Abstract
Polo-like kinases are essential cell cycle regulators that are conserved from yeast to humans. Unlike higher eukaryotes, who express multiple Polo-like kinase family members that perform many important functions, budding yeast express only a single Polo-like kinase, Cdc5, which is the homologue of mammalian cell cycle master regulator Polo-like kinase 1. Cdc5 is a fascinating multifaceted protein that is programmed to target its many substrates in a timely, sequential manner to ensure proper cell cycle progression. Over the years, many lessons about Polo-like kinase 1 have been learned by studying Cdc5 in budding yeast. Cdc5 has been well documented in regulating mitotic entry, chromosome segregation, mitotic exit, and cytokinesis. Cdc5 also plays important roles during cell division after DNA damage. Here, we briefly review the many functions of Cdc5 and its regulation in the absence and presence of DNA damage.
Keywords: Polo-like kinase 1, Plk1, Cdc5, mitosis, DNA damage, cell cycle
Introduction
Polo-like kinase 1 (Plk1) is an essential mitotic regulator that is highly conserved among eukaryotes including yeast, C. elegans, fruit fly, zebra fish, mouse, and humans (Archambault and Glover 2009; de Carcer et al. 2011). Polo was first reported in Drosophila neuroblasts with circular metaphase chromosome arrangements, which corresponded to mutations in the polo gene (Sunkel and Glover 1988). Strikingly, treating human cells with a Plk1 inhibitor results in an identical circular chromosome phenotype (Takaki et al. 2008). The conservation among Plk1 homologs can be further appreciated by the finding that human PLK1 cDNA, when expressed from a yeast vector, can restore the viability of a budding yeast Polo depletion strain (Lee and Erikson 1997).
All Plk1 homologs are composed of an N-terminal Ser/Thr catalytic kinase domain and a C-terminal non-catalytic polo-box domain (PBD), which is made up of two separate polo-box motifs (Figure 1). The polo boxes, as well as the sequence directly N-terminal to them, act together as a single phosphoserine/threonine-binding module. Crystal structure studies of the PBD have discovered that the two polo boxes fold together to form a pincer-like structure (Elia et al. 2003). The PBD regulates the subcellular localization of Plk1 and is required for Polo to bind to its substrates. Indeed, mutations in the PBD prevent Polo from binding to its substrates, which can result in severe cell cycle defects (Elia et al. 2003; Ratsima et al. 2011; Song et al.2000). Budding yeast express a single Plk1 homolog Cdc5 whose structure and functions are highly conserved with higher eukaryotes. Here, we briefly review the functions and regulation of Cdc5 during the cell cycle in the absence and presence of DNA damage.
Figure 1. Key motifs found inside Cdc5.
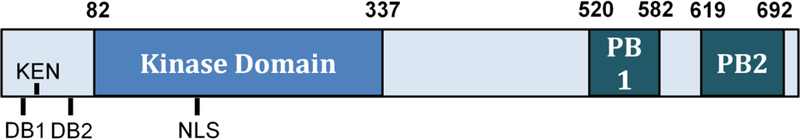
Cdc5 consists of a kinase domain in its N-terminal half, and a polo-box domain (PBD) towards its C-terminus. The destruction sequences (two Destruction Boxes and a KEN motif), which are recognized by APCCdh1 for proteasome-mediated degradation in G1 phase, lie in the N-terminus. The Cdc5 kinase domain, through which Cdc5 phosphorylates its substrates, contains a nuclear localization signal. Residue T242 is phosphorylated by Cdk1, which is required for the kinase activity of Cdc5. Phosphorylation of T238 is required for full kinase activity and for recruitment of Cdc5 to SPBs after DNA-damage. The L251W mutation is found in the adaptation-defective cdc5-ad allele. The Polo Box Domain (PBD), made up of two polo boxes, is required for Cdc5 binding to its substrates. Mutating W517/V518/L530 to F/A/A (cdc5-FAA) prevents Cdc5 from localizing to its substrates and is lethal (Song et al. 2000). The cdc5–16 (W517F H641A K643M) PBD mutant cannot bind to substrates that have undergone a priming phosphorylation event; however, this mutant still allows for cell viability.
Cdc5 activation and inactivation
Cdc5 starts to be expressed in S-phase and its protein levels remain high until the end of cell division (Charles et al. 1998; Cheng et al. 1998; Shirayama et al. 1998). Cdc5’s protein kinase activity depends on phosphorylation by cyclin-dependent kinase 1 (Cdk1), a master regulator of cell cycle control (Benanti 2016; Bloom and Cross 2007; Mortensen et al. 2005). Cdc5 contains one strict Cdk1 consensus site (S/T-P-X-K/R) and four minimal sites (S/T-P). Of these sites, mutating the minimal site T242 rendered cells inviable (Mortensen et al. 2005). Indeed, cdc5-T242A has no kinase activity, indicating that Cdk1-dependent phosphorylation of T242 is required for Cdc5 kinase activity. An additional Cdk1-dependent phosphorylation site, T70, is required for activation of the mitotic exit network (Rodriguez-Rodriguez et al. 2016). Cdc5 contains a conserved phosphorylation site, T238, which is phosphorylated by Aurora A in human cells (Macurek et al. 2008; Rawal et al. 2016; Seki et al. 2008). Phosphorylation at this site in budding yeast is not essential for cell viability, however, cdc5-T238A has reduced kinase activity and DNA damage checkpoint adaptation defects, as discussed later in this review (Rawal et al. 2016; Rodriguez-Rodriguez et al. 2016). The kinase activity of Cdc5 remains high throughout mitosis (Cheng et al. 1998).
After mitosis and cytokinesis have been completed, the Anaphase Promoting Complex (APC) subunit Cdh1 targets Cdc5 for proteasome-mediated degradation (Charles et al. 1998; Cheng et al. 1998; Shirayama et al. 1998). Cdc5 has three known degron sequences in its N-terminus (Figure 1) (Arnold et al. 2015; Charles et al. 1998; Shirayama et al. 1998). Ablation of a KEN destruction box (Pfleger and Kirschner 2000) results in much more robust stabilization of Cdc5 in G1 than mutating the two evolutionarily conserved DB destruction boxes (Arnold et al. 2015).
Functions of Cdc5 during mitosis
Cdc5 performs a vast variety of functions in mitosis (Figure 2). Biochemical studies have identified hundreds of Cdc5 substrates, many of which remain to be fully characterized (Snead et al. 2007). Inactivation of Cdc5 results in a large-budded telophase arrest (Charles et al. 1998; Hartwell et al. 1973; Jaspersen et al. 1998), suggesting that the only essential functions of Cdc5 occur later during cell division. Below, we summarize the known functions of Cdc5 during the different mitotic stages.
Figure 2. Overview of Cdc5 functions in key cell cycle pathways.
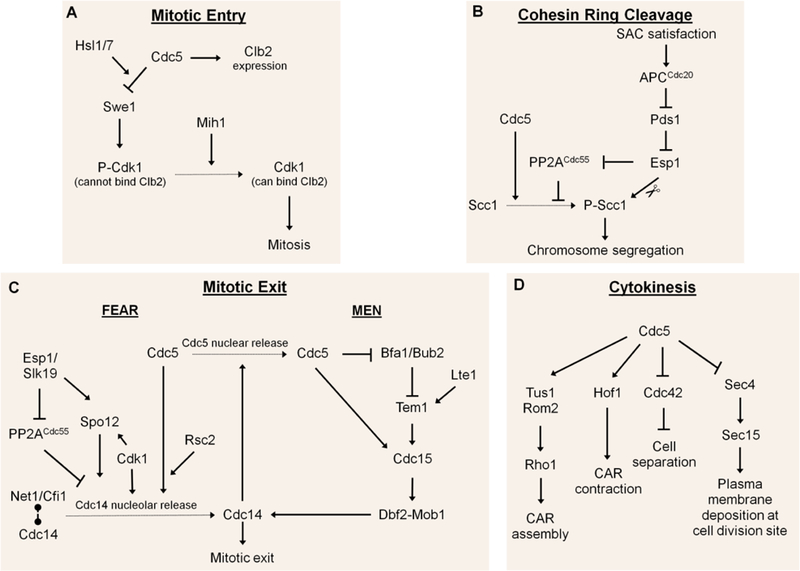
A) To initiate mitotic entry, Cdc5 promotes expression of mitotic cyclin Clb2 and inactivates Swe1 kinase. Swe1 phosphorylates Cdk1 and prevents Cdk1 from interacting with Clb2. At G2/M, Swe1 gets phosphorylated by Cdc5. Hsl1/7 are required for efficient phosphorylation of Swe1 by Cdc5. This phosphorylation of Swe1 targets it for degradation. Mih1 dephosphorylates Cdk1 and allows Cdk1 to bind Clb2 and trigger entry into mitosis.
B) In metaphase, Cdc5 phosphorylates cohesin kleisin subunit Scc1. This phosphorylation is antagonized by PP2ACdc55, which is down-regulated by Esp1 once the spindle assembly checkpoint (SAC) (Wang et al. 2014) has been satisfied. Esp1 cleaves phosphorylated Scc1, allowing sister chromatid separation.
C) Mitotic exit in budding yeast is driven by Cdc14 phosphatase, which antagonizes Cdk1 activity. Prior to anaphase, Cdc14 is sequestered in the nucleolus, spatially separated from its nuclear and cytoplasmic substrates, by anchoring protein Net1/Cfi1. In early anaphase, phosphorylation of Net1/Cfi1 by Cdk1 and Cdc5 triggers the release of Cdc14 to the nucleus (FEAR). FEAR is also controlled by other components including Esp1, Slk19, and Spo12. The second mitotic exit pathway, the MEN, is triggered in the cytoplasm by the Tem1 GTPase, which is negatively regulated by the Bfa1/Bub2 GAP and positively regulated by the Lte1 GEF. When Cdc14 is released from the nucleus by FEAR, it triggers the release of Cdc5 from the nucleus to the cytoplasm. In the cytoplasm, Cdc5 activates the MEN by phosphorylating the Bfa1/Bub2 and Cdc15. MEN promotes the release of Cdc14 from the nucleus to the cytoplasm, further driving mitotic exit.
D) Cdc5 controls multiple aspects of cytokinesis. Cdc5 phosphorylates Rho1 GEFs Tus1 and Rom2, which promotes contractile actin ring (CAR) assembly. Cdc5 promotes the recruitment of F-BAR protein Hof1 to the bud neck to trigger CAR contraction. Cdc5 also promotes cell separation by inhibiting Cdc42 and regulates the cell size at the bud neck by preventing Sec4 from interacting with its effector protein Sec15.
Mitotic entry
Cdc5 promotes timely mitotic entry (Nakashima et al. 2008; Sakchaisri et al. 2004), acting as a negative regulator of the Swe1 protein kinase and as a positive regulator of cyclin Clb2 expression (Figure 2A). Before mitosis, the phosphorylation of Cdk1 on tyrosine 19 by the kinase Swe1 (budding yeast WEE1 homolog) prevents Cdk1 from interacting specifically with Clb2 but not with the G1 cyclins Cln1–3 or S-phase cyclins Clb5 and Clb6 (Booher et al. 1993; Hu et al. 2008). In G2, the protein kinase Hsl1 and its associating protein Hsl7 help recruit Swe1 and Cdc5 to the bud neck (Asano et al. 2005; Sakchaisri et al. 2004). Cdc5 phosphorylates Swe1, targeting it for ubiquitin-mediated degradation (Asano et al. 2005). At least two other kinases, Cdk1 and the PAK homolog Cla4, also phosphorylate Swe1 to target it for degradation (Sakchaisri et al. 2004; Sia et al. 1998). The phosphatase Mih1 subsequently dephosphorylates Cdk1 on Y19, allowing it to interact with Clb2 and drive mitosis (Russell et al. 1989; Sia et al. 1996).
Cdc5 also promotes mitotic entry by acting within the nucleus, and nuclear exclusion of Cdc5 in G2/M results in delayed mitotic entry (Botchkarev et al. 2014; Nakashima et al. 2008). Cdc5 phosphorylates the Mcm1-Fkh2-Ndd1 transcription factor complex required for Clb2 expression (Darieva et al. 2006). Thus, it is probable that nuclear import of Cdc5 is required for mitotic entry by regulating the expression of Clb2.
Maintenance of shape of mitotic nucleus
Very interesting recent work revealed that Cdc5 plays a role in regulating the shape of the nucleus during mitosis (Walters et al. 2014). Nuclei of medium-budded cells undergo a space-restricted nuclear envelope expansion in the vicinity of the nucleolus resulting in the formation of “flares.” Cdc5 was identified as one of four proteins, alongside Fas1, Fas2, and Acc1, to be required for this expansion of the nuclear envelope. A cdc5 mutant, cdc5-nf, is specifically defective in forming nuclear flares at 34˚C but does not appear to have any other mitotic defects. Because other known cdc5 temperature-sensitive mutants, as well as auxin-inducible degradation of Cdc5, fail to form these nuclear envelope flares during nocodazole arrest, it is likely that Cdc5 kinase activity is required for nuclear flare formation.
Cohesin ring cleavage
A key nuclear function of Cdc5 is to promote the efficient cleavage of the cohesin ring subunit Scc1 by the Esp1 protease (separase) (Figure 2B) (Alexandru et al. 2001; Ciosk et al. 1998; Hornig and Uhlmann 2004). Through metaphase, sister chromatids are held together by the cohesin ring, which is composed of the subunits Scc1/Mcd1, Scc3, Smc1, and Smc3 (Gruber et al. 2003). When the mitotic spindle microtubules capture all of the sister chromatids by their kinetochores, spindle assembly checkpoint proteins activate the APC subunit Cdc20, an E3 ligase whose function is required for anaphase onset (Hoyt et al. 1991; Hwang et al. 1998). Among other substrates, Cdc20 targets Pds1 (securin) for degradation (Cohen-Fix et al. 1996; Visintin et al. 1997), allowing Esp1 to cleave Scc1 and allow sister chromatids to segregate faithfully to the two daughter cells in anaphase (Uhlmann et al. 1999).
During mitosis, Cdc5 binds to chromatin in the cohesin-associated regions (Mishra et al. 2016; Rossio et al. 2010). Cdc5 phosphorylates specifically chromatin-bound Scc1 on ten residues (Alexandru et al. 2001; Hornig and Uhlmann 2004). Cdc5-phosphorylated Scc1 can be dephosphorylated by the protein phosphatase 2A (PP2A) regulatory subunit B Cdc55 (Yaakov et al. 2012). Since PP2ACdc55 is inhibited by Esp1 in anaphase, it is likely that PP2ACdc55 down-modulates the Cdc5-dependent phosphorylation level of Scc1 until anaphase onset to prevent precocious Scc1 cleavage by low levels of Esp1 activity prior to anaphase (Figure 2B) (Yaakov et al. 2012). At the metaphase-to-anaphase transition, Cdc5 is required to remove cohesin from chromatin, which is important for faithful chromosome segregation, since cells expressing the cdc5–99 mutant, which is defective in mitotic exit, maintain cohesin on chromatin in anaphase and have chromosome segregation defects (Mishra et al. 2016). In line with this, it was recently reported that a different point mutant of Cdc5, cdc5-T238A, has an increased chromosome loss rate and that Cdc5 acts in parallel with Sgs1 to maintain genome integrity (Rawal et al. 2016). Cdc5 also helps maintain genome integrity during mitosis by contributing to the degradation of the DNA replication firing factor Sld2 (Reusswig et al. 2016).
Mitotic Exit
In anaphase, Cdc5 triggers two branches of the mitotic exit cascade – Cdc Fourteen Early Anaphase Release (FEAR) and the Mitotic Exit Network (MEN) (Figure 2C) (Stegmeier and Amon 2004). The main goal of both pathways is the nucleolar release and activation of Cdc14, an essential phosphatase in budding yeast, which drives mitotic exit in part by inactivating Cdk1 and dephosphorylating Cdk1 substrates (Machin et al. 2016; Stegmeier and Amon 2004; Wurzenberger and Gerlich 2011). From G1 phase through metaphase, Cdc14 is kept sequestered in the nucleolus by the anchoring protein Net1, also known as Cfi1 (Shou et al. 1999; Visintin et al. 1999). By keeping Cdc14 in the nucleolus, Cfi1/Net1 prevents Cdc14 from accessing its substrates in the nucleus and in the cytoplasm. At anaphase onset, as a result of the FEAR network, which is controlled by many components including Esp1, Slk19, Spo12, PP2ACdc55 (D’Amours and Amon 2004; Queralt et al. 2006; Stegmeier et al. 2002; Tomson et al. 2009), Net1 becomes phosphorylated by Cdk1 and Cdc5 (Figure 2C) (Azzam et al. 2004; Rodriguez-Rodriguez et al. 2016; Shou et al. 2002; Yoshida and Toh-e 2002). This phosphorylation event liberates Cdc14 from Net1. Cdc5 physically interacts with FEAR network proteins Esp1 and Slk19, as well as with Cdc14 (Rahal and Amon 2008; Stegmeier et al. 2002). The RSC chromatin-remodeling complex accessory subunit Rsc2, is also required for Net1 phosphorylation and physically interacts with the PBD of Cdc5, suggesting that Rsc2 can promote Cdc5 activity towards Net1 (Rossio et al. 2010). However, it has been argued that Cdc5, instead of directly phosphorylating Net1, primarily functions in FEAR by stimulating degradation of the Cdk1-inhibitory kinase Swe1 (Liang et al. 2009). Thus, although the molecular mechanism of Cdc5 involvement in FEAR has been debated, it is well-accepted that Cdc5 is important for Cdc14 activation.
When Cdc14 is released from Cfi1/Net1-dependent anchoring in the nucleolus, it can gain access to its substrates in the nucleus and cytoplasm to promote mitotic exit. Cdc5 also promotes chromosome condensation in anaphase by targeting condensin components Brn1, Ycg1, and Ycs4 (St-Pierre et al. 2009), and mitotic spindle elongation (Park et al. 2008; Roccuzzo et al. 2015). It was recently shown that Cdc5 targets Sfi1, a component of the yeast centrosomes or spindle pole bodies (SPBs) (Jaspersen and Winey 2004), to prevent untimely SPB duplication until the daughter cells segregated the nuclei and exited from mitosis (Elserafy et al. 2014).
In late anaphase, Cdc5 targets the Bfa1/Bub2 GTPase-activating protein (GAP) complex on the cytoplasmic surface of daughter-facing SPB (Geymonat et al. 2003; Gruneberg et al. 2000; Gryaznova et al. 2016; Hu et al. 2001; Pereira et al. 2000). This phosphorylation event is important for activation of the second mitotic exit cascade, the MEN. The MEN is controlled by the activation status of the Tem1 GTPase (Bardin and Amon 2001; Lee et al. 2001; Shirayama et al. 1994). Prior to anaphase, Tem1 is kept inactive by the Bfa1/Bub2 GAP complex (Geymonat et al. 2002; Pereira et al. 2000). When the spindle is correctly oriented and the cell is ready to exit from mitosis, Cdc5 phosphorylates Bfa1 on the cytoplasmic surface of daughter-facing SPB, a modification that is required and sufficient to inactivate the Bfa1/Bub2 (Geymonat et al. 2003; Gruneberg et al. 2000; Gryaznova et al. 2016; Hu et al. 2001; Pereira et al. 2000). Tem1, no longer under influence of Bfa1/Bub2 GAP activity, becomes activated by the Lte1 guanine exchange factor (GEF) in the daughter cell (Bardin et al. 2000; Pereira et al. 2000). This activation allows Tem1 to trigger the MEN through its substrate Cdc15 kinase (Asakawa et al. 2001; Jaspersen et al. 1998; Shirayama et al. 1994; Visintin and Amon 2001). Cdc5 phosphorylates Bfa1 on four additional residues to maintain Bfa1 localization to the daughter SPB and away from the mother SPB in anaphase (Kim et al. 2012). Cdc5 also directly phosphorylates Cdc15 independently of Tem1 GTPase activity (Rock and Amon 2011). Thus, Cdc5 targets both Bfa1 and Cdc15 to activate MEN.
Cytokinesis
Cdc5 controls multiple events that contribute to cytokinesis (Figure 2D). Contractile actin ring (CAR) assembly is a hallmark of cytokinesis in yeast and mammalian cells (Wloka and Bi 2012). The Rho1 GTPase regulates CAR assembly in yeast (Tolliday et al. 2002). Cdc5 promotes CAR assembly by phosphorylating the Rho1 GEFs Tus1 and Rom2, thereby targeting their localization to the bud neck (Yoshida et al. 2006). Phosphorylation of Tus1 and Rom2 by Cdc5 is required for CAR assembly and for activation of Rho1 at the bud neck (Yoshida et al. 2006). Additionally, Cdc5 inhibits activity of the Cdc42 GTPase, likely via interaction with the Cdc42 GAPs Bem2 and Bem3 (Atkins et al. 2013). This inhibition of Cdc42 is required for cytokinesis and cell separation. Cdc5 also promotes recruitment of the F-BAR protein Hof1 to the actomyosin ring, where Hof1 in turn promotes actomyosin ring contraction and membrane ingression (Meitinger et al. 2011). Cdc5 also phosphorylates Sec4 at the bud neck during cytokinesis, which blocks Sec4 interaction with its downstream effector Sec15 and prevents the addition of new plasma membrane to the bud neck, thus regulating cell size at the bud neck (Lepore et al. 2016).
Regulation of Cdc5 localization during the cell cycle
It is remarkable that a single Cdc5 protein can know where to target each of its substrates in a timely manner to promote mitotic progression. One important mechanism through which Cdc5 binds to many of its substrates is via priming phosphorylation as reviewed elsewhere (Lee et al. 2005). Another key layer of Cdc5 regulation lies in its ability to change localization during the cell cycle (Figure 3, Table 1).
Figure 3. Cdc5 localization during the cell cycle.
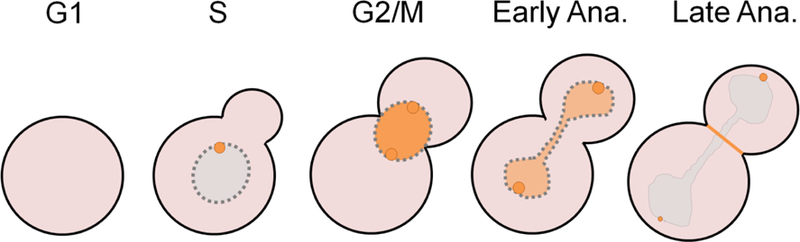
Cdc5 starts to accumulate at the SPB in S-phase. Following SPB duplication, in G2, Cdc5 accumulates in the nucleus and decorates the nuclear surface of SPBs. Cdc5 remains in the nucleus and at both SPBs in early anaphase. Later in anaphase, Cdc5 is released from the nucleus to the cytoplasm. In late anaphase, Cdc5 localizes strongly at the cytoplasmic side of the daughter SPB, and weakly at the mother SPB, as well as at the bud neck. After cytokinesis is complete, Cdc5 gets targeted by APCCdh1 for degradation in G1 of the following cell cycle.
Table 1.
Known regulators of Cdc5 localization during the cell cycle
| Cell cycle stage | Regulator | Function |
|---|---|---|
| G2/M | Cnm67, Nud1, Bbp1 | Contribute to Cdc5 SPB localization |
| TORC1, PP2APph21/22, Srp1, Kap95 | Nuclear import of Cdc5 | |
| Anaphase | Cdc14 | Required to release of Cdc5 from the nucleus to the cytoplasm, to release Cdc5 from its G2/M SPB population, and for Cdc5 localization to the bud neck |
| Bfa1 | Required for Cdc5 localization to daughter SPB | |
| G1 | APCCdh1 | Targets Cdc5 for degradation |
Cdc5 localization during the cell cycle was first reported by (Charles et al. 1998; Cheng et al. 1998; Shirayama et al. 1998; Song et al. 2000). Cdc5 accumulates in the nucleus in G2-phase and remains strongly nuclear through early anaphase. Analysis of the Cdc5 amino acid sequence revealed two putative nuclear localization signal (NLS) motifs: 58KKKR and 171KRRK; however, only mutation of the latter resulted in de-localization of Cdc5 from the nucleus (Nakashima et al. 2008). This Cdc5 NLS is conserved among polo kinases (Nakashima et al 2008). Nuclear import of Cdc5 is regulated by TORC1, PP2A catalytic subunits Pph21/22, and karyopherin complex subunits Srp1 and Kap95 (Nakashima et al. 2008). This nuclear import of Cdc5 is important for mitotic entry (Botchkarev et al. 2014; Nakashima et al. 2008). Cdc5 gets released back from the nucleus to the cytoplasm in late anaphase in a Cdc14-dependent manner (Botchkarev et al. 2014). Blocking nuclear release of Cdc5 in anaphase results in defects in mitotic exit (Botchkarev et al. 2014). The mechanism through which Cdc14 triggers the release of Cdc5 from the nucleus in anaphase is still unclear, and a nuclear export sequence (NES) within Cdc5 remains to be defined. It has been reported that the nuclear Cdc7-Dbf4 protein complex, whose function is required for the regulation of DNA replication (Sclafani 2000), physically interacts with Cdc5 and thereby prevents Cdc5-dependent MEN activation prior to anaphase (Chen and Weinreich 2010; Miller et al. 2009).
Cdc5 also undergoes change in localization at the SPBs during the cell cycle. Cdc5 starts to localize at the SPB in S-phase, when there is only one SPB present in the cell (Botchkarev et al. 2014; Lee et al. 2005). Following SPB duplication in G2-phase, Cdc5 localizes to the nuclear surface of both mother-facing and daughter-facing SPBs (mSPB and dSPB respectively) (Botchkarev et al. 2017). Indeed, in G2/M, Cdc5 can bind to nuclear SPB component Spc110 and kinetochore proteins Slk19, Cse4, and Tid3 (Rahal and Amon 2008; Snead et al. 2007). Likewise, blocking nuclear import of Cdc5 in karyopherin mutants srp1–35 and kap95–14 prevented the localization of Cdc5 to the SPBs at least in small and medium budded cells (Nakashima et al. 2008). When overexpressed, Cdc5 was detected at both nuclear and cytoplasmic SPB surfaces using immuno-electron microscopy (Park et al. 2004a).
In anaphase, Cdc14 promotes the release of Cdc5 from the inner plaque of the mSPB and dSPB (Botchkarev et al. 2017). Following Cdc5 translocation to the cytoplasm in anaphase (Botchkarev et al. 2014), Cdc5 localizes to the outer plaque of the dSPB in a Bfa1-dependent manner (Botchkarev et al. 2017) and continues to decorate the dSPB until the end of the cell cycle (Botchkarev et al. 2017; Botchkarev et al. 2014; Cheng et al. 1998; Maekawa et al. 2007; Shirayama et al. 1998; Song et al. 2000). Cdc5 localization to the SPBs likely depends on the phosphorylation of its SPB substrates by priming kinases. The cdc5–16 “pincer” mutant, which contains three point mutations in its C-terminal polo-box domain that render it unable to bind phospho-primed substrates (Elia et al. 2003; Ratsima et al. 2011), cannot localize to the SPBs during mitosis (Botchkarev et al. 2014; Park et al. 2004b; Ratsima et al. 2011). In the absence of SPB components Cnm67, Nud1, and Bbp1, Cdc5 becomes partially re-localized away from the SPBs (Park et al. 2004a; Park et al. 2004b).
Cdc5 localizes to the bud neck during G2 (Sakchaisri et al. 2004; Song et al. 2000) as well as in anaphase (Botchkarev et al. 2014; Meitinger et al. 2011; Sakchaisri et al. 2004; Song et al. 2000). Cdc5 localization to the bud neck in anaphase is thought to be required for cytokinesis and regulation of membrane deposition (Lepore et al. 2016; Meitinger et al. 2011). Cdc5 signal from the bud neck disappears prior to actomyosin ring contraction (Meitinger et al. 2011).
Cdc5 functions and regulation after DNA damage
Cdc5 plays important functions in the cellular response to DNA damage events. When a cell suffers from a DNA damage event, the DNA damage response signaling cascade becomes activated (Harrison and Haber 2006). The main functions of the DNA damage response are to arrest the cell cycle in G2/M phase and to repair the DNA damage event. If the DNA damage cannot be repaired, after a prolonged G2/M arrest, the cells can resume their cell cycle progression and continue dividing in the presence of the DNA damage. This event is termed adaptation.
The first hint that Cdc5 is involved in regulating the DNA damage checkpoint came from the discovery that an L251W point mutant of Cdc5, termed cdc5-ad, is defective in adaptation to DNA damage (Toczyski et al. 1997). Upon DNA damage, cells expressing cdc5-ad remain permanently arrested as dumbbell cells and exhibit a continued hyperphosphorylation of Rad53, a measure of an activated DNA damage checkpoint (Pellicioli et al. 2001). The cdc5-ad mutation is not simply loss of function of Cdc5, because it retains kinase activity (Rawal et al. 2016) and co-expression of Cdc5 and cdc5-ad in diploid cells does not fully suppress the cdc5-ad adaptation defect (Dotiwala et al. 2007; Vidanes et al. 2010). Therefore, it is highly likely that cdc5-ad is a semi-dominant allele; but its precise role in maintaining G2/M arrest after DNA damage is not clear.
Other work suggests that the DNA damage checkpoint down-regulates Cdc5. The temperature-sensitive cdc13–1 allele triggers DNA damage checkpoint-induced G2/M arrest, by the loss of protection of telomeres (Lin and Zakian 1996; Weinert et al. 1994). When Cdc13 is inactivated, Cdc5 has reduced kinase activity compared to nocodazole-induced G2/M arrest (Cheng et al. 1998; Zhang et al. 2009). Rad53 is important to down-regulate Cdc5 kinase activity after DNA damage (Zhang et al. 2009), although Cdc5 substrates Swe1 and Scc1 can still be phosphorylated (Ratsima et al. 2016). But Cdc5 in turn regulates Rad53. Overexpression of Cdc5 significantly shortens DNA damage-induced G2/M arrest and promotes dephosphorylation/inactivation of Rad53 (Donnianni et al. 2010; Vidanes et al. 2010). Rad53 dephosphorylation is mediated in part by the Ptc2 and Ptc3 phosphatases (Leroy et al. 2003), but Cdc5 can still promote Rad53 dephosphorylation in a ptc2∆ ptc3∆ strain (Vidanes et al. 2010). It is possible that Cdc5 acts through the PP4 phosphatase, Pph3, which also regulates Rad53 (O’Neill et al. 2007).
Cdc5 accumulates in the nucleus after DNA damage in cdc13–1. Bypassing DNA damage-induced G2/M arrest by inactivating Rad53 is sufficient to reduce the nuclear localization of Cdc5 and increase the phosphorylation of Cdc5’s cytoplasmic mitotic exit substrate Bfa1 (Valerio-Santiago et al. 2013). Consistent with this finding, we have shown that Cdc5 localizes to the nuclear surface of SPBs, spatially separated from Bfa1, during DNA damage-induced G2/M arrest (Botchkarev et al. 2017). Thus, nuclear sequestration of Cdc5 after DNA damage away from its cytoplasmic substrate Bfa1 may be a mechanism that contributes to a prolonged G2/M arrest after DNA damage. A new Cdc5 adaptation defective mutant was recently identified, cdc5-T238A, which has reduced kinase activity, is defective in SPB localization after an irreparable DSB, and its recruitment to the SPBs correlates with the timing of adaptation (Rawal et al. 2016). It was also shown that artificial sequestration of Cdc5 to the SPB inner plaque after DNA damage is sufficient to promote adaptation (Ratsima et al. 2016). It is likely that Cdc5 has important nuclear and cytoplasmic roles in adaptation.
Concluding thoughts
In the four decades since the isolation of the first cdc5 mutant by Lee Hartwell and colleagues (Hartwell et al. 1973), and in almost three decades since discovery of Polo kinase by Claudio Sunkel and David Glover (Sunkel and Glover 1988), Polo-like kinases have been established as critical multifaceted regulators of cell division in many eukaryotes (Archambault and Glover 2009; Lee et al. 2005). Looking forward, many more functions and mechanisms of Cdc5 regulation are likely to be uncovered. Since Polo kinases are highly overexpressed in many types of cancer (Degenhardt and Lampkin 2010; Strebhardt and Ullrich 2006), lessons learned about budding yeast Cdc5 functions and regulation may be applicable to solving cancer problems. In fact, Cdc5 studies have extended to even as far as the involvement of Plk2 in Parkinson’s disease (Wang et al. 2012). Thus, due to its conservation with higher eukaryotes and simple genetics, the budding yeast remains an excellent model system to study Polo-like kinases, their regulation, and their various functions in many different health and disease contexts.
Acknowledgements
We are grateful to Satoshi Yoshida, in whose lab V.B. initiated his study of Cdc5. Work was supported by National Institutes of Health Grant GM61766 to J.E.H.
Literature cited
- Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K (2001) Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast Cell 105:459–472 [DOI] [PubMed] [Google Scholar]
- Archambault V, Glover DM (2009) Polo-like kinases: conservation and divergence in their functions and regulation Nat Rev Mol Cell Biol 10:265–275 10.1038/nrm2653 [DOI] [PubMed] [Google Scholar]
- Arnold L, Hockner S, Seufert W (2015) Insights into the cellular mechanism of the yeast ubiquitin ligase APC/C-Cdh1 from the analysis of in vivo degrons Mol Biol Cell 26:843–858 10.1091/mbc.E14-09-1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Yoshida S, Otake F, Toh-e A (2001) A novel functional domain of Cdc15 kinase is required for its interaction with Tem1 GTPase in Saccharomyces cerevisiae Genetics 157:1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano S et al. (2005) Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast EMBO J 24:2194–2204 10.1038/sj.emboj.7600683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins BD, Yoshida S, Saito K, Wu CF, Lew DJ, Pellman D (2013) Inhibition of Cdc42 during mitotic exit is required for cytokinesis J Cell Biol 202:231–240 10.1083/jcb.201301090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam R et al. (2004) Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus Science 305:516–519 10.1126/science.1099402 [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Amon A (2001) Men and sin: what’s the difference? Nat Rev Mol Cell Biol 2:815– 826 10.1038/35099020 [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A (2000) A mechanism for coupling exit from mitosis to partitioning of the nucleus Cell 102:21–31 [DOI] [PubMed] [Google Scholar]
- Benanti JA (2016) Create, activate, destroy, repeat: Cdk1 controls proliferation by limiting transcription factor activity Curr Genet 62:271–276 10.1007/s00294-015-0535-5 [DOI] [PubMed] [Google Scholar]
- Bloom J, Cross FR (2007) Multiple levels of cyclin specificity in cell-cycle control Nat Rev Mol Cell Biol 8:149–160 10.1038/nrm2105 [DOI] [PubMed] [Google Scholar]
- Booher RN, Deshaies RJ, Kirschner MW (1993) Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins EMBO J 12:3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VV Jr., Garabedian MV, Lemos B, Paulissen E, Haber JE (2017) The budding yeast Polo-like kinase localizes to distinct populations at centrosomes during mitosis Mol Biol Cell 10.1091/mbc.E16-05-0324 [DOI] [PMC free article] [PubMed]
- Botchkarev VV Jr., Rossio V, Yoshida S (2014) The budding yeast Polo-like kinase Cdc5 is released from the nucleus during anaphase for timely mitotic exit Cell Cycle 13:3260–3270 10.4161/15384101.2014.953882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO (1998) The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae Curr Biol 8:497–507 [DOI] [PubMed] [Google Scholar]
- Chen YC, Weinreich M (2010) Dbf4 regulates the Cdc5 Polo-like kinase through a distinct non-canonical binding interaction J Biol Chem 285:41244–41254 10.1074/jbc.M110.155242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Hunke L, Hardy CF (1998) Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase cdc5p Mol Cell Biol 18:7360–7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K (1998) An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast Cell 93:1067–1076 [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D (1996) Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p Genes Dev 10:3081–3093 [DOI] [PubMed] [Google Scholar]
- D’Amours D, Amon A (2004) At the interface between signaling and executing anaphase--Cdc14 and the FEAR network Genes Dev 18:2581–2595 10.1101/gad.1247304 [DOI] [PubMed] [Google Scholar]
- Darieva Z et al. (2006) Polo kinase controls cell-cycle-dependent transcription by targeting a coactivator protein Nature 444:494–498 10.1038/nature05339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carcer G, Manning G, Malumbres M (2011) From Plk1 to Plk5: functional evolution of polo-like kinases Cell Cycle 10:2255–2262 10.4161/cc.10.14.16494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt Y, Lampkin T (2010) Targeting Polo-like kinase in cancer therapy Clin Cancer Res 16:384–389 10.1158/1078-0432.CCR-09-1380 [DOI] [PubMed] [Google Scholar]
- Donnianni RA et al. (2010) Elevated levels of the polo kinase Cdc5 override the Mec1/ATR checkpoint in budding yeast by acting at different steps of the signaling pathway PLoS Genet 6:e1000763 10.1371/journal.pgen.1000763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotiwala F, Haase J, Arbel-Eden A, Bloom K, Haber JE (2007) The yeast DNA damage checkpoint proteins control a cytoplasmic response to DNA damage Proc Natl Acad Sci U S A 104:11358–11363 10.1073/pnas.0609636104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia AE et al. (2003) The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain Cell 115:83–95 [DOI] [PubMed] [Google Scholar]
- Elserafy M et al. (2014) Molecular mechanisms that restrict yeast centrosome duplication to one event per cell cycle Curr Biol 24:1456–1466 10.1016/j.cub.2014.05.032 [DOI] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Smith SJ, Wheatley E, Rittinger K, Johnston LH, Sedgwick SG (2002) Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1 J Biol Chem 277:28439–28445 10.1074/jbc.M202540200 [DOI] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Walker PA, Johnston LH, Sedgwick SG (2003) In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5 J Biol Chem 278:14591–14594 10.1074/jbc.C300059200 [DOI] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K (2003) Chromosomal cohesin forms a ring Cell 112:765– 777 [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E (2000) Nud1p links astral microtubule organization and the control of exit from mitosis EMBO J 19:6475–6488 10.1093/emboj/19.23.6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryaznova Y, Koca Caydasi A, Malengo G, Sourjik V, Pereira G (2016) A FRET-based study reveals site-specific regulation of spindle position checkpoint proteins at yeast centrosomes Elife 5 10.7554/eLife.14029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JC, Haber JE (2006) Surviving the breakup: the DNA damage checkpoint Annu Rev Genet 40:209–235 10.1146/annurev.genet.40.051206.105231 [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Mortimer RK, Culotti J, Culotti M (1973) Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants Genetics 74:267–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig NC, Uhlmann F (2004) Preferential cleavage of chromatin-bound cohesin after targeted phosphorylation by Polo-like kinase EMBO J 23:3144–3153 10.1038/sj.emboj.7600303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT (1991) S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function Cell 66:507–517 [DOI] [PubMed] [Google Scholar]
- Hu F, Gan Y, Aparicio OM (2008) Identification of Clb2 residues required for Swe1 regulation of Clb2-Cdc28 in Saccharomyces cerevisiae Genetics 179:863–874 10.1534/genetics.108.086611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Wang Y, Liu D, Li Y, Qin J, Elledge SJ (2001) Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints Cell 107:655–665 [DOI] [PubMed] [Google Scholar]
- Hwang LH et al. (1998) Budding yeast Cdc20: a target of the spindle checkpoint Science 279:1041–1044 [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO (1998) A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae Mol Biol Cell 9:2803–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M (2004) The budding yeast spindle pole body: structure, duplication, and function Annu Rev Cell Dev Biol 20:1–28 10.1146/annurev.cellbio.20.022003.114106 [DOI] [PubMed] [Google Scholar]
- Kim J, Luo G, Bahk YY, Song K (2012) Cdc5-dependent asymmetric localization of bfa1 fine-tunes timely mitotic exit PLoS Genet 8:e1002450 10.1371/journal.pgen.1002450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Erikson RL (1997) Plk is a functional homolog of Saccharomyces cerevisiae Cdc5, and elevated Plk activity induces multiple septation structures Mol Cell Biol 17:3408–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Park JE, Asano S, Park CJ (2005) Yeast polo-like kinases: functionally conserved multitask mitotic regulators Oncogene 24:217–229 10.1038/sj.onc.1208271 [DOI] [PubMed] [Google Scholar]
- Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH (2001) Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5 Curr Biol 11:784–788 [DOI] [PubMed] [Google Scholar]
- Lepore D, Spassibojko O, Pinto G, Collins RN (2016) Cell cycle-dependent phosphorylation of Sec4p controls membrane deposition during cytokinesis J Cell Biol 214:691–703 10.1083/jcb.201602038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy C, Lee SE, Vaze MB, Ochsenbein F, Guerois R, Haber JE, Marsolier-Kergoat MC (2003) PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break Mol Cell 11:827–835 [DOI] [PubMed] [Google Scholar]
- Liang F, Jin F, Liu H, Wang Y (2009) The molecular function of the yeast polo-like kinase Cdc5 in Cdc14 release during early anaphase Mol Biol Cell 20:3671–3679 10.1091/mbc.E08-10-1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA (1996) The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo Proc Natl Acad Sci U S A 93:13760–13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin F, Quevedo O, Ramos-Perez C, Garcia-Luis J (2016) Cdc14 phosphatase: warning, no delay allowed for chromosome segregation! Curr Genet 62:7–13 10.1007/s00294-015-0502-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macurek L et al. (2008) Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery Nature 455:119–123 10.1038/nature07185 [DOI] [PubMed] [Google Scholar]
- Maekawa H, Priest C, Lechner J, Pereira G, Schiebel E (2007) The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal J Cell Biol 179:423–436 10.1083/jcb.200705197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G (2011) Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis Genes Dev 25:875–888 10.1101/gad.622411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Gabrielse C, Chen YC, Weinreich M (2009) Cdc7p-Dbf4p regulates mitotic exit by inhibiting Polo kinase PLoS Genet 5:e1000498 10.1371/journal.pgen.1000498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK et al. (2016) Polo kinase Cdc5 associates with centromeres to facilitate the removal of centromeric cohesin during mitosis Mol Biol Cell 27:2286–2300 10.1091/mbc.E16-01-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen EM, Haas W, Gygi M, Gygi SP, Kellogg DR (2005) Cdc28-dependent regulation of the Cdc5/Polo kinase Curr Biol 15:2033–2037 10.1016/j.cub.2005.10.046 [DOI] [PubMed] [Google Scholar]
- Nakashima A et al. (2008) The yeast Tor signaling pathway is involved in G2/M transition via polo-kinase PLoS One 3:e2223 10.1371/journal.pone.0002223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill BM, Szyjka SJ, Lis ET, Bailey AO, Yates JR 3rd, Aparicio OM, Romesberg FE (2007) Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage Proc Natl Acad Sci U S A 104:9290–9295 10.1073/pnas.0703252104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ et al. (2008) Requirement for the budding yeast polo kinase Cdc5 in proper microtubule growth and dynamics Eukaryot Cell 7:444–453 10.1128/EC.00283-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ et al. (2004a) Requirement for Bbp1p in the proper mitotic functions of Cdc5p in Saccharomyces cerevisiae Mol Biol Cell 15:1711–1723 10.1091/mbc.E03-07-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE et al. (2004b) Novel functional dissection of the localization-specific roles of budding yeast polo kinase Cdc5p Mol Cell Biol 24:9873–9886 10.1128/MCB.24.22.9873-9886.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE (2001) Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest Mol Cell 7:293–300 [DOI] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E (2000) The Bub2p spindle checkpoint links nuclear migration with mitotic exit Mol Cell 6:1–10 [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW (2000) The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1 Genes Dev 14:655–665 [PMC free article] [PubMed] [Google Scholar]
- Queralt E, Lehane C, Novak B, Uhlmann F (2006) Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast Cell 125:719–732 10.1016/j.cell.2006.03.038 [DOI] [PubMed] [Google Scholar]
- Rahal R, Amon A (2008) The Polo-like kinase Cdc5 interacts with FEAR network components and Cdc14 Cell Cycle 7:3262–3272 10.4161/cc.7.20.6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratsima H, Ladouceur AM, Pascariu M, Sauve V, Salloum Z, Maddox PS, D’Amours D (2011) Independent modulation of the kinase and polo-box activities of Cdc5 protein unravels unique roles in the maintenance of genome stability Proc Natl Acad Sci U S A 108:E914–923 10.1073/pnas.1106448108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratsima H, Serrano D, Pascariu M, D’Amours D (2016) Centrosome-Dependent Bypass of the DNA Damage Checkpoint by the Polo Kinase Cdc5 Cell Rep 14:1422–1434 10.1016/j.celrep.2016.01.014 [DOI] [PubMed] [Google Scholar]
- Rawal CC, Riccardo S, Pesenti C, Ferrari M, Marini F, Pellicioli A (2016) Reduced kinase activity of polo kinase Cdc5 affects chromosome stability and DNA damage response in S. cerevisiae Cell Cycle 15:2906–2919 10.1080/15384101.2016.1222338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusswig KU, Zimmermann F, Galanti L, Pfander B (2016) Robust Replication Control Is Generated by Temporal Gaps between Licensing and Firing Phases and Depends on Degradation of Firing Factor Sld2 Cell Rep 17:556–569 10.1016/j.celrep.2016.09.013 [DOI] [PubMed] [Google Scholar]
- Roccuzzo M, Visintin C, Tili F, Visintin R (2015) FEAR-mediated activation of Cdc14 is the limiting step for spindle elongation and anaphase progression Nat Cell Biol 17:251– 261 10.1038/ncb3105 [DOI] [PubMed] [Google Scholar]
- Rock JM, Amon A (2011) Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit Genes Dev 25:1943–1954 10.1101/gad.17257711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez JA, Moyano Y, Jativa S, Queralt E (2016) Mitotic Exit Function of Polo-like Kinase Cdc5 Is Dependent on Sequential Activation by Cdk1 Cell Rep 15:2050– 2062 10.1016/j.celrep.2016.04.079 [DOI] [PubMed] [Google Scholar]
- Rossio V et al. (2010) The RSC chromatin-remodeling complex influences mitotic exit and adaptation to the spindle assembly checkpoint by controlling the Cdc14 phosphatase J Cell Biol 191:981–997 10.1083/jcb.201007025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P, Moreno S, Reed SI (1989) Conservation of mitotic controls in fission and budding yeasts Cell 57:295–303 [DOI] [PubMed] [Google Scholar]
- Sakchaisri K et al. (2004) Coupling morphogenesis to mitotic entry Proc Natl Acad Sci U S A 101:4124–4129 10.1073/pnas.0400641101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA (2000) Cdc7p-Dbf4p becomes famous in the cell cycle J Cell Sci 113 ( Pt 12):2111–2117 [DOI] [PubMed] [Google Scholar]
- Seki A, Coppinger JA, Jang CY, Yates JR, Fang G (2008) Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry Science 320:1655– 1658 10.1126/science.1157425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Toh EA (1994) The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase Mol Cell Biol 14:7476–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K (1998) The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae EMBO J 17:1336–1349 10.1093/emboj/17.5.1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W et al. (2002) Cdc5 influences phosphorylation of Net1 and disassembly of the RENT complex BMC Mol Biol 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W et al. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex Cell 97:233–244 [DOI] [PubMed] [Google Scholar]
- Sia RA, Bardes ES, Lew DJ (1998) Control of Swe1p degradation by the morphogenesis checkpoint EMBO J 17:6678–6688 10.1093/emboj/17.22.6678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia RA, Herald HA, Lew DJ (1996) Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast Mol Biol Cell 7:1657–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead JL et al. (2007) A coupled chemical-genetic and bioinformatic approach to Polo-like kinase pathway exploration Chem Biol 14:1261–1272 10.1016/j.chembiol.2007.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Grenfell TZ, Garfield S, Erikson RL, Lee KS (2000) Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures Mol Cell Biol 20:286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J et al. (2009) Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity Mol Cell 34:416–426 10.1016/j.molcel.2009.04.013 [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Amon A (2004) Closing mitosis: the functions of the Cdc14 phosphatase and its regulation Annu Rev Genet 38:203–232 10.1146/annurev.genet.38.072902.093051 [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A (2002) Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase Cell 108:207–220 [DOI] [PubMed] [Google Scholar]
- Strebhardt K, Ullrich A (2006) Targeting polo-like kinase 1 for cancer therapy Nat Rev Cancer 6:321–330 10.1038/nrc1841 [DOI] [PubMed] [Google Scholar]
- Sunkel CE, Glover DM (1988) polo, a mitotic mutant of Drosophila displaying abnormal spindle poles J Cell Sci 89 ( Pt 1):25–38 [DOI] [PubMed] [Google Scholar]
- Takaki T, Trenz K, Costanzo V, Petronczki M (2008) Polo-like kinase 1 reaches beyond mitosis--cytokinesis, DNA damage response, and development Curr Opin Cell Biol 20:650–660 10.1016/j.ceb.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Toczyski DP, Galgoczy DJ, Hartwell LH (1997) CDC5 and CKII control adaptation to the yeast DNA damage checkpoint Cell 90:1097–1106 [DOI] [PubMed] [Google Scholar]
- Tolliday N, VerPlank L, Li R (2002) Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis Curr Biol 12:1864–1870 [DOI] [PubMed] [Google Scholar]
- Tomson BN, Rahal R, Reiser V, Monje-Casas F, Mekhail K, Moazed D, Amon A (2009) Regulation of Spo12 phosphorylation and its essential role in the FEAR network Curr Biol 19:449–460 10.1016/j.cub.2009.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K (1999) Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1 Nature 400:37–42 10.1038/21831 [DOI] [PubMed] [Google Scholar]
- Valerio-Santiago M, de Los Santos-Velazquez AI, Monje-Casas F (2013) Inhibition of the mitotic exit network in response to damaged telomeres PLoS Genet 9:e1003859 10.1371/journal.pgen.1003859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidanes GM, Sweeney FD, Galicia S, Cheung S, Doyle JP, Durocher D, Toczyski DP (2010) CDC5 inhibits the hyperphosphorylation of the checkpoint kinase Rad53, leading to checkpoint adaptation PLoS Biol 8:e1000286 10.1371/journal.pbio.1000286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Amon A (2001) Regulation of the mitotic exit protein kinases Cdc15 and Dbf2 Mol Biol Cell 12:2961–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus Nature 398:818–823 10.1038/19775 [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A (1997) CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis Science 278:460–463 [DOI] [PubMed] [Google Scholar]
- Walters AD et al. (2014) The yeast polo kinase Cdc5 regulates the shape of the mitotic nucleus Curr Biol 24:2861–2867 10.1016/j.cub.2014.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S et al. (2012) alpha-Synuclein disrupts stress signaling by inhibiting polo-like kinase Cdc5/Plk2 Proc Natl Acad Sci U S A 109:16119–16124 10.1073/pnas.1206286109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jin F, Higgins R, McKnight K (2014) The current view for the silencing of the spindle assembly checkpoint Cell Cycle 13:1694–1701 10.4161/cc.29027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH (1994) Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair Genes Dev 8:652–665 [DOI] [PubMed] [Google Scholar]
- Wloka C, Bi E (2012) Mechanisms of cytokinesis in budding yeast Cytoskeleton (Hoboken) 69:710–726 10.1002/cm.21046 [DOI] [PubMed] [Google Scholar]
- Wurzenberger C, Gerlich DW (2011) Phosphatases: providing safe passage through mitotic exit Nat Rev Mol Cell Biol 12:469–482 10.1038/nrm3149 [DOI] [PubMed] [Google Scholar]
- Yaakov G, Thorn K, Morgan DO (2012) Separase biosensor reveals that cohesin cleavage timing depends on phosphatase PP2A(Cdc55) regulation Dev Cell 23:124–136 10.1016/j.devcel.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Kono K, Lowery DM, Bartolini S, Yaffe MB, Ohya Y, Pellman D (2006) Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis Science 313:108–111 10.1126/science.1126747 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Toh-e A (2002) Budding yeast Cdc5 phosphorylates Net1 and assists Cdc14 release from the nucleolus Biochem Biophys Res Commun 294:687–691 10.1016/S0006-291X(02)00544-2 [DOI] [PubMed] [Google Scholar]
- Zhang T, Nirantar S, Lim HH, Sinha I, Surana U (2009) DNA damage checkpoint maintains CDH1 in an active state to inhibit anaphase progression Dev Cell 17:541–551 10.1016/j.devcel.2009.09.006 [DOI] [PubMed] [Google Scholar]


