Abstract
Treatment of glioblastoma (GBM) remains a challenge using conventional chemotherapy, such as temozolomide (TMZ), and is often ineffective as a result of drug resistance. We have assessed a novel nitrone-based agent, OKN-007, and found it to be effective in decreasing tumor volumes and increasing survival in orthotopic GBM xenografts by decreasing cell proliferation and angiogenesis and increasing apoptosis. In this study, we assessed combining OKN-007 with TMZ in vivo in a human G55 GBM orthotopic xenograft model and in vitro in TMZ-resistant and TMZ-sensitive human GBM cell lines. For the in vivo studies, magnetic resonance imaging was used to assess tumor growth and vascular alterations. Percent animal survival was also determined. For the in vitro studies, cell growth, IC50 values, RNA-seq, RT-PCR, and ELISA were used to assess growth inhibition, possible mechanism-of actions (MOAs) associated with combined OKN-007 + TMZ versus TMZ alone, and gene and protein expression levels, respectively. Microarray analysis of OKN-007–treated rat F98 glioma tumors was also carried out to determine possible MOAs of OKN-007 in glioma-bearing animals either treated or not treated with OKN-007. OKN-007 seems to elicit its effect on GBM tumors via inhibition of tumorigenic TGF-β1, which affects the extracellular matrix. When combined with TMZ, OKN-007 significantly increases percent survival, decreases tumor volumes, and normalizes tumor blood vasculature in vivo compared to untreated tumors and seems to affect TMZ-resistant GBM cells possibly via IDO-1, SUMO2, and PFN1 in vitro. Combined OKN-007 + TMZ may be a potentially potent treatment strategy for GBM patients.
Introduction
High-grade gliomas are deadly, diffuse tumors that are highly vascularized and are resistant to apoptosis. Gliomas are the most common malignant primary brain tumors [1]. Glioblastoma (GBM), a grade IV glioma, accounts for the majority of all gliomas and is the most common of all malignant central nervous system tumors in adults [1]. GBMs have the highest incidence rate (3.2 per 100,000 population) and the highest number of cases of all malignant tumors with 12,760 cases projected for 2018 [1]. Unfortunately, the 5-year survival rate for glioblastomas is 5.5% [1], and there are no current effective therapies for this devastating disease. Current standard-of-care (SOC) for GBM involves surgical resection and follow-up treatments with radiotherapy and chemotherapeutic drugs such as temozolomide (TMZ), as well as bevacizumab, a monoclonal antibody targeting vascular endothelial growth factor (VEGF). Although TMZ is considered to be one of the most effective chemotherapeutic agents to prolong GBM patient survival [2], these SOC treatments, including TMZ, have not provided any long-term survival, and reoccurrence is very common. The recurrence for high-grade gliomas, particularly GBM, is due to difficulty in achieving a gross surgical section coupled with resistance to chemo- (mainly TMZ) [3], [4] and radiotherapies [5], [6]. Glioma-initiating cells that have stem-like properties have been linked with resistance to therapy and progression [7], [8].
From previous studies, it is well known that O6-methylguanine-DNA methyltransferase (MGMT) [9], N-methylpurine DNA glycosylase (MPG; also known as alkylpurine-DNA-N-glycosylase or APNG) [10], and hypoxia inducible factor 1α (HIF-1α) [11] are all associated with TMZ-resistance in GBM. Drug resistance against TMZ is mediated by increasing MGMT levels [2]. More specifically, it has been known for some time that MGMT promoter methylation is a predictive biomarker associated with alkylating chemotherapy, particularly TMZ [7]. There are several other mechanisms-of-action (MOAs) that have been recently associated with TMZ-resistance. For instance, it is known that if HIF-1α is downregulated, then a GBM cell becomes more TMZ sensitive [11]. Hedgehog (Hh) signaling, which is deregulated in GBM, is also associated with TMZ resistance (U87-MG, T98G) [12]. It has also been recently shown that connexins, particularly gap junction protein connexin43 (Cx43), play a role in the microenvironment of malignant gliomas and are capable in rendering GBM cells (U251) chemotherapeutic resistant, such as that elicited by TMZ [2], [13]. Activation of the PI3K pathway has also been recently found to be associated with TMZ resistance [14]. Recently, it was established that the sex-determining region Y (SRY)-box9 protein (SOX9) expression is associated with poor GBM patient prognosis and TMZ resistance (U87, U251) [15]. Another group recently found that stearoyl-coenzyme A desaturase 1 (SCD1) is upregulated in TMZ-resistant GBM cells [16]. It was also determined recently that X-ray repair cross-complementing 3 is involved with in TMZ-resistance by promoting DNA double-strand break repair [17]. Another group recently reported that long noncoding RNA (LncRNA) tumor suppressor candidate 7 expression is negatively associated with TMZ resistance in U87TR GBM cells [18]. The upregulation of MSH6, a key component of DNA mismatch repair, has also been recently found to be associated with TMZ resistance in GBM [19]. In U87 cells treated with TMZ, key candidate proteins and pathways associated with TMZ resistance include 28 downregulated proteins, of which 9 proteins (DHX9, HNRNPR, RPL3, HNRNPA3, SF1, DDX5, EIF5B, BTF3, and RPL8) were identified as key candidate proteins involved in ribosome and spliceosome pathways, and highly associated with GBM patient prognosis [20]. It should be noted that many of the above studies were done in GBM cells in vitro.
Regarding previously published possible treatment strategies, some approaches have included inhibiting NF-κB, a transcriptional regulator of MGMT, to reduce MGMT levels [7]; silencing SOX9, a transcriptional factor expressed in most solid tumors and a key regulator of glioma stem cells, and its target PDK1 (pyruvate dehydrogenase kinase 1) [8]; combining Akt and SCD1 inhibition [16]; and silencing LncRNA-H19 decreases TMZ resistance (TMZ-resistant cell lines: U251TMZ, M059JTMZ) by suppressing epithelial-mesenchymal transition via the Wnt/β-catenin pathway [21]; to suppress TMZ-resistance glioma cell growth. Again, in many cases, these studies were done in vitro.
We have previously demonstrated that a novel antiglioma agent, OKN-007 [OKlahoma Nitrone; disodium 4-(tert-butyl-imino) methyl) benzene-1,3-disulfonate N-oxide)], is able to decrease cell proliferation and angiogenesis, as well as increase apoptosis [22], [23]. Regarding angiogenesis, our group established that OKN-007 specifically inhibits HIF-1α protein expression [22]. In reference to angiogenesis, it was also established by our group that OKN-007 can specifically reduce vascular endothelial growth factor receptor 2 (VEGFR2) protein expression [24], and in addition reduce free radical levels [25], using molecular-targeted magnetic resonance imaging (MRI) probes for both of these biomarkers.
Related with this study, our group decided to assess whether OKN-007, if administered with TMZ, could have a synergistic effect for both treatments. We conducted an in vivo orthotopic xenograft GBM study to assess animal survival and effect on tumor volume reduction, as well as an effect on vascular perfusion. In addition, we also investigated the possible MOAs associated with OKN-007 treatment when combined to TMZ in both TMZ-resistant and TMZ-sensitive human GBM cells in vitro using qPCR and ELISA methods for determining HIF-1α, MGMT, and MPG gene and protein levels, respectively. In addition, RNA-seq was used to further elucidate the MOA regarding gene expression associated with combined OKN and TMZ treatment compared to TMZ alone in both TMZ-sensitive and TMZ-resistant GBM cell lines. Assessment of OKN-007 regarding its effect on cell migration was also studied in vitro using microfluidic chambers. The MOA of OKN-007 in a rodent GBM model was also further characterized with microarray, RT-PCR, and ELISA assessments.
Materials and Methods
In Vivo Studies
Rodents and Treatments
Animal studies were conducted in accordance to the OMRF Institutional Animal Care and Use Committee policies, which follow NIH guidelines. For the F98 rat glioma cell implantation model, F98 cells (105 in 10-μl volume) were intracerebrally implanted with a stereotaxic device (2 mm lateral and 2 mm anterior to the bregma and at a 3 mm depth) in a total of 15 Fischer 344 rats (male 200-250 g). The animals were divided into two groups once tumors reached 10-20 mm3 in volume (as determined by MRI): OKN-007 treated (n = 8) and untreated (UT) (n = 7) groups. Rats were treated until tumors reached 200-250 mm3 in volume or for a total of 4-6 weeks. OKN-007 was administered in the drinking water (18 mg/kg; 0.018% w/v). For the G55 GBM cell implantation model, 2-month-old male nude mice (Hsd:Athymic Nude-Foxn1nu mice; Harlan Inc., Indianapolis, IN) were implanted intracerebrally with human G55 xenograft cells (1 × 106) per ml suspended in 4 μl in cell culture media of 1% agarose solution. Once tumors reached 10-15 mm3 (determined via MRI), mice were treated either with OKN-007 in the drinking water (150 mg/kg; 0.20% w/v for a 20 g mouse) daily or with TMZ (30 mg/kg) via gavage every 3 days. Mice were treated until the tumors reached 100-150 mm3 or for a total of 4-6 weeks. For both rodent studies, OKN-007 was dissolved in water and made fresh every 2 days. Water bottles were weighed, and the amount of OKN-007 consumed per rodent was determined. No significant deviation was observed in the volume of liquid uptake of OKN-007 in these rodents. The average intake of OKN-007 was approximately 10 mg/kg/day/rat [22] or 140-150 mg/kg/day/mouse. TMZ was dissolved in 5% DMSO and 5% solutol-15 in sterile saline and administered via gavage. All groups were stratified to ensure that tumor sizes were similar before initiation of treatment.
MRI
MRI experiments were performed on a Bruker Bio-spec 7.0-T/30-cm horizontal-bore magnet imaging system. Animals were immobilized by using 1.5%-2.5% isoflurane and 0.8 L/min O2 and placed in a 72-mm quadrature volume coil for signal transmission, and either a surface rat-head or mouse-head coil was used for signal reception. T2-weighted morphological imaging was obtained with a slice thickness of 0.5 mm and a field of view of 4 × 5 cm2 for rats or 2 × 2 cm2 for mice, with an approximate in-plane resolution of 150 μm for rats and 80 μm for mice and with a repetition time of 3000 milliseconds and an echo time of 63 milliseconds for a total acquisition time of 13 minutes. Tumor volumes were calculated from 3D MRI slices rendered MRI datasets using Amira v5.6.0 (FEI) [9], [10], [11]. Tumor volumes were transposed from morphological image data sets. Comparative tumor volumes were obtained at the same time as the mean maximum tumor volumes for untreated tumor-bearing mice at days 19–22 [26].
Perfusion imaging
In order to assess microvascular alterations associated with tumor capillaries, the perfusion imaging method, arterial spin labeling, was used as previously described [26]. Perfusion maps were obtained on a single axial slice of the brain located on the point of the rostrocaudal axis where the tumor had the largest cross section. Five regions of interest (ROIs) were manually outlined around the tumor, and appropriate ROIs were also taken from the contralateral side of the brain for comparison purposes. To calculate the differences in relative cerebral blood flow (rCBF) values, tumor rCBF values were obtained at late (days 18-26 following intracerebral implantation of cells for untreated mice) and early (days 10-13 following cell implantation) tumor stages and normalized to rCBF values in the contralateral brain region of corresponding animals.
RNA Isolation and Preparation
For the rat F98 glioma study, all rats were euthanized after the last MRI examination. The brain of each animal was removed and snap frozen in liquid nitrogen before storage in a −80°C freezer. Total RNA from all tumor tissues from all treatment groups was purified with an RNeasy Mini Kit (Qiagen) and quantified by spectrophotometry (Nanodrop). cDNA was synthesized using SuperScript IV Reverse Transcriptase Kit (Invitrogen).
Microarray Analysis
The Illumina TotalPrep RNA Amplification Kit was used for labeling cRNA (Ambion, Austin, TX), as previously described [27]. Four × four treated/untreated samples were profiled using Affymetrix RaGene-1_0-st-v1 microarrays. Exon-level summarized measures were quantile normalized and tested for differential expression using Significance Analysis of Microarrays [28] at a false discovery rate (FDR) <40% and fold-change <1.5. Functional enrichment analysis was performed using Ingenuity Pathways Analysis (IPA) (Ingenuity Systems, www.ingenuity.com).
Histology and Immunohistochemistry (IHC)
All mice were euthanized after the last MRI examination. Perfusion fixation (10% neutral buffered formalin administered via a tail vein injection) was used on anesthetized (Isoflurane) mice, and whole brain of each animal was removed, further preserved in 10% neutral buffered formalin, and processed routinely. Paraffin-embedded tissues were sectioned in 5-μm sections, mounted on super frost plus glass slides, stained with hematoxylin and eosin (H&E), and examined by light microscopy. IHC was done to establish TGFβ1 levels by staining tissue samples with anti-TGFβ1 antibody (rabbit anti-TGFβ1, cat. no. 250876, 1 mg/ml, ABBIOTEC, San Diego, CA). For TGFβ1 IHC, sections were incubated in an antigen retrieval solution (citrate buffer, pH 6, Vector Laboratories, Burlingame CA) for 20 minutes in a rice steamer followed by a 20-minute cool down in deionized water.
Statistical analysis
Survival curves were analyzed using Kaplan–Meier curves. Tumor volumes, changes in normalized rCBF, and tumor blood volumes were analyzed and compared by two-way ANOVA with multiple comparisons. Data were represented as mean ± SD, and P values of *.05, **.01, ***.001, and ****.0001 were considered statistically significant. For microarray data, random variance t statistics for each gene were used [29].
In Vitro Studies
Cells and Culture Media
Most GBM cells were obtained from the American Tissue Culture Collection (ATCC; Manassas, VA, USA) [U-138-ATCC (CRL-HTB-16) glioblastoma, LN-18-ATCC (CRL-2610) glioblastoma, LN-229-ATCC (CRL-2611) glioblastoma, and T-98G-ATCC (CRL-1690)]. U-251 GBM cells were obtained from Sigma-Aldrich (N#09063001 also known before as U-373 MG (ATCC HTB-17)). G55 cells were obtained from Dr. Michael Sughrue, who was a resident at University of California San Francisco [originally obtained from C. David James (Department of Neurological Surgery, UCSF, CA, USA), who characterized the cells].
Cells were cultured in Dulbecco's modified Eagle's medium (Gibco BRL, Crand Island, NY) supplemented with 10% fetal bovine serum (Gibco) in a standard humidified incubator at 37°C under 5% CO2.
IC50 Concentrations: Protocol for TMZ Sensitivity Determination
The sensitivities of the six glioma cell lines to TMZ were evaluated from the concentrations required for 50% growth inhibition (IC50; also known as GI50) in comparison with untreated controls [8]. Briefly, cells were plated at 1 × 104 cells per well in 24-well, flat-bottomed plates and incubated with medium for 24 hours. The cells were subsequently washed twice with medium and incubated further with fresh medium (control) or medium containing 0.1-1000 μM of TMZ. For each plate that contained growth medium with ±TMZ, there was a plate with medium containing 1 mM OKN. After exposure to the various concentrations of TMZ for 72 hours, cells were detached by trypsinization and the numbers counted. The experiments were repeated at least four times at each concentration.
For varying concentrations of OKN-007, percent of cell viability for human GBM cells treated with either TMZ alone (500 μM) or TMZ combined with OKN (500 μM, 1 mM or 2 mM) was calculated. The G55 cells used for the OKN-007 varying concentration component were low-passage cells (TMZ resistant) determined from the IC50 values.
RNA Preparation
To avoid contributions from artificial sources in the experimentally measured expression patterns, each cell line was grown in four independent cultures, and the entire process was carried out independently on mRNA extracted from each culture.
Cell lines LN-18 and LN-229 were subjected to an assessment of their gene expression profile for four groups: cells, cells with TMZ, cells with TMZ-OKN combined treatment, and cells with OKN. The extracted total RNA was purified with an RNeasy Mini Kit (Qiagen), quantified by spectrophotometry (Nanodrop).
Quantification of mRNA by Real-Time Quantitative RT-PCR for HIF-1a, MPG, and MGMT
Total RNA from all cell lines with all treatments was purified with an RNeasy Mini Kit (Qiagen) and quantified by spectrophotometry (Nanodrop). cDNA was synthesized using SuperScript IV Reverse Transcriptase Kit (Invitrogen).
Target gene mRNA was amplified and measured by Bio-Rad CFX96 Real Time System. Gene expression was determined using the SYBR Select Master Mix (Applied Biosystems). Fluorescence signals, which are proportional to the concentration of the PCR product, are measured at the end of each cycle and immediately displayed on a computer screen, permitting real-time monitoring of the PCR. The reaction is characterized by the point during cycling when amplification of PCR products is first detected, rather than the amount of PCR product accumulated after a fixed number of cycles. The higher the starting quantity of the template, the earlier a significant increase in fluorescence is observed. The threshold cycle is defined as the fractional cycle number at which fluorescence passes a fixed threshold above the baseline. Fluorescence data were converted into cycle threshold measurements exported to Microsoft Excel. Glyceraldehyde-3-phosphatase dehydrogenase (GAPDH) mRNA expression levels were used as the quantitative internal control. For precise quantification, the mRNA expression level of each sample was normalized using the expression of the GAPDH gene.
All primers were synthesized by Integrated DNA Technologies.
The primers used were:
HIF-1α: F 5′-GTCGGACAGCCTCACCAAACAGAGC-3′ and R 5′-GTTAACTTGATCCAAAGCTCTGAG-3′. For MGMT, F 5′-GGGTCTGCACGAAATAAAGC-3′, R 5′-CTCCGGACCTCCGAGAAC-3′ [6], 5′-GTC CTA GTC CGG CGA CTT CC-3′, and 5′-CTT GTC TGG GCA GGC CCT TTG C-3′ were used to amplify 603-bp transcripts of MPG [9].
ELISA for HIF-1a, MPG, and MGMT
Protein expressions were assessed in all six glioma cell lines for all four groups of treatment (cell alone, cells with TMZ, cells with OKN, cells with combined OKN and TMZ)) as described above.
Cell Lysates
Cells were lysed before assaying. In brief, cells were washed by cold PBS gently, detached with trypsin, and collected by centrifugation at 1000×g for 5 minutes. Then, cells were washed three times in cold PBS and then suspended in fresh lysis buffer. Lysates were centrifuged at 1500×g for 10 minutes at 2°C-8°C to remove cellular debris.
The assay is based on the sandwich ELISA principle. Each well of the supplied microtiter plate has been precoated with a target specific capture antibody. Standards or samples are added to the wells, and the target antigen binds to the capture antibody. Unbound standard or sample is washed away. A biotin-conjugated detection antibody is then added which binds to the captured antigen. Unbound detection antibody is washed away. An avidin–horseradish peroxidase (HRP) conjugate is then added which binds to the biotin. Unbound avidin-HRP conjugate is washed away. A TMB substrate is then added which reacts with the HRP enzyme resulting in color development. A sulfuric acid stop solution is added to terminate color development reaction, and then the optical density (OD) of the well is measured at a wavelength of 450 nm ± 2 nm. The OD of an unknown sample can then be compared to an OD standard curve generated using known antigen concentrations in order to determine its antigen concentration. The antigen concentration determined from ELISA was then normalized to the total protein concentration of each cell lysate in order to have comparison between groups. ELISA kits for MPG and for HIF1a were purchased from CLOUD-CLONE Corp., and kits for human MGMT were from LifeSpan Biosciences, Inc.
RNA-seq
Prior to RNA-seq analysis, quality control measures were implemented. Concentration of RNA was ascertained via fluorometric analysis on a Thermo Fisher Qubit fluorometer. Overall quality of RNA was verified using an Agilent Tapestation instrument. Following initial QC steps, sequencing libraries were generated using the Lexogen Quantseq FWD library prep kit according to the manufacturer’s protocol. Briefly, the first strand of cDNA was generated using 5′-tagged poly-T oligomer primers. Following RNase digestion, the second strand of cDNA was generated using 5′-tagged random primers. A subsequent PCR step with additional primers added the complete adapter sequence to the initial 5′ tags, added unique indices for demultiplexing of samples, and amplified the library. Final libraries for each sample were assayed on the Agilent Tapestation for appropriate size and quantity. These libraries were then pooled in equimolar amounts as ascertained via fluorometric analyses. Final pools were absolutely quantified using qPCR on a Roche LightCycler 480 instrument with Kapa Biosystems Illumina Library Quantification reagents. Sequencing was performed on an Illumina Nextseq 500 instrument with High Output chemistry and 75-bp single-ended reads.
Raw sequencing files were processed with bbduk (decontamination using Kmers) [30] trimming of poly A tails and adaptor sequences. Fastqc [31] and multiQC [32] were used to check the quality of the resulting fastq files. High-quality scores (phred scores) of 33-36 were present in all samples with 9.3 ± 1.3 million reads each. Sorted bam files aligned with Tophat2 [33] to the GRCh38 genome were then provided to two separate pipelines in parallel in order to analyze the robust responses from each package.
Counts and differential gene expression were obtained with R using the ‘GenomicAlignments’ function ‘summarizeOverlaps’ and the negative binomial generalized linear modeling package DESeq2 [34].
Cell Migration
For the migration study, six-well chambers with polydimethylsiloxane (PDMS) microchannels inside were coated with 10 μg/ml of laminin (Sigma-Aldrich) in each well. G55 cells were seeded (50 × 103) in 100 μl and supplemented with 2 ml of media in each well. The chambers were incubated in the incubator at 37°C with 5% CO2 until scheduled time points. Some chambers were treated with OKN-007 (1 mM), TMZ (500 μM), or OKN-007 combined with TMZ, and for each treatment group, one well was left untreated as a control. Images of the cells inside the microchannels were taken using an Olympus CK40 inverted microscope (Japan) under 10×, and the distances of the same cells traveled were measured at 22, 28, and 46 hours postseeding; cell migration speed (μm/h) was calculated. Each treatment was repeated at least three times, and data are shown as mean ± S.D.
Statistical Analysis
RT-PCR gene and ELISA protein levels, and cell migrations were analyzed and compared by two-way ANOVA with multiple comparisons. RNA-seq data were analyzed using FDR <0.05 from the Benjamini-Hochberg FDR values provided by DESeq2 [34], [35], [36]. Data were represented as mean ± SD, and P values of *.05, **.01, ***.001, and ****.0001 were considered statistically significant.
Results
In Vivo G55 Orthotopic Xenograft GBM Model
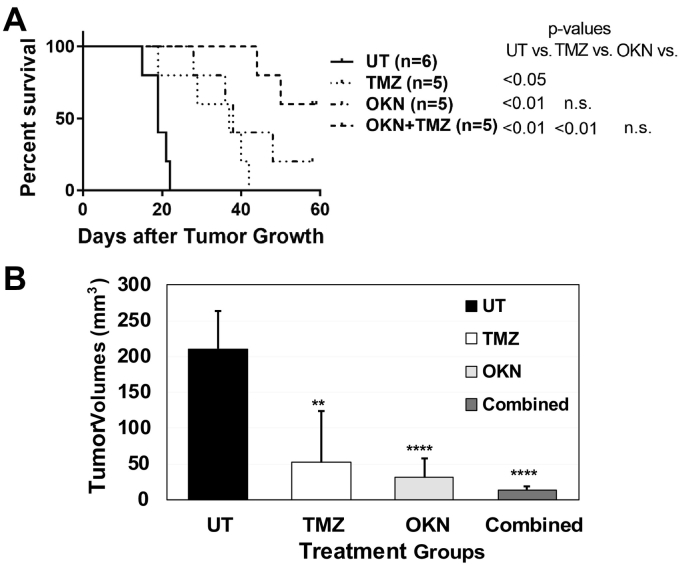
The percent animal survival data indicated that 60% of the combined therapy (OKN-007 + TMZ)–treated mice remained alive 60 days following tumor detection and treated for over 50 days (Figure 1A). One of the OKN-007-treated mice (20% of the mice treated) was also alive 60 days following tumor detection. Statistical analysis indicated that all of the treated mice (either OKN-007 alone, TMZ alone, or combined therapy) were found to have a significant increase in percent survival when compared to untreated (UT) G55 glioma-bearing mice. It was also found that the combined therapy mice had a significantly longer survival than TMZ-treated mice. However, statistical analysis indicated that there was no significance between the OKN-007–treated and combined therapy mice even though 60% of mice survived for the combined group compared to 20% for OKN-007 treatment alone.
Figure 1.
(A) Percent animal survival Kaplan-Meier curve for UT and TMZ-, OKN-007–, and combined OKN-007 and TMZ–treated G55 glioma-bearing mice. All treatment groups were found to have a significantly higher survival (P < .05 or more) compared to UT G55-glioma-bearing mice. The combined treatment group was found to have a significantly higher percent survival (P < .01) than the TMZ-treated group. (B) In vivo tumor volumes (mm3) obtained at days 19-22 following MRI detection of tumors (>5 mm3). All treatment groups were found to have significantly lower tumor volumes (P < .01 or more) compared to the UT group. N = 5 for each treatment group.
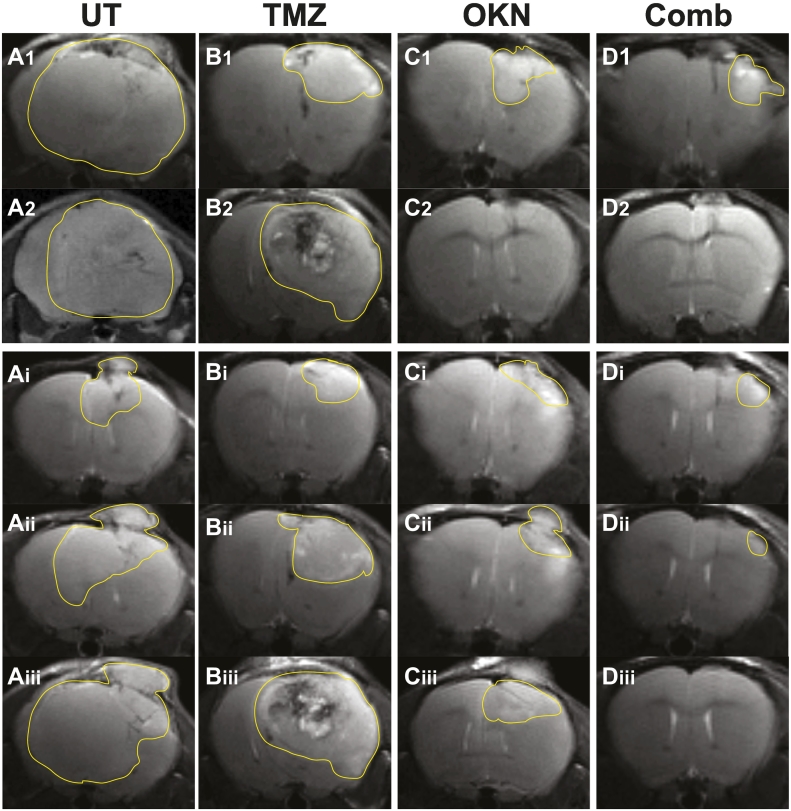
The tumor volumes were compared at the same time period when the UT mice were euthanized, i.e., tumor volumes reached 150 mm3 or larger (at 19-22 days following tumor detection). At 19-22 days following tumor detection, mice that were treated with either TMZ, OKN-007 or combined therapy were all found to have significantly decreased tumor volumes when compared to UT mice (Figure 1B). None of the treated mice were found to significantly differ from each other in tumor volumes; however, the combined therapy had the lowest tumor volume mean when compared to either TMZ- or OKN-007–treated mice. Representative MR images depicting tumors (mid-tumor regions) are also shown for each treatment group investigated (Figure 2). Representative tumors for all treatment groups as they developed from close to tumor detection until the last time points are also shown (Figure 2, A-D panels i-iii).
Figure 2.
Representative MR images depicting tumors in the midtumor region (maximal tumor) for (A) UT or each treatment group [(B) TMZ, (C) OKN-007 (OKN), or (D) combined OKN-007 and TMZ (Comb)]. Tumors are highlighted with a faint line to depict tumor boundaries. Images in panels “1” or “2” are multiple examples in treatment groups A-D, at 19-22 days following initial tumor detection, depicting either consistency in the UT group or variability in the treatment groups. There were no detectable tumors in images C2 or D2 for the OKN-007– or combined-therapy groups following treatment at this time-point range. Images in panels “i” are early detection of tumors at 3-6 days following initial detection. Images in panels “ii” for groups A-C are tumors at 10-12 days following initial detection. The tumor in panel Dii for the combined treatment group is 45 days following initial detection. Images in panels “iii” for groups A-C are tumors at 15-22 days following initial detection at the last imaging time point (representative midsurvival). The tumor in panel Diii for the combined treatment group is 52 days following initial detection, where no tumor was detected.
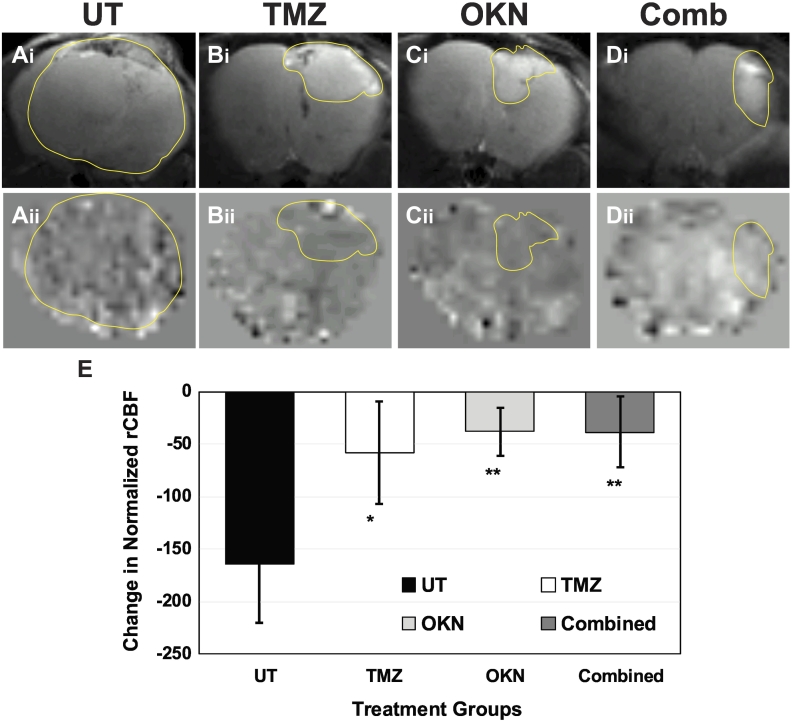
Normalized differences in tumor rCBF were found to significantly decrease in all treated mice compared to UT mice (Figure 3E). There were no significant differences between treated groups due to the small number of animals per group. Although not significant, it appears that both the combined therapy and OKN-007–treated groups seemed to have more normalized (less change in normalized rCBF) perfusion rates in their tumors compared to TMZ treatment. Representative morphological MR images (Figure 3, A-Di) and their corresponding perfusion maps (Figure 3, A-Dii) are shown for each treatment group.
Figure 3.
Vascular changes as depicted by perfusion MRI (rCBF in tumors at 21-22 days following tumor detection minus rCBF at initial tumor detection) in normalized (normalized to contralateral or normal brain tissue) rCBF values for (A) UT, (B) TMZ, (C) OKN, or (D) combined treated G55 glioma-bearing mice. Images at the top panel labeled “i” are representative T2-weighted morphological MR images for each treatment group, whereas the images labeled “ii” are representative perfusion maps for each treatment group. (E) Quantitative assessment of a change in normalized rCBF in UT and TMZ-, OKN-, or combined therapy–treated G55 glioma-bearing mice. All treatment groups had a significantly lower (P < .05 or more) change in normalized rCBF compared to the UT group. N = 5 for each treatment group.
In Vitro GBM Cell Study
From the in vitro GBM cell growth curves, it was established from the TMZ-sensitive cells (U251, LN229) that most of the cells were killed more than 50% with TMZ concentrations of 100 μM or less (Table 1). For the TMZ-resistant GBM cells (T98, LN18, U138, G55), the effect of combined therapy was substantial at TMZ concentrations less than 100 μM. The efficacy of combined therapy was significant.
Table 1.
IC50 Values for Human GBM Cells Treated with Either TMZ Alone (0, 0.1, 1, 10, 100, or 1000 μM) or TMZ Combined with OKN (1 mM)
| Cell line | TMZ, μM | TMZ, μM + OKN-007 |
|---|---|---|
| TMZ resistant | ||
| G55 (low) | 567.4 | 63.4 |
| U138 | 448.1 | 195.5 |
| LN18 | 773.7 | 31.9 |
| T98 | 438.3 | 295.4 |
| T98 (2) | 447.2 | 138.9 |
| TMZ sensitive | ||
| LN229 | 107.5 | Less 1 (<1) |
| LN229 (2) | ~100 | Less 1 (<1) |
| U251 | 176.5 | 8.0 |
| G55 (high) | 94.3 | 37.9 |
G55 GBM cells studied were both low- (<10 passages) and high- (>30 passages) passage cells. G55 low-passage cells were found to be TMZ resistant, whereas high-passage cells were more TMZ sensitive. LN229 and T98 GBM cell IC50 determinations were repeated.
A concentration of 1 mM OKN was found to be just as effective in reducing cell viabilities compared to the 2-mM concentration for some cells (LN229, U138, G55) (Table 2). In the other cells (U251, T98, LN18), 2 mM OKN was found to be slightly more effective in reducing cell viabilities compared to the 1-mM concentration (Table 2).
Table 2.
Percent of Cell Viability for Human GBM Cells Treated with Either TMZ Alone (500 μM) or TMZ Combined with OKN (500 μM, 1 mM or 2 mM)
| Cell line | TMZ, 500 μM | OKN, TMZ, both 500 μM | OKN, 1 mM; TMZ, 500 μM | OKN, 2 mM; TMZ, 500 μM |
|---|---|---|---|---|
| TMZ resistant | ||||
| G55 | 78.7 | 67.8 | 49.6 | 46.2 |
| U138 | 87.4 | 85.4 | 77.6 | 81.7 |
| LN18 | 61.7 | 78.9 | 60.6 | 41.1 |
| T98 | 78.0 | 68.3 | 56.1 | 37.2 |
| TMZ sensitive | ||||
| LN229 | 68.5 | 56.4 | 26.9 | 28.2 |
| U251 | 38.1 | 40.5 | 27.0 | 22.2 |
G55 low-passage TMZ-resistant cells were used.
HIF-1α, MGMT, and MPG
RT-PCR (Table 3) indicated that HIF-1α gene fold-changes had increased in all TMZ-treated and OKN-007 + TMZ–treated cells. OKN-007 increased HIF-1α gene fold-changes in all cells, except T98 cells, which had decreased gene fold-changes. MGMT gene fold-change is decreased in LN18 cells treated with either TMZ or OKN-007. OKN-007 decreased MGMT gene fold-change in T98 cells. MGMT gene fold-changes are increased in U251 cells following either TMZ, OKN-007, or combined treatment. MGMT gene fold-changes are increased slightly in LN18 combined treatment. MPG gene fold-changes are increased in most cells following TMZ treatment; in OKN-007–treated G55 and U138 cells; and in G55, LN18, and U138 cells treated with OKN-007 combined with TMZ. There were decreases in MPG gene fold-changes with OKN-007-treatment for LN18, T98, U251, and LN229 cells and also for T98 and LN229 cells treated with OKN-007 + TMZ.
Table 3.
RT-PCR Gene Fold-Changes for HIF-1α, MGMT, and MPG in TMZ-Resistant and TMZ-Sensitive Human GBM Cells Treated with Either TMZ, OKN, or Combined OKN + TMZ, or Untreated (UT)
| HIF-1α |
MGMT |
MPG |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cells | UT | TMZ | OKN | Comb | UT | TMZ | OKN | Comb | UT | TMZ | OKN | Comb |
| TMZ resistant | ||||||||||||
| G55 | 1.00 | 1.85 | 1.20 | 2.10 | - | - | - | - | 1.06 | 2.53 | 4.52 | 3.58 |
| LN18 | 1.03 | 2.11 | 1.35 | 2.76 | 1.06 | 0.77 | 0.42 | 1.26 | 1.00 | 2.95 | 0.60 | 5.40 |
| U138 | 1.00 | 2.05 | 2.93 | 3.17 | - | - | - | - | 1.13 | 7.14 | 11.10 | 12.70 |
| T98 | 1.00 | 1.72 | 0.58 | 1.56 | 1.00 | 1.07 | 0.54 | 1.15 | 1.10 | 1.22 | 0.62 | 0.73 |
| TMZ sensitive | ||||||||||||
| U251 | 1.00 | 1.48 | 1.59 | 1.56 | 1.00 | 1.32 | 1.22 | 1.24 | 1.10 | 1.11 | 0.81 | 1.16 |
| LN229 | 1.00 | 1.41 | 1.09 | 2.03 | - | - | - | - | 1.10 | 1.22 | 0.62 | 0.73 |
N = 2 for each cell treatment group, i.e., no statistical analyses was done. No substantial fold-changes were detected where there is a dash (−) indicated.
ELISA established protein levels (Table 4) indicated that HIF-1α was elevated in most cells treated with TMZ or combined treatment. OKN-007 decreased HIF-1α levels slightly in U138 cells, whereas in all other cells, this protein was slightly elevated in this treatment group. MGMT was elevated in G55, T98, and U251 cells treated with TMZ or combined treatment. OKN-007 slightly decreased MGMT in G55 and LN18 cells. MPG was mainly only elevated in U251 cells treated with TMZ or OKN-007 combined with TMZ. OKN-007 decreased MPG levels in LN229 cells. Specifically, protein levels in Table 4 indicate that, regarding HIF-1α, TMZ and combined treatments resulted in significantly higher levels, compared to UT cells, for G55, T98, and U251 cells, whereas OKN treatment had higher levels in T98 and U251 cells. HIF-1α levels were significantly higher in LN229 cells compared to TMZ-treated cells alone. Regarding MGMT, TMZ and combined treatments had significantly higher levels, compared to UT cells, for G55, T98, and U251 cells. MGMT levels were higher in combined treatment LN229 cells compared to either UT cells or TMZ-treated cells. Regarding MPG, TMZ and combined treatments resulted in significantly higher levels, compared to UT cells, for G55 and U251 cells; OKN treatment had higher levels in G55 cells; and TMZ alone had higher levels in U138 cells.
Table 4.
ELISA Protein Level (ng/mg Cell Lysate) Changes for HIF-1α, MGMT, and MPG in TMZ-Resistant and TMZ-Sensitive Human GBM Cells Treated with Either TMZ, OKN, or Combined OKN + TMZ, or Untreated (UT)
| Cells | HIF-1α |
MGMT |
MPG |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UT | TMZ | OKN | Comb | UT | TMZ | OKN | Comb | UT | TMZ | OKN | Comb | |
| TMZ resistant | ||||||||||||
| G55 | 0.14 ±0.02 |
1.08 ±0.04 *** |
0.19 ±0.06 |
1.13 ±0.25 *** |
0.37 ±0.10 |
1.19 ±0.13 *** |
0.31 ±0.06 |
1.11 ±0.11 *** |
1.15 ±0.09 |
1.50 ±0.22 * |
1.41 ±0.10 ** |
1.77 ±0.32 ** |
| LN18 | 0.76 ±0.06 |
0.83 ±0.07 |
0.92 ±0.14 |
0.90 ±0.36 |
1.23 ±0.22 |
1.28 ±0.16 |
0.98 ±0.07 |
1.31 ±0.15 |
2.18 ±0.36 |
2.27 ±0.25 |
2.38 ±0.24 |
2.20 ±0.32 |
| U138 | 0.40 ±0.10 |
0.84 ±0.35 |
0.33 ±0.05 |
0.44 ±0.12 |
0.77 ±0.25 |
0.98 ±0.12 |
0.75 ±0.04 |
0.93 ±0.19 |
1.30 ±0.03 |
1.50 ±0.15 * |
1.22 ±0.15 |
1.30 ±0.32 |
| T98 | 0.42 ±0.05 |
0.77 ±0.26 * |
0.58 ±0.05 ** |
0.59 ±0.05 ** |
0.62 ±0.10 |
1.76 ±0.22 *** |
0.84 ±0.19 |
1.57 ±0.55 * |
3.04 ±0.59 |
3.82 ±0.69 |
3.61 ±0.62 |
3.27 ±0.40 |
| TMZ sensitive | ||||||||||||
| U251 | 0.30 ±0.04 |
1.41 ±0.68 * |
0.91 ±0.21 ** |
0.91 ±0.19 *** |
0.92 ±0.30 |
1.44 ±0.24 * |
1.00 ±0.21 |
1.44 ±0.28 * |
2.70 ±0.40 |
4.99 ±0.67 ** |
2.77 ±0.09 |
4.09 ±0.57 ** |
| LN229 | 0.68 ±0.21 |
0.74 ±0.25 |
0.66 ±0.21 |
1.14 ±0.19 *† |
0.67 ±0.08 |
0.75 ±0.09 |
0.69 ±0.06 |
0.89 ±0.06 *† |
2.34 ±0.41 |
1.93 ±0.34 |
1.88 ±0.29 |
2.23 ±0.21 |
N = 4 for each cell treatment group. For HIF-1α: TMZ and combined treatments resulted in significantly higher levels, compared to UT cells, for G55, T98, and U251 cells, whereas OKN treatment had higher levels in T98 and U251 cells. HIF-1α levels were significantly higher in LN229 cells compared to TMZ-treated cells alone. For MGMT: TMZ and combined treatments had significantly higher levels, compared to UT cells, for G55, T98, and U251 cells. MGMT levels were higher in combined treatment LN229 cells compared to either UT cells or TMZ-treated cells. For MPG: TMZ and combined treatments resulted in significantly higher levels, compared to UT cells, for G55 and U251 cells; OKN treatment had higher levels in G55 cells, and TMZ alone had higher levels in U138 cells. *P < .05, **P < .01, ***P < .001, †P < .05. *,**, or ***Significant differences between UT and treated cells. †Significant differences between TMZ and combined treatments.
RNA-seq Data
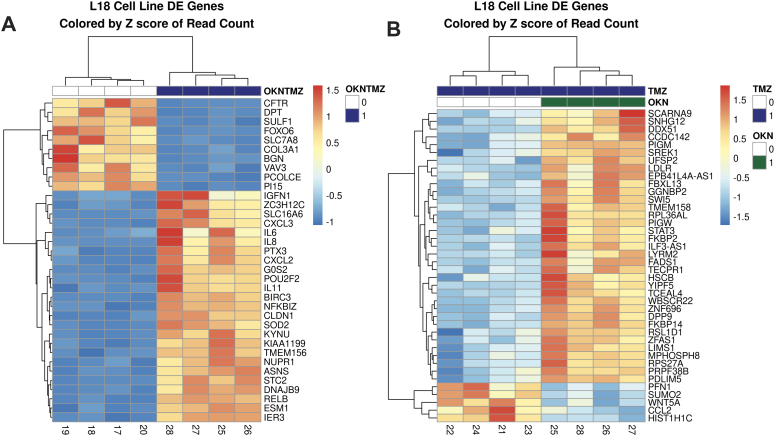
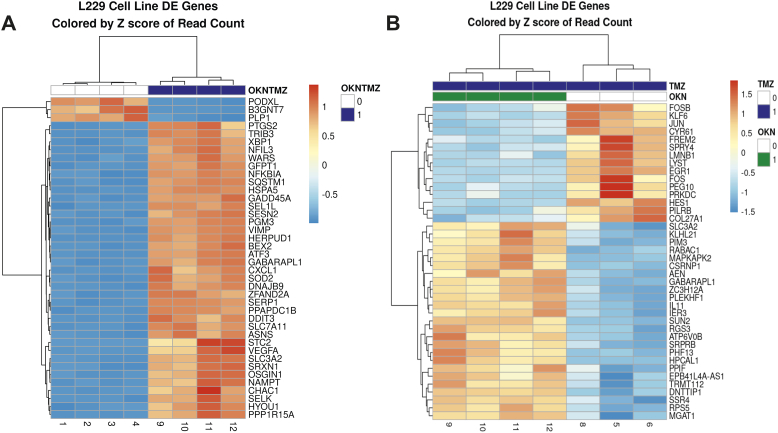
Figure 4A shows that the TMZ-resistant LN18 cells had 25 upregulated genes and 10 down-regulated genes when comparing the combined TMZ + OKN group to UT cells, whereas in Figure 4B, there were 37 upregulated genes and 3 genes downregulated in the TMZ + OKN group compared to TMZ alone group.
Figure 4.
Gene fold-changes when comparing: (A) TMZ + OKN to UT treatment groups or (B) TMZ + OKN to TMZ-alone treatment groups in LN18 GBM cells.
Figure 5A shows that the TMZ-sensitive LN229 cells had 37 upregulated genes and 3 downregulated genes when comparing the combined TMZ + OKN group to UT cells, whereas in Figure 5B, there were 21 upregulated genes and 19 genes downregulated in the TMZ + OKN group compared to TMZ alone group.
Figure 5.
Gene fold-changes when comparing: (A) TMZ + OKN to UT treatment groups or (B) TMZ + OKN to TMZ-alone treatment groups in LN229 GBM cells.
In Vitro Cell Migration Study
G55 GBM cell migration studies indicated that the cell migration speed for OKN-treated cells was significantly decreased at 22 and 46 hours following treatment compared to UT cells (Figure 6). Cells treated with TMZ were found to be significantly decreased at 28 and 46 hours following treatment compared to UT cells. It was also found that combined OKN + TMZ treatments significantly decreased cell migration at 22 and 28 hours following treatment compared to TMZ alone.
Figure 6.
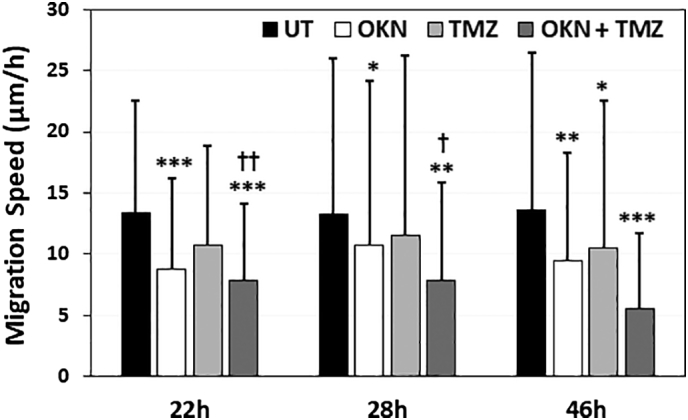
Migration speeds of G55 GBM cells in PDMS microchannels coated with laminin, either UT or treated with either OKN-007 (OKN), TMZ, or both OKN + TMZ at 22, 28, and 46 hours posttreatment. Chambers were treated with OKN-007 (1 mM), TMZ (500 μM), or OKN-007 combined with TMZ. There was a significant difference between UT cells (*) and those treated with OKN or OKN + TMZ at 22 and 28 hours and with all treatment groups at 46 hours. There was a significant difference between TMZ and OKN + TMZ groups (†) at 22 and 28 hours. *(†) P < .05, **(††) P < .01, *** P < .001. n = 62-68 for UT, n = 62-68 for TMZ-, 87-99 for OKN-007–treated, and 189-200 combined G55 cells, from repeated measures, where ‘n’ is the number of migrating cells counted. Values indicated are means ± S.D.
Microarray Data from Rat F98 Gliomas Either UT or Treated with OKN-007
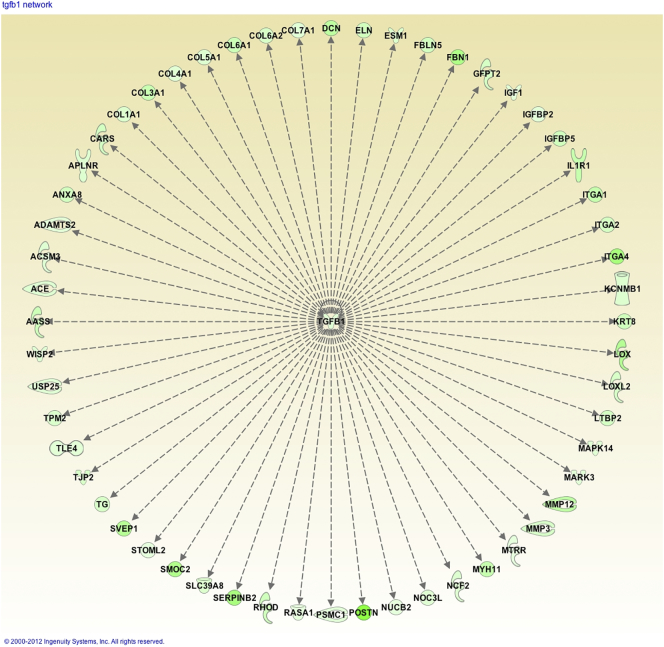
Microarray analysis identified predominant downregulation of genes following OKN treatment. A total of 384 genes had at least 1 exon significantly downregulated, while only 3 were upregulated (Supplementary Table). Major downregulated gene pathways affected include TGFβ1, PDGFBB, P38 MAPK, NFκB, some MMPs (particularly MMP12), DCN (decorin), SERPINB2, LUM, lipopolysaccharide-binding protein (LBP), and several collagens (see Supplementary File 1). Pathway analysis indicated that OKN-treated F98 tumors downregulated several genes associated with the extracellular matrix (ECM) (e.g., collagen and MMP genes), all with a connection to TGFβ1 (see Supplementary File 2). Upstream regulator analysis identified TGFβ1 as the most significant inhibited upstream regulator, controlling 57 downregulated genes (Figure 7). TGFβ1 was itself downregulated nearly two-fold.
Figure 7.
TGFβ1 as a master regulator of 57 downregulated genes by OKN-007 in rat F98 treated gliomas versus untreated tumors (Upstream Regulator Analysis, IPA). OKN-007 downregulates 57 genes including collagens, MMP12 (tissue remodeling), SERPINB2 (serpin peptidase inhibitor), and IGFBP5 (insulin-like growth factor binding protein).
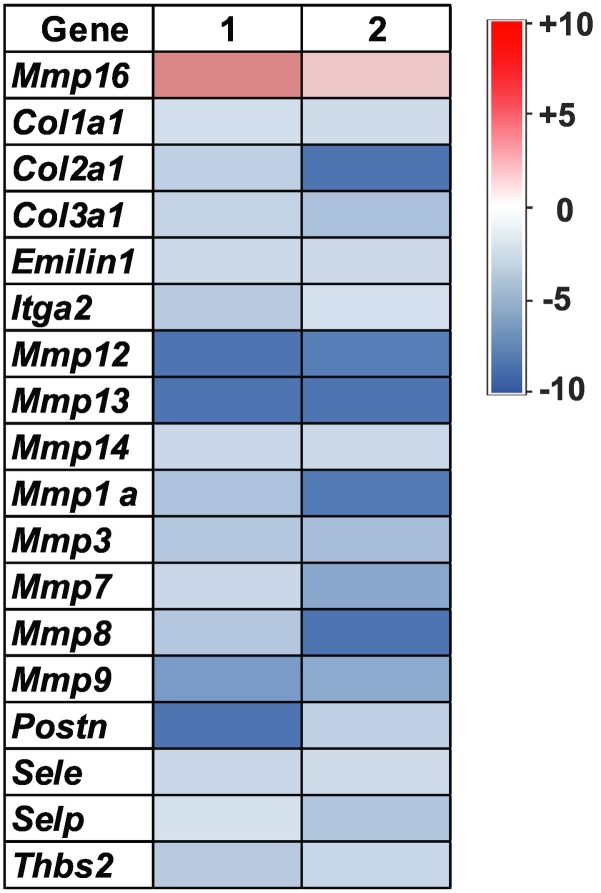
RT-PCR was able to confirm that several ECM genes were downregulated with OKN-treatment compared to UT F98 tumors (Figure 8).
Figure 8.
RT-PCR of RNA isolated from F98 untreated and OKN-007–treated tumors with +10- to −10-fold changes in gene expressions, particularly associated with collagen and MMP-associated genes.
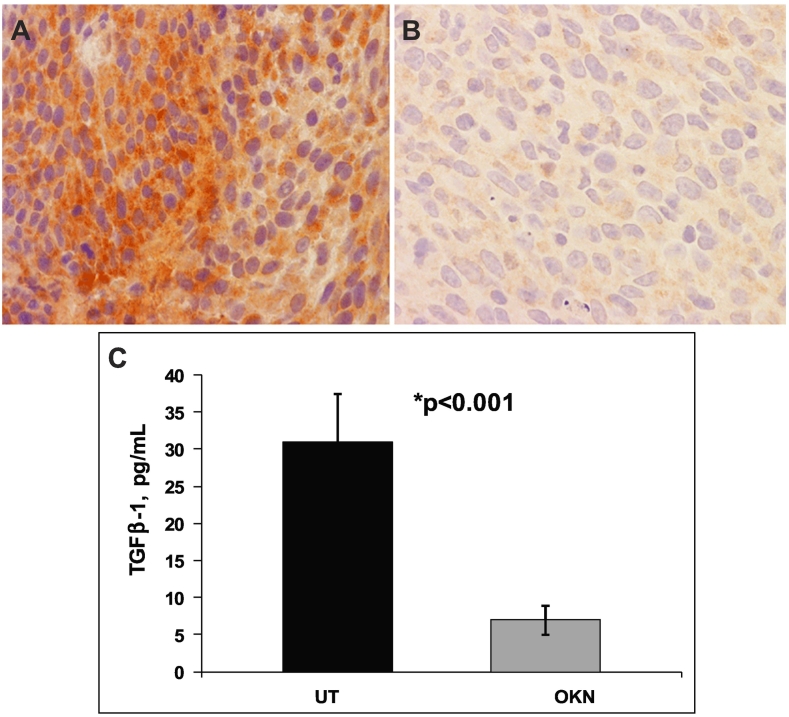
Immunohistochemistry showed that TGFβ1 protein levels were substantially reduced in OKN-treated F98 gliomas compared to UT tumors (Figure 9, A and B). The decrease in TGFβ1 protein levels was also confirmed using ELISA, indicating a significantly decreased (P < .001) level of TGFβ1 protein expression in OKN-treated F98 gliomas compared to UT tumors (Figure 9C).
Figure 9.
TGFβ1 IHC of rat F98 orthotopic tumors (midtumor region) either (A) untreated or (B) treated with OKN-007. (C) ELISA TGFβ1 protein levels (pg/ml) in UT or OKN-007 (OKN)–treated from F98 tumor tissue lysates.
Discussion
When OKN-007 is combined with TMZ, it can be effective against both TMZ-sensitive and TMZ-resistant GBM cells in vitro, synergistically decrease tumor volumes, as well as increase animal survival and normal vascularization in vivo in a G55 orthotopic xenograft GBM model.
We were able to establish that OKN-007 when combined with TMZ can augment the effect of TMZ in TMZ-sensitive GBM cells, as well as render TMZ-resistant GBM cells more sensitive to TMZ and/or the effect of OKN-007 on tumor cell growth. We were also able to establish that OKN-007, by itself, can downregulate several genes associated with the ECM through TGFβ1 and also be effective against cell migration. We previously found that OKN-007 can effectively inhibit cell proliferation; decrease HIF-1α and VEGFR2, which are both associated with angiogenesis; and increase apoptosis [22].
Combined OKN-007 and TMZ treatment in the in vivo G55 xenograft study indicated an enhanced % survival (Figure 1A) and decreased tumor volumes (Figure 1B) with minimal variability when compared to single-agent treatments. Of particular interest, more animals continued to survive with the combined treatment group compared to single-agent treatment groups. There was a large variability in the reduction of tumor volumes for the TMZ treatment group (Figures 1B and 2B). Vascular changes, as measured by perfusion MRI, indicated that OKN-007 (either alone or in combined treatment with TMZ) was possibly responsible for returning tumor vascularization to more normal perfusion rates (Figure 3E).
Other TMZ combined therapies that have been recently investigated include silencing GLI1, which is associated with Hedgehog signaling and specifically affected glioma-like stem cells (U87-MG, T98G) [12]; inhibiting Wnt/β-catenin signaling, which downregulates the expression of aldehyde dehydrogenase isoform 3A1 (ALDH3A1) [37]; using a miR-519a mimic, where miR-519a functions as a tumor suppressor by targeting the signal transducer and activator of transcription 3 (STAT3)–mediated autophagy, and promoting TMZ-induced autophagy (U87-MG/TMZ) [38]; inhibiting PI3K to sensitize GBM cells to TMZ [14]; inhibiting the SOX9/CA9 (carbonic anhydrase 9)-mediated oncogenic pathway to enhance TMZ sensitivity [15]; and co-delivery of TMZ and siRNA targeting the BCL-2 gene using a folate-conjugated triblock copolymer (Fa-PEG-PEI-PCL, Fa-PEC) of poly(ε-caprolactone) (PCL), poly(ethylenimine) (PEI), and poly(ethylene glycol) (PEG) nanocarrier construct [39]. All of the therapies combined with TMZ either are used to augment the effect of TMZ and/or effect TMZ-resistant GBM cells.
For the in vitro IC50 component of the study, for all cells, OKN-007 had a substantial effect in reducing the TMZ IC50 concentration so that the GBM cells all became TMZ sensitive (or more TMZ sensitive if they already were) (Table 1). There should be a cautionary note regarding assessing TMZ resistance with the G55 cells. We established that a low-passage G55 cell line is actually TMZ resistant, whereas high-passage G55 (>30 passages) may become TMZ sensitive. For the in vivo G55 xenograft data, we used a low-passage G55 cell line (<10 passages), which was a similar passage to those used for our in vitro studies.
From the varying concentrations of OKN-007, it was concluded that a concentration of 1 mM OKN was just as effective in reducing cell viabilities compared to the 2-mM concentration for some cell lines (e.g., LN229, U138, G55), whereas for other cell lines (e.g., U251, T98, LN18), 2 mM OKN was slightly more effective in reducing cell viabilities compared to the 1-mM concentration (Table 2), further validating the use of the 1-mM OKN-007 concentration for the IC50 value determinations with varying concentrations of TMZ (shown in Table 1).
At the gene level (Table 3), for TMZ-treated cells, HIF-1α gene fold-changes increase in all cells (compared to untreated cells). This is similar to what has already been reported by Fisher et al. (2007) where TMZ treatment of U251-MG and U87-MG resulted in the upregulation of HIF-1α [4]. Combined TMZ + OKN treatment results in some cells increasing HIF-1α gene fold-changes more so than TMZ alone (e.g., LN229, LN18, U138, and G55), but overall, there does not seem to be any substantial decreasing effect on HIF-1α with combined treatment. Possibly, OKN-007, when combined with TMZ, may be augmenting the effect of TMZ regarding the modulation of HIF-1α. There is noted reduced HIF-1α gene expression due to combined treatment in T98, but it was not significant. OKN alone seems to only decrease HIF-1α gene fold-changes in the T98 GBM cells compared to untreated cells. HIF-1α gene fold-changes in OKN treatment for U138 and U251 seem to still be increased (compared to untreated cells). Unlike protein levels, there is no reduction in HIF-1α by OKN-007 at the gene level. There was a reduction effect of OKN-007 only in T98 cells. HIF-1α gene fold-changes may differ in a more hypoxic environment, such as in a tumor (in vivo), compared to the normoxic conditions in cells.
Regarding protein levels in the TMZ-resistant LN18 GBM cell line and the TMZ-sensitive LN229 cell line (Table 4), TMZ seems to elevate HIF-1α in all cells, elevates MGMT in most cells (except LN229), and elevates MPG in only T98 and U251 cells. OKN-007 for most cells seems to not affect HIF-1α protein (compared to untreated cells), OKN-007 by itself seems to not effect protein levels of MGMT (compared to untreated cells, except LN18), and MPG is still elevated in T98 cells with OKN treatment alone. Of interest, HIF-1a was induced by TMZ in U251, U138, and T98 cells and then decreased by OKN-007. Combined TMZ + OKN seems to keep HIF-1α elevated (except for U138 cells), MGMT protein levels were not substantially changed with combined TMZ + OKN treatment, and MPG is still elevated in U251 cells with combined TMZ + OKN treatment, whereas other cells are not substantially affected (from either TMZ, OKN, or combined TMZ + OKN treatments). Specifically, in U251, T98, and G55 cells, the level of MGMT increased due to TMZ, but the treatment of OKN-007 did not inhibit the expression of MGMT.
There were only modest increases in MGMT gene fold-changes for TMZ alone or combined TMZ + OKN in only three cell lines (U251, T98, LN18) that had detectable levels. OKN treatment alone slightly increased MGMT gene fold-changes in U251 cells but conversely decreased MGMT gene fold-changes in LN18 and T98 cells (compared to untreated cells). Regarding OKN, these results may indicate that OKN alone may decrease MGMT gene expression, which may decrease resistant for GBM cells.
MPG gene fold-changes (unlike the protein levels) in TMZ treated cells were elevated more than two-fold in three cell lines (G55, LN18 and U138). With combined therapy, four cell lines (G55, LN229, LN18, and U138) had elevated MPG gene fold-changes (compared to untreated cells). Of interest, OKN treatment alone had continued elevated MPG gene fold-changes for two cell lines (G55 and U138) and some decreases for two other cell lines (LN18, T98) (compared to untreated cells). There were no decreases in MPG by OKN-007 that were observed.
It does not appear that OKN-007 affects TMZ-resistance through either MGMT or MPG.
From the RNA-seq data for the TMZ-resistant LN18 GBM cells (Figure 4), when comparing combined TMZ + OKN to UT treatments (Figure 4A), interesting downregulated genes include DPT-TRAMP that enhances TGF-β1 activity, FOXO6 (elevated expression correlates with progression and prognosis in gastric cancer [40]), COL3A1 (overexpression associated with bladder tumor progression [41]), BGN (plays a role in enhancing vessel formation and tumor cell migration [42]), and VAV3 (implicated in pancreatic cancer development [43]), and interesting upregulated genes include NFKBIZ (promotes apoptosis [44]), SOD2 (superoxide dismutase antioxidant [45]), and ASNS (responsive to cellular stress [46]). When comparing combined TMZ + OKN treatment to TMZ alone (Figure 4B), interesting upregulated genes are RNF149 (stress sensor gene that amplifies p53 response to arrest cell cycle [47]), IDO-1 (involved in human gliomas [48]), and SLC14a2 (endogenous transmembrane protein upstream-of-mTORC2 (UT2) negatively regulates activation of STAT3 [49]). Interesting downregulated genes include SUMO2 (overexpression of SUMO in conditions such as brain ischemia and hypoxia could increase cell survival, whereas the knockdown of SUMO expression has proven to be toxic to cells and associated with TGFβ1 in resistant glioma cells) [50], HIST1H1C/H1.2 (knockdown of histone HIST1H1C inhibits high glucose–induced inflammation and cell toxicity [51]), and PFN1 (profiling-1 phosphorylation is associated with tumor aggressiveness in human glioma) [52].
For RNA-seq data for the TMZ-sensitive LN229 cells (Figure 5), when comparing combined TMZ + OKN to UT treatments (Figure 5A), an interesting downregulated gene includes PODXL (a marker for gastric cancer [53]), and interesting upregulated genes include TRIB3 (induces apoptosis and reverses resistance to chemotherapy in hepatic carcinoma cells [54]) and CHAC1 (enhances glioma apoptosis [55]). When comparing combined TMZ + OKN treatment to TMZ alone (Figure 5B), interesting downregulated genes are IGR1 (associated with shorter disease-free survival in patients whose tumors are ER positive and HER2 positive [56]; IGF1R signaling pathway is very relevant in drug resistance [57]), XIST (increased level associated with shorter survival and poorer prognosis [58]), and PRKDC (prognostic biomarker for chemoresistance in breast cancer patients [59]). Interesting upregulated genes include ZC3H12A (crucial negative regulator of inflammation [60]) and RN7SK (potential antiproliferative and tumor-suppressive function [61]).
It appears that combined OKN-007 and TMZ-treatment affects different genes in TMZ-resistant or TMZ-sensitive GBM cells, which will need to be further pursued.
From further bioinformatics analysis, FOS has the most robust downregulation, which can be observed as an OKN effect across all cell lines and all treatment conditions (TMZ, OKN, and TMZ + OKN), as well as when the data are separated into each treatment condition or cell line separately. FOS has also been observed to be sufficient for decreasing cell viability and sensitizing glioblastoma to DNA damage via radiation, so it is possible that this may be helping TMZ cause DNA damage as well [6]. MGMT had nonsignificant changes in many of the conditions; however, for LN229, the log2 fold-change was ~3 with a P value of ~.057.
Other MOAs that could be associated with TMZ-resistance and that combined OKN + TMZ therapy may be affecting include TGFβ1 [50], [62] and also possibly Akt [63] and macrophages [64]. In our microarray assessment of rat F98 gliomas treated with OKN, TGFβ1 played a major role in the MOA for OKN efficacy by downregulating 57 genes that are commonly linked to TGFβ1. Perhaps, in combined OKN + TMZ therapy, OKN is affecting TGF-β1 and eliciting a response on TMZ-resistant cells.
OKN-007 was also found to significantly decrease G55 cell migration rates both when administered as a single agent and when combined with TMZ (Figure 6). Also, it was found that the combined treatment was found to significantly decrease cell migration rates compared to TMZ alone.
From microarray analysis of OKN-007–treated versus untreated rat F98 glioma RNA, it was established that downregulated genes mainly included members of integrin and collagen families, which are enriched in the ECM and cell adhesion (Figure 7; Supplementary 1). Cell viability of tumor tissues treated with OKN-007 was decreased, whereas cell death was increased via the downregulation of ANGPT2, DLL4, HPX, IGF1, and TGFβ1 genes. “Angiogenesis” was also decreased in OKN-007–treated samples. Several immune response genes were notably downregulated, with LBP being the most downregulated (Supplementary 1). TGFβ1, the last exon of which was downregulated, was the master regulator of 57 genes all downregulated by OKN-007 (Figure 7). TGFβ1 participates in nearly all processes mentioned above and as such may be considered as the main upstream regulator downregulated by OKN-007.
In addition to integrins and collagens, there is a notable group of ECM glycoproteins, such as lumican (LUM), fibrillin1 and 5 (FBN1), laminin (LAMA2). All of them are downregulated; however, matrix metallopeptidase 3 (MMP3), an enzyme degrading all aforementioned proteins, ADAMTS9 and PRSS2, other peptidases, are also downregulated. CD248, expressed in vascular endothelial cells of malignant tumors but not in normal blood vessels, is downregulated. That suggests that ECM maintenance and remodeling are active in untreated glioma samples and are normalized by OKN-007 treatment.
TGFβ1 and a member of TGF-β proteins latent transforming growth factor β binding protein (LTBP2) regulating cell adhesion and migration among other functions are also downregulated. Similarly, other cell adhesion–related molecules, such as POSTN (periostin) and LUM (lumican), are downregulated. F11r (F11 receptor), an immunoglobulin superfamily gene member and an important regulator of cell adhesion, cell-cell interaction, and formation of tight junction, is also downregulated. Collectively, it is suggestive that cell adhesion is downregulated. These findings could also be supportive regarding the cell migration inhibition properties of OKN-007.
LBP is the strongest downregulated immune-related gene, and the majority of its exons are downregulated. This gene is involved in the acute-phase immunologic response to gram-negative 8 bacterial infections. DMBT1 (deleted in malignant brain tumors 1) plays a role in the interaction of tumor cells and the immune system. Cytokine receptor IL1R1, oncostatin M receptor, which heterosimerizes with IL6 and IL31AR, interferon (alpha, beta and omega) receptor 1, and F11 receptors are all downregulated.
Overall, processes overrepresented by downregulated genes appear to be brought back to a normal state, in contrast with untreated glioma samples, where they are aberrantly regulated. For example, the single-exon CD248 gene is highly expressed in malignant tumors (and in the untreated glioma samples from this study). CD248 was downregulated in OKN-007–treated samples, suggesting a reversal of the tumor cells back to a normal state.
RT-PCR assessment of untreated and OKN-007–treated F98 rat glioma RNA samples for ECM genes (Figure 8) confirmed that predominantly collagen and MMP genes were mainly downregulated by OKN-007, although MMP16 seemed to be upregulated. Downregulation of ECM genes would also hinder cell migration.
IHC and ELISA were used to confirm that TGFβ1 protein expression was decreased with OKN-007 treatment in the F98 rat glioma model (Figure 9).
A proposed MOA schematic for OKN-007 is depicted in Figure 10, indicating that OKN-007 affects the TGF-β1 pathway, possibly through LTBP2, as suggested from the microarray data. The main effect of OKN-007, based on microarray, RT-PCR, ELISA, and IHC data, is that this compound mainly affects the TGF-β1 pathway. Our group previously found that OKN-007 directly reduces free radicals (which are included in ROS) using immunospin trapping in conjunction with in vivo molecular-targeted MRI [25]. In addition, our group established that OKN-007 decreases cell proliferation and angiogenesis, and increases apoptosis [22]. From this study, it was determined that OKN-007 decreases cell migration. The microarray and RT-PCR data also indicate that OKN-007 mainly affects ECM-related genes.
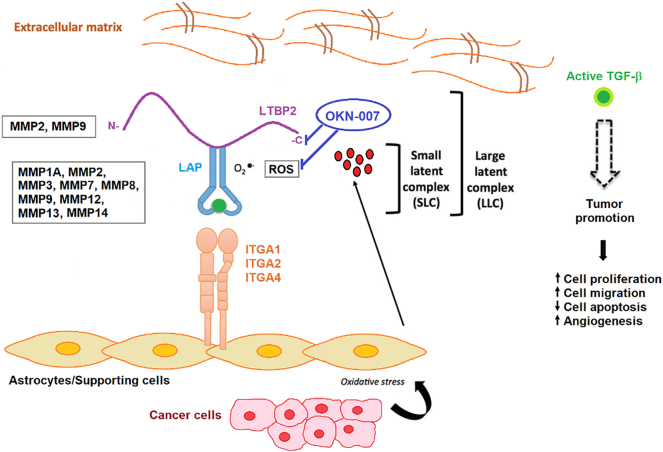
Figure 10.
Stromal activators of TGF-β in the tumor microenvironment. MMP2 and MMP9 proteolytically cleave LTBP, thereby releasing latent TGF-β from the ECM. MMP1A, MMP2, MMP3, MMp7, MMP8, MMP9, MMP12, MMP13, and MMP14 activate latent TGF-β via proteolytic cleavage of the latency-associated peptide (LAP), while integrins expressed on astrocytes (ITGA1, 2 and 4) bind to the large latent complex (LLC) and activate latent TGF-β through MMP-dependent cleavage of LAP. Integrins (ITGA1, 2 and 4) bind to the LLC and induce conformational changes in the latent complex via contractile action from activated astrocytes. ROS produced by activated astrocytes via the induction of oxidative stress from adjacent cancer cells can lead to the oxidation of the LAP domain and induce allosteric changes that release mature TGF-β from LAP. The mature (active) form of TGF-β can then bind to its cognate receptor and exert its tumor-promoting and tumor-suppressive properties. Dashed arrow indicates recruitment of the mature TGF-β protein to its cognate receptor. Modified from Costanza et al. (2017) [74]. Based on microarray and RT-PCR data from the rat F98 glioma model, comparing untreated to OKN-007–treated tumor tissue, OKN-007 is thought to act on LTBP (see Supplementary File 1, Supplementary File 2), as well as reactive oxygen species (ROS) [25]. LTBP2, MMP1A, MMP2, MMP3, MMP7, MMP8, MMP9, MMP12, MMP13, and MMP14 were all found to be downregulated in microarray and/or RT-PCR data from the F98 glioma study.
Stromal activators of TGF-β in the tumor microenvironment. MMP2 and MMP9 proteolytically cleave LTBP, thereby releasing latent TGF-β from the ECM. MMP1A, MMP2, MMP3, MMp7, MMP8, MMP9, MMP12, MMP13, and MMP14 activate latent TGF-β via proteolytic cleavage of the latency-associated peptide (LAP), while integrins expressed on astrocytes (ITGA1, 2 and 4) bind to the large latent complex (LLC) and activate latent TGF-β through MMP-dependent cleavage of LAP. Integrins (ITGA1, 2 and 4) bind to the LLC and induce conformational changes in the latent complex via contractile action from activated astrocytes. ROS produced by activated astrocytes via the induction of oxidative stress from adjacent cancer cells can lead to the oxidation of the LAP domain and induce allosteric changes that release mature TGF-β from LAP. The mature (active) form of TGF-β can then bind to its cognate receptor and exert its tumor-promoting and tumor-suppressive properties. Dashed arrow indicates recruitment of the mature TGF-β protein to its cognate receptor. Modified from Costanza et al. (2017) [74]. Based on microarray and RT-PCR data from the rat F98 glioma model, comparing untreated to OKN-007–treated tumor tissue, OKN-007 is thought to act on LTBP (see Supplementary Files 1 and 2), as well as reactive oxygen species (ROS) [25]. LTBP2, MMP1A, MMP2, MMP3, MMP7, MMP8, MMP9, MMP12, MMP13, and MMP14 were all found to be downregulated in microarray and/or RT-PCR data from the F98 glioma study.
Our data indicate that OKN-007, from in vivo experiments, seems to affect some of the molecular mechanisms associated with astrocytic activation, including affecting (1) the expression of the TGF-β receptor on the plasma membrane of astrocytes during activation and (2) a hypoxic insult, which leads to the stabilization and activation of HIF-1α [65]. These processes lead to gene transcription that affects apoptosis regulation, proliferation, migration, and angiogenesis [65].
OKN-007 has also been shown to be a heparin sulfate sulfatase Sulf2 inhibitor in hepatocellular carcinoma and thought to mediate tumor suppression via TGFβ1/SMAD2 and Hedgehog/GLI1 signaling [66]. Heparan sulfate proteoglycans (HSPGs) have also been associated with tumor drug resistance, and targeting HSPGs may be an effective approach to enhance anticancer chemotherapy efficacy and reduce drug resistance [67]. For example, it has been suggested that Sulf2 may play a key role in sorafenib susceptibility and resistance in liver cancer and that targeting Sulf2 may help sorafenib treatment in liver cancer patients [68]. In our own group, we were able to establish that OKN-007 was able to significantly decrease Sulf2 protein levels (P < .05) in a pediatric GBM patient-derived xenograft (PDX) model (IC-3752GBM) in athymic mice [69]. In that study, we did not look at gene levels of Sulf2. It has however been shown by Yue et al. (2013) that TGF-β1 induced the expression of Sulf2 in vitro in A549 cells, adenocarcinoma cells derived from type II alveolar epithelial cells [70]. It is possible that Sulf2 is a downstream effect following OKN-007 inhibition of the TGF-β1 pathway; however, our F98 rat glioma microarray or RT-PCR data do not present any supportive evidence that Sulf2 is affected by OKN-007 at the gene level in this glioma model.
As gliomas are highly invasive tumors, we also used microfluidic chambers to see if OKN-007 not only had a role in inhibiting cell proliferation, angiogenesis, and increasing apoptosis [22] but also has a role in inhibiting glioma cell migration/invasion [71]. We showed that OKN-007 significantly decreased migration velocity. Glioma cell invasion is dependent on the interaction between the glioma cells and ECM components such as fibronectin, collagen IV, tenascin-C, and fibronectin that stimulate different downstream migration pathways [72]. Perhaps OKN-007 is affecting cell migration via its effect on the ECM (including collagen, LAMA2, and MMP genes), where microarray and RT-PCR data support this hypothesis.
As TGF-β1 plays a major role in TMZ-resistance [50], [62], OKN-007 may actually be affecting TMZ-resistance by targeting TGF-β1. Additional supportive data in the LN18 RNA-seq data show that SUMO2, which is associated with TGF-β1 TMZ resistance, is also knocked down when OKN is combined with TMZ (compared to TMZ alone).
Based on the in vitro combined OKN-007 + TMZ treatment studies, for future studies, the following biomarkers, HIF-1α, TGF-β1, c-FOS, PFN-1, and SUMO2, all need to be further pursued as potential key molecular components that may be involved in the role that OKN-007 may play in synergistically affecting TMZ resistance when combined with TMZ. From studies in our group and those done by others, Sulf2 may also be an interesting biomarker to evaluate. For future studies, it would also be informative to assess OKN-007 with a panel of PDX GBM cells with known genetic alterations rather than extensively passaged GBM cell lines.
It should be noted that in previous studies for U251 and U87 cell lines, gene expression profiles generated from tissue culture were significantly different from those generated from a subcutaneous (s.c.) implanted tumor, which was significantly different from those grown intracerebrally (i.c.). The disparity between the i.c gene expression profiles and those generated from s.c. xenografts suggests that whereas an in vivo growth environment modulates gene expression, orthotopic growth conditions induce a different set of modifications [73]. This may imply the importance of using an appropriate orthotopic in vivo model to correctly represent the tumor microenvironment.
Conclusions
OKN-007 is an interesting antiglioma agent, which not only can be effective on its own, by targeting the tumorigenic TGF-β1 pathway via downregulation of key genes associated with the ECM and cell invasion, but also can elicit an effect on TMZ-resistant GBM cells/tissues. The MOA associated with combined OKN-007 + TMZ therapy does not seem to occur via either HIF-1α, MGMT, or MPG. OKN-007 itself did decrease some of these genes and proteins for some of the GBM cells, but the majority of GBM cells were either not affected or had elevated levels, with both OKN-007 or combined treatment groups. In vitro RNA-seq analysis provided some insights to other possible MOAs regarding how OKN-007 may play a role on TMZ resistance when combined with TMZ, and these will need to be further pursued in more detail in vivo taking into consideration the ECM and tumor microenvironment. Combining OKN-007 with TMZ seems like a promising therapeutic strategy that may prevent TMZ-resistant GBM cells from proliferating and migrating, as well as possibly extending the effect of TMZ.
The following are the supplementary data related to this article.
Microarray data of genes either up- or downregulated from rat F98 glioma-bearing tumor RNA samples that were treated with OKN-007 (T) and compared to those that were untreated (U).
Ingenuity Pathways Analysis (IPA) of genes and associated pathways from microarray data of genes either up- (red) or down-regulated (green) from rat F98 glioma-bearing tumor RNA samples that were treated with OKN-007 (T) and compared to those that were untreated (U).
Acknowledgements
We would like to thank Dr. Bart Frank (OMRF Microarray Core) for conducting the initial microarray analysis for the F98 rat glioma study.
Footnotes
Funding: Research reported in this publication was supported in part by the Oklahoma Medical Research Foundation (R.A.T.), Oblato, Inc. (R.A.T.), and the National Cancer Institute Cancer Center Support Grant P30CA225520 awarded to the University of Oklahoma Stephenson Cancer Center regarding use of the Tissue Pathology Share Resource. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Declaration of interest: R. A. T. has an issued patent on the use of OKN-007 in gliomas and a patent application regarding using combined OKN-007 and TMZ treatment for gliomas. Oblato, Inc., is currently conducting the phase I clinical trials in human GBM patients for OKN-007 as a single therapeutic agent and combined with TMZ.
References
- 1.Ostrom Q.T., Gittleman H., Liao P., Vecchione-Koval T., Wolinsky Y., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro Oncol. 2017;19:v1–v88. doi: 10.1093/neuonc/nox158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai SW, Huang BR, Liu YS, Lin HY, Chen CC, Tsai CF, Lu DY, Lin C. Differential characterization of temozolomide-resistant human glioma cells. Int J Mol Sci. 2018;19(1) doi: 10.3390/ijms19010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi J, Dong B, Zhou P, Guan W, Peng Y. Functional network analysis of gene-phenotype connectivity associated with temozolomide. Oncotarget. 2017;8(50):87554–87567. doi: 10.18632/oncotarget.20848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher T, Galanti G, Lavie G, Jacob-Hirsch J, Kventsel I, Zeligson S, Winkler R, Simon AJ, Amariglio N, Rechavi G. Mechanisms operative in the antitumor activity of temozolomide in glioblastoma multiforme. Cancer J. 2007;13:335–344. doi: 10.1097/PPO.0b013e318157053f. [DOI] [PubMed] [Google Scholar]
- 5.Furnari F.B., Fenton T., Bachoo R.M., Mukasa A., Stommel J.M., Stegh A., Hahn W.C., Ligon K.L., Louis D.N., Brennan C. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 6.Liu ZG, Jiang G, Tang J, Wang H, Feng G, Chen F, Tu Z, Liu G, Zhao Y, Peng MJ. c-Fos over-expression promotes radioresistance and predicts poor prognosis in malignant glioma. Oncotarget. 2016;7(40):65946. doi: 10.18632/oncotarget.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Happold C, Stojcheva N, Silginer M, Weiss T, Roth P, Reifenberger G, Weller M. Transcriptional control of O6-methyguanine DNA methyltransferase expression and temozolomide resistance in glioblastoma. J Neurochem. 2018;144(6):780–790. doi: 10.1111/jnc.14326. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Xu X, Liu N, Cheng Y, Jin W, Zhang P, Wang X, Yang H, Liu H, Tu Y. SOX9-PDK1 axis is essential for glioma stem cell self-renewal and temozolomide resistance. Oncotarget. 2017;9(1):192–204. doi: 10.18632/oncotarget.22773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao YH, Wang ZF, Cao CJ, Weng H, Xu CS, Li K, Li JL, Lan J, Zeng XT, Li ZQ. The clinical significance of O6-methylguanine-DNA-methyltransferase promoter methylation status in adult patients with glioblastoma: a meta-analysis. Front Neurol. 2018;9:127. doi: 10.3389/fneur.2018.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang JB, Svilar D, Trivedi RN, Wang XH, Goellner EM, Moore B, Hamilton RL, Banze LA, Brown AR, Sobol RW. N-methylpurine DNA glycosylase and DNA polymerase beta modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro Oncol. 2011;13(5):471–486. doi: 10.1093/neuonc/nor011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang JH, Ma ZX, Huang GH, Xu QF, Xiang Y, Li N, Sidlauskas K, Zhang EE, Lv SQ. Downregulation of HIF-1a sensitizes U251 glioma cells to the temozolomide (TMZ) treatment. Exp Cell Res. 2016;343(2):148–158. doi: 10.1016/j.yexcr.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Melamed JR, Morgan JT, Ioele SA, Gleghorn JP, Sims-Mourtada J, Day ES. Investigating the role of Hedgehog/GLI1 signaling in glioblastoma cell response to temozolomide. Oncotarget. 2018;9(43):27000–27015. doi: 10.18632/oncotarget.25467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grek CL, Sheng Z, Naus CC, Sin WC, Gourdie RG, Ghatnekar GG. Novel approach to temozolomide resistance in malignant glioma: connexin43-directed therapeutics. Curr Opin Pharmacol. 2018;41:79–88. doi: 10.1016/j.coph.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas B, Klinger V, Keksel C, Bonigut V, Kiefer D, Caspers J, Walther J, Wos-Maganga M, Weickhardt S, Rohn G. Inhibition of the PI3K but not the MEK/ERK pathway sensitizes human glioma cells to alkylating drugs. Cancer Cell Int. 2018;18:69. doi: 10.1186/s12935-018-0565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Wang Z, Liu N, Cheng Y, Jin W, Zhang P, Wang X, Yang H, Liu H, Zhang Y. Association between SOX9 and CA9 in glioma, and its effects on chemosensitivity to TMZ. Int J Oncol. 2018;53(1):189–202. doi: 10.3892/ijo.2018.4382. [DOI] [PubMed] [Google Scholar]
- 16.Dai S, Yan Y, Xu Z, Zeng S, Qian L, Hou L, Li X, Sun L, Gong Z. SCD1 confers temozolomide resistance to human glioma cells via the Akt/GSK3β/β-catenin signaling axis. Front Pharmacol. 2018;8:960. doi: 10.3389/fphar.2017.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roos WP, Frohnapfel L, Quiros S, Ringel F, Kaina B. XRCC3 contributes to temozolomide resistance of glioblastoma cells by promoting DNA double-strand break repair. Cancer Lett. 2018;424:119–126. doi: 10.1016/j.canlet.2018.03.025. [DOI] [PubMed] [Google Scholar]
- 18.Shang C, Tang W, Pan C, Hu X, Hong Y. Long non-coding RNA TUSC7 inhibits temozolomide resistance by targeting miR-10a in glioblastoma. Cancer Chemother Pharmacol. 2018;81(4):671–678. doi: 10.1007/s00280-018-3522-y. [DOI] [PubMed] [Google Scholar]
- 19.Sun Q, Pei C, Li Q, Dong T, Xing W, Zhou P, Gong Y, Zhen Z, Gao Y, Xiao Y. Up-regulation of MSH6 is associated with temozolomide resistance in human glioblastoma. Biochem Biophys Res Commun. 2018;496(4):1040–1046. doi: 10.1016/j.bbrc.2018.01.093. [DOI] [PubMed] [Google Scholar]
- 20.Yi GZ, Xiang W, Feng WY, Chen ZY, Li YM, Deng SZ, Guo ML, Zhao L, Sun XG, He MY. Identification of key candidate proteins and pathways associated with temozolomide resistance in glioblastoma based on subcellular proteomics and bioinformational analysis. Biomed Res Int. 2018;2018 doi: 10.1155/2018/5238760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia L, Tian Y, Chen Y, Zhang G. The silencing of LncRNA-H19 decreases chemoresistance of human glioma cells to temozolomide by suppressing epithelial-mesenchymal transition via the Wnt/β-catenin pathway. Onco Targets Ther. 2018;11:313–321. doi: 10.2147/OTT.S154339. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Towner RA, Gillespie DL, Schwager A, Saunders DG, Smith N, Njoku CE, Krysiak RS, III, Larabee C, Iqbal H, Floyd RA. Regression of glioma growth in F98 and U87 rat glioma models by the nitrone OKN-007. Neuro Oncol. 2013;15:330–340. doi: 10.1093/neuonc/nos337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coutinho de Souza P, Balasubramanian K, Njoku C, Smith N, Gillespie DL, Schwager A, Abdullah O, Ritchey JW, Fung K-M, Saunders D. OKN-007 decreases tumor necrosis and tumor cell proliferation and increases apoptosis in a preclinical F98 rat glioma model. J Magn Reson Imaging. 2015;42:1582–1591. doi: 10.1002/jmri.24935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coutinho de Souza P, Smith N, Pody R, He T, Njoku C, Silasi-Mansat R, Lupu F, Meek B, Chen H, Dong Y. OKN-007 decreases VEGFR-2 levels in a preclinical GL261 mouse glioma model. Am J Nucl Med Mol Imaging. 2015;5(4):363–378. [PMC free article] [PubMed] [Google Scholar]
- 25.Coutinho de Souza P, Smith N, Atolagbe O, Ziegler J, Nijoku C, Lerner M, Ehrenshaft M, Mason RP, Meek B, Plafker SM. OKN-007 decreases free radical levels in a preclinical F98 rat glioma model. Free Radic Biol Med. 2015;87:157–168. doi: 10.1016/j.freeradbiomed.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziegler J, Pody R, Coutinho de Souza P, Evans B, Saunders D, Smith N, Mallory S, Njoku C, Dong Y, Chen H. ELTD1, an effective anti-angiogenic target for gliomas: preclinical assessment in mouse GL261 and human G55 xenograft glioma models. Neuro Oncol. 2017;19(2):175–185. doi: 10.1093/neuonc/now147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffitts J, Tesiram Y, Reid GE, Saunders D, Floyd RA, Towner RA. In vivo MRS assessment of altered fatty acyl unsaturation in liver tumor formation of a TGF alpha/c-myc transgenic mouse model. J Lipid Res. 2009;50:611–622. doi: 10.1194/jlr.M800265-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright GW, Simon R. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 30.Bushnell B. BBTools software package. 2014. http://sourceforgenet/projects/bbmap
- 31.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. [Google Scholar]
- 32.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 36.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suwala AK, Koch K, Rios DH, Aretz P, Uhlmann C, Ogorek I, Felsberg J, Reifenberger G, Kohrer K, Deenen R. Oncotarget. 2018;9(32):22703–22716. doi: 10.18632/oncotarget.25210. Inhibition of Wnt/beta-catenin signaling downregulates expression of aldehyde dehydrogenase isoform 3A1 (ALDH3A1) to reduce resistance against temozolomide in glioblastoma in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Chen L, Li JJ, Zhou Q, Huang A, Liu WW, Wang K, Gao L, Qi ST, Lu YT. miR-519a enhances chemosensitivity and promotes autophagy in glioblastoma by targeting STAT3/Bcl2 signaling pathway. J Hematol Oncol. 2018;11(1):70. doi: 10.1186/s13045-018-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng Y, Huang J, Xiao H, Wu T, Shuai X. Codelivery of temozolomide and siRNA with polymeric nanocarrier for effective glioma treatment. Int J Nanomedicine. 2018;13:3467–3480. doi: 10.2147/IJN.S164611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang JH, Tang HS, Li XS, Zhang XL, Yang XZ, Zeng LS, Ruan Q, Huang YH, Liu GJ, Wang J. Elevated FOXO6 expression correlates with progression and prognosis in gastric cancer. Oncotarget. 2017;8(19):31682–31691. doi: 10.18632/oncotarget.15920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan L, Shu B, Chen L, Qian K, Wang Y, Qian G, Zhu Y, Cao X, Xie C, Xiao Y. Overexpression of COL3A1 confers a poor prognosis in human bladder cancer identified by co-expression analysis. Oncotarget. 2017;8(41):70508–70520. doi: 10.18632/oncotarget.19733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myren M, Kirby DJ, Noonan ML, Maeda A, Owens RT, Ricard-Blum S, Kram V, Kilts TM, Young MF. Biglycan potentially regulates angiogenesis during fracture repair by altering expression and function of endostatin. Matrix Biol. 2016;52-54:141–150. doi: 10.1016/j.matbio.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuboi M, Taniuchi K, Furihata M, Naganuma S, Kimura M, Watanabe R, Shimizu T, Saito M, Dabanaka K, Hanazaki K. Vav3 is linked to poor prognosis of pancreatic cancers and promotes the motility and invasiveness of pancreatic cancer cells. Pancreatology. 2016;16(5):905–916. doi: 10.1016/j.pan.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Okamoto K, Iwai Y, Oh-hora M, Yamamoto M, Morio T, Aoki K, Ohya K, Jetten AM, Akira S, Muta T. IκBξ regulates TH17 development by cooperating with ROR nuclear receptors. Nature. 2010;464:1381–1387. doi: 10.1038/nature08922. [DOI] [PubMed] [Google Scholar]
- 45.Wang W, Jin Y, Zeng N, Ruan Q, Qian F. SOD2 facilitates the antiviral innate immune response by scavenging reactive oxygen species. Viral Immunol. 2017;30(8):582–589. doi: 10.1089/vim.2017.0043. [DOI] [PubMed] [Google Scholar]
- 46.Balasubramanian MN, Butterworth EA, Kilberg MS. Asparagine synthesase: regulation by cell stress and involvement in tumor biology. Am J Physiol Endocrinol Metab. 2013;304(8):E789–E799. doi: 10.1152/ajpendo.00015.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nosrati N, Kapoor NR, Kumar V. DNA damage stress induces the expression of ribosomal protein S27a gene in a p53-dependent manner. Gene. 2015;559(1):44–51. doi: 10.1016/j.gene.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 48.Adams S, Teo C, McDonald KL, Zinger A, Bustamante S, Lim CK, Sundaram G, Braidy N, Brew BJ, Guillemin GJ. Involvement of the kynurenine pathway in human glioma pathophysiology. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0112945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee D, Wang YH, Kalaitzidis D, Ramachandran J, Eda H, Sykes DB, Raje N, Scadden DT. Endogenous transmembrane protein UT2 inhibits pSTAT3 and suppresses hematological malignancy. J Clin Invest. 2016;126(4):1300–1310. doi: 10.1172/JCI84620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshino A, Ogino A, Yachi K, Ohta T, Fukushima T, Watanabe T, Katayama Y, Okamoto Y, Naruse N, Sano E. Gene expression profiling predicts response to temozolomide in malignant gliomas. Int J Oncol. 2010;36(6):1367–1377. doi: 10.3892/ijo_00000621. [DOI] [PubMed] [Google Scholar]
- 51.Wang W, Wang Q, Wan D, Sun Y, Wang L, Chen H, Liu C, Petersen RB, Li J, Xue W. Histone HIST1H1C/H1.2 regulates autophagy in the development of diabetic retinopathy. Autophagy. 2017;13(5):941–954. doi: 10.1080/15548627.2017.1293768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu C, Tu Y, Yuan J, Mao X, He S, Wang L, Fu G, Zong J, Zhang Y. Aberrant expression of N-methylpurine-DNA glycosylase influences patient survival in malignant gliomas. J Biomed Biotechnol. 2012;2012:760679. doi: 10.1155/2012/760679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Zhu Z, Sheng J, Yu Z, Yao B, Huang K, Zhou L, Qui Z, Huang C. miR-509-3-5P inhibits the invasion and lymphatic metastasis by targeting PODXL and serves as a novel prognostic indicator for gastric cancer. Oncotarget. 2017;8(31):34867–34883. doi: 10.18632/oncotarget.16802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Zhu D, Hou L, Hu B, Xu M, Meng X. TRB3 reverses chemotherapy resistance and mediates crosstalk between endoplasmic reticulum stress and AKT signaling pathways in MHCC97H human hepatocellular carcinoma cells. Oncol Lett. 2018;15(1):1343–1349. doi: 10.3892/ol.2017.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen PH, Shen WL, Shih CM, Ho KH, Cheng CH, Lin CW, Lee CC, Liu AJ, Chen KC. The CHAC1-inhibited Notch3 pathway is involved in temozolomide-induced glioma cytotoxicity. Neuropharmacology. 2017;116:300–314. doi: 10.1016/j.neuropharm.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 56.McDermott MSJ, Canonici A, Ivers L, Browne BC, Madden SF, O'Brien NA, Crown J, O'Donovan N. Dual inhibition of IGF1R and ER enhances response to trastuzumab in HER2 positive breast cancer cells. Int J Oncol. 2017;50(6):2221–2228. doi: 10.3892/ijo.2017.3976. [DOI] [PubMed] [Google Scholar]
- 57.Yuan J, Yin Z, Tao K, Wang G, Gao J. Function of insulin-like growth factor 1 receptor in cancer resistance to chemotherapy. Oncol Lett. 2018;15(1):41–47. doi: 10.3892/ol.2017.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene innon-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478(2):811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 59.Sun G, Yang L, Dong C, Ma B, Shan M, Ma B. PRKDC regulates chemosensitivity and is a potential prognostic and predictive marker of response to adjuvant chemotherapy in breast cancer patients. Oncol Rep. 2017;37(6):3536–3542. doi: 10.3892/or.2017.5634. [DOI] [PubMed] [Google Scholar]
- 60.Wawro M, Kochan J, Krzanik S, Jura J, Kasza A. Intact NYN/PIN-like domain is crucial for the degradation of inflammation-related transcripts by ZC3H12D. J Cell Biochem. 2017;118(3):487–498. doi: 10.1002/jcb.25665. [DOI] [PubMed] [Google Scholar]
- 61.Bazi Z, Bertacchi M, Abasi M, Mohammadi-Yeganeh S, Soleimani M, Wagner N, Ghanbarian H. Rn7SK small nuclear RNA is involved in neuronal differentiation. J Cell Biochem. 2018;119(4):3174–3182. doi: 10.1002/jcb.26472. [DOI] [PubMed] [Google Scholar]
- 62.Wang Z, Wang K, Wang R, Liu X. SUMOylation regulates TGF-β1/Smad4 signaling in-resistant glioma cells. Anticancer Drugs. 2017;29:136–144. doi: 10.1097/CAD.0000000000000578. [DOI] [PubMed] [Google Scholar]
- 63.Fan Y, Potdar AA, Gong Y, Eswarappa SM, Donnola S, Lathia JD, Hambardzumyan D, Rich JN, Fox PL. Profilin-1 phosphorylation directs angiocrine expression and glioblastoma progression through HIF-1α accumulation. Nat Cell Biol. 2014;16(5):445–456. doi: 10.1038/ncb2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeng H, Yang Z, Xu N, Liu B, Fu Z, Lian C, Guo H. Connective tissue growth factor promotes temozolomide resistance in glioblastoma through TGF-β1-dependent activation of Smad/ERK signaling. Cell Death Dis. 2017;8(6) doi: 10.1038/cddis.2017.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang C, Rahimpour S, Yu ACH, Lonser RR, Zhuang Z. Regulation and dysregulation of astrocyte activation and implications in tumor formation. Cell Mol Life Sci. 2013;70:4201–4211. doi: 10.1007/s00018-013-1274-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng X, Gai X, Han S, Moser CD, Hu C, Shire AM, Floyd RA, Roberts LR. The human sulfatase 2 inhibitor 2,4-disulfonylphenyl-tert-butylnitrone (OKN-007) has an antitumor effect in hepatocellular carcinoma mediated via suppression of TGFB1/SMAD2 and Hedgehog/GLI1 signaling. Genes Chromosomes Cancer. 2013;52(3):225–236. doi: 10.1002/gcc.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lanzi C, Zaffaroni N, Cassinelli G. Targeting heparin sulfate proteoglycans and their modifying enzymes to enhance anticancer chemotherapy efficacy and overcome drug resistance. Curr Med Chem. 2017;24:1–27. doi: 10.2174/0929867324666170216114248. [DOI] [PubMed] [Google Scholar]
- 68.Yoon S, Lee E-J, Choi J-H Chung T, Kim DY, Im J-Y, Bae M-H, Kwon J-H, Kim H-H, Kim HC, Park YN. Recapitulation of pharmacogenomics data reveals that invalidation of SULF2 enhance sorafenib susceptibility in liver cancer. Oncogene. 2018 doi: 10.1038/s41388-018-0291-3. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 69.Coutinho de Souza P, Mallory S, Smith N, Saunders D, Li X-N, McNall-Knapp RY, Fung K-M, Towner RA. Inhibition of pediatric glioblastoma tumor growth by the anti-cancer agent OKN-007 in orthotopic mouse xenografts. PLos One. 2015;10(8) doi: 10.1371/journal.pone.0134276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yue X, Lu J, Auduong L, Sides MD, Lasky JA. Overexpression of Sulf2 in idiopathic pulmonary fibrosis. Glycobiology. 2013;23(6):709–719. doi: 10.1093/glycob/cwt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szopa W, Burley TA, Kramer-Marek G, Kaspera W. Diagnostic and therapeutic biomarkers in glioblastoma: current status and future perspectives. Biomed Res Int. 2017;2017 doi: 10.1155/2017/8013575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Demuth T, Berens ME. Molecular mechanisms of glioma cell migration and invasion. J Neuro-Oncol. 2004;70(2):217–228. doi: 10.1007/s11060-004-2751-6. [DOI] [PubMed] [Google Scholar]
- 73.Camphausen K, Purow B, Sproul M, Scott T, Ozawa T, Deen DF, Tofilon PJ. Influence of in vivo growth on human glioma cell line gene expression: Convergent profiles under orthotopic conditions. Proc Natl Acad Sci U S A. 2005;102(23):8287–8292. doi: 10.1073/pnas.0502887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Costanza B, Umelo IA, Bellier J, Castronovo V, Turtoi A. Stromal modulators of TGF-β in cancer. J Clin Med. 2017;6:7. doi: 10.3390/jcm6010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microarray data of genes either up- or downregulated from rat F98 glioma-bearing tumor RNA samples that were treated with OKN-007 (T) and compared to those that were untreated (U).
Ingenuity Pathways Analysis (IPA) of genes and associated pathways from microarray data of genes either up- (red) or down-regulated (green) from rat F98 glioma-bearing tumor RNA samples that were treated with OKN-007 (T) and compared to those that were untreated (U).