Abstract
For the past 60 years Caulobacter spp. have been commonly attributed an aquatic and oligotrophic lifestyle yet are not uncommon in nutrient-rich or soil environments. This study evaluates the environmental and ecological associations of Caulobacter to reconcile past evidence, largely limited to culturing and microscopy, with currently available metagenomic and genomic data. The distribution of Caulobacter species and their characteristic adhesion-conferring genes, holdfast (hfaAB), were determined using collections of 10,641 16S rRNA gene libraries (196 studies) and 2625 shotgun metagenomes (190 studies) from a range of terrestrial and aquatic environments. Evidence for ecotypic variation was tested in 26 genomes sourced from soil, rhizosphere, plant, groundwater, and water. Caulobacter were, on average, fourfold more relatively abundant in soil than in aquatic environments, and abundant in decomposing wood, compost, and particulate matter (in air and water). Caulobacter holdfast genes were 35-fold more abundant in soils than aquatic environments. Ecotypic differences between soil and aquatic Caulobacter were evident in the environmental associations of several species and differences in genome size and content among isolates. However, most abundant species were common to both environments, suggesting populations exist in a continuum that was evident in the re-analysis of studies on the temporal dynamics of, and sources of bacterioplankton to, lakes and rivers. This study provides a new perspective on the ecological profile of Caulobacter, demonstrating that members of this genus are predominantly soil-borne, possess an overlooked role in plant matter decomposition and a dependency on water-mediated dispersal.
Introduction
The earliest descriptions of Caulobacter date back to the turn of the 20th century when biologists first observed bacteria with stalked, or ‘prosthecate,’ cell morphology [1–3]. Their ability to outlive other bacterial organoheterotrophs in nutrient depleted, long-term incubations and their frequent isolation from aquatic environments led to their characterization as oligotrophic and aquatic [1, 4]. Subsequent studies of the aquatic isolate Caulobacter crescentus CB15 (syn. C. vibrioides) supported this characterization and revealed physiological adaptations to low nutrient conditions that included optimizing cell surface area via stalk elongation [5, 6] and an uncommon configuration of outer membrane transporters [7, 8]. Their ability to adhere to surfaces using holdfast protein and their dimorphic lifecycle have also been interpreted as adaptations to aquatic, oligotrophic environments [8, 9]. The widespread acceptance of this ecophysiological profile is apparent in the general lack of supporting citation when referred to in the literature [6, 8, 10–15].
The first isolations of Caulobacter from soil date back to Nemec and Bystricky [16] and Poindexter [4], but were treated as exceptional given that all prior Caulobacter isolates were from oligotrophic, freshwater sources [1–3, 17–19]. Caulobacter have since been isolated from nutrient-rich environments, like wastewater [20] and the rhizosphere [21, 22], and observed in high abundance in pulp mill waste lagoons [23]. Yet, in general, the ecology and function of Caulobacter from non-aquatic or nutrient-rich environments has received little attention. With the rise of metagenomics, one can find a patchwork of cultivation-independent evidence for the presence of Caulobacter in soils and their role in decomposition, including the degradation of cellulose [24–26], lignin [27, 28], and polyaromatic hydrocarbons [29, 30]. Their role in decomposition is supported by the capacity of C. crescentus CB15 to grow on lignocellulose-degradation by-products, like xylose and vanillin [31–34], and the capacity of other species to grow on cellulose [35]. This evidence raises questions about the prevalence and ecology of Caulobacter in soils and the extent to which soil and aquatic strains differ in physiology and genetics (i.e., ecotypic variation).
A reliance on water, or moist conditions, is likely to be a defining characteristic of Caulobacter ecology in terrestrial environments. The number of culturable Caulobacter significantly decreased when soil was dried and increased when soil was agitated in water prior to culturing [5, 36]. Similarly, the deleterious effect of prolonged exposure to dry conditions was evident in the decline of soil Caulobacter populations in the decades following timber harvesting [37] and in a soil warming experiment [38]. The reduction in Caulobacter populations in drier soil is likely due to attrition from the inability of irreversibly bound cells to disperse and colonize new resources. The importance of water-mediated dispersal was evident in the rapid appearance of Caulobacter in overlaying water when river sediments were re-wet, surpassed only by fast-growing Bacillus spp. [39]. The potential increase in dispersal of Caulobacter during periods of wetness raises the possibility that sizable populations may wash from terrestrial into aquatic environments. The potential for this process to occur is supported by their membership in the stable autochthonous community in alpine groundwater [40]. The vagile life stage of Caulobacter is expected to have important ramifications for their soil ecology which have yet to be examined.
Several lines of evidence exist for the current ecophysiological profile of Caulobacter, yet the most heavily cited and influential ecological research was conducted by Poindexter in the mid-20th century (Figure S1) and remains largely untested by the tools of modern molecular ecology. The present study aims to explore the ecology of Caulobacter via a meta-analysis of publicly available 16S rRNA gene amplicon libraries, whole-shotgun metagenomes and genomes. The study represents the first cultivation-independent environmental survey of Caulobacter and seeks to (i) test the assumption that Caulobacter predominate in aquatic, nutrient-poor environments, (ii) determine the extent of ecotypic variation in Caulobacter populations between soil and aquatic environments, and (iii) to examine the role of water-mediated dispersal in Caulobacter ecology. The results have the potential to change perspectives on the involvement of Caulobacter spp. in soil processes, such as organic matter cycling, soil aggregation (given their adhesive properties), and possibly plant-microbe interactions.
Methods
Environmental survey of Caulobacter 16S rRNA genes
16S rRNA gene amplicon libraries (‘targeted locus’ metagenomes) were obtained from the NCBI using the BioProject portal with NCBI taxonomy IDs corresponding to metagenomes from soil (410658), phyllosphere (662107), sediment (749907; 412755; 556182), aquatic (1169740), freshwater (449393), marine (408172), and gut sources (749906; 408170; 410661; 1202446; 1510822; 1436733; 506599). All BioProjects with greater than three 16S rRNA gene sequencing libraries and sufficient metadata to identify the source and sampling location were included (Table S1). All experimentally manipulated samples were discarded or used separately in targeted analyses if the study design was relevant to the objectives of the present study. A study of contaminant bacterial DNA present in four commonly used DNA extraction kits [41] was included to rule out systemic bias in sequencing libraries. The complete list of BioProject metadata can be found in Supplementary Data, including an account of which samples in each project failed to meet the acceptance criteria. The following environments were represented: lake, glacier, groundwater, pond, river, estuary, marine, wetland, grassland, forest, plant, agricultural, compost, canyon, tundra, shrubland, alpine, atmosphere, host-associated, urban, and built. The following sample sources were represented: sediment, water, particle-attached (aqueous), biofilm, air, soil, phyllosphere, rhizosphere, wood, plastic, reagents, ice, feces, rumen, and gut (details in Table S1).
Sequencing libraries were quality filtered (‘trim.seqs’; q-scoreavg = 30) and classified using mothur [42], with its implementation of the RDP Classifier [25], against the Greengenes database (database gg_13_8_99; August 2013). This dataset served for all taxonomy-based analyses of relative abundance. Libraries with fewer than 500 quality processed reads were discarded. All sequences classified to the family Caulobacteraceae were subsequently assigned to operational taxonomic units (OTUs) at a similarity threshold of 99% using ‘closed-reference OTU picking’ in QIIME [43] with the SILVA tree as reference, which included a total of 442 Caulobacter sequences (‘SILVA_128_SSURef’; downloaded 20 July 2017). This form of OTU selection was necessary for integrating 16S rRNA amplicon libraries spanning different variable regions. All counts were normalized to total counts per thousand reads, a measure of relative abundance, for each library. All analyses can be reproduced using data and scripts available in Supplementary Data package. This includes the targeted analysis of certain datasets used in case studies examining the association of Caulobacter with low-nutrient environments and water-mediated dispersal.
Environmental survey of holdfast proteins in shotgun metagenomes
All publicly available metagenomes hosted on IMG/ER [44] were queried using the ‘Find Functions’ tool with KEGG orthology numbers for holdfast gene (hfa) subunit A (K13585) and B (K13586), which are necessary for Caulobacter surface adherence [45]. The total counts per metagenome, sample metadata, and amino acid sequences were downloaded for all shotgun metagenomes present in IMG/ER as of 31 May 2017. Metagenomes originated from a diverse set of environmental sources from across North American and the Pacific and Atlantic Oceans (Figure S2). The ‘Ecosystem Category’ was used to distinguish between ‘terrestrial’, ‘aquatic’, and ‘plant’ (plant-associated) environments and ‘Ecosystem Type’ was used as a more detailed descriptor of sample sources. All hfaA (n = 8614) and hfaB (n = 26,340) sequences were classified to genus level based on diamond BLAST [46] protein homology searches against a database containing all hfaAB present in genomes from IMG/ER (110 genomes). Classifications were designated based on the top BLAST hit with >80% similarity across 90% length of gene. All counts were normalized to counts per million bases for each metagenome assembly.
Comparative genomics
All publicly available Caulobacter genomes were downloaded using the pyani script ‘genbank_get_genomes_by_taxon.py’ with the NCBI taxonomy ID 75 on 31 May 2017 [47]. Three metagenome-assembled genomes and one genome from an unclassified member of Caulobacteraceae (‘PMMR1’) were discarded. C. crescentus NA1000 was also discarded, since it is an engineered variant of C. crescentus CB15. The unpublished genome of a Caulobacter strain (‘iso597’) isolated from forest soil by VanInsberghe et al., [48] was included in analyses and is now publicly available via IMG/ER (Taxon ID 2524614525). Of the total 26 genomes analyzed, four were derived from single-cell genome amplification and the remainder from characterized isolates. Comparison of genome content was based on reciprocal best BLAST hits computed with GET_HOMOLOGUES [49]. Phylogenetic relatedness was determined by BLAST-based average nucleotide identity (ANI) calculations with pyani and by multi-locus sequence alignments using the ‘insert genome into tree’ tool on KBase (details in Supplementary Methods [50]). Oligotrophic traits were assessed using the methodology and genomic signatures (COGs) presented in Lauro et al., [51]. Functional gene annotations were based on homology to clusters of orthologous groups (COGs) provided by IMG/ER. The annotation of carbohydrate-active enzymes (CAZymes) was based on diamond BLASTx protein homology searches against a local CAZy database (downloaded 19 August 2015). The following glycosyl hydrolase families were deemed endoglucanases involved in cellulose degradation: 5, 6, 7, 8, 9, 12, 26, 44, 45, 48, 51, 61, 74, 81, and 131 [52–60]. ORF prediction was performed with Prodigal ([61]; v. 2.6.2) prior to BLAST searches. Genes with >60% identity were attributed homologous function.
Bioinformatics
Statistics were performed using R (v. 3.3.1, R Core Team, 2016) with a general dependency on the following packages: reshape2, ggplot2, plyr [62–64], Hmisc [65], and phyloseq [66]. Complete linkage, hierarchical clustering was performed on ANI distances and the Bray-Curtis dissimilarity of shared genomic content using ‘vegdist’ from the R-package vegan [67]. A tanglegram was prepared from the clustering data with ‘dendextend’ [68]. A maximum likelihood phylogenetic tree of lovK, a photo-responsive gene regulator [15] was built using MEGA6 [69] with the Jones–Taylor–Thornton substitution model and uniform substitution rate.
Results
Environmental distribution of Caulobacter in 16S rRNA gene libraries
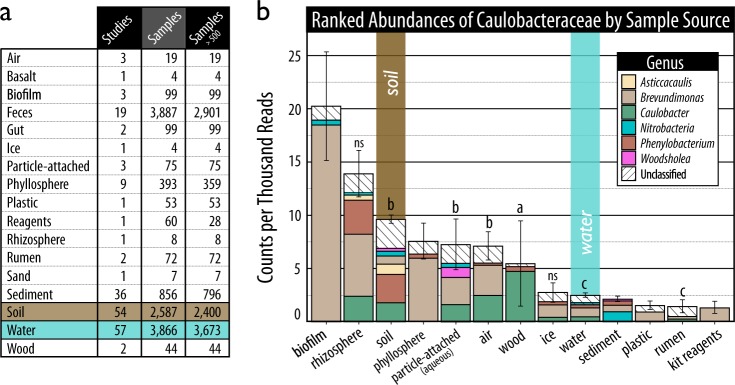
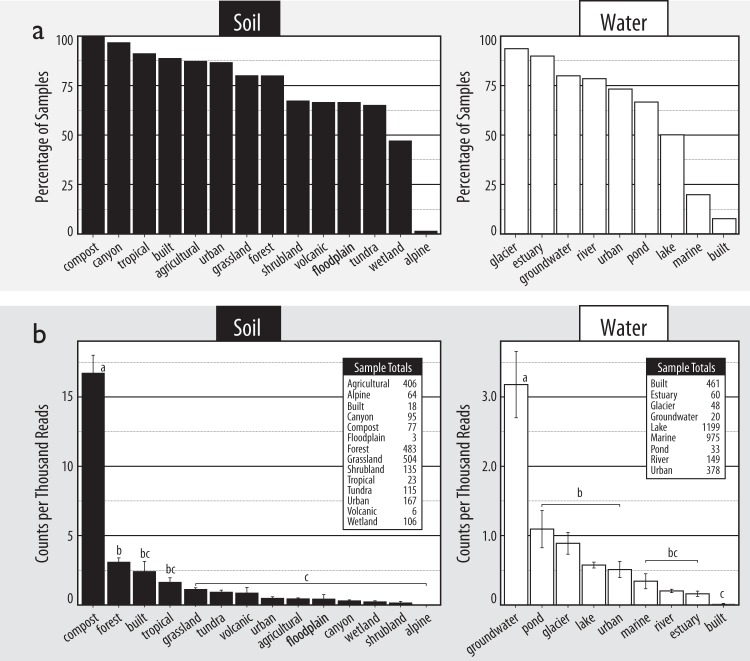
To test whether Caulobacter predominate in aquatic and/or oligotrophic environments, their relative abundance was calculated in 10,641 16S rRNA gene libraries from 196 studies spanning a variety of terrestrial and aquatic habitats (Fig. 1a). Not only were Caulobacter significantly more abundant in soils (on average 0.17% of sequences per library) than aquatic (0.04%) environments (Fig. 1b; Mann–Whitney; U3224 = 5.5 × 106; p ≈ 0), they were more frequently detected in soil samples, on average in 76 vs. 41% of samples, respectively (Fig. 2a). Caulobacter were found at greatest relative abundances in habitats not associated with nutrient limitation, evident in the fact three of the top five samples originated from decomposing wood (17.1% of total reads; BioProject ID: PRJNA205418), compost (6.3%; PRJEB7318) and buried wood in a boreal peat forest (3.9%; PRJEB669). In types of water and soil environments, Caulobacter were found at their highest relative abundances in groundwater and compost, respectively (Fig. 2b). Close relatives to Caulobacter also exhibited associations with soils, including members of Asticcacaulis and Phenylobacterium (Fig. 1a). Caulobacter were absent from sequences identified as contaminants in DNA extraction kits, demonstrating the lack of potential systemic methodological bias (Fig. 1a).
Fig. 1.
The ranked relative abundance of 16S rRNA genes classified to genera in the family Caulobacteraceae in (a) sequencing libraries from a variety of environmental samples (b) averaged and plotted as stacked bar plots. In (a), the total number of samples are shown before and after removing those with fewer than 500 quality-filtered sequences. In (b), the lettering denotes statistically supported differences in the relative abundance of specifically Caulobacter (Tukey HSD; p < 0.001). Error bars represent the standard error of counts of all Caulobacteraceae
Fig. 2.
The environmental distribution of Caulobacter in 16S rRNA gene libraries from sources of soil and water samples ranked by (a) the percentage of libraries in which Caulobacter were detected and (b) relative abundance. In (b), tables show the total number of samples from each environment. Lettering denotes statistically supported differences in the relative abundance of Caulobacter (Tukey HSD; p < 0.05). Error bars represent standard error
Environmental distribution of holdfast genes
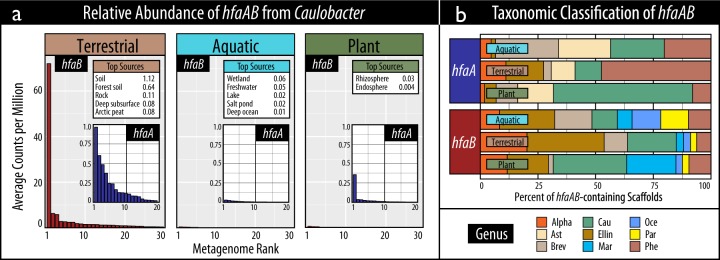
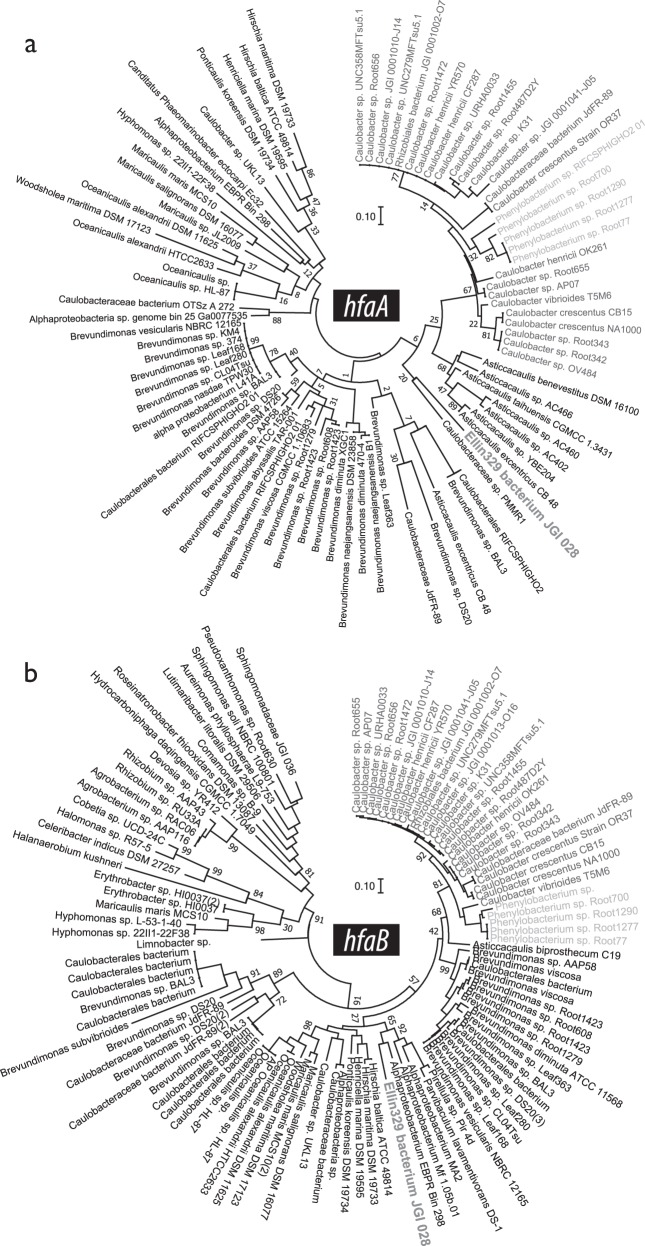
The relative abundance of holdfast (hfaAB) encoded by Caulobacter was also greatest in terrestrial environments, supporting the 16S rRNA gene-based evidence (Fig. 3a). A total of 2625 out of an available 8058 shotgun metagenome assemblies from 190 studies contained at least one predicted holdfast gene (Table S1) and the relative abundances of hfaA and hfaB were concordant (Spearman’s ρ = 0.9; p < 0.001). On average, the highest relative abundances of Caulobacter hfaAB occurred in soil metagenomes (0.46 counts per million bp assembly) and was significantly greater than in aquatic metagenomes (0.01 cpm; Mann–Whitney; U63 = 16,052; p = 0.001). The sources where the highest relative abundances of hfaAB occurred were unspecified soil, wetland and the rhizosphere for each environment (Fig. 3a). The greatest proportion of hfaB were classified to the candidate alphaproteobacterial order ‘Ellin329’ (31%), followed by Caulobacter (25%). For hfaA, this trend was only observed in terrestrial environments (Fig. 3b). All holdfast-encoding taxa were members of the Alphaproteobacteria and hfaAB encoded by Caulobacter were phylogenetically distinct, grouping closest to hfaAB from Phenylobacterium (Fig. 4). A more phylogenetically diverse set of taxa encoded hfaB compared to hfaA. The relative abundance of hfaA and hfaB from all taxa were on average 12- and 21-fold more abundant in terrestrial versus aquatic metagenomes, respectively.
Fig. 3.
An overview of the composition of holdfast genes, hfaA and hfaB, encoded by Caulobacter in terrestrial, aquatic and plant-associated samples according to (a) the ranked relative abundance of gene copies in assembled shotgun metagenomes, and (b) taxonomic classifications. In (a), tables show the top five environmental sources where the greatest relative abundances occur. In (b), the taxonomic abbreviations stand for the following: Alpha (unclassified Alphaproteobacteria), Ast (Asticcacaulis), Brev (Brevundimonas), Cau (Caulobacter), Ellin (Ellin329), Mar (Maricaulis), Oce (Oceanicaulis), Par (Parvibaculum), and Phe (Phenylobacterium)
Fig. 4.
The phylogeny of all (a) HfaA and (b) HfaB present in genomes available through IMG/ER (n = 93 and 110, respectively). Each maximum likelihood tree is based on the Jones–Taylor–Thornton substitution model and was bootstrapped 100 times
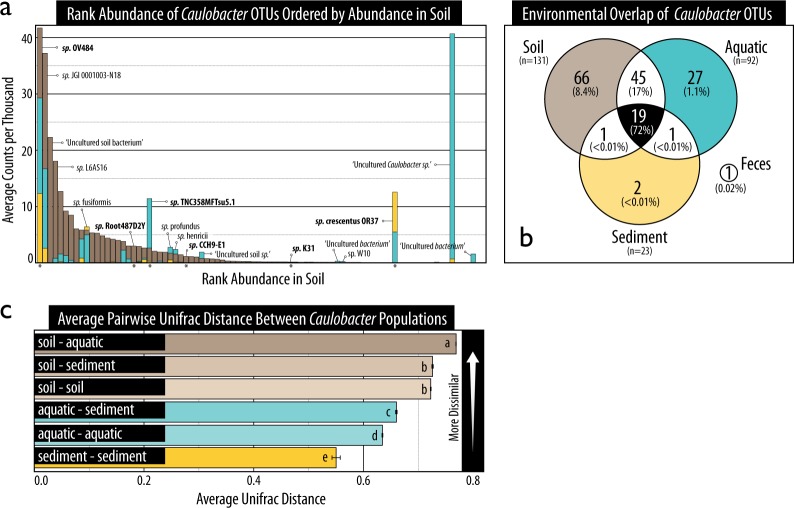
Ecotypic variation in Caulobacter OTUs
The environmental distributions of Caulobacter species were evaluated to identify ecotypic distinctions between soil and aquatic populations. A subset of Caulobacter species were exclusive to either aquatic and sediment or soil environments (Fig. 5a, b). Of the 131,600 sequences classified to the family Caulobacteraceae, ~18% were assigned to 162 Caulobacter OTUs at 99% similarity. Two highly abundant OTUs (ranked 2nd and 7th, respectively) were exclusive to aquatic and sediment samples: an uncultured Caulobacter sp. (cloned from a freshwater lake; JF275033) and C. crescentus OR37 (isolated from groundwater). The fourth-ranked OTU (‘uncultured soil bacterium’; FQ658960) as well as seven of the top 20 OTUs were exclusive to soils (Fig. 5a). Ecotypic differences were also apparent at the population level, where beta-diversity differed most between soil and aquatic habitats based on Unifrac phylogenetic distance (Fig. 5c). Beta-diversity was significantly higher among soils than among sediment or aquatic environments, suggesting a considerable degree of diversity within soil populations. Similarly, soils had the highest OTU richness (131 observed species) compared with aquatic (92) and sediment sources (23). Despite clear distinction in environmental distribution among a subset of OTUs, the majority of Caulobacter sequences (72%) were assigned to 19 OTUs found in all three environments (Fig. 5b). The two most abundant of these were related to C. sp. OV484 (plant-root isolate) and C. sp. JGI 0001003-N18 (unknown origin). Notably, no sequences formed an OTU based on the C. crescentus CB15 reference sequence present in the SILVA database.
Fig. 5.
An OTU-based account of the composition of Caulobacter populations in soil, sediment and aquatic environments according to (a) their ranked relative abundance, (b) the overlap in OTUs shared among the three sample sources, and (c) the average Unifrac phylogenetic distance. In (a), bars are superimposed (i.e., not stacked). The species name of abundant or common OTUs have been provided and those represented by genomes analyzed in this study have been bolded and marked with an asterisk. In (b), the percentage of sequences represented by all overlapping OTUs is shown in brackets beneath the number of overlapping OTUs. In (c), lettering denotes statistically supported differences in average pairwise distances (Tukey HSD; p < 0.05). Error bars represent standard error. OTUs had a minimum of 99% similarity to reference sequences in SILVA
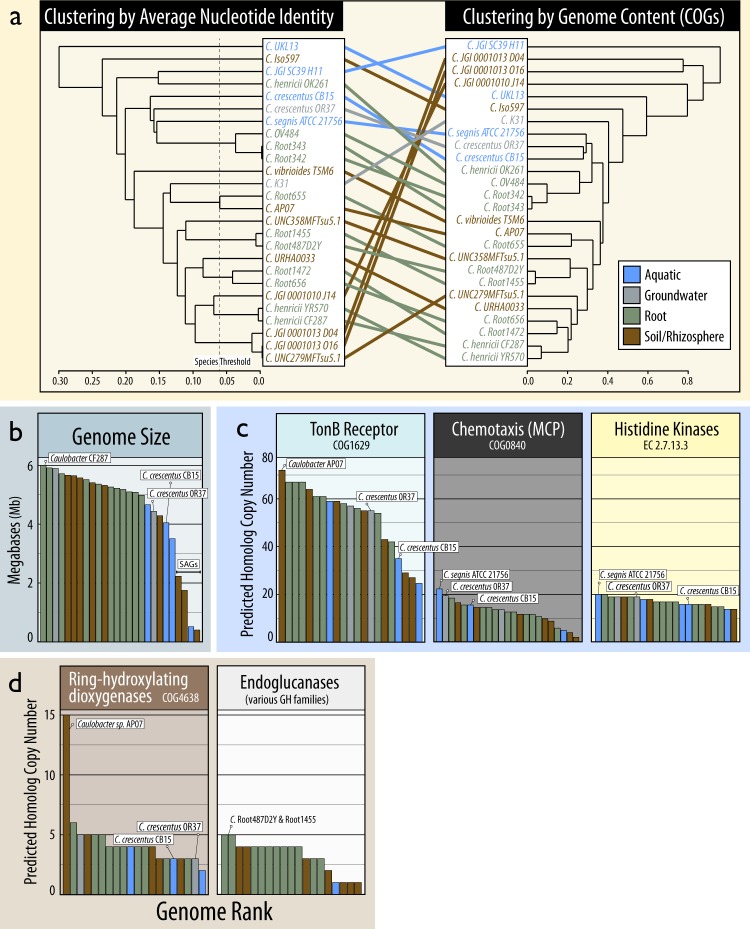
Ecotypic variation in Caulobacter genomes
Signatures of ecotypic variation were assessed based on phylogeny and functional gene content in 22 Caulobacter isolate genomes and four single-amplified genomes (SAGs) from 14 different studies. Genomes were sourced from bulk soil (3 genomes), rhizosphere soil (6), plant-root (11), aquatic (4), and groundwater (2) (Table S3). Aquatic isolates and the groundwater isolate, C. crescentus OR37, consistently grouped according to phylogeny and functional gene content based on multi-locus sequence alignments (Figure S3), ANI, COG profiles (Fig. 6a) and CAZyme content (Figure S4). Notably, sequences assigned to an OTU based on the C. crescentus OR37 16S rRNA gene were exclusive to aquatic and sediment habitats (Fig. 5a), suggesting it may represent an aquatic/groundwater ecotype. Genomes from aquatic isolates and C. crescentus OR37 tended to be smaller than those isolated from root, bulk soil, or rhizosphere (Fig. 6b) and were substantially under-represented in genes encoding endoglucanases (Fig. 6d). Isolates from bulk soil also encoded a greater number of ring-hydroxylating dioxygenases, including several ring-cleaving families (COG3565, COG3805, and COG5517) exclusive to two of the three soil isolates: C. vibrioides T5M6 and C. iso597, and the groundwater isolate (C. sp. K31). These differences corroborated the existence of Caulobacter ecotypes differentiated largely by aquatic and groundwater versus soil habitats. The four SAGs were outliers in genome size and gene content, indicating a poor quality of assembly.
Fig. 6.
Comparisons of Caulobacter genomes based on (a) their phylogenetic and functional relatedness, (b) genome size, (c) abundance of transport, chemotaxis, and signaling genes identified as signatures of oligotrophy in Caulobacter [8], and (d) catabolic genes commonly involved in decomposing lignocellulose. In a the phylogenetic relatedness is based on average nucleotide identity (1 – ANI) and functional relatedness is based on Bray-Curtis dissimilarity of COG profiles. The ANI threshold associated with separate species are indicated (Konstantinidis and Tiedje, 2005). The dendrograms in (a) were ordered using the ladderize function. In (b–d), the rank location of the type strain, C. crescentus CB15, and the closely related C. crescentus OR37 are displayed (when present) and the genome(s) with the highest number of copies of each gene family
Genetic signatures of oligotrophy were used to test whether such traits contribute to ecotypic variation in terrestrial and aquatic isolates. Proposed Caulobacter-specific signatures of oligotrophy, such as an overabundance of histidine kinases, methyl-accepting chemotaxis proteins (MCPs) and TonB-dependent outer membrane proteins [8], did not consistently differentiate aquatic from terrestrial genomes, though MCPs were highest in an aquatic isolate and C. crescentus OR37 (Fig. 6c). 16S rRNA gene copy number ranged between 1 and 7 (µrrn = 1.8) and did not significantly differ between aquatic (µrrn = 1.6) and terrestrial (µrrn = 1.4) isolates. Aquatic isolate genomes and C. crescentus OR37 grouped together based on a set of genes common to oligotrophic bacteria [51], yet did not exhibit a semblance to the prototypical pattern of oligotrophy, which clustered with the soil isolate C. segnis iso597 (Figure S5). The same group of aquatic/groundwater isolates clustered based on the phylogenetic relatedness of lovK (Figure S6), a gene responsible for the light-induced production of holdfast [15], though lovK was conserved in 23 of the 26 genomes, including all soil isolates, suggesting potential photo-responsiveness is not exclusive to aquatic ecotypes.
Case studies of Caulobacter ecology
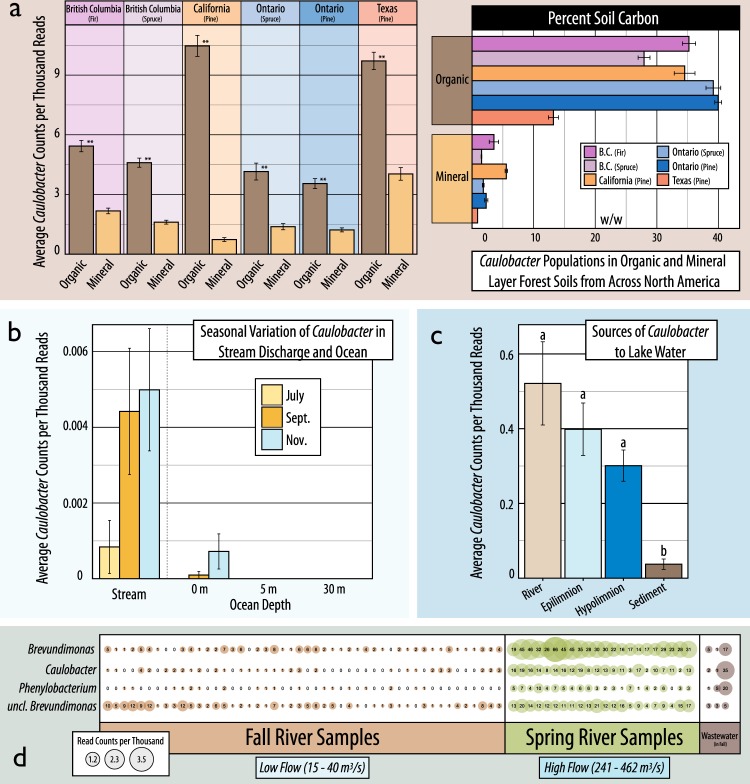
Several studies in the collection of 16S rRNA gene libraries were used to assess the distribution of Caulobacter across terrestrial, riverine, lake, and ocean samples, supporting the potential role of water-mediated dispersal. In a transect from upland coastal forest soils to adjoining stream and ocean water, Caulobacter were observed at higher relative abundances in streams during periods of high precipitation (Fall season) compared to drier summer months, with populations in the adjacent ocean water mirroring the temporal dynamics (Fig. 7b). In a survey of bacterial river populations, Caulobacter were also at their peak relative abundance at Spring thaw when river flow was greatest (Fig. 7d). In a study of lakes, Caulobacter were found at highest relative abundance in tributaries and decreased with increasing depth from the epilimnion to lake sediment (Fig. 7c). In the aforementioned coastal study, identical Caulobacter sequences (i.e. ~100% similarity) were detected in adjacent soil, stream and ocean (Figure S7b) with two of the top ten most abundant phylotypes common to all three habitats (~14% of total Caulobacter reads). However, the largest Caulobacter populations were exclusive to soil (~75%), and, to a lesser extent, stream water (~9%), while none were exclusive to ocean water. The importance of soil moisture regime was apparent in the reduced Caulobacter populations in a long-term drying experiment in agricultural soil (Figure S7c). However, in a second case study, the relative abundance of Caulobacter did not significantly differ between heated moist soils versus heated dry soils (not shown; PRJNA287307).
Fig. 7.
A compilation of several studies that demonstrate the associations of Caulobacter with organic matter rich soils and the spatial and temporal dynamics of populations in aquatic systems. In (a), the relative abundance of Caulobacter is contrasted between organic (5 cm deep) and mineral layer (5–20 cm) forest soils from across North America (data from ref. [26]). In (b), the seasonal differences in the relative abundance of Caulobacter in stream and ocean water (at stream discharge) are shown during drier Summer and rainier Fall months on the coast of British Columbia (PRJEXXXX). In (c), the relative abundance of Caulobacter are shown in sources of bacterioplankton to lake systems (PRJNA263673). In (d), the relative abundance of Caulobacter in the Grand River (Ontario, Canada) are shown during Fall and Spring flows at various locations (x-axis), including wastewater discharge streams (PREJEXXXX)
Several studies provided information regarding the association of Caulobacter with soil nutrient conditions and decomposition. In a comprehensive study of over 700 forest soils [26], Caulobacter populations were significantly more abundant in the organic layer than mineral layer soils, which contained significantly less organic carbon (t-test; p < 0.001; Fig. 7a). In a study targeting agricultural soil decomposers, Caulobacter exhibited characteristics of other trophic generalists, like Pseudomonas spp., in their capacity to metabolize a variety of organic compounds, including xylose, cellulose, vanillin, fatty acids, oxalate, and amino acids (Figure S7a; [70]). Fifteen out of the 196 amplicon libraries had data for organic matter or carbon content, though none exhibited significant correlation with Caulobacter relative abundances.
Discussion
The molecular-based meta-analysis presented here demonstrates that Caulobacter populations are more common and abundant in soil than aquatic environments. The results were consistent in both phylogenetic gene marker and shotgun metagenomic datasets, where canonical Caulobacter holdfast genes were targeted. The use of relative abundance data to support this conclusion is justifiable given that microbial biomass is, on average, far greater in soil compared to aquatic environments [71–75]. Therefore, it is reasonable to equate higher relative abundance with higher total abundance. Other potential methodological biases would likely have underestimated relative abundance in soils, since the use of metagenome assemblies rather than raw reads favors aquatic populations with lower diversity and better assembly [76, 77]. Similarly, normalizing hfaAB counts to total assembly size would inflate estimates of gene counts in populations with smaller average genomes sizes, such as aquatic populations [78]. The results, therefore, offer a comprehensive and robust new perspective on the scope and scale of terrestrial Caulobacter populations.
The long-standing lack of recognition for soil Caulobacter populations may be attributed to a combination of factors that likely biased early research. When Caulobacter were first described, aquatic communities were routinely sampled by submersing glass slides in water, then characterizing the observable populations with microscopy [1, 5, 79]. The capacity of Caulobacter to strongly adhere to surfaces would have favored their attachment to slides and their readily distinguishable stalked, vibrioid morphology, and striking rosette formations [2] would have improved the likelihood of identification. Conversely, their irreversible attachment to surfaces would have complicated detection in soils, hindering efforts to dislodge and culture or visualize cells in an era limited to these methods. The association with oligotrophic aquatic environments may also result from their proclivity to bind to surfaces, since Caulobacter are less likely to occur in an unbound state where concentrations of suspended organic matter are high. Indeed, an inverse relationship between the number of Caulobacter and concentrations of aqueous particulate matter has been previously reported [5, 36], and was supported in the current dataset by the high-relative abundance of Caulobacter in particle-attached metagenomes (air and aqueous). It should come as less of a surprise, then, that Caulobacter were found in greater abundances in soil and other terrestrial environments when the harsher extraction methods used to recover DNA from environmental sources, like bead beating, liberated the material of surface-bound cells.
The putative role of Caulobacter in plant matter decomposition was apparent in one of the earliest descriptions of the genus, where cellulose and chitin were each used to obtain enrichment cultures [1], as well as early genomic analysis [8] and in vitro characterizations [31, 32, 34, 35]. The results of the present study are the first to demonstrate the strength of their association with environments where decomposition is a primary process. The highest average relative abundances of Caulobacter were found in compost and forest soils, and two of the top five individual samples came from separate studies on decomposing wood (PRJNA205418 & PRJEB669). The abundance of hfaAB was similarly high in forest soils, surpassed only by the unspecified aggregated ‘soil’ category. Their predominance in forest soils is noteworthy given the accumulation therein of lignified plant matter, which is consistent with the enrichment of Caulobacter in lignin-degrading experiments in temperate and tropical forest soils [27, 28] and in co-culture with wood-rot fungi [80, 81]. The capacity for lignin-degradation was supported by the presence of ring-cleaving dioxygenase genes in two of the three soil isolate genomes. This newly recognizable role in decomposition implies an adaptive benefit for surface adhesion, possibly aiding the colonization of insoluble plant polymers while ensuring proximity to the by-products of extra-cellular catabolism. This possibility is strengthened by the high proportion of holdfast encoded by the candidate alphaproteobacterial order Ellin329, which are abundant in peatlands and can degrade xylan and cellulose [82].
The existence of aquatic and terrestrial ecotypes was supported by the species-level distributions of Caulobacter and by differences in genomic composition. These ecotypes lead one to reject the hypothesis that aquatic populations are wholly comprised of allochthonous persister cells derived from terrestrial run-off. The Caulobacter species that were exclusive to aquatic and sediment habitats matched a clone from a German freshwater lake [83] and a heavy-metal tolerant groundwater isolate C. crescentus OR37 [84]. The phylogenetic relatedness and similarity in genome content between OR37 and C. crescentus CB15 corroborates the use of CB15 as a model aquatic ecotype, though not ideal given its general absence from 16S rRNA gene libraries. Aquatic genomes were notably smaller than those from terrestrial sources, a trait common in oligotrophic and aquatic bacteria [85]. However, the content of aquatic genomes did not closely match previously proposed traits of bacterial oligotrophs [51], suggesting that we do not have sufficient or specific understanding of relevant gene markers, or that soil and aquatic populations share adaptations for oligotrophy. There were a greater number of ecotypes in soil than aquatic environments (66 vs. 27 OTUs, respectively), with the highest ranking ecotype first identified in a study on PAH-degradation in soil [86]. Expanding the collection of ecotype genomes identified here will greatly improve the power to resolve traits associated with aquatic versus soil and oligotroph versus decomposer ecophysiology.
The prevalence of Caulobacter species common to both soil and aquatic habitats provided a degree of support for the hypothesis that a continuum exists between the two environments driven, in part, by water-mediated dispersal. The largest Caulobacter populations from any aqueous environment occurred in groundwater metagenomes, suggesting groundwater outflow may act as a source to aquatic systems, concordant with previous observations in alpine groundwater systems [40]. Patterns of seasonal abundance further supported the possibility that a proportion of Caulobacter are allochthonous to lakes and rivers, given peak abundances occurred at times of heavier precipitation and run-off. These results conform with previous observations of higher Caulobacter cell counts in wetter Spring months in lakes [87] and rivers [88]. In the former case, Caulobacter were purportedly replenished from sediments from lake mixing [87], yet, the relatively low overall abundance in sediment versus river metagenomes in the present study, suggest the source is more likely riverine or terrestrial run-off. Based on these observations, one might conceive of Caulobacter’s oligotrophic traits as adaptations to the transient conditions experienced during dispersal. To characterize Caulobacter as facultative oligotrophs, or oligotolerant, better reconciles their oligotrophic traits with their relatively fast growth rates in nutrient-rich media [4, 89, 90], inhibition of holdfast production in low-nutrient conditions [91] and their high-relative abundances in wastewater [20], forest soil and compost. These largely circumstantial observations require additional quantitative evidence to determine the extent that aquatic populations of Caulobacter are replenished by water-borne dispersal from terrestrial habitats.
Conclusions
This study supports a shift away from the aquatic oligotroph paradigm and invites a broader consideration of the types of interactions that can occur between Caulobacter cell and environment. There are many ways in which oligotrophy, surface-attachment and a dimorphic lifecycle could confer fitness in both aquatic and terrestrial environments. This is evident in the variety of stimuli that regulate holdfast expression during Caulobacter’s cell cycle, such as nutrient depletion [91, 92], light-exposure [15], or surface contact [93]. The hypothesis that oligotolerance aids Caulobacter during water-mediated dispersal remains to be tested. Yet, it is clear we hold a naive view of the range of environments or processes where facultative oligotrophy is advantageous. Similarly, the prevalence of holdfast genes in soil from sources other than Caulobacter (an average of 10–20-fold more than aquatic environments) suggests we lack understanding about the range of life strategies for a holdfast-producers, the most apparent strategy relating to the digestion of insoluble plant fibers. By establishing the presence of soil Caulobacter populations, this study raises several important questions about their role in organic matter cycling and soil aggregation, and even their potential role in the rhizosphere, as recently evidenced [94]. For these question, and others, the apparent ease of culturing Caulobacter and their distinguishable cell morphology will aid research efforts now that we know to look.
Electronic supplementary material
Acknowledgements
This research was enabled by computational support from WestGrid (www.westgrid.ca) and Compute Canada (Calcul Canada; www.computecanada.ca) with funding from a NSERC graduate scholarship and University of British Columbia Four Year Doctoral Fellowship. Several analyses were made possible through data sharing by Dr. Colleen Kellog, Sara Coyotzi, Dr. Josh Neufeld, Dr. Nicholas Youngblut, Dr. Daniel Buckley, Dr. David VanInsberghe and Dr. William W. Mohn. Assistance in revising the manuscript was provided by Dr. Nagissa Mahmoudi.
Compliance with ethical standards
Conflict of interest
The author declares that they have no conflicts of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0257-z) contains supplementary material, which is available to authorized users.
References
- 1.Henrici AT, Johnson DE. Studies of freshwater bacteria: II. stalked bacteria, a new order of schizomycetes. J Bacteriol. 1935;30:61. doi: 10.1128/jb.30.1.61-93.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones M. A peculiar microorganism showing rosette formation. Zentr Bakteriol Parasitenk Abt II. 1905;14:459–63. [Google Scholar]
- 3.Loeffler F. Weitere Untersuchungen uber die Beizung und Farbung der Geisseln bei den Bakterien. Cent Bakteriol Parasitenkd. 1890;7:625–39. [Google Scholar]
- 4.Poindexter JS. Biological properties and classification of the Caulobacter group. Bacteriol Rev. 1964;28:231. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poindexter JS. The Caulobacters: ubiquitous unusual bacteria. Microb Rev. 1981;45:123–79. doi: 10.1128/mr.45.1.123-179.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner JK, Setayeshgar S, Sharon LA, Reilly JP, Brun YV. A nutrient uptake role for bacterial cell envelope extensions. Proc Natl Acad Sci USA. 2006;103:11772–77. doi: 10.1073/pnas.0602047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denancé N, et al. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS ONE. 2007;2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen J, et al. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci USA. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abraham WR, Rohde M, Bennasar A. The Family Caulobacteraceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes. Berlin: Springer-Verlag; 2014. pp. 179–205. [Google Scholar]
- 10.Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Entcheva-Dimitrov P, Spormann AM. Dynamics and control of biofilms of the oligotrophic bacterium Caulobacter crescentus. J Bacteriol. 2004;186:8254–66. doi: 10.1128/JB.186.24.8254-8266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benz R, Jones MD, Younas F, Maier E, Modi N, Mentele R, et al. OmpW of Caulobacter crescentus functions as an outer membrane channel for cations. PLoS ONE. 2015;10:e0143557. doi: 10.1371/journal.pone.0143557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landt SG, Lesley JA, Britos L, Shapiro L. CrfA, a small noncoding RNA regulator of adaptation to carbon starvation in Caulobacter crescentus. J Bacteriol. 2010;192:4763–75. doi: 10.1128/JB.00343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boutte CC, Crosson S. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment. Mol Microbiol. 2011;80:695–714. doi: 10.1111/j.1365-2958.2011.07602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci USA. 2007;104:18241–46. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemec P, Bystricky V. Peculiar morphology of some microorganisms accompanying diatomaceae preliminary report. J Gen Appl Microbiol. 1962;8:121–9. [Google Scholar]
- 17.Houwink AL. Caulobacter versus Bacillus spec. div. Nature. 1951;168:654. doi: 10.1038/168654b0. [DOI] [PubMed] [Google Scholar]
- 18.Omeliansky V. A new bacillus: Bacillus jagellatus. Zh Mikrobiol Epidemiol Immunobiol. 1914;1:24. [Google Scholar]
- 19.Bowers LE, Weaver R, Grula EA, Edwards O. Studies on a strain of Caulobacter from water I.: isolation and identification as Caulobacter vibrioides Henrici and Johnson with Emended Description. J Bacteriol. 1954;68:194. doi: 10.1128/jb.68.2.194-200.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacRae JD, Smit J. Characterization of caulobacters isolated from wastewater treatment systems. Appl Environ Microbiol. 1991;57:751–8. doi: 10.1128/aem.57.3.751-758.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moya G, Yan ZF, Won K, Yang JE, Wang QJ, Kook M, et al. Caulobacter hibisci sp. nov., isolated from rhizosphere of Hibiscus syriacus L.(Mugunghwa flower) IJSEM. 2017;67:3167–73. doi: 10.1099/ijsem.0.002021. [DOI] [PubMed] [Google Scholar]
- 22.Sun LN, Yang ED, Hou XT, Wei JC, Yuan ZX, Wang WY. Caulobacter rhizosphaerae sp. nov., a stalked bacterium isolated from rhizosphere soil. IJSEM. 2017;67:1771–6. doi: 10.1099/ijsem.0.001860. [DOI] [PubMed] [Google Scholar]
- 23.Stanley PM, Ordal EJ, Staley JT. High numbers of prosthecate bacteria in pulp mill waste aeration lagoons. Appl Env Microbiol. 1979;37:1007–11. doi: 10.1128/aem.37.5.1007-1011.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verastegui Y, Cheng J, Engel K, Kolczynski D, Mortimer S, Lavigne J, et al. Multisubstrate isotope labeling and metagenomic analysis of active soil bacterial communities. mBio. 2014;5:e01157–14. doi: 10.1128/mBio.01157-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilhelm RC, Cardenas E, Maas K, Leung H, McNeil L, Berch S, et al. Biogeography and organic matter removal shape long-term effects of timber harvesting on forest soil microbial communities. ISME J. 2017;11:2552–68. doi: 10.1038/ismej.2017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeAngelis KM, Allgaier M, Chavarria Y, Fortney JL, Hugenholtz P, Simmons B, et al. Characterization of trapped lignin-degrading microbes in tropical forest soil. PLoS ONE. 2011;6:e19306. doi: 10.1371/journal.pone.0019306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilhelm RC, Singh R, Eltis L, Mohn WW. Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. (in review, May 23rd, 2018). [DOI] [PMC free article] [PubMed]
- 29.Corgié S, Beguiristain T, Leyval C. Spatial distribution of bacterial communities and phenanthrene degradation in the rhizosphere of Lolium perenne L. Appl Environ Microbiol. 2004;70:3552–7. doi: 10.1128/AEM.70.6.3552-3557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yergeau E, Sanschagrin S, Beaumier D, Greer CW. Metagenomic analysis of the bioremediation of diesel-contaminated Canadian high arctic soils. PLoS ONE. 2012;7:e30058. doi: 10.1371/journal.pone.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee D, Bourquin A. Metabolism of aromatic compounds by Caulobacter crescentus. J Bacteriol. 1987;169:1993–6. doi: 10.1128/jb.169.5.1993-1996.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hottes AK, Meewan M, Yang D, Arana N, Romero P, McAdams HH, Stephens C. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J Bacteriol. 2004;186:1448–61. doi: 10.1128/JB.186.5.1448-1461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Presley GN, Payea MJ, Hurst LR, Egan AE, Martin BS, Periyannan GR. Extracellular gluco-oligosaccharide degradation by Caulobacter crescentus. Microbiol. 2014;160:635–45. doi: 10.1099/mic.0.072314-0. [DOI] [PubMed] [Google Scholar]
- 34.Thanbichler M, Iniesta AA, Shapiro L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acid Res. 2007;35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song N, Cai HY, Yan ZS, Jiang HL. Cellulose degradation by one mesophilic strain Caulobacter sp. FMC1 under both aerobic and anaerobic conditions. Bioresour Technol. 2013;131:281–7. doi: 10.1016/j.biortech.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Krasil’nikov NA, Belyaev SS. Distribution of Caulobacter in certain soils. Mikrobiologiya. 1967;36:1083–6. [PubMed] [Google Scholar]
- 37.Wilhelm RC, Cardenas E, Leung H, Szeitz A, Jensen LD, Mohn WW. Long-term enrichment of stress-tolerant cellulolytic soil populations following timber harvesting evidenced by multi-omic stable isotope probing. Front Microbiol. 2017;8:537. doi: 10.3389/fmicb.2017.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pold G, Melillo JM, DeAngelis KM. Two decades of warming increases diversity of a potentially lignolytic bacterial community. Front Microbiol. 2015;6:480. doi: 10.3389/fmicb.2015.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fazi S, Amalfitano S, Piccini C, Zoppini A, Puddu A, Pernthaler J. Colonization of overlaying water by bacteria from dry river sediments. Environ Microbiol. 2008;10:2760–72. doi: 10.1111/j.1462-2920.2008.01695.x. [DOI] [PubMed] [Google Scholar]
- 40.Farnleitner AH, Wilhartitz I, Ryzinska G, Stadler H, Burscher MM, Hornek R, et al. Bacterial dynamics in spring water of alpine karst aquifers indicates the presence of stable autochthonous microbial endokarst communities. Environ Microbiol. 2005;7:1248–1259. doi: 10.1111/j.1462-2920.2005.00810.x. [DOI] [PubMed] [Google Scholar]
- 41.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffat MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87–98. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75:7537–41. [DOI] [PMC free article] [PubMed]
- 43.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markowitz VM, Chen IA, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucl Acid Res. 2012;40(D1):D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole JL, Hardy GG, Bodenmiller D, Toh E, Hinz A, Brun YV. The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk. Mol Microbiol. 2003;49:1671–83. doi: 10.1046/j.1365-2958.2003.03664.x. [DOI] [PubMed] [Google Scholar]
- 46.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 47.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8:12–24. [Google Scholar]
- 48.VanInsberghe D, Hartmann M, Stewart GR, Mohn WW. Isolation of a substantial proportion of forest soil bacterial communities detected via pyrotag sequencing. Appl Environ Microbiol. 2013;79:2096–8. doi: 10.1128/AEM.03112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79:7696–701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arkin AP, Stevens RL, Cottingham RW, Maslov S, Henry CS, Dehal P. et al. The DOE Systems Biology Knowledgebase (KBase). bioRxiv. (2016). 10.1101/096354.
- 51.Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, et al. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA. 2009;106:15527–33. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen JC, Brown K, et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochem. 2010;49:3305–16. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 53.Hasper AA, Dekkers E, van Mil M, van de Vondervoort PJI, de Graaff LH. EglC, a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Appl Environ Microbiol. 2002;68:1556–60. doi: 10.1128/AEM.68.4.1556-1560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lafond M, Navarro D, Haon M, Couturier M, Berrin JG. Characterization of a broad-specificity β-Glucanase acting on β-(1,3)-, β-(1,4)-, and β-(1,6)-Glucans that defines a new glycoside hydrolase family. Appl Environ Microbiol. 2012;78:8540–6. doi: 10.1128/AEM.02572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petersen L, Ardèvol A, Rovira C, Reilly PJ. Mechanism of cellulose hydrolysis by inverting GH8 endoglucanases: a QM/MM metadynamics study. J Phys Chem B. 2009;113:7331–9. doi: 10.1021/jp811470d. [DOI] [PubMed] [Google Scholar]
- 56.Boyce A, Walsh G. Characterisation of a novel thermostable endoglucanase from Alicyclobacillus vulcanalis of potential application in bioethanol production. Appl Microbiol Biotechnol. 2015;99:7515–25. doi: 10.1007/s00253-015-6474-8. [DOI] [PubMed] [Google Scholar]
- 57.Vlasenko E, Schülein M, Cherry J, Xu F. Substrate specificity of family 5, 6, 7, 9, 12, and 45 endoglucanases. Bioresour Technol. 2010;101:2405–11. doi: 10.1016/j.biortech.2009.11.057. [DOI] [PubMed] [Google Scholar]
- 58.Warner CD, Go RM, García-Salinas C, Ford C, Reilly PJ. Kinetic characterization of a glycoside hydrolase family 44 xyloglucanase/endoglucanase from Ruminococcus flavefaciens FD-1. Enzyme Microb Technol. 2011;48:27–32. doi: 10.1016/j.enzmictec.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Brumm PJ, Hermanson S, Gowda K, Xie D, Mead DA. Clostridium thermocellum Cel5L-cloning and characterization of a new, thermostable GH5 cellulase. Int J Biochem Res Rev. 2015;6:62. [Google Scholar]
- 60.Yagüe E, Béguin P, Aubert JP. Nucleotide sequence and deletion analysis of the cellulase-encoding gene celH of Clostridium thermocellum. Gene. 1990;89:61–67. doi: 10.1016/0378-1119(90)90206-7. [DOI] [PubMed] [Google Scholar]
- 61.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wickham H. Reshaping data with the reshape Package. J Stat Soft. 2007;21:1–20. [Google Scholar]
- 63.Wickham H. The split-apply-combine strategy for data analysis. J Stat Soft. 2011;40:1–29. [Google Scholar]
- 64.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag; 2009. [Google Scholar]
- 65.Harrell FE, Dupont C. Hmisc: Harrell Miscellaneous. R package version 3.17-1. (2015). https://cran.r-project.org/web/packages/Hmisc/Hmisc.pdf
- 66.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2014;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB et al. vegan: Community Ecology Package. R package version 1.15-4. (2015). http://CRAN.R-project.org/package=vegan
- 68.Galili T. dendextend: an R package for visualizing, adjusting, and comparing trees of hierarchical clustering. Bioinformatics. 2015;15:3718–3720. doi: 10.1093/bioinformatics/btv428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tamura K, Stecher G, Peterson P, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Youngblut ND, Barnett SE, Buckley DH. HTSSIP: an R package for analysis of high throughput sequencing data from nucleic acid stable isotope probing (SIP) experiments. PLoS ONE. 2018;13:e0189616. doi: 10.1371/journal.pone.0189616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cole JJ, Pace ML, Caraco NF, Steinhart GS. Bacterial biomass and cell size distributions in lakes: more and larger cells in anoxic waters. Limnol Oceanogr. 1993;38:1627–1632. [Google Scholar]
- 72.Frostegård Aring, Bååth E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fertil Soils. 1996;22:59–65. [Google Scholar]
- 73.Simon M, Cho BC, Azam F. Significance of bacterial biomass in lakes and the ocean: comparison to phytoplankton biomass and biogeochemical implications. Mar Ecol Prog Ser. 1992;86:103–110. [Google Scholar]
- 74.Van EsF, Meyer-Reil LA. Biomass and metabolic activity of heterotrophic marine bacteria. In: Alexander M, editor. Advances in microbial ecology. Berlin: Springer; 1982. pp. 111–170. [Google Scholar]
- 75.Vance E, Brookes P, Jenkinson D. An extraction method for measuring soil microbial biomass carbon. Soil Biol Biochem. 1987;19:703–707. [Google Scholar]
- 76.Torsvik V, Øvreås L, Thingstad TF. Prokaryotic diversity--magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 77.van der Walt AJ, Van Goethem MW, Ramond J-B, Makhalanyane TP, Reva O, Cowan DA. Assembling metagenomes, one community at a time. bioRxiv. 2017;120154. [DOI] [PMC free article] [PubMed]
- 78.Raes J, Korbel J, Lercher M, von Mering C, Bork P. Prediction of effective genome size in metagenomic samples. Genome Biol. 2007;8:R10. doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stove J, Stanier R. Cellular differentiation in stalked bacteria. Nature. 1962;196:1189–1192. [Google Scholar]
- 80.Folman LB, Klein Gunnewiek PJA, Boddy L, De Boer W. Impact of white-rot fungi on numbers and community composition of bacteria colonizing beech wood from forest soil. FEMS Microbiol Ecol. 2008;63:181–191. doi: 10.1111/j.1574-6941.2007.00425.x. [DOI] [PubMed] [Google Scholar]
- 81.Valaskova V, de Boer W, Klein Gunnewiek PJA, Pospisek M, Baldrian P. Phylogenetic composition and properties of bacteria coexisting with the fungus Hypholoma fasciculare in decaying wood. ISME J. 2009;3:1218–1221. doi: 10.1038/ismej.2009.64. [DOI] [PubMed] [Google Scholar]
- 82.Harbison AB, Carson MA, Lamit LH, Basiliko N, Bräuer SL. A novel isolate and widespread abundance of the candidate alphaproteobacterial order (Ellin 329), in southern Appalachian peatlands. FEMS Microbiol Lett. 2016;363:fnw151. doi: 10.1093/femsle/fnw151. [DOI] [PubMed] [Google Scholar]
- 83.Jogler M, Siemens H, Chen H, Bunk B, Sikorski J, Overmann J. Identification and targeted cultivation of abundant novel freshwater sphingomonads and analysis of their population substructure. Appl Environ Microbiol. 2011;77:7355–7364. doi: 10.1128/AEM.05832-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Utturkar SM, Bollmann A, Brzoska RM, Klingeman DM, Epstein SA, Palumbo AV, et al. Draft genome sequence for Caulobacter sp. Strain OR37, a bacterium tolerant to heavy metals. Genome Announc. 2013;1:e00322–13. doi: 10.1128/genomeA.00322-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giovannoni SJ, Thrash JC, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2015;8:1553–1565. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin F, Torelli S, Le Paslier D, Barbance A, Martin-Laurent F, Bru D, et al. Betaproteobacteria dominance and diversity shifts in the bacterial community of a PAH-contaminated soil exposed to phenanthrene. Environ Pollut. 2012;162:345–353. doi: 10.1016/j.envpol.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 87.Staley JT, Konopka A, Dalmasso J. Spatial and temporal distribution of Caulobacter spp. in two mesotrophic lakes. FEMS Microbiol Lett. 1987;45:1–6. [Google Scholar]
- 88.Staley JT. Incidence of prosthecate bacteria in a polluted stream. Appl Microbiol. 1971;22:496–502. doi: 10.1128/am.22.4.496-502.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Campos M, Surovtsev IV, Kato S, Paintdakhi A, Beltran B, Ebmeier SE, et al. A constant size extension drives bacterial cell size homeostasis. Cell. 2014;159:1433–1446. doi: 10.1016/j.cell.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koch A. Oligotrophs versus copiotrophs. BioEssays. 2001;23:657–661. doi: 10.1002/bies.1091. [DOI] [PubMed] [Google Scholar]
- 91.Feibig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH, Crosson S. A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet. 2014;10:e1004101. doi: 10.1371/journal.pgen.1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Curtis PD. Stalk formation of Brevundimonas and how it compares to Caulobacter crescentus. PLoS ONE. 2017;12:e0184063. doi: 10.1371/journal.pone.0184063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. Surface contact stimulates the just‐in‐time deployment of bacterial adhesins. Mol Microbiol. 2012;83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhalnina K, Louie KB, Hao Z, Mansoori N, Nunes da Rocha U, Shi S, et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol. 2018;3:470–480. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.