.2 g of plant material (fresh weight) was frozen and ground with mortar and pestle. Ground plant material was extracted with 10 mL of methanol for 1 h at 37 °C in a glass vial. Plant methanol extract was dried under nitrogen gas in a separate glass vial. Dried plant methanol extract was resuspended in water (10 mL) and partitioned with hexane (2 × 10 mL) and ethyl acetate (2 × 10 mL), and subsequently extracted with n-butanol (10 mL). The n-butanol extract was dried in vacuo and resuspended in 2 mL of methanol for LC-MS analysis. Peptide extracts were subjected to high-resolution MS analysis with the following LC-MS parameters: LC: Phenomenex Kinetex 2.6-μm C18 reverse-phase, 100-Å, 150 × 3 mm LC column; LC gradient: solvent A, 0.1% formic acid; solvent B, acetonitrile (0.1% formic acid); 0–2 min, 5% B; 2–23 min, 5–95% B; 23–25 min, 95% B; 25–30 min, 5% B; 0.5 mL/min; MS: positive ion mode; full MS: resolution, 70,000; mass range, 425–1,250 m/z; dd-MS2 (data-dependent MS/MS): resolution, 17,500; loop count, 5; collision energy, 15–35 eV (stepped); dynamic exclusion, 1 s. LC-MS data were analyzed with QualBrowser in the Thermo Xcalibur software package (version 3.0.63; Thermo Scientific).
For comparative chemotyping of lyciumin concentrations in different plant tissues, see SI Appendix, Supporting Text.
Lyciumin Genome Mining.
Prediction of lyciumin genotypes.
For prediction of lyciumin precursor genes in a plant genome, LbaLycA homologs were searched by tblastn search in the 6-frame translated genome sequence [JGI Phytozome, version 12.1, and prerelease genomes (13)] or by blastp of Refseq protein sequences (NCBI genomes; SI Appendix, Table S4). In addition, annotated BURP domains were identified by “BURP domain” keyword search (13). All identified BURP domain proteins from a plant genome were then searched for lyciumin core peptide sequences with the search criteria of a glutamine and proline as the first and second amino acid, respectively, in the core peptide sequence and a tryptophan at the eighth position of the core peptide sequence. A BURP domain protein, which contained one or multiple sequences matching these lyciumin core peptide criteria, was a candidate lyciumin precursor peptide and, thus, its gene a predicted lyciumin genotype in the target plant genome.
To complement missing core peptide sequences from a lyciumin precursor gene with a sequence gap in the potato genome (PGSC0003DMG400047074), a Russett potato tuber transcriptome (NCBI SRA, SRR5970148) was assembled by Trinity (version 2.4) (40) and rnaSPAdes (version 1.0, kmer 25,75) (41). Precursor peptide transcripts with missing core peptide sequences were searched in both de novo transcriptome assemblies by LbaLycA tblastn search.
Prediction of lyciumin chemotypes.
A lyciumin structure was predicted from a putative lyciumin core peptide sequence by transformation of the glutamine at the first position to a pyroglutamate and formation of a covalent bond between the indole nitrogen of the tryptophan at the eighth position with the α-carbon of the residue at the fourth position by loss of two hydrogens (SI Appendix, Fig. S10).
Lyciumin chemotyping.
LC-MS data of peptide extracts from a predicted lyciumin producing plant was analyzed for lyciumin mass signals by (i) parent mass search (base peak chromatogram of calculated [M+H]+ of predicted lyciumin structure, Δm = 5 ppm), (ii) fragment mass search of pyroglutamate-proline-b ion in MS/MS data (C10H13N2O3+, 209.09207 m/z, Δm = 5 ppm), and (iii) iminium ion mass search of specific amino acids of predicted structure in MS/MS data (for example, pyroglutamate iminium ion [M+H]+ 84.04439 m/z). Putative mass signals of predicted lyciumin structures were confirmed by MS/MS data analysis with QualBrowser in the Thermo Xcalibur software package (version 3.0.63; Thermo Scientific).
Cloning of Lyciumin Precursor Gene StuBURP from Solanum tuberosum.
Tuber sprout tissue was removed from a sprouting potato tuber and total RNA was extracted with the Qiagen RNeasy Plant Mini kit. cDNA was prepared from sprout total RNA with SuperScript III First-Strand Synthesis System (Invitrogen). A de novo transcriptome was assembled from a Russett potato RNA-seq dataset (NCBI SRA, SRR5970148) and transcripts homologous to target lyciumin precursor PGSC0003DMG400047074 were used to design cloning primers (StuBURP-pEAQ-fwd, TGCCCAAATTCGCGACCGGTATGGAGTTGCATCACCAATA; StuBURP-pEAQ-rev, CCAGAGTTAAAGGCCTCGAGTTAGTTTTCAGCCACTTGAAGAACTG) for amplification of precursor peptide gene StuBURP with Phusion High-Fidelity DNA polymerase (New England Biolabs). StuBURP was cloned into pEAQ-HT (16), which was linearized by restriction enzymes AgeI and XhoI, by Gibson cloning assembly (New England Biolabs) (55). Cloned StuBURP was sequenced by Sanger sequencing from pEAQ-HT-StuBURP.
Lyciumin Engineering in Nicotiana benthamiana.
Predicted lyciumin precursor Sali3-2 (Glyma.12G217400) was synthesized as an IDT gBlock with a 5′-adapter (tgcccaaattcgcgaccggt) and a 3′-adapter (ctcgaggcctttaactctgg) for Gibson assembly (5). pEAQ-HT was digested by AgeI and XhoI restriction enzymes, and the Sali3-2 gBlock was cloned into the digested pEAQ-HT with Gibson Assembly Master Mix (55). pEAQ-HT-Sali3-2 was verified by Sanger sequencing and transformed into Agrobacterium tumefaciens LBA4404 for heterologous expression as described above. Constructs for lyciumin engineering were Sali3-2 mutants of its core peptide sequence (Fig. 4B). Sali3-2 mutants were synthesized as gBlocks and cloned into pEAQ-HT for heterologous expression in N. benthamiana as described above. Chemotyping of infiltrated N. benthamiana leaves for lyciumins was done as described above. For gene sequences, please see SI Appendix, Supporting Text.
Phylogenetic Analysis of Lyciumin Precursor Peptides.
Please see SI Appendix, Supporting Text.
Purification and Structure Elucidation of Lyciumins.
Please see SI Appendix, Supporting Text.
Gene Expression Analysis of Characterized Lyciumin Precursors.
Please see SI Appendix, Supporting Text.
Glutamine Cyclotransferase Coexpression Assays with LbaLycA in Nicotiana benthamiana.
Please see SI Appendix, Supporting Text.
LbaLycA-Based Lyciumin Production in Nicotiana benthamiana in Comparison with Source Plant Extraction.
Please see SI Appendix, Supporting Text.
Supplementary Material
Acknowledgments
We thank Frank C. Schroeder (Cornell University) and Li-Jun Ma (University of Massachusetts, Amherst) for helpful discussion and for assisting with a number of exploratory experiments related to this study. We thank the Tomato Genetics Resource Center (University of California, Davis) for providing seeds of S. lycopersicum Heinz 1706-BG, and Dong Wang (University of Massachusetts, Amherst) for providing seeds of M. truncatula. R.D.K. is a Howard Hughes Medical Institute postdoctoral fellow of the Life Sciences Research Foundation. This work was supported by grants from the Thome Foundation, the Pew Scholars Program in the Biomedical Sciences, the Searle Scholars Program, and the Family Larsson Rosenquist Foundation.
Footnotes
Conflict of interest statement: R.D.K. and J.-K.W. have filed a patent application on heterologous production and diversification of branched cyclic peptides using the biosynthetic system discovered in this study. J.-K.W. is a co-founder, a member of the Scientific Advisory Board, and a shareholder of DoubleRainbow Biosciences, which develops biotechnologies related to natural products.
This article is a PNAS Direct Submission.
Data deposition: The gene sequences reported in this paper have been deposited in the GenBank database [accession nos. MH124242 (LbaLycA), MH124243 (LbaQC), and MH124244 (StuBURP)]; the Lycium barbarum root transcriptome reported in this paper has been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. SRR6896657); and the LC-MS datasets reported in this paper have been deposited in the Global Natural Products Social Molecular Networking–MassIVE database (accession nos. MSV000082522–MSV000082557).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813993115/-/DCSupplemental.
References
- 1.Li FS, Weng JK. Demystifying traditional herbal medicine with modern approach. Nat Plants. 2017;3:17109. doi: 10.1038/nplants.2017.109. [DOI] [PubMed] [Google Scholar]
- 2.Tu Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat Med. 2011;17:1217–1220. doi: 10.1038/nm.2471. [DOI] [PubMed] [Google Scholar]
- 3.Rishton GM. Natural products as a robust source of new drugs and drug leads: Past successes and present day issues. Am J Cardiol. 2008;101:43D–49D. doi: 10.1016/j.amjcard.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Drewry DH, Macarron R. Enhancements of screening collections to address areas of unmet medical need: An industry perspective. Curr Opin Chem Biol. 2010;14:289–298. doi: 10.1016/j.cbpa.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 5.Medema MH, Fischbach MA. Computational approaches to natural product discovery. Nat Chem Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M, et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziemert N, Alanjary M, Weber T. The evolution of genome mining in microbes—a review. Nat Prod Rep. 2016;33:988–1005. doi: 10.1039/c6np00025h. [DOI] [PubMed] [Google Scholar]
- 8.Kersten RD, et al. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim E, Moore BS, Yoon YJ. Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nat Chem Biol. 2015;11:649–659. doi: 10.1038/nchembio.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazio GC, Xu R, Matsuda SP. Genome mining to identify new plant triterpenoids. J Am Chem Soc. 2004;126:5678–5679. doi: 10.1021/ja0318784. [DOI] [PubMed] [Google Scholar]
- 11.Huang AC, et al. Unearthing a sesterterpene biosynthetic repertoire in the Brassicaceae through genome mining reveals convergent evolution. Proc Natl Acad Sci USA. 2017;114:E6005–E6014. doi: 10.1073/pnas.1705567114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kautsar SA, Suarez Duran HG, Blin K, Osbourn A, Medema MH. plantiSMASH: Automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 2017;45:W55–W63. doi: 10.1093/nar/gkx305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodstein DM, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matasci N, et al. Data access for the 1,000 plants (1KP) project. Gigascience. 2014;3:17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anarat-Cappillino G, Sattely ES. The chemical logic of plant natural product biosynthesis. Curr Opin Plant Biol. 2014;19:51–58. doi: 10.1016/j.pbi.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sainsbury F, Thuenemann EC, Lomonossoff GP. pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J. 2009;7:682–693. doi: 10.1111/j.1467-7652.2009.00434.x. [DOI] [PubMed] [Google Scholar]
- 17.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craik DJ, Daly NL, Bond T, Waine C. Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 19.Craik DJ, et al. Ribosomally-synthesised cyclic peptides from plants as drug leads and pharmaceutical scaffolds. Bioorg Med Chem. 2018;26:2727–2737. doi: 10.1016/j.bmc.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Mylne JS, et al. Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat Chem Biol. 2011;7:257–259. doi: 10.1038/nchembio.542. [DOI] [PubMed] [Google Scholar]
- 21.Condie JA, et al. The biosynthesis of Caryophyllaceae-like cyclic peptides in Saponaria vaccaria L. from DNA-encoded precursors. Plant J. 2011;67:682–690. doi: 10.1111/j.1365-313X.2011.04626.x. [DOI] [PubMed] [Google Scholar]
- 22.Gruber CW, et al. A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J Biol Chem. 2007;282:20435–20446. doi: 10.1074/jbc.M700018200. [DOI] [PubMed] [Google Scholar]
- 23.Saska I, et al. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J Biol Chem. 2007;282:29721–29728. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen GK, et al. Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat Chem Biol. 2014;10:732–738. doi: 10.1038/nchembio.1586. [DOI] [PubMed] [Google Scholar]
- 25.Barber CJ, et al. The two-step biosynthesis of cyclic peptides from linear precursors in a member of the plant family Caryophyllaceae involves cyclization by a serine protease-like enzyme. J Biol Chem. 2013;288:12500–12510. doi: 10.1074/jbc.M112.437947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chekan JR, Estrada P, Covello PS, Nair SK. Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants. Proc Natl Acad Sci USA. 2017;114:6551–6556. doi: 10.1073/pnas.1620499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan NH, Zhou J. Plant cyclopeptides. Chem Rev. 2006;106:840–895. doi: 10.1021/cr040699h. [DOI] [PubMed] [Google Scholar]
- 28.Mohimani H, Pevzner PA. Dereplication, sequencing and identification of peptidic natural products: From genome mining to peptidogenomics to spectral networks. Nat Prod Rep. 2016;33:73–86. doi: 10.1039/c5np00050e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yahara S, et al. Structures of anti-ace and anti-renin peptides from lycii-radicis cortex. Tetrahedron Lett. 1989;30:6041–6042. [Google Scholar]
- 30.Yahara S, et al. Cyclic peptides, acyclic diterpene glycosides and other compounds from Lycium chinense Mill. Chem Pharm Bull (Tokyo) 1993;41:703–709. doi: 10.1248/cpb.41.703. [DOI] [PubMed] [Google Scholar]
- 31.Morita H, Yoshida N, Takeya K, Itokawa H, Shirota O. Configurational and conformational analyses of a cyclic octapeptide, lyciumin A, from Lycium chinense Mill. Tetrahedron. 1996;52:2795–2802. [Google Scholar]
- 32.Morita H, Suzuki H, Kobayashi J. Celogenamide A, a new cyclic peptide from the seeds of Celosia argentea. J Nat Prod. 2004;67:1628–1630. doi: 10.1021/np049858i. [DOI] [PubMed] [Google Scholar]
- 33.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 34.Hattori J, Boutilier KA, van Lookeren Campagne MM, Miki BL. A conserved BURP domain defines a novel group of plant proteins with unusual primary structures. Mol Gen Genet. 1998;259:424–428. doi: 10.1007/s004380050832. [DOI] [PubMed] [Google Scholar]
- 35.Ding X, Hou X, Xie K, Xiong L. Genome-wide identification of BURP domain-containing genes in rice reveals a gene family with diverse structures and responses to abiotic stresses. Planta. 2009;230:149–163. doi: 10.1007/s00425-009-0929-z. [DOI] [PubMed] [Google Scholar]
- 36.Boutilier KA, et al. Expression of the BnmNAP subfamily of napin genes coincides with the induction of Brassica microspore embryogenesis. Plant Mol Biol. 1994;26:1711–1723. doi: 10.1007/BF00019486. [DOI] [PubMed] [Google Scholar]
- 37.Bassüner R, et al. Abundant embryonic mRNA in field bean (Vicia faba L.) codes for a new class of seed proteins: cDNA cloning and characterization of the primary translation product. Plant Mol Biol. 1988;11:321–334. doi: 10.1007/BF00027389. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi-Shinozaki K, Shinozaki K. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet. 1993;238:17–25. doi: 10.1007/BF00279525. [DOI] [PubMed] [Google Scholar]
- 39.Zheng L, Heupel RC, DellaPenna D. The beta subunit of tomato fruit polygalacturonase isoenzyme 1: Isolation, characterization, and identification of unique structural features. Plant Cell. 1992;4:1147–1156. doi: 10.1105/tpc.4.9.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang MC, Zou Y, Watanabe K, Walsh CT, Tang Y. Oxidative cyclization in natural product biosynthesis. Chem Rev. 2017;117:5226–5333. doi: 10.1021/acs.chemrev.6b00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Y, et al. Expression of a vacuole-localized BURP-domain protein from soybean (SALI3-2) enhances tolerance to cadmium and copper stresses. PLoS One. 2014;9:e98830. doi: 10.1371/journal.pone.0098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ragland M, Soliman KM. Global Environmental Biotechnology. Springer Science; London: 1997. A molecular approach to understanding aluminum tolerance in soybean (Glycine max L) pp. 125–138. [Google Scholar]
- 45.Cheng S, et al. 10KP: A phylodiverse genome sequencing plan. Gigascience. 2018;7:1–9. doi: 10.1093/gigascience/giy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Field B, Osbourn AE. Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science. 2008;320:543–547. doi: 10.1126/science.1154990. [DOI] [PubMed] [Google Scholar]
- 47.Mylne JS, et al. Cyclic peptides arising by evolutionary parallelism via asparaginyl-endopeptidase-mediated biosynthesis. Plant Cell. 2012;24:2765–2778. doi: 10.1105/tpc.112.099085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flühe L, et al. The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat Chem Biol. 2012;8:350–357. doi: 10.1038/nchembio.798. [DOI] [PubMed] [Google Scholar]
- 49.Schramma KR, Bushin LB, Seyedsayamdost MR. Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink. Nat Chem. 2015;7:431–437. doi: 10.1038/nchem.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Costa MMR, et al. Molecular cloning and characterization of a vacuolar class III peroxidase involved in the metabolism of anticancer alkaloids in Catharanthus roseus. Plant Physiol. 2008;146:403–417. doi: 10.1104/pp.107.107060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sterjiades R, Dean JF, Eriksson KE. Laccase from sycamore maple (Acer pseudoplatanus) polymerizes monolignols. Plant Physiol. 1992;99:1162–1168. doi: 10.1104/pp.99.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davin LB, et al. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science. 1997;275:362–366. doi: 10.1126/science.275.5298.362. [DOI] [PubMed] [Google Scholar]
- 53.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Priyam A, et al. 2015. Sequenceserver: A modern graphical user interface for custom BLAST databases. bioRxiv:10.1101/033142. Preprint, posted November 27, 2015.
- 55.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.