Abstract
Double-stranded (ds) RNAs play essential roles in many processes of cell metabolism. The knowledge of three-dimensional (3D) structure, stability, and flexibility of dsRNAs in salt solutions is important for understanding their biological functions. In this work, we further developed our previously proposed coarse-grained model to predict 3D structure, stability, and flexibility for dsRNAs in monovalent and divalent ion solutions through involving an implicit structure-based electrostatic potential. The model can make reliable predictions for 3D structures of extensive dsRNAs with/without bulge/internal loops from their sequences, and the involvement of the structure-based electrostatic potential and corresponding ion condition can improve the predictions for 3D structures of dsRNAs in ion solutions. Furthermore, the model can make good predictions for thermal stability for extensive dsRNAs over the wide range of monovalent/divalent ion concentrations, and our analyses show that the thermally unfolding pathway of dsRNA is generally dependent on its length as well as its sequence. In addition, the model was employed to examine the salt-dependent flexibility of a dsRNA helix, and the calculated salt-dependent persistence lengths are in good accordance with experiments.
Introduction
RNAs play a pervasive role in gene regulation and expression. In addition to single-stranded (ss) RNAs such as mRNAs and tRNAs, double-stranded (ds) RNAs are widespread in cells and are involved in a variety of biological functions (1, 2, 3). For examples, small noncoding dsRNAs can play a critical role in mediating neuronal differentiation (4), dsRNA segments of special lengths can inhibit the translation of mRNA molecules into proteins through attaching to mRNAs (5, 6), and dsRNAs of more than 30 basepair (bp) length can be key activators of the innate immune response against viral infections (7). Generally, dsRNAs realize their biological functions through becoming partially melted or changing their conformations (2, 3, 4, 5, 6, 7, 8, 9). Furthermore, the interchain interactions in stabilizing structures of dsRNAs are very sensitive to the environment (e.g., temperature and ion conditions) (10, 11, 12, 13, 14). Thus, a full understanding of dsRNA-mediated biology would require the knowledge of three-dimensional (3D) structures, structural stability, and flexibility of dsRNAs in ion solutions.
The 3D structures of RNAs, including dsRNAs, can be measured by several experimental methods such as x-ray crystallography, NMR spectroscopy, and cryo-electron microscopy. However, it is still technically challenging and expensive to experimentally derive 3D structures of RNAs at high resolution, and the RNA structures deposited in the Protein Data Bank (PDB) are still limited (15). Therefore, as complementary methods, some computational models have been developed in recent years, aiming to predict RNA 3D structures in silico (16, 17, 18, 19, 20, 21, 22, 23). The fragment assembly models (24, 25, 26, 27, 28, 29, 30, 31) such as MC-Fold/MC-Sym pipeline (24), 3dRNA (25, 26, 27), RNAComposer (28), and Vfold3D (29, 30) can successfully predict 3D structures of RNAs, including even large RNAs, at fast speed; however, these methods are generally based on given secondary structures and the limited known RNA 3D structures deposited in the PDB. Although the fragment assembly method of fragment assembly of RNA (FARNA) (31) can predict 3D structures for RNAs from sequences, it can only be efficient for small RNAs because of its full-atomic resolution. In parallel ways, some coarse-grained (CG) models (32, 33, 34, 35, 36, 37, 38, 39, 40, 41) such as iFold (42), SimRNA (43), HiRE-RNA (44), and RACER (45, 46) have been proposed to predict 3D structures for RNAs with medium length from their sequences based on knowledge-based statistical potentials and/or experiential parameters. However, these existing 3D-structure prediction models seldom make quantitative predictions for thermodynamic stability and flexibility of RNAs.
Simultaneously, some models have been employed to predict the thermodynamics of RNAs. Vfold2D/VfoldThermal (29, 30) with involving thermodynamic parameters can make reliable predictions on the free-energy landscape of RNAs, including pseudoknots, at the secondary structure level. The model proposed by Denesyuk and Thirumalai (47, 48) can predict the thermodynamics of small RNAs well, whereas such a structure-based (Gö-like) model could not predict 3D structures of RNAs solely from the sequences. Although other models such as iFold (42), HiRE-RNA (44), oxRNA (49), and NARES-2P (50, 51) may give melting curves of RNAs, there is still a lack of extensive experimental validation for these models.
Furthermore, RNAs are highly charged polyanionic molecules, and RNA structure and stability are generally sensitive to solution ion conditions, especially multivalent ions such as Mg2+ (8, 10, 11, 12, 13, 14). The role of ions in RNA structure and stability—especially the role of Mg2+, which is generally beyond the mean field descriptions (52, 53)—is seldom involved in the existing 3D-structure prediction models. To predict the 3D structures and stability of RNAs in ion solutions, we have developed a CG model with implicit electrostatic potential (54, 55), and the model has been validated through making reliable predictions on 3D structures and stability of RNA hairpins and pseudoknots as well as the ion effect on their stability. However, the model of the previous version was developed to predict the 3D structure and stability of ssRNA and cannot be directly employed to simulate the folding of RNAs with multiple strands because the translational entropy change of RNA strands during structure formation was not taken into account in the model. Unlike ssRNAs, the structure folding and stability of RNAs with multiple strands would be strongly dependent on strand concentration, which can make predictions on folding of dsRNAs very inefficient, especially for predicting dsRNA stability at (low) experimental strand concentration.
In this work, we further developed our previous three-bead CG model for ssRNAs to predict the 3D structure and stability of dsRNAs from their sequences by involving strand translations associated with strand concentration. For predicting stability of dsRNAs, we involved a correction of finite size effect (56) for dsRNA with two strands in a simulation box and introduced a technique of transforming melting curves of dsRNAs at high strand concentrations to those at low strand concentrations (57, 58) to promote the simulation efficiency of the model. Furthermore, an implicit structure-based electrostatic potential is introduced to capture the effect of ions such as Mg2+ on 3D structures and stability of dsRNAs. As compared with the extensive experimental data, this model can predict the 3D structures, stability, and flexibility of various dsRNAs with high accuracy, and the effects of monovalent/divalent ions on the stability and flexibility of dsRNAs can be well captured by this model. Additionally, our further analyses show that the thermally unfolding pathway of dsRNA is dependent on not only its length but also its sequence.
Methods
CG structure model and energy function
To reduce the complexity of nucleotides, in our CG model, one nucleotide is represented by three beads: the phosphate bead (P), sugar ring bead (C), and base bead (N) (54, 55). The P and C beads are placed at the P and C4′ atom positions, and the base bead (N) is placed at N9 atom position for purine or N1 for pyrimidine; see Fig. 1. The three beads are treated as van der Waals spheres with the radii of 1.9, 1.7, and 2.2 Å, respectively (54, 55).
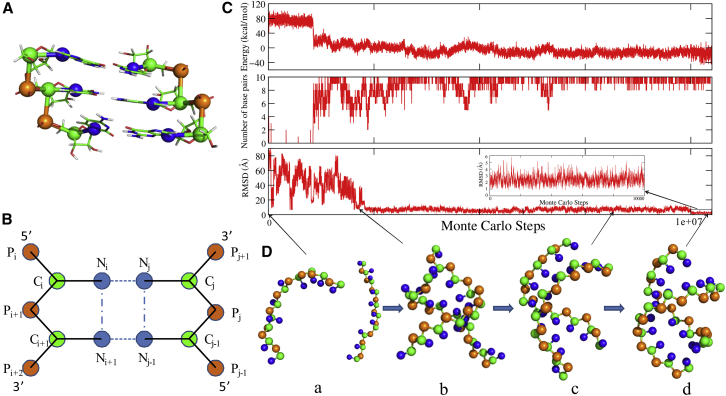
Figure 1.
(A) The CG representation for one fragment of dsRNA superposed on an all-atom representation. The beads of P (orange) and C (green) are located at the P atom in the phosphate group and the C4′ atom in the sugar ring, respectively. The beads of N (blue) are located at the N9 atom position for purine or N1 atom position for pyrimidine. (B) A schematic representation for pseudobonds (solid lines), base pairing (dashed lines), and base stacking (dash-dotted line) in our model. (C and D) An illustration for the folding process of a typical dsRNA (PDB: 2JXQ) in our model. (C) The system energy (top), number of basepairs (middle), and the RMSD (bottom) along the simulated annealing MC simulation. The inset is the zoomed RMSD of the structure in the refinement procedure at the end of the simulation. (D) The four typical conformation states in the folding procedure. The structures are shown with PyMol (http://www.pymol.org). To see this figure in color, go online.
The potential energy of a CG dsRNA is composed of two parts, bonded potential Ubonded and nonbonded potential Unonbonded (54, 55):
| (1) |
The bonded potential Ubonded represents the energy associated with pseudocovalent bonds between contiguous CG beads within any single chain, which includes bond-length energy Ub, bond-angle energy Ua, and dihedral-angle energy Ud:
| (2) |
The initial parameters of these potentials were derived from the statistical analysis on the available 3D structures of RNA molecules in the PDB (http://www.rcsb.org/pdb/home/home.do), and two sets of parameters Parahelical and Paranonhelical were provided for stems and single strands/loops, respectively. Note that only Paranonhelical is used in the folding process and both Parahelical and Paranonhelical are used in structure refinement (54, 55). The nonbonded potential Unonbonded in Eq. 1 includes the following five components:
| (3) |
Ubp is the base-pairing interaction between Watson-Crick (G-C and A-U) and wobble (G-U) basepairs. Ubs and Ucs are sequence-dependent base stacking and coaxial stacking interactions between two neighbor basepairs and between two neighbor stems, respectively. The strengths of Ubs and Ucs were derived from the combined analysis of available thermodynamic parameters and Monte Carlo (MC) simulations (54, 55). Uexc represents the excluded volume interaction between two CG beads, and it is modeled by a purely repulsive Lennard-Jones potential.
The last term Uel in Eq. 3 is a structure-based electrostatic energy for an RNA, which is newly refined in the model to better capture the contribution of monovalent and divalent ions to RNA 3D structures. The electrostatic potential is treated as a combination of Debye-Hückel approximation and the counterion condensation theory (52, 53, 54, 55),
| (4) |
where rij is the distance between the i-th and j-th P beads, each of which carries a unit negative charge (−e). lD is the Debye length of ion solution. is the permittivity vacuum, and ε is the effective temperature-dependent dielectric constant of water (54, 55). The reduced negative charge Qi on the i-th P bead is given by
| (5) |
where fi is the fraction of ion neutralization. In this model, beyond the assumption of uniform distribution of binding ions along RNA chain in our previous model, fi is dependent on RNA 3D structure and includes the contributions of monovalent and divalent ions
| (6) |
Here, x and 1 − x represent the contribution fractions from monovalent and divalent ions, which can be derived from the tightly bound ion model (55, 59, 60, 61, 62, 63). is the binding fraction of ν-valent ions and is given by
| (7) |
where N is the number of P beads. represents the average charge-neutralization fraction of ions, and the counterion condensation theory gives that (52, 53, 54, 55) , where b is the average charge spacing on RNA backbone and lB is the Bjerrum length. ϕi in Eq. 7 is the electrostatic potential at the i-th P bead and can be approximately calculated by
| (8) |
Equations 5, 6, 7, and 8 show that the structure-based reduced charge fraction Qi needs to be obtained through an iteration process: 1) use ion-neutralization fraction to calculate fi through Eq. 6, where the initial ; 2) calculate Qi through Eq. 5 and substitute Qi in Eq. 8 to calculate ϕi; 3) calculate (renew) of ν-valent ions through Eq. 7; 4) repeat 1–3 until becomes converged. See more details in the Supporting Materials and Methods.
The detailed descriptions of the CG energy function as well as the parameters for the potentials in Eqs. 1, 2, and 3 can be found in the Supporting Materials and Methods.
Simulation algorithm
To effectively avoid the traps in local energy minima, the MC simulated annealing algorithm is used to sample conformations for dsRNA at given monovalent/divalent ion conditions. Based on the sequence of dsRNA, two initial random CG chains can be generated and be separately placed in a cubic box, the size of which is determined by concentration of ssRNA. Generally, the simulation of a dsRNA system with a given ion condition is performed from a high temperature (e.g., 110°C) to the target temperature (e.g., room/body temperature). At each temperature, the conformations of the dsRNA are sampled by intrastrand pivot moves and interstrand translation/rotation through the Metropolis algorithm until the system reaches enough equilibrium. In this process, the newly refined electrostatic potential Uel is involved (see Eq. 4), and Uel can only be obtained after an iterative process for Qi. In practice, Uel is renewed over every 20 MC steps, and generally, Uel can be obtained through ∼4 times iterations for converged Qi. Thus, the increase in computation cost due to involving the newly refined Uel is negligible compared to the whole simulation cost. The equilibrium conformations of the system at each temperature can be saved to obtain 3D structures and structural properties of the dsRNA at each temperature.
Calculation of melting temperature
The stability of dsRNAs generally depends on strand concentration due to the contribution of translation entropy of melted ssRNA chains (64). However, for dsRNA with low strand concentrations (e.g., 0.1 mM in experiments), a very long simulation time is generally required to reach equilibrium for the dsRNA system. To make our calculation efficient, the simulations for dsRNAs can be performed at relatively high strand concentrations (44, 65). To avoid the boundary effect, we generally took a suitable high strand concentration (e.g., 10 mM for dsRNA ≤6 bp and 1 mM for dsRNA >6 bp). Correspondingly, the volume of dsRNAs can become negligible compared to the simulation boxes, e.g., dsRNAs of 6 and 14 bp only occupy the simulation boxes with side length of ∼70 and ∼150 Å by ∼2 and ∼0.4% in volume, respectively. Based on the equilibrium conformations at each temperature T, the fraction Φ(T) of unfolded state characterized as a completely dissociated ss chain can be obtained at T. Because the small system of the simulation (two strands in a simulational box) can lead to a significant finite size effect (56), the predicted Φ(T) needs to be further corrected to the fraction θh(T) of unfolded state at the high bulk strand concentration (56):
| (9) |
where a = 1 and 2 for non-self-complementary and self-complementary sequences, respectively (56). Afterwards, based on θh(T) at the high strand concentration, the fraction θ(T) of unfolded state at an experimental strand concentration Cs (e.g., ∼0.1 mM) can be calculated by (57, 58)
| (10) |
where . Finally, the fractions θ(T) of unfolded state can be fitted to a two-state model to obtain the melting temperature Tm (54, 55, 57),
| (11) |
where dT is an adjustable parameter. More details about the calculation of melting temperature are given in the Supporting Materials and Methods. For long dsRNAs whose unfolding can be a non-two-state transition, we still used the above formulas to estimate their melting temperatures, in analogy to related experiment (66).
Results and Discussion
In this section, our model was first employed to predict 3D structures of extensive dsRNAs in monovalent/divalent ion solutions. Afterwards, the model was used to predict stability of extensive dsRNAs and the effects of monovalent/divalent ions and further to analyze the thermally unfolding pathway of various dsRNAs. Finally, the model was employed to examine the salt-dependent flexibility of a dsRNA helix. Our predictions and analyses were extensively compared with the available experiments and existing models.
Structure predictions for dsRNAs in ion solutions
Two sets of available dsRNAs were used in this work on 3D structure prediction. One set includes 16 dsRNAs whose structures were determined by x-ray experiments (defined as the x-ray set), and the other set contains 10 dsRNAs whose structures were determined by NMR experiments in ion solutions (defined as the NMR set). The PDB codes as well as the descriptions of the dsRNAs in two sets are shown in Tables 1 and S3, respectively. In the following, because of the lack of ionic conditions for dsRNAs in the x-ray set, we first made the predictions on 3D structures for 26 dsRNAs in the x-ray set and NMR set at high salt (e.g., 1 M Na+), i.e., assuming dsRNAs are nearly fully neutralized and consequently ignoring electrostatic contribution to the predicted structures. Afterwards, we further employed our model to predict 3D structures for dsRNAs in NMR set at their respective experimental ion conditions to examine whether the model with structure-based electrostatic potential can improve 3D structure predictions for dsRNAs in ion solutions.
Table 1.
10 dsRNAs in the NMR Set for Structure Prediction at Respective Salt Conditions
| PDB | Descriptiona | Length (nt) | [Na+]/[Mg2+] mM/mMb | Mean/Minimal RMSDs (Å)c |
|
|---|---|---|---|---|---|
| In 1M [Na+] | In Experimental Solution | ||||
| 2GM0 | I | 70 | 250/0.1 | 7.8/3.5 | 6.1/3.1 |
| 2M1O | H | 14 | 80/0.1 | 2.4/1.1 | 2.0/0.7 |
| 1TUT | I | 22 | 80/3 | 3.9/2.2 | 3.2/1.7 |
| 2KYD | H | 40 | 150/10 | 3.8/1.2 | 3.7/1.2 |
| 2D1A | I | 78 | 50/0 | 7.4/4.1 | 6.9/3.8 |
| 2DD1 | I | 20 | 90/0 | 3.7/1.5 | 3.3/1.5 |
| 2K7E | I | 24 | 110/0 | 2.8/1.2 | 2.4/1.3 |
| 2LX1 | I | 22 | 90/0 | 5.6/3.3 | 4.8/2.4 |
| 2JXQ | H | 20 | 60/0 | 2.1/0.8 | 2.0/0.8 |
| 1F5G | I | 20 | 80/0 | 3.2/1.9 | 2.8/1.7 |
H stands for dsRNAs of complementary duplex, and I stands for dsRNAs with internal loop.
The experimental ion conditions for structure determination by NMR method.
The RMSDs are calculated over all three CG beads of predicted structures by our model from the corresponding atoms of the native structures.
Structure predictions for dsRNAs at 1M [Na+]
For 26 dsRNAs in the x-ray set and NMR set, the 3D structures were predicted from sequences with strand concentration of 1 mM at high salt concentration (e.g., 1 M Na+), regardless of possible ion effects. In the following, we used a paradigm dsRNA (PDB: 2JXQ; shown in Table 1) to show the structure predicting process of dsRNA with our model, which is shown in Fig. 1 C. First, the energy of the system reduces with the decrease of temperature (from 100°C to room temperature), and the dsRNA folds into native-like structures (e.g., structure c in Fig. 1 D) from an initial random configuration (e.g., structure a in Fig. 1 D). Second, a further structure refinement (∼1.2 × 107 MC steps) is performed at the target temperature (e.g., room temperature), in which the last predicted structure from the annealing process is taken as input and the parameters Paranonhelical of bonded potentials are replaced by Parahelical for the base-paired regions to better capture the geometry of helical stems (54, 55). Finally, an ensemble of refined 3D structures (∼10,000 structures) can be obtained over the last ∼1 × 106 MC steps, and these structures can be evaluated by the root mean-square deviation (RMSD) values calculated over all the beads in predicted structures from the corresponding atoms in the native structures in the PDB (67). As shown in Figs. 1 C and 2, for the dsRNA of PDB: 2JXQ, the mean RMSD (the averaged value over the refined structures) and the minimal RMSD (from the structure closest to the native one) are 2.1 and 0.8 Å, respectively.
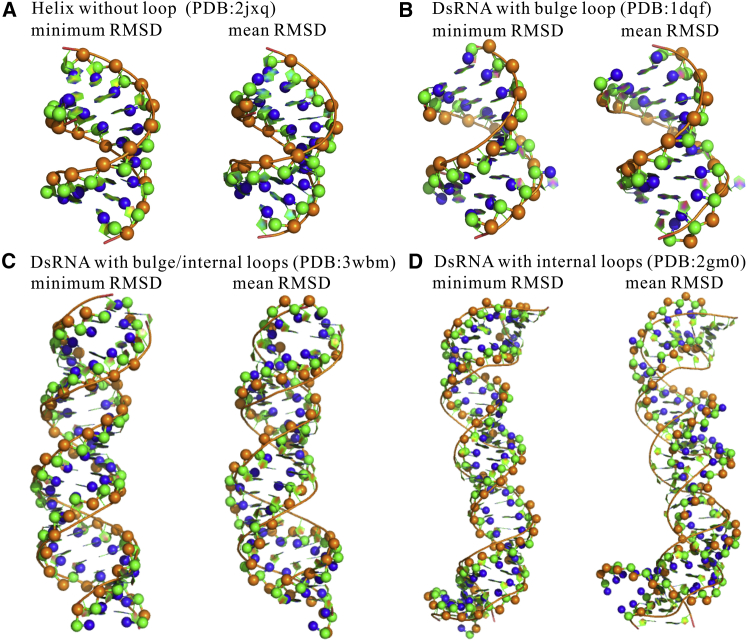
Figure 2.
The predicted 3D structures (ball-stick) with mean/minimal RMSDs in comparison with the corresponding native structures (cartoon) for four typical dsRNAs. (A) dsRNA helix without loop (PDB: 2JXQ) with mean/minimal RMSDs of 2.0 Å/0.8 Å. (B) dsRNA containing a bulge loop (PDB: 1DQF) with mean/minimal RMSDs of 2.5 Å/1.6 Å. (C) dsRNA containing two bulge loops and two internal loops (PDB: 3WBM) with mean/minimal RMSDs of 5.4 Å/2.3 Å. (D) dsRNA containing four internal loops (PDB: 2GM0) with mean/minimal RMSDs of 6.1 Å/3.1 Å. The structures are shown with PyMol (http://www.pymol.org). To see this figure in color, go online.
Following the above process, the 3D structures of 26 dsRNAs, including 15 dsRNAs with bulge/internal loops, were predicted by this model with an overall mean RMSD of ∼3.3 Å and an overall minimal RMSD of ∼1.8 Å; see Figs. 2, 3, and Table S1. This shows that our model with coaxial/base stacking can reliably capture the 3D shapes of various dsRNAs, including those with bulge/internal loops. For 13 dsRNAs with internal loops, the overall mean RMSD is ∼4.2 Å, which is slightly larger than that (∼3.3 Å) of all the 26 dsRNAs. This is because large internal loops generally contain noncanonical basepairs, which is ignored in this model. For example, the dsRNA of PDB: 3WBM contains two internal loops with several noncanonical basepairs to keep the helix more continuous than the predicted one; see Fig. 2 C.
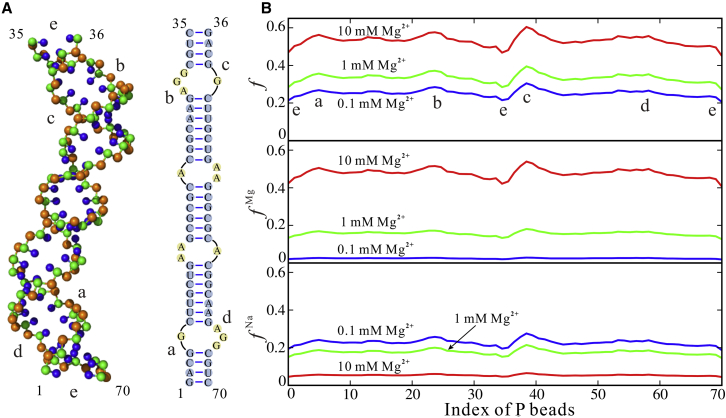
Figure 3.
(A) The predicted 3D structure (left, shown with PyMol) and secondary structure (right) of dsRNA (PDB: 2GM0). (B) The calculated ion-charge-neutralization fractions along P beads of the predicted structure of the dsRNA at different ion conditions; see Eq. 6. f, the total ion-neutralization fractions (top); fMg, neutralization fractions of Mg2+ (middle); fNa, neutralization fractions of Na+ (bottom). The blue, green, and red lines denote the cases of the dsRNA in 150 mM Na+ solutions mixed with 0.1, 1, and 10 mM Mg2+, respectively. The two bent regions labeled with (a, d) and (b, c) induced by the internal loops correspond to the ion-neutralization-fraction peaks in (B), and two helical ends of the dsRNA labeled with (e) correspond to the ion-neutralization-fraction troughs in (B). To see this figure in color, go online.
Structure predictions for dsRNAs in respective ion solutions
Because RNA structures can be strongly influenced by ions (10, 11, 12, 13, 14), we introduced the structure-based electrostatic potential in our model to improve the 3D structure prediction for dsRNAs at the respective ion conditions. In the following, we first examined the structure-based electrostatic potential through the charge-neutralization fractions fNa and fMg of Na+ and Mg2+ along with an example dsRNA (PDB: 2GM0) in mixed Na+/Mg2+ solutions. As shown in Fig. 3 B, fNa and fMg appear dependent on Na+/Mg2+ concentrations as well as dsRNA structures: 1) as Mg2+ concentration increases, fMg increases and fNa decreases because of the competition between Mg2+ and Na+ in binding to an RNA and the lower binding-entropy penalty for Mg2+ at higher Mg2+ concentration (10, 11, 12, 13, 14, 59, 60, 61, 62, 63); 2) fNa and fMg are larger at bent regions and appear less at two ends, which is attributed to the higher charge density of P beads at bending regions and lower charge density of P beads at two ends of the dsRNA. Therefore, the newly refined electrostatic potential (Eqs. 4, 5, 6, 7, and 8) can capture the structure-based ion binding and the competitive binding between Na+ ions and Mg2+ ions to dsRNAs.
To examine whether the involvement of the implicit structure-based electrostatic potential (Eqs. 4, 5, 6, 7, and 8) and corresponding ion conditions can improve 3D structure prediction for dsRNAs, we further predicted the 3D structures for 10 dsRNAs in the NMR set at their respective experimental ion conditions. For the 10 dsRNAs, the overall mean RMSD between predicted structures in ion solutions and the native structures is ∼3.7 Å, which is visibly smaller than that (∼4.3 Å) of the predictions at 1 M [Na+]; see Table 1. This suggests that the inclusion of structure-based electrostatic potential can account for ionic conditions for RNAs and the implementation of the corresponding experimental ion conditions can improve the predictions on 3D structures for dsRNAs in ion solutions. Furthermore, Table 1 also shows that such improvement appears more pronounced for the dsRNAs with internal loops and for the ion conditions containing Mg2+, e.g., the mean RMSDs of the dsRNA of 2GM0 and 1TUT decrease from 7.8 and 3.9 Å to 6.1 and 3.2 Å, respectively. This indicates that the newly refined electrostatic potential can effectively involve the RNA structure information and the effect of ions such as Mg2+. In addition, we have further predicted 3D structures for dsRNAs in NMR set using our model in the previous version and for several ssRNAs including hairpins and pseudoknots involved in (55, 68) using our model; see Table S4. The comparisons between predictions from our model and its previous version show a visible improvement (∼0.4 Å in mean RMSD) on predicting RNA structures, including both dsRNAs and ssRNAs (55, 69). This indicates that the inclusion of the newly refined structure-based electrostatic potential can better capture RNA structures at their respective ion conditions.
Comparisons with other models
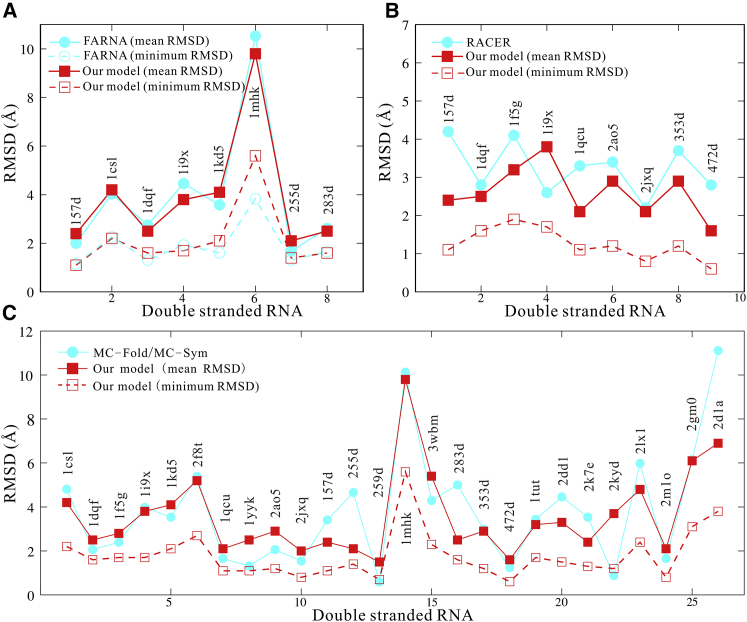
To further examine our model, we made extensive comparisons with three existing RNA-structure-prediction models: FARNA (31), RACER (45, 46), and MC-Fold/MC-Sym pipeline (24). FARNA is a fragment-assembly model with high resolution for small RNAs (31). As shown in Fig. 4 A, the average mean RMSD (∼3.9 Å) from our model is very slightly smaller than that (∼4.1 Å) from FARNA. Afterwards, we made the comparison with a newly developed CG model, RACER (45). As shown in Fig. 4 B, the mean RMSD of the predictions from our model (∼2.6 Å) is slightly smaller than that (∼3.2 Å) from RACER. Furthermore, we made the extensive comparison with MC-Fold/MC-Sym pipeline (24), a well-established RNA 2D/3D structure prediction model with a web server (http://www.major.iric.ca/MC-Pipeline). We used the web server of MC-Fold/MC-Sym pipeline to predict the structures of all the dsRNAs involved in our structure prediction and chose the top predicted structure to make comparisons with our model. As shown in Fig. 4 C, the average mean RMSD (∼3.3 Å) of the predictions from our model is slightly smaller than that (∼3.8 Å) of the top structure from MC-Fold/MC-Sym pipeline. Therefore, the above comparisons show that our model can be reliable in predicting the 3D structures of dsRNAs. Beyond 3D structure predictions, our model can also predict the stability and flexibility of dsRNAs in ion solutions. It should be noted that the computational cost of our model is comparable with that of RACER (46) but much higher than that of the fragment-assembly methods such as MC-Fold/MC-Sym (24) because our model and RACER both involved conformation sampling, unlike fragment-assembly methods. For example, for dsRNA with 20 nt, it takes ∼3 h for our model to predict its 3D structure with one thread on the early generation Intel i7 central processing unit and takes ∼10 min by MC-Fold/MC-Sym on its server. However, compared with the existing models, our model can also predict the 3D structures at different temperatures and can estimate thermodynamic stability for dsRNAs simultaneously.
Figure 4.
The comparisons of the predicted 3D structures between our model and the existing models: (A) FARNA (31), (B) RACER (45, 46), and (C) MC-Fold/MC-Sym pipeline (24). The RMSDs of structures predicted by FARNA are calculated over the C4′ atom (31). The RMSDs of structures predicted by RACER, MC-Fold/MC-Sym pipeline, and our model are calculated over all CG beads (24, 45). The data of FARNA and RACER are taken from (31) to (45), respectively. To see this figure in color, go online.
Stability of dsRNAs in ion solutions
Stability of dsRNAs with various sequences
As described in Methods, for dsRNA with a given strand concentration, the melting curve as well as the melting temperature Tm can be calculated with our model. For example, for the sequence (CGCG)2, the melting curve of the dsRNA with a high strand concentration of 10 mM can be predicted based on the fractions of unfolded state at different temperatures, and the melting curve as well as the melting temperature Tm of the dsRNA at low experimental strand concentration (0.1 mM) can be calculated through Eqs. 9, 10, and 11; see Fig. 5, A and B. As shown in Fig. 5, A and B, the predicted Tm of the sample sequence (CGCG)2 with experimental strand concentration of 0.1 mM is ∼19.5°C, which agrees well with the corresponding experimental value (∼19.3°C). Furthermore, we further predicted the thermodynamic stability of 22 dsRNAs (from 4-bp to 14 bp) with various complementary sequences; see Table 2. Here, dsRNAs are assumed in solutions of 1 M [Na+] to solely examine the stabilities of dsRNAs of various sequence and make comparisons with extensive experimental data (58, 66, 70, 71, 72). As shown in Table 2, the Tm values of extensive dsRNAs from our model are in good agreement with the corresponding experimental data with the mean deviation ∼1.3°C and maximal deviations <2.5°C. Such agreement indicates that the sequence-dependent base-pairing and base-stacking interactions in our model can capture the stability of dsRNAs of extensive sequences and different lengths well (58, 66, 70, 71, 72).
Figure 5.
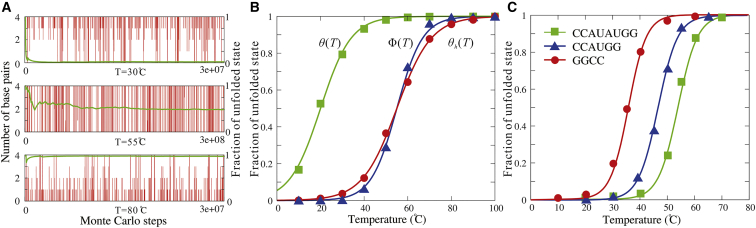
(A) The time evolution of the number of basepairs (vertical or red lines) and the average fractions of unfolded state (transverse or green lines) for the sample dsRNA of (CGCG)2 at 30°C (top), 55°C (middle), and 80°C (bottom). (B) The fractions of unfolded state as functions of temperature for the dsRNA of (CGCG)2. Symbols show the predicted data at different temperatures: blue triangle, at high strand concentration (10 mM); red circle, corrected value of θh(T) by Eq. 9; and green square, at experimental strand concentration (0.1 mM) derived by Eq. 10. Lines are the fitted melting curve to the predicted data through Eq. 11. More details can be found in the Supporting Materials and Methods. (C) The fractions of unfolded states for three dsRNAs with 0.1 mM strand concentration as functions of temperature. Symbols are the predicted data and lines are the fitted curves for the three dsRNAs through Eq. 11. To see this figure in color, go online.
Table 2.
The Melting Temperature Tm Values for 20 dsRNAs in 1 M [Na+] Solution
| Sequencea | Length (bp) | Expt. (°C)d | Pred. (°C) | Deviation (°C) | |
|---|---|---|---|---|---|
| 1 | CGCG | 4 | 19.9 | 19.5 | −0.4 |
| 2 | GGCC | 4 | 34.3 | 35.1 | +0.8 |
| 3 | GCGC | 4 | 26.6 | 28.1 | +1.5 |
| 4 | CCGG | 4 | 27.2 | 26.6 | −0.6 |
| 5 | CGCGCG | 6 | 57.8 | 58.5 | +0.7 |
| 6 | CCGCGG | 6 | 59.8 | 61.2 | +1.4 |
| 7 | CCAUGG | 6 | 46.4 | 45.3 | −1.1 |
| 8 | GUCGAC | 6 | 45.3 | 46.8 | +1.5 |
| 9 | AGCGCU | 6 | 50.2 | 51.1 | +0.9 |
| 10 | UCAUGA | 6 | 27.2 | 25.7 | −1.5 |
| 11 | GAGGAG | 6 | 50.9 | 52.0 | +1.1 |
| 12 | AACUAGUU | 8 | 45.7 | 45.5 | −0.2 |
| 13 | ACUUAAGU | 8 | 40.3 | 41.3 | +1.0 |
| 14 | ACCUUUGG | 8 | 56.3 | 54.8 | −1.5 |
| 15 | GCCAUGGC | 8 | 71.4 | 69.5 | −1.9 |
| 16 | GCUGCGAC | 8 | 67.9 | 65.8 | −2.1 |
| 17 | CCAUAUGG | 8 | 57.0 | 59.1 | +2.1 |
| 18 | GAACGUUC | 8 | 52.3 | 53.6 | +1.3 |
| 19 | AAGGUUGGAA | 10 | 66.5 | 64.1 | −2.4 |
| 20 | AUUGGAUACAAAb | 12 | 55.4 | 58.1 | +2.7 |
| 21 | AAAAAAAUUUUUUUc | 14 | 33.2 | 35.0 | +1.8 |
| 22 | CCUUGAUAUCAAGG | 14 | 76.7 | 74.3 | −2.4 |
Thermally unfolding pathways of dsRNAs
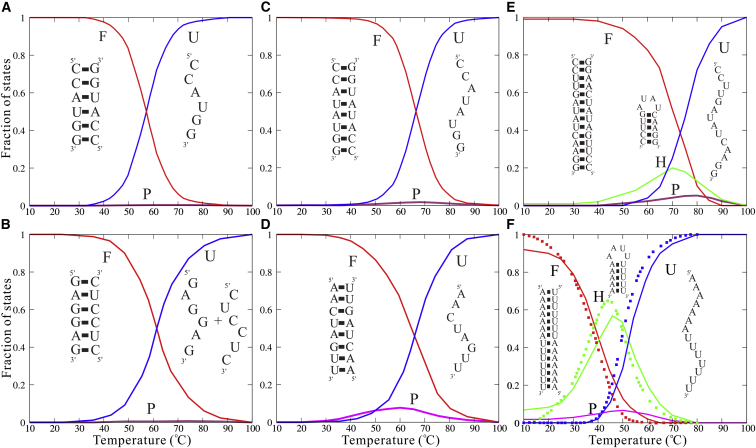
Because intermediate states of RNAs can be important to their functions (1, 2, 3, 10, 68, 69), we made further analyses of thermally unfolding pathways for different dsRNAs. To distinguish the possible different states of dsRNAs at different temperatures in our simulations, all the states for dsRNA were more detailedly divided into unfolded state (U, two disassociated single strands), possible hairpin state (H, with at least one hairpin), folded helix state (F, with the formation of all basepairs except for the two end ones), and partially folded helix state (P, other conformations besides U, F, and H states).
As shown in Fig. 6, the unfolding pathways of dsRNAs are dependent on their length as well as sequences. For short sequences (≤6 bp), dsRNAs undergo the standard two-state melting transitions and there are almost no intermediate states such as P and H states (see Fig. 6, A and B), which is consistent with the previous experiments (66). As chain length increases to ∼8 bp, P states begin to appear and can become visible at ∼Tm; see Fig. 6, C and D. Figs. 6, C and D also show that the unfolding pathways of dsRNAs with the same chain length but different sequences would be slightly different, e.g., the fraction of P state of (AACUAGUU)2 with end A-U basepairs is slightly higher than that of (CCAUAUGG)2 with end G-C basepairs. This is because the unstable end A-U basepairs can induce more notable P states than stable end G-C basepairs; see Fig. S1.
Figure 6.
The fractions of F (folded, red lines), P (partially folded, pink lines), H (hairpin, green lines), and U (unfolded, blue lines) states as functions of temperature for the unfolding of (CCAUGG)2 (A), (GAGGAG, CUCCUC) (B), (CCAUAUGG)2 (C), (AACUAGUU)2 (D), (CCUUGAUAUCAAGG)2 (E), and (AAAAAAAUUUUUUU)2 (F). The full symbols in (F) are the corresponding experimental data from (71). The insets illustrate the typical predicted secondary structures for F, U, and H states. To see this figure in color, go online.
For dsRNAs with more than 10 bp, their thermally unfolding pathways become more complex and interesting because their ss chain may fold to hairpin structures. As shown in Fig. 6, E and F for the dsRNA of (CCUUGAUAUCAAGG)2 and (AAAAAAAUUUUUUU)2, the fractions of F state are near unity at low temperature. As temperature increases, the dsRNAs begin to melt and H states would form from the melted ssRNAs with the maximal fractions of ∼0.2 and ∼0.5 for the two dsRNAs, respectively. At higher temperatures, the dsRNAs become almost completely melted as the U state. Notably, as shown in Fig. 6 F, the unfolding pathway of (AAAAAAAUUUUUUU)2 predicted by our model is very close to the corresponding experiments (66, 71), suggesting that the melting pathways of dsRNAs can be well captured by our analyses with this model. The difference on unfolding pathways between the two dsRNAs is attributed to the different sequences, i.e., G-C content, especially at two ends. Specifically, the fractions of states follow the order of for (CCUUGAUAUCAAGG)2 at ∼70°C, whereas for (AAAAAAAUUUUUUU)2 at ∼45°C, such an order becomes . To reveal what determines the order, we calculated the stability for the states with Mfold (73). We found that the order of state fractions is in agreement with that of state stability. For example, the formation free energies for F, H, and P states are ∼−0.5, ∼0.2, and ∼1.5 kcal/mol for (CCUUGAUAUCAAGG)2 at ∼70°C, respectively. For (AAAAAAAUUUUUUU)2 at ∼45°C, the formation free energies for H, F, and P states are ∼−0.8, ∼−0.1, and ∼0.6 kcal/mol. This indicates that the unfolding pathway of dsRNA is dependent on the stability of possible states.
Although unfolding of long dsRNAs can be non-two-state transitions, in analogy to experiments (66), we can still estimate their melting temperatures by assuming strands completely disassociated state as U state (66); see Eq. 11 in Methods.
Stability of dsRNAs with bulge/internal loops
Beyond the dsRNAs with complementary sequences shown above, the stability of other eight dsRNAs with bulge/internal loops was examined by our model. As shown in Table 3, for the dsRNAs with single/double-bulge loops of different loop lengths (sequences 1–6) and the dsRNAs with internal loops (sequences seven and eight), the mean deviation between the predicted Tm values and the experimental data (74, 75, 76, 77, 78, 79, 80) is ∼2.6°C, which indicates that our model with the coaxial stacking potential can roughly estimate the stability of dsRNAs with bulge/internal loops. However, such predictions, especially for dsRNAs with long bulge/internal loops, are not as precise as those for dsRNAs without loops. The detailed comparisons with experimental data show that the predicted Tm values for the dsRNAs with a one-nucleotide (nt) bulge loop are slightly higher than experimental data, whereas our model underestimates the stability of dsRNAs with longer bulge loops, which may suggest that the coaxial stacking potential Ucs involved in our model may slightly overestimate the coaxial interaction strength while underestimating the coaxial interaction range. For the dsRNAs with an internal loop (e.g., AA/AA or AAA/AAA), our model underestimates their stability, which may be attributed to the ignorance of noncanonical basepairs in this model (80).
Table 3.
The Melting Temperatures Tm for Eight dsRNAs with Bulge/Internal Loop in 1 M Na+ Solution
| Sequencea | Descriptionb | Experiment (°C)c | Prediction (°C) | Deviation (°C) | |
|---|---|---|---|---|---|
| 1 | UGACGCUCA, ACUG GAGU | B | 42.2 | 45.0 | +2.8 |
| 2 | GACUAUGUC, CUGA ACAG | B | 34.2 | 36.1 | +1.9 |
| 3 | GACUAGUGUC, CUGA ACAG | B | 31.9 | 29.5 | −2.4 |
| 4 | GACUCAUCCUG, CUGG GGAC | B | 40.7 | 37.9 | −2.8 |
| 5 | GCAGUUCACG, CG CAAG GC | B | 35.5 | 36.8 | +1.3 |
| 6 | CGAGUAC CG, GC CAUGAGC | B | 43.6 | 41.5 | −2.1 |
| 7 | CGC AA GCG, GCG AA CGC | I | 35.5 | 33.2 | −2.3 |
| 8 | CGCAAAGGC, GCGAAACCG | I | 38.5 | 33.1 | −5.4 |
Effects of monovalent and divalent ions
The thermal stability of RNA molecules is generally sensitive to the ionic conditions (10, 11, 12, 13, 14, 59, 60, 61, 62, 63). Particularly, Mg2+ is efficient in neutralizing the negative charges on the RNA molecule and generally plays an important role in RNA folding (14, 59, 60, 61, 62, 63, 81, 82, 83, 84). However, most of the existing structure prediction models cannot quantitatively predict the stability of dsRNAs in ion solutions, especially in the presence of Mg2+. Here, we employed our model to examine the stability for dsRNAs over a wide range of monovalent and divalent ion concentrations.
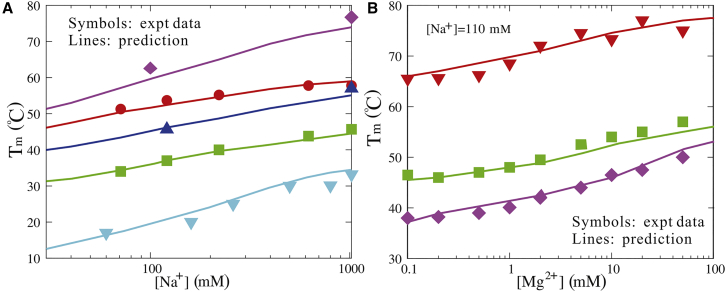
First, we examined the effect of monovalent ions on the stability of dsRNAs. As shown in Fig. 7 A, for five dsRNAs with different sequences and lengths, the predicted melting temperature Tm values from our model agree well with the experimental data (58, 66, 70, 71, 72, 84), with a mean deviation <2°C over the wide range of [Na+]. As [Na+] increases from 10 mM to 1 M, Tm values of the dsRNAs obviously increase, which is attributed to lower ion-binding entropy penalty and stronger ion neutralization for basepair formation at higher [Na+]; see also Fig. S1. Furthermore, Fig. 7 A shows that the [Na+]-dependence of Tm is stronger for longer dsRNAs. This is because basepair formation of longer dsRNAs causes larger buildup of negative charges and consequently causes stronger [Na+]-dependent ion binding.
Figure 7.
(A) The predicted melting temperatures Tm and corresponding experimental data (58, 66, 70, 71, 72, 84) as functions of [Na+] for sequences CCUUGAUAUCAAGG, CGCGCG, CCAUAUGG, AACUAGUU, and AAAAAAAUUUUUUU (from top to bottom). (B) The predicted melting temperatures Tm and corresponding experimental data (84) as functions of [Mg2+] for sequences CCUUGAUAUCAAGG, CCAUAUGG, and CCAUGG (from top to bottom). Note that the solutions contain 110 mM Na+ as background (84). To see this figure in color, go online.
Second, we examined the stability of dsRNAs in mixed monovalent and divalent ion solutions. As shown in Fig. 7 B, for three different dsRNAs, the predicted Tm values are in good accordance with the experimental data over the wide range of [Mg2+] (84). Fig. 7 B also shows that there are three ranges in Tm-[Mg2+] curves: 1) at low [Mg2+] (relatively to [Na+]), the stability of the dsRNAs is dominated by the background [Na+] and Tm values of the dsRNAs are almost the same as the corresponding pure [Na+]; 2) with the increase of [Mg2+], Mg2+ ions begin to play a role and Tm increases correspondingly; and 3) when [Mg2+] becomes very high (relatively to [Na+]), the stability is dominated by Mg2+. Furthermore, it is shown that Mg2+ is very efficient in stabilizing dsRNAs. Even in the background of 110 mM Na+, ∼1 mM Mg2+ begins to enhance the stability of dsRNAs, and 10 mM Mg2+ (+110 mM background Na+) can achieve the similar stability to 1 M Na+ for dsRNAs; see sequences of CCUUGAUAUCAAGG and CCAUAUGG in Fig. 7, A and B. This is attributed to the high ionic charge of Mg2+ and the consequent efficient role in stabilizing dsRNAs (58, 59, 60, 61, 62, 63, 81, 82, 83, 84).
Flexibility of dsRNA in ion solutions
DsRNAs generally are rather flexible in ion solutions because of their polymeric nature, and the flexibility is extremely important for their biological functions. Additionally, dsRNA flexibility is highly dependent on solution ion conditions (85, 86, 87, 88, 89, 90, 91, 92, 93). In this section, we further employed our model to examine the flexibility of a 40-bp RNA helix in ion solutions. The sequence of the dsRNA helix, which is selected according to a previous study (91), is 5′-CGACUCUACGGAAGGGCAUCCUUCGGGCAUCACUACGCGC-3′, with 57% CG content in its central 30-bp segment and the other chain being fully complementary to it. First, we predicted the 3D structures for the dsRNA helix from the sequence at 25°C, and afterwards, we performed further simulations for the dsRNA helix at various ion conditions based on the predicted structures. Enough conformations at equilibrium at each ion condition were used to analyze the salt-dependent flexibility of the dsRNA helix; see Fig. S2.
Structure fluctuation of dsRNA in ion solutions
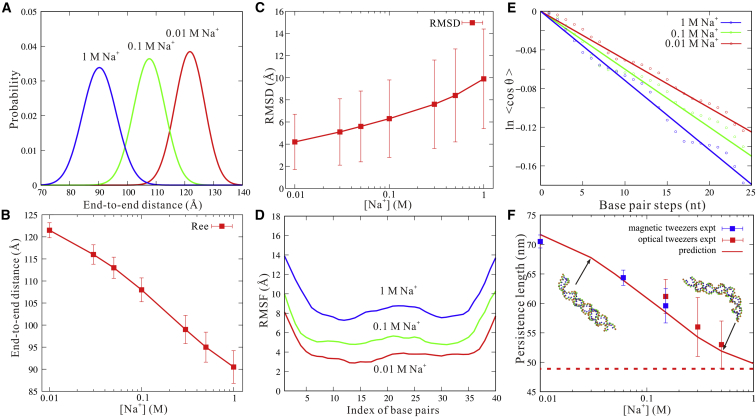
In the following, we first examined the structure fluctuation of the dsRNA helix through calculating end-to-end distance, RMSD variance, and root mean-square fluctuation (RMSF) at different [Na+] values (85). As [Na+] increases, the end-to-end distance of the dsRNA helix decreases, e.g., from ∼125 Å at 10 mM [Na+] to ∼90 Å at 1 M [Na+], and simultaneously, the variance of end-to-end distance increases; see Fig. 8, A and B. This indicates the stronger bending conformations and the higher bending fluctuation for the dsRNA helix at higher ion concentrations, which are attributed to the stronger ion neutralization on P bead charges and consequently the reduced electrostatic repulsion due to bending (86, 87, 88, 89, 90, 91, 92, 93, 94). The RMSD variance of the dsRNA helix at different [Na+] values calculated based on the conformation-averaged reference structure also indicates that the dsRNA helix would become more flexible with the increase of [Na+]; see Fig. 8 C. To examine local structure fluctuation, we further calculated the RMSF of the centers of each basepair of the dsRNA helix at different [Na+] values. As shown in Fig. 8 D, the RMSF increases as [Na+] increases from 0.01 to 1 M, which is because the stronger ion binding and charge neutralization on P beads enable the larger fluctuation of basepairs along the helix (94). Additionally, end effect contributes to an extra increase of RMSF at the two helical ends (94).
Figure 8.
(A) The distributions of end-to-end distance for the 40-bp dsRNA helix at 1, 0.1, and 0.01 M [Na+], respectively. (B) The mean end-to-end distances of the 40-bp dsRNA helix as a function of [Na+]. Here, the error bars denote the variance for the end-to-end distances. (C) The RMSD of the 40-bp dsRNA helix calculated based on the conformation-averaged reference structure of the respective simulation as a function of [Na+]. Here, the error bars denote the variances for the RMSDs. (D) The RMSF of basepairs along the 40-bp dsRNA helix from simulated ensembles at 1, 0.1, and 0.01 M [Na+] values. (E) The fitting curves for lp through Eq. 12 for the 40-bp dsRNA helix at 1, 0.1, and 0.01 M [Na+] values. (F) The predicted persistence length lp (lines) for the 40-bp dsRNA helix as a function of [Na+]. Blue squares, experimental data from magnetic tweezers method (93); red squares, experimental data from optical tweezers method (93). The dashed line in (F) shows the predicted lp assuming all P beads are electrically neutral. To see this figure in color, go online.
Persistence length of dsRNA helix in ion solutions
Generally, the flexibility of a polymer can be described by its persistence length lp (95), and lp can be calculated by the following (96):
| (12) |
where and and are the first and i-th bond-direction vectors, respectively. b in Eq. 12 is the average bond length. According to Eq. 12, lp of the dsRNA helix can be obtained through modeling the dsRNA helix as a bead chain composed of the central beads of basepairs; see Fig. 8 E. To avoid the end effect (94), the first and last five basepairs were excluded in our calculations and the bond vectors are selected as those over every five continuous basepairs (91).
As shown in Fig. 8 F, the persistence lengths of the 40-bp dsRNA helix at different [Na+] values predicted by our model are in quantitative agreement with the corresponding experimental data (93). For example, the deviation of lp between prediction and experiments is less than ∼2 nm over the wide range of [Na+]. As [Na+] increases from 0.01 to 1 M, lp of the dsRNA helix decreases from ∼70 to ∼50 nm. This is because more binding ions neutralize the negative P bead charges on the dsRNA helix more strongly and can reduce the electrostatic bending repulsion along the strands more strongly, causing stronger bending flexibility at high [Na+]. Additionally, we have taken out the representative structures at 0.1 mM [Na+] and have calculated the ion-charge-neutralization fractions along the structures. As shown in Fig. S3, the ion-charge-neutralization fraction changes as RNA structure is changed, especially in the bending regions.
Conclusions
Knowledge of the 3D structures and thermodynamic properties of dsRNAs are crucial for understanding their biological functions. In this work, we have further developed our previous CG model by introducing a structure-based electrostatic potential and employed the model to predict 3D structures, stability, and flexibility of dsRNAs in monovalent/divalent ion solutions. Our predictions were extensively compared with experimental data, and the following conclusions have been obtained:
-
1)
Our model can predict 3D structures from sequences for extensive dsRNAs with/without bulge/internal loops in monovalent/divalent ion solutions well with overall mean RMSD <3.5 Å, and the involvement of the structure-based electrostatic potential and corresponding experimental ion conditions generally improves the structure predictions with smaller RMSDs for dsRNAs in ion solutions.
-
2)
Our model can make good predictions for the stability for dsRNAs with extensive sequences over wide ranges of monovalent/divalent ion concentrations with mean deviation <2°C, and our analyses show that the thermally unfolding pathway of dsRNA is dependent on its length as well as its sequence.
-
3)
Our model can well capture the salt-dependent flexibility of dsRNAs, and the predicted salt-dependent persistence lengths are in good accordance with experiments.
Although our predictions agree well with the extensive experimental data on 3D structure, stability and flexibility of dsRNAs, there are still several limitations in our model. First, despite the fact that the structure-based electrostatic potential can efficiently capture the effects of monovalent/divalent ions on the structure, stability, and flexibility of dsRNAs, the model was not examined for RNAs with more complex structures, and the model cannot consider concrete ion distribution and specific ion binding around an RNA. Further development of this model may need to involve the effect of ions through an implicit-explicit combined treatment for ions (54). Second, the model only involves canonical and wobble basepairs (A-U, G-C, and G-U) and ignores noncanonical basepairs because over ∼90% basepairs in the ds stems of natural RNAs are canonical ones (97). However, noncanonical basepairs are usually found in RNAs of complex structures, e.g., in interhelical junction loops (98), and for those dsRNAs with bulge/internal loops, the ignorance of noncanonical basepairs may (only) slightly affect the predictions on the structure and stability (97). The noncanonical basepairs can also be possibly involved in the model for predicting RNAs of complex structures at the level of three CG beads through two treatments: 1) derive the (local) bond energy parameters for noncanonical basepairs based on the corresponding structures in the PDB and involve them in Eqs. S3–S5 (54); or 2) given Turner’s nearest-neighbor thermodynamic parameters for noncanonical base stacking (99), derive the basepair/stacking parameters for noncanonical basepairs based on the corresponding structures in the PDB (54) and involve these parameters in Eqs. S7–S9. Finally, our model is a CG model, and it is still necessary to rebuild all-atom structures based on predicted CG ones. Nevertheless, our model can predict the 3D structures, stability, and flexibility of dsRNAs over the wide ranges of monovalent/divalent ion concentrations well and can be a good basis for further development for a predictive model with higher accuracy.
Author Contributions
Z.-J.T., L.J., and Y.-Z. S. designed the research. L.J., Y.-Z.S., and C.-J.F. performed the simulation. Z.-J.T., L.J., and Y.-L.T. analyzed the data. L.J., Y.-Z.S., and Z.-J.T. wrote the manuscript. All authors discussed the result and reviewed the manuscript.
Acknowledgments
We are grateful to Professors Shi-Jie Chen (University of Missouri), Xiangyun Qiu (George Washington University), Jian Zhang (Nanjing University) and Wenbing Zhang (Wuhan University) for valuable discussions.
This work was supported by grants from the National Science Foundation of China (11575128, 11605125, and 11774272). Parts of the numerical calculation in this work were performed on the supercomputing system in the Supercomputing Center of Wuhan University.
Editor: Tamar Schlick.
Footnotes
Lei Jin and Ya-Zhou Shi contributed equally to this work.
Supporting Materials and Methods, three figures, and four tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)31006-3.
Supporting Material
References
- 1.Watson J.D. Pearson/Benjamin Cummings; San Francisco, CA: 2008. Molecular Biology of the Gene. [Google Scholar]
- 2.Tinoco I., Jr., Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 3.Li P.T., Vieregg J., Tinoco I., Jr. How RNA unfolds and refolds. Annu. Rev. Biochem. 2008;77:77–100. doi: 10.1146/annurev.biochem.77.061206.174353. [DOI] [PubMed] [Google Scholar]
- 4.Kuwabara T., Hsieh J., Gage F.H. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- 5.Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 6.Meister G., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 7.Akira S., Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 8.Chen S.J. RNA folding: conformational statistics, folding kinetics, and ion electrostatics. Annu. Rev. Biophys. 2008;37:197–214. doi: 10.1146/annurev.biophys.37.032807.125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mustoe A.M., Brooks C.L., Al-Hashimi H.M. Hierarchy of RNA functional dynamics. Annu. Rev. Biochem. 2014;83:441–466. doi: 10.1146/annurev-biochem-060713-035524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Draper D.E., Grilley D., Soto A.M. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 11.Lipfert J., Doniach S., Herschlag D. Understanding nucleic acid-ion interactions. Annu. Rev. Biochem. 2014;83:813–841. doi: 10.1146/annurev-biochem-060409-092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodson S.A. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Draper D.E. Folding of RNA tertiary structure: linkages between backbone phosphates, ions, and water. Biopolymers. 2013;99:1105–1113. doi: 10.1002/bip.22249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koculi E., Hyeon C., Woodson S.A. Charge density of divalent metal cations determines RNA stability. J. Am. Chem. Soc. 2007;129:2676–2682. doi: 10.1021/ja068027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose P.W., Prlić A., Burley S.K. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017;45:D271–D281. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sim A.Y., Minary P., Levitt M. Modeling nucleic acids. Curr. Opin. Struct. Biol. 2012;22:273–278. doi: 10.1016/j.sbi.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao Z., Westhof E. RNA Structure: advances and assessment of 3D structure prediction. Annu. Rev. Biophys. 2017;46:483–503. doi: 10.1146/annurev-biophys-070816-034125. [DOI] [PubMed] [Google Scholar]
- 18.Schlick T., Pyle A.M. Opportunities and challenges in RNA structural modeling and design. Biophys. J. 2017;113:225–234. doi: 10.1016/j.bpj.2016.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun L.Z., Zhang D., Chen S.J. Theory and modeling of RNA structure and interactions with metal ions and small molecules. Annu. Rev. Biophys. 2017;46:227–246. doi: 10.1146/annurev-biophys-070816-033920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somarowthu S. Progress and current challenges in modeling large RNAs. J. Mol. Biol. 2016;428:736–747. doi: 10.1016/j.jmb.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y.Z., Wu Y.Y., Tan Z.J. RNA structure prediction: progress and perspective. Chin. Phys. B. 2014;23:078701. [Google Scholar]
- 22.Cragnolini T., Derreumaux P., Pasquali S. Ab initio RNA folding. J. Phys. Condens. Matter. 2015;27:233102. doi: 10.1088/0953-8984/27/23/233102. [DOI] [PubMed] [Google Scholar]
- 23.Zhou H.X. Theoretical frameworks for multiscale modeling and simulation. Curr. Opin. Struct. Biol. 2014;25:67–76. doi: 10.1016/j.sbi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parisien M., Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y., Huang Y., Xiao Y. Automated and fast building of three-dimensional RNA structures. Sci. Rep. 2012;2:734. doi: 10.1038/srep00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Zhao Y., Xiao Y. 3dRNAscore: a distance and torsion angle dependent evaluation function of 3D RNA structures. Nucleic Acids Res. 2015;43:e63. doi: 10.1093/nar/gkv141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Mao K., Xiao Y. Optimization of RNA 3D structure prediction using evolutionary restraints of nucleotide-nucleotide interactions from direct coupling analysis. Nucleic Acids Res. 2017;45:6299–6309. doi: 10.1093/nar/gkx386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popenda M., Szachniuk M., Adamiak R.W. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao S., Chen S.J. Physics-based de novo prediction of RNA 3D structures. J. Phys. Chem. B. 2011;115:4216–4226. doi: 10.1021/jp112059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X., Zhao P., Chen S.J. Vfold: a web server for RNA structure and folding thermodynamics prediction. PLoS One. 2014;9:e107504. doi: 10.1371/journal.pone.0107504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das R., Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc. Natl. Acad. Sci. USA. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyeon C., Thirumalai D. Capturing the essence of folding and functions of biomolecules using coarse-grained models. Nat. Commun. 2011;2:487. doi: 10.1038/ncomms1481. [DOI] [PubMed] [Google Scholar]
- 33.Jonikas M.A., Radmer R.J., Altman R.B. Coarse-grained modeling of large RNA molecules with knowledge-based potentials and structural filters. RNA. 2009;15:189–199. doi: 10.1261/rna.1270809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boudard M., Barth D., Cohen J. GARN2: coarse-grained prediction of 3D structure of large RNA molecules by regret minimization. Bioinformatics. 2017;33:2479–2486. doi: 10.1093/bioinformatics/btx175. [DOI] [PubMed] [Google Scholar]
- 35.Kim N., Laing C., Schlick T. Graph-based sampling for approximating global helical topologies of RNA. Proc. Natl. Acad. Sci. USA. 2014;111:4079–4084. doi: 10.1073/pnas.1318893111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain S., Schlick T. F-RAG: generating atomic coordinates from RNA graphs by fragment assembly. J. Mol. Biol. 2017;429:3587–3605. doi: 10.1016/j.jmb.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J., Bian Y., Wang W. RNA fragment modeling with a nucleobase discrete-state model. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2012;85:021909. doi: 10.1103/PhysRevE.85.021909. [DOI] [PubMed] [Google Scholar]
- 38.Bian Y., Zhang J., Wang W. Free energy landscape and multiple folding pathways of an H-type RNA pseudoknot. PLoS One. 2015;10:e0129089. doi: 10.1371/journal.pone.0129089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J., Zhang J., Wang W. Structure prediction of RNA loops with a probabilistic approach. PLoS Comput. Biol. 2016;12:e1005032. doi: 10.1371/journal.pcbi.1005032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uusitalo J.J., Ingólfsson H.I., Faustino I. Martini coarse-grained force field: extension to RNA. Biophys. J. 2017;113:246–256. doi: 10.1016/j.bpj.2017.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sieradzan A.K., Makowski M., Liwo A. A general method for the derivation of the functional forms of the effective energy terms in coarse-grained energy functions of polymers. I. Backbone potentials of coarse-grained polypeptide chains. J. Chem. Phys. 2017;146:124106. doi: 10.1063/1.4978680. [DOI] [PubMed] [Google Scholar]
- 42.Ding F., Sharma S., Dokholyan N.V. Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA. 2008;14:1164–1173. doi: 10.1261/rna.894608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boniecki M.J., Lach G., Bujnicki J.M. SimRNA: a coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res. 2016;44:e63. doi: 10.1093/nar/gkv1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cragnolini T., Derreumaux P., Pasquali S. Coarse-grained simulations of RNA and DNA duplexes. J. Phys. Chem. B. 2013;117:8047–8060. doi: 10.1021/jp400786b. [DOI] [PubMed] [Google Scholar]
- 45.Xia Z., Bell D.R., Ren P. RNA 3D structure prediction by using a coarse-grained model and experimental data. J. Phys. Chem. B. 2013;117:3135–3144. doi: 10.1021/jp400751w. [DOI] [PubMed] [Google Scholar]
- 46.Bell D.R., Cheng S.Y., Ren P. Capturing RNA folding free energy with coarse-grained molecular dynamics simulations. Sci. Rep. 2017;7:45812. doi: 10.1038/srep45812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denesyuk N.A., Thirumalai D. Coarse-grained model for predicting RNA folding thermodynamics. J. Phys. Chem. B. 2013;117:4901–4911. doi: 10.1021/jp401087x. [DOI] [PubMed] [Google Scholar]
- 48.Hori N., Denesyuk N.A., Thirumalai D. Salt effects on the thermodynamics of a frameshifting RNA pseudoknot under tension. J. Mol. Biol. 2016;428:2847–2859. doi: 10.1016/j.jmb.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Šulc P., Romano F., Louis A.A. A nucleotide-level coarse-grained model of RNA. J. Chem. Phys. 2014;140:235102. doi: 10.1063/1.4881424. [DOI] [PubMed] [Google Scholar]
- 50.He Y., Maciejczyk M., Liwo A. Mean-field interactions between nucleic-acid-base dipoles can drive the formation of a double helix. Phys. Rev. Lett. 2013;110:098101. doi: 10.1103/PhysRevLett.110.098101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He Y., Liwo A., Scheraga H.A. Optimization of a nucleic acids united-RESidue 2-point model (NARES-2P) with a maximum-likelihood approach. J. Chem. Phys. 2015;143:243111. doi: 10.1063/1.4932082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manning G.S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q. Rev. Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 53.Hayes R.L., Noel J.K., Onuchic J.N. Generalized Manning condensation model captures the RNA ion atmosphere. Phys. Rev. Lett. 2015;114:258105. doi: 10.1103/PhysRevLett.114.258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi Y.Z., Wang F.H., Tan Z.J. A coarse-grained model with implicit salt for RNAs: predicting 3D structure, stability and salt effect. J. Chem. Phys. 2014;141:105102. doi: 10.1063/1.4894752. [DOI] [PubMed] [Google Scholar]
- 55.Shi Y.Z., Jin L., Tan Z.J. Predicting 3D structure, flexibility, and stability of RNA hairpins in monovalent and divalent ion solutions. Biophys. J. 2015;109:2654–2665. doi: 10.1016/j.bpj.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ouldridge T.E., Louis A.A., Doye J.P. Extracting bulk properties of self-assembling systems from small simulations. J. Phys. Condens. Matter. 2010;22:104102. doi: 10.1088/0953-8984/22/10/104102. [DOI] [PubMed] [Google Scholar]
- 57.Borer P.N., Dengler B., Uhlenbeck O.C. Stability of ribonucleic acid double-stranded helices. J. Mol. Biol. 1974;86:843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- 58.Tan Z.J., Chen S.J. RNA helix stability in mixed Na+/Mg2+ solution. Biophys. J. 2007;92:3615–3632. doi: 10.1529/biophysj.106.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan Z.J., Chen S.J. Electrostatic correlations and fluctuations for ion binding to a finite length polyelectrolyte. J. Chem. Phys. 2005;122:44903. doi: 10.1063/1.1842059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan Z.J., Chen S.J. Salt contribution to RNA tertiary structure folding stability. Biophys. J. 2011;101:176–187. doi: 10.1016/j.bpj.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan Z.J., Chen S.J. Predicting ion binding properties for RNA tertiary structures. Biophys. J. 2010;99:1565–1576. doi: 10.1016/j.bpj.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang F.H., Wu Y.Y., Tan Z.J. Salt contribution to the flexibility of single-stranded nucleic acid offinite length. Biopolymers. 2013;99:370–381. doi: 10.1002/bip.22189. [DOI] [PubMed] [Google Scholar]
- 63.Xi K., Wang F.H., Tan Z.J. Competitive binding of Mg2+ and Na+ ions to nucleic acids: from helices to tertiary structures. Biophys. J. 2018;114:1776–1790. doi: 10.1016/j.bpj.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Privalov P.L., Crane-Robinson C. Translational entropy and DNA duplex stability. Biophys. J. 2018;114:15–20. doi: 10.1016/j.bpj.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao S., Chen S.J. Free energy landscapes of RNA/RNA complexes: with applications to snRNA complexes in spliceosomes. J. Mol. Biol. 2006;357:292–312. doi: 10.1016/j.jmb.2005.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xia T., SantaLucia J., Jr., Turner D.H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 67.Parisien M., Cruz J.A., Major F. New metrics for comparing and assessing discrepancies between RNA 3D structures and models. RNA. 2009;15:1875–1885. doi: 10.1261/rna.1700409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y., Gong S., Zhang W. The thermodynamics and kinetics of a nucleotide base pair. J. Chem. Phys. 2016;144:115101. doi: 10.1063/1.4944067. [DOI] [PubMed] [Google Scholar]
- 69.Shi Y.Z., Jin L., Tan Z.J. Predicting 3D structure and stability of RNA pseudoknots in monovalent and divalent ion solutions. PLoS Comput. Biol. 2018;14:e1006222. doi: 10.1371/journal.pcbi.1006222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakano S., Fujimoto M., Sugimoto N. Nucleic acid duplex stability: influence of base composition on cation effects. Nucleic Acids Res. 1999;27:2957–2965. doi: 10.1093/nar/27.14.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hickey D.R., Turner D.H. Solvent effects on the stability of A7U7p. Biochemistry. 1985;24:2086–2094. doi: 10.1021/bi00329a042. [DOI] [PubMed] [Google Scholar]
- 72.Chen Z., Znosko B.M. Effect of sodium ions on RNA duplex stability. Biochemistry. 2013;52:7477–7485. doi: 10.1021/bi4008275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tomcho J.C., Tillman M.R., Znosko B.M. Improved model for predicting the free energy contribution of dinucleotide bulges to RNA duplex stability. Biochemistry. 2015;54:5290–5296. doi: 10.1021/acs.biochem.5b00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crowther C.V., Jones L.E., Serra M.J. Influence of two bulge loops on the stability of RNA duplexes. RNA. 2017;23:217–228. doi: 10.1261/rna.056168.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Znosko B.M., Silvestri S.B., Serra M.J. Thermodynamic parameters for an expanded nearest-neighbor model for the formation of RNA duplexes with single nucleotide bulges. Biochemistry. 2002;41:10406–10417. doi: 10.1021/bi025781q. [DOI] [PubMed] [Google Scholar]
- 77.Murray M.H., Hard J.A., Znosko B.M. Improved model to predict the free energy contribution of trinucleotide bulges to RNA duplex stability. Biochemistry. 2014;53:3502–3508. doi: 10.1021/bi500204e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen G., Znosko B.M., Turner D.H. Factors affecting thermodynamic stabilities of RNA 3 x 3 internal loops. Biochemistry. 2004;43:12865–12876. doi: 10.1021/bi049168d. [DOI] [PubMed] [Google Scholar]
- 79.Longfellow C.E., Kierzek R., Turner D.H. Thermodynamic and spectroscopic study of bulge loops in oligoribonucleotides. Biochemistry. 1990;29:278–285. doi: 10.1021/bi00453a038. [DOI] [PubMed] [Google Scholar]
- 80.SantaLucia J., Jr., Kierzek R., Turner D.H. Stabilities of consecutive A.C, C.C, G.G, U.C, and U.U mismatches in RNA internal loops: evidence for stable hydrogen-bonded U.U and C.C.+ pairs. Biochemistry. 1991;30:8242–8251. doi: 10.1021/bi00247a021. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Z.L., Wu Y.Y., Tan Z.J. Divalent ion-mediated DNA-DNA interactions: a comparative study of triplex and duplex. Biophys. J. 2017;113:517–528. doi: 10.1016/j.bpj.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Denesyuk N.A., Thirumalai D. How do metal ions direct ribozyme folding? Nat. Chem. 2015;7:793–801. doi: 10.1038/nchem.2330. [DOI] [PubMed] [Google Scholar]
- 83.Leipply D., Draper D.E. Effects of Mg2+ on the free energy landscape for folding a purine riboswitch RNA. Biochemistry. 2011;50:2790–2799. doi: 10.1021/bi101948k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Serra M.J., Baird J.D., Westhof E. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. RNA. 2002;8:307–323. doi: 10.1017/s1355838202024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hagerman P.J. Flexibility of RNA. Annu. Rev. Biophys. Biomol. Struct. 1997;26:139–156. doi: 10.1146/annurev.biophys.26.1.139. [DOI] [PubMed] [Google Scholar]
- 86.Bao L., Zhang X., Tan Z.J. Flexibility of nucleic acids: from DNA to RNA. Chin. Phys. B. 2016;25:018703. [Google Scholar]
- 87.Li J., Wijeratne S.S., Kiang C.H. DNA under force: mechanics, electrostatics, and hydration. Nanomaterials (Basel) 2015;5:246–267. doi: 10.3390/nano5010246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sutton J.L., Pollack L. Tuning RNA flexibility with helix length and junction sequence. Biophys. J. 2015;109:2644–2653. doi: 10.1016/j.bpj.2015.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen H., Meisburger S.P., Pollack L. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc. Natl. Acad. Sci. USA. 2012;109:799–804. doi: 10.1073/pnas.1119057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Drozdetski A.V., Tolokh I.S., Onufriev A.V. Opposing effects of multivalent ions on the flexibility of DNA and RNA. Phys. Rev. Lett. 2016;117:028101. doi: 10.1103/PhysRevLett.117.028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bao L., Zhang X., Tan Z.J. Understanding the relative flexibility of RNA and DNA duplexes: stretching and twist-stretch coupling. Biophys. J. 2017;112:1094–1104. doi: 10.1016/j.bpj.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang X., Bao L., Tan Z.J. Radial distribution function of semiflexible oligomers with stretching flexibility. J. Chem. Phys. 2017;147:054901. doi: 10.1063/1.4991689. [DOI] [PubMed] [Google Scholar]
- 93.Herrero-Galán E., Fuentes-Perez M.E., Arias-Gonzalez J.R. Mechanical identities of RNA and DNA double helices unveiled at the single-molecule level. J. Am. Chem. Soc. 2013;135:122–131. doi: 10.1021/ja3054755. [DOI] [PubMed] [Google Scholar]
- 94.Wu Y.Y., Bao L., Tan Z.J. Flexibility of short DNA helices with finite-length effect: from base pairs to tens of base pairs. J. Chem. Phys. 2015;142:125103. doi: 10.1063/1.4915539. [DOI] [PubMed] [Google Scholar]
- 95.Kebbekus P., Draper D.E., Hagerman P. Persistence length of RNA. Biochemistry. 1995;34:4354–4357. doi: 10.1021/bi00013a026. [DOI] [PubMed] [Google Scholar]
- 96.Ullner M., Woodward C.E. Orientational correlation function and persistence lengths of flexible polyelectrolytes. Macromolecules. 2002;35:1437–1445. [Google Scholar]
- 97.Roy A., Panigrahi S., Bhattacharyya D. Structure, stability, and dynamics of canonical and noncanonical base pairs: quantum chemical studies. J. Phys. Chem. B. 2008;112:3786–3796. doi: 10.1021/jp076921e. [DOI] [PubMed] [Google Scholar]
- 98.Leontis N.B., Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Turner D.H., Mathews D.H. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2010;38:D280–D282. doi: 10.1093/nar/gkp892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.