Abstract
Cyanobacteria are photosynthetic organisms responsible for ~25% of the organic carbon fixation on earth. A key step in carbon fixation is catalyzed by ribulose bisphosphate carboxylase/oxygenase (RuBisCO), the most abundant enzyme in the biosphere. Applying Zernike phase contrast electron cryo-tomography and automated annotation, we identified individual RuBisCO molecules and their assembly intermediates leading to the formation of carboxysomes inside Syn5 cyanophage infected cyanobacteria Synechococcus WH8109 cells. Surprisingly, more RuBisCO molecules were found to be present as cytosolic free-standing complexes or clusters than as packaged assemblies inside carboxysomes. Cytosolic RuBisCO clusters and partially assembled carboxysomes identified in the cell tomograms support a concurrent assembly model involving both the protein shell and the enclosed RuBisCO. In mature carboxysomes, RuBisCO is neither randomly nor strictly icosahedrally packed within protein shells of variable sizes. A time averaged molecular dynamics simulation showed a semi-liquid probability distribution of the RuBisCO in carboxysomes and correlated well with carboxysome subtomogram averages. Our structural observations reveal the various stages of RuBisCO assemblies, which could be important for understanding cellular function.
Keywords: RuBisCO, Carboxysome biogenesis, Zernike phase contrast cryo-electron tomography, Convolutional neural network-based annotation, Molecular dynamics simulation
Introduction
The ability of photosynthetic organisms to fix inorganic CO2 from the atmosphere into organic carbon is fundamental to the maintenance of life on earth. Carbon enters the biosphere through the Calvin-Benson-Bassham cycle of autotrophic organisms such as plants, algae and bacteria. The most important enzyme responsible for carbon fixation is ribulose bisphosphate carboxylase/oxygenase (RuBisCO), which is the most abundant enzyme in the biosphere. RuBisCO catalyzes the reaction of CO2 with a five-carbon molecule, ribulose-1,5-bisphosphate (RuBP), and the breakdown of the resulting six-carbon molecule into two molecules of 3-phosphoglycerate (3PGA). In cyanobacteria, RuBisCO accounts for 2 to 10 percent of the total cell protein. Despite its pivotal role in carbon fixation, RuBisCO is an inefficient enzyme due to its slow kinetics and poor discrimination between CO2 and O2 substrates. When cyanobacteria first evolved, O2 concentration in the atmosphere was not nearly as significant. However, the current higher atmospheric O2 concentration and RuBisCO’s high affinity for O2 compromise its carbon fixation activity, leading to an energy-depleting and unproductive process called photorespiration [1,2], To improve the efficiency of carbon fixation in an oxygen-rich atmosphere, marine cyanobacteria have developed a sophisticated CO2 concentrating mechanism (CCM) by sequestering essential carbon fixing enzymes, including RuBisCO, in a specialized polyhedral cellular compartment known as the carboxysome [3-5]. Found in cyanobacteria and chemoautotrophic bacteria, carboxysomes are icosahedral microcompartments whose structure is reminiscent of icosahedral viral capsids. Major shell proteins form a proteinaceous molecular layer, perforated with pores that facilitate the selective entry of the carbon fixation substrates, bicarbonate and RuBP, and exit of the product, 3PGA, but inhibit the entry of O2 or leakage of CO2 [6-11]. By enclosing RuBisCO in a proteinaceous shell, the cells produce a more localized and concentrated supply of CO2 [3, 5, 12-19].
RuBisCO complexes differ in their molecular forms. Although all RuBisCO enzymes are characterized by their signature multimeric composition, the number of large subunits and the presence or absence of small subunits provide the structural distinction between two major forms [12, 20-28] that are involved in carbon fixation. Form I RuBisCO, a hexadecameric enzyme with a core composed of eight large subunits and eight small subunits, is the most prevalent form in cyanobacteria, chemoautotrophs, and higher order plants [29]. Form II RuBisCO is composed of multimers of large subunits with only a 30% amino acid sequence homology to its form I equivalent [30]. Enzymatic activity analyses and structural studies suggested that intra-carboxysomal RuBisCO may not account for the total amount of RuBisCO [31]. However, the relative distribution and organization of RuBisCO in cytosol vs those enclosed in carboxysomes had not been fully characterized.
Cryo-electron tomography (CryoET) is an imaging technique that is well-suited for visualization of individual macromolecules and their interacting partners inside a cell [32-36]. We used a 200 keV cryo-electron microscope equipped with Zernike phase optics to investigate the distribution and structures of RuBisCO inside cyanobacteria both as free-standing enzyme molecules and in their various stages of assembly during carboxysome biogenesis. The Zernike phase plate substantially enhances the image contrast, as shown previously [37-39], In this study, we used cyanobacteria Synechococcus sp. WH8109 cells as a model system. The cells, infected with cyanophage Syn5 prior to being prepared for tomographic imaging, revealed detailed structural features of intracellular macromolecular complexes.
Because of its central role in carbon fixation, RuBisCO has long been a focus for genetic engineering. Improving its carboxylation efficiency and enhancing substrate specificity would directly improve plant or bacterial productivity. Novel insights into RuBisCO structure and distribution and carboxysome biogenesis in cells can serve as a basis for future genetic applications.
Results
1. Intracellular RuBisCO exists both as cytosolic free-standing complexes and as packaged assemblies in carboxysomes.
Cyanobacterium Synechococcus sp. WH8109 is widely distributed in the world’s oceans. Its complete genome sequence has been determined and it grows robustly to high cell densities under laboratory conditions [40]. The Synechococcus sp. WH8109 cells cultured in a laboratory are usually between 0.6 μm and 1.6 μm in size. They possess internal thylakoid membranes embedded with photosynthetic complexes and light harvesting pigments, phosphate inclusions, and carbon fixing carboxysomes. Infection by phages causes cells to become leaky and consequently thinner than their uninfected counterparts, resulting in better image contrast in the electron microscope. The extent of cell membrane rupture can be controlled by monitoring phage infection progression. We have reconstructed 56 WH8109 cell tomograms, 9 uninfected and 47 infected at various time points post Syn5 infection. The number of carboxysomes in infected cells does not appear to be correlated with infection progression [37], as evidenced by the average carboxysome count per cell in the infected and uninfected cells. In our analysis of RuBisCO distribution and carboxysome structures in this paper, data from infected cells are used.
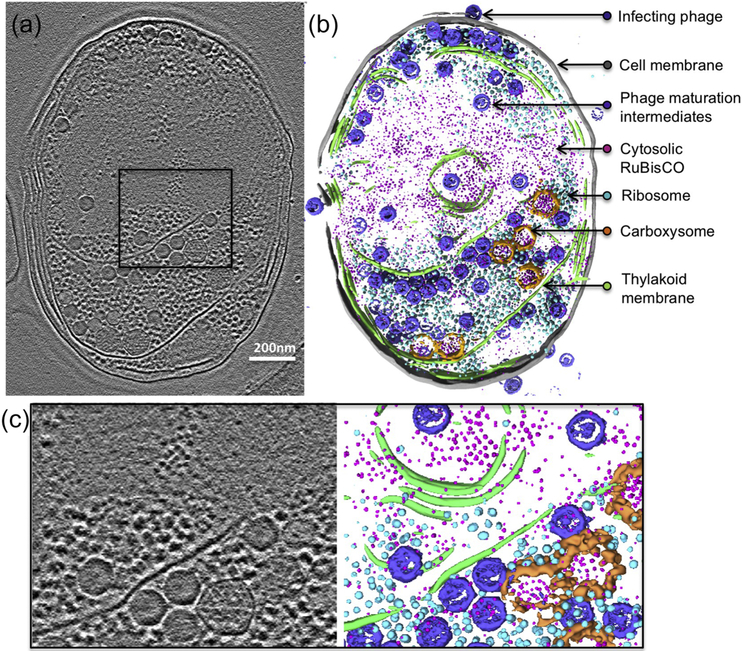
Because of the relatively high image contrast with a Zernike phase plate in the electron microscope, we can easily recognize RuBisCO, either free-standing or in association with carboxysomes at different stages of assembly, from nascent to mature (Fig. 1a). However, there are many other subcellular structures present in these tomograms that often obscure views of RuBisCO. Manual picking or segmentation of these features of interest requires immense human effort and is subject to bias [41, 42]. In order to circumvent these challenges, we employed a convolutional neural network algorithm trained on subcellular features of interest to detect them automatically in the tomograms (Fig. 1b, Sup. Movie 1) [43]. These features include the cell membrane, thylakoid membrane, ribosomes, cyanophage maturation intermediates, and RuBisCO existing as both free-standing units and assembled in carboxysomes.
Figure 1. Zernike phase contrast cryoET enables direct visualization of subcellular structures in Synechococcus sp. WH8109 cells.
(a) A 54Å slab taken from a WH8109 cell tomogram. (b) Volume rendering of the entire cell tomogram shown in (a). Carboxysome shell is segmented and colored in orange. RuBisCO complexes are colored in magenta. (c) Zoom-in view of the region boxed in (a).
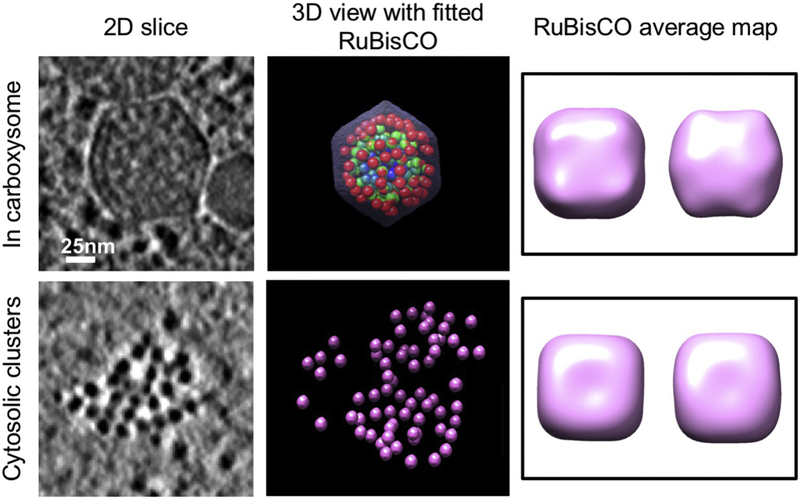
RuBisCO complexes in WH8109 cell tomograms are roughly spherical to cuboidal with a diameter of 13 nm and often exhibit a hollow center in slice views. To validate these automatically annotated densities, we performed subtomogram averaging of both putative cytosolic free-standing RuBisCO clusters and carboxysome-enclosed RuBisCO complexes. Atomic structures for biochemically purified RuBisCO from Synechococcus PCC6301 had been determined by X-ray crystallography [44]. The RuBisCO complexes from this cyanobacterial species are hexadecameric with 8 large subunits located at the corners of a rough cube, forming a core surrounded by 8 small subunits. Given that we imaged whole cells, our data collection was not aimed at high resolution, so the subtomogram averages of the automatically annotated RuBisCO exhibit ~40Å resolution (Fig. 2), providing information on the shape of the enzyme with limited structural details. Nevertheless, the overall sizes and shapes of the subtomogram averages from automatically annotated cytosolic free-standing RuBisCO agree well with subtomogram average of the (presumably authentic) intra-carboxysomal RuBisCO, confirming that the automatic annotated subcellular densities are indeed RuBisCO complexes. Note that training of the neural network applied only information from the tomogram densities themselves, while the atomic structure of RuBisCO was not used.
Figure 2. Applying automated neural-network based annotation and subtomogram analysis to study RuBisCO distribution and packing in individual carboxysomes.
Left: slice views of a carboxysome and a cytosolic RuBisCO cluster; Middle: isosurface views of a carboxysome and of a RuBisCO cluster. The carboxysome is radially colored with blue representing the innermost layer of RuBisCO, green, representing the middle layer, and red to represent the outermost layer. Right: top and side views of RuBisCO subtomogram averages from carboxysomes (top) and from cytosolic clusters (bottom).
The free-standing RuBisCO complexes were often found in the central region of the cell surrounded by the thylakoid, in isolation or in clusters (Fig. 1). Sizes of RuBisCO clusters range from small aggregates of less than ten RuBisCO complexes, to larger assemblies of over thirty RuBisCO complexes (Fig. 2). Some of these clusters form organized 2D or 3D repeating structures, while others exhibit less structural rigidity with less order and looser packing. In the annotated tomograms, there is an average of 160 RuBisCO within each carboxysome. The number of cytosolic free-standing RuBisCO that were automatically annotated was estimated to be in the range of several thousand, indicating that the number of cytosolic RuBisCO complexes is much higher than that of carboxysome-enclosed complexes (Sup. table 1). Moreover, the ratio of cytosolic to carboxysome RuBisCO from our analysis is likely an underestimate due to the high confidence threshold we imposed for our automated annotation. In our neural-network based annotation, cytosolic RuBisCO located at top, bottom or edges of the tomogram that may have been affected more by the missing wedge or other reconstruction artifacts were all excluded from final quantification analysis. Indeed, quantitative Western blotting of the large subunit of RuBisCO complex (rbcL), revealed that the total amount of RuBisCO large subunit in exponential phase cells, including both in hexadecameric RuBisCO complexes and free, isolated subunits, is at least an order of magnitude higher (Sup Fig. 1). Based on our analyses, there are definitely many more cytosolic “free” RuBisCO than are enclosed in carboxysomes.
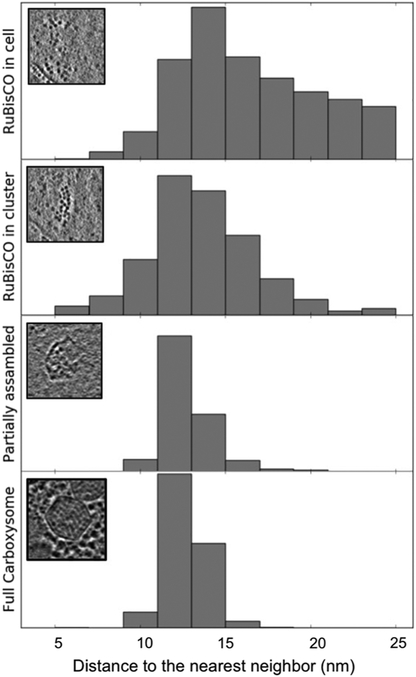
In order to measure the degree of association in RuBisCO assemblies, we compared the distance between nearest neighbors, a parameter indicating how tightly the RuBisCO complexes are associated. In freestanding RuBisCO clusters, defined as 3 or more RuBisCO molecules within 30 nm to each other, the distance from the center of a RuBisCO molecule to the center of its nearest neighbor is measured and plotted. The distribution has the same histogram peak in their nearest-neighbor spacing as for those enclosed inside carboxysomes, whether partially or fully assembled (Fig. 3). The similar value for the mode in the nearest-neighbor distance distributions of cytosolic and of carboxysome RuBisCO suggested that RuBisCO has an intrinsic propensity to self-assemble in the cell, and thus may play a functional role in carboxysome biogenesis. The progressively narrower distribution of nearest neighbor spacing observed in Figure 3 could simply indicate the spatial constraints imposed by the shell. From our tomograms, we could not distinguish if the RuBisCO clusters are assembly intermediates before RuBisCO acquires the shell or temporary products of carboxysome disassembly or dynamic RuBisCO recycling.
Figure 3. Comparison of distance to the nearest neighbors of RuBisCO complexes inside WH8109 cells.
The insets are representative clusters or particles in each class.
2. RuBisCO packing in carboxysome shows probability distributions constrained by size and shape of the shell
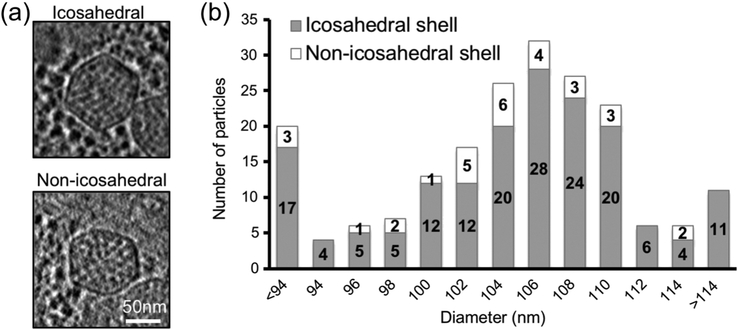
RuBisCO molecules were observed in either partially or completely assembled carboxysomes (Fig. 4). Some of the carboxysome intermediates may represent the early stage of biogenesis, since only a small portion of the shell is complete. These intermediates usually have a less defined shape relative to that of fully assembled carboxysomes. Particles in the intermediate or late stage of assembly, including those with a shell that is at least half assembled, usually displayed a more defined shape and shell angularity (Fig. 5). This is most likely attributed to incorporation of multiple pentameric units into the shell, which define the vertices [3, 8, 9, 45]. In Syn5-infected Synechococcus sp. W8109 cell tomograms, the number of carboxysomes varies from one to nine, with an average of 4.7 per cell. The intracellular carboxysomes appear to be in close proximity to the thylakoid, but no associations or direct interactions were detected. Of the 198 carboxysome subvolumes extracted from 44 cell tomograms, 168 (85%) were found to exhibit icosahedral symmetry in their shell, defined by i) displaying obvious pointed vertices when icosahedral symmetry was applied along the symmetry axes, and ii) showing stronger cross-correlation to an icosahedral shell model than to a spherical model. The asymmetric particles are either more elongated in one direction compared to a canonical icosahedron or characterized by either irregular shapes or lack of angularity (Fig. 4). The direction of the elongation has no correlation to that of the missing wedge.
Figure 4. Carboxysome size distribution and the numbers of icosahedral and non-icosahedral particles in each size group.
(a) Central slabs of 54 Å of representative icosahedral and non-icosahedral particles. (b) Carboxysome size distribution histogram including both non-symmetric and icosahedral particles in each size group. Intracellular carboxysome subtomograms were classified into various size groups based on their diameters by radial density plotting.
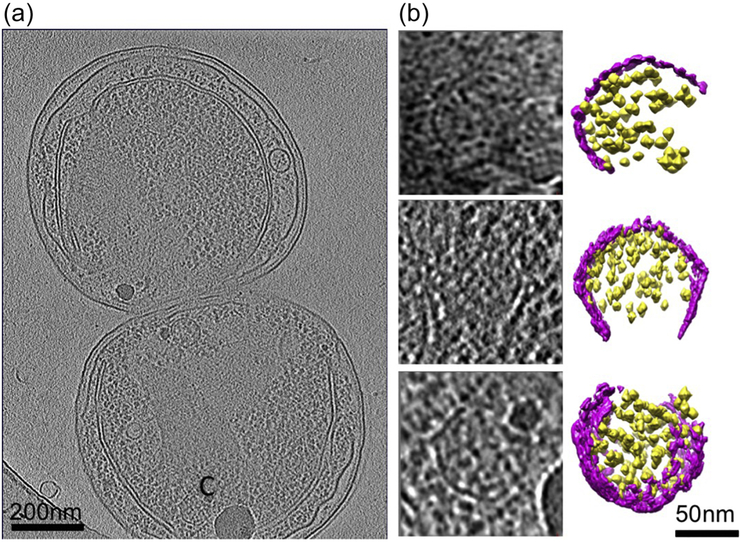
Figure 5. Partially assembled carboxysomes identified in Synechococcus WH8109 cell tomograms.
(a) Section view of a tomogram of a cell that has just completed cell division showing a partially assembled carboxysome labeled by “C”. (b) Slice and color isosurface views of partially assembled carboxysomes. The carboxysomes were arranged from particles with a small number of RuBisCO and shell units, probably representing particles in the early stage of carboxysome biogenesis, to particles that are close to completion of assembly. Magenta: shell; yellow: RuBisCO.
The diameter of the intracellular carboxysomes, defined as two times the distance between the center and the peak corresponding to the outer shell layer in a 1D radial density plot of the particle, follows a Gaussian distribution. The dominant size group has a diameter of 106 nm (N= 32 particles). In our WH8109 cell tomograms, we observed carboxysomes with sizes outside the ranges observed from purified samples [46, 47], suggesting the purification process might have preferentially enriched the dominant size classes. Therefore, purified samples may not represent the full range of sizes that are present under physiological conditions inside the cell [48].
RuBisCO in intracellular carboxysomes often were shown to line up on regular lattices in sectional views (Fig. 4a). Radial density plots of averaged maps of various size groups clearly showed RuBisCO molecules are arranged in three or four concentric layers residing under the carboxysome shell layer (Sup Fig. 2). To study RuBisCO complex packing in carboxysomes, we generated three-dimensional RuBisCO distribution models for individual carboxysomes (Fig. 2, Sup. Movie 2). The average number of RuBisCO in individual carboxysome particles from different size groups increases with diameter, from 150 in the 104 nm size group to 180 in the 110 nm size group, approximately linearly with respect to volume. Tomography and subtomogram averaging of purified carboxysomes were unable to reach a resolution that allow delineation of the shell subunits [46, 47], so it remains uncertain whether the size distribution of the shell is due to differences in triangulation number of the shell unit packing or some other form of shell protein conformational variation as seen in the capsid lattice of HIV and T4 phage [49, 50].
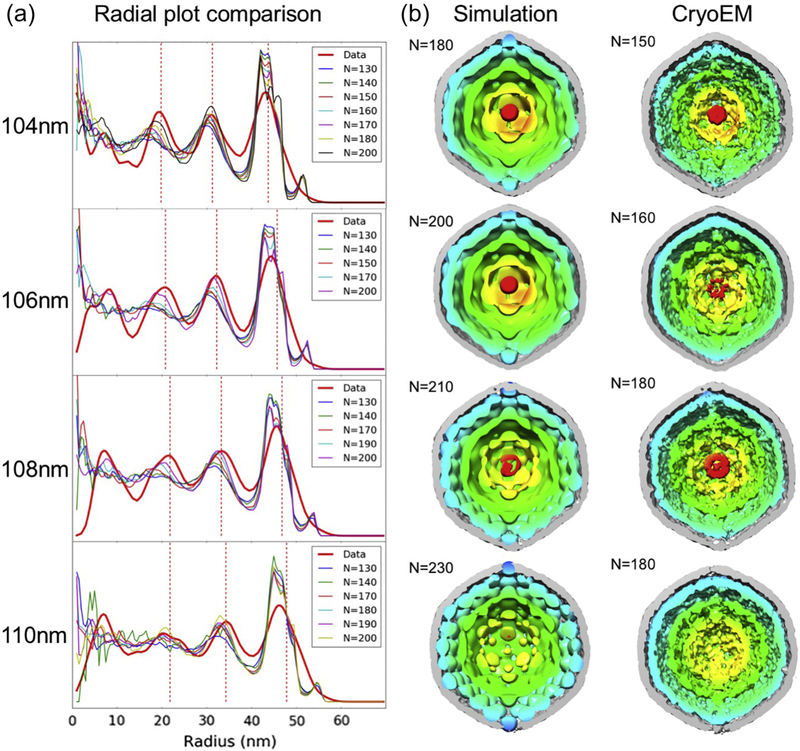
To examine the nature of RuBisCO packing in carboxysomes, we conducted molecular dynamics simulations by imposing the shape and size of the shell from cryoET subtomogram density maps as constraints and using the numbers of RuBisCO complexes enclosed as simulation input variables (Fig. 6). Each RuBisCO was modeled as a single sphere. The interaction potential was a Lennard-Jones 6-12 potential with a well depth of 1 kJ/mol. The time averages of 100 microsecond molecular dynamics simulation snapshots yielded a probability distribution of the RuBisCO locations in the carboxysome. The time averages of initially randomly arranged RuBisCO molecules within these icosahedral shells of defined diameters were then compared to our subtomogram averages from the corresponding carboxysome size groups (Fig. 6). The 1D radial density plot of simulation time averages showed the same characteristic layered packing with the position of the outermost layer maintaining a 7 nm distance from the shell, which is consistent with the radius of the RuBisCO complexes. The positions of inner layers appear to be more sensitive to the variation in RuBisCO numbers, populating the center as the total number of RuBisCO increases (Fig. 6). We also calculated the number of RuBisCO that “best” fit into the shell by identifying the time average simulation map having the highest 1D radial plot cross-correlation score with the subtomogram averages for each size group from experimental data (Fig. 6). Because no specific RuBisCO self-association potential term was included in the Molecular Dynamics calculation, the number of RuBisCO molecules required to give the best 1D correlation with the subtomogram averages is slightly higher than that actually found in these averages. Putting more RuBisCO molecules than actually present into the Molecular Dynamics simulation therefore partially compensated for the lack of such an attraction term. Along with our observations of RuBisCO self-association in the cytosol and as quantitated in Figure 3, this data suggests that inter-RuBisCO interaction is present in RuBisCO packing within carboxysomes [51, 52].
Figure 6. Molecular dynamic simulation vs experimental subtomogram averages of carboxysomes.
(a) Radial density plots of individual carboxysome simulations with variable numbers of RuBisCO (N) enclosed were compared to those of experimental subtomogram averages (red line). The red dashed lines denote the locations of the density peaks in the radial density plots. The cross sectional views of best matching simulation maps were shown with corresponding subtomogram average maps in (b).
Discussion
Individual RuBisCO complexes and their distribution inside the cyanobacteria have not been studied in detail by cryoET, partly because the thickness of the cell and the low image contrast intrinsic in cellular tomograms obscure recognition of individual enzymes inside the cell. Images taken with the Zernike phase contrast electron microscope showed an improved contrast compared to conventional imaging and enabled direct recognition and identification of individual RuBisCO complexes, free in cytosol or enclosed in carboxysomes, from cell tomograms. The capability of visualizing individual enzymes inside a cell opens the opportunity to derive the assembly process of large macromolecules and assemblies like the carboxysome by cryoET, image analysis, and annotation methods.
Biochemical evidence suggested that there is cytosolic RuBisCO activity [31, 53]. Our cryoET and quantitative Western analysis support the notion that RuBisCO in the cytosol may account for the observed cytosolic carbon fixation activity. An interesting question is why there are more cytosolic RuBisCO than those in the carboxysomes. One possibility is that a high amount of RuBisCO in the cytosol may be required in order to achieve assembly, and the cytosolic pool represents a necessary cost. Another possibility is a genuine structural and functional difference between the cytosolic and carboxysomal RuBisCO. Previous studies suggested that in some proteobacterial species, cytosolic form IAq RuBisCOs exhibit a higher affinity and specificity for CO2 even in the presence of O2 [53]. During periods of high dissolved inorganic carbon (DIC) and NH3-limited conditions, cells favor upregulated production of the cytosolic form RuBisCO because of its higher sensitivity to fluctuating inorganic carbon concentrations compared to the carboxysomal counterpart [54].
Regulation of RuBisCO expression is not well understood in cyanobacteria. It may be possible that, like proteobacteria, Synechococcus WH8109 cells also possess multiple forms of RuBisCOs, an adaptation that confers flexibility with carbon sequestration and fixation in volatile environments where CO2 and O2 levels constantly fluctuate [53]. At our current resolution and with the number of tomograms we have collected, we will not be able to distinguish if the cytosolic RuBisCO clusters are structurally different compared to the ones that are incorporated into carboxysomes. Furthermore, we also cannot rule out the possibility that the cytosolic clusters are products of carboxysome disassembly. Tracking structural elements during carboxysome assembly or disassembly will require integration of genetic manipulation, and use of biochemical and biophysical tools.
Carboxysome biogenesis appeared to be a relatively rare or rapid event because we observe, on average, less than 1 partially assembled particle per cell tomogram. Often, these partially assembled particles were located near mature carboxysomes, suggesting the carboxysome biogenesis may be a spatially-confined system, in which the major structural and functional proteins are translated locally or are transported to these locations for more efficient assembly.
There are two major types of carboxysomes, α-carboxysomes and β-carboxysomes, that differ in gene organization and evolutionarily distinct forms of protein components [55]. While oceanic cyanobacteria such as Synechococcus WH8109 possess predominantly α-carboxysomes containing RuBisCO form 1A, the β-carboxysome type encapsulating RuBisCO form 1B is distributed mainly in freshwater cyanobacteria. Although functional similarity is conserved between both carboxysome types, they differ in protein composition and the process by which RuBisCO enzymes are packaged into the carboxysomes. The β-carboxysomes are characterized by their sophisticated internal structure facilitated by scaffolding proteins, whereas α-carboxysomes appear less ordered and smaller in size with thinner shell [56, 57]. In the biogenesis of beta-carboxysomes, RuBisCO molecules first assemble into large preformed pre-carboxysomes. The subsequent integration of the shell defines these preformed structures into mature carboxysomes [13, 56, 57]. Through live cell, time-lapse fluorescent microscopy, it has been shown that the carboxysome biogenesis in the β-cyanobacteria, S. elongatus PCC 7942, occurs preferentially at the site of preexisting carboxysomes and requires a RuBisCO foci prior to colocalization of shell proteins [7]. The concurrent assembly of carboxysomes in Synechococcus sp. WH8109 cells contrasts with the biogenesis of beta-carboxysomes [7, 48]. The presence and abundance of cytosolic RuBisCO clusters (Sup Fig. 3a, b) in WH8109 cell tomograms suggested that they might play some role in initiation of carboxysome assembly. These cytosolic RuBisCO clusters may recruit shell protein subunits to form partially assembled carboxysomes that we observed in our tomograms (Sup Fig. 3c). The interactions between the first layer of RuBisCO complexes with the shell protein subunits might seed the addition of shell subunits to deposit and envelop the RuBisCO complexes. Addition of new RuBisCO complexes to the core and recruitment of shell protein subunits continue until the shell seals up to form a complete carboxysome particle (Sup Fig. 3d). The shell protein subunits may play a role in carboxysome size and morphology determination.
Carboxysome shell protein subunits had been shown to self-assemble into icosahedral cages in vitro [8, 9]. We did not observe such empty carboxysome shells within cyanobacteria WH8109 cells. This could be partially due to “cell crowdedness” that affects shell protein interaction kinetics. In α-carboxysomes, an 850 amino acid protein CsoS2 is shown to be critical for carboxysome assembly [57-59]. In our study, we were unable to detect if this protein is involved in RuBisCO self-clustering or aggregation. Future work on the presence or absence of RuBisCO clusters in CsoS2 deletion mutants would reveal possible functional roles of CsoS2 in self-clustering of RuBisCO complexes.
Materials and Methods
Cell culture
The Synechococcus sp. WH8109 cells were grown in gas dispersion bottles in artificial sea water (ASW) with continuous aeration [40] as described previously. Cell cultures at exponential phase were infected with Syn5 phage at a multiplicity of infection (MOI) of 5 to produce the thinner cells [37]. At 65 to 75 minutes post-infection, cells were centrifuged at 8500 g for 5 minutes. The cell pellet was gently resuspended in fresh ASW medium and concentrated 100-fold for subsequent cryoET imaging.
Tomographic tilt series acquisition and reconstruction
An aliquot of 3.5 μl concentrated infected or uninfected WH8109 cell samples were mixed with 10 nm gold fiducial markers and applied to Quantifoil holey grids (Quantifoil, Germany) and plunge frozen using a Vitrobot Mark VI (Thermo Fisher Scientific). The frozen, hydrated samples were imaged in a JEM2200FS electron microscope (JEOL, Japan). This electron microscope has an in-column energy filter (the slit was set to 20eV) and an airlock system to allow insertion of Zernike phase plates [38, 60-62]. The illumination setting used was: spot size 1; condenser aperture = 70 μm; objective aperture = 60 μm.
Tilt series of WH8109 cells were collected under low dose conditions on a Gatan 4k×4k CCD camera (Gatan, Inc.) at 25,000× microscope magnification and defocus close to 0. The image pixel size was calibrated to be 4.52 Å/pixel. Typically, a tilt series ranges from −60° to 60° at 3° step increments. The accumulated dose for each tilt series was 40-50 electrons/Å2. IMOD [63] was used to align tilt series and to reconstruct tomograms.
Automated RuBisCO annotation and nearest neighbor analysis
Free-standing RuBisCO in WH8109 cell tomograms were semi-automatically annotated using a convolutional neural-network based tool in EMAN2 [43, 64, 65]. RuBisCO in partial and fully assembled carboxysome are more difficult to annotate reliably using the same procedure due to their lower contrast. Therefore, we determined the coordinates of carboxysome RuBisCO complexes by calculating the cross-correlation of shell-subtracted carboxysome subtomograms with a missing wedge corrupted RuBisCO model. This model has a diameter of 13 nm, the size of a RuBisCO molecule. In the calculation of the number of carboxysome-enclosed RuBisCO in a cell, the average number of RuBisCO in carboxysomes was used. RuBisCO complexes outside the cells were manually removed from the result. Distance between RuBisCO molecule and its nearest neighbor were calculated from the coordinates of each RuBisCO. A RuBisCO was considered to be in a cluster when it had more than two RuBisCO complexes within a 30 nm radius.
RuBisCO Subtomogram averaging
Subvolumes of the annotated RuBisCO were extracted from WH8109 cell tomograms using EMAN2. Reference-free 2D class averaging was performed with the 2D projections of the subvolumes using the EMAN2 single particle reconstruction protocol. Good 2D class averages were used to generate a 3D initial model of the RuBisCO (Sup. Fig. 4). Subvolumes of RuBisCO were aligned and averaged using the EMAN2 single particle tomography tools. Only RuBisCO molecules in clusters were used to generate the final average. D4 symmetry was applied in the subtomogram averaging.
Quantification of rbcL by Western Blot
Synechococcus sp. WH8109 cell growth curve was plotted by measuring cell growth (OD750 on a Denovix DS-11 FX+ spectrophotometer/fluorometer) and cell count (on a Beckman Coulter Counter) vs the culture time. From the growth curve, we established a strong correlation (R2 = 0.99) between the OD750 reading and the cell density. Synechococcus sp. WH8109 cells growing at exponential phase were harvested for quantitative Western blot. The cell density (cell count / ml) was calculated based on OD reading of the culture using the growth curve as a reference. Cells were then lysed by freeze and thaw cycles. An aliquot of 2.7 μl of cell lysate samples containing 4 μg of total protein were loaded on a mini protein TGX stain-free gel (4-20%) under denaturing conditions (65°C water bath for 5 min). Samples from three independent cell cultures were loaded. The blotted bands were immunodetected with a specific anti-rabbit rbcL antibody and subsequently by an HRP-conjugated chicken anti-rabbit secondary antibody. Western blotting images were obtained using a chemiluminescence (ECL detection solution) and visualized by ChemiDoc MP Imaging System. Quantification of rbcL was done using a rbcL standard purchased from Agrisera. Number of RuBisCO large subunits per cell was calculated based on cell density, volume of cell culture collected, total volume of cell lysate, and volume of sample loaded to the gel.
Molecular dynamic simulation
In the present work, we model each RuBisCO as a Lennard-Jones sphere with a diameter of 13 nm corresponding to the crystal structure of cyanobacterial RuBisCO. A small attraction (<kT) showed no tendency for the system to spontaneously associate. The icosahedral carboxysome shell potential was adjusted to allow the position of the outermost layer of RuBisCO to be 7 nm from the shell as discussed above. The simulations were performed for 100 μs or 100 million time steps in the microcanonical ensemble at 300 K (with variation less than ±1 K) with a 1 ps time step. Coordinates were sampled with 500,000 total snapshots recorded at 200 step intervals. The time averaged densities were not symmetry averaged. Most packing densities were found to be liquid-like, while the highest densities explored a glassy state. The software used for the Molecular Dynamic simulation was the Extended System Program from the Pettitt group.
Carboxysome subvolume classification and averaging
Subvolumes of carboxysomes were extracted from WH8109 tomograms manually. These subvolumes were subjected to a symmetry search algorithm (e2symsearch.py from EMAN2 [65]) to align the subvolumes to their symmetry axis, if present. After the alignment, a spherical radial plot (average density in spherical shells of different radii) as well as an icosahedral radial plot (average density in icosahedral shells of different radius) was calculated for each carboxysome. Carboxysomes with a higher density peak in the icosahedral radial plot than the spherical radial plot (i.e. more similar to an icosahedron than to a sphere) were classified as icosahedral, while others were considered non-icosahedral. Carboxysome subtomograms with identified icosahedral symmetry were then aligned to a common icosahedral orientation.
For the carboxysome subvolumes found to have icosahedral symmetry, the radii were calculated by a 1D radial density plot. The icosahedral carboxysome subvolumes were then further classified into 11 size groups with diameters ranging from 94 nm to 114 nm. In each size group, a subtomogram average of the corresponding size group is achieved by averaging particles that are aligned to their icosahedral symmetry axis.
To compare with simulation results, radial plot of averaged density map from each size group is generated and compared to corresponding radial plots of simulated maps with different RuBisCO numbers. Peak positions of the three outermost RuBisCO layers of the radial plot were used to measure the similarity between experimental data and simulation results. For each size group, simulation result with peak positions that correlates best with the average density map is defined as the best fit.
Visualization
Visualizations of the cell tomograms and averaged carboxysome maps of various size groups were done in Chimera [66].
Supplementary Material
Highlights.
RuBisCO catalyzes a key step in carbon fixation in cyanobacteria.
CryoET identified RuBisCO as free-standing molecules and as packed in carboxysomes.
Carboxysome assembles by concurrent recruitment of RuBisCO clusters and the shell.
Spatial dispositions of RuBisCO may be related to its physiological conditions.
Acknowledgements
This research was supported by grants from National Institutes of Health (P41GM103832, R01GM037657 and R01GM080139) and Robert Welch Foundation (Q1242 and H-0037). The density maps of carboxysome subtomogram averages of 104 nm, 106 nm, 108 nm and 110 nm size groups have been deposited in the EMDB under accession codes EMD-7854, EMD-7855, EMD-7856 and EMD-7857 respectively. We thank Orly Levitan for providing rbcL antibody standard for Western blot, and Jennifer Jiang and Nikhita Nambiar for editing of the manuscript.
Abbreviations
- RuBisCO
ribulose bisphosphate carboxylase/oxygenase
- CCM
carbon concentration mechanism
- CryoET
cryo-electron tomography
- RuBP
ribulose-1,5-bisphosphate
- 3PGA
3-phosphoglycerate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Grotzschel S, de Beer D. Effect of oxygen concentration on photosynthesis and respiration in two hypersaline microbial mats. Microb Ecol. 2002;44:208–16. [DOI] [PubMed] [Google Scholar]
- [2].Hackenberg C, Huege J, Engelhardt A, Wittink F, Laue M, Matthijs HC, et al. Low-carbon acclimation in carboxysome-less and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology. 2012;158:398–413. [DOI] [PubMed] [Google Scholar]
- [3].Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, et al. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–6. [DOI] [PubMed] [Google Scholar]
- [4].Zarzycki J, Axen SD, Kinney JN, Kerfeld CA. Cyanobacterial-based approaches to improving photosynthesis in plants. J of Exp Bot. 2013;64:787–98. [DOI] [PubMed] [Google Scholar]
- [5].Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–22. [DOI] [PubMed] [Google Scholar]
- [6].Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA. The structure of CcmP, a tandem bacterial microcompartment domain protein from the beta-carboxysome, forms a subcompartment within a microcompartment. J Biol Chem. 2013;288:16055–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen AH, Robinson-Mosher A, Savage DF, Silver PA, Polka JK. The bacterial carbon-fixing organelle is formed by shell envelopment of preassembled cargo. PLoS One. 2013;8:e76127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dryden KA, Crowley CS, Tanaka S, Yeates TO, Yeager M. Two-dimensional crystals of carboxysome shell proteins recapitulate the hexagonal packing of three-dimensional crystals. Protein Sci. 2009;18:2629–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, et al. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–8. [DOI] [PubMed] [Google Scholar]
- [10].Kinney JN, Axen SD, Kerfeld CA. Comparative analysis of carboxysome shell proteins. Photosynth Res. 2011;109:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, et al. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol. 2009;392:319–33. [DOI] [PubMed] [Google Scholar]
- [12].Espie GS, Kimber MS. Carboxysomes: cyanobacterial RubisCO comes in small packages. Photosynth Res. 2011;109:7–20. [DOI] [PubMed] [Google Scholar]
- [13].Kerfeld CA, Melnicki MR. Assembly, function and evolution of cyanobacterial carboxysomes. Curr Opin Plant Biol. 2016;31:66–75. [DOI] [PubMed] [Google Scholar]
- [14].Klanchui A, Cheevadhanarak S, Prommeenate P, Meechai A. Exploring Components of the CO2-Concentrating Mechanism in Alkaliphilic Cyanobacteria Through Genome-Based Analysis. Comput Struct Biotechnol J. 2017;15:340–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Montgomery BL, Lechno-Yossef S, Kerfeld CA. Interrelated modules in cyanobacterial photosynthesis: the carbon-concentrating mechanism, photorespiration, and light perception. J Exp Bot. 2016;67:2931–40. [DOI] [PubMed] [Google Scholar]
- [16].Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot. 2008;59:1441–61. [DOI] [PubMed] [Google Scholar]
- [17].Price GD, Pengelly JJ, Forster B, Du J, Whitney SM, von Caemmerer S, et al. The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J Exp Bot. 2013;64:753–68. [DOI] [PubMed] [Google Scholar]
- [18].Mangan N, Brenner M. Systems analysis of the CO2 concentrating mechanism in cyanobacteria. Elife. 2014:e02043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rosgaard L, de Porcellinis AJ, Jacobsen JH, Frigaard NU, Sakuragi Y. Bioengineering of carbon fixation, biofuels, and biochemicals in cyanobacteria and plants. J Biotechnol. 2012;162:134–47. [DOI] [PubMed] [Google Scholar]
- [20].Hansen S, Vollan VB, Hough E, Andersen K. The crystal structure of rubisco from Alcaligenes eutrophus reveals a novel central eight-stranded beta-barrel formed by beta-strands from four subunits. J Mol Biol. 1999;288:609–21. [DOI] [PubMed] [Google Scholar]
- [21].Kellogg E, Juliano N. The structure and function of RuBisCO and their implications for systematic studies. Am J Bot. 1997;84:413. [PubMed] [Google Scholar]
- [22].Matsumura H, Mizohata E, Ishida H, Kogami A, Ueno T, Makino A, et al. Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate. J Mol Biol. 2012;422:75–86. [DOI] [PubMed] [Google Scholar]
- [23].Menon BB, Dou Z, Heinhorst S, Shively JM, Cannon GC. Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RubisCO species. Plos One. 2008;3:e3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Okano Y, Mizohata E, Xie Y, Matsumura H, Sugawara H, Inoue T, et al. X-ray structure of Galdieria Rubisco complexed with one sulfate ion per active site. FEBS Lett. 2002;527:33–6. [DOI] [PubMed] [Google Scholar]
- [25].Taylor TC, Andersson I. Structural transitions during activation and ligand binding in hexadecameric Rubisco inferred from the crystal structure of the activated unliganded spinach enzyme. Nat Struct Biol. 1996;3:95–101. [DOI] [PubMed] [Google Scholar]
- [26].Taylor TC, Backlund A, Bjorhall K, Spreitzer RJ, Andersson I. First crystal structure of Rubisco from a green alga, Chlamydomonas reinhardtii. J Biol Chem. 2001;276:48159–64. [DOI] [PubMed] [Google Scholar]
- [27].Valegard K, Hasse D, Andersson I, Gunn LH. Structure of Rubisco from Arabidopsis thaliana in complex with 2-carboxyarabinitol-1,5-bisphosphate. Acta Crystallogr D Struct Biol. 2018;74:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Satagopan S, Chan S, Perry LJ, Tabita FR. Structure-function studies with the unique hexameric form II ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Rhodopseudomonas palustris. J Biol Chem. 2014;289:21433–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Joshi J, Mueller-Cajar O, Tsai YC, Hartl FU, Hayer-Hartl M. Role of small subunit in mediating assembly of red-type form I Rubisco. J Biol Chem. 2015;290:1066–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev. 2007;71:576–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Beudeker RF, Cannon GC, Kuenen JG, Shively JM. Relations between d-ribulose-1,5-bisphosphate carboxylase, carboxysomes and CO2 fixing capacity in the obligate chemolithotroph Thiobacillus neapolitanus grown under different limitations in the chemostat. Arch Microbiol. 1980;124:185–9. [Google Scholar]
- [32].Baumeister W Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr Opin Struct Biol. 2002;12:679–84. [DOI] [PubMed] [Google Scholar]
- [33].Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–13. [DOI] [PubMed] [Google Scholar]
- [34].Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–65. [DOI] [PubMed] [Google Scholar]
- [35].Li Z, Jensen GJ. Electron cryotomography: a new view into microbial ultrastructure. Curr Opin Microbiol. 2009;12:333–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gan L, Jensen GJ. Electron tomography of cells. Q Rev Biophys. 2012;45:27–56. [DOI] [PubMed] [Google Scholar]
- [37].Dai W, Fu C, Raytcheva D, Flanagan J, Khant HA, Liu X, et al. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature. 2013;502:707–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dai W, Fu C, Khant HA, Ludtke SJ, Schmid MF, Chiu W. Zernike phase-contrast electron cryotomography applied to marine cyanobacteria infected with cyanophages. Nat Protoc. 2014;9:2630–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dai W, Schmid MF, King JA, Chiu W. Identifying the assembly pathway of cyanophage inside the marine bacterium using electron cryo-tomography. Microbial cell. 2014;1:45–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pope WH, Weigele PR, Chang J, Pedulla ML, Ford ME, Houtz JM, et al. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a "horned" bacteriophage of marine Synechococcus. J Mol Biol. 2007;368:966–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hecksel CW, Darrow MC, Dai W, Galaz-Montoya JG, Chin JA, Mitchell PG, et al. Quantifying variability of manual annotation in cryo-electron tomograms. Microsc Microanal. 2016;22:487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Luengo I, Darrow MC, Spink MC, Sun Y, Dai W, He CY, et al. SuRVoS: super-region volume segmentation workbench. J Struct Biol. 2017;198:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen M, Dai W, Sun SY, Jonasch D, He CY, Schmid MF, et al. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat Methods. 2017;14:983–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Newman J, Gutteridge S. The X-ray structure of Synechococcus ribulose-bisphosphate carboxylase/oxygenase-activated quaternary complex at 2.2-Å resolution. Journal Biol Chem. 1993;268:25876–86. [PubMed] [Google Scholar]
- [45].Tanaka S, Sawaya MR, Phillips M, Yeates TO. Insights from multiple structures of the shell proteins from the beta-carboxysome. Protein Sci. 2009;18:108–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, et al. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol. 2007;372:764–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, et al. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol. 2006;364:526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ. Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells. J Mol Biol. 2010;396:105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rao VB, Black LW. Structure and assembly of bacteriophage T4 head. Virol J. 2010;7:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Roy H, Kupferschmid M, Bell JA. Theory of chaperonin action: Inertial model for enhancement of prokaryotic rubisco assembly. Protein science : a publication of the Protein Society. 1992;1:925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and Mg-ATP. Nature. 1989;342:884. [DOI] [PubMed] [Google Scholar]
- [53].Badger MR, Bek EJ. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot. 2008;59:1525–41. [DOI] [PubMed] [Google Scholar]
- [54].Mangiapia M, Usf M, Brown TW, Chaput D, Haller E, Harmer TL, et al. Proteomic and Mutant Analysis of the CO2 Concentrating Mechanism of Hydrothermal Vent Chemolithoautotroph Thiomicrospira crunogena. J Bacteriol. 2017;199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rae BD, Long BM, Badger MR, Price GD. Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol Mol Biol Rev. 2013;77:357–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell. 2013;155:1131–40. [DOI] [PubMed] [Google Scholar]
- [57].Cai F, Dou Z, Bernstein SL, Leverenz R, Williams EB, Heinhorst S, et al. Advances in understanding carboxysome sssembly in Prochlorococcus and Synechococcus implicate CsoS2 as a critical component. Life (Basel). 2015;5:1141–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kinney JN, Salmeen A, Cai F, Kerfeld CA. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J Biol Chem. 2012;287:17729–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Baker SH, Lorbach SC, Rodriguez-Buey M, Williams DS, Aldrich HC, Shively J. The correlation of the gene csoS2 of the carboxysome operon with two polypeptides of the carboxysome in Thiobacillus neapolitanus. Arch Microbiol. 1999;172:233. [DOI] [PubMed] [Google Scholar]
- [60].Rochat RH, Liu X, Murata K, Nagayama K, Rixon FJ, Chiu W. Seeing the portal in herpes simplex virus type 1 B capsids. J Virol. 2011;85:1871–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Murata K, Liu X, Danev R, Jakana J, Schmid MF, King J, et al. Zernike phase contrast cryo-electron microscopy and tomography for structure determination at nanometer and subnanometer resolutions. Structure. 2010;18:903–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Danev R, Glaeser RM, Nagayama K. Practical factors affecting the performance of a thin-film phase plate for transmission electron microscopy. Ultramicroscopy. 2009;109:312–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kremer JR, Mastronarde DN, McIntosh JR. Computer visulasualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–6. [DOI] [PubMed] [Google Scholar]
- [64].Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. [DOI] [PubMed] [Google Scholar]
- [65].Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. [DOI] [PubMed] [Google Scholar]
- [66].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.