Significance
Using an interactomic approach, we have identified the nuclear receptor REV-ERBα as a O-GlcNAc transferase (OGT) protein partner. REV-ERBα protects cytoplasmic OGT from proteasomal degradation and facilitates cytosolic and nuclear protein O-GlcNAcylation while REV-ERα ligands decreased cytoplasmic OGT activity. REV-ERBα thus exerts pleiotropic activities through OGT, coordinating signal transduction, epigenomic programming, and transcriptional response in the liver.
Keywords: O-GlcNAcylation, REV-ERBα, metabolism, epigenomics, signal transduction
Abstract
The nuclear receptor REV-ERBα integrates the circadian clock with hepatic glucose and lipid metabolism by nucleating transcriptional comodulators at genomic regulatory regions. An interactomic approach identified O-GlcNAc transferase (OGT) as a REV-ERBα−interacting protein. By shielding cytoplasmic OGT from proteasomal degradation and favoring OGT activity in the nucleus, REV-ERBα cyclically increased O-GlcNAcylation of multiple cytoplasmic and nuclear proteins as a function of its rhythmically regulated expression, while REV-ERBα ligands mostly affected cytoplasmic OGT activity. We illustrate this finding by showing that REV-ERBα controls OGT-dependent activities of the cytoplasmic protein kinase AKT, an essential relay in insulin signaling, and of ten-of-eleven translocation (TET) enzymes in the nucleus. AKT phosphorylation was inversely correlated to REV-ERBα expression. REV-ERBα enhanced TET activity and DNA hydroxymethylated cytosine (5hmC) levels in the vicinity of REV-ERBα genomic binding sites. As an example, we show that the REV-ERBα/OGT complex modulates SREBP-1c gene expression throughout the fasting/feeding periods by first repressing AKT phosphorylation and by epigenomically priming the Srebf1 promoter for a further rapid response to insulin. Conclusion: REV-ERBα regulates cytoplasmic and nuclear OGT-controlled processes that integrate at the hepatic SREBF1 locus to control basal and insulin-induced expression of the temporally and nutritionally regulated lipogenic SREBP-1c transcript.
O-GlcNAcylation is a posttranslational modification (PTM) targeting both cytoplasmic and nuclear proteins (1), affecting their cellular sublocalization, stability, and/or activity (2). O-GlcNAcylation involves a biosynthetic arm, the hexosamine biosynthetic pathway (HBP), and a catalytic arm acting through two enzymes with opposite activities, namely O-GlcNAc transferase (OGT) catalyzing N-acetylglucosamine (GlcNAc) transfer from UDP-GlcNAc to polypeptides, and O-GlcNAcase (OGA/MGEA5), which removes this sugar moiety (3). As generation of the rate-limiting UDP-GlcNAc requires glucose (Glc), glutamine, gluconeogenic or ketogenic amino acids, fatty acids, uridine, and ATP as precursors, OGT activity reflects the cellular metabolic status (2). OGT modifies hundreds of proteins, including proteins involved in the regulation of metabolism, such as carbohydrate-responsive element-binding protein (ChREBP) (4). OGT also couples metabolism to epigenomic control of transcription. As an example, when integrated into a host cell factor 1 (HCF-1)/OGT complex, PGC1α is O-GlcNAcylated and is protected from ubiquitinylation and degradation (5). In addition, OGT interacts with the corepressor Sin3a and ten-eleven translocation (TET methylcytosine hydroxylases) family members to O-GlcNAcylate RNA polymerase II and histone H2B (6). Hence, OGT imposes a metabolic control over transcriptional regulation.
The cyclically expressed nuclear receptors REV-ERBα and REV-ERBβ are key transcription factors involved in the regulation of the circadian clock machinery and of metabolism. Genetic ablation of Rev-erbα and/or Rev-erbβ influences adipocyte differentiation, lipid, cholesterol, and glucose metabolism and leads to cardiometabolic abnormalities (7). REV-ERBα has a prominent regulatory role in hepatic metabolism by acting as a transcriptional repressor regulated by small synthetic ligands and heme, its natural ligand (8). REV-ERBα represses gene expression by recruiting a HDAC3/NCoR corepressor complex in a ligand-dependent manner, and liver Hdac3 or Ncor gene deletion partially phenocopies that of Rev-erbα (9, 10). Recent data suggest that REV-ERBα regulates liver metabolic gene expression through tethering to HNF6, whereas clock genes are regulated through direct DNA binding (11). REV-ERBα, through its interaction with HSP90, affects the stability of the glucocorticoid receptor (12). REV-ERBα thus increases its functional versatility by engaging in various protein–protein interactions. However, little is known about other REV-ERBα interaction partners and associated functions. In this study, we have identified OGT as a binding partner of REV-ERBα, and we have characterized the functional consequences of this partnership on integrated hepatic intracellular signaling.
Results
REV-ERBα Interacts with OGT.
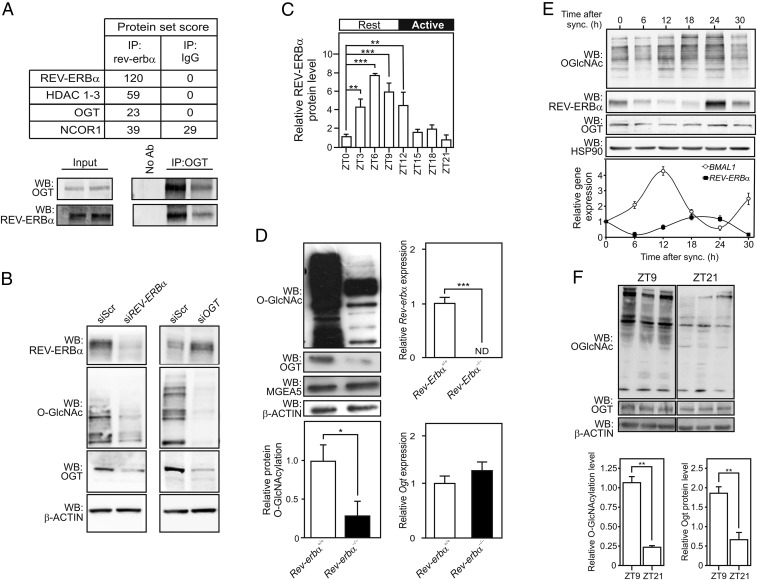
To identify novel REV-ERBα−interacting proteins, nuclear extracts from human hepatoma HepG2 cells were cross-linked then immunoprecipitated using a REV-ERBα−specific antibody. This procedure followed by nano LC-MS/MS (RIME; ref. 13) identified several nuclear REV-ERBα−interacting proteins (Fig. 1A, Upper and SI Appendix, Table S1), including the known REV-ERBα corepressor complex NCOR1/HDAC3 as well as OGT. The REV-ERBα/OGT interaction was confirmed by co-IP of native protein complexes (Fig. 1A, Lower). Considering the instrumental roles of both proteins in controlling liver metabolism, we investigated the functional role(s) of the REV-ERBα/OGT interaction.
Fig. 1.
REV-ERBα interacts with OGT and controls O-GlcNAcylation. (A) Most-relevant proteins identified by RIME in HepG2 cells extract (Upper). Validation of the REV-ERBα/OGT interaction by anti-OGT coimmunoprecipitation (IP) of HepG2 whole cellular extract followed by anti-OGT and anti-REV-ERBα WB (n = 2; Lower). (B) O-GlcNAcylation levels in cellular extracts from HepG2 cells depleted from REV-ERBα or OGT mRNAs. (C) Time-dependent REV-ERBα protein expression level in mouse livers. (D) O-GlcNAcylation, OGT, and OGA proteins levels in protein extracts from WT or Rev-erbα−/−mouse livers at ZT6. Rev-erbα and Ogt mRNA levels were assessed by RT-qPCR. ND, not detectable. (E) O-GlcNAcylation, REV-ERBα, and OGT protein levels (Upper) and REV-ERBα and BMAL1 relative gene expression (Lower) in synchronized U2OS cells. (F) O-GlcNAcylation and OGT protein levels in mouse livers at ZT9 and ZT21. Results are expressed as mean ± SEM, and values were compared by a two-way ANOVA followed by a Bonferroni post hoc test (C) or a t test (D and F). *P < 0.05, **P < 0.01, ***P < 0.001.
REV-ERBα Regulates Cellular O-GlcNAcylation Levels.
We first monitored protein O-GlcNAcylation levels in HepG2 cells by Western blotting (WB) with an anti-O-GlcNAc antibody (RL2; Fig. 1B) at 25 mM Glc, a condition increasing protein O-GlcNAcylation (SI Appendix, Fig. S1A). siRNA-mediated knockdown of REV-ERBα or of OGT equally decreased protein O-GlcNAcylation and OGT levels (Fig. 1B). As REV-ERBα knockdown did not affect OGT mRNA expression (SI Appendix, Fig. S1B), we concluded that the REV-ERBα protein level controls OGT protein stability. REV-ERBα protein expression fluctuates in a circadian manner in mouse liver reaching the highest expression at ZT6–ZT12 (Fig. 1C and SI Appendix, Fig. S1C). Since REV-ERBα also interacts with OGT in liver extracts (SI Appendix, Fig. S1D), we examined whether O-GlcNAcylation regulation by REV-ERBα occurs in vivo. Protein O-GlcNAcylation was compared at ZT6 in wild-type and Rev-erbα−deficient mice. Protein O-GlcNAcylation and OGT protein expression were drastically reduced in Rev-erbα−/− livers (Fig. 1D and SI Appendix, Fig. S1E), whereas Ogt mRNA steady-state levels were not altered in this genetic background (Fig. 1D).
REV-ERBα mRNA and protein expression cycles in synchronized human osteosarcoma U2OS cells. Maximal expression of REV-ERBα protein was observed in serum-shocked U2OS cells 24 h after synchronization and inversely correlated with Bmal1 expression, a cognate direct REV-ERBα target gene (Fig. 1E). OGT and protein O-GlcNAcylation levels peaked 24 h after synchronization, thus following REV-ERBα protein expression and activity as monitored by Western blot and BMAL1 expression, respectively, and exhibited a trough 30 h after the serum shock, corresponding to the nadir of REV-ERBα expression (Fig. 1E). Of note, OGT and O-GlcNAcylation fluctuations were not observed in an U2OS cell subclone in which REV-ERBα protein expression was abrogated through CrispR/Cas9-mediated gene inactivation (SI Appendix, Fig. S1F, Left and SI Appendix, Fig. S1G). Increased REV-ERBα, OGT, and protein O-GlcNAcylation levels were also detected in mouse livers at ZT9 compared with ZT21, corresponding to maximal and minimal liver REV-ERBα protein expression in vivo, respectively (Fig. 1 C and F). Taken together, these data show that the REV-ERBα protein level controls OGT protein level and/or activity. As REV-ERBβ displays overlapping functions with REV-ERBα (7, 9), we assessed whether REV-ERBβ also regulates protein O-GlcNAcylation in HepG2 cells. Knockdown of REV-ERBβ expression affected neither OGT expression nor cellular protein O-GlcNAcylation levels (SI Appendix, Fig. S1 A and B). Furthermore, CrispR/Cas9-mediated REV-ERBβ gene ablation did not affect the circadian modulation of OGT and O-GlcNAcylation. (SI Appendix, Fig. S1F, Right and SI Appendix, Fig. S1G), showing that the observed OGT protein stabilization is REV-ERBα−specific.
Whether a reciprocal regulation occurs between REV-ERBα and OGT was assessed (SI Appendix, Fig. S2). 6-diazo-5-oxo-1-norleucine (DON) is a chemical inhibitor of glucosamine-fructose-6-phosphate aminotransferase, which alters UDP-GlcNAc–dependent pathways including OGT-catalyzed O-GlcNAcylation. DON efficiently blunted the glucose-induced expression of liver pyruvate kinase known to be an OGT-dependent, ChREBP-mediated process (4). In contrast, glucose and/or DON treatments did not modify endogenous REV-ERBα transcriptional activity (SI Appendix, Fig. S2A) despite an observed stabilization of the REV-ERBα protein by DON. The ability of REV-ERBα to recruit the transcriptional corepressor NCOR1, as assessed in a mammalian two-hybrid assay, was neither altered by the OGA/MGEA5-specific inhibitor Thiamet G nor by the OGT activator glucosamine (SI Appendix, Fig. S2B). Immunoprecipitated REV-ERBα submitted to Click-it chemistry, which substitutes O-GlcNAc moieties with azido-modified galactose and allows subsequent detection with alkyne-derived markers, did not provide evidence for REV-ERBα O-GlcNAcylation (SI Appendix, Fig. S2C). The absence of REV-ERBα O-GlcNAcylation was confirmed by mass spectrometry analysis of REV-ERBα immunopurified from HepG2 cells. Therefore, REV-ERBα transcriptional activity is not sensitive to HBP modulation and REV-ERBα is not detectably O-GlcNAcylated in the studied conditions.
REV-ERBα Ligand Binding Interferes with O-GlcNAcylation.
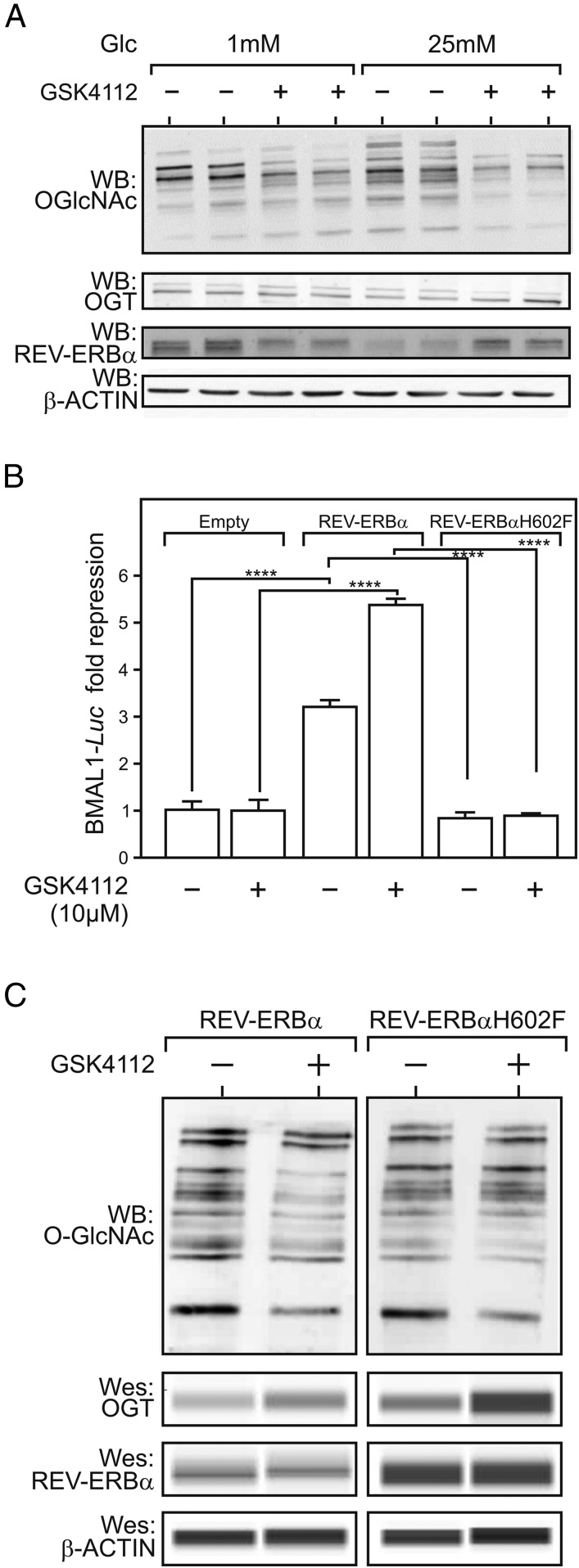
We next assessed whether ligand binding to REV-ERBα affects protein O-GlcNAcylation. We first interfered with intracellular heme levels (typically around 2–4 µM, ref. 14) in HepG2 cells by knocking down the expression of the rate-limiting heme synthesis enzyme ALAS1 (SI Appendix, Fig. S3A). This resulted in a modest but detectable decrease in heme level (SI Appendix, Fig. S3B) and in increased cellular protein O-GlcNAcylation (SI Appendix, Fig. S3C). This result was consistent with the effect of succinylacetone, a potent inhibitor of heme synthesis, which also increased protein O-GlcNAcylation at both low and high glucose concentrations without significantly altering REV-ERBα protein levels (SI Appendix, Fig. S3D). As treatment of HepG2 cells with the heme precursor delta-aminolevulinic acid did not reproducibly induce major changes in protein O-GlcNAcylation probably due to an precise regulation of intracellular heme level (SI Appendix, Fig. S3D), we treated cells with the high affinity, heme-mimicking REV-ERBα synthetic agonist GSK4112 (Fig. 2A). This compound efficiently blunted protein O-GlcNAcylation at low and high glucose concentrations, again irrespectively of REV-ERBα protein levels (Fig. 2A). To ensure that this effect was mediated by REV-ERBα ligation, we compared the ability of wild-type REV-ERBα to regulate cellular protein O-GlcNAcylation to that of a heme binding-crippled REV-ERBα mutant. Heme binding to REV-ERBα (Kd 2–4 µM, ref. 15) is prevented by mutating H602 into F (16). REV-ERBα H602F displayed a decreased transcriptional repressive activity (Fig. 2B) when overexpressed in REV-ERBα−negative HEK293 cells (SI Appendix, Fig. S3E). Treatment with the REV-ERB agonist GSK4112 blunted protein O-GlcNAcylation in the presence of wild-type REV-ERBα−expressing cells, but not in H602F-expressing cells (Fig. 2C). Thus, REV-ERBα ligation decreases protein O-GlcNAcylation without affecting OGT level as opposed to REV-ERBα deficiency. Taken together, this suggests that REV-ERBα controls both OGT activity and activity.
Fig. 2.
REV-ERBα agonists reduce protein O-GlcNAcylation. (A) Protein O-GlcNAcylation, OGT, and REV-ERBα levels after treatment of HepG2 cells with the synthetic REV-ERBα agonist GSK4112 (10 µM). (B) REV-ERBα repressive activity determined by a transactivation assay using a Bmal-luc reporter gene, expression vectors coding for either wild-type or mutated REV-ERBα (WT, H602F) or empty vector (empty) transfected into HEK293 cells treated or not with 10 µM GSK4112. (C) O-GlcNAcylation, OGT, and REV-ERBα protein levels in HEK293 whole-cell extracts after transfection with REV-ERBα WT or H602F and treatment or not with 10 µM GSK4112 at 25 mM Glc. Results are expressed as mean ± SEM, and values were compared by a two-way ANOVA followed by a Tukey post hoc test, ****P < 0.0001. Wes, Simple Western (ProteinSimple).
REV-ERBα Modulates OGT Activity in both Cytoplasmic and Nuclear Compartments.
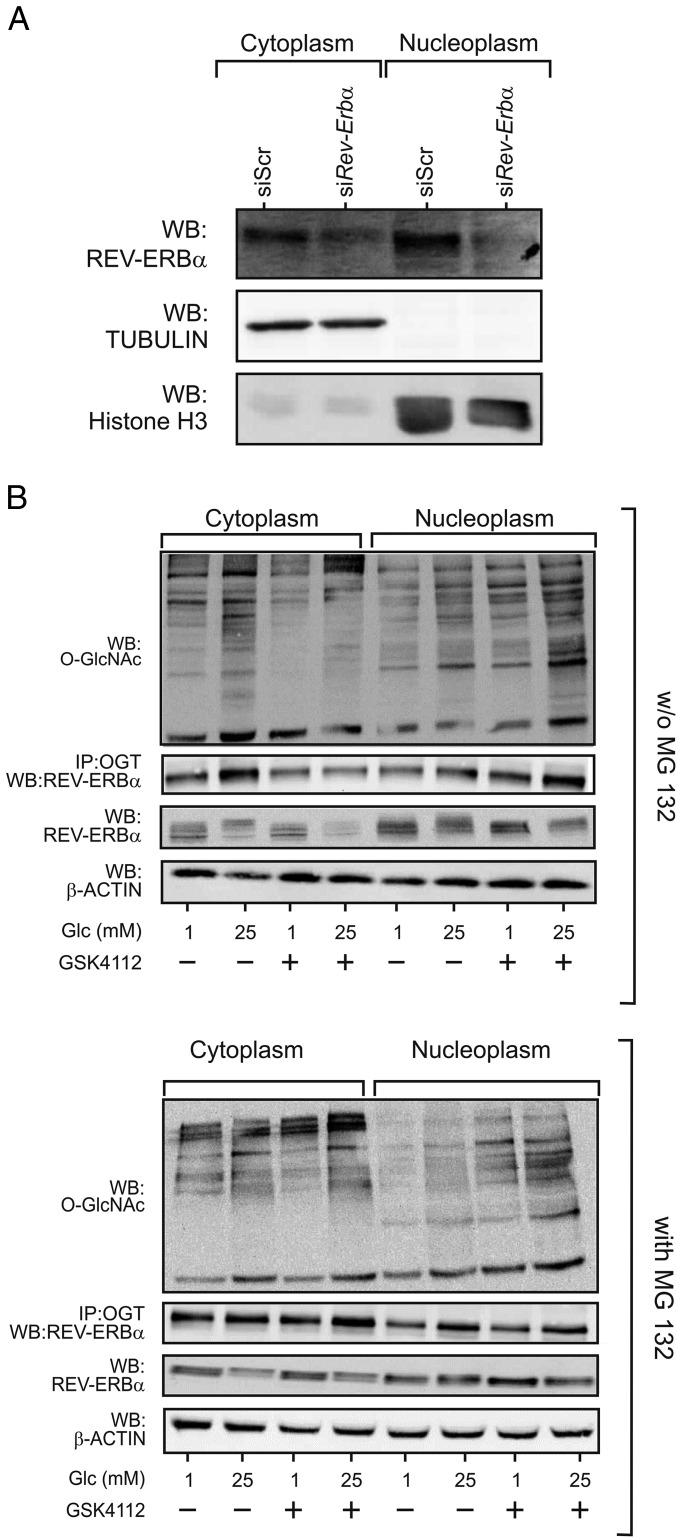
As we confirmed that OGT and REV-ERBα are located in both the cytoplasmic and nuclear compartments (refs. 2 and 17, Fig. 3 A and B, and SI Appendix, Fig. S4A), thus possibly undergoing distinct cellular fates, we investigated the possibility that cytoplasmic and nuclear OGT could be regulated differentially by REV-ERBα. We observed that OGT degradation, hence decreased total protein O-GlcNacylation, triggered by REV-ERBα protein depletion was prevented upon 26S proteasome complex inhibition by MG132 (carbobenzoxy-Leu-Leu-leucinal) (SI Appendix, Fig. S4B). In contrast, the ligand-induced decrease in cellular protein O-GlcNAcylation was not MG132-sensitive (SI Appendix, Fig. S4C). Cell fractionation showed that REV-ERBα protein level decreased in the cytoplasm and increased in the nuclear compartment upon ligand treatment (Fig. 3B, Upper and SI Appendix, Fig. S4A), in agreement with a previous study (18). We thus tested the possibility that the altered REV-ERBα subcellular distribution triggered by GSK4112 selectively modifies OGT activity in the cytoplasmic and nucleoplasmic compartments (Fig. 3B). GSK4112 treatment blunted the glucose-induced O-GlcNAcylation of cytoplasmic proteins. In contrast, O-GlcNAcylation of nucleoplasmic proteins increased in these conditions, thus concomitant with ligand-induced nuclear accumulation of REV-ERBα and with increased interaction of nuclear OGT with REV-ERBα. Proteasome inhibition prevented ligand-induced cytoplasmic REV-ERBα depletion and abolished the ligand-induced decrease of cytoplasmic REV-ERBα/OGT interaction and O-GlcNAcylation levels (Fig. 3B). However, MG132 did not influence protein O-GlcNAcylation in the nuclear compartment, indicating that nuclear OGT is insensitive to proteasomal degradation (Fig. 3B). Taken together, these data suggest that ligand treatment triggers REV-ERBα cytoplasmic depletion, hence negatively impacting protein O-GlcNAcylation in a proteasome-dependent manner, while promoting its accumulation in the nucleus and interaction with OGT, hence increasing nuclear OGT activity.
Fig. 3.
REV-ERBα interaction with cytoplasmic OGT prevents its proteasomal degradation. (A) REV-ERBα is detected in both the cytoplasm and nucleus of target cells. Cytoplasmic and nuclear fractions from HepG2 cells were analyzed by WB for their REV-ERBα content. (B) Cytoplasmic and nuclear protein O-GlcNAcylation and REV-ERBα protein levels were determined by WB. The REV-ERBα/OGT interaction was analyzed by OGT IP followed by REV-ERBα WB analysis. HepG2 cells were cultured at 1 or 25 mM Glc and treated with 10 µM GSK4112 with (Lower) or without (Upper) 5 µM MG132.
REV-ERBα−Dependent O-GlcNAcylation Targets Cytoplasmic and Nuclear Structural and Effector Proteins.
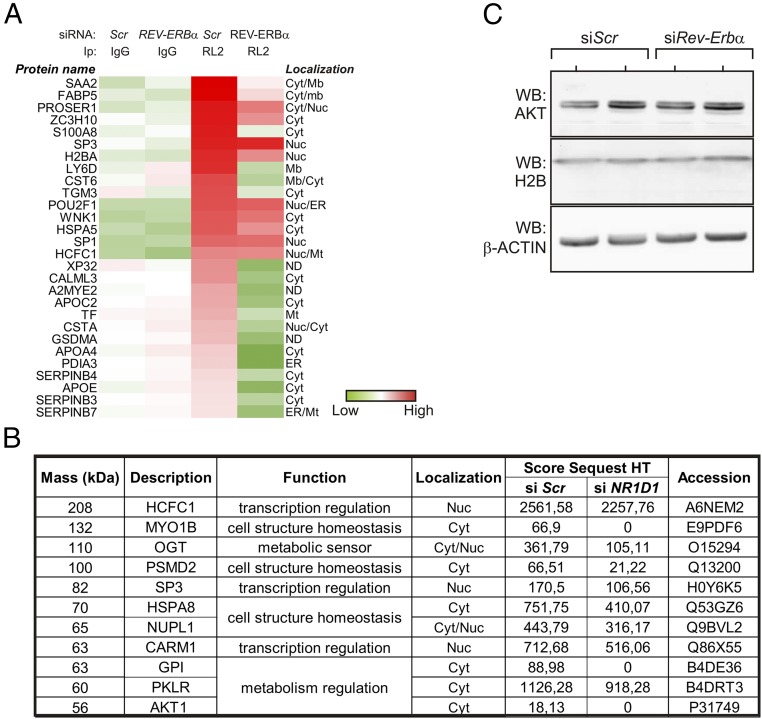
Since data above point at REV-ERBα protein expression and subcellular localization as a set point of protein O-GlcNAcylation, we identified cellular proteins whose O-GlcNAcylation is potentially controlled by REV-ERBα by label-free mass spectrometry. HepG2 cellular extracts were immunoprecipitated with either a nonspecific IgG (Ctrl IgG) or the anti-O-GlcNAc antibody RL2, after transfection with a negative control siRNA (Ctrl) or an anti-REV-ERBα siRNA (Fig. 4A). This approach identified 281 proteins present in at least one condition, among which 71 were specifically enriched upon RL2 IP. Twenty-eight proteins were less represented in immunoprecipitates from REV-ERBα–depleted cells (Fig. 4A), including several previously identified O-GlcNAcylated proteins with structural, signaling, or transcriptional functions such as histone H2B, the transcription factors SP1 and SP3, HCFC1/VP16-accessory protein, and the protein kinase WNK1 (17, 19, 20). To detect lower expressed putative OGT targets, immunoprecipitates were also resolved by SDS/PAGE and areas of the gel ranging from 50 kDa to 170–200 kDa were analyzed by LC-MS/MS after tryptic digestion (Fig. 4B and SI Appendix, Table S2). Top ranking proteins again included HCFC1 and SP3, and interestingly other proteins such as OGT itself, the nuclear receptor coactivator CARM1, and AKT1. Although this approach could also potentially identify proteins with a decreased steady-state level in REV-ERBα−depleted cells or interacting with O-GlcNAcylated proteins, many of them have previously been described as bona fide OGT targets and O-GlcNAcylated polypeptides (17, 21, 22). As an example, both AKT and H2B were similarly expressed in naive and REV-ERBα−depleted cells, suggesting that OGT activity does not affect their stability (Fig. 4C). Taken together, these analyses show that REV-ERBα expression likely regulates the O-GlcNAcylation level of multiple cytoplasmic and nuclear proteins or of associated proteins with important regulatory functions (SI Appendix, Table S2).
Fig. 4.
REV-ERBα impacts on protein O-GlcNAcylation in both cytoplasmic and nuclear compartments. (A) Label-free mass spectrometry analysis of O-GlcNAcylated proteins. Control (IgG) or O-GlcNAcylated protein-enriched (RL2) immunoprecipitates from siRNA (Scr or REV-ERBα)-transfected HepG2 cells were analyzed by label-free mass spectrometry analysis. (B) LC-MS/MS analysis of O-GlcNAcylated proteins. HepG2 cells were treated like in A, and immunoprecipitates were fractionated by SDS/PAGE before mass spectrometry analysis. (C) Cellular abundance of AKT and H2B in siRNA-transfected HepG2 cells. HepG2 cells were cultured at 25 mM Glc. Cyt, cytoplasm; ER, endoplasmic reticulum; Mb, plasma membrane; Mt, mitochondria; Nuc, nucleoplasm.
Cytoplasmic OGT Regulates AKT Phosphorylation in a REV-ERBα−Dependent Manner.
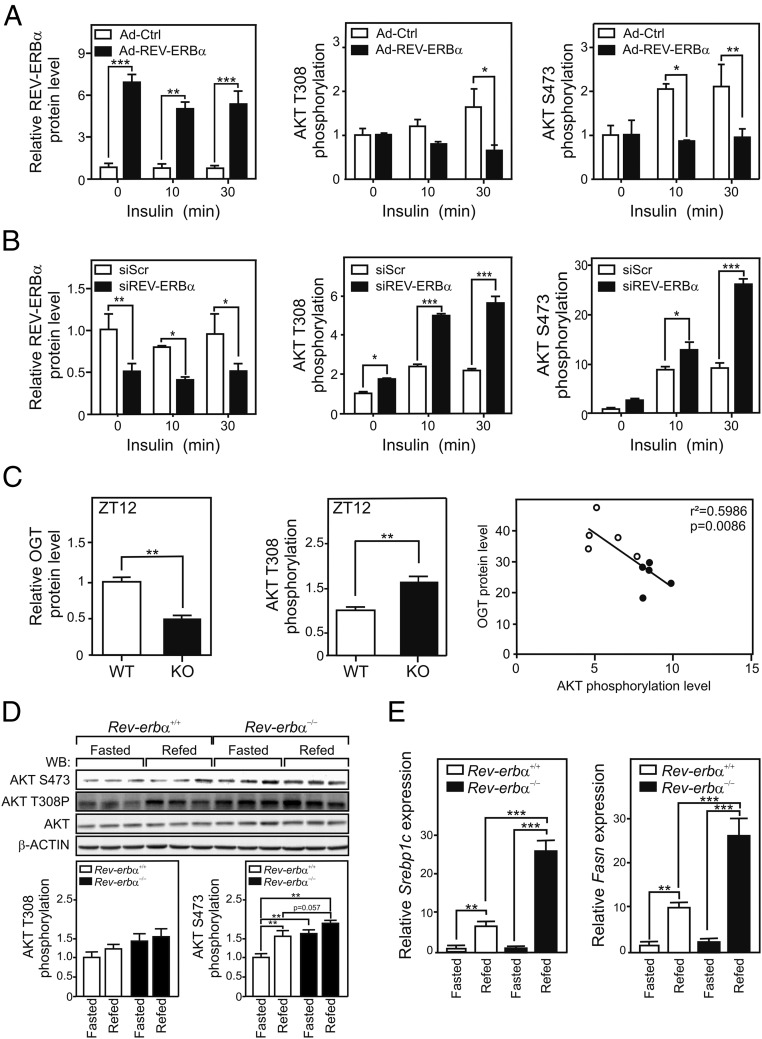
In response to insulin, AKT T308 and S473 are phosphorylated by PDK1 and mTORC2, respectively (23). O-GlcNAcylation impairs AKT phosphorylation and inhibits insulin signaling in mouse liver (22, 24). Since REV-ERBα overexpression increased AKT O-GlcNAcylation in HepG2 cells as assessed by Click-It chemistry (SI Appendix, Fig. S5A), we further tested whether REV-ERBα controls AKT phosphorylation. Overexpression of REV-ERBα through adenoviral transduction (Fig. 5A and SI Appendix, Fig. S5 B and C) increased OGT protein level and cellular protein O-GlcNAcylation levels (SI Appendix, Fig. S5B). In parallel, insulin-stimulated AKT phosphorylation decreased at both S473 and T308 upon REV-ERBα overexpression (Fig. 5A and SI Appendix, Fig. S5C). Conversely, REV-ERBα knockdown increased AKT phosphorylation (Fig. 5B and SI Appendix, Fig. S5D). To assess whether REV-ERBα−dependent OGT stabilization also controls AKT phosphorylation in vivo, OGT protein and AKT phosphorylation levels were analyzed in liver extracts from ad libitum-fed Rev-erbα+/+ or Rev-erbα−/− mice. Rev-erbα deficiency reduced OGT protein levels in mouse liver (Fig. 5C and SI Appendix, Fig. S5E) as described above (Fig. 1E). Both circadianly (ZT0) and genetically induced Rev-erbα depletion (Rev-erbα−/−) were associated with increased AKT T308 phosphorylation, which correlated with OGT protein levels (Fig. 5C and SI Appendix, Fig. S5E). The expression of the lipogenic transcription factor Srebp1c in mice is positively (auto)regulated through the insulin/AKT pathway and, after S1P/S2P-mediated processing, controls the expression of genes coding for lipid biosynthetic enzymes such as Scd and Fasn (25). Basal AKT phosphorylation at S473, a process known to be insulin-dependent (26), was significantly higher in fasted Rev-erbα−/− liver, and T308 phosphorylation showed a similar trend. Similarly, phosphorylation levels were higher in refed Rev-erbα−/− liver (Fig. 5D). In Rev-erbα−/− mouse liver, the response to refeeding also translated into increased hepatic expression of Srebp1c and of its target gene Fasn when compared its Rev-erbα+/+ counterpart (Fig. 5E). These findings thus suggest that REV-ERBα also acts nontranscriptionally on this signaling axis by controlling AKT O-GlcNAcylation.
Fig. 5.
REV-ERBα expression level alters AKT phosphorylation. (A) REV-ERBα protein level, T308, and S473 AKT phosphorylation were quantified in HepG2 cellular extracts after transduction with control (LacZ, open bars) or REV-ERBα (filled bars) adenoviruses and treated with 60 nM insulin for the indicated times. (B) HepG2 cells were transfected with siRNA (Scr and REV-ERBα) and treated with 60 nM insulin for the indicated times. REV-ERBα, AKT T308, and S473 phosphorylation were quantified. (C) OGT (Left) and phospho-AKT T308 (Center) hepatic content. The correlation analysis in ad libitum-fed WT (n = 5) and Rev-erbα KO (n = 5) mice at ZT12 (Right) was performed with data from SI Appendix, Fig. S5E. (D) Hepatic P-AKT T308 and AKT protein levels (ZT6) from WT or Rev-erbα KO mice after a 30-h fast (Fasted) or refed for 6 h after fasting (Refed). (E) Relative Srebp1c (Left) and Fasn (Right) gene expression in livers (ZT6) from WT and Rev-erbα KO mice fasted or refed as described in D. Data are expressed as mean ± SEM. The statistical significance was assessed by a two-way ANOVA followed by a Bonferroni post hoc test (A, B, and E) or a t test (C). *P < 0.05, **P < 0.001, and ***P < 0.001.
TET Activity and Methylcytosine Hydroxylation Are REV-ERBα−Dependent.
The label-free mass spectroscopy analysis also identified H2B as a REV-ERBα−dependent O-GlcNAcylated protein (Fig. 4). Histone H2B O-GlcNAcylation is catalyzed by chromatin-bound OGT through interaction with TET oxidases (TET), which have recently emerged as major epigenomic players by regulating cytosine hydroxymethylation (27). Reciprocally, OGT O-GlcNAcylates TET enzymes and alters their enzymatic properties through ill-defined mechanisms (21). We therefore tested whether REV-ERBα impacts on TET activity through OGT.
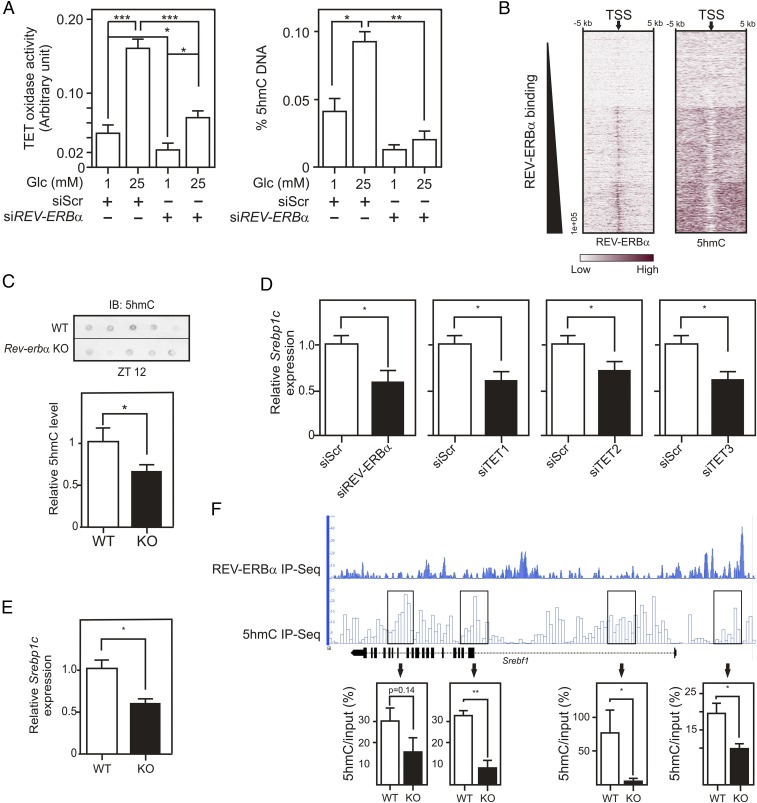
As described (28), glucose significantly increases TET enzymatic activity in HepG2 cells (Fig. 6A). REV-ERBα knockdown blunted Glc-induced TET enzymatic activity (Fig. 6A). REV-ERBα depletion did not reduce nuclear OGT protein content but reduced its enzymatic activity as total nuclear protein O-GlcNAcylation decreased by ∼20% (SI Appendix, Fig. S6 A and B) without affecting TET protein levels (SI Appendix, Fig. S6C). In line, global cytosine hydroxymethylation of the HepG2 cell genome was significantly decreased upon REV-ERBα knockdown at both 1 and 25 mM Glc (Fig. 6A). GSK4112 treatment increased TET enzymatic activity at 1 mM Glc but was unable to do so at 25 mM Glc, probably reflecting a saturation of the system (SI Appendix, Fig. S6D). REV-ERBα overexpression increased cytosine hydroxymethylation, and this response were clearly blunted by OSMI-1, a cell permeable OGT inhibitor (ref. 29 and SI Appendix, Fig. S6 E and F). REV-ERBα is thus an important determinant of TET activity and controls global 5hmC levels in an OGT-dependent manner in vitro.
Fig. 6.
Nuclear REV-ERBα controls nuclear OGT and TET activities and affects DNA hydroxymethylation. (A) TET oxidase activities (Left) and 5hmC levels (Right) in HepG2 nuclear protein extracts or in genomic DNA, respectively. Cells were transfected with siRNA (control or REV-ERBα) and incubated at 1 or 25 mM Glc. (B) Heatmaps of REV-ERBα and 5hmC signal intensities in clusterized Gencode TSS. Gencode TSS (arrow) were aligned and extended 5 kb on each side. (C) 5hmC levels in Rev-erbα+/+ (WT) and Rev-erbα−/− (KO; n = 5–6) mouse livers. (D) Relative SREBP1C gene expression determined by RT-qPCR on siRNA (Scr, REV-ERBα, TET1, TET2, or TET3)-transfected HepG2 cells. (E) Relative Srebp1c gene expression in liver from Rev-erbα+/+ or Rev-erbα−/− mice (ZT12). (F) Representation of REV-ERBα chromatin occupancy and hydroxymethylated region localization at the mouse hepatic Srebf1 locus (Upper). Srebf1 hydroxymethylation was quantified by hMeDIP-qPCR (Bottom) in hepatic genomic DNA from Rev-erbα+/+ (n = 2) or Rev-erbα−/− mice (n = 2). Histogram represents mean ± SEM. The statistical significance of differences was assessed by a two-way ANOVA followed by a Bonferroni post hoc test (A) or by a t test. *P < 0.05, **P < 0.01, ***P < 0.001.
TET/OGT complexes are mostly targeted to promoter regions through interaction of TET with DNA (19). We thus investigated whether REV-ERBα genomic binding overlaps with 5hmC content in mouse liver. Using previously published data for C57BL/6 mouse liver (30), we confirmed that 5hmC localizes to genomic regions neighboring transcription start sites (TSS; SI Appendix, Fig. S7A). Mapping REV-ERBα genomic binding sites in C57BL/6 mouse liver (31) in the vicinity (±5 kb) of TSS showed a direct correlation between REV-ERBα binding and 5hmC density (Fig. 6B and SI Appendix, Fig. S7B). 5hmC levels were measured in liver from Rev-erbα+/+ and Rev-erbα−/− in mice (Fig. 6C). Rev-erbα−/− mouse DNA showed a decreased 5hmC content compared with DNA from wild-type littermates while the genetic background did not affect TET protein levels (SI Appendix, Fig. S7C). Taken together, these observations show that REV-ERBα impacts on nuclear OGT activity and increases DNA 5hmC level.
In addition to nutritional regulation, Srebp1c expression undergoes diurnal variation, with a peak occurring at ZT14–18, which is imposed in part by REV-ERBα, whose knockout decreases Srebp1c expression (32, 33). We first tested whether REV-ERBα regulates SREBP1C expression in a cell-autonomous manner. REV-ERBα deficiency in HepG2 cells decreased SREBP1C basal expression (Fig. 6D). TET1, TET2, or TET3 knockdown (Fig. 6D and SI Appendix, Fig. S7D) led to a significantly decreased expression of SREBP1C (Fig. 6D). Thus, TETs sustain gene expression at this locus in vitro. Similarly, Rev-erbα deficiency decreased hepatic Srebp1c gene expression in ad libitum-fed mice (Fig. 6E). REV-ERBα bound to genomic regions in the vicinity or within the Srebf1 gene, and an increased 5hmC density was observed around REV-ERBα binding sites (Fig. 6F, Upper). Interestingly, 5hmC density at these sites decreased in Rev-erbα−/− livers, thereby paralleling the decreased Srebp1c gene expression in this genetic background (Fig. 6F, Lower).
In conclusion, these data show that REV-ERBα controls TET oxidase activity, thereby controlling epigenomic marking at the Srebf1 locus whose basal expression is regulated through the REV-ERBα/OGT/TET axis.
Discussion
The regulatory pathways controlling OGT activity and expression are not yet fully understood. OGT is regulated by the UDP-GlcNAc pool, which fluctuates with the varying availability of nutrients such as glucose, glutamate, and free fatty acids. In addition, various PTMs modulate OGT activity, localization, substrate selectivity, or stability such as O-GlcNAcylation itself, phosphorylation, or ubiquitinylation (2). OGT-protein partners, such as MYPT1 and CARM1, affect OGT activity or substrate selectivity (34). Reciprocally, OGT regulates the activity of numerous proteins, including circadian clock proteins (35).
In this study, we identify a mechanism regulating OGT stability and activity. We show that REV-ERBα, but not REV-ERBβ with which it shares only 53% amino acid sequence homology, interacts with cytoplasmic and nuclear OGT. REV-ERBα itself is however not a substrate for OGT, and its transcriptional properties are not affected by the HBP/OGT pathway. A synthetic REV-ERBα ligand favored REV-ERBα cytoplasmic depletion and nuclear accumulation and, consequently, induced cytoplasmic proteasomal OGT degradation, thereby compromising protein O-GlcNAcylation in this cellular compartment while increasing nuclear OGT activity. Collectively, this suggests that REV-ERBα unexpectedly acts as a regulator of protein stability, therefore expanding the repertoire of its biological properties beyond that of a transcriptional regulator.
Mass spectrometry analysis identified O-GlcNAcylated proteins or proteins bound to them, among which AKT was an interesting target as a key regulator of insulin signaling. AKT phosphorylation is reduced upon OGT overexpression, hence inducing insulin resistance (36) and REV-ERBα (over)expression impinges on AKT phosphorylation in vitro. OGT protein level is reduced in Rev-erbα−/− mouse liver and correlates with increased AKT phosphorylation, thus potentially impacting on hepatic insulin signaling. Interestingly, insulin signaling is a circadian-regulated phenomenon, as mice display decreased insulin signaling at ZT7 (37), corresponding to the Rev-erbα expression zenith (Fig. 1C). In addition, AKT phosphorylation peaks at night in mouse liver when Rev-erbα expression is lowest (38, 39). Insulin signaling follows a circadian rhythm in humans, with a low insulin responsiveness being observed in the evening when REV-ERBα is expressed (40, 41). These data are in line with our observation that physiologically (ZT0) or genetically induced (Rev-erbα−/− mice) REV-ERBα depletion reduces liver OGT protein content and favors AKT phosphorylation, hence leading to increased insulin responsiveness. In the presence of REV-ERBα (ZT6–12 in mouse liver), cytoplasmic OGT is stabilized and AKT phosphorylation is decreased, hence providing a molecular basis for the observed reduced insulin signaling. Thus, circadian modulation of insulin signaling could occur, at least partly, through the REV-ERBα−dependent control of cytoplasmic OGT and of its target AKT.
The REV-ERBα/OGT interaction also occurs in the nucleus. Contrasting with cytoplasmic OGT, nuclear OGT protein level is not modulated by REV-ERBα expression levels. However, OGT activity is reduced upon REV-ERBα down-regulation, as suggested by decreased H2B O-GlcNAcylation in siRev-erbα-transfected HepG2 cells. The impact of nuclear REV-ERBα on OGT activity was investigated by monitoring TET1/2/3 oxidase activity, which were described as glucose-sensitive and OGT-sensitive enzymes in ES cells (19, 28, 42). The REV-ERBα−dependant increase of TET activity and DNA hydroxymethylation are also OGT-dependent. Reduced expression of REV-ERBα decreases nuclear protein O-GlcNAcylation levels and blunts glucose-induced TET activity and DNA hydroxymethylation in HepG2 cells. These findings, showing that OGT contributes to increased TET activity and 5hmC content, contrast with the reported decreased protein stability of O-GlcNAcylated TET1 in ES cells (43) and increased nuclear export of O-GlcNAcylated TET3 in HeLa and HEK cells (44). It is therefore likely that TET family members, which are expressed in hepatocyte cell lines and mouse liver, are regulated in a cell/tissue-specific manner.
Comparison of the mouse liver REV-ERBα cistrome to the 5hmC genomic landscape (30, 31) revealed that high-density 5hmC regions localized in the vicinity of strong REV-ERBα DNA binding sites. Conversely, weak REV-ERBα binding sites associated to weakly hydroxymethylated regions, suggesting a direct relationship between REV-ERBα genomic binding site occupancy and DNA hydroxymethylation. No specific enrichment in HNF6-dependent or in HNF6-independent REV-ERBα binding sites (11, 45) was observed in our analysis. This suggested that REV-ERBα binding correlates with the 5hmC pattern regardless of its DNA binding mode. Furthermore, Rev-erbα KO mice display a globally reduced liver DNA hydroxymethylation (∼35%).
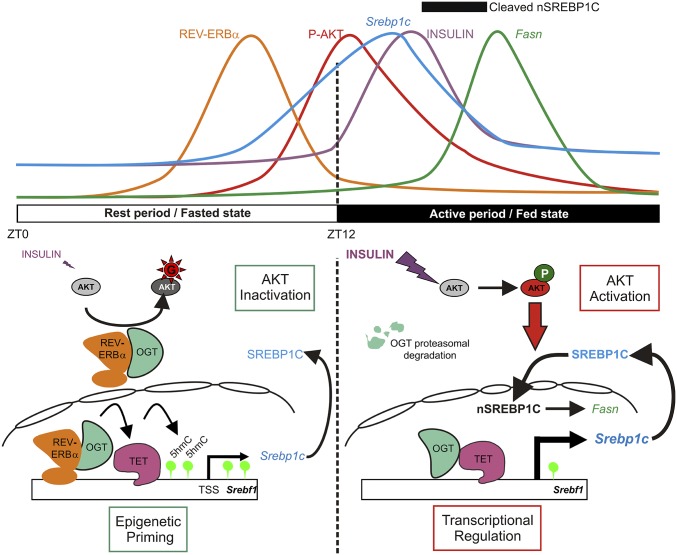
Taken together, these data indicate that the REV-ERBα/OGT functional interaction impinges on cytosine hydroxymethylation both in vitro and in vivo. Predicting the transcriptional outcome of this process at the cellular level remains difficult, as several groups reported that TET1 DNA binding is not only associated with gene activation but also with gene repression (30, 46). However, our observation that TET1/2/3 regulate SREBP1C expression suggests that REV-ERBα regulates this important lipogenic gene by altering epigenomic modifications at this locus. Since Srebp1c expression is not HDAC3-sensitive (47), and since REV-ERBα is not associated to HDAC3 or HNF6 at this locus (10, 11, 31), REV-ERBα may act as a positive regulator of Srebp1c expression. Whether this property actually extends beyond the control of Srebp1c expression awaits further investigation, but this nevertheless demonstrates that REV-ERBα acts as a sophisticated regulator of metabolic flexibility by interconnecting temporally and spatially hormonal signals, metabolism, and circadian rhythm. Thus, during the rest period, when insulin level is low and REV-ERBα is high, the REV-ERBα/OGT complex could act at different levels. In the cytoplasm, the REV-ERBα/OGT complex would block the AKT signaling pathway and, in parallel, will activate TET enzymes in the nucleus to promote an epigenomic state favoring SREBP1C basal expression (Fig. 7). This original REV-ERBα−dependant regulation could prime the liver for a faster response to insulin signaling at the start of the active phase. During the active feeding period, during which REV-ERBα is not expressed and when insulin peaks, cytoplasmic OGT is degraded, allowing the full activation of the AKT signaling pathway (Fig. 7). Increased DNA methylation was reported in mouse liver during the active period (48), which could result from the observed reduction of TET activity in the absence of REV-ERBα. As methylation is most often a dynamic modification characterizing inactive chromatin, these reciprocal epigenomic modifications may be involved in the circadian regulation of hepatic lipogenesis.
Fig. 7.
REV-ERBα/OGT participates to the control of circadian metabolic flexibility. During the rest period (from ZT0 to ZT12), insulin level is low and cytoplasmic REV-ERBα stabilizes OGT protein, which prevents AKT phosphorylation. In the nucleus, REV-ERBα binds to and activates OGT, favoring TETs enzyme activity. The REV-ERBα−dependent TETs activation results in DNA hydroxymethylation of Srebf1 locus enhancing Srebp1c basal expression. This epigenetic mechanism could prime the liver, allowing an immediate response to feeding. During the active period (from ZT12 to ZT24), the cytoplasmic OGT, which is not protected anymore by REV-ERBα, is degraded by the proteasome. In the nucleus, OGT and TET activities decrease in the absence of REV-ERBα. In parallel, insulin level increases in response to the food intake. In this context, the liver can fully respond to insulin, triggering the maturation of nSREBP1c followed by Fasn expression induction. Data were adapted from our and several other studies (32, 33, 38, 39).
Experimental Procedures
Animal Experimentation.
Animals were handled in accordance with institutional guidelines and approved by the Comité d’Ethique en Expérimentation Animale du Nord-Pas de Calais (C2EA-75). All mice (12-wk-old) were housed in a 12 h/12 h light/dark cycle for 2 wk before experimentation and fed ad libitum on a chow diet (A04, Safe Diets) with free access to drinking water. Wild-type C57BL/6J mice were from Charles River Laboratories. Rev-erbα KO and their wild-type control littermates were bred at Institut Pasteur de Lille animal facility as described (49). Mice were killed by cervical dislocation, and livers were snap-frozen in liquid nitrogen and stored at −80 °C until use. Samples were thawed only once as protein O-GlcNAcylation was found to be unstable upon multiple freeze-thawing cycles.
Chemicals.
Glucosamine, MG132, OGT inhibitors (DON and azaserine), the OGA inhibitor Thiamet G, δ-aminolevulinic acid, and succinyl acetone were purchased from Sigma-Aldrich. Compounds were used at indicated concentrations (see figure legends).
Cell Culture.
HepG2 cells, derived from a human hepatocellular carcinoma, were purchased from ATCC (HB8065) and grown in DMEM containing 25 mM Glc supplemented with 10% FBS, 1 mM nonessential amino acids, 1 mM sodium pyruvate, and 1% penicillin/streptomycin in a humidified incubator with 5% CO2 at 37 °C. Treatments were performed as described in DMEM containing 4 mM glutamine (Life Technologies-Gibco-BRL) supplemented with 10% dextran/charcoal-treated FBS at indicated Glc concentrations. Human embryonic kidney (HEK) 293 cells were purchased from ATCC (CRL-1573) and maintained at 37 °C under 5% CO2 in DMEM containing 25 mM Glc supplemented with 10% FBS and 1% penicillin/streptomycin. The U2OS cell line (HTB-96; ATCC) derived from human osteosarcoma was grown in McCoy 5A medium containing 17 mM Glc supplemented with 10% FBS and 1% penicillin/streptomycin.
The U2OS rev-erbα−/− clone was generated using the CRISPR-paired nickase technology (Sigma-Aldrich). Briefly, cells were transfected with two RNA guide-encoding vectors (pU6-gRNA HSL0002457008 and HSL0002457018) and Cas9-D10A nicking nuclease mutant encoding vector (pCMV-Cas9-D10A) using the JetPEI transfection reagent according to manufacturer recommendations. Clone selection was done by limiting dilution and screened by WB.
Statistical Analyses.
Raw data were analyzed using GraphPad Prism 7.0. Results are expressed as mean ± SEM (n = 3–6 for in vitro experiments, n values are indicated in figure legends for in vivo experiments), and groups were compared using either a t test or ANOVA followed by a post hoc test as indicated in figure legends. Specific analyses are described in corresponding experimental procedure sections.
All other information is available in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by Agence Nationale de la Recherche Grants OGlcRev and ANR-10-LABX-46, Fondation pour la Recherche Médicale Grant Equipe labellisée FRM 2015 DEQ20150331724, European Foundation for the Study of Diabetes/Lilly European Diabetes Research Program, European Commission EuRhythDia FP7-health Grant 278397, and French Proteomic Infrastructure Grant ANR-10-INBS-08–03. B.S. is a recipient of Advanced European Council Grant 694717. O-GlcNAcylated protein identification was done at “Plateforme Analyses Glycoconjugués” (FR3688 FRABio, CNRS, Université de Lille).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805397115/-/DCSupplemental.
References
- 1.Levine ZG, Walker S. The biochemistry of O-GlcNAc transferase: Which functions make it essential in mammalian cells? Annu Rev Biochem. 2016;85:631–657. doi: 10.1146/annurev-biochem-060713-035344. [DOI] [PubMed] [Google Scholar]
- 2.Bond MR, Hanover JA. O-GlcNAc cycling: A link between metabolism and chronic disease. Annu Rev Nutr. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vocadlo DJ. O-GlcNAc processing enzymes: Catalytic mechanisms, substrate specificity, and enzyme regulation. Curr Opin Chem Biol. 2012;16:488–497. doi: 10.1016/j.cbpa.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 4.Guinez C, et al. O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes. 2011;60:1399–1413. doi: 10.2337/db10-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruan HB, et al. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 2012;16:226–237. doi: 10.1016/j.cmet.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis BA, Hanover JA. O-GlcNAc and the epigenetic regulation of gene expression. J Biol Chem. 2014;289:34440–34448. doi: 10.1074/jbc.R114.595439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant D, et al. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbα. ACS Chem Biol. 2010;5:925–932. doi: 10.1021/cb100141y. [DOI] [PubMed] [Google Scholar]
- 9.Bugge A, et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Z, et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell. 2013;52:769–782. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. HNF6 and Rev-erbα integrate hepatic lipid metabolism by overlapping and distinct transcriptional mechanisms. Genes Dev. 2016;30:1636–1644. doi: 10.1101/gad.281972.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okabe T, et al. REV-ERBα influences the stability and nuclear localization of the glucocorticoid receptor. J Cell Sci. 2016;129:4143–4154. doi: 10.1242/jcs.190959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammed H, et al. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11:316–326. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- 14.Ryter SW, Tyrrell RM. An HPLC method to detect heme oxygenase activity. Curr Protoc Toxicol. 2001;Chapter 9:Unit 9.6. doi: 10.1002/0471140856.tx0906s05. [DOI] [PubMed] [Google Scholar]
- 15.Hering Y, et al. Development and implementation of a cell-based assay to discover agonists of the nuclear receptor REV-ERBα. J Biol Methods. 2018;5:e94. doi: 10.14440/jbm.2018.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin L, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 17.Ha C, Lim K. O-GlcNAc modification of Sp3 and Sp4 transcription factors negatively regulates their transcriptional activities. Biochem Biophys Res Commun. 2015;467:341–347. doi: 10.1016/j.bbrc.2015.09.155. [DOI] [PubMed] [Google Scholar]
- 18.Li T, et al. Novel role of nuclear receptor Rev-erbα in hepatic stellate cell activation: Potential therapeutic target for liver injury. Hepatology. 2014;59:2383–2396. doi: 10.1002/hep.27049. [DOI] [PubMed] [Google Scholar]
- 19.Vella P, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: Direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci USA. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dehennaut V, Leprince D, Lefebvre T. O-GlcNAcylation, an epigenetic mark. Focus on the histone code, TET family proteins, and polycomb group proteins. Front Endocrinol (Lausanne) 2014;5:155. doi: 10.3389/fendo.2014.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruan HB, Singh JP, Li MD, Wu J, Yang X. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol Metab. 2013;24:301–309. doi: 10.1016/j.tem.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Qian K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017;18:452–465. doi: 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azzout-Marniche D, et al. Insulin effects on sterol regulatory-element-binding protein-1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem J. 2000;350:389–393. [PMC free article] [PubMed] [Google Scholar]
- 26.Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delatte B, Deplus R, Fuks F. Playing TETris with DNA modifications. EMBO J. 2014;33:1198–1211. doi: 10.15252/embj.201488290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhliwayo N, Sarras MP, Jr, Luczkowski E, Mason SM, Intine RV. Parp inhibition prevents ten-eleven translocase enzyme activation and hyperglycemia-induced DNA demethylation. Diabetes. 2014;63:3069–3076. doi: 10.2337/db13-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortiz-Meoz RF, et al. A small molecule that inhibits OGT activity in cells. ACS Chem Biol. 2015;10:1392–1397. doi: 10.1021/acschembio.5b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomson JP, et al. DNA immunoprecipitation semiconductor sequencing (DIP-SC-seq) as a rapid method to generate genome wide epigenetic signatures. Sci Rep. 2015;5:9778. doi: 10.1038/srep09778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Martelot G, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilardi F, et al. CycliX Consortium Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet. 2014;10:e1004155. doi: 10.1371/journal.pgen.1004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung WD, Sakabe K, Housley MP, Dias WB, Hart GW. O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J Biol Chem. 2008;283:33935–33941. doi: 10.1074/jbc.M806199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaasik K, et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 2013;17:291–302. doi: 10.1016/j.cmet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 37.Shi SQ, Ansari TS, McGuinness OP, Wasserman DH, Johnson CH. Circadian disruption leads to insulin resistance and obesity. Curr Biol. 2013;23:372–381. doi: 10.1016/j.cub.2013.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robles MS, Humphrey SJ, Mann M. Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab. 2017;25:118–127. doi: 10.1016/j.cmet.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Jouffe C, et al. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11:e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrasco-Benso MP, et al. Human adipose tissue expresses intrinsic circadian rhythm in insulin sensitivity. FASEB J. 2016;30:3117–3123. doi: 10.1096/fj.201600269RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinkhasov BB, Selyatinskaya VG, Astrakhantseva EL, Anufrienko EV. Circadian rhythms of carbohydrate metabolism in women with different types of obesity. Bull Exp Biol Med. 2016;161:323–326. doi: 10.1007/s10517-016-3406-2. [DOI] [PubMed] [Google Scholar]
- 42.Yang H, et al. TET-catalyzed 5-methylcytosine hydroxylation is dynamically regulated by metabolites. Cell Res. 2014;24:1017–1020. doi: 10.1038/cr.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi FT, et al. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013;288:20776–20784. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Q, et al. Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked β-N-acetylglucosamine transferase (OGT) J Biol Chem. 2014;289:5986–5996. doi: 10.1074/jbc.M113.524140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science. 2015;348:1488–1492. doi: 10.1126/science.aab3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papazyan R, et al. Physiological suppression of lipotoxic liver damage by complementary actions of HDAC3 and SCAP/SREBP. Cell Metab. 2016;24:863–874. doi: 10.1016/j.cmet.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maekawa F, et al. Diurnal expression of Dnmt3b mRNA in mouse liver is regulated by feeding and hepatic clockwork. Epigenetics. 2012;7:1046–1056. doi: 10.4161/epi.21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woldt E, et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19:1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.