Abstract
The light-harvesting chlorophyll a/b binding proteins (LHCP) belong to a large family of membrane proteins. They form the antenna complexes of photosystem I and II and function in light absorption and transfer of the excitation energy to the photosystems. As nuclear-encoded proteins, the LHCPs are imported into the chloroplast and further targeted to their final destination—the thylakoid membrane. Due to their hydrophobicity, the formation of the so-called ‘transit complex’ in the stroma is important to prevent their aggregation in this aqueous environment. The posttranslational LHCP targeting mechanism is well regulated through the interaction of various soluble and membrane-associated protein components and includes several steps: the binding of the LHCP to the heterodimeric cpSRP43/cpSRP54 complex to form the soluble transit complex; the docking of the transit complex to the SRP receptor cpFtsY and the Alb3 translocase at the membrane followed by the release and integration of the LHCP into the thylakoid membrane in a GTP-dependent manner. This review summarizes the molecular mechanisms and dynamics behind the posttranslational LHCP targeting to the thylakoid membrane of Arabidopsis thaliana.
Keywords: LHCP, CpSRP, Transit complex, Alb3, CpFtsY, Thylakoid membrane
Introduction and overview of LHCP transport to the thylakoid membrane
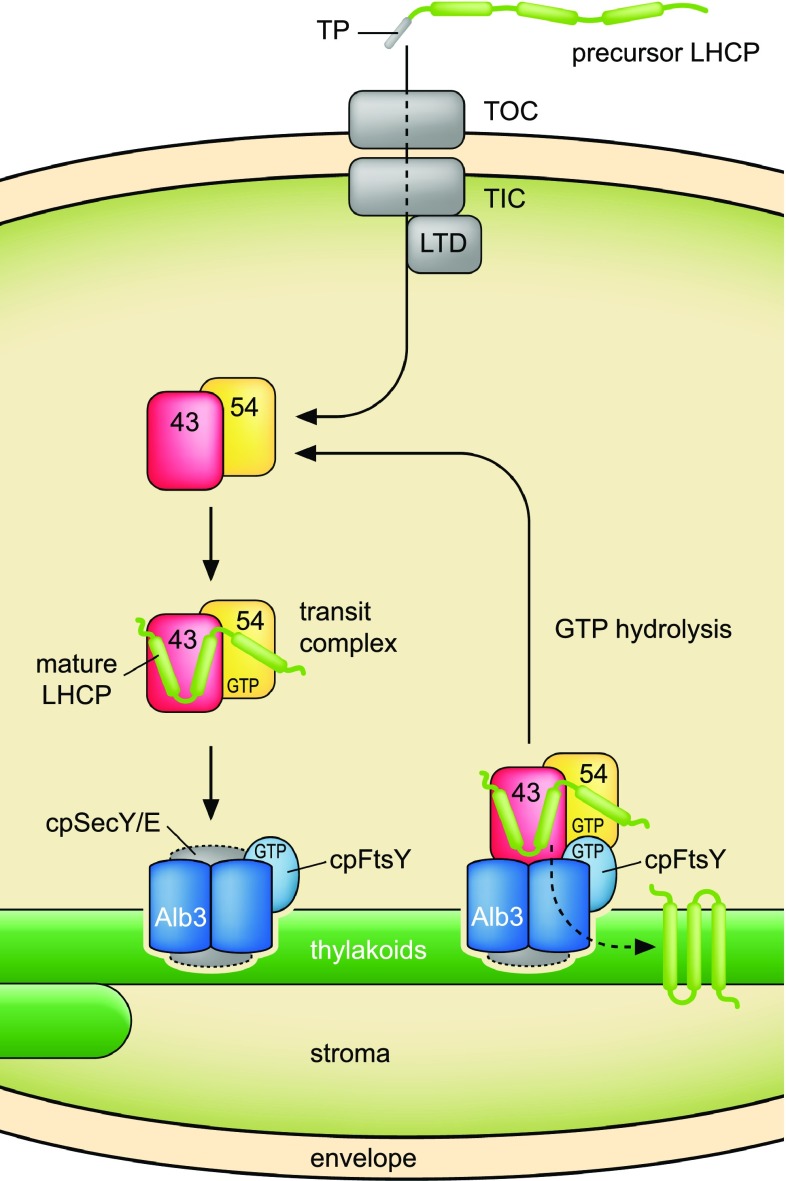
The capture of light energy is essential for biomass production through photosynthesis. In organisms ranging from green algae to vascular plants, photosystems I and II are associated with antenna complexes that consist of the light-harvesting chlorophyll a/b binding proteins (LHCPs) and are specialized for the harvesting and transfer of energy to the photosystems. LHCPs are integral thylakoid membrane proteins with three membrane-spanning regions and represent the most abundant proteins in this membrane system. LHCPs are encoded in the nucleus, translated in the cytosol, and targeted to the chloroplast via N-terminal transit sequences. Upon import into the chloroplast, which is mediated by two translocons in the outer and inner envelope membrane (TOC/TIC) (Jarvis 2008; Paila et al. 2015; Bölter and Soll 2016; Sjuts et al. 2017), the transit sequence is cleaved off (Richter and Lamppa 1999) (Fig. 1). The question of how the LHCPs are translocated through the stroma and subsequently inserted and assembled in the thylakoid membrane has been a subject of study for approximately three decades. In early studies, it was shown that a proteinaceous stromal factor is required for the formation of a soluble, stable ~ 120 kDa LHCP intermediate termed the transit complex, which traverses the stroma before thylakoid insertion (Fulsom and Cline 1988; Cline et al. 1989; Reed et al. 1990; Payan and Cline 1991). This factor was later identified as the so-called chloroplast signal recognition particle (cpSRP), which is located in the stromal fraction of the chloroplast (Li et al. 1995; Schünemann et al. 1998; Klimyuk et al. 1999). The cpSRP complex of higher plants is well characterized; it consists of two subunits, the conserved 54 kDa GTPase cpSRP54 and a unique chloroplast-specific 43 kDa protein, cpSRP43 (Franklin and Hoffman 1993; Schünemann et al. 1998; Klimyuk et al. 1999) (Fig. 1). CpSRP54 is homologous to cytosolic eukaryotic SRP54 and to the prokaryotic 54 homolog (Ffh) (Franklin and Hoffman 1993; Li et al. 1995), which are required for cotranslational protein transport to the endoplasmic reticulum and the plasma membrane, respectively (Akopian et al. 2013; Saraogi and Shan 2014; Voorhees and Hegde 2016). Consistent with the previous finding of a soluble LHCP intermediate, it has been demonstrated that complex formation between cpSRP and LHCP prevents aggregation of the hydrophobic LHCP in the aqueous milieu of the stroma and maintains it in an insertion-competent stage (Schünemann et al. 1998; Yuan et al. 2002; Goforth et al. 2004). The handover of the LHCP from the TOC/TIC import translocon to the cpSRP complex involves the ankyrin-repeat protein LTD (LHCP translocation defect), which is able to interact with the Tic machinery, LHCP, and cpSRP (Ouyang et al. 2011) (Fig. 1). Although cpSRP is sufficient to keep LHCP soluble and in an insertion-competent stage, the insertion of LHCPs into the thylakoid membrane requires additional factors. They comprise (i) the thylakoid membrane-associated SRP receptor cpFtsY (Kogata et al. 1999; Tu et al. 1999; Yuan et al. 2002), which is a homolog of the eukaryotic SRP receptor SRα and the prokaryotic FtsY, (ii) GTP, which is hydrolyzed by the SRP GTPases cpSRP54 and cpFtsY (Akopian et al. 2013) and (iii) the integral thylakoid membrane translocase Alb3 (albino 3) (Moore et al. 2000) (Fig. 1). Alb3 is a homolog of the bacterial YidC and mitochondrial Oxa proteins, which mediate the insertion, assembly, and folding of membrane proteins in the plasma membrane and inner mitochondrial membrane, respectively (Dünschede and Schünemann 2011; Wang and Dalbey 2011; Saller et al. 2012; Hennon et al. 2015).
Fig. 1.
LHCPs are targeted to the thylakoid membrane via the posttranslational cpSRP-dependent transport pathway. LHCPs are imported posttranslationally into chloroplasts via the TOC/TIC translocon in the outer and inner envelope membrane. After import into the stroma, the transit peptide is cleaved off and the LHCPs are forwarded to the cpSRP complex by LTD. The transit complex consisting of cpSRP43, cpSRP54, and LHCP traverses the stroma and docks to the thylakoid membrane via interaction with cpFtsY and the Alb3 insertase. Alb3/cpFtsY are associated with the cpSecY translocase, which is, however, most likely not involved in the insertion process. GTP hydrolysis catalyzed by the SRP GTPases cpSRP54 and cpFtsY drives the dissociation of protein components
In this review, we summarize the molecular details of the individual steps of posttranslational cpSRP-dependent LHCP transport in plants, including cpSRP43/cpSRP54 heterodimerization, cpSRP/LHCP transit complex formation, docking of the transit complex at the thylakoid membrane, and insertion of LHCP into the membrane. We also discuss aspects of the regulation and dynamics of the transport machinery. For information on the evolution of this transport system and on the overlapping function of cpSRP pathway components in the cotranslational transport of plastid-encoded proteins, we refer to previous reviews (Henry et al. 2007; Richter et al. 2010; Ziehe et al. 2017).
Formation of the cpSRP43/54 heterodimer in Arabidopsis thaliana
The chloroplast-specific cpSRP43 is a multidomain protein that consists of three chromodomains (CD1, CD2, CD3) and four ankyrin repeats (Ank1-Ank4) (Klimyuk et al. 1999; Goforth et al. 2004; Stengel et al. 2008). The N-terminal region of cpSRP43 harbors the first chromodomain (CD1), which is followed by 4 ankyrin repeats (Ank1-4) and two additional chromodomains (CD2, CD3) in the C-terminus (Fig. 2b). The second cpSRP subunit, cpSRP54, consists of an N-terminal N domain, a central G domain with GTPase activity and a methionine-rich M domain in the C-terminus (Franklin and Hoffman 1993) (Fig. 2c). In 2008, Stengel et al. published the first crystal structure of cpSRP43 (CD1-Ank4), revealing a unique arrangement of the chromodomains and the ankyrin repeats (Stengel et al. 2008) (Table 1). The crystal structure shows the characteristic helix-turn-helix motifs of Ank2 and Ank3 and the elongated nature of the Ank1 and Ank4 helices. CD1 is composed of three antiparallel β-sheets and a vertical α-helix that is oriented in the direction of the first ankyrin helix. Overall, the crystal structure reveals the elongated horseshoe character of the CD1-Ank4 region that is typical of ankyrin-repeat proteins.
Fig. 2.
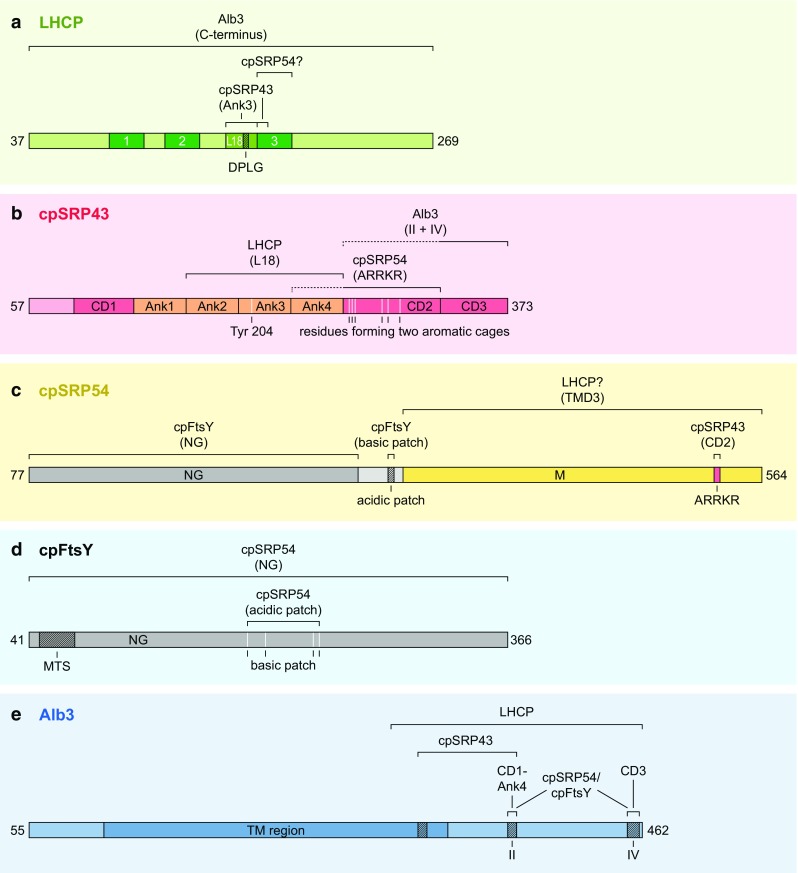
Scheme of protein–protein interactions within the posttranslational cpSRP transport pathway. The mature forms of the proteins involved in posttranslational LHCP transport and their functional domains are shown. Amino acid positions on the left and right correspond to the Arabidopsis thaliana proteins, except of LHCP, which refers to Lhcb1 of Pisum sativum. The diverse interaction partners and the interacting domains are indicated. Further details and references are given in the main text. a LHCP harbors three transmembrane domains (dark green, 1–3). The L18 region containing the crucial DPLG motif, which is responsible for cpSRP43 (Ank3) binding, is located between the second and third transmembrane domains. The binding region for cpSRP43 extends into LHCP’s transmembrane domain three. It is discussed if cpSRP54 binds to transmembrane domain three of LHCP. Furthermore, there is a direct interaction between Alb3’s C-terminus and LHCP. b CpSRP43 comprises three chromodomains (CD1-CD3, red) and four ankyrin repeats (Ank1-Ank4, orange). The Ank2-Ank4 region with the conserved Y204 binds the LHCP L18 peptide. The interaction with the ARRKR motif of cpSRP54 is accomplished via a twinned aromatic cage located in CD2, which is formed by six residues. A stimulating effect of Ank4 for cpSRP54/cpSRP43 complex formation was demonstrated (depicted by dashed line). CpSRP43 binds to the Alb3’s C-terminus (motifs II and IV) via CD2-3, whereby CD3 plays the major role (dashed and solid lines, respectively). c CpSRP54 is composed of a N-terminal NG domain (gray) and a C-terminal M domain (yellow) connected by a linker region (light gray). CpSRP54 binds to cpSRP43 with its C-terminal ARRKR motif (red) within the M domain. The NG domain binds to the homologous domain in cpFtsY. An acidic patch (dark shaded, E313; D314; E316; D317) next to the M domain forms an additional interaction site for cpFtsY. The M domain possibly also binds to the third transmembrane domain of LHCP. d Like cpSRP54, cpFtsY comprises a NG domain. A membrane targeting sequence (MTS, dark shaded) is located close to the N-terminus. As mentioned above cpFtsY interacts with cpSRP54 via its NG domain. Additionally, it contains a basic patch (K191; K193; K203; R204; K235; K236; K 240) as counterpart for cpSRP54’s acidic patch to provide an interaction via the complementary charged regions. e Alb3 is predicted to have five transmembrane domains which are summarized and depicted as transmembrane (TM) region (dark blue). CpSRP43 binds to a motif within the TM region, motif II and motif IV. Further data indicate that the binding to motif II and motif IV is mediated by CD1-Ank4 and CD3, respectively. The Alb3 C-terminus also binds the cpSRP54/cpFtsY complex, whereby the binding interface is probably provided by motifs II and IV. Additionally, LHCPs bind to the C-terminal region of Alb3
Table 1.
Crystal structures of proteins and protein complexes of the posttranslational cpSRP pathway
| Macromolecule | Fragment | Organism | PDB number | References |
|---|---|---|---|---|
| cpSRP43 | ||||
| cpSRP43 CD3 | Residue: 319–368; His-tag cleaved off | A. thaliana | 5E4X | Horn et al. (2015) |
| cpSRP43 CD1-Ank4 | Residue: 85–267; His-tag cleaved off | A. thaliana | 3DEO | Stengel et al. (2008) |
| cpSRP43/cpSRP54 | ||||
| cpSRP43∆CD3 complexed with RRKRp | cpSRP43 residue: 85–318; His-tag cleaved off | A. thaliana | 3UI2 | Holdermann et al. (2012) |
| cpSRP54 residue: 528–540; His-tag cleaved off | A. thaliana | |||
| cpSRP43/LHCP | ||||
| cpSRP43 CD1-Ank4 complexed with L18 | Residue: 85–267; His-tag cleaved off | A. thaliana | 3DEP | Stengel et al. (2008) |
| cpSRP43/Alb3 | ||||
| cpSRP43 CD2-CD3 fused via thioredoxin to Alb3 motif IV | Thioredoxin residue: 3-109; His-tag | E. coli | 5E4W | Horn et al. (2015) |
| cpSRP43 residue: 265–369 | A. thaliana | |||
| Alb3 residue: 453–461; GST-tag cleaved off | A. thaliana | |||
| cpFtsY | ||||
| cpFtsY | Residue: 80–383; His-tag | P. patens | 4AK9 | Träger et al. (2012) |
| cpFtsY | Residue: 24–112; thioredoxin | A. thaliana | 2OG2 | Chandrasekar et al. (2008) |
| cpFtsY | Residue: 65–366; His-tag cleaved off | A. thaliana | 3B9Q | Stengel et al. (2007) |
| cpSRP54/cpFtsY | ||||
| cpSRP54 complexed with cpFtsY | cpSRP54 residue: 77–371; His-tag | A. thaliana | 5L3R | Wild et al. (2016) |
| cpFtsY residue: 80–366; His-tag | A. thaliana | |||
All available crystal structures and the corresponding PDB numbers are listed. Additional information about the crystallized fragments and the corresponding organism (A. thaliana, Arabidopsis thaliana; P. patens, Physcomitrella patens; E. coli, Escherichia coli) is given
CpSRP43/cpSRP54 complex formation was intensively studied by several groups. Initially, the cpSRP54 M domain was identified as the main binding region for cpSRP43 (Jonas-Straube et al. 2001; Groves et al. 2001; Goforth et al. 2004). Later, a 10-residue segment within the C-terminal tail region of cpSRP54M (RRKRp10) was shown to be important for cpSRP43 binding. This segment contains the conserved positively charged cpSRP43 binding motif ARRKR comprising residues 535–539 of cpSRP54 (Funke et al. 2005; Dünschede et al. 2015) (Fig. 2c). The formation of the cpSRP complex is mainly accomplished by the interaction of the ARRKR motif with cpSRP43-CD2. CD2 consists of three-stranded antiparallel β-sheets with a perpendicular α-helix and thus has the characteristic chromodomain architecture (Holdermann et al. 2012) (Table 1). In contrast to CD1, which is tightly connected to the ankyrin repeats, CD2 does not participate in any tertiary interactions with the N-terminal domains of cpSRP43 and is therefore more flexible. Within CD2 are located residues that form two aromatic cages that together present the binding interface for cpSRP54 (Holdermann et al. 2012) (Fig. 2b). Cage 1, which is formed by E268, W291 and D293, recognizes R537 of the cpSRP43 binding motif, whereas R536 is bound by the second aromatic cage, which consists of F267, Y269, and H304. Further detailed study revealed that the RRKRp10 peptide binds at the interface between CD2 and Ank4 and that in this complex CD2 is more closely positioned to Ank4 compared to free cpSRP43 (Holdermann et al. 2012). The importance of the Ank4 region for the cpSRP43/cpSRP54 interaction was also indicated by the observation that the affinity of binding of full-length cpSRP43 to RRKRp10 (Kd 0.39 µM) is significantly increased in comparison to that of cpSRP43 CD2 (Holdermann et al. 2012). Notably, full-length cpSRP54 and cpSRP54M bind cpSRP43 with even higher affinity (Kd 2–95 nM) (Hermkes et al. 2006; Gao et al. 2015; Ziehe et al. 2016) (Table 2), suggesting that additional regions of cpSRP54 are required to support high-affinity cpSRP complex formation. Little is known about the dynamics of the formation of this complex in vivo, but current data indicate that most if not all of the cpSRP43 is complexed with cpSRP54 in the stroma (Schünemann et al. 1998; Klimyuk et al. 1999).
Table 2.
Kd values of protein–protein interactions within the posttranslational cpSRP-pathway of Arabidopsis thaliana
| Interaction | Method | Kd [µM] | References |
|---|---|---|---|
| cpSRP43/Alb3 | |||
| 43/Alb3 C-terminus | FA | 12–18 | Liang et al. (2016) |
| 43/Alb3 C-terminus | ITC | 5.1 | Horn et al. (2015) |
| 43/Alb3 C-terminus | ITC | 9.7–13.2 | Falk et al. (2010); Falk and Sinning (2010a, 2010b) |
| 43/Alb3 C-terminus | ITC | 0.094 | Lewis et al. (2010) |
| cpSRP43/LHCP | |||
| 43/LHCP | LS | 0.17/1.5 | Liang et al. (2016) |
| 43/LHCP + 54M | 0.26 | ||
| 43/LHCP | FA | 0.14–0.3 | Jaru-Ampornpan et al. (2010) |
| 43/L18 | ITC | 0.322 | Gao et al. (2015) |
| 43/L18 + RRKRp10 | 0.107 | ||
| 43/L18 | ITC | 1.17 | Stengel et al. (2008) |
| 43/L11 | FA | 0.022 | Liang et al. (2016) |
| 43/L11 + Alb3 C-terminus | 0.011 | ||
| cpSRP43/cpSRP54 | |||
| 43/54 | MST | 0.05 | Ziehe et al. (2016) |
| 43/54 | FCS | 0.095 | Gao et al. (2015) |
| 43/54M | SPR | 0.0025 | Hermkes et al. (2006) |
| cpSRP54/cpFtsY | |||
| 54/FtsY - PG | FRET | 2.34 | Chandrasekar and Shan (2017) |
| 54/FtsY + PG | 0.13 | ||
| 54/FtsY* | FRET | 0.77 | Chandrasekar et al. (2017) |
| 54NG/FtsY* | 12 | ||
Enlisted are the dissociation constants (Kd values) of protein complexes of the posttranslational cpSRP-pathway. Additionally, the methods by which the Kd values were determined as well as possible variations of the experimental settings are shown
FA fluorescence anisotropy, FCS fluorescence correlation spectroscopy, FRET forster resonance energy transfer, ITC isothermal titration calorimetry, LA light scattering, MST microscale thermophoresis, SPR surface plasmon resonance spectroscopy
* measured in the presence of 0.01 % Nikkol (mimicking of the effect of lipids)
CpSRP binds LHCP to form a soluble LHCP transport intermediate, the transit complex
As described in the introduction, the LHC proteins are bound by the cpSRP complex in a way that maintains their solubility and insertion competence. Several studies have aimed to identify the intermolecular contacts between LHCP and the cpSRP subunits within the transit complex as summarized below.
Using various LHCP truncation constructs, an 18-residue-long binding site between the second and third transmembrane domains, L18 (VDPLYPGGSFDPLGLADD), and a hydrophobic region following the L18 motif were shown to be crucial for transit complex formation with cpSRP (DeLille et al. 2000) (Fig. 2a). The L18 motif harboring the DPLG sequence is conserved among LHCPs (Stengel et al. 2008; Barros and Kühlbrandt 2009) and therefore seems to be an important feature of members of this protein family. Tu et al. identified cpSRP43 as the binding partner for the L18 motif, while a direct interaction of cpSRP54 with LHCP was not detected (Tu et al. 2000). Further studies mapped the binding interface between LHCP and cpSRP via a pepscan analysis and confirmed the cpSRP43/L18 interaction (Groves et al. 2001). Cross-linking studies with pea Lhcb1 and cpSRP43 or a cpSRP complex revealed the presence of direct contacts between the L18 motif of Lhcb1 and the first part of TMD3 with cpSRP43; no contacts between Lhcb1 and cpSRP54 were detected (Cain et al. 2011). The structure of the cpSRP43/L18 complex was resolved by Stengel et al. (2008) (Table 1). CpSRP43 forms two predominantly hydrophobic grooves on its concave surface. L18 binds to groove 1, which is formed by ankyrin repeats 2–4. The DPLG motif is compactly folded and wraps around Y204 of Ank3 (Fig. 2b) and it was shown that mutations in the DPLG motif or in Y204 of cpSRP43 impair the cpSRP43/L18 interaction (Stengel et al. 2008). Studies to quantitatively analyze the interaction of cpSRP43 with the L18 region of LHCP reported dissociation constants (Kd) ranging from 22 nM to 1.17 µM (Table 2). While the interaction between cpSRP43 and LHCP has been unambiguously proven, the question whether cpSRP54 contacts LHCP directly is less clear. As mentioned above, binding of cpSRP54 to LHCP was not observed in some studies, but other studies have reported evidence for cpSRP54/LHCP interaction. Initial reports revealed that cpSRP54 binds to residues within the third transmembrane domain (High et al. 1997; Groves et al. 2001) (Fig. 2c), emphasizing the importance of this transmembrane domain for transit complex formation (DeLille et al. 2000). Recently, it has been demonstrated that the absence of cpSRP54 or mutations within the M domain of cpSRP54 impair the formation of the cpSRP/LHCP transit complex (Dünschede et al. 2015; Henderson et al. 2016). Although the foregoing studies indicate that cpSRP54 plays an important role in transit complex formation, its precise contribution remains unclear. Apparently, it is not essential for the formation of soluble LHCP as it was shown that cpSRP43 alone acts as an ATP-independent chaperone for LHCP and is sufficient to maintain its solubility (Falk and Sinning 2010b; Jaru-Ampornpan et al. 2010). Therefore, cpSRP54 probably acts as an optimizing element that maintains the transit complex in an ideal insertion-competent state, thereby rendering the transport process more efficient (see also below in ‘Regulation and dynamics of the transport machinery’).
Docking of the transit complex at the membrane and LHCP insertion
The cpSRP receptor cpFtsY binds peripherally to thylakoid membranes, and biochemical and genetic data prove that cpFtsY is linked to LHCP insertion (Kogata et al. 1999; Tu et al. 2000; Yuan et al. 2002; Tzvetkova-Chevolleau et al. 2007; Marty et al. 2009). Similar to cpSRP54, cpFtsY contains an NG domain that is necessary for GTP binding and hydrolysis (Fig. 2d). Crystal structures of various plant cpFtsY proteins reveal the characteristic four helix bundle within the N domain and the five G motifs within the G domain (Stengel et al. 2007; Chandrasekar et al. 2008; Träger et al. 2012) (Table 1). Tethering of cpFtsY to the membrane is mediated via an amphipathic helix located at the N-terminus (Stengel et al. 2007; Marty et al. 2009) (Fig. 2d). Within this region, two conserved phenylalanine residues, F48 and F49, are crucial for membrane binding, and it was demonstrated that cpFtsY is only functional in LHCP insertion when it is attached to the thylakoid membrane (Marty et al. 2009). CpFtsY is able to bind cpSRP54 and complex formation is established by interaction between the homologous NG domains of the two proteins (Jaru-Ampornpan et al. 2007; Stengel et al. 2007; Chandrasekar et al. 2008; Wild et al. 2016) (Table 1). Interestingly, recent data indicate that the M domain of cpSRP54 accelerates and stabilizes cpSRP54/cpFtsY complex formation via interaction between a positively charged cluster in the G domain of cpFtsY and a negatively charged cluster within the M domain of cpSRP54 (Jaru-Ampornpan et al. 2007; Chandrasekar et al. 2017) (Table 2). Furthermore, complex formation between cpFtsY and cpSRP54 is considerably stimulated by anionic phospholipids (Chandrasekar and Shan 2017) (Table 2).
In addition to cpFtsY, the integral thylakoid membrane protein Alb3 is involved in LHCP insertion. The characterization of Alb3 as the responsible insertase is based on the results of several studies that demonstrated specific inhibition of LHCP insertion by anti-Alb3 antibodies (Moore et al. 2000, 2003), and a direct interaction of Alb3 with components of the cpSRP transport pathway (Moore et al. 2003; Bals et al. 2010; Falk et al. 2010; Lewis et al. 2010; Dünschede et al. 2011; Walter et al. 2015; Chandrasekar and Shan 2017) (see also below). Consistently, the alb3 null mutant in Arabidopsis thaliana displays an albino phenotype (Sundberg et al. 1997). Similar to bacterial YidC, the crystal structure of which was recently solved (Kumazaki et al. 2014a, b), Alb3 is predicted to contain five conserved transmembrane helices and a structurally disordered C-terminus protruding into the stroma of the chloroplast (Falk et al. 2010) (Fig. 2e). Blue native PAGE indicates that Alb3 can form dimers (Dünschede et al. 2011) (Fig. 1).
Several studies have described a direct interaction between Alb3 and cpSRP43 (Bals et al. 2010; Falk et al. 2010; Lewis et al. 2010; Dünschede et al. 2011; Liang et al. 2016). Two positively charged binding motifs within the C-terminus of Alb3, motif II (Dünschede et al. 2011; Falk et al. 2010) and motif IV (Falk et al. 2010), are important for cpSRP43 binding (Fig. 2e). Structural data revealed that motif IV binds to cpSRP43 CD3 (Horn et al. 2015) (Table 1). Biochemical data point to an interaction of motif II and cpSRP43 CD1-Ank4 (Liang et al. 2016). Furthermore, a binding site within the transmembrane region of Alb3 was described (Dünschede et al. 2011) (Fig. 2e). These data led to the conclusion that the transit complex is recruited to Alb3 via cpSRP43/Alb3 interaction. This docking model was further supported by the finding that cpSRP43 alone is able to keep LHCPs soluble (Falk and Sinning 2010b; Jaru-Ampornpan et al. 2010) and by the results of Tzvetkova-Chevolleau et al., who postulated an alternative LHCP transport pathway in Arabidopsis thaliana that bypasses cpFtsY and cpSRP54 but still requires cpSRP43 for LHCP targeting (Tzvetkova-Chevolleau et al. 2007). The latter authors demonstrated that the ffc/cpftsy double-knockout mutant lacking functional cpSRP54 and cpFtsY has a less severe phenotype and accumulates more LHCPs than the cpftsy single-knockout mutant. Therefore, these data provide support for an LHCP transport mechanism that depends on an efficient interaction between Alb3 and cpSRP43. However, the dissociation constant of the cpSRP43/Alb3 C-terminus interaction was described inconsistently. Whereas a Kd of ~ 90 nM, indicating high-affinity binding, was reported by Lewis et al. (2010), other reports point to a rather weak, transient interaction (Kd 5–18 µM) (Falk et al. 2010; Falk and Sinning 2010a; Horn et al. 2015; Liang et al. 2016) (Table 2). Notably, the affinity of cpSRP43 for full-length Alb3 has not been determined yet. Therefore, the contribution of the Alb3/cpSRP43 interaction to the recruitment of the transit complex to the membrane remains unclear.
Other data support the existence of an alternative LHCP targeting mode in which the transit complex recruitment to Alb3 is accomplished primarily via an interaction between Alb3 and the cpSRP54/cpFtsY complex. The studies of Moore et al (2003) indicate that Alb3 can bind the cpSRP54/cpFtsY complex even in the absence of cpSRP43 and LHCP, and a recent study reported that Alb3 C-terminus binds the cpSRP54/cpFtsY complex with an affinity in the submicromolar range (Chandrasekar and Shan 2017). Additional data suggested that motifs II and IV within Alb3 C-terminus, which are responsible for the cpSRP43 interaction, might also play a role in binding the cpSRP54/cpFtsY complex (Chandrasekar and Shan 2017) (Fig. 2e).
Various studies have demonstrated that LHCP insertion is Alb3-dependent and independent of the thylakoid membrane cpSecY/E translocase (Mori et al. 1999; Moore et al. 2003). However, a direct association between Alb3 and the cpSecY translocase has been shown by coimmunoprecipitation experiments, double immunogold labeling and cross-linking studies, while there is no clear evidence for the presence of an uncomplexed pool of Alb3 (Klostermann et al. 2002). The Alb3/cpSecY translocase association was confirmed by Moore et al. (2003) who showed in diverse precipitation analyses that a stabilized complex consisting of cpFtsY and cpSRP can precipitate Alb3 and cpSecY from solubilized thylakoid membranes. Interestingly, recent data obtained in comigration and coimmunoprecipitation analyses of solubilized thylakoid membrane complexes indicate that cpFtsY and Vipp1 are additional components of the Alb3/cpSecY-containing complex in the thylakoid membrane (Walter et al. 2015). Therefore, it seems possible that the transit complex docks to a preformed cpFtsY/Alb3/cpSecY complex in the thylakoid membrane; however, the cpSec translocase does not appear to be involved in contact formation or the insertion process (Fig. 1).
Regulation and dynamics of the transport machinery
The nucleotide requirement for LHCP integration was examined by in vitro reconstitution assays in two main studies. Initially, Hoffman and Franklin (1994) showed that GTP is the only nucleotide required for integration and demonstrated an inhibitory effect of non-hydrolyzable analogs of GTP (Hoffman and Franklin 1994). The requirement for GTP hydrolysis in LHCP insertion was confirmed by Yuan et al. (2002). Notably, this study also described a stimulatory effect of ATP and of the non-hydrolyzable analog AMP-PNP, indicating that a yet unknown ATP-binding protein might be involved in LHCP integration. GTP is not required for formation of the transit complex (Yuan et al. 2002); however, it is important for triggering the GTPase cycle of the cpSRP54/cpFtsY complex at the membrane (Jaru-Ampornpan et al. 2007, 2009), which is a multistep process comprising assembly of the GTP-loaded cpSRP54/cpFtsY complex, reciprocal GTPase activation and dissociation of the complex. Interestingly, within the GTPase cycle, the cpSRP54/cpFtsY assembly step plays a crucial role in LHCP insertion, and GTPase activation enhances the insertion efficiency to some extent (Nguyen et al. 2011). Molecular dynamic simulations indicate that binding of GTP to cpFtsY is an important step in cpSRP54/cpFtsY complex formation because it induces conformational changes in cpFtsY that favor the formation of a complex with cpSRP54 (Yang et al. 2011), which is a kinetically fast interaction (Jaru-Ampornpan et al. 2007). In recent years, several mechanisms that regulate the GTPase activity of the individual SRP GTPases and of the cpSRP54/cpFtsY complex have been described. GTPase assays using the soluble recombinant cpSRP54/cpFtsY complex were used to demonstrate that cpSRP43 and the C-terminus of Alb3 stimulate GTP hydrolysis by the complex and that the stimulatory effect of Alb3 C-terminus is strictly coupled to the presence of cpSRP43 (Goforth et al. 2004; Lewis et al. 2010). A regulatory effect of Alb3 C-terminus on GTP hydrolysis by the cpSRP54/cpFtsY complex was also described by Chandrasekar and Shan (2017). However, in that case Alb3 C-terminus had an inhibitory, cpSRP43-independent effect on GTP hydrolysis, which led to the hypothesis that this negative regulation might enable positioning of the transit complex on the translocase and transfer of LHCP to Alb3 before GTP hydrolysis occurs. The inconsistent findings are probably due to the use of different experimental conditions. Chandrasekar and Shan (2017) performed GTPase assays in the presence of PG liposomes and reported that the regulatory effect of Alb3 C-terminus on GTPase activation is dependent on the presence of anionic phospholipids (Chandrasekar and Shan 2017). The role of lipids in regulating the GTPase cycle is further supported by the finding that liposomes stimulate the basal GTP hydrolysis rate of cpFtsY (Marty et al. 2009).
Although knowledge of the dynamics of the transport machinery is rather limited, some of the mechanisms (besides regulation of the GTPase cycle) involved in coordinating the order of events have recently been elucidated. A central role is assigned to cpSRP43 because it shows high interdomain dynamics, a feature that probably enables it to undergo flexible interactions with its several binding partners (Gao et al. 2015) (Fig. 2b). The binding of cpSRP54 to cpSRP43 reduces the flexibility of cpSRP43 (Gao et al. 2015) and induces a conformational change (Liang et al. 2016) that results in an enhanced binding affinity of cpSRP43 to LHCP (three to sixfold) (Gao et al. 2015; Liang et al. 2016). The affinity between the activated cpSRP43 and the L18 motif of LHCP was determined to be in the range of 100–300 nM (Gao et al. 2015; Liang et al. 2016) (Table 2). The release of LHCP from cpSRP is triggered upon interaction of cpSRP43 with the insertase Alb3, as it was shown that the addition of recombinant Alb3 C-terminus dissociates soluble cpSRP43/LHCP complexes (Lewis et al. 2010; Liang et al. 2016) and that this effect is coupled to the presence of the cpSRP43 binding motifs II and IV in Alb3 C-terminus (Liang et al. 2016). Furthermore, it was observed that Alb3 C-terminus weakens the interaction between cpSRP43 and the RRKR10p peptide, leading to the hypothesis that this might contribute to the release and transfer of LHCP to the insertase (Falk et al. 2010; Horn et al. 2015).
The thylakoid membrane as the site of cpSRP-dependent LHCP insertion and pigment loading
Approximately 30 years ago, it was demonstrated in in vitro experiments that LHCP is inserted into thylakoid membranes but not into envelope membranes (Cline 1986). In later studies, the in vitro insertion assay of LHCP into thylakoids has been extended and successfully used by several groups to study the molecular details of this pathway (see above and Kuttkat et al. 1995). The thylakoid membrane as the site of cpSRP-dependent LHCP insertion is further supported by the exclusive localization of the Alb3 translocase in thylakoid membrane (Gerdes et al. 2006). In vivo data support the important role of cpSRP-dependent LHCP transport; the ffc/chaos double-knockout mutant, which lacks cpSRP54 and cpSRP43, showed pale green leaves due to the loss of 85% of its chlorophyll as well as a strong decrease in the levels of most LHCPs and greatly reduced number of thylakoids (Amin et al. 1999; Hutin et al. 2002). For a detailed summary of the phenotypes of cpSRP pathway mutants, we refer to previous review articles (Schünemann 2004; Henry et al. 2007; Richter et al. 2010). The biogenesis of stable LHC complexes in the thylakoid membrane requires assembly with pigments (Kuttkat et al. 1995, 1997; Plumley and Schmidt 1995; Tanaka and Tanaka 2011). Because the soluble LHCP/cpSRP transit complex forms in the absence of pigments and in vitro experiments have demonstrated that inserted LHCP is complexed with pigments and assembled in trimers (Kuttkat et al. 1995), it is very likely that pigment loading occurs at the site of insertion in the thylakoid membrane. This is consistent with the finding that the chlorophyll (chl) b-deficient Arabidopsis thaliana cao-1 mutant can efficiently import LHCPs, while stable assembly with PSII is affected (Nick et al. 2013). Furthermore, studies using a chl b-deficient Chlamydomonas reinhardtii mutant point to an interconnection between pigment synthesis and LHCP biogenesis that occurs at the thylakoid membrane (Plumley and Schmidt 1995). However, Reinbothe et al. (2006) observed severely impaired LHCP import into chloroplasts from a chl b-deficient mutant of Arabidopsis thaliana (Reinbothe et al. 2006), and studies with Chlamydomonas reinhardtii mutants showed that the absence of chl b led to an accumulation of LHCPs in the cytosol and the vacuole (Park and Hoober 1997). Based on that evidence, a model of LHCII assembly was hypothesized in which chl b is incorporated into LHCP in the envelope membrane during import. Non-pigment-loaded LHCP would reenter the cytosol for degradation (Hoober et al. 2007). The transfer of the pigment-loaded LHCII from the envelope to the thylakoid was hypothesized to be mediated by vesicles. Indeed, in recent years, there has been increasing evidence for the presence of a vesicle transport system in chloroplasts. However, the question of whether or not proteins are transported in addition to lipids has not yet clearly been answered (Karim et al. 2014; Karim and Aronsson 2014; Lindquist et al. 2016). Tanz et al. (2012) suggested a vesicle-based transport of LHCPs predominantly in cotyledons. As described above, the ffc/chaos mutant shows a severely compromised phenotype but still contains residual levels of LHCPs, indicating that at least some members of the LHCP family can be transported in a cpSRP43/54-independent manner in these plants. Considering that the upregulation of stromal chaperones such as ClpC plays a role in compensating for the absence of cpSRP54 in ffc mutants (Rutschow et al. 2008) and that yeast mutants compensate for the loss of the SRP-pathway by a reduced growth rate and induction of heat shock proteins (Mutka and Walter 2001), it is likely that the cpSRP double mutant uses similar strategies to adapt to the loss of cpSRP. Taken together, the model of vesicle-mediated transport of (pigment-loaded) LHCP remains speculative, at least for chloroplasts of higher plants, whereas the current in vitro and in vivo data indicate that cpSRP-dependent LHCP transport, which includes insertion and pigment loading at the thylakoid membrane, plays a primary role in LHCP biogenesis in higher plants.
Conclusions
In the last decades, considerable progress has been made in understanding the molecular details of cpSRP/Alb3-dependent LHCP transport to the thylakoid membrane. However, several central issues need to be investigated in the future to get a more complete picture of LHCP transport. To further decipher this mechanism, it is important to get more structural information about single components and protein complexes. Here, it will be especially challenging to elucidate the structure of the transit complex, the Alb3 insertase, and finally the docking complex. In addition, little is known about the spatiotemporal coordination of LHCP transport, insertion, and pigment delivery/assembly.
Acknowledgements
We thank Klaus Hagemann for his excellent graphical contribution.
References
- Akopian D, Shen K, Zhang X, Shan SO. Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin P, Sy DA, Pilgrim ML, Parry DH, Nussaume L, Hoffman NE. Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol. 1999;121:61–70. doi: 10.1104/pp.121.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bals T, Dünschede B, Funke S, Schünemann D. Interplay between the cpSRP pathway components, the substrate LHCP and the translocase Alb3: an in vivo and in vitro study. FEBS Lett. 2010;584:4138–4144. doi: 10.1016/j.febslet.2010.08.053. [DOI] [PubMed] [Google Scholar]
- Barros T, Kühlbrandt W. Crystallisation, structure and function of plant light-harvesting Complex II. Biochim Biophys Acta. 2009;1787:753–772. doi: 10.1016/j.bbabio.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Bölter B, Soll J. Once upon a time–chloroplast protein import research from infancy to future challenges. Mol Plant. 2016;9:798–812. doi: 10.1016/j.molp.2016.04.014. [DOI] [PubMed] [Google Scholar]
- Cain P, Holdermann I, Sinning I, Johnson AE, Robinson C. Binding of chloroplast signal recognition particle to a thylakoid membrane protein substrate in aqueous solution and delineation of the cpSRP43-substrate interaction domain. Biochem J. 2011;437:149–155. doi: 10.1042/BJ20110270. [DOI] [PubMed] [Google Scholar]
- Chandrasekar S, Shan SO. Anionic phospholipids and the Albino3 translocase activate signal recognition particle-receptor interaction during light-harvesting Chlorophyll a/b-binding protein targeting. J Biol Chem. 2017;292:397–406. doi: 10.1074/jbc.M116.752956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar S, Chartron J, Jaru-Ampornpan P, Shan SO. Structure of the chloroplast signal recognition particle (SRP) receptor: domain arrangement modulates SRP-receptor interaction. J Mol Biol. 2008;375:425–436. doi: 10.1016/j.jmb.2007.09.061. [DOI] [PubMed] [Google Scholar]
- Chandrasekar S, Sweredoski MJ, Sohn CH, Hess S, Shan SO. Co-evolution of two GTPases enables efficient protein targeting in an RNA-less chloroplast signal recognition particle pathway. J Biol Chem. 2017;292:386–396. doi: 10.1074/jbc.M116.752931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K. Import of proteins into chloroplasts. Membrane integration of a thylakoid precursor protein reconstituted in chloroplast lysates. J Biol Chem. 1986;261:14804–14810. [PubMed] [Google Scholar]
- Cline K, Fulsom DR, Viitanen PV. An imported thylakoid protein accumulates in the stroma when insertion into thylakoids is inhibited. J Biol Chem. 1989;264:14225–14232. [PubMed] [Google Scholar]
- DeLille J, Peterson EC, Johnson T, Moore M, Kight A, Henry R. A novel precursor recognition element facilitates posttranslational binding to the signal recognition particle in chloroplasts. Proc Natl Acad Sci USA. 2000;97:1926–1931. doi: 10.1073/pnas.030395197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dünschede B, Schünemann D. The role of Alb3 and its homologs in the insertion and assembly of thylakoid membrane proteins. Endocyto Cell Res. 2011;21:19–25. [Google Scholar]
- Dünschede B, Bals T, Funke S, Schünemann D. Interaction studies between the chloroplast signal recognition particle subunit cpSRP43 and the full-length translocase Alb3 reveal a membrane-embedded binding region in Alb3 protein. J Biol Chem. 2011;286:35187–35195. doi: 10.1074/jbc.M111.250746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dünschede B, et al. Chloroplast SRP54 was recruited for posttranslational protein transport via complex formation with chloroplast SRP43 during land plant evolution. J Biol Chem. 2015;290:13104–13114. doi: 10.1074/jbc.M114.597922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Sinning I. The C terminus of Alb3 interacts with the chromodomains 2 and 3 of cpSRP43. J Biol Chem. 2010;285:le25–26. doi: 10.1074/jbc.L110.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Sinning I. cpSRP43 is a novel chaperone specific for light-harvesting chlorophyll a,b-binding proteins. J Biol Chem. 2010;285:21655–21661. doi: 10.1074/jbc.C110.132746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Ravaud S, Koch J, Sinning I. The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J Biol Chem. 2010;285:5954–5962. doi: 10.1074/jbc.M109.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin AE, Hoffman NE. Characterization of a chloroplast homologue of the 54-kDa subunit of the signal recognition particle. J Biol Chem. 1993;268:22175–22180. [PubMed] [Google Scholar]
- Fulsom DR, Cline K. A Soluble Protein Factor is Required in Vitro for Membrane Insertion of the Thylakoid Protein, pLAHCP. Plant Physiol. 1988;88:1146–1153. doi: 10.1104/pp.88.4.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke S, Knechten T, Ollesch J, Schünemann D. A unique sequence motif in the 54-kDa subunit of the chloroplast signal recognition particle mediates binding to the 43-kDa subunit. J Biol Chem. 2005;280:8912–8917. doi: 10.1074/jbc.M409992200. [DOI] [PubMed] [Google Scholar]
- Gao F, et al. Regulation of structural dynamics within a signal recognition particle promotes binding of protein targeting substrates. J Biol Chem. 2015;290:15462–15474. doi: 10.1074/jbc.M114.624346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes L, et al. A second thylakoid membrane-localized Alb3/OxaI/YidC homologue is involved in proper chloroplast biogenesis in Arabidopsis thaliana. J Biol Chem. 2006;281:16632–16642. doi: 10.1074/jbc.M513623200. [DOI] [PubMed] [Google Scholar]
- Goforth RL, et al. Regulation of the GTPase cycle in post-translational signal recognition particle-based protein targeting involves cpSRP43. J Biol Chem. 2004;279:43077–43084. doi: 10.1074/jbc.M401600200. [DOI] [PubMed] [Google Scholar]
- Groves MR, Mant A, Kuhn A, Koch J, Dubel S, Robinson C, Sinning I. Functional characterization of recombinant chloroplast signal recognition particle. J Biol Chem. 2001;276:27778–27786. doi: 10.1074/jbc.M103470200. [DOI] [PubMed] [Google Scholar]
- Henderson RC, et al. Domain organization in the 54-kDa subunit of the chloroplast signal recognition particle. Biophys J. 2016;111:1151–1162. doi: 10.1016/j.bpj.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennon SW, Soman R, Zhu L, Dalbey RE. YidC/Alb3/Oxa1 Family of Insertases. J Biol Chem. 2015;290:14866–14874. doi: 10.1074/jbc.R115.638171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry R, Goforth RL, Schünemann D. The chloroplast SRP/FtsY and Alb3 in the integration into the thylakoid membrane. In: Ross ED, Koehler C, Tamanoi F, editors. The enzymes, molecular machines involved in protein transport across cellular membranes. Cambridge: Academic Press; 2007. [Google Scholar]
- Hermkes R, Funke S, Richter C, Kuhlmann J, Schünemann D. The alpha-helix of the second chromodomain of the 43 kDa subunit of the chloroplast signal recognition particle facilitates binding to the 54 kDa subunit. FEBS Lett. 2006;580:3107–3111. doi: 10.1016/j.febslet.2006.04.055. [DOI] [PubMed] [Google Scholar]
- High S, et al. Chloroplast SRP54 interacts with a specific subset of thylakoid precursor proteins. J Biol Chem. 1997;272:11622–11628. doi: 10.1074/jbc.272.17.11622. [DOI] [PubMed] [Google Scholar]
- Hoffman NE, Franklin AE. Evidence for a stromal GTP requirement for the integration of a chlorophyll a/b-binding polypeptide into thylakoid membranes. Plant Physiol. 1994;105:295–304. doi: 10.1104/pp.105.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdermann I, Meyer NH, Round A, Wild K, Sattler M, Sinning I. Chromodomains read the arginine code of post-translational targeting. Nat Struct Mol Biol. 2012;19:260–263. doi: 10.1038/nsmb.2196. [DOI] [PubMed] [Google Scholar]
- Hoober JK, Eggink LL, Chen M. Chlorophylls, ligands and assembly of light-harvesting complexes in chloroplasts. Photosynth Res. 2007;94:387–400. doi: 10.1007/s11120-007-9181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn A, Hennig J, Ahmed YL, Stier G, Wild K, Sattler M, Sinning I. Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction. Nat Commun. 2015;6:8875. doi: 10.1038/ncomms9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin C, Havaux M, Carde JP, Kloppstech K, Meiherhoff K, Hoffman N, Nussaume L. Double mutation cpSRP43–/cpSRP54—is necessary to abolish the cpSRP pathway required for thylakoid targeting of the light-harvesting chlorophyll proteins. Plant J. 2002;29:531–543. doi: 10.1046/j.0960-7412.2001.01211.x. [DOI] [PubMed] [Google Scholar]
- Jaru-Ampornpan P, Chandrasekar S, Shan SO. Efficient interaction between two GTPases allows the chloroplast SRP pathway to bypass the requirement for an SRP RNA. Mol Biol Cell. 2007;18:2636–2645. doi: 10.1091/mbc.e07-01-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaru-Ampornpan P, Nguyen TX, Shan SO. A distinct mechanism to achieve efficient signal recognition particle (SRP)-SRP receptor interaction by the chloroplast srp pathway. Mol Biol Cell. 2009;20:3965–3973. doi: 10.1091/mbc.e08-10-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaru-Ampornpan P, Shen K, Lam VQ, Ali M, Doniach S, Jia TZ, Shan SO. ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat Struct Mol Biol. 2010;17:696–702. doi: 10.1038/nsmb.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 2008;179:257–285. doi: 10.1111/j.1469-8137.2008.02452.x. [DOI] [PubMed] [Google Scholar]
- Jonas-Straube E, Hutin C, Hoffman NE, Schünemann D. Functional analysis of the protein-interacting domains of chloroplast SRP43. J Biol Chem. 2001;276:24654–24660. doi: 10.1074/jbc.M100153200. [DOI] [PubMed] [Google Scholar]
- Karim S, Aronsson H. The puzzle of chloroplast vesicle transport—involvement of GTPases. Front Plant Sci. 2014;5:472. doi: 10.3389/fpls.2014.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S, et al. A novel chloroplast localized Rab GTPase protein CPRabA5e is involved in stress, development, thylakoid biogenesis and vesicle transport in Arabidopsis. Plant Mol Biol. 2014;84:675–692. doi: 10.1007/s11103-013-0161-x. [DOI] [PubMed] [Google Scholar]
- Klimyuk VI, et al. A chromodomain protein encoded by the arabidopsis CAO gene is a plant- specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell. 1999;11:87–99. doi: 10.1105/tpc.11.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klostermann E, Droste Gen Helling I, Carde JP, Schünemann D. The thylakoid membrane protein ALB3 associates with the cpSecY-translocase in Arabidopsis thaliana. Biochem J. 2002;368:777–781. doi: 10.1042/bj20021291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogata N, Nishio K, Hirohashi T, Kikuchi S, Nakai M. Involvement of a chloroplast homologue of the signal recognition particle receptor protein, FtsY, in protein targeting to thylakoids. FEBS Lett. 1999;447:329–333. doi: 10.1016/S0014-5793(99)00305-1. [DOI] [PubMed] [Google Scholar]
- Kumazaki K, et al. Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci Rep. 2014;4:7299. doi: 10.1038/srep07299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazaki K, et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature. 2014;509:516–520. doi: 10.1038/nature13167. [DOI] [PubMed] [Google Scholar]
- Kuttkat A, Grimm R, Paulsen H. Light-harvesting chlorophyll a/b-binding protein inserted into isolated thylakoids binds pigments and is assembled into trimeric light-harvesting complex. Plant Physiol. 1995;109:1267–1276. doi: 10.1104/pp.109.4.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuttkat A, Edhofer I, Eichacker LA, Paulsen H. Light-harvesting chlorophyll a/b-binding protein stably inserts into etioplast membranes supplemented with Zn-pheophytin a/b. J Biol Chem. 1997;272:20451–20455. doi: 10.1074/jbc.272.33.20451. [DOI] [PubMed] [Google Scholar]
- Lewis NE, et al. A dynamic cpSRP43-Albino3 interaction mediates translocase regulation of chloroplast signal recognition particle (cpSRP)-targeting components. J Biol Chem. 2010;285:34220–34230. doi: 10.1074/jbc.M110.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Henry R, Yuan J, Cline K, Hoffman NE. A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc Natl Acad Sci USA. 1995;92:3789–3793. doi: 10.1073/pnas.92.9.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang FC, Kroon G, McAvoy CZ, Chi C, Wright PE, Shan SO. Conformational dynamics of a membrane protein chaperone enables spatially regulated substrate capture and release. Proc Natl Acad Sci USA. 2016;113:E1615–E1624. doi: 10.1073/pnas.1524777113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist E, Solymosi K, Aronsson H. Vesicles Are persistent features of different plastids. Traffic. 2016;17:1125–1138. doi: 10.1111/tra.12427. [DOI] [PubMed] [Google Scholar]
- Marty NJ, et al. The membrane-binding motif of the chloroplast signal recognition particle receptor (cpFtsY) regulates GTPase activity. J Biol Chem. 2009;284:14891–14903. doi: 10.1074/jbc.M900775200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Harrison MS, Peterson EC, Henry R. Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J Biol Chem. 2000;275:1529–1532. doi: 10.1074/jbc.275.3.1529. [DOI] [PubMed] [Google Scholar]
- Moore M, Goforth RL, Mori H, Henry R. Functional interaction of chloroplast SRP/FtsY with the ALB3 translocase in thylakoids: substrate not required. J Cell Biol. 2003;162:1245–1254. doi: 10.1083/jcb.200307067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Summer EJ, Ma X, Cline K. Component specificity for the thylakoidal Sec and Delta pH-dependent protein transport pathways. J Cell Biol. 1999;146:45–56. doi: 10.1083/jcb.146.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutka SC, Walter P. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol Biol Cell. 2001;12:577–588. doi: 10.1091/mbc.12.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TX, Chandrasekar S, Neher S, Walter P, Shan SO. Concerted complex assembly and GTPase activation in the chloroplast signal recognition particle. Biochemistry. 2011;50:7208–7217. doi: 10.1021/bi200742a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick S, Meurer J, Soll J, Ankele E. Nucleus-encoded light-harvesting chlorophyll a/b proteins are imported normally into chlorophyll b-free chloroplasts of Arabidopsis. Mol Plant. 2013;6:860–871. doi: 10.1093/mp/sss113. [DOI] [PubMed] [Google Scholar]
- Ouyang M, et al. LTD is a protein required for sorting light-harvesting chlorophyll-binding proteins to the chloroplast SRP pathway. Nat Commun. 2011;2:277. doi: 10.1038/ncomms1278. [DOI] [PubMed] [Google Scholar]
- Paila YD, Richardson LGL, Schnell DJ. New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J Mol Biol. 2015;427:1038–1060. doi: 10.1016/j.jmb.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Hoober JK. Chlorophyll synthesis modulates retention of apoproteins of light-harvesting complex II by the chloroplast in Chlamydomonas reinhardtii. Physiol Plantarum. 1997;101:135–142. doi: 10.1034/j.1399-3054.1997.1010118.x. [DOI] [Google Scholar]
- Payan LA, Cline K. A stromal protein factor maintains the solubility and insertion competence of an imported thylakoid membrane protein. J Cell Biol. 1991;112:603–613. doi: 10.1083/jcb.112.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumley GF, Schmidt GW. Light-harvesting Chlorophyll a/b Complexes: interdependent pigment synthesis and protein assembly. Plant Cell. 1995;7:689–704. doi: 10.1105/tpc.7.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JE, Cline K, Stephens LC, Bacot KO, Viitanen PV. Early events in the import/assembly pathway of an integral thylakoid protein. Eur J Biochem. 1990;194:33–42. doi: 10.1111/j.1432-1033.1990.tb19423.x. [DOI] [PubMed] [Google Scholar]
- Reinbothe C, et al. A role for chlorophyllide a oxygenase in the regulated import and stabilization of light-harvesting chlorophyll a/b proteins. Proc Natl Acad Sci USA. 2006;103:4777–4782. doi: 10.1073/pnas.0511066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Lamppa GK. Stromal processing peptidase binds transit peptides and initiates their ATP-dependent turnover in chloroplasts. J Cell Biol. 1999;147:33–44. doi: 10.1083/jcb.147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter CV, Bals T, Schünemann D. Component interactions, regulation and mechanisms of chloroplast signal recognition particle-dependent protein transport. Eur J Cell Biol. 2010;89:965–973. doi: 10.1016/j.ejcb.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Rutschow H, Ytterberg AJ, Friso G, Nilsson R, van Wijk KJ. Quantitative proteomics of a chloroplast SRP54 sorting mutant and its genetic interactions with CLPC1 in Arabidopsis. Plant Physiol. 2008;148:156–175. doi: 10.1104/pp.108.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saller MJ, Wu ZC, de Keyzer J, Driessen AJ. The YidC/Oxa1/Alb3 protein family: common principles and distinct features. Biol Chem. 2012;393:1279–1290. doi: 10.1515/hsz-2012-0199. [DOI] [PubMed] [Google Scholar]
- Saraogi I, Shan SO. Co-translational protein targeting to the bacterial membrane. Biochim Biophys Acta. 2014;1843:1433–1441. doi: 10.1016/j.bbamcr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schünemann D. Structure and function of the chloroplast signal recognition particle. Curr Genet. 2004;44:295–304. doi: 10.1007/s00294-003-0450-z. [DOI] [PubMed] [Google Scholar]
- Schünemann D, Gupta S, Persello-Cartieaux F, Klimyuk VI, Jones JDG, Nussaume L, Hoffman NE. A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc Natl Acad Sci USA. 1998;95:10312–10316. doi: 10.1073/pnas.95.17.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjuts I, Soll J, Bolter B. Import of soluble proteins into chloroplasts and potential regulatory mechanisms. Front Plant Sci. 2017;8:168. doi: 10.3389/fpls.2017.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel KF, Holdermann I, Wild K, Sinning I. The structure of the chloroplast signal recognition particle (SRP) receptor reveals mechanistic details of SRP GTPase activation and a conserved membrane targeting site. FEBS Lett. 2007;581:5671–5676. doi: 10.1016/j.febslet.2007.11.024. [DOI] [PubMed] [Google Scholar]
- Stengel KF, Holdermann I, Cain P, Robinson C, Wild K, Sinning I. Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science. 2008;321:253–256. doi: 10.1126/science.1158640. [DOI] [PubMed] [Google Scholar]
- Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, Coupland G. ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell. 1997;9:717–730. doi: 10.1105/tpc.9.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A. Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim Biophys Acta. 2011;1807:968–976. doi: 10.1016/j.bbabio.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Tanz SK, et al. The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LHCB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings. Plant J. 2012;69:743–754. doi: 10.1111/j.1365-313X.2011.04833.x. [DOI] [PubMed] [Google Scholar]
- Träger C, et al. Evolution from the prokaryotic to the higher plant chloroplast signal recognition particle: the signal recognition particle RNA is conserved in plastids of a wide range of photosynthetic organisms. Plant Cell. 2012;24:4819–4836. doi: 10.1105/tpc.112.102996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu CJ, Schuenemann D, Hoffman NE. Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J Biol Chem. 1999;274:27219–27224. doi: 10.1074/jbc.274.38.27219. [DOI] [PubMed] [Google Scholar]
- Tu CJ, Peterson EC, Henry R, Hoffman NE. The L18 domain of light-harvesting chlorophyll proteins binds to chloroplast signal recognition particle 43. J Biol Chem. 2000;275:13187–13190. doi: 10.1074/jbc.C000108200. [DOI] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T, et al. Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell. 2007;19:1635–1648. doi: 10.1105/tpc.106.048959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Hegde RS. Toward a structural understanding of co-translational protein translocation. Curr Opin Cell Biol. 2016;41:91–99. doi: 10.1016/j.ceb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Walter B, Hristou A, Nowaczyk MM, Schünemann D. In vitro reconstitution of co-translational D1 insertion reveals a role of the cpSec-Alb3 translocase and Vipp1 in photosystem II biogenesis. Biochem J. 2015;468:315–324. doi: 10.1042/BJ20141425. [DOI] [PubMed] [Google Scholar]
- Wang P, Dalbey RE. Inserting membrane proteins: The YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim Biophys Acta. 2011;1808:866–875. doi: 10.1016/j.bbamem.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Wild K, et al. Structural basis for conserved regulation and adaptation of the signal recognition particle targeting complex. J Mol Biol. 2016;428:2880–2897. doi: 10.1016/j.jmb.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Yang MJ, Pang XQ, Zhang X, Han KL. Molecular dynamics simulation reveals preorganization of the chloroplast FtsY towards complex formation induced by GTP binding. J Struct Biol. 2011;173:57–66. doi: 10.1016/j.jsb.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Yuan J, Kight A, Goforth RL, Moore M, Peterson EC, Sakon J, Henry R. ATP stimulates signal recognition particle (SRP)/FtsY-supported protein integration in chloroplasts. J Biol Chem. 2002;277:32400–32404. doi: 10.1074/jbc.M206192200. [DOI] [PubMed] [Google Scholar]
- Ziehe D, Dünschede B, Zenker M, Funke S, Nowaczyk MM, Schünemann D. The Chloroplast SRP systems of chaetosphaeridium globosum and physcomitrella patens as intermediates in the evolution of SRP-dependent protein transport in higher plants. PLoS ONE. 2016;11:e0166818. doi: 10.1371/journal.pone.0166818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehe D, Dünschede B, Schünemann D. From bacteria to chloroplasts: evolution of the chloroplast SRP system. Biol Chem. 2017;398:653–661. doi: 10.1515/hsz-2016-0292. [DOI] [PubMed] [Google Scholar]