Abstract
Chemotaxis-mediated motility enables Campylobacter jejuni to navigate through complex environmental gradients and colonize diverse niches. C. jejuni is known to possess several methyl accepting chemotaxis proteins (MCPs), also called transducer-like proteins (Tlps). While the role of some of the Tlps in chemotaxis has been identified, their regulation and role in virulence is still not very clear. Here, we investigated the contribution of Tlp2 to C. jejuni chemotaxis, stress survival and colonization of the chicken gastrointestinal tract. The Δtlp2 deletion mutant showed decreased chemotaxis toward aspartate, pyruvate, inorganic phosphate (Pi), and iron (FeSO4). Transcriptional analysis of tlp2 with a promoter fusion reporter assay revealed that the tlp2 promoter (Ptlp2) was induced by Pi and iron, both in the ferrous (Fe2+) and ferric form (Fe3+). RT-PCR analysis using overlapping primers indicated that the phoX gene, located immediately downstream of tlp2, is co-transcribed with tlp2. A transcription start site was identified at 53 bp upstream of the tlp2 start codon. The Δtlp2 mutant showed decreased colonization of the chicken gastrointestinal tract. Collectively, our findings revealed that the tlp2 plays a role in C. jejuni pathogenesis and colonization in the chicken host and its expression is regulated by iron.
Keywords: transducer like protein, chemotaxis, iron, regulation, promoter
Introduction
Foodborne gastrointestinal illness caused by a gram negative bacterium, Campylobacter jejuni, has seen a surge in incidence in the recent years (CDC, 2013). In the United States, Food and Drug Administration (FDA) has placed Campylobacter species in the list of “qualifying pathogens” capable of posing a serious public health risk (Food and Drug Administration and HHS, 2014). The prevalence and transmission of Campylobacter can be attributed to its widespread colonization in the gastrointestinal tract of farm animals, especially chickens (Hermans et al., 2012). It is well established that C. jejuni employs motility and chemotaxis to colonize the avian and mammalian gastrointestinal tract (Yao et al., 1994; Hendrixson and DiRita, 2004; Young et al., 2007; Hermans et al., 2011; Chandrashekhar et al., 2015, 2017). Directional motility in C. jejuni is mediated by the chemotaxis system, composed of chemoreceptors and other core signal transduction proteins (Lertsethtakarn et al., 2011).
Transducer like proteins (Tlps) are the key components involved in sensing environmental signals through chemotaxis or energy taxis in C. jejuni. (Marchant et al., 2002; Vegge et al., 2009; Korolik, 2010; Tareen et al., 2010; Reuter and van Vliet, 2013; Rahman et al., 2014). Amino acids (aspartate, glutamate and serine), organic acid salts (succinate, isocitrate, and formate), bile and mucin are chemoattractants for C. jejuni (Hugdahl et al., 1988; Hartley-Tassell et al., 2010; Tareen et al., 2010). C. jejuni Tlps have been classified into three groups (A-C), based on sequence analysis and structural homology (Marchant et al., 2002; Chandrashekhar et al., 2017). The C. jejuni Tlp2 (CJJ81176_0180) is a group A transducer-like protein (Marchant et al., 2002) with transmembrane domains, a periplasmic ligand binding domain and a cytoplasmic signaling domain. BLAST analysis of the predicted amino acid sequence of Tlp2 shows greatest homology to C. jejuni Tlp3 and Tlp4 (60% identity). The cytoplasmic signaling domain is identical to Tlp3 but the periplasmic domain shows only 38% identity with Tlp3 (Rahman et al., 2014). An earlier study in C. jejuni NCTC11168 strain revealed that tlp2 deletion mutant exhibited no chemotaxis and invasion defects (Vegge et al., 2009). However, recent evidence indicates that tlp2 is one of the most abundantly expressed tlps in mice infected with C. jejuni NCTC 11168-O (Day et al., 2012), thus emphasizing the significance of understanding the role of Tlp2 in C. jejuni pathophysiology. This warranted us to further investigate the role of C. jejuni Tlp2 in chemotaxis, virulence, and host colonization.
Iron is an essential nutrient and a cofactor for proteins involved in cellular metabolism, enzyme catalysis, and sensing extracellular and intracellular signals (Lill, 2009). The bioavailability of iron in the host and environment (10−18–10−24M) being lower than the minimum requirement for bacterial growth (10−7M), makes iron a key player in the host-pathogen interaction (Braun and Hantke, 2003). Chemotaxis toward iron has been studied in Shewanella oneidensis and the magnetotactic bacteria Geobacter metallireducens (Childers et al., 2002; Bencharit and Ward, 2005). In these bacteria, the chemotactic response to iron is due to the fact that it serves as an insoluble electron acceptor (Childers et al., 2002; Bencharit and Ward, 2005; Harris et al., 2010). Knowledge about the role of iron as an electron acceptor in C. jejuni, chemotaxis toward iron and/or regulation of Tlp genes by iron in C. jejuni is still scarce. However, a study in Helicobacter pylori indicated that tlpB, chemoreceptor for sensing bicarbonate and arginine, is induced by iron through a fur-independent mechanism (Ernst et al., 2005). Interestingly, a recent study in C. jejuni has identified that tlp genes (Cj0262c and Cj1110c) are regulated by iron and/or Ferric uptake regulator (Fur) protein (Butcher et al., 2012). The study also revealed that cj0145 (phoX), a gene located immediately downstream of tlp2, is induced in the presence of iron although the specific mechanism of regulation is still unexplored (Butcher et al., 2012).
Here we investigated the role tlp2 in C. jejuni chemotaxis, in-vitro virulence and colonization of the chicken gastrointestinal tract. We provide evidence that iron regulates chemotaxis in C. jejuni and tlp2 contributes to in-vivo colonization of the chicken gastrointestinal tract. The findings of this study not only highlight the significance of tlp2 in C. jejuni pathogenesis but also elaborate on the complex mechanism by which iron regulates the chemotaxis in C. jejuni through Tlp2.
Materials and Methods
Bacterial Strains, Media and Growth Conditions
Bacterial strains and plasmids used in this study are described in Table 1. C. jejuni used in this study are derivatives of strain 81–176 (WT) (Korlath et al., 1985) and NCTC 11168. C. jejuni strains were grown on Mueller-Hinton media (MH; Oxoid, Hampshire, United Kingdom) under microaerophilic conditions [(85% N2 (v/v), 10% CO2 (v/v) and 5% O2 (v/v)] in a DG250 Microaerophilic Workstation (Microbiology International, Frederick, Maryland, United States) at 42°C. E. coli DH5α was used for plasmid propagation and cloning purposes and was routinely cultured on Luria-Bertani (LB) medium at 37°C overnight. Growth media was supplemented with appropriate antibiotics; chloramphenicol (10 μg/ml for Campylobacter; 20 μg/ml for E. coli) and kanamycin (30 μg/ml for Campylobacter; 50 μg/ml for E. coli) as required.
Table 1.
Bacterial strains and plasmids used in this study.
| Strains | Relevant description | Source/Reference |
|---|---|---|
| C. jejuni 81–176 WT | Wild type strain of C. jejuni | Dr. Qijing Zhang |
| Δtlp2 | C. jejuni 81–176 derivative with deletion in tlp2 gene; tlp2::kan | This study |
| Δtlp2-cm | C. jejuni 81–176 derivative with deletion in tlp2 gene; tlp2::cm | This study |
| tlp2 comp | C. jejuni 81–176 tlp2 mutant complemented with wild type copy of tlp2 on pRY112 | This study |
| C. jejuni NCTC11168Δfur | C. jejuni NCTC11168 derivative with deletion in fur gene; fur::tet | Dr. Jun Lin |
| WT Ptlp2-pMW10 | C. jejuni 81–176 WT reporter strain carrying Ptlp2-pMW10 | This study |
| Δtlp2 Ptlp2-pMW10 | C. jejuni 81–176 Δtlp2 strain carrying Ptlp2-pMW10 | This study |
| Δfur Ptlp2-pMW10 | C. jejuni 81–176 Δfur strain carrying Ptlp2-pMW10 | This study |
| WT Pint-pMW10 | C. jejuni 81–176 WT reporter strain carrying Ptlp2-phoXint-pMW10 | This study |
| E. coli DH5α | E. coli strain used for cloning | Invitrogen |
| Plasmids | ||
| pZero-1 | Cloning vector for making suicide vector; Zeo | Invitrogen |
| pUC4K | Source plasmid for kanamycin resistance gene; Kan | Amersham |
| pUC4C | Source plasmid for chloramphenicol resistance gene; Cm | This study |
| pMW10 | Promoter shuttle vector; pMW10, Kan | Wosten et al., 1998 |
| pRY112 | E.coli-Campylobacter shuttle vector for complementation; Cm | Yao et al., 1993 |
| pRK2013 | Helper plasmid for complementation; Kan | Grabowska et al., 2011 |
| pZero1-tlp2 | pZero-1 containing the upstream and downstream sequences of tlp2; Zeo | This study |
| pZero1-Δtlp2-kan | pZero1-tlp2 with tlp2 gene replaced by the pUC4K kan gene through inverse PCR; Zeo, Kan | This study |
| pZero1-Δtlp2-cm | pZero1-tlp2-kan where kanwas replaced by the chloramphenicol gene; Zeo, Cm | This study |
| Ptlp2-pMW10 | pMW10 carrying the tlp2 promoter; Kan | This study |
| Pint-pMW10 | pMW10 carrying the intergenic region between tlp2 and phoX; Kan | This study |
Cm, chloramphenicol resistance; Kan, kanamycin resistance; Zeo, Zeocin resistance.
Generation and Complementation of tlp2 Mutant
Recombinant DNA techniques were performed as per standard procedures (Sambrook et al., 1989). C. jejuni tlp2 mutant was created by double crossover allelic exchange method as previously described (Rajashekara et al., 2009). Oligonucleotides used in the present study were synthesized from Integrated DNA Technologies (Skokie, IL, United States) and are listed in Table 2. Briefly, the gene of interest (tlp2) plus ∼ 1 kb flanking DNA was amplified by PCR from C. jejuni strain 81–176 genome. The purified PCR products were ligated into zeocin-resistant pZErO-1 (zero background cloning vector) (Invitrogen, Carlsbad, CA, United States), and the ligation product was transformed into Library Efficiency DH5α E. coli competent cells (Invitrogen) to generate the plasmid pZErO1-tlp2. The whole plasmid except the target gene was amplified by inverse PCR. Purified inverse PCR products were ligated either to a kanamycin resistant cassette (from pUC4K) or a chloramphenicol resistance cassette (from pUC4C), and the resulting suicide vector was electroporated into C. jejuni. Transformants were selected on MH agar supplemented with chloramphenicol or kanamycin. Individual clones were confirmed for deletion of the target gene by PCR. The tlp2 mutant with kanamycin resistance was used in all the assays; except for reporter studies, in which case tlp2 mutant with chloramphenicol resistance was used as reporter plasmid carries kanamycin resistance.
Table 2.
Oligonucleotide primers used in this study.
| Name | Sequence |
|---|---|
| Primers for gene deletion | |
| tlp 2F | ATATATGGTACCTTGCTACTAGTAT TTTGTTC |
| tlp 2R | AATTAACTCGAGCATAACCTTGT GGTACTATA |
| tlp 2F inv | ATATATGGATCCGAGAACA TGGTAGAGGCTTT |
| tlp 2R inv | ATATATGGATCCCCAGCATC TCTAAAATTCTT |
| Complementation primers | |
| tlp 2 comp F | AATGAAGTCGACAAATTATA ACGATATTAAGC |
| tlp 2 comp R | AATTAAGGTACCAAAACCTTT TCTTCTTAACA |
| Primers for RT-PCR and (q)RT-PCR | |
| Intergenic region P-180/181 (P1) F | ATAGCGTAGCTCAAATTGAT |
| Intergenic region P-180/181 (P1) R | AAGCATAGCAGCACTTAAAT |
| tlp2 control (P2) F | TGCAAATCTTGCTAAAACTA |
| tlp2 control (P2) R | GTCCAAATTCATCATTGCTT |
| phoX control (P3) F | GCTATGGATTTAACAAAACT |
| phoX control (P3) R | GTTAAACTGTCCTACATACA |
| phosR F | GCAAACATAATCATCACAACCAC |
| phosR R | GAGAGCAAGGATACAAAGAAGC |
| pstS F | CCTTATACAAACTGGAATCAAATC |
| pstS R | GACACATCACTCATTACAAGC |
| pstC F | CGCTTATGCTTTAGGTATGAC |
| pstC R | GCTGCCATCACCACTATC |
| Primers for promoter fusion reporter studies and primer extension assay | |
| tlp2_PF_F | ACA TTG ACA TCC CGG GTA TTT GCA GC |
| tlp2_PF_R | AAT CAG TGA GAT CTT CAA TTT TAC GC |
| CJJ81-180_PE_temp_F | GGG GGC AAA ATA ACA TTG ACA TCT AGA G |
| CJJ81-180_PE_temp_R | GCA TCT TGA CTA TCT AAC TGT TCT ATA GG |
| CJ81-180_PE_R1 | ACC TAA AAT TAT CAA ACA CAC TAC TGC G |
| CJJ81-180_PE_R2 | TAA TTT ATT TCA GCA TTC ACA ACT TCA TG |
| CJJ81-181_PE_temp_F | AAA CTG CAG GTA TCA CTC AAA TCA ATG |
| CJJ81-181_PE_temp_R | ACC TAG CAA ATC CTT ATC CTT AAG C |
| CJJ81-181_PE_R1 | TTG CAA AAA AAG CCA CCA TAG AAC C |
| CJJ81-181_PE_R2 | TTA AAA CCT TTG CTT CAT AAC CTT GTG G |
The complemented strain was created by amplifying coding regions of tlp2 along with its potential promoter region by PCR using primers indicated in Table 2. The resulting fragment was cloned into SalI-KpnI digested pRY112 (Yao et al., 1993) and the complementation plasmid was introduced into the Δtlp2 deletion mutant by biparental conjugation as described (Miller et al., 2000). Transconjugants were selected on MH agar supplemented with kanamycin and chloramphenicol and the resulting complementation strain was designated tlp2 comp as listed in Table 1.
Chemotaxis Assay
To quantify chemotaxis, we adapted a modified capillary chemotaxis assay that quantitatively measures bacterial tactic responses (Mazumder et al., 1999; Cerda et al., 2003). The assay was previously used for quantifying chemotaxis in subsurface microaerophilic bacteria including Campylobacter (Mazumder et al., 1999; Chandrashekhar et al., 2015) and other Epsilonproteobacteria, such as H. pylori (Cerda et al., 2003, 2011). Briefly, C. jejuni wild type (WT), Δtlp2 mutant and the complemented strains were grown microaerobically at 42°C for 18 h on MH agar and resuspended in chemotaxis buffer (Phosphate Buffered Saline, PBS or Normal Saline, pH 7.4) and OD600 was adjusted to 0.5. A 100 μl volume of a solution of the compounds [All compounds at 0.1M except Pi (Inorganic Ventures, Christiansburg, VA, United States) at 1 mM and FeSO4 (Sigma) at 0.1 mM] to be tested for chemotaxis response (buffer alone served as control) was aspirated through a 22 G stainless-steel needle (0.254 mm diameter × 20 mm long) into a 1 ml tuberculin syringe. The 0.1 M concentration of the compounds was selected based on previous studies and a series of preliminary experiments that showed that 100 mM resulted in the strongest chemotaxis response (Vegge et al., 2009; Tareen et al., 2010). A 100 μl of the OD600 adjusted bacterial suspension was drawn into a 200 μl disposable pipette tip and the needle-syringe system was fitted to the pipette tip in such a way that the needle was immersed into the bacterial suspension. The system was positioned horizontally and incubated at 42°C for 1 h. The needle-syringe system was then separated from the bacterial suspension containing pipette tip and contents of the syringe were 10-fold serially diluted in chemotaxis buffer, plated onto MH agar plates and incubated at 42oC under microaerophilic conditions to determine colony-forming units (CFUs). Relative Chemotaxis Ratio (RCR) toward a test compound was ascertained as a ratio between the numbers of bacteria entering the test needle-syringes to those in the control needle-syringes. A test compound was considered as an attractant if the RCR was ≥ 2 (Mazumder et al., 1999). Results were expressed as the mean of three independent assays. A mutant was considered deficient in chemotaxis toward a substrate if both the corresponding RCR value was significantly < 2 (P < 0.05) and the CFU of the mutants were significantly lower (P < 0.05) than those of the wildtype. A C. jejuni 81–176 cheY mutant which is incapable of directional movement (negative control) (Yao et al., 1997) and 0.1% porcine gastric mucin (positive control; Sigma) were also used to evaluate the integrity of the assay. To test the response to repellents, C. jejuni cultures were mixed with a repellent and the bacteria that entered the syringe, which in this instance contained only buffer, to escape the repellent were quantified as described above. To account for any methodological bias, capillary chemotaxis results were further verified by using the disk method (Vegge et al., 2009) for selected compounds.
Determination of the tlp2 Transcriptional Levels With Reporter Gene Assays
The partial coding region of tlp2 and the upstream region was amplified with tlp2-PF_F (SmaI) and tlp2-PF_R (BglII) primers and cloned into pMW10, a shuttle vector for E. coli and C. jejuni, containing a promoterless lacZ gene (Wosten et al., 1998). The plasmid was mobilized into C. jejuni WT and Δtlp2::Cm strains by electroporation. The Δtlp2::Cm strain was used for reporter studies since pMW10 carries kanamycin resistance. TheΔtlp2::Cm was generated as described above. β-Galactosidase assay was performed with C. jejuni strains harboring the tlp2 promoter (Ptlp2)-lacZ transcriptional fusion construct, as described previously (Wosten et al., 1998). To examine the effect of Pi and iron on tlp2 transcription, reporter strains were incubated in MOPS and MEMα (Life technologies, Invitrogen) supplemented with Pi and FeSO4 or FeCl3 (Iron), respectively.
Additionally, reporter fusions were also created for the intergenic region between tlp2 (Cjj81176_0180) and phoX (Cjj81176_181) to determine any potential promoter in the intergenic region. The intergenic region was amplified with specific primers listed in Table 2 and cloned into pMW10 using the BamHI-XbaI sites. Reporter gene assays were carried out as described above.
Reporter gene assays were also carried out in the C. jejuni 81–176 Δfur mutant. C. jejuni 81–176 Δfur mutant was created by natural transformation of WT containing the (Ptlp2)-lacZ transcriptional fusion construct with genomic DNA from C. jejuni NCTC11168 Δfur mutant as described previously (Jeon et al., 2008; Gangaiah et al., 2009). Briefly, 1 ml of C. jejuni WT reporter strain was resuspended to an OD600 of 0.5. Approximately, 5 μg of genomic DNA from C. jejuni NCTC11168 Δfur mutant was added and incubated for 4 h microaerobically. The bacteria were plated on MH plates supplemented with appropriate antibiotics and incubated microaerobically at 42oC for 48 h. The deletion of the fur gene in 81–176 was confirmed by PCR.
RNA Extraction and Reverse Transcriptase Overlapping PCR
Briefly, C. jejuni WT grown overnight in MH agar plate was scraped and resuspended to an OD600 of 0.05 in MEM-α or MH broth and grown up to mid log phase (6 h), respectively. Total RNA was extracted using RNeasy Mini Kit (Qiagen) and quantified using NanoDrop ND-2000c spectrophotometer (Wilmington, DE, United States). cDNA synthesized using SuperScript® III First-Strand Synthesis SuperMix (Invitrogen), was used as a template for PCR with a set of overlapping primers for the tlp2, phoX, and the 135 base pairs intergenic region between tlp2 and phoX. (Table 2).
Primer Extension Assay
Primer extension assay was performed as described previously (Kim et al., 2011). Briefly, C. jejuni WT strain was grown for 6 h (mid-log phase) with shaking in MH broth at 42oC and harvested by centrifugation at 10,000 × g for 5 min. Total RNA was purified with TRIzol (Invitrogen) according to the manufacturer’s instructions. Purified RNA was resuspended in sterile distilled RNase-free water, and the RNA concentration was determined by measuring the OD of the solution at 260 and 280 nm using NanoVue (GE Healthcare). A portion (10 pmol) of the PE_R primer was labeled with 32P at the 5′ end by 10 U of T4 polynucleotide kinase (Invitrogen) and 80 μCi of [γ-32P] dATP for 30 min at 37°C. The labeling mixture was heated at 70°C for 10 min and purified with MicroSpin G-25 columns (GE Healthcare). The γ-32P-end-labeled primer (0.5 pmol) was co-precipitated with 15 μg of total RNA by the addition of sodium acetate and absolute ethanol. The pellet was washed with 75% ethanol, dried at room temperature, and resuspended in 20 μl of 250 mM KCl, 2 mM Tris (pH 7.9), and 0.2 mM EDTA. The mixture was heated to 65°C and then was allowed to cool to room temperature for 1 h. After annealing, 50 μl of reaction solution containing 5 μg of actinomycin D, 700 μM deoxynucleoside triphosphates, 10 mM MgCl2, 5 mM DTT, 20 mM Tris (pH 7.6), 30 U of RNasin (Promega), and 150 U of Superscript® III reverse transcriptase (Invitrogen) was added. The mixture was incubated at 42°C for 70 min and treated with 100 U of RNase T1 (Invitrogen) at 37°C for 15 min. The sample was ethanol precipitated after addition of 1.4 μl of 5M NaCl with 2.5 volumes of absolute ethanol and then washed with 75% ethanol. Sample was resuspended with 6 μl of formamide dye and 4 μl of Tris-EDTA (pH 8.0) buffer and then denatured at 90°C for 3 min. The samples were resolved on 6% polyacrylamide-8M urea gels, and the reverse transcription signals were analyzed by using BAS 2500 (Fuji Film). Primers, CJJ81176_180_PE and CJJ81176_181_PE (Table 2) were used for sequencing the upstream regions of tlp2 and phoX, for transcription start site with a SequiTherm EXCELII DNA sequencing system (Epicenter).
Alkaline Phosphatase Assay
PhoX activity was determined as described previously (Drozd et al., 2011). Briefly, WT, Δtlp2, and the tlp2 comp strains were grown overnight on MH plates with appropriate antibiotics. The cultures were gently scraped, washed and resuspended in MEM and incubated at 42°C microaerobically with shaking for 2 h. Cultures were then centrifuged for 10 min at 7000 × g and supernatant was removed. Cells were gently washed with 50 mM MOPS buffer (pH 7.4) (Sigma) and incubated with shaking at 42oC for 2 h following which, OD600 readings were taken. Cells were pelleted and resuspended in PNPP buffer containing 2 mM p-nitrophenyl phosphate (PNPP; Sigma) and incubated at 37°C. OD measurements at 550 nm and 420 nm were taken, and the phosphatase activity was calculated as described previously (Wosten et al., 2006). The assay was performed a total of three times with duplicate samples in each assay. Additionally, effect of iron on PhoX activity was assessed by supplementation of FeSO4 at 40 μM concentration in MOPS buffer.
Nutrient Downshift Assay
The role of tlp2 in C. jejuni survival under nutrient downshift was assessed using MEM-α as described previously (Gangaiah et al., 2010). Briefly, mid-log-phase cultures of WT, Δtlp2, and the tlp2 comp strains were pelleted, washed twice and resuspended in MEM-α with OD600 adjusted to 0.05. The bacterial suspensions were incubated microaerobically at 42°C with shaking. Samples were taken over time, serially diluted (10-fold) in MEM-α media and plated on MH agar for determining CFU. The experiment was performed three times and the average for each time point was taken.
Quantitative Reverse Transcriptase PCR (qRT-PCR) Analysis of Phosphate Uptake Genes
The C. jejuni WT and Δtlp2 cultures were assessed for changes in expression of phosphate uptake genes (phosR, pstC, and pstS) (Wosten et al., 2006). Briefly, C. jejuni WT and Δtlp2 strains were grown to mid-log phase in MEM-α microaerobically, with shaking at 42oC. Total RNA was extracted using RNeasy Mini Kit (Qiagen) and cDNA was synthesized using SuperScript® III First-Strand Synthesis SuperMix (Invitrogen). RNA and cDNA concentrations and purity were determined using NanoDrop ND-2000c spectrophotometer (Wilmington, DE, United States). Quantitative RT-PCR was performed with a SensiMixPlus SYBR RT-PCR kit (Quantace, Norwood, MA, United States) in a Mastercycler ep realplex2 thermal cycler (Eppendorf, Westbury, NY, United States). Gene specific primers (Table 2) used in this analysis have been described previously (Drozd et al., 2014). The relative levels of expression of target genes were normalized to 16S rRNA gene expression of the same strain. The relative fold changes in gene expression was calculated using the comparative threshold cycle (CT) method to yield fold-difference in transcript level compared to WT (Livak and Schmittgen, 2001). The qRT-PCR was performed a total of three times with duplicate samples in each assay.
Invasion and Intracellular Survival Assays
Invasion and intracellular survival of C. jejuni WT and Δtlp2 mutant in INT 407 cell line (human embryonic intestine cells, ATCC CCL 6) was assessed as described previously (Kassem et al., 2012). Briefly, mid-log phase grown bacterial cells were collected by centrifugation (5,000 × g, 10 min), washed twice with MEM containing 1% (v/v) FBS and resuspended in MEM. INT 407 cells (1.4 × 105 per well) in MEM with 10% (v/v) fetal bovine serum (FBS) were seeded in 24-well tissue culture plate and incubated for 18 h at 37°C with 5% CO2. INT 407 cells were infected with multiplicity of infection (MOI) 100 for invasion and intracellular survival assays and incubated for 3 h at 37°C. Following 3 h of incubation with bacteria; cells were treated with gentamicin (150 μg/ml) and incubated for additional 2 h. After 2 h of incubation, the infected cells were rinsed three times with MEM, lysed with 0.1% (v/v) Triton-X 100, serially diluted in MEM and plated on MH agar. The percent invasion was calculated as follows: (no. of CFU recovered after lysis of INT 407 cells/CFU added to each well) × 100.
To assess survival of C. jejuni WT and Δtlp2 mutant in INT 407 cell line, following 2 h of gentamicin treatment, the infected cells were washed with MEM three times and covered with MEM containing gentamicin (10 μg/ml) and incubated for 24 h at 37°C. After 24 h of incubation, infected cells were washed with MEM, lysed and plated as described above. In parallel, we also cultured the supernatant of gentamicin treated monolayers to ensure the quality of the gentamicin protection assay.
Chicken Colonization Assay
Chicken colonization study was performed as described previously (Gangaiah et al., 2009). Briefly, 3 day-old specific pathogen free chickens (n = 6 for each group) were obtained from a local hatching facility (Food Animal Health Research Program, OARDC, Wooster, OH, United States). Campylobacter free chickens were inoculated orally with 104 CFU of the C. jejuni WT and Δtlp2 mutant strain in 200 μl of PBS (pH 7.4). Chickens were euthanized after 7 days post-inoculation and ceca, duodenum, jejunum, liver, spleen and bursa were collected aseptically, weighed, homogenized, serially diluted in PBS (pH 7.4) and plated on appropriate MH agar containing Campylobacter selective supplement with or without kanamycin to determine colony forming units (CFU). Plates were incubated at 42°C microaerobically and CFUs per gram of tissues were determined.
Statistical Analysis
Statistical significance of data generated in this study was determined using two tailed Student’s t-test. Results of the promoter fusion assay were statistically analyzed using one way Anova with Dunett’s multiple comparison posttests. Data from the chicken colonization experiment was analyzed using the Mann Whitney test. P ≤ 0.01 or 0.05 (α level) was considered statistically significant.
Results
The Δtlp2 Mutant Is Defective in Chemotaxis Toward Aspartate, Pyruvate, Pi and Iron
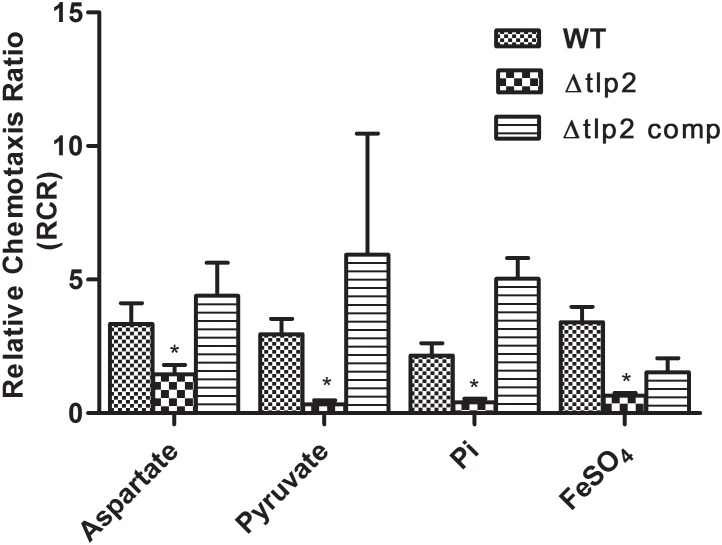
To assess the role of tlp2 in C. jejuni chemotaxis, a deletion mutant was constructed with the coding region of tlp2 being replaced with kanamycin resistance gene. Syringe capillary chemotaxis assays were performed to determine the chemotactic activity of C. jejuni WT, the Δtlp2 mutant and the complemented strains toward different substrates (Table 3). Substrates with RCR values > 2 and < 0.1 were considered as chemo attractants and repellants, respectively, for WT C. jejuni (Hugdahl et al., 1988; Cerda et al., 2003; Chandrashekhar et al., 2015). In addition, capillary assay showed strong chemotaxis of C. jejuni toward 0.1% porcine gastric mucin (RCR = 9.0), while a non-motile cheY mutant had an RCR below the detection limit (∼0) for some of the known attractants (Chandrashekhar et al., 2015). Compared to the WT, the Δtlp2 mutant was defective in chemotaxis toward aspartate (P = 0.0292) and pyruvate (P = 0.0010) with RCR values < 2 (Figure 1) (RCR values: aspartate: 3.81 for the WT and 1.45 for the Δtlp2 mutant; pyruvate: 2.96 for the WT and 0.33 for the Δtlp2 mutant) (Cerda et al., 2003). Even though the tlp2 mutant showed RCR values less than 2 for isocitrate, succinate and propionate; they were not statistically significant (Table 3). Interestingly, Δtlp2 mutant also showed a chemotaxis defect toward Pi and iron (FeSO4), compared to WT (Figure 1). We observed that FeSO4, but FeCl3.6H20 (ferric iron source) and (NH4)2SO4 (sulfate source) were not a chemoattractant for C. jejuni, based on the RCR indices for these compounds (FeCl3.6H20: RCR of 1.40 and (NH4)2SO4: RCR of 0.40) (Table 3). The chemotaxis defect was restored to WT levels in the complemented strain; however, chemotaxis toward iron was partially restored in the complemented strain (Figure 1). Chemotaxis results were also confirmed by assessing chemotaxis using the disk method for selected substrates such as aspartate and pyruvate (data not shown).
Table 3.
RCR valuesa for the WT and tlp2 mutant for all compounds tested.
| Chemicals tested | WT | Δtlp2 |
|---|---|---|
| Aspartate | 3.81 ± 0.32 | 1.45 ± 0.35 |
| L-glutamine | 4.19 ± 0.77 | 2.93 ± 0.38 |
| L-serine | 2.04 ± 0.46 | 1.99 ± 0.82 |
| Fumarate | 9.32 ± 1.45 | 2.52 ± 0.79 |
| Isocitrate | 2.51 ± 0.41 | 1.5 ± 0.65b |
| Formate | 4.41 ± 0.13 | 2.36 ± 1.05 |
| Succinate | 3.73 ± 1.2 | 1.2 ± 0.70b |
| Pyruvate | 2.96 ± 0.566 | 0.33 ± 0.15 |
| Propionate | 2.97 ± 1.51 | 1.35 ± 0.30b |
| Inorganic phosphate | 2.15 ± 0.46 | 0.4 ± 0.15 |
| Deoxycholic acid | <0.1 | <0.1 |
| Cholic acid | <0.1 | <0.1 |
| FeSO4 | 3.40 ± 0.58 | 0.66 ± 0.10 |
| FeCl3.6H20a | 1.4 ± 0.35 | NT |
| (NH4)2SO4a | 0.1 | NT |
The results show the means and standard errors of three independent experiments. An RCR value of 2 or above indicates chemotaxis toward the test chemical (Mazumder et al., 1999). aThe RCR value of the WT strain was <2.0, hence the compounds were not chemoattractants for C. jejuni 81–176 and were therefore not tested (NT) for chemotaxis with the mutant. bThe RCR of these compounds are lesser than 2 for the tlp2 mutant, however, not significant statistically P > 0.05.
FIGURE 1.
The Δtlp2 mutant was significantly defective in chemotaxis toward aspartate, pyruvate, inorganic phosphate and ferrous sulfate. Chemotaxis was determined using the capillary method (Chandrashekhar et al., 2015) where an RCR-value of 2 or above indicates chemotaxis toward the test chemical. This graph represents only those compounds (out of 15 compounds) for which a defect in chemotaxis was observed in the Δtlp2 mutant. The complete results for chemotaxis toward all 15 compounds tested are listed in Table 3. ∗P ≤ 0.05. The results show the means and standard errors of three independent experiments.
Iron Induces tlp2 Promoter (Ptlp2) Activity
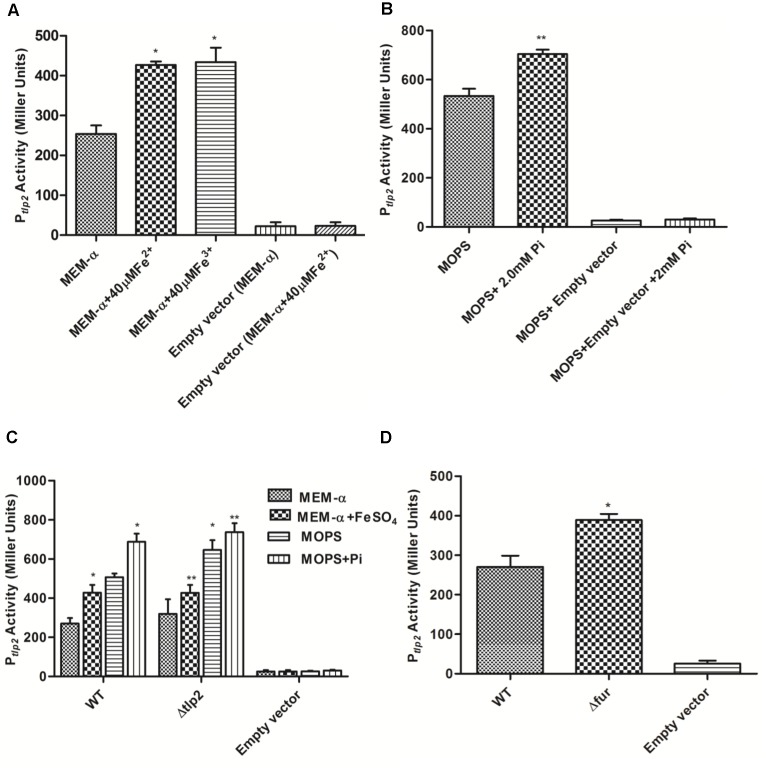
Decreased chemotaxis toward iron observed in the Δtlp2 mutant encouraged us to investigate the tlp2 expression under different growth conditions. The level of tlp2 transcription was quantified with β-galactosidase assays in presence of metal ions, such as Fe2+, Fe3+, Cu2+, Ca2+, Mg2+ and Zn2+ in MEM-α that does not contain these metals (van Vliet et al., 1998; Kim et al., 2011). Assay was performed in the presence of 20 μM CuCl2, 40 μM FeSO4, 40 μM FeCl3, 40 μM MnCl2, and 10 μM ZnCl2. Since MEM-α media already has Ca2+ (1.8 mM) and Mg2+ (0.8 mM), we did not supplement the media with these two metal ions. Iron in both ferrous (FeSO4) and ferric (FeCl3) forms induced tlp2 expression at 40 μM concentrations (Figure 2A), whereas other metals had no effect on the level of tlp2 transcription (Supplementary Figure S1A). For Fe2+, 40 μM was used based on the dose response assay (Supplementary Figure S1B) which showed best result at this concentration (Supplementary Figure S1B). Concentrations of iron as low as 5 μM FeSO4 also significantly induced tlp2 expression (Supplementary Figure S1B).
FIGURE 2.
β-galactosidase activity of Campylobacter jejuni WT carrying Ptlp2-lacZ transcriptional fusion construct. (A) β-galactosidase activity in the absence (uninduced) and presence of 40 μM FeSO4 or FeCl3(H2O)6 (induced) added to MEM-α. (B) β-galactosidase activity in the absence (uninduced) and presence of 2mM Pi (induced) added to MOPS. (C) β-galactosidase activity of the Ptlp2-lacZ fusion assays in the Δtlp2::Cm mutant in the presence or absence of 40 μM FeSO4 in MEM-α and in the presence or absence of 2mM Pi in MOPS. (D) β-galactosidase activity of the Ptlp2-lacZ fusion in the Δfur mutant in MEM-α. The cells were incubated for 8 h before carrying out the assay. The results show the means and standard deviations of three independent experiments. ∗P < 0.05 where each group is compared with the WT reporter strain that is not induced (MEM-α or MOPS) and ∗∗P < 0.05 where each group is compared with the WT that is induced (with FeSO4 or Pi).
Similarly, the activity of Ptlp2 was investigated in the presence of Pi due to the observed chemotaxis defect toward Pi (Table 3 and Figure 1). MOPS buffer was used as a low phosphate medium for the incubation of the C. jejuni reporter strains (Gangaiah et al., 2009). The concentration of Pi added to MOPS buffer ranged from 1 to 3 mM but Ptlp2 was most significantly induced in the presence of 2 and 3 mM of Pi (Figure 2B and Supplementary Figure S1C). Further, Ptlp2 activity in the tlp2 deletion mutant was also studied to assess the effect of the gene product on its promoter activity. Since pMW10 shuttle vector has a kanamycin resistant cassette, we created a Δtlp2 mutant with a chloramphenicol resistant cassette. We found that the tlp2 expression was also induced in the Δtlp2 mutant in the presence of Pi and FeSO4 similar to WT (Figure 2C). Even though the Ptlp2 activity in Δtlp2 mutant was higher than the WT both in the presence or absence of Pi and Fe, the difference was not statistically significant. These results suggest that tlp2 transcription is independent of Tlp2 protein levels in the cell.
Ferric uptake regulator protein (Fur) plays an important role in C. jejuni iron homeostasis (van Vliet et al., 1998). In addition, iron and Fur are shown to regulate tlp genes (Cj0262c; Tlp4 and Cj1110c; Tlp8) in C. jejuni (Butcher et al., 2012). We, therefore, investigated if tlp2 expression is regulated by Fur. Interestingly, the Ptlp2 activity was increased in a Δfur mutant of C. jejuni 81–176 in MEM-α (Figure 2D). These observations revealed a role for fur in the regulation of tlp2 expression. Since a fur mutation derepresses genes involved in iron acquisition in C. jejuni (Holmes et al., 2005), there will be over-accumulation of iron in the fur mutant. The increased levels of intracellular levels may increase the Ptlp2 activity in the fur mutant.
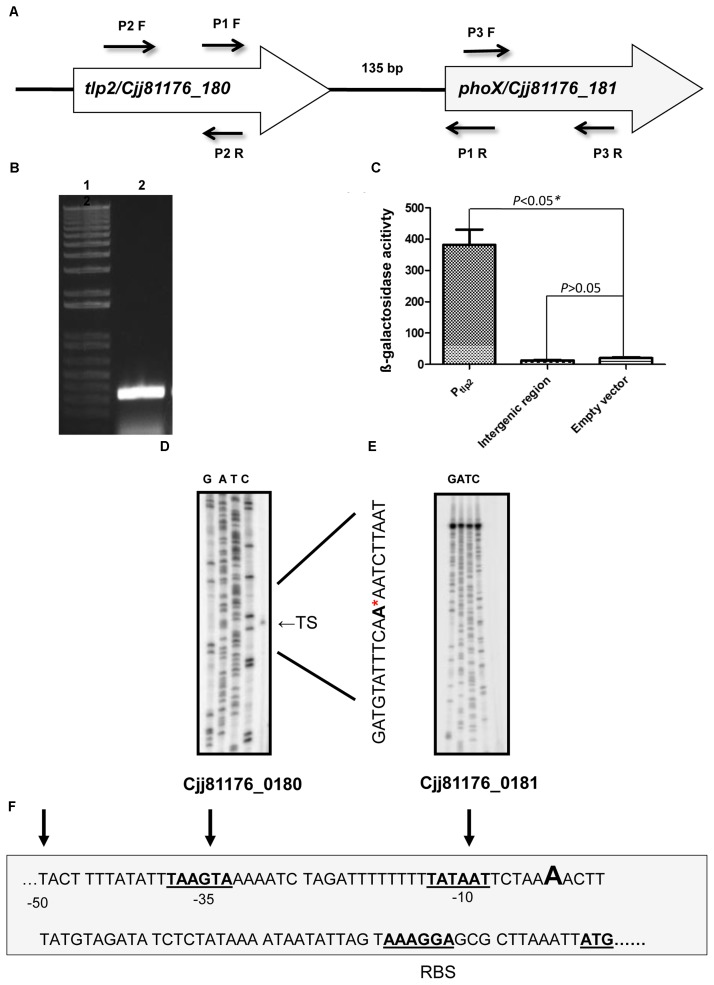
The tlp and phoX Genes Are Co-transcribed
The tlp2 gene (CJJ81176_180) is located upstream to phoX (CJJ81176_181) in the same orientation with an intergenic region of 135 base pairs (Figure 3A). Additionally, a previous study indicated that phoX gene (Cj0145), located immediately downstream of tlp2, is induced by iron and was also enriched in the CjFur ChIP-chip assay (Butcher et al., 2012). As the Δtlp2 mutant is defective in chemotaxis toward iron (Fe) and Pi, and the tlp2 transcription is modulated by iron and Pi, we hypothesized that these two genes may be co-transcribed. To test this, total RNA was extracted from WT grown in MEM-α and analyzed by RT-PCR using primers designed to amplify a flanking region of the two genes. The results indicated that tlp2 and phoX are co-transcribed (Figure 3B). In addition, an amplicon was also observed when WT was grown in nutrient rich MH broth (data not shown). These results implied that phoX is co-transcribed with tlp2 under the conditions tested in this study.
FIGURE 3.
(A) Genetic organization of tlp2. The tlp2 gene (cjj81176_180) is located upstream of the phoX gene (Cjj81176_181) which encodes the alkaline phosphatase (PhoX) enzyme. The tlp2 and phoX genes are separated by a 135 bp intergenic region. (B) Reverse Transcriptase overlapping PCR showing co-transcription of tlp2 and phoX. Intergenic region was amplified with primer pair P1 F (forward) and R (reverse) in WT strain grown in MEM-α. Primers for the tlp2 (P2 F and R) and phoX (P3 F and R) genes were included as control regions as well (data not shown). (C) The Pint-lacZ fusion showed no β-galactosidase activity compared to the Ptlp2–lacZ fusion in the WT strain. Determination of the transcriptional start site for tlp2 (D) and phoX (E) by a primer extension assay. Only one transcriptional start site is seen upstream to tlp2, designated TS (Transcriptional Start) indicated with an arrowhead on the right and by the ∗ in the sequence. No transcriptional start site was found in the region upstream to phoX (F): The −10 and −35 elements of the Ptlp2 are underlined and the ribosomal binding site is indicated as RBS.
Further, an intergenic region between tlp2 and phoX was fused to the promoterless lacZ gene to confirm that the promoter activity observed was specific to Ptlp2. The reporter strains did not show any promoter activity; the promoter activity in the β-galactosidase assay was similar to that of the negative control (empty plasmid) (Figure 3C). Similar findings were observed when media were supplemented with iron or Pi (data not shown), confirming that there was no promoter in the intergenic region between tlp2 and phoX under these tested conditions.
Furthermore, a primer extension analysis revealed a single transcription start site (TS) upstream to the tlp2 gene (Figures 3D,F). The TS is located 53 bp upstream of the tlp2 start codon with a ribosomal binding site located 12 bp upstream from the start codon. The −10 region was identified with the first T of the TATA box located 59 bp upstream of the start codon. Consistent with the results above, no transcription start site was observed in the 135 bp intergenic region between tlp2 and phoX (Figure 3E). This result indicates that tlp2 and phoX genes constitute an operon, and the transcription of phoX is dependent on the tlp2 promoter and they are co-transcribed.
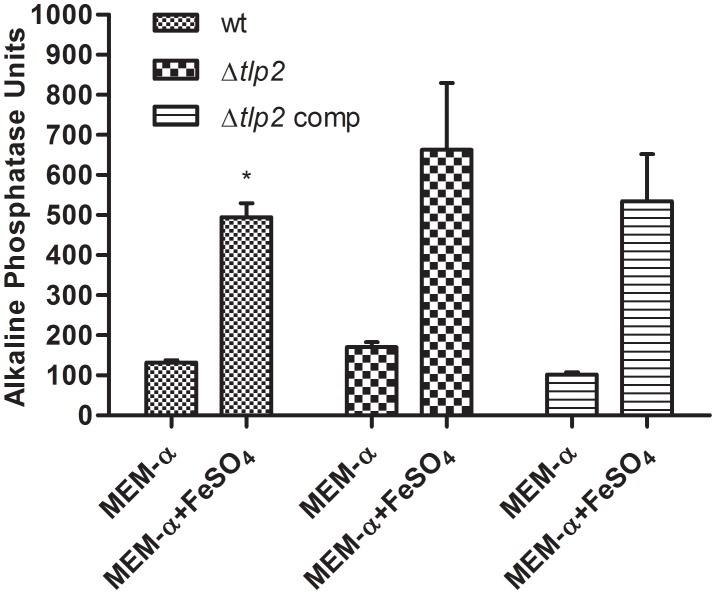
Alkaline Phosphatase Activity Is Increased in the Presence of Iron
A study investigating the regulatory potential of Fur of C. jejuni identified that phoX is activated by iron (Butcher et al., 2012). Therefore, the PhoX activity of C. jejuni WT was evaluated in MEM-α supplemented with 40 μM FeSO4. The PhoX activity of the WT strain increased approximately four-fold in the presence of iron (Figure 4). Similarly, a higher PhoX activity was also observed in the Δtlp2 mutant in MEM-α in the presence of iron similar to the WT (Figure 4); however, this increase was not significant (P ≤ 0.09). In the complemented strain, the PhoX activity was similar to the WT with or without iron (Figure 4). These results suggest that iron upregulates the PhoX activity in C. jejuni and potentially intersects the phosphate utilization pathway of C. jejuni.
FIGURE 4.
C. jejuni WT and Δtlp2 strains shows a higher PhoX activity in the presence of iron (40 μM FeSO4 ). The bacterial cultures grown in MEM-α in the presence or absence of 40 μM FeSO4. The data is a mean of 3 independent trials. ∗P ≤ 0.05.
Deletion of tlp2 Affected Nutrient Stress Survival
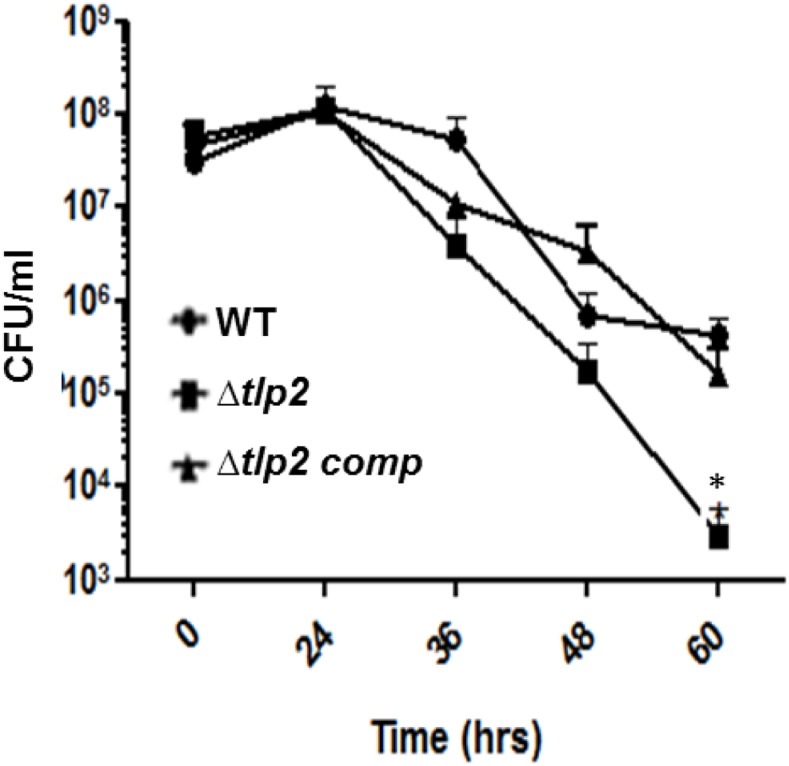
The effect of a tlp2 mutation on stress survival was monitored by comparing the growth of the Δtlp2 mutant strain to the WT C. jejuni in nutrient-limited conditions. The Δtlp2 mutant did not display any growth defect when grown in nutrient-rich MH broth (data not shown); however, the tlp2 mutant on transition from nutrient rich MH broth to nutrient deficient MEM (without glutamine) exhibited survival defects in the late stationary phase especially 36 h and onward. The survivability of the tlp2 mutant strain was decreased by one and more than two orders of magnitude at 36 and 60 h, respectively, as compared to the WT (P < 0.05) (Figure 5).
FIGURE 5.
Survival of Δtlp2 mutant under nutrient downshift. Survival under nutrient stress was assessed by resuspending MH agar grown bacterial cultures in MEM-α, and determining CFU at different time points. Each data point represents the mean ± SE of 3 independent experiments. ∗P ≤ 0.05.
Deletion of tlp2 Affected Intracellular Survival in Intestinal Epithelial Cells
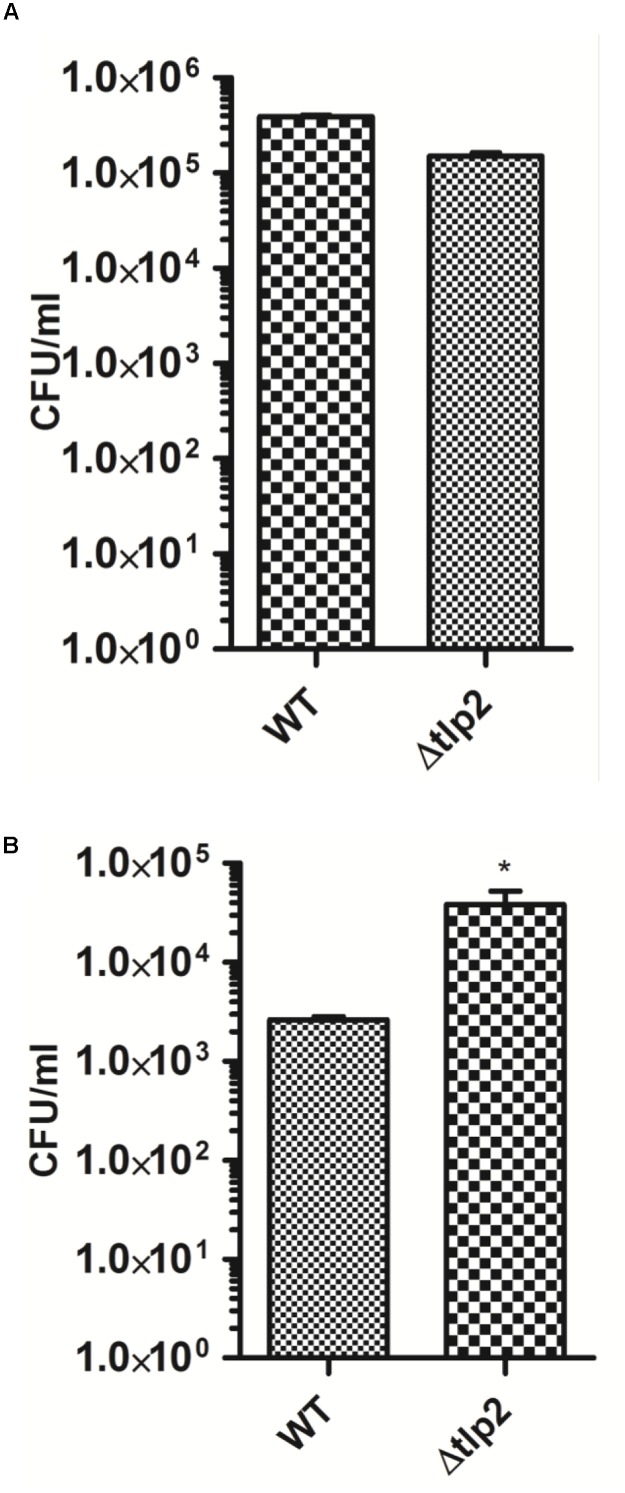
The consequence of tlp2 deletion on virulence-associated traits of C. jejuni was evaluated by the ability of Δtlp2 mutant to invade and survive within the human intestinal epithelial INT 407 cells (Candon et al., 2007). The Δtlp2 demonstrated similar invasion in INT 407 cells; however, the Δtlp2 mutant showed a higher intracellular survival, with almost 2 logs more bacteria recovered compared to the WT (Figures 6A,B).
FIGURE 6.
The Δtlp2 mutant exhibits no invasion defect (A) and increased intracellular survival (B) in INT 407 human intestinal cells. Bacteria were infected at an MOI of 100 for both invasion (2 h of gentamicin treatment) and intracellular survival assays (24 h of further incubation). Each data point represents the mean ± SE of 3 independent experiments. ∗P ≤ 0.05.
The Δtlp2 Mutant Is Defective in Colonization of the Chicken Gastrointestinal Tract
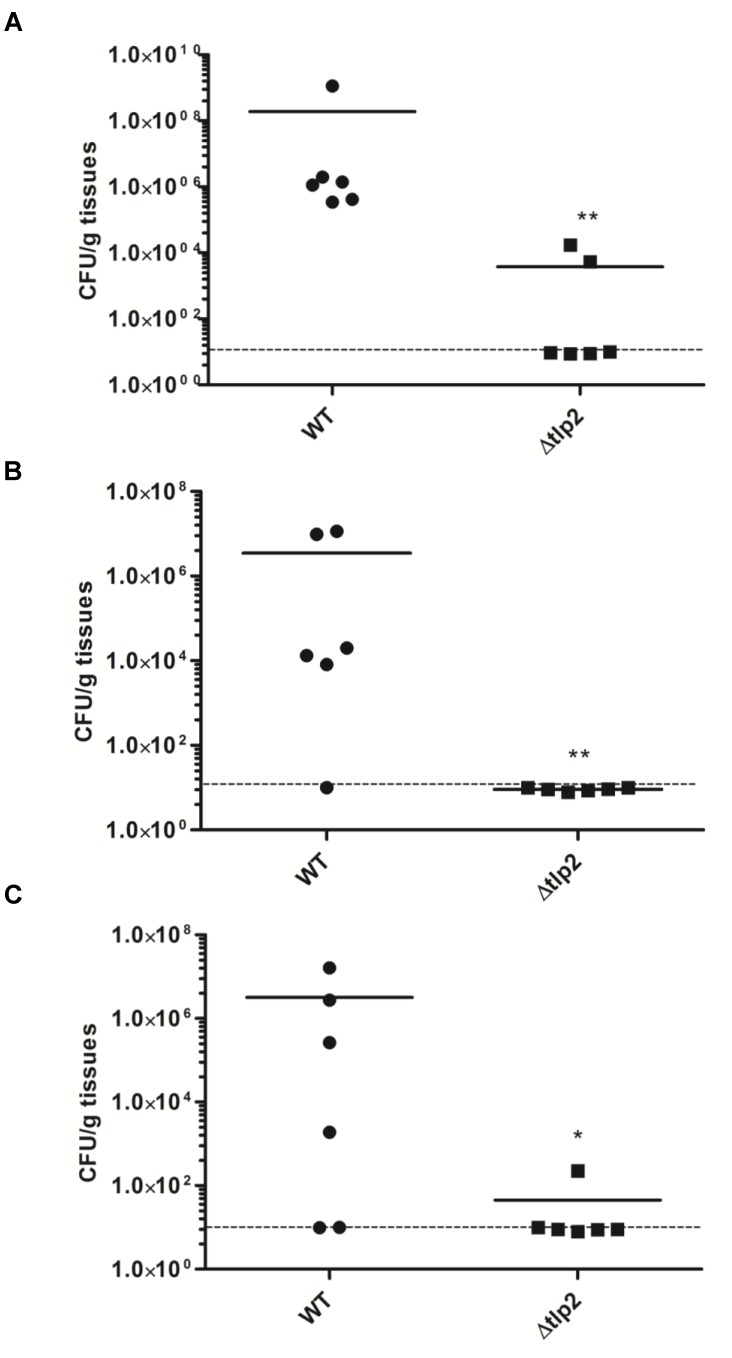
To investigate the role of Tlp2 in colonization of C. jejuni, we investigated the colonization of Δtlp2 mutant and WT in different segments of the chicken intestine. The Δtlp2 mutant and WT were inoculated into 3-day old chicks orally (104 CFU/chicken), and bacterial burden was analyzed after 7 days of infection. Colonization of the C. jejuni WT strain in the chicken gastrointestinal tract (cecum and duodenum/jejunum) ranged from 4 × 107 to 2 × 108 CFU per gram of tissue; while in the Δtlp2 mutant varied from 2 × 102 to 8 × 103 CFU per gram of tissue. The Δtlp2 mutant showed a 4–5 logs decrease in cecal colonization compared to the WT (Figure 7A). The Δtlp2 mutant was not detected in the duodenum and colonization of the jejunum was also reduced by almost 4 logs (Figures 7B,C). However, the liver, spleen, and bursa showed no colonization by C. jejuni WT and Δtlp2 mutant. These findings suggest that the tlp2 is essential for achieving optimal colonization in the proximal and distal segments of the gastrointestinal tract, including the cecum.
FIGURE 7.
Colonization of the Δtlp2 mutant in chicken gastrointestinal tract. The Δtlp2 mutant exhibited a significant defect in colonization of the (A) cecum (B) duodenum and (C) jejunum, compared to the WT. Each data point represents CFU/g of tissue. ∗P ≤ 0.05 and ∗∗P ≤ 0.01 Dotted line represents the detection limit.
Discussion
In this study, we characterized the role of tlp2 in chemotaxis, stress survival, and colonization of the chicken gut. Our results indicated that tlp2 is involved in chemotaxis toward aspartate, pyruvate, Pi, and iron. Promoter fusion assays revealed that iron, in the ferrous and ferric form induces the tlp2 promoter activity. Iron is essential for C. jejuni colonization in the host as it is one of the limiting nutrients sequestered away from the pathogen by the host and the bioavailability of iron in the intestine is not very well understood (Naikare et al., 2006).
Predicted domain structure of the Tlp2 in the SMART database (Schultz et al., 1998) revealed a single periplasmic Cache_1 (Ca2+ channels and chemotaxis receptors) domain (Anantharaman and Aravind, 2000) and a cytoplasmic MCP signaling domain. Cache domain is found in the extracellular or periplasmic portions of chemoreceptors from Gram-positive and Gram-negative bacteria, and is associated with sensing of small molecules (Anantharaman and Aravind, 2000). The cache domains of Pseudomonas aeruginosa and Vibrio cholerae have been associated with chemotaxis toward amino acids (Nishiyama et al., 2012). The Cache domain is responsible for interaction with multiple ligands and thereby chemotaxis (Tasneem et al., 2005). Δtlp2 mutant shows decreased chemotaxis toward aspartate, pyruvate, iron and Pi. Additionally, Tlp2 shows 38% identity with the periplasmic region of the multiple ligand binding Tlp3 (Ccml) of C. jejuni, possessing a single cache domain which can potentially bind to multiple ligands with varying affinity (Rahman et al., 2014).
Studies in S. oneidensis and G. metallireducens report chemotaxis toward iron in the ferrous form (Childers et al., 2002; Bencharit and Ward, 2005). Iron is a redox active metal, and chemotaxis to iron suggests bacterial movement through reduced metal gradients toward potential electron acceptors (oxidized ferric form). Our observations in the WT (C. jejuni 81–176) strain show that it is chemotactic toward ferrous iron. The chemotactic response of C. jejuni toward iron (Fe2+) can be explained as bacterial adaptation to the assimilatory requirement for iron, as it is an important constituent of iron sulfur proteins and other cellular processes (Bencharit and Ward, 2005). Comparably, the chemotactic response of H. pylori toward a metal ion (zinc) has been primarily attributed to the mechanism of nutrient acquisition by bacteria (Sanders et al., 2013).
A study in C. jejuni, employing CjFur ChIP-chip analysis, identified cj0145 (phoX) as a novel gene in the Fur regulon in C. jejuni, which is activated by iron (Butcher et al., 2012). Much in line with the study above, we found in our study that PhoX activity in the WT is significantly increased in the presence of iron. A recent study on the Pseudomonas fluorescens PhoX revealed that iron is a cofactor required for enzyme activity, additionally implying that the bioavailability of iron affects bacterial phosphate uptake (Yong et al., 2014). Although a similar mechanism for increased PhoX activity in C. jejuni in the presence of iron can be envisioned, a further biochemical investigation on C. jejuni PhoX is needed to identify the precise role of iron in its enzymatic activity.
The sensing, uptake and utilization of inorganic phosphate in prokaryotes enables their ability to withstand conditions of phosphate deprivation. The Pi sensing or taxis has been studied in bacterial pathogens such as Enterococcus cloacae and P. aeruginosa under conditions of phosphate starvation, with two chemotactic transducers identified for Pi taxis in P. aeruginosa. The Pho regulon and the phosphate uptake system regulate Pi taxis in both bacteria (Kusaka et al., 1997; Wu et al., 2000). C. jejuni being an enteric pathogen is subjected to its survival under low phosphate conditions in the chicken gastrointestinal tract. While the uptake and utilization of Pi in C. jejuni through the two-component PhosS/PhosR operon has been previously described (Wosten et al., 2006), nothing is known about Pi taxis in this microaerophile. In our study, C. jejuni WT is chemotactic toward Pi, whereas the Δtlp2 mutant displayed a decreased chemotaxis. The decreased cellular availability of Pi in the tlp2 mutant was accompanied by an upregulation of the phosR (response regulator of Pho regulon) and the genes for phosphate uptake (pstC and pstS) which is normally induced in response to Pi limitation (Supplementary Figure S2) (Wosten et al., 2006). Additionally, the tlp2 mutant’s decreased survival under nutrient mediated stress (Figure 5) can be attributed to the Pi limiting conditions created due to decreased Pi taxis. Earlier studies have indicated that survival under low-nutrient stress is regulated by PPK1 mediated synthesis of poly-P from Pi (Candon et al., 2007; Gangaiah et al., 2009).
PhoX hydrolyzes phospho-organic compounds to Pi, a preferred phosphate source and a building block for poly-P in C. jejuni (Candon et al., 2007; Drozd et al., 2011). PhoX in C. jejuni is activated by the PhosS-PhosR two component system, under phosphate limiting conditions (Wosten et al., 2006). However, what remains to be investigated is whether PhosR also regulates the tlp2 promoter activity in C. jejuni. The tlp2 gene is located upstream to phoX in C. jejuni, and our investigation of tlp2 transcriptional organization revealed that both genes are transcribed together from a single promoter (Ptlp2) located upstream to tlp2. These findings however, contradict a previous finding in C. jejuni 81116, where phoX was shown to be transcribed by a promoter located in the intergenic region of tlp2 and phoX, when C. jejuni was grown in a chemically defined medium (Wosten et al., 2006). However, we could not observe any promoter activities in the intergenic region using a primer extension assay under our experimental conditions (Figure 3). Further, both strains possess 135 bps intergenic region between tlp2 and phoX; however, showed 95.5% sequence similarity. Therefore, the disparity could be due to the different media and strains used in the two different studies.
The Δtlp2 mutant exhibited an increased intracellular survival in INT 407 cell monolayer than the WT strain. The group A Tlps 1, 4, 7 and 10 but not Tlp2, have been shown to play a role in C. jejuni invasion of human intestinal epithelial cells (Vegge et al., 2009; Hartley-Tassell et al., 2010; Tareen et al., 2010). C. jejuni is known to survive within epithelial cells and can be viable for up to 24 h (Watson and Galan, 2008). Studies have also indicated a role for iron acquisition in C. jejuni intracellular survival (Naikare et al., 2006). This therefore piqued our interest in identifying a role for a Tlp involved in chemotaxis toward iron, in C. jejuni survival within host cells. The results of our study showed that the deletion of tlp2 increased intracellular recovery of C. jejuni. This was in contrary to our belief that deletion of tlp2 would decrease the survival of C. jejuni in host cells, due to the decreased chemotaxis toward iron. It must however, be noted that intracellular C. jejuni undergo a metabolic reprogramming which affects their survival within epithelial cells (Svensson et al., 2009; Liu et al., 2012). The increased intracellular survival in the Δtlp2 mutant may indicate a dysregulation of cellular process which warrants further investigation.
The role of tlp2 in tissue specific colonization of the chicken gastrointestinal tract was investigated. Mutation in tlp2 resulted in a colonization defect in the cecum, with a more profound reduction seen in the duodenum and jejunum. Catabolism of amino acids such as aspartate and serine are essential for C. jejuni colonization of the avian gut (Guccione et al., 2008), as reflected by the tlp1 mutant (aspartate chemoreceptor), which was severely impaired in colonization of the chicken ceca (Hartley-Tassell et al., 2010). The tlp2 mutant demonstrated a decreased chemotaxis toward aspartate, which might explain the reduced colonization. The utilization of glutamine, glutathione and asparagine in C. jejuni 81–176 is associated to tissue-specific colonization of the murine intestine (Hofreuter et al., 2008). However, it is not known if the ability to metabolize these nutrients also supports tissue specific colonization in the chicken intestinal tract. Additionally, chemotaxis toward pyruvate and fumarate mediated by Tlp9 represents energy taxis in C. jejuni. Energy taxis is an essential driving force for C. jejuni for establishment during colonization of the host (Vegge et al., 2009). The chicken cecum represents an iron and phosphate limiting environment for C. jejuni and iron acquisition is known to be essential for C. jejuni colonization of the chicken (Naikare et al., 2006). It is not surprising to see that Δtlp2 mutant, defective in chemotaxis toward Pi and iron, is also defective in colonization of the chicken cecum, duodenum, and jejunum. These findings clearly indicate that tlp2 contributes to C jejuni interaction with host cells, which is an important determinant for C jejuni pathogenesis and colonization of the chicken gastrointestinal tract.
In summary, the present study identifies a role for tlp2 in C. jejuni chemotaxis, stress survival and colonization of the chicken gastrointestinal tract. Further, our findings indicate that iron regulates tlp2. The tlp2 mutant was also defective in chemotaxis to Pi and showed increased PhoX activity. This suggests a possible cross-talk between iron and phosphate regulatory pathways, which needs further investigation. In addition, the increased PhoX activity in the presence of iron seen in C. jejuni indicates that iron may reduce the bioavailability of phosphate. Our findings in this study suggest a basis for future biochemical characterization of PhoX in C. jejuni.
Ethics Statement
Animal experiments were conducted according to the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The animal studies were approved by the Institutional Animal Care and Use Committee (IACUC), the Ohio State University. Chickens were housed at the Food Animal Health Research Program Animal Care Facility, which is fully accredited by AAALAC and the animals were supervised by a senior veterinarian. Infectious agents were administered using manual restraint for less than one minute to minimize distress. Before necropsy, chickens were euthanized by carbon dioxide inhalation. This method is consistent with the recommendations of the panel on euthanasia of the American Veterinary Medical Association and by the Ohio State University Institutional Laboratory Animal Care and Use Committee.
Author Contributions
GR and KC designed the experiments. KC, SH, BJ, and SR performed the experiments and collected the data. KC, GR and VS analyzed the data. KC, GR, VS, and BJ wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer YK and handling Editor declared their shared affiliation.
Acknowledgments
We thank Dr. Juliette Hanson for assistance with chicken colonization studies. We acknowledge Dr. Jun Lin (The University of Tennessee) for providing the C. jejuni NCTC11168 fur mutant strain and Dr. John Gunn (Ohio State University) for critically reading the manuscript.
Footnotes
Funding. This work was supported by the funds from Ohio Agricultural Research and Development Center (OARDC); The Ohio State University. KC was supported by a fellowship from the Indian Council of Agricultural Research (ICAR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02674/full#supplementary-material
References
- Anantharaman V., Aravind L. (2000). Cache - a signaling domain common to animal Ca(2 + )-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 25 535–537. 10.1016/S0968-0004(00)01672-8 [DOI] [PubMed] [Google Scholar]
- Bencharit S., Ward M. J. (2005). Chemotactic responses to metals and anaerobic electron acceptors in Shewanella oneidensis MR-1. J. Bacteriol. 187 5049–5053. 10.1128/JB.187.14.5049-5053.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V., Hantke K. (2003). “Mechanisms of bacterial iron transport,” in Microbial Transport System, ed. Winkelmann G. (Weinheim: Wiley-VCH Verlag GmBH & Co; ), 289–311. [Google Scholar]
- Butcher J., Sarvan S., Brunzelle J. S., Couture J. F., Stintzi A. (2012). Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc. Natl. Acad. Sci. U.S.A. 109 10047–10052. 10.1073/pnas.1118321109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candon H. L., Allan B. J., Fraley C. D., Gaynor E. C. (2007). Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. J. Bacteriol. 189 8099–8108. 10.1128/JB.01037-1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. (2013). “Incidence and trends of infection with pathogens transmitted commonly through food — foodborne diseases active surveillance network, 10 u.s. sites, 1996–2012,” in Morbidity and Mortality Weekly Report, ed. Charlotte K. (Washington, DC: U.S. Government PrintingOffice; ). [PMC free article] [PubMed] [Google Scholar]
- Cerda O., Rivas A., Toledo H. (2003). Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol. Lett. 224 175–181. 10.1016/S0378-1097(03)00423-3 [DOI] [PubMed] [Google Scholar]
- Cerda O. A., Nunez-Villena F., Soto S. E., Ugalde J. M., Lopez-Solis R., Toledo H. (2011). tlpA gene expression is required for arginine and bicarbonate chemotaxis in helicobacter pylori. Biol. Res. 44 277–282. /S0716-97602011000300009 [PubMed] [Google Scholar]
- Chandrashekhar K., Gangaiah D., Pina-Mimbela R., Kassem I. I., Jeon B. H., Rajashekara G. (2015). Transducer like proteins of Campylobacter jejuni 81-176: role in chemotaxis and colonization of the chicken gastrointestinal tract. Front. Cell. Infect. Microbiol. 5:46. 10.3389/fcimb.2015.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekhar K., Kassem I. I., Rajashekara G. (2017). Campylobacter jejuni transducer like proteins: chemotaxis and beyond. Gut Microbes 8 323–334. 10.1080/19490976.2017.1279380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers S. E., Ciufo S., Lovley D. R. (2002). Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature 416 767–769. 10.1038/416767a. [DOI] [PubMed] [Google Scholar]
- Day C. J., Hartley-Tassell L. E., Shewell L. K., King R. M., Tram G., Day S. K., et al. (2012). Variation of chemosensory receptor content of Campylobacter jejuni strains and modulation of receptor gene expression under different in vivo and in vitro growth conditions. BMC Microbiol. 12:128. 10.1186/1471-2180-12-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozd M., Chandrashekhar K., Rajashekara G. (2014). polyphosphate-mediated modulation of Campylobacter jejuni biofilm growth and stability. Virulence 5 680–690. 10.4161/viru.34348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozd M., Gangaiah D., Liu Z., Rajashekara G. (2011). Contribution of TAT system translocated PhoX to Campylobacter jejuni phosphate metabolism and resilience to environmental stresses. PLoS One 6:e26336. 10.1371/journal.pone.0026336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst F. D., Bereswill S., Waidner B., Stoof J., Mader U., Kusters J. G., et al. (2005). Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151(Pt 2), 533–546. 10.1099/mic.0.27404-27400 [DOI] [PubMed] [Google Scholar]
- Food and Drug Administration and HHS. (2014). Establishing a list of qualifying pathogens under the food and drug administration safety and innovation act: final rule. Fed. Regist. 79 32464–32481. [PubMed] [Google Scholar]
- Gangaiah D., Kassem I. I, Liu Z., Rajashekara G. (2009). Importance of polyphosphate kinase 1 for Campylobacter jejuni viable-but-nonculturable cell formation, natural transformation, and antimicrobial resistance. Appl. Environ. Microbiol. 75 7838–7849. 10.1128/AEM.01603-1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaiah D., Liu Z., Arcos J., Kassem I. I, Sanad Y., Torrelles J. B., et al. (2010). Polyphosphate kinase 2: a novel determinant of stress responses and pathogenesis in Campylobacter jejuni. PLoS One 5:e12142. 10.1371/journal.pone.0012142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowska A. D., Wandel M. P., Lasica A. M., Nesteruk M., Roszczenko P., Wyszynska A., et al. (2011). Campylobacter jejuni dsb gene expression is regulated by iron in a Fur-dependent manner and by a translational coupling mechanism. BMC Microbiol. 11:166. 10.1186/1471-2180-11-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E., Leon-Kempis Mdel R., Pearson B. M., Hitchin E., Mulholland F., van Diemen P. M., et al. (2008). Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 69 77–93. 10.1111/j.1365-2958.2008.06263.x [DOI] [PubMed] [Google Scholar]
- Harris H. W., El-Naggar M. Y., Bretschger O., Ward M. J., Romine M. F., Obraztsova A. Y., et al. (2010). Electrokinesis is a microbial behavior that requires extracellular electron transport. Proc. Natl. Acad. Sci. U.S.A. 107 326–331. 10.1073/pnas.0907468107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley-Tassell L. E., Shewell L. K., Day C. J., Wilson J. C., Sandhu R., Ketley J. M., et al. (2010). Identification and characterization of the aspartate chemosensory receptor of Campylobacter jejuni. Mol. Microbiol. 75 710–730. 10.1111/j.1365-2958.2009.07010.x [DOI] [PubMed] [Google Scholar]
- Hendrixson D. R., DiRita V. J. (2004). Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52 471–484. 10.1111/j.1365-2958.2004.03988.x [DOI] [PubMed] [Google Scholar]
- Hermans D., Pasmans F., Messens W., Martel A., Van Immerseel F., Rasschaert G., et al. (2012). Poultry as a host for the zoonotic pathogen Campylobacter jejuni. Vector Borne Zoonotic Dis. 12 89–98. 10.1089/vbz.2011.0676 [DOI] [PubMed] [Google Scholar]
- Hermans D., Van Deun K., Martel A., Van Immerseel F., Messens W., Heyndrickx M., et al. (2011). Colonization factors of Campylobacter jejuni in the chicken gut. Vet. Res. 42:82. 10.1186/1297-9716-42-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreuter D., Novik V., Galan J. E. (2008). Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 4 425–433. 10.1016/j.chom.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Holmes K., Mulholland F., Pearson B. M., Pin C., McNicholl-Kennedy J., Ketley J. M., et al. (2005). Campylobacter jejuni gene expression in response to iron limitation and the role of fur. Microbiology 151(Pt 1), 243–257. 10.1099/mic.0.27412-27410 [DOI] [PubMed] [Google Scholar]
- Hugdahl M. B., Beery J. T., Doyle M. P. (1988). Chemotactic behavior of Campylobacter jejuni. Infect. Immun. 561560–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon B., Muraoka W., Sahin O., Zhang Q. (2008). Role of cj1211 in natural transformation and transfer of antibiotic resistance determinants in Campylobacter jejuni. Antimicrob. Agents Chemother. 52 2699–2708. 10.1128/aac.01607-1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem I. I., Khatri M., Esseili M. A., Sanad Y. M., Saif Y. M., Olson J. W., et al. (2012). Respiratory proteins contribute differentially to Campylobacter jejuni’s survival and in vitro interaction with hosts’ intestinal cells. BMC Microbiol. 12:258. 10.1186/1471-2180-12-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Hwang S., Ryu S., Jeon B. (2011). Regulation of perR expression by iron and PerR in Campylobacter jejuni. J. Bacteriol. 193 6171–6178. 10.1128/JB.05493-5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlath J. A., Osterholm M. T., Judy L. A., Forfang J. C., Robinson R. A. (1985). A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152 592–596. 10.1093/infdis/152.3.592 [DOI] [PubMed] [Google Scholar]
- Korolik V. (2010). Aspartate chemosensory receptor signalling in Campylobacter jejuni. Virulence 1 414–417. 10.4161/viru.1.5.12735 [DOI] [PubMed] [Google Scholar]
- Kusaka K., Shibata K., Kuroda A., Kato J., Ohtake H. (1997). Isolation and characterization of Enterobacter cloacae mutants which are defective in chemotaxis toward inorganic phosphate. J. Bacteriol. 179 6192–6195. 10.1128/jb.179.19.6192-6195.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertsethtakarn P., Ottemann K. M., Hendrixson D. R. (2011). Motility and chemotaxis in Campylobacter and Helicobacter. Annu. Rev. Microbiol. 65 389–410. 10.1146/annurev-micro-090110-102908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R. (2009). Function and biogenesis of iron-sulphur proteins. Nature 460 831–838. 10.1038/nature08301 [DOI] [PubMed] [Google Scholar]
- Liu X., Gao B., Novik V., Galan J. E. (2012). Quantitative proteomics of intracellular Campylobacter jejuni reveals metabolic reprogramming. PLoS Pathog 8:e1002562. 10.1371/journal.ppat.1002562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Marchant J., Wren B., Ketley J. (2002). Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol. 10 155–159. 10.1016/S0966-842X(02)02323-5 [DOI] [PubMed] [Google Scholar]
- Mazumder R., Phelps T. J., Krieg N. R., Benoit R. E. (1999). Determining chemotactic responses by two subsurface microaerophiles using a simplified capillary assay method. J. Microbiol. Methods 37 255–263. 10.1016/S0167-7012(99)00072-X [DOI] [PubMed] [Google Scholar]
- Miller W. G., Bates A. H., Horn S. T., Brandl M. T., Wachtel M. R., Mandrell R. E. (2000). Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66 5426–5436. 10.1128/AEM.66.12.5426-5436.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naikare H., Palyada K., Panciera R., Marlow D., Stintzi A. (2006). Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74 5433–5444. 10.1128/IAI.00052-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama S., Suzuki D., Itoh Y., Suzuki K., Tajima H., Hyakutake A., et al. (2012). Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect. Immun. 80 3170–3178. 10.1128/IAI.00039-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman H., King R. M., Shewell L. K., Semchenko E. A., Hartley-Tassell L. E., Wilson J. C., et al. (2014). Characterisation of a multi-ligand binding chemoreceptor ccml (tlp3) of Campylobacter jejuni. PLoS Pathog 10:e1003822. 10.1371/journal.ppat.1003822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajashekara G., Drozd M., Gangaiah D., Jeon B., Liu Z., Zhang Q. (2009). Functional characterization of the twin-arginine translocation system in Campylobacter jejuni. Foodborne Pathog Dis. 6 935–945. 10.1089/fpd.2009.0298 [DOI] [PubMed] [Google Scholar]
- Reuter M., van Vliet A. H. (2013). Signal balancing by the CetABC and CetZ chemoreceptors controls energy taxis in Campylobacter jejuni. PLoS One 8:e54390. 10.1371/journal.pone.0054390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular cloning: a laboratory manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Sanders L., Andermann T. M., Ottemann K. M. (2013). A supplemented soft agar chemotaxis assay demonstrates the Helicobacter pylori chemotactic response to zinc and nickel. Microbiology 159(Pt 1), 46–57. 10.1099/mic.0.062877-62870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95 5857–5864. 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson S. L., Davis L. M., MacKichan J. K., Allan B. J., Pajaniappan M., Thompson S. A., et al. (2009). The CprS sensor kinase of the zoonotic pathogen Campylobacter jejuni influences biofilm formation and is required for optimal chick colonization. Mol. Microbiol. 71 253–272. 10.1111/j.1365-2958.2008.06534.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareen A. M., Dasti J. I., Zautner A. E., Gross U., Lugert R. (2010). Campylobacter jejuni proteins Cj0952c and Cj0951c affect chemotactic behaviour towards formic acid and are important for invasion of host cells. Microbiology 156(Pt 10), 3123–3135. 10.1099/mic.0.039438-39430 [DOI] [PubMed] [Google Scholar]
- Tasneem A., Iyer L. M., Jakobsson E., Aravind L. (2005). Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 6:R4. 10.1186/gb-2004-6-1-r4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet A. H., Wooldridge K. G., Ketley J. M. (1998). Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180 5291–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegge C. S., Brondsted L., Li Y. P., Bang D. D., Ingmer H. (2009). Energy taxis drives Campylobacter jejuni toward the most favorable conditions for growth. Appl. Environ. Microbiol. 75 5308–5314. 10.1128/AEM.00287-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R. O., Galan J. E. (2008). Campylobacter jejuni survives within epithelial cells by avoiding delivery to lysosomes. PLoS Pathog 4:e14. 10.1371/journal.ppat.0040014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten M. M., Boeve M., Koot M. G., van Nuenen A. C., van der Zeijst B. A. (1998). Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten M. M., Parker C. T., van Mourik A., Guilhabert M. R., van Dijk L., van Putten J. P. (2006). The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol. Microbiol. 62 278–291. 10.1111/j.1365-2958.2006.05372.x [DOI] [PubMed] [Google Scholar]
- Wu H., Kato J., Kuroda A., Ikeda T., Takiguchi N., Ohtake H. (2000). Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J. Bacteriol. 182 3400–3404. 10.1128/JB.182.12.3400-3404.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R., Alm R. A., Trust T. J., Guerry P. (1993). Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130 127–130. 10.1016/0378-1119(93)90355-7 [DOI] [PubMed] [Google Scholar]
- Yao R., Burr D. H., Doig P., Trust T. J., Niu H., Guerry P. (1994). Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14 883–893. 10.1111/j.1365-2958.1994.tb01324.x [DOI] [PubMed] [Google Scholar]
- Yao R., Burr Donald H., Guerry P. (1997). CheY-mediated modulation of Campylobacter jejuni virulence. Mol. Microbiol. 23 1021–1031. 10.1046/j.1365-2958.1997.2861650.x [DOI] [PubMed] [Google Scholar]
- Yong S. C., Roversi P., Lillington J., Rodriguez F., Krehenbrink M., Zeldin O. B., et al. (2014). A complex iron-calcium cofactor catalyzing phosphotransfer chemistry. Science 345 1170–1173. 10.1126/science.1254237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K. T., Davis L. M., Dirita V. J. (2007). ‘Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5 665–679. 10.1038/nrmicro1718 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.