Abstract
The understanding of the mechanisms of antibiotic resistance development are fundamental to alert and preview beforehand, the large scale dissemination of resistance to antibiotics, enabling the design of strategies to prevent its spread. The mecA-mediated methicillin resistance conferring resistance to broad-spectrum β-lactams is globally spread in staphylococci including hospitals, farms and community environments, turning ineffective the most widely used and efficient class of antibiotics to treat staphylococcal infections. The use of whole genome sequencing (WGS) technologies at a bacterial population level has provided a considerable progress in the identification of key steps that led to mecA-mediated β-lactam resistance development and dissemination. Data obtained from multiple studies indicated that mecA developed from a harmless core gene (mecA1) encoding the penicillin-binding protein D (PbpD) from staphylococcal species of animal origin (S. sciuri group) due to extensive β-lactams use in human created environments. Emergence of the resistance determinant involved distortion of PbpD active site, increase in mecA1 expression, addition of regulators (mecR1, mecI) and integration into a mobile genetic element (SCCmec). SCCmec was then transferred into species of coagulase-negative staphylococci (CoNS) that are able to colonize both animals and humans and subsequently transferred to S. aureus of human origin. Adaptation of S. aureus to the exogenously acquired SCCmec involved, deletion and mutation of genes implicated in general metabolism (auxiliary genes) and general stress response and the adjustment of metabolic networks, what was accompanied by an increase in β-lactams minimal inhibitory concentration and the transition from a heterogeneous to homogeneous resistance profile. Nowadays, methicillin-resistant S. aureus (MRSA) carrying SCCmec constitutes one of the most important worldwide pandemics. The stages of development of mecA-mediated β-lactam resistance described here may serve as a model for previewing and preventing the emergence of resistance to other classes of antibiotics.
Keywords: β-lactams resistance, Staphylococcus sciuri, staphylococcal cassette chromosome mec (SCCmec), methicillin-resistant Staphylococcus aureus (MRSA), whole genome sequencing
Introduction
Antimicrobial resistance threatens the effective prevention and treatment strategies of an increasing range of bacterial infections. In 21st century we are facing the real possibility that minor injuries and common infections can lead to death. A detailed understanding of the evolutionary processes occurring in nature that lead to resistance development is thus essential for anticipating its emergence and to restrain its spread.
One of the best models of resistance development is the emergence of methicillin resistance in staphylococci not only due the fact that it is extremely well documented, but mainly because it gave rise to methicillin-resistant Staphylococcus aureus (MRSA) pandemics – presently a major public health concern (Oliveira et al., 2001, 2002; Lindsay, 2013; Otto, 2013).
Due to their high efficacy and low toxicity, β-lactams are the most widely used class of antibiotics (Shahid et al., 2009). They inhibit bacterial cell wall biosynthesis through irreversible binding to the traspeptidase domain of penicillin-binding proteins (PBPs) (Ghuysen, 1991, 1994).
Staphylococcus species have a broad distribution in nature and consist of large populations. They are common commensals of the skin and mucous membranes of humans and animals (Kloos, 1980, 1986, 1997) and are ubiquitously recovered from the environment (Huijbers et al., 2015). Although during most of its existence they live as mere colonizers, when the skin and mucous membranes barrier of their host is impaired and the host is immunocompromized staphylococci may arise as important pathogens. Among all staphylococcal species, S. aureus, is considered to be the most pathogenic, being associated to a myriad of infections ranging from mild skin infections to life-threatening diseases (Crossley and Archer, 1997).
The major driving force for the emergence of β-lactams resistance in staphylococci was the continuous exposure to β-lactams in multiple environments: in soils where they had to co-exist with penicillin-producing fungi; in production animal farms wherein large amounts of β-lactam antibiotics were used as food additives (National Research Council, 1980; Castanon, 2007), and during treatment of bacterial infections (Westh et al., 2004).
All MRSA contain a copy of an exogenous mec gene that codifies for PBPs with low affinity for β-lactams (mecA, mecB, mecC, and mecD) (Hartman and Tomasz, 1984; Harrison et al., 2013; Gomez-Sanz et al., 2015; Schwendener et al., 2017; Becker et al., 2018; Schwendener and Perreten, 2018), In this review we will focus on the evolution and emergence of methicillin resistance mediated by mecA which encodes an extra PBP, PBP2A (Hartman and Tomasz, 1984) with a low binding affinity to virtually all β-lactams. In the presence of β-lactams antibiotics the transpeptidase domain of all native PBPs is inactivated, but bacteria containing mecA continue to synthesize cell wall as a result of the cooperation between transpeptidase domain of the PBP2A and the transglycosylase domain of the native staphylococcal PBP2 (Pinho et al., 2001). The few β-lactam to which mecA does not confer resistance include ceftobiprole and ceftaroline which are active against MRSA (Entenza et al., 2002; Ishikawa et al., 2003) and penicillin G, ampicillin and amoxicillin which are active against penicillinase-negative MRSA strains (a minority, nowadays).
Several efforts have been made to clarify the origin of mecA-mediated resistance to β-lactams in staphylococci and the use of state-of-the-art WGS technology has provided unprecedented advances (see Table 1). Nevertheless, the precise steps that led to β-lactam resistance development and dissemination are still not totally clear and are a matter of speculation.
Table 1.
Insights into β-lactam resistance development provided by WGS.
| mecA evolution |
| • mecA homologs are ubiquitous in S. fleurettii and S. vitulinus |
| •mecA homologue is present in the native location In S. fleurettii and S. vitulinus |
| • mecA and mecA2 in S. vitulinus do not provide resistance to β-lactams |
| •mecA homologs diversification begun with the use of antibiotics (1940s) |
| •rnecB and mecC were Identified |
| SCCmec evolution |
| •mec complex and SCC elements evolved in parallel in different chromosomal locations |
| • mecA homologs native location in S. sciuri species group is 200 Kb from orfX |
| •mecA and mecR2 originated from S. sciuri homologs |
| • mecRl/mecl were added to mecA to form the mec complex in S. fleurettii or S. vitulinus |
| • The last donors of J1 region and ccr to SCCmec were S. sciuri |
| • The last donors of mec complex, J2 and J3 regions to SCCmec were S. vitulinus/S. fleurettii |
| • Origin of SCC and SCCrnec is probably S. sciuri |
| •mecA was probably integrated into a SCC in 5. sciuri |
| • SCCmec III was probably the most ancient SCCmec type |
| Expression of β-lactam resistance |
| • Genetic basis of hetero-to-homo resistance conversion |
| – Tandem amplification of SCCmec |
| – Mutations in relA and rpoB |
| • Mechanisms of β-lactam resistance development in S. sciuri and S. vitulinus |
| – Alterations in mecAl/mecA2 promoter |
| – Alterations in PbpD structure |
| – SCCmec acquisition |
History of β-Lactams and β-Lactam Resistance
Penicillin, a natural antibacterial compound produced by fungi, was first discovered in 1928 by Alexander Fleming (Fleming, 1929). However, due to low production yield, instability of the compound and problems in purification it was only later, in 1941, that penicillin was used as an antibiotic to treat human bacterial infections. The necessity to treat sick and wounded soldiers in the Second World War promoted the mass production of penicillin, and in 1945 this antibiotic was already used routinely in human clinical practice (Aminov, 2017).
Based on studies showing that penicillin is a growth promoter of chickens, pigs and livestock (National Research Council, 1980; Castanon, 2007), in 1951, the Food and Drug Administration (FDA) also approved the use of penicillin in animals (Hao et al., 2014). Nowadays, penicillin and other β-lactams continue to be used in many countries in food production animals not only to enhance animal growth, but also to treat infections and as a prophylactic (Hao et al., 2014). In fact, recent surveillance studies in Europe indicate that 25% of all antibiotic consumption (in mg/PCU) in veterinary setting relate to penicillin (report SE, 2010/2015), much of which are used for non-therapeutic purposes in chickens, cattle, and swine, compared with just a small quantity used for clinical treatments (World Organization for Animal Health, 2016).
A natural consequence of penicillin exposure was the development of antibiotic resistance. In fact, in 1942, only 2 years after the introduction of penicillin into clinical practice, the first penicillin-resistant S. aureus emerged in a hospital (Barber and Rozwadowska-Dowzenko, 1948) and shortly after (1960s) were also disseminated in the community (Rountree and Freeman, 1955), reaching around 80%. Penicillin resistance emerged due to the acquisition of β-lactamases that were able to hydrolyze and inactivate penicillin. Further developments to overcome resistance to penicillin included the synthesis of penicillinase-resistant penicillins, such as methicillin in 1960. However, due to its amazing adaptative power, S. aureus that were resistant to methicillin and to all β-lactams emerged right after its first use in the treatment of bacterial infections, through the acquisition of mecA (Jevons, 1961). This event has lead to the emergence of methicillin-resistant S. aureus (MRSA) strains and to one of the most important bacterial pandemics in hospitals worldwide (Oliveira et al., 2002; Grundmann et al., 2006). The MRSA rates in hospitals increased then, exponentially, reaching extremely high levels (above 60%) in the 1990s, mainly in Southern European countries (Deurenberg and Stobberingh, 2008; Chambers and Deleo, 2009; Lindsay, 2013). Similar to what was observed before for penicillin, this was followed by a wave of MRSA emergence in the community (Herold et al., 1998; Continuous Discharge Certificate [CDC], 1999; Vandenesch et al., 2003; Vlack et al., 2006), causing infections in otherwise healthy persons. Community-associated MRSA (CA-MRSA) are nowadays endemic in the community in specific countries like United States and here they have also become major multidrug resistant hospital clones (Moran et al., 2006; Otto, 2013). Additionally, resistance to β-lactams has expanded into farm environments wherein specific MRSA clones, like ST398 have become frequent colonizers of production animals and of humans in contact with them (Armand-Lefevre et al., 2005; Voss et al., 2005; Fluit, 2012).
As was observed for β-lactams, the emergence of drug resistance has been described following the introduction of each new antimicrobial class. The recent awareness by political authorities of the problem of antimicrobial resistance has lead to actions toward the banning of antimicrobial use in animals (European Union [EU], 2003; Food and Drug Administration [FDA], 2018). However, rules controlling antimicrobial use in animals have been applied mainly in Europe (National Research Council, 1980; World Organization for Animal Health, 2016), and still some of the antimicrobials, like penicillins, used to treat human disease continue to be heavily used in animals in European countries (Grave et al., 2012).
The Structural Element of β-Lactam Resistance: the mecA Gene
β-lactams target the PBPs, involved in the synthesis of peptidoglycan, the major structural component of the bacterial cell wall. In particular, PBPs catalyze the main reactions involved in the polymerization of peptidoglycan, namely the elongation of glycan strands (transglycosilation) and the cross-linking between stem peptides of different glycan strands (transpeptidation) (Macheboeuf et al., 2006). Binding of β-lactams to native PBPs turns them inactive, what prevents peptidoglycan synthesis and bacterial growth (Waxman and Strominger, 1983). This reaction involves the break of the β-lactam ring amide bond and acylation of the PBPs, which gives rise to a serine ester-linked acyl derivative that is extremely stable and has a low rate of deacylation.
The mecA gene encodes a high-molecular weight class B PBP, called PBP2A (Hartman and Tomasz, 1984), which contains two domains, the C-terminal domain which is known to have a transpeptidation function, and a N-terminal domain to which no function has been attributed, the so called non-binding (NB) domain. Resistance is provided by the fact that this extra PBP has a lower efficiency of acylation by β-lactams, which is believed to result from a lower affinity for these compounds (Fuda et al., 2004) and from a slow rate of acylation. Resolution of PBP2A crystal structure showed that the poor acylation rate observed is due to the presence of a distorted active site, provided by a higher flexibility of the NB domain and by differences in regions close to the active site groove in the transpeptidase domain (Lim and Strynadka, 2002). Furthermore, the position of Ser403 was considered crucial for the effective nucleophilic attack of the β-lactam ring, which leads to acylation of the protein (Lim and Strynadka, 2002).
The Mobile Element Carrying mecA: SCCmec
The mecA gene is carried in a mobile genetic element called staphylococcal cassette chromosome mec (SCCmec) (International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements [IWG-SCC] et al., 2009). SCCmec is delimited by distinctive terminal inverted and direct repeats formed upon SCCmec insertion (DR-left downstream orfX, attL; DR-right at SCCmec end, attR) in a single chromosomal location, in the 3′ end of orfX, a RNA methyltransferase that is localized near the origin of replication (Boundy et al., 2013). This mobile genetic element is composed of two central elements, the mec complex containing mecA and intact and deleted forms of its regulators (mecI, mecR1) and the ccr complex composed of cassette chromosme recombinases (ccr) involved in its mobility (Katayama et al., 2000). The remaining portions of SCCmec, are composed of non-essential components, namely additional metal and antibiotic resistance genes carried by transposons and plasmids, as well as genes of unknown function, which are named J regions. The J3 region is located between orfX and mec complex, the J2 region is flanked by mec complex and the ccr complex and the J1 region between ccr complex and the right extremity of the element (see Figure 1). More recently, mecR2, coding for an anti-repressor of mecA was described to exist downstream mecI, which together with mecI and mecR1 constitute an unusual three-component arrangement (Arede et al., 2012). So far as many as thirteen different structural types of SCCmec have been described in S. aureus1 (Ito et al., 2001, 2004; Ma et al., 2002; Oliveira et al., 2006; Berglund et al., 2008; Zhang et al., 2009; Garcia-Alvarez et al., 2011; Li et al., 2011; Wu et al., 2015; Baig et al., 2018) that range between 20 and 70 Kb. The different types of SCCmec correspond to different combinations of mec complex class (A-E), according to the presence/absence of regulatory genes and insertion sequences, and ccr allotypes (ccrAB and ccrC).
FIGURE 1.
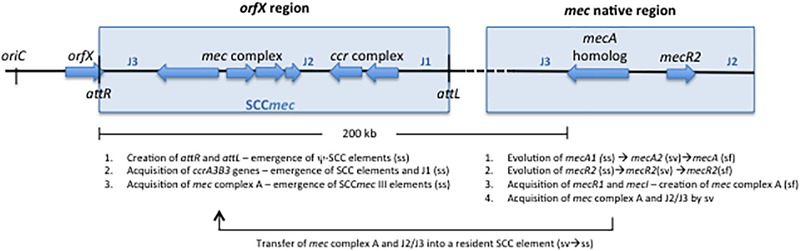
Schematic representation of the genomic events possibly associated with the evolution and assembly of SCCmec III occurring in the orfX and mec native region of species of Staphylococcus sciuri group. The basic structure of SCCmec in orfX is shown, including attL, attR, mec complex, ccr complex, J1, J2, and J3 regions. The elements present in the native region in species of the S. sciuri group (S. sciuri, S. vitulinus, and S. fleurettii) are shown, including a mecA homolog, the mecR2, J2 and J3 regions. S. sciuri: ss; S. vitulinus: sv; S. fleurettii: sf.
SCCmec is believed to have been acquired a limited number of times in S. aureus (Robinson and Enright, 2003), but acquisition of this element appears to provide a major advantage to bacteria mainly in the hospital environment. In fact, the acquisition of different types of SCCmec by methicillin-susceptible S. aureus (MSSA) of diverse genetic backgrounds gave rise to several MRSA pandemics over time (Chambers and Deleo, 2009), namely the Iberian (ST247-I), Brazilian (ST239-III), New York-Japan (ST5-II), EMRSA16 (ST36-II), EMRSA15 (ST22-IV), Berlin (ST45-IV), United States 300 (ST8-IV), and ST398-V (Deurenberg and Stobberingh, 2009). In contrast to S. aureus, in coagulase-negative staphylococci (CoNS), there is not a clear association between SCCmec and specific genetic backgrounds (Rolo et al., 2012). However, it is still not clear if this derives from the fact that SCCmec was acquired a higher number of times or from a higher instability of SCCmec structure in these species. Nevertheless, specific SCCmec types appear to be more common to certain CoNS species. SCCmec type I and VI are more common in Staphylococcus hominis (Bouchami et al., 2012), SCCmec III in Staphylococcus sciuri group of species (Rolo et al., 2017a), SCCmec IV in S. epidermidis (Miragaia et al., 2007), and SCCmec V in S. haemolyticus (Bouchami et al., 2010).
The true breadth of genetic diversity of SCCmec elements is still unknown. While in S. aureus this structure has been shown to be relatively stable, in CoNS, such as S. epidermidis and S. haemolyticus, a high genetic diversity has been described (Miragaia et al., 2007). This might be due to the fact that CoNS species have a high recombination rate (Miragaia et al., 2007), an enhanced ability to acquire and maintain exogenous genetic material or because these elements have been acquired earlier by these species than by S. aureus. In the era of WGS where detailed information on the entire genomes of thousands of staphylococci are being gathered, the number of types and subtypes of SCCmec have increased exponentially1, and this challenges the traditional criteria and methodologies that were previously defined to classify SCCmec types, mainly based on PCR. A web-tool, SCCmecFinder, able to identify all SCCmec element types (I to XIII) and SCCmec IV and V subtypes was recently developed to classify SCCmec types based on WGS data (Kaya et al., 2018). The characterization of the SCCmec elements is based on two different gene prediction approaches to achieve correct annotation.
The SCC Elements
Although due to its clinical relevance, SCCmec is the most popular element, several other SCC elements not carrying the mec complex (SCC) or either mec complex and ccr complex (pseudo-SCC) have been identified at the orfX site. These elements can carry diverse genes relevant for staphylococcal survival and virulence, namely conferring heavy metal resistance genes (Chongtrakool et al., 2006) providing capsule production (Luong et al., 2002), cell-wall biosynthesis (Mongkolrattanothai et al., 2004), restriction/modification functions or immune protection (Holden et al., 2004). These elements can be found alone in the chromosome or in tandem with SCCmec or other SCC elements, being in this case named composite islands (CI). Examples of such CIs include the SCCmec III-SCCmer from the pandemic MRSA Brazilian clone (Chongtrakool et al., 2006) and the SCCmec IV-ACME from the USA300 CA-MRSA clone (Shore et al., 2011b). Although many studies have described the structure and contents of these elements, few studies have addressed their true contribution for staphylococcal virulence or fitness (Diep et al., 2008).
Besides being inserted at the exact same location as SCCmec, SCC and pseudo-SCC elements have been described to have regions of homology with SCCmec (Katayama et al., 2003a) (see Table 2), suggesting that their evolutionary history is related. Still, until recently the nature of their relatedness remained elusive.
Table 2.
Nucleotide identity (%) of SCC elements found in S. sciuri, S. vitulinus and S. fleurettii with S. aureus SCCmec.
| SCC element | Species | ccr type | mec complex | Homology with S. aureus SCCmec region (%) |
|---|---|---|---|---|
| SCC11/01 | S. sciuri | ccrA5B5 | – | J1: IVa (46), SCCmec S. xylosus (mecC) (59) |
| J2: III (67–96), IX (79–93) | ||||
| J3: IX (69–91) | ||||
| SCCK6-15937 | S. sciuri | ccrA5Bnew | – | J1:III(90), V(79) |
| J2: III (95) | ||||
| J3: IX (99) | ||||
| ψ-SCCK6-10930 | S. sciuri | – | Jl: V (65) | |
| J3: III (55–80) | ||||
| SCCmec III-like B | S. sciuri | ccrA3B5 | A | J2: II (99–100), III (60–100). IX (73) J3: III (100) |
| SCCmec III-like A | S. sciuri | ccrA3B5 | A | J1: X (96), III (90–100) |
| J2: III (38–100) | ||||
| J3: IV (42) | ||||
| SCCK116 | S. sciuri | ccrC | – | J2: II (90), III (91–98), V (73–80) |
| SCC-CISS27-I SCC-CISS27 - II | S. sciuri | ccrA1B3 ccrA1B3 | J2: III(77), X(90) | |
| Jl: IX (77), XI (73) | ||||
| J2: II (94), IV (62–79), IX (73–97) | ||||
| SCC402567 | S. fleuretti | ccrAnewB5 | – | J2: IX (75), III (77–96) |
| SCC11683 | S. vitulinus | ccrABnew | – | J1: IX (56–95) J2: IX (97) |
| SCC401946 | S. vitulinus | ccrABnew | – | J1: IX (76), V (82) J3: IV (51) |
Transfer of SCC Elements
SCCmec with exactly the same nucleotide sequence were found in different staphylococcal strains and species, suggesting that this element is frequently transferred among staphylococci (Shore et al., 2011b). However, the mechanisms of SCC and SCCmec insertion and excision from the chromosome as well as the mechanism of transfer is still elusive and it is likely that several different mechanisms are involved.
When grown in antibiotic free medium SCCmec can be excised from the chromosome, a reaction catalyzed by the cassette chromosome recombinases (Ccr) (Katayama et al., 2000). Immediately after excision, SCCmec circularizes into an extrachromosomal intermediate form and the attSCC sites are created in the chromosome (attB) and the intermediate circular form (attS). The created attSCC sites, work like recognition sites for the Ccr enzymes for a later integration into the chromosome. During SCCmec excision and insertion orfX remains always intact (Katayama et al., 2000).
Electrophoretic mobility shift assay (EMSA) showed that Ccr enzymes recognize a minimum of 14-bp sequence in the terminal sequence of the orfX that contains always the conserved sequence (TATCATAA), which is also found in SCCmec (attS). However, DNA sequences flanking the att sites were shown to also have a role on the frequency and efficiency of SCCmec insertion (Wang et al., 2012). This sequence specificity was thus probably important in the epidemiology of SCCmec acquisition by staphylococci and might partly explain the association of some SCCmec to specific genetic backgrounds or staphylococcal species (Oliveira et al., 2002; Miragaia et al., 2007; Bouchami et al., 2010, 2012; Rolo et al., 2017a).
According to studies of translational fusions of the ccr promoter with green fluorescent protein the ccr activity and associated SCCmec excision is a bistable process occurring only in a small fraction of cells within the population (Stojanov et al., 2013). The fate of the extrachromosomal circularized SCC elements after they are formed is still a mystery. SCC elements have once been regarded as non-replicative, due to the absence of a replication origin, but recent crystallographic studies have provided evidence that SCCmec elements encode an active MCM-like helicase (Mir-Sanchis et al., 2016), suggesting that they can eventually replicate in the cytoplasm before being transferred, but further studies are needed to confirm these observations.
The transfer of SCCmec have been successfully achieved in the laboratory by several different genetic mechanisms. In particular susceptible strains in contact with phage lysates containing this mobile genetic element were shown to become resistant to β-lactams and SCCmec elements were successfully packaged into bacteriophage capsides (Cohen and Sweeney, 1970; Maslanova et al., 2013; Scharn et al., 2013; Chlebowicz et al., 2014; Haaber et al., 2016). Additionally, transfer of SCCmec was also achieved through natural transformation, upon induction of SigH in very specific laboratory growth conditions (Morikawa et al., 2012). Moreover, a chromosomally encoded and laboratory-constructed derivative of SCCmec was captured on a conjugative plasmid and transferred by filter-mating into different S. aureus and S. epidermidis recipients (Ray et al., 2016). Also, evidence of possible transfer of SCCmec by conjugation was the finding of a mecA homologue within a plasmid of Macrococcus caseolyticus, a species phylogenetically related to Staphylococcus (Tsubakishita et al., 2010a).
In spite of the huge effort of the scientific community in elucidating the mechanism of SCCmec transfer, many of the studies described occurred in very artificial conditions and it remains to be clarified which mechanism(s) are actually more frequent in vivo.
The Origin of the Methicillin Resistance Determinant – mecA Evolution in S. sciuri Group
Early studies, based on structural nucleotide identity, have proposed that the mecA gene originated from recombination between a PBP from E. coli (Song et al., 1987) or from Enterococcus hirae (Archer and Niemeyer, 1994) with a β-lactamase encoding gene. A theory that was later supported by the finding by WGS of another mec allotype (mecC) as part of a class E mec complex, containing blaZ (mecI-mecR1-mecC-blaZ) in Macrococcus caseolyticus (Tsubakishita et al., 2010a) and S. xylosus (Harrison et al., 2013).
Other lines of evidence suggest that mecA originated from native PBPs of species of the Staphylococcus sciuri group – a primordial phylogenetic clade, including S. sciuri, Staphylococcus fleurettii, Staphylococcus vitulinus, Staphylococcus lentus, and Staphylococcus stepanovicci (Schleifer et al., 1983; Couto et al., 1996; Zhou et al., 2008; Antignac and Tomasz, 2009; Hauschild et al., 2010; Tsubakishita et al., 2010b), which most important ecological niches are the soil and skin and mucous membranes of wild and production animals.
The use of WGS on a large collection of isolates belonging to the S. sciuri group revealed the presence of homologues of S. aureus mecA with different levels of homology that were ubiquitous within some of the species (Couto et al., 1996; Wu et al., 1996; Hiramatsu et al., 2013) (see Figure 2A). However, in contrast to mecA in S. aureus, mecA homologs in S. sciuri group species were all located approximately 200 Kb apart from orfX (native location) between mva and xyl operons, outside any SCC element (Rolo et al., 2017b) (see Figure 2B). The results suggest that mecA has been transmitted vertically during the early stages of staphylococcal speciation (see Figure 2A; Step 1, Figure 3).
FIGURE 2.
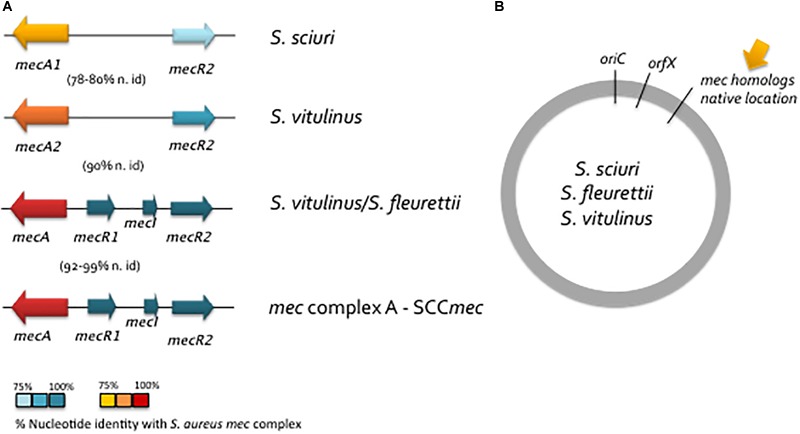
Schematic representation of the structure of mec native region in S. sciuri group species. (A) Structure of mec native region in S. sciuri, S. vitulinus and S. fleurettii in comparison to mec complex A in S. aureus. Colors indicate the level of identity of the mec complex A from S. aureus with the corresponding region in S. sciuri species group as depicted in Figure legend. (B) Location of the mec native region in the S. sciuri group of species.
FIGURE 3.
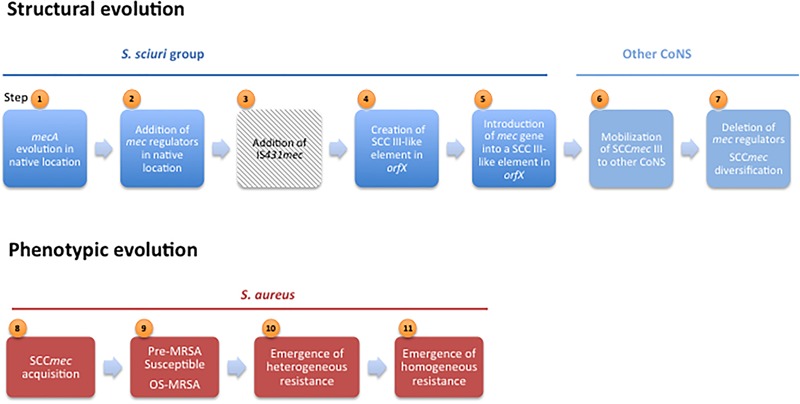
Steps in structural and phenotypic evolution of mecA-mediated β-lactam resistance.
Although the primary function of these mecA precursors was probably related to cell wall synthesis and not to antimicrobial resistance, the complete evolution from a native PBP into a resistance determinant appears to have been a stepwise process that occurred within this group of species. S. sciuri carries the most ancestral form of mecA (mecA1) which has 85% homology in nucleotide sequence with S. aureus mecA; S. vitulinus harbors an intermediary form (mecA2) with 94% homology and all S. fleurettii and some S. vitulinus have a mecA form that is almost identical to that of S. aureus mecA (mecAf, mecAv; 99% homology) (Rolo et al., 2017b; Tsubakishita et al., 2010b) (see Figure 2 and Table 3). In S. lentus and S. stepanovicii so far no mecA homolog has been described, (Tsubakishita et al., 2010b; Calazans-Silva et al., 2014), but no extensive and detailed study was ever performed in these two species.
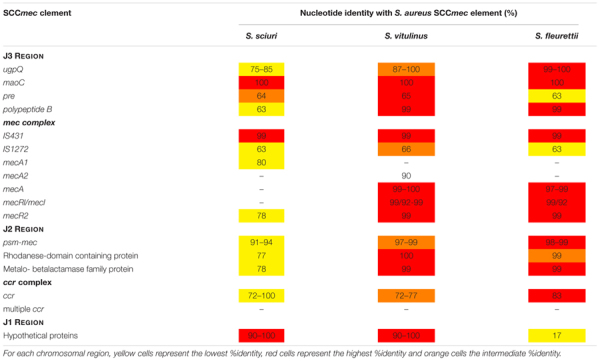
Table 3.
Nucleotide identity (%) of chromosoomal regions of S. sciuri, S. fleurettii and S. vitulinus with those found within S. aureus SCCmec.
 |
The only mecA homologue that confers resistance to β-lactams is the mecA in S. fleuretti. The same mecA homologue in S. vitulinus and the mecA1 and mecA2 found in S. sciuri and S. vitulinus, respectively, do not confer resistance to β-lactams in the great majority of strains (Couto et al., 1996; Wu et al., 1998; Monecke et al., 2012; Rolo et al., 2017b). However, S. sciuri and S. vitulinus strains exhibiting β-lactam resistance have been reported (Couto et al., 2003; Tsubakishita et al., 2010b; Rolo et al., 2017b). Recent work by Rolo et al. (2017b) wherein a large collection of S. sciuri and S. vitulinus were analyzed by WGS showed that β-lactam resistance in these species emerged multiple times during evolution and was driven mainly by the contact with human created environments, namely with the beginning of the use of antibiotics in production animals and humans. The mechanisms of resistance development in these two species included: (i) the structural diversification of the non-binding domain of native PBPs which altered the structure of the active site and exposure of ser403; (ii) mutations and insertion of IS256 in the promoters of mecA homologs that were associated to an increased expression of the proteins; (iii) acquisition of SCCmec. Additionally, like in S. aureus, the bacterial genetic background plays an important role in the expression of β-lactam resistance in the S. sciuri group of species, since the exact same gene allele was found associated to both susceptible and resistant strains (Rolo et al., 2017b).
Additional evidence supporting that S. sciuri mecA1 was the evolutionary precursor of S. aureus mecA, include the fact that this gene could be recruited to express methicillin resistance in S. sciuri after stepwise exposure to methicillin (Wu et al., 2001). Moreover, the activated copy of S. sciuri mecA1 was, similarly, able to restore methicillin resistance phenotype, when transduced into methicillin-susceptible S. aureus (MSSA), conferring high level, homogeneous and broad-spectrum β-lactam resistance (Wu et al., 2001). Furthermore, the S. sciuri mecA1 when transduced into MSSA was shown to act exactly like S. aureus mecA, being controlled by S. aureus mecA regulators (mecI and mecR1) and its product (PBP4) taking part in cell wall biosynthesis, producing a peptidoglycan typical of methicillin-resistant S. aureus (Antignac and Tomasz, 2009).
Besides mecA, other mec genes have been identified that are associated with β-lactam resistance, namely mecB and mecD in Macrococcus caseolyticus (Baba et al., 2009; Schwendener et al., 2017; Schwendener and Perreten, 2018) and mecC in S. aureus (Garcia-Alvarez et al., 2011; Shore et al., 2011a), S. xylosus (Harrison et al., 2013), S. sciuri carnaticus (Harrison et al., 2014), and S. stepanovicii (Loncaric et al., 2013). The mecB and mecD are the most distant from S. aureus mecA, having, respectively, a nucleotide identity with mecA that is equal or lower than 62%, whereas mecC has 69% nucleotide sequence identity. All mec forms confer resistance to β-lactams to their natural hosts and their introduction into a susceptible S. aureus genetic background was able to provide a resistance phenotype, confirming that they should encode a PBP with low-affinity to β-lactams that participates in cell wall synthesis (Baba et al., 2009; Kim et al., 2012). Both mecB and mecC were carried within mobile genetic elements structurally similar to SCCmec that were inserted in the orfX region (SCCmec XI, SCCmec IX-like) (Gomez-Sanz et al., 2015) and mecB was additionally found within a plasmid in M. caseolyticus (Baba et al., 2009; Tsubakishita et al., 2010a). The mecD gene is carried within a resistance island (McRImecD-1, McRImecD-2) that is inserted in 3′ end of the rpsI gene. Besides mecD this island contains genes for an integrase of the tyrosine recombinase family, but does not resemble either SCC elements or mecB-carrying mobile genetic elements (Schwendener et al., 2017; Schwendener and Perreten, 2018). However, none of the mecB, mecC or mecD was found within the native location (200 Kb apart from orfX).
The exact evolutionary link between mecA, mecB, mecC and mecD forms is still undetermined. Among all mec genes, mecA is apparently, the most successful in Staphylococcus. The mecB was recently found within a plasmid in a single S. aureus human carriage strain belonging to ST7 (Becker et al., 2018) and mecC has been limited to only a few S. aureus clonal lineages (CC130 and ST425) and four Staphylococcus species (S. sciuri, S. xylosus, S. stepanovicci, and S. aureus) (Harrison et al., 2013, 2014; Loncaric et al., 2013; Becker et al., 2014; Semmler et al., 2016). MRSA harboring mecC are believed to have a zoonotic origin and although they were reported in several different countries, they have been rarely observed in human infection (Becker et al., 2014). However, surveillance of dissemination of these mec genes should not be disregarded, since antibiotic use and the consequent selective pressure could drive fast evolutionary leaps that can lead to their precipitous spread.
Stages in the Evolution of SCCmec
Most of the efforts have been focused on the clarification of the origin and evolution of the β-lactam resistance determinant (mecA). Much less information is available regarding the evolution of SCCmec, the mobile element carrying mecA, which is responsible for the worldwide spreading of β-lactam resistance among staphylococci. SCCmec is a mosaic-like element that was described to contain multiple transposable elements, plasmids and insertion sequences in J regions (Ito et al., 2001), a genetic environment that per si can promote and facilitate genetic variation and recombination, what has been hindering the reliable tracing of their phylogeny.
The characterization of the native location of mecA homologs, the SCC insertion site and the genetic background of a large collection of isolates belonging to S. sciuri group by comparative genomics showed that SCC elements and mecA and flanking regions evolved in parallel in these species in these two distinct chromosomal locations (Rolo et al., 2017b).
Assembly of the mec Complex in the Native Location
The mecA homologs flanking genes in the native location were found to be the same as those flanking mecA inside SCCmec, encompassing the J2 and J3 regions (Rolo et al., 2017b) (see Table 3). Moreover, as for native mecA homologs, the level of homology of their flanking genes (psm-mec and ugpQ) with the same genes in SCCmec from MRSA, varied according to the phylogeny, wherein those of S. fleuretti were the most similar and those of S. sciuri were the most distant (Rolo et al., 2017b). The results suggest that the first stage of SCCmec evolution included the evolution of mecA homologs and their neighbor genes in the native location (Step 1, Figure 3). This was followed by the creation of the mec complex (Step 2, Figure 3). The mecR2 was the first regulator to be added in S. sciuri at the native location near mecA1, since the most ancient precursor of mecR2 was found in this species. This gene organization was preserved along phylogeny and became ubiquitous in S. fleurettii and S. vitulinus (Tsubakishita et al., 2010b). Addition of mecR1 and mecI happened later, after the evolution of the ancestral mecA1 into mecA was complete, as demonstrated by the lack of these regulators in S. sciuri and their occurrence in the native location of S. fleurettii and S. vitulinus near mecA (Tsubakishita et al., 2010b; Rolo et al., 2017b). These finding came to reconcile previous controversies, suggesting that although mecA1 was the original precursor of mecA, S. fleuretti/S. vitulinus were probably the last donors of the mec complex to give rise to SCCmec. The addition of IS431 element probably occurred later, after mecA, regulators and neighboring regions were mobilized into a SCC element located in the orfX region (Step 3, Figure 3). Alternatively, it could have been added during their mobilization, as it was never detected in the native location in any of the strains tested.
Acquisition and expression of mecA in species in which this gene is not native imposes a fitness cost to bacteria (Ender et al., 2004). For this reason, the step of addition of regulators with subsequent mecA repression appears to have been particularly crucial in the maintenance of the gene in new host species (Katayama et al., 2003b) and thus in mecA dissemination. In fact, some of the first methicillin resistant staphylococci, the so-called pre-MRSA (Step 9, Figure 3) (Hiramatsu et al., 1992; Kuwahara-Arai et al., 1996), contained intact regulators and a susceptible phenotype.
Studies wherein the mec complex region was characterized in MRSA and MR-CoNS revealed that although mecI and the 3′ end of mecR1 are deleted in a great proportion of contemporary clinical strains (Katayama et al., 2001), the 5′ end portion of mecR1 as well as a copy of IS431 downstream mecA (IS431-R) are conserved in every strain. Moreover, deletion of mecI and mecR1 promoted by IS431 was accomplished in vitro upon selection with methicillin in a S. haemolyticus with a mec complex type A (Suzuki et al., 1993). These observations are in accordance with the view that the mec regulators and the IS431-R together with mecA were once the original components of the mec region DNA and that deletion of the regulators occurred at a later time in evolution (Step 7, Figure 3). On the other hand, the similarity of nucleotide sequence in regions located upstream of the mec complex in four different mec complex classes, suggests that deletion of the mec regulators must have occurred after the establishment of the prototypic mec complex A in a SCCmec element (Katayama et al., 2001).
Thus the assembly of mec complex A seems to be the first step of genetic evolution, followed by its establishment in SCCmec and subsequent deletion of the regulators to originate the different mec complex classes (mec complex B, C, D, E).
SCC Element Evolution in the orfX Region
Analysis of the S. sciuri orfX region showed that SCC elements most probably originated in S. sciuri and were assembled from housekeeping genes located in this region (Step 4, Figure 3), as evidenced by the finding of the same housekeeping genes either outside or within SCC elements (Rolo et al., 2017a) – a finding not observed in the other species of the S. sciuri group. Additionally, in S. sciuri the most ancestral forms of cassette chromosome recombinases (ccr) and the highest genetic diversity were found, including almost all ccr allotypes described in S. aureus (Rolo et al., 2014). Interestingly, it was also in this species that a SCCmec type III-like was found with high homology simultaneously with the mec complex and J2 region of S. aureus SCCmec type III and the J1 region and an ancestral form of ccrAB3 of S. sciuri SCC non-mec (Rolo et al., 2017a) (see Table 2). The results suggest that SCCmec III originated in S. sciuri, probably through the integration of the mec complex and J2 region from S. vitulinus/S. fleuretti into a resident SCC non-mec carrying ccrAB3 (Step 5, Figure 3). However, the mechanism that mobilized the mec complex from S. vitulinus/S. fleuretti to an SCC in S. sciuri is still not known. Once formed SCCmec III probably disseminated to other CoNS species, namely S. epidermidis and S. aureus ST239, wherein SCCmec III was found to be prevalent (Miragaia et al., 2007; Harris et al., 2012) (Step 6, Figure 3).
Although it is apparent that the origin of SCCmec type III is S. sciuri, the source of the remaining SCCmec types elements remains unclear. In contrast to S. sciuri, which carried a high diversity of ccr allotypes, methicillin susceptible CoNS species belonging to more recent clades in the phylogeny of staphylococci, which include Staphylococcus epidermidis, Staphylococcus haemolyticus and Staphylococcus hominis, were particularly enriched in a specific allotype of ccr. The ccrAB2 was found to be common in S. epidermidis (Miragaia et al., 2007), ccrAB1 in S. hominis (Bouchami et al., 2012) and ccrC in S. haemolyticus (Bouchami et al., 2010), which is coincidently the same type of ccr carried by the most frequent SCCmec in these species. It is thus tempting to speculate that each SCCmec type can result from the integration of mec complex, probably through recombination, into a resident SCC element in these species. This hypothesis is supported by the identification of SCC non-mec elements carrying different ccrAB types in S. aureus and CoNS with regions of high homology with known SCCmec types (Katayama et al., 2003a). However, the enrichment of certain SCCmec types in particular species may also derive from the described specificity of the different types of Ccr enzymes as described above (Wang et al., 2012).
SCCmec Diversification
The next stage of SCCmec evolution that is believed to be still ongoing includes the diversification and dissemination of the SCCmec element among the staphylococcal population (Step 7, Figure 3). The existence of similar regions among different SCCmec types, like the J1 region in SCCmec type II and IV or the mec complex B between SCCmec type I and IV (Chongtrakool et al., 2006) suggest that the different SCCmec types are related.
The involvement in the diversification process of species, such as S. epidermidis, S. hominis and S. haemolyticus is apparent. Besides being reservoirs of specific types of SCCmec and ccr allotypes, these species harbor a huge number of non-described SCCmec types (Wisplinghoff et al., 2003; Miragaia et al., 2007; Bouchami et al., 2010), evidencing their key role in the current diversification of SCCmec.
Moreover, factors associated to hospital environment appear to be driving the diversification and acquisition of SCCmec in species like S. epidermidis (Rolo et al., 2012). One of the factors related to the clinical setting that might be triggering SCCmec diversification is the use of antibiotics, namely β-lactams and vancomycin, which were already shown to promote the expression of recombinases (Higgins et al., 2009). The excision of SCC elements promoted by ccr overexpression may create new opportunities of recombination between different elements within the same strain, giving rise to new SCCmec structures.
The Importance of Genetic Background for the Acquisition of SCCmec and for the Expression of β-Lactam Resistance in S. aureus
The transfer of SCCmec from CoNS to S. aureus was probably a subsequent step (Step 8). Several lines of evidence suggest that acquisition and expression of mecA by S. aureus was a complex process involving multiple genetic and metabolic alterations. The construction of a Tn551 transposon library in the background of the MRSA strain COL and subsequent screening for a decreased level of methicillin resistance has identified several factors (fem or auxiliary genes) that, together with the mecA gene, are crucial for the expression of high-level and homogeneous resistance to methicillin (de Lencastre and Tomasz, 1994) (Step 11, see Figure 3). Although having a substantial impact on oxacillin resistance, these genes are not directly implicated in the expression of mecA, but they are mainly involved in cell wall metabolism and stress response (de Lencastre and Tomasz, 1994). However, these studies were performed in a single MRSA strain (COL) and these same genes appear to have different contributions to β-lactams resistance in other S. aureus genetic backgrounds (Memmi et al., 2008; Figueiredo et al., 2014), suggesting that expression of β-lactam resistance is extremely complex and that auxiliary genes in different MRSA strains might be different or use different mechanisms. Additionally, whether the identified auxiliary genes in COL also contribute to β-lactam resistance expression in other Staphylococcus species is unknown.
What appears to hold true is that not all S. aureus appear to have the same ability to accommodate mecA. The existence of a host barrier was evidenced by the finding that SCCmec was acquired by a limited number of S. aureus genetic backgrounds (e.g., ST239, ST45, ST22, ST8, ST5) (Robinson and Enright, 2003) while other genetic backgrounds despite being successful, like ST121, were rarely observed carrying mecA (Rao et al., 2015). Furthermore, when a recombinant plasmid, carrying intact mecA, was introduced into strains that have never experienced the presence of mecA, they were unable to maintain or express β-lactam resistance, a phenomena not observed when the same assay was performed in MSSA strains from which SCCmec has been excised (Katayama et al., 2003b). Interestingly, either the presence of β-lactamase (blaR1-blaI) or mecA regulatory genes (mecR1-mecI), which control mecA expression, allowed the maintenance and expression of plasmid-carried mecA in the naïve genetic background (Katayama et al., 2003b), which is indicative that besides the genetic backgrounds the repression of mecA was important for the acquisition and stability of mecA in staphylococci. Actually, the so-called pre-MRSA although carrying mecA, showed a susceptible phenotype, which was shown to result from mecI-mediated repression of mecA transcription (Kuwahara-Arai et al., 1996) (Step 9, Figure 3). The integration of newly acquired genes into the recipient metabolic network is a complex mechanism that frequently represents a large fitness cost for bacteria. The presence of the regulators will probably work as safeguard mechanisms that will silence the newly acquired gene and avoid potentially harmful consequences of its expression in the new bacterial host, while it is still not adapted (Ochman et al., 2000; Navarre et al., 2006).
A different phenomenon supporting the importance of genetic background for the expression of β-lactam resistance is the emergence of the so-called oxacillin susceptible MRSA (OS-MRSA) (Step 9, Figure 3), strains that like pre-MRSA carry mecA and do not express β-lactam resistance, but that in contrast do not carry mecI (SCCmec IV or V) (Giannouli et al., 2010; Andrade-Figueiredo and Leal-Balbino, 2016; Phaku et al., 2016). OS-MRSA have been recently described as a cause of infections in humans (Andrade-Figueiredo and Leal-Balbino, 2016) and have been also isolated from animals (Phaku et al., 2016). Functional and genomic analysis of OS-MRSA and MRSA strains identified mutations in femA, a known auxiliary gene, as the possible cause of the observed decreased resistance to β-lactams (Giannouli et al., 2010; Phaku et al., 2016). Although being described many years after the emergence of pre-MRSA, the exact date of OS-MRSA emergence is uncertain. Actually, since for several decades detection of MRSA in many hospitals was based in purely phenotypic approaches, OS-MRSA may have passed unnoticed. It could be that OS-MRSA correspond to strains that have recently acquired mecA and that have developed alternative mechanisms to compensate for the cost of acquisition of an exogenous gene.
Altogether, data suggest that for acquiring and maintaining mecA, S. aureus strains had to adapt its genetic background, compensated for mecA/SCCmec fitness cost, or were already intrinsically equipped for it. Still it remains to be clarified which genetic determinants and mechanism are involved in this adaptation process.
Homogeneous and Heterogeneous Expression of Resistance to Methicillin
The genetic alterations in genetic backgrounds and associated metabolic alterations described above to have occurred upon SCCmec acquisition were frequently paralleled by alterations in the cell population profile of β-lactam resistance expression. Clinical MRSA isolates, when cultured, frequently exhibit a low level of methicillin resistance, but contain subpopulations of bacteria displaying very high levels of resistance to this antibiotic, a feature called heterogeneous resistance (Tomasz et al., 1991) (Step 10, Figure 3). Exposure of the hetero-MRSA strains to β-lactam antibiotics originates mutant strains in which all cells are uniformly highly resistant to β-lactams, named homogeneous methicillin resistance (Step 11, Figure 3) (Tomasz et al., 1991). Both hetero and homo resistance phenotypes can be found in clinical MRSA isolates, but they appear to correspond to two different and sequential evolutionary stages of β-lactams resistance expression. However, the molecular basis of the emergence of heterogeneous resistance and of the heterogeneous-to-homogeneous conversion is not totally understood and appears to derive by multiple different mechanism, of which only a few have yet been identified.
Genetic analysis of colonies within the highly resistant subpopulation of a heteregeneous MRSA strains, showed that high resistance was associated to the deletion of lytH, encoding a putative lytic enzyme homologous to a N-acetylmuramyl-L-alanine amidase (Fujimura and Murakami, 1997). But other mutations have been identified to provide the same type of phenotype, like mutations in mecI or in its promoter (Kondo et al., 2001). More recently, the comparison of the whole genome of strains selected from high and low level resistant subpopulations identified, in highly resistant strains, two additional mutations in relA, which is involved in the synthesis of (p)ppGppas, an effector of the stringent stress response to many environmental and genetic changes (Mwangi et al., 2013).
Ryffel et al. (1990) and Berger-Bachi and Rohrer (2002) first hypothesized that the heterogeneous-to-homogeneous conversion of methicillin resistance results from a spontaneous chromosomal mutation that is not linked to mecA. Kondo et al. (2001) showed by in vitro trans-complementation studies that hmrA and hmrB, which encode a putative aminohydrolase and an acyl carrier protein, respectively, were responsible for the conversion of the heterogenous profile (eagle type) of the N315 pre-MRSA strain into a uniformly highly resistant MRSA strain. Almost 20 years later a study wherein the whole genome of hetero-MRSA strain (N315) and its derivative homogeneously resistant strain selected by imipenem exposure, were compared confirmed that they differed in a single non-synonymous mutation in rpoB, encoding the RNA polymerase β subunit (Aiba et al., 2013). Furthermore more recently, WGS revealed that tandem amplification of the SCCmec near its integration site was another alternative mechanism driving the heterogenous-to-homogeneous conversion (Gallagher et al., 2017).
Concluding Remarks
The development of mecA-mediated resistance to β-lactams was induced by human use of β-lactam antibiotics both to treat human infections and feed additives and involved several key genetic events: (1) the evolution of a native gene into a resistance determinant occurring at the native location; (2) the evolution of the SCC elements occurring at the orfX region; (3) integration of the mec complex and neighboring regions into a SCC element; (4) the adaptation of the host bacteria genetic background; (5) dissemination of SCCmec among staphylococci colonizing animals; (6) dissemination of SCCmec among staphylococci colonizing both animals and humans. Strikingly, most of the events that lead to β-lactam resistance development have occurred within the group of the most primitive animal-related Staphylococcus species isolated from production animals or human infection, suggesting it was a bacterial survival strategy against the human use of antimicrobials. The jump of SCCmec from animal to human-associated Staphylococcus species, like S. aureus, was a key event leading to several worldwide pandemics.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was financially supported by: Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER funds through COMPETE2020 - Programa Operacional Competitividade e Internacionalização (POCI) and by national funds through FCT – Fundação para a Ciência e a Tecnologia; ONEIDA project (LISBOA-01-0145-FEDER-016417) co-funded by FEEI – “Fundos Europeus Estruturais e de Investimento” from “Programa Operacional Regional Lisboa 2020” and by national funds from FCT – “Fundação para a Ciência e a Tecnologia; project PTDC/FIS-NAN/0117/2014 and project PTDC/CVT-CVT/29510/2017 from Fundação para a Ciência e Tecnologia (FCT) and project EXPOSE Ref. 02/SAICT/2016, funded by Portugal 2020, projetos de Investigação Cientìfica e Desenvolvimento Tecnológico (IC&DT), Programa Operacional Regional do Norte e de Lisboa.
References
- Aiba Y., Katayama Y., Hishinuma T., Murakami-Kuroda H., Cui L., Hiramatsu K. (2013). Mutation of RNA polymerase beta-subunit gene promotes heterogeneous-to-homogeneous conversion of beta-lactam resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 57 4861–4871. 10.1128/AAC.00720-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminov R. (2017). History of antimicrobial drug discovery: major classes and health impact. Biochem. Pharmacol. 133 4–19. 10.1016/j.bcp.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Andrade-Figueiredo M., Leal-Balbino T. C. (2016). Clonal diversity and epidemiological characteristics of Staphylococcus aureus: high prevalence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with clinical isolates in Brazil. BMC Microbiol. 16:115. 10.1186/s12866-016-0733-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antignac A., Tomasz A. (2009). Reconstruction of the phenotypes of methicillin-resistant Staphylococcus aureus by replacement of the staphylococcal cassette chromosome mec with a plasmid-borne copy of Staphylococcus sciuri pbpD gene. Antimicrob. Agents Chemother. 53 435–441. 10.1128/AAC.01099-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer G. L., Niemeyer D. M. (1994). Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 2 343–347. 10.1016/0966-842X(94)90608-4 [DOI] [PubMed] [Google Scholar]
- Arede P., Milheirico C., de Lencastre H., Oliveira D. C. (2012). The anti-repressor MecR2 promotes the proteolysis of the mecA repressor and enables optimal expression of beta-lactam resistance in MRSA. PLoS Pathog. 8:e1002816. 10.1371/journal.ppat.1002816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armand-Lefevre L., Ruimy R., Andremont A. (2005). Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg. Infect. Dis. 11 711–714. 10.3201/eid1105.040866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Kuwahara-Arai K., Uchiyama I., Takeuchi F., Ito T., Hiramatsu K. (2009). Complete genome sequence of Macrococcus caseolyticus strain JCSCS5402, [corrected] reflecting the ancestral genome of the human-pathogenic staphylococci. J. Bacteriol. 191 1180–1190. 10.1128/JB.01058-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig S., Johannesen T. B., Overballe-Petersen S., Larsen J., Larsen A. R., Stegger M. (2018). Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 61 74–76. 10.1016/j.meegid.2018.03.013 [DOI] [PubMed] [Google Scholar]
- Barber M., Rozwadowska-Dowzenko M. (1948). Infection by penicillin-resistant staphylococci. Lancet 2 641–644. 10.1016/S0140-6736(48)92166-7 [DOI] [PubMed] [Google Scholar]
- Becker K., Ballhausen B., Kock R., Kriegeskorte A. (2014). Methicillin resistance in Staphylococcus isolates: the “mec alphabet” with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. Int. J. Med. Microbiol. 304 794–804. 10.1016/j.ijmm.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Becker K., van Alen S., Idelevich E. A., Schleimer N., Seggewiss J., Mellmann A., et al. (2018). Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg. Infect. Dis. 24 242–248. 10.3201/eid2402.171074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bachi B., Rohrer S. (2002). Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178 165–171. 10.1007/s00203-002-0436-0 [DOI] [PubMed] [Google Scholar]
- Berglund C., Ito T., Ikeda M., Ma X. X., Soderquist B., Hiramatsu K. (2008). Novel type of staphylococcal cassette chromosome mec in a methicillin-resistant Staphylococcus aureus strain isolated in Sweden. Antimicrob. Agents Chemother. 52 3512–3516. 10.1128/AAC.00087-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchami O., Ben Hassen A., de Lencastre H., Miragaia M. (2010). High prevalence of mec complex C and ccrC is independent of SCCmec type V in Staphylococcus haemolyticus. Eur. J. Clin. Microbiol. Infect. Dis. 31 605–614. 10.1007/s10096-011-1354-3 [DOI] [PubMed] [Google Scholar]
- Bouchami O., Ben Hassen A., de Lencastre H., Miragaia M. (2012). Molecular epidemiology of methicillin-resistant Staphylococcus hominis (MRSHo): low clonality and reservoirs of SCCmec structural elements. PLoS One 6:e21940. 10.1371/journal.pone.0021940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boundy S., Safo M. K., Wang L., Musayev F. N., O’Farrell H. C., Rife J. P., et al. (2013). Characterization of the Staphylococcus aureus rRNA methyltransferase encoded by orfX, the gene containing the staphylococcal chromosome Cassette mec (SCCmec) insertion site. J. Biol. Chem. 288 132–140. 10.1074/jbc.M112.385138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calazans-Silva A. C., Medeiros P. T., Araujo D. M., Carvalho B. O., Coelho I. S., Coelho S. M., et al. (2014). Genetic analysis of mecA gene and detection of homologue pbpD in Staphylococcus sciuri group. Braz. J. Microbiol. 45 651–655. 10.1590/S1517-83822014000200038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon J. I. (2007). History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. 86 2466–2471. 10.3382/ps.2007-00249 [DOI] [PubMed] [Google Scholar]
- Chambers H. F., Deleo F. R. (2009). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7 629–641. 10.1038/nrmicro2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowicz M. A., Maslanova I., Kuntova L., Grundmann H., Pantucek R., Doskar J., et al. (2014). The Staphylococcal Cassette Chromosome mec type V from Staphylococcus aureus ST398 is packaged into bacteriophage capsids. Int. J. Med. Microbiol. 304 764–774. 10.1016/j.ijmm.2014.05.010 [DOI] [PubMed] [Google Scholar]
- Chongtrakool P., Ito T., Ma X. X., Kondo Y., Trakulsomboon S., Tiensasitorn C., et al. (2006). Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 50 1001–1012. 10.1128/AAC.50.3.1001-1012.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S., Sweeney H. M. (1970). Transduction of methicillin resistance in Staphylococcus aureus dependent on an unusual specificity of the recipient strain. J. Bacteriol. 104 1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Continuous Discharge Certificate [CDC] (1999). Four pediatric deaths from community-acquired methicillin resistant Staphylococcus aureus - Minnesota and North Dakota, 1997-1999. MMWR 48 707–710. [PubMed] [Google Scholar]
- Couto I., de Lencastre H., Severina E., Kloos W., Webster J. A., Hubner R. J., et al. (1996). Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb. Drug Resist. 2 377–391. 10.1089/mdr.1996.2.377 [DOI] [PubMed] [Google Scholar]
- Couto I., Wu S. W., Tomasz A., de Lencastre H. (2003). Development of methicillin resistance in clinical isolates of Staphylococcus sciuri by transcriptional activation of the mecA homologue native to s. J. Bacteriol. 185 645–653. 10.1128/JB.185.2.645-653.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley K. B., Archer G. L. (1997). The Staphylococci in Human Disease. London: Churchill Livingstone. [Google Scholar]
- de Lencastre H., Tomasz A. (1994). Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 38 2590–2598. 10.1128/AAC.38.11.2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenberg R. H., Stobberingh E. E. (2008). The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8 747–763. 10.1016/j.meegid.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Deurenberg R. H., Stobberingh E. E. (2009). The molecular evolution of hospital- and community-associated methicillin-resistant Staphylococcus aureus. Curr. Mol. Med. 9 100–115. 10.2174/156652409787581637 [DOI] [PubMed] [Google Scholar]
- Diep B. A., Stone G. G., Basuino L., Graber C. J., Miller A., des Etages S. A., et al. (2008). The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 197 1523–1530. 10.1086/587907 [DOI] [PubMed] [Google Scholar]
- Ender M., McCallum N., Adhikari R., Berger-Bachi B. (2004). Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob. Agents Chemother. 48 2295–2297. 10.1128/AAC.48.6.2295-2297.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entenza J. M., Hohl P., Heinze-Krauss I., Glauser M. P., Moreillon P. (2002). BAL9141, a novel extended-spectrum cephalosporin active against methicillin-resistant Staphylococcus aureus in treatment of experimental endocarditis. Antimicrob. Agents Chemother. 46 171–177. 10.1128/AAC.46.1.171-177.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Union [EU] (2003). REGULATION (EC) No 1831/2003 OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL on additives for use in animal nutrition. Off. J. Eur. Union. L 268/29 1–30. (accessed Dec 30, 2005). [Google Scholar]
- Figueiredo T. A., Ludovice A. M., Sobral R. G. (2014). Contribution of peptidoglycan amidation to beta-lactam and lysozyme resistance in different genetic lineages of Staphylococcus aureus. Microb. Drug Resist. 20 238–249. 10.1089/mdr.2014.0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. (1929). Classics in infectious diseases: on the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae by Alexander Fleming, Reprinted from the British Journal of Experimental Pathology 10:226-236, 1929. Rev. Infect. Dis. 2 129–139. 10.1093/clinids/2.1.129 [DOI] [PubMed] [Google Scholar]
- Fluit A. C. (2012). Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 18 735–744. 10.1111/j.1469-0691.2012.03846.x [DOI] [PubMed] [Google Scholar]
- Food and Drug Administration [FDA] (2018). Antimicrobial Resistance. Avilable at: http://www.fda.gov/animalveterinary/safetyhealth/antimicrobialresistance/default.htm [Google Scholar]
- Fuda C., Suvorov M., Vakulenko S. B., Mobashery S. (2004). The basis for resistance to beta-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 279 40802–40806. 10.1074/jbc.M403589200 [DOI] [PubMed] [Google Scholar]
- Fujimura T., Murakami K. (1997). Increase of methicillin resistance in Staphylococcus aureus caused by deletion of a gene whose product is homologous to lytic enzymes. J. Bacteriol. 179 6294–6301. 10.1128/jb.179.20.6294-6301.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A., Coughlan S., Black N. S., Lalor P., Waters E. M., Wee B., et al. (2017). Tandem amplification of the staphylococcal cassette chromosome mec element can drive high-level methicillin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 61:e00869-17. 10.1128/AAC.00869-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alvarez L., Holden M. T., Lindsay H., Webb C. R., Brown D. F., Curran M. D., et al. (2011). Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 11 595–603. 10.1016/S1473-3099(11)70126-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuysen J. M. (1991). Serine beta-lactamases and penicillin-binding proteins. Annu. Rev. Microbiol. 45 37–67. 10.1146/annurev.mi.45.100191.000345 [DOI] [PubMed] [Google Scholar]
- Ghuysen J. M. (1994). Molecular structures of penicillin-binding proteins and beta-lactamases. Trends Microbiol. 2 372–380. 10.1016/0966-842X(94)90614-9 [DOI] [PubMed] [Google Scholar]
- Giannouli S., Labrou M., Kyritsis A., Ikonomidis A., Pournaras S., Stathopoulos C., et al. (2010). Detection of mutations in the FemXAB protein family in oxacillin-susceptible mecA-positive Staphylococcus aureus clinical isolates. J. Antimicrob. Chemother. 65 626–633. 10.1093/jac/dkq039 [DOI] [PubMed] [Google Scholar]
- Gomez-Sanz E., Schwendener S., Thomann A., Gobeli Brawand S., Perreten V. (2015). First staphylococcal cassette chromosome mec containing a mecB-carrying gene complex independent of transposon Tn6045 in a Macrococcus caseolyticus isolate from a canine infection. Antimicrob. Agents Chemother. 59 4577–4583. 10.1128/AAC.05064-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grave K., Greko C., Kvaale M. K., Torren-Edo J., Mackay D., Muller A., et al. (2012). Sales of veterinary antibacterial agents in nine European countries during 2005-09: trends and patterns. J. Antimicrob. Chemother. 67 3001–3008. 10.1093/jac/dks298 [DOI] [PubMed] [Google Scholar]
- Grundmann H., Aires-de-Sousa M., Boyce J., Tiemersma E. (2006). Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368 874–885. 10.1016/S0140-6736(06)68853-3 [DOI] [PubMed] [Google Scholar]
- Haaber J., Leisner J. J., Cohn M. T., Catalan-Moreno A., Nielsen J. B., Westh H., et al. (2016). Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat. Commun. 7:13333. 10.1038/ncomms13333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H., Cheng G., Iqbal Z., Ai X., Hussain H. I., Huang L., et al. (2014). Benefits and risks of antimicrobial use in food-producing animals. Front. Microbiol. 5:288 10.3389/fmicb.2014.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. R., Feil E. J., Holden M. T., Quail M. A., Nickerson E. K., Chantratita N., et al. (2012). Evolution of MRSA during hospital transmission and intercontinental spread. Science 327 469–474. 10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison E. M., Paterson G. K., Holden M. T., Ba X., Rolo J., Morgan F. J., et al. (2014). A novel hybrid SCCmec-mecC region in Staphylococcus sciuri. J. Antimicrob. Chemother. 69 911–918. 10.1093/jac/dkt452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison E. M., Paterson G. K., Holden M. T., Morgan F. J., Larsen A. R., Petersen A., et al. (2013). A Staphylococcus xylosus isolate with a new mecC allotype. Antimicrob. Agents Chemother. 57 1524–1528. 10.1128/AAC.01882-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. (1984). Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild T., Stepanovic S., Zakrzewska-Czerwinska J. (2010). Staphylococcus stepanovicii sp. nov., a novel novobiocin-resistant oxidase-positive staphylococcal species isolated from wild small mammals. Syst. Appl. Microbiol. 33 183–187. 10.1016/j.syapm.2010.03.004 [DOI] [PubMed] [Google Scholar]
- Herold B. C., Immergluck L. C., Maranan M. C., Lauderdale D. S., Gaskin R. E., Boyle-Vavra S., et al. (1998). Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279 593–598. 10.1001/jama.279.8.593 [DOI] [PubMed] [Google Scholar]
- Higgins P. G., Rosato A. E., Seifert H., Archer G. L., Wisplinghoff H. (2009). Differential expression of ccrA in methicillin-resistant Staphylococcus aureus strains carrying staphylococcal cassette chromosome mec type II and IVa elements. Antimicrob. Agents Chemother. 53 4556–4558. 10.1128/AAC.00395-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu K., Asada K., Suzuki E., Okonogi K., Yokota T. (1992). Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 298 133–136. 10.1016/0014-5793(92)80039-J [DOI] [PubMed] [Google Scholar]
- Hiramatsu K., Ito T., Tsubakishita S., Sasaki T., Takeuchi F., Morimoto Y., et al. (2013). Genomic basis for methicillin resistance in Staphylococcus aureus. Infect. Chemother. 45 117–136. 10.3947/ic.2013.45.2.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M. T., Feil E. J., Lindsay J. A., Peacock S. J., Day N. P., Enright M. C., et al. (2004). Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U.S.A. 101 9786–9791. 10.1073/pnas.0402521101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers P. M., Blaak H., de Jong M. C., Graat E. A., Vandenbroucke-Grauls C. M., de Roda Husman A. M. (2015). Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ. Sci. Technol. 49 11993–12004. 10.1021/acs.est.5b02566 [DOI] [PubMed] [Google Scholar]
- International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements [IWG-SCC], Ito, T. K., Hiramatsu, D. C., Oliveira, H., de Lencastre, K., Zhang, H., et al. (2009). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53 4961–4967. 10.1128/AAC.00579-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Matsunaga N., Tawada H., Kuroda N., Nakayama Y., Ishibashi Y., et al. (2003). TAK-599, a novel N-phosphono type prodrug of anti-MRSA cephalosporin T-91825: synthesis, physicochemical and pharmacological properties. Bioorg. Med. Chem. 11 2427–2437. 10.1016/S0968-0896(03)00126-3 [DOI] [PubMed] [Google Scholar]
- Ito T., Katayama Y., Asada K., Mori N., Tsutsumimoto K., Tiensasitorn C., et al. (2001). Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45 1323–1336. 10.1128/AAC.45.5.1323-1336.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Ma X. X., Takeuchi F., Okuma K., Yuzawa H., Hiramatsu K. (2004). Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48 2637–2651. 10.1128/AAC.48.7.2637-2651.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevons M. P. (1961). Celbenin-resistant’ staphylococci. Br. Med. J. 1 124–125. 10.1136/bmj.1.5219.124-a [DOI] [Google Scholar]
- Katayama Y., Ito T., Hiramatsu K. (2000). A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44 1549–1555. 10.1128/AAC.44.6.1549-1555.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y., Ito T., Hiramatsu K. (2001). Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-Mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin- resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45 1955–1963. 10.1128/AAC.45.7.1955-1963.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y., Takeuchi F., Ito T., Ma X. X., Ui-Mizutani Y., Kobayashi I., et al. (2003a). Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 185 2711–2722. 10.1128/JB.185.9.2711-2722.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y., Zhang H. Z., Hong D., Chambers H. F. (2003b). Jumping the barrier to beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 185 5465–5472. 10.1128/JB.185.18.5465-5472.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H., Hasman H., Larsen J., Stegger M., Johannesen T. B., Allesoe R. L., et al. (2018). SCCmecFinder, a Web-Based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 3:e00612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Milheirico C., Gardete S., Holmes M. A., Holden M. T., de Lencastre H., et al. (2012). Properties of a novel PBP2A protein homolog from Staphylococcus aureus strain LGA251 and its contribution to the beta-lactam-resistant phenotype. J. Biol. Chem. 287 36854–36863. 10.1074/jbc.M112.395962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos W. (1986). “Ecology of human skin,” in Coagulase-Negative Staphylococci eds Mardh P.-A., Schleifer K. H. (Stockholm: Almqvist & Wiksell; ). [Google Scholar]
- Kloos W. (1997). “Taxonomy and systematics of staphylococci indigenous to humans,” in The Staphylococci in Human Disease ed. Crossley K. B. (New York, NY: Churchill Livingstone; ) 113–137. [Google Scholar]
- Kloos W. E. (1980). Natural populations of the genus Staphylococcus. Annu. Rev. Microbiol. 34 559–592. 10.1146/annurev.mi.34.100180.003015 [DOI] [PubMed] [Google Scholar]
- Kondo N., Kuwahara-Arai K., Kuroda-Murakami H., Tateda-Suzuki E., Hiramatsu K. (2001). Eagle-type methicillin resistance: new phenotype of high methicillin resistance under mec regulator gene control. Antimicrob. Agents Chemother. 45 815–824. 10.1128/AAC.45.3.815-824.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara-Arai K., Kondo N., Hori S., Tateda-Suzuki E., Hiramatsu K. (1996). Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP 2’ production. Antimicrob. Agents Chemother. 40 2680–2685. 10.1128/AAC.40.12.2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Skov R. L., Han X., Larsen A. R., Larsen J., Sorum M., et al. (2011). Novel types of staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 55 3046–3050. 10.1128/AAC.01475-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D., Strynadka N. C. (2002). Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 9 870–876. [DOI] [PubMed] [Google Scholar]
- Lindsay J. A. (2013). Hospital-associated MRSA and antibiotic resistance-what have we learned from genomics? Int. J. Med. Microbiol. 303 318–323. 10.1016/j.ijmm.2013.02.005 [DOI] [PubMed] [Google Scholar]
- Loncaric I., Kubber-Heiss A., Posautz A., Stalder G. L., Hoffmann D., Rosengarten R., et al. (2013). Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J. Antimicrob. Chemother. 68 2222–2225. 10.1093/jac/dkt186 [DOI] [PubMed] [Google Scholar]
- Luong T. T., Ouyang S., Bush K., Lee C. Y. (2002). Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 184 3623–3629. 10.1128/JB.184.13.3623-3629.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. X., Ito T., Tiensasitorn C., Jamklang M., Chongtrakool P., Boyle-Vavra S., et al. (2002). Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46 1147–1152. 10.1128/AAC.46.4.1147-1152.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheboeuf P., Contreras-Martel C., Job V., Dideberg O., Dessen A. (2006). Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30 673–691. 10.1111/j.1574-6976.2006.00024.x [DOI] [PubMed] [Google Scholar]
- Maslanova I., Doskar J., Varga M., Kuntova L., Muzik J., Maluskova D., et al. (2013). Bacteriophages of Staphylococcus aureus efficiently package various bacterial genes and mobile genetic elements including SCCmec with different frequencies. Environ. Microbiol. Rep. 5 66–73. 10.1111/j.1758-2229.2012.00378.x [DOI] [PubMed] [Google Scholar]
- Memmi G., Filipe S. R., Pinho M. G., Fu Z., Cheung A. (2008). Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 52 3955–3966. 10.1128/AAC.00049-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miragaia M., Thomas J. C., Couto I., Enright M. C., de Lencastre H. (2007). Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing (MLST) data. J. Bacteriol. 189 2540–2552. 10.1128/JB.01484-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir-Sanchis I., Roman C. A., Misiura A., Pigli Y. Z., Boyle-Vavra S., Rice P. A. (2016). Staphylococcal SCCmec elements encode an active MCM-like helicase and thus may be replicative. Nat. Struct. Mol. Biol. 23 891–898. 10.1038/nsmb.3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke S., Muller E., Schwarz S., Hotzel H., Ehricht R. (2012). Rapid microarray-based identification of different mecA alleles in Staphylococci. Antimicrob. Agents Chemother. 56 5547–5554. 10.1128/AAC.00574-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolrattanothai K., Boyle S., Murphy T. V., Daum R. S. (2004). Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob. Agents Chemother. 48 1823–1836. 10.1128/AAC.48.5.1823-1836.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran G. J., Krishnadasan A., Gorwitz R. J., Fosheim G. E., McDougal L. K., Carey R. B., et al. (2006). Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355 666–674. 10.1056/NEJMoa055356 [DOI] [PubMed] [Google Scholar]
- Morikawa K., Takemura A. J., Inose Y., Tsai M., Nguyen Thi le T., Ohta T., et al. (2012). Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog. 8:e1003003. 10.1371/journal.ppat.1003003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi M. M., Kim C., Chung M., Tsai J., Vijayadamodar G., Benitez M., et al. (2013). Whole-genome sequencing reveals a link between beta-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb. Drug Resist. 19 153–159. 10.1089/mdr.2013.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (1980). Committee to Study the Human Health Effects of Subtherapeutic Antibiotic Use in Animal Feeds. Antibiotics In Animal Feeds, National Research Council (US) Committee to Study the Human Health Effects of Subtherapeutic Antibiotic Use in Animal Feeds The Effects on Human Health of Subtherapeutic Use of Antimicrobials in Animal Feeds. Washington, DC: National Academies Press. [PubMed] [Google Scholar]
- Navarre W. W., Porwollik S., Wang Y., McClelland M., Rosen H., Libby S. J., et al. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313 236–238. 10.1126/science.1128794 [DOI] [PubMed] [Google Scholar]
- Ochman H., Lawrence J. G., Groisman E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405 299–304. 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- Oliveira D. C., Milheirico C., de Lencastre H. (2006). Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob. Agents Chemother. 50 3457–3459. 10.1128/AAC.00629-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira D. C., Tomasz A., de Lencastre H. (2001). The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7 349–361. 10.1089/10766290152773365 [DOI] [PubMed] [Google Scholar]
- Oliveira D. C., Tomasz A., de Lencastre H. (2002). Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2 180–189. 10.1016/S1473-3099(02)00227-X [DOI] [PubMed] [Google Scholar]
- Otto M. (2013). Community-associated MRSA: what makes them special? Int. J. Med. Microbiol. 303 324–330. 10.1016/j.ijmm.2013.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phaku P., Lebughe M., Strauss L., Peters G., Herrmann M., Mumba D., et al. (2016). Unveiling the molecular basis of antimicrobial resistance in Staphylococcus aureus from the Democratic Republic of the Congo using whole genome sequencing. Clin. Microbiol. Infect. 22:644 e1–644.e5. [DOI] [PubMed] [Google Scholar]
- Pinho M. G., de Lencastre H., Tomasz A. (2001). An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. U.S.A. 98 10886–10891. 10.1073/pnas.191260798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Q., Shang W., Hu X., Rao X. (2015). Staphylococcus aureus ST121: a globally disseminated hypervirulent clone. J. Med. Microbiol. 64 1462–1473. 10.1099/jmm.0.000185 [DOI] [PubMed] [Google Scholar]
- Ray M. D., Boundy S., Archer G. L. (2016). Transfer of the methicillin resistance genomic island among staphylococci by conjugation. Mol. Microbiol. 100 675–685. 10.1111/mmi.13340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- report SE (2010/2015). Sales of veterinary antimicrobial agents in 30 European countries in 2015∗. [Google Scholar]
- Robinson D. A., Enright M. C. (2003). Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47 3926–3934. 10.1128/AAC.47.12.3926-3934.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolo J., de Lencastre H., Miragaia M. (2012). Strategies of adaptation of Staphylococcus epidermidis to hospital and community: amplification and diversification of SCCmec. J. Antimicrob. Chemother. 67 1333–1341. 10.1093/jac/dks068 [DOI] [PubMed] [Google Scholar]
- Rolo J., de Lencastre H., Miragaia M. (2014). High frequency and diversity of cassette chromosome recombinases (ccr) in methicillin-susceptible Staphylococcus sciuri. J. Antimicrob. Chemother. 69 1461–1469. 10.1093/jac/dku028 [DOI] [PubMed] [Google Scholar]
- Rolo J., Worning P., Nielsen J. B., Bowden R., Bouchami O., Damborg P., et al. (2017a). Evolutionary origin of the staphylococcal cassette chromosome mec (SCCmec). Antimicrob. Agents Chemother. 61:e02302-16. 10.1128/AAC.02302-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolo J., Worning P., Nielsen J. B., Sobral R., Bowden R., Bouchami O., et al. (2017b). Evidence for the evolutionary steps leading to mecA-mediated beta-lactam resistance in staphylococci. PLoS Genet. 13:e1006674. 10.1371/journal.pgen.1006674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree P. M., Freeman B. M. (1955). Infections caused by a particular phage type of Staphylococcus aureus. Med. J. Aust. 42 157–161. [PubMed] [Google Scholar]
- Ryffel C., Tesch W., Birch-Machin I., Reynolds P. E., Barberis-Maino L., Kayser F. H., et al. (1990). Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Gene 94 137–138. 10.1016/0378-1119(90)90481-6 [DOI] [PubMed] [Google Scholar]
- Scharn C. R., Tenover F. C., Goering R. V. (2013). Transduction of staphylococcal cassette chromosome mec elements between strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 57 5233–5238. 10.1128/AAC.01058-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer K. H., Geyer U., Kilpper-Balz R., Devriese L. A. (1983). Elevation of Staphylococcus sciuri subsp. lentus (Kloos et al.) to species status: Staphylococcus lentus (Kloos et al.) comb. nov. Syst. Appl. Microbiol. 4 382–387. 10.1016/S0723-2020(83)80022-8 [DOI] [PubMed] [Google Scholar]
- Schwendener S., Cotting K., Perreten V. (2017). Novel methicillin resistance gene mecD in clinical Macrococcus caseolyticus strains from bovine and canine sources. Sci. Rep. 7:43797. 10.1038/srep43797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendener S., Perreten V. (2018). The integrase of the Macrococcus caseolyticus resistance island mecD (McRImecD) inserts DNA site-specifically into Staphylococcus and Bacillus chromosomes. Mol. Microbiol. 10.1111/mmi.14112 [DOI] [PubMed] [Google Scholar]
- Semmler T., Harrison E. M., Lubke-Becker A., Ulrich R. G., Wieler L. H., Guenther S., et al. (2016). A look into the melting pot: the mecC-harboring region is a recombination hot spot in Staphylococcus stepanovicii. PLoS One 11:e0147150. 10.1371/journal.pone.0147150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid M., Sobia F., Singh A., Malik A., Khan H. M., Jonas D., et al. (2009). Beta-lactams and beta-lactamase-inhibitors in current- or potential-clinical practice: a comprehensive update. Crit. Rev. Microbiol. 35 81–108. 10.1080/10408410902733979 [DOI] [PubMed] [Google Scholar]
- Shore A. C., Deasy E. C., Slickers P., Brennan G., O’Connell B., Monecke S., et al. (2011a). Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 55 3765–3773. 10.1128/AAC.00187-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore A. C., Rossney A. S., Brennan O. M., Kinnevey P. M., Humphreys H., Sullivan D. J., et al. (2011b). Characterization of a novel arginine catabolic mobile element (ACME) and staphylococcal chromosomal cassette mec composite island with significant homology to Staphylococcus epidermidis ACME type II in methicillin-resistant Staphylococcus aureus genotype ST22-MRSA-IV. Antimicrob. Agents Chemother. 55 1896–1905. 10.1128/AAC.01756-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. D., Wachi M., Doi M., Ishino F., Matsuhashi M. (1987). Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 221 167–171. 10.1016/0014-5793(87)80373-3 [DOI] [PubMed] [Google Scholar]
- Stojanov M., Sakwinska O., Moreillon P. (2013). Expression of SCCmec cassette chromosome recombinases in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. J. Antimicrob. Chemother. 68 749–757. 10.1093/jac/dks494 [DOI] [PubMed] [Google Scholar]
- Suzuki E., Kuwahara-Arai K., Richardson J. F., Hiramatsu K. (1993). Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob. Agents Chemother. 37 1219–1226. 10.1128/AAC.37.6.1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A., Nachman S., Leaf H. (1991). Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 35 124–129. 10.1128/AAC.35.1.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubakishita S., Kuwahara-Arai K., Baba T., Hiramatsu K. (2010a). Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob. Agents Chemother. 54 1469–1475. 10.1128/AAC.00575-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubakishita S., Kuwahara-Arai K., Sasaki T., Hiramatsu K. (2010b). Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob. Agents Chemother. 54 4352–4359. 10.1128/AAC.00356-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenesch F., Naimi T., Enright M. C., Lina G., Nimmo G. R., Heffernan H., et al. (2003). Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9 978–984. 10.3201/eid0908.030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlack S., Cox L., Peleg A. Y., Canuto C., Stewart C., Conlon A., et al. (2006). Carriage of methicillin-resistant Staphylococcus aureus in a Queensland Indigenous community. Med. J. Aust. 184 556–559. [DOI] [PubMed] [Google Scholar]
- Voss A., Loeffen F., Bakker J., Klaassen C., Wulf M. (2005). Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11 1965–1966. 10.3201/eid1112.050428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Safo M., Archer G. L. (2012). Characterization of DNA sequences required for the CcrAB-mediated integration of staphylococcal cassette chromosome mec, a Staphylococcus aureus genomic island. J. Bacteriol. 194 486–498. 10.1128/JB.05047-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D. J., Strominger J. L. (1983). Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu. Rev. Biochem. 52 825–869. 10.1146/annurev.bi.52.070183.004141 [DOI] [PubMed] [Google Scholar]
- Westh H., Zinn C. S., Rosdahl V. T. (2004). An international multicenter study of antimicrobial consumption and resistance in Staphylococcus aureus isolates from 15 hospitals in 14 countries. Microb. Drug Resist. 10 169–176. 10.1089/1076629041310019 [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H., Rosato A. E., Enright M. C., Noto M., Craig W., Archer G. L. (2003). Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 47 3574–3579. 10.1128/AAC.47.11.3574-3579.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Organization for Animal Health (2016). OIE Annual report on the use of antimicrobial agents in animals. Better Understanding of The Global Situation. OIE Annual report. [Google Scholar]
- Wu S., de Lencastre H., Tomasz A. (1998). Genetic organization of the mecA region in methicillin-susceptible and methicillin-resistant strains of Staphylococcus sciuri. J. Bacteriol. 180 236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Piscitelli C., de Lencastre H., Tomasz A. (1996). Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb. Drug Resist. 2 435–441. 10.1089/mdr.1996.2.435 [DOI] [PubMed] [Google Scholar]
- Wu S. W., de Lencastre H., Tomasz A. (2001). Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J. Bacteriol. 183 2417–2424. 10.1128/JB.183.8.2417-2424.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Li F., Liu D., Xue H., Zhao X. (2015). Novel type XII Staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase, CcrC2. Antimicrob. Agents Chemother. 59 7597–7601. 10.1128/AAC.01692-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., McClure J. A., Elsayed S., Conly J. M. (2009). Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53 531–540. 10.1128/AAC.01118-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Antignac A., Wu S. W., Tomasz A. (2008). Penicillin-binding proteins and cell wall composition in beta-lactam-sensitive and -resistant strains of Staphylococcus sciuri. J. Bacteriol. 190 508–514. 10.1128/JB.01549-07 [DOI] [PMC free article] [PubMed] [Google Scholar]


