Cancer often arises in preneoplastic fields of histologically normal appearance. These fields have been extensively studied in ulcerative colitis, a cancer predisposing inflammatory bowel disease. Here, we review the field effect in ulcerative colitis and its utility to improve early cancer detection.
Abstract
Cumulative evidence indicates that a significant proportion of cancer evolution may occur before the development of histological abnormalities. While recent improvements in DNA sequencing technology have begun to reveal the presence of these early preneoplastic clones, the concept of ‘premalignant field’ was already introduced by Slaughter more than half a century ago. Also referred to as ‘field effect’, ‘field defect’ or ‘field cancerization’, these terms describe the phenomenon by which molecular alterations develop in normal-appearing tissue and expand to form premalignant patches with the potential to progress to dysplasia and cancer. Field effects have been well-characterized in ulcerative colitis, an inflammatory bowel disease that increases the risk of colorectal cancer. The study of the molecular alterations that define these fields is informative of mechanisms of tumor initiation and progression and has provided potential targets for early cancer detection. Herein, we summarize the current knowledge about the molecular alterations that comprise the field effect in ulcerative colitis and the clinical utility of these fields for cancer screening and prevention.
Introduction
Cancers evolve through an iterative process of mutation, selection and clonal expansion (1). Most tumor types take multiple years to develop, during which time preneoplastic cells clonally expand and progressively acquire the molecular alterations necessary to allow them to fully escape growth control checkpoints and invade surrounding tissues. Precisely how much of this process occurs prior to the development of morphologically recognizable malignancy is unknown, but it has been estimated that cancer cells accumulate about half of their mutational load before tumor initiation (2). This implies that a substantial proportion of a cancer’s development entails precancerous clonal evolution within histologically normal-appearing tissue.
During the last several years, new and extremely sensitive DNA sequencing technologies have begun to directly reveal the presence of these early clones (3,4). However, the notion that preneoplastic changes precede cancer was recognized more than a half century ago. Prior to even the modern understanding of DNA as the genetic material, Slaughter et al. proposed the concept of field cancerization to describe “preconditioned epithelium activated over an area in which multiple cell groups undergo a process of irreversible change towards cancer”. This preneoplastic condition was originally described in oral carcinoma on the basis of subtle morphological abnormalities in surrounding tissue. It was postulated that this abnormal “field” underlied the relatively common finding of multiple synchronous primary tumors and the high frequency of local recurrence in head and neck squamous cell cancers, despite apparent complete tumor resection (5).
Although Slaughter and colleagues recognized field cancerization as an important clinical phenomenon, they lacked a mechanistic explanation. The following decades revealed examples of field cancerization associated with many other epithelial cancers and demonstrated that the fields could be characterized by defined molecular aberrations present in histologically normal tissue (6). Contemporary molecular tools have provided important insights into the basis of the cancer-prone phenotype of these fields. In a variety of examples (6,7), peritumoral fields were shown to encompass populations of clonally related cells that bear some, but not all, of the genetic changes of the tumor itself. The relative ease with which a second tumor or tumor relapse can occur within such a field simply reflects the relatively low evolutionary hurdle for one of these partially dysregulated cells to acquire the last molecular change(s) needed for full-blow malignancy.
While the specific nature of the fields, and the molecular mechanisms that initiate them, are likely to be different in different tissue types, the field concept illustrates the general notion of multistep carcinogenesis, wherein ancestral populations of preneoplastic cells can both precede and co-exist with a cancer (6). Such a field effect may be thought of as an early stage of the neoplastic process where selected mutant cells can incrementally enhance growth properties. Because these preneoplastic populations are often morphologically normal in appearance and may be very small, they frequently go undetected in sporadic tumors. Several preneoplastic diseases, however, often exhibit large preneoplastic fields and offer an excellent opportunity to study the initial stages of tumor development. Ulcerative colitis (UC) and Barrett’s esophagus (BE) are among the best characterized. Fields can expand several centimeters in BE (8,9) and practically the entire length of the colon (~150 cm) in UC (10). In both diseases, chronic inflammation generates extensive damage to epithelial cells, leading to increased cell replication and/or direct DNA damage. Subsequent mutations that alter growth control genes enable clonal expansions, which result in patches of cells that share identical mutations. In some cases, a single genetic change can be found in all cells within an entire field many centimeters in size, indicating a single clonal founder cell. In other cases, multiple independent clones with distinct genetic signatures evolve simultaneously in response to the inflammatory environment.
A further advantage of the study of these preneoplastic diseases is that, in addition to non-dysplastic fields and overt cancer, intermediate degrees of dysplasia are routinely found during endoscopic surveillance. These intermediate stages help delineate with even finer precision the multistep sequence of tumor progression and provide a unique longitudinal window of opportunity to study early cancer and its patterns of evolution (11,12).
In this review, we focus on UC as a model of inflammation-mediated tumorigenesis in which the field effect has been extensively characterized. The search terms used to query the literature include field effect, field defect and field cancerization in UC. We prioritized a broad discussion on the concept of field effect and its implications as opposed to a detailed recollection of articles describing precancerous fields. We first describe the disease’s epidemiology, histologic sequence of neoplastic progression and the types of molecular alterations that have, thus far, been identified in these fields. We then discuss the clinical implications of these fields with a special focus on their use as biomarkers of early or imminent cancer. Field effects have been identified in a growing variety of cancers including: breast (13,14), head and neck (15), bladder (16,17), colorectal (18), gastric (19–21), prostate (22,23), lung (24,25), skin (26–28), liver (29), ovarian (30) and cervical (31). Although UC is responsible for only a small fraction of the global burden of colon cancer, we posit that the lessons learned from studying tumor progression in this unique disease can be applied to the understanding, prevention and clinical management of many other malignancies associated with field effects.
Colorectal cancer risk in UC
Ulcerative colitis is one of the two major types of inflammatory bowel disease and is characterized by uninterrupted stretches of chronic inflammation of the colon mucosa. It affects roughly one million patients in the United States and its prevalence is increasing worldwide (32,33). The cause of UC remains to be fully determined, but a preponderance of evidence suggests that it is the result of a complex interaction between a dysregulated host immune system, the gut microbiome and diet (34–38). A significant aspect of the management of UC is that it elevates the risk of colorectal cancer (CRC) (39) and cancer-related deaths, although improvements in surveillance methods appear to have decreased both the incidence (40) and mortality (41,42) of CRC in UC in recent years. The increased risk of CRC is attributable to multiple aspects of chronic inflammation and immune dysregulation (43,44). Patients whose inflammation is more severe (45) and more extensive (39,46) are more likely to develop CRC. Other risk factors include prolonged disease duration (46–49), concurrent diagnosis of primary sclerosing cholangitis, an autoimmune disorder of the biliary system (50–52), a family history of sporadic CRC (44,53,54), early age of UC onset (55–57) and extent of dysplasia (58).
Colorectal cancer development in the setting of UC differs from that of sporadic CRC in several respects (59). Histologically, adenomatous polyps typically precede sporadic CRC whereas UC-associated CRC (UC-CRC) often arises from flat dysplasia. Sporadic CRC is believed to be initiated by mutations in APC, followed by mutations in KRAS and TP53 (60), although it is currently appreciated that this progression does not necessary follow a linear sequence (61). In UC, TP53 mutations appear to be the initiating mutation in most lesions, although mutations in KRAS have also been identified as a founder event in a minority of cases (62). Recently, next generation sequencing-based studies confirmed a higher frequency of TP53 mutations and lower frequency of APC and KRAS mutations in UC-CRC compared to sporadic CRC (63,64). Epidemiologically, the mean age for CRC development in the general population is 64 (65) versus 43 years for UC-CRC (66). In addition, the prognosis of UC-CRC is poorer than sporadic CRC, although it is unclear if this reflects the tumor biology itself, the average stage of disease at diagnosis, or other health challenges faced by UC patients (67) including those related to immunosuppressive therapies.
A common trait between sporadic and UC-associated CRC appears to be the pivotal role of an abnormal intestinal microbiota as an initiating mechanism. In the last 10 years, a large body of evidence has accumulated linking CRC with a dysbiotic gut microbiota and dysregulated immunity, both in the context of sporadic CRC and inflammatory bowel diseases (68–70). A dysbiotic microbiota contributes to tumor progression directly by generating reactive metabolites and carcinogens, and indirectly by disrupting the epithelial cell barrier in the host (70). This causes local intolerance to antigens of normal flora and leads to dysregulation of the adaptive and innate immune response and subsequent chronic inflammation (68). Diet appears to be an important contributor in this process, as it has a major influence on the gut flora and is transformed into metabolites that can have protective or promoting roles in tumor progression (71). In the case of patients with UC, genetic predisposition might contribute to a dysbiotic gut flora, causing the extensive chronic inflammation characteristic of this disease (68) and increasing the risk of tumor progression through the extensive cell proliferation required in repeated cycles of wound and repair (12,72).
The sequence of tumor progression in UC
Tumor progression in UC is clinically described as a multistep process defined by increasing degrees of histological abnormalities, progressing from no dysplasia, to low-grade dysplasia, high-grade dysplasia and finally cancer (Figure 1). Although often represented linearly and sequentially, it is important to recognize that tumor evolution is usually branched, not linear (73) and, in the case of UC tumorigenesis, not every dysplastic stage may be observed. This sequence might also occur independently in multiple locations in the colon. It is well established that in UC patients, multiple areas of the colon can simultaneously develop dysplastic changes and that independent synchronous cancers can evolve in parallel within these fields (74).
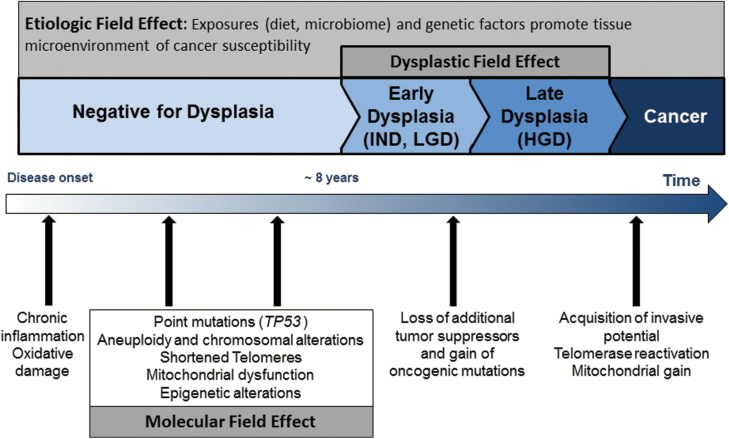
Figure 1.
Proposed model of carcinogenesis in UC. This model integrates the etiological, molecular and dysplastic field effects with known cellular and molecular events that contribute to the different stages of carcinogenesis in UC. The arrow indicates the temporal direction of dysplastic progression and the color gradient reflects the increasing risk of cancer at the different stages of the process. Cancer arises within dysplastic and/or molecular fields. Molecular fields precede dysplastic fields and are more extensive, thus offering an excellent opportunity for precancer detection.
Recent mounting evidence has led to the appreciation that clinical patterns of progression to CRC in UC may differ based on the age of disease onset. UC has two peaks of incidence: early onset occurs between 25 and 35 years of age while late onset arises between 55 and 65 years of age (55,75). Clinical studies have recognized for some time that patients with late onset tend to have less extensive disease and a lower risk of CRC development (76–78). Interestingly, Brackmann et al. (57,58) reported that among UC patients with CRC, those with late age of onset tended to develop cancer without widespread dysplasia. Conversely, patients with early age of onset typically exhibited extensive dysplasia at CRC diagnosis. The presence of extensive dysplasia was also associated with longer disease duration prior to CRC development, higher probability of presenting with active inflammation (57) and worse CRC prognosis (58). These findings suggested that the development of CRC in patients with early onset disease is related to long exposure to inflammation and subsequent development of dysplasia. However, in patients with late onset of disease, CRC might arise independently of observable dysplasia or, alternatively, tumors might be fast growing and displace their original localized dysplastic fields in a clonal sweep. Our group has identified molecular evidence further supporting fundamental differences between early and late onset disease. We demonstrated that UC-cancer patients with early onset of disease have extensive fields of molecular abnormalities throughout their colons compared to UC-cancer patients who have late onset of disease. Specifically, large clonal populations with shortened telomeres could be found in multiple non-dysplastic areas of the colon of early onset UC Progressors (patients with high-grade dysplasia or cancer), but this was almost never the case in the Progressors who had late onset of UC (79). While more research is needed to better distinguish these two modes of CRC progression in UC, their recognition as distinct entities has important clinical implications. Since patients with late onset of disease appear to develop cancer without a widespread field effect, these patients might benefit from partial colon resection instead of full colectomy, which is the current standard-of-care. The ability to safely use segmental resection of the colon in older UC patients would spare them significant morbidity.
Molecular alterations characterize preneoplastic fields in UC
The field effect was originally described based on regions of tissue sharing histological abnormalities (5), but the concept was later broadened to include clonal molecular abnormalities in otherwise histologically normal-appearing tissue, as it was recognized that multiple molecular changes produce Slaughter’s original observation (6). More recently, the term has been used more broadly to describe an “etiologic” field effect, which takes into consideration the contribution of environmental and genetic factors that produce cancer susceptibility (80). The genetic component of UC is well established as well as the critical role of an altered microbiome, which deregulates immunity and triggers inflammation. While the presence of these factors is not indicative of cancer progression, they contribute to the neoplastic process by producing a predisposing microenvironment (80). Figure 1 illustrates the three conceptual types of field effects—etiological, molecular and morphological—in the context of UC cancer progression. These fields represent increasing levels of cancer susceptibility that are operative in the colons of patients with UC, each level contributing to the development of the next. However, only molecular and dysplastic fields are indicative of an underlying preneoplastic process. Here we focus on molecular fields due to their importance to understand tumor progression and their potential for early cancer detection.
In this section, we first summarize the genetic alterations that have provided evidence of clonal field effects in UC. These alterations include somatic point mutations, chromosomal alterations and passenger mutations in polyguanine tracts. We then describe a second set of molecular alterations that define preneoplastic fields in UC without an implicit assumption of clonality. These include telomere shortening, mitochondrial alterations and epigenetic changes. Some of these alterations have provided meaningful clues in terms of the underlying molecular mechanisms that may drive tumor evolution in UC and for a more extensive review on that topic the readers are referred to a recent excellent article by Choi et al. (12). Of note, many of the molecular alterations discussed here, both clonal and non-clonal, harbor potential as UC cancer biomarkers. However, our goal is not the description of the biomarker value of each alteration, which has been previously done by us and others (81–83), but the review of the evidence for a field effect based on those alterations and the discussion of the clinical applicability of those fields for optimal cancer surveillance.
Clonal alterations
Point mutations
Mutations in TP53 are the most common and best characterized single nucleotide variants in UC-associated preneoplastic fields. TP53 mutations (84) and loss of heterozygosity (84,85) occur early in UC neoplastic development. UC patients with TP53 mutations in non-dysplastic biopsies are four times more likely to progress to dysplasia and cancer (86). Additionally, there is a strong correlation between mutations in highly conserved regions of TP53 and the histological progression from low-grade dysplasia to cancer in UC patients (82). In an early study by our group, we examined alterations in TP53 by fluorescence in situ hybridization (FISH) in order to characterize the spatial pattern of these mutational events in individual cells within crypts (87). A detailed analysis of multiple crypts demonstrated that most TP53 FISH abnormalities are shared by all the crypts within a colonic region, indicating monoclonality. The observation of the same TP53 alterations in the two branches of a crypt in fission strongly suggested that clonal expansion of mutated cells occurs by crypt fission and provided direct observation of how clonal fields propagate themselves in UC. In a later study, Leedham et al. (62) identified several molecular alterations, including TP53 and KRAS mutations, in individual, microdissected dysplastic crypts and adjacent non-dysplastic crypts. In one UC Progressor case, the same founding mutation was found in spatially separated tumors 14 cm apart from each other and in the nondysplastic surrounding tissue, clearly demonstrating field cancerization (62). The authors also found clonally disparate tumors in another UC Progressor, supporting the idea that a common etiologic risk factor, i.e. inflammation, has sufficient carcinogenic potential to facilitate the emergence of multiple synchronous clonal fields throughout the colon.
Although it is rapidly becoming the new technical standard, we are aware of only two studies that have yet applied Next Generation Sequencing to the study of somatic mutations in UC, and both were limited to the interrogation of tumors, rather than of preneoplastic fields (63,64). While these reports provide a useful baseline from which to compare the mutational landscape of sporadic versus UC-associated CRC, additional knowledge remains to be generated from applying this technology to a comprehensive study of the spatial and temporal pattern of mutation accumulation in preneoplastic fields.
Aneuploidy and chromosomal alterations
For more than 25 years, aneuploidy has been recognized as an early occurring alteration in UC carcinogenesis, often found even before the appearance of dysplasia (88,89). The presence of aneuploid fields is associated with a higher risk of progression to dysplasia (90), histological grade (90), disease duration (90) and the presence of primary sclerosing cholangitis (91). More sensitive technologies, including comparative genomic hybridization (92) and FISH, revealed that chromosomal alterations occur early in UC tumorigenesis, often preceding histologically defined dysplasia (93) and affecting the entirety of the colon (10). The relative timing and frequency of numerical chromosomal alterations in UC differs significantly from those of sporadic CRC, supporting the conclusion that neoplastic progression follows distinct pathways in these diseases (93). More recently, CGH-array studies have demonstrated that in UC Progressors, chromosomal alterations can be found in distant normal-appearing biopsies (94) and the same alteration can be shared by multiple biopsies spanning most of the length of the colon (95). This indicates that the field effect can be very large and raises the fundamental question of how these clones propagate. The fields also appeared to be graded in nature, as copy number alterations increased in frequency and magnitude with proximity to dysplasia.
While the mechanisms for localized clonal expansions have been well characterized (12), it is unclear how a clone can generate the extensive pancolonic fields described above. Clones originate from stem cells that expand to occupy the whole crypt via niche succession and then crypts laterally expand by crypt-fission to generate monoclonal patches (12). Niche succession might occur by neutral drift (96), but the expansion of certain mutations beyond a crypt appears to involve selection (97,98). This process can generate clones of >10 cm in size (62), but it appears unlikely that the same process would extent throughout the whole organ. Alternative hypotheses are convergent evolution and long-distance stem cell migration aimed at mucosal healing, as proposed in Choi et al. (12). Further investigation in this area is highly needed and would greatly benefit from more advanced methods of lineage detection, as explained in the next section.
Clonal expansions detected by passenger mutations
The ability to recognize clonal expansions requires the presence and identification of one or more genetic changes that identically mark the clone’s progeny as related to each other, yet distinct from adjacent cells. This lineage marker is typically a putative molecular driver of the clonal outgrowth. The challenge is, however, that just as with tumors, multiple genetic changes can drive the clonal expansion. These changes vary from one person to another, and even among different clones within a single individual. Thus, many of these fields might be undetectable if only screening for known driver mutations. An alternative approach is to focus on passenger mutations. As cells divide, replication errors produce mutations, the vast majority of which are functionally neutral and offer no selective advantage or disadvantage (7). These mutations are carried in the daughter cells as ‘passenger’ mutations and offer a much larger repertoire of somatic variants to enable the detection of clonal fields and elucidate lineage relationships.
Our group pioneered an approach for clone detection based on passenger mutations in polyguanine tracts (PolyG), which are highly mutable repetitive DNA sequences interspersed throughout the genome (99,100). We demonstrated that extensive clonal fields defined by PolyG mutations were present in histologically normal colonic biopsies of UC Progressors, but were almost entirely absent in UC Non-progressors (patients without dysplasia). These fields were composed of thousands of cells that appeared microscopically normal, but had aberrantly proliferated from an original precursor cell, indicating the presence of an ‘occult’ process of neoplastic evolution. In a later study, these clonal fields were only found in patients with early onset of disease, suggesting that alternative pathways of progression without extensive clonal expansions might be operative in patients that develop UC later in life (100).
Other molecular alterations identified in preneoplastic fields
Telomere shortening
Telomere shortening is another well-documented, early event in UC tumorigenesis. While it is not informative about clonality, telomere shortening has provided insight about the extent of the field effect and mechanisms of tumor progression in UC (101–106). The colonic epithelium of patients with UC has shorter telomeres than age-matched non-UC patients (102,107). This shortening appears to occur within the first 8 years of disease duration (102) which, interestingly, coincides with the time at which clinical risk of CRC for UC patients increases. This suggests that the onset of cancer may depend upon telomeres becoming critically short. Telomere shortening occurs diffusely in the colonic epithelium of UC patients (102), especially in those with more severe clinical phenotypes (108). Additionally, telomere shortening is more common in biopsies closer to dysplasia (103) and is more extreme in UC Progressors compared to Non-progressors (103,105,106).
Mitochondrial dysfunction
Growing evidence indicates that epithelial cell mitochondrial dysfunction is present in UC (109), although it remains unclear whether it is a cause or consequence of the disease and whether it contributes to cancer progression. The role of mitochondria in cancer, in general, is controversial. A prevailing hypothesis was that mutations in mitochondrial DNA were somehow advantageous to tumors and clonally expanded into fields under positive selection (110). This was proposed to be the basis of the Warburg effect—the observation that most cancers use glycolysis for energy production (111–113). This idea has been challenged, however, by more recent studies arguing that many cancers do, in fact, rely on oxidative phosphorylation and mitochondrial function (114–116) and that mitochondrial DNA mutations either accumulate randomly and clonally expand as passengers without selective pressure, or are selected against (117–119).
In UC, genetic (120), proteomic (37,121–123) and metabolic (124–126) studies have identified mitochondrial alterations in colonic biopsies, both in non-dysplastic mucosa and in UC-associated cancers. Unfortunately, some results are contradictory and the relevance of these findings is still unclear. Our group previously demonstrated that in UC Progressors, the levels of cytochrome C oxidase subunit I (COX), a protein of complex I of the electron transport chain, decreased with proximity to dysplasia, indicating the presence of a gradient field effect. COX staining was completely absent in some dysplastic areas but, remarkably, was typically high in high-grade dysplasia and cancer (127). These results are concordant with mitochondrial dysfunction as a feature of UC colonic epithelium (109), but suggest that function might be restored later in progression to allow for the metabolic demands of cancer cells (116).
The mechanisms that trigger these processes are unknown. Field effects that include mitochondrial dysfunction can sometimes arise from clonal expansions of mitochondrial DNA mutations through crypt conversion and crypt fission, as previously characterized in the aging colon (128). Additionally, PGC1α, the master regulator of mitochondrial biogenesis, might mediate mitochondrial dysfunction. PGC1α expression is decreased in the intestinal epithelium of patients with UC. Notably, in mice, its deletion confers susceptibility to colitis, whereas restoration of the protein ameliorates the disease and restores mitochondrial integrity (129). PGC1α also offers a potential link between telomeres and mitochondria: in telomerase knockout mice, shortened telomeres trigger mitochondrial dysfunction via TP53 and PGC1α signaling (130,131). This intriguing connection deserves further investigation in UC tumorigenesis, especially since both alterations exhibit a similar pattern of initial dysfunction followed by later recovery.
Epigenetic changes
Chronic inflammation is well known to play a role in the progression of cancer—UC-mediated CRC being only one of many examples (132). As noted above, the inflammatory state can mutate DNA and lead to disruption of growth control genes as well as accelerate mutation acquisition by increasing cell turnover. Additionally, chronic inflammation can epigenetically alter epithelial cells without a clonal relationship (132). Gloria et al. (133) first identified DNA methylation changes in the setting of UC by measuring the incorporation of labeled methyl groups into DNA. They demonstrated that UC colonic DNA is globally hypomethylated compared to normal controls and suggested that epigenetic changes in UC colonic mucosa contribute to cancer progression. Since then, epigenetic changes have been more extensively studied as precursor lesions in UC. It has been observed that histone modification genes are overexpressed in UC and that the level of overexpression correlates with both disease extent and duration (134). In non-dysplastic, but inflamed, UC tissue, hypomethylation of bivalent H3K27me3-associated promoters facilitates the upregulation of cancer progression associated genes, including those associated with cell movement, death, survival and proliferation. These inflammation-induced changes create a field of susceptibility that might predispose to cancer progression (135). Additionally, both dysplastic and normal-appearing UC Progressor epithelium features hypermethylation at CpG islands, similar to what is seen with aging, and might contribute to increased susceptibility (136–138). Other studies have observed significantly higher methylation in genes associated with UC inflammation from UC Progressor non-dysplastic tissues when compared with UC Non-progressor tissue (139). Collectively, these findings in preneoplastic fields are consistent with the epigenetic alterations found in CRC, in which both global hypomethylation and regional hypermethylation are observed (140). Thus, the current view is that epigenetic alterations play a role in the development and progression of UC (141). These alterations might create an epigenetic field effect as a result of the clonal expansion of stem cells that carry epigenetic changes or non-clonally as a result of environmental exposures and inflammation (132,141,142).
Model of cancer progression in UC: accelerated colon aging?
The molecular alterations that define preneoplastic fields in UC are remarkably similar to the changes that occur in normal, aging tissue. Based on this, our group proposed the idea that UC can be thought of as a disease of accelerated colon aging. In normal individuals, telomere length declines progressively with age (81), but in UC the rate of decline is accelerated, especially within the first 8 years after disease diagnosis (102). On average, individuals with UC at age 40 carry colonocyte telomeres as short as non-UC individuals at age 60. This finding fits with the epidemiological observation that CRC develops at a mean age of 64 in the general population, but a mean age of 43 in UC patients—about 20 years earlier (66).
Similarly, chromosomal alterations (96,143), mitochondrial loss (144,145) and DNA methylation (146) changes have been reported in both normal aging colon and preneoplastic fields in UC. In the normal colon, these age-related alterations are attributed to the increased load of somatic mutations and molecular damage over time. While somatic mutations can lead to clonal fields through biased competition and expansion of mutated intestinal stem cells (147), extensive molecular damage can lead to non-clonal fields of cancer predisposing alterations, such as telomere shortening caused by oxidative damage (148).
We have made an effort to integrate the findings described above in our proposed model of carcinogenesis in UC (Figure 1). We postulate that in UC, genetic susceptibility and an altered microbiome and immunity lead to chronic inflammation and concomitant increased cell turnover and oxidative damage. This accelerates the rate of mutation accumulation and molecular damage, thus effectively producing accelerated aging of the colon. When telomeres become critically short, uncapped chromosome ends trigger a DNA damage response (149). In the presence of proficient TP53, this response results in the activation of cellular senescence; however, if TP53 is mutated, cells bypass senescence and cell division continues, producing preneoplastic fields in which dysfunctional telomeres trigger end-to-end fusions and chromosomal instability (106). This instability and the progressive exhaustion of telomeres eventually leads cells to crisis and death (150). During this process, however, additional genetic and epigenetic alterations accumulate and might disrupt other tumor suppressors genes and activate proto-oncogenes. One of the many consequences of this can be mutational or epigenetic reactivation of telomerase. Reactivated telomerase rescues the cells from crisis, allows further proliferation under the drive of mutated oncogenes, and eventually results in the development of invasive malignancy (151). At that step cancer cells might also acquire proficient mitochondria to cover the metabolic needs to rapid growth (116).
Field effect implications: opportunities for early cancer detection
Preneoplastic fields are an indication of an emerging neoplastic process and, therefore, they could be used to improve cancer detection in UC. The current cancer surveillance system is based on colonoscopic screening for dysplasia and it is likely to be one of the factors contributing to the decrease in UC-associated CRC in recent years (43). However, this approach has limited sensitivity (152), fails to detect every patient at risk (153), and it is time consuming and expensive. A more efficient and sensitive system would be highly desirable, especially in view of the worldwide overall increase in UC incidence (154).
The American Gastroenterology Association recommends that colonoscopic surveillance should begin 8–10 years after disease diagnosis and should occur every 1–2 years depending on dysplasia findings (155,156). Until recently, the guidelines included collection of at least 32 random quadrant biopsies to achieve 90% sensitivity for histological identification of dysplasia (90). However, gastroenterologists’ non-adherence to colonoscopy guidelines, patients’ non-compliance to the surveillance plan, and a lack of agreement between pathologists upon histological assessment reduce the efficacy of this surveillance approach (56,157,158). Over the last decade, the superiority of chromoendoscopy (CE) for the detection of dysplasia has been established (159–161). Whereas standard colonoscopic methods rely on the use of white light to visualize areas of dysplasia, CE uses dyes that stain the mucosa, increasing the contrast and the sensitivity to find dysplasia. CE is the method currently recommended for colonoscopic surveillance (162), but it is still limited by the requisite detection of morphological changes that are visible by endoscopy.
As described above, cumulative evidence demonstrates that molecular changes precede the morphological changes associated with dysplasia. Thus, focusing on the identification of preneoplastic fields may prove to be the most sensitive approach for identifying patients at risk of CRC progression. Figure 2 illustrates the potential use of the field effect to improve cancer colonoscopic surveillance in patients with UC. The premise is that CE with targeted biopsies is performed, in accordance to current recommendations (162), but in addition, two biopsies are collected at each colon segment for molecular analysis of field effects. The extension of the fields, as well as the degree of molecular alterations, might reflect the likelihood of cancer progression and could potentially be used to personalize colonoscopic surveillance, even in the absence of dysplastic findings. This hypothesis is based on the fact that cancer is a probabilistic process that depends not only on the rate of mutation, but also on the number of cells at risk (163). Thus, patients with pancolonic and multifocal fields might benefit from closer monitoring as compared with patients with localized, unifocal fields. Given the vast heterogeneity of genetic changes that can lead to UC-CRC (64), traditional genetic markers such as TP53 mutations are unlikely to be universal detectors of preneoplastic fields. Passenger mutations, such as indels in PolyG tracts, overcome this problem and might offer a promising solution for the identification of preneoplastic progression in UC. This information could be integrated into predictive statistical models to identify patients at risk. In addition, studies of the field effect and its role in early cancer detection in UC could be supported by the use of mathematical modeling to quantify the extent and behavior of these fields and identify at-risk patients (164,165). Such computational approaches have been successfully applied to predict the size, shape and distribution of fields (164), to predict cancer risk in Barrett’s esophagus (166) and head and neck premalignant lesions (167), and to evaluate the prognostic value of putative biomarkers (165).
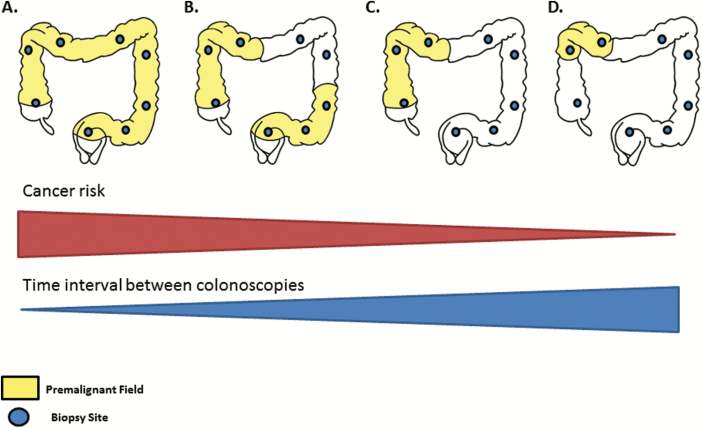
Figure 2.
Implications of the field effect on UC colonoscopic surveillance. By analyzing several screening biopsies procured along the colon, field effects can be identified and could possibly be used to predict cancer progression risk. The colon diagrams represent four clinical scenarios in which dysplasia may not be detected by chromoendoscopy but in which screening biopsies could identify molecular fields: (A) pancolonic field, (B) multifocal fields, (C) extensive field and (D) localized field. Cancer is a probabilistic process that depends on the rate of mutation and the number of cells at risk. Thus, we postulate that large, multifocal fields are likely to be associated with higher risk of cancer progression compared to small, unifocal fields. Studies with large numbers of patients are needed to validate this model, quantify the progression risk accordingly, and integrate it with other known cancer risk factors in UC. If such approach was developed, it would facilitate tailoring the time interval between colonoscopies to the level of risk for each patient.
Moving forward, large studies are required to establish the value of molecular fields for cancer detection and prediction in a clinical setting. Fortunately, large repositories of archival UC biopsies already exist, which include multiple longitudinal colonoscopies for each patient, each containing multiple random colonic biopsies. Such repositories are an excellent resource for determining the biomarker potential of molecular fields for cancer prediction.
Conclusions
Here, we have discussed the concept of the field effect, the molecular alterations that define these fields and their implications for early cancer detection in UC. Preneoplastic fields precede cancer progression in UC and have been characterized through the analysis of TP53 mutations, chromosomal alterations, telomere shortening, mitochondrial dysfunction and epigenetic alterations. Biologically, these fields reflect the early events that lead to tumor progression in UC and enable accurate spatial and temporal characterization of in vivo tumor evolution. Clinically, preneoplastic fields provide an opportunity to improve early cancer detection in UC and to personalize colonoscopic surveillance. Beyond its applications in UC, the study of the field effect is highly relevant to current efforts to understand ‘precancer’ in order to improve early detection and prevention of cancer (168).
Funding
JJS R01CA160674, T32HL007093; TAB R01CA160674. RAR R01CA181308, R01CA160674.
Conflict of Interest Statement: JJS is a founder and equity holder of TwinStrand Biosciences.
Abbreviations
- BE
Barrett’s esophagus
- CE
chromoendoscopy
- CRC
colorectal cancer
- FISH
fluorescence in situ hybridization
- PolyG
polyguanine tract
- UC
ulcerative colitis
References
- 1. Nowell P.C. (1976)The clonal evolution of tumor cell populations. Science, 194, 23–28. [DOI] [PubMed] [Google Scholar]
- 2. Tomasetti C., et al. (2013)Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc. Natl. Acad. Sci. USA, 110, 1999–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martincorena I., et al. (2015)Somatic mutation in cancer and normal cells. Science, 349, 1483–1489. [DOI] [PubMed] [Google Scholar]
- 4. Krimmel J.D., et al. (2016)Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc. Natl. Acad. Sci. USA, 113, 6005–6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Slaughter D.P., et al. (1953)Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer, 6, 963–968. [DOI] [PubMed] [Google Scholar]
- 6. Braakhuis B.J., et al. (2003)A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res., 63, 1727–1730. [PubMed] [Google Scholar]
- 7. Salk J.J., et al. (2010)Passenger mutations as a marker of clonal cell lineages in emerging neoplasia. Semin. Cancer Biol., 20, 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brabender J., et al. (2005)The molecular signature of normal squamous esophageal epithelium identifies the presence of a field effect and can discriminate between patients with Barrett’s esophagus and patients with Barrett’s-associated adenocarcinoma. Cancer Epidemiol. Biomarkers Prev., 14, 2113–2117. [DOI] [PubMed] [Google Scholar]
- 9. Prevo L.J., et al. (1999)p53-mutant clones and field effects in Barrett’s esophagus. Cancer Res., 59, 4784–4787. [PubMed] [Google Scholar]
- 10. Rabinovitch P.S., et al. (1999)Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer Res., 59, 5148–5153. [PubMed] [Google Scholar]
- 11. Reid B.J. (2010)Early events during neoplastic progression in Barrett’s esophagus. Cancer Biomark., 9, 307–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choi C.R., et al. (2017)Clonal evolution of colorectal cancer in IBD. Nat. Rev. Gastroenterol. Hepatol., 14, 218–229. [DOI] [PubMed] [Google Scholar]
- 13. Rivenbark A.G., et al. (2012)Field cancerization in mammary carcinogenesis - Implications for prevention and treatment of breast cancer. Exp. Mol. Pathol., 93, 391–398. [DOI] [PubMed] [Google Scholar]
- 14. Heaphy C.M., et al. (2009)Mammary field cancerization: molecular evidence and clinical importance. Breast Cancer Res. Treat., 118, 229–239. [DOI] [PubMed] [Google Scholar]
- 15. Jaiswal G., et al. (2013)Field cancerization: concept and clinical implications in head and neck squamous cell carcinoma. J. Exp. Ther. Oncol., 10, 209–214. [PubMed] [Google Scholar]
- 16. Cheng L., et al. (2010)The origins of urothelial carcinoma. Expert Rev. Anticancer Ther., 10, 865–880. [DOI] [PubMed] [Google Scholar]
- 17. Höglund M. (2007)Bladder cancer, a two phased disease?Semin. Cancer Biol., 17, 225–232. [DOI] [PubMed] [Google Scholar]
- 18. Amaro A., et al. (2016)Molecular evolution of colorectal cancer: from multistep carcinogenesis to the big bang. Cancer Metastasis Rev., 35, 63–74. [DOI] [PubMed] [Google Scholar]
- 19. Maeda M, et al. (2016)Mechanisms for the induction of gastric cancer by Helicobacter pylori infection: aberrant DNA methylation pathway. Gastric Cancer. 20 (Suppl 1), 8–15. [DOI] [PubMed] [Google Scholar]
- 20. Rugge M., et al. (2013)Precancerous lesions in the stomach: from biology to clinical patient management. Best Pract. Res. Clin. Gastroenterol., 27, 205–223. [DOI] [PubMed] [Google Scholar]
- 21. Graham T.A., et al. (2011)Field cancerization in the GI tract. Future Oncol., 7, 981–993. [DOI] [PubMed] [Google Scholar]
- 22. Nonn L., et al. (2009)Evidence for field cancerization of the prostate. Prostate, 69, 1470–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Squire J.A., et al. (2011)Prostate cancer as a model system for genetic diversity in tumors. Adv. Cancer Res., 112, 183–216. [DOI] [PubMed] [Google Scholar]
- 24. Beane J., et al. (2017)Detecting the presence and progression of premalignant lung lesions via airway gene expression. Clin. Cancer Res., 23, 5091–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gomperts B.N., et al. (2011)Evolving concepts in lung carcinogenesis. Semin. Respir. Crit. Care Med., 32, 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stern R.S., et al. (2002)p53 mutation in nonmelanoma skin cancers occurring in psoralen ultraviolet a-treated patients: evidence for heterogeneity and field cancerization. J. Invest. Dermatol., 119, 522–526. [DOI] [PubMed] [Google Scholar]
- 27. Verkouteren J.A.C., et al. (2017)Epidemiology of basal cell carcinoma: scholarly review. Br. J. Dermatol., 177, 359–372. [DOI] [PubMed] [Google Scholar]
- 28. Merkel E.A., et al. (2017)Malignant melanoma of sun-protected sites: a review of clinical, histological, and molecular features. Lab. Invest., 97, 630–635. [DOI] [PubMed] [Google Scholar]
- 29. Utsunomiya T., et al. (2014)Specific molecular signatures of non-tumor liver tissue may predict a risk of hepatocarcinogenesis. Cancer Sci., 105, 749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kobayashi H., et al. (2017)The conceptual advances of carcinogenic sequence model in high-grade serous ovarian cancer. Biomed. Rep., 7, 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chu T.Y., et al. (1999)Monoclonality and surface lesion-specific microsatellite alterations in premalignant and malignant neoplasia of uterine cervix: a local field effect of genomic instability and clonal evolution. Genes. Chromosomes Cancer, 24, 127–134. [DOI] [PubMed] [Google Scholar]
- 32. Kappelman M.D., et al. (2013)Recent trends in the prevalence of Crohn’s disease and ulcerative colitis in a commercially insured US population. Dig. Dis. Sci., 58, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hanauer S.B. (2006)New lessons: classic treatments, expanding options in ulcerative colitis. Colorectal Dis., 8 (Suppl 1), 20–24. [DOI] [PubMed] [Google Scholar]
- 34. Palmieri O., et al. (2015)Genome-wide pathway analysis using gene expression data of colonic mucosa in patients with inflammatory bowel disease. Inflamm. Bowel Dis., 21, 1260–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ye B.D., et al. (2016)Genetic variation in IBD: progress, clues to pathogenesis and possible clinical utility. Expert Rev. Clin. Immunol., 12, 1091–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grivennikov S.I., et al. (2010)Immunity, inflammation, and cancer. Cell, 140, 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hsieh S.Y., et al. (2006)Comparative proteomic studies on the pathogenesis of human ulcerative colitis. Proteomics, 6, 5322–5331. [DOI] [PubMed] [Google Scholar]
- 38. Payne C.M., et al. (2008)Hydrophobic bile acids, genomic instability, Darwinian selection, and colon carcinogenesis. Clin. Exp. Gastroenterol., 1, 19–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dyson J.K., et al. (2012)Colorectal cancer in inflammatory bowel disease: what is the real magnitude of the risk?World J. Gastroenterol., 18, 3839–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Castaño-Milla C., et al. (2014)Systematic review with meta-analysis: the declining risk of colorectal cancer in ulcerative colitis. Aliment. Pharmacol. Ther., 39, 645–659. [DOI] [PubMed] [Google Scholar]
- 41. Söderlund S., et al. (2009)Decreasing time-trends of colorectal cancer in a large cohort of patients with inflammatory bowel disease. Gastroenterology, 136, 1561–7; quiz 1818. [DOI] [PubMed] [Google Scholar]
- 42. Herrinton L.J., et al. (2012)Incidence and mortality of colorectal adenocarcinoma in persons with inflammatory bowel disease from 1998 to 2010. Gastroenterology, 143, 382–389. [DOI] [PubMed] [Google Scholar]
- 43. Garg S.K., et al. (2016)Risk of cancer in inflammatory bowel disease: going up, going down, or still the same?Curr. Opin. Gastroenterol., 32, 274–281. [DOI] [PubMed] [Google Scholar]
- 44. Loftus E.V., Jr (2004)Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology, 126, 1504–1517. [DOI] [PubMed] [Google Scholar]
- 45. Itzkowitz S.H., et al. (2004)Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am. J. Physiol. Gastrointest. Liver Physiol., 287, G7–17. [DOI] [PubMed] [Google Scholar]
- 46. Levin B. (1999)Risk of cancer in ulcerative colitis. Gastrointest. Endosc., 49(3 Pt 2), S60–S62. [DOI] [PubMed] [Google Scholar]
- 47. Lakatos L., et al. (2006)Risk factors for ulcerative colitis-associated colorectal cancer in a Hungarian cohort of patients with ulcerative colitis: results of a population-based study. Inflamm. Bowel Dis., 12, 205–211. [DOI] [PubMed] [Google Scholar]
- 48. Beaugerie L., et al. (2012)Clinical, serological and genetic predictors of inflammatory bowel disease course. World J. Gastroenterol., 18, 3806–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Madanchi M., et al. (2016)Malignancies in patients with inflammatory bowel disease: a single-centre experience. Digestion, 94, 1–8. [DOI] [PubMed] [Google Scholar]
- 50. Zheng H.H., et al. (2016)Increased risk of colorectal neoplasia in patients with primary sclerosing cholangitis and inflammatory bowel disease: a meta-analysis of 16 observational studies. Eur. J. Gastroenterol. Hepatol., 28, 383–390. [DOI] [PubMed] [Google Scholar]
- 51. Brentnall T.A., et al. (1996)Risk and natural history of colonic neoplasia in patients with primary sclerosing cholangitis and ulcerative colitis. Gastroenterology, 110, 331–338. [DOI] [PubMed] [Google Scholar]
- 52. Shetty K., et al. (1999)The risk for cancer or dysplasia in ulcerative colitis patients with primary sclerosing cholangitis. Am. J. Gastroenterol., 94, 1643–1649. [DOI] [PubMed] [Google Scholar]
- 53. Askling J., et al. (2001)Colorectal cancer rates among first-degree relatives of patients with inflammatory bowel disease: a population-based cohort study. Lancet, 357, 262–266. [DOI] [PubMed] [Google Scholar]
- 54. Velayos F.S., et al. (2006)Predictive and protective factors associated with colorectal cancer in ulcerative colitis: a case-control study. Gastroenterology, 130, 1941–1949. [DOI] [PubMed] [Google Scholar]
- 55. Jess T., et al. (2012)Decreasing risk of colorectal cancer in patients with inflammatory bowel disease over 30 years. Gastroenterology, 143, 375–81.e1; quiz e13. [DOI] [PubMed] [Google Scholar]
- 56. Lutgens M.W., et al. (2013)Declining risk of colorectal cancer in inflammatory bowel disease: an updated meta-analysis of population-based cohort studies. Inflamm. Bowel Dis., 19, 789–799. [DOI] [PubMed] [Google Scholar]
- 57. Brackmann S., et al. (2009)Two distinct groups of colorectal cancer in inflammatory bowel disease. Inflamm. Bowel Dis., 15, 9–16. [DOI] [PubMed] [Google Scholar]
- 58. Brackmann S., et al. (2010)Widespread but not localized neoplasia in inflammatory bowel disease worsens the prognosis of colorectal cancer. Inflamm. Bowel Dis., 16, 474–481. [DOI] [PubMed] [Google Scholar]
- 59. Neumann H., et al. (2011)Cancer risk in IBD: how to diagnose and how to manage DALM and ALM. World J. Gastroenterol., 17, 3184–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cho K.R., et al. (1992)Genetic alterations in the adenoma–carcinoma sequence. Cancer, 70(6 Suppl), 1727–1731. [DOI] [PubMed] [Google Scholar]
- 61. Sprouffske K., et al. (2011)Accurate reconstruction of the temporal order of mutations in neoplastic progression. Cancer Prev. Res. (Phila)., 4, 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Leedham S.J., et al. (2009)Clonality, founder mutations, and field cancerization in human ulcerative colitis-associated neoplasia. Gastroenterology, 136, 542–50.e6. [DOI] [PubMed] [Google Scholar]
- 63. Yaeger R., et al. (2016)Genomic alterations observed in colitis-associated cancers are distinct from those found in sporadic colorectal cancers and vary by type of inflammatory bowel disease. Gastroenterology, 151, 278–287.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Robles A.I., et al. (2016)Whole-exome sequencing analyses of inflammatory bowel disease-associated colorectal cancers. Gastroenterology, 150, 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Amersi F., et al. (2005)Colorectal cancer: epidemiology, risk factors, and health services. Clin. Colon Rectal Surg., 18, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Eaden J.A., et al. (2001)The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut, 48, 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ou B., et al. (2016)Survival of colorectal cancer in patients with or without inflammatory bowel disease: a meta-analysis. Dig. Dis. Sci., 61, 881–889. [DOI] [PubMed] [Google Scholar]
- 68. DuPont A.W., et al. (2011)The intestinal microbiota and chronic disorders of the gut. Nat. Rev. Gastroenterol. Hepatol., 8, 523–531. [DOI] [PubMed] [Google Scholar]
- 69. Zhu Y., et al. (2011)Gut microbiota and probiotics in colon tumorigenesis. Cancer Lett., 309, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Arthur J.C., et al. (2011)The struggle within: microbial influences on colorectal cancer. Inflamm. Bowel Dis., 17, 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. O’Keefe S.J. (2016)Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol., 13, 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Romano M., et al. (2016)From inflammation to cancer in inflammatory bowel disease: molecular perspectives. Anticancer Res., 36, 1447–1460. [PubMed] [Google Scholar]
- 73. Gerlinger M., et al. (2014)Cancer: evolution within a lifetime. Annu. Rev. Genet., 48, 215–236. [DOI] [PubMed] [Google Scholar]
- 74. Harpaz N., et al. (2013)Precancerous lesions in inflammatory bowel disease. Best Pract. Res. Clin. Gastroenterol., 27, 257–267. [DOI] [PubMed] [Google Scholar]
- 75. Baars J.E., et al. (2012)Age at diagnosis of inflammatory bowel disease influences early development of colorectal cancer in inflammatory bowel disease patients: a nationwide, long-term survey. J. Gastroenterol., 47, 1308–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Charpentier C, et al. (2014)Natural history of elderly-onset inflammatory bowel disease: a population-based cohort study. Gut., 63, 423–432. [DOI] [PubMed] [Google Scholar]
- 77. Quezada S.M., et al. (2012)Association of age at diagnosis and ulcerative colitis phenotype. Dig. Dis. Sci., 57, 2402–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hou J.K., et al. (2016)Characteristics and behavior of elderly-onset inflammatory bowel disease: a multi-center US study. Inflamm. Bowel Dis., 22, 2200–2205. [DOI] [PubMed] [Google Scholar]
- 79. Salk J.J., et al. (2013)Clonal expansions and short telomeres are associated with neoplasia in early-onset, but not late-onset, ulcerative colitis. Inflamm. Bowel Dis., 19, 2593–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lochhead P., et al. (2015)Etiologic field effect: reappraisal of the field effect concept in cancer predisposition and progression. Mod. Pathol., 28, 14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Risques R.A., et al. (2006)Cancer surveillance in inflammatory bowel disease: new molecular approaches. Curr. Opin. Gastroenterol., 22, 382–390. [DOI] [PubMed] [Google Scholar]
- 82. Thorsteinsdottir S., et al. (2011)Pathogenesis and biomarkers of carcinogenesis in ulcerative colitis. Nat. Rev. Gastroenterol. Hepatol., 8, 395–404. [DOI] [PubMed] [Google Scholar]
- 83. Chen R., et al. (2016)Biomarkers for colitis-associated colorectal cancer. World J. Gastroenterol., 22, 7882–7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Brentnall T.A., et al. (1994)Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology, 107, 369–378. [DOI] [PubMed] [Google Scholar]
- 85. Burmer G.C., et al. (1992)Neoplastic progression in ulcerative colitis: histology, DNA content, and loss of a p53 allele. Gastroenterology, 103, 1602–1610. [DOI] [PubMed] [Google Scholar]
- 86. Lashner B.A., et al. (1999)Evaluation of the usefulness of testing for p53 mutations in colorectal cancer surveillance for ulcerative colitis. Am. J. Gastroenterol., 94, 456–462. [DOI] [PubMed] [Google Scholar]
- 87. Chen R., et al. (2005)The initiation of colon cancer in a chronic inflammatory setting. Carcinogenesis, 26, 1513–1519. [DOI] [PubMed] [Google Scholar]
- 88. Porschen R., et al. (1992)DNA aneuploidy in Crohn’s disease and ulcerative colitis: results of a comparative flow cytometric study. Gut, 33, 663–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lindberg J.O., et al. (1999)DNA aneuploidy as a marker of premalignancy in surveillance of patients with ulcerative colitis. Br. J. Surg., 86, 947–950. [DOI] [PubMed] [Google Scholar]
- 90. Rubin C.E., et al. (1992)DNA aneuploidy in colonic biopsies predicts future development of dysplasia in ulcerative colitis. Gastroenterology, 103, 1611–1620. [DOI] [PubMed] [Google Scholar]
- 91. Holzmann K., et al. (2001)Flow cytometric and histologic evaluation in a large cohort of patients with ulcerative colitis: correlation with clinical characteristics and impact on surveillance. Dis. Colon Rectum, 44, 1446–1455. [DOI] [PubMed] [Google Scholar]
- 92. International HIVCS et al. (2010)The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science, 330, 1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Aust D.E., et al. (2000)Chromosomal alterations in ulcerative colitis-related and sporadic colorectal cancers by comparative genomic hybridization. Hum. Pathol., 31, 109–114. [DOI] [PubMed] [Google Scholar]
- 94. Bronner M.P., et al. (2010)Array-based comparative genomic hybridization in ulcerative colitis neoplasia: single non-dysplastic biopsies distinguish progressors from non-progressors. Mod. Pathol., 23, 1624–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lai L.A., et al. (2012)Pan-colonic field defects are detected by CGH in the colons of UC patients with dysplasia/cancer. Cancer Lett., 320, 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kang H., et al. (2013)Direct measurements of human colon crypt stem cell niche genetic fidelity: the role of chance in non-Darwinian mutation selection. Front. Oncol., 3, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Martincorena I., et al. (2013)Non-random mutation: the evolution of targeted hypermutation and hypomutation. Bioessays, 35, 123–130. [DOI] [PubMed] [Google Scholar]
- 98. Preston S.L., et al. (2003)Bottom-up histogenesis of colorectal adenomas: origin in the monocryptal adenoma and initial expansion by crypt fission. Cancer Res., 63, 3819–3825. [PubMed] [Google Scholar]
- 99. Boyer J.C., et al. (2002)Sequence dependent instability of mononucleotide microsatellites in cultured mismatch repair proficient and deficient mammalian cells. Hum. Mol. Genet., 11, 707–713. [DOI] [PubMed] [Google Scholar]
- 100. Salk J.J., et al. (2009)Clonal expansions in ulcerative colitis identify patients with neoplasia. Proc. Natl. Acad. Sci. USA, 106, 20871–20876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. O’Sullivan J., et al. (2006)Telomere length in the colon declines with age: a relation to colorectal cancer?Cancer Epidemiol. Biomarkers Prev., 15, 573–577. [DOI] [PubMed] [Google Scholar]
- 102. Risques R.A., et al. (2008)Ulcerative colitis is a disease of accelerated colon aging: evidence from telomere attrition and DNA damage. Gastroenterology, 135, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Risques R.A., et al. (2011)Ulcerative colitis-associated colorectal cancer arises in a field of short telomeres, senescence, and inflammation. Cancer Res., 71, 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Brentnall T.A. (2003)Molecular underpinnings of cancer in ulcerative colitis. Curr. Opin. Gastroenterol., 19, 64–68. [DOI] [PubMed] [Google Scholar]
- 105. Friis-Ottessen M., et al. (2014)Telomere shortening correlates to dysplasia but not to DNA aneuploidy in longstanding ulcerative colitis. BMC Gastroenterol., 14, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. O’Sullivan J.N., et al. (2002)Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat. Genet., 32, 280–284. [DOI] [PubMed] [Google Scholar]
- 107. Kinouchi Y., et al. (1998)Telomere shortening in the colonic mucosa of patients with ulcerative colitis. J. Gastroenterol., 33, 343–348. [DOI] [PubMed] [Google Scholar]
- 108. Tahara T., et al. (2015)Telomere length in non-neoplastic colonic mucosa in ulcerative colitis (UC) and its relationship to the severe clinical phenotypes. Clin. Exp. Med., 15, 327–332. [DOI] [PubMed] [Google Scholar]
- 109. Novak E.A., et al. (2015)Mitochondrial dysfunction in inflammatory bowel disease. Front. Cell Dev. Biol., 3, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Chatterjee A., et al. (2011)Mitochondrial subversion in cancer. Cancer Prev. Res. (Phila)., 4, 638–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sánchez-Aragó M., et al. (2010)Selection of cancer cells with repressed mitochondria triggers colon cancer progression. Carcinogenesis, 31, 567–576. [DOI] [PubMed] [Google Scholar]
- 112. Larman T.C., et al. ; Cancer Genome Atlas Research Network. (2012)Spectrum of somatic mitochondrial mutations in five cancers. Proc. Natl. Acad. Sci. USA, 109, 14087–14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Yu M. (2012)Somatic mitochondrial DNA mutations in human cancers. Adv. Clin. Chem., 57, 99–138. [DOI] [PubMed] [Google Scholar]
- 114. Ward P.S., et al. (2012)Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell, 21, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Smolková K., et al. (2011)Waves of gene regulation suppress and then restore oxidative phosphorylation in cancer cells. Int. J. Biochem. Cell Biol., 43, 950–968. [DOI] [PubMed] [Google Scholar]
- 116. Zong W.X., et al. (2016)Mitochondria and Cancer. Mol. Cell, 61, 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ju Y.S., et al. (2014)Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife, 3, e02935. doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pereira L., et al. (2012)Somatic mitochondrial DNA mutations in cancer escape purifying selection and high pathogenicity mutations lead to the oncocytic phenotype: pathogenicity analysis of reported somatic mtDNA mutations in tumors. BMC Cancer, 12, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Stewart J.B., et al. (2015)Simultaneous DNA and RNA mapping of somatic mitochondrial mutations across diverse human cancers. PLoS Genet., 11, e1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Nishikawa M., et al. (2005)Accumulation of mitochondrial DNA mutation with colorectal carcinogenesis in ulcerative colitis. Br. J. Cancer, 93, 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Brentnall T.A., et al. (2009)Proteins that underlie neoplastic progression of ulcerative colitis. Proteomics Clin. Appl., 3, 1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chen R., et al. (2014)Up-regulation of mitochondrial chaperone TRAP1 in ulcerative colitis associated colorectal cancer. World J. Gastroenterol., 20, 17037–17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. May D., et al. (2011)Investigating neoplastic progression of ulcerative colitis with label-free comparative proteomics. J. Proteome Res., 10, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Santhanam S., et al. (2012)Mitochondrial electron transport chain complex dysfunction in the colonic mucosa in ulcerative colitis. Inflamm. Bowel Dis., 18, 2158–2168. [DOI] [PubMed] [Google Scholar]
- 125. Santhanam S., et al. (2007)Impairment of mitochondrial acetoacetyl CoA thiolase activity in the colonic mucosa of patients with ulcerative colitis. Gut, 56, 1543–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sifroni K.G., et al. (2010)Mitochondrial respiratory chain in the colonic mucosal of patients with ulcerative colitis. Mol. Cell. Biochem., 342, 111–115. [DOI] [PubMed] [Google Scholar]
- 127. Ussakli C.H., et al. (2013)Mitochondria and tumor progression in ulcerative colitis. J. Natl. Cancer Inst., 105, 1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Greaves L.C., et al. (2006)Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc. Natl. Acad. Sci. USA, 103, 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cunningham K.E., et al. (2016)Peroxisome proliferator-activated Receptor-γ coactivator 1-α (PGC1α) protects against experimental murine colitis. J. Biol. Chem., 291, 10184–10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Sahin E., et al. (2011)Telomere dysfunction induces metabolic and mitochondrial compromise. Nature, 470, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sahin E., et al. (2012)Axis of ageing: telomeres, p53 and mitochondria. Nat. Rev. Mol. Cell Biol., 13, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Chiba T., et al. (2012)Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation. Gastroenterology, 143, 550–563. [DOI] [PubMed] [Google Scholar]
- 133. Glória L., et al. (1996)DNA hypomethylation and proliferative activity are increased in the rectal mucosa of patients with long-standing ulcerative colitis. Cancer, 78, 2300–2306. [DOI] [PubMed] [Google Scholar]
- 134. Gerçeker E., et al. (2015)Never in mitosis gene A-related kinase 6 and aurora kinase A: new gene biomarkers in the conversion from ulcerative colitis to colorectal cancer. Oncol. Rep., 34, 1905–1914. [DOI] [PubMed] [Google Scholar]
- 135. Hahn M.A., et al. (2014)Loss of the polycomb mark from bivalent promoters leads to activation of cancer-promoting genes in colorectal tumors. Cancer Res., 74, 3617–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Issa J.P., et al. (2001)Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res., 61, 3573–3577. [PubMed] [Google Scholar]
- 137. Kim B.N., et al. (2005)Methylation and expression of p16INK4 tumor suppressor gene in primary colorectal cancer tissues. Int. J. Oncol., 26, 1217–1226. [PubMed] [Google Scholar]
- 138. Kang K., et al. (2016)A genome-wide methylation approach identifies a new hypermethylated gene panel in ulcerative colitis. Int. J. Mol. Sci., 17(8), 1291. doi: 10.3390/ijms17081291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Garrity-Park M.M., et al. (2016)A biomarker panel to detect synchronous neoplasm in non-neoplastic surveillance biopsies from patients with ulcerative colitis. Inflamm. Bowel Dis., 22, 1568–1574. [DOI] [PubMed] [Google Scholar]
- 140. Jones P.A., et al. (2007)The epigenomics of cancer. Cell, 128, 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ventham N.T., et al. (2013)Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology, 145, 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Luo Y., et al. (2014)Field cancerization in the colon: a role for aberrant DNA methylation?Gastroenterol. Rep. (Oxf)., 2, 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hsieh J.C., et al. (2013)Large chromosome deletions, duplications, and gene conversion events accumulate with age in normal human colon crypts. Aging Cell, 12, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Greaves L.C., et al. (2010)Defects in multiple complexes of the respiratory chain are present in ageing human colonic crypts. Exp. Gerontol., 45, 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Greaves L.C., et al. (2012)Comparison of mitochondrial mutation spectra in ageing human colonic epithelium and disease: absence of evidence for purifying selection in somatic mitochondrial DNA point mutations. PLoS Genet., 8, e1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kaz A.M., et al. (2014)Patterns of DNA methylation in the normal colon vary by anatomical location, gender, and age. Epigenetics, 9, 492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Snippert H.J., et al. (2014)Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep., 15, 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. von Zglinicki T. (2002)Oxidative stress shortens telomeres. Trends Biochem. Sci., 27, 339–344. [DOI] [PubMed] [Google Scholar]
- 149. d’Adda di Fagagna F., et al. (2003)A DNA damage checkpoint response in telomere-initiated senescence. Nature, 426, 194–198. [DOI] [PubMed] [Google Scholar]
- 150. Shay J.W. (2016)Role of telomeres and telomerase in aging and cancer. Cancer Discov., 6, 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. DePinho R.A. (2000)The age of cancer. Nature, 408, 248–254. [DOI] [PubMed] [Google Scholar]
- 152. Fornaro R., et al. (2016)Colorectal cancer in patients with inflammatory bowel disease: the need for a real surveillance program. Clin. Colorectal Cancer, 15, 204–212. [DOI] [PubMed] [Google Scholar]
- 153. Mooiweer E., et al. ; Dutch Initiative on Crohn’s and Colitis. (2015)Incidence of interval colorectal cancer among inflammatory bowel disease patients undergoing regular colonoscopic surveillance. Clin. Gastroenterol. Hepatol., 13, 1656–1661. [DOI] [PubMed] [Google Scholar]
- 154. Molodecky N.A., et al. (2012)Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology, 142, 46–54.e42. [DOI] [PubMed] [Google Scholar]
- 155. Farraye F.A., et al. (2010)AGA technical review on the diagnosis and management of colorectal neoplasia in inflammatory bowel disease. Gastroenterology, 138, 746–774. [DOI] [PubMed] [Google Scholar]
- 156. Yashiro M. (2014)Ulcerative colitis-associated colorectal cancer. World J. Gastroenterol., 20, 16389–16397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Zisman T.L., et al. (2012)Prospective study of the progression of low-grade dysplasia in ulcerative colitis using current cancer surveillance guidelines. Inflamm. Bowel Dis., 18, 2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Ullman T.A. (2005)Colonoscopic surveillance in inflammatory bowel disease. Curr. Opin. Gastroenterol., 21, 585–588. [DOI] [PubMed] [Google Scholar]
- 159. Wu L., et al. (2012)The diagnostic accuracy of chromoendoscopy for dysplasia in ulcerative colitis: meta-analysis of six randomized controlled trials. Colorectal Dis., 14, 416–420. [DOI] [PubMed] [Google Scholar]
- 160. Subramanian V., et al. (2011)Meta-analysis: the diagnostic yield of chromoendoscopy for detecting dysplasia in patients with colonic inflammatory bowel disease. Aliment. Pharmacol. Ther., 33, 304–312. [DOI] [PubMed] [Google Scholar]
- 161. Marion J.F., et al. ; Chromoendoscopy Study Group at Mount Sinai School of Medicine. (2016)Chromoendoscopy is more effective than standard colonoscopy in detecting dysplasia during long-term surveillance of patients with colitis. Clin. Gastroenterol. Hepatol., 14, 713–719. [DOI] [PubMed] [Google Scholar]
- 162. Soetikno R., et al. (2016)Paradigm shift in the surveillance and management of dysplasia in inflammatory bowel disease (West). Dig. Endosc., 28, 266–273. [DOI] [PubMed] [Google Scholar]
- 163. Nunney L., et al. (2015)Peto’s paradox and the hallmarks of cancer: constructing an evolutionary framework for understanding the incidence of cancer. Philos. Trans. R Soc. Lond. B Biol. Sci., 370(1673). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Foo J., et al. (2014)Multifocality and recurrence risk: a quantitative model of field cancerization. J. Theor. Biol., 355, 170–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Dhawan A., et al. (2016)A computational modeling approach for deriving biomarkers to predict cancer risk in premalignant disease. Cancer Prev. Res. (Phila)., 9, 283–295. [DOI] [PubMed] [Google Scholar]
- 166. Curtius K., et al. (2016)A molecular clock infers heterogeneous tissue age among patients with Barrett’s Esophagus. PLoS Comput. Biol., 12, e1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Ryser M.D., et al. (2016)Quantifying the dynamics of field cancerization in tobacco-related head and neck cancer: a multiscale modeling approach. Cancer Res., 76, 7078–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Spira A., et al. (2017)Precancer atlas to drive precision prevention trials. Cancer Res., 77, 1510–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]