Abstract
Although many advanced biosensing techniques have been purposed for cytokine profiling, there are no clinically available methods that integrate high-resolution immune cell monitoring and in situ multiplexed cytokine detection together in a biomimetic tissue microenvironment. The primary challenge arises due to the lack of suitable label-free sensing techniques and difficulty for sensor integration. In this work, we demonstrated a novel integration of a localized-surface plasmon resonance (LSPR)-based biosensor with a biomimetic microfluidic ‘adipose-tissue-on-chip’ platform for an in-situ label-free, high-throughput and multiplexed cytokine secretion analysis of obese adipose tissue. Using our established adipose-tissue-on-chip platform, we were able to monitor the adipose tissue initiation, differentiation, maturation and simulate the hallmark formation of crown-like structures (CLS) during pro-inflammatory stimulation. With integrated antibody-conjugated LSPR barcode sensor arrays, our platform enables simultaneous multiplexed measurements of pro-inflammatory (IL-6, and TNF-α) and anti-inflammatory (IL-10, and IL-4) cytokines secreted by the adipocytes and macrophages. As a result, our adipose-tissue-on-chip platform is capable of identifying stage-specific cytokine secretion profiles from a complex milieu during obesity progression, highlighting its potential as a high-throughput preclinical readout for personalized obesity treatment strategies.
Introduction
The adipose tissue of obese patients is characterized as a tissue microenvironment composed of adipose tissue macrophages, precursor and hypertrophic adipocytes, and other immune cells that predominantly produce pro-inflammatory cytokines, such as IL-6, IL-1β, and TNF-α, for chronic, low-grade inflammation.1,2 Adipose tissue inflammation is a critical risk factor of obesity and is linked to the development of complications like Type 2 Diabetes Mellitus, cardiovascular diseases and certain cancers. Pathologically, inflammation in obese adipose tissue has been defined as the accumulation of immune cells, specifically macrophages, around a necrotizing adipocyte, an entity known as a crown-like structure (CLS).3,4 There is a limited understanding of obesity-associated inflammation in adipose tissue microenvironment during disease progression. Previous studies of adipose tissue inflammation have mostly focused on ex vivo studies of CLS using adipose tissues isolated from mouse models or patient surgical waste,3,4 or characterizing the immunogenicity of adipocyte cell lines without a local, chronic inflammatory milieu.5,6 Hence, there is a demand for a platform that can report the spatiotemporal dynamics of cell cytokine secretion profiles during adipocyte differentiation and CLS formation and resolution in an adipose tissue microenvironment.
Cytokines are key immune regulators of inflammation and form complex immune-body cytokine networks for intercellular communications among immune cells and parenchymal cells of organs and tissues.7 Precisely quantifying cytokine secretions from immune and body cells remains a critical challenge due to their small molecule size, extremely low concentrations, and transient alterations during cell-cell signaling in the extracellular milieu.8,9 The enzyme-linked immunosorbent assay (ELISA) is widely used and regarded as a “gold standard” method for cytokine profiling. Conventional ELISA assays rely on fluoresce labeling, long term sample incubation and time-consuming manipulation processes, which greatly limit the real-time, multiplex cytokine measurements and the ability to monitor cell functions in a dynamic manner under niche-specific conditions.10 Recently, a few label-free detection strategies such as mechanical,11,12 electrochemical,13–16 optical,17–19 or surface plasma based biosensors,20–24 have been developed for real-time profiling multiplexed cytokines in biological fluid samples, such as serum,13 urine, or saliva13. However, tools to measure cytokine secretion dynamics and the associated cellular immune functions within immune-body tissue microenvironments remain in infancy, owing to difficulties in measurement setup for signal acquisition, and sensor integration.
Recent advances in lab-on-a-chip technologies have allowed the integration of cytokine biosensors and on-chip cell culture possible.25 For example, Luchansky et al.26 and Oh et al.27 have demonstrated platforms capable of dynamic and multiplexed T cell cytokine secretion profiling using integrated microfluidic devices with silicon micro-ring resonator sensor and nanoplasmonic gold nanoparticle sensor, respectively. However, these methodologies still cannot achieve on-chip, in situ measurements from cells due to laborious sample preparation steps of separating supernatants from cell culture. Others have shown microfluidic biosensing devices capable of on-chip immune cell isolation and integrated with28–30 or AlphaLISA31,32 sensors for detecting cytokine secretions of live cells. However, until now, there is still a lack of technologies that can achieve label-free, dynamic, and multiplexed cytokine profiling to monitor cellular functions in situ in a biomimetic tissue environment. Importantly, the need of sampling for these detection technologies will affect the original concentrations of cytokines released by the cells, compromising the ability to continuously monitor cell secretion profiles for extended culture periods (during cell differentiation and polarization). This limitation becomes even more pronounced when studying on-chip bio-mimic system with small volume of medium contained in a microfluidic chip (< 100 μL).
In this work, we demonstrate, for the first time, a novel, integrated microfluidic ‘adipose-tissue-on-chip’ platform for in situ, high-throughput, multiplexed analysis of cytokine secretion dynamics within a biomimetic adipose tissue microenvironment. Our biomimetic ‘adipose-tissue-on-chip’ platform permits a real-time, chronological observation of the dynamic stages of adipocyte differentiation and CLS formation. Simultaneously, we equipped a label-free localized surface plasmon resonance (LSPR)-based nanoplasmonic biosensor10,28,33,34 to the biomimetic adipose tissue microenvironment for in situ cytokine monitoring of adipose tissue inflammation. Multiplexed measurements of cytokine concentrations were achieved with a unique dark-field imaging scheme using antibody-conjugated gold nanorod (AuNR) LSPR nanobiosensor barcode microarrays.27,35 This on-chip immunoassay technology affords a wide dynamic range (~10−10,000 pg/mL) of cytokine detection and requires a low operating sample volume (down to ~1 μL) and short assay time (~30 min). More importantly, our label-free, imaging-based LSPR immunoassay is ideal for an in situ, dynamic monitoring of proinflammatory [tumor-necrosis-factor alpha (TNF-α) and interleukin-6 (IL-6)] and anti-inflammatory [interleukin-10 (IL-10) and interleukin-4 (IL-4)] cytokine secretions from the adipose tissue inflammation. Our platform eliminates the need of sampling by avoiding complex culture media separation procedures, which allows on-chip culture and continuous monitoring without perturbing cell/tissue microenvironments. Using our platform, we can quantitatively characterize a time-course functional response of adipocytes under inflammatory stimulation, as well as the immunophenotype switch of macrophages in an inflammatory adipose tissue microenvironment. This may facilitate the future development of new prognostic tools useful for personalized treatment strategies that address problems in obesity and its complications.
Results and Discussion
Integrated Adipose-Tissue-on-Chip Nanoplasmonic Biosensing Platform Design
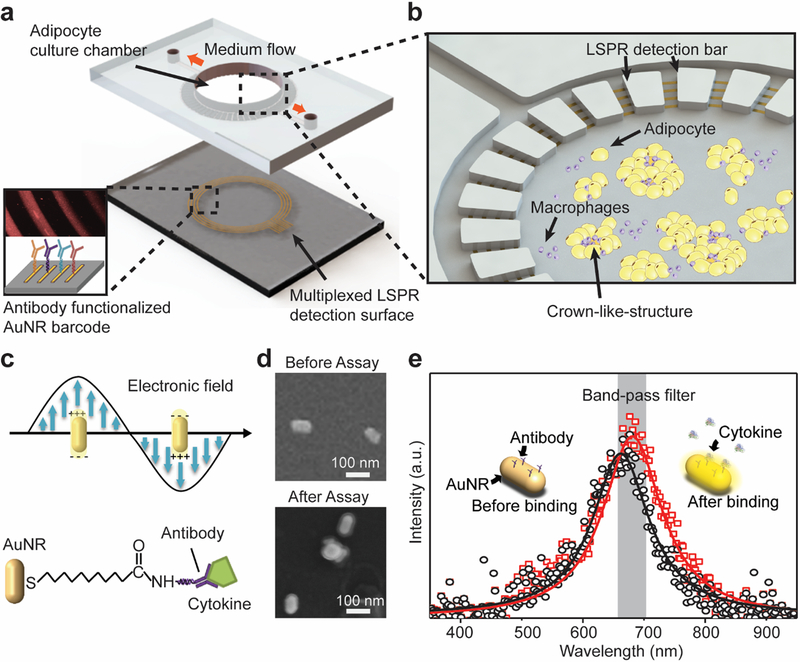
The integrated ‘adipose-tissue-on-chip’ used in this study consists of two components: a central adipose tissue culture chamber and a peripheral nanoplasmonic LSPR sensing array (Figure 1a& Figure S1). A polydimethylsiloxane (PDMS) layer with an incubation chamber (4-mm central diameter) was fabricated and bound to cover glass for adipose-tissue-on-chip culture. It has been considered that the formation of CLS composed of dead adipocytes surrounded by macrophages is a defining hallmark of obesity and adipose tissue dysfunction.36 Therefore, in this study, we presented a biomimetic culture model using murine adipocytes and macrophages, which are the predominant cells that are present in CLS and mediate inflammation in obese adipose tissue to form a biomimetic adipose tissue microenvironment. 3T3L1 cells, a widely-used precursor adipocyte line, and J774A.1 (J7) mouse macrophages were used and cultured in the central chamber at 37°C and 5% CO2 conditions for 15 days for on-chip formation of CLSs of obese adipose tissue (Figure 1b).
Figure 1. Adipose-tissue-on-chip sensing platform for an in situ multiplexed analysis of adipose tissue inflammation.
(a) Schematic of the integrated LSPR optofluidic platform device. The bottom layer is an AuNR-patterned barcode for multiplexed LSPR detection. The top PDMS layer provides a cell culturing chamber for adipocyte culturing and observation for on-chip CLS formation with introduction of macrophages. (b) Enlarged schematic showing the adipocyte culture chamber is surrounded by multiple microchannels connected to LSPR cytokine detection barcode arrays. The circular LSPR sensing arrays are align covered under the microchannel patterns surrounding the cell culturing chamber. The adipocytes and macrophages culture medium supernatant is then diffused into the microchannels and interact with the AuNR LSPR sensors. (c) Principle of LSPR-based biosensing with cytokine-binding events on AuNRs. (d) SEM images of individual AuNR biosensors immobilized on glass before and after immunoassay. After cytokine assay, the AuNR surfaces show a protein-coated layer. (e) Principle of dark-field intensity imaging of LSPR nanoplasmonic biosensor microarrays, where the binding between targeted antigen and sensing surface induces spectral redshift and intrinsic intensity enhancement of AuNR scattering light. Measuring intensity changes with a band-pass filter enable us to quantify the analyte concentration in samples.
To build up a complete profile of immune cell cytokine secretion, we integrated a LSPR ‘barcode’ sensing array of four circular AuNR stripes aligned perpendicular to the microchannel (1000 μm long, 200 μm wide) patterns surrounding the adipose tissue chamber for cytokine detection (Figure S1). Each LSPR AuNR detection stripe has a width of 100 μm and an inter-bar gap of 100 μm and functionalized with different cytokine detection antibodies on another glass layer using a one-step microfluidic patterning method (Figure S1d&S2). The assembled device allows cell culture medium to diffuse from the adipose tissue chamber into the microchannels and interact with the AuNR barcodes for a “label-free” cytokine detection of the adipose tissue microenvironment (Figure 1b and Figure S1c). The integrated microfluidic adipose-tissue-on-chip coupled with LSPR dark-field imaging permitted us to conduct a functional immunoassay for an in-situ, multiplex monitoring of a group of immune cells secreting cytokines from the adipose tissue microenvironment. Such a device can be readily used to provide predictive information and mechanistic insights for unraveling the complex, adaptive nature of obese adipose tissue in a proinflammatory milieu under macrophage recruitment.
Cytokine Detection Standard Curve Acquisition and Validation with ELISA.
LSPR is a plasmonic phenomenon that arises around nanoscale structures or nanoparticles of a conductive metal (AuNRs in this study) when light is illuminated on its surface (Figure 1c&d).37 When the incident light frequency matches the natural frequency of electron oscillation of the AuNRs, it generates significantly enhanced near-field LSPR “hot spots” at the corners of the nanoparticles and results in an increased number of scattered photons in far-field, which are sensitive to the condition changes in the vicinity of metal nanoparticles. Cytokine binding events lead to changes in the near-field refractive index around AuNRs, shifting the intensity and phase of the extinction spectra (Figure 1e) and, thus, increasing dark-field LSPR image brightness. Applying this LSPR mechanism, we established a microfluidic dark-field imaging platform for rapid, precise, and dynamic cytokine measurements (Figure S3a).
The layout and preparation of LSPR AuNR strips are shown in Figure 1a and Figure S2 (see details in Materials and Methods). Aqueous cetrimonium bromide (CTAB) coated AuNRs (NanoSeedz Ltd.) with average lengths of 80±5 nm and widths of 40±3 nm were used as the sensing elements and uniformly distributed on the glass substrate with an interparticle distance >100 nm to avoid plasmonic coupling between adjacent particles (Figure 1d). Before the detection, we first functionalized each of the four AuNR strips with cytokine-specific antibodies targeting pro-inflammatory (TNF-α and IL-6)) and anti-inflammatory (IL-10 and IL-4) cytokines using standard EDC/NHS chemistry (see details in Materials and Methods, Figure 1a, and Figure S2). After the cytokine binding, a ~10 nm thick protein layer coating on the AuNRs was observed, which further confirms the binding of antibody/cytokine molecules to the sensor surface. We also monitored the spectral red-shift at the end of each bio-conjugation step during the AuNR surface functionalization and after the measurement of 10 ng/mL of IL-6 using a spectrophotometer attached to microscope (Figure S3b and Figure 1e). In our measurement, we observed the resonance peak wavelength of the extinction spectrum of our AuNRs on the glass substrate was shifted from 662 nm to around 681 nm, confirming the cytokine binding events to the sensor surface.
The integrated nanoplasmonic biosensing platform was then mounted on a dark-field microscopy stage for rapid, high-sensitivitiy, and dynamic cytokine detection (Figure 2a and Figure S3a; see details in Materials and Methods). The scattering spectrum can be spatially mapped across the surface to obtain a LSPR image. An increase in the surface antigen-antibody binding thus increases the brightness of the LSPR image. The LSPR images of the AuNR nanoplasmonic biosensor microarrays were captured in real time using a 10× objective lens, a dark-field condenser, an electron multiplying charge coupled device (EMCCD) through a band-pass filter (674−686 nm) (Figure 2a and Figure S3a). The captured LSPR images were then processed by a customized MATLAB program to selectively detect the intensity changes of the LSPR images in a microfluidic channel resulting from the cytokine surface binding to AuNRs (Figure 1a).
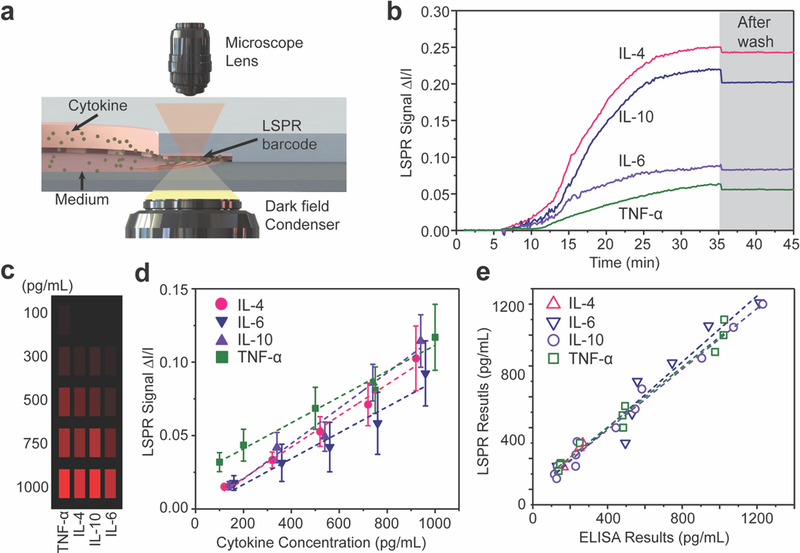
Figure 2. Label-free and multiplex nanoplasmonic measurement of cell-secreted cytokines.
(a) Schematic of LSPR signal detection on a dark-field microscopy stage. (b) Real-time LSPR signals during the multiplex cytokine detection. The gray area shows the LSPR signal after washing with PBS. (c) Mapping of intensity variations at LSPR microarray sensing spots for four different types of cytokines at different concentrations. (d) Calibration curves of purified IL-4, IL-6. IL-10, and TNF-α obtained from LSPR nanoplasmonic biosensor microarray chip. Each sample were measured ten times. Error bar: standard error. (e) Correlation between data obtained from independent LSPR microarray and gold standard ELISA experiments using the same samples.
Prior to multiplexed analyte detection, we performed parallel measurements of different cytokines using the LSPR sensing units on the microfluidic chip. Standard curves were acquired for each cytokine to determine the assay time, dynamic range and limit of detection (LOD) of the sensors. First, we measured real-time cytokine signal variations associated with analyte surface binding in a multiplex scheme with a mixture of four target cytokines suspended in phosphate buffered saline (PBS) solution. In the cytokine solution, a different concentration level was assigned to cytokines IL-4, IL-6, IL-10 and TNF-α, at 3000, 1000, 500, and 250 pg/mL, respectively. We loaded the cytokine mixture into one of the microfluidic channels of the LSPR microarray device and subsequently measured the time-course signal variation from the sensor spots (Figure 2b). Analyte-antibody binding reached equilibrium within 30 min after the introduction of the cytokine mixture, as evidenced by signal plateaus. The rapid analyte binding kinetics allowed the assay to be performed with a very short incubation time, without adversely affecting the on-chip cell culture. Such a time-saving detection tool for multiplexed cytokine profiling can be performed at the point of interest during adipose tissue-on-a-chip culture. The signal intensity was only reduced by ~ 8% after washing with PBS to remove any nonspecific bound molecules. We then spiked a PBS solution with purified IL-4, IL-6, IL-10 and TNF-α with known concentrations from 100 to 1000 pg/mL and quantified the percentage of scattering intensity change induced by the specific binding of target analyte cytokines to antibody-conjugated AuNR microarrays. Figure 2c shows the mapping of intensity variations at our LSPR microarray sensing spots for the four different types of cytokines at varying concentration. We recorded intensity values of LSPR sensing spots before (I) and after (I + ΔI) sample incubation and plotted standard curves showing the fractional intensity shift ΔI/I as a function of cytokine concentrations (Figure 2d).
We further compared readouts from the LSPR nanoplasmonic biosensor with those acquired by the “gold standard” ELISA (Biolegend) (Figure 2e). The PBS solutions spiked with different concentrations of cytokines were tested using both the LSPR biosensors and ELISA. We found an excellent correlation between the two types of measurements across a wide dynamic range with slope values of 1.069, 0.982, 1.305, 0.976 and R2 of 0.918, 0.985, 0.983, 0.979 for cytokines IL-6, TNF-α, IL-4 and IL-10, correspondingly. Therefore, we validated the accuracy of our LSPR nanoplasmonic biosensors. It should be noted that typical ELISA measurements require at least 100 μL of sample medium for each cytokine detection. Thus, a total of at least 400 μL media is required to measure 4 types of cytokines. However, the in-vitro adipose-tissue-on-chip in this study only requires a few microliter of samples, which is inaccessible by conventional ELISA.
On-chip Adipose Tissue Cell Culture
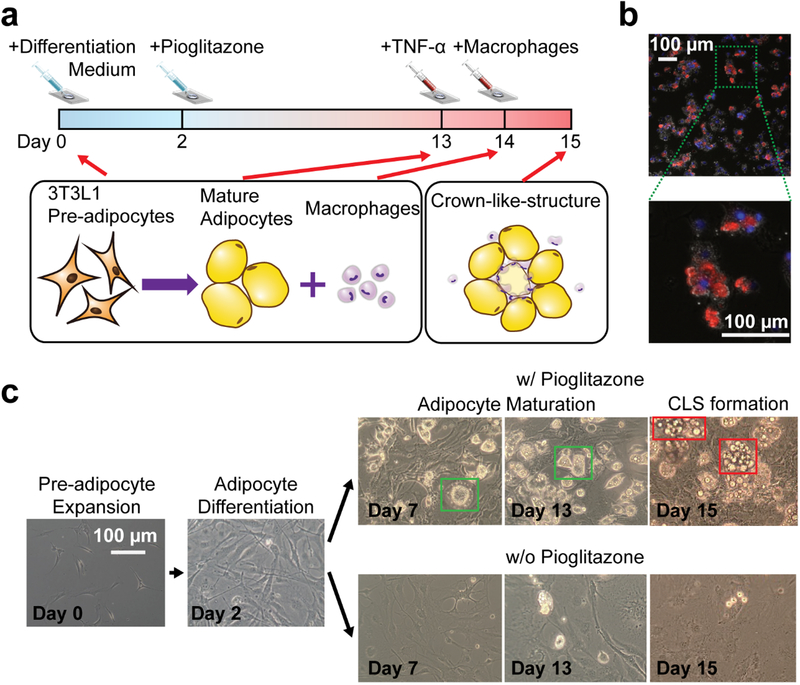
To form a biomimetic adipose tissue microenvironment, 3T3L1 precursor adipocyte cells and J7 mouse macrophages were used and cultured in the adipose tissue chamber for 15 days (Figure 3a). We quantitatively characterized time-course responses of adipocytes during maturation and inflammatory stimulation and obtained a dynamic correlation between the cytokine secretion profile undergo adipocyte differentiation and CLS formation (Figure 3b) within the adipose tissue microenvironment. Initially, 1,000 3T3L1 pre-adipocytes were loaded into the culturing chamber (Day 0) and treated with differentiation medium (see Materials and Methods) for 2 days to initial adipocyte differentiation. Following that, the adipocytes were continuously cultured in maintenance medium for another 11 days. Pioglitazone is recognized to activate PPAR alpha to differentiate pre-adipocytes into mature adipocytes. Thus, we accelerated the maturation of adipocytes in our in vitro system by adding 0.2 μM of pioglitazone into the maintenance medium on Day 2.5 As shown in Figure 3c, 3T3L1 cells treated with pioglitazone exhibited the unique adipocyte morphology, evidenced by increasing accumulation of lipid drops and many signet ring appearances of adipose cells.38
Figure 3. On-chip adipocyte culture, differentiation, inflammatory stimulation, and CLS formation.
(a) Schematic of the on-chip adipocyte culture, differentiation, inflammatory stimulation, and CLS formation process. After the 3T3L1 pre-adipocytes were loaded to the adipocyte culture chamber, the cell culture and expansion medium was replaced with differentiation medium to initial adipocyte differentiation, and denoted the date as Day 0. To enhance adipocyte differentiation, 0.2 μM of pioglitazone was added into the maintenance medium on Day 2. The adipocytes were then continuously cultured in maintenance medium with pioglitazone for another 11 days. On Day 13, 2ng/mL of TNF-α was added into the medium, stimulating an inflammatory microenvironment. The macrophages were added on Day 14. Subsequently, the formation of CLSs were observed on Day 15. (b) Representative fluorescent image of the adipocytes, macrophages, and CLS on chip. The nucleus of adipocytes was stained by DAPI (blue color) and the macrophages were stained by iNOS M1 macrophage marker (red color). The enlarged image clearly shows a CLS with M1 macrophages surrounding adipocyte cells. (c) Optical microscope images of the on-chip adipocyte differentiation and CLS formation process. The 3T3L1 cells treated with pioglitazone exhibited the unique mature adipocyte morphology, evidenced by increasing accumulation of lipid drops and many signet ring appearances of adipose cells.
After Day 13, 2 ng/mL of purified TNF-α were added to the cell culture chamber to trigger the pro-inflammatory response from the adipocytes as well as to promote the necrosis of adipocytes, mimicking the dysfunctional adipose tissue with chronic inflammation caused by obesity. After a 24-hour incubation, the medium with TNF-α was removed and replaced by fresh maintenance medium on Day 14. Subsequently, different densities of J7 macrophages (0, 500, and 1000 cells, denoted as 0×Mac, 1×Mac and 2×Mac in Figure 4) were introduced into three different devices to imitate different levels of macrophage infiltration, respectively. After fifteen days of adipocyte differentiation, maturation, and inflammatory stimulation, bio-mimic CLSs consisting of macrophages encircling a necrotizing adipocyte3,4 were successfully observed in our microfluidic chip, validating adipose tissue inflammation in the adipose tissue microenvironment. Figure 3b presents fluorescent image of the adipocytes, macrophages, and CLS on the chip after cell fixing and staining on Day 15. Macrophages were stained with iNOS M1 macrophage marker (red color) and the nucleus of adipocyte were stained with DAPI (blue color). Clearly, there are a number of CLS signature by clusters of macrophages circulating adipocytes. We confirmed macrophages in CLS are mostly polarized to M1 pro-inflammatory phenotype.39 This observation confirmed that we have successfully created an inflammatory adipose tissue. The polarization of macrophages also concurs with results from in vivo observations from obese human and mice.36,40 Moreover, we can directly observe the CLS on-chip even without fluorescent staining, owning to the unique appearance of CLS (Figure 3c). We observed that the density of CLS positively correlated with an increasing number of J7 macrophages (Figure S4), indicating that the number of recruited macrophages may correlate to the level of inflammation in the adipose tissue.
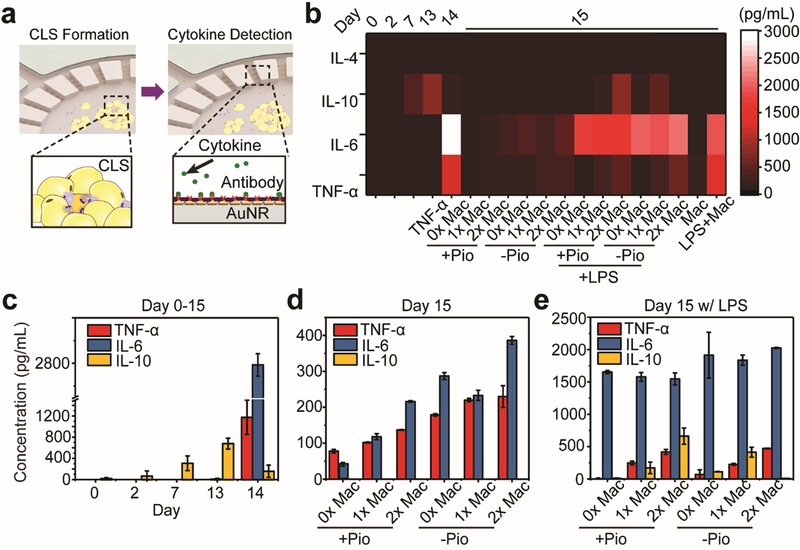
Figure 4. On-chip Multiplexed Cytokine Secretion Profile Measurement.
(a) Schematic of on-chip CLS formation and cell-secreted cytokine detection. (b) A heat map visualization representing the concentrations of IL-4, IL-6, IL-10 and TNF-α at different stages of adipose-tissue-on-chip, with different concentration of macrophages and with/without different reagents such as Pio and LPS. (c) Cytokine secretion profiles of 3T3L1 adipocytes during the 13-day differentiation process. The adipocytes exhibited an increasing trend of IL-10 before Day 13. After adding a pro-inflammatory stimulation of 2 ng/mL TNF- α, IL-10 levels were suppressed and IL-6 levels were raised significantly to high levels. (d) Comparison of cytokine secretion profiles of adipose-tissue-on-chip after adding different densities of J7 macrophages in the medium with (+Pio) or without (-Pio) pioglitazone. (e) Cytokine secretion profiles of the adipose-tissue-on-chip after pro-inflammatory LPS stimulation in the medium with (+Pio) or without (-Pio) pioglitazone. From (c)-(e) the results were measured from at least 3 samples in same condition. Error bar: Standard error.
On-chip Cytokine Secretion Profile Measurement from the Adipocytes and Adipose Tissue Macrophages
During obesity progression, adipose tissue macrophages and hypertrophic adipocytes produce a pro-inflammatory milieu of cytokines, such as IL-6 and TNF-α, for facilitating chronic low-grade inflammation.1,2 Therefore, an in situ multiplexed cytokine profiling is a key to monitoring the status changes of adipose tissue inflammation from initiation to a chronic stage, as indicated by the CLS formation. To obtain the multiplexed adipocyte cytokine secretion profile from the adipose tissue cells and macrophages, we prepared LSPR sensing arrays on glass slides with each AuNR barcode functionalized by antibodies targeting IL-4, IL-6, IL-10, or TNF-α, respectively (Figure 4a). After each cytokine measurement, we peeled off the glass slide deposited with AuNRs, and replenished the incubation chamber with fresh cell culture medium. We quantitatively measured the four cytokine secretion levels in the adipose-tissue-on-chip on Day 0 (expansion medium), Day 2 (differentiation medium), Day 7 and Day 13 (maintenance medium + pioglizatone), Day 14 (maintenance medium + TNF-α), and Day 15 (maintenance medium ± pioglizatone + J7 macrophages). The results were plotted as a heat map ‘barcode” visual, as shown in Figure 4b. The heat map sensing barcode provides a multiparametric and spatiotemporal analysis of the immune status of the adipose tissue microenvironment. It is noteworthy that anti-inflammatory cytokines, especially IL-4 were nearly undetectable. However, pro-inflammatory cytokines were observed predominantly in concentrations on Day 14 and Day 15, confirming that a pro-inflammatory microenvironment was formed after TNF-α stimulation.
We first compared adipocyte cytokine secretion profiles during the differentiation process (from Day 0–13) to after TNF-α stimulation (Day 14). The secretion of anti-inflammatory cytokine, IL-10, was the most prominent in healthy developing 3T3-L1 adipocytes under low density macrophage culture or pioglitazone conditions. In Figure 4c, we observed elevated secretions of IL-10 but negligible levels of other secretory cytokines from Day 0–13, confirming the healthy state of the adipocytes. Pioglitazone has previously been reported to suppress adipose tissue inflammation41 and can be attributed to the observed increasing levels of anti-inflammatory cytokine IL-10. On Day 14 after a 24-hr stimulation with 2 ng/mL of TNF-α, adipocytes exhibited a significant change from anti-inflammatory into an inflammatory phenotype as evidenced by the elevated levels of IL-6 and suppressed expressions of IL-10, similar to the obesity-induced changes in the adipose tissue microenvironment. On Day 15 after removing TNF-α-conditioned medium, adipocyte without J7 macrophages continued expressing pro-inflammatory cytokines but non-detectable levels of anti-inflammatory cytokines (Figure 4d), suggesting a low-grade inflammatory milieu in the adipose-tissue-on-chip, confirming that we have successfully produced a biomimetic, obesity-induced, and inflammatory adipose tissue microenvironment. We then compared the cytokine secretion profiles of the adipose-tissue-on-chip after adding different densities of J7 macrophages. IL-6 and TNF-α levels have been observed to increase with the number of J7 macrophages introduced into the chip, indicating the macrophages were mainly polarized into the inflammatory, M1 phenotype, concurring with our previous immunofluorescence staining and analysis results (Figure 3b) and other studies on obese adipose tissue macrophages. Juxtaposed with the observation of increasing CLS density (Figure S4), our results correlate CLS formation to the level of adipose tissue inflammation. Devices using medium without pioglitazone expressed higher levels of pro-inflammatory cytokines after D14, confirming the suppressive effect of pioglitazone on adipose tissue inflammation. These results suggest that CLS formation is cell and microenvironment-dependent and can be modulated under in vitro conditions to recapitulate aspects of adipose tissue inflammation in real-time.
It has been well known that lipopolysaccharide (LPS) can initiate an acute pro-inflammatory response of macrophages and promote the secretion of pro-inflammatory cytokines TNF-α and IL-6.42 Thus, to validate our sensing platform can map phenotype changes as a response to inflammatory milieu alterations, we stimulated J7 macrophages with LPS (100 ng/mL) both off-chip and on-chip and subsequently quantified cytokine secretion profiles. First, we pre-stimulated J7 macrophages (density ~ 5×104 cells/mL) with 100 ng/mL of LPS for 24 hrs in T75 flasks, then collected 50 μL of supernatant samples from the J7 macrophage culture medium and loaded it into our microfluidic LSPR biosensor device for cytokine detection. We also collected control samples from the J7 cell culture medium without LPS stimulation for comparison. Significant increases in TNF-α and IL-6 were observed in the supernatants collected from LPS-stimulated J7 macrophages, in consistent with previous studies (Figure S5).43,44
Then, we performed an on-chip LPS stimulation by simultaneously adding 100 ng/mL of LPS and loading J7 macrophages in the adipose-tissue-on-chip cell culture chamber on Day 14. On-chip multiplexed cytokine secretion profiles were measured on Day 15. Unlike the low-grade inflammation with IL-6 and TNF-α around 200–300 pg/mL in the non-stimulated chips, LPS stimulation created a high-grade inflammation in the adipose-tissue-on-chip, displaying heightened secretion levels (> 1500 pg/mL) of IL-6. Interestingly, we also observed distinct IL-10 levels after LPS stimulation. This observation is consistent with those previously reported where LPS challenge prompts the expression of anti-inflammatory cytokines like IL-10 in multiple types of cells, such as 3T3L1 adipocytes6, microglia45, Kupffer cells46, etc. Our integrated nanoplasmonic adipose-on-chip has demonstrated to be a reliable platform to monitor dynamic multiplexed cytokine secretion profiles of cells in a biomimetic adipose tissue microenvironment.
Conclusion
Although many cytokine sensing techniques have been developed, they have been hindered by the complexity to troubleshoot signal acquisition, sensor integration, low sensitivity, or lack of multiplexity. Thus, this has stressed an urgent need to equip novel label-free cytokine biosensors to profile the dynamic in situ cytokine secretion behaviors of a complex biomimetic tissue microenvironment. In this work, we demonstrated a novel integration of a LSPR biosensor with a biomimetic microfluidic adipose-tissue-on-chip platform to quantitatively characterize the multiplexed cytokine secretion patterns of adipose tissue cells altered by pro-inflammatory stimulations. Particularly, we were able to easily observe the appearance of the adipocytes and the on-chip formation of the CLS in our microfluidic biomimetic adipose tissue microenvironment chip. With integrated antibody-conjugated AuNR LSPR barcode arrays, our platform enables simultaneous multiplexed measurements of pro-inflammatory (IL-6, and TNF-α) and anti-inflammatory (IL-10, and IL-4) cytokines secreted by the adipocytes and macrophages. Our cytokine secretion assay was rapid (< 30 min), sensitive (~ 20 pg/mL), and easy to implement for multiplexed and real-time detection. The multiplexed time-course cytokine secretion data obtained from this work enabled us to monitor the dynamic, stage-specific responses of adipocytes and encircling macrophages during differentiation and pro-inflammatory stimulation in the adipose tissue microenvironment. To the best of our knowledge, this study is the first to integrate a label-free, multiplexed biosensor into a complex biomimetic tissue microenvironment and quantitatively characterize the dynamic cytokine secretion behaviors in a spatiotemporal manner. We envision this integrated ‘adipose-tissue-on-chip’ biosensing platform will not only be applied as a precise drug-efficacy screening and prognostic tool for personalized treatment regimens and risk prevention against obesity but can also be adapted to other biomimetic microfluidic cultures to drive future research in precision immunology for personalized medicine.
MATERIALS AND METHODS
Microfluidic device fabrication.
Poly-dimethylsiloxane (PDMS) was used to frame the microfluidic device to reconstruct a biomimetic adipose tissue microenvironment in vitro. Master molds were first fabricated on silicon substrates by employing photolithography with SU-8 negative photoresist (2025, Microchem). The master molds were silanized with trichloro(1H,1H,2H,2H-perfluorooctyl) silane (448931, Sigma-Aldrich) vapor overnight in vacuum desiccation to facilitate subsequent release of PDMS from the molds. A PDMS precursor (Sylgard-184, Dow Corning) was prepared by mixing a PDMS curing agent with the base (wt:wt = 1:10), and poured onto the molds, and cured overnight in a 60 °C oven. Two separate fully cured PDMS structures with microfluidic channels were fabricated using different molds: one for patterning the circulated AuNRs biosensor barcodes on a glass substrate and the other for forming the on-chip culturing and cytokine detection layer (Figure S1).
Assembly of adipose-tissue-on-chip device and LSPR cytokine detection layer
To assemble the integrated culture and detection platform, we first permanently bound a clean 18 cm × 18 cm glass cover slide on the top side of the PDMS microfluidic device. The device was then sterilized by exposing it under UV light for 5 mins. Subsequently, the device was inverted, producing a 4 mm diameter cylindrical chamber with a sealed bottom. The adipocytes and macrophages were then cultured on-chip in a single chamber in a cell culture incubator. Before taking each multiplexed cytokine measurement, a fresh glass substrate with patterned AuNR sensing array was temperately bound to the PDMS microfluidic device. The circular AuNR sensing array and microfluidic chamber were aligned under an optical microscope. After bonding, the whole device was then inverted to allow conditioned culture media to diffuse into the microchannels and AuNR sensing array for LSPR cytokine detection. After each measurement, the AuNR patterned glass substrate was peeled off, and the device was put back to the incubator to for on-chip cell culture.
Cell Culture.
Mouse 3T3L1 precursor adipocyte cell line (ATCC) and J774A.1 macrophages (ATCC) were cultured separately in pre-adipocyte expansion medium containing 90% of Dulbecco’s Modified Eagle’s Medium (DMEM, sigma Al) supplemented with 10% bovine serum (Gibco) and 1% penicillin/streptomycin (Gibco) in T75 flasks. Both cell lines were grown in a 37 °C and 5% CO2 incubator to reach 80% confluence for any further experimentation.
On-chip adipocyte differentiation, maturation and inflammatory stimulation.
The culture chamber was incubated in 1 mg/mL fibronectin solution for 1 hour for helping the adhesion of cells on the surface of glass. After removing fibronectin and washing, pre-adipocytes and 35μL of cell culture medium were added into the chamber for the on-chip culture experiment. After the 3T3L1 pre-adipocytes were loaded to the adipocyte culture chamber (1000 cells per device) and cultured for 2 days, the pre-adipocyte expansion medium was replaced with differentiation medium to initial adipocyte differentiation, and denoted the date as Day 0. The differentiation medium contains 90% DMEM (Caisson), 10% FBS (Gibco), 1% penicillin/streptomycin (Gibco), 1.0 μg/mL Insulin (Gibco), 1.0 μM Dexamethasone, and 0.5 mM Methylisobutylxanthine (IBMX) to chemically induce the differentiation of the 3T3L1 cells. To enhance adipocyte differentiation, the differentiation medium was replaced with a pioglitazone (0.2 μM; Alfa Aesar) supplemented adipocyte maintenance medium on Day 2. The adipocyte maintenance medium contains 90% DMEM (Caisson), 10% FBS (Gibco), 1% penicillin/streptomycin (Gibco), 1.0 μg/mL Insulin (Gibco). The adipocytes were then continuously cultured in adipocyte maintenance medium with pioglitazone for another 11 days with the culture medium replaced every 2−3 days. On Day 13, 2ng/mL of TNF-α (ebioscience) was added into the adipocyte maintenance medium, stimulating an inflammatory microenvironment for 24 hours. On Day 14, the medium with TNF-α were replaced by normal adipocyte maintenance medium with 0.2 μM of pioglitazone. Different amount of J7 macrophages (0, 500, and 1000 cells, denoted as 0×Mac, 1×Mac and 2×Mac) were simultaneously introduced into three different devices to mimic different level of inflammation. The 3T3L1 adipocytes and J7 macrophages were co-cultured in the devices for another 24 hours before taking cytokine measurement at the final stage. A control experiment following the same procedure but without addition of pioglitazone was performed and compared. All cells were incubated at 37 °C with 5% CO2 and 100% humidity in a CO2 Cell Culture Incubator.
LSPR Nanoplasmonic Biosensor Microarray Fabrication.
The fabrication of LSPR nanoplasmonic biosensor microarray is shown as schematic in Figure S2. A piranha-cleaned glass substrate was first oxygen-plasma treated at 20 W for 120 s. Then, a colloidal solution suspending positively charged CTAB-coated AuNRs (Nanoseedz, Hong Kong) were flown into the PDMS microfluidic patterning channels (Figure S1d) covered by the plasma treated glass substrate. The surface of the glass substrate was negatively charged. The AuNRs were immobilized onto the glass substrate by means of electrostatic interactions and formed parallel sensor array patterns. Subsequently, 1 mM of 11-Mercaptoundecanoic acid (Sigma-Aldrich) was dissolved in 10% ethanol, loaded into the microfluidic patterning channels, and incubated overnight to functionalize the AuNR surfaces with, which replaced CTAB through a ligand exchange process. 0.4 M EDC (1-ethyl-3-[3-(dimethylamino)propyl] carbodiimide hydrochloride, Thermo Scientific) and 0.6 M NHS (N-hydroxysuccinimide, Thermo Scientific) were mixed at a 1:1 volume ratio in 0.1 M MES (1-ethyl-3-[3-(dimethylamino)propyl] carbodiimide hydrochloride, Thermo Scientific) solution. The EDC/NHS/MES solution was loaded to the same microfluidic channels and incubated for 30 min to activate the ligand. This was followed by antibody coating of the AuNR sensor patters that involved loading of probe antibodies (anti-mouse IL-4, IL-6, IL-10, TNF-α, ebioscience, USA) in PBS solution at a concentration of 50 μg/mL into individual patterning channels. Subsequently, 3% BSA in deionized water solution was loaded through the channels and incubated for 20 min for sensor surface passivation to eliminate nonspecific binding of biomolecules. At the end of every incubation step above, the sensor surfaces were thoroughly washed using deionized water, and any excessive solution and unbound molecules were removed.
LSPR Dark-Field Imaging for Cytokine Detection.
Before each multiplexed cytokine measurement, a freshly made glass substrate with AuNR LSPR biosensor microarray chip was temperately bonded onto the side of culture chamber with the microchannel patterns. The alignment of the circular AuNR arrays and the microchannels were operated using an optical microscope. The device was then reversed to have the cell culture medium diffuse into the microchannels during the LSPR measurement. After the measurement, the AuNR patterned glass substrate was peeled off, and the device was sent back to incubator to continue the on-chip cell culture. The LSPR biosensor microarray chip was mounted on the motorized stage of an upright microscope (Axio Imager 2, Zeiss) to position the on-chip sensing spot at ease and to automate the signal scanning. A dark-field condenser (NA = 1.45) was closely placed to the backside of the glass substrate (the opposite side of the AuNR-deposited sensor side) using lens oil. The light scattered from the AuNR nanoplasmonic biosensor arrays was collected using a 10× objective lens under the chip and then filtered by a band-pass filter (674−686 nm, Semrock). This light signal was collected by an electron multiplying CCD (EMCCD, Photometrics) camera (Figure S3). The captured LSPR images were then processed by a customized MATLAB program to selectively detect the intensity changes of the LSPR images in a microfluidic channel resulting from the cytokine surface binding to AuNRs. Image analysis was performed using our customized MATLAB code.
Spectral Shift Measurement of Biomolecule Binding using Spectrophotometer.
The scattering spectrum of the AuNRs during each step of biomolecule binding were measured using a fiber spectrometer (Ocean Optics. UR2000). The spectrometer was mounted onto the camera C-mount port of the dark-field upright microscope (Axio Imager 2, Zeiss) to check the dark-field scattering spectrum of the AuNR substrate. The spectral shift at each bio-conjugation step was monitored (Figure S3).
Cytokine Measurement with ELISA Immunoassays.
The validation of cytokine measurement was carried out by measuring PBS solutions spiked with different concentrations of cytokines (mouse IL-4, IL-6, IL-10, TNF-α, ebioscience, USA) using both LSPR microarrays and the gold-standard ELISA. ELISA kits (Mouse IL-4 ELISA MAX™ Deluxe, Mouse IL-6 ELISA MAX™ Deluxe, Mouse IL-10 ELISA MAX™ Deluxe, and Mouse TNF-α ELISA MAX™ Deluxe, Biolegend) were used in this study according to manufacturer’s protocols. Briefly, cytokine solutions or cell culture supernatants were collected as samples. Capture antibody solution was added into all wells of a 96-well plate provided in the kit, the plate was subsequently sealed and incubated at 4°C overnight. After washing and blocking, 100 μL of each standards and samples were added into the wells and were incubated for 2 hours at room temperature. The wells were then incubated with detection antibody for 1 hour, followed by washing and incubation with HRP-labeled Avidin for 30 minutes. TMB mixture (1:1) was then applied to visualize chemiluminescence for 20 minutes in the dark. Then, 100 μL of 2N sulfuric acid solution was added in each well to stop the reaction. The reading of ELISA results was carried out using a plate reader (SpectraMax i3, Molecular Devices) by reading the absorbance at 450 nm within 15 mins after adding the stop solution.
Immunofluorescence Staining and Analysis
We fixed and stained the cells cultured on the chip after day 15. Cells were fixed with 4% paraformaldehyde (15711, Electron Microscopy Sciences) for 30 min, permeabilized with 0.3% Triton X- 100 (11332481001, Sigma-Aldrich) for 10 min, and then blocked with 3% bovine serum for 1 hr on ice to eliminate nonspecific binding. Specifically, we stained M1 phenotype macrophages by incubating the chip with iNOS primary antibodies (NB300–605, Novus Biologicals, 5 μg/mL) for 1 hr, and then visualized with Alexa Fluor 555 conjugated goat anti-mouse IgG secondary antibodies (Invitrogen, 5 mg/mL). All cell nucleus was stained with DAPI. Fluorescent images were obtained using an inverted microscope (Zeiss Axio Observer.Z1) equipped with a digital CMOS camera (ORCA-Flash4.0 LT Digital CMOS camera, Hamamatsu Photonics) and a 10x objective.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge financial supports from the National Science Foundation (CBET 1701322 to W. C., CBET 1701363 to P.C.), the National Institute of Health (NIH/NIBIB 1R21EB025406–01A1 to W. C.), the New York University Global Seed Grant, and NYU Clinical and Translational Science Institute Collaborative Translational Pilot Award (NIH/NCATS 1UL1 TR001445). J.O.A. was supported by the Sackler Foundation and the Helmsley Center for Disorders of the Digestive System, both at Rockefeller University, during the initial phases of this work.
REFERENCES
- 1.Dalmas E, Clément K and Guerre-Millo M, Trends Immunol, 2011, 32, 307–314. [DOI] [PubMed] [Google Scholar]
- 2.Zeyda M and Stulnig TM, Immunol. Lett, 2007, 112, 61–67. [DOI] [PubMed] [Google Scholar]
- 3.Morris PG, Hudis CA, Giri D, Morrow M, Falcone DJ, Zhou XK, Du B, Brogi E, Crawford CB, Kopelovich L, Subbaramaiah K and Dannenberg AJ, Cancer Prev. Res. (Phila), 2011, 4, 1021–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cinti S, Mitchell G, Barbatelli G, Murano I, Ceresi E, Faloia E, Wang S, Fortier M, Greenberg AS and Obin MS, J. Lipid Res, 2005, 46, 2347–55. [DOI] [PubMed] [Google Scholar]
- 5.Haka AS, Barbosa-Lorenzi VC, Lee HJ, Falcone DJ, Hudis CA, Dannenberg AJ and Maxfield FR, J. Lipid Res, 2016, 57, 980–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lira FS, Rosa JC, Cunha CA, Ribeiro EB, Oller do Nascimento C, Oyama LM and Mota JF, Lipids Health Dis, 2011, 10, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frankenstein Z, Alon U and Cohen IR, Biol. Direct, 2006, 1, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu G, Qi M, Hutchinson MR, Yang G and Goldys EM, Biosens. Bioelectron, 2016, 79, 810–821. [DOI] [PubMed] [Google Scholar]
- 9.Schenk T, Irth H, Marko-Varga G, Edholm L-E, Tjaden U and van der Greef J, J. Pharm. Biomed. Anal, 2001, 26, 975–985. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, Huang N-T, Chung M-T, Cornell TT and Kurabayashi K, Adv. Drug Deliv. Rev, 2015, 95, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Hurk R and Evoy S, Sensors Actuators B Chem, 2013, 176, 960–965. [Google Scholar]
- 12.Dutta P, Sanseverino J, Datskos PG and Sepaniak MJ, NanoBiotechnology, 2005, 1, 237–244. [Google Scholar]
- 13.Pui T-S, Agarwal A, Ye F, Huang Y and Chen P, Biosens. Bioelectron, 2011, 26, 2746–2750. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Tuleouva N, Ramanculov E and Revzin A, Anal. Chem, 2010, 82, 8131–8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pui TS, Kongsuphol P, Arya SK and Bansal T, Sensors Actuators B Chem, 2013, 181, 494–500. [Google Scholar]
- 16.Kongsuphol P, Ng HH, Pursey JP, Arya SK, Wong CC, Stulz E and Park MK, Biosens. Bioelectron, 2014, 61, 274–279. [DOI] [PubMed] [Google Scholar]
- 17.Luchansky MS and Bailey RC, J. Am. Chem. Soc, 2011, 133, 20500–20506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandal S, Goddard JM and Erickson D, Lab Chip, 2009, 9, 2924. [DOI] [PubMed] [Google Scholar]
- 19.Luchansky MS and Bailey RC, Anal. Chem, 2010, 82, 1975–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inci F, Filippini C, Baday M, Ozen MO, Calamak S, Durmus NG, Wang S, Hanhauser E, Hobbs KS, Juillard F, Kuang PP, Vetter ML, Carocci M, Yamamoto HS, Takagi Y, Yildiz UH, Akin D, Wesemann DR, Singhal A, Yang PL, Nibert ML, Fichorova RN, Lau DT-Y, Henrich TJ, Kaye KM, Schachter SC, Kuritzkes DR, Steinmetz LM, Gambhir SS, Davis RW and Demirci U, Proc. Natl. Acad. Sci, 2015, 112, E4354–E4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y-C, Chiang C-Y, Li C-H, Chang T-C, Chiang C-S, Chau L-K, Huang K-W, Wu C-W, Wang S-C and Lyu S-R, Analyst, 2013, 138, 4599. [DOI] [PubMed] [Google Scholar]
- 22.Huang T, Nallathamby PD and Xu X-HN, J. Am. Chem. Soc, 2008, 130, 17095–17105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang C-Y, Hsieh M-L, Huang K-W, Chau L-K, Chang C-M and Lyu S-R, Biosens. Bioelectron, 2010, 26, 1036–1042. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Perdiguero J, Retolaza A, Bujanda L and Merino S, Talanta, 2014, 119, 492–497. [DOI] [PubMed] [Google Scholar]
- 25.Corrie SR and Plebanski M, Mol. Immunol, 2018, 98, 28–35. [DOI] [PubMed] [Google Scholar]
- 26.Luchansky MS and Bailey RC, J. Am. Chem. Soc, 2011, 133, 20500–20506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh BR, Chen P, Nidetz R, McHugh W, Fu J, Shanley TP, Cornell TT and Kurabayashi K, ACS Sensors, 2016, 1, 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh B-R, Huang N-T, Chen W, Seo JH, Chen P, Cornell TT, Shanley TP, Fu J and Kurabayashi K, ACS Nano, 2014, 8, 2667–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Ota S, Guo B, Ryu J, Rhodes C, Xiong Y, Kalim S, Zeng L, Chen Y, Teitell MA and Zhang X, Nano Lett, 2011, 11, 3431–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Soler M, Özdemir CI, Belushkin A, Yesilköy F and Altug H, Lab Chip, 2017, 17, 2208–2217. [DOI] [PubMed] [Google Scholar]
- 31.Chen W, Huang N-T, Oh B, Lam RHW, Fan R, Cornell TT, Shanley TP, Kurabayashi K and Fu J, Adv. Healthc. Mater, 2013, 2, 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang N-T, Chen W, Oh B-R, Cornell TT, Shanley TP, Fu J and Kurabayashi K, Lab Chip, 2012, 12, 4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Y, Chen P, Chung MT, Nidetz R, Park Y, Liu Z, McHugh W, Cornell TT, Fu J and Kurabayashi K, Nano Lett, 2017, 17, 2374–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng YB, Kiraly B, Weiss PS and Huang TJ, Nanomedicine, 2012, 7, 751–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen P, Chung MT, McHugh W, Nidetz R, Li Y, Fu J, Cornell TT, Shanley TP and Kurabayashi K, ACS Nano, 2015, 9, 4173–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wentworth JM, Naselli G, Brown WA, Doyle L, Phipson B, Smyth GK, Wabitsch M, O’Brien PE and Harrison LC, Diabetes, 2010, 59, 1648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haes AJ, Haynes CL, McFarland AD, Schatz GC, Van Duyne RP and Zou S, MRS Bull, 2005, 30, 368–375. [Google Scholar]
- 38.Church C, Horowitz M and Rodeheffer M, Adipocyte, 2012, 1, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui X, Morales R-TT, Qian W, Wang H, Gagner J-P, Dolgalev I, Placantonakis D, Zagzag D, Cimmino L, Snuderl M, Lam RHW and Chen W, Biomaterials, 2018, 161, 164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lumeng CN, Bodzin JL and Saltiel AR, J. Clin. Invest, 2007, 117, 175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Permana PA, Zhang W, Wabitsch M, Fischer-Posovszky P, Duckworth WC and Reaven PD, Am. J. Physiol. Endocrinol. Metab, 2009, 296, E1076–84. [DOI] [PubMed] [Google Scholar]
- 42.Yücel G, Zhao Z, El-Battrawy I, Lan H, Lang S, Li X, Buljubasic F, Zimmermann W-H, Cyganek L, Utikal J, Ravens U, Wieland T, Borggrefe M, Zhou X-B and Akin I, Sci. Rep, 2017, 7, 2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seow V, Lim J, Iyer A, Suen JY, Ariffin JK, Hohenhaus DM, Sweet MJ and Fairlie DP, J. Immunol, 2013, 191, 4308–16. [DOI] [PubMed] [Google Scholar]
- 44.Cui X, Morales R-TT, Qian W, Wang H, Gagner J-P, Dolgalev I, Placantonakis D, Zagzag D, Cimmino L, Snuderl M, Lam RHW and Chen W, Biomaterials, 2018, 161, 164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henry CJ, Huang Y, Wynne AM and Godbout JP, Brain. Behav. Immun, 2009, 23, 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knoll P, Schlaak J, Uhrig A, Kempf P, zum Büschenfelde K-HM and Gerken G, J. Hepatol, 1995, 22, 226–229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.