Abstract
Three oxidative products of 5-methylcytosine (5mC) occur in mammalian genomes. We evaluated if these cytosine modifications in a CG dinucleotide altered DNA binding of four B-HLH homodimers and three heterodimers to the E-Box motif CGCAG|GTG. We examined 25 DNA probes containing all combinations of cytosine in a CG dinucleotide and none changed binding except for carboxylation of cytosine (5caC) in the strand CGCAG|GTG. 5caC enhanced binding of all examined B-HLH homodimers and heterodimers, particularly the Tcf3|Ascl1 heterodimer which increased binding ~10-fold. These results highlight a potential function of the oxidative products of 5mC, changing the DNA binding of sequence-specific transcription factors.
Keywords: Carboxylation, E-Box motif, CG dinucleotide, Basic-helix-loop-helix, DNA binding, Tcf3|Ascl1 heterodimer
Introduction
In mammals, ~60–80% of the cytosines in the CG dinucleotide are methylated in somatic cells, particularly in the CG poor regions of the genome [1]. The biological consequences of 5mC in the CG dinucleotide vary [2] [3] [4]. Methylation can inhibit the DNA binding of transcription factors (TFs) involved in housekeeping functions like ETS (CCGGAA), SP1 (CCCGCC), and NRF-1 (CGCCTGCG) [5] suggesting a mechanistic link between hypermethylation of CG islands and gene suppression that is observed in some cancers [6]. Alternatively, CG dinucleotide methylation can increase DNA binding of TFs [7] resulting in repression [8] and/or activation of nearby genes [9]. For example, C/EBP family members preferentially bind methylated DNA sequences and are critical for activation of tissue specific promoters during differentiation [7].
Recently, the TET family of dioxygenases was identified that iteratively oxidize 5mC to 5-hydroxymethylcytosine (5hmC), then 5-formylcytosine (5fC), and finally 5-carboxylcytosine (5caC) [10]. Both 5fC and 5caC can be removed by mammalian thymine DNA glycosylase (TDG) and replaced with cytosine (C) to complete the demethylation of 5mC, which occurs when cells differentiate. The abundance of different cytosine forms varies dramatically within cells and between cell types suggesting a potential biological function [10] [11] [12].
The effect of 5hmC, 5fC, and 5caC on DNA binding of TFs is only now being investigated [13]. In the present study, we used the Electrophoretic mobility shift assay (EMSA) to examine the DNA binding of four B-HLH homodimers and three heterodimers to 25 double-stranded DNA 28-mers (dsDNA) containing the E-Box 8-mer CGCAG|GTG with different cytosine forms of the CG dinucleotide and observe that 5caC enhances DNA binding. These results were confirmed circular dichroism (CD) thermal denaturation.
Material and methods
Protein binding microarrays
The 40,000 feature array design consists of 60-mer DNAs, 35-bps are unique DNA sequences connected to a common 25-bp sequence used for double stranding [14]. CG dinucleotides were enzymatically methylated and the effect on DNA binding was determined [9].
In-vitro transcription & translation
Protein synthesis was performed using in-vitro translation kit (PureExpress, NEB) and the resulting reaction was diluted to a ratio of 1:5 with CD buffer (150 mM KCl, 12.5 mM K2HPO4-KH2PO4, pH 7.4, 1 mM DTT, 0.25 mM EDTA). 2 µL of diluted reaction mixture containing Tcf3|Ascl1 heterodimer was used in EMSA assays described below.
DNA oligonucleotides
Twenty single-stranded DNA 28-mer (ssDNA) cartridge-purified oligonucleotides were purchased from W.M. Keck Oligonucleotide Synthesis Facility at Yale to examine two DNA sequences. Five 28-mer DNAs (CTGACCGATACGCAG|GTGCCTGACTGAC) termed the sense strand (a) contained different versions of the cytosine in bold (C, 5mC, 5hmC, 5fC, 5caC). The strong E-Box motif is underlined and the center of the dyad is marked. Five 28-mer DNAs termed the anti-sense strand (b) (GTCAGTCAGGCAC|CTGCGTATCGGTCAG) contained different versions of the cytosine in bold. The weak E-box 28-mer on the sense-strand (a) is CTGACCCATACGCAA|ATGTCTGACTGAC. The anti-sense strand was end-labeled with γ-32P ATP (specific activity 5000 Ci/mmol, MP Biomedicals) using T4 polynucleotide kinase (NEB), and was purified by ProbeQuant G-50 micro column (GE Healthcare Biosciences). dsDNA probes were generated by annealing the labeled anti-sense strand and unlabeled sense strand.
EMSA
The binding of B-HLH proteins to 25 dsDNAs with all possible modifications of cytosine in a CG dinucleotide was analyzed by EMSA [15]. For Tcf3|Ascl1 heterodimer made by IVT, 2 µL of diluted protein was used. For EMSA with purified B-HLH domains, 10 µM dimer was heated at 65°C for 15 min in the presence of 1 mM DTT, followed by cooling at room temperature for 5 min. Protein dimers and 32P-labeled dsDNA (7 pM) were then added to the EMSA binding buffer (CD buffer containing 0.5 mg/mL BSA, 10% glycerol, 0.02µg/µL poly dIdC, 10 mM MgCl2) in a final volume of the reaction 20 µL.
Protein expression and purification
The DNA binding B-HLH domains of Tcf3, Tcf4, Tcf12, and Ascl1 were expressed from a T7 expression vector named pT5 plasmid [16] in E. coli BL21 DE3 (LysE) cells. Cells were grown, induced, and collected by centrifugation at +4°C for 15 min at 6000×g. The pellet was resuspended in 4 mL lysis buffer (50 mM Tris-HCl, pH 8.0; 1 mM EDTA; 1 mM DTT; 0.2 mM PMSF), frozen on dry ice and lysed at room temperature in the presence of 1.3 M KCl. The lysate was centrifuged at 30,000 rpm for 30 min in the Beckman L8-80 ultracentrifuge in the 60Ti rotor. The pellet was brought to 4 M urea, sonicated, heated at 65°C for 15 min, and centrifuged at 5400×g for 10 min [17]. The supernatant was dialyzed to a low salt buffer (20 mM Tris-HCl, pH 8.0, 10 mM KCl, 1 mM EDTA, 1 mM DTT, 0.2 mM PMSF) using the Amicon Ultra-15 column (catalog # UFC901024, EMD Millipore), and then loaded to a SP Sepharose column (catalog # 17-0729-01, GE Healthcare Biosciences). The protein was then eluted off the column using 300 mM and 1,000 mM KCl and purified by HPLC. Tcf4 has a C-terminal His tag (HHHHHH) and Ascl1 has a C-terminal Flag φ10 tag (MDYKDDDDKHMASMTGGQQMGRDP). The amino acid sequences for the proteins are.
Tcf3:MGHMVHRPWIQDEVLSLEEKDLRDRERRMANNARERVRVRDINEAFRELGRMCQ LHLKSDKAQTKLLILQQAVQVILGLEQQVRERNLNPKAACGGRTRIVSAHNSENEL
Tcf4:MGNNDDEDLTPEQKAEREKERRMANNARERLRVRDINEAFKELGRMVQLHLKSD KPQTKLLILHQAVAVILSLEQQVRERNLNPKAACLKRREEELHHHHHH
Tcf12:MGSTNEDEDLNPEQKIEREKERRMANNARERLRVRDINEAFKELGRMCQLHLKS EKPQTKLLILHQAVAVILSLEQQVRERNLNPKAACLKRREEEL
Ascl1:MASGFGYSLPQQQPAAVARRNERERNRVKLVNLGFATLREHVPNGAANKKMSK VETLRSAVEYIRALQQLLDEHDAVSAAFQAGVLSPELMDYKDDDDKHMASMTGGQQMGRDP
CD spectroscopy
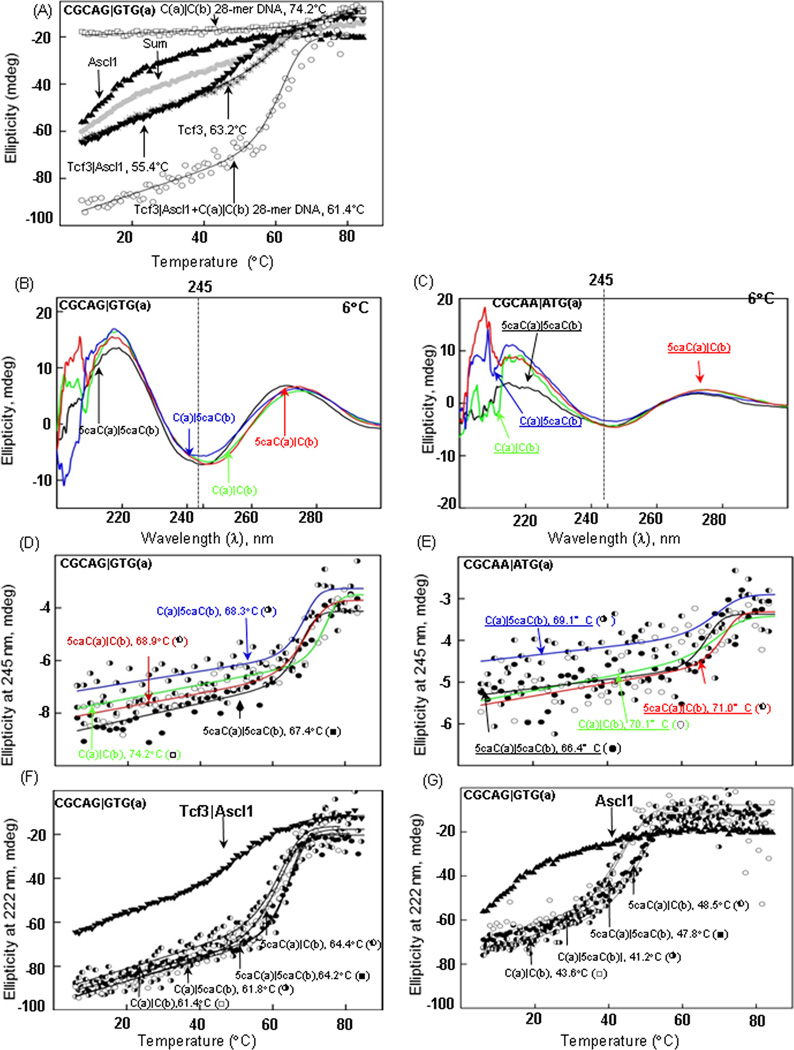
CD spectroscopy was performed using a Jasco J-720 spectropolarimeter and thermal denaturation curves were fitted [18]. The sum line in figure 3A is twice the concentration.
Figure 3. (A) Circular dichroism spectra and thermal denaturation of DNA, Protein, and DNA-Protein complex.
CD at 222 nm of the thermal stability of 2 µM Tcf3 homodimer (✳), Ascl1 homodimer (▲), and Tcf3Ascl1 heterodimer in the absence (▼), or presence of dsDNA 28-mer containing the unmodified E-Box CGCAG|GTG (○). The grey circles show the sum of the Tcf3 and Ascl1 thermal denaturation curves. Thermal denaturation at 245 nm of 2 µM dsDNA 28-mer containing CGCAG|GTG (□). (B–C) CD spectra from 200 nm to 300 nm at 6°C for four dsDNAs that vary 5caC in the CG dinucleotide (2 µM) containing strong E-box (CGCAG|GTG) and weak E-Box (CGCAA|ATG). (D–E) Thermal stability of the four dsDNA 28-mers described in B monitored by circular dichroism at 245 nm. A fitted curve to a two-state transition is shown. (F–G) Thermal stability of 2 µM Tcf3|Ascl1heterodimer and Ascl1 homodimer monitored at 222 nm in the absence (▼) or presence of the four DNAs described above.
Crystal structure of transcription factor E47 (Tcf3) homodimer
The image of the X-ray structure of the E47 homodimer bound to DNA [19] was generated using the program Chimera http://www.cgl.ucsf.edu/chimera/.
Results
Protein binding microarrays
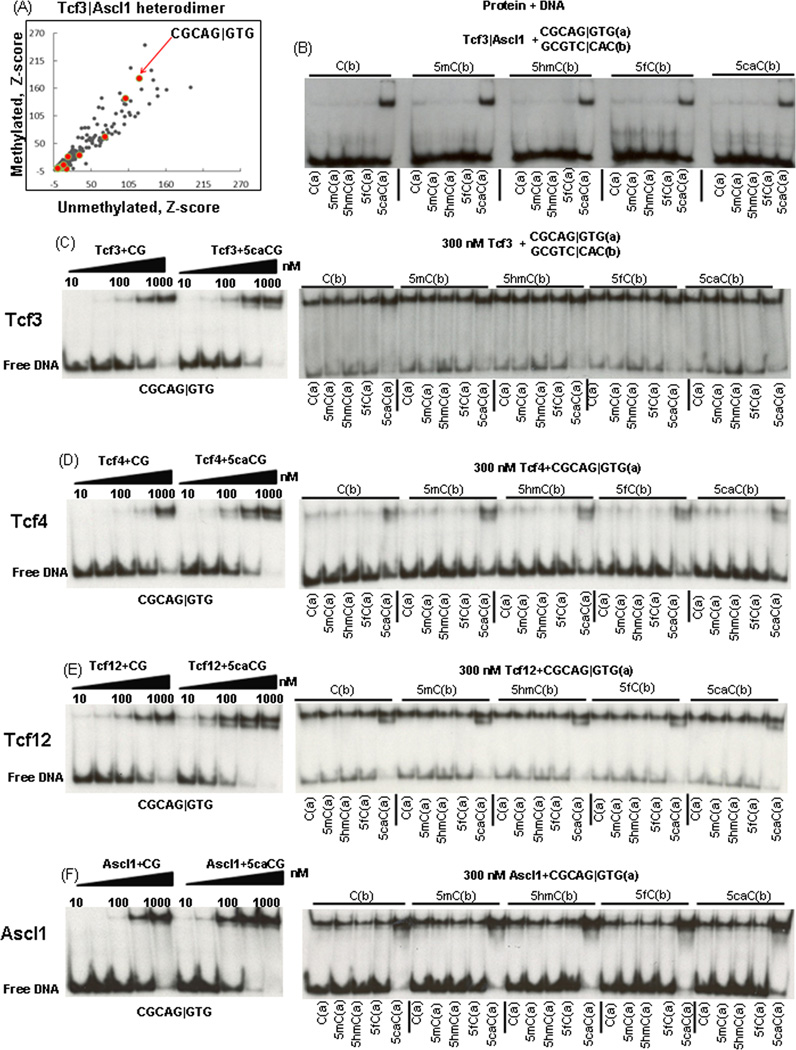
We used protein binding microarrays [14] to determine the DNA binding specificities of the Tcf3|Ascl1 heterodimer binding to unmethylated and enzymatically methylated CG dinucleotides using Agilent microarrays containing 40,000 features. The Tcf3|Ascl1 heterodimer bound the E-box motif 8-mer CGCAG|GTG well when both cytosines in the CG dinucleotide were either unmethylated (C) or 5mC (Figure 1A). Methylation of a CG dinucleotide in the center of E-Box (CGCAC|GTG) inhibits binding (Table1).
Figure 1. 8-mers bound by Tcf3|Ascl1 heterodimer and EMSA with pure Tcf3 B-HLH domain.
A) Z-scores for 8-mer DNA binding by the Tcf3|Ascl1 heterodimer calculated from protein binding arrays that contained either unmethylated or methylated cytosine in the CG dinucleotide [9]. All 16 E-Box sequences CGCAN|NTG are in red. B) EMSA of Tcf3 & Ascl1 mixture produced by in-vitro transcription translation reaction binding to 25 dsDNA 28-mers containing CGCAG|GTG with different chemical forms of the CG dinucleotide. The heterodimer only binds when 5caC is in the CG dinucleotide in the 8-mer CGCAN|NTG. EMSA with pure Tcf3 B-HLH domain: C) a half-log dilution from 1,000 nM to 10 nM for the Tcf3 homodimer binding two DNA 28-mers showing preferential binding to DNA containing 5caCs in the CG dinucleotide. The right panel shows 300 nM Tcf3 homodimer binding to 25 DNAs with different cytosine forms of the CG dinucleotide. D) Tcf4, E) Tcf12, and F) Ascl1.
Table 1.
Z-scores for the E-Box motifs (CGCAN|NTG) bound by Tcf3|Ascl1 heterodimer on methylated and unmethylated arrays.
| E-Box motif 8-mer | Unmethylated | Methylated |
|---|---|---|
| CGCAG|GTG | 120.7 | 180.0 |
| CGCAC|CTG | 101.0 | 140.0 |
| CGCAG|CTG | 70.5 | 63.7 |
| CGCA|GATG | 32.9 | 27.4 |
| CGCAT|CTG | 15.8 | 24.2 |
| CGCAC|ATG | 9.4 | 8.2 |
| CGCAT|ATG | 7.7 | 5.6 |
| CGCAT|GTG | 5.6 | 3.7 |
| CGCAA|GTG | 1.5 | 2.2 |
| CGCAA|CTG | 2.3 | 2.0 |
| CGCAG|TTG | 3.3 | 1.4 |
| CGCAA|TTG | 0.3 | 1.2 |
| CGCAC|TTG | 1.7 | 1.0 |
| CGCAA|ATG | 0.6 | 0.7 |
| CGCAC|GTG | 13.8 | 0.2 |
| CGCAT|TTG | 0.7 | 0.2 |
In vitro translated Tcf3&Ascl1 proteins binding 25 different dsDNA with modified CG dinucleotides
The five ssDNA 28-mers (CTGACCGATACGCAG|GTGCCTGACTGAC) with different cytosines were annealed with the complementary ssDNA to make 25 dsDNAs with different chemical forms of the CG dinucleotide. Figure 1B is an EMSA with 25 DNAs shows that the Tcf3|Ascl1 mixture bound five DNAs and all contain 5caC for the C in bold (CGCAG|GTG). Other cytosine modifications did not affect dramatically DNA binding.
Four B-HLH homodimers binding 25 dsDNAs
To quantify the contribution of 5caC to Tcf3|Ascl1 binding, we used pure B-HLH domains. Figure 1C–F presents an EMSA using two DNAs, unmodified DNA and DNA with two 5caCs in the CG dinucleotide in CGCAG|GTG. A half-log dilution from 1,000 nM to 10 nM of four B-HLH homodimers shows 5caC increases binding of all four homodimers with Ascl1 showing the largest increase in binding by ~6-fold. Next, we examined homodimer binding to 25 dsDNAs with different CG dinucleotides. All four homodimers at 300 nM preferentially bound the five DNAs containing 5caC in the CG dinucleotide in CGCAG|GTG (Table 2).
Table 2.
~Kd ranges for B-HLH homodimers and heterodimers binding dsDNA 28-mers with the E-Box CGCAG|GTG, as estimated by EMSA. Fold increase in binding caused by 5caC.
| B-HLH domains | CGCAG|GTG Kd (nM) |
~fold increase in binding caused by 5caC |
|---|---|---|
| Tcf3 | 300–1,000 | ~2 |
| Tcf4 | 300–1,000 | ~3 |
| Tcf12 | 100–300 | ~4 |
| Ascl1 | 300–1,000 | ~6 |
| Tcf3|Ascl1 | 10–30 | ~10 |
| Tcf4|Ascl1 | 60–200 | ~6 |
| Tcf12|Ascl1 | 60–200 | ~10 |
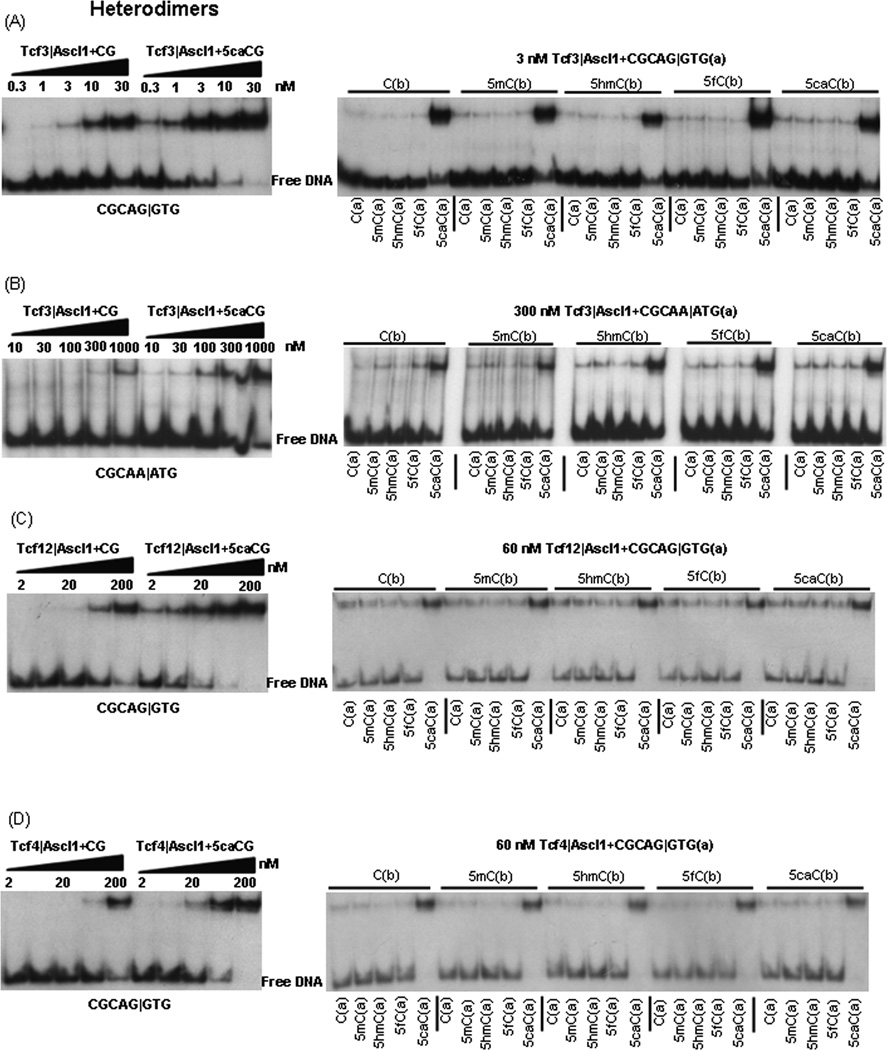
Tcf3|Ascl1 heterodimer binds CGCAG|GTG ~10-fold better when the CG contains 5caC
We next examined an equimolar mixture of purified Tcf3 and Ascl1 B-HLH domains binding to modified CG dinucleotides. Figure 2A presents an EMSA of the Tcf3|Ascl1 heterodimer with a half-log dilution from 30 nM to 0.3 nM binding to the two DNAs described previously, unmodified and 5caC containing DNA. The mixture of Tcf3 and Ascl1 binds better (Kd=10–30 nM) than either the Tcf3 homodimer (Kd=300–1,000 nM) or the Ascl homodimer (Kd=300–1,000 nM) indicating that the mixture is forming Tcf3|Ascl1 heterodimers as expected [20]. Two 5caCs in the CG dinucleotide increases DNA binding ~10-fold to a Kd between 1 and 3 nM. We next examined 3 nM Tcf3|Ascl1 heterodimer binding to 25 dsDNAs. Only DNA containing 5caC in the cytosine in bold CGCAG|GTG (a) is well bound. A modest inhibition of binding is observed with 5hmC in the CGCAG|GTG 8-mer.
Figure 2. Tcf3|Ascl1, Tcf4|Ascl1, and Tcf12|Ascl1 heterodimers binding modified CGs in two 8-mer: CGCAG|GTG and CGCAA|ATG.
A) EMSA showing a half-log dilution from 30 nM to 0.3 nM for the Tcf3|Ascl1 heterodimer binding two dsDNA 28-mers containing CGCAG|GTG, unmodified and containing 5caC in the CG dinucleotide. The right panel shows 3 nM Tcf3|Ascl1 heterodimer binding to 25 DNAs (see Figure 1). (B) EMSA showing a dilution from 1,000 nM to 10 nM for the Tcf3|Ascl1 heterodimer binding two dsDNA 28-mers containing a weak E-Box (CGCAA|ATG). The right panel shows binding of 300 nM Tcf3|Ascl1 heterodimer to 25 DNAs. (C) EMSA showing a dilution from 200 nM to 2 nM for the Tcf12|Ascl1 heterodimer binding two dsDNAs 28-mers containing CGCAG|GTG. The right panel shows 60 nM Tcf12|Ascl1 heterodimer binding to 25 dsDNAs. D) Tcf4|Ascl1.
The Tcf3|Ascl1 heterodimer binds the weak E-box CGCAA|ATG ~10-fold better when the CG contains 5caC
Tcf3|Ascl1 binding the weak E-Box CGCAA|ATG (Table 1) is enhanced by 5caC. Figure 2B presents an EMSA with a half-log dilution from 1,000 nM to 10 nM of the Tcf3|Ascl1 heterodimer binding unmodified and 5caC containg DNA. Binding to CGCAA|ATG is ~ 100-fold weaker than CGCAG|GTG. Again, 5caC in the CG dinucleotide is bound ~10-fold better. When 300 nM of protein was used, of the 25 dsDNAs that were examined, only the 5 probes containing 5caC for the cytosine in bold (CGCAA|ATG) are well bound.
The Tcf4|Ascl1 and Tcf12|Ascl1 heterodimers
Figure 2C–D presents an EMSA of Tcf12|Ascl1and Tcf4|Ascl1 heterodimers from 200 nM to 2 nM binding the two DNA probes with CGCAG|GTG discussed previously. Both Tcf12|Ascl1 and Tcf4|Ascl1 mixtures bind unmodified DNA better than the either homodimer indicating heterodimer formation. 5caC increases binding of Tcf12|Ascl1 and Tcf4|Ascl1 heterodimers ~10-fold and ~6-fold respectively. Tcf12|Ascl1 and Tcf4|Ascl1 heterodimers binding to 25 dsDNAs show binding to the five DNAs containing 5caC in CGCAG|GTG)
CD spectra and stability of the Tcf3|Ascl1 heterodimer bound to 4 dsDNAs
CD spectroscopy was also used to examine DNA binding. Thermal stability of Tcf3, Ascl1, and Tcf3|Ascl1 mixture was measured at 222 nm to determine the α-helical content of the dimers (Figure 3A). With heating, the Tcf3 homodimer cooperatively looses ellipticity at 222 nm as observed for other B-HLH homodimers [21] [22] with a Tm of 63.2 °C. The denaturation is well fit using a two-state model of α-helical dimers becoming unhelical monomers. The Ascl1 homodimer is less stable and we do not observe a low temperature baseline. The mixture of Tcf3 and Ascl1 has a Tm of 55.4 °C which is more than the sum of the two homodimer denaturations suggesting heterodimer formation. Addition of DNA increased both the ellipticity and stability of Tcf3|Ascl1 heterodimer. The Tcf3|Ascl1 heterodimer bound to DNA is less stable than dsDNA alone suggesting that the loss of ellipticity at 222 nm is not a consequence of the melting of the DNA but that the heterodimer denatures upon heating in the presence of dsDNA, a wavelength were the ellipticity of DNA does not change when dsDNA is denatured [7].
CD spectra and thermal stability of DNA containing 5caC
Next, we determined if 5caC in a CG dinucleotide changes the stability of both strong (CGCAG|GTG) and weak E-Box (CGCAA|ATG) motif dsDNA. The CD spectra from 200 nm to 300 nm of 4 dsDNAs at 6°C, unmodified cytosine, 5caC on one strand, and 5caC on both strands, is similar (Figure 3B and 3C), with a minimum at 245 nm and maximum between 270 nm and 280 nm, traits of B-form DNA [23]. Thermal stability at 245 nm shows that, for both strong and weak E-Box motif the unmodified DNA is most stable of the 4 DNAs, denaturing at 74°C and 70°C respectively. The two DNAs containing one 5caC in the strong E-Box are less stable, (68°C and 69°C) while the DNA with two 5caC is the least stable (67°C) (Table 3, Figure 3 D). Similarly, for weak E-Box the two DNAs with one 5caC are less stable and the DNA with two 5caC is the least stable, as seen for the 28-mers with CGCAG|GTG in the center (Figure 3E, Table 4). The thermal denaturation of Tcf3|Ascl1 heterodimer bound to dsDNA containing CGCAA|ATG did not produce a clear two-state transition indicative of poor binding as observed with EMSA.
Table 3.
The CD thermal denaturation monitored at 245 nm for four dsDNA 28-mers containing CGCAG|GTG (a) where the two cytosines in the CG dinucleotide are either C or 5caC. Thermal denaturation of Tcf3|Ascl1 heterodimer and Ascl1 homodimer monitored at 222 nm bound to four DNAs.
| CGCAG|GTG (°C) | |||
|---|---|---|---|
| DNA (E-Box) | DNA (°C±S.E.) |
DNA+Tcf3|Ascl1 (°C±S.E.) |
DNA+ Ascl1 (°C±S.E.) |
| C(a)|C(b) (○) | 74.2±0.27 | 61.4±0.15 | 43.6±0.65 |
| C(a)|5caC(b) (◑) | 68.3±0.27 | 61.8±0.18 | 41.2±0.51 |
| 5caC(a)|C(b) (◐) | 68.9±0.30 | 64.4±0.18 | 48.5±0.38 |
| 5caC(a)|5caC(b) (●) | 67.4±0.28 | 64.2±0.07 | 47.8±0.15 |
Table 4.
The CD thermal denaturation monitored at 245 nm for four dsDNA 28-mers containing CGCAA|ATG (a) where the cytosine in the CG dinucleotide are either C or 5caC.
| DNA (E-Box) | CGCAA|ATG (°C) | |
|---|---|---|
| Exp # 1 (°C±S.E.) |
Exp # 2 (°C±S.E.) |
|
| C(a)|C(b) (○) | 70.1±1.32 | 72.9±0.75 |
| C(a)|5caC(b) (◑) | 69.1±1.04 | 69.1±1.18 |
| 5caC(a)|C(b) (◐) | 71.0±0.55 | 69.9±0.76 |
| 5caC(a)|5caC(b) (●) | 66.4±0.50 | 67.3±0.60 |
CD spectra and stability of the Tcf3|Ascl1 heterodimer bound to modified dsDNAs
The thermal stability of the Tcf3|Ascl1 heterodimer monitored at 222 nm is greater when bound to the two DNAs containing 5caC in the CG dinucleotide in CGCAG|GTG (Tm = 64.4 and 64.2°C) compared to the two DNAs containing cytosine (Tm = 61.4 and 61.8°C) (Figure 3F, Table 3). The thermal stability of the Ascl1 homodimer shows similar traits, stability is higher when bound to two DNAs that contain 5caC in the CG dinucleotide in CGCAG|GTG (Tm = 48.5 and 47.8°C) compared to the two DNA containing unmodified cytosine (Tm= 43.6 and 41.2°C) (Figure 3G, Table 3).
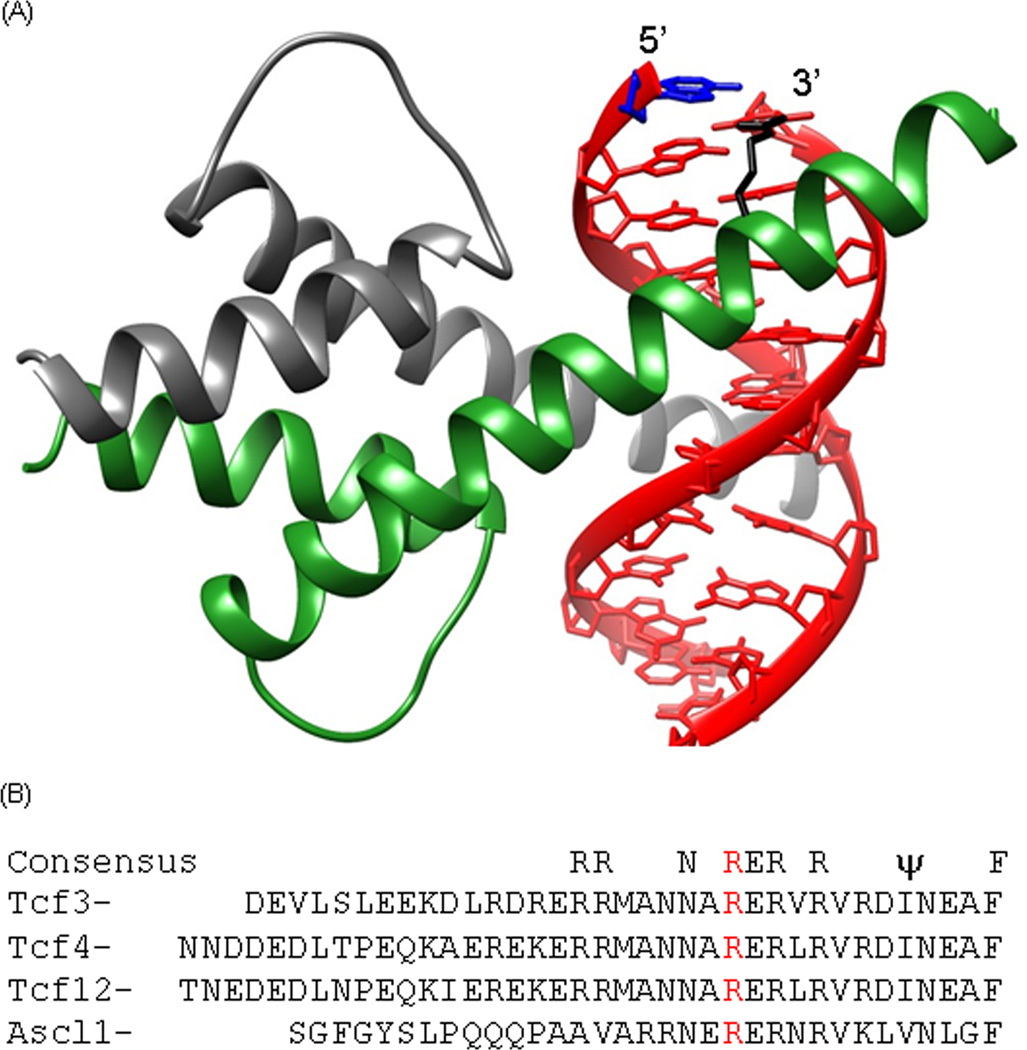
Crystal structure of transcription factor E47 (Tcf3) homodimer bound to DNA
Most crystal structures of B-HLH domains bound to DNA do not show a protein interaction with the base in the position analogous to the 5caC [24]. However, the E47 (Tcf3) homodimer bound to E-Box DNA shows an arginine interacting with an adenine (Figure 4A) that is in the same position as the 5caC that increases Tcf3|Ascl1 binding [19] suggesting a direct protein-DNA interaction. Figure 4B presents the amino acid sequence of DNA binding region of the four B-HLH proteins used in this study. The arginine in the E47 homodimer structure that interacts with the base 5-bp from the center of the dyad is conserved in four B-HLH proteins examined and may explain preferentially bind of 5caC.
Figure 4. Crystal structure of transcription factor E47 (Tcf3) homodimer bound to DNA.
The DNA is in red. One monomer is in grey, the second monomer is in green, the adenine in the same position as the critical 5caC is in blue and the invariant arginine is in black. B) Amino acid sequence for the DNA regions of the B-HLH transcription factors [28] of Tcf3, Tcf4, Tcf12, and Ascl1. The invariant arginine that interacts with the adenine in the E47 homodimer|DNA complex is in red.
Discussion
Three oxidative products of 5mC have recently been identified in mammalian genomes and their biological significance is being investigated [11]. Their abundance varies in tissues [10] suggesting they are regulated intermediates with the potential to have biological functions. A potential function of the three oxidative products of 5mC is to change the sequence-specific DNA binding of TFs. We used EMSA and CD spectroscopy to examine the effect of five cytosine nucleotides (C, 5mC, 5hmC, 5fC, and 5caC) on binding of 4 B-HLH homodimers and 3 B-HLH heterodimers both binding to DNA. We examined 25 DNAs containing different 11 combinations of modified C on the two Cs in the CG dinucleotide of the “flank” of the E-box 8-mer CGCAG|GTG. When the cytosine in bold is carboxylated (5caC), binding in increased for all 4 homodimers and 3 heterodimers. The Tcf3|Ascl1 heterodimer showed the strongest preferential of ~10-fold, more than either homodimer. However, we do not know which monomer in the Tcf3|Ascl heterodimer is binding 5caC. The heterodimer could exist as an ensemble of two states, one where theTcf3 monomer is interacting with 5caC and the second where the Ascl1 monomer is interacting with 5caC. Thus, changing the monomer which is ~ 20 angstroms away from 5caC can change the preferential binding of the second monomer in the dimer. Elucidating the allosterical mechanisms acting over long distances would be interesting to unravel.
B-HLH TFs recognize E-box sequences (CAN|NTG) [20] (for clarity, we place a vertical line in the center of B-HLH dyad). Tcf3 (aka E12, E47) heterodimerizes with different B-HLH proteins in different tissues in mouse [25] and human [20]. For example, Tcf3 heterodimerizes with myoD to drive muscle differentiation [26] and NeuroD to drive neuron differentiation [27].
There are over 60 members of the B-HLH family of transcription factors or proteins dimerize as homodimers and heterodimers and binds to E-Box like sequences (CAN|NTG) [20]. Some B-HLH members, e.g. Myc|Max heterodimers, drive cell growth [21] while other members, e.g. Tcf3|MyoD heterodimers, drive cell differentiation [28]. The various cytosine modifications in E-Box motifs may add an additional layer to DNA binding specificity of this family of proteins. We propose that arginine in the E47 homodimer (Tcf3) that is interacting with the same base as the critical 5caC may mediate the preferential binding to 5caC [19]. This arginine is conserved in Tcf family members and their dimerization partners but not for the B-HLH proteins involved in cell growth like Myc and Max. Potentially when 5caC is produced during the demethylation of tissue specific enhancers [3], Tcf3 and its various heterodimer partners bind these DNA sequences and shift the cell toward differentiation and away from B-HLH dimers involved in cell growth that do not have the arginine hypothesized to bind 5caC.
The abundance of oxidation products of 5mC in cells can be modulated by either activating TET enzymes or inactivating thymine DNA glycosylase (TDG)-mediated base excision repair [29]. Determining if B-HLH proteins bind to 5caC containing CGCAN|NTG in cells is difficult because their occurrences in genome are rare [10]. If experimental systems can be identified where these modification are more abundant, it may become feasible. Methods have been developed to determine the occurrence of 5hmC [30] and 5fC [31] at single CG dinucleotide resolution. Development of methods to determine 5caC in the genome at CG dinucleotide resolution is necessary. In summary, some B-HLH proteins preferentially bind the E-Box motif (CAN|NTG) with a CG dinucleotide on the flank when it contains 5caC.
Highlights.
B-HLH proteins bind the E-Box 8-mer CGCAN|NTG better to carboxylcytosine in the CG dinucleotide.
The Tcf3|Ascl1 heterodimer preferentially binds to 5caC better than either homodimer.
Increased binding of 5caC to B-HLH proteins was confirmed by circular dichroism thermal denaturation.
5caC increased Tcf3|Ascl1 heterodimer binding ~10-fold
5hmC and 5fC did not change binding.
Acknowledgements
We thank Peter Schuck, NIBIB and Grzegorz Piszczek, NHLBI, NIH for advice on CD spectroscopy.
Funding
This work is supported by the intramural research project of National Cancer Institute, NIH, Bethesda, USA.
Abbreviations
- C
Cytosine
- B-HLH
Basic-helix-loop-helix
- E-Box
Enhancer Box
- Tcf3
Transcription factor 3
- Tcf4
Transcription factor 4
- Tcf12
Transcription factor 12
- Ascl1
Achaete-scute homolog 1
- 5mC
5-methylcytosine
- 5hmC
5-hydroxymethylcytosine
- 5fC
5-formylcytosine
- 5caC
5-carboxylcytosine
- EMSA
electrophoretic mobility shift assay, double-stranded DNA (dsDNA), single-stranded DNA (ssDNA)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bird A, Taggart M, Frommer M, et al. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 2.Vinson C, Chatterjee R, Fitzgerald P. Transcription factor binding sites and other features in human and Drosophila proximal promoters. Subcell Biochem. 2011;52:205–222. doi: 10.1007/978-90-481-9069-0_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chatterjee R, Vinson C. CpG methylation recruits sequence specific transcription factors essential for tissue specific gene expression. Biochim Biophys Acta. 2012;1819:763–770. doi: 10.1016/j.bbagrm.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinson C, Chatterjee R. CG methylation. Epigenomics. 2012;4:655–663. doi: 10.2217/epi.12.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rozenberg JM, Shlyakhtenko A, Glass K, et al. All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues. BMC Genomics. 2008;9:67. doi: 10.1186/1471-2164-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rishi V, Bhattacharya P, Chatterjee R, et al. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci U S A. 2010;107:20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Toh H, Sasaki H, et al. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26:2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mann IK, Chatterjee R, Zhao X J, et al. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res. 2013;23:988–997. doi: 10.1101/gr.146654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue A, Shen L, Dai Q, et al. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spruijt CG, Gnerlich F, Smits AH, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Lam KN, van Bakel H, Cote AG, et al. Sequence specificity is obtained from the majority of modular C2H2 zinc-finger arrays. Nucleic Acids Res. 2011;39:4680–4690. doi: 10.1093/nar/gkq1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinson CR, Hai T, Boyd SM. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev. 1993;7:1047–1058. doi: 10.1101/gad.7.6.1047. [DOI] [PubMed] [Google Scholar]
- 16.Ahn S, Olive M, Aggarwal S, et al. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olive M, Krylov D, Echlin DR, et al. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- 18.Krylov D, Mikhailenko I, Vinson C. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. EMBO J. 1994;13:2849–2861. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellenberger T, Fass D, Arnaud M, et al. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 20.Murre C, Bain G, van Dijk MA, et al. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 21.Krylov D, Kasai K, Echlin DR, et al. A general method to design dominant negatives to B-HLHZip proteins that abolish DNA binding. Proc Natl Acad Sci U S A. 1997;94:12274–12279. doi: 10.1073/pnas.94.23.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rishi V, Gal J, Krylov D, et al. SREBP-1 dimerization specificity maps to both the helix-loop-helix and leucine zipper domains: use of a dominant negative. J Biol Chem. 2004;279:11863–11874. doi: 10.1074/jbc.M308000200. [DOI] [PubMed] [Google Scholar]
- 23.Kypr J, Kejnovska I, Renciuk D, et al. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma PC, Rould MA, Weintraub H, et al. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 25.Lazorchak A, Jones ME, Zhuang Y. New insights into E-protein function in lymphocyte development. Trends Immunol. 2005;26:334–338. doi: 10.1016/j.it.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Weintraub H, Davis R, Tapscott M S, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 27.Farah MH, Olson JM, Sucic HB, et al. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- 28.Vinson CR, Garcia KC. Molecular model for DNA recognition by the family of basic-helix-loop-helix-zipper proteins. New Biol. 1992;4:396–403. [PubMed] [Google Scholar]
- 29.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu M, Hon GC, Szulwach KE, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song CX, Szulwach KE, Dai Q, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]