Abstract
Killer-cell immunoglobulin-like receptor (KIR) genes are inherited as haplotypes. They are expressed by NK cells and linked to outcomes of infectious diseases and pregnancy in humans. Understanding how genotype relates to phenotype is difficult because of the extensive diversity of the KIR family. Indeed, high-resolution KIR genotyping and phenotyping in single NK cells in the context of disease association is lacking. Here, we describe a new method to separate NK cells expressing allotypes of the KIR2DL1 gene carried by the KIR A haplotype (KIR2DL1A) from those expressing KIR2DL1 alleles carried by the KIR B haplotype (KIR2DL1B). We find that in KIR AB heterozygous individuals, different KIR2DL1 allotypes can be detected both in peripheral blood and in uterine single NK cells. Using this new method, we demonstrate that both blood and uterine NK cells co-dominantly express KIR2DL1A and KIR2DL1B allotypes, but with a predominance of KIR2DL1A variants, which associate with enhanced NK cell function. In a case-control study of pre-eclampsia, we show that KIR2DL1A, not KIR2DL1B, associates with increased disease risk. This method will facilitate our understanding of how individual KIR2DL1 allelic variants affect NK cell function and contribute to disease risk.
Introduction
Natural Killer (NK) cells are important effectors in immune responses to viruses and tumours (1). Increasingly, tissue resident NK cells are described which defy the classical paradigm of NK cells as ‘killers’ (Reviewed in (2)). Rather, they fulfil tissue specific roles such as interacting with placental cells during early pregnancy (3). NK cell function is tightly controlled by continuously integrating signals from activating and inhibitory receptors including the Killer-cell immunoglobulin-like receptors (KIR). This polymorphic gene family is expressed predominantly by NK cells and to a lesser extent T cells. Both activating and inhibitory KIR exist, some of which bind HLA class I ligands to regulate NK functional responses. In addition, inhibitory KIR that bind cognate HLA class I ligands act with other inhibitory receptors such as CD94/NKG2A, to tune the reactive potential of NK cells to their environment; a dynamic process known as education (4). Because both KIR and HLA are highly polymorphic, this diversity creates a spectrum of NK cell function both within and between individuals. More than 200 studies demonstrate disease association to variants of KIR and their HLA ligands (Reviewed in (5)). These include associations with outcomes in infectious diseases such as HIV/AIDS (6, 7) and hepatitis (8), complications of pregnancy such as pre-eclampsia (9–11) and the outcome of immunotherapy in cancer patients (12). However, the complexity of both the KIR and HLA gene families presents an obstacle to understanding how these genetic associations contribute to pathogenesis.
The diversity of the KIR system arises from several sources. Firstly, KIR genes are inherited as 2 haplotypes, classified based on gene content. The KIR A haplotype has fewer genes with mainly inhibitory KIR, whereas KIR B haplotypes contain additional activating KIR. In this way, individuals will differ by the restricted suite of KIR genes they possess. This is compounded by the fact that the same KIR gene can be found on multiple haplotypes. Therefore, copy numbers of specific KIR genes can also vary. A further source of variation is the extensive KIR allelic polymorphism, for example over 100 alleles have been described for KIR3DL1/S1 (IPD-KIR database). In addition to the genetic diversity, KIR expression is stochastically regulated by two opposing sense and antisense promoters (13). Thus, in a population of NK cells there is variegated expression of KIR or none at all. Moreover, KIR genes are located within the leukocyte receptor complex on chromosome 19 and segregate independently from their HLA ligands encoded on chromosome 6. As a result, individuals may not have all the cognate ligands for their particular KIR repertoire; this will affect the education status of their NK cells. The polymorphic nature of the KIR/HLA system complicates linking genotype to phenotype in the context of disease association studies.
The inhibitory receptor KIR2DL1 recognises HLA-C allotypes bearing a C2 epitope (C2+HLA-C), defined by lysine at position 80. A number of disease associations studies have linked KIR2DL1 with outcome in cancer and transplantation (14) (15) (16). However, some of the strongest associations come from disorders of pregnancy. For example, mothers with two KIR A haplotypes (KIR AA genotype) are at increased risk of disorders of placentation if the fetus carries a C2 epitope inherited from the father (9, 11). Conversely, mothers with a KIR B haplotype (containing activating KIR2DS1 that can also bind C2) are at low risk, but instead these mothers have an increased risk of delivering a large baby (17). When the fetus is homozygous for alleles encoding a C1 epitope (C1+HLA-C) the mother’s KIR genotype has no effect, so C2 is the crucial fetal ligand. These results suggest that binding of the maternal inhibitory KIR2DL1 to trophoblast C2+HLA-C increases the risk of pregnancy disorders, whereas activating KIR2DS1 promotes fetal growth. Functional experiments in mice support the idea that receptor/ligand interactions leading to strong NK inhibition impede fetal growth (18) and there is good evidence for natural selection against strong inhibitory C2-specific human KIR2DL1 variants (19).
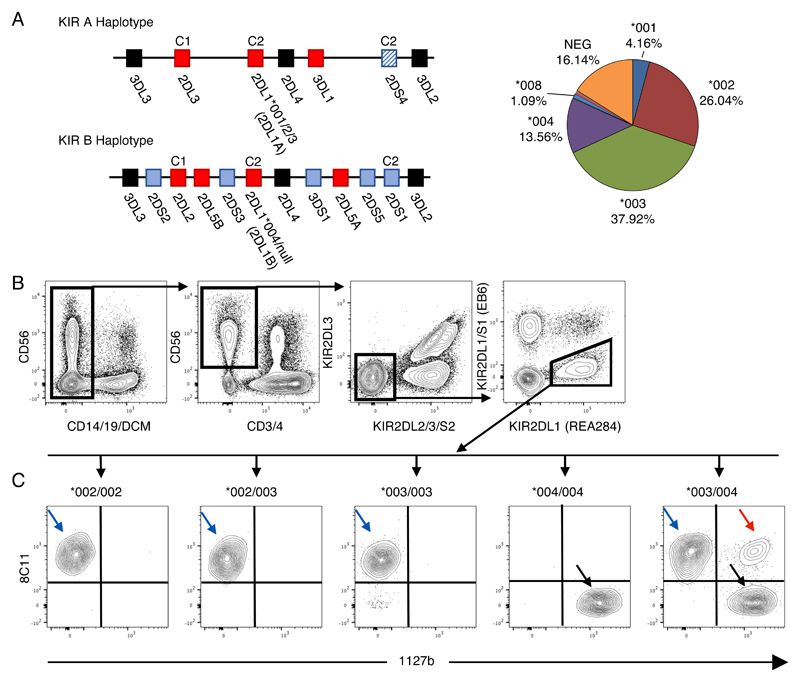
Currently 26 KIR2DL1 alleles have been identified which can be grouped according to which haplotypes they tend to segregate onto (20). Those typically found on the KIR A haplotype in European populations are KIR2DL1*003, *002 or *001, denoted hereon as KIR2DL1A. The dominant allele on the KIR B haplotype is KIR2DL1*004, designated as KIR2DL1B (Figure 1A). These alleles vary in frequency across populations (Figure 1A) and show differences in expression levels of both RNA and protein (15, 21–23). Moreover, functional studies have shown that KIR2DL1 allotypes bind C2+HLA-C allotypes with variable affinities, form an hierarchy of inhibition in response to their cognate ligand and differ in their ability to respond to missing self (24–26). However, most of these studies have been restricted to using in vitro systems, cell lines or donors homozygous for the alleles of interest. Thus, it is unclear how these data translate into functional differences on primary NK cells. In particular, it is not known whether KIR2DL1 allotypes are co-dominantly expressed by NK cells or if they confer different educating signals when presented with identical HLA-C environments.
Figure 1. A new strategy to identify different KIR2DL1 allotypes in single NK cells.
A) Representative KIR A and B Haplotypes. Cognate HLA-C ligands are depicted above their receptor. Framework genes (present in all common haplotypes) are shown in black, activating KIR in blue and inhibitory KIR in red. In European populations the KIR2DL1 alleles on the A haplotype are KIR2DL1*001, *002 or *003. The KIR2DL1 allele on the B haplotype is almost exclusively KIR2DL1*004. Allele frequencies in the cohort in this study are depicted in the pie chart. B) Cryopreserved PBMCs from donors typed for KIR2DL1 alleles were stained. Gating strategy to identify KIR2DL2/3/S2neg 2DS1neg 2DL1pos NK cells in blood. The clone names of specific mAbs recognizing KIR2DL1 are included in parentheses on selected FACS plots to clarify how they were used to gate on KIR2DL1 single positive cells. C) Results for donors with different KIR2DL1 alleles are shown. Using antibody clones 8C11 and 1127b, three subsets were identified in donors heterozygous for KIR2DL1*003 and *004: *003 single positive cells (003sp, blue arrow), *004 single positive cells (004sp, black arrows), and *003/*004 double positive cells (003004dp, red arrow).
A major stumbling block to studying the effects of allelic variation of KIR on NK cell function and its contribution to disease has been the lack of antibodies specific for individual KIR receptors. As a result, anti-KIR antibodies often display cross reactivity for the products of several KIR genes. This property has been exploited to study expression of different KIR2DL3 allotypes in heterozygotes, but has thus far not been possible for KIR2DL1 (27, 28). Here, we have identified a staining combination of KIR-specific antibodies to separate NK cells expressing KIR2DL1A and KIR2DL1B allotypes within the same individual. We find that KIR2DL1A allotypes are preferentially expressed by peripheral blood NK (pbNK) cells and KIR2DL1Apos NK cells respond more strongly in missing self assays than KIR2DL1Bpos cells from the same individual and his or her HLA background. In a case-control study, we show that the risk of developing pre-eclampsia is associated with the number of KIR2DL1A, but not KIR2DL1B gene copy number. Finally, we study the expression of KIR2DL1 allotypes on the tissue resident population of NK cells that are likely to be driving this association in pregnancy. Similar to their peripheral blood counterparts, we find that in decidual NK cells, the KIR2DL1 positive niche is dominated by expression of KIR2DL1A allotypes.
Methods
Cohorts and genotyping
Genomic DNA was obtained from three case/control UK Caucasian cohorts with pre-eclamptic patients and one prospective cohort: controls (n=679), pre-eclamptics (n=693). Informed written consent was obtained from the subjects and the corresponding ethical approvals are: Cambridge Research Ethics Committee (reference numbers 01/197, 05/Q0108/367, 07/H0308/163), GOPEC (Derb reference number 09/H0401/66), London multiregional ethical committee (MREC No. 05/MRE02/20). Pre-eclampsia was defined by the clinical appearance of hypertension and proteinuria. For samples used for phenotyping and functional analysis, genomic DNA was isolated from decidual tissue and digested with proteinase K and RNase A (Roche) in combination with tissue lysis and protein precipitation buffers (Qiagen), prior to precipitation of DNA with isopropanol. For blood samples, Genomic DNA was isolated using the QIAamp DNA Mini Blood Kit (Qiagen). KIR genes presence/absence and HLA-C1/C2 genotyping for the three previously published pre-eclamptic cohorts and the families was performed by PCR-SSP as described previously (9, 10, 29). The two remaining cohorts, the prospective cohort of controls from Kiel, Germany and a cohort of pre-eclamptic patients, were typed for maternal KIR genes by multiplexed quantitative PCR (30). Genotyping of KIR2DL1 alleles was performed by pyrosequencing targeting exons 1, 4, 5, 7, and 9 (31).
KIR genotype analysis
KIR haplotype regions were defined following the KIR2011 Workshop recommendations. Briefly the centromeric A region (cA) was defined by the presence of KIR2DL3 and KIR2DL1, the centromeric B region (cB) by KIR2DS2 and KIR2DL2, the telomeric A region (tA) by KIR3DL1 and KIR2DS4, telomeric B region (tB) by KIR3DS1 and KIR2DS1.
Primary tissue
Samples were obtained from three different sources. Firstly, previously characterized cryopreserved PBMCs from healthy donors were kindly provided by Karl-Johan Malmberg, Karolinska Institutet. Secondly, peripheral blood and matched decidua were obtained from women undergoing elective terminations of first trimester pregnancies. Mononuclear cells were isolated by enzymatic digestion of decidual tissue as described previously (32) and cryopreserved. Thirdly, peripheral blood was purchased from the NHS Blood and Transplant unit and PBMC were cryopreserved. Ethical approval for this study was obtained from the Cambridge Research Ethics Committee (study 04/Q0108/23) and Karolinska Institute ethics review board with all participants supplying fully informed consent.
Cell lines
YTS cells were stably transfected with a pcDNA3.1 plasmid encoding a single KIR2DL1 allele (either *001, *003 or *004). These cells were obtained from Pippa Kennedy, Manchester University. Cells were grown in RPMI medium (10% FCS) under selection with G418 (1.5 mg/ml).
Flow cytometry of NK cells
Directly conjugated antibodies used for staining are listed in Table1. Where required, antibodies were biotinylated using the Fluoreporter Mini-biotin-XX protein labelling kit (Life Technologies) and detected using streptavidin-Qdot 605 (Invitrogen/Thermofisher). Viability was assessed through staining with LIVE/DEAD Aqua (Invitrogen/Thermofisher). Cells were thawed in complete medium (RPMI 1640 medium, antibiotics, 10% Fetal Calf Serum), counted and distributed at approximately 1e6 cells/well in 200ul FACS Wash (FW, 1xPBS, 2% Fetal Calf Serum, 2mM EDTA). Cells were then stained for 1hr at 4C unless otherwise stated in 30ul of cocktail. KIR2DS1 was detected as previously described (33). After staining with a secondary cocktail containing LIVE/DEAD Aqua and streptavidin-Qdot 605 for 30 minutes at 4C, cells were fixed in 1% paraformaldehyde for 10 minutes at room temperature. Finally, fixed cells were resuspended in FW and analysed on a LSR Fortessa (BD Biosciences). The antibody 8C11 only bound KIR2DL1pos cells in donors carrying the alleles KIR2DL1*002 and/or *003. The antibody 1127b only bound KIR2DL1pos cells in donors carrying the allele KIR2DL1*004 (Figure 1)
Table 1. Antibodies for flow cytometry.
| Antigen | Clone | Fluorophore | Supplier |
|---|---|---|---|
| CD3 | UCHT1 | PE-Cy5 | Beckman Coulter |
| CD4 | OKT4 | PE-Cy5 | BioLegend |
| CD14 | M5E2 | BV510 | Biosciences |
| CD19 | HIB19 | BV510 | Biosciences |
| CD56 | N901 | PE-Dazzle | BioLegend |
| KIR2DL1 | REA284 | PAC-Vio770 | Miltenyi |
| KIR2DL1/S1 | EB6 | PE-Cy7 | Beckman Coulter |
| KIR2DL2/3/S2 | GL183 | PE-Cy5.5 | Beckman Coulter |
| or | CH-L | BB515 | Biosciences |
| KIR2DL3 | 180701 | FITC | R&D Systems |
| KIR3DL1 | DX9 | BV421 | BioLegend |
| NKG2A | Z199 | APC | Beckman Coulter |
| KIR2DS1/L1 | 1127b | PE | R&D Systems |
| KIR2DL1/L2/3/S2 | 8C11 | Biotin | C. Retiére (gift) |
| CD107a | H4A3 | BV650 | BioLegend |
Functional assays
PBMCs were thawed and rested overnight in complete medium. 5e5 PBMCs were co-cultured with 5e4 K562 target cells for 6 hours in 96-well U bottom plates at 37C. After 1 hour of incubation, Golgi Plug (BD Biosciences) and Golgi Stop (BD Biosciences) were added. This method is adapted from Bryceson et al 2010 (34). Cells were then stained and fixed as described above.
Statistics
Categorical data was analysed using the chi-square and Fisher's exact test with two-tailed mid-p adjustment and Student’s t-test for continuous data. The magnitude of the effect was estimated by odds ratios (OR) and their 95% confidence intervals (CI). For comparisons of matched groups, a repeated measures one-way ANOVA with Tukey’s multiple comparisons test was performed. Statistical analyses were largely performed using PRISM (GraphPad Software Inc.) and the open source statistical package R (www.r-project.org). A P-value of ≤ 0.05 was considered to be statistically significant.
Results
A new strategy to identify different KIR2DL1 allotypes in single NK cells
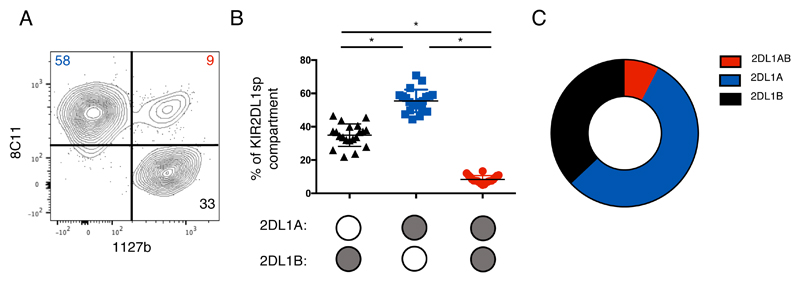
To identify antibodies that can distinguish KIR2DL1 allotypes, we used a panel of anti-KIR antibodies to stain YTS cells transfected with individual FLAG tagged KIR2DL1A (KIR2DL1*001,003) and KIR2DL1B (KIR2DL1*004) alleles (Figure S1). Two antibodies were identified that recognised selectively either KIR2DL1A allotypes (mAb 8C11), or KIR2DL1B, (mAb 1127b). The binding profile for 8C11 was consistent with the predictions made by David et al. (35). Both antibodies also cross react with other KIR (Table S1) and thus we designed a panel that would be able to distinguish specific KIR2DL1 allotypes on primary NK cells. Cryopreserved PBMCs from donors typed for KIR2DL1 alleles were then stained. KIR2DL1*001 donors are lacking due to their rarity but, because, KIR2DL1*001 and *002 do not differ in their extracellular domains, results obtained for KIR2DL1*002 can be used as a proxy for KIR2DL1*001. Due to antibody cross reactivity, NK cells expressing KIR2DL2/3, KIR2DS2 and KIR2DS1 were gated out of the analysis (Figure 1B). As expected from our findings on YTS cells, 8C11 only recognises KIR2DL1A and not KIR2DL1B on PBMCs; conversely 1127b binds only KIR2DL1B and not KIR2DL1A allotypes (Figure 1C). As further confirmation, KIR2DL1A single positive (sp), KIR2DL1Bsp and KIR2DL1AB double positive (dp) subsets were sorted from a KIR2DL1*003/004 heterozygous donor and KIR2DL1 transcripts amplified by RT-PCR. The sequences of the KIR2DL1 allele-specific transcripts in each subset matched the protein data (data not shown). In addition, to ensure that there was no steric hindrance of the antibodies, we permuted the order of antibody staining (data not shown). For KIR2DL1A/B heterozygous donors, our gating strategy allowed us to measure the percentage of cells expressing each allotype in a subset of KIR2DL1 positive cells (KIR2DL2/3/S2neg,2DS1neg,2DL1pos pbNK cells) (Figure 2A). Expression of KIR2DL1A dominates in all donors, with the mean proportion of KIR2DL1Asp pbNK cells observed being 56% in comparison to 35% for KIR2DL1B cells and only 8% for KIR2DL1ABdp cells (n = 20) (Figure 2B,C). These results show that, for the first time, we are able to visualise expression of two different KIR2DL1 allotypes present on KIR A and KIR B haplotypes within the same heterozygous individual. Moreover, although there is co-dominant expression, KIR2DL1A is expressed by the majority of NK cells.
Figure 2. KIR2DL1A allotypes are preferentially expressed by KIR2DL1sp pbNK cells.
Cryopreserved PBMCs from KIR2DL1A/B heterozygotes were stained as in Figure 1, (n=20). Cells were gated to identify KIR2DL2/3/S2neg 2DS1neg 2DL1pos NK cells (KIR2DL1sp subset). A) KIR2DL1sp subset stained with 8C11 and 1127b in a donor heterozygous for KIR2DL1A/B. B) Proportions of KIR2DL1A single positive, KIR2DL1B single positive and KIR2DL1AB double positive subsets are shown as percentages of the KIR2DL1 subset (mean +/- SD), summarised in the donut plot in panel C). Filled circles in panel B indicate pbNK cells expressing either 2DL1A, or 2DL1B, or both allotypes together.
* P < 0.0001; [RM one-way ANOVA, Tukey’s multiple comparisons test]
KIR2DL1A single positive NK cells are more responsive than KIR2DL1B single positive NK cells
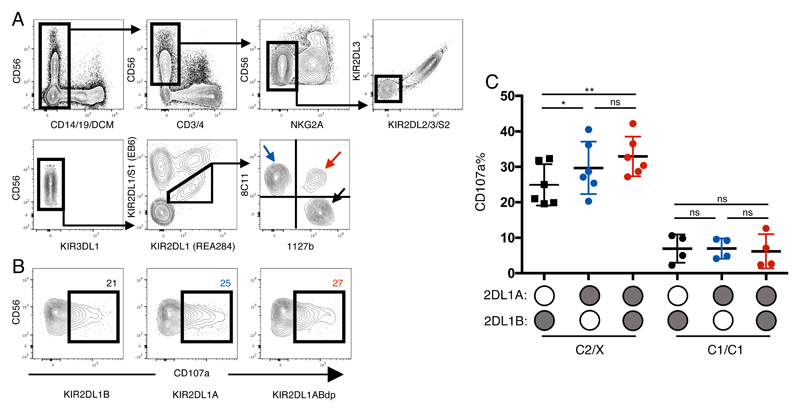
Although previous attempts to link KIR2DL1 alleles to different NK functions have been limited, KIR2DL1*004 was found to be hypofunctional in the context of missing self (26). However, this compared responses between donors with and without KIR2DL1*004, hence with different HLA backgrounds. We measured missing self responses of pbNK to the standard HLA class I-deficient K562 cell line to study how different KIR2DL1 allotypes within the same donor educate NK cells to alter their responsiveness. To rule out the influence of other inhibitory receptors, these were excluded using a flow cytometry gating strategy from individuals heterozygous for KIR2DL1A and KIR2DL1B. Thus, we focused on NKG2Aneg KIR2DL2/3/S2neg KIR3DL1neg KIR2DS1neg KIR2DL1pos NK cells (Figure 3A). Donors were further stratified by whether they possess a C2+HLA-C allele, the cognate ligand for KIR2DL1. Although both KIR2DL1A and KIR2DL1B are able to educate NK cells (Figure 3B,C), there is a significantly higher missing self response in KIR2DL1Asp compared with KIR2DL1Bsp NK cells. Moreover, an additive response in NK cells expressing both KIR2DL1 alleles can be seen compared with KIR2DL1B but not KIR2DL1A. All three KIR2DL1 subsets were similarly hyporesponsive in C1/C1 donors. These results show that KIR2DL1A allotypes are better educators of NK cells than KIR2DL1B allotypes from the same donor.
Figure 3. KIR2DL1A single positive NK cells are more responsive than KIR2DL1B single positive NK cells.
Cryopreserved PBMCs were co-cultured with K562 and degranulation was assessed by staining with anti-CD107a antibody (n=10). All donors are heterozygous for KIR2DL1A and B alleles. A) NKG2Aneg KIR2DL2/3/S2neg 3DL1neg 2DS1neg 2DL1pos NK cells were identified. KIR2DL1Asp (blue arrow), KIR2DL1Bsp (black arrow) and KIR2DL1ABdp (red arrow); B) Representative CD107a degranulation FACS plots for allele specific subsets from the same donor. From Left to Right: KIR2DL1Bsp, KIR2DL1Asp, KIR2DL1ABdp; C) Degranulation data (mean +/- SD) for KIR2DL1 allele specific subsets is shown for 10 donors. Donors are stratified by HLA-C status as mothers homozygous for HLA-C with C1 epitope (C1/C1) and those with at least one C2 epitope (C2/X).
*P < .05; **P < .01; ns, not significant [RM one-way ANOVA, Tukey’s multiple comparisons test]
KIR2DL1A allotypes and gene copy number associate with greater risk of pre-eclampsia
Having established that there are both phenotypic and functional differences between KIR2DL1 allotypes, we wanted to assess their impact in the context of risk of disease. Previous genetic studies have shown that mothers with two KIR A haplotypes are at increased risk of pre-eclampsia or fetal growth restriction, if the fetus carries a C2 epitope (9). These results suggest that binding of KIR2DL1 to C2+HLA-C on placental extravillous trophoblast (ETV) cells leads to strong inhibition of decidual NK cells (dNK) and compromised placental development. To understand how different KIR2DL1 alleles contribute to the risk of pregnancy disorders, we analysed a case/control cohort comparing 693 pre-eclamptic and 679 control pregnancies. When mothers lacking KIR2DS1 are classified according to KIR2DL1 genotype, the presence of KIR2DL1A was strongly associated with an increased risk of pre-eclampsia compared to mothers who lacked KIR2DL1A (P = 0.0006, Odds Ratio (OR) = 1.46 [1.18-1.80]) (Table 2). In contrast KIR2DS1-ve mothers who carry KIR2DL1B, were not at increased risk (P = 0.328, OR = 0.85 [0.6- 1.18]). This effect is seen even without correcting for fetal HLA-C genotype (Table 2).
Table 2. The presence of KIR2DL1A but not KIR2DL1B, in the absence of KIR2DS1, confers increased risk of pre-eclampsia.
| Controls (n=679) |
Pre-eclamptic patients (=693) |
||||||
|---|---|---|---|---|---|---|---|
| N | F(%) | N | F(%) | P-value | OR | 95% CI | |
|
KIR2DL1A carriersa (KIR2DS1 negative) |
352 | 51.8% | 423 | 61.0% | 0.0006 | 1.46 | 1.18-1.80 |
|
Othersb (KIR2DL1A negative and/or KIR2DS1 positive) |
327 | 48.2% | 270 | 39.0% | |||
|
KIR2DL1B carriersc (KIR2DS1 negative) |
84 | 12.4% | 74 | 10.7% | 0.328 | 0.85 | 0.61-1.18 |
|
Othersd (KIR2DL1B negative and/or KIR2DS1 positive) |
594 | 87.6% | 618 | 89.3% | |||
defined as KIR2DL1A+ KIR2DS1- donors, with or without KIR2DL1B present.
KIR2DL1A negative and/or KIR2DS1 positive includes: KIR2DL1A- KIR2DS1-, KIR2DL1A-KIR2DS1+, and KIR2DL1A+ KIR2DS1+ donors, with or without KIR2DL1B present.
defined as KIR2DL1B+ KIR2DS1- donors, with or without KIR2DL1A present
KIR2DL1B negative and/or KIR2DS1 positive includes: KIR2DL1B- KIR2DS1-, KIR2DL1B- KIR2DS1+, and KIR2DL1B+ KIR2DS1+ donors, with or without KIR2DL1A present.
Having identified that women who possess KIR2DL1A alleles are more at risk than those with other KIR2DL1 genotypes we next addressed the effect of increasing the number of copies of KIR2DL1A. Mothers were grouped according to whether they had 0, 1 or 2 copies of KIRDL1A or KIR2DL1B. We found that increasing the copy number of KIR2DL1A was associated with increased risk of developing pre-eclampsia (P = 0.006). The increasing OR as KIR2DL1A copy number increases, indicates an additive genetic risk model (Table 3). The increased risk associated with KIR2DL1A copy number remained even when KIR2DS1 was included as a covariate. In contrast, increasing copy number of KIR2DL1B had the opposite effect, apparently lowering the risk of pre-eclampsia as KIR2DL1B copy number increased although this trend did not reach statistical significance (p = 0.26, OR = 0.68 [0.32-1.34]). Taken together, the results in Tables 2 and 3 show that KIR2DL1A alleles are associated with a greater risk of pre-eclampsia than KIR2DL1B alleles.
Table 3. KIR2DL1A confers more risk of pre-eclampsia as copy number increases.
| Controls | Pre-eclamptic patients | ||||||
|---|---|---|---|---|---|---|---|
| N | F(%) | N | F(%) | P-value | OR | 95% CI | |
| KIR2DL1A CNV | |||||||
| 0 | 135 | 10.6% | 56 | 7.8% | 0.006 | 1 | |
| 1 | 539 | 42.3% | 282 | 39.4% | 1.26 | 0.90-1.79 | |
| 2 | 599 | 47.1% | 377 | 52.7% | 1.52 | 1.09-2.14 | |
| KIR2DL1B CNV | |||||||
| 0 | 961 | 75.5% | 553 | 77.3% | 0.261 | 1 | |
| 1 | 284 | 22.3% | 151 | 21.1% | 0.92 | 0.74-1.15 | |
| 2 | 28 | 2.2% | 11 | 1.5% | 0.68 | 0.32-1.34 | |
CNV is copy number variation of KIR2DL1A or KIR2DL1B alleles. The value can be 0, 1 or 2 copies in this regression analysis.
Preferential expression of KIR2DL1A allotypes in tissue resident decidual NK cells
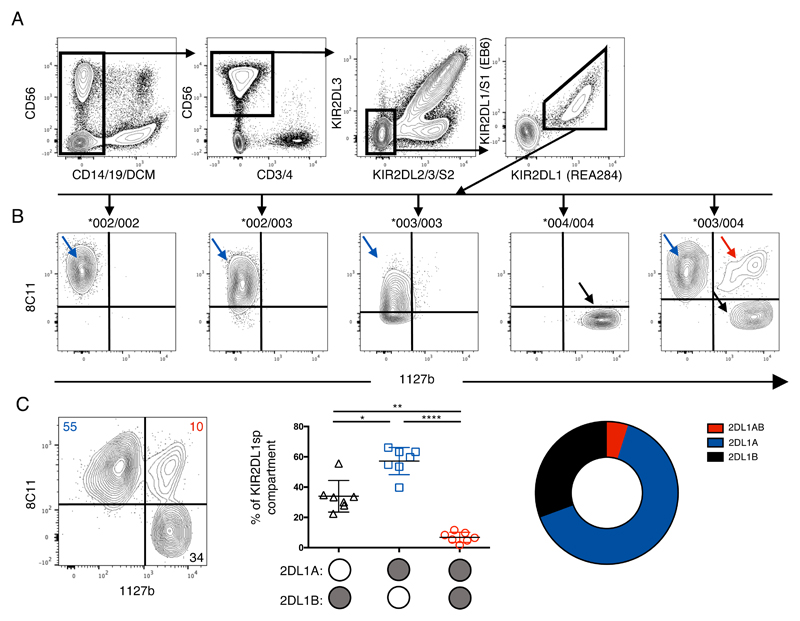
Our findings show that KIR2DL1A compared to KIR2DL1B alleles are associated with the risk of developing pre-eclampsia and with both phenotypic and functional differences in pbNK cells. However, it is decidual NK (dNK) cells, present in the lining of the pregnant uterus (decidua), rather than pbNK that are likely to be responsible for the biological effects on placental development. Early in pregnancy, dNK cells account for ~70% of the decidual leukocytes (36). They interact with surrounding maternal cells in the decidua and with the invading fetal trophoblast cells that express HLA-C molecules. The receptor expression profiles of dNK cells are quite different than those of pbNK cells (37, 38) with a greater proportion of dNK cells expressing KIR2DL1 compared to pbNK cells from the same donor (39). Education of decidual NK cells is also regulated differently from that of pbNK cells (40). We compared the expression of KIR2DL1A and KIR2DL1B allotypes on dNK cells in heterozygous individuals in a similar analysis to Figures 1 and 2. dNK cells express both KIR2DL1A and KIR2DL1B allotypes and co-dominant expression of both allotypes is observed in heterozygotes (Figure 4A, 4B). In accordance with the results of pbNK cells (Figure 2), for KIR2DL1A/B heterozygous donors, there is preferential expression of KIR2DL1A allotypes (57%) in KIR2DL1sp dNK cells (n= 7) (Figure 4C) compared to 34% for KIR2DL1Bsp and 7% for KIR2DL1ABdp. When comparing matched NK cells from blood and decidua, the expression patterns of the KIR2DL1 allotypes were very similar (Figure S2). Taken together these results indicate that the mechanism governing the relative frequency of KIR2DL1A/B allele expression is similar in pbNK cells and in dNK cells.
Figure 4. Preferential expression of KIR2DL1A allotypes in tissue resident decidual NK cells.
Cryopreserved decidual mononuclear cells from donors typed for KIR2DL1 alleles were stained to identify dNK cells expressing KIR2DL1 allotypes (n=7). A) Gating strategy to identify KIR2DL2/3/S2neg 2DS1neg 2DL1pos dNK cells; B) dNK cells from donors typed for KIR2DL1 alleles were stained and gated as in (A). Results for donors with different KIR2DL1 alleles are shown above each chart; C) 2D FACS plot is representative of biallelic KIR2DL1 expression in dNK cells from a donor heterozygous for KIR2DL1*003 and *004. Proportions of KIR2DL1A single positive, KIR2DL1B single positive and KIR2DL1AB double positive subsets are shown as percentages of the KIR2DL1 subset (mean +/- SD), and summarised in the donut plot.
*P < .05; **P < .01; *** P < .0001; ns, not significant [RM one-way ANOVA, Tukey’s multiple comparisons test]
Discussion
We have developed a method to identify allotype-specific NK-cell subsets for the polymorphic receptor KIR2DL1. Using this method, we show that alleles associated with either KIR haplotype, KIR2DL1A and KIR2DL1B, are both expressed in heterozygous individuals, with KIR2DL1A prevailing both phenotypically and functionally in both peripheral and uterine NK cells. Because KIR2DL1A are associated with an increased risk to develop pre-eclampsia, this result could help to focus on patients at higher risk.
The lack of KIR specific antibodies has limited studies of individual KIR. This problem is amplified when studying KIR alleles because alleles can differ by a handful of residues. More recently, by combining cross-reactive anti-KIR antibodies it has been possible to achieve improved resolution. For example, KIR2DL3*005 allotypes can be identified from other KIR2DL3 allotypes using an unexpected cross reactivity of the anti-KIR2DL1/S1 clone, EB6 (28). By exploiting the cross reactivity of the anti-KIR antibodies, 8C11 and 1127b (35), we are able to visualise expression of two different KIR2DL1 alleles carried on KIR A and KIR B haplotypes within the same heterozygous individual.
The surface expression of a particular KIR can be affected by allelic polymorphism, promoter differences, copy number variation, HLA ligand background and HCMV status (21, 41–44). In donors homozygous for KIR2DL1 alleles, the most common KIR2DL1B allele in Europeans, KIR2DL1*004, is expressed by significantly fewer NK cells than the most common KIR2DL1A allele, KIR2DL1*003 (21). In accordance with this, and although there is bi-allelic expression of KIR2DL1 allotypes, KIR2DL1Asp NK cells dominated the KIR2DL1sp NK cell niche in our KIR2DL1A/B heterozygous donors.
Using this approach, the effects of KIR2DL1 alleles on NK function can, for the first time, be compared within the same individual. In this scenario, NK cells expressing different KIR2DL1 allotypes have been educated in identical HLA class I environments. By reducing variation attributed to inter-individual differences we have been able to assess the functional capabilities of KIR2DL1 allotypes. KIR2DL1A proved to be the strongest educator when compared to KIR2DL1B in missing self assays, in agreement with previous data which shows KIR2DL1*004 to be hyporesponsive in comparison to KIR2DL1 alleles in other donors (26). In contrast to education by KIR2DL3 alleles which was found to be non-additive with respect to degranulation (27), our data suggests that there is an additive effect, with a significant difference in responsiveness between cells expressing KIR2DL1B alone, compared to KIR2DL1ABdp pbNK cells in donors with the C2 epitope that binds both KIR2DL1A and KIR2DL1B allotypes.
From our data it is not clear whether the differences in function of these alleles are attributable to differences in ligand binding avidity or in the quality or quantity of signalling. Binding of Fc fusion proteins of KIR2DL1A allotypes to microbeads coated with C2+HLA-C is only slightly higher than for KIR2DL1*004 Fc (24). Notably, the significant differences in binding of each KIR2DL1 Fc protein to different C2+HLAC allotypes, emphasises the importance of comparing functional effects of KIR2DL1 alleles in the same HLA-C background. Following ligand binding, KIR2DL1 proteins also differ in their signalling capacity. KIR2DL1A alleles commonly found in Europeans have an arginine residue at position 245, whilst the KIR2DL1*004 allele on the B haplotype has a cysteine in this position (C245). When KIR2DL1A alleles transfected into YT-Indy cells bind C2 ligands, they recruit more of the tyrosine phosphatase SHP-2, than those with C245, resulting in enhanced inhibitory signalling and longer surface expression (25). We now show these functional differences are apparent even in primary NK cells, educated in the same HLA-C background.
Genes encoding KIR receptors and their HLA ligands are associated with pregnancy disorders. Homozygosity for the KIR A haplotype (KIR AA) in the mother is more frequently associated with the disorders known as the Great Obstetrical Syndromes (GOS), including pre-eclampsia, recurrent miscarriage and fetal growth restriction (9, 10, 17, 29). This effect is strongest when the fetus carries a C2 epitope, strongly implicating KIR2DL1/C2 receptor ligand interactions. We now show that KIR2DL1A but not KIR2DL1B increases the risk of developing pre-eclampsia in a dose dependent manner, particularly when KIR2DS1, the antagonist for KIR2DL1, is absent. This suggests binding of KIR2DL1B with a C2 epitope on fetal extravillous trophoblast does not contribute to risk of GOS. Due to small numbers in each group, we were not able to compare the risk of preeclampsia in KIR2DS1- mothers carrying only KIR2DL1B, with those lacking a KIR2DL1 gene. The only precedent for allele specific associations with pregnancy disorders is confined to an activating KIR. Certain African KIR2DS5 alleles are associated with decreased risk of pre-eclampsia (11). Our finding of an association of specific KIR2DL1 alleles in pre-eclampsia is the first report of allele specific effects for an inhibitory KIR on pregnancy outcome.
To further understand how allelic variants can drive disease progression we analysed the NK cell population that is physiologically relevant to pre-eclampsia, namely dNK cells and show that like the blood compartment, KIR2DL1A is preferentially expressed by KIR2DL1sp dNK cells. Alleles of a gene coding for an inhibitory KIR may mediate their effects via two different mechanisms – NK-cell education and inhibition. Through calibrating the functional potential of dNK cells, education by the mother’s HLA-C could affect the ability of dNK cells to regulate trophoblast invasion. For example, genetic evidence suggests that for pregnancies in which the fetus is C2+HLA-C, mothers who are KIR AA and C2+HLA-C are at lower risk of developing pre-eclampsia than KIR AA and C1C1+HLA-C mothers (10). This is consistent with the idea that maternal KIR2DL1pos dNK cells are educated by C2+HLA-C prior to pregnancy. Alternatively, if excessive NK cell inhibition occurs through KIR AA dNK cells and fetal C2+HLA-C on invading trophoblast during pregnancy, this may lead to reduced remodelling of the uterine vasculature and poorer pregnancy outcomes in both humans and mice (reviewed in (45)) (18). There exists a hierarchy of inhibition which places common KIR2DL1A above KIR2DL1B in response to 721.221 cells expressing the C2+HLA-C allele Cw6 (25). This model would predict increased inhibition when dNK expressing KIR2DL1A encounters fetal trophoblast expressing C2+HLA-C compared to dNK expressing KIR2DL1B. The ability to stain both KIR2DL1 allotypes within NK cells from the same genetic background will, for the first time, permit dissection of both the effects of maternal HLA-C on dNK education, and whether KIR2DL1 allotypes generate different responses to fetal HLA-C.
Genetic studies that analyse KIR allelic variation are increasingly common (7, 11, 14, 46). Of equal importance, is the need to develop methods that allow us to characterise how these allelic variants contribute to functional differences at the cellular level and ultimately cause pathology. Allelic variability of other KIR genes such as KIR3DL1, KIR2DL2/3 and KIR3DL2 has an impact on phenotype and function in the context of infection and autoimmunity (8, 47, 48). For example, KIR3DL1 polymorphisms associate with delayed HIV progression and predict patient survival in response to immunotherapy for neuroblastoma (7, 12). Evidence that functional differences between KIR2DL1 alleles are important and subject to natural selection comes from recent population studies. These show the frequency of C2 is inversely correlated with the KIR A haplotype (19). Our data suggests that pre-eclampsia may contribute to this selective pressure to reduce the KIR A frequency and the strongly inhibitory KIR2DL1 alleles it carries. We have developed a method that will allow assessment of phenotype and function of KIR2DL1 allele-specific subpopulations of cells within an individual. This will facilitate our understanding of how KIR2DL1 receptor polymorphisms affect cellular function and ultimately contribute to disease progression.
Supplementary Material
Acknowledgements
We thank the members of the Colucci and the Moffett labs for suggestions and discussions and for the support of the Centre for trophoblast Research, Cambridge. We are grateful to Diane Moore and all the staff and patients of the Rosie maternity Hospital, without whom this work would not have been possible. We thank Karl-Johan Malmberg for providing PBMCs from healthy donors, Pippa Kennedy and Dan Davis, for providing YTS cells transfected with KIR2DL1 allele constructs, Anne-Ferguson-Smith for help with the pyrosequencing, and Gordon Smith, for access to the POPS cohort. Christiane Kling kindly provided DNA from control patients from a prospective recurrent miscarriage cohort and where available, from their male partners. Ryan Bjordhal from R&D systems, kindly provided monoclonal antibody 1127b for evaluation.
Grant support: This work was funded by the Wellcome Trust (Grant 200841/Z/16/Z to FC and AM), the Medical Research Council (Grant MR/P001092/1 to AS), the National Institutes of Health (Grant NIH U01 AI090905 to PJN and R01 AI17892 to PP) and the Cambridge NIHR BRC Cell Phenotyping Hub (to FC). The POPS study is funded by the Women’s Health theme of the NIHR Cambridge Biomedical Research Centre and the Stillbirth and Neonatal Death Society (SANDS). OH was supported by a MedImmune-Cambridge PhD fellowship. OC was supported by a CTR Next Generation Fellowship.
Footnotes
Authors’ contribution
OH designed and executed experiments, analysed data and wrote manuscript
OC designed and executed experiments, analysed data and wrote manuscript
MAI designed experiments, analysed data and edited manuscript
CR and CK provided essential resources
TV executed experiments
PN, HH, designed and executed experiments and analysed data
PP provided resources and edited manuscript
J Jayaraman, J Traherne, J Trowsdale, designed and executed experiments and analysed data
HG provided resources and edited manuscript
MI provided resources
AM designed experiments, analysed data and edited manuscript
AS designed experiments, analysed data and wrote manuscript
FC designed experiments, analysed data and wrote manuscript
References
- 1.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 2.Björkström NK, Ljunggren H-G, Michaëlsson J. Vol. 16. Nature Publishing Group; 2016. Emerging insights into natural killer cells in human peripheral tissues; pp. 310–320. [DOI] [PubMed] [Google Scholar]
- 3.Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, Prus D, Cohen-Daniel L, Arnon TI, Manaster I, Gazit R, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med. 2006;12:1065–1074. doi: 10.1038/nm1452. [DOI] [PubMed] [Google Scholar]
- 4.Goodridge JP, Önfelt B, Malmberg K-J. Newtonian cell interactions shape natural killer cell education. Immunol Rev. 2015;267:197–213. doi: 10.1111/imr.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Béziat V, Hilton H, Norman PJ, Traherne JA. Deciphering the KIR system at super-resolution for NK and T cell biology. Immunology. 2016 doi: 10.1111/imm.12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin MP, Gao X, Lee J-H, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 7.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 9.Hiby SE, Walker JJ, O'shaughnessy KM, Redman CWG, Carrington M, Trowsdale J, Moffett A. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200:957–965. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiby SE, Apps R, Sharkey AM, Farrell LE, Gardner L, Mulder A, Claas FH, Walker JJ, Redman CC, Morgan L, Tower C, et al. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J Clin Invest. 2010;120:4102–4110. doi: 10.1172/JCI43998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakimuli A, Chazara O, Hiby SE, Farrell L, Tukwasibwe S, Jayaraman J, Traherne JA, Trowsdale J, Colucci F, Lougee E, Vaughan RW, et al. A KIR B centromeric region present in Africans but not Europeans protects pregnant women from pre-eclampsia. Proc Natl Acad Sci USA. 2015;112:845–850. doi: 10.1073/pnas.1413453112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forlenza CJ, Boudreau JE, Zheng J, Le Luduec JB, Chamberlain E, Heller G, Cheung NKV, Hsu KC. KIR3DL1 Allelic Polymorphism and HLA-B Epitopes Modulate Response to Anti-GD2 Monoclonal Antibody in Patients With Neuroblastoma. Journal of Clinical Oncology. 2016:1–13. doi: 10.1200/JCO.2015.64.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson SK. Probabilistic bidirectional promoter switches: noncoding RNA takes control. Mol Ther Nucleic Acids. 2014;3:e191. doi: 10.1038/mtna.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bari R, Rujkijyanont P, Sullivan E, Kang G, Turner V, Gan K, Leung W. Effect of donor KIR2DL1 allelic polymorphism on the outcome of pediatric allogeneic hematopoietic stem-cell transplantation. J Clin Oncol. 2013;31:3782–3790. doi: 10.1200/JCO.2012.47.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babor F, Manser AR, Fischer JC, Scherenschlich N, Enczmann J, Chazara O, Moffett A, Borkhardt A, Meisel R, Uhrberg M. KIR ligand C2 is associated with increased susceptibility to childhood ALL and confers an elevated risk for late relapse. Blood. 2014;124:2248–2251. doi: 10.1182/blood-2014-05-572065. [DOI] [PubMed] [Google Scholar]
- 16.Dutta A, Saikia N, Phookan J, Baruah MN, Baruah S. Association of killer cell immunoglobulin-like receptor gene 2DL1 and its HLA-C2 ligand with family history of cancer in oral squamous cell carcinoma. Immunogenetics. 2014;66:439–448. doi: 10.1007/s00251-014-0778-1. [DOI] [PubMed] [Google Scholar]
- 17.Hiby SE, Apps R, Chazara O, Farrell LE, Magnus P, Trogstad L, Gjessing HK, Carrington M, Moffett A. Maternal KIR in Combination with Paternal HLA-C2 Regulate Human Birth Weight. The Journal of Immunology. 2014;192:5069–5073. doi: 10.4049/jimmunol.1400577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieckbusch J, Gaynor LM, Moffett A, Colucci F. MHC-dependent inhibition of uterine NK cells impedes fetal growth and decidual vascular remodelling. Nat Commun. 2014;5:3359. doi: 10.1038/ncomms4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nemat-Gorgani N, Hilton HG, Henn BM, Lin M, Gignoux CR, Myrick JW, Werely CJ, Granka JM, Möller M, Hoal EG, Yawata M, et al. Different Selected Mechanisms Attenuated the Inhibitory Interaction of KIR2DL1 with C2+ HLA-C in Two Indigenous Human Populations in Southern Africa. J Immunol. 2018;200:2640–2655. doi: 10.4049/jimmunol.1701780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, Parham P. Polymorphic HLA-C Receptors Balance the Functional Characteristics of KIR Haplotypes. J Immunol. 2015;195:3160–3170. doi: 10.4049/jimmunol.1501358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunphy SE, Guinan KJ, Chorcora CN, Jayaraman J, Traherne JA, Trowsdale J, Pende D, Middleton D, Gardiner CM. 2DL1, 2DL2 and 2DL3 all contribute to KIR phenotype variability on human NK cells. Genes Immun. 2015;16:301–310. doi: 10.1038/gene.2015.15. [DOI] [PubMed] [Google Scholar]
- 22.McErlean C, Gonzalez AA, Cunningham R, Meenagh A, Shovlin T, Middleton D. Differential RNA expression of KIR alleles. Immunogenetics. 2010;62:431–440. doi: 10.1007/s00251-010-0449-9. [DOI] [PubMed] [Google Scholar]
- 23.Schonberg K, Sribar M, Enczmann J, Fischer JC, Uhrberg M. Analyses of HLA-C-specific KIR repertoires in donors with group A and B haplotypes suggest a ligand-instructed model of NK cell receptor acquisition. Blood. 2011;117:98–107. doi: 10.1182/blood-2010-03-273656. [DOI] [PubMed] [Google Scholar]
- 24.Hilton HG, Norman PJ, Nemat-Gorgani N, Goyos A, Hollenbach JA, Henn BM, Gignoux CR, Guethlein LA, Parham P. Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer Population. PLoS Genet. 2015;11:e1005439–19. doi: 10.1371/journal.pgen.1005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N, Holladay M, Rooney B, Leung W. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine245. Blood. 2009;114:5182–5190. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beziat V, Traherne JA, Liu LL, Jayaraman J, Enqvist M, Larsson S, Trowsdale J, Malmberg KJ. Influence of KIR gene copy number on natural killer cell education. Blood. 2013;121:4703–4707. doi: 10.1182/blood-2012-10-461442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falco M, Romeo E, Marcenaro S, Martini S, Vitale M, Bottino C, Mingari MC, Moretta L, Moretta A, Pende D. Combined Genotypic and Phenotypic Killer Cell Ig-Like Receptor Analyses Reveal KIR2DL3 Alleles Displaying Unexpected Monoclonal Antibody Reactivity: Identification of the Amino Acid Residues Critical for Staining. The Journal of Immunology. 2010;185:433–441. doi: 10.4049/jimmunol.0903632. [DOI] [PubMed] [Google Scholar]
- 29.Hiby SE, Regan L, Lo W, Farrell L, Carrington M, Moffett A. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Human Reproduction. 2008;23:972–976. doi: 10.1093/humrep/den011. [DOI] [PubMed] [Google Scholar]
- 30.Jiang W, Johnson C, Jayaraman J, Simecek N, Noble J, Moffatt MF, Cookson WO, Trowsdale J, Traherne JA. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. 2012;22:1845–1854. doi: 10.1101/gr.137976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norman PJ, Hollenbach JA, Nemat-Gorgani N, Guethlein LA, Hilton HG, Pando MJ, Koram KA, Riley ME, Abi-Rached L, Parham P. Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS Genet. 2013;9:e1003938. doi: 10.1371/journal.pgen.1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Male V, Gardner L, Moffett A. Isolation of Cells from the Feto-Maternal Interface. John Wiley & Sons, Inc; Hoboken, NJ, USA: 2001. 313–7.40.11. [DOI] [PubMed] [Google Scholar]
- 33.Fauriat C, Ivarsson MA, Ljunggren H-G, Malmberg K-J, Michaëlsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood. 2010;115:1166–1174. doi: 10.1182/blood-2009-09-245746. [DOI] [PubMed] [Google Scholar]
- 34.Bryceson YT, Fauriat C, Nunes JM, Wood SM, Björkström NK, Long EO, Ljunggren H-G. Functional analysis of human NK cells by flow cytometry. Methods Mol Biol. 2010;612:335–352. doi: 10.1007/978-1-60761-362-6_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David G, Morvan M, Gagne K, Kerdudou N, Willem C, Devys A, Bonneville M, Folléa G, Bignon J-D, Retière C. Discrimination between the main activating and inhibitory killer cell immunoglobulin-like receptor positive natural killer cell subsets using newly characterized monoclonal antibodies. Immunology. 2009;128:172–184. doi: 10.1111/j.1365-2567.2009.03085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King A, Balendran N, Wooding P, Carter NP, Loke YW. CD3- leukocytes present in the human uterus during early placentation: phenotypic and morphologic characterization of the CD56++ population. Dev Immunol. 1991;1:169–190. doi: 10.1155/1991/83493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Male V, Sharkey A, Masters L, Kennedy PR, Farrell LE, Moffett A. The effect of pregnancy on the uterine NK cell KIR repertoire. Eur J Immunol. 2011;41:3017–3027. doi: 10.1002/eji.201141445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivarsson MA, Stiglund N, Marquardt N, Westgren M, Gidlöf S, Bjorkstrom NK. Composition and dynamics of the uterine NK cell KIR repertoire in menstrual blood. Mucosal Immunol. 2017;10:322–331. doi: 10.1038/mi.2016.50. [DOI] [PubMed] [Google Scholar]
- 39.Xiong S, Sharkey AM, Kennedy PR, Gardner L, Farrell LE, Chazara O, Bauer J, Hiby SE, Colucci F, Moffett A. Maternal uterine NK cell–activating receptor KIR2DS1 enhances placentation. J Clin Invest. 2013;123:4264–4272. doi: 10.1172/JCI68991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharkey AM, Xiong S, Kennedy PR, Gardner L, Farrell LE, Chazara O, Ivarsson MA, Hiby SE, Colucci F, Moffett A. Tissue-Specific Education of Decidual NK Cells. J Immunol. 2015;195:3026–3032. doi: 10.4049/jimmunol.1501229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, Parham P. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. The Journal of Immunology. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic Control of Variegated KIR Gene Expression: Polymorphisms of the Bi-Directional KIR3DL1 Promoter Are Associated with Distinct Frequencies of Gene Expression. PLoS Genet. 2008;4:e1000254. doi: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Béziat V, Liu LL, Malmberg J-A, Ivarsson MA, Sohlberg E, Björklund AT, Retière C, Sverremark-Ekström E, Traherne J, Ljungman P, Schaffer M, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. 2013;121:2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Béziat V, Dalgard O, Asselah T, Halfon P, Bedossa P, Boudifa A, Hervier B, Theodorou I, Martinot M, Debré P, Björkström NK, et al. CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur J Immunol. 2012;42:447–457. doi: 10.1002/eji.201141826. [DOI] [PubMed] [Google Scholar]
- 45.Moffett A, Colucci F. Uterine NK cells: active regulators at the maternal-fetal interface. J Clin Invest. 2014;124:1872–1879. doi: 10.1172/JCI68107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahn RS, Moslehi H, Martin MP, Abad-Santos M, Bowcock AM, Carrington M, Liao W. Inhibitory KIR3DL1 alleles are associated with psoriasis. Br JDermatol. 2016;174:449–451. doi: 10.1111/bjd.14081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Augusto DG, O'Connor GM, Lobo-Alves SC, Bass S, Martin MP, Carrington M, McVicar DW, Petzl-Erler ML. Pemphigus is associated with KIR3DL2 expression levels and provides evidence that KIR3DL2 may bind HLA-A3 and A11 in vivo. Eur J Immunol. 2015;45:2052–2060. doi: 10.1002/eji.201445324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bari R, Thapa R, Bao J, Li Y, Zheng J, Leung W. KIR2DL2/2DL3-E35 alleles are functionally stronger than -Q35 alleles. Scientific Reports. 2016;6:1625. doi: 10.1038/srep23689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.