Abstract
In the past few years, the study of therapeutic RNA nanotechnology has expanded tremendously to encompass a large group of interdisciplinary sciences. It is now evident that rationally designed programmable RNA nanostructures offer unique advantages in addressing contemporary therapeutic challenges such as distinguishing target cell types and ameliorating disease. However, to maximize the therapeutic benefit of these nanostructures, it is essential that we understand the immunostimulatory aptitude of such tools and identify potential complications. We present a set of 16 nanoparticle platforms that are highly configurable. These novel nucleic acid-based polygonal platforms are programmed for controllable self-assembly from RNA and/or DNA strands via canonical Watson-Crick interactions. We demonstrate that the immunostimulatory properties of these particular designs can be tuned to elicit the desired immune response or lack thereof. To advance our current understanding of the nanoparticle properties that contribute to the observed immunomodulatory activity and establish corresponding designing principles, we conducted QSAR (quantitative structure-activity relationships) modeling. The results demonstrate that molecular weight, together with melting temperature and half-life, strongly predict the observed immunomodulatory activity. This framework provides the fundamental guidelines necessary for the development of a new library of nanoparticles with predictable immunomodulatory activity.
Keywords: RNA nanotechnology, RNA and DNA nanoparticles, RNA polygons, Self-assembly, Immunology, QSAR
Introduction
The field of nanotherapeutics is exponentially growing due to the ability of nanoparticles to overcome many of the limitations noted for traditional small and macromolecular drugs. Nanotechnology is increasingly used in drug delivery due to the unique physical and chemical properties of nanoparticles, such as hydrophobicity, size, surface charge, and the presence of targeting moieties. These properties can overcome barriers that commonly limit the efficacy of traditional small and macromolecular drugs. The development of therapeutic nucleic acids (TNAs) that have rapidly evolved from conventional (e.g., siRNAs) to nanotechnology-formulated concepts (e.g., siRNAs incorporated into liposomes) and to the more controllable new generation of nano-TNAs represents one such example of utilizing benefits of nanotechnology for improving the quality of traditional therapeutics. These nano-TNAs rely on rationally designed nucleic acids (RNA, DNA or their chemical analogs) to engineer well-defined, fully programmable, and self-assembling nanoparticles, in which the nucleic acids serve as both a carrier and an active pharmaceutical ingredient [1–23]. One major limitation to the clinical use of conventional TNAs is their immunostimulatory properties including the induction of cytokines, chemokines, and type I and II interferons. Translational considerations for nano-TNA have been discussed before, and among other areas, include an understanding of the immunological properties[24]. The ability to predict the effects of the nano-TNAs on the immune system would allow maximizing their therapeutic index. For example, diseases of the central nervous system (CNS) are extremely difficult to treat due to the highly selective permeability of the blood brain barrier. Moreover, the neuroinflammation, which has been implicated in degenerative CNS pathologies such as multiple sclerosis, Alzheimer’s disease, and Parkinson’s disease[25], may also arise from delivery of therapeutic agents with undesirable immunostimulation into the brain, and therefore may counteract the efficacy of these drugs. Nano-TNAs have the potential to overcome both of these barriers. Their delivery to the brain can be achieved by previously characterized carriers, such as bolaamphiphiles[26–30], and rational design of nano-TNAs can help avoid the undesirable neuroinflammation. Therefore, it is necessary to characterize the immunomodulatory effects of nano-TNAs on resident CNS cells such as microglia and astrocytes that are essential for the initiation and progression of immune responses in the CNS [31] through the production of various inflammatory cytokines[32–34]. Earlier studies by our research groups and others revealed that while some types of nano-TNAs do not induce an immunological response, the immunogenicity of other assemblies strongly depends on their connectivity and composition[4, 35–37]. Recently, we introduced a novel design strategy that allows for the simple and efficient construction of RNA nanoparticles[38]. The assemblies, exemplified by nano-triangles, are solely based on Watson-Crick interactions and therefore, can be made not only of RNAs but also of DNAs and even RNA and DNA mixtures. The alterations in composition significantly affect the thermodynamic and chemical stabilities of nanoparticles as well as their immunological properties.
In the present study, we expanded the library of nanoparticles from triangle to hexagon with the same connectivity rules and assessed the effect of the nano-TNA size and composition on their immunomodulatory activity in human glia-like cells. For these purposes, RNA triangles (~75 kDa) were compared to RNA tetragons (~100 kDa), RNA pentagons (~125 kDa), and RNA hexagons (~150 kDa). Similarly, we compared corresponding DNA polygons and two types of different RNA/DNA hybrids for each polygon. We demonstrate that nucleic acid polygons primarily stimulate an interferon response in contrast to a damaging inflammatory cytokine response. Additionally, we report that nucleic acid composition significantly alters the amount of type I interferons release by microglia-like cells. Together these data suggest that nano-TNAs may be specifically engineered to minimize detrimental inflammatory responses while promoting beneficial host immunity. Finally, to establish a set of design rules that allow engineering of nucleic acid-based polygons with predicted immunological activities for further confirmative biological screening experiments, we applied a QSAR modeling technique to the experimental dataset generated for 16 polygons.
Methods
Assembly of polygons and their characterization.
The nucleic acid sequences for construction of a polygon library containing 16 candidates were computationally designed using 2D folding programs including Mfold[39] and NUPACK[40–43]. The nucleic acid strands encoding the composition of polygons are listed in the supporting information. All DNAs were purchased from IDT (idtdna.com) and all RNA strands were produced from PCR-amplified DNA templates using in vitro run-off transcription. Briefly, synthetic DNAs coding for the sequence of the designed RNA were amplified by PCR using primers containing the T7 RNA polymerase promoter. Resulting DNA templates were transcribed with T7 RNA polymerase. Transcription was performed in 80 mM HEPES-KOH, pH 7.5; 2.5 mM spermidine; 50 mM DTT; 25 mM MgCl2; 5 mM NTPs; 0.2 μM of DNA templates, and “home-made” T7 RNA polymerase ~100 units/μL. Transcription was stopped with RQ1 DNase. Transcribed RNAs were purified with a denaturing urea gel electrophoresis (PAGE) (15% acrylamide, 8M urea). The RNAs were eluted from gel slices overnight at 4°C into 1 × TBE buffer containing 300 mM NaCl. After precipitating the RNA in 2.5 volumes of 100% ethanol, samples were rinsed with 90% ethanol, vacuum dried, and dissolved in double-deionized water.
The polygons were assembled one pot from an equimolar mixture of nucleic acid strands (1 μM) in 1 × TMS buffer (80 mM Tris-HCl pH = 8.0, 100 mM NaCl, 5 mM MgCl2) with subsequent heating and cooling processes (annealing) from 80 °C to 4 °C in 20 min. All assemblies were tested with 7% native-PAGE and/or 3 % agarose gels. Native-PAGE ran for 1 hour at 4 °C at a constant 90 V and then were stained with ethidium bromide before imaging with the Bio-Rad ChemiDoc MP system.
3D modeling.
3D models of each RNA polygon were built using Discovery Studio Visualizer[44]. The energy minimization was applied for structural refinement of each polygon, using the ff10 force field and the Amber12 molecular dynamics package[14, 45].
Atomic force microscopy (AFM).
Assembled RNA polygons (5 μL of 50 nM stock) were deposited on APS modified mica, incubated for ~2 min and air dried, as described previously. AFM visualization was performed using a MultiMode AFM Nanoscope IV system (Bruker Instruments, Santa Barbara, CA) in tapping mode. The images were recorded with a 1.5 Hz scanning rate using a TESPA-300 probe from Bruker with a resonance frequency of 320 kHz and spring constant of about 40 N/m. Images were processed by the FemtoScan Online software package (Advanced Technologies Center, Moscow, Russia)[46, 47].
Dynamic Light Scattering (DLS).
The average hydrodynamic radii for assembled polygons (at 1 μM final concentration) were measured in a micro-cuvette (Starna Cells, Inc) using Zetasizer nano-ZS (Malvern Instrument, LTD). All measurements were done at room temperature according to instrumentation protocol.
Degradation assay in fetal bovine serum (FBS).
The experiment was conducted by incubation of nucleic acid polygons (1 μM) in an aqueous 2% (v/v) FBS solution at 37 °C, and aliquots (10 μL) were collected at 1, 5, 10, 20, 40, 60, and 90 min. Aliquots were immediately snap-frozen on dry ice to prevent any further degradation by nucleases presented in FBS. The collected samples were analyzed by a 7% native PAGE. We used ImageJ software to evaluate the fractions of remaining polygons by integrating the intensities of the bands corresponding to NPs. Integration areas for each time point were compared to the integration area for the control polygon of the same concentration in the absence of FBS. Plots were generated using OriginPro 8 Software where the remaining fraction (%) of polygons was plotted against FBS exposure time (min). An exponential decay function was used to fit data points following F(t) = F0 * e(−t/τ), where F(t) and F0 are the fractions at time t and at initial time 0 respectively; τ is exponential decay time constant.
Equilibrium dissociation constant (KD) measurements.
To measure the apparent KD for polygon assemblies, titration experiments were carried out. For this, fixed concentrations (10 nM) of IR-700 conjugated dT2 or rT2 strands were titrated with various concentrations (0.01 nM – 1000 nM) of the corresponding triangle, tetragon, pentagon or hexagon short side oligonucleotides. For instance, for DNA triangle, 10 nM IR-700 dT2 were mixed with mixtures of unlabeled [dT1, dT3, dT4] at 0.1, 0.5, 1, 5, 10, 50, 100, 500, and 1000 nM. The samples were annealed and analyzed with a 7% native-PAGE. The quantified polygon fractions (f) were plotted versus the total concentration (Ct) of the polygons. Non-linear sigmoidal curve fitting was applied to the data from two independent experiments using Origin 8.0 software. The general equilibrium equation for multi-strand nucleic acid components was used according to
where n = numbers of oligonucleotide strands: triangle n = 4, tetragon n = 5, pentagon n = 6, hexagon n = 7. The bands corresponding to polygons were quantified using ImageJ software. The yield for each polygon was calculated by dividing the corresponding quantified value for triangles by the total sum of the values for all monomers, dimers, and trimers present in the lane.
Structural integrity of polygons associated with Lipofectamine 2000 (L2K).
To ensure that all polygons remain intact during the transfection experiments, polygons (at 1 μM) were incubated with 2 μL of L2K at 25 °C for 30 minutes. Polygon/L2K complexes (4 μL) were then mixed with 2 μL of Triton X-100 (Sigma Aldrich) for an additional 30 minutes at 25 °C. All samples were analyzed by 7% native-PAGE and visualized by AFM (Figure 2C and supporting S3).
Figure 2:
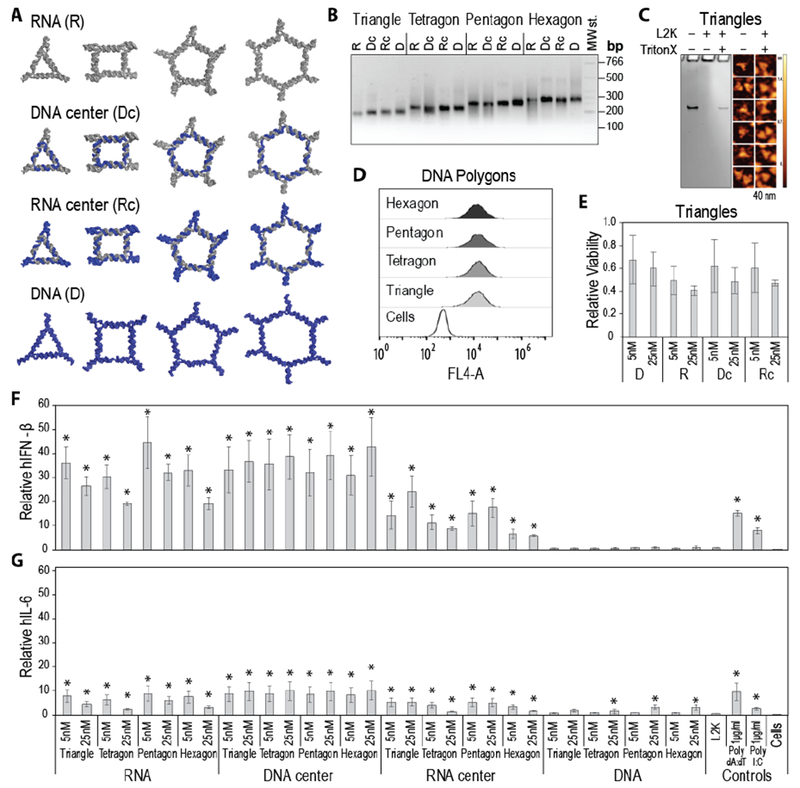
Cell culture experiments with programmable polygons. Human microglia-like cell lines were transfected with polygons at a final concentration of 5 nM and 25 nM. (A) 3D models of tested polygons with RNA strands shown in grey and DNA strands in blue. (B) Assemblies all polygons visualized by agarose gel. “MW st.” denotes the low molecular weight DNA ladder (NEB). (C) Structural integrity of polygons associated with Lipofectamine 2000 (L2K) confirmed by the release studies with Triton X100. The results are analyzed by native-PAGE and visualized by AFM. (D) Relative cellular uptakes assessed by flow cytometry and (E) cell viability assays of polygons transfected with L2K. Polygons tested without L2K showed negligible change in cell viability (data not shown). (F-G) 24 hours post transfection with 16 RNA, DNA and RNA/DNA polygons, cell supernatants were collected and levels of IFN-β (F) and IL-6 (G) production were assessed by specific-capture ELISA. In C, E, and F, results were normalized to transfection reagent alone treated cells (L2K) and presented as the mean +/− SEM. Statistically significant results are indicated with asterisks (p value < 0.05).
Transfections.
The human microglia-like cell line, hμglia or hHμ, was a generous gift from the laboratory of Dr. Jonathan Karn (Case Western Reserve University)[48]. Primary human microglia cells purchased from ScienCell were immortalized using SV40 and hTert antigens and sorted for the microglial/macrophage cell marker CD11b. These cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 5% FBS and 100 U/ml penicillin-100 μg/ml streptomycin at 37° C with 5% CO2. The human astrocyte-like cell line, U87 MG (ATCC HTB-14) was grown in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10 % FBS, 1 mM sodium pyruvate, and 100 U/ml penicillin with 100 μg/ml streptomycin at 37 °C 5 % CO2. Transfection of U87 MG cells and hHμ cells was conducted using Lipofectamine 2000 (L2K) (Invitrogen). Polygons or the positive controls poly dA:dT naked and poly I:C naked (Invivogen) were pre-incubated with L2K and Opti-MEM medium prior to transfection. Cells were transfected with polygon/L2K complexes at a final concentration of polygons of 5 nM or 25 nM or with positive control/L2K complexes at a final concentration of 1ug/ml. Media used for transfection was either DMEM supplemented with 5% FBS or EMEM supplemented with 10% FBS and 1 mM sodium pyruvate. Four hours post transfection the cell culture media was changed to media additionally supplemented with 100 U/ml penicillin with 100 μg/ml streptomycin. The cell supernatants were collected for further analysis twenty-four hours post transfection.
Cell Viability:
hHμ cells were plated in a 96 well plate at 5,000 cells per well (six wells per each sample) and polygons were transfected at a final concentration of either 5 nM or 25 nM. The cells were then incubated for four hours at 37° C and 5% CO2 and the transfection media was replaced with the fresh one. Twenty-four hours post transfection, 20 μL of CellTiter-Blue (Promega) was added to each well and incubated for 2.5 hours. Absorbance was measured at 490 nM using a Tecan Ultra (Tecan) plate reader and normalized to solutions transfected only with L2K.
Relative uptake efficiencies in hHμ:
hHμ wells were plated in a 24 well plate at 50,000 cells per well and polygons tagged with IR-700 were transfected. The cells were then incubated in the solution for four hours at 37 °C and 5% CO2 prior to media change. Twenty-four hours post transfection, cells were treated with Cell Dissociation Buffer (Gibco) and analyzed using a BD Accuri C6 Flow Cytometer. Untreated cells were used as control.
Enzyme-linked Immunosorbent Assay (ELISA).
Specific capture ELISAs were performed to quantify concentrations of human IL-6, IL-8, and IFN-β as previously described by our laboratory[49]. A commercially available ELISA kit was used to measure IL-8 (R&D Systems). The IL-6 ELISA was conducted using a rat anti-human IL-6 capture antibody (BD Pharmingen) and a biotinylated rat anti-human IL-6 detection antibody (BD Pharmingen). The IFN-β ELISA was carried out using a polyclonal rabbit anti-human IFN-β capture antibody (Abcam) and a biotinylated polyclonal rabbit anti-human IFN-β detection antibody (Abcam). Bound antibody was detected using streptavidin-horseradish peroxidase (BD Biosciences) followed by the addition of tetramethylbenzidine (TMB) substrate. H2SO4 was used to stop the reaction and absorbance was measured at 450 nm. Dilutions of recombinant cytokines for IL-6 and IFN-β (BD Pharmingen, Abcam) were used to generate a standard curve. The concentration of each cytokine was determined by extrapolation of absorbances in the study samples to that in the standard curve prepared from known concentrations of the relevant cytokine.
Statistics.
Experimental results were normalized to the L2K alone treated control and presented as the mean +/− SEM. Statistical significance was determined using a Student’s two-tailed t-test conducted with GraphPad Prism Software. A P-value of less than 0.05 was considered to be statistically significant.
Limulus Amoebocyte Lysate (LAL) assay.
The LAL assay was utilized to assess preparation contamination with the bacterial endotoxin, lipopolysaccharide. The polygons were tested at several dilutions according to a standardized procedure described earlier (https://ncl.cancer.gov/sites/default/files/protocols/NCL_Method_STE-1.2.pdf)[2]. Controls included the addition of known quantities of an endotoxin standard to nanoparticle samples to rule out potential nanoparticle interference with the assay. Reported values are from dilutions that demonstrated acceptable spike recovery and did not interfere with the assay.
Quantitative Structure-Activity Relationship (QSAR) modeling.
Data set:
In this study, 16 polygonal nanoparticles (both RNA- and DNA-based) were used for the construction of quantitative structure-activity relationships (QSAR). Three types of immune responses were identified based on the levels of IFN-β, IL-6, and IL-8 release experimentally measured from hHμ cells, and we used these activity values for QSAR modeling. The physicochemical properties and immune-response activities (observed and predicted) of studied polygons are presented in Supporting Tables S2 and S3. The polygon sequences can be found in the supplemental materials (Supporting Table S1).
QSAR approach.
For the development of the QSAR model we used two types of descriptors: physicochemical properties of constructed nanoparticles and sequence-based descriptors generated by Word2vec[50] approach as well as Random Forest (RF)[51] technique for model building.
Descriptors.
To generate the sequence-based descriptors we have used the Word2vec approach implemented in the KNIME analytic platform[52]. Word2vec is a two-layer neural network which is trained to reconstruct the linguistic contexts of words. As input Word2vec uses text and as an output, it produces a continuous vector space where semantically similar words are mapped to nearby points. Thus, sequences of nanoparticles were transformed into the vectors of real numbers of ten dimensions using nucleotides as the words. In additional to sequence-based descriptors, we also used the six physicochemical properties of constructed nanoparticles: molecular weight, GC content (%), diameter (nm), Tm (°C), decay time (min) and KD (nM).
Machine learning method
RF.
For the development of QSAR models, we used the RF implemented in the KNIME analytic platform[53], which is a modern and predictive machine learning approach. RF is an ensemble of decision trees and more trees reduce the variance. The classification from each tree can be thought of as a vote; the most votes determine the classification. The regression output was calculated as a mean value of all trees. Each tree was grown as the following: A random sample of nanoparticles (67%) was selected from the initial modeling set as the training set for the current tree. Not selected samples were used as a test set called an out-of-bag (OOB), which typically is 33% of initial modeling data. The randomly selected descriptors from the training set were used to split the nodes in the tree. Each tree is grown until it reaches the maximum tree depth parameter. The internal model evaluation was done according to the performance with the OOB set. To construct the best RF model, the following parameters were considered during a 5-fold cross validation procedure (5-fold CV): the number of trees (100), and the number of descriptors (16).
Model construction and validation.
To estimate the predictivity of the developed models we used 5-fold external cross-validation procedure (5-fold CV)[54]. During this procedure the initial data set was randomly divided into 5 parts. Four parts were used as the training set for model building and the remaining part was used as the test set for the assessment of external predictive accuracy. In additional, the Y-randomization (shuffling of the dependent values) was performed during 5-fold CV to assure that the accuracy of the model was not obtained due to chance correlations.
Evaluation of the model prediction accuracy.
To estimate the accuracy of prediction the following statistical parameters were calculated:
-
1)Determination coefficient
where , is predicted value for each particular object, is average activity value from the training set, and n is the number of objects in the training set. -
2)Root mean square error
where , is predicted value for each particular compound and n is the number of objects in the training set.
Results and Discussion
We constructed four types of equilateral polygons that can self-assemble from single-stranded longer central and shorter side strands. All polygons, while being different in size, number of sides, and the total number of strands entering their assemblies, have minimal variance in their sequence signatures (Figure 1A). For example, polygons with n number of sides would share the sequences with n-1 polygons but have an extra short side strand and an elongated central strand in their assembly. Polygons assembled via one-pot assembly were extensively analyzed by gels (Figures 1, 2B, and supporting S1) with the average yield estimated to be greater than 90%. The type of polygon was determined by the sequence of the longer strand. Thus the assembly of the particular shape is guided by the addition of the corresponding central strand to a mixture of all short RNAs. The structural evaluation of the assemblies by AFM and DLS provide additional evidence of formation of the designed polygon structures (Figure 1). AFM studies revealed that the shapes of resulting polygons were similar to their computed 3D models. The hydrodynamic diameters of polygons in aqueous solution were measured to be ~15 nm, ~16 nm, ~20 nm, and ~24 nm for triangles, tetragons, pentagons, and hexagons, respectively. These experimental results were in agreement with the predicted sizes.
Figure 1:
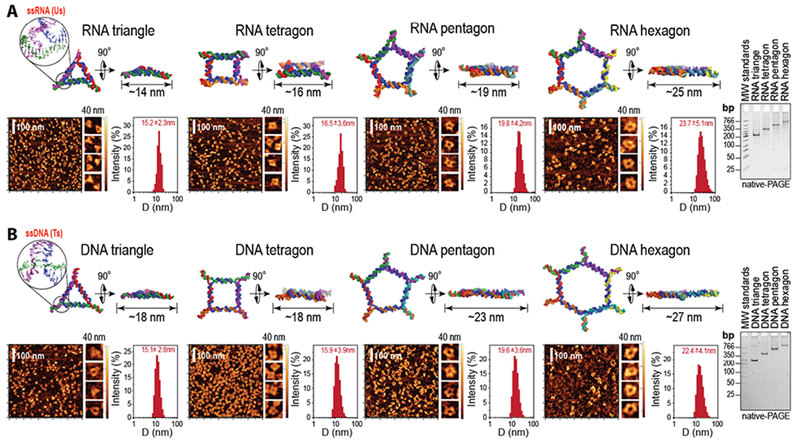
Programmable nucleic acid RNA (A) and DNA (B) polygons. Each panel presents energy minimized 3D models of RNA and DNA nanoparticles (identical sequences are colored the same), with corresponding AFM images, hydrodynamic radii measured by DLS (presented as +/− SEM), and ethidium bromide total staining native-PAGE results. “MW standards” denote the low molecular weight DNA ladder (NEB) used as the size marker.
Due to the design principles that rely only on canonical Watson-Crick interactions, polygons have an ability to efficiently assemble not only from RNAs but also from DNAs as well as from different ratios of RNA and DNA strands. This composition flexibility offers a rapid, convenient, and cost effective way to engineer different polygons with tunable physicochemical properties dictated by the nature of RNA and DNA. For example, by using various combinations of just 20 different RNA and DNA strands (10 each), it becomes possible to easily assemble a total of 240 unique RNA, DNA, and RNA/DNA hybrid polygons (16 triangles, 32 tetragons, 64 pentagons, and 128 hexagons). To show the feasibility of this approach, we have synthesized 16 polygons (Figure 2) made of all RNAs, all DNAs, only central strand RNA, and only central strand DNA and then further extensively characterized their properties and tested their immunogenicity. For all polygons, relative sizes (D), melting temperatures (Tm), dissociation constants (KD), and stabilities in blood serum (τ) were measured and the results are summarized in Supporting Table S2.
Prior to the immunological studies, the levels of endotoxin in prepared samples were assessed. Endotoxin is a component of the cell wall of Gram-negative bacteria and is a common contaminant in biotechnology and nanotechnology therapeutics[55]. Common sources of endotoxin are laboratory glassware, spatulas used to weigh out reagents, water, commercially available enzymes and oligonucleotides. Autoclaving kills bacteria, but does not eliminate endotoxin. Likewise, water purification systems remove ions, but not endotoxin. It is common knowledge in the area of nanotechnology that as much as 30-50% of nanoparticles fail during preclinical stage due to endotoxin contamination. Since endotoxins are potent immunostimulants, which may induce production of proinflammatory cytokines, we tested polygons for the presence of this contaminant using LAL assay. The level of endotoxin in all tested samples was below 0.05 EU/mL of 10 nM stock, which corresponds to less than 5 pg/mL concentration in our in vitro assays. The results are shown in Supporting Table S1. These levels of endotoxin are insufficient to elicit significant production of pro-inflammatory cytokines by glial cells.
Currently, one of the primary limitations in the translation of TNAs to the clinic is the stimulation of both off-target effects and immunotoxicity. In addition, treatment of CNS diseases is especially difficult due to an inability of therapies to cross the blood brain barrier and the sensitivity of the CNS to inflammatory damage. The TNAs discussed in this study can be complexed with lipid-based carriers that permit delivery to target cells within the CNS. Importantly, the carrier does not alter the structure of polygons (Figure 2C and supporting Figure S3). In the CNS, glial cells are key initiators of immune responses. Glial cells use a variety of cell surface, endosomal, and cytosolic receptors to sense pathogen-associated molecular patterns (PAMPs) such as nucleic acids. Due to the complexing of TNAs with lipid-based carriers, we predict TNAs will be identified by endosomal and/or cytosolic nucleic acid sensors. Therefore, in order to determine the immunomodulatory activity of TNA polygons that differ in their nucleic acid composition, human microglia-like cells and astrocyte-like cells were transfected side-by-side with 16 different polygons or positive controls and inflammatory mediator release was determined by specific capture ELISA (Figures 2 and S4). The positive controls selected were poly dA:dT, a synthetic analog of B-DNA recognized by DNA sensors; and poly (I:C) a synthetic dsRNA polymer recognized by RNA sensors. We observed no significant release of IFN-β or IL-6 from astrocyte-like cells transfected with nano-TNAs (data not shown). Interestingly, transfection of microglia-like cells with 12 out of the 16 polygons resulted in significant IFN-β release with minimal IL-6 or IL-8 production compared to the transfection reagent alone control. We observed no statistically significant difference in the release of I L-6 and I L-8 for polygons compared to our positive controls poly dA:dT and poly (I:C). These data suggest that these polygons primarily promote an interferon response rather than a damaging inflammatory cytokine production. The cytokine responses to these polygons did not show a clear dose dependency, potentially due to ligand saturation effects or reductions in cell viability resulting from greater activation and terminal differentiation of these cells, although it should be noted that such reductions were not statistically significant (Figure 2E).
Additionally, we observed that the nucleic acid composition of the TNA polygons significantly affected the release of inflammatory mediators. We observed that the polygons composed exclusively of RNA, or those that had central strand of RNA or DNA, stimulated a robust 10 to 40-fold increase in IFN-β responses compared to transfection reagent alone control. Additionally, compared to our positive controls, RNA and DNA center polygons induced a significant increase in the release of IFN-β further indicating the potency of these polygons as an interferon stimulus. In contrast, polygons composed exclusively of DNA do not stimulate significant IFN-β release above our transfection reagent alone control and induced significantly less release of IFN-β compared to our positive controls. These data suggest an RNA composition is required to stimulate an interferon response. Additionally, our data indicate DNA polygons are more immunologically quiescent compared to other polygon compositions and our positive controls. Interestingly, we observed a trend in inflammatory mediator responses attributable to polygon type for nano-TNAs composed exclusively of RNA, or TNAs composed of DNA with an RNA center. For these compositions, triangle and pentagon structures tended to simulate more IFN-β release compared to tetragon and hexagon compositions, suggesting a role for polygon type in cytokine production. However, further investigations will be required to definitively establish the role of polygon type in immune mediator release. Overall, these results hold promise for the development of these novel polygon nano-TNAs for clinical use given that their nucleic acid composition may dictate the cytokine response of the recipient. For example, all polygons composed solely of DNA elicit minimal inflammatory cytokine responses, thus avoiding the negative effects often associated with nanoparticle delivery, while RNA polygons may have use as adjuvants due to their ability to stimulate interferon responses. As such, these nano-TNAs provide an opportunity to engineer specific therapies for a variety of medical purposes. The effects shown in Figure 2 were not limited to the use of just Lipofectamine 2000. Experiments conducted using two other transfection reagents showed similar trends in cytokine release in response to polygon administration (Supporting Figure S5).
To construct a model that allows engineering of nucleic acid-based polygons with predicted immunological activities for further biological screening, we applied QSAR modeling technique to the experimental dataset of the 16 polygons (Figure 3). The two types of descriptors together with Random Forest were used for developing of QSAR models. To investigate the informativeness of the descriptors we built three separate models. The first model was based only on the physicochemical properties. The second model was based only on the sequence-based descriptors and the third model was based on both physicochemical properties together with sequence-based descriptors. The performance of each model was evaluated by 5-fold CV procedure (Supporting Figure S6). The statistical characteristics of the generated models are presented in Table 1.
Figure 3:
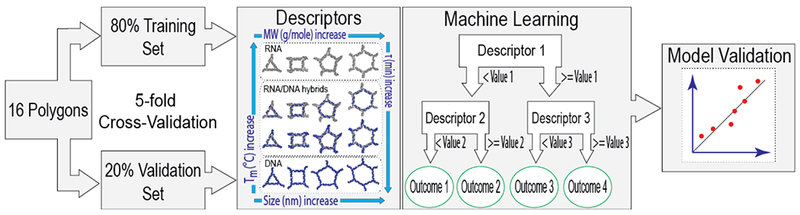
Schematic representation of quantitative structure-activity relationship (QSAR) modeling used in this project.
Table 1.
Model accuracy estimated during 5-fold cross validation procedure.
| IFN-β | IL-6 | IL-8 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Descriptors | R2 | RMSE | R2y-rand | R2 | RMSE | R2y-rand | R2 | RMSE | R2y-rand |
| physicochemical (PC) | 0.728 | 7.806 | −0.318 | 0.803 | 1.333 | −0.252 | 0.669 | 0.486 | −0.358 |
| sequence-based (Seq) | 0.392 | 11.66 | −0.649 | 0.353 | 2.415 | −0.674 | 0.539 | 0.574 | −0.706 |
| PC and Seq | 0.696 | 8.225 | −0.540 | 0.682 | 1.694 | −0.469 | 0.565 | 0.558 | −0.614 |
As shown in Table 1, the models constructed using two types of descriptors together demonstrated strong predictive power. All models showed negative determination coefficients after y-randomization procedures, proving the lack of correlations by chance. The models based only on sequence descriptors showed poor predictivity for IFN-β and IL-6 responses, and moderate predictivity for IL-8. The best predictive accuracy was obtained using only the physicochemical descriptors. These results are not surprising since the developed polygons have little variation across their sequences. However, the sequence information still showed some effect in the models, especially for I L-8 responses, and might play a significant role in predicting the behavior of polygons with more diverse shapes and structures. Thus, to predict immune responses to novel structure-diverse polygons, the models based on both descriptor types, as well as models based only on physicochemical descriptors, should be applied to predict nano-TNA immunomodulatory activity.
As mentioned above, the best prediction results were obtained using the physicochemical descriptors. Besides yielding good accuracy, these types of descriptors allow mining for clear interpretations of the developed models. Since the RF algorithm was used to build the QSAR models, the contribution of each descriptor into the tree-based model can be readily calculated. Thus, for each descriptor, we calculated an importance value as a ratio of the number of models, which used the descriptor as a split of the tree to the number of times the descriptor was the candidate for splitting. The sum of the importance values for all descriptors was scaled to 100% for comparison purposes. The results obtained are presented in Table 2.
Table 2.
Descriptor’s contribution into the Random Forest models. Bold and italic font represents the most important descriptors.
| IFN-β | IL-6 | IL-8 | |
|---|---|---|---|
| MW | 20.54% | 19.29% | 19.38% |
| GC content | 11.94% | 5.96% | 6.98% |
| Size (diameter) | 2.99% | 2.98% | 5.99% |
| Tm, °C | 24.01% | 25.04% | 21.88% |
| τ, min | 31.30% | 30.38% | 31.38% |
| KD, nM | 9.23% | 16.36% | 14.39% |
Table 2 shows that MW, Tm, and τ provide the major contributors to the RF models across all immune response activities. These important descriptors have interesting relationships with biological activities. It can be seen that DNA-based nanoparticles that have lower Tm values also induce low immune responses. Interestingly, the low decay time (30 min and less) for most of the nanoparticles corresponded to higher immune-response values. Although clear relationships between the physicochemical properties of nanoparticles and biological activities were discovered during modeling, future extension of the data set will improve the predictivity of these models and increase the confidence level in result interpretation.
Conclusion
In conclusion, we have developed novel nano-TNA platforms that are highly reconfigurable in both their physicochemical and immunological properties. These nano-TNAs can be specifically designed with the desired size, shape, melting temperature, enzymatic decay rates, and immunomodulatory activity. Our data strongly indicate that the nucleic acid composition of the nano-TNAs, specifically the combination of RNA to DNA, determines many of the physicochemical and immunological properties. Our data indicates that the properties of RNA and DNA determine stability factors including Tm and τ. In the present study, we have focused on defining the role of polygon type and composition in the initiation of immune responses, but further studies will be required to determine the influence of lipid-based carriers on the delivery of nano-TNAs and the initiation of polygon-induced immune responses in order to develop these agents for clinical use. We have concluded that polygons with an RNA composition stimulate a robust interferon response and minimal inflammatory cytokine release. In contrast, assemblies composed solely of DNA stimulate minimal interferon and inflammatory cytokine release. Additionally, we observed a trend for polygon type to contribute to the robustness of the immunological response. While our current data focuses on defining the role of polygon type and composition in initiating an immune response using one transfection reagent for delivery, preliminary experiments conducted using two additional transfection reagents display similar trends in the cytokine profile released in response to polygon transfection (Supporting Figure S6). These results suggest that the conclusions from our current data could be applied to additional carriers. However, future studies are necessary to fully investigate the role of lipid-based carriers in delivery and initiation of polygon induced immune responses in order to translate these nano-TNAs to clinical use. Interestingly, by developing QSAR models we were able to demonstrate that the physicochemical properties of the nano-TNAs, which are determined by the ratio of RNA and DNA, are the best predictors of immunological activity. Specifically, MW, Tm, and τ predict nano-TNA immunomodulatory activity. The QSAR models have also allowed for the generation of a library of nano-TNAs and their predicted immunological activity. Most importantly, the construction of this library provides a set of design principles for nano-TNAs. These design principles allow engineering nano-TNAs with specific physicochemical and immunological properties for desired medical applications. The flexibility of designing nano-TNAs with differing RNA and DNA composition as well as polygon type provides the potential for efficient and cost-effective development of an extensive library of nano-TNAs with an array of physicochemical and immunological properties. Studies are ongoing to identify the molecular mechanisms, such as cytosolic nucleic acid sensors, that underlie the immune responses of these cells to our nano-TNAs. The ability to tailor these key nano-TNA properties to therapeutic applications brings us one step closer to a more personalized medical treatment plan for patients for a myriad of diseases and conditions.
Supplementary Material
Acknowledgements
The research was supported by UNCC Department of Chemistry start-up funds to KAA, UNCC Faculty Research Grant to KAA and IM, research grant to KAA and IM from UNCC Center for Biomedical Engineering and Science (CBES), and by BSU Department of Chemistry start-up funds and Indiana Academy of Science grant #G9000602A to EFK. MAD discloses that this project was funded in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. AVZ discloses that this project was funded in part with Intramural Research Program, National Center for Advancing Translational Sciences, National Institutes of Health (1ZIATR000058-02). Authors would like to thank Dr. Alexander Lushnikov and Dr. Alexey Krasnoslobodtsev for performing AFM imaging of the polygons at the Nanoimaging core facility at the University of Nebraska Medical Center. We also thank Morgan Chandler (UNCC) for critical reading and helpful discussion.
Footnotes
Competing Financial Interests
The authors declare that they have no competing financial interests.
Supporting Information
Supplementary information accompanies this paper. All RNA and DNA sequences, supporting figures S1-6, and supporting tables S1-3 are shown in supplementary information.
REFERENCES
- 1.Jasinski D; Haque F; Binzel DW; Guo P, ACS Nano 2017. DOI 10.1021/acsnano.6b05737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonin KA; Grabow WW; Walker FM; Bindewald E; Dobrovolskaia MA; Shapiro BA; Jaeger L, Nat Protoc 2011, 6 (12), 2022–2034. DOI 10.1038/nprot.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afonin KA; Kasprzak WK; Bindewald E; Kireeva M; Viard M; Kashlev M; Shapiro BA, Acc. Chem. Res 2014, 47 (6), 1731–1741. DOI 10.1021/ar400329z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afonin KA; Viard M; Kagiampakis I; Case CL; Dobrovolskaia MA; Hofmann J; Vrzak A; Kireeva M; Kasprzak WK; KewalRamani VN; Shapiro BA, ACS Nano 2015, 9 (1), 251–9. DOI 10.1021/nn504508s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Afonin KA; Viard M; Koyfman AY; Martins AN; Kasprzak WK; Panigaj M; Desai R; Santhanam A; Grabow WW; Jaeger L; Heldman E; Reiser J; Chiu W; Freed EO; Shapiro BA, Nano letters 2014, 14 (10), 5662–71. DOI 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afonin KA; Viard M; Martins AN; Lockett SJ; Maciag AE; Freed EO; Heldman E; Jaeger L; Blumenthal R; Shapiro BA, Nature nanotechnology 2013, 8 (4), 296–304. DOI 10.1038/nnano.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo P, Nature nanotechnology 2010, 5 (12), 833–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng L; Li SK; Liu H; Liu CY; LaSance K; Haque F; Shu D; Guo P, Pharmaceutical research 2014, 31 (4), 1046–58. DOI 10.1007/s11095-013-1226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jasinski DL; Khisamutdinov EF; Lyubchenko YL; Guo P, ACS Nano 2014, 8 (8), 7620–9. DOI 10.1021/nn502160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rychahou P; Haque F; Shu Y; Zaytseva Y; Weiss HL; Lee EY; Mustain W; Valentino J; Guo P; Evers BM, ACS Nano 2015, 9 (2), 1108–16. DOI 10.1021/acsnano.5b00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu D; Li H; Shu Y; Xiong G; Carson WE; Haque F; Xu R; Guo P, ACS Nano 2015. DOI 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shu Y; Haque F; Shu D; Li W; Zhu Z; Kotb M; Lyubchenko Y; Guo P, RNA 2013, 19 (6), 767–77. DOI 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart JM; Viard M; Subramanian HK; Roark BK; Afonin KA; Franco E, Nanoscale 2016, 8 (40), 17542–17550. DOI 10.1039/c6nr05085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinbrecher T; Latzer J; Case DA, J Chem Theory Comput 2012, 8 (11), 4405–4412. DOI 10.1021/ct300613v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JB; Hong J; Bonner DK; Poon Z; Hammond PT, Nat Mater 2012, 11 (4), 316–22. DOI 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roh YH; Lee JB; Shopsowitz KE; Dreaden EC; Morton SW; Poon Z; Hong J; Yamin I; Bonner DK; Hammond PT, ACS Nano 2014, 8 (10), 9767–80. DOI 10.1021/nn502596b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H; Lee JS; Lee JB, Sci Rep 2016, 6, 25146 DOI 10.1038/srep25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H; Lytton-Jean AK; Chen Y; Love KT; Park AI; Karagiannis ED; Sehgal A; Querbes W; Zurenko CS; Jayaraman M; Peng CG; Charisse K; Borodovsky A; Manoharan M; Donahoe JS; Truelove J; Nahrendorf M; Langer R; Anderson DG, Nature nanotechnology 2012, 7 (6), 389–93. DOI 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H; Lytton-Jean AKR; Chen Y; Love KT; Park AI; Karagiannis ED; Sehgal A; Querbes W; Zurenko CS; Jayaraman M; Peng CG; Charisse K; Borodovsky A; Manoharan M; Donahoe JS; Truelove J; Nahrendorf M; Langer R; Anderson DG, Nature nanotechnology 2012, 7 (6), 389–393. DOI 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X; Xu Y; Yu T; Clifford C; Liu Y; Yan H; Chang Y, Nano letters 2012, 12 (8), 4254–9. DOI 10.1021/nl301877k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuller VJ; Heidegger S; Sandholzer N; Nickels PC; Suhartha NA; Endres S; Bourquin C; Liedl T, ACS Nano 2011, 5 (12), 9696–702. DOI 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 22.Kumar V; Palazzolo S; Bayda S; Corona G; Toffoli G; Rizzolio F, Theranostics 2016, 6 (5), 710–25. DOI 10.7150/thno.14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rangnekar A; Nash JA; Goodfred B; Yingling YG; LaBean TH, Molecules 2016, 21 (2). DOI 10.3390/molecules21020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobrovolskaia MA, DNA and RNA Nanotechnology 2016, 3 (7), 1–10. DOI https://doi.org/10.1515/rnan-2016-0001. [Google Scholar]
- 25.Chen WW; Zhang X; Huang WJ, Mol Med Rep 2016, 13 (4), 3391–6. DOI 10.3892/mmr.2016.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grinberg S; Linder C; Heldman E, Crit Rev Oncog 2014, 19 (3-4), 247–60. [DOI] [PubMed] [Google Scholar]
- 27.Gupta K; Afonin KA; Viard M; Herrero V; Kasprzak W; Kagiampakis I; Kim T; Koyfman AY; Puri A; Stepler M; Sappe A; KewalRamani VN; Grinberg S; Linder C; Heldman E; Blumenthal R; Shapiro BA, J Control Release 2015, 213, 142–151. DOI 10.1016/j.jconrel.2015.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim T; Afonin KA; Viard M; Koyfman AY; Sparks S; Heldman E; Grinberg S; Linder C; Blumenthal RP; Shapiro BA, Mol Ther Nucleic Acids 2013, 2, e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popov M; Abu Hammad I; Bachar T; Grinberg S; Linder C; Stepensky D; Heldman E, European journal of pharmaceutics and biopharmaceutics : official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.V 2013, 85 (3 Pt A), 381–9. DOI 10.1016/j.ejpb.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Popov M; Grinberg S; Linder C; Waner T; Levi-Hevroni B; Deckelbaum RJ;Heldman E, J Control Release 2012, 160 (2), 306–14. DOI 10.1016/j.jconrel.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 31.Ransohoff RM; Brown MA, J Clin Invest 2012, 122 (4), 1164–71. DOI 10.1172/JCI58644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perry VH; Holmes C, Nat Rev Neurol 2014, 10 (4), 217–24. DOI 10.1038/nrneurol.2014.38. [DOI] [PubMed] [Google Scholar]
- 33.Ramesh G; MacLean AG; Philipp MT, Mediators Inflamm 2013, 2013, 480739 DOI 10.1155/2013/480739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sofroniew MV, Nat Rev Neurosci 2015, 16 (5), 249–63. DOI 10.1038/nrn3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afonin KA; Desai R; Viard M; Kireeva ML; Bindewald E; Case CL; Maciag AE; Kasprzak WK; Kim T; Sappe A; Stepler M; Kewalramani VN; Kashlev M; Blumenthal R; Shapiro BA, Nucleic acids research 2014, 42 (3), 2085–2097. DOI 10.1093/nar/gkt1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halman JR; Satterwhite E; Roark B; Chandler M; Viard M; Ivanina A; Bindewald E; Kasprzak WK; Panigaj M; Bui MN; Lu JS; Miller J; Khisamutdinov EF; Shapiro BA; Dobrovolskaia MA; Afonin KA, Nucleic acids research 2017. DOI 10.1093/nar/gkx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khisamutdinov EF; Li H; Jasinski DL; Chen J; Fu J; Guo P, Nucleic acids research 2014, 42 (15), 9996–10004. DOI 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bui MN; Brittany Johnson M; Viard M; Satterwhite E; Martins AN; Li Z; Marriott I; Afonin KA; Khisamutdinov EF, Nanomedicine 2017. DOI 10.1016/j.nano.2016.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuker M, Nucleic acids research 2003, 31 (13), 3406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zadeh JN; Steenberg CD; Bois JS; Wolfe BR; Pierce MB; Khan AR; Dirks RM; Pierce NA, J Comput Chem 2010, 32 (1), 170–3. DOI 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 41.Dirks RM; Lin M; Winfree E; Pierce NA, Nucleic acids research 2004, 32 (4), 1392–403. DOI 10.1093/nar/gkh291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfe BR; Pierce NA, ACS Synth Biol 2015, 4 (10), 1086–100. DOI 10.1021/sb5002196. [DOI] [PubMed] [Google Scholar]
- 43.Zadeh JN; Wolfe BR; Pierce NA, J Comput Chem 2011, 32 (3), 439–52. DOI 10.1002/jcc.21633. [DOI] [PubMed] [Google Scholar]
- 44.Dassault Systèmes BIOVIA DSME, Release 2017, San Diego: Dassault Systèmes, 2016. [Google Scholar]
- 45.Case DA; Cheatham TE 3rd; Darden T; Gohlke H; Luo R; Merz KM Jr.; Onufriev A; Simmerling C; Wang B; Woods RJ, J Comput Chem 2005, 26 (16), 1668–88. DOI 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shlyakhtenko LS; Gall AA; Lyubchenko YL, Methods in molecular biology 2013, 931, 295–312. DOI 10.1007/978-1-62703-056-4_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shlyakhtenko LS; Gall AA; Filonov A; Cerovac Z; Lushnikov A; Lyubchenko YL, Ultramicroscopy 2003, 97 (1-4), 279–87. DOI 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 48.Garcia-Mesa Y; Jay TR; Checkley MA; Luttge B; Dobrowolski C; Valadkhan S; Landreth GE; Karn J; Alvarez-Carbonell D, J Neurovirol 2017, 23 (1), 47–66. DOI 10.1007/s13365-016-0499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chauhan VS; Sterka DG Jr.; Gray DL; Bost KL; Marriott I, Journal of immunology 2008, 180 (12), 8241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mikolov T; Chen K; Corrado G; Dean J, ArXiv13013781 Cs 2013. [Google Scholar]
- 51.Breiman L, Mach Learn 2001, 45 (1), 5–32. [Google Scholar]
- 52.Warr WA, J Comput Aided Mol Des 2012, 26 (7), 801–4. DOI 10.1007/s10822-012-9577-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zakharov AV; Peach ML; Sitzmann M; Filippov IV; McCartney HJ; Smith LH; Pugliese A; Nicklaus MC, Future Med Chem 2012, 4 (15), 1933–44. DOI 10.4155/fmc.12.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tropsha A, Mol Inform 2010, 29 (6-7), 476–88. DOI 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 55.Dobrovolskaia MA, J Control Release 2015, 220 (Pt B), 571–83. DOI 10.1016/j.jconrel.2015.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


