Significance
We do not fully understand how behavior evolves. Here we investigate the genomic basis of bower building among Lake Malawi cichlid fishes. Males construct bowers of two major types, pits and castles, to attract females in mating displays. Thousands of genetic variants are strongly associated with divergence in bower behavior. Remarkably, F1 hybrids of pit-digging and castle-building species perform sequential construction of first pit and then castle bowers. Analysis of brain gene expression in hybrids showed behavior-dependent allele-specific expression with preferential expression of pit-digging alleles during pit digging and castle-building alleles during castle building. Our results suggest that behaviors evolve via complex genetic architectures featuring cis-regulatory differences whose effects on gene expression are specific and context dependent.
Keywords: Malawi cichlids, bower building, cis-regulatory evolution, genome sequencing
Abstract
Many behaviors are associated with heritable genetic variation [Kendler and Greenspan (2006) Am J Psychiatry 163:1683–1694]. Genetic mapping has revealed genomic regions or, in a few cases, specific genes explaining part of this variation [Bendesky and Bargmann (2011) Nat Rev Gen 12:809–820]. However, the genetic basis of behavioral evolution remains unclear. Here we investigate the evolution of an innate extended phenotype, bower building, among cichlid fishes of Lake Malawi. Males build bowers of two types, pits or castles, to attract females for mating. We performed comparative genome-wide analyses of 20 bower-building species and found that these phenotypes have evolved multiple times with thousands of genetic variants strongly associated with this behavior, suggesting a polygenic architecture. Remarkably, F1 hybrids of a pit-digging and a castle-building species perform sequential construction of first a pit and then a castle bower. Analysis of brain gene expression in these hybrids showed that genes near behavior-associated variants display behavior-dependent allele-specific expression with preferential expression of the pit-digging species allele during pit digging and of the castle-building species allele during castle building. These genes are highly enriched for functions related to neurodevelopment and neural plasticity. Our results suggest that natural behaviors are associated with complex genetic architectures that alter behavior via cis-regulatory differences whose effects on gene expression are specific to the behavior itself.
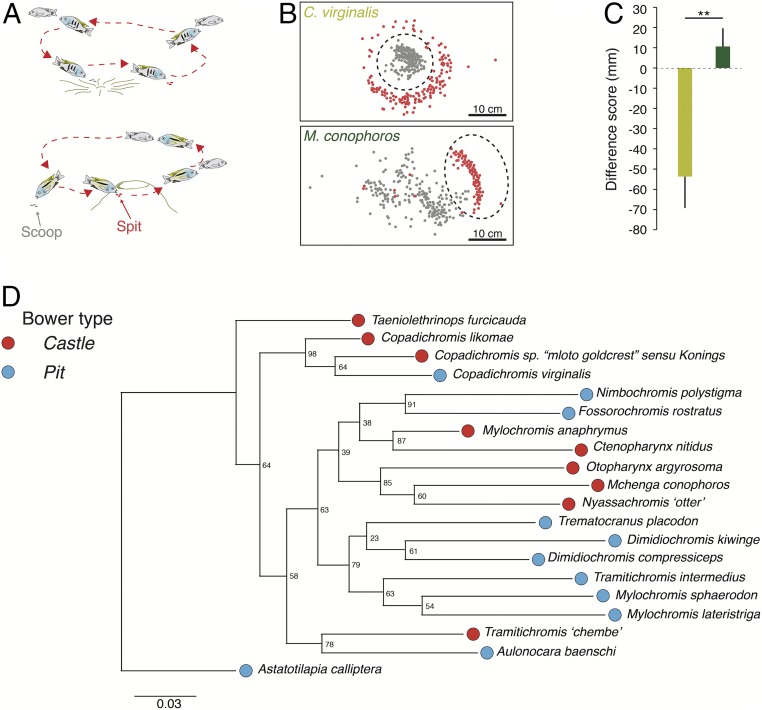
Understanding behavioral evolution requires identifying the genetic and regulatory architectures encoding neural development and function. To characterize the evolution of a complex social behavior, we focused on the remarkable bower-building feats performed by ∼200 cichlid fish species in Lake Malawi that live on sandy substrate. Bowers are species-specific sand structures that serve as signals in male–male competition and female mate choice (1). Malawi cichlid species build two basic bower types: (i) pits, which are depressions that resemble nests in the sand, and (ii) castles, which resemble miniature volcanoes (2). Bower building requires highly repetitive activity in which males perform hundreds of scoop–spit bouts with their mouths per hour, interspersing construction with the courtship of females and aggressive encounters with conspecific males (Fig. 1A) (2, 3). To dig pits, males collect sand from the center of the pit and spit it elsewhere, while to build castles, males gather sand from elsewhere and spit it in a targeted location (Fig. 1 B and C and Movie S1). Pit and castle bower types are distributed widely across the Malawi cichlid sand-dweller phylogeny, suggesting that parallel evolution and/or hybridization may be responsible (4). Furthermore, bower building is innate. Naive males born in an aquarium, who have experienced neither sand nor other males, perform species-specific bower behavior when housed with sand and gravid females.
Fig. 1.
Bower building. (A) Characteristic behavioral patterns associated with pit digging (Upper) and castle bower building (Lower). (B) Average locations of scoops (gray) and spits (red) during bower-building trials in the pit-digging species C. virginalis and the castle-building species M. conophoros. Consensus locations of the bowers are indicated by dashed black circles. (C) Results of a Student’s t test (two-tailed, P = 0.006) comparing difference score (in millimeters; mm) between C. virginalis (pit digging; yellow) and M. conophoros (castle building; green). (**P < 0.01.) (D) Maximum-likelihood phylogeny from genome-wide variants of the species sequenced in this study. Numbers at nodes are bootstrap support values.
Results
Extensive Genetic Differences Exist Between Pit-Digging and Castle-Building Species.
To identify genetic variants associated with bower building, we sequenced the genomes of 20 male individuals from 20 sand-dwelling Lake Malawi cichlid species: 11 pit-digging species and nine castle-building species (SI Appendix, Table S1). Species in these two groups construct either pits or castles despite differences within the groups in color pattern, ecology, feeding mode, and other characteristics (2). Sequence reads from each species (mean coverage ∼25×) were aligned using the Malawi cichlid reference genome (4) and were mapped to Malawi linkage groups (LGs) (Methods). A maximum-likelihood phylogeny based on variant sites (Fig. 1D) is consistent with repeated evolution of pit-digging and castle-building behavior in our sampled species.
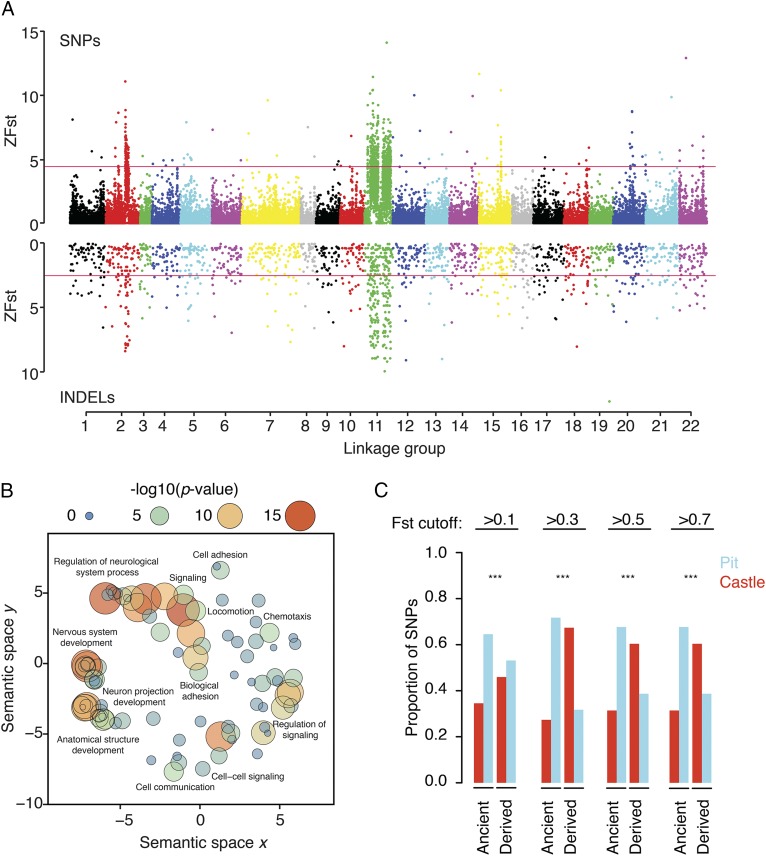
Like other recently evolved species flocks (5), East African cichlids share genetic polymorphisms because of incomplete lineage sorting and hybridization (6–9). Therefore, we used both population-based and phylogeny-based analytical approaches to understand genomic correlates of bower building. We applied the population-based fixation index (FST) to identify genomic regions differentiating pit-digging vs. castle-building species. We observed ∼15.5 million SNPs and ∼130,000 insertion/deletions (indels) in the sample set; 1.5% of variable sites were notably divergent between pit-digging vs. castle-building groups (FST >0.2, compared with 0.08 genome-wide mean). We compared patterns of FST divergence across the genome (Fig. 2A) with a population-structure–corrected genome-wide association study on bower behavior and found that the two are strongly correlated (SI Appendix, Fig. S1).
Fig. 2.
Genome-wide divergence associated with bower building. (A) Manhattan plot of genome-wide ZFST for SNPs and indels between pit-digging and castle-building species. (B) Semantic similarity of GO biological process terms enriched for high-FST variants. (C) Bar plot of SNP proportions per FST cutoff for ancestral and derived SNPs. SNPs in which castle-building species possess the alternate allele are colored red; those in which pit-digging species possess the alternate allele are colored blue. ***P < 0.001, Fisher’s exact test.
We next identified outlier 10-kb regions based on mean FST of SNPs [10% false-discovery rate (FDR), FST >0.2] and individual indels (10% FDR, FST >0.1) (Fig. 2A). Outlier regions were observed on every LG, but peaks on LG2 and LG11 are striking for their size (1.3 Mb and 6 Mb, respectively) and consistency across SNP and indel data. Broad peaks of differentiation on LG2 and LG11 could be caused by structural changes [e.g., inversions (10)] associated with bower behavior or with other traits such as male sex determination (11). However, sex-determination systems are not known for these species, and we could not identify structural variants that could explain the broad peaks of genetic differentiation on LG2 or LG11 (Methods) (12, 13).
Characterizing Variants Associated with Bower Type.
We hypothesized that if high-FST regions across the genome are the product of selection on bower building or associated behavioral traits, then they should be enriched near genes involved in brain function and development. To test this, we examined 1,563 genes located within 25 kb of an outlier 10-kb region (∼30% of these genes are located on LGs 2 and 11) for functional enrichment (14). This gene set was significantly enriched for tissue types, pathways, human disorders, and phenotypes associated with brain development and behavior, including axon guidance, synaptic transmission, autism spectrum disorder, and spatial learning (Fig. 2B and Dataset S1). Together, these analyses identify genomic regions and genetic variants associated with bower behavior and demonstrate that genes near these variants are strikingly enriched for putative functions in brain and behavior.
To assess the role of ancestral variation in differentiation of pit-digging vs. castle-building behaviors, we classified SNPs as either “new” if they were only found in sand-dwellers or “ancient” if they were also found in the genomes of non–sand-dwellers, including species from other African rift lakes. For new variants found only in sand-dweller species, we marked alleles as derived if they were not shared with the rock-dweller Metriaclima zebra reference genome (SI Appendix, Fig. S2A). Such standing genetic variation has been recruited for rapid adaptation in sticklebacks and other cichlid fish species (8, 15). We used odds ratios (ORs) from a Fisher’s exact test comparing SNP-level allele counts to infer whether pit digging and castle building associated preferentially with ancestral or derived alleles (SI Appendix, Fig. S2B). We found that among more genetically diverged SNPs, castle-building species tended to have derived alleles (P < 0.0001), while pit-digging species were enriched for ancestral alleles (P < 0.0001) (Fig. 2C). Indeed, increasing FST was related to greater divergence in ORs between derived and ancestral alleles (SI Appendix, Fig. S2C). Furthermore, when comparing the overall distribution of FST measures, 9% of all SNPs were ancient, but in high-FST variants (FST ≥0.2) this proportion was elevated to 20% (χ2 test, P value < 2.2e-16) (SI Appendix, Fig. S3). These data suggest that both standing genetic variation and derived alleles shared by castle-building species contribute to the overall genetic architecture of bower behavior.
Allele Sharing Among Bower-Building Species May Be Due to Introgression.
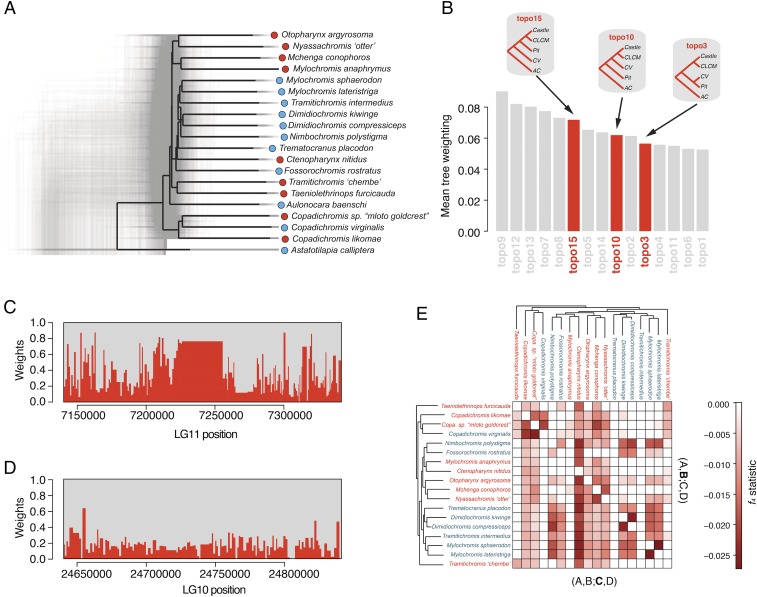
The above observations, along with low bootstrap support values for some nodes on the whole-genome maximum-likelihood phylogeny (Fig. 1D), suggest that sand-dweller genomes may have been subject to evolutionary processes leading to species tree contraventions, such as incomplete lineage sorting and introgression. To test this, we constructed maximum-likelihood phylogenies from nonoverlapping windows of 10,000 SNPs (1,927 in total) (16). The resulting local phylogenies demonstrate the presence of a variety of tree topologies (Fig. 3A). Using TWISST (17), a method that measures the “weights” of various tree topologies genome-wide, we found moderate to strong support for a variety of trees, including several that group species by bower phenotype (Fig. 3B and SI Appendix, Fig. S4 A and B). Furthermore, support for trees that group by bower phenotype varied across LGs with an extremely strong increase in weights on LGs 2 and 11, reflecting the strong genetic divergence seen via FST in these regions (Fig. 3 C and D and SI Appendix, Fig. S4C).
Fig. 3.
Complex phylogenetic relationships among sand-dwelling Malawi cichlids. (A) We plotted 1,927 phylogenies resulting from nonoverlapping 10,000 SNP windows using DensiTree. The consensus phylogeny produced by DensiTree is colored black. (B) Bar plot of mean genome-wide weightings for the 15 tree topologies tested with Twisst. Trees grouping clades by bower phenotype (topologies 15, 10, and 3) are highlighted. See SI Appendix, Fig. S4 for visualizations of all 15 topologies. (C) An example of a stacked plot of topology weightings along a region of LG11 with strong support for groupings by phenotype. (D) An example of a stacked plot of a mixed-weight region on LG10. (E) Heatmap of the most significant f4 values. Species in the x and y axes were either B or C in the form f4(A,B;C, A. calliptera). Names of pit-digging and castle-building species are in blue and red font, respectively. A darker red square indicates more signal of gene flow between the species pairs in the respective row and column.
We next tested for signals of admixture by comparing the observed similarity in allele frequencies among species pairs with those expected given their phylogenetic relatedness (as established by comparisons with pairs of outgroup species) (18). We found many species pairs with signals of gene flow that were stronger than expected by chance, as evidenced by significantly negative admixture statistics (Methods and Fig. 3E). Similarly, using TreeMix (19), we found that analyses of admixture scenarios incorporating all species in our dataset also supported multiple admixture events, although the predicted number and specifics of these events could not be confidently estimated (SI Appendix, Fig. S5). We also observed that signals of admixture varied by genomic location and in many cases recapitulated regions of high divergence (SI Appendix, Fig. S6) (20). Given these species’ high degrees of relatedness and the potential of natural selection acting on standing genetic variation, it is difficult to ascertain the importance of introgression in the evolution of bower building. Nonetheless, taken together these patterns suggest that gene flow has occurred across the sand-dwelling clade and may have impacted variants important for bower building. Our observations of complex evolutionary histories reflecting both segregation of ancestral polymorphism and gene flow between species are consistent with findings from other recent studies of African cichlid genome-wide divergence (8, 20). Specific hypotheses of gene flow between bower-building species could be tested by additional population and geographic sampling (21).
Bower Building Is Associated with Context-Dependent, Allele-Specific Expression.
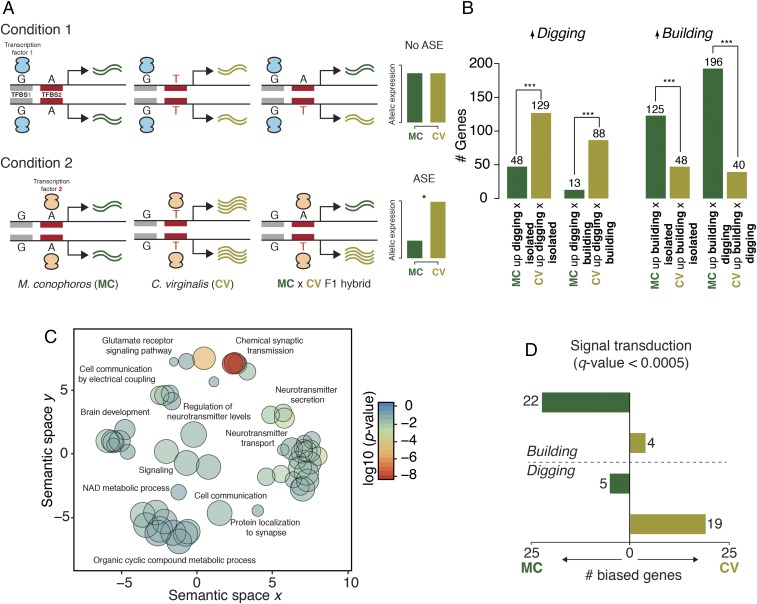
Behavioral traits can be associated with rapid transcriptional changes and distinct neurogenomic states (22), so we next asked how gene expression was activated in the brains of pit-digging vs. castle-building cichlids. To do this, we assayed whole-brain gene expression by RNA-sequencing (RNA-seq) in interspecific hybrid males to measure allele-specific expression (ASE), an approach that can identify cis-regulatory divergence between closely related species (23, 24). We crossed the pit-digging Copadichromis virginalis (CV; sire) with the castle-builder Mchenga conophoros (MC; dam) based on previous laboratory observations confirming the viability of this cross. Remarkably, CV × MC F1 hybrids produced an unusual intermediate “pit-castle” bower by carrying out parental behaviors in sequence: First, a pit is excavated for several days to weeks followed by a transition to the construction of a castle (SI Appendix, Fig. S7). This observation suggests that both pit-digging and castle-building behavioral control circuits are functional in the F1 male brain. We took advantage of this sequential bower construction to compare brain RNA-seq data from CV × MC F1 hybrid males during three behavioral contexts: pit digging (n = 2), castle building (n = 2), or in isolation without conspecifics or sand (control; n = 2) (Fig. 4A). Given that the terminology for behaviors observed in the hybrids overlaps with that used for the pure-species genomic comparisons above, we hereafter denote F1 hybrid behavior with the suffix “-phase.” Sequences were aligned using the M. zebra reference genome, and ASE was measured from gene-level read counts.
Fig. 4.
Behaviorally dependent allele-specific expression. (A) Cartoon representation of allele-specific expression under different contexts. In context 1, the sequence in the transcription factor binding site (TFBS1) is identical in the two alleles, leading to an expected ∼50:50 allelic ratio in the F1 hybrid. In context 2, there is a variant between the species in TFBS2 leading to ASE in the F1 hybrid. (B) Bar plots indicating the distribution of significantly differentially biased genes across building and digging contexts. Significance was calculated using a Fisher’s exact test; ***P < 5 × 10−5. (C) Semantic similarity of GO biological process terms enriched for genes with diffASE. Node size is the log of the size of the category represented. Nodes are colored by the log10 P value of the enrichment. (D) An example of a result from a sign test comparing context-dependent allele-specific expression (signal transduction; Reactome pathway R-HSA-162582).
We found the presence of ASE in the brain transcriptomes of fish in all three behavioral contexts. We identified 621 genes with significant ASE across replicates in at least one behavioral context (Bonferroni-corrected P < 0.05) (Dataset S2). Because many of these genes may be unrelated to bower building, we reasoned that differential ASE (diffASE) across contexts—e.g., cases in which one allele is more highly expressed than the other only during the pit-phase or castle-phase—would enrich for those involved in the behavior. We found robust variation in the number of CV- and MC-biased genes between behavioral contexts that, surprisingly, reflected a pattern of species bias (Fig. 4B). Specifically, significantly more genes are MC-biased during the castle-phase than during the pit-phase (counts: 196 MC-biased genes, 40 CV-biased genes; Fisher’s exact P < 7.45 × 10−19) or than in isolation (counts: 125 MC-biased genes, 48 CV-biased genes; Fisher’s exact P = 1.37 × 10−7), while significantly more genes are CV-biased during the pit-phase than during the castle-phase (counts: 13 MC-biased genes, 88 CV-biased genes; Fisher’s exact P = 3.03 × 10−11) and during the pit-phase compared with isolation (counts: 48 MC-biased genes, 129 CV-biased genes; Fisher’s exact P = 2.58 × 10−8). Furthermore, we identified a number of individual genes with “discordant ASE” because their direction of allelic bias switched between behavioral contexts (SI Appendix, Fig. S8 A and B). Notable examples of this phenomenon include the genes atp1b4, an ion pump with brain-specific expression in fish (25) (digging: CV allele 2.98-fold higher expression; building: MC allele 10.83-fold higher expression) and dgcr8, a core component in microRNA biogenesis that is required for inhibitory synaptic function (26) (digging: CV allele 2.55-fold higher; building: MC allele 2.75-fold higher) (SI Appendix, Fig. S8 C and D; full results are given in Dataset S2).
These patterns of differential allelic expression in F1 animals indicate an unexpected amount of dynamic genomic regulation associated with behavior. Notably we observed that, within the same brain containing alleles from both parental genomes, castle-building cis-regulatory elements are specifically activated during the castle-phase, and vice versa for the pit-phase. These results add to a growing body of literature illustrating context-dependent transcriptional response with experience or changes in behavior (27, 28) and further suggest that evolutionary differences between species in relation to brain function and behavior may arise from variation in such context-dependent regulation of gene expression.
Context- and Lineage-Specific Induction Identifies Behavior-Dependent Genes and Pathways.
We reasoned that if the context-dependent ASE observed above is biologically relevant, then genes with shared patterns of allelic induction across contexts should be enriched for similar functional roles. To this end we first identified genes with significant diffASE—varying ratios between CV and MC alleles dependent on context—between at least two of the three behavioral contexts (435 genes) (Methods). We performed gene-set enrichment analysis using all other genes detected in at least one of the three contexts (9,703 genes) as background. This analysis identified a number of enriched categories spanning various aspects of neural structure and function (Dataset S3). For example, genes with diffASE were significantly enriched in a number of neural-specific Reactome pathways such as transmission across chemical synapses (R-HSA-888590; q-value = 8.13 × 10−6) and GABA synthesis, release, reuptake and degradation (R-HSA-112315; q-value = 5.38 × 10−7). Enriched gene ontology (GO) biological processes categories were predominately related to synaptic function, neurotransmitter regulation and signaling, and ion transport and binding (Fig. 4C and Dataset S3), while cellular component sets included structures and loci important for neuronal function such as clathrin-sculpted vesicles (GO:0060198; q-value = 1.06 × 10−5) and postsynaptic densities (GO:0014069; q-value = 3.46 × 10−5).
We further refined these tests by identifying genes that displayed significant differential induction of one or both alleles during building or digging behaviors (building up-regulated genes, n = 171; digging up-regulated genes, n = 174) (Methods). These genes differ from those with diffASE in that the comparison of interest is the expression of individual alleles across rather than within contexts. While diffASE is ascertained by identifying different ratios between alleles, differential induction is more similar to traditional differential expression tests in that it is concerned with the expression of individual alleles across contexts. Analyzing these genes might then provide insight into the divergence of context-dependent regulation of alleles between CV and MC. To assay the roles of genes with differential induction, we performed gene-set enrichment tests as above. Genes up-regulated during building were enriched for neurotransmitter release (R-HSA-112310; q-value = 4.70 × 10−2) and ion channel transport (R-HSA-983712; q-value = 1.97 × 10−7). Similarly, digging-induced genes were associated with ion homeostasis (R-HSA-5578775; q-value = 2.90 × 10−2) and chemical synaptic transmission (GO:0007268; q-value = 3.20 × 10−2). These results indicate that genes with differential ASE and induction are coherently enriched for specific neural processes in comparison with the rest of the transcriptome, adding evidence in support of the idea that context-dependent gene regulation may support distinct neural states related to behavior.
We extended this analysis to explore the role of lineage-specific differences in gene regulation via consideration of independent regulation of CV and MC alleles across behavior states. To do so, we applied a sign test to the directionality of CV and MC alleles between the pit- and castle-phases (Dataset S2) (24, 29). We compared the allelic counts of individual genes across phases (pit vs. castle), avoiding a bias for the sampled tissue as opposed to a typical GO enrichment test which would compare the complete focal gene list with a background. We found several hierarchically organized gene sets that matched a pattern of significant differential induction of CV and MC alleles between digging and building (Dataset S3). The identified gene sets were largely involved in cell signaling and communication (an example is plotted in Fig. 4D). These observations suggest that lineage-specific selection may have played a role in producing differential regulation of neural-signaling genes in CV and MC related to their species-specific bower behavior.
Bower-Associated SNPs and cis-Regulatory Variation.
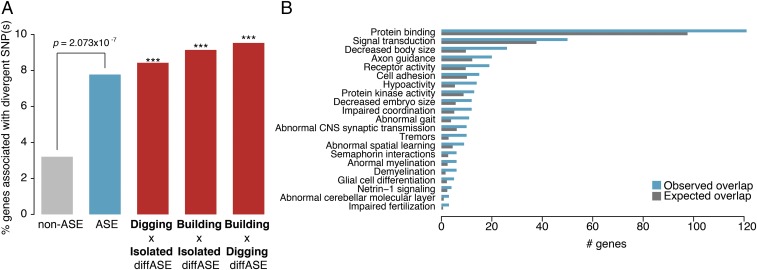
Finally, we asked the extent to which genomic divergence among bower-building species is associated with cis-regulatory changes inferred from the CV × MC intercross. If natural selection had acted on regulatory variants associated with pit digging and castle building, it would result in enrichment of highly differentiated SNPs proximal to genes that display ASE. Indeed, we found that ASE genes were significantly enriched near high-FST SNPs (48 of 621 ASE genes vs. 332 of 1,0221 non-ASE genes; Fisher’s exact test, P = 2.07 × 10−7) (Fig. 5A). Furthermore, this enrichment increases when considering those genes displaying diffASE (Fig. 5A). We also detected over 30 pathways that showed significant overlap between ASE and high-FST gene lists (Bonferroni-corrected P < 0.05; hypergeometric test) (Fig. 5B). As in previous analyses of ASE and FST alone, many of these pathways are involved in neurodevelopment, neuroplasticity, and behavioral regulation, suggesting concordance between patterns of genetic and regulatory divergence among bower-building species.
Fig. 5.
Intersection of genome-wide SNPs and ASE. (A) Bar plot comparing the number of ASE, non-ASE, and diffASE (across all contexts) genes associated with highly divergent SNPs between pit-digging and castle-building species. ***P < 1 × 10−4, Fisher’s exact test comparing genes overlapping SNPs and genes not overlapping SNPs. (B) Categories in which the observed amount of overlap (blue) between genes associated with highly divergent SNPs and with ASE is significantly greater than expected (gray).
Discussion
By combining genome sequencing across many closely related species with analysis of ASE in the brains of behaving hybrid animals, we provide a genome-wide view of how a complex behavior has evolved. Our results suggest that the evolution of bower building was associated with polygenic selection on ancient and new genetic variants that regulate genes involved in neural activity and synaptic plasticity in specific behavioral contexts. The observation of context-dependent ASE associated with sequential pit-digging and castle-building behavior in F1 males suggests how cis-regulatory divergence across many genes may combine to produce the evolutionary divergence of a complex behavior.
The elevated FST values at thousands of variants among the 20 diverse bower-building species examined revealed that bower building is associated with a complex but phylogenetically consistent genetic architecture. The observation that these sites are both ancestral (polymorphisms shared with species outside of Lake Malawi) and derived parallels similar findings from studies of Malawi cichlids (6, 8, 20) and other recently evolved species flocks such as sticklebacks (15) and finches (5). Notably, we found that castle-building species tended to possess derived variants at more genetically diverged sites, suggesting that, at least from a genomic perspective, castle building is the younger, derived behavior. The observation that the species believed to be similar to the common ancestor of Malawi cichlids, Astatotilapia calliptera, digs pits and is positioned at the base of the Lake Malawi phylogeny lends credence to this idea (Fig. 1A). The phylogenetic distribution of bower building suggests that repeated instances of selection, be it on standing variation or introgressed alleles, may have acted to differentially fix the genetic architecture associated with castle bowers in a number of sand-dwelling species. That we detect potential genomic signatures of gene flow supports the notion that introgression may have played a role in the propagation of the derived castle-building behavior among sand-dwelling cichlid species. The importance of gene flow across species boundaries has been highlighted before in cichlid fish adaptive evolution (8, 30), but bower building represents a special case of this general phenomenon, as the behavior is sex specific and unlikely to increase male survival. It will be interesting to test specific hypotheses of gene flow between particular bower-building species using more targeted sampling and genetic methods.
Given the large regions of increased genetic divergence identified on LGs 2 and 11, it is intriguing to consider the possibility that there may be “supergene”-like elements underlying bower building similar to those that have been found to be associated with other animal behaviors such as male reproductive morphs in the ruff (31, 32) or insect social organization (33). Our observation that F1 hybrids of a cross between pit-digging and castle-building species can build both structures and do so in a mutually exclusive, sequential fashion may further support the notion that the varying bower types require genomic regions working in a modular and independent fashion. Such a finding would not be without precedent among the cichlids of Lake Malawi. The orange-blotch (OB) coloration phenotype found among >20 rock-dwelling species is associated with a tightly linked genomic locus resembling a supergene in its size and inheritance and has arisen independently at least three times (34).
Our approach to sequence the genomes of individuals from pit-digging vs. castle-building species has identified numerous genetic variants associated with bower behavior, not unlike the genetic architecture of other complex traits, including human neurological disorders (35). Integrating genome sequencing with RNA-seq from F1 hybrid brains pairs our strategy with the complexity of the bower trait. Our results fit the general pattern observed in other complex traits: numerous genetic associations with the phenotype (36) and an expectation that many of these variants exert their effects via context-dependent cis regulation of gene expression (37, 38). Models like Malawi cichlids may thus occupy a “sweet spot” in complexity, combining a rich genetic, evolutionary, and phenotypic profile with tractable biology that could yield insights into the origins of behavioral diversity. Of utmost importance will be continued efforts toward correct phenotypic categorization of such complex traits. In this study, we decided to characterize bower building into two qualitative categories and used this definition to perform genome-wide tests across 20 diverse species. However, our species cohort may share other traits correlated with bower type, adding noise to our measures of genetic association, or may represent more than two behavioral strategies (although this scenario seems unlikely, given the behavioral and genetic findings presented here). In general, the use of bower building as a model complex trait will benefit from careful work on the roles of ontogeny, intraspecific variation, and behavioral variability in the regulation of this behavior.
The extensive, context-dependent transcriptomic divergence associated with bower building in F1 hybrids provides intriguing insights into the regulatory basis of behavioral evolution. For example, it appears that bower building is defined by modularity at multiple levels of biological organization. The transition in CV × MC F1 hybrids from pit to castle bowers may be considered as a shift between distinct behavioral modules associated with distinct behavioral patterns reflective of the respective parental species. The finding that these phases are associated with distinct transcriptomic states suggests that the pit and castle alleles function modularly based on behavioral context, best reflected by the significant number of genes displaying discordant ASE across these behavioral modules. This is in opposition to other scenarios in which regulatory divergence might be static across behavioral conditions or minimal in the context of brain function and behavior. Instead, the transcriptomic states associated with the pit- and castle-phases in F1 hybrids appear to be more similar to those found between tissues in an organism, arising from potentially distinct regulatory or epigenomic cellular environments. In this case, given the genomic and evolutionary signatures identified by the whole-genome analyses, it appears that these differences in regulation are at least partly due to extensive sequence-level variation in a number of functionally related regulatory elements associated with behavior. Notably, this may suggest that the genome harbors regulatory loci specifically involved in the dynamic coordination of behavior and brain function analogous to well-known genetic modulators of morphology and that these loci underlie the evolution of behavioral diversity.
Methods
Bower Behavioral Measurements.
Individual adult, reproductive subject males (C. virginalis, n = 4 and M. conophoros, n = 4) were each housed with one to three adult, reproductive stimulus females of the same species in 43.2 × 91.4 × 40.6 cm (160-L) glass aquariums maintained on a 12-h:12-h light:dark cycle., A tray (Newbury Black Poly Saucer, SA1412BK; Dynamic Design) 5.1 cm deep and 35.6 cm in diameter was placed in each tank and was filled with sand (Sahara Sand, 00254; CaribSea, Inc.). All animal experiments were approved by the Georgia Tech Institutional Animal Care and Use Committee (protocol no. A11034) and by the Stanford University Animal Care and Use Committee (protocol no. APLAC-28757). For each subject, 90- to 120-min videos were recorded between 3–8 h after lights-on during periods of high bower-building activity using a GoPro camera (Hero4 Silver, CHDHY-401; GoPro) housed in a waterproof compartment (Clear Standard Housing, AHSRH401; GoPro) and placed top-down directly above the sand-filled tray. The behavior of male subjects was scored using The Observer XT 12 software (Noldus) according to the following definitions: “scooping” was defined as opening the mouth and collecting sand, and “spitting” was defined as expulsion of sand from the mouth. For each individual, screenshots were captured at the precise moment of every scoop and spit event, and these screenshots were exported to Paint (Microsoft) to extract spatial coordinates of the mouth for every scoop and spit event.
Bower-building difference scores were calculated to measure the spatial dispersion of scoops compared with the spatial dispersion of spits for each subject male. To calculate the difference score for each subject, we first determined the coordinates for each subject’s average scoop location and average spit location. Using these coordinates, we calculated the absolute distance from the average spit location to each individual spit location and from the average scoop location to each individual scoop location. To generate a quantitative metric of spatial dispersion, these distances were then averaged, yielding two distance scores for each subject, one for scoops, and one for spits. To compare the spatial dispersion of scoops vs. spits, the difference between these distance scores (scoop distance score minus spit distance score) was calculated for each subject, thus providing an estimate of differences in spatial patterns of scooping and spitting sand for each animal. A two-tailed Student’s t test was used to compare these difference scores between species.
Genome Sequencing, Alignment, and Variant Identification.
We chose diverse representative species of pit-digging and castle-building groups, selected from multiple genera across the phylogenetic tree of sand-dwellers (2). Genomic DNA was extracted from fin clips of 20 individuals collected in Lake Malawi (11 pit diggers and 9 castle builders) (SI Appendix, Table S1) using the Qiagen DNeasy kit (catalog no. 69504; Qiagen).
Libraries were constructed following the Illumina TruSeq DNA library-preparation protocol. Paired-end sequencing (2 × 100) was performed on the Illumina Hi-Seq 2500 system at the Georgia Institute of Technology. Raw sequence reads were quality controlled using the NGS QC Toolkit (39). Quality control was performed as follows: First, raw reads with an average PHRED quality score below 20 were removed. The remaining reads were further trimmed of low-quality bases at the 3′ end. Quality-control reads for each of the genomes were aligned to the new M. zebra reference genome (4) using bwa-mem (version 0.7.4) and default parameters (40). We used Picard Tools (https://broadinstitute.github.io/picard/) to mark PCR duplicates. Sequences were mapped to 87% of the reference genome on average, with mean coverage of 24.31×. Variant discovery and filtering were performed using HaplotypeCaller within the Genome Analysis Toolkit (GATK) program according to GATK best-practices recommendations (41–43).
Deletions were identified using modifications to a previously published approach (44). Candidate deletions were first identified using chimeric reads as identified by Burrows–Wheeler Aligner (40) (i.e., reads that included the SA tag) in which each alignment mapped to the same contig and the same strand. These reads were used to infer the breakpoints and insertion sequence of a candidate deletion. Candidate deletions that were also present in the sequencing of M. zebra were excluded as likely errors in the reference sequence. Each candidate deletion was then genotyped in each of the pit-digging and castle-building species by collecting all the reads with primary alignments that fell within 10 bp of the candidate deletion and with a mapping-quality score greater than 10. These reads were realigned to both the reference and the candidate deletion sequence using a striped Smith–Waterman alignment from the scikit-bio Python library. Reads were classified as reference, mutant, or undetermined based on their mapping score to the two alleles. Deletions were categorized as homozygous mutant if 80% or more of the reads were categorized as mutant, as homozygous reference if 80% or more of the reads were identified as reference, and as heterozygote if they fell in between. Candidate deletions that were reference homozygous in all species were filtered from the dataset.
Tests of Genetic Divergence and Enrichment.
We excluded sites with more than 50% missing genotypes from the whole-genome sequencing data. We calculated FST per variant and in 10-kb windows using the –weir-fst-pop parameter from the VCFtools program (45) with the flags -fst-window-size 0 for individual sites and -fst-window-size 10,000 for 10-kb windows. Nucleotide divergence was calculated using VCFtools with the -window-pi 10,000 flag. Thresholds were estimated using R-package fdr-tool (46). FST values were converted to a normalized scale for visualizing these data on LGs (Fig. 2A) using Fisher’s Z-transformation. To compare FST with a population-structure–controlled genome-wide association analysis, we employed GEMMA v0.96 (47). We first computed a kinship matrix using the filtered sites from which FST was calculated and then performed the association test on bower type using a linear mixed model (-lmm flag) factoring in relatedness via the kinship matrix.
SNP and indel variants were annotated using the SnpEff (4.3i) program (48) and were analyzed for functional enrichment using GeneAnalytics (geneanalytics.genecards.org) (14). Using the binomial distribution, this algorithm tests the null hypothesis that there is no functional overrepresentation. A resulting score is presented for each match in the form of a −log2 transformed P value corrected for multiple comparisons via the FDR method. Scores are arranged into three significance categories: high (Padj <0.0001), medium (Padj <0.05), and low (Padj >0.05). Semantic similarity plots for significantly enriched categories were produced with REVIGO (49).
Identifying Structural Variants.
To predict structural variants on a genome-wide basis, we used breakdancer-max (1.1) (12) with default parameters. As read-pair mismatches in size (pairs are farther away than expected) and direction (pairs in the same orientation) can be used to identify inversions, we also used the Integrated Genome Browser (13) to manually validate predicted structural variants from breakdancer-max, particularly those on LGs 2 and 11. Thousands of structural variants were predicted for each species, but none consistently associated with the broad FST peaks on LGs 2 and 11.
Improved Genome Annotation.
In the original National Center for Biotechnology Information (NCBI) release of the latest M. zebra reference genome (4), 15,361 of 26,490 predicted protein-coding genes were annotated as hypothetical or without orthologs. To improve this annotation, we identified orthologs for genes in two additional ways: (i) using a phylogenetic method using TreeFam, a curated database of phylogenetic trees of animal genes, and (ii) via reciprocal blast against the human genome and five fish genomes (50). The final annotation merged orthologous genes identified by both methods; 1,900 hypothetical genes remain.
Assigning SNPs and Genome Contigs to Linkage Maps.
We used Chromonomer (1.03) (51) to anchor the gap-filled “M_zebra_UMD1” assembly (4) to LGs using two different genetic maps, both generated via traditional F2 crosses and genotyped with restriction site-associated DNA sequencing (RAD-seq). First, a genetic map from 160 F2 individuals from an M. zebra × M. mbenjii cross resulting in 834 markers in 22 LGs and spanning 1,933 cM (52) was used to anchor the M_zebra_UMD1 assembly. This initial anchored assembly was subsequently reanchored with Chromonomer using a second genetic map. The second genetic map was generated by genotyping 268 F2 individuals from a Labeotropheus fuelleborni × Pseudotropheus tropheops “red cheek” cross, resulting in 946 markers in 24 LGs and spanning 1,453.3 cM (53). BWA mem (version 0.7.12-r1044) (54) was used in both Chromonomer runs to create the input SAM file by aligning respective map-marker sequences to the appropriate assembly or intermediate assembly. A minimum of two markers was required to anchor a contig to a particular LG. The resulting FASTA file of the anchored M_zebra_UMD1 assembly was used for subsequent analysis.
Phylogenetic Analysis.
A maximum-likelihood phylogeny was constructed with the variant data using the SNPhylo pipeline (55). Default parameters were used with an additional flag -M 0.5.
Ancestral Allele Reconstruction.
Pairwise whole-genome alignments of Neolamprologus brichardi (a species belonging to an older radiation from Lake Tanganyika), A. burtoni (a riverine species found in East Africa around Lake Tanganyika), and Pundamilia nyererei (a species from a recent radiation in Lake Victoria) were each constructed against the latest Lake Malawi cichlid genome, M. zebra, using the last alignment algorithm (A. burtoni against M. zebra, P. nyererei against M. zebra, N. brichardi against M. zebra) (56). The generated .maf files from the alignment were converted to sam format using the maf-convert script within the last alignment package. Resulting sam files were converted to bam format via SAMtools (57) and were used to add ancestral allele information into the vcf file obtained from variant discovery.
Detection of Ancient/Derived Allele Enrichment Among Pit-Digging and Castle-Building Species.
To assess biases in the presence of ancient (polymorphic within and outside of Lake Malawi) and derived (polymorphic only among sand-dwelling species) alleles among pit-digging and castle-building species, we first intersected our SNP-level FST measurements with the lists of ancient and derived SNPs as identified through the ancestral allele reconstruction methods outlined above. We then calculated a P value and OR on allele counts at each SNP using a Fisher exact test. We assessed both the degree of divergence at each SNP via FST values and the direction of divergence through the ORs from the Fisher’s exact test. For example, at derived SNPs, an OR <1 indicated that the castle-building species tended to possess the derived allele, while an OR >1 indicated a bias toward the derived allele for the pit-diggers. For ancient SNPs, an OR <1 indicated that the castle-building species tended to possess the non-M. zebra allele (recall that variants were called in relation to the M. zebra reference genome), while an OR >1 indicated that pit-digging species tended to possess the non-M. zebra allele.
To assess systematic differences in the possession of derived or ancient variants between pit-digging and castle-building species, we performed Fisher’s exact tests at various FST thresholds. These tests were applied to both the proportion of SNPs with an OR >1 and OR <1 for the derived and ancient lists (as seen in Fig. 2C) in addition to comparing the mean ORs at various P value thresholds (as seen in SI Appendix, Fig. S2C).
Analyses of Gene Flow and Incomplete Lineage Sorting.
Maximum-likelihood phylogenies were produced for 10-kb genomic bins using the python function phyml_sliding_windows.py created by Simon Martin (available at https://github.com/simonhmartin/genomics_general). This resulted in 1,927 trees that were subsequently visualized using DensiTree v2.2.5 (16).
We used TWISST (17) to analyze the distribution of phylogenetic topologies across genomic windows. To do so the .vcf file containing genotype information for the pit-digging and castle-building species was filtered to include only biallelic sites in which all species possessed genotypes. Indels were also removed. The function phyml_sliding_windows.py was then used to create maximum-likelihood phylogenies from these variants in windows containing 50 informative SNPs. TWISST was then run on these trees, testing for variation in the tree topologies in the following five clades:
Clade 1 (pit diggers): Trematrocranus placodon, Dimidiochromis kiwinge, Dimidiochromis compressiceps, Tramitichromis intermedius, Mylochromis sphaerodon, Mylochromis lateristriga
Clade 2 (C. virginalis): C. virginalis
Clade 3 (Copadichromis castle builders): Copadichromis sp. “mloto goldcrest,” Copadichromis likomae
Clade 4 (castle builders): Mylochromis anaphrymus, Ctenopharynx nitidus, Otopharynx argyrosoma, M. conophoros, Nyassachromis sp. “otter”
Clade 5 (A. calliptera): A. calliptera
Four Population Tests.
TreeMix was run on genotype counts using the settings –k 2000 and –m 1,2,4,6,8,10 to assess support for admixture events among bower-building species. We extended this analysis by computing the f4 statistic for every possible four-population combination using the fourpop function in TreeMix with the setting –k 500. P values were calculated for every four-population comparison from the reported z-scores and adjusted using Bonferroni correction. To use A. calliptera as an outgroup in the detection of possible gene flow among sand-dwellers, results for just the combination (A,B; C, A. calliptera) were extracted. This led to 14,536 comparisons, of which 3,706 had significant (Bonferroni-corrected P < 0.05) f4 statistics (Fig. 3E and Dataset S4). The most significant comparisons for each species pair in which the species were (A,B;C,D) and (A,B;C,D) were then collected.
We used the fd statistic to identify patterns of possible introgression across the genome. The fd statistic function is similar to f4 in that it compares genotype frequencies between four populations; however, whereas f4 is a genome-wide measure, fd can be calculated locally within genomic windows and therefore allows the detection of genomic regions with particularly strong signals of introgression19. We calculated fd using the python function ABBABABAwindows.py created by Simon Martin (available at https://github.com/simonhmartin/genomics_general) over 10-kb windows containing at least 50 informative SNPs for the four population groups identified as most significant from analyses of the f4 statistic.
RNA-Seq Library Construction.
To assess the allele-specific expression, whole brains were obtained from the following animals:
One CV male; digging (sire of all analyzed F1 hybrids)
Two CV × M. conophoros (MC) F1 hybrids; digging
Two CV × MC F1 hybrids; building
Two CV × MC F1 hybrids; isolated
For these experiments individual males were housed with three to five conspecific females and were allowed to develop territories and initiate bower construction. We confirmed that bower behavior was being performed reliably (i.e., was consistent for >24 h), and on the evening before the experiment we separated focal males from females using a transparent divider and flattened the bower. At lights-on the next morning the barrier was removed, and the males’ behavior was observed. Males were killed via decapitation 30 min after the initiation of consistent behavior and, to prevent mRNA degradation, brains were dissected into RNAlater (catalog no. AM7020; Thermo Fisher) less than 10 min after the animal was killed. Whole brains were homogenized in TRIzol (catalog no. 12183555; Thermo Fisher) using a pestle. RNA was isolated using a Qiagen RNeasy mini kit (catalog no. 74104; Qiagen). RNA-seq libraries were constructed using Illumina TruSeq kits following the manufacturer’s protocols. All libraries were sequenced as multiplexed samples in one lane of an Illumina HiSeq 2000 system. Two biological replicates per context were chosen for analyzing ASE following previously published methods (24) that found a similar sample size was sufficient for reliably detecting allelic biases from RNA-seq data.
RNA-Seq Alignments and SNP Calling.
RNA-seq read quality was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Illumina adapters were removed using SeqPrep (https://github.com/jstjohn/SeqPrep). We obtained the M. zebra genome assembly and annotations from NCBI RefSeq (assembly accession: GCF_000238955.2; Assembly name: M_zebra_UMD1). RNA-seq reads were aligned to the M. zebra genome using STAR 2.4 (58) with the options –SortedByCoordinate, –outSAMattributes MD NH NM, and –clip5pNbases 6.
Read groups were added with AddOrReplaceReadGroups.jar in Picard Tools 1.92 (https://github.com/broadinstitute/picard). The resulting bam files were sorted using SAMtools (57). Duplicate reads were marked using MarkDuplicates.jar in Picard Tools. We then applied GATK 3.3 indel realignment and duplicate removal and performed SNP and indel discovery using UnifiedGenotyper following the suggested GATK best practices (41–43).
We filtered the resulting .vcf files to identify all heterozygous sites in the F1 hybrid samples with quality scores >30. We also produced a list of all homozygous sites in the CV parental sample with quality scores >30. To allow proper phasing of heterozygous sites, we filtered the F1 hybrid list to include only sites that intersected with the CV homozygous SNPs.
Detection and Quantification of ASE.
The M. zebra reference genome was masked at high-confidence heterozygous sites using the perl script MaskReferencefromBED.pl (https://github.com/TheFraserLab/ASErhttp://github.com/TheFraserLab/ASEr). To control for reference bias, all hybrid samples were then realigned to this masked reference using the same STAR options as above. Duplicates were marked using MarkDuplicates.jar in Picard Tools, and the bam files were sorted using Sam Tools. The SNP-level ASE was then calculated with the python script CountSNPASE.py (https://github.com/TheFraserLab/ASEr).
All downstream ASE analyses were conducted using R version 3.2.3 (59). After SNP-level ASE was calculated, we filtered for more than five counts per allele for every gene within each sample. To conduct gene-level analyses, the subset of SNPs that met this expression cutoff were then summed without normalization into gene-level counts using the gene coordinates in the ref_M_zebra_UMD1_top_level.gff3 annotation (NCBI M. zebra annotation release 102). For all genes in each sample, ASE was calculated by taking the log2 ratio of the gene-level CV allele counts over the gene-level MC allele counts. After alleles were filtered and summed into genes, we investigated the distribution of ASE ratios across all genes for each sample. Distributions consistently skewed toward either allele could be potentially indicative of biases in read alignment or other technical artifacts. Analysis of ASE ratio distributions showed each to be roughly normal and centered around a log2 ratio of zero, indicating a lack of evidence for strong bias toward either species’ allele across samples (SI Appendix, Fig. S9).
We next calculated the significance of ASE per gene. Since allelic counts from RNA-seq data are prone to overdispersion, we identified significant ASE using a beta-binomial test comparing the CV and MC counts at each gene with the R package MBASED (60) (1- sample analysis; default parameters; run for 1,000,000 simulations). The resulting P values were adjusted for multiple tests using Bonferroni correction. For a gene to be considered significant within a context, we required that both replicates possess Bonferroni All P values <0.05 and that the direction of ASE (either CV biased or MC biased) was the same between replicates.
Identifying diffASE.
DiffASE and differential allelic induction were identified using the two-sample analysis in MBASED (60) (default parameters; run for 1,000,000 simulations) which, as in the one-sample analysis used to assay ASE, employs a beta-binomial model of read counts to control for over dispersion. MBASED was used to compare all possible pairings of the three behavioral contexts (digging, building, and isolated). This produced the pairings: digging × building, digging × isolated, and building × isolated. Furthermore, since MBASED tests for significance in only one of the two contexts at a time, we also ran all three comparisons in their reciprocal directions (building × digging, isolated × digging, and isolated × building). To identify diffASE, the expression of the CV and MC alleles within each context were compared (i.e., CV allelecontext1 vs. MC allelecontext1 compared with CV allelecontext2 vs. MC allelecontext2). Significant cases represent scenarios in which the ratios between the alleles diverge between contexts (as represented in Fig. 4A). Differential induction was assayed by testing for significant variation between CV and MC alleles across contexts, using the ratio of each allele to itself for the test of significance (i.e., CV allelecontext1 vs. CV allelecontext2 compared with MC allelecontext1 vs. MC allelecontext2).
The resulting P values for each pairing were combined across replicates using Fisher’s method and were adjusted with Bonferroni correction. We then compared these combined and adjusted P values across the reciprocal contexts (e.g., digging × isolated vs. isolated × digging) and selected the lowest P value for downstream analyses.
Gene Set Enrichment Tests.
For the enrichment analyses of diffASE genes and genes with allelic induction in building and digging, the corresponding lists were produced as well as background sets corresponding to all other genes detected in one of the three behavioral contexts. Gene enrichments were calculated with PANTHER (61) and filtered for a q-value <0.05. The allelic induction lists were produced in a method similar to those for diffASE by using MBASED, but instead of comparing alleles within contexts the analysis was run on alleles across contexts (e.g., comparing CV allele expression in digging vs. building). The resulting P values would then reflect the differential induction of each allele across pairs of contexts. For the gene set enrichment tests, only genes that had significant induction (Bonferroni-corrected P value <0.05) of at least one allele in either the building or digging contexts alone were selected. Semantic similarity plots for significantly enriched categories were produced with REVIGO (45).
Lineage-specific variation was assayed by comparing the number of genes enriched within gene sets that possessed differential allelic induction of either the CV or MC alleles during building or digging. To do so a .gmt file containing gene sets from the human Kyoto Encyclopedia of Genes and Genomes, GO, Molecular Signatures Database, National Cancer Institute, Institute of Bioinformatics, NetPath, HumanCyc, Reactome, and Panther databases was downloaded from the Bader laboratory website (download.baderlab.org/EM_Genesets/) in April 2018. Genes were then assigned to categories, and a Fisher’s exact test was performed on a 2 × 2 contingency table in which the rows represented digging and building and the columns were the CV and MC alleles. To limit the number of tests performed, we ran the Fisher’s exact test on each ontology independently and required each gene set to have 20 genes represented. The resulting P values were then adjusted for multiple tests using Bonferroni correction.
Supplementary Material
Acknowledgments
We thank Bosco Rusua and Richard Zatha for assistance with fieldwork in Malawi, Shweta Biliya and Fred Vannberg for help with the genome sequencing, and Tom Kocher and members of the J.T.S., R.D.F., and H.B.F. laboratories for critical comments. This project was supported by NIH Grants R01GM101095 (to J.T.S. and R.D.F.), R01NINDS034950 (to R.D.F.), 2R01GM097171-05A1 (to H.B.F.), and R21AG050304 and R01GM114170 (to P.T.M.). R.A.Y. was supported by the Stanford Bio-X Bowes Fellowship and a Stanford Center for Computational, Evolutionary, and Human Genomics Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All sequencing data in support of the findings of this study have been deposited in the Gene Expression Omnibus (accession no. GSE97490) and in the National Center for Biotechnology Information BioProject databank (accession no. PRJNA380805). Code for the allele-specific expression pipeline is available at https://github.com/TheFraserLab/ASEr.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810140115/-/DCSupplemental.
References
- 1.Keenleyside MHA. Cichlid Fishes: Behavior, Ecology and Evolution. 1st Ed Chapman and Hall; London: 1991. [Google Scholar]
- 2.York R, et al. Evolution of bower building in Lake Malawi cichlid fish: Phylogeny, morphology, and behavior. Front Ecol Evol. 2015;3:18. [Google Scholar]
- 3.Magalhaes IS, Croft GE, Joyce DA. Altering an extended phenotype reduces intraspecific male aggression and can maintain diversity in cichlid fish. PeerJ. 2013;1:e209. doi: 10.7717/peerj.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conte MA, Kocher TD. An improved genome reference for the African cichlid, Metriaclima zebra. BMC Genomics. 2015;16:724. doi: 10.1186/s12864-015-1930-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamichhaney S, et al. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature. 2015;518:371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 6.Loh YHE, et al. Comparative analysis reveals signatures of differentiation amid genomic polymorphism in Lake Malawi cichlids. Genome Biol. 2008;9:R113. doi: 10.1186/gb-2008-9-7-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loh YHE, et al. Origins of shared genetic variation in African cichlids. Mol Biol Evol. 2013;30:906–917. doi: 10.1093/molbev/mss326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brawand D, et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meier JI, et al. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat Commun. 2017;8:14363. doi: 10.1038/ncomms14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parnell NF, Streelman JT. Genetic interactions controlling sex and color establish the potential for sexual conflict in Lake Malawi cichlid fishes. Heredity (Edinb) 2013;110:239–246. doi: 10.1038/hdy.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen K, et al. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freese NH, Norris DC, Loraine AE. Integrated genome browser: Visual analytics platform for genomics. Bioinformatics. 2016;32:2089–2095. doi: 10.1093/bioinformatics/btw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-Ari Fuchs S, et al. GeneAnalytics: An integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS. 2016;20:139–151. doi: 10.1089/omi.2015.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones FC, et al. Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouckaert RR. DensiTree: Making sense of sets of phylogenetic trees. Bioinformatics. 2010;26:1372–1373. doi: 10.1093/bioinformatics/btq110. [DOI] [PubMed] [Google Scholar]
- 17.Martin SH, Van Belleghem SM. Exploring evolutionary relationships across the genome using topology weighting. Genetics. 2017;206:429–438. doi: 10.1534/genetics.116.194720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin SH, Davey JW, Jiggins CD. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol. 2015;32:244–257. doi: 10.1093/molbev/msu269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malinsky M, et al. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science. 2015;350:1493–1498. doi: 10.1126/science.aac9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mims MC, Darrin Hulsey C, Fitzpatrick BM, Streelman JT. Geography disentangles introgression from ancestral polymorphism in Lake Malawi cichlids. Mol Ecol. 2010;19:940–951. doi: 10.1111/j.1365-294X.2010.04529.x. [DOI] [PubMed] [Google Scholar]
- 22.Robinson GE, Fernald RD, Clayton DF. Genes and social behavior. Science. 2008;322:896–900. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittkopp PJ, Kalay G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2011;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- 24.Fraser HB, et al. Systematic detection of polygenic cis-regulatory evolution. PLoS Genet. 2011;7:e1002023. doi: 10.1371/journal.pgen.1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestov NB, et al. Evolution of Na,K-ATPase beta m-subunit into a coregulator of transcription in placental mammals. Proc Natl Acad Sci USA. 2007;104:11215–11220. doi: 10.1073/pnas.0704809104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu R, et al. Loss of microRNAs in pyramidal neurons leads to specific changes in inhibitory synaptic transmission in the prefrontal cortex. Mol Cell Neurosci. 2012;50:283–292. doi: 10.1016/j.mcn.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandrasekaran S, et al. Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc Natl Acad Sci USA. 2011;108:18020–18025. doi: 10.1073/pnas.1114093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hrvatin S, et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci. 2018;21:120–129. doi: 10.1038/s41593-017-0029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orr HA. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics. 1998;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irisarri I, et al. Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nat Commun. 2018;9:3159. doi: 10.1038/s41467-018-05479-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Küpper C, et al. A supergene determines highly divergent male reproductive morphs in the ruff. Nat Genet. 2016;48:79–83. doi: 10.1038/ng.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamichhaney S, et al. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax) Nat Genet. 2016;48:84–88. doi: 10.1038/ng.3430. [DOI] [PubMed] [Google Scholar]
- 33.Schwander T, Libbrecht R, Keller L. Supergenes and complex phenotypes. Curr Biol. 2014;24:R288–R294. doi: 10.1016/j.cub.2014.01.056. [DOI] [PubMed] [Google Scholar]
- 34.Roberts RB, Ser JR, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 2009;326:998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science. 2015;349:1489–1494. doi: 10.1126/science.aaa8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: From polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moyerbrailean GA, et al. High-throughput allele-specific expression across 250 environmental conditions. Genome Res. 2016;26:1627–1638. doi: 10.1101/gr.209759.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel RK, Jain M. NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auwera GA, et al. From FastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGrath PT, et al. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature. 2011;477:321–325. doi: 10.1038/nature10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danecek P, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strimmer K. fdrtool: A versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 47.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramakrishnan Varadarajan A, Mopuri R, Streelman JT, McGrath PT, Grath P. Genome-wide protein phylogenies for four African cichlid species. BMC Evol Biol. 2018;18:1. doi: 10.1186/s12862-017-1072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amores A, et al. A RAD-tag genetic map for the platyfish (Xiphophorus maculatus) reveals mechanisms of karyotype evolution among teleost fish. Genetics. 2014;197:625–641. doi: 10.1534/genetics.114.164293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Quin CT, Drilea AC, Conte MA, Kocher TD. Mapping of pigmentation QTL on an anchored genome assembly of the cichlid fish, Metriaclima zebra. BMC Genomics. 2013;14:287. doi: 10.1186/1471-2164-14-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albertson RC, et al. Genetic basis of continuous variation in the levels and modular inheritance of pigmentation in cichlid fishes. Mol Ecol. 2014;23:5135–5150. doi: 10.1111/mec.12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997.
- 55.Lee TH, Guo H, Wang X, Kim C, Paterson AH. SNPhylo: A pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics. 2014;15:162. doi: 10.1186/1471-2164-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.R Core Team 2016. R: A Language and Environment for Statistical Computing (R Found Stat Comput, Vienna), Version R.3.4.3.
- 60.Mayba O, et al. MBASED: Allele-specific expression detection in cancer tissues and cell lines. Genome Biol. 2014;15:405. doi: 10.1186/s13059-014-0405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomas PD, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.