Abstract
In contrast to responses against infectious challenge, T cell responses induced via adjuvanted subunit vaccination are dependent on interleukin-27 (IL-27). We show that subunit vaccine–elicited cellular responses are also dependent on IL-15, again in contrast to the infectious response. Early expression of interferon regulatory factor 4 (IRF4) was compromised in either IL-27– or IL-15–deficient environments after vaccination but not infection. Because IRF4 facilitates metabolic support of proliferating cells via aerobic glycolysis, we expected this form of metabolic activity to be reduced in the absence of IL-27 or IL-15 signaling after vaccination. Instead, metabolic flux analysis indicated that vaccine-elicited T cells used only mitochondrial function to support their clonal expansion. Loss of IL-27 or IL-15 signaling during vaccination resulted in a reduction in mitochondrial function, with no corresponding increase in aerobic glycolysis. Consistent with these observations, the T cell response to vaccination was unaffected by in vivo treatment with the glycolytic inhibitor 2-deoxyglucose, whereas the response to viral challenge was markedly lowered. Collectively, our data identify IL-27 and IL-15 as critical to vaccine-elicited T cell responses because of their capacity to fuel clonal expansion through a mitochondrial metabolic program previously thought only capable of supporting quiescent naïve and memory T cells.
INTRODUCTION
The most robust and durable vaccine platforms use attenuated infectious agents, against which both T and B cell memory can last for many decades (1, 2). Unfortunately, not all infectious agents can be attenuated for the purposes of vaccination (e.g., HIV, hepatitis C virus, and tuberculosis), mandating the use of adjuvanted subunit vaccines to promote protective immunity. Infectious model organisms, such as lymphocytic choriomeningitis virus or Listeria monocytogenes (LM), have been used extensively in the laboratory for studying the molecular and cellular underpinnings of robust T cell immunity (3). The immunological factors and pathways central to the cellular response against model organisms are reasonably assumed to be the same factors and pathways that will be central to subunit vaccine–induced immunity. For example, interleukin-12 (IL-12) is well documented by Curtsinger and Mescher to play a critical role as a “signal 3 cytokine” supporting maximal T cell differentiation and survival (4). IL-12 is also important in the generation of short-lived effector cells (SLECs) during primary infectious challenge (5), a subset important for the elimination of the primary infection and the eventual resolution of the response to resting memory. Findings such as these have encouraged the pursuit of formulations that induce the same inflammatory components produced during the infectious process for use as vaccine adjuvants.
Most of the vaccine adjuvants tested to date are effective at augmenting antibody responses, but their capacity to facilitate cellular immunity is typically orders of magnitude lower than attenuated infectious agents (6, 7). Although numerous factors are likely to be involved, a key difference between vaccine adjuvant administration and infectious challenge is that the inflammation induced by adjuvants is generally less potent and resolves more quickly (8). The degree to which downstream adaptive immunity is dependent on this inflammation calls into question the transferability of the rules governing infection-elicited immunity to vaccine adjuvant–elicited immunity. An excellent example of this is the vastly divergent roles played by IL-27 in immune responses to adjuvanted vaccines versus natural infection. IL-27 is a cytokine closely related to IL-6, IL-12, and IL-23 and has been linked to the inhibition of cell-mediated immunity in the context of autoimmunity and infectious disease (9, 10). In contrast to the reports of inhibitory effects of IL-27 in response to infectious challenges, CD4+ and CD8+ T cell responses to a broad range of adjuvanted subunit vaccines are highly dependent on T cell–intrinsic IL-27 signaling, which mediates the prolonged survival of T cells in response to vaccination (7). Thus, the vast majority of data on IL-27 derived from infectious model systems did not predict it to be a major determinant of vaccine-elicited cellular immunity.
A major focus of T cell biology in recent years has been the identification and manipulation of the metabolic pathways that fuel T cell clonal expansion, memory formation, and long-term survival (11). The metabolic program used by T cells is dynamic, changing depending on the activation state and differentiation status of the T cell. Whereas naïve T cells largely generate adenosine 5′-triphosphate (ATP) via oxidative phosphorylation, one common feature of activated T cells is an early switch to aerobic glycolysis, or so-called “Warburg metabolism,” in which these rapidly dividing cells chiefly convert glucose to lactate rather than feeding pyruvate into the Krebs cycle within the mitochondria (12). However, quiescent memory T cells do not rely on aerobic glycolysis but instead exhibit high rates of fatty acid β-oxidation in a metabolic program characterized by high mitochondrial spare respiratory capacity (SRC) (13, 14). The reason usually given for why rapidly dividing cells switch between these two forms of metabolism is their high demand for biomass generation (12, 15). CD8+ T cells can divide as often as once every 2 to 4 hours (16), a rate once thought to be catastrophic to cellular survival. Aerobic glycolysis, while producing far less ATP than oxidative phosphorylation, produces a number of metabolic intermediates that serve as precursors for amino acid, lipid, and nucleotide synthesis (12). It is believed that mitochondrial function, though superior at generating ATP, fails to produce sufficient levels of these precursors to double the mass of a T cell during peak clonal expansion (17). Given our increasing appreciation for the disparate factors governing T cell responses to infection and vaccination (8), whether vaccine-elicited cells follow this same paradigm, or whether their metabolic program is distinctly shaped by their unique inflammatory environment, remains to be determined.
The data that we present here not only identify a number of factors in addition to IL-27, which function distinctly in vaccine-elicited immunity versus infection-elicited immunity, but also identify the metabolic program fueling the clonal expansion and survival of T cells responding to subunit vaccination. In addition to IL-27, we show that the magnitude of vaccine-elicited CD8+ T cell immunity is highly dependent on IL-15, Tbet, and Eomes. This is again in contrast to T cell responses to infectious challenge, where the loss of any of these factors changes the long-term fate of the T cells but leaves early CD8+ T cell proliferation and survival largely unaffected (18–23). Last, our data show that these cytokine and transcriptional events are necessary to support a metabolic program fueled not by aerobic glycolysis but through mitochondrial function. Collectively, our data broaden the conclusion that divergent biological mechanisms guide subunit vaccine–elicited immunity from those derived from infectious challenge.
RESULTS
IL-15 is required for the CD8+ T cell response to adjuvanted subunit vaccination but not to infectious challenge
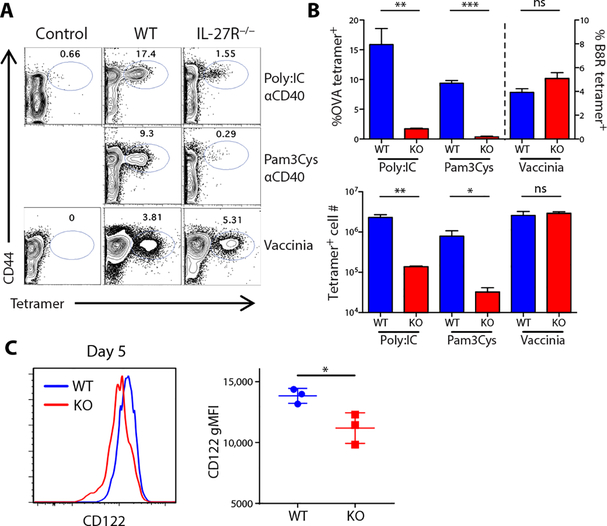
We previously showed that a combination adjuvant consisting of an anti-CD40 antibody and a Toll-like receptor (TLR) agonist could induce CD4+ and CD8+ T cell responses, in both mice (7, 24) and primates (25), on par with those observed to infectious challenges such as LM or vaccinia virus (VV). CD8+ T cell responses to this vaccination are dependent on IL-27R (Fig. 1, A and B) in a T cell–intrinsic manner (7). This is observable as a decrease in either the percentage or total numbers of antigen-specific T cells (Fig. 1B) and contrasts with the response to infectious challenge with VV, which is fully intact in both wild-type (WT) and IL-27R−/− hosts (Fig. 1, A and B). Furthermore, this IL-27 dependency is not limited to the use of combined adjuvants but is also a critical requirement for the T cell response to single-adjuvant vaccinations (7). These results highlight the principle that T cell responses to subunit vaccines and infections exhibit different activation requirements to facilitate maximal responses.
Fig. 1. IL-27 is required for the CD8+ T cell response to adjuvanted subunit vaccination but not to infectious challenge.
(A and B) Tetramer staining on splenic CD8+ T cells from control (unimmunized), C57BL/6J, and IL-27R−/− mice immunized with poly (I:C)/αCD40/OVA or Pam3Cys/αCD40/OVA or infected with 5 × 106 PFU of VV-WR at 7 dpi. ns, not significant. (C) Tetramer-positive T cells were analyzed by flow cytometry for CD122 (IL-2 receptor β-chain) expression 5 days after immunization with poly (I:C)/αCD40/OVA. Data indicate means ± SEM, n ≥ 3 mice per group, representative of five experiments.
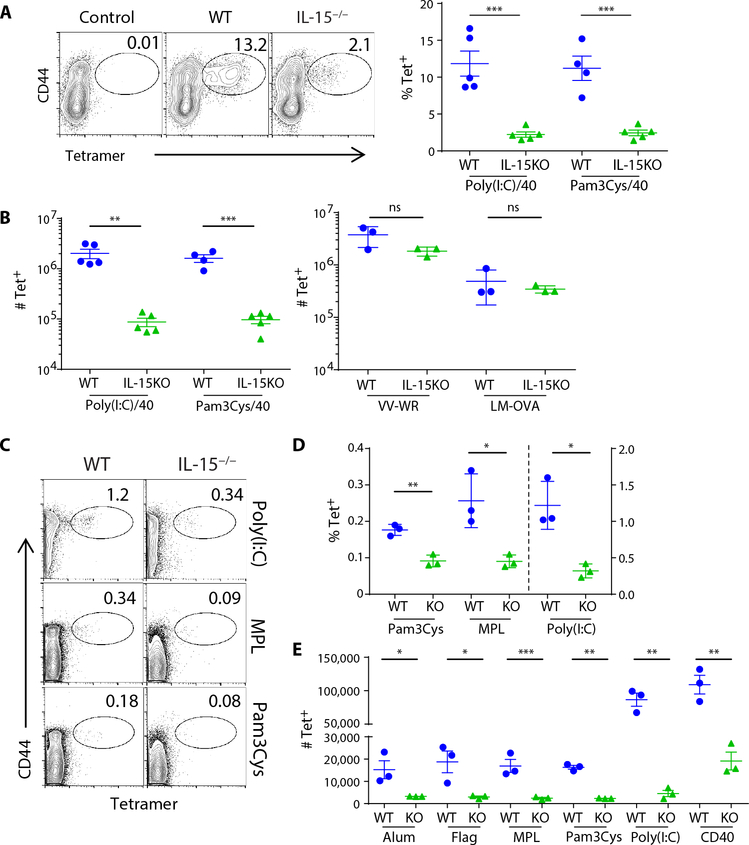
In our previous report, we concluded that IL-27 must be facilitating the survival of the T cells beyond day 3 after vaccination (7). Closer investigation of the cells at day 5, when IL-27R−/− T cells are undergoing attrition and WTT cells are expanding, revealed subs -tantially reduced levels of CD122 expression in the IL-27R−/− T cells (Fig. 1C). CD122 is the β-chain for both IL-2 and IL-15 signaling, suggesting decreased sensitivity of these cells to IL-2 and/or IL-15. Although IL-15 controls cell fate decisions between effector and memory lineages and promotes memory cell homeostasis (19, 21), primary CD8+ T cell responses to infectious challenge are independent of this cytokine (18, 22, 23). However, the loss of CD122 on IL-27R−/− T cells, coupled with the fact that vaccine-elicited T cells already demonstrated a differential dependency on one cytokine (IL-27) compared with infections, led us to hypothesize that IL-15 plays a critical role in vaccine-elicited immunity. Consistent with this hypothesis, there was a substantial deficit in the primary CD8+ T cell response to subunit vaccination in the IL-15−/− host (Fig. 2, A and B). Given this profound impact on the primary T cell response, as well as the established role for IL-15 in maintaining memory CD8+ T cell populations, the memory pool 30 days after subunit vaccination was unsurprisingly similarly compromised in the IL-15−/− host as well (fig. S1). As documented previously, IL-15−/− hosts mount a primary T cell response comparable with WT in response to infectious challenge with either ovalbumin (OVA)–expressing LM (LM-OVA) or VV (Fig. 2B). This dependency on IL-15 was broadly applicable to a host of single adjuvants as well (Fig. 2, C to E), similar to our observations with IL-27 (7). These data indicated that the CD8+ T cell response to a broad spectrum of subunit vaccine adjuvants depends on two cytokines, IL-27 and IL-15, which previous infectious models did not predict.
Fig. 2. IL-15 is required for the generation of the primary CD8+ T cell response to vaccination but not to infectious challenge.
(A and B) The abundance of splenic antigen-specific T cells was determined for WT or IL-15−/− mice 7 days after vaccination with poly(I:C)/αCD40/OVA or Pam3Cys/αCD40/OVA. The percentage of CD8+ T cells that were tetramer-positive (A) and the total number of tetramer-positive T cells (B) are shown for cells responding to immunization, VV-WR, or LM-OVA. (C to E) Similarly, tetramer staining was performed in mice responding to specified single adjuvants plus OVA 7 dpi. Data indicate means ± SEM, n ≥ 3 mice per group, representative of four (A and B) and three (C, D, and E) experiments. Tet, tetramer; Flag, flagellin.
Tbet and Eomes are required for maximal CD8+ T cell responses to adjuvanted subunit vaccination
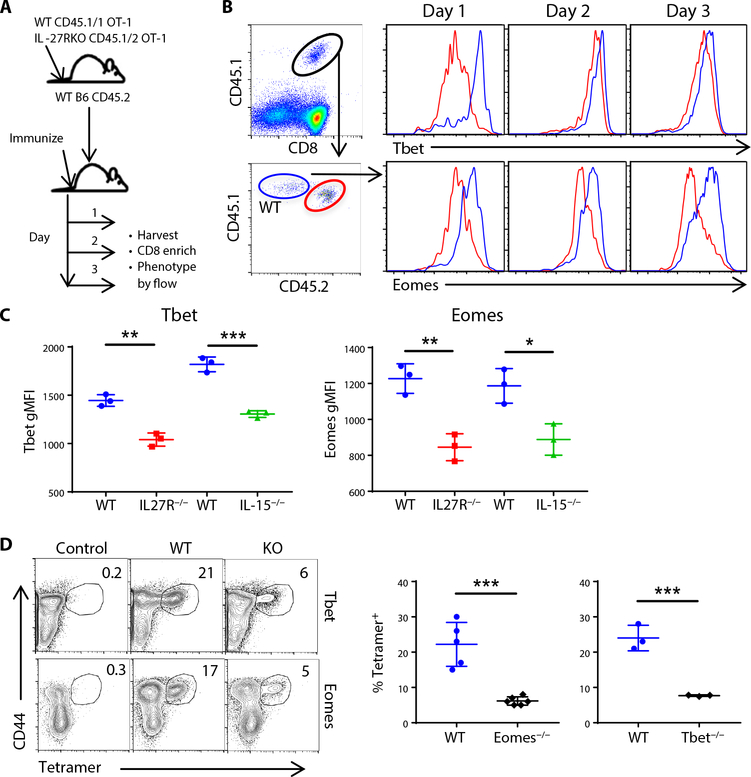
To help study early events after vaccination, we used a coadoptive transfer model, transferring small numbers of congenically marked WT and IL-27R−/− OT-1 T cells [T cell receptor (TCR) transgenic T cells that are OVA-specific] into a naïve recipient (Fig. 3A) and compared their responses at early time points after vaccination. We initially examined the expression of Tbet and Eomes (Fig. 3B) because they are two transcription factors important in T cell differentiation and fate determination, are known to influence CD122 expression, and are downstream of both IL-27 (26–28) and IL-15 (29, 30) signaling. Both Tbet and Eomes expression were substantially reduced in the IL-27R−/− T cells compared with the WT (Fig. 3B), observable as soon as day 1 after vaccination and remaining low through at least day 3 (Fig. 3, B and C). This was not unique to IL-27R−/− T cells, because WTT cells transferred into an IL-15−/− host demonstrated a similar reduction in both Tbet and Eomes during the early phases of T cell activation (Fig. 3C). This expression profile was somewhat unexpected given the documented roles of these transcription factors primarily as driving long-term fate decisions, not early T cell activation and expansion (20). We therefore determined the requirement for each factor by immunizing Tbet−/− or Eomesfl/fl/dLCK-cre mice and compared their T cell responses with WT controls as described above. As with T cells deficient in IL-27 or IL-15 signaling, T cells deficient in either Tbet or Eomes were substantially compromised in their response to combined adjuvant subunit vaccination (Fig. 3D). Thus, much like the cytokines that induce them, these transcription factors play a more critical role in the early activation and expansion of T cells after vaccination than their documented roles after infectious challenge.
Fig. 3. Tbet and Eomes are required for maximal CD8+ T cell responses to adjuvanted subunit vaccination.
(A to C) Congenically marked OT-1 T cells from WT and IL-27R−/− backgrounds were cotransferred into C57BL/6J mice and harvested for flow cytometric analysis 1, 2, and 3 days after vaccination. (A) OT-1 cotransfer method schematic. (B) Flow plots show the gating strategy for WT and IL-27R−/− OT-1 cells and representative Tbet and Eomes staining. (C) The MFI of Tbet and Eomes at day 3 after immunization in WT/IL-27R−/− OT-1 cotransfers and in single transfers of WT OT-1 cells transferred into WT mice or IL-15−/− mice. (D) Tetramer staining was performed on CD8+ T cells in control (unimmunized), C57BL/6J (WT), Tbet KO, or Eomes KO (Eomesfl/fl × dLck-Cre) mice 7 dpi. Data shown are means ± SEM, n ≥ 3 mice per group, representative of three (for Tbet−/−) and four (for Eomesfl/fl × dLck-Cre) experiments.
IL-27 and IL-15 increase IRF4 expression, a required transcription factor for CD8+ T cell responses to adjuvanted subunit vaccination
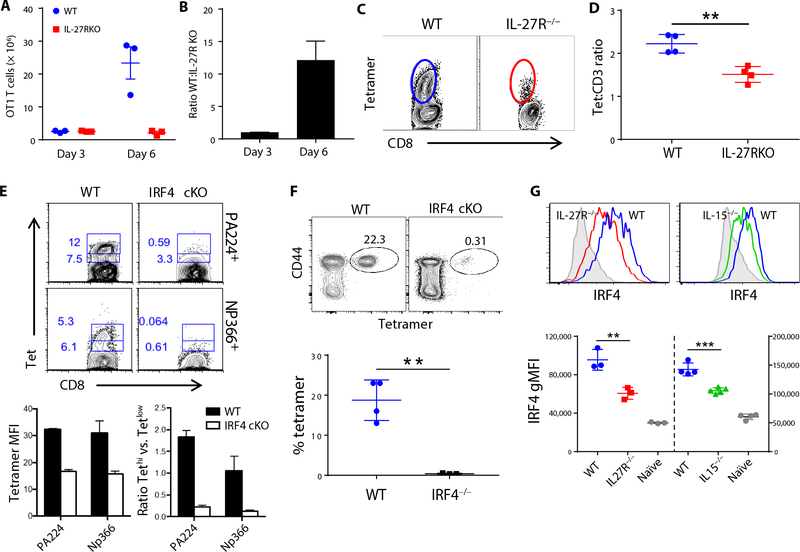
After vaccination, IL-27R−/− T cells initially expand commensurate to their WT counterparts, achieving a 1:1 ratio on day 3 [Fig. 4, A and B, and (7)]. However, by day 6, the ratio is >10:1 in favor of the WT cells (Fig. 4, A and B), a phenomenon also observed in the ratio of WT to IL-15−/− T cells (fig. S2). Carboxyfluorescein diacetate succinimidyl ester labeling and Ki67 staining revealed (respectively) that neither the number of divisions nor duration of proliferation was different between WT and IL-27R−/− T cells (7). This is consistent with a role for IL-27 in mediating the survival of the T cells late after antigen recognition, a function strikingly reminiscent of that described for interferon regulatory factor 4 (IRF4). IRF4 is a transcription factor known to be important in the differentiation and maintenance of a proliferating T cell’s energy consumption (31, 32). In response to infection, IRF4−/− T cells initially proliferate identical to WT T cells but then fail to accumulate at later time points because of their compromised survival. We observed further similarities between IRF4−/− and IL-27R−/− responses in the affinity of the surviving T cells after vaccination/infectious challenge, respectively. IL-27R−/− T cells show greatly reduced tetramer staining [Fig. 4C and (7)], a proxy for TCRs with reduced affinity for peptide:major histocompatibility complex, at late times after vaccination. This difference holds when using a tetramer/CD3 geometric mean fluorescence intensity (gMFI) ratio to correct for any differences in surface levels of TCR [Fig. 4D and (7)]. Likewise, the few IRF4−/− T cells surviving after challenge with influenza also display reduced binding to tetramer compared with controls (Fig. 4E). Given these similarities, we investigated whether the vaccine-elicited response also required IRF4. Using the T cell conditional IRF4−/− (IRF4 cKO) host (32), we found that the vaccine- elicited response was almost completely ablated in the absence of IRF4 expression (Fig. 4F). This indicates that, unlike the other factors described above, IRF4 is a required transcription factor for mediating T cell responses to either infection or subunit vaccination. Furthermore, the expression of this critical transcription factor is influenced by IL-27 and/or IL-15 in the context of vaccination. Three days after vaccination, IRF4 was substantially reduced in both IL-27R−/− and IL-15−/− hosts (Fig. 4G), indicating a dependency on these cytokines for maximal IRF4. IRF4 levels were also significantly lower in cells responding to infection compared with vaccination (fig. S3) where it is known to be linked to the intensity of TCR stimulation (32, 33). However, IRF4 was recently shown to also be influenced by IL-2 and IL-15 in vitro (34), consistent with the results presented here, confirming this connection between IRF4 and IL-15 in an in vivo setting. Therefore, unlike Tbet and Eomes, IRF4 represents an important nexus between vaccine- and infection-elicited responses.
Fig. 4. IL-27 and IL-15 influence IRF4 expression, a required transcription factor for the T cell responses to subunit vaccination.
(A and B) C57BL/6J mice received 5 × 103 purified OT-1 T cells 1 day before vaccination. Total numbers of WT and IL-27R−/− OT-1 cells were analyzed at 3 and 6 days after vaccination (A), and the ratio of WT to IL-27R−/− OT-1 cells was determined for each time point (B). (C and D) Endogenous CD8+ T cells were analyzed for Kb-SIINFEKL tetramer staining 7 days after immunizing C57BL/6J mice (C), and the ratio of tetramer MFI:CD3ε MFI was determined for each of four mice per group (D). (E) C57BL/6J and IRF4 cKO (Irf4fl/fl × CD8α-Cre) mice were challenged with influenza virus and tetramer-stained 7 dpi. Representative plots show distribution of low and high tetramer staining. Graphs show MFI of tetramer on tetramer-positive events (left) and the ratio of tetramer-high versus tetramer-low events (right) in WT and IRF4 cKO mice. (F) C57BL/6J and IRF4 cKO mice were vaccinated and tetramer-stained 7 dpi. Representative plots show the percentage of tetramer staining cells out of total CD8+ T cells. The graph below shows means ± SEM, n = 3 to 4 mice per group. (G) IRF4 staining on tetramer-positive cells from WT, IL-15−/−, and IL-27R−/− mice 3 days after immunization. Data shown are means ± SEM, n = 3 mice per group, representative of three (A and B), five (C and D), two (E and F), and four (G) experiments.
Vaccine-induced T cell responses display a metabolic program characterized by mitochondrial function, not aerobic glycolysis
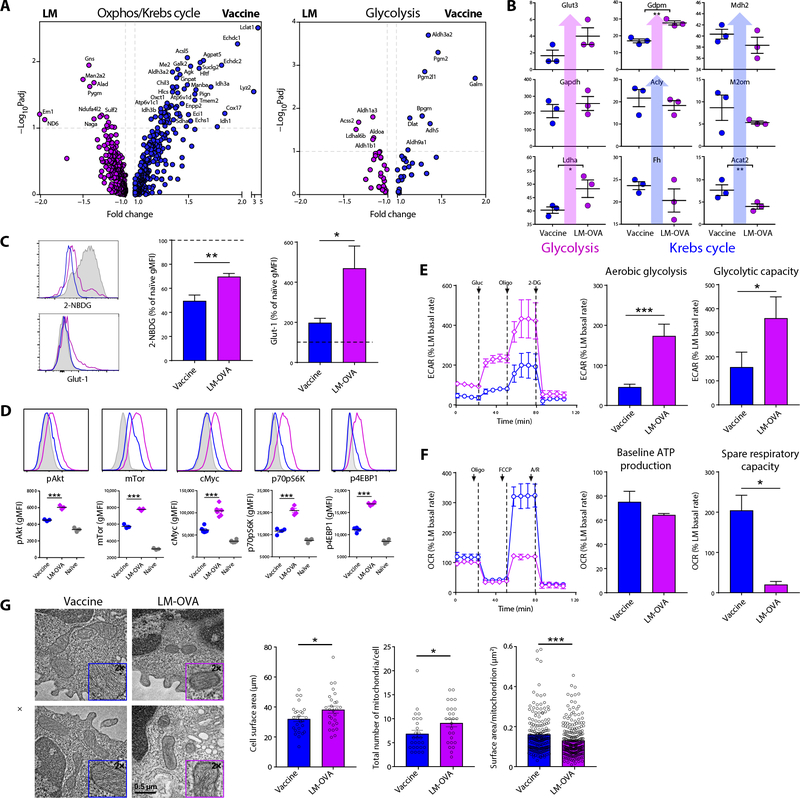
Numerous genes induced by IRF4 are involved in glycolysis and mitochondrial function (31, 35), and the loss of IRF4 leads predominantly to an early loss of glycolytic function in the T cell (31, 35). This role in metabolism is consistent with a requirement for IRF4 specifically in the survival of primary effector cells, because they are known to be heavily dependent on aerobic glycolysis (11). In contrast, memory T cells support their metabolic demands primarily by mitochondrial fatty acid oxidation (14) and are thus less dependent on IRF4. We therefore anticipated that vaccine-induced T cells from WT hosts would display robust, IRF4-mediated glycolytic function, whereas T cells from IL-27–or IL-15–deficient settings would show a substantial loss of glycolytic function, thereby explaining their loss of survival after vaccination. However, transcriptional analyses suggested a reprogramming of T cells responding to vaccination in favor of mitochondrial function, not aerobic glycolysis. OVA-specific T cells responding to vaccination or LM-OVA challenge were isolated by flow sorting and differential gene expression determined by Affymetrix gene array analysis (fig. S4). We filtered the gene expression data using a lower fold change threshold with the rationale that cellular metabolism is a complex process regulated by hundreds of gene products such that smaller fold changes in a large number of related genes would be expected to have a significant biological impact. Whereas genes associated with glycolysis showed no substantial pattern of differential expression, genes associated with the Krebs cycle and oxidative phosphorylation predominated in T cells responding to subunit vaccination (Fig. 5A), revealing a broader underlying genetic program directed not toward glycolysis but to mitochondrial function. These unexpected findings were confirmed at the protein level, showing increases in glycolytic enzymes in T cells responding to LM-OVA but increases in Krebs cycle enzymes in the vaccine-elicited T cells (Fig. 5B). Flow cytometric analysis was consistent with these findings, with vaccine-elicited T cells showing marked reductions in signaling elements critical to glycolytic metabolism such as AKT, the mammalian target of rapamycin (mTOR), cMyc, pS6K, and p4EBP1 (Fig. 5C) (11). In line with previous ex vivo studies (36), glucose uptake [as demonstrated by 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose (2-NBDG) fluorescence] was reduced in both infection- and vaccine-elicited T cells compared with naïve CD8+ T cells (Fig. 5D); nevertheless, glucose uptake in vaccine-elicited T cells was significantly reduced compared with that in infection-elicited T cells. Moreover, they showed lower expression of the glucose transporter Glut-1 (Fig. 5D). To validate these findings at the metabolic level, we performed experiments using the Seahorse metabolic flux analyzer. We initially measured both extracellular acidification rates (ECARs; a measure of aerobic glycolysis) and oxygen consumption rates (OCRs; a measure of mitochondrial function) of WT T cells responding to vaccination or infection with LM-OVA. As expected, T cells responding 5 days after LM challenge were engaged in aerobic glycolysis, in terms of both baseline aerobic glycolysis and glycolytic capacity (Fig. 5E). However, in agreement with the analysis above, subunit vaccine–derived WT T cells showed markedly reduced glycolysis for both of these parameters (Fig. 5E). Instead, these cells had elevated mitochondrial function, showing an SRC orders of magnitude higher than that observed in LM-elicited T cells and similar to what is seen in memory T cells (Fig. 5F). Cell surface marker analyses also indicate that these cells are predominantly programmed toward memory, with most expressing low killer cell lectin-like receptor G1 (KLRG1) and high IL-7 receptor-α (CD127) (fig. S5). To follow up on the functional differences that we observed, we used transmission electron microscopy (TEM) to evaluate mitochondrial morphology. Mitochondrial fission has been associated with highly glycolytic effector T cells, whereas fusion has been observed in memory T cells (37). TEM analysis revealed that cells responding to vaccination were slightly smaller and had fewer mitochondria, and notably, that those mitochondria were larger than their infection counterparts (Fig. 5G). These phenotypes are consistent with mitochondrial fission occurring after infection and mitochondrial fusion occurring after vaccination. Together, these data indicate that CD8+ T cells develop a memory- like metabolic phenotype very early after vaccination.
Fig. 5. Vaccine-induced T cell responses display a metabolic program characterized by mitochondrial function, not aerobic glycolysis.
(A) Seven days after immunization or infection with LM-OVA, relative RNA expression was determined for antigen-specific T cells sorted from C57BL/6J mice. Differentially expressed genes (adjusted P value false discovery rate (FDR) < 0.1 and fold change of 1.1 or greater) were filtered for Gene Ontology Consortium (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway association with electron transport chain, oxidative phosphorylation (Oxphos), tricarboxylic acid cycle and metabolic process, or glycolysis. Gene expression in T cells responding to LM challenge or combined adjuvant vaccine immunization T cells was determined by Affymetrix gene arrays (as described in Materials and Methods) using Transcriptome Analysis Console Software (Thermo Fisher Scientific). All differentially expressed genes (significant between vaccine- and LM-OVA–responding T cells) were next determined with a fold change cutoff at ±1.1. (B) Proteomics analyses performed on OT-1 T cells purified 5 dpi, showing proteins important to glycolysis and the Krebs cycle in vaccine-treated cells versus LM-OVA. (C) Glut-1 expression and 2-NBDG uptake on antigen-specific endogenous CD8+ T cells were tetramer-stained 5 dpi as a percentage of CD44− naïve CD8+ T cell MFI. Data are combined from two experiments, where n = 6 to 7 mice per group. (D) Representative flow plots and average gMFI of pAkt, mTor, cMyc, p70pS6K, and 4EBP1 on OT-1 T cells 5 days after vaccination (blue), infection (magenta), or in CD44− endogenous CD8+ T cells (gray). Data shown are means ± SEM, representative of ≥3 experiments. (E and F) Meta bolic flux was performed on splenic OT-1 T cells purified 5 dpi with LM-OVA, or αCD40/poly(I:C)/OVA, respectively. (E) ECAR was measured at baseline, and after injection with glucose, oligomycin, and 2-DG, at indicated time points, from which baseline aerobic glycolysis and glycolytic capacity were determined. Data shown are combined from four separate experiments, n = 4 per group, where each n value equals the average of all the biological replicates from one of the four experiments. The total numbers of mice included were 22 for LM-OVA and 26 for vaccine. (F) OCR was measured at baseline and after injection of oligomycin, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), and antimycin and rotenone (A/R). From this, baseline ATP production and SRC were determined. Data shown are combined from three separate experiments, n = 3 per group, where each n value equals the average of all the biological replicates from one of the three experiments. Ten mice (LM-OVA) or 14 (vaccine) mice were included. Ratio paired t tests were used to determine significance for (E) and (F). (G) OT-1 T cells were purified 5 dpi and analyzed by TEM. Representative TEM images are shown. Scale bar, 0.5 μm. Cell surface area and the total number of mitochondria were quantified from 27 cells per group. The surface area of each mitochondrion from each cell was also quantified.
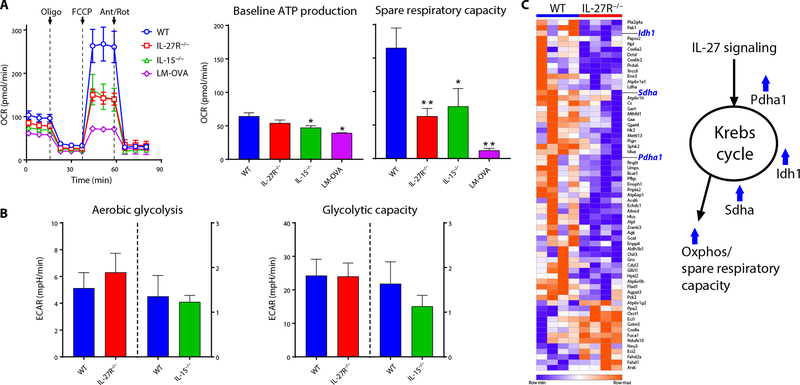
These data were in contrast to our initial expectations, suggesting instead that IL-27 and IL-15 were needed to support mitochondrial function, not glycolysis, in the vaccine-induced response. In keeping with this new prediction, T cells responding in the absence of IL-27 or IL-15 signaling had substantially reduced mitochondrial function (Fig. 6A) with no compensatory increase in glycolytic function (Fig. 6B). RNA sequencing (RNA-seq) analysis provided substantial transcriptional support for these observations (Fig. 6C and fig. S6); transcript levels of several key Krebs cycle enzymes were significantly lower in IL-27R−/− T cells compared with WT, including pyruvate dehydrogenase (Pdha), isocitrate dehydrogenase (Idh1), and succinate dehydrogenase (Sdha). Collectively, these data indicate that whereas T cell responses to infection display the canonical signatures of being fueled by aerobic glycolysis, T cells responding to subunit vaccination show little glycolytic activity. Instead, they display robust mitochondrial function dependent on IL-27 and IL-15. It is important to note that the T cells at the time point of our analysis are still performing cellular division every 4 to 6 hours. Elevated mitochondrial function is well documented for memory T cells (14), but this is after the cells have exited the process of clonal expansion and thus no longer require the metabolic intermediates derived from aerobic glycolysis for generating reducing power and biomass. Although glycolysis was previously shown to be unnecessary to support in vitro activation and proliferation (38), our in vivo demonstration that vaccine-elicited T cells appear to be using mitochondrial function to support such marked clonal expansion is unprecedented.
Fig. 6. Vaccine-induced augmentation of SRC is dependent on IL-27 and IL-15.
WT OT-1 T cells were adoptively transferred into C57BL/6J or IL-15−/− recipients (WT, IL-15−/−), or IL-27R−/− OT-1 T cells were transferred into C57BL/6J recipients (IL-27R−/−) and vaccinated 1 day later. Alternatively, WT OT-1 T cells were transferred into C57BL/6J recipients and infected 1 day later (LM-OVA). OCR and ECAR were measured ex vivo on purified OT-1 cells 5 dpi (A and B). Ant/Rot, antimycin and rotenone. Baseline ATP production, SRC, aerobic glycolysis, and glycolytic capacity were determined as in Fig. 5. RNA-seq on WT and IL-27R−/− OT-1 T cells responding to subunit vaccination was performed as described in Materials and Methods (C). The resulting gene list was filtered for all differentially expressed genes (adjusted P value FDR < 0.1) with a fold change of 1.15 or greater and that are associated with the GO metabolic pathways metabolic process, trixarboxylic acid cycle, and/or electron transport chain (GO:0008152, GO:0006099, and GO:0022900). Heat map was generated using Morpheus (Broad Institute). For OCR, data are combined from two experiments, where n = 6 mice per group (all vaccination groups) and n = 3 mice (infection), and values are means ± SEM. For ECAR, data are representative from a total of three experiments each, where n = 7 (WT versus IL-27R−/−) or n = 3 (WT versus IL-15−/−).
Glycolytic blockade reduces T cell response to infection but not to subunit vaccination
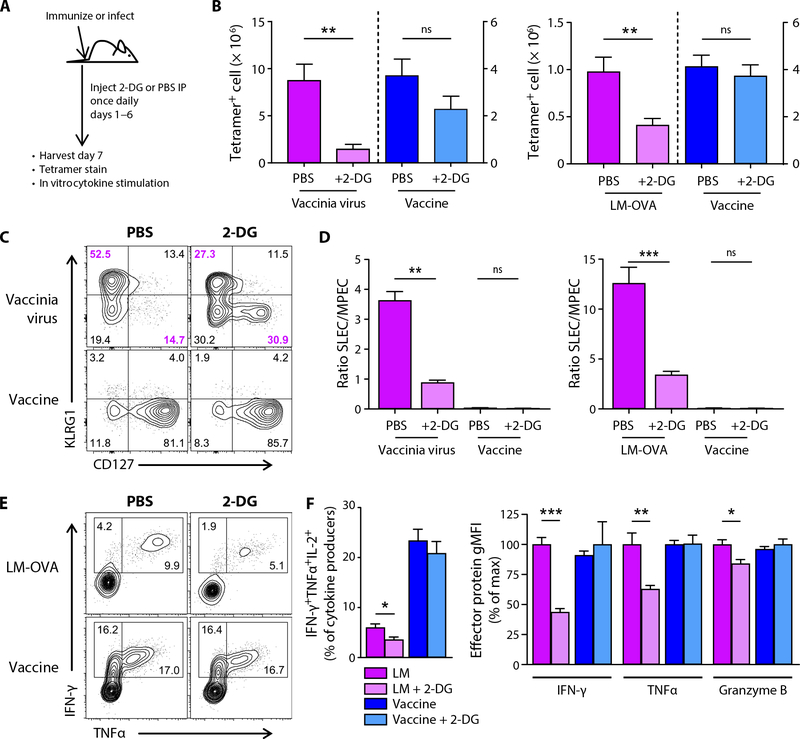
These data suggest that the energy and biomass demands for T cells responding to subunit vaccination are met largely by mitochondrial function. To specifically address this, we challenged mice with either subunit vaccination or infection, with or without concomitant injection of 2-deoxyglucose (2-DG), a competitive inhibitor for the glycolytic process. Mice were treated with 2-DG on days 1 to 6 after initial vaccination/viral challenge, and the peak of the T cell response was measured at day 7 (Fig. 7A). As expected, 2-DG treatment markedly reduced the magnitude of the T cell response to VV or LM-OVA challenge (Fig. 7B), confirming the glycolytic requirement for the clonal expansion of infection-specific responses. In contrast, 2-DG treatment failed to have any meaningful impact on the response to vaccination (Fig. 7B). Previous data showed that inhibition of glycolysis with rapamycin facilitates a skewing of a virus- specific response toward memory phenotype cells (39) and that glucose withdrawal inhibits cellular function, specifically interferon-γ (IFN-γ) production (38). We therefore investigated whether 2-DG had the same influence on the infection- and vaccine-specific responses. Similar to rapamycin, treatment with 2-DG coincided with a decrease in the percentage of CD127−KLRG1+ SLECs and a corresponding increase in CD127+KLRG1− memory precursor effector cells (MPECs) 7 days after viral or bacterial challenge (Fig. 7, C and D). In contrast, the T cells responding to vaccination display an overwhelmingly MPEC phenotype early, culminating in the vast majority of the T cells being memory phenotype at the peak of the response, a phenotype on which 2-DG had no impact (Fig. 7, C and D) (40). Consistent with previous reports on the importance of glycolysis for maximal cytokine production (38) and lytic activity (41), glycolysis blockade with 2-DG also resulted in reduced polyfunctional cytokine production after infection with LM-OVA (Fig. 7, E and F) and reduced production of IFN-γ, tumor necrosis factor–α (TNFα), and granzyme B (Fig. 7F). However, much like the proliferative response, neither cytokine production nor granzyme expression by vaccine- elicited cells was affected. These data are again consistent with the conclusion that the magnitude, phenotype, and function of the T cell response to vaccination are minimally affected by glycolysis and its downstream effects. Collectively, these studies confirm that vaccine- elicited T cell responses derive their metabolic support largely from the mitochondria, a function that requires the participation of IL-27 and IL-15, thereby explaining why the vaccine-elicited response is dependent on these cytokines.
Fig. 7. Glycolytic blockade reduces T cell response to infection but not to subunit vaccination.
WT C57BL/6J mice were either immunized, infected with 1 × 107 PFU of VV-OVA, or infected with 2 × 103 CFU of LM-OVA. Starting 1 day later, mice were injected intraperitoneally (IP) with PBS or 2-DG daily. (A) Experimental schematic. (B) Spleens were harvested at 7 dpi, and tetramer staining was performed for Kb-TSYKFESV (B8R) or Kb-SIINFEKL (OVA) tetramer-positive cells in VV-WR–infected and vaccinated mice (left) or Kb-SIINFEKL tetramer-positive cells in LM-OVA–infected or vaccinated mice (right). Shown are means ± SEM from a representative experiment (VV) or two experiments combined (LM-OVA) of ≥3 experiments each, where n = 4 to 5 mice per group. The relative percentages of SLEC and MPEC subsets were assessed in tetramer-positive events (C), and the ratios of the number of SLECs to the number of MPECs were determined (D). Splenocytes were also stained 7 dpi for granzyme B and for intracellular cytokines after 5 hours of stimulation with SIINFEKL peptide (E). (F) Percentage of IFN-γ+TNFα+IL-2+ within all CD8+ T cells, making any one or more of these cytokines (left). Relative quantity of IFN-γ, TNFα, or granzyme B produced by CD8+ T cells, making the respective effector proteins normalized to percentage of max for LM or vaccine, respectively, for each protein (right). Percentages shown (C and E) are the averages from four to five mice per group.
DISCUSSION
Since Otto Warburg first described the fermentation of glucose in cancer cells in the presence of sufficient levels of oxygen to maintain oxidative phosphorylation (42), scientists have noted the so-called Warburg effect in rapidly dividing cell types, including in T cells. Various hypotheses have attempted to explain why cells transition toward aerobic glycolysis, though none have been universally accepted. Despite its inefficiency at producing ATP, aerobic glycolysis may meet the energy demands of tumors and effector T cells because nutrients (including glucose) are abundant owing to constant exposure to circulating blood, such that even inefficient production of ATP still meets the cells’ energy needs (12). In addition, ATP may not even be a limiting factor, given that the energy requirements of rapidly dividing cells may be lower than quiescent cells, not higher (43). That said, whether rapid division per se requires Warburg metabolism remains unclear. A leading explanation for why rapidly dividing cells use aerobic glycolysis is that glucose catabolism is necessary for meeting the increased demand for biomass (12, 44). Multiple biosynthetic pathways branch off from glycolysis, including the pentose phosphate and serine biosynthesis pathways, which contribute to the generation of nucleotides and amino acids. The final conversion of pyruvate to lactate is said to be important because of its production of β-nicotinamide adenine dinucleotide (NAD+), a limiting factor for additional glycolysis (12, 15). However, as Liberti and Locasale point out, the production of lactate is mutually exclusive with bio-mass generation—the carbons contained within glucose leave the cell when lactate is excreted (17). Furthermore, the shunting of glucose- derived pyruvate into the Krebs cycle allows for the generation of amino acids not accessible from early glycolytic intermediates alone. Thus, although it is clear that some tumor cells require glucose for their high growth rates and most produce large quantities of lactate, no claim can reasonably be made that rapid cellular proliferation demands aerobic glycolysis.
Our studies of vaccine-elicited T cells provide an example of cells undergoing clonal expansion in vivo independent of aerobic glycolysis. Metabolic flux assays confirmed that infection induced high rates of aerobic glycolysis in T cells, whereas vaccination induced a level of aerobic glycolysis barely above the limit of detection. Vaccination instead imbues T cells with a memory T cell–like metabolic program characterized by a high SRC that is unusual in its dependence on IL-27 and IL-15. Inhibiting glycolysis by in vivo administration of 2-DG had no effect on the generation of a robust CD8+ T cell response to vaccination while severely limiting the magnitude of the response to infection (Fig. 7). Restricting glycolysis significantly skewed the phenotype of infection-elicited T cells away from SLECs toward MPECs, consistent with previous observations (39, 45, 46), but unsurprisingly left the overwhelmingly MPEC phenotype of vaccine-elicited T cells unaffected. Using glucose-free media, Chang and colleagues demonstrated that glycolysis was not required for the activation and rapid proliferation of T cells in vitro but was necessary to support the development of effector functions (38). However, we (7, 24, 25, 40) and others (47–49) have extensively documented the robust effector function of T cells derived from the combined adjuvant method of vaccination used here. This indicates that the previously observed dependency of effector function on glycolysis is either an in vitro observation or one that does not apply to vaccine-elicited responses. The data collectively call into question the existing dogma that aerobic glycolysis in rapidly dividing cells is critical to fuel the anabolic processes necessary for their proliferation and differentiation, and further underscore the divergence in T cell programming adopted by cells responding to infection and vaccination. Recent data from Goldrath and colleagues revealed that mitochondrial function could be uncoupled from the formation of T cell memory (50), and the data that we present here show an analogous decoupling of aerobic glycolysis from the metabolic demands of primary T cell clonal expansion.
We have used a broad array of vaccine adjuvants in the present work and therefore propose that our conclusions are broadly applicable to most adjuvants in current use. New adjuvants may be identified in the future that induce cellular immunity through a mechanism of action divergent from the cytokine and metabolic dependencies that we have herein uncovered. Furthermore, we have yet to confirm or deny the IL-27 and IL-15 dependency of vaccine- elicited cellular immunity in higher primates, and therefore, some degree of species- related differences may also be found. That said, the implications of these data are significant because they apply to immuno-oncology and therapeutic vaccination. Because the success of the current checkpoint blockade immunotherapies [e.g., PD-1 (programmed cell death-1) or CTLA-4 (cytotoxic T-lymphocyte associated protein 4)] depends on their capacity to induce robust and enduring T cell function (51), the addition of vaccine strategies that can elicit T cells capable of functioning in the highly suppressive tumor microenvironment would be of obvious therapeutic benefit. T cell function is suppressed in the tumor microenvironment, in part, because these high rates of aerobic glycolysis by the tumor cells deprive effector T cells of their primary fuel source (52, 53). In addition, T cells within the tumor display a significant loss of mitochondrial mass and function, further contributing to antitumor immune dysfunction (54, 55). However, and directly related to our findings here, antitumor T cell function could be substantially enhanced by increasing mitochondrial function and biogenesis through ectopic PGC1α (PPARγ coactivator 1α) expression (54) or by specifically increasing fatty acid catabolism by pre-treatment of transferred T cells with fenofibrate (55). Here, we reveal that subunit vaccine–elicited T cells already have increased mitochondrial function with little dependence on glucose, indicating that they may be well suited for promoting antitumor immunity, given the notoriously low glucose availability in the tumor microenvironment.
The precise reason(s) for these extreme differences in phenotype and metabolic dependency are not fully clear, though it is reasonable to hypothesize that differences in the degree and duration of inflammation are responsible. Tbet and Blimp1 expression are downstream of inflammatory factors and are well established as drivers of SLEC formation (56, 57). Given the lack of SLEC formation in response to vaccination (fig. S5 and Fig. 7), one might predict that vaccine-elicited T cells show reduced Blimp1 and Tbet expression. Curiously, at the peak of the vaccine response, the expression of both Blimp1 (40) and Tbet (fig. S7) are actually higher than that induced by infection. This suggests that an MPEC phenotype is acquired in response to vaccination despite high levels of these transcription factors. Reiner and colleagues recently showed an association between phosphatidylinositol 3-kinase (PI3K)/AKT activity and skewing toward an effector cell fate (58). Because PI3K/AKT is a signaling component of a variety of inflammatory mediators (59), the levels of inflammation present during infection would be expected to skew toward SLECs through PI3K/AKT activation of mTOR (60) and phosphorylation of forkhead box protein O1 (61), exporting it from the nucleus where it helps to maintain a commitment to a memory cell fate through Tcf7 expression. Our data are consistent with these previous findings, with LM-elicited T cells expressing substantially higher levels of pAKT and mTOR (Fig. 5). Likewise, high levels of inflammation are largely absent after adjuvant administration; our recent use of an IL-27p28-GFP reporter host revealed that production of this cytokine so critical to vaccine-elicited T cell immunity was only produced in the first 6 to 18 hours after vaccination (62). It may be that this limited amount of inflammation does not induce sufficient PI3K/AKT activation to support an effector cell phenotype or metabolic fate, instead driving a memory phenotype and metabolic program that require IL-27 and IL-15.
Last, it is worth emphasizing the growing list of factors with differential functions in the T cell response to subunit vaccination or infection, a list that now includes IL-15, Tbet, and Eomes in addition to IL-27. The unusual dependence of the vaccine-elicited T cell response on these factors, and the relative indifference with which the infection-elicited response proceeds in their absence, further supports the assertion that basic immune mechanisms derived from infectious biology overlap poorly with signals critical for subunit vaccine–derived cellular immunity (8). These data suggest that approaching subunit vaccination, at least as it pertains to the generation of T cell immunity, as an area of immunological study distinct and independent from infectious biology may better advance our development of vaccines and adjuvants capable of generating clinically relevant and therapeutic T cell immunity.
MATERIALS AND METHODS
Study design
Broadly, the goal of these studies was to identify cellular and molecular mechanisms by which a robust vaccine-elicited cellular response is generated and to what degree those mechanisms are held uniquely or in common from a response instigated by an infectious agent. This was accomplished by evaluating either the endogenous CD8+ T cell response or the response of a small number of transferred congenically marked transgenic T cells, at various time points after either subunit vaccination or infectious challenge. Comparisons between the response to vaccination and infection in WT hosts and the response to vaccination in WT and genetically modified hosts formed the basis for the conclusions drawn.
Experimental models
All experiments involving mice were conducted following protocols approved by the University of Colorado Denver Institutional Animal Care and Use Committee (IACUC) or the Mayo Clinic IACUC according to guidelines provided by the Association for Assessment and Accreditation of Laboratory Animal Care. Female C57BL/6J, congenic B6 Ly5.1 (CD45.1), CD45.2+ OVA-specific TCR-transgenic (OT-1), and Tbet−/− (B6.129S6-Tbx21tm1Glm/J) mice were obtained from the Jackson Laboratory. IL-27Rα−/− mice were provided by Genentech. IL-15−/− (C57BL/6NTac-IL15tm1Imx N5) mice were obtained from Taconic. Eomesfl/fl mice were provided by E. J. Wherry (20). dLck-Cre mice were provided by M. J. Bevan, originally from N. Killeen (63). Irf4fl/fl CD8α-Cre (32) mice were bred and maintained at the Mayo Clinic. CD45.2+ and CD45.1+ OVA-specific TCR- transgenic (OT-1), IL-27Rα−/− CD45.1+ OT-1, and Eomesfl/fl dLCK-Cre mice were bred and maintained at the University of Colorado Denver. Mice used were aged 6 to 12 weeks old, group-housed at a temperature range of 21° to 23°C, with a 14-hour on and 10-hour off light cycle, and fed an irradiated standard diet (2920X; Envigo).
Infections, immunizations, and treatments
Female mice were challenged via tail vein injections with either 2000 colony-forming units (CFU) per mouse LM expressing whole OVA (LM-OVA) or 5 × 106 plaque-forming units (PFU) per mouse VV Western Reserve (VV-WR) strain or the same strain expressing OVA (VV-OVA). For influenza virus infection, WT (IRF4fl/fl) and IRF4 conditional knockout (cKO; CD8-cre IRF4fl/fl) mice were infected with influenza A/PR8/34 strain (~200 PFU per mouse). Influenza was inoculated in anesthetized mice through intranasal route as described before (64). Data were analyzed in lung CD8+ T cells at day 9 after influenza infection. Mice were immunized via tail vein injection or intraperitoneal injection with the indicated innate receptor agonist, with or without αCD40 antibody clone FGK4.5 (BioXcell), and 100 to 150 μg of whole chicken OVA (Sigma). OVA protein was detoxified by phase separation (65) and was lipopolysaccharide- free as determined by a limulus assay. The following adjuvant doses were used for immunizations: Poly I:C/αCD40 (80 μg/40 μg), Pam3Cys/αCD40 (25 μg/50 μg), Poly I:C (50 μg), Pam3Cys (25 μg), flagellin (15 μg), monophosphoryl lipid A (MPL; 40 μg), and Alum (50 μg). Treatment with 2-DG (1 g/kg) or vehicle control [phosphate- buffered saline (PBS)] was administered by daily intraperitoneal injection at 1 to 6 days post-infection/immunization (dpi).
Statistical analysis
GraphPad Prism (version 7.0c, GraphPad) was used for all statistical analyses. Experiments were performed independently at least twice with a minimum of three mice per group. Figure legends detail number of experimental replicates and n values. Unless noted, data shown are means ± SEM. Significance was defined by unpaired Student’s t tests unless specified. *P ≤ 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Acknowledgments:
We thank J. Bourne and the Electron Microscopy Center for assistance with the preparation and imaging of samples on the TEM. We thank E. Kitten and the Human Immune Monitoring Shared Resource for help with cell sorting. We also thank the Mass Spectrometry–based Proteomics facility and M. Dzieciatkowska for technical assistance with proteomics analyses.
Funding: This work was supported by National Institute of Allergy and Infectious Diseases training grant T32-AI074491–09 (J.K.) and NIH grants AI066121 (R.M.K.) and AI126899 (R.M.K. and C.A.H.).
Footnotes
SUPPLEMENTARY MATERIALS
immunology.sciencemag.org/cgi/content/full/3/27/eaas9822/DC1
Methods
Fig. S1. In response to subunit vaccination, the formation of T cell memory is substantially reduced in an IL-15−/− host.
Fig. S2. IL-15 is required to support survival of vaccine-elicited T cells, not for their initial expansion to antigenic challenge.
Fig. S3. Vaccine-elicited T cells express levels of IRF4 substantially higher than in T cells responding to viral challenge.
Fig. S4. Vaccine- and infection-elicited T cells express both overlapping and unique gene sets when compared with naïve T cells.
Fig. S5. Vaccine-elicited T cells express predominantly a central memory phenotype as compared with T cells responding to infection.
Fig. S6. Global gene expression differences and representative gene sets in WT versus IL-27R−/− T cells responding to subunit vaccination.
Fig. S7. Tbet expression in vaccine-elicited T cells from WT mice is elevated compared with WT T cells responding to virus infection.
Table S1. Raw data.
Competing interests: R.M.K. is a founder of ImmuRx Inc., a vaccine company for which intellectual property is based on the combined TLR agonist/anti-CD40 immunization platform.
REFERENCES AND NOTES
- 1.Amanna IJ, Carlson NE, Slifka MK, Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med 357, 1903–1915 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Amanna IJ, Slifka MK, Crotty S, Immunity and immunological memory following smallpox vaccination. Immunol. Rev 211, 320–337 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Bosio CM, MacLeod M, Marrack P, Kedl RM, Utility of mouse models in vaccine design and development, in Vaccinology: Principles and Practice, Morrow WJW, Sheikh NA, Schmidt CS, Davies DH, Eds. (Wiley-Blackwell, 2012), pp. 94–109. [Google Scholar]
- 4.Curtsinger JM, Mescher MF, Inflammatory cytokines as a third signal for T cell activation. Curr. Opin. Immunol 22, 333–340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM, Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffman RL, Sher A, Seder RA, Vaccine adjuvants: Putting innate immunity to work. Immunity 33, 492–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pennock ND, Gapin L, Kedl RM, IL-27 is required for shaping the magnitude, affinity distribution, and memory of T cells responding to subunit immunization. Proc. Natl. Acad. Sci. U.S.A 111, 16472–16477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennock ND, Kedl JD, Kedl RM, T cell vaccinology: Beyond the reflection of infectious responses. Trends Immunol 37, 170–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.C. A. Hunter, R. Kastelein, Interleukin-27: Balancing protective and pathological immunity. Immunity 37, 960–969 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida H, Hunter CA, The immunobiology of interleukin-27. Annu. Rev. Immunol 33, 417–443 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Buck MD, O’Sullivan D, Pearce EL, T cell metabolism drives immunity. J. Exp. Med 212, 1345–1360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vander Heiden MG, Cantley LC, Thompson CB, Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang L-S, Jones RG, Choi Y, Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Windt GJW, Everts B, Chang C-H, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL, Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lunt SY, Vander Heiden MG, Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol 27, 441–464 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Yoon H, Kim TS, Braciale TJ, The cell cycle time of CD8+ T cells responding in vivo is controlled by the type of antigenic stimulus. PLOS ONE 5, e15423 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberti MV, Locasale JW, The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci 41, 211–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manjunath N, Shankar P, Wan J, Weninger W, Crowley MA, Hieshima K, Springer TA, Fan X, Shen H, Lieberman J, von Andrian UH, Effector differentiation is not prerequisite for generation of memory cytotoxic T lymphocytes. J. Clin. Invest 108, 871–878 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubinstein MP, Lind NA, Purton JF, Filippou P, Best JA, McGhee PA, Surh CD, Goldrath AW, IL-7 and IL-15 differentially regulate CD8+ T-cell subsets during contraction of the immune response. Blood 112, 3704–3712 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, DeJong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL, Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science 321, 408–411 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sprent J, Turnover of memory-phenotype CD8+ T cells. Microbes Infect 5, 227–231 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Becker TC, Wherry EJ, Boone D, Murali-Krishna K, Antia R, Ma A, Ahmed R, Interleukin 15 is required for proliferative renewal of virus-specific memory CD8 T cells. J. Exp. Med 195, 1541–1548 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yajima T, Yoshihara K, Nakazato K, Kumabe S, Koyasu S, Sad S, Shen H, Kuwano H, Yoshikai Y, IL-15 regulates CD8+ T cell contraction during primary infection. J. Immunol 176, 507–515 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Ahonen CL, Doxsee CL, McGurran SM, Riter TR, Wade WF, Barth RJ, Vasilakos JP, Noelle RJ, Kedl RM, Combined TLR and CD40 triggering induces potent CD8+ T cell expansion with variable dependence on type I IFN. J. Exp. Med 199, 775–784 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson EA, Liang F, Lindgren G, Sandgren KJ, Quinn KM, Darrah PA, Koup RA, Seder RA, Kedl RM, Loré K, Human anti-CD40 antibody and poly IC:LC adjuvant combination induces potent T cell responses in the lung of nonhuman primates. J. Immunol 195, 1015–1024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hibbert L, Pflanz S, de Waal Malefyt R, Kastelein RA, IL-27 and IFN-α signal via Stat1 and Stat3 and induce T-Bet and IL-12Rβ2 in naive T cells. J. Interferon Cytokine Res 23, 513–522 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Kamiya S, Owaki T, Morishima N, Fukai F, Mizuguchi J, Yoshimoto T, An indispensable role for STAT1 in IL-27-induced T-bet expression but not proliferation of naive CD4+ T cells. J. Immunol 173, 3871–3877 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Mayer KD, Mohrs K, Reiley W, Wittmer S, Kohlmeier JE, Pearl JE, Cooper AM, Johnson LL, Woodland DL, Mohrs M, Cutting edge: T-bet and IL-27R are critical for in vivo IFN-γ production by CD8 T cells during infection. J. Immunol 180, 693–697 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL, Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol 6, 1236–1244 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Klose CSN, Blatz K, d’Hargues Y, Hernandez PP, Kofoed-Nielsen M, Ripka JF, Ebert K, Arnold SJ, Diefenbach A, Palmer E, Tanriver Y, The transcription factor T-bet is induced by IL-15 and thymic agonist selection and controls CD8αα+ intraepithelial lymphocyte development. Immunity 41, 230–243 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, Nutt SL, Kallies A, The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol 14, 1155–1165 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Yao S, Buzo BF, Pham D, Jiang L, Taparowsky EJ, Kaplan MH, Sun J, Interferon regulatory factor 4 sustains CD8+ T cell expansion and effector differentiation. Immunity 39, 833–845 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nayar R, Enos M, Prince A, Shin H, Hemmers S, Jiang J.-k., Klein U, Thomas CJ, Berg LJ, TCR signaling via Tec kinase ITK and interferon regulatory factor 4 (IRF4) regulates CD8+ T-cell differentiation. Proc. Natl. Acad. Sci. U.S.A 109, E2794–E2802 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang S, Shen Y, Pham D, Jiang L, Wang Z, Kaplan MH, Zhang G, Sun J, IRF4 modulates CD8+ T cell sensitivity to IL-2 family cytokines. Immunohorizons 1, 92–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahnke J, Schumacher V, Ahrens S, Käding N, Feldhoff LM, Huber M, Rupp J, Raczkowski F, Mittrücker H-W, Interferon regulatory factor 4 controls TH1 cell effector function and metabolism. Sci. Rep 6, 35521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bengsch B, Johnson AL, Kurachi M, Odorizzi PM, Pauken KE, Attanasio J, Stelekati E, McLane LM, Paley MA, Delgoffe GM, Wherry EJ, Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8+ T cell exhaustion. Immunity 45, 358–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buck MD, O’Sullivan D, Klein Geltink RI, Curtis JD, Chang C-H, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJW, Chen Q, Huang SC-C, O’Neill CM, Edelson BT, Pearce EJ, Sesaki H, Huber TB, Rambold AS, Pearce EL, Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang C-H, Curtis JD, Maggi LB Jr., Faubert B, Villarino AV, O’Sullivan D, Huang SC-C, van der Windt GJW, Blagih J, Qiu J, Weber JD, Pearce EJ, Jones RG, Pearce EL, Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R, mTOR regulates memory CD8 T-cell differentiation. Nature 460, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards LE, Haluszczak C, Kedl RM, Phenotype and function of protective, CD4-independent CD8 T cell memory. Immunol. Res 55, 135–145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mah AY, Rashidi A, Keppel MP, Saucier N, Moore EK, Alinger JB, Tripathy SK, Agarwal SK, Jeng EK, Wong HC, Miller JS, Fehniger TA, Mace EM, French AR, Cooper MA, Glycolytic requirement for NK cell cytotoxicity and cytomegalovirus control. JCI Insight 2, e95128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warburg O, Wind F, Negelein E, The metabolism of tumors in the body. J. Gen. Physiol 8, 519–530 (1927). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Epstein T, Xu L, Gillies RJ, Gatenby RA, Separation of metabolic supply and demand: Aerobic glycolysis as a normal physiological response to fluctuating energetic demands in the membrane. Cancer Metab 2, 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boroughs LK, DeBerardinis RJ, Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol 17, 351–359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Araki K, Youngblood B, Ahmed R, The role of mTOR in memory CD8+ T-cell differentiation. Immunol. Rev 235, 234–243 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, Roychoudhuri R, Palmer DC, Muranski P, Karoly ED, Mohney RP, Klebanoff CA, Lal A, Finkel T, Restifo NP, Gattinoni L, Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest 123, 4479–4488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho H-I, Celis E, Optimized peptide vaccines eliciting extensive CD8 T-cell responses with therapeutic antitumor effects. Cancer Res 69, 9012–9019 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hailemichael Y, Dai Z, Jaffarzad N, Ye Y, Medina MA, Huang X-F, Dorta-Estremera SM, Greeley NR, Nitti G, Peng W, Liu C, Lou Y, Wang Z, Ma W, Rabinovich B, Sowell RT, Schluns KS, Davis RE, Hwu P, Overwijk WW, Persistent antigen at vaccination sites induces tumor-specific CD8+ T cell sequestration, dysfunction and deletion. Nat. Med 19, 465–472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, Franci C, Cheung TK, Fritsche J, Weinschenk T, Modrusan Z, Mellman I, Lill JR, Delamarre L, Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Phan AT, Doedens AL, Palazon A, Tyrakis PA, Cheung KP, Johnson RS, Goldrath AW, Constitutive glycolytic metabolism supports CD8+ T cell effector memory differentiation during viral infection. Immunity 45, 1024–1037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen DS, Mellman I, Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Ho P-C, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui Y-C, Cui G, Micevic G, Perales JC, Kleinstein SH, Abel ED, Insogna KL, Feske S, Locasale JW, Bosenberg MW, Rathmell JC, Kaech SM, Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho P-C, Kaech SM, Reenergizing T cell anti-tumor immunity by harnessing immunometabolic checkpoints and machineries. Curr. Opin. Immunol 46, 38–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, Ferris RL, Delgoffe GM, The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 701–703 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Kurupati R, Liu L, Zhou XY, Zhang G, Hudaihed A, Filisio F, Giles-Davis W, Xu X, Karakousis GC, Schuchter LM, Xu W, Amaravadi R, Xiao M, Sadek N, Krepler C, Herlyn M, Freeman GJ, Rabinowitz JD, Ertl HCJ, Enhancing CD8+ T cell fatty acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy. Cancer Cell 32, 377–391.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stelekati E, Shin H, Doering TA, Dolfi DV, Ziegler CG, Beiting DP, Dawson L, Liboon J, Wolski D, Ali M-AA, Katsikis PD, Shen H, Roos DS, Haining WN, Lauer GM, Wherry EJ, Bystander chronic infection negatively impacts development of CD8+ T cell memory. Immunity 40, 801–813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xin A, Masson F, Liao Y, Preston S, Guan T, Gloury R, Olshansky M, Lin J-X, Li P, Speed TP, Smyth GK, Ernst M, Leonard WJ, Pellegrini M, Kaech SM, Nutt SL, Shi W, Belz GT, Kallies A, A molecular threshold for effector CD8+ T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat. Immunol 17, 422–432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin W-HW, Adams WC, Nish SA, Chen Y-H, Yen B, Rothman NJ, Kratchmarov R, Okada T, Klein U, Reiner SL, Asymmetric PI3K signaling driving developmental and regenerative cell fate bifurcation. Cell Rep 13, 2203–2218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, Tsatsanis C, Akt signaling pathway in macrophage activation and M1/M2 polarization. J. Immunol 198, 1006–1014 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Finlay D, Cantrell DA, Metabolism, migration and memory in cytotoxic T cells. Nat. Rev. Immunol 11, 109–117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hess Michelini R, Doedens AL, Goldrath AW, Hedrick SM, Differentiation of CD8 memory T cells depends on Foxo1. J. Exp. Med 210, 1189–1200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kilgore AM, Welsh S, Cheney EE, Chitrakar A, Blain TJ, Kedl BJ, Hunter CA, Pennock ND, Kedl RM, IL-27p28 production by XCR1+ dendritic cells and monocytes effectively predicts adjuvant-elicited CD8+ T cell responses. Immunohorizons 2, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang DJ, Wang Q, Wei J, Baimukanova G, Buchholz F, Stewart AF, Mao X, Killeen N, Selective expression of the Cre recombinase in late-stage thymocytes using the distal promoter of the Lck gene. J. Immunol 174, 6725–6731 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Sun J, Madan R, Karp CL, Braciale TJ, Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat. Med 15, 277–284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aida Y, Pabst MJ, Removal of endotoxin from protein solutions by phase separation using Triton X-114. J. Immunol. Methods 132, 191–195 (1990). [DOI] [PubMed] [Google Scholar]
- 66.Haluszczak C, Akue AD, Hamilton SE, Johnson LDS, Pujanauski L, Teodorovic L, Jameson SC, Kedl RM, The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J. Exp. Med 206, 435–448 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang H, Lei R, Ding S-W, Zhu S, Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Contreras-López O, Moyano TC, Soto DC, Gutiérrez RA, Step-by-step construction of gene co-expression networks from high-throughput arabidopsis RNA sequencing data. Methods Mol. Biol 1761, 275–301 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao Y, Smyth GK, Shi W, The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hochberg Y, Benjamini Y, More powerful procedures for multiple significance testing. Stat. Med 9, 811–818 (1990). [DOI] [PubMed] [Google Scholar]
- 73.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, Xia J, MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46, W486–W494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reisz JA, Wither MJ, Dzieciatkowska M, Nemkov T, Issaian A, Yoshida T, Dunham AJ, Hill RC, Hansen KC, D’Alessandro A, Oxidative modifications of glyceraldehyde 3-phosphate dehydrogenase regulate metabolic reprogramming of stored red blood cells. Blood 128, e32–e42 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.