Abstract
Doublet and triplet microtubules are essential and highly stable core structures of centrioles, basal bodies, cilia, and flagella. In contrast to dynamic cytoplasmic microtubules, their luminal surface is coated with regularly arranged microtubule inner proteins (MIPs). However, the protein composition and biological function(s) of MIPs remain poorly understood. Using genetic, biochemical, and imaging techniques, we identified Tetrahymena RIB72A and RIB72B proteins as ciliary MIPs. Fluorescence imaging of tagged RIB72A and RIB72B showed that both proteins colocalize to Tetrahymena cilia and basal bodies but assemble independently. Cryoelectron tomography of RIB72A and/or RIB72B knockout strains revealed major structural defects in the ciliary A-tubule involving MIP1, MIP4, and MIP6 structures. The defects of individual mutants were complementary in the double mutant. All mutants had reduced swimming speed and ciliary beat frequencies, and high-speed video imaging revealed abnormal highly curved cilia during power stroke. Our results show that RIB72A and RIB72B are crucial for the structural assembly of ciliary A-tubule MIPs and are important for proper ciliary motility.
INTRODUCTION
Centrioles, basal bodies, cilia, and flagella are microtubule assemblies that are critical for fundamental cellular functions, including centrosome biogenesis, assembly and organization of the microtubule cytoskeleton, cell sensing and signaling by primary cilia, and propelling cells or generating fluid flow across tissues (Satir, 2017). Defects in ciliary assembly or function lead to a diverse array of human diseases, such as developmental disorders, numerous types of cancer, and ciliary diseases collectively known as ciliopathies. The latter can cause birth defects that affect development of the brain (microcephaly), skeleton, and heart (e.g., situs inversus), polycystic kidney disease, Bardet-Biedl syndrome, and primary ciliary dyskinesia (Fliegauf et al., 2007; Satir, 2017).
The core structure of cilia/flagella and basal bodies is a highly conserved, cylindrical arrangement of nine doublet or triplet microtubules, respectively. Whereas singlet cytoplasmic microtubules are usually characterized by dynamic instability, doublet and triplet microtubules form a highly stable scaffold, which serves as a platform for a plethora of associated proteins. A common structural feature shared by doublets, triplets, and other highly stable microtubules, such as the subpellicular and ventral disk microtubules of the pathogens Plasmodium and Giardia, respectively, is the presence of a unique class of microtubule-associated proteins, called microtubule inner proteins (MIPs) that are regularly distributed along the luminal surface of the microtubules (Nicastro et al., 2006, 2011; Maheshwari et al., 2015; Ichikawa et al., 2017; Kirima and Oiwa, 2017). The three-dimensional structures and periodicities of MIPs have been best described in doublet microtubules (DMTs) of motile cilia where they assemble into a complex arrangement of at least six conserved MIP structures (Nicastro et al., 2006, 2011; Ichikawa et al., 2017) (Figure 1B). However, the protein composition, interactions, and biological function(s) of these and other MIPs have remained elusive.
FIGURE 1:

Schematic models showing the general organization of MIPs in the doublet microtubules and predicted structural motifs of T. thermophila RIB72A and RIB72B. (A) Simplified cross-sectional view of a cilium with nine DMTs surrounding the central pair complex (CPC); the viewing direction is from the proximal cilium end. Outer and inner dynein arms (ODA, IDA), and the nexin-dynein regulatory complex (N-DRC) connect neighboring DMTs, whereas the radial spokes (RS) connect to the CPC. One DMT is boxed and shown larger in B. (B) Schematic of a DMT with known axonemal MIPs and other microtubule-associated structures in cross-sectional view. Note that MIP5 in the B-tubule is not shown, as it is too small to be resolved by conventional cryo-ET. (C) Domain structures of T. thermophila RIB72A and RIB72B as predicted by NCBI BLAST, showing three DM-10 domains of unknown function for both proteins, and for RIB72A a C-terminal EF-hand motif. At, A-tubule; Bt, B-tubule; IJ, inner junction; M, membrane.
Using genetic, biochemical, and imaging techniques, including cryoelectron tomography (cryo-ET), we characterized two MIP candidates, RIB72A and RIB72B, in the motile cilia of Tetrahymena thermophila. Motile cilia typically contain a [9+2] axonemal core with nine DMTs surrounding a central pair (CP) of singlet microtubules (Figure 1A) (Carbajal-Gonzalez et al., 2013). Each DMT consists of an A-tubule and a B-tubule, with 13 and 10 protofilaments (PFs), respectively (Witman et al., 1978) (Figure 1B). Outer and inner dynein motors, and complexes that regulate the activity of the dyneins, such as the nexin–dynein regulatory complex and radial spokes, are arranged regularly along the A-tubules to generate the oscillatory movement of cilia and flagella (Gibbons, 1965; Porter and Sale, 2000; Heuser et al., 2009). The 72-kDa ribbon-associated protein RIB72 was first identified as a protein associated with the “hyperstable protofilament-ribbon,” a structure left after Sarkosyl extraction of sea urchin sperm flagella (Hinchcliffe and Linck, 1998). Subsequent studies showed that RIB72 is conserved from algae to humans and tightly associates with the flagellar DMTs—possibly on the luminal side—of the unicellular algae Chlamydomonas reinhardtii (Ikeda et al., 2003). Mutations in the human RIB72 homologue, hsEFHC1, cause juvenile myoclonic epilepsy (Suzuki et al., 2004). A common feature of RIB72 homologues are three DM10 domains that are important for ciliary localization (Zhao et al., 2016) and in many cases a C-terminal, Ca2+-binding EF-hand motif (Zhao, 2015) (Figure 1C).
Here, consistent with previous findings that localized RIB72 to Chlamydomonas flagella (Ikeda et al., 2003), we detected two RIB72 proteins, RIB72A and RIB72B, in the basal bodies and cilia of Tetrahymena. Individual and double-knockout mutant cells showed slower swimming speed and abnormal ciliary waveform. A null mutant-wildtype comparison using cryo-ET combined with subtomogram averaging revealed severe structural defects of the A-tubule MIP1, -4, and -6. This shows that RIB72A and RIB72B are required for the assembly of a large subset of MIP structures that may play an important role in proper ciliary motility.
RESULTS
RIB72A and RIB72B both localize to basal bodies and cilia in Tetrahymena
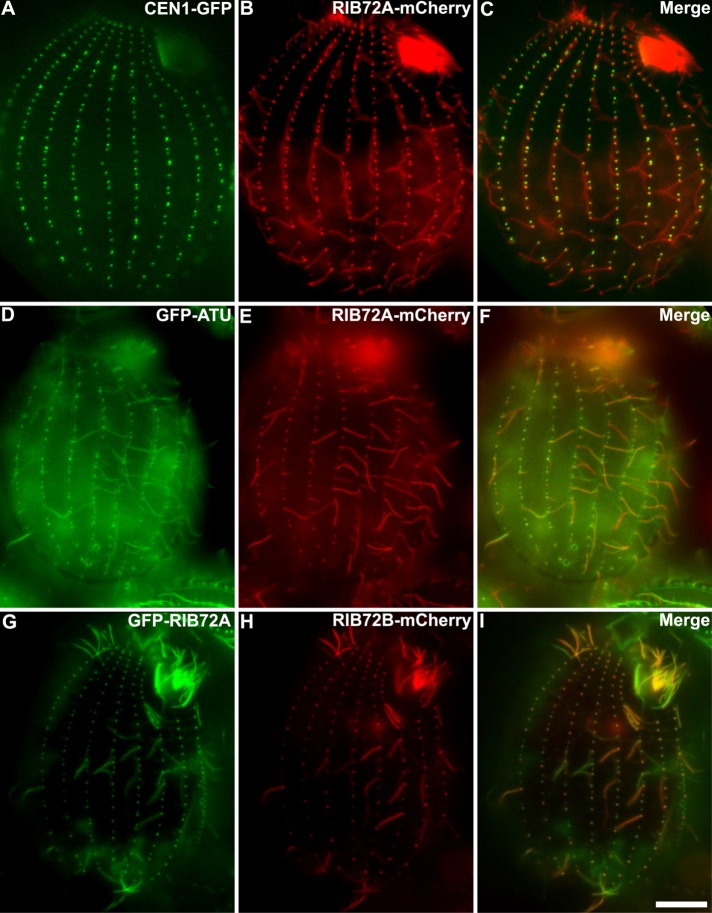
There are two genes encoding RIB72 in the genome of T. thermophila: RIB72A TTHERM_00143690) and RIB72B (TTHERM_00584850) (Eisen et al., 2006). Previous proteomics studies revealed RIB72A protein as a basal body component in Tetrahymena (Kilburn et al., 2007). We expressed RIB72A-mCherry fusion protein in Tetrahymena cells coexpressing either the basal body marker centrin (GFP-CEN1) or α-tubulin (GFP-ATU1). Fluorescence microscopy revealed that RIB72A-mCherry localized to basal bodies (Figure 2, A–C) and along the entire ciliary length (Figure 2, D–F).
FIGURE 2:
Fluorescence images reveal that Tetrahymena RIB72A and RIB72B colocalize to cilia and basal bodies. Fluorescence light microscopy images of fixed Tetrahymena cells expressing RIB72A or RIB72B tagged by fluorescent proteins along with basal body and microtubule markers. (A–C) RIB72A-mCherry (B) coexpressed with the basal body marker GFP-CEN1 (A) shows colocalization at basal bodies (C) in a WT background. (D–F) RIB72A-mCherry (E) coexpressed with the microtubule marker GFP-ATU1 (D) showing colocalization along the ciliary length (F) in a WT background. (G–I) Coexpressed GFP-RIB72A (G) and RIB72B-mCherry (H) showing colocalization at basal bodies and cilia (I) in a WT background. Cilia displaying signal from a single tagged Rib72 protein moved during imaging as live cells were used to limit fixation artifacts. Scale bar: 10 μm.
We constructed Tetrahymena knockout (KO) strains lacking RIB72A, RIB72B, or both genes. GFP-RIB72A expressed in the RIB72B-KO background localized to both cilia and basal bodies (Supplemental Figure S1A), suggesting that RIB72A assembles in these organelles independently of RIB72B. Also, RIB72B-mCherry localized to both cilia and basal bodies in the absence of RIB72A (Supplemental Figure S1, A and B). Coexpression of GFP-RIB72A and RIB72B-mCherry showed colocalization of the two proteins in most but not all cilia suggesting that RIB72A and RIB72B coexist in at least a subset of cilia (Figure 2, G–I).
Loss of RIB72A and/or RIB72B decreases swimming speed and phagocytosis rate in Tetrahymena
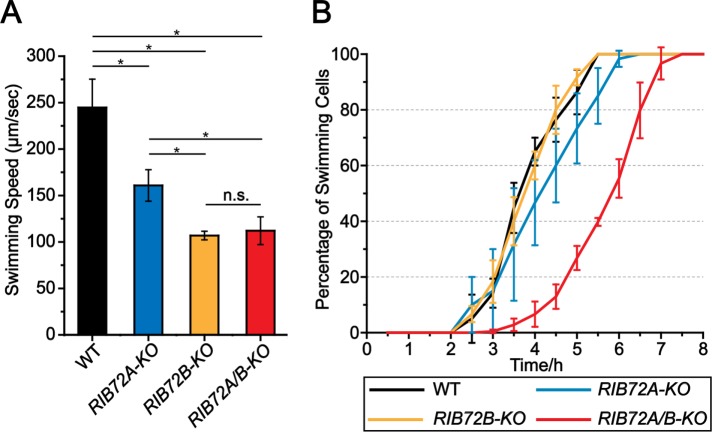
The swimming speeds of RIB72A-KO (∼160 μm/s) and RIB72B-KO (∼106 μm/s) cells were reduced to 2/3 and half of the wild-type (WT) velocity (245 μm/s), respectively (Figure 3A). The swimming velocity of the double-knockout RIB72A/B-KO (∼112 μm/s) was about as low as that of RIB72B-KO (Figure 3A).
FIGURE 3:
Gene knockouts of RIB72A and RIB72B decrease swimming velocity of Tetrahymena cells. (A) Loss of RIB72A or RIB72B decreases swimming velocity as compared with WT (100%): RIB72A-KO 66%, RIB72BKO 44%, RIB72A/B-KO 46% (n = 20 cells per stain). Error bars represent standard deviations and asterisks indicate statistical significance at p < 0.01 (one-way analysis of variance [ANOVA]). n.s. stands for nonsignificant, p > 0.05. (B) The percentage of cells recovering motility (>25 μm/s) after deciliation was recorded.
Tetrahymena use oral cilia to move particles into the oral groove for feeding through phagocytosis. We conducted a phagocytosis assay by adding India ink as “food” to the growth media and scoring the number of ink-containing food vacuoles per cell. The food vacuoles were smaller and less numerous in both RIB72B-KO and RIB72A/B-KO (Supplemental Figure S1, C–F). These results show that RIB72A and especially RIB72B are important for the functionality of oral cilia.
Following deciliation by a pH shock, all strains, WT and the three KO mutants, were able to regenerate cilia as evidenced by the recovery of cell motility, but RIB72A-KO and especially the double mutant RIB72A/B-KO restored motility at a slower rate than WT (Figure 3B), indicating that RIB72A-KO and double-knockout cells either assemble slower or fewer cilia following deciliation.
High-speed video imaging reveals abnormal waveform and decreased beating frequency in the three RIB72A/B knockout strains
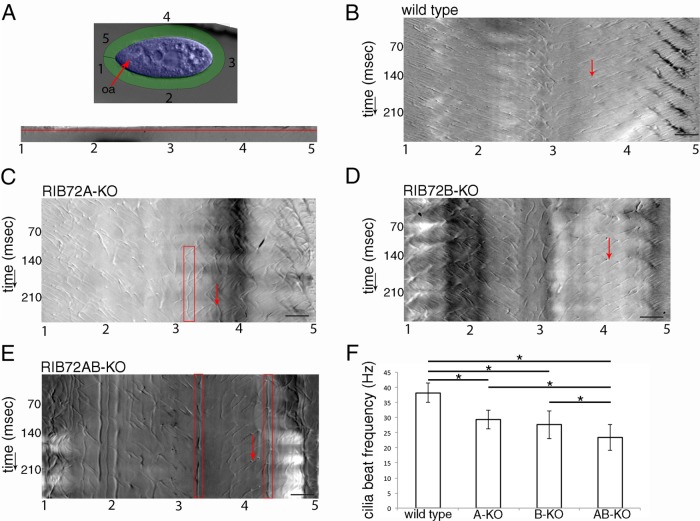
To analyze cilia beating in live cells, we collected high-speed videos of forward-swimming cells that were partially restrained inside microfluidic channels (Supplemental Videos S1–S5). Using an image processing workflow that was originally developed for Paramecium (Funfak et al., 2015), we extracted video segments that contain the zone of beating cilia around the entire cell circumference as outlined in Figure 4A. These videos were used to produce chronographs along a single line positioned slightly above the cell body surface (Figure 4, B–E; also see Supplemental Figure S2 for technical details). In the chronograph of a wild-type cell, short gray-scale diagonal lines represent traces of individual cilia as they undergo their power stroke (Figure 4B). In the recovery stroke, the cilia are too close to the cell surface to be visible (Supplemental Figure S2). The chronograph shows the time (y-axis) that it takes for a cilium occupying a particular position on the cell surface (x-axis) to pass through the field of view (examples are shown by the red arrows in Figure 4, B–E). Wild-type cilia were uniformly active along the entire cell surface and were beating with an average frequency of 38 Hz (±3.2, n = 45 cilia, 9 cells) (Figure 4, B and F). When a cilium stalls, this produces a vertical or wavy line on the chronograph (Supplemental Figure S2). The RIB72A-KO chronogram contained a few vertical or wavy lines consistent with stalled or pivoting cilia (red box in Figure 4C), while such cilia were not apparent in the RIB72B-KO chronograph (Figure 4D). The stalled cilia were very prominent in the double-knockout chronograph (red boxes in Figure 4E). In the single and double knockouts, the cilia that were not stalled showed reduced beat frequencies. The RIB72A-KO and RIB72B-KO cilia beat at 29 and 28 Hz, respectively (23 and 27% reduction compared with wild type) and the RIB72A/B-KO cilia beat at 23 Hz (39% reduction). The inverse slope of each diagonal line in the chronograph represents the speed of power stroke. The power stroke in the double-knockout cilia was 43% slower as compared with the wild type (Supplemental Figure S3A). Thus, the reduced beat frequency in RIB72A/B-KO is at least partly due to the slower movement of the cilium during the power stroke.
FIGURE 4:
Chronographs based on high-speed video imaging show that loss of either RIB72A or RIB72B or both proteins strongly affects the motility of cilia. (A) Outline of the image analysis workflow, based on Funfak et al. (2015) as applied here to Tetrahymena (see Supplemental Figure S2 for details). As shown in the top panel, the first frame of a video was used to mark the cell body (blue) and cilia (green) regions. Numbers correspond to the positions along the cell circumference starting at the cell’s anterior tip and going around the cell in a counterclockwise direction. The bottom panel shows a single frame of a movie of an extracted and unwrapped cilia region. Numbers 1–5 correspond to the positions marked around the cell circumference as shown in the top panel. The red line shows the position which was plotted over time to produce the chronograph seen in B. (B–E) Chronographs of the ciliary zones from wild-type (B) (Supplemental Video S1), single (C, D) (Supplemental Videos S2 and S3) and double-knockout cells (E) (Supplemental Video S4). The numbers on the x-axis correspond to the positions around the cell circumference as indicated in A; the y-axis represents time. Red arrows show examples of single beat duration measurements as distances between two consecutive diagonal gray-scale lines representing the same cilium in consecutive beat cycles. Note the presence of stalled or pivoting cilia indicated by red boxes in C and E. (F) Average beat frequencies in the wild-type (38.1 + 3.2 Hz, n cilia = 45, n cells = 9), RIB72A-KO (29.3 + 3.1 Hz, n cilia = 48, n cells = 8), RIB72B-KO (27.6 + 4.6 Hz, n cilia = 64, n cells = 9), and RIB72AB-KO cells (23.3 + 4.3 Hz, n cilia = 50, n cells = 8) cells. Error bars represent standard deviations, and asterisks indicate statistical significance at p < 0.001 (one-way ANOVA). Abbreviations: oa, oral apparatus. Scale bars are 10 microns.
Video S1.
Video S1. A wild‐type Tetrahymena cell swimming inside a microfluidic channel, recorded at 2000 frames/sec. (Stills appear in Figure 4 and Figure S3).
Video S2.
Video S2. A Tetrahymena RIB72A‐KO cell swimming inside a microfluidic channel, recorded at 2000 frames/sec. (Stills appear in Figure 4).
Video S3.
Video S3. A Tetrahymena RIB72B‐KO cell swimming inside a microfluidic channel, recorded at 2000 frames/sec. (Stills appear in Figure 4).
Video S4.
Video S4. A Tetrahymena RIB72A/B‐KO cell swimming inside a microfluidic channel, recorded at 2000 frames/sec. (Stills appear in Figure 4 and Figure S3).
Video S5.
Video S5. A Tetrahymena RIB72A/B‐KO cell swimming inside a microfluidic channel, recorded at 2000 frames/sec. Red arrow marks a cilium with a kink near the distal region.
The metachronal waves were visible near the anterior end of the wild-type chronograph, as dark diagonal lines (Figure 4B) (Funfak et al., 2015). The metachronal waves were visible but less frequent in the single KO strains but completely absent in the double-knockout cells (Figure 4, B–E). Thus, the loss of RIB72A/B also affects the coordination of beating between cilia. An analysis of the successive frames of extracted cilia videos for the double-knockout cells revealed abnormalities in the shape of individual cilia. Specifically, during the power stroke, wild-type cilia remained relatively straight, while some of the RIB72A/B-KO cilia appear excessively curved (Supplemental Figure S3B). Interestingly, the curving occurred halfway into the power stroke and in the opposite direction to the bend that normally forms during the recovery stroke. Furthermore, some RIB72A/B-KO cilia had a kink near the distal tip (Supplemental Video S5). We conclude that the loss of RIB72A and RIB72B reduces the beat frequency, increases the incidence of cilium stalling, reduces the metachronal activity, and alters the shape of the cilium during the power stroke.
Cryo-ET reveals defects in the MIP1, MIP4, and MIP6 structures in RIB72A-KO and RIB72B-KO and additive defects in the double knockout
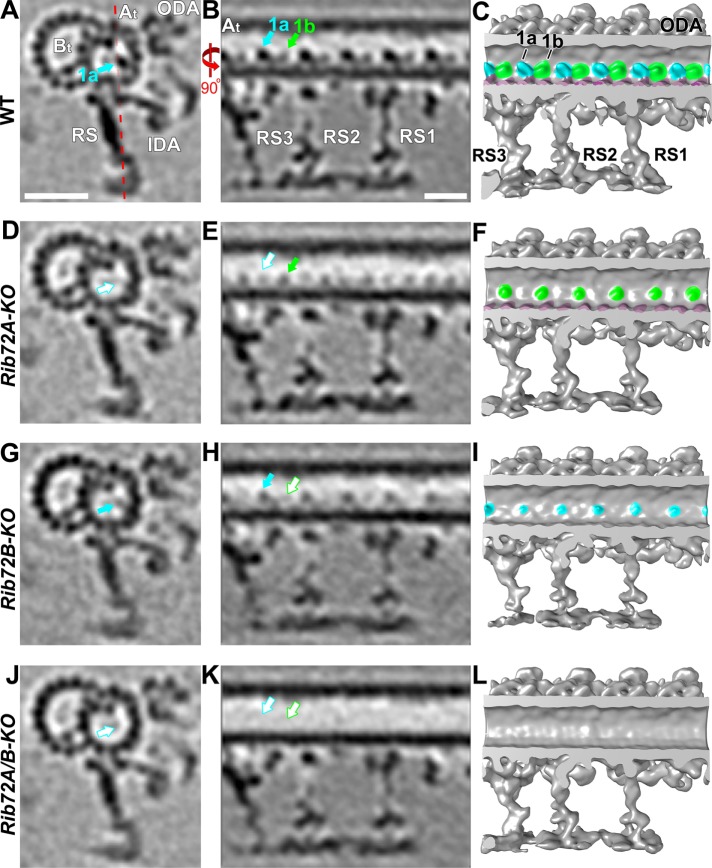
We performed cryo-ET and subtomogram averaging of the 96-nm axonemal repeats and compared the three-dimensional structures among WT, RIB72A-KO, RIB72B-KO, and RIB72A/B-KO to determine the effects of RIB72A and/or RIB72B removal on the ciliary structure. The axonemal microtubules themselves (DMTs and CPC) and all axonemal structures that bind to the exterior of the microtubules, such as the dyneins and regulatory complexes, appeared fully assembled in all four strains (Supplemental Figure S4). In contrast, the mutant averages revealed clear defects in several MIP structures on the luminal side of the A-tubule (Figures 5–7). Previous cryo-ET studies named the major MIP densities in ciliary DMTs as MIP1-6 based on their association with certain subsets of DMT PFs (Figure 1B). Absence of RIB72A and RIB72B led to defects in different subcomplexes of MIP structures, namely of MIP1 (PF A5-A7) (Figure 5), MIP4 (PF A10-A13) (Figure 6), and MIP6 (PF A1-4) (Figure 7) (summarized in Table 1). In the double mutant RIB72A/B-KO these defects were additive and thus most severe (Figures 5–7) (summarized in Table 1).
FIGURE 5:
Losses of RIB72A and RIB72B affect the MIP1 structure differently. (A–L) Averaged tomographic slices (A, B, D, E, G, H, J, K) and isosurface renderings (C, F, I, L) in cross-sectional (A, D, G; from proximal) and longitudinal views (B, C, E, F, H, I; proximal on the right) of Tetrahymena axonemal repeat units of RIB72A-KO (D-F), RIB72B-KO (G–I), and the double mutant (J–L) showed different defects in the MIP1a (blue) and/or MIP1b (green) structure compared with WT (A–C). The RIB72A-KO mutant is missing MIP1a (open blue arrow) but not MIP1b (solid green arrow); the RIB72B-KO mutant lacks MIP1b (open green arrow) but not MIP1a (solid blue arrow). The double KO shows both defects, that is, MIP1a and MIP1b are missing (open blue and green arrows). Red dotted line in A indicates slice position of longitudinal tomographic slices, for example, in B. Other labels: At, A-tubule; Bt, B-tubule; IDA, inner dynein arms; ODA, outer dynein arms; RS, radial spokes. Scale bars: 25 nm (A, valid also for D, G), 16 nm (B, valid also for E, H).
FIGURE 6:
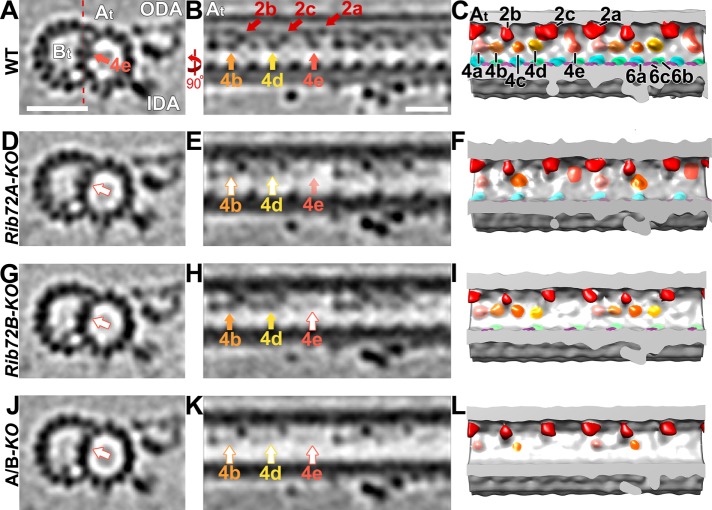
Loss of RIB72A/B affects multiple MIP4 subunits. (A–L) Tomographic slices (A, B, D, E, G, H, J, K) and isosurface renderings (C, F, I, L) in cross-sectional (A, D, G, J; from proximal) and longitudinal views (B, C, E, F, H, I, K, L; proximal on the left) of Tetrahymena axonemal repeat units. RIB72A-KO (D-F), RIB72B-KO (G–I), and the double mutant (J–L) have various defects in the MIP4 structures (4a, rose; 4b, light orange; 4c, dark orange; 4d, yellow; 4e tomato) as compared with WT (A–C); missing/reduced structures are indicated by open arrows outlined by the respective colors for MIP4 subunits. The RIB72A mutant has defects in the MIP4b (light orange arrow), MIP4d (yellow arrow), and a slight defect in MIP4e (tomato arrow). The RIB72B mutant has defects only in the MIP4e subunit and no others. The defects in the double KO were additive, that is, MIP4b, 4d, and 4e are missing. Red dotted line in A indicates slice position of longitudinal tomographic slices, for example, in B. Other labels: At, A-tubule; Bt, B-tubule; IDA, inner dynein arms; ODA, outer dynein arms; RS, radial spoke; red, MIP2a/b. Scale bars: 25 nm (A, valid also for D, G, J), 16 nm (B, valid also for E, H, K).
FIGURE 7:
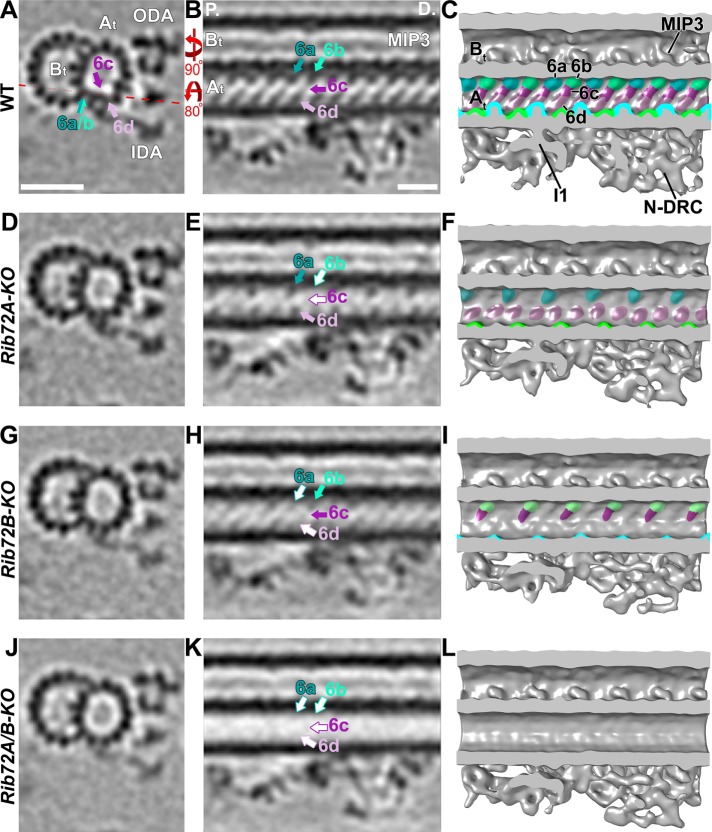
Loss of RIB72A/B affects the MIP6 structure and shows that MIP6 is composed of multiple subunits. (A–L) Tomographic slices (A, B, D, E, G, H, J, K) and isosurface renderings (C, F, I, L) in cross-sectional (A, D, G, J; from proximal) and longitudinal bottom view (B, C, E, F, H, I, K, L; proximal on the left) of Tetrahymena axonemal repeat units. RIB72A-KO (D–F), RIB72B-KO (G–I), and the double mutant (J–L) each have defects in the MIP6 structures (6a, teal; 6b, aqua; 6c, purple; 6d, mauve) as compared with WT (A–C); missing structures are indicated by open arrows outlined by the respective colors for MIP6 subunits. The RIB72A mutant lacked MIP6b (aqua arrow) and MIP6c (purple arrow), whereas the RIB72B mutant was missing only MIP6a (teal arrow) and MIP6d (mauve arrow). The defects in the double KO were additive, that is, all four MIP6 subunits were missing. Red dotted line in A indicates slice position of longitudinal tomographic slices, for example, in B. Other labels: At, A-tubule; Bt, B-tubule; I1, I1 dynein; IDA, inner dynein arms; MIP3; N-DRC, nexin dynein regulatory complex; ODA, outer dynein arms. Scale bars: 25 nm (A, valid also for D, G, J), 16 nm (B, valid also for E, H, K).
TABLE 1:
Summary of MIPs present or absent in WT and here studied mutants.
| MIP components (MW) | Strain | |||
| WT | RIB72A-KO | RIB72B-KO | RIB72A/B-KO | |
| 1a (~55) | + | – | + | – |
| 1b (~20) | + | + | – | – |
| 4b (~20) | + | – | + | – |
| 4d (~20) | + | – | + | – |
| 4e (~25) | + | ± | – | – |
| 6a (~25) | + | + | – | – |
| 6b (~18) | + | – | + | – |
| 6c (~15) | + | – | + | – |
| 6d (~20) | + | + | – | – |
+ indicates that the component is present; ± indicates that the component reduced; – indicates that the component is lacking. Molecular weight is indicated in parentheses (in kDa).
MIP1 consists of a larger MIP1a and an intervening shorter MIP1b subunit, and together the MIP1a/b minimum repeat has a 16-nm periodicity (Figure 5, A–C). The density corresponding to MIP1a was missing in RIB72A-KO, whereas RIB72B-KO lacked MIP1b (Figure 5, D–I), and the double mutant RIB72A/B-KO lost the entire density of MIP1 (Figure 5, J–L). From the subtomogram averages, we estimated the molecular weight of the subunit MIP1a to be ∼50 kDa and that of MIP1b ∼20 kDa.
MIP4 consists of multiple densities that are attached to the A-tubule-luminal side of the DMT midpartition (PFs A10-A13), which was also shown to represent most of the hyperstable PF-ribbon (Nicastro et al., 2011; Linck et al., 2014). Here, we could distinguish five subdensities of the Tetrahymena MIP4, namely MIP4a-e, which showed an overall minimum repeat periodicity of 48 nm along PF A12 (MIP4a-d) and PF A11-12 (MIP4e) (Figure 6C). Based on the tomographic data, the sizes of these MIP4 subdensities were estimated to be ∼20 kDa each (Figure 6, A–C). The structural defects of MIP4 were relatively mild in RIB72B-KO axonemes that lacked only MIP4e (Figure 6, G–I). In contrast, the defects were similarly strong in RIB72A-KO and RIB72A/B-KO that were missing MIP4b and 4d and were reduced (Figure 6, D–F) or missing MIP4e (Figure 6, J–L), respectively. The densities of the A-tubule MIP2 along PF A8-A12 remained completely unchanged in the mutants (Figure 6).
MIP6 was not originally described but visible in higher-resolution single-particle cryo-EM reconstruction spanning PFs A1-4 with 8-nm periodicity (Maheshwari et al., 2015). In our studies, by comparing the structural defects in RIB72A-KO, RIB72B-KO, and RIB72A/B-KO to wildtype axonemes, we found that MIP6 was composed of at least four different subunits, namely MIP6a–d, that ranged in size from ∼15 to 25 kDa each (Figure 7). MIP6a and 6b alternated along PF A1 with an overall periodicity of 16 nm, whereas the MIP6c and 6d densities formed a continuous ridge that spanned PFs A2-3 with an overall periodicity of 8 nm (Figure 7, A–C). The structural defects in the single KO mutants were complementary, that is, RIB72A-KO was missing MIP6b and MIP6c (Figure 7, D–F), whereas RIB72B-KO lacked MIP6a and MIP6d (Figure 7, G–I). Similarly to MIP1, the defects in the double mutant RIB72A/B-KO were additive, that is, the entire MIP6 structure was absent (Figure 7, J–L).
RIB72B-GFP rescue shows additional density in the area of MIP4
To test whether RIB72B protein was a structural component of the RIB72B-dependent MIP-complex or required only for its assembly, we imaged axonemes from a RIB72B-KO rescue-strain that expressed N-terminally tagged RIB72B-GFP. The swimming speed of the RIB72B-GFP rescue strain was similar to that of WT (Supplemental Figure S1H), and all structural MIP defects of RIB72B-KO (MIPs 1b, 4e, 6a, and 6d) were rescued to resemble the WT structure (Figure 8). Moreover, the resolution of the subtomogram averages was sufficiently high to visualize the additional density corresponding to GFP (27 kDa) at the inner surface of the A-tubule PF A12 and close to the proximal end of MIP4e, demonstrating that RIB72B is a structural component of the RIB72B-dependent MIP-complex with its C-terminal domain likely located at MIP4e (Figure 8, F and L). We also attempted to visualize GFP-tagged RIB72A; however, in the studied strains neither full structural rescue of the RIB72AKO nor GFP-tagged RIB72A was observed in the axonemal averages.
FIGURE 8:
RIB72BKO axonemes rescued with RIB72B-GFP show no structural defects and an additional tag density. (A–L) Tomographic slices (A–F) and isosurface renderings (G–L) in longitudinal views (in A, D, G, J proximal is on the right; in all remaining panels proximal is on the left) of Tetrahymena axonemal repeat units. The axonemal average from RIB72B-KO rescued with RIB72B-GFP (D-F, J-L) showed the same structures as seen in WT (A-C, G-I), that is, the mutant defects were rescued. Specifically, the loss of MIPs 1b, 4e, 6a, and 6d in the RIB72B-KO mutant (as seen in Figures 5H, empty green arrow; 6H, empty tomato arrow; and 7, G–I, empty teal and mauve arrows, respectively) is rescued by expression of RIB72B-GFP (green arrow and structure in D and J, tomato arrow and structure in L and F, and teal and mauve arrows and structures in E and K, respectively) and resemble the WT structures (green arrow in A, tomato arrow in C, and teal arrow in B, respectively). In addition, the extra density of the GFP-tag was visible in the vicinity of MIP4e in the rescued strain (green circles and densities in F and L) that was not observed in WT (compared with C and I). Slice positions of the tomographic slices are the same as in Figures 5 (for MIP1), 6 (for MIP4), and 7 (for MIP6); MIP structures are colored as follows: MIP1a blue, MIP1b green, MIP4a rose, MIP4b light orange, MIP4c dark orange, MIP4d yellow, MIP4e tomato, MIP6a teal, MIP6b aqua, MIP6c purple, MIP6d mauve. Other labels: At, A-tubule; Bt, B-tubule; I1, I1 dynein; IDA, inner dynein arms; MIP3; N-DRC, nexin dynein regulatory complex; ODA, outer dynein arms; RS, radial spokes; GFP, green fluorescent protein. Scale bar: 16 nm (valid A–F).
DISCUSSION
RIB72A and RIB72B are conserved MIPs required for the assembly of two independent MIP supramolecular complexes inside the ciliary A-tubule
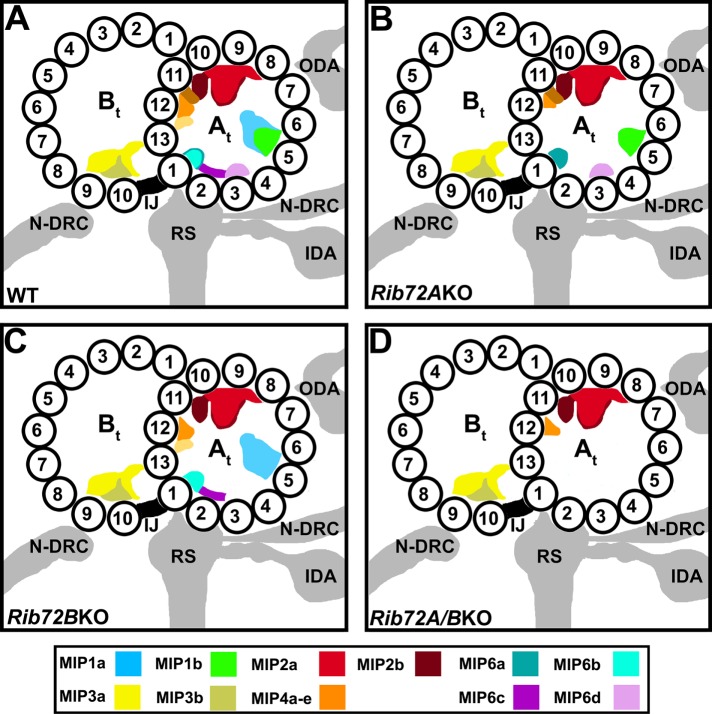
Our subtomogram averages clearly showed that several A-tubule MIP structures were missing or reduced in the absence of RIB72A and/or RIB72B. In addition, our localization of the additional GFP-tag density at PF A12 in the RIB72B-GFP rescue strain demonstrated that RIB72B itself is a component of the RIB72B-dependent MIP-complex. This is consistent with previous studies, such as 1) identification of RIB72 as a component of the hyperstable ribbon extracted from Sarkosyl-treated axonemes (Patel-King et al., 2002; Linck et al., 2014), 2) negative-staining immuno-EM with anti-RIB72 antibodies that labeled partially splayed or degraded A-tubules more frequently than intact A-tubules (Ikeda et al., 2003), and 3) a more recent cryo-immunogold-ET study with anti-SpRIB74 (the RIB72 homologue in the sea urchin Strongylocentrotus purpuratus) that detected gold labels along the side of the hyperstable ribbon facing the A-tubule lumen, again suggesting that spRIB74 localizes to the luminal surface of PF A11-12-13-1 (Linck et al., 2014). However, previous studies were hampered by limited efficiency of antibody labeling and reduced resolution of negative staining immuno-EM, the long labels with primary and secondary antibodies (20–25 nm length) and by the reduced structure preservation due to the need for partial disassembly of the DMTs to allow antibody access to the luminal microtubule surface. Here we provided direct visual evidence that RIB72B of Tetrahymena is a MIP bound to the luminal surface of the A-tubule, and that absence of RIB72A and/or RIB72B causes severe structural MIP defects. These data suggest that RIB72A and RIB72B are crucial for the stable assembly of two independent supramolecular MIP complexes that together cover the luminal surface of about half of the A-tubule diameter. The RIB72A-dependent and RIB72B-dependent MIP supracomplexes were complementary and composed of [MIPs 1a, 4b, 4d, (4e), 6b, 6c] and [MIPs 1b, 4e, 6a, 6d], respectively (Table 1 and Figure 9).
FIGURE 9:
Summary schematics of MIP locations in the DMTs of WT (A), RIB72A-KO (B), RIB72B-KO (C), and RIB72A/B-KO (D) cilia. The structural comparison between the averaged DMTs of WT and mutant axonemes revealed that loss of RIB72A causes the loss of MIPs 1a (blue) 4b (light orange), 4d (yellow), 4e (tomato) (partially missing), 6b (aqua), and 6c (purple). The absence of RIB72B results in the loss of MIPs 1b (green), 4e (tomato), 6a (teal), and 6d (mauve). The defects in the double knockouts are additive. Coloring of all MIPs is shown in the color legend. Other labels: At, A-tubule; Bt, B-tubule; IDA, inner dynein arms; IJ, inner junction; N-DRC, nexin dynein regulatory complex; ODA, outer dynein arms; RS, radial spokes; 1–13, PF numbers.
The observed structural defects in each single and the double KO mutant seemed to be larger than the molecular size of a single copy of the 72-kDa RIB72A and/or 61-kDa RIB72B proteins. Based on the missing densities in our subtomogram averages of RIB72A/B-KO, we estimated the total molecular weight of the RIB72A-dependent and RIB72B-dependent MIP supracomplexes combined to be ∼240 kDa. This is nearly twice of the sum of RIB72A and RIB72B (133 kDa), suggesting either that multiple copies of each RIB72 proteins are involved or that additional, unidentified proteins contribute to these MIP supracomplexes. A recent biochemical and immunogold labeling study suggested that the EF-hand containing protein FAP85 is an A-tubule MIP and may be associated with the MIP1 structure in Chlamydomonas flagella (Kirima and Oiwa, 2017).
RIB72A and RIB72B likely form two independent MIP supramolecular complexes inside the A-tubule of basal bodies
Using cryo-ET, we showed that both RIB72A and RIB72B are axonemal MIPs. However, our fluorescence light microscopy localized both proteins not only to the ciliary length but also to the basal bodies. Previous fluorescence light microscopy of labeled RIB72 homologues in other species, such as Chlamydomonas, also demonstrated RIB72 localization along the length of axonemes but not to basal bodies (Ikeda et al., 2003). This suggests that both proteins are also MIPs in the basal bodies, which is consistent with a previous study that identified the Tetrahymena proteins, RIB72A/Bbc73 and RIB72B/Bbc60, in the basal body proteome (Kilburn et al., 2007). Although previous cryo-ET studies of basal bodies and centrioles were limited in their resolution, because of technical challenges due to their shorter length and accessibility/need for isolation from the cell body, some MIP densities were also observed in the triplet microtubules of these organelles (Guichard et al., 2012; Li et al., 2012). Together this suggests that at least some, if not all, MIP structures described for ciliary DMTs, including the highly conserved RIB72A-dependent and RIB72B-dependent MIP supracomplexes in the A-tubule, are also assembled in the A- and B-tubules of basal bodies.
Possible functional role(s) of ciliary MIPs
The overall structures and periodicities of MIPs inside DMTs appears conserved from algae to human (Nicastro et al., 2011; Lin et al., 2014), suggesting that the biological function(s) of MIPs in motile cilia is conserved. Previous studies have hypothesized various functional roles for ciliary MIPs, such as stabilization of DMT, anchoring of axonemal complexes to the outer surface, and regulation of beat frequency and/or ciliary waveform (Nicastro et al., 2011; Linck et al., 2014; Ichikawa et al., 2017), but so far little experimental evidence exists to test these hypotheses. Here we have studied ciliary functions by phenotypically comparing wild-type and Rib72A/B-KO mutants, including swimming velocity, phagocytosis efficiency, and ciliary waveform and frequency. Our results showed that the MIP defects—even though all other structures seemed to have fully assembled in the axoneme—caused abnormal ciliary motility. Specifically, high-speed video imaging of beating cilia revealed decreased beating frequency and unusual waveform with occasional distal cilia kinking, demonstrating that RIB72A and RIB72B are important for the proper ciliary beating.
MIPs are a common feature of highly stable microtubules that experience mechanical stress. In contrast to regular cytoplasmic microtubules, DMTs are highly stable and cold resistant (Andrieux et al., 2002). In motile cilia, the DMTs provide a stable scaffold for many associated complexes while being exposed to vigorous bending with ∼40 Hz frequency, that is, the bending direction changes every ∼8 ms. In the malaria-causing pathogen Plasmodium, the MIPs are proposed to stabilize the subpellicular microtubules to provide the sporozoites with a highly elastic, yet stable, cytoskeleton that remains intact while the cells undergo extreme bending as they glide and squeeze through tissue barriers (Cyrklaff et al., 2007). Attachment of the diarrhea-causing parasite Giardia lamblia to the host’s intestinal epithelium is mediated by a suction-type mechanism caused by the ventral disk, a large spiral-shaped array of stable and MIP-containing microtubules (Schwartz et al., 2012). The microtubule-stabilizing drug taxol and its derivatives also bind to the inside of the microtubule wall (Nogales et al., 1999). Thus, the inner scaffold formed by MIPs could add physical interactions that strengthen tubulin dimer and protofilament coherence, thereby increasing microtubule stability and maybe regulating microtubule elasticity.
However, despite severe MIP defects, especially in the double RIB72A/B-KO mutant, we did not observe large-scale DMT defects by cryo-ET in inactive (ATP-free) isolated axonemes, such as partially depolymerized microtubules or holes in PFs, neither in the raw tomograms nor the subtomogram averages. It is possible that smaller defects in the microtubule lattice were present but averaged out in the subtomogram averages. However, imaging of actively beating mutant cilia revealed abnormally high-curved cilia during the power stroke and in the double KO there were occasional kinks (Suplemental Figure S3B) that could be a sign of a broken axoneme. Smaller axonemal pieces that were broken off would likely get lost during axoneme isolation. The abnormal curvature and kinks in mutant cilia suggest that RIB72A and RIB72B probably contribute to the stability and possibly the elasticity of the axoneme in actively beating cilia.
Another possible function of MIPs could be regulation during cilia movement. One proposed regulatory mode is the interaction of MIPs with axonemal complexes on the outside of DMTs, contributing to their regulation. In fact, protease digestion experiments in which a ∼10-kDa fragment was cleaved from Chlamydomonas RIB72 suggested that this fragment was likely to reside on the outer microtubule surface and was therefore accessible to protease (Ikeda et al., 2003; Ichikawa et al., 2017). This fragment could interact with associated axonemal complexes, such as the regulatory I1 dynein or the calmodulin and spoke associated complex (CSC), and through these interactions regulate cilia motility. Importantly, a recent high-resolution study showed MIPs projecting through the DMT and interacting with proteins on the exterior of the DMT (Ichikawa et al., 2017). Interestingly, the EF-hand calcium-binding domain at the C-terminus of RIB72A is highly conserved among various species (Ikeda et al., 2003). In Chlamydomonas, the flagella show a Ca2+-dependent waveform conversion, and the sensitivity of RIB72 to trypsin digestion depends on the Ca2+ concentration (Ikeda et al., 2003). Axoneme curvature has also been proposed as a signal that switches dynein activity on/off on opposite sides of the flagellum to generate the undulating motion typical of cilia (Satir and Matsuoka, 1989). If the MIPs change the stability/elasticity properties of the DMTs, then this could also result in delayed or lack of proper signaling to coordinate dynein activities in beating cilia. Combined with our high-speed imaging results showing abnormal beating waveform and frequencies of MIP-depleted cilia, including stalling and pivoting, RIB72A and RIB72B might also be important for signaling and regulation of ciliary motility.
We demonstrated that RIB72A and RIB72B are ciliary MIPs that are required for proper ciliary motility. These proteins may contribute to DMT stability and elasticity and possibly to the regulation of ciliary beating.
MATERIALS AND METHODS
Construction of gene knockout strains
Native expression of C-terminal fusion proteins Rib72a-mCherry and Rib72b-mCherry were made using the p4T2-1-NEO2-mCherry vector. In brief, two regions of homology were cloned into the vector flanking the NEO2 gene, which encodes resistance to paromomycin. Homology regions consist of 1 kb of DNA from the 3′ end of the RIB72A or RIB72B gene, terminating at the stop codon and 1 kb of DNA from the 3′UTR 100 base pairs after the stop codon of RIB72A or RIB72B. Rib72a-mCherry and Rib72b-mCherry were also generated with resistance to cyclohexamide. In these strains the NEO2 coding sequence was replaced by the RPL38A coding sequence, which confers resistance to cyclohexamide. Native expression of Rib72a-GFP was generated using the same strategy described above with the mCherry coding sequence replaced with the GFP coding sequence. The RIB72A null strain was also generated as described above except the upstream homology region was replaced with 1 kb of homologous DNA from the 5′UTR of RIB72A.
Exogenous expression rescue constructs for N-terminal fusion proteins GFP-Rib72a and GFP-Rib72b were made using the pBS-MTT-GFP-gtw plasmid. In brief, cDNA from RIB72A or RIB72B was generated by reverse transcription PCR (RT-PCR) using SuperScriptII reverse transcriptase (Invitrogen, Grand Rapids, NY). The cDNA of these genes was cloned into the pENTR4 Gateway Entry Vector (Invitrogen) then shuttled into the pBS-MTT-GFP-gtw plasmid through gateway cloning (Invitrogen). This plasmid integrates into the genome at the RPL29 locus and its MTT promoter allows for inducible control of expression by using CdCl2.
The above-described constructs were linearized and introduced to wild-type Tetrahymena cells by biolistic bombardment. Only the Rib72b rescue construct was integrated into a non-wild-type strain background, RIB72BΔ cells. Plasmid DNA integrated directly into the genome through homologous recombination. Successful integration of plasmid DNA was identified by resistance to either paromomycin or cyclohexamide drug.
The Tetrahymena strains lacking one or both of the RIB72 homologues were constructed by gene disruptions (initially in the micronucleus) using homologous DNA recombination and biolistic bombardment (Cassidy-Hanley et al., 1997; Hai and Gorovsky, 1997) as previously described in detail (Dave et al., 2009) using mating strains CU428 and B2068 (obtained from the Tetrahymena Stock Center, Cornell University). RIB72A (TTHERM_00143690) was interrupted by insertion of neo3 cassette (Shang et al., 2002), while RIB72B (TTHERM_00584850) was disrupted by neo4 (Mochizuki, 2008). Single- and double-knockout homozygous heterokaryons were obtained by crosses as previously described (Dave et al., 2009). Genotyping of heterokaryon progeny during crosses was performed by PCR of genomic DNA of outcross progeny using primers that detect a junction between the targeted gene and neo.
The primers used for construction of targeting plasmids and testing for a loss of the targeted sequences are as follows: Amplification of homology arms for knocking out RIB72A were 5′- ATTAGGGCCCAGTGATTAAAACACAGACAAA-3′ and 5′-ATTACCGCGGAGGCTCCCATAAATGATTA-3′; 5′-TTTATATCGATTATTTTAATTGGCAGAGAGT-3′ and 5′-ATTAGAGCTCATTGATAGGATTGTTGTGAT-3′. Amplification of homology arms for knocking out RIB72B were 5′-TTTTGGGCCCTAATTGGTGGTCCTACTGG-3′ and 5′-AAAATCGATACCTTTAGGCTATCCAATCT-3′;
5′-AAACCGCGGAAGCAATCTAAAAGCCATAGT-3′ and 5′-TTTGAGCTCGTATTAAACT TAGCATTGAATC-3′. Primers to test for the loss of targeted RIB72A sequence were 5′-TCTAGGAAAGTGTTGTTGAA-3′ and 5′-ATAGTAATTGAGACCACCTT-3′. Primers to test for the loss of targeted RIB72B sequence were 5′-TTTGAGCTCGTATTAAACT TAGCATTGAATC-3′ and 5′-ATGTTCCTCCAG TTGTCTAT-3′.
Fluorescence microscopy
Generation of the fluorescent fusion Tetrahymena strains was achieved as previously described in detail (Winey et al., 2012). Briefly, plasmid DNA was introduced to cells by biolistic bombardment, and transformation was mediated by homologous recombination (Cassidy-Hanley et al., 1997; Hai and Gorovsky, 1997). Bbc73 was tagged with mCherry at its C-terminus and integrated at its endogenous locus under control of its native promoter. This fluorescent fusion protein was coexpressed with N-terminal GFP fusions of Cen1 (Vonderfecht et al., 2012), ATU1 or Bbc60 were expressed under control of an inducible promoter (Shang et al., 2002). Fluorescence images were acquired at room temperature using an Eclipse Ti inverted microscope (Nikon, Japan) equipped with a CFI Plan Apo VC 60 × H numerical aperture 1.4 objective (Nikon, Japan) and a CoolSNAP HQ2 charge-coupled device camera (Photometrics, Tuscon, AZ). MetaMorph Imaging software (Molecular Devices, Sunnyvale, CA) was used to collect images and generate the projection images. For live cell imaging, cells were grown in 2% super proteose peptone (SPP) medium, washed once with 10 mM Tris-HCl, pH 7.4, pelleted, and placed on microscope slides (VWR, Radnor, PA) (Gorovsky et al., 1975). In strains where gene expression was under control of an inducible MTT1 promotor, 0.5 μg/ml concentration of CdCl2 was used. For immunofluorescence, cells were fixed using either 3% formaldehyde or 70% ethanol and then placed on poly-l-lysine–treated slides. Both the centrin-1 antibody 20H5 and anti-mouse Alexa488 were diluted 1:1000 in phosphate-buffered saline (PBS) + 1% bovine serum albumin (BSA). Primary antibody incubation was done at 4°C overnight, and secondary antibody incubation was done for 1 h at room temperature. Cells were washed in PBS + 0.1% BSA five times after each antibody incubation. Finally, cells were mounted in Citifluor (Ted Pella, Redding, CA) and imaged.
Swim speed assay
Tetrahymena cells were grown at 30°C in SPP media (2% protease peptone, 0.1% yeast extract, 0.2% glucose, 0.003% FeCl3) to mid–log phase, ∼2 × 105 cells/ml. Cells were then placed on a slide and imaged using differential interference contrast (DIC) imaging with a 20× objective at 10 frames/s on a Nikon Ti microscope (Nikon, Japan) using NIS Elements software (Nikon). ImageJ with the ImageJ software plugin MTrackJ (E. Meijering) was used to track and quantify the movement of Tetrahymena cells. Twenty cells were quantified per condition, and the experiment was performed in triplicate.
Reciliation assay
Tetrahymena cells were grown in 2% SPP medium to mid–log phase, ∼2 × 105 cells/ml. Deciliation was performed as described previously (Calzone and Gorovsky, 1982). Briefly, cells were placed in starvation media (10 mM Tris-HCl, pH 7.4) for 24 h at 30°C. Cells were then placed in deciliation medium (10% Ficoll 400, 10 mM NaOAc, 10 mM EDTA, 10 mM CaCl2, pH 4.2), and cilia were mechanically sheared by passage through an 18.5-gauge syringe. Cells were then placed in regeneration medium (15 mM Tris-HCl, 2.0 mM CaCl2, pH 7.95) and allowed to regenerate cilia at 30°C. Cells were imaged every 30 min as described above in the swim speed assay. Twenty cells from each time point were scored for forward movement (>25 μm/s), and the percentage of the 20 cells that showed forward motility was recorded. The experiment was completed in triplicate.
Phagocytosis assay
Tetrahymena cells were incubated with 0.1% India ink in 2% SPP media at 30°C. At 0.5, 2, 4.5, and 7.5 h after the India ink was added, cells were washed in 10 mM Tris buffer (pH 7.5) and fixed with 4% glutaraldehyde (Sigma-Aldrich). Cells were mounted on slides and the average number of food vacuoles per cell was quantified (n = 10 cells) using phase contrast imaging with a 60× oil immersion objective on an upright light microscope (Nikon Eclipse LV, Japan) equipped with a scientific complementary metal-oxide semiconductor (sCMOS) camera (Andor, UK).
High-speed video analysis of cilia in swimming Tetrahymena cells
Observations of cilia in actively swimming Tetrahymena cells were performed and data were quantified as described previously for Paramecium tetraurelia (Funfak et al., 2015), with minor modifications. A highly concentrated cell suspension (in 2% SPP medium) was injected into the inlet of a microfluidic channel (33 μm in height and 50 μm in width) using a controllable pressure source (Fluigent). The pressures between the inlet and outlet of the channel were equilibrated to prevent residual flow. DIC imaging of forward-swimming cells was done on an inverted microscope (Nikon TE2000), using a 60× objective lens. Videos were recorded on a Photron 1024PCI camera at 2000 frames per second. Video image processing and quantification was done exactly as described (Funfak et al., 2015). Matlab (The MathWork, Natick, MA) along with the CR Toolbox (Barbacci et al., 2014) were used to mark the cell body and the zone of beating cilia in the first frame of the movie. The cell body and ciliary zone boundaries was adjusted every 30–50 frames to track the position of the moving cell. The segment of the video containing cilia was digitally unwrapped, producing a rectangle where all ciliary bases are aligned horizontally, generating an “extracted cilia video.” ImageJ was used to plot the gray-scale over time at a line positioned within the ciliary zone but away from the cell surface, creating a space-time diagram (chronograph). This produces an image containing a number of short diagonal lines that correspond to traces of individual cilia. On the time (y) axis, successive diagonal lines corresponding to the same cilium can be identified based on their vertical position and shape. The beat frequency was quantified by measuring the distance (on the y-axis) between diagonal lines corresponding to the same cilium. The speed of the power stroke corresponded to the inverse slope of a diagonal line. See Supplemental Figure S2 for additional information about the methodology used to quantify ciliary motility.
Cryosample preparation and cryo-ET
Cryosamples were prepared and imaged as previously described (Nicastro et al., 2006, 2011). Briefly, axonemes were isolated from cultured Tetrahymena cells using the pH-shock method. When the pH of the buffer solution was rapidly lowered to 4.3, the cells released their intact cilia, which were collected by centrifugation. Cilia were then demembranated with detergent (IGPAL 630; Sigma-Aldrich) and the axonemes collected by centrifugation. Colloidal gold solution (10 nm; Sigma–Aldrich) was applied to Quantifoil grids R2/2 Holey carbon 400 mesh Quantifoil grids (Quantifoil MicroTools GmbH, Germany) and allowed to air-dry. The amounts of 3 μl of sample and 1 μl of 10-fold-concentrated BSA-coated 10-nm gold solution (Iancu et al., 2006) were added to the grid, and excess fluid was blotted with filter paper before the grid was rapidly plunge frozen in liquid ethane with a homemade manual plunge freezing device. This generates a layer of vitreous ice with embedded axonemes. Frozen grids were stored in liquid nitrogen until used.
For cryo-ET, frozen grids were loaded into a cryo-holder (Gatan, Eindhoven, Netherlands) and inserted into a Tecnai F30 transmission electron microscope (FEI, Hillsboro, OR) equipped with a field emission gun, GIF2000 post column energy filter (Gatan, Eindhoven, Netherlands), and a 2048 × 2048 pixel charge-coupled device (CCD) camera (Gatan, Eindhoven, Netherlands). Single-axis tilt series were recorded using the microscope control software SerialEM (Mastronarde, 2005) with the TEM operated at 300 kV in low-dose mode. The sample was tilted from −65 to +65° in increments of 2.0°. The total accumulated electron dose was limited to ∼100 e/Å2. The defocus was −8 μm, and the energy filter was operated in zero loss mode with a 20-eV slit width. The magnification was 13,000× with a final pixel size of 1.077 nm.
Image processing
The software program IMOD (Kremer et al., 1996) was used for fiducial alignment and tomogram reconstruction using weighted back projection. Subtomogram averaging was performed using the PEET software (Nicastro et al., 2006; Heumann et al., 2011) to average the 96-nm axonemal repeat units from three-dimensional reconstructed Tetrahymena axonemes. The resolution of the DMT averages was estimated using the 0.5 criterion of the Fourier shell correlation method (Supplemental Table 1). The Chimera package (Pettersen et al., 2004) was used for three-dimensional visualization and surface rendering.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chen Xu for training and management of the electron microscopes in the Louise Mashal Gabbay Cellular Visualization Facility at Brandeis University (Waltham, MA). This work was supported by funding from the National Institutes of Health (GM083122 to D.N., GM074746 and GM127571 to M.W., and GM089912 to J.G.) and the National Science Foundation (MBC-033965 to J.G.). P.L. was supported by a predoctoral fellowship from the American Heart Association. The three-dimensional averaged structures have been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-7806, EMD-7811, EMD-7805, and EMD-7807.
Abbreviations used:
- A t
A-tubule
- B t
B-tubule
- cryo-ET
cryoelectron tomography
- DMT
doublet microtubule
- IDA
inner dynein arms
- MIP
microtubule inner protein
- N-DRC
nexin-dynein regulatory complex
- ODA
outer dynein arms.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-06-0405) on August 22, 2018.
REFERENCES
- Andrieux A, Salin PA, Vernet M, Kujala P, Baratier J, Gory-Faure S, Bosc C, Pointu H, Proietto D, Schweitzer A, et al. (2002). The suppression of brain cold-stable microtubules in mice induces synaptic defects associated with neuroleptic-sensitive behavioral disorders. Genes Dev , 2350–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacci A, Diener J, Hémon P, Adam B, Donès N, Reveret L, Moulia B. (2014). A robust videogrametric method for the velocimetry of wind-induced motion in trees. Agric Meteorol , 220–229. [Google Scholar]
- Calzone FJ, Gorovsky MA. (1982). Cilia regeneration in Tetrahymena. A simple reproducible method for producing large numbers of regenerating cells. Exp Cell Res , 471–476. [DOI] [PubMed] [Google Scholar]
- Carbajal-Gonzalez BI, Heuser T, Fu X, Lin J, Smith BW, Mitchell DR, Nicastro D. (2013). Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton , 101–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. (1997). Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics , 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyrklaff M, Kudryashev M, Leis A, Leonard K, Baumeister W, Menard R, Meissner M, Frischknecht F. (2007). Cryoelectron tomography reveals periodic material at the inner side of subpellicular microtubules in apicomplexan parasites. J Exp Med , 1281–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave D, Wloga D, Sharma N, Gaertig J. (2009). DYF-1 Is required for assembly of the axoneme in Tetrahymena thermophila. Eukaryot Cell , 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. (2006). Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol , e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H. (2007). When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell , 880–893. [DOI] [PubMed] [Google Scholar]
- Funfak A, Fisch C, Abdel Motaal HT, Diener J, Combettes L, Baroud CN, Dupuis-Williams P. (2015). Paramecium swimming and ciliary beating patterns: a study on four RNA interference mutations. Integ Biol , 90–100. [DOI] [PubMed] [Google Scholar]
- Gibbons IR. (1965). An effect of adenosine triphosphate on the light scattered by suspensions of cilia. J Cell Biol , 707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky MA, Yao MC, Keevert JB, Pleger GL. (1975). Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol , 311–327. [DOI] [PubMed] [Google Scholar]
- Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, Brune A, Ishikawa T, Sachse C, Gonczy P. (2012). Cartwheel architecture of Trichonympha basal body. Science , 553. [DOI] [PubMed] [Google Scholar]
- Hai B, Gorovsky MA. (1997). Germ-line knockout heterokaryons of an essential alpha-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc Natl Acad Sci USA , 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann JM, Hoenger A, Mastronarde DN. (2011). Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J Struct Biol , 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. (2009). The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol , 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Linck RW. (1998). Two proteins isolated from sea urchin sperm flagella: structural components common to the stable microtubules of axonemes and centrioles. J Cell Sci (Pt 5), 585–595. [DOI] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE. (2006). Electron cryotomography sample preparation using the Vitrobot. Nat Protoc , 2813–2819. [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Liu D, Kastritis PL, Basu K, Hsu TC, Yang S, Bui KH. (2017). Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins. Nat Commun , 15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Brown JA, Yagi T, Norrander JM, Hirono M, Eccleston E, Kamiya R, Linck RW. (2003). Rib72, a conserved protein associated with the ribbon compartment of flagellar A-microtubules and potentially involved in the linkage between outer doublet microtubules. J Biol Chem , 7725–7734. [DOI] [PubMed] [Google Scholar]
- Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, Yates JR, 3rd, Winey M. (2007). New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol , 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirima J, Oiwa K. (2017). Flagellar-associated protein FAP85 is a microtubule inner protein that stabilizes microtubules. Cell Struct Funct , 1–14. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol , 71–76. [DOI] [PubMed] [Google Scholar]
- Li S, Fernandez JJ, Marshall WF, Agard DA. (2012). Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J , 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Yin W, Smith MC, Song K, Leigh MW, Zariwala MA, Knowles MR, Ostrowski LE, Nicastro D. (2014). Cryo-electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat Commun , 5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linck R, Fu X, Lin J, Ouch C, Schefter A, Steffen W, Warren P, Nicastro D. (2014). Insights into the structure and function of ciliary and flagellar doublet microtubules: tektins, Ca2+-binding proteins, and stable protofilaments. J Biol Chem , 17427–17444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari A, Obbineni JM, Bui KH, Shibata K, Toyoshima YY, Ishikawa T. (2015). alpha- and beta-tubulin lattice of the axonemal microtubule doublet and binding proteins revealed by single particle cryo-electron microscopy and tomography. Structure , 1584–1595. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol , 36–51. [DOI] [PubMed] [Google Scholar]
- Mochizuki K. (2008). High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene , 79–83. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Fu X, Heuser T, Tso A, Porter ME, Linck RW. (2011). Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc Natl Acad Sci USA , E845–E853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. (2006). The molecular architecture of axonemes revealed by cryoelectron tomography. Science , 944–948. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. (1999). High-resolution model of the microtubule. Cell , 79–88. [DOI] [PubMed] [Google Scholar]
- Patel-King RS, Benashski SE, King SM. (2002). A bipartite Ca2+-regulated nucleoside-diphosphate kinase system within the Chlamydomonas flagellum. The regulatory subunit p72. J Biol Chem , 34271–34279. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem , 1605–1612. [DOI] [PubMed] [Google Scholar]
- Porter ME, Sale WS. (2000). The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J Cell Biol , F37–F42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. (2017). CILIA: before and after. Cilia , 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P, Matsuoka T. (1989). Splitting the ciliary axoneme: implications for a “switch-point” model of dynein arm activity in ciliary motion. Cell Motil Cytoskeleton , 345–358. [DOI] [PubMed] [Google Scholar]
- Schwartz CL, Heumann JM, Dawson SC, Hoenger A. (2012). A detailed, hierarchical study of Giardia lamblia’s ventral disc reveals novel microtubule-associated protein complexes. PLoS One , e43783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky MA. (2002). A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc Natl Acad Sci , 3734–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Delgado-Escueta AV, Aguan K, Alonso ME, Shi J, Hara Y, Nishida M, Numata T, Medina MT, Takeuchi T, et al. (2004). Mutations in EFHC1 cause juvenile myoclonic epilepsy. Nat Genet , 842–849. [DOI] [PubMed] [Google Scholar]
- Vonderfecht T, Cookson MW, Giddings TH, Jr, Clarissa C, Winey M. (2012). The two human centrin homologues have similar but distinct functions at Tetrahymena basal bodies. Mol Biol Cell , 4766–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Stemm-Wolf AJ, Giddings TH, Jr, Pearson CG. (2012). Cytological analysis of Tetrahymena thermophila. Methods Cell Biol , 357–378. [DOI] [PubMed] [Google Scholar]
- Witman GB, Plummer J, Sander G. (1978). Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J Cell Biol , 729–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2015). The Multiple Functions of EFHC1 in Tetrahymena and Xenopus. PhD thesis. University of Colorado, Boulder. http://libraries.colorado.edu/record=b8507521~S3. [Google Scholar]
- Zhao Y, Shi J, Winey M, Klymkowsky MW. (2016). Identifying domains of EFHC1 involved in ciliary localization, ciliogenesis, and the regulation of Wnt signaling. Dev Biol , 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.