Significance
In aerobic organisms, the terminal enzymes of the respiratory chain such as cytochrome c oxidase receive electrons and reduce molecular oxygen to water. Heme A is an essential cofactor for these oxidases and constitutes the redox-active sites and the proton-transfer pathways. Heme A is synthesized by heme A synthase (HAS). The molecular architecture and reaction mechanism is unknown. In this work, we determined the crystal structure of HAS from Bacillus subtilis in the apo form at high resolution. The structure in the holo form was also constructed using the crystal structure. These structures provide some clues for the heme A biosynthetic process, including substrate binding manner and catalytic residues.
Keywords: crystal structure, heme A biosynthesis, membrane protein, CtaA, formylation reaction
Abstract
Heme A is an essential cofactor for respiratory terminal oxidases and vital for respiration in aerobic organisms. The final step of heme A biosynthesis is formylation of the C-8 methyl group of heme molecule by heme A synthase (HAS). HAS is a heme-containing integral membrane protein, and its structure and reaction mechanisms have remained unknown. Thus, little is known about HAS despite of its importance. Here we report the crystal structure of HAS from Bacillus subtilis at 2.2-Å resolution. The N- and C-terminal halves of HAS consist of four-helix bundles and they align in a pseudo twofold symmetry manner. Each bundle contains a pair of histidine residues and forms a heme-binding domain. The C-half domain binds a cofactor-heme molecule, while the N-half domain is vacant. Many water molecules are found in the transmembrane region and around the substrate-binding site, and some of them interact with the main chain of transmembrane helix. Comparison of these two domain structures enables us to construct a substrate-heme binding state structure. This structure implies that a completely conserved glutamate, Glu57 in B. subtilis, is the catalytic residue for the formylation reaction. These results provide valuable suggestions of the substrate-heme binding mechanism. Our results present significant insight into the heme A biosynthesis.
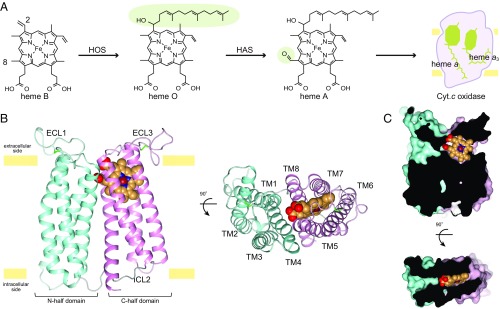
Heme A is a kind of iron porphyrin compound with one formyl group and one hydroxyethylfarnesyl group (Fig. 1A). This molecule plays a key role in the reduction of molecular oxygen and forms the proton pump pathway of cytochrome c oxidase and quinol oxidase, which are the terminal enzymes of the aerobic respiratory chain (1–3). Heme A is synthesized by two enzymatic reactions from heme B. In the first reaction of heme A biosynthesis, the C-2 vinyl group of heme B is converted to a hydroxyethylfarnesyl group to form heme O by heme O synthase (HOS) (4). In the second reaction, the C-8 methyl group of heme O is converted to a formyl group to form heme A by heme A synthase (HAS) (5, 6). HAS is found in a wide range of organisms which carry out aerobic respiration, and is termed Cox15p in eukaryotes and CtaA in archaea and bacteria (6, 7). HAS is an integral membrane protein and located in the bacterial cytoplasmic membrane and mitochondrial inner membrane.
Fig. 1.
Overall structure of BsHAS. (A) Scheme of heme A biosynthesis. The hydroxyethylfarnesyl group and the formyl group are shaded in green. (B) Overall structure of BsHAS. The structure is a ribbon drawing as viewed from within the membrane and the extracellular side. The N-half domain is shown in cyan and C-half is shown in pink. The heme molecule is shown as spheres. (C) Heme-binding site. A section of the surface representation of BsHAS is shown.
Although HAS is essential for aerobic organisms, only limited information is available regarding its structure and catalytic mechanism. HAS has little sequence homology with other proteins, and its structure is unknown. Low-spin heme b is detected from HAS and thus is considered to be a cofactor (8, 9). Sequence analyses and membrane topology analysis using fusion proteins indicate that HAS has eight transmembrane helices with both the N and C termini exposed on the cytoplasmic side of the membrane (6, 8, 10, 11). HAS is classified into two types according to both the phylogeny and key features of the amino acid sequence (12) (SI Appendix, Fig. S1). Type-1 HAS is characterized by the pairs of cysteine residues, while type-2 HAS lacks the cysteine residues. The catalytic residues are still unknown, while several amino acid residues are completely conserved in the Cox15p/CtaA family. Biochemical studies for HAS are limited due to the lack of in vitro enzyme assay systems, and the activity has not been reported for any preparation of purified sample (7). HAS from Bacillus subtilis (BsHAS), which belongs to type-1 HAS, has been relatively well studied (5, 8, 13–15). Various variants have been produced by site-directed mutagenesis based on the sequence conservation to specify the catalytically important residues (10, 16, 17) (SI Appendix, Table S1). However, the interpretation of the results of mutational analyses is ambiguous, since structural information is absent. In addition, mutations of HAS have been linked to mitochondrial diseases (18, 19), Alzheimer’s disease (20), and various cancers (COSMIC; https://cancer.sanger.ac.uk/cosmic and ref. 21). HAS is also involved in the infectivity of some protozoan parasites, such as Trypanosoma cruzi (22). Accordingly, HAS could be a potential target for new drugs. Therefore, the atomic structures are desired for overcoming the current stagnation.
Results
Overall Structure.
Here we report the crystal structure of BsHAS at 2.2-Å resolution (SI Appendix, Figs. S2 and S3 and Tables S2 and S3). BsHAS has eight transmembrane helices (TM1–TM8) and two long extracellular loop regions (ECL1 and ECL3) (Fig. 1B and SI Appendix, Fig. S4). The N-half (TM1–TM4) and C-half (TM5–TM8) domains are connected by an intracellular loop (ICL2). Each domain is formed from four-helix bundles and is organized in counterclockwise order as viewed from the extracellular side. Each four-helix bundle contains a pair of conserved histidine residues and forms a heme-binding site. The C-half domain has one heme B molecule, while a cavity exists in the N-half domain (Fig. 1C). Thus, we concluded that the C-half domain is the cofactor-heme binding domain and the N-half domain is the substrate-heme binding domain. Consequently, the crystal structure is assigned as the substrate-free state.
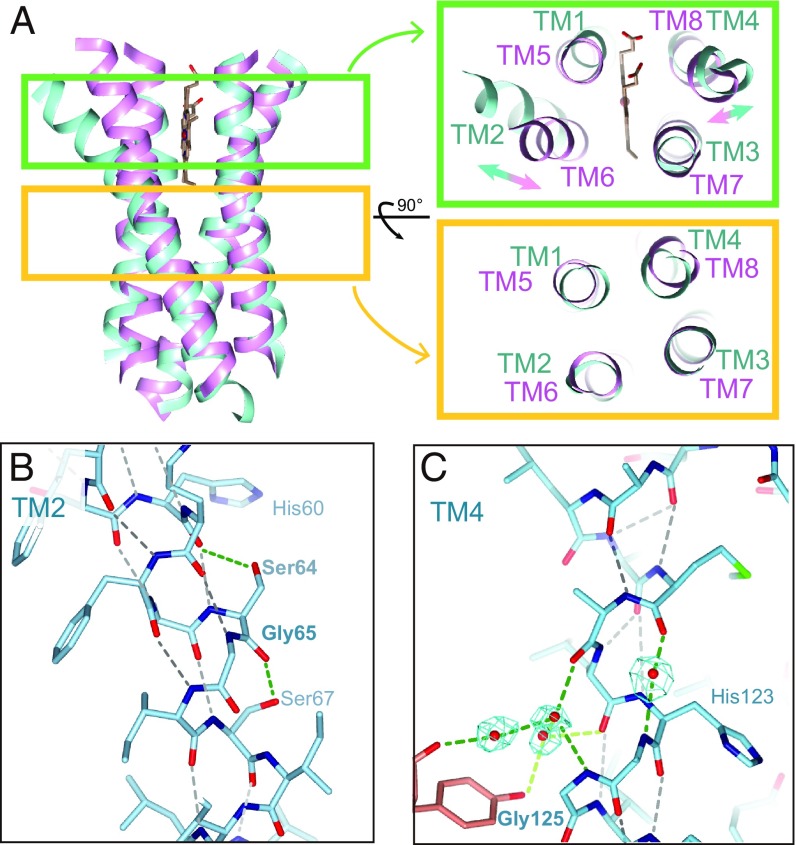
Eight-transmembrane HAS has been considered to arise from tandem gene duplication of ancestral four-transmembrane HAS (8, 23). Consistent with this, the structures of the N- and C-half domains are related by a pseudo twofold symmetry with an axis running perpendicular to the membrane. The transmembrane regions of these domains superimpose well, with a root mean square deviation (r.m.s.d.) of 2.3 Å over the main chain atoms (Fig. 2A). However, there are relatively large differences between TM2–TM6 and TM4–TM8. TM2 is kinked at a highly conserved glycine residue (Gly65 in B. subtilis) with its extracellular end shifted outward by ∼5 Å (Fig. 2B). In TM6, which is the C-half domain counterpart of TM2, the corresponding residue of Gly65 is Leu221 and the helix is straight. In TM4, the hydrogen-bonding pattern is broken by interaction between the backbone and water molecules, and the helix is slightly kinked (Fig. 2C). The two pairs of cysteine residues in ECL1 and ECL3 form disulfide bridges, respectively, as confirmed by the anomalous difference Fourier map (SI Appendix, Fig. S5). Bacterial HAS activity was shown to be strictly dependent on molecular oxygen, and an oxygen supply is necessary for the heme A synthesis (13, 24). In contrast, an isotopic labeling study suggested that the source of the oxygen atom in the formyl group of product heme A is not derived from molecular oxygen but rather from water molecules in type-1 HAS (14). This would be true in type-2 HAS. The necessity of molecular oxygen for type-1 and type-2 HAS activation has been studied only in vivo; it is not clear that molecular oxygen acts on HAS directly. Thus, the origin of the molecular oxygen requirement is unknown. On the other hand, as for type-1 HAS, the formation of disulfide bridges by oxidation may be one of the factors of the molecular oxygen requirement.
Fig. 2.
Pseudo twofold symmetric structure. (A) Structural superimposition of transmembrane helices of the N (cyan)- and C (pink)-half domains. (B) The helical secondary structure breaking region of TM2. The hydrogen bonds in the main chain are indicated by black dashes and the others are shown with green dashes. (C) TM4. The omit map for waters is shown in cyan (4.0σ).
Cofactor Heme-Binding Domain.
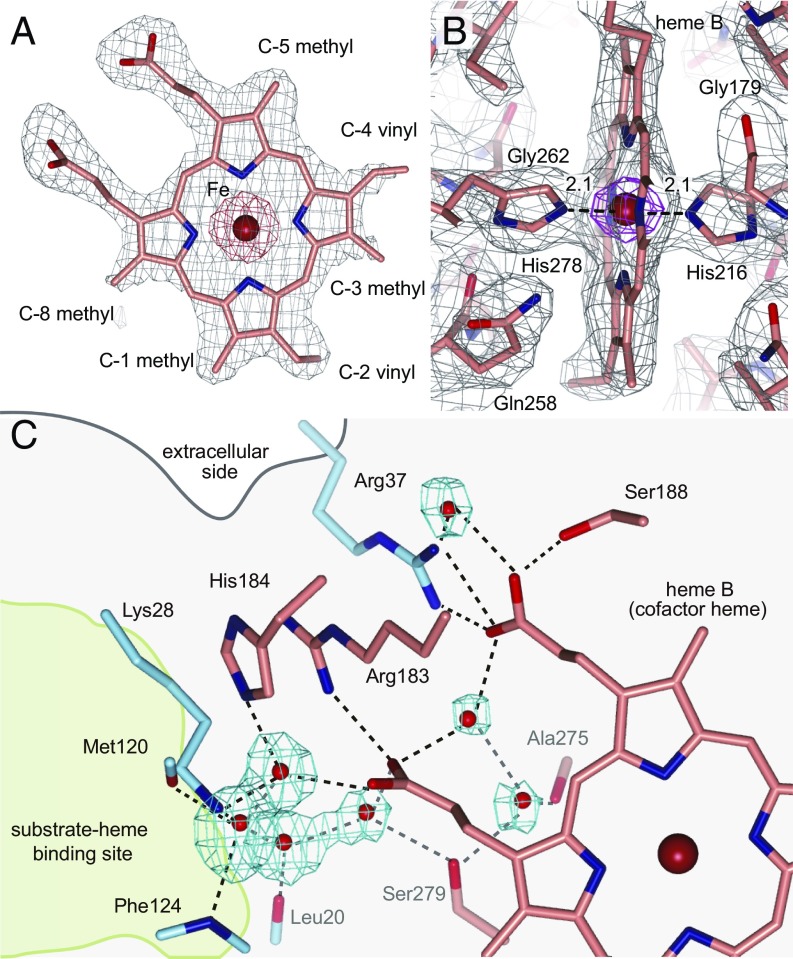
The two sides of the macrocycle plane of hemes are distinguishable between β-face and α-face. The conformation and orientation of cofactor-heme B is clearly defined from the electron density map (Fig. 3 A and B). The conserved His278 and His216, respectively, bind to the β-face and α-face of heme b as axial ligands (Fig. 3B). This bis-histidine hexa-coordination of the heme iron is consistent to the low-spin heme as indicated with EPR spectroscopy (9). Many water molecules were found in the cavity in the transmembrane region of BsHAS (Fig. 3C). These water molecules form clusters, and interact with both the cofactor-heme b and the substrate-heme binding site through hydrogen bonds. A part of this hydrogen-bonding network reaches the hydrophobic region of the lipid bilayer. These water molecules and the hydrogen-bonding network may be associated with heme A synthesis in terms of the conformational change of helices. In this case, water molecules interact with the main chain of transmembrane helices and propionic group of heme molecule.
Fig. 3.
Cofactor-heme binding site. (A) Omit map of the cofactor-heme for determination of its orientation and conformation. The heme b is viewed from the α-face (back side). The omit map is shown for the whole heme molecule (gray mesh, 1.5σ; red mesh, 10.0σ). (B) Environment of the cofactor-heme b. The 2mFobs − DFcalc electron density map is shown (gray mesh, 1.0σ; magenta mesh, 8.0σ). (C) Hydrogen bond network. Water molecules are shown as red spheres with an omit map in cyan (4.0σ), and amino acid residues included in the hydrogen bond network are shown as sticks. The dotted lines indicate hydrogen bonds.
Structural Comparison.
A structural comparison based on the C-half domain of HAS using the Dali server (25) resulted in ∼2,500 proteins with Z score ≥2 in the PDB25 database. However, most of these are soluble proteins. The heme-containing membrane protein with the highest structural similarity was bovine mitochondrial cytochrome c oxidase [PDB ID code 1OCC (26), Z score = 8.9, r.m.s.d. = 2.7 Å for 119 residues, identity = 8%], followed by superoxide oxidase from Escherichia coli [PDB ID code 5OC0 (27), Z score = 8.8, r.m.s.d. = 3.7 Å for 125 residues, identity = 7%], bacterial caa3-type cytochrome c oxidase from Thermus thermophilus [PDB ID code 2YEV (28), Z score = 8.3, r.m.s.d. = 2.8 Å for 116 residues, identity = 8%] and cytochrome bc1 complex from Rhodobacter sphaeroides [PDB ID code 5KKZ (29), Z score = 7.5, r.m.s.d. = 2.9 Å for 126 residues, identity = 8%].
Although bovine cytochrome c oxidase is the most similar structure in heme-containing membrane protein, the portion of similar structure to BsHAS is not heme-binding domain (SI Appendix, Fig. S6A). Moreover, the similar structure is only a part of the subunit III in cytochrome c oxidase. The result is almost the same in the caa3 oxidase from T. thermophilus. While the superoxide oxidase (SOO) has the second-highest Z score, the overall structure of SOO is very similar to the C-half domain of BsHAS except for some differences (SI Appendix, Fig. S6B). SOO has two heme molecules, while C-half domain of BsHAS contains one heme molecule. The long extracellular loop exists between the third and fourth helix in SOO, while BsHAS has the long loop between the first and second helix. SOO oxidizes superoxide anion using ubiquinone as the electron acceptor (27). However, the reaction mechanism of SOO is also unclear and the mechanismic relationship between SOO and HAS is unknown. In the case of cytochrome b of cytochrome bc1 complex, a part of the heme-binding domain is similar to BsHAS (SI Appendix, Fig. S6C). In the present structure of BsHAS, the highly conserved glutamine residue (Gln258 in B. subtilis) interacts perpendicularly with the macrocycle plane of the cofactor-heme b. A similar close interaction between glutamine and heme b in BsHAS is also observed in the cytochrome bc1 complex (SI Appendix, Fig. S6C, Right). HAS and these proteins have different functions from each other, although the participation of electrons in the reactions is a common feature. Thus, the result of the structure comparison does not provide any clues for the reaction mechanism of HAS.
Substrate Heme-Binding Domain.
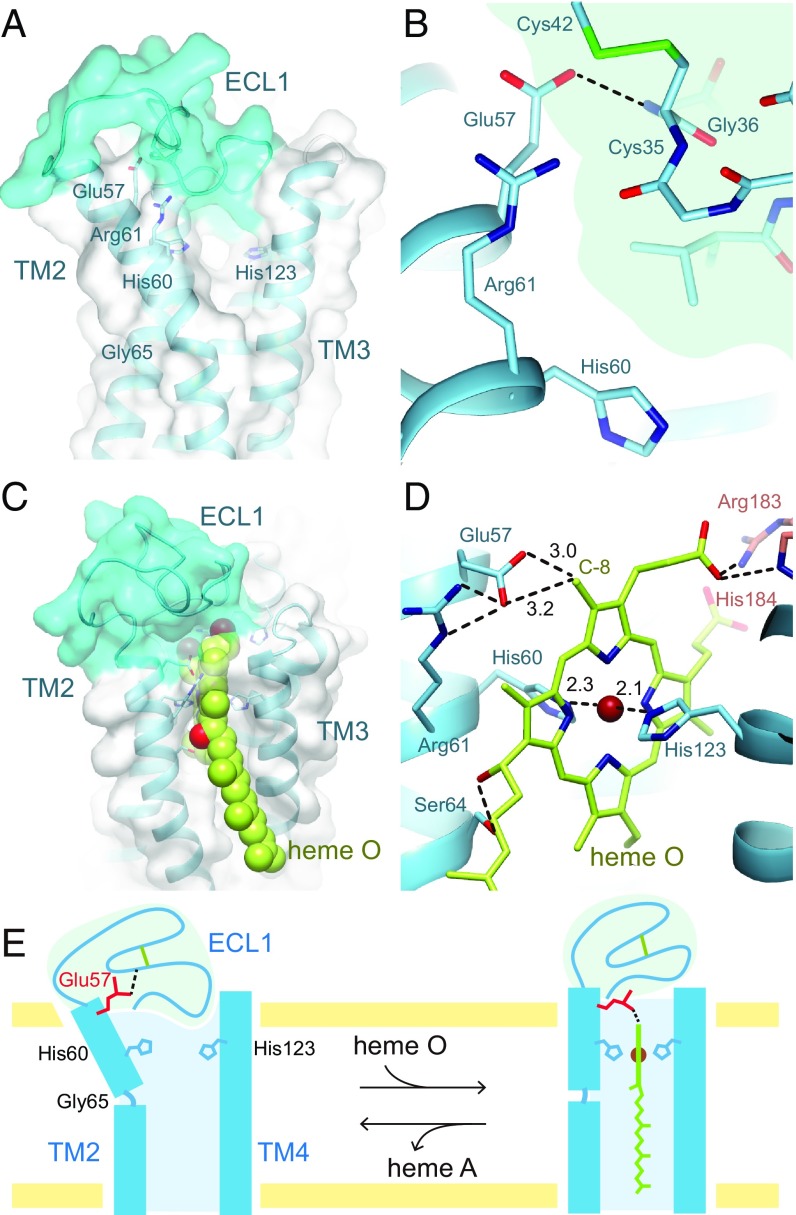
In the substrate heme-binding domain, ECL1 plugs the substrate-binding site instead of the heme molecule (Fig. 4A). The distance between His60 and His123, which are considered to be the axial ligands for substrate heme O, is ∼6.5 Å due to the bending of TM2. Since the spectroscopic study of heme a in a product-bound cytochrome ba form of BsHAS suggests bis-histidine ligation (9), the His Nε2–His Nε2 distance is expected to decrease with structural changes of TM2 and TM4 in the substrate-binding state. Since the hydrogen-bonding pattern of the backbone in the vicinity of the ligand histidine residue is deviated from the regular pattern (Fig. 2 B and C), it is reasonable to assume that TM2 and TM4 are flexible and can change their conformations upon heme O binding. A completely conserved glutamate residue (Glu57 in B. subtilis) in TM2 forms a hydrogen bond with the main chain of ECL1 (Fig. 4B). On the other hand, Gln213 in TM6, which is the counterpart of Glu57, interacts with Arg217 and locates near the cofactor-heme b (SI Appendix, Fig. S3F). While the present structure is in the substrate-free state, the sequence similarities between the N and C halves allowed us to generate a plausible structural model for the substrate-binding state (Fig. 4 C and D and Dataset S1). This model was constructed from the substrate-free state structure with bending TM2 inwards by ∼15° at Gly65. As a result, Glu57 locates near the C-8 methyl group of the substrate-heme O. In addition, the Ser64 and hydrophobic residues in the cytoplasmic half of TM2 and TM3 interact with the OH and polyprenyl groups of the 2-hydroxyethylfarnesyl moiety, respectively. The substrate-binding model strongly suggests that Glu57 is the catalytic residue for the formylation reaction.
Fig. 4.
Substrate-heme binding site. (A) Position of ECL1 and transmembrane helices. The molecular surface of ECL1 is colored cyan. (B) Interaction in the substrate-heme binding site. The ECL1 region is shaded in pale cyan. Glu57 interacts with the main chain of Gly36 in ECL1. (C) A proposed model of the substrate-heme binding state viewed from within the membrane. The heme O molecule is shown as spheres. (D) The catalytic site of the substrate-heme binding state in the proposed model. (E) Substrate heme O binding and conformational changes of ECL1 and TM2.
Discussion
ECL1 completely plugs the substrate-heme binding site in the crystal structure and moves away from this site in the substrate-binding state. In consideration of the above, we proposed the following scheme for substrate binding (Fig. 4E). In the substrate-free state, TM2 is kinked outward and the expanded substrate-heme binding site is plugged with ECL1. First, the polyprenyl group of heme O interacts with the hydrophobic residues of TM2 and TM3, and then the OH group of the hydroxyethylfarnesyl moiety contacts Ser64. Subsequently, the porphyrin ring of heme O is tugged to the substrate-heme binding site, and the Fe atom interacts with Nε2 atoms of His60 and His123. As a result, TM2 is transformed into a straight helix. Thus, ECL1 is moved from the transmembrane region and Glu57 can interact with the C-8 methyl group of heme O.
The substrate-binding model strongly suggests that the conserved glutamate residue (Glu57 in B. subtilis) is the catalytic residue. Thus, it is plausible that the formylation reaction goes through ester cross-links of the C-8 methyl group of heme O with the carboxylate group of Glu57, which was proposed by Brown et al. (14). In this reaction scheme, molecular oxygen binds to iron of the substrate-heme. However, the substrate-heme has bis-histidine hexa-coordination as with cofactor-heme b. Therefore, it cannot be assumed that the molecular oxygen binds to the substrate-heme iron. Moreover, the shortest distance between the two heme porphyrin rings is ∼13 Å in the substrate-binding model, while this can be shorter between the substituents on the two hemes. Thus, electrons can be transferred directly between the two heme molecules, since the maximum distance for electron transfer is 14 Å in the protein milieu (30). Accordingly, we propose the modified reaction scheme, in which the cofactor-heme b participates in the formylation reaction (SI Appendix, Fig. S7). The contribution of cofactor-heme b to the reaction is supported by the mutation analysis which shows that the H216M variant of BsHAS accumulates an alcohol form of heme O (heme I) (16). However, the electron carrier molecules are still unknown and further studies are needed to elucidate the details of the reaction mechanism.
In this paper, we report the structure of BsHAS at high resolution. This structure enables us to reexamine the interpretations of previous mutational analyses and design experiments on HAS from a novel point of view. Although there are still many unanswered questions, our work should provide a number of clues for further studies on heme A biosynthesis.
Methods
Sequence Alignment.
Amino acid sequence alignments were generated using CLUSTALW (31) and corrected manually. All sequences were obtained from the Uniprot database (B. subtilis: P12946; Geobacillus stearothermophilus: A0A087LHZ1; Aeropyrum pernix: Q9YBA3; R. sphaeroides: Q3IXW9; Saccharomyces cerevisiae: P40086; Homo sapiens: Q7KZN9). The A. pernix sequence was repeated twice.
Overexpression and Purification.
HAS from B. subtilis was expressed and purified as described previously (10) with some modifications. The BsHAS was introduced into the pET15b vector with an N-terminal His tag and a thrombin cleavage site, and expressed in E. coli BL21(DE3)/pLysS. For purification, cells were resuspended in 50 mM Tris⋅HCl (pH 8.0), 1× protease inhibitor mixture (Nacalai Tesque) and 0.5 mg⋅mL−1 lysozyme, and disrupted by sonication. Isolated membranes were solubilized at 1.2% sucrose monolaurate (SM-1200) (SML; Mitsubishi-Kagaku Foods) and 50 mM Na-Hepes (pH 7.4) at ∼277 K. The mixture was immediately centrifuged for 40 min at 185,000 × g. The supernatant was applied twice to an Ni-NTA superflow (Qiagen) column, and the protein was eluted with a buffer containing 50 mM Na-Hepes (pH 7.4), 300 mM NaCl, 150 mM imidazole, and 0.1% SML. After removal of the His tag by thrombin, the resultant major fragment was purified on a HiLoad 16/60 Superdex 200 column (GE Healthcare) in a buffer containing 50 mM Na-Hepes (pH 7.4), 300 mM NaCl, and 0.1% SML, and the peak fractions were collected for crystallization.
Spectroscopic Measurements.
The UV-Vis spectra of the purified enzyme in 50 mM Na-Hepes (pH 7.4) and 0.1% SML was measured with a V-630 UV-Vis spectrophotometer (JASCO) in a range from 350 nm to 650 nm at a room temperature of ∼298 K. The reduced samples were prepared by adding sodium ascorbate at a final concentration of 100 mM.
Crystallization and X-Ray Data Collection.
Ultrafiltration was used to replace the detergent with dodecyl-β-d-maltoside (β-DDM; Anatrace), and the purified HAS was concentrated to ∼20 mg⋅mL−1. The protein solution was reconstituted into lipidic cubic phase (LCP) by mixing with molten monoolein (Sigma-Aldrich) with a coupled syringe in a ratio of 40/60 (wt/wt) protein/lipid. Crystallizations were set up by transferring 0.5 μL of the protein-laden mesophase and 15 μL of a precipitant solution onto a glass against 300 μL of the precipitant solution. Crystals appeared within ∼1 wk in a precipitant solution containing 30% (vol/vol) PEG600, 100 mM NaCl, 100 mM Li2SO4, and 100 mM Na-Hepes (pH 7.0) and grew to full size in 1 mo. Heavy atom derivatives were prepared by the soaking method using a saturated concentration of p-chloromercuribenzoic acid (PCMB) and potassium tetrachloroplatinate for 2 h and 24 h, respectively. Diffraction data were collected at BL41XU (proposal nos. 2013A1869, 2013B1363, 2014A1365, 2016B2709, and 2017B2728 to K.T.) and BL44XU (proposal nos. 2014A6950, 2015A6543, 2016A6642, and 2017A6572 to K.M.) of SPring-8. The wavelength of X-rays was set to 1.0 Å for native and heavy atom derivative datasets, 1.74 Å for Fe anomalous datasets, and 1.75 Å for S detection datasets. Crystals were cooled by cryostream coolers at cryogenic temperatures of 15 K at BL41XU and 40 K at BL44XU. Each dataset was obtained from a single crystal, respectively. The space group was determined to be H3 with 1 molecule per asymmetric unit and the crystals were twinned with twin fractions of 3–50%. The data were processed with the HKL2000 program (32) and the XDS program (33). The resolution limit was defined from CC1/2 ∼50% for the native dataset (34) and I/σ(I) ∼2.0 for the other datasets. Datasets with twin ratios lower than 10% were employed for further analyses.
Structure Determination.
Initial experimental phases were obtained via single-wavelength anomalous dispersion experiments (SAD), thereby utilizing the anomalous scattering of the iron in the intrinsic heme at a resolution of 4.8 Å using the SOLVE program (35), and subsequent density modification was performed with the RESOLVE program (36). This Fe-SAD phase was used for the heavy atom search. Subsequently, multiple isomorphous replacement with anomalous scattering (MIRAS) was performed at a resolution of 4.2 Å using the SOLVE program. The X-ray diffraction data for Fe anomalous detection were used as the native data for MIRAS. The phases were improved by multicrystal averaging using the MIRAS phase with the DMMulti program (37) in the CCP4 suite (38). This multicrystal averaging was performed by using several datasets which have very different unit cell parameters and low isomorphism. The density-modified phases were good enough to visualize the membrane-spanning helices (SI Appendix, Fig. S3A). The crystal lattice is composed of stacks of 2D membrane protein layers and belongs to the type I membrane crystal (39). The initial model was built manually with the XtalView program (40). The model was constructed by autotracing with the ARP/wARP program (41) using high-resolution native data and modified manually with the XtalView and COOT (42) programs. The refinement calculations were performed with the CNS (43) and PHENIX (44) programs. The final model contains 308 residues (−5–303), one heme B molecule, 32 waters, one Cu2+ ion, three SO42− ions, and 21 monoolein molecules. The composition of metal ion was determined by X-ray fluorescence spectroscopy. The Rwork and Rfree factors in the resolution range from 30 Å to 2.2 Å were 20.6% and 22.9%, respectively (SI Appendix, Table S3). Residues in favored, allowed and disallowed regions in the Ramachandran plot account for 96.74%, 2.93%, and 0.33% of total residues, respectively, based on analysis with the Molprobity program (45). All figures were generated using the Pymol program (46).
Model Building of Substrate-Heme Binding State.
To create an initial model of the substrate-heme binding state, the heme O molecule was manually placed in the substrate-heme binding site and backbone torsion angles of Gly65 were corrected to form a straight helix with the XtalView program. In addition, side-chain torsion angles of Glu57 and Arg61 were corrected in an analogous manner to Gln213 and Arg217 in the C-half domain. The conformations of the model were energetically optimized by the molecular dynamics using the model_anneal.inp script and model_minimize.inp of the CNS program.
Supplementary Material
Acknowledgments
We thank Dr. K. Kita (Nagasaki University), Dr. Y. Hanazono, and Messrs K. Hirasawa and M. Ono (Kyoto University) for their contributions to the initial steps of this work; Messrs Y. Tai and H. Dao (Kyoto University) for their contributions to the manuscript preparation; and the BL41XU beamline staff and BL44XU beamline staff of SPring-8 for their help with the X-ray data collections. This work was supported by the Photon and Quantum Basic Research Coordinated Development Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (K.M.) and the Japan Society of Promotion of Science (JSPS) KAKENHI Grant 15J02413 (to S.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6A2J for native dataset and 6IED for S-anomalous dataset).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813346115/-/DCSupplemental.
References
- 1.Tsukihara T, et al. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 2.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 3.Yoshikawa S, Shimada A. Reaction mechanism of cytochrome c oxidase. Chem Rev. 2015;115:1936–1989. doi: 10.1021/cr500266a. [DOI] [PubMed] [Google Scholar]
- 4.Saiki K, Mogi T, Anraku Y. Heme O biosynthesis in Escherichia coli: The cyoE gene in the cytochrome bo operon encodes a protoheme IX farnesyltransferase. Biochem Biophys Res Commun. 1992;189:1491–1497. doi: 10.1016/0006-291x(92)90243-e. [DOI] [PubMed] [Google Scholar]
- 5.Svensson B, Lübben M, Hederstedt L. Bacillus subtilis CtaA and CtaB function in haem A biosynthesis. Mol Microbiol. 1993;10:193–201. doi: 10.1111/j.1365-2958.1993.tb00915.x. [DOI] [PubMed] [Google Scholar]
- 6.Mogi T. Biosynthesis and role of heme O and heme A. In: Kadish K, Gulard R, Smith K, editors. The Porphyrin Handbook. Academic; Cambridge, MA: 2003. pp. 157–181. [Google Scholar]
- 7.Hederstedt L. Heme A biosynthesis. Biochim Biophys Acta. 2012;1817:920–927. doi: 10.1016/j.bbabio.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 8.Svensson B, Hederstedt L. Bacillus subtilis CtaA is a heme-containing membrane protein involved in heme A biosynthesis. J Bacteriol. 1994;176:6663–6671. doi: 10.1128/jb.176.21.6663-6671.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svensson B, Andersson KK, Hederstedt L. Low-spin heme A in the heme A biosynthetic protein CtaA from Bacillus subtilis. Eur J Biochem. 1996;238:287–295. doi: 10.1111/j.1432-1033.1996.0287q.x. [DOI] [PubMed] [Google Scholar]
- 10.Mogi T. Probing structure of heme A synthase from Bacillus subtilis by site-directed mutagenesis. J Biochem. 2009;145:625–633. doi: 10.1093/jb/mvp017. [DOI] [PubMed] [Google Scholar]
- 11.Throne-Holst M. 2001. Synthesis of hemes found in heme-copper oxidases of Bacillus subtilis. PhD thesis (Department of Microbiology, Lund University, Lund, Sweden)
- 12.He D, Fu C-J, Baldauf SL. Multiple origins of eukaryotic cox15 suggest horizontal gene transfer from bacteria to jakobid mitochondrial DNA. Mol Biol Evol. 2016;33:122–133. doi: 10.1093/molbev/msv201. [DOI] [PubMed] [Google Scholar]
- 13.Brown KR, Allan BM, Do P, Hegg EL. Identification of novel hemes generated by heme A synthase: Evidence for two successive monooxygenase reactions. Biochemistry. 2002;41:10906–10913. doi: 10.1021/bi0203536. [DOI] [PubMed] [Google Scholar]
- 14.Brown KR, Brown BM, Hoagland E, Mayne CL, Hegg EL. Heme A synthase does not incorporate molecular oxygen into the formyl group of heme A. Biochemistry. 2004;43:8616–8624. doi: 10.1021/bi049056m. [DOI] [PubMed] [Google Scholar]
- 15.Morrison MS, Cricco JA, Hegg EL. The biosynthesis of heme O and heme A is not regulated by copper. Biochemistry. 2005;44:12554–12563. doi: 10.1021/bi050893d. [DOI] [PubMed] [Google Scholar]
- 16.Hederstedt L, Lewin A, Throne-Holst M. Heme A synthase enzyme functions dissected by mutagenesis of Bacillus subtilis CtaA. J Bacteriol. 2005;187:8361–8369. doi: 10.1128/JB.187.24.8361-8369.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewin A, Hederstedt L. Heme A synthase in bacteria depends on one pair of cysteinyls for activity. Biochim Biophys Acta. 2016;1857:160–168. doi: 10.1016/j.bbabio.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Bugiani M, Tiranti V, Farina L, Uziel G, Zeviani M. Novel mutations in COX15 in a long surviving Leigh syndrome patient with cytochrome c oxidase deficiency. J Med Genet. 2005;42:e28. doi: 10.1136/jmg.2004.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alfadhel M, et al. Infantile cardioencephalopathy due to a COX15 gene defect: Report and review. Am J Med Genet A. 2011;155A:840–844. doi: 10.1002/ajmg.a.33881. [DOI] [PubMed] [Google Scholar]
- 20.Vitali M, et al. Analysis of the genes coding for subunit 10 and 15 of cytochrome c oxidase in Alzheimer’s disease. J Neural Transm (Vienna) 2009;116:1635–1641. doi: 10.1007/s00702-009-0324-8. [DOI] [PubMed] [Google Scholar]
- 21.Forbes SA, et al. COSMIC: High-resolution cancer genetics using the Catalogue of Somatic Mutations in Cancer. Curr Protoc Hum Genet. 2016;91:10.11.1–10.11.37. doi: 10.1002/cphg.21. [DOI] [PubMed] [Google Scholar]
- 22.Merli ML, Cirulli BA, Menéndez-Bravo SM, Cricco JA. Heme A synthesis and CcO activity are essential for Trypanosoma cruzi infectivity and replication. Biochem J. 2017;474:2315–2332. doi: 10.1042/BCJ20170084. [DOI] [PubMed] [Google Scholar]
- 23.Lewin A, Hederstedt L. Compact archaeal variant of heme A synthase. FEBS Lett. 2006;580:5351–5356. doi: 10.1016/j.febslet.2006.08.080. [DOI] [PubMed] [Google Scholar]
- 24.Hannappel A, Bundschuh FA, Ludwig B. Characterization of heme-binding properties of Paracoccus denitrificans Surf1 proteins. FEBS J. 2011;278:1769–1778. doi: 10.1111/j.1742-4658.2011.08101.x. [DOI] [PubMed] [Google Scholar]
- 25.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 27.Lundgren CAK, et al. Scavenging of superoxide by a membrane-bound superoxide oxidase. Nat Chem Biol. 2018;14:788–793. doi: 10.1038/s41589-018-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyons JA, et al. Structural insights into electron transfer in caa3-type cytochrome oxidase. Nature. 2012;487:514–518. doi: 10.1038/nature11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esser L, et al. Hydrogen bonding to the substrate is not required for Rieske iron-sulfur protein docking to the quinol oxidation site of complex III. J Biol Chem. 2016;291:25019–25031. doi: 10.1074/jbc.M116.744391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowtan K. ‘dm’: An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsl Prot Crystallogr. 1994;31:34–38. [Google Scholar]
- 38.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Michel H. Crystallization of membrane protein. Trends Biochem Sci. 1983;8:56–59. [Google Scholar]
- 40.McRee DE. XtalView/Xfit–A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 41.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delano WL. 2002. The PyMOL Molecular Graphics System (Schrödinger, LLC), Version 1.5.0.5.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.